Abstract
Purpose
Adiponectin is an insulin-sensitizing and anticarcinogenic hormone that is encoded by a gene on chromosome 3. Here, we analyzed the expression of adiponectin and its receptor Adipor1 in primary uveal melanoma (UM) with regard to the monosomy-3 status and clinical factors, as well as the physiological response of UM cells to adiponectin.
Methods
Immunohistochemistry was performed on the primary UM of 34 patients. Circulating melanoma cells (CMC) were isolated by immunomagnetic enrichment. Monosomy-3 was evaluated by Immuno-FISH. Gene expression was analyzed using the RNAseq data of The Cancer Genome Atlas study. Cultures of choroidal melanocytes and UM were established from the samples of two patients. The proliferative potential of the UM cell lines Mel-270 and OMM-2.5 was determined by immunocytochemistry, immunoblotting, cell cycle analysis, nucleolar staining, and adenosine triphosphate (ATP) levels.
Results
UM with monosomy-3 exhibited a lower immunoreactivity for adiponectin and Adipor1, which was associated with monosomy-3-positive CMC and the development of extraocular growth or metastases. Both proteins were more abundant in the irradiated tumors and present in the cultured cells. Gene expression profile indicated the impairment of adiponectin-mediated signaling in the monosomy-3 tumors. Adiponectin induced a significant decline in the ATP levels, Ki-67 expression, cells in the G2/M phase, and nucleolar integrity in UM cultures.
Conclusions
Adiponectin deficiency appears to enhance the metastatic potential of the UM cells with monosomy-3 and the termination of tumor dormancy. Counteracting insulin resistance and improving the serum adiponectin levels might therefore be a valuable approach to prevent or delay the UM metastases.
Keywords: uveal melanoma, monosomy-3, insulin resistance, adiponectin, Adipor1, circulating melanoma cells (CMC), nucleoli, tumor dormancy
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults, with an annual incidence rate of approximately six to seven cases per million people. Despite the advances in diagnosis and local tumor control, up to 50% of the patients may develop metastases predominantly in the liver within 5 to 10 years. Because of the lack of efficient therapies, UM metastasis results in an average survival time of less than 6 months after diagnosis.1–3
The most significant prognostic factor for UM is the loss of one copy of chromosome 3 (monosomy-3 [M3]) in the primary tumor cells, with the occurrence of metastatic disease almost exclusively in the patients having this condition.4–6 The latency in the development of metastases suggests that the UM cells have already started to disseminate at the time of initial diagnosis and remained dormant in the new microenvironment until the conditions became favorable for their growth.1,7,8 Because the metastases of UM occur mainly through the hematogenous route,9 dissemination of the primary tumor cells into peripheral blood is a prerequisite.7,10,11 Circulating melanoma cells (CMC) seem to arise early in the disease and are found in almost all patients with UM.8,12–14 Further differentiation such as the M3 status can therefore provide valuable information for the early identification of patients at high metastatic risk.15 Knowledge of the pathophysiological factors that favor the survival of M3-positive CMC and their micrometastases over such long timeframes would also be indispensable for the development of a preventive therapy.
A novel study has recently reported the association of insulin resistance with a more aggressive course of UM. In this study, patients with UM or choroidal nevus had significantly lower levels of the insulin-sensitizing hormone adiponectin in their serum compared with control subjects. The UM patients who developed systemic metastases also exhibited a significant decline in serum adiponectin compared with the patients who had no metastases during the follow-up. The authors therefore suggested that insulin resistance, which was associated with lower serum adiponectin levels, may promote a proangiogenic and antiapoptotic environment, contributing toward a more aggressive clinical course.16 The decrease in the body's sensitivity to insulin has indeed gained more attention in cancer research lately, because of its association with a higher risk for development of various tumors.17–19 However, insulin resistance is not widely recognized as an established prognostic factor for UM yet, and the molecular mechanisms underlying its involvement in the aggressive course of this disease are not known.
Adiponectin is a peptide hormone that can enhance insulin sensitivity mainly by inhibiting the release of glucose from the liver. Accordingly, the levels of circulating adiponectin are reduced in people with insulin resistance, type 2 diabetes, and obesity. Initially, adiponectin was considered to be produced exclusively by the mature adipocytes. However, later studies have demonstrated its expression at modest levels in several other tissues including human skeletal muscle, liver, cardiomyocytes, and brain.20–23 Interestingly, the serum levels of adiponectin are inversely correlated to the risk for development of obesity-associated malignancies such as colon, renal, endometrial, and postmenopausal breast cancers. Experimental studies have also demonstrated that adiponectin can exert anticarcinogenic effects by both directly binding the adiponectin receptors on tumor cells and limiting cell proliferation, as well as indirectly by improving insulin sensitivity and reducing inflammation in the affected tissues.23,24 Remarkably, the Adipoq gene encoding human adiponectin (NCBI Gene ID: 9370) is located on chromosome 3q27.3, a locus that has already been linked to type-2 diabetes and metabolic syndrome.20,23,24 Moreover, the nuclear peroxisome proliferator-activated receptor gamma (PPARG, Gene ID: 5468), which promotes the expression and secretion of adiponectin, and the adaptor protein APPL1 (Gene ID:26060), which directly interacts with the adiponectin receptors, are also encoded by genes on chromosome 3p25.2 and 3p14.3, respectively.25–28 However, it is not known yet whether the UM cells express adiponectin or its receptors, whether the presence of M3 leads to a decrease in tumor adiponectin levels, and whether adiponectin induces a physiological response in the UM cells.
To address these questions, we initially performed an immunohistochemical analysis of adiponectin and its major receptor Adipor124 on the primary tumors of 34 UM patients who underwent operation in our clinic. To validate these findings in a larger and independent cohort, we examined the mRNA expression levels of the major components of the adiponectin-mediated signaling in the RNAseq data of The Cancer Genome Atlas (TCGA) Study.29,30 We could also establish cultures from the primary tumor of two of our UM patients to visualize the adiponectin protein with regard to the M3 status in intact cells by an Immuno-FISH assay. In addition, we analyzed the outcomes of adiponectin treatment on the proliferative potential of the well-characterized UM cell lines Mel-270 and OMM-2.5, which were derived from the primary tumor and liver metastases, respectively, of a UM patient.31,32 Our results demonstrated significantly lower levels of the adiponectin protein and Adipor1 in the primary tumors having a higher extent of M3, which was associated with the prevalence of M3 in the CMC of these patients and the development of metastases or extraocular growth. In contrast, the irradiated tumors exhibited more adiponectin and Adipor1 compared with the native samples. The basal levels of adiponectin were significantly lower in the UM cells with M3 that were cultured in normal medium with serum. Treatment of the UM cell lines with adiponectin could in turn suppress the cell-cycle progression and proliferation, together with the impairment of adenosine triphosphate (ATP) levels and nucleolar integrity. Our results therefore provide novel insights into the possible mechanisms underlying the higher metastatic potential of the UM cells with M3 and the first evidence to the protective role of adiponectin in the maintenance of UM dormancy.
Methods
Patient Selection
A total of 34 consecutive patients with clinically localized UM presenting at the Department of Ophthalmology, University of Lübeck, Germany, between December 2009 and January 2018 were enrolled in the study. UM was diagnosed with clinical and ultrasound examination performed by a specialized ophthalmologist. The study was authorized by the local ethic committee of the University of Lübeck (File number: 10-200) and conforms to the guidelines of the Declaration of Helsinki as revised in Tokyo and Venice. All patients received an explanation about the nature and possible consequences of the study and gave informed consent before their inclusion.
To evaluate the metastatic status, patients received liver function tests (alkaline phosphatase, aspartate aminotransferase, alanine aminotransferase, bilirubin), ultrasound of the abdomen, as well as the computer tomography of the chest and abdomen. Standardized A and B scans (I3 eyecubed System-ABD; Ellex Inc, Sacramento, CA, USA) and ultrasound biomicroscopy (VuMax II; Sonomed Inc, NY, USA) were obtained to evaluate the size of the tumor and the exact intraocular localization and to document ciliary body involvement.
Analysis of Gene Expression in the UM Cohort of the TCGA Study
Data validation was performed by analyzing the mRNA expression of the genes of interest in the UM cohort of the TCGA study, which enrolled 80 patients.29,30 Gene expression data were downloaded from the Xena platform of the University of California Santa Cruz (https://xena.ucsc.edu/).33 The graphical summaries of the genomic alterations, clinical factors, and gene expression heatmaps were generated using the cBioPortal resource (https://www.cbioportal.org/).34,35 Kaplan-Meier curves were constructed using the UCSC Xena platform.
Immunohistochemistry (IHC) on the Primary Tumors
Samples of the primary tumor (n = 38) were obtained from 34 UM patients after biopsy, resection, or enucleation, fixed in 4.5% formalin, and embedded in paraffin. Sections with a thickness of 6 µm were collected onto Superfrost Ultra-Plus slides. Deparaffinization was performed in three changes of xylol and two changes of absolute ethanol, followed by a graded series of ethanol descending from 90% to 50% for 5 minutes each, and a final wash in triple distilled water for 5 minutes. Antigen retrieval was performed by incubating the deparaffinized sections in preheated 10 mmol/L sodium citrate buffer, pH 6.0, with 0.025% Tween 20 for 20 minutes in a steam-cooker. The slides were cooled to room temperature, rinsed three times for 10 minutes in phosphate-buffered saline solution (PBS), and incubated with sterile-filtered blocking buffer (3% bovine serum albumin (BSA) in 10 mmol/L Tris-HCl, pH 7.5, 120 mmol/L KCl, 20 mmol/L NaCl, 5 mmol/L ethylenediamine tetra-acetic acid (EDTA), 0.1% Triton X-100) supplemented with 5% goat serum for 30 minutes at room temperature. Sections were then incubated with the rabbit primary antibodies against adiponectin (Abcam, Cambridge, UK; ab62551; 1:150 dilution in blocking buffer), Adipor1 (Abcam; ab126611; 1:50 dilution in blocking buffer), or BAP1 (Abcam; ab199396; 1:20 dilution in blocking buffer) overnight at 4°C. The negative controls were incubated with the blocking buffer alone. After rinsing the sections three times for 5 minutes with PBS, sections were incubated in freshly prepared 3% hydrogen peroxide solution in PBS for 15 minutes, rinsed twice with PBS, and incubated with horseradish peroxidase (HRP)–conjugated goat anti rabbit secondary antibodies (Jackson Immunoresearch, Cambridgeshire, UK; 111-035-003; 1:250 in blocking buffer) for 1 hour. Sections were rinsed three times for 5 minutes with PBS followed by 10 minutes’ incubation with the HRP green substrate that was freshly prepared by mixing 90 µL of HRP-Green chromogen with 1 mL of HRP-Green buffer as instructed by the manufacturer (42 Life Sciences, Bremerhaven, Germany) and washing for 5 minutes in triple-distilled water. Nuclei were counterstained with nuclear fast red for 10 minutes. The slides were then briefly rinsed in triple-distilled water for 1 minute, dehydrated for 30 seconds each in a graded series of ethanol (75%–96% 2 × 100%) followed by two times for 5 minutes in xylol, and mounted in nonaqueous medium.
Analysis of IHC Intensity by Image Deconvolution
For each patient, images of the entire tumor area were acquired under 200× magnification by light microscopy (Leica, Wetzlar, Germany). Image deconvolution was performed using the Fiji software (version 2.0.0-rc-54/1.51g) to separate the layers of nuclei, pigmentation, and IHC reaction with minimal overlap and background. The parameters for the deconvolution were optimized by initially selecting the regions of interest that exhibited only the pure color of each layer. The red (R), green (G), and blue (B) values that we could optimize for the deconvolution of our IHC samples were as follows: Magenta for nuclei (R1: 0.482, G1: 0.719, B1: 0.501); green for IHC (R2: 0.776, G2: 0.501, B2: 0.382); brown for the pigmentation (R3: 0.446, G3: 0.616, B3: 0.649). The gray value of the IHC layer was inverted so that the regions with a strong immunoreactivity would appear lighter and acquire higher pixel intensities compared with the background. The tumor area was then circumscribed on the original image, and the integrated density (area × intensity) of the selected region was determined by redirecting the measurement to the inverted IHC image. Deconvolution was performed by these user-defined values on all the images of a tumor. The sum of the integrated densities was then divided by the total area to determine the mean IHC-intensity for each tumor. The mean intensity of the negative control was subtracted from the intensity of all tumors. The total number of images quantified for adiponectin and Adipor1 were 2272 and 2568, respectively.
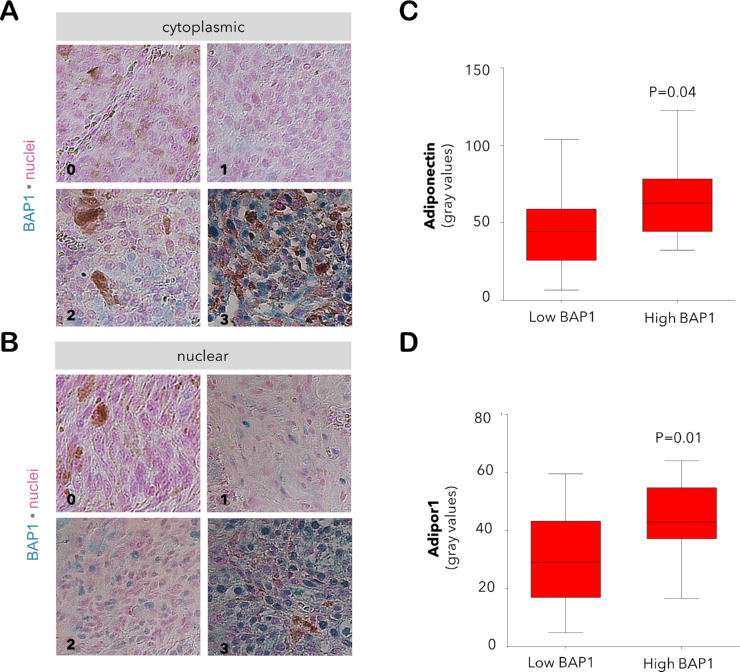
Scoring of BAP1 Intensity
Images of the entire tumor area were acquired under 200× magnification by light microscopy (Leica). The immunoreactivity for BAP1 in the cytoplasm and nucleus was graded as described36 using a scale of 0 to 3 (0: positive staining in less than 10% of cells; 1: positive staining in 11%–33% of cells; 2: positive staining in 34%–66% of cells; 3: positive staining in more than 67% of cells). Tumors with the grades of 0 and 1 were collectively defined as “low BAP1” samples whereas the tumors with the grades of 2 and 3 were assigned to the “high BAP1” group.
Immunomagnetic Enrichment of CMC
CMC were isolated from whole blood within 3 hours of blood collection as described.14 Isolated cells were processed as cytospins and stored at −20°C until use.
Immuno-FISH
Frozen cytospins that were aged for 24 hours at room temperature were processed for Immuno-FISH as described,15 using the primary mouse anti-melanoma-associated chondroitin sulfate proteoglycan (Abcam; ab78284; 1/5 in IHC-blocking buffer) or rabbit anti-adiponectin (Abcam, 1/10 dilution in blocking buffer) antibodies and Alexa 488-conjugated goat anti-mouse (ThermoFisher, Waltham, MA, USA; A-11001; 1:100 in blocking buffer) or (ab150077, 1:100 in blocking buffer). The incubation with the secondary antibodies and all the subsequent steps for post-fixation, denaturation, and hybridization were performed under protection from light. The CEP3 probe (Abbott GmbH, Wiesbaden, Germany) was prepared as instructed by the manufacturer and applied on to the cells at a volume of 4 µL/cytospin. The nuclei were counterstained with 0.5 µg/mL DAPI in PBS for 10 minutes. Cells were mounted in Mowiol, and analyzed by fluorescence microscopy (Leica DMI 6000 B) using the appropriate filter sets (A4: Ex: 360/40, Em: 470/40 nm; L5: Ex: 460/40, Em: 527/30 nm; Cy3: Ex: 545/30, Em: 610/75 nm). Images were acquired using a monochrome digital camera (DFC 350 FX; Leica) attached to the microscope and the Leica Application Software (Advanced Fluorescence 2.3.0, build 5131). Intensity of the adiponectin immunoreactivity in the cells was quantified using the Fiji software by circumscribing the cytoplasm (omitting the nucleus) and correcting for background by subtracting the mean intensity of the negative controls.
For the Immuno-FISH analysis on the paraffin sections of primary tumor samples, sections were deparaffinized, processed for antigen retrieval, and incubated in blocking buffer as described above for IHC, followed by the overnight incubation with monoclonal rabbit antibodies against Melan-A (Abcam, ab210546; 1:100 in blocking buffer) at 4°C and Alexa 488-conjugated goat anti-rabbit secondary antibodies (Abcam, 1:100 in blocking buffer) for 1 hour under protection from light. The post-fixation, denaturation, hybridization, washing, and counterstaining of the nuclei were then performed as described,15 with the extension of hybridization to 72 hours and using 10 µL probe/cm2.
Assessment of M3
The copy number of chromosome 3 in the tumor samples was determined in the overlay images of cells positive for Melan-A. Signals on the retina of 3 patients were also quantified as a positive control for diploidy. Cells with only one distinct signal or a doublet signal (two signals that were separated by a distance of less than twice the diameter of a signal) were defined as being M3-positive. Quantification of the signals was performed by determining both the percentage of M3-positive cells within an area and the chromosomal index as described.37 For the former method, the percentage of cells with M3 and two or more signals within a given area was calculated in a minimum of 203 nonoverlapping nuclei except for one patient with a very small sample (n = 88 quantified nuclei). The chromosomal index was determined by counting the total hybridization signals and dividing by the number of nuclei in a given area. The median results of both quantifications were calculated. Tumors that had a percentage of M3-cells that was equal to or above the median or a chromosomal index equal to or below the median were given the score 1. Only the tumors that received the score 1 for both parameters were classified as high M3 tumors. Samples that received the score 0 with both methods were classified as low M3 and the tumors that received the score 1 with only one method were classified as intermediate M3. For the subsequent statistical analysis, the low and intermediate M3 tumors were combined and defined as “low M3 tumors.”
Culture of Human Choroidal Melanocytes
Isolation of human choroidal melanocytes from adult eyes was performed as described38 with slight modifications. Briefly, the enucleation samples of two patients who underwent operation in our clinic were cut into halves toward the optic nerve, and the choroid was removed together with the retinal pigment epithelial (RPE) layer from the sclera. The isolated choroid-RPE tissue was rinsed twice for 10 minutes in Hank's buffered saline solution (HBSS) with penicillin/streptomycin and incubated in 0.05% Trypsin-EDTA in HBSS for 1 hour at 37°C to release the RPE cells. The choroid samples were then rinsed with HBSS and incubated in 0.25% Trypsin/EDTA overnight at 4°C. The isolated cells were passed through a 70-µm cell strainer, spun in a centrifuge at 200g for 5 minutes, and seeded in uncoated T25 tissue culture flasks prefilled with normal UM medium (RPMI 1640 [Biochrom, Berlin, Germany] supplemented with 10% fetal bovine serum, 2 mmol/L L-Glutamine, 100 units/mL penicillin, 100 µg/mL streptomycin). The remaining choroid tissue was incubated further with 1 U/mL dispase in DMEM/F-12 (StemCell Technologies, Köln, Germany) at 37°C for 3 hours, with the collection of the dissociated cells every hour. The collected cells were rinsed as described above and pooled with the initially seeded cells in the T25 flasks. Cells were allowed to grow for 3 to 4 days, after which the medium was replaced twice weekly. Subculturing or harvesting the cells for the experiments was performed by trypsinization for 8 to 10 minutes followed by centrifugation at 300g for 8 minutes. Cells in the passages 1 and 2 were used for the experiments.
Culture of the Primary UM Cells
UM cultures were established from the enucleation samples of two patients. Approximately one-half of each tumor sample was fixed in formalin, embedded in paraffin and processed for IHC. The remaining tissue was washed two times in cell culture-grade 1× PBS (Biochrom) with penicillin/streptomycin and dissociated into single cells using a neural tissue dissociation kit with papain (Miltenyi Biotec, Bergisch Gladbach, Germany) following the manufacturer's instructions. The cells were seeded into a 25-cm2 culture flask and grown in UM medium at 37°C with 5% CO2 with the replacement of medium by approximately 80% every 2 to 3 days. Harvesting the cells for subculturing or the experiments was performed by incubation in dispase (1 U/mL in DMEM/F-12) at 37°C until cell detachment was observed under a microscope. Cytospins for Immuno-FISH were prepared by spinning the cells on to Superfrost Ultra Plus slides (1000–3000 cells/slide) at 135g for 8 minutes using a cytocentrifuge (Tharmac, Waldsolms, Germany) followed by air-drying overnight and storing at −20°C. Cells in passage 2 were used for the experiments.
Culture of UM Cell Lines
The primary (Mel-270) and metastatic (OMM-2.5) UM cell lines, which have been characterized extensively,31,32 were kindly provided by Martine J. Jager (Leiden University Medical Center, Leiden, The Netherlands). Cells were grown under normoxic conditions at 37°C with 5% CO2 in UM medium and passaged twice weekly by trypsinization.
For the experiments, cells at passages 21 to 29 were collected by trypsinization for 2 to 3 minutes, seeded on to 8-well slides (Sarstedt, Nümbrecht, Germany, 104 cells/well), 96-well plates (8.103 cells/well), 48-well plates (8.104 cells/well), or 6-well plates (2.105 cells/well) and allowed to attach overnight. For the treatments involving serum deprivation, cells were washed twice for 10 minutes with serum-free medium before introducing the test medium. Recombinant human adiponectin (Peprotech, Hamburg, Germany; 450-24) was reconstituted in sterile triple-distilled water. Cells were incubated in normal UM medium with 10% fetal bovine serum (FBS) or serum free medium ± adiponectin (30 µg/mL) for 1 day.
Immunocytochemistry
Cells grown in 8-well slides were fixed in 2% paraformaldehyde (PFA)-PBS for 10 minutes followed by 4% PFA-PBS for a further 10 minutes, rinsed three times in PBS and incubated with the IHC-blocking buffer for 30 minutes. The primary antibodies against Melan-A (1:200), HMB-45 (Abcam; ab787; 1:200), p-Mel (Santa Cruz, Heidelberg, Germany; sc-377325, 1:200), MITF (Merck, Darmstadt, Germany; AB4139, 1:200), adiponectin (1:200), Ki-67 (Abcam; ab15580; 1:500), and YAP (Abcam; ab52771; 1:200) were diluted in the IHC-blocking buffer. For the detection of Adipor1 (1:200) and Adipor2 (Abcam; ab189446; 1:50) on the cell surface, blocking the cells and dilution of the antibodies were performed using 3% BSA-PBS. The cells were incubated with the primary antibodies overnight at 4°C followed by the Cy3-conjugated anti-rabbit (Jackson Immunoresearch; 111-165-003; 1:800) or Alexa 488-conjugated anti-mouse antibodies (1:100) in the corresponding blocking buffer for 1 hour at room temperature protected from light. Nuclei were counterstained with DAPI (0.5 µg/mL in PBS) for 10 minutes, and the cells mounted in Mowiol were visualized by fluorescence microscopy (Leica). Images were acquired with a monochrome digital camera using the Advanced Fluorescence software (Leica).
The mean intensity of the nuclear Ki-67 staining was measured with a semi-automated method using the Fiji software. The non-overlapping nuclei that lie completely within a DAPI-image were selected automatedly by adjusting the threshold and analyzed by redirecting the measurement to the Ki-67 image. The mean intensity of the cells that were visually recognized as negative in the Ki-67 image was taken as the cutoff. The percentage of (Ki67-positive)-cells with a mean intensity above the cut-off value was calculated on a minimum of 205 cells per group.
Cell-Cycle Analysis
Analysis of cell cycle phases was performed by measuring the integrated density of DNA in the fluorescence microscopy images of the immunostainings as described39 using the Fiji software. A minimum of 525 nonoverlapping nuclei that completely lie within the image were quantified for each group. Histograms of DNA content in the cell population were generated to visualize the distribution of the integrated densities. The cutoff values for the cell cycle phases were established from the histogram of the cells that were treated with normal medium supplemented with 10% FBS. The percentages of cells within each phase was then calculated for all the groups and presented as the mean ± SD of 3 or 4 independent experiments for the OMM-2.5 and Mel-270 cells, respectively.
Silver Staining of AgNOR
NOR silver staining was performed as described40 with slight modifications. Briefly, cells grown in 8-well slides were fixed with 2% PFA-PBS for 10 minutes, rinsed briefly in PBS and distilled water, and incubated in a freshly prepared solution of one volume of 2% gelatin in 1% formic acid and two volumes of 50% silver nitrate for 12 minutes at 37°C. The slides were rinsed three times for 1 minute in triple-distilled water, mounted in Mowiol, and observed by light microscopy (Leica) at a magnification of 200×. Quantification of the AgNOR area was performed in a minimum of 202 nuclei per group using the Fiji software in a semi-automated manner, by converting the images to 16-bit, background subtraction, setting the threshold for the selection of nucleoli, automated quantification of the total nucleolar area, and the division of this value by the total nuclei number in the analyzed region.
Immunoblotting
The Mel-270 and OMM-2.5 cells that were grown in 48- or 6-well-plates were washed twice with ice-cold PBS, placed on ice, and lysed in ice-cold cell lysis buffer (50 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 2 mmol/L EDTA, 1% NP-40 [v/v], 0.1% SDS, 0.5% sodium deoxycholate, 50 mmol/L NaF) supplemented with protease (Sigma-Aldrich, Munich, Germany; 1/100) and phosphatase inhibitors (Sigma-Aldrich, 1/100) immediately before use (50 µL/well for 48-well plates, 100 µL/well for 6-well plates) for 15 minutes under gentle agitation on a shaker. Samples of primary tumors obtained from two patients were freshly frozen in liquid nitrogen, whereas the other halves of the tissues were fixed in formalin and processed for IHC. Frozen tissues were disrupted and homogenized for 2 minutes in the lysis buffer using the Tissue Lyser LT (Qiagen, Hilden, Germany), followed by an incubation for 15 minutes on ice under gentle agitation. Cell and tissue lysates were cleared by centrifugation at 12,000g, 4°C for 20 minutes (Sigma 2-16PK; Hettich, Tuttlingen, Germany). The supernatants were collected and stored at −80°C if not used immediately. Protein concentration was determined by the bicinchoninic acid (BCA) assay (ThermoFisher; 23225). Electrophoresis was performed by mixing the samples with an equal volume of 2X Laemmli buffer, denaturing at 95°C to 99°C for 5 minutes and running in 4% to 10% TGX stain-free polyacrylamide gels (Bio-Rad, Munich, Germany) under denaturing and nonreducing conditions with 10 µg of protein/well. After electrophoresis, protein loading in the gels was visualized using the ChemiDoc MP stain-free imaging (Bio-Rad) by placing the gels directly on to the imaging tray with 2 to 3 mL distilled water underneath and activating with ultraviolet light for 5 minutes. The gels were then equilibrated in blotting buffer (48 mmol/L Tris, 39 mmol/L Glycine, 10% methanol [v/v], 0.04% SDS [w/v]) for 10 minutes and transferred onto methanol-activated polyvinylidene fluoride (PVDF) membranes by semi-dry blotting (Biotec-Fischer, Reiskirchen, Germany) at a constant current of 0.8 mA/cm2 for 1 hour. Protein transfer on to the membranes was visualized by the ChemiDoc MP stain-free system. Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline solution with 0.1% Tween-20 for 1 hour under gentle agitation followed by the incubation with rabbit primary antibodies against adiponectin (1:500), Ki-67 (1:2000), YAP (1:1000), Adipor1 (1:250), Melan-A (1:5000) or GAPDH (Abcam, ab9485, 1:2000) diluted in blocking buffer overnight at 4°C. After three washes of 5 minutes with Tris-buffered saline solution with 0.1% Tween-20, the membranes were incubated with the StarBright Blue 700 goat anti-rabbit antibodies (Bio-Rad;12004161; Exmax/Emmax = 470 nm/700 nm, 1:1000 in blocking buffer) or HRP-conjugated goat anti-rabbit antibodies (1:1000 in in blocking buffer) for 1 hour at room temperature. The membranes were washed as described above, and signal detection by enhanced chemiluminescence was performed by incubating the blots treated with HRP-conjugated antibodies for 2 minutes in a mixture of 10 mL of 0.025% (w/v) Luminol - 0.1 mol/L Tris-HCl (pH 8.6) with 3 µL 30% H2O2 and 1 mL 0.11% p-hydroxycoumaric acid–dimethyl sulfoxide (w/v) that was prepared shortly before use. Imaging of the enhanced chemiluminescence and fluorescent blots was performed by using the ChemiDoc MP system. The gray values of the fluorescent blot were inverted to obtain dark signals against a clear background.
ATP Assay
ATP levels were measured by a luminescence assay as instructed by the manufacturer (Abcam, ab 113849). Briefly, the Mel-270 and OMM-2.5 cells were seeded into 96-well plates at a density of 8000 cells/well (100 µL/well), allowed to attach overnight, rinsed twice with serum-free medium, and incubated with or without 30 µg/mL adiponectin in serum-free medium in triplicates for 8 to 10 hours. The kit reagents were brought to room temperature approximately 30 minutes before use. The assay is based on the lysis of cells followed by the incubation with a substrate that emits light in the presence of ATP. Because the lysis buffer is highly alkaline, protein quantification is not recommended by the manufacturer as a control for equal cellular material. Phase contrast images of the center of each well were therefore acquired using an inverse microscope (Leica) at 50× magnification as a control for cell density, which was quantified using the Fiji software. Cells were lysed in the detergent solution (50 µL/well) by shaking vigorously on an orbital shaker at 600 to 700 revolutions per minute (rpm) for 5 minutes, followed by the incubation with the substrate (50 µl/well) for 5 minutes at 600 to 700 rpm under light protection. A group of control wells was incubated without the substrate. The plates were left steady for 10 minutes under light protection for dark adaptation and luminescence was measured using a multi-mode microplate reader (SpectraMax i3x; Molecular Devices, Munich, Germany). After background subtraction, the mean luminescence of three wells from each group was normalized to the mean cell density of the corresponding group. Results were expressed as percentage, taking the values of the untreated group as 100%. Data represent the mean ± SEM of three independent experiments.
Statistical Analysis
The data sets were analyzed using the NCSS statistical software (Version 8.0.13; NCSS, Kaysville, UT, USA) under Windows 7. Logistic regression analysis was performed by taking the absence of a given condition as the reference (baseline) value. Mann-Whitney U test was performed to evaluate the association of gene expression with the binary variables. Kruskal-Wallis test was applied when the parameters could be categorized into three or more subgroups. Barnard's exact test was performed to analyze the proportion of categorical variables with respect to the development of metastases or extraocular growth. Data from cell culture experiments and the association of metastases with the continuous variables were analyzed by non-paired two-sided t-testing, assuming equal variance among the samples. P values less than 0.05 were considered as significant.
Results
Lower Levels of the Adiponectin and Adipor1 Proteins in the Primary UM Samples with Prevalent M3
The expression of adiponectin and Adipor1 at the protein level was initially analyzed in 38 primary UM samples obtained from 34 patients who underwent operation in our clinic. Two samples from different time points (biopsy, endoresection, or enucleation) were available from four of these patients. Liver metastases were present in one patient at the time of presentation, whereas all the remaining patients were diagnosed with primary UM without metastases or extraocular growth. Irradiation was performed before the acquisition of 20 of these samples. The clinical data of the patients and the histologic characteristics of these tumors are presented in Table 1.
Table 1.
Baseline Characteristics of the UM Patients and Tumor Samples Analyzed for Protein Expression
| Patients (n = 34) | |
|---|---|
| Sex | |
| Female | 16 (47.1%) |
| Male | 18 (52.9%) |
| Age in years, mean (SD) | 66.88 (11.6) |
| Tumor location | |
| Ciliary body | 2 (5.9%) |
| Choroid | 32 (94.1%) |
| Tumor size in mm, Mean (SD) | |
| Base 1 | 12.12 (4.1)* |
| Base 2 | 12.53 (4.6)* |
| Prominence | 7.58 (4.3)* |
| Tumor samples (n = 38) | |
| Sample acquisition | |
| Biopsy | 2 (5.3%) |
| Resection | 15 (39.5%) |
| Enucleation | 21 (55.3%) |
| Irradiation | |
| No | 18 (47.4%) |
| Yes | 20 (52.6%) |
| Prominent cell type | |
| Spindle | 5 (13.2%) |
| Mixed | 20 (52.6%) |
| Epithelioid | 13 (34.2%) |
| M3-positive cells | |
| Median | 30.0% |
| Min–Max | 8.8%–85.5% |
| Chromosome 3 index | |
| Median | 1.0 |
| Min–Max | 0.7–1.6 |
| M3 status | |
| Low | 13 (34.2%) |
| Intermediate | 9 (23.7%) |
| High | 16 (42.1%) |
| M3 status in non-irradiated tumors | 18 |
| Low | 4 (22.2%) |
| Intermediate | 3 (16.7%) |
| High | 11 (61.1%) |
| M3 status in irradiated tumors | 20 |
| Low | 9 (45.0%) |
| Intermediate | 6 (30.0%) |
| High | 5 (25.0%) |
SD, Standard deviation; M3, monosomy-3.
Data missing from one patient.
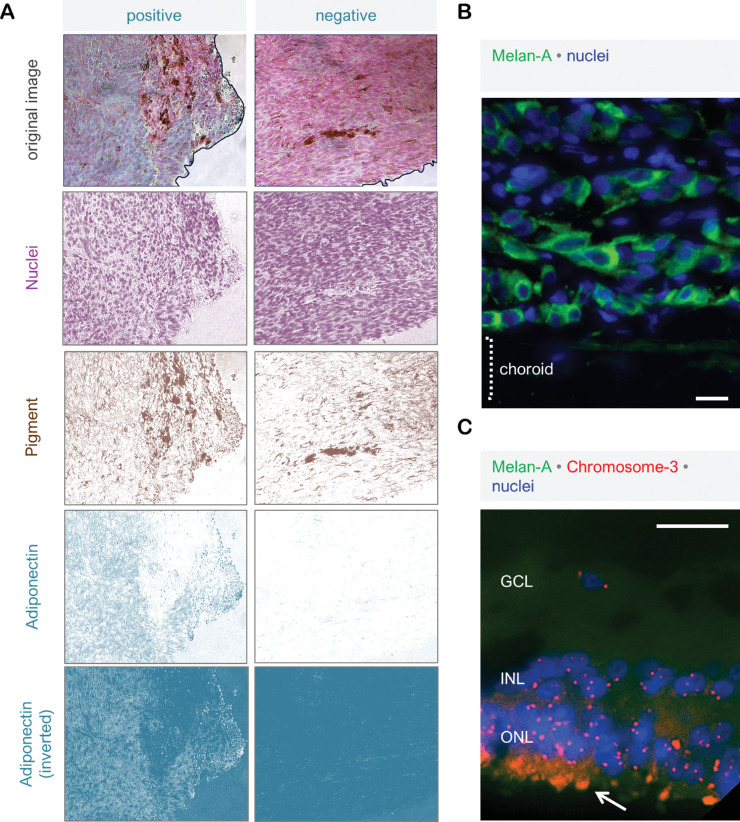
Immunohistochemistry (IHC) for adiponectin and Adipor1 on the paraffin sections of these samples revealed positive signals in all of the tumors. However, the staining intensity exhibited a very nonuniform pattern within some tumors, which complicated the subjective scoring of such samples. For a representative and reproducible quantification of the immunoreactivity, we therefore acquired the images of the entire tumor area from all the samples under 200x magnification and developed an objective quantification approach by deconvoluting the images, so that the IHC-signals would be separated from the nuclear staining and pigmentation with minimal overlap and background. The integrated density (Intensity × Area) of the IHC-layer was then measured on all the images of a given tumor and divided by the entire tumor area to determine the mean intensity of each sample (Fig. 1a). The extent of M3 in these tumors was analyzed by Immuno-FISH, which was optimized for the detection of chromosome-3 in the cells specifically expressing the melanoma marker Melan-A (Figs. 1b and 1c).
Figure 1.
Objective quantification of the immunohistochemical staining and the assessment of M3 status. (a) IHC on the paraffin sections of primary UM was performed using HRP-conjugated secondary antibodies and the HRP-green substrate, which yields a blue-green reaction product (left panel). The right panel demonstrates the negative control of the same tumor, which was incubated without the primary antibodies. Nuclei were counterstained with nuclear fast red. For each patient, light microscopy images of the entire tumor area were acquired under 200 × magnification. Image deconvolution was then performed using the Fiji software to separate the layers of nuclei, pigmentation, and IHC reaction with minimal overlap and background. The gray value of the IHC layer (as demonstrated for adiponectin) was inverted so that the regions exhibiting a stronger immunoreactivity would appear lighter and acquire higher pixel intensities compared to the background. The tumor area was then circumscribed on the original image (as demonstrated by the black lines), and the integrated density (area × intensity) of the selected region was determined by redirecting the measurement to the inverted IHC image. This approach enabled the measurement of IHC intensity even in the samples that exhibited a very weak immunoreactivity, such as the negative control in the right panel. The mean IHC-intensities of the circumscribed areas in the positive and negative stainings were measured as 44.728 and 1.975 gray values, respectively, using this method. (b) Melan-A was selected as a suitable marker for the distinction of UM borders from the adjacent tissues by fluorescence microscopy. This marker was used in the subsequent Immuno-FISH assays to ensure that the nuclei that were quantified for chromosome 3 belonged to the UM cells. (c) Immuno-FISH for chromosome 3 was also performed on the retinal tissue of three enucleation samples as a positive control for diploid cells. Arrow indicates the autofluorescence in photoreceptor outer segments. GCL, ganglion cell layer; INL, inner nerve layer; ONL, outer nerve layer. Scale bars in (b) and (c) = 25 µm.
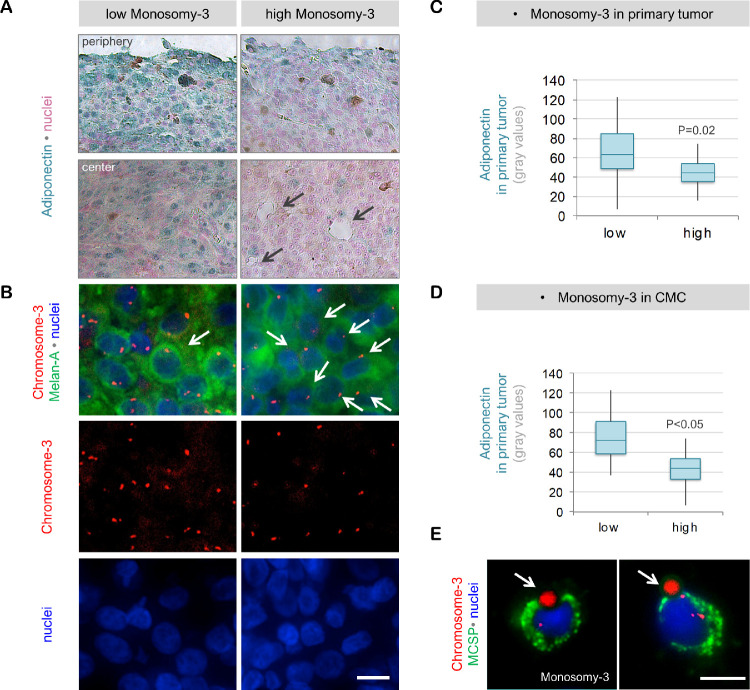
Quantification of the IHC for adiponectin demonstrated the presence of this protein at significantly lower levels in the tumors that were classified as having a high degree of M3 (n = 16), compared to the tumors with a low to moderate degree of M3, which were collectively termed as “low M3” hereafter (n = 22; P = 0.02, please see the Methods for a detailed description of the M3 classification). CMC analysis was performed in 33 of the 34 patients and yielded a positive result for 90,9% (n = 30 of 33) of the patients. For analyzing the association of CMC parameters with the expression of adiponectin or Adipor1 in the patients with two tumor samples (n = 4), only the values quantified in the first tumor sample were taken. The Immuno-FISH assay failed to reveal clear signals for chromosome 3 in the CMC of two patients. Of the remaining cases (n = 31), patients with a higher prevalence of M3 in their CMC (n = 15) also had significantly less adiponectin in their primary tumor compared with the patients who were CMC negative or had a lower percentage of CMC with M3 (n = 16; P < 0.05; Fig. 2). In contrast, the irradiated tumors (n = 20) exhibited a stronger immunoreactivity for adiponectin (median 64.4; mean ± SD 69.0 ± 23.6) compared to the native samples (n = 18; median 42.2; mean ± SD 43.6 ± 19.3; P = 0.01; Fig. 2a).
Figure 2.
Adiponectin levels in the primary UM samples with regard to the M3 status. (a) IHC for adiponectin demonstrated a strong and relatively homogenous distribution of this protein in both the margins and the center of a primary UM, which was classified as having „low M3“ (obtained from Patient 3, male, 67 years). In contrast, the “high M3” tumor (derived from patient 14, female, 48 years) exhibited significantly less adiponectin in the periphery, which decreased further toward the center. The adiponectin-positive cells in the center of this latter tumor, which constituted less than 10% of the cell population, were mainly detected in the vicinity of blood vessels (indicated by the black arrows). Patient 3 received stereotactic radiotherapy 17 months before undergoing endoresection, whereas patient 14 underwent enucleation without irradiation. Patient 14 developed liver metastases within 2 years, whereas no metastases or extraocular growth were detected in Patient 3 during the follow-up time of 9 years. (b) Immuno-FISH assay demonstrating the copy number of chromosome 3 in the Melan-A positive cells of these tumors. Note the lower number of chromosome 3 signals despite the higher nuclear density in the “high M3” tumor. Examples of cells with M3 are indicated by the white arrows in both tumors. (c) Quantification of adiponectin in the entire tumor area with respect to the M3 status. Tumors with a higher percentage of M3 positive cells (n = 16) had significantly lower levels of adiponectin compared to the low M3 tumors (n = 22) as determined by logistic regression analysis. (d) Patients with a higher percentage of M3 in their CMC (n = 15) also had significantly less adiponectin in their primary tumor compared with the patients with lower levels of M3-positive CMC (n = 16) as determined by logistic regression. (e) Examples of some of the CMC detected in patient 14 at the time of diagnosis with primary UM. Arrows indicate the immunobeads. No CMC were detected in patient 3. Scale bars in (b) and (e) = 10 µm. MCSP, Melanoma-associated chondroitin sulfate proteoglycan.
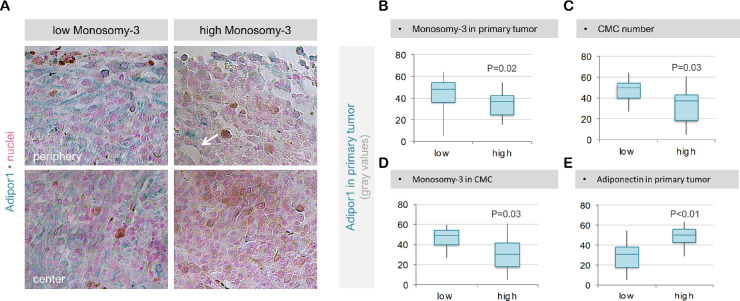
Expression of Adipor1 followed a very similar pattern to adiponectin, being detected at significantly less levels in the high M3 tumors (n = 16) compared to the low M3 samples (n = 22; P = 0.02). Patients with a higher number of CMC or prevalence of M3 in their CMC (n = 15) also exhibited a weaker Adipor1 expression in their primary tumor compared with the patients who had lower CMC parameters (n = 16; P < 0.03). The levels of Adipor1 were highly correlated with the abundance of adiponectin in the corresponding primary tumor (Fig. 3). Irradiated tumors (n = 20) exhibited a stronger immunoreactivity for Adipor1 (median 44.0; mean ± SD 46.0 ± 10.7 gray values) compared to the native samples (n = 18; median 33.3; mean ± SD 32.8 ± 15.2 gray values; P = 0.01; Fig. 3a).
Figure 3.
Expression of Adipor1 in primary UM samples with regard to the M3 status. (a) The irradiated, low M3 tumor of patient 3 exhibited a strong and mainly homogenous expression of Adipor1 in both the periphery and center. In contrast, Adipor1 was detected at significantly weaker levels in the periphery and was almost absent in the center of the nonirradiated, high M3 tumor of patient 14. Arrow indicates stronger Adipor1 expression in the proximity of a blood vessel in the latter tumor. Quantification of Adipor1 intensity in the entire tumor area demonstrated significantly lower levels of this protein in the patients who had a high extent of M3 in their primary tumor (b), a higher number of CMC (c), and a higher percentage of CMC with M3 (d). Adipor1 expression correlated positively with the adiponectin levels in the corresponding primary UM (e). The P values in (b–e) were determined by logistic regression analysis.
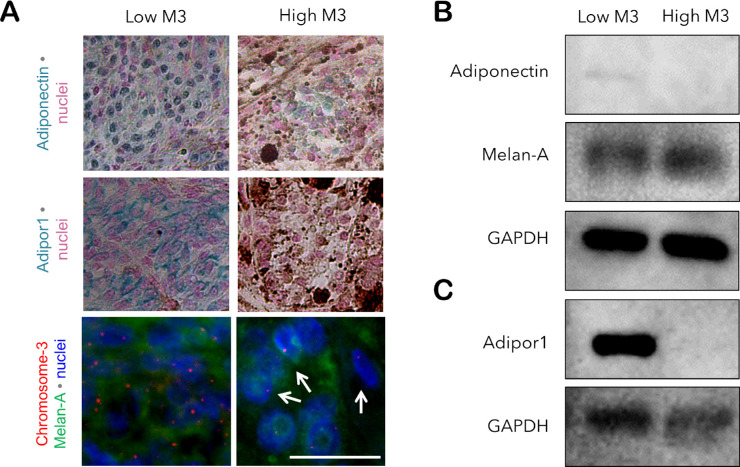
To validate this differential expression pattern, we have also performed immunoblotting for adiponectin and Adipor1 on the lysates of freshly frozen tumor samples that were available from two patients with low versus high M3. The remaining halves of the tissues were fixed in formalin and processed for IHC and Immuno-FISH. The purity of the tissue lysates for melanoma cells was verified by an additional immunoblotting for Melan-A, which revealed strong signals in both tumors. Adiponectin could be detected as a very faint band at a molecular weight of approximately 62 kDa only in the low M3 tumor. Likewise, the Adipor1 protein was highly abundant as an approximately 45- to 50-kDa band in the lysate of the low M3 sample, whereas the high M3 tumor revealed no signals (Fig. 4).
Figure 4.
Validation of the differential protein levels of adiponectin and Adipor1 with regard to the M3 status by immunoblotting. Freshly frozen tissue was available from two samples that were classified as having low versus high M3 (patients 5 and 6; female; age: 53 and 69 years, respectively). The remaining halves of the tissues were fixed in formalin and processed for immunostaining and immuno-FISH. Both patients underwent irradiation before the acquisition of the tumor samples. (A) IHC for adiponectin and Adipor1 demonstrated a reduction in the levels of both proteins in the high M3 sample. Arrows indicate several cells with M3 detected by immuno-FISH. Immunoblotting of the lysates of these tumors also revealed the stronger presence of (B) adiponectin and (C) Adipor1 in the low M3 sample. Expression of Melan-A was analyzed as a positive control for the abundance of melanoma cells in the tissue lysates. Patient 6 with the high M3 sample developed metastases within 5 years, whereas no metastases were detected in patient 5 with the low M3-tumor during the follow-up time of 8 years. Scale bar = 25 µm.
Expression of the Adiponectin and Adipor1 Proteins with Regard to BAP1
To further verify the association of adiponectin and Adipor1 deficiency with the malignant potential of UM, we have evaluated the levels of these proteins with respect to the BAP1 status in 37 of the primary tumors of our patients, whereas one sample with low M3 was omitted due to the insufficient material amount. The immunoreactivity for BAP1 was highly correlated with the M3 status of the corresponding tumors. Cytoplasmic BAP1 was abundant in 16 of the 21 samples with low M3 (76.2%) compared to the expression of this protein in seven of the 16 tumors with high M3 (43.8%, P = 0.04, Pearson's Chi-Square test). Likewise, the nuclear BAP1 was present in 11 of the 21 tumors with low M3 (52.4%) as opposed to its detection in two of the 16 samples with high M3 (12.5%, P = 0.01, Pearson's Chi-Square test). The levels of adiponectin and Adipor1 were also reduced in the tumors with a lower cytoplasmic BAP1 (P < 0.05, Mann-Whitney U test), whereas no significant difference was observed between the expression of these proteins and nuclear BAP1 (Fig. 5).
Figure 5.
Expression of adiponectin and Adipor1 with regard to the BAP1 levels in the primary tumor of our UM patients. The immunoreactivity for BAP1 in the (A) cytoplasm and (B) nucleus was graded at a scale of 0–3 (0: positive staining in less than 10% of cells; 1: positive staining in 11%–33% of cells; 2: positive staining in 34%–66% of cells; 3: positive staining in more than 67% of cells). Images were acquired under 200x magnification. The immunoreactivity for (C) adiponectin and (D) Adipor1 was significantly reduced in the tumors with lower cytoplasmic levels of BAP1 (n = 14 and n = 23 samples with low versus high BAP1, respectively; Mann-Whitney U test).
Inverse Association of the Adiponectin and Adipor1 Protein Levels in the Primary UM with the Prevalence of M3 in the CMC and the Metastatic Risk
Association of the adiponectin and Adipor1 protein levels in the primary UM with the risk of developing metastases or extraocular growth was analyzed in 26 of the 34 patients, who had a follow-up time of 2 to 9 years, whereas the remaining eight patients were excluded because of their shorter follow-up time. Adiponectin and Adipor1 were detected at significantly lower levels in the patients who have developed metastases or extraocular growth (n = 13; P = 0.02 and P = 0.04, respectively). The prevalence of M3 was significantly higher in both the primary tumor and the CMC of the patients with metastases or extraocular growth (P < 0.01). No significant association was observed between the metastatic risk and other clinical parameters including the age, gender, affected eye, tumor size, optic nerve invasion, ciliary body invasion, irradiation, as well as the presence and number of CMC (Table 2).
Table 2.
Association of the Metastases/Extraocular Growth With Clinical Parameters and Adiponectin/Adipor1 Protein Levels (Follow-Up Time: 2–9 Years)
| All Patients | No Metastases | Metastases/Extraocular | ||
|---|---|---|---|---|
| (N = 26) | (N = 13) | Growth (N = 13) | P * | |
| Age at diagnosis, median (min, max) | 68 (39–83) | 67 (53–82) | 69 (39–83) | 0.51 |
| Sex, no. (%) | 0.85 | |||
| Female | 13 (50.0) | 6 (46.2) | 7 (53.9) | |
| Male | 13 (50.0) | 7 (53.9) | 6 (46.2) | |
| Eye, no. (%) | 0.56 | |||
| Right | 12 (46.2) | 7 (53.9) | 5 (38.5) | |
| Left | 14 (53.9) | 6 (46.2) | 8 (61.5) | |
| Tumor size in mm, median (min, max) | ||||
| Base 1 | 13.1 (1.4–18.6) | 12.2 (5.0–18.6) | 13.6 (1.4–17.5) | 0.63 |
| Base 2 | 13.0 (1.4–21.8) | 12.8 (6.0–21.8) | 14.4 (1.4–19.0) | 0.94 |
| Prominence | 7.7 (0.5–13.5) | 7.5 (0.5–12.9) | 8.2 (0.9–13.5) | 0.79 |
| Optic nerve invasion, no. (%) | 0.68 | |||
| No | 23 (88.5) | 12 (92.3) | 11 (84.6) | |
| Yes | 3 (11.5) | 1 (7.7) | 2 (15.4) | |
| Ciliary body invasion, no. (%) | ||||
| No | 24 (92.3) | 11 (84.6) | 13 (100) | 0.22 |
| Yes | 2 (7.7) | 2 (15.4) | 0 (0) | |
| Irradiation, no. (%) | 0.53 | |||
| No | 11 (42.3) | 4 (30.8) | 6 (46.2) | |
| Yes | 15 (57.7) | 9 (69.2) | 7 (53.9) | |
| CMC-positive, no. (%) | 0.22 | |||
| No | 2 (7.7) | 2 (15.4) | 0 (0) | |
| Yes | 24 (92.3) | 11 (84.6) | 13 (100) | |
| CMC-No. / 50 mL blood, median (min, max) | 8.2 (0-51) | 4 (0-51) | 10 (3.8-37.5) | 0.62 |
| M3 in CMC, No. (%) | 0.02 | |||
| No | 10 (38.5) | 8 (61.5) | 2 (15.4) | |
| Yes | 16 (61.5) | 5 (38.5) | 11 (84.6) | |
| % of CMC with M3, median (min, max) | 20.9 (0-100) | 0 (0-33.3) | 37.5 (11.8-100) | 0.00 |
| M3 in primary tumor, no. (%) | ||||
| Low | 8 (30.8) | 8 (61.5) | 0 (0.0) | 0.00 |
| Intermediate | 5 (19.2) | 3 (23.1) | 2 (15.4) | 0.74 |
| High | 13 (50.0) | 2 (15.4) | 11 (84.6) | 0.00 |
| Cytoplasmic BAP1 protein, no. (%) | 0.02 | |||
| High (grade 2+3) | 15 (60.0)† | 10 (83.3)† | 5 (38.5) | |
| Low (grade 0+1) | 10 (40.0) | 2 (16.7) | 8 (61.5) | |
| Nuclear BAP1 protein, no. (%) | 0.01 | |||
| High (grade 2+3) | 8 (32.0)† | 7 (58.3)† | 1 (7.7) | |
| Low (grade 0+1) | 17 (68.0) | 5 (41.7) | 12 (92.3) | |
| Adiponectin in primary tumor, median (min, max) | 52.8 (6.6-122.8) | 78.2 (6.6-122.8) | 46.8 (15.8-74.3) | 0.02 |
| Adipor1 in primary tumor, median (min, max) | 41.8 (4.8-64.0) | 52.2 (4.8-60.7) | 36.9 (15.3-64.0) | 0.04 |
CMC: Circulating melanoma cells; M3: Monosomy-3.
P values for the numerical variables and the proportions of the categorical variables were determined by the non-paired t-test and Barnard's exact test, respectively.
Data missing from one patient.
Validation of Gene Expression in the Independent UM Cohort of the TCGA Study
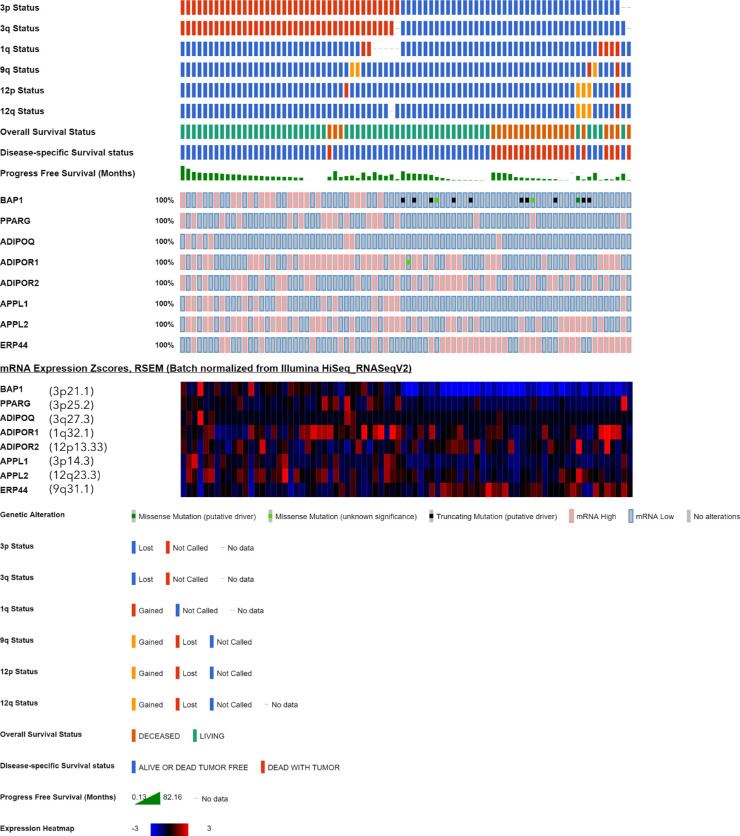
To verify our above findings, we also analyzed the expression of adiponectin, Adipor1, Adipor2, and PPARG at the mRNA level with respect to the prognostic factors and survival using the RNAseq data of the TCGA study from 80 UM patients. In addition, we included the genes Appl1 (Locus: 3p14.3) and Appl2 (Locus: 12q23.3), which encode the primary downstream effectors of the adiponectin receptors,27,28 as well as the Erp44 gene (Locus: 9q31.1), which functions as a negative regulator of adiponectin secretion.41
The baseline values of the clinical and histologic factors of the TCGA UM-cohort are summarized in Table 3. Complete information for the copy number of chromosome 3p and 3q was available for 77 of the 80 patients in the cBioPortal resource, whereas the remaining three patients (TCGA-V4-A9EW, TCGA-VD-A8KH, TCGA-WC-A88A) were excluded from the M3 analysis because of partially or completely missing data on this parameter. The heatmap for mRNA levels with regard to the M3 status and the copy number of the other chromosomes that harbor the genes of interest demonstrated a differential expression pattern for the majority of the selected genes. The expression of adiponectin remained within the 1-SD range of the mean value for most of the tumors, with higher z-scores detected in a limited number of cases mainly in the disomy 3 group. In contrast, the expression of PPARG, Adipor1, APPL1, and APPL2 was considerably lower, whereas the levels of ERP44 were higher in the M3 group (Fig. 6).
Table 3.
Baseline Characteristics of the UM Cohort of the TCGA Study (n = 80 Patients).
| Age in years (n = 80), median (range) | 61.5 (22.0-86.0) |
| Sex (n = 80) | |
| Female | 35 (43.8%) |
| Male | 45 (56.3%) |
| Eye color (n = 30) | |
| Brown | 15 (50.0%) |
| Green | 6 (20.0%) |
| Blue | 9 (30.0) |
| Tumor LBD in mm (n = 74), median (range) | 16.8 (10.0-23.6) |
| Tumor height in mm (n = 76), median (range) | 11.0 (4.4-16.0) |
| AJCC clinical stage (n = 80) | |
| IIA and -B | 36 (45.0%) |
| IIIA-C | 40 (50.0%) |
| IV | 4 (5.0%) |
| Extrascleral extension (n = 80) | |
| No | 71 (88.8%) |
| Yes | 9 (11.3%) |
| Chromosome 3 status (n = 77) | |
| M3 absent | 38 (49.4%) |
| M3 present | 39 (50.6%) |
| BAP1 expression (n = 80), median (range) | 19.5 (15.9-21.7) |
| Cell type (n = 80) | |
| Spindle | 43 (53.8%) |
| Spindle/Epithelioid | 14 (17.5%) |
| Epithelioid/Spindle | 7 (8.8%) |
| Epithelioid | 16 (20.0%) |
| Closed loops (n = 80) | |
| Absent | 38 (47.5%) |
| Present | 42 (52.5%) |
| Mitotic count (n = 80) | |
| 0–5 | 63 (78.8%) |
| >5–10 | 11 (13.8%) |
| >11 | 6 (7.5%) |
| Tumor infiltrating lymphocytes density (n = 80) | |
| Mild | 71 (88.8%) |
| Moderate | 7 (8.8%) |
| Heavy | 2 (2.5%) |
| Tumor infiltrating macrophages density (n = 80) | |
| Mild | 40 (50.0%) |
| Moderate | 29 (36.3%) |
| Heavy | 11 (13.8%) |
| Pigmentation (n = 80) | |
| Mild | 39 (48.8%) |
| Moderate | 27 (33.8%) |
| Heavy | 14 (17.5%) |
| Progression (n = 80) | |
| Censored | 50 (62.5%) |
| Progression | 30 (37.5%) |
| Metastases (n = 80) | |
| No | 54 (67.5%) |
| Yes | 26 (32.5%) |
| Time to metastases (n = 80) | |
| No metastases | 54 (67.5%) |
| >2 years | 7 (8.8%) |
| 1–2 years | 7 (8.8%) |
| <1 year | 12 (15.0%) |
| Disease specific survival (n = 78) | |
| Alive or dead as a result of other causes | 58 (74.4%) |
| Dead as a result of UM metastases | 20 (25.6%) |
Percentages are rounded to one digit and may not be amounting to 100.
LBD: largest basal diameter; AJCC: American Joint Committee on Cancer; M3: Monosomy 3. Information on the chromosome 3 status and tumor progression was retrieved from the cBioPortal database. The remaining parameters were obtained from the study of Robertson et al.29
Figure 6.
Expression of the major genes involved in adiponectin-mediated signaling in the UM cohort of the TCGA study (n = 80 patients). Each column represents a tumor sample, which was clustered according to the copy number of chromosome 3p and 3q (red: normal, blue: loss) in the uppermost two rows. Tumors with the loss of both 3p and 3q were considered as having M3, whereas these data were incomplete for n = 3 samples. The copy number of chromosomes 1q, 9q, and 12, which harbor several of the genes of interest, as well as the survival status are also demonstrated in the upper group of rows, whereas the middle rows indicate the presence of genetic alterations. The lower rows construct the expression heatmap, which was generated from the mRNA z-scores, with blue and red representing mRNA levels that were up to three standard deviations lower or higher than the mean, respectively, whereas black indicates an expression at the mean. The loci of the analyzed genes are indicated in parentheses next to the gene symbol in the heatmap. Expression of BAP1 was included as a reference.
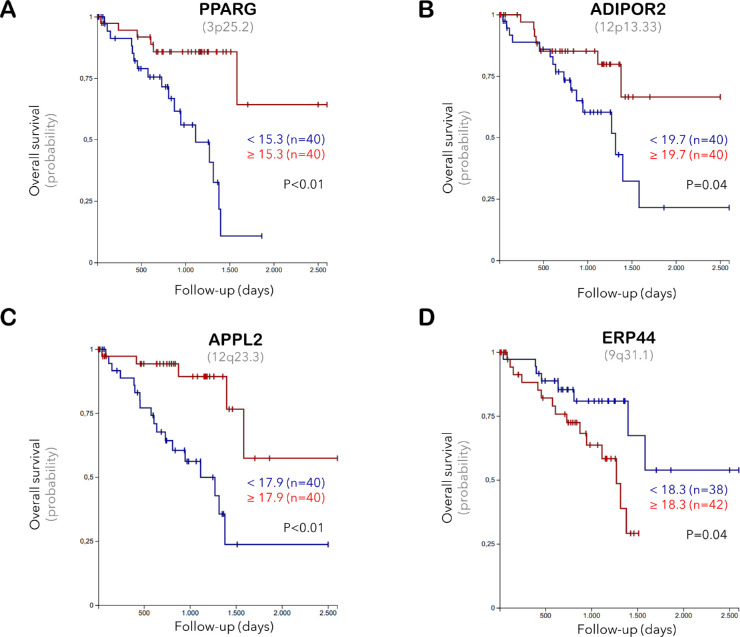
The association of gene expression with tumor characteristics and prognosis was analyzed based on the median expression of each gene. The adiponectin mRNA was detected at very low levels in the entire TCGA cohort (median 0.0, range 0.0–9.7). Although the expression range of adiponectin was slightly decreased in the patients having primary tumors with M3, an epithelioid morphology, lower BAP1 levels, closed loops, higher mitotic count, advanced clinical stage, extrascleral extension, metastases, shorter time to metastases, and worse survival rate, we did not detect a significant association between the mRNA levels of adiponectin and any of the prognostic factors. In contrast, the expression of PPARG, Adipor1, APPL1, and APPL2 was notably reduced in the M3-tumors (P < 0.05). Lower PPARG levels also correlated with an increased tumor thickness, advanced clinical stage, extrascleral extension, reduced BAP1 expression, epithelioid structure, and an adverse prognosis. The decrease in Adipor1 levels was associated with the BAP1 deficiency (P = 0.03) and tumor progression (P = 0.01) in spite of the higher expression of Adipor1 in the patients with blue eye color (n = 9 of 30 patients with available information, P = 0.01). Tumors with a moderate to heavy pigmentation and infiltration with macrophages exhibited lower levels of APPL1 and APPL2 (P < 0.01). The expression of APPL2 was also negatively correlated with the development of metastases and UM-related death, whereas the abundance of ERP44 was increased in the M3 tumors and associated with a more epithelioid structure, presence of closed loops, and worse prognosis (P < 0.05, Supplementary Table 1). The overall survival rate was reduced for the patients exhibiting lower levels of PPARG, Adipor2, and APPL2, as well as a higher ERP44 expression in their primary tumors (P < 0.05, Fig. 7).
Figure 7.
Kaplan-Meier curves demonstrating the probability of overall survival with respect to the mRNA expression of (A) PPARG, (B) Adipor2, (C), APPL2, and (D) ERP44 in the primary UM samples of the TCGA study. The median expression level of each gene was taken as the cutoff value. Gene loci are indicated in parentheses underneath the gene symbol.
Detection of Adiponectin and its Receptors in the Cultures of Human Choroidal Melanocytes and UM Cells
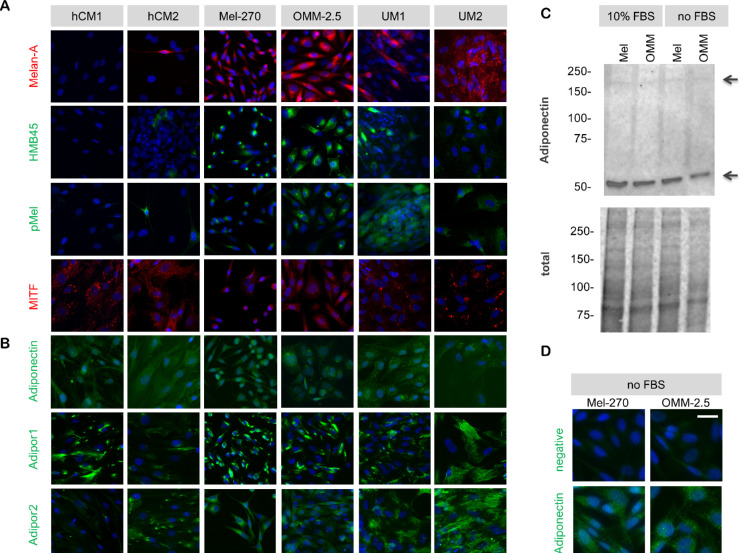
To analyze the basal levels of adiponectin and its receptors, we also established cultures from human choroidal melanocytes (hCM), as well as the high M3 tumor samples of two of our patients. We also included the well-established UM cell lines Mel-270 and OMM-2.5. The hCM1 cells were generated from a patient who had no history of ocular tumors and underwent enucleation because of perforation, whereas the hCM2 cells were derived from the tumor-free choroidal tissue of a patient with posterior UM (male, 56 years). The hCM1 cells would therefore be expected to be more representative of the normal melanocytes whereas the hCM2 culture would be likely to contain transformed cells.
Culture purity was evaluated by a panel of melanocyte and melanoma-specific markers. The hCM1 cells exhibited a strong expression of the melanocyte differentiation protein MITF but were negative for HMB45, Melan-A, and p-Mel. In contrast, the MITF expression appeared slightly lower in the hCM2 cells, while the latter markers could be detected in 5-10% of the cell population, suggesting that the hCM2 culture was less differentiated and consisted of some activated/transformed melanocytes. The UM cultures generated in our laboratory (UM1 and UM2) exhibited a strong immunoreactivity for Melan-A and p-Mel, with an uneven HMB45 expression pattern and moderate to low levels of MITF. The Mel-270 and OMM-2.5 cells were both strongly positive for Melan-A and HMB-45, but expressed weaker levels of p-Mel and MITF (Fig. 8a).
Figure 8.
Expression of adiponectin and its receptors in the cultured human choroidal melanocytes (hCM) and UM cells. (a) Culture purity was evaluated by the levels of melanocyte- and melanoma-specific markers. The hCM1 cells were derived from the choroid of a patient without an ocular tumor, whereas the hCM2 cells were obtained from the tumor-free choroidal region of a patient with posterior UM. The UM1 and UM2 cells were isolated from the primary tumors of patients 19 and 14, respectively, in our study. The donor of UM1 cells (male, 82 years) had posterior UM which was classified as a high M3 tumor. UM2 cells were derived from the high M3 tumor of the patient (female, 48 years), who was referred to in the Figures 2 and 3, as well. (b) Expression of adiponectin, as well as its receptors Adipor1 and Adipor2 in the cells grown in normal medium with 10% FBS. Expression of the receptors on the cell surface was analyzed by immunocytochemistry on non-permeabilized cells. Adipor1 could be detected on all the cell types whereas the expression of Adipor2 was considerably stronger in the UM cultures. Scale bar = 50 µm. (c) Immunoblotting under non-reducing conditions demonstrated the expression of adiponectin in the whole lysates of Mel-270 and OMM-2.5 cells that were either grown in normal medium with 10% FBS or serum-deprived for 2 days. The total protein loading in the lanes was detected by stain-free gel imaging. (d) Immunocytochemical analysis of adiponectin levels in the Mel-270 and OMM-2.5 cells that were serum-deprived for 2 days. The negative controls were incubated without the primary antibody. Scale bar = 25 µm.
Adiponectin was detected at a moderate level in all of these cells whereas the expression of its receptors exhibited cell-type specific patterns. Adipor1 was detected on the surface of all cell types while the Adipor2 isoform was almost absent from the hCM1 cells but strongly expressed on the hCM2 cells and all of the UM cultures (Fig. 8b). Adiponectin was also present in the whole lysates of Mel-270 and OMM-2.5 cells that were grown in normal medium or serum deprived for 2 days, mainly as a dimer with a molecular weight of approximately 62 kDa. The hexamer isoform of adiponectin, which has a molecular weight of approximately 180 kDa, was detected at very low levels (Figs. 8c and 8d).
Lower Basal Levels of Adiponectin in the Cultured UM Cells with M3
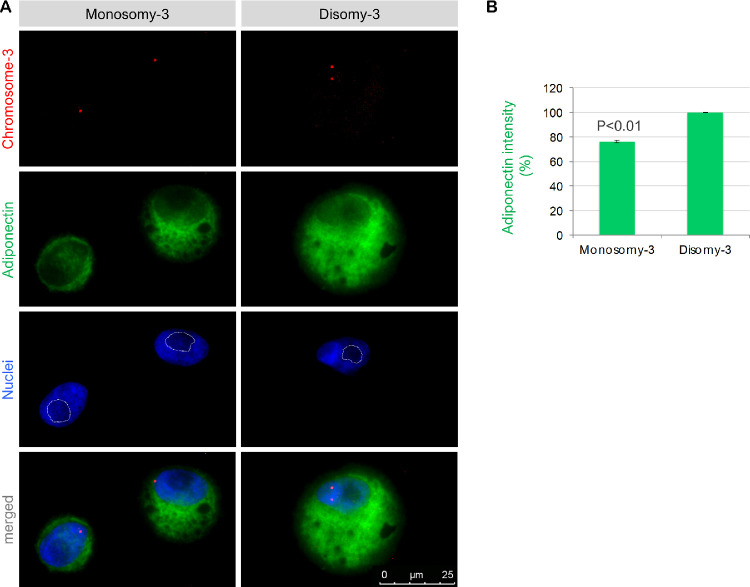
To gain further insight into the outcomes of M3 on the basal adiponectin levels, we performed an Immuno-FISH assay for the adiponectin protein and chromosome 3 on the untreated UM1 and UM2 cells that were grown in normal medium with 10% FBS. This condition was selected to simulate the normal environment that the CMC would be exposed to. Our results demonstrated the presence of adiponectin at approximately 22% lower levels in the UM cells with M3 (P = 0.03). Notably, the nucleoli of the M3-positive cells tended to occupy a larger area, as estimated by the darker regions in the nuclear staining with DAPI (Fig. 9).
Figure 9.
Basal adiponectin levels in the cultured UM cells with M3 versus disomy-3. (a) Immuno-FISH was performed for the codetection of adiponectin and chromosome 3 on the cytospins of intact UM1 and UM2 cells (both at passage 2). Images presented were acquired from the UM1 cells. Notice the larger nucleolar area (which appear darker in the nuclear stainings and were marked by the white lines) in the cells with M3. (b) The mean gray value of the adiponectin staining was quantified by circumscribing the cytoplasmic region (excluding the nuclei) in a total of 112–180 nonoverlapping UM2 and UM1 cells, respectively, with clear signals for chromosome 3. The number of M3-positive cells that could be quantified was 43–69 for the UM2 and UM1 cells, respectively. The intensity of the cells with disomy-3 was taken as 100%. Data represent the mean ± standard deviation of the measurements for the UM1 and UM2 cells. P value was determined by two-sided t-test assuming equal variance.
Induction of a More Quiescent Phenotype in the UM Cells Exposed to a Physiologically Relevant Concentration of Adiponectin
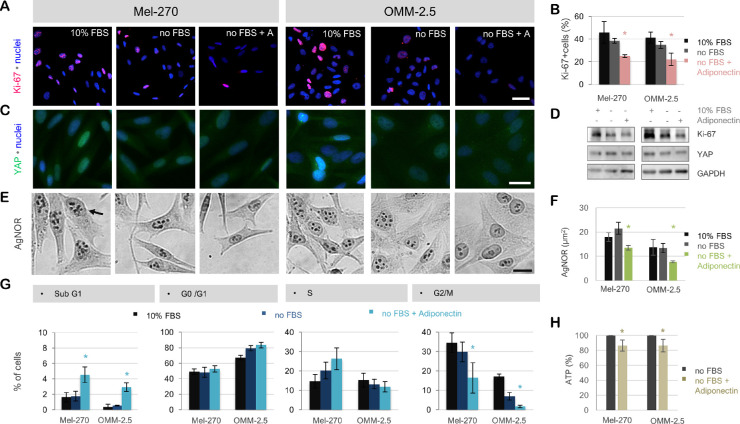
To determine the response of the UM cells to adiponectin, the Mel-270 and OMM-2.5 cells were incubated with a physiological concentration of adiponectin at 30 µg/mL under serum deprivation for 1d. Adiponectin could significantly suppress the proliferation marker Ki-67 in both cell types, together with a decrease in the percentage of cells in the G2/M phase and a slight accumulation of cells in the Sub G1 phase. This effect was also associated with a gradual decline in the expression of YAP and the area occupied by the silver-binding nucleolar organizer regions (AgNOR) after 1d, as well as an approximately 15% decrease in ATP levels after 8 to 10 hours (P < 0.05, Fig. 10).
Figure 10.
Adiponectin induces a more quiescent phenotype in the UM cells by suppressing proliferation and energy production. (a) Immunostaining for the proliferation marker Ki-67 on the Mel-270 and OMM-2.5 cells after a 1-day incubation in normal medium with 10% FBS, serum-free medium (no FBS), or serum-free medium with 30 µg/ml Adiponectin (no FBS + A). Scale bar = 50 µm. (b) Quantification of the Ki-67 immunostaining was performed on a minimum of 205 nuclei per group. Cells with a mean Ki-67 intensity that was above the cutoff value of negative cells were defined as being positive. Data represent the mean ± SEM of n = 3 independent experiments. *P < 0.05 compared to the corresponding no FBS group as determined by the two-sided t-test. (c) YAP-immunostaining demonstrating a gradual decline in the levels of this protein in response to serum deprivation ± adiponectin. The intensity of the nuclear staining also underwent a progressive decrease, suggesting the impairment of DNA duplication. Scale bar = 25 µm. (d) Immunoblotting for Ki-67 and YAP in the whole cell lysates after a 1-day exposure to the indicated treatments. Images are representative of n = 3 experiments. (e) Silver staining of nucleolar organizer regions (AgNOR, indicated by the arrow), demonstrating the reduction in the number and area of the nucleolar structures in response to serum deprivation + Adiponectin. Scale bar = 25 µm. (f) Quantification of the AgNOR area. Data represent the mean ± SEM of n = 3 experiments with a minimum of n = 202 quantified nuclei per group. *P < 0.05 compared to the corresponding no FBS group as determined by the two-sided t-test. (g) Cell-cycle analysis after a 1-day exposure to the indicated treatments. Adiponectin could mainly impair the progression of UM cells into the G2/M phase and induce a slight but significant accumulation of the cells in the Sub G1 phase. The Mel-270 and OMM-2.5 cells also differed in their cell-cycle rates, with the former cell type apparently requiring more time for the G2/M phase, as demonstrated by the higher percentage of untreated Mel-270 cells in the G2/M phase and the lower cell density of this group compared with the untreated OMM-2.5 cells in panel (a). Data represent the mean ± SEM of n = 3 independent experiments. *P < 0.05 compared to the no FBS group of the corresponding cell as determined by the two-sided t-test. (h) ATP levels in the serum deprived cells treated with or without adiponectin for 8 to 10 hours as measured by a luminescence assay. The values of untreated cells were taken as 100%. Data represent the mean ± SEM of n = 3 independent experiments with triplicate wells for each group. *P < 0.05 compared to the no FBS group of the corresponding cell as determined by the two-sided t-test.
Discussion
The development of efficient therapies against UM has been sadly impeded by the insufficient knowledge on the molecular mechanisms that underlie the peculiar potential of the disseminated UM cells to remain dormant for many years before forming highly aggressive metastases mainly in the liver. It has also remained largely unknown how the presence of M3, the most important prognostic factor for UM metastases, is involved in the termination of UM dormancy and whether some pathophysiological factors aggravate the course of events. A very important clue on this subject was recently provided by the study of Sevim and Kiratli,16 which reported the association of insulin resistance, characterized by the lower serum levels of adiponectin, with an unfavorable prognosis in UM. Remarkably, adiponectin is a hormone with anti-carcinogenic activities and encoded by a gene on chromosome 3.23,24 In this study, we therefore evaluated whether adiponectin and its major receptor Adipor1 are present in the UM cells, whether M3 alters the tumor adiponectin levels, and how the UM cells respond to adiponectin treatment.
Our results demonstrated significantly lower levels of the adiponectin protein and Adipor1 in the primary UMs which had a higher prevalence of M3. Likewise, our validation analysis using the TCGA study demonstrated a gene expression profile that indicated the impairment of the adiponectin-mediated signaling in the M3 tumors, which also correlated with several prognostic factors and overall survival. The extent of Adipor1 expression was also very consistent with the amount of the adiponectin protein in the corresponding tumor, suggesting that the UM cells with adiponectin deficiency may be retracting the Adipor1 and already entering a state of local insulin resistance. However, we could not distinguish whether the adiponectin in the paraffin sections of our primary UM samples represented the tumor-derived adiponectin or the systemic hormone that might have bound on to these cells. Adiponectin is indeed the most abundant serum adipokine that constitutes 0.01% or 3 to 30 µg/mL of plasma protein.42,43 In some tumors with a very weak immunoreactivity for adiponectin and Adipor1, we could identify individual cells with a strong staining mainly in the proximity of the blood vessels (Figs. 2 and 3), suggesting that these cells might have received a high amount of circulating adiponectin and retained their Adipor1 expression. However, the tumors with a low percentage of M3 usually exhibited an intense and mostly uniform immunoreactivity for adiponectin even in the poorly vascularized central regions, indicating that the UM cells may be able to produce their own adiponectin. Consistently, we could detect the adiponectin protein mainly as the dimer isoform in the lysates of Mel-270 and OMM-2.5 cells that were serum-deprived for 2 days. Our findings also demonstrate a mild but significant adiponectin deficiency by approximately 22% in the UM cells with M3 that were cultured in normal medium with serum supplementation in order to simulate the environment that the CMC would be exposed to. Additional studies on the expression of adiponectin and its receptors at the mRNA level are needed to gain more insight into the influence of M3 on the adiponectin production capacity of UM cells. An Immuno-FISH assay for the adiponectin protein and chromosome 3 on the paraffin sections of UM samples, as well as serum-deprived cultures, might also be very informative to determine the levels of this hormone concurrently with the copy number of chromosome 3.
The lower levels of adiponectin and Adipor1 in our primary UM samples were significantly associated with a higher percentage of M3 in the CMC of these patients and the risk of developing metastases or extraocular growth within the follow-up time of 2 to 9 years. These findings suggest that the impairment of adiponectin signaling in the primary tumor might be augmenting the metastatic potential of the disseminated cells. Consistent with this hypothesis, we could also provide the first proof to the direct antiproliferative effect of adiponectin on the UM cell lines Mel-270 and OMM-2.5 as demonstrated by the downregulation of Ki-67, decrease in the percentage of cells in the G2/M phase, and a slight but significant accumulation of cells in the Sub G1 phase in response to adiponectin at its maximal plasma concentration of 30 µg/mL. This more quiescent state was also associated with the reduction of ATP levels and the area of the so-called AgNOR proteins in the nucleoli of the UM cell lines in our study. The AgNOR proteins, which can be selectively stained by silver methods, have proven to be very reliable indicators of nucleolar function, and their amount was directly related to the rapidity of cell proliferation in validated human cancer cell lines.40 Our findings therefore suggest that after dissemination, CMC exposed to considerably higher levels of adiponectin may be shifting to a more dormant status. The gradual decline in the serum adiponectin levels, as observed in the case of insulin resistance, would then weaken this suppression over time. The CMC with M3 or their micrometastases might be recovering more easily from this dormancy, possibly because of their own adiponectin deficiency and receptor retraction, which deserves further investigation.
Remarkably, the increase in nucleolar size is one of the established prognostic factors which has shown a positive correlation with the malignancy of UM and is a feature of the epithelioid UM cells with an aggressive metastatic potential. However, this factor has received less consideration in clinical practice, probably because of its laborious measurement.6,44,45 The nucleolus is a prominent, highly specialized, and dynamic compartment in the nucleus, where the production of ribosomes, the protein translation machinery of cells, occurs. The nucleoli organize around the chromosomes that contain ribosomal DNA repeats, enabling the transcription of genes for ribosomal RNAs. The assembly of ribosomal subunits and their transport into the cytoplasm are also regulated in the nucleolus. In addition, the nucleolus serves as a reservoir for “dormant” proteins involved in cell cycle and DNA damage response.46–48
Ribosome biosynthesis is reported to be the most energy-consuming process in human cells, which is persistently hyperactivated in cancer. Accordingly, almost all types of cancer cells exhibit an increase in the size or number of nucleoli, whereas cellular stressors or senescence induce the nucleolar shrinkage.46–50 However, the factors that account for the larger nucleolar size in the malignant UM cells have remained unknown so far. In this study, the nuclear staining of the UM cultures we have generated indicated the presence of larger nucleoli in the cells with M3 (Fig. 5), which remains to be validated by quantifying specific nucleoli markers. We could also provide the first direct evidence to the involvement of adiponectin deficiency in the malignant UM phenotype with abundant nucleoli by demonstrating a significant impairment of nucleolar structure and ATP levels after the treatment of the cell lines Mel-270 and OMM-2.5 with adiponectin. Our findings therefore suggest that the UM cells can no longer produce sufficient energy to afford high levels of nucleoli in response to adiponectin by yet unknown mechanisms. Interestingly, the irradiated tumors in our study also contained significantly higher levels of adiponectin and Adipor1, further supporting the role of adiponectin in the energetic collapse of these cells. Adiponectin indeed exerts its downstream effects by mainly activating the AMP-Kinase (AMPK), which is the major energy sensor in cells.24,51 AMPK responds to an increase in the ratio of AMP/ATP by initiating a cascade of catabolic events that diverts the cells into a quiescent, energy-saving mode.51,52 Interestingly, the inhibition of mutant GNAQ signaling could induce the AMPK-dependent autophagy of UM cells,53 providing further support to the central role of AMPK in the cellular adaptation efforts to cope with energy starvation. Likewise, the AMPK activator aminoimidazole carboxamide ribonucleotide could significantly suppress the proliferation of UM cells.54 Moreover, AMPK could directly phosphorylate the Hippo pathway effector YAP at multiple sites and inhibit its transcriptional activity in non-melanoma cells.55 The YAP-mediated signaling has indeed received more attention in the past years as a downstream regulator of the overactivated GNAQ/GNA11 pathway in UM cells.56–58 In this study, we have also observed a slight decline in YAP levels in the UM cells treated with adiponectin. Future studies focusing particularly on the activities of AMP-Kinase and YAP might therefore provide a deeper insight into the molecular mechanisms of adiponectin action in UM.
The association of M3 with lower adiponectin levels may be primarily due to the loss of the genes encoding adiponectin and its major transcriptional regulator PPARG, which both map to chromosome 3.20,25,26 PPARG may also be involved in the expression of the adiponectin receptors as indicated by the upregulation of the Adipor2 mRNA and protein in the hepatocytes that were treated with the PPAR-gamma agonist rosiglitazone.59 Moreover, the expression of the Erp44 gene, which encodes a negative regulator of adiponectin secretion, was elevated in the M3 tumors and significantly associated with a worse prognosis. The ERP44 protein functions as a chaperone that can prevent the secretion of adiponectin by retaining this peptide hormone in the early secretory compartment of the endoplasmic reticulum.41 Remarkably, the cytoplasmic BAP1 was recently found to be localized predominantly in the endoplasmic reticulum fractions of fibroblasts.60 In this study, we have also detected higher Erp44 expression in the BAP1 negative tumors of the TCGA cohort (P < 0.01, Supplementary Table 1). However, we could not find any information on whether BAP1 interacts with ERP44 and whether the loss of BAP1 contributes to the increased expression of ERP44 in the M3 tumors, which remain to be investigated. Since the Erp44 gene maps to the 9q31.1 locus, which does not undergo clinically relevant copy number alterations in the UM patients, the upregulation of ERP44 in the tumors with a higher metastatic risk further signifies the impairment of adiponectin signaling as an important factor that may be aggravating the course of UM. The significant decline in the mRNA levels of Adipor1 in the M3 tumors may in turn be a secondary response to the reduction of secreted adiponectin, which may be exerting autocrine or paracrine effects on the UM cells that need to be examined in detail in future studies.
A limitation of our study is the assessment of M3 status in the paraffin sections only by Immuno-FISH without the further validation using alternative techniques. By performing Immuno-FISH with a centromeric probe for chromosome 3 alone, we were not able to detect the partial deletions on this chromosome or an isodisomy and might have overestimated the pure disomy-3 in some cases. However, FISH on paraffin sections might also lead to the underestimation of the diploid status due to the possible truncation of nuclei particularly when the sections have a thickness of 4 µm or less.37,61 In our study, we have used 6-µm sections to decrease the risk of obtaining false-positive results for M3. Our positive controls for the diploidy of the retina also supported the adequacy of this approach. Moreover, our Immuno-FISH assay enabled us to detect chromosome 3 simultaneously with the melanoma marker Melan-A and ensure that the quantified nuclei belonged to the tumor cells rather than the tumor infiltrating or endothelial cells. Because of the very small size of some of the tumor samples and partly due to our attempts for establishing UM cultures with M3, we did not have sufficient material to analyze further chromosomal aberrations, either. Primary UMs indeed exhibit frequent alterations on chromosomes 1, 6, and 8, as well. The gain of 8q, which is also associated with a reduced survival rate, occurs mostly in combination with M3. Likewise, the loss of 6q or 1p increase the metastatic risk, whereas the gain of 6p exists almost in a mutually exclusive pattern with M3 and is a strong predictor of good prognosis in the absence of other chromosomal anomalies.6 Conducting subsequent studies on the levels of adiponectin and its receptors with regard to these aberrations might therefore provide additional insight into the behavior of the heterogenous UMs under insulin resistance.
Interestingly, the high frequency of M3 and its inverse relation to the 6p gain in UM cells suggest that these aberrations are the consequences of some (patho-)physiological conditions rather than being random events. Chromosomes are indeed not randomly distributed in the nucleus but tend to occupy distinct territories.62–65 Remarkably, the three-dimensional FISH analysis of human fibroblasts has shown that one copy of chromosome-3 and -8 appear adjacent to each other in both poles of the nucleus, whereas a segment of chromosome-6 resides between one copy of chromosome-3 and -1.66 This constellation, which remains to be analyzed in the uveal melanocytes and UM cells, might in turn provide a novel insight into the peculiar cytogenetics of UM. During the proliferation of UM cells, the spatial conditions in the nucleus may be favoring the missegregation of either chromosome-3 or -6p, but not both of them concurrently, directing one of the daughter cells to an aggressive phenotype. However, it is still unknown, which factors can trigger the large scale chromosomal aberrations in UM cells in such an intriguingly systematic manner and whether it would be possible to divert the proliferating UM cells to the more benign genotype with the 6p gain by modulating some external events. Interestingly, the nucleoli can act as a spatial organizer in the nucleus, keeping distinct chromosomal segments separated from each other.67 It would be therefore very exciting to analyze whether the large nucleoli form such a physical barrier during the condensation of chromosomes in the nonquiescent UM cells, which later hinders the coexistence of the fatal M3 with the favorable 6p gain in the same nucleus. It would also be very interesting to determine whether adiponectin can prevent the generation of further UM cells with M3 by disrupting the nucleolar integrity as a novel therapy approach to maintain the existing tumors in a more benign genotype and quiescent state.
The quiescence of disseminated UM cells over many years has also remained an enigmatic subject, with the factors enabling this process and the conditions that result in the termination of tumor dormancy still being elusive.1,7,8 However, a deeper insight into the actions of adiponectin might provide valuable clues to these events. As stated earlier, one of the direct mechanisms that accounts for the insulin-sensitizing effects of adiponectin is its inhibitory action on the release of glucose from the liver cells.24 Yet, it is not known, whether the metastasized UM cells with M3 (and with a possible shortage of endogenous adiponectin) can consume the adiponectin present in the liver and impair the normal adiponectin signaling within the liver. Although such an adiponectin consumption may initially maintain the UM cells in a more quiescent state in the liver, the tumor cells could be promoting the release of glucose from the hepatic cells and exploiting the liver for sustaining their own energy supply in this manner. The liver indeed serves as the body's major glucose reservoir, helping to maintain the blood sugar levels by regulating the storage and release of glucose depending on the body's needs. The liver is also responsible for the release of more than 80% of the glucose after an overnight fasting.21,68 With these features, the liver represents a very attractive seeding soil for the tumor cells that need a fast and continuous access to the glucose reserves. As the systemic adiponectin levels further decline under insulin resistance, the liver cells may no longer be receiving sufficient adiponectin to prevent the glucose release or maintain the UM micrometastases quiescent, which would induce the detrimental imbalance towards the termination of UM dormancy. The tendency of the UM (macro)metastases to grow so rapidly to a lethal extent in the liver1 also indicates that the cells with M3 may have an inherent ability to benefit from a sustained glucose supply from the liver cells via the mechanism suggested above, which urgently deserves further investigation.
In response to insulin resistance, complementary physiological mechanisms are activated to maintain the blood glucose levels in the normal range. Accordingly, signaling proteins with an insulin-mimetic activity, such as the insulin-like growth factor-1 (IGF-1) become more available in the circulation.17,69–71 IGF-1 is indeed considered to be the major chemoattractant for UM metastasis, because it is mainly released from the liver cells.69 Insulin resistance might therefore be aggravating the course of UM by not only creating an adiponectin-deficient environment, but also promoting the bioavailability of IGF-1. Interestingly, the circulating levels of adiponectin are lower whereas IGF-1 levels are higher in elderly men compared with women,72,73 suggesting that the sex-specific differences of these hormones might be one of the reasons underlying the slight predominance and worse prognosis of UM in the male patients,74,75 which needs to be analyzed in more detail with regard to insulin resistance. Similar to the UM micrometastases, insulin resistance also follows a rather stealthy course, remaining usually undetected for many years until the subtle systemic changes it induces accumulate to a pathological extent that manifests itself as metabolic disorders.76 It may be therefore highly advantageous to inform the patients with UM or melanocytic lesions on the adverse effects of insulin resistance and immediately take preventive measures. Because the insulin resistance is associated with many comorbidities such as hypertension or hyperlipidemia and increases the risk of developing other severe conditions including cardiovascular disease and numerous malignancies,17–19,76 lifestyle changes to avoid insulin resistance and adiponectin deficiency would also be expected to improve the overall well-being of the UM patients. The dietary and lifestyle modifications that could increase the levels of circulating adiponectin in humans involve a low glycemic load-diet, fiber supplementation, daily intake of fish or omega-3 supplements, moderately intense aerobic exercise, and a Mediterranean diet with the consumption of fruits, deep yellow vegetables, nuts, and coffee,24,77–79 whereas smoking was associated with lower adiponectin levels.79,80 In severe cases of adiponectin deficiency, therapeutic interventions with the type-2 diabetes drug Metformin, which could exert an inhibitory effect on cultured UM cells (de Andrade BDM et al., IOVS 2017;58:ARVO E-Abstract 3964), may also be beneficial to improve the circulating adiponectin levels,81 although there is to our knowledge no study reporting the efficacy of such drugs in UM patients. We therefore believe that further research on the adiponectin-mediated signaling events in UM cells is urgently needed and holds the great promise of opening new avenues not only toward the better understanding of the pathophysiology of UM but also the development of efficient therapies.
In conclusion, our findings provide novel insights into the mechanisms that may be contributing to the highly aggressive metastatic potential of the UM cells with M3 and further support the critical role of insulin resistance in aggravating the course of this disease. Taking immediate actions against insulin resistance and improving the levels of circulating adiponectin by lifestyle changes or therapeutic interventions might therefore be a valuable and easy to consider approach to prevent or delay the UM metastases. This would also improve the overall life quality and offer a new hope for the patients who are affected by this devastating disease.
Supplementary Material
Acknowledgments
The authors thank Christine Örün and Anna Kraus for excellent technical assistance.
Supported by Werner Und Klara Kreitz-Stiftung, Kiel, Germany, Grant F279097.
Disclosure: A. Tura, None; C. Thieme, None; A. Brosig, None; H. Merz, None; M. Ranjbar, None; S. Vardanyan, None; H. Zuo, None; T. Maassen, None; V. Kakkassery, None; S. Grisanti, None
References
- 1. Nichols EE, Richmond A, Daniels AB. Micrometastatic dormancy in uveal melanoma: a comprehensive review of the evidence, mechanisms, and implications for future adjuvant therapies. Int Ophthalmol Clin. 2017; 57: 1–10. [DOI] [PubMed] [Google Scholar]
- 2. Aronow ME, Topham AK, Singh AD. Uveal melanoma: 5-year update on incidence, treatment, and survival (SEER 1973-2013). Ocul Oncol Pathol. 2018; 4: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blum ES, Yang J, Komatsubara KM, Carvajal RD. Clinical management of uveal and conjunctival melanoma. Oncology (Williston Park). 2016; 30: 29–32,34–43,48. [PubMed] [Google Scholar]
- 4. Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jöckel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996; 347: 1222–1225. [DOI] [PubMed] [Google Scholar]
- 5. Scholes AG, Damato BE, Nunn J, Hiscott P, Grierson I, Field JK. Monosomy 3 in uveal melanoma: correlation with clinical and histologic predictors of survival. Invest Ophthalmol Vis Sci. 2003; 44: 1008–1011. [DOI] [PubMed] [Google Scholar]
- 6. Kaliki S, Shields CL, Shields JA. Uveal melanoma: estimating prognosis. Indian J Ophthalmol. 2015; 63: 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco PL, Lim LA, Miyamoto C, Burnier MN. Uveal melanoma dormancy: an acceptable clinical endpoint? Melanoma Res. 2012; 22: 334–340. [DOI] [PubMed] [Google Scholar]
- 8. Ossowski L, Aguirre-Ghiso JA.. Dormancy of metastatic melanoma. Pigment Cell Melanoma Res. 2010; 23: 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ardjomand N, Komericki P, Langmann G, et al.. Lymph node metastases arising from uveal melanoma. Wien Klin Wochenschr. 2005; 117: 433–435. [DOI] [PubMed] [Google Scholar]
- 10. Wöll E, Bedikian A, Legha SS. Uveal melanoma: natural history and treatment options for metastatic disease. Melanoma Res. 1999; 9: 575–581. [PubMed] [Google Scholar]
- 11. Torres V, Triozzi P, Eng C, et al.. Circulating tumor cells in uveal melanoma. Future Oncol. 2011; 7: 101–109. [DOI] [PubMed] [Google Scholar]
- 12. Callejo SA, Antecka E, Blanco PL, Edelstein C, Jr Burnier MN. Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye (Lond). 2007; 21: 752–759. [DOI] [PubMed] [Google Scholar]
- 13. Schuster R, Bechrakis NE, Stroux A, et al.. Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma. Clin Cancer Res. 2007; 13: 1171–1178. [DOI] [PubMed] [Google Scholar]
- 14. Tura A, Lüke J, Merz H, et al.. Identification of circulating melanoma cells in uveal melanoma patients by dual-marker immunoenrichment. Invest Ophthalmol Vis Sci. 2014; 55: 4395–4404. [DOI] [PubMed] [Google Scholar]
- 15. Tura A, Merz H, Reinsberg M, et al.. Analysis of monosomy-3 in immunomagnetically isolated circulating melanoma cells in uveal melanoma patients. Pigment Cell Melanoma Res. 2016; 29: 583–589. [DOI] [PubMed] [Google Scholar]
- 16. Sevim DG, Kiratli H.. Serum adiponectin, insulin resistance, and uveal melanoma: clinicopathological correlations. Melanoma Res. 2016; 26: 164–172. [DOI] [PubMed] [Google Scholar]
- 17. Arcidiacono B, Iiritano S, Nocera A, et al.. Insulin resistance and cancer risk: an overview of the pathogenetic mechanisms. Exp Diabetes Res. 2012; 2012: 789174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Djiogue S, Nwabo Kamdje AH, Vecchio L, et al.. Insulin resistance and cancer: the role of insulin and IGFs. Endocr Relat Cancer. 2013; 20: R1–R17. [DOI] [PubMed] [Google Scholar]
- 19. Orgel E, Mittelman SD.. The links between insulin resistance, diabetes, and cancer. Curr Diab Rep. 2013; 13: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006; 116: 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Combs TP, Marliss EB.. Adiponectin signaling in the liver. Rev Endocr Metab Disord. 2014; 15: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thundyil J, Pavlovski D, Sobey CG, Arumugam TV. Adiponectin receptor signalling in the brain. Br J Pharmacol. 2012; 165: 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katira A, Tan PH.. Evolving role of adiponectin in cancer-controversies and update. Cancer Biol Med. 2016; 13: 101–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dalamaga M, Diakopoulos KN, Mantzoros CS. The role of adiponectin in cancer: a review of current evidence. Endocr Rev. 2012; 33: 547–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Greene ME, Blumberg B, McBride OW, et al.. Isolation of the human peroxisome proliferator activated receptor gamma cDNA: expression in hematopoietic cells and chromosomal mapping. Gene Expr. 1995; 4: 281–299. [PMC free article] [PubMed] [Google Scholar]
- 26. Maeda N, Takahashi M, Funahashi T, Kihara S, Nishizawa H, Kishida K, et al.. PPARgamma ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001; 50: 2094–2099. [DOI] [PubMed] [Google Scholar]
- 27. Deepa SS, Dong LQ.. APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab . 2009; 296: E22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Achari AE, Jain SK.. Adiponectin, a therapeutic target for obesity, diabetes, and endothelial dysfunction. Int J Mol Sci . 2017; 18. pii: E1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robertson AG, Shih J, Yau C, et al.. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2018; 33: 151. [DOI] [PubMed] [Google Scholar]
- 30. Jager MJ, Brouwer NJ, Esmaeli B. The Cancer Genome Atlas Project: an integrated molecular view of uveal melanoma. Ophthalmology. 2018; 125: 1139–1142. [DOI] [PubMed] [Google Scholar]
- 31. Jager MJ, Magner JA, Ksander BR, Dubovy SR. Uveal melanoma cell lines: where do they come from? (An American Ophthalmological Society Thesis). Trans Am Ophthalmol Soc. 2016; 114: T5. [PMC free article] [PubMed] [Google Scholar]
- 32. Griewank KG, Yu X, Khalili J, et al.. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 2012; 25: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu J, Li M, Zhang Y. Long noncoding RNA HOXA-AS2 regulates the expression of SCN3A by sponging miR-106a in breast cancer. J Cell Biochem . 2019; 120: 14465–14475. [DOI] [PubMed] [Google Scholar]
- 34. Gao J, Aksoy BA, Dogrusoz U, et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal . 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cerami E, Gao J, Dogrusoz U, et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov . 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szalai E, Wells JR, Ward L, Grossniklaus HE. Uveal melanoma nuclear BRCA1-associated protein-1 immunoreactivity is an indicator of metastasis. Ophthalmology . 2018; 125: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sandinha MT, Farquharson MA, Roberts F. Identification of monosomy 3 in choroidal melanoma by chromosome in situ hybridisation. Br J Ophthalmol. 2004; 88: 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu DN, McCormick SA, Ritch R, Pelton-Henrion K. Studies of human uveal melanocytes in vitro: isolation, purification and cultivation of human uveal melanocytes. Invest Ophthalmol Vis Sci. 1993; 34: 2210–2219. [PubMed] [Google Scholar]
- 39. Roukos V, Pegoraro G, Voss TC, Misteli T. Cell cycle staging of individual cells by fluorescence microscopy. Nat Protoc. 2015; 10: 334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Derenzini M, Trerè D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998; 152: 1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 41. Hampe L, Radjainia M, Xu C, et al.. Regulation and quality control of adiponectin assembly by endoplasmic reticulum chaperone ERp44. J Biol Chem. 2015; 290: 18111–18123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng Y, Scherer PE.. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann N Y Acad Sci. 2010; 1212: E1–E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gu HF. Biomarkers of adiponectin: plasma protein variation and genomic DNA polymorphisms. Biomark Insights. 2009; 4: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moshari A, McLean IW.. Uveal melanoma: mean of the longest nucleoli measured on silver-stained sections. Invest Ophthalmol Vis Sci. 2001; 42: 1160–1163. [PubMed] [Google Scholar]
- 45. Miyamoto C, Balazsi M, Bakalian S, Fernandes BF, Jr Burnier MN. Uveal melanoma: ocular and systemic disease. Saudi J Ophthalmol. 2012; 26: 145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strunnikov AV. A case of selfish nucleolar segregation. Cell Cycle. 2005; 4: 113–117. [DOI] [PubMed] [Google Scholar]
- 47. Lindström MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene. 2018; 37: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shaw P, Doonan J.. The nucleolus. Playing by different rules? Cell Cycle. 2005; 4: 102–105. [DOI] [PubMed] [Google Scholar]
- 49. Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta. 2014; 1842: 802–816. [DOI] [PubMed] [Google Scholar]
- 50. Pfister AS. Emerging role of the nucleolar stress response in autophagy. Front Cell Neurosci. 2019; 13: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ke R, Xu Q, Li C, Luo L, Huang D. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol Int. 2018; 42: 384–392. [DOI] [PubMed] [Google Scholar]
- 52. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012; 13: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ambrosini G, Musi E, Ho AL, de Stanchina E, Schwartz GK. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol Cancer Ther. 2013; 12: 768–776. [DOI] [PubMed] [Google Scholar]
- 54. Al-Moujahed A, Nicolaou F, Brodowska K, et al.. Uveal melanoma cell growth is inhibited by aminoimidazole carboxamide ribonucleotide (AICAR) partially through activation of AMP-dependent kinase. Invest Ophthalmol Vis Sci. 2014; 55: 4175–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang W, Xiao ZD, Li X, et al.. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015; 17: 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu FX, Luo J, Mo JS, et al.. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014; 25: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jager MJ, Dogrusöz M, Woodman SE. Uveal melanoma: identifying immunological and chemotherapeutic targets to treat metastases. Asia Pac J Ophthalmol (Phila). 2017; 6: 179–185. [DOI] [PubMed] [Google Scholar]
- 58. Field MG, Harbour JW.. GNAQ/11 mutations in uveal melanoma: is YAP the key to targeted therapy? Cancer Cell. 2014; 25: 714–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun X, Han R, Wang Z, Chen Y. Regulation of adiponectin receptors in hepatocytes by the peroxisome proliferator-activated receptor-gamma agonist rosiglitazone. Diabetologia . 2006; 49: 1303–1310. [DOI] [PubMed] [Google Scholar]
- 60. Amelio I. Genes versus environment: cytoplasmic BAP1 determines the toxic response to environmental stressors in mesothelioma. Cell Death Dis . 2017; 8: e2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rauser S, Weis R, Braselmann H, et al.. Significance of HER2 low-level copy gain in Barrett's cancer: implications for fluorescence in situ hybridization testing in tissues. Clin Cancer Res. 2007; 13: 5115–5123. [DOI] [PubMed] [Google Scholar]
- 62. Cremer T, Cremer C.. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001; 2: 292–301. [DOI] [PubMed] [Google Scholar]
- 63. Williams RR. Transcription and the territory: the ins and outs of gene positioning. Trends Genet. 2003; 19: 298–302. [DOI] [PubMed] [Google Scholar]
- 64. Guasconi V, Souidi M, Ait-Si-Ali S. Nuclear positioning, gene activity and cancer. Cancer Biol. Ther. 2005; 4: 134–138. [DOI] [PubMed] [Google Scholar]
- 65. Dostie J, Bickmore WA.. Chromosome organization in the nucleus - charting new territory across the Hi-Cs. Curr Opin Genet Dev. 2012; 22: 125–131. [DOI] [PubMed] [Google Scholar]
- 66. Speicher MR, Carter NP.. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005; 6: 782–792. [DOI] [PubMed] [Google Scholar]
- 67. Schöfer C, Weipoltshammer K. Nucleolus and chromatin. Histochem Cell Biol. 2018; 150: 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gerich JE. Role of the kidney in normal glucose homeostasis and in the hyperglycaemia of diabetes mellitus: therapeutic implications. Diabet Med. 2010; 27: 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Binoux M. The IGF system in metabolism regulation. Diabete Metab. 1995; 21: 330–337. [PubMed] [Google Scholar]
- 70. Clemmons DR. Metabolic actions of insulin-like growth factor-I in normal physiology and diabetes. Endocrinol Metab Clin North Am. 2012; 41: 425–443, vii–viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cohen DH, LeRoith D.. Obesity, type 2 diabetes, and cancer: the insulin and IGF connection. Endocr Relat Cancer. 2012; 19: F27–45. [DOI] [PubMed] [Google Scholar]
- 72. Song HJ, Oh S, Quan S, et al.. Gender differences in adiponectin levels and body composition in older adults: Hallym aging study. BMC Geriatr. 2014; 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chigogora S, Zaninotto P, Kivimaki M, Steptoe A, Batty GD. Insulin-like growth factor 1 and risk of depression in older people: the English Longitudinal Study of Ageing. Transl Psychiatry. 2016; 6: e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD. Uveal melanoma: epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol. 2017:11: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zloto O, Pe'er J, Frenkel S. Gender differences in clinical presentation and prognosis of uveal melanoma. Invest Ophthalmol Vis Sci. 2013; 54: 652–656. [DOI] [PubMed] [Google Scholar]
- 76. Singh B, Saxena A.. Surrogate markers of insulin resistance: a review. World J Diabetes. 2010; 1: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Silva FM, de Almeida JC, Feoli AM. Effect of diet on adiponectin levels in blood. Nutr Rev. 2011; 69: 599–612. [DOI] [PubMed] [Google Scholar]
- 78. Tsukinoki R, Morimoto K, Nakayama K. Association between lifestyle factors and plasma adiponectin levels in Japanese men. Lipids Health Dis. 2005; 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Esfahani M, Movahedian A, Baranchi M, Goodarzi MT. Adiponectin: an adipokine with protective features against metabolic syndrome. Iran J Basic Med Sci. 2015; 18: 430–442. [PMC free article] [PubMed] [Google Scholar]
- 80. Kotani K, Hazama A, Hagimoto A, et al.. Adiponectin and smoking status: a systematic review. J Atheroscler Thromb. 2012; 19: 787–794. [DOI] [PubMed] [Google Scholar]
- 81. Su JR, Lu ZH, Su Y, et al.. Relationship of serum adiponectin levels and metformin therapy in patients with type 2 diabetes. Horm Metab Res. 2016; 48,92–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.