Abstract
Purpose
To investigate the relationship of growth in drusen size with genetic susceptibility and adherence to the alternate Mediterranean diet.
Methods
Participants in this analysis had complete ocular, genetic, and dietary data with mean follow-up time of 10.2 years in the Age-Related Eye Disease database. Maximal drusen size was graded on an ordinal scale and two-step progression was determined. A genetic risk score using variants associated with advanced AMD and derived from a stepwise regression model yielded 11 variants in 8 genes. Adherence to the alternate Mediterranean diet was assessed using a nine-component score based on intake of vegetables, fruits, legumes, whole cereals, fish, meat, nuts, alcohol, and monounsaturated-to-saturated fatty acids ratio. Multivariate Cox proportional hazards models were used.
Results
Among 3023 eligible eyes, 19% had drusen growth. In the stepwise selection, common and rare risk alleles for CFH Y402H, CFH rs1410996, CFH R1210C, C3 R102G, C3 K155Q, and ARMS2/HTRA1, as well as VEGF-A, TIMP3, NPLOC4, and HSPH1 variants were significantly associated with 2-step progression in drusen size, and the C2 E318D protective allele conferred decreased risk, adjusting for other covariates. A higher genetic risk score conferred a higher risk (hazard ratio per 1-unit increase, 2.68; 95% confidence interval, 2.23–3.23; P < 0.001), and a medium/high adherence to alternate Mediterranean diet score (4–9) tended to lower risk (hazard ratio, 0.83; 95% confidence interval, 0.68–0.99; P = 0.049), adjusting for all covariates.
Conclusions
Genetic susceptibility was independently related to drusen growth. A Mediterranean-style diet with healthful nutrient-rich foods (fruits, vegetables, legumes and fish), may reduce enlargement of drusen, the hallmark of AMD.
Keywords: mediterranean diet, drusen progression, genetics, nutrition, AMD
AMD is the leading cause of visual impairment among the elderly population in industrialized countries.1 Advanced forms of AMD lead to central vision loss. Early AMD and intermediate forms, asymptomatic or associated with minimal vision loss, affect between 15% and 25% of the population aged 75 to 85 years.2 These early stages are characterized by the presence of drusen and pigment alterations in the macular region, which usually precede geographic atrophy and neovascular disease, the two advanced subtypes causing visual loss. Drusen—yellow deposits between the retinal pigment epithelium and Bruch's membrane—are the major hallmarks of AMD and are heterogeneous with regard to shape, size, location, and extent.3–6
Many studies have investigated factors associated with progression from nonadvanced to advanced AMD. This transition is multifactorial7 and is influenced by demographic, behavioral, environmental, phenotypic, and genetic factors, including age, smoking,8–10 and nutrition,11–14 as well as several genetic variants.7,15–18 However, information is sparse related to factors associated with drusen size progression, even though early AMD with drusen increases the risk of developing more advanced disease.9,19,20
Further exploration of increasing drusen size over time and how such progression is influenced by genetic and modifiable factors would be helpful to identify higher risk subtypes among the millions of people world-wide diagnosed with nonadvanced stages of AMD. This knowledge could also increase understanding of the biology of drusen growth and could lead to preventive measures before the development of visually disabling forms of AMD. In this study, we assessed the association of drusen size progression measured on an ordinal scale with common and rare AMD genetic variants and diet quality.
Methods
Study Population
Data were derived from the Age-Related Eye Disease Study (AREDS) database. The original study was a multicenter, randomized clinical trial to assess the effect of antioxidant and mineral supplements on the risk of AMD and cataracts.21 The four treatment groups were antioxidants, zinc, antioxidants and zinc combined, or placebo. The protocol was approved by a data and safety monitoring committee and by each institutional review board for the participating ophthalmic centers. Participants were aged 55 to 80 years at baseline and were required to have at least one eye with a visual acuity of no worse than 20/32. Examinations were performed at baseline, at the 2-year visit, and annually thereafter during 13 years of follow-up. The mean follow-up time was 10.2 years. Informed consent was obtained from participants before enrollment, and all research followed the tenets of the Declaration of Helsinki.
Procedures
Data on demographic factors, environmental exposures, and habitual diet were obtained. Retinal photographs were taken according to a standardized protocol by certified photographers using certified cameras.22 Trained fundus graders, masked to clinical and phenotypic information, ascertained signs of AMD from annual stereoscopic color images using a standardized protocol at a single reading center.
Study Participants
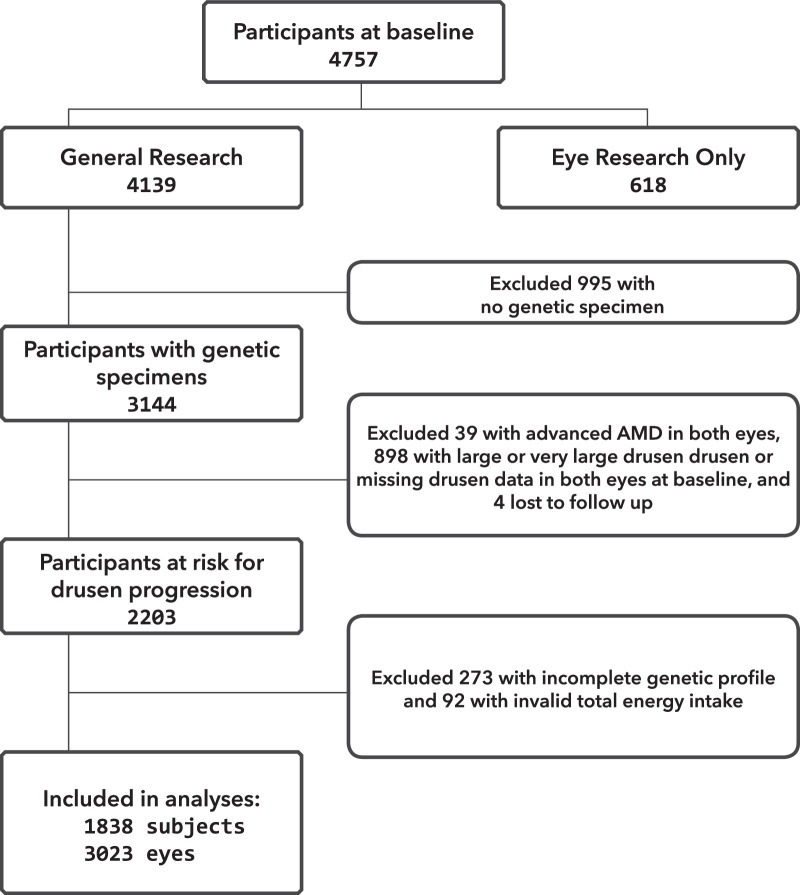
Data were accessed from NIH Database of Genotypes and Phenotypes. Figure 1 illustrates the selection procedures for participants included in the present study. Among the 4757 participants at baseline, 618 participants consented only to “eye research” and, for these participants, phenotype and genetic data could not be linked and they were not included in the analyses. Among the remaining 4139 participants who consented to “general research,” we excluded 995 participants for lack of a genetic specimen. Of the 3144 participants with a genetic specimen, 4 were removed from the dataset owing to lack of follow-up information. Thirty-nine participants with advanced AMD in both eyes at baseline and 898 participants with the highest drusen sizes (drusen grade 3 or 4, as defined below) or missing drusen data in both eyes at baseline were also removed. Furthermore, an additional 365 participants were excluded from the dataset: 273 because of incomplete genotyping information (genotyping rate of <100% for the variants evaluated), and 92 because of an invalid total energy intake (valid total energy intake range is 600–3200 kcal for women and 600–4200 kcal for men). Complete data, including AMD grade at baseline and at least one follow-up visit, dietary data, and demographic, behavioral, and genetic covariates were available for 1838 participants and 3023 eyes at risk for drusen progression. Most of our sample (96%) was Caucasian.
Figure 1.
Flowchart showing study sample selection.
Definition of Drusen Size Progression
Advanced AMD was defined by neovascular AMD, central or noncentral geographic atrophy, or both advanced forms together. Drusen grades based on measured maximum size within the grid among eyes without advanced AMD were defined as follows: 0, no drusen or questionable drusen; 1, small drusen (<63 μm); 2, intermediate drusen (63–124 μm); 3, large drusen (125–249 μm); and 4, very large drusen (≥250 μm). Eye-specific progression of maximum drusen size according to this ordinal scale was defined as an eye advancing at least two grades during the study period. Follow-up for this analysis ended when an eye progressed two grades over the study period (progression from grade 0 to grade 2, or from grade 1 to grade 3, or from grade 2 to grade 4). Eyes that had no indication of drusen size progression were censored if and when they reached advanced AMD or at the last follow-up visit. Eyes with advanced AMD or highest drusen sizes (drusen grade 3 or 4) at baseline, and therefore unable to progress 2 grades, were excluded from the analysis.
Dietary Data
Data on food consumption were collected at enrollment with a validated, self-administered, semiquantitative 90-item, food frequency questionnaire based on the National Cancer Institute Health Habits and History Questionnaire (version 2.1). Participants were asked to report how often, on average, they consumed each food or beverage item during the past year. Consumption was classified into nine levels from “never or less than one per month” to “six or more per day.” For each item, average serving size was recorded as “small,” “medium, or “large” with respect to standard examples. The University of Minnesota Nutrition Coordinating Center Food Composition Database (version 31) was used with the estimated quantity of nutrient intake and DietSys software (version 3.0; Block Dietary Data Systems, Berkeley, CA) to derive individual nutrient values for each questionnaire item. The instrument was validated through a telephone-administered 24-hour dietary recall at 3 and 6 months after enrollment in a sample of 197 randomly selected participants.
We used these raw diet data to create an alternate Mediterranean Diet (aMeDi) score, which is a validated score widely used to estimate adherence to the Mediterranean diet in the US population.23 We previously determined that this score was related to progression from nonadvanced to advanced AMD,13 and this finding was later confirmed in a European population.14 This score is a slightly modified version of the original Mediterranean Diet score published by Trichopoulou et al.24 and is more representative of the dietary patterns observed in the US population. The original Mediterranean diet does not distinguish between whole and refined grains and includes dairy products rather than nuts. The aMeDi score includes nine components: vegetables (excluding potatoes), fruits, legumes, whole grain, nuts, fish, red and processed meat, alcohol consumption, and the monounsaturated fatty acid to saturated fatty acid ratio. Daily intake of each food or beverage group was calculated as the sum of the number of medium servings consumed per day. For each component hypothesized to benefit health, one point was given if intake was above the sex-specific median, and zero otherwise. For alcohol, one point was given to men if their consumption was between 2.7 and 12.2 g/d and women if their consumption was between 1.0 and 4.2 g/d. These cutoffs, corresponding to the third quartile of distribution of total alcohol consumption in this population, were chosen to represent mild to moderate consumption. For components presumed to be detrimental to health, one point was given if intake was below the sex-specific median, and zero otherwise. Total aMeDi score was calculated by adding the scores (zero or one point) for each food category for each participant. Scores range from zero (nonadherence) to nine (perfect adherence).
Demographic and Behavioral Covariates
Baseline age, sex, education, smoking, body mass index (BMI; kg/m2), AREDS treatment group, multivitamin supplement use, and ocular characteristics were included. Pack-years was defined as the product of smoking duration (years) by average packs of cigarettes smoked per day. The median value among ever smokers was 20 pack-years, and smoking status was classified as follows: never, less than 20 pack-years, and 20 pack-years or more. AREDS treatment was defined as antioxidant alone, zinc alone, antioxidant plus zinc group, or placebo. Multivitamin use was defined as never for participants who reported never having taken a multivitamin supplement and who were not taking Centrum during the study, and was defined as ever for participants who declared that they had taken a multivitamin supplement in the past or at present, or were taking Centrum during the study.
Genotype Data
DNA samples were purchased and we assessed genetic variants associated with advanced AMD using array-based and gene sequencing platforms, as previously described.17,25–27 All single nucleotide polymorphisms had a high genotype call rate (>98%). Variants in this study represented common and rare variants in all known genetic pathways: complement factor H (CFH) Y402H (rs1061170); CFH rs1410996; CFH R1210C (rs121913059); CFH N1050Y (rs35274867); complement factor I (CFI) (rs10033900); complement factor B (CFB) R32Q (rs641154); complement component 2 (C2) E318D (rs9332739); complement component 3 (C3) R102G (rs2230199); C3 K155Q (rs147859257); complement component 9 (C9) (rs34882957); vascular endothelial growth factor A (VEGFA) (rs943080); transforming growth factor, beta receptor I (TGFBR1) (rs334353); hepatic lipase (LIPC) (rs10468017); cholesteryl ester transfer protein (CETP) (rs3764261); ATP binding cassette subfamily A member 1 (ABCA1) (rs1883025); apolipoprotein C1 (APOC1) (rs4420638); apolipoprotein H (APOH) (rs1801689); age-related maculopathy susceptibility 2 (ARMS2/HTRA1) (rs10490924); Pellino E3 ubiquitin protein ligase family member 3 (PELI3) (rs145732233); TNF receptor superfamily member 10a (TNFRSF10A) (rs3278062); solute carrier family 16 member 8 (SLC16A8) (rs8135665); paired immunoglobin-like type 2 receptor beta and alpha (PILRB/PILRA) (rs11769700); transmembrane protein 97 (TMEM97) (rs704); Collagen type VIII alpha 1 chain (COL8A1) (rs13095226); collagen type IV alpha 3 chain (COL4A3 (rs11884770); chymotrypsinogen B1 (CTRB1) (rs8056814); A disintegrin-like and metalloprotease with thrombospondin type 1 motif 9 (ADAMTS9) (rs6795735); tissue inhibitor of metalloproteinases inhibitor 3 (TIMP3) (rs9621532); DNA repair protein RAD51 paralog B (RAD51B) (rs8017304); nuclear protein localization 4 homolog/tetraspanin 10 (NPLOC4/TSPAN10) (rs9895741); and heat shock protein family H (HSPH1/B3GALTL) (rs9542236).
Statistical Analysis
The associations between baseline demographic, behavioral, and ocular characteristics with drusen size progression were determined using Cox proportional hazards models with the eye as the unit of analysis, with estimation of hazard ratios (HR) and 95% confidence intervals (CI). Comparisons were adjusted for age and sex.
For analysis of progression of maximum drusen size, we used survival analysis methodology with the eye as the unit of analysis. The associations of each single nucleotide polymorphism with drusen size progression were analyzed using Cox proportional hazards models (PROC PHREG with the covariate aggregate option in SAS 9.4, SAS Institute Inc., Cary, NC), adjusting for correlation of drusen progression in the 2 eyes. In the multivariate models we adjusted for age (≤64, 65–to 74, and >74 years), sex, education (high school or less, high school or more), smoking (never smoker, <20, and ≥20 pack-years), BMI (<25.0, 25.0–29.9, and ≥30 kg/m2), AREDS treatment (four treatment categories), multivitamin supplement use (never, ever), and maximum drusen size category at baseline in each eye.
To select the genetic variants most highly related to increase in drusen size, a stepwise regression method was applied with a significance level with a P value of 0.05 or less for entry and a P value of 0.10 or greater for remaining in the model.17 Eight genetic variants were retained and a composite genetic risk score was calculated using the sum of the regression coefficients multiplied by the corresponding number of risk alleles and summed over these genetic variants, similar to previous analyses.17,28
Participants were classified according to categories of the aMeDi score.13 The aMeDi score was categorized as low (0–3) or medium-high (4–9). The association of aMeDi score with drusen size progression was analyzed using the methodology explained above and models were further adjusted for total energy intake (kilocalories per day).
Interactions between the genetic risk score (classified as below the median, greater than or equal to the median) and aMeDi score were analyzed using the fully adjusted model described above. P values of less than 0.05 were considered statistically significant for all analyses. All statistical analyses were performed using SAS software, version 9.4.
Results
The incidence of drusen progression is shown in Supplementary Figure S1. The mean follow-up time was 10.2 years (range, 3–13 years), and 82.4% had 10 years or more of follow-up time and 1.7% had 5 years or less. Among 3023 eyes, 8% had two-step drusen growth at 5 years, and 19% had drusen growth at 10 years, based on the Kaplan-Meier survival analysis. Baseline demographic, behavioral, and ocular characteristics for eyes which were progressors or nonprogressors, adjusted for age and sex, are shown in the Supplementary Table S1. Older age, smoking, and drusen size at baseline were associated with higher risk of drusen progression (Ptrend < 0.0001, Ptrend = 0.049 and Ptrend < 0.0001, respectively). Sex, education, BMI, AREDS treatment, and multivitamin use did not differ significantly between progressing and nonprogressing eyes.
The associations between drusen growth and genetic susceptibility are displayed in Table 1. The risk alleles for CFH Y402H, CFH rs1410996, C3 R102G, ARMS2/HTRA1, and HSPH1 rs9542236 variants were significantly associated with an increased risk of drusen size progression, and the protective alleles for C2 E318D and ABCA1 conferred a significant decreased risk of progression, after adjustment for age, sex, education, smoking, BMI, drusen size grade at baseline, AREDS treatment, and multivitamin supplement use.
Table 1.
Associations Between Drusen Size Progression and Genetic Susceptibility
| Progressors | Nonprogressors | Model 1* | Model 2† | |||
|---|---|---|---|---|---|---|
| n (%) | n (%) | HR (95% CI) | P Value‡ | HR (95% CI) | P Value‡ | |
| 587 (19.4) | 2436 (80.6) | |||||
| Complement pathway | ||||||
| CFH Y402H: rs1061170 | <0.001 | 0.11 | ||||
| TT | 179 (30.5) | 949 (39.0) | Ref | |||
| CT | 268 (45.7) | 1098 (45.0) | 1.28 (1.04–1.58) | 1.24 (0.94–1.64) | ||
| CC | 140 (23.8) | 389 (16.0) | 1.73 (1.36–2.2) | 1.28 (0.9–1.82) | ||
| CFH: rs1410996 | <0.001 | 0.02 | ||||
| TT | 80 (13.6) | 430 (17.7) | Ref | |||
| CT | 229 (39.0) | 1148 (47.1) | 1.07 (0.81–1.41) | 1.01 (0.7–1.46) | ||
| CC | 278 (47.4) | 858 (35.2) | 1.64 (1.24–2.16) | 1.44 (0.97–2.15) | ||
| CFH R1210C: rs121913059 | 0.25 | <0.001 | ||||
| CC | 584 (99.5) | 2431 (99.8) | Ref | |||
| TC | 3 (0.5) | 5 (0.2) | 2.37 (0.54–10.37) | 4.26 (1.69–10.74) | ||
| CFH N1050Y: rs35274867 | 0.15 | 0.15 | ||||
| AA | 578 (98.5) | 2363 (97.0) | Ref | |||
| TA/TT | 9 (1.5) | 73 (3.0) | 0.59 (0.28–1.22) | 0.52 (0.2–1.4) | ||
| CFI: rs10033900 | 0.08 | 0.19 | ||||
| CC | 137 (23.3) | 666 (27.3) | Ref | |||
| CT | 305 (52.0) | 1186 (48.7) | 1.23 (0.99–1.53) | 1.23 (0.97–1.57) | ||
| TT | 145 (24.7) | 584 (24.0) | 1.25 (0.97–1.62) | 1.24 (0.94–1.64) | ||
| CFB R32Q: rs641153 | 0.71 | 0.64 | ||||
| CC | 583 (99.3) | 2413 (99.1) | Ref | |||
| CT/TT | 4 (0.7) | 23 (0.9) | 0.80 (0.24–2.63) | 0.80 (0.23–2.75) | ||
| C2 E318D: rs9332739 | 0.001 | <0.001 | ||||
| GG | 561 (95.6) | 2212 (90.8) | Ref | |||
| CC/CG | 26 (4.4) | 224 (9.2) | 0.48 (0.3–0.75) | 0.43 (0.26–0.7) | ||
| C3 R102G: rs2230199 | <0.001 | <0.001 | ||||
| CC | 315 (53.7) | 1564 (64.2) | Ref | |||
| CG | 219 (37.3) | 782 (32.1) | 1.30 (1.08–1.57) | 1.37 (1.114–1.68) | ||
| GG | 53 (9.0) | 90 (3.7) | 2.26 (1.65–3.1) | 2.35 (1.67–3.33) | ||
| C3 K155Q: rs147859257 | 0.22 | 0.06 | ||||
| TT | 575 (98.0) | 2409 (98.9) | Ref | |||
| GT | 12 (2.0) | 27 (1.1) | 1.66 (0.74–3.73) | 2.08 (0.92–4.68) | ||
| C9 P167S: rs34882957 | 0.64 | 0.34 | ||||
| GG | 576 (98.1) | 2399 (98.5) | Ref | |||
| AG | 11 (1.9) | 37 (1.5) | 1.18 (0.6–2.33) | 1.16 (0.56–2.41) | ||
| Angiogenesis pathway | ||||||
| VEGFA: rs943080 | 0.09 | 0.05 | ||||
| CC | 113 (19.3) | 589 (24.2) | Ref | |||
| CT | 319 (54.3) | 1211 (49.7) | 1.29 (1.02–1.64) | 1.29 (1.00–1.67) | ||
| TT | 155 (26.4) | 636 (26.1) | 1.27 (0.97–1.66) | 1.33 (0.98–1.77) | ||
| TGFBR1: rs334353 | 0.54 | 0.75 | ||||
| TT | 332 (56.6) | 1403 (57.6) | Ref | |||
| GT | 210 (35.8) | 874 (35.9) | 0.99 (0.82–1.2) | 1.03 (0.83–1.26) | ||
| GG | 45 (7.6) | 159 (6.5) | 1.20 (0.85–1.7) | 1.02 (0.68–1.53) | ||
| Lipid pathway | ||||||
| LIPC: rs10468017 | 0.57 | 0.32 | ||||
| CC | 308 (52.5) | 1259 (51.7) | Ref | |||
| TC | 244 (41.6) | 988 (40.6) | 0.83 (0.55–1.25) | 0.80 (0.52–1.24) | ||
| TT | 35 (5.9) | 189 (7.7) | 1.01 (0.84–1.21) | 0.96 (0.79–1.17) | ||
| CETP: rs3764261 | 0.26 | 0.20 | ||||
| CC | 244 (41.6) | 1087 (44.6) | Ref | |||
| AC | 279 (47.5) | 1106 (45.4) | 1.10 (0.91–1.33) | 1.17 (0.96–1.44) | ||
| AA | 64 (10.9) | 243 (10.0) | 1.15 (0.85–1.56) | 1.29 (0.93–1.79) | ||
| ABCA1: rs1883025 | 0.02 | 0.07 | ||||
| CC | 354 (60.3) | 1315 (54.0) | Ref | |||
| TC | 205 (34.9) | 952 (39.1) | 0.85 (0.70–1.03) | 0.87 (0.71–1.07) | ||
| TT | 28 (4.8) | 169 (6.9) | 0.68 (0.44–1.03) | 0.73 (0.46–1.13) | ||
| APOC1/APOE: rs4420638 | 0.25 | 0.98 | ||||
| AA | 434 (73.9) | 1720 (70.6) | Ref | |||
| GA | 153 (26.1) | 716 (29.4) | 0.89 (0.72–1.09) | 1.01 (0.82–1.26) | ||
| APOH: rs1801689 | 0.99 | 0.31 | ||||
| AA | 547 (93.2) | 2278 (93.5) | Ref | |||
| CA/CC | 40 (6.8) | 158 (6.5) | 1.00 (0.72–1.4) | 1.23 (0.86–1.75) | ||
| Immune inflammatory pathway | ||||||
| ARMS2/HTRA1 A69S: rs10490924 | <0.001 | <0.001 | ||||
| GG | 294 (50.1) | 1497 (61.5) | Ref | |||
| GT | 251 (42.7) | 803 (33.0) | 1.53 (1.27–1.84) | 1.61 (1.31–1.97) | ||
| TT | 42 (7.2) | 136 (5.5) | 1.40 (0.98–2.01) | 1.27 (0.87–1.85) | ||
| PELI3 A307V:rs145732233 | 0.83 | 0.82 | ||||
| CC | 583 (99.3) | 2414 (99.1) | Ref | |||
| TC | 4 (0.7) | 22 (0.9) | 0.87 (0.23–3.29) | 0.86 (0.23–3.26) | ||
| TNFRSF10A: rs13278062 | 0.46 | 0.57 | ||||
| TT | 149 (25.4) | 640 (26.3) | Ref | |||
| GT | 294 (50.1) | 1247 (51.2) | 1.03 (0.82–1.28) | 1.05 (0.83–1.35) | ||
| GG | 143 (24.5) | 549 (22.5) | 1.10 (0.86–1.42) | 1.08 (0.82–1.43) | ||
| SLC16A8: rs8135665 | 0.48 | 0.83 | ||||
| CC | 385 (65.6) | 1575 (64.7) | Ref | |||
| TC | 182 (31.1) | 774 (31.8) | 0.95 (0.78–1.16) | 1.03 (0.84–1.27) | ||
| TT | 20 (3.3) | 87 (3.5) | 0.86 (0.50–1.46) | 0.78 (0.42–1.43) | ||
| PILRB/PILRA: rs11769700 | 0.21 | 0.23 | ||||
| TT | 359 (61.2) | 1544 (63.4) | Ref | |||
| CT | 207 (35.3) | 795 (32.6) | 1.16 (0.96–1.41) | 1.15 (0.93–1.42) | ||
| CC | 21 (3.5) | 97 (4.0) | 1.04 (0.62–1.77) | 1.06 (0.57–1.94) | ||
| TMEM97/VTN: rs704 | 0.55 | 0.82 | ||||
| AA | 143 (24.4) | 588 (24.1) | Ref | |||
| AG | 296 (50.4) | 1155 (47.4) | 1.14 (0.91–1.42) | 1.20 (0.94–1.53) | ||
| GG | 148 (25.2) | 693 (28.5) | 0.93 (0.72–1.2) | 0.98 (0.74–1.3) | ||
| Extracellular matrix | ||||||
| COL8A1 P195L: rs13095226 | 0.56 | 0.69 | ||||
| TT | 488 (83.1) | 1985 (81.5) | Ref | |||
| CT/CC | 99 (16.9) | 451 (18.5) | 0.93 (0.74–1.18) | 0.95 (0.73–1.24) | ||
| COL4A3: rs11884770 | 0.67 | 0.90 | ||||
| CC | 300 (51.1) | 1291 (53.0) | Ref | |||
| TC | 245 (41.7) | 935 (38.4) | 0.93 (0.65–1.31) | 0.84 (0.55–1.29) | ||
| TT | 42 (7.2) | 210 (8.6) | 1.12 (0.93–1.35) | 1.16 (0.95–1.43) | ||
| CTRB1: rs8056814 | 0.37 | 0.29 | ||||
| GG | 482 (82.1) | 1944 (79.8) | Ref | |||
| AA/AG | 105 (17.9) | 492 (20.2) | 0.90 (0.72–1.13) | 0.88 (0.68–1.13) | ||
| ADAMTS9/ADAMTS9-AS2: rs6795735 | 0.77 | 0.88 | ||||
| CC | 172 (29.3) | 727 (29.8) | Ref | |||
| TC | 297 (50.6) | 1163 (47.7) | 1.09 (0.89–1.35) | 1.12 (0.89–1.39) | ||
| TT | 118 (20.1) | 546 (22.5) | 0.94 (0.73–1.23) | 0.97 (0.72–1.3) | ||
| TIMP3: rs9621532 | 0.06 | 0.01 | ||||
| AA | 516 (87.9) | 2200 (90.3) | Ref | |||
| CA/CC | 71 (12.1) | 236 (9.7) | 1.29 (0.99–1.69) | 1.41 (1.05–1.88) | ||
| DNA repair/protein binding | ||||||
| RAD51B: rs8017304 | 0.44 | 0.85 | ||||
| AA | 240 (40.9) | 972 (39.9) | Ref | |||
| AG | 272 (46.3) | 1101 (45.2) | 0.87 (0.65–1.16) | 0.90 (0.66–1.24) | ||
| GG | 75 (12.8) | 363 (14.9) | 1.01 (0.83–1.22) | 1.07 (0.87–1.32) | ||
| NPLOC4/TSPAN10: rs9895741 | 0.19 | 0.09 | ||||
| GG | 240 (40.9) | 1067 (43.8) | Ref | |||
| AG | 259 (44.1) | 1037 (42.6) | 1.08 (0.89–1.32) | 1.17 (0.94–1.45) | ||
| AA | 88 (15.0) | 332 (13.6) | 1.18 (0.91–1.54) | 1.22 (0.91–1.64) | ||
| HSPH1/B3GALTL: rs9542236 | 0.02 | 0.02 | ||||
| TT | 177 (30.1) | 811 (33.3) | Ref | |||
| CT | 287 (48.9) | 1226 (50.3) | 1.09 (0.89–1.34) | 1.09 (0.87–1.36) | ||
| CC | 123 (21.0) | 399 (16.4) | 1.39 (1.08–1.79) | 1.40 (1.06–1.86) | ||
Model 1: HRs calculated using the Cox proportional hazards model adjusting for age, sex, education, smoking, BMI, drusen size grade at baseline, AREDS treatment and multivitamin supplement use. Each gene was analyzed separately. Eyes were used as the unit of analysis.
Model 2: HRs calculated using the Cox proportional hazards model adjusting for age, sex, education, smoking, BMI, drusen size grade at baseline, AREDS treatment, multivitamin supplement use, and all genetic variants. Eyes were used as the unit of analysis.
P for trend
n (%) refers to the number of eyes.
After further adjustments for all genes together in the same model (Table 1, model 2), higher risk of two-step drusen enlargement was associated with common and rare variants in CFH: rs1410996 and CFH R1210C, and variants in C3, VEGFA, ARMS2/HTRA1, TIMP3, and HSPH1/B3GALTL. The rare C3 variant, K155Q, conferred borderline risk (P = 0.06). A lower risk of progression was seen for the protective variant in the gene C2, but ABCA1 was no longer significant (P = 0.07), although the effect size was similar in both models (HR, 0.68–0.73). The other genetic variants were not associated with the progression in drusen size. The magnitude of the effects for the risk genotypes ranged from an increased risk of progression of drusen size of 27% for ARMS2/HTRA1 (HR, 1.27), to more than a four-fold higher risk of drusen size progression for the rare CFH R1210C variant (HR, 4.26). C2 was associated with a reduction in risk of 57% (HR, 0.43).
Table 2 displays the stepwise model of genetic factors related to drusen size progression. HRs represent risk per allele. After applying the stepwise selection criteria, 11 genetic variants in 8 genes were selected for inclusion in the model. These include CFH rs1061170 (Y402H), CFH rs1410996, CFH rs121913059 (R1210C), C2 rs9332739 (E318D), C3 rs2230199 (R102G), C3 K115Q, VEGFA, ARMS2/HTRA1 rs10490924, TIMP3 rs9621532, NPLOC4/TSPAN10, and HSPH1/B3GALTL rs9542236. HRs ranged from 1.16 to 5.85 for the risk variants, and the HR was 0.44 for the protective variant. These genetic variants, independently associated with drusen size progression, are in the complement, angiogenesis, immune, inflammatory, extracellular matrix, and DNA repair/protein binding pathways.
Table 2.
Stepwise Model of Genetic Factors Associated with Drusen Size Progression
| Drusen Size Progression | ||
|---|---|---|
| HR (95% CI)* | P Value | |
| Complement pathway | ||
| CFH Y402H: rs1061170 | 1.22 (1.02–1.43) | 0.03 |
| CFH: rs1410996 | 1.22 (1.01–1.48) | 0.04 |
| CFH R1210C: rs121913059 | 5.85 (2.44–14.06) | <0.001 |
| C2 E318D: rs9332739 | 0.44 (0.28–0.69) | <0.001 |
| C3 R102G: rs2230199 | 1.45 (1.26–1.67) | <0.001 |
| C3 K155Q: rs147859257 | 2.19 (0.98–4.88) | 0.05 |
| Angiogenesis pathway | ||
| VEGFA: rs943080 | 1.19 (1.04–1.36) | 0.01 |
| Immune inflammatory pathway | ||
| ARMS2/HTRA1 A69S: rs10490924 | 1.34 (1.17–1.54) | <0.001 |
| Extracellular matrix | ||
| TIMP3: rs9621532 | 1.52 (1.16–2.01) | 0.003 |
| DNA repair/protein binding | ||
| NPLOC4/TSPAN10: rs9895741 | 1.16 (1.02–1.32) | 0.02 |
| HSPH1/B3GALTL: rs9542236 | 1.21 (1.06–1.39) | 0.005 |
HRs for drusen size progression represent risk per allele, and are adjusted for age, sex, education, smoking, BMI, drusen size grade at baseline, AREDS treatment, multivitamin supplement use. Eyes were used as the unit of analysis.
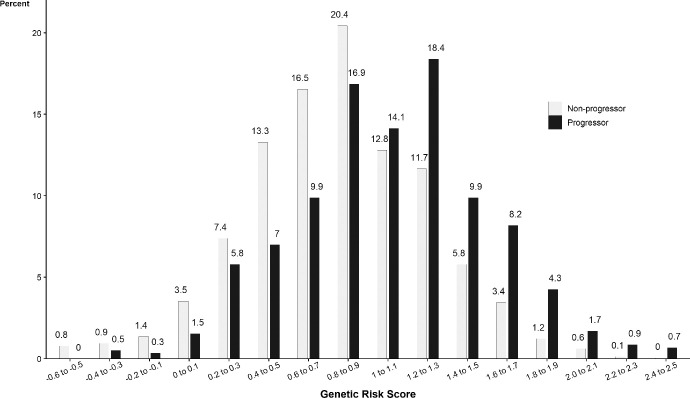
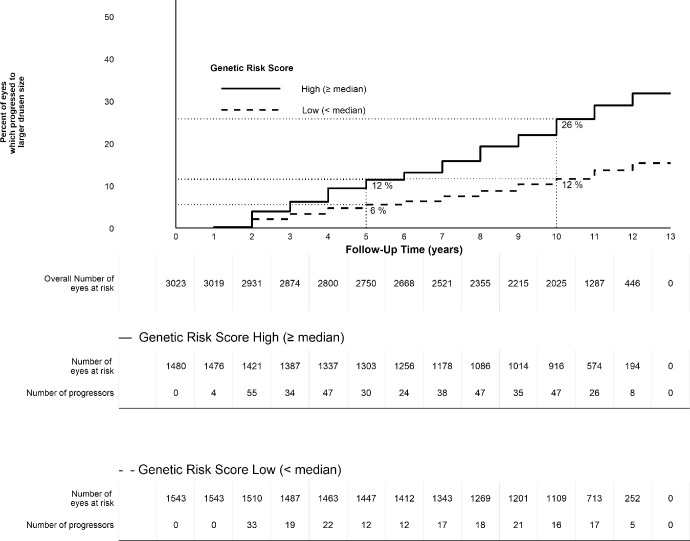
Table 3 displays the associations between drusen size progression, genetic susceptibility, and aMeDi score after adjustment for baseline demographic, behavioral, and ocular characteristics, as well as both genetic susceptibility and aMeDi diet in the same model. Genetic susceptibility and adherence to aMeDi diet were independently associated with drusen progression. Although smoking tended to be associated with drusen growth when adjusted only for age and sex, it was not significantly related in this multivariate model. The calculated genetic risk score ranged from –0.60 to 2.54. We rounded off the estimates and plotted their distribution for eyes that progressed and those that did not progress (Fig. 2). Eyes that progressed to larger drusen size tended to have higher genetic risk scores and nonprogressors tended to have lower genetic risk scores, although there was overlap. Eyes of participants with a higher genetic risk score had about a 2.7-fold increased risk per 1-unit increase in the score (HR, 2.68; 95% CI, 2.23–3.23; P < 0.001). Figure 3 shows the Kaplan-Meier curves for the probability of drusen progression depending on the category of the genetic risk score. For eyes of participants with a genetic risk score of less than the median, the probability of progression was 6% at 5 years and 12% at 10 years. For eyes of participants in the median or higher genetic risk score, the rates were higher: the 5-year rate of progression was 12% and the 10-year rate was 26%. A medium/high adherence to the aMeDi score with four or more components above the median tended to lower risk for drusen progression (HR for medium/high adherence 0.83; 95% CI, 0.68–0.99; P = 0.049). No significant interaction between genetic risk score and aMeDi score was detected.
Table 3.
Association Between Drusen Size Progression, Genetic Susceptibility, and aMeDi Score
| Progressors | Nonprogressors | |||
|---|---|---|---|---|
| Mean (SD) | Mean (SD) | HR (95% CI)* | P Value | |
| Genetic Risk score† | 1.06 (0.49) | 0.81 (0.45) | 2.68 (2.23–3.23) | <0.001 |
| aMeDi Score | n (%) | n (%) | ||
| Low (0–3) | 212 (21) | 796 (79) | Ref | |
| Medium–high (4–9) | 375 (19) | 1640 (81) | 0.83 (0.68–0.99) | 0.049 |
HRs calculated using Cox proportional hazards model adjusting for all covariates: age, sex, education, smoking, BMI, drusen size grade at baseline, total energy intake, AREDS treatment, multivitamin supplement use, genetic risk score (continuous), aMeDi score. For genetic score, HR is the change for a 1-unit increase. Eyes were used as the unit of analysis.
Gene score includes: 11 variants in 8 genes: CFH Y402H, CFH rs1410996, CFH R1210C, C2, C3 R102G, C3 K155Q, ARMS2/HTRA1 A69S, TIMP3, NPLOC4/TSPAN10, VEGFA and HSPH1/B3GALTL. Genetic risk score used as a continuous variable is based on the stepwise model presented in Table 2.
n (%) refers to the number of eyes.
Figure 2.
Percent of eyes with drusen progression (2 steps) or no drusen progression during the follow-up time, according to the Genetic Risk Score.
Figure 3.
Kaplan-Meier curves for the probability of eyes progressing to larger drusen size (2 steps) by follow-up time according to high or low Genetic Risk Score. The survival estimates were obtained from the Kaplan-Meier survival curves.
Discussion
Drusen are the major clinical signs of early AMD. Although the association between progression to advanced AMD and genetic variants is well-established, little is known about the impact of genetics and diet on drusen size progression. This study adds new information about earlier stages of AMD by assessing the influence of genes and a healthy diet on two-step drusen size progression over 13 years in a large cohort, controlling for demographic, behavioral, and ocular factors. When adjusting for all covariates and selecting the most highly associated genetic variants, risk alleles in seven genes increased the rate of drusen progression, and protective alleles in one gene significantly decreased the risk of drusen progression. Participants with a higher polygenic risk score were more likely to progress to a larger drusen size. Our study also highlights for the first time that higher adherence to the Mediterranean diet score (aMeDi score) was associated with a decreased risk of drusen size progression adjusting for genetic and other variables. Genetic susceptibility and adherence to aMeDi score were independently associated with drusen progression.
A previous analysis showed an association between genes and transition among consecutive stages of AMD based on the Clinical Age-Related Maculopathy Grading System with a scale of 1 to 5 from no AMD to advanced stages.6,19 The incidence of medium drusen among persons without any AMD at baseline was associated with CFH rs1061170 and ARMS2.20 A longitudinal analysis of a small subgroup of the AREDS sample (N = 75) did not find an association between a genetic score, including 19 common risk variants, and progression of drusen based on color photos assessed with an automated algorithm, possibly owing to low power.29 We have previously shown that carriers of the rare variant CFH R1210C have a higher drusen burden in the macular and extra-macular locations,26,30 but no previous studies have explored the impact of this high impact genetic variant or the high-risk rare C3 variant31 on drusen size progression. Both of these rare variants were shown to be associated with drusen growth in this prospective study.
Our study reports new findings and expands on previous results in several ways. We explored prospective associations of drusen size progression determined by a 2-unit increase in measured drusen size on an ordinal scale. We analyzed genetic variants including both common and rare variants, together with a healthy dietary pattern over 13 years. We evaluated a large cohort with a large number of drusen size progressors. These results highlight the role of genetics in the transition to larger drusen for both common and rare variants in various pathways: the complement pathway–CFH R1210C, C3 K155Q, and C2, and genetic variants in the angiogenesis, immune, and inflammatory pathway; extracellular matrix; and DNA repair pathways. Furthermore, we created a genetic risk score incorporating the most highly related variants after adjusting for other covariates, and demonstrated that participants with a higher polygenic risk score were at higher risk to have a two-step increase in drusen size, independent of other factors.
Our results support the impact of the complement pathway in the development of drusen. Dysregulation owing to genetic variants leading to uncontrolled activation of the alternative complement pathway could have an impact at the level of Bruch's membrane, and could lead to an impaired ability to dampen immune complex deposition. Factor H protein has been found as a molecular constituent of drusen.32,33 Results are consistent with our previous cross-sectional study in a different population, which showed that common variants in CFH are related to higher drusen area and volume based on automatic OCT measurements.34 The high-impact, rare CFH R1210C mutation compromises C-terminal factor H function, and this variant has been shown to be associated with a high drusen burden and earlier age of onset of AMD.26,30 Although rare, the CFH R1210C mutation was most highly associated with enlargement of drusen in these analyses, with almost a six-fold increase in the risk for drusen growth.
The common C3 R102G single nucleotide polymorphism influenced drusen growth and is reported to have functional consequences on the C3 protein. This variant generates the fast and slow electrophoretic haplotypes of C3 (C3F and C3S) showing a differential capacity to bind monocyte complement receptor C3F, which is the risk variant for AMD, and has been previously reported as associated with other autoimmune-mediated conditions.35–37 Drusen contain C3 and its activation products membrane attack complex. The higher impact rare C3 mutation, K155Q, results in resistance to proteolytic inactivation by CFH and CFI, implicating loss of C3 protein regulation and excessive alternative complement activation in AMD pathogenesis.31 This variant also tended to be associated with drusen enlargement. Evidence suggests that local inflammation and activation of the complement cascade contribute to the pathogenesis of drusen and AMD.
The classical pathway of the complement system may be implicated in drusen progression, because the protective allele for the C2 R102G variant was associated with a decreased risk of drusen progression. This variant is also known to be associated with a decreased risk of advanced AMD.27,38
Other than CFH and C3, the other genetic variant most significantly associated with drusen growth is the common variant in the gene locus ARMS2/HTRA1. In contrast with the complement pathway, the gene products of ARMS2/HTRA1 and their functional properties as well as the role of this gene locus in development of drusen and drusen size progression are less well-understood. These two genes at the chromosome 10q26 locus are in high linkage disequilibrium, which means that their association is higher than would be expected if unlinked or associated randomly, and it is not yet certain which gene product leads to AMD pathology. However, it is clear that this biologic pathway plays a role in nonadvanced stages of AMD, as shown in the current study of progression in drusen size, and previous studies with different study designs.19 Furthermore, the variant in ARMS2/HTRA1 was recently shown to be independently associated with drusen area and volume measured on OCT, as was CFH.34 Several studies have proposed that the loss of ARMS2 protein owing to this mutation may implicate mitochondrial dysfunction, which in turn may increase oxidative stress in the RPE layer.39,40 A study in 2017 found that ARMS2 was expressed in human monocytes and microglia cells and facilitated removal of cellular debris by local complement activation; the A69S variant resulted in ARMS2 deficiency, which could impair the removal of cellular debris at Bruch's membrane, possibly leading to the development of drusen.41 However, another study in 2017 presented support for the gene HTRA1 as the risk factor for AMD, based on protein expression in eye tissues.42
The VEGFA gene, which relates to the function of angiogenesis, cell growth, and vascular permeability, is associated with advanced AMD.43 However, the mode of action of VEGFA on drusen progression has not yet been explored. TIMP3 is a tissue inhibitor of metalloproteinases-3, an extracellular matrix protein. A mutation in this gene has been shown to disrupt the RPE membrane44 and in a study on induced pluripotent stem cell-derived RPE, a TIMP3 mutation was associated with deposits in the RPE membrane.45 Potential roles in drusen development for the genes in the DNA repair/protein-binding pathways, NPLOC4/TSPAN10 and HSPH1/B3GALTL, have not yet been explored.
In a previous report,46 the ABCA1 gene in the lipoprotein pathway was implicated in early AMD. ABCA1 protein has been shown to be expressed in the retina and RPE,47 and has also been synthetized and expressed in macrophages, which are the predominant cells involved in creating the progressive plaque lesions of atherosclerosis.48,49 It is possible that functional variants regulating the expression level of ABCA1 promote cholesterol efflux, decrease the activation of the inflammatory pathway in subretinal drusen, and result in less drusen accumulation. However, although the ABCA1 variant was associated with drusen progression individually, it was not retained in the stepwise model as a major factor in these analyses.
Although individual genetic variants and the genetic risk score were more significantly associated with drusen progression in these analyses, diet quality also tended to modify risk. The protective effects of eating a Mediterranean-type diet have been shown for different outcomes, such as overall mortality,50 cardiovascular events,51,52 cognitive decline,53 and more recently vision impairment, diabetic retinopathy,54 and AMD.13,14,55,56 Two of the AMD reports are prospective studies showing that this type of diet decreased the rate of progression from nonadvanced to advanced AMD—one in the US population reported in 201513 and the other confirming these results in a European cohort published in 2019.14 The biological basis of this healthy diet is based on the protective effects of a diet rich in nutrient-rich foods: fish providing polyunsaturated omega-3 fatty acids, and plant foods such as fruits, vegetables, legumes, olive oil, as well as nuts, providing antioxidant and anti-inflammatory vitamins and minerals such as vitamin B9, vitamin E, zinc, lutein, and zeaxanthin.7,11,12,57–59 Our previous study showed that higher levels of serum antioxidants, including vitamin C, lutein, and zeaxanthin, as well as higher fish intake were associated with lower levels of C-reactive protein, a marker for systemic inflammation.59
The dietary questionnaire was validated through a telephone-administrated 24-hour dietary recall on a subsample, as in other analyses of food frequency questionnaires. The same instrument was used for all individuals, providing rankings of dietary intake that are useful in association analyses. We used a well-known and validated method to assess diet and created a dietary pattern score. Our aMeDi score was developed by using sex-specific thresholds to better account for differences between men and women. Other strengths include standardized data collection, a long follow-up time, a large number of drusen size progressors, and data available for major dietary, and other behavioral confounders. Our analyses accounted for time to progression in each eye and included genetic variants in all pathways related to AMD. Dietary assessment was conducted before drusen progression, which decreased the likelihood of reverse causation and minimized the potential for dietary changes as a result of worsening disease. The phenotypic grading of drusen size was assigned without knowledge of risk factors or genotype. Our findings are applicable to an older Caucasian population in the US and may be relevant for similar populations in other countries.
In conclusion, this study presents new information which shows that genetic variants and a genetic risk score involving several biologic pathways significantly influenced a two-step increase in drusen growth, and that higher adherence to a Mediterranean-type diet may be associated with a reduced risk of drusen size progression over time. Modifiable factors such as a healthy dietary pattern could play a role in delaying progression of earlier stages of AMD, in addition to the known role of these healthy behaviors on progression to more advanced disease. Genetic predictors and diet quality were independently related to an increase in drusen size. Knowledge of nature and nurture, genetic susceptibility, and modifiable factors, may identify participants at a greater risk of early disease progression who could benefit from preventive measures.7
Supplementary Material
Acknowledgments
Supported in part by NIH R01-EY011309, R01-EY028602, R01-EY022445; American Macular Degeneration Foundation, Northampton, Massachusetts; and The Macular Degeneration Center of Excellence, University of Massachusetts Medical School, Department of Ophthalmology and Visual Sciences, Worcester, Massachusetts.
Disclosure: B.M.J. Merle, None; B. Rosner, None; J.M. Seddon, Laboratoires Thea (C), Gemini Therapeutics (R)
Amended May 18, 2020: Reference #39 was a duplicate of #34. It has been removed and the subsequent references and in-text citations renumbered.
References
- 1. Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012; 379: 1728–1738. [DOI] [PubMed] [Google Scholar]
- 2. Wong WL, Su X, Li X, et al.. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014; 2: e106–e116. [DOI] [PubMed] [Google Scholar]
- 3. Abdelsalam A, Del Priore L, Zarbin MA. Drusen in age-related macular degeneration: pathogenesis, natural course, and laser photocoagulation-induced regression. Surv Ophthalmol. 1999; 44: 1–29. [DOI] [PubMed] [Google Scholar]
- 4. Bird AC, Bressler NM, Bressler SB, et al.. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995; 39: 367–374. [DOI] [PubMed] [Google Scholar]
- 5. Klein R, Davis MD, Magli YL, Segal P, Klein BE, Hubbard L. The Wisconsin age-related maculopathy grading system. Ophthalmology. 1991; 98: 1128–1134. [DOI] [PubMed] [Google Scholar]
- 6. Seddon JM, Sharma S, Adelman RA. Evaluation of the clinical age-related maculopathy staging system. Ophthalmology. 2006; 113: 260–266. [DOI] [PubMed] [Google Scholar]
- 7. Seddon JM. Macular degeneration epidemiology: nature-nurture, lifestyle factors, genetic risk, and gene-environment interactions – the Weisenfeld Award Lecture. Invest Ophthalmol Vis Sci. 2017; 58: 6513–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996; 276: 1141–1146. [PubMed] [Google Scholar]
- 9. Saunier V, Merle BMJ, Delyfer M-N, et al.. Incidence of and risk factors associated with age-related macular degeneration: four-year follow-up from the ALIENOR Study. JAMA Ophthalmol. 2018; 136: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gopinath B, Liew G, Flood VM, Joachim N, Burlutsky G, Mitchell P. Combined influence of poor health behaviours on the prevalence and 15-year incidence of age-related macular degeneration. Sci Rep. 2017; 7: 4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seddon JM, Ajani UA, Sperduto RD, et al.. Dietary carotenoids, vitamins A, C, and E, and advanced age-related macular degeneration. Eye Disease Case-Control Study Group. JAMA. 1994; 272: 1413–1420. [PubMed] [Google Scholar]
- 12. Seddon JM, Cote J, Rosner B. Progression of age-related macular degeneration: association with dietary fat, transunsaturated fat, nuts, and fish intake. Arch Ophthalmol. 2003; 121: 1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merle BMJ, Silver RE, Rosner B, Seddon JM. Adherence to a Mediterranean diet, genetic susceptibility, and progression to advanced macular degeneration: a prospective cohort study. Am J Clin Nutr. 2015; 102: 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merle BMJ, Colijn JM, Cougnard-Grégoire A, et al.. Mediterranean diet and incidence of advanced age-related macular degeneration: the EYE-RISK Consortium. Ophthalmology. 2019; 126: 381–390. [DOI] [PubMed] [Google Scholar]
- 15. Yu Y, Wagner EK, Souied EH, et al.. Protective coding variants in CFH and PELI3 and a variant near CTRB1 are associated with age-related macular degeneration†. Hum Mol Genet. 2016; 25: 5276–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fritsche LG, Igl W, Bailey JNC, et al.. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016; 48: 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seddon JM, Rosner B.. Validated prediction models for macular degeneration progression and predictors of visual acuity loss identify high-risk individuals. Am J Ophthalmol. 2019; 198: 223–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delcourt C, Delyfer M-N, Rougier M-B, et al.. ARMS2 A69S polymorphism and the risk for age-related maculopathy: the ALIENOR study. Arch Ophthalmol. 2012; 130: 1077–1078. [DOI] [PubMed] [Google Scholar]
- 19. Yu Y, Reynolds R, Rosner B, Daly MJ, Seddon JM. Prospective assessment of genetic effects on progression to different stages of age-related macular degeneration using multistate Markov models. Invest Ophthalmol Vis Sci. 2012; 53: 1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joachim NDL, Mitchell P, Kifley A, Wang JJ. Incidence, progression, and associated risk factors of medium drusen in age-related macular degeneration: findings from the 15-year follow-up of an Australian cohort. JAMA Ophthalmol. 2015; 133: 698–705. [DOI] [PubMed] [Google Scholar]
- 21. Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001; 119: 1417–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Age-Related Eye Disease Study Research Group. The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study Report Number 6. Am J Ophthalmol. 2001; 132: 668–681. [DOI] [PubMed] [Google Scholar]
- 23. Fung TT, Rexrode KM, Mantzoros CS, Manson JE, Willett WC, Hu FB. Mediterranean diet and incidence of and mortality from coronary heart disease and stroke in women. Circulation. 2009; 119: 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trichopoulou A, Kouris-Blazos A, Wahlqvist ML, et al.. Diet and overall survival in elderly people. BMJ. 1995; 311: 1457–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seddon JM, Reynolds R, Yu Y, Rosner B. Three new genetic loci (R1210C in CFH, variants in COL8A1 and RAD51B) are independently related to progression to advanced macular degeneration. PloS One. 2014; 9: e87047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raychaudhuri S, Iartchouk O, Chin K, et al.. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011; 43: 1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maller J, George S, Purcell S, et al.. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006; 38: 1055–1059. [DOI] [PubMed] [Google Scholar]
- 28. Seddon JM, Silver RE, Kwong M, Rosner B. Risk prediction for progression of macular degeneration: 10 common and rare genetic variants, demographic, environmental, and macular covariates. Invest Ophthalmol Vis Sci. 2015; 56: 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoffman JD, van Grinsven MJJP, Li C, et al.. Genetic association analysis of drusen progression. Invest Ophthalmol Vis Sci. 2016; 57: 2225–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferrara D, Seddon JM.. Phenotypic characterization of complement factor H R1210C rare genetic variant in age-related macular degeneration. JAMA Ophthalmol. 2015; 133: 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seddon JM, Yu Y, Miller EC, et al.. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013; 45: 1366–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bora NS, Gobleman CL, Atkinson JP, Pepose JS, Kaplan HJ. Differential expression of the complement regulatory proteins in the human eye. Invest Ophthalmol Vis Sci. 1993; 34: 3579–3584. [PubMed] [Google Scholar]
- 33. Johnson PT, Betts KE, Radeke MJ, Hageman GS, Anderson DH, Johnson LV. Individuals homozygous for the age-related macular degeneration risk-conferring variant of complement factor H have elevated levels of CRP in the choroid. Proc Natl Acad Sci U S A. 2006; 103: 17456–17461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seddon JM, Dossett JP, Widjajahakim R, Rosner B. Association between perifoveal drusen burden determined by OCT and genetic risk in early and intermediate age-related macular degeneration. Invest Ophthalmol Vis Sci. 2019; 60: 4469–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007; 39: 1200–1201. [DOI] [PubMed] [Google Scholar]
- 36. Yates JRW, Sepp T, Matharu BK, et al.. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007; 357: 553–561. [DOI] [PubMed] [Google Scholar]
- 37. Zerbib J, Richard F, Puche N, et al.. R102G polymorphism of the C3 gene associated with exudative age-related macular degeneration in a French population. Mol Vis. 2010; 16: 1324–1330. [PMC free article] [PubMed] [Google Scholar]
- 38. Gold B, Merriam JE, Zernant J, et al.. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006; 38: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanda A, Chen W, Othman M, et al.. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007; 104: 16227–16232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakai S, Honda S, Matsumiya W, Miki A, Nakamura M. ARMS2 variants may predict the 3-year outcome of photodynamic therapy for wet age-related macular degeneration. Mol Vis. 2017; 23: 514–519. [PMC free article] [PubMed] [Google Scholar]
- 41. Micklisch S, Lin Y, Jacob S, et al.. Age-related macular degeneration associated polymorphism rs10490924 in ARMS2 results in deficiency of a complement activator. J Neuroinflammation. 2017; 14: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liao S-M, Zheng W, Zhu J, et al.. Specific correlation between the major chromosome 10q26 haplotype conferring risk for age-related macular degeneration and the expression of HTRA1. Mol Vis. 2017; 23: 318–333. [PMC free article] [PubMed] [Google Scholar]
- 43. Yu Y, Bhangale TR, Fagerness J, et al.. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011; 20: 3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jacobson SG, Cideciyan AV, Bennett J, Kingsley RM, Sheffield VC, Stone EM. Novel mutation in the TIMP3 gene causes Sorsby fundus dystrophy. Arch Ophthalmol. 2002; 120: 376–379. [DOI] [PubMed] [Google Scholar]
- 45. Galloway CA, Dalvi S, Hung SSC, et al.. Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc Natl Acad Sci U S A. 2017; 114: E8214–E8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Y, Reynolds R, Fagerness J, Rosner B, Daly MJ, Seddon JM. Association of variants in the LIPC and ABCA1 genes with intermediate and large drusen and advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011; 52: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Duncan KG, Hosseini K, Bailey KR, et al.. Expression of reverse cholesterol transport proteins ATP-binding cassette A1 (ABCA1) and scavenger receptor BI (SR-BI) in the retina and retinal pigment epithelium. Br J Ophthalmol. 2009; 93: 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmitz G, Kaminski WE, Porsch-Ozcürümez M, et al.. ATP-binding cassette transporter A1 (ABCA1) in macrophages: a dual function in inflammation and lipid metabolism? Pathobiol J Immunopathol Mol Cell Biol. 1999; 67: 236–240. [DOI] [PubMed] [Google Scholar]
- 49. Tang C, Liu Y, Kessler PS, Vaughan AM, Oram JF. The macrophage cholesterol exporter ABCA1 functions as an anti-inflammatory receptor. J Biol Chem. 2009; 284: 32336–32343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trichopoulou A, Costacou T, Bamia C, Trichopoulos D. Adherence to a Mediterranean diet and survival in a Greek population. N Engl J Med. 2003; 348: 2599–2608. [DOI] [PubMed] [Google Scholar]
- 51. Trichopoulou A, Bamia C, Trichopoulos D. Mediterranean diet and survival among patients with coronary heart disease in Greece. Arch Intern Med. 2005; 165: 929–935. [DOI] [PubMed] [Google Scholar]
- 52. Martínez-González MÁ, Corella D, Salas-Salvadó J, et al.. Cohort profile: design and methods of the PREDIMED study. Int J Epidemiol. 2012; 41: 377–385. [DOI] [PubMed] [Google Scholar]
- 53. Samieri C, Grodstein F, Rosner BA, et al.. Mediterranean diet and cognitive function in older age. Epidem. 2013; 24: 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sala-Vila A, Díaz-López A, Valls-Pedret C, et al.. Dietary marine ω-3 fatty acids and incident sight-threatening retinopathy in middle-aged and older individuals with type 2 diabetes: prospective investigation from the PREDIMED Trial. JAMA Ophthalmol. 2016; 134: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 55. Raimundo M, Mira F, Cachulo M da L, et al.. Adherence to a Mediterranean diet, lifestyle and age-related macular degeneration: the Coimbra Eye Study – report 3. Acta Ophthalmol (Copenh). 2018; 96: e926–e932. [DOI] [PubMed] [Google Scholar]
- 56. Hogg RE, Woodside JV, McGrath A, et al.. Mediterranean Diet Score and its association with age-related macular degeneration: the European Eye Study. Ophthalmology. 2017; 124: 82–89. [DOI] [PubMed] [Google Scholar]
- 57. Reynolds R, Rosner B, Seddon JM. Dietary omega-3 fatty acids, other fat intake, genetic susceptibility, and progression to incident geographic atrophy. Ophthalmology. 2013; 120: 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Merle BMJ, Silver RE, Rosner B, Seddon JM. Dietary folate, B vitamins, genetic susceptibility and progression to advanced nonexudative age-related macular degeneration with geographic atrophy: a prospective cohort study. Am J Clin Nutr. 2016; 103: 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seddon JM, Gensler G, Klein ML, Milton RC. C-reactive protein and homocysteine are associated with dietary and behavioral risk factors for age-related macular degeneration. Nutrition. 2006; 22: 441–443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.