Abstract
Background
Protein Disulfide Isomerase (PDI) proteins are part of the Thioredoxin (TRX) protein superfamily. PDIs are involved in the formation and rearrangement of disulfide bonds between cysteine residues during protein folding in the endoplasmic reticulum (ER) and are implicated in stress response pathways.
Methods
Eight children from four consanguineous families residing in distinct geographies within the Middle East and Central Asia were recruited for study. All probands showed structurally similar microcephaly with lissencephaly (microlissencephaly) brain malformations. DNA samples from each family underwent whole exome sequencing (WES), assessment for repeat expansions, and confirmatory segregation analysis.
Results
An identical homozygous variant in TMX2 (c.500G>A), encoding Thioredoxin-related transmembrane protein 2, segregated with disease in all four families. This variant changed the last coding base of exon six, and impacted mRNA stability. All patients presented with microlissencephaly, global developmental delay, intellectual disability, and epilepsy. While TMX2 is an activator of cellular C9ORF72 repeat expansion toxicity, patients showed no evidence of C9ORF72 repeat expansions.
Conclusion
The TMX2 c.500G>A allele associates with recessive microlissencephaly, and patients show no evidence of C9ORF72 expansions. TMX2 is the first PDI implicated in a recessive disease, suggesting a protein isomerization defect in microlissencephaly.
Keywords: TMX2, thioredoxin, ER stress, microlissencephaly, protein disulfide isomerase
INTRODUCTION
Lissencephaly (LIS, lissos means smooth in Greek) refers to a smooth surface of the cerebral cortex.[1] Previously termed ‘type I’ and ‘type II’ to refer to the absence or presence of coexistent microcephaly,[1] respectively, recent lissencephaly molecular classification has identified eight recurrently mutated genes in recessive forms of disease: RELN (MIM:600514), NDE1 (MIM:609449), LAMB1 (MIM:150240), KATNB1 (MIM:602703), CDK5 (MIM:123831), TMTC3 (MIM:617218).[2–7] Primary microcephaly (MPCH) refers to reduced head size with reduced cerebral volume; and at the more severe end of the spectrum, there is often a reduction in the complexity of the folding of the cerebral cortex, termed ‘simplified gyral pattern.’[8] Therefore, most cases with microcephaly show only mild disruption of the gyral folding on brain MRI, but at the severe end of microcephaly spectrum the cerebral cortex can show gyri paucity, and can be difficult to distinguish from forms of lissencephaly. A distinct entity termed ‘microlissencephaly’ describes the combination of lissencephaly with microcephaly.[9] Of the recessive causes for lissencephaly only a few show notable additional microcephaly: RELN, NDE1, KATNB1, and a few chromosomal deletion syndromes.[2, 3, 5, 10] This suggests the existence of genetic conditions in which the degree of lissencephaly is greater than would be expected from the degree of microcephaly alone, supporting the use of the term microlissencephaly for appropriate conditions.
Some genes linked to microcephaly with lissencephaly, such as TMTC3, are implicated in the endoplasmic reticulum (ER) stress response, which is activated upon misfolding of secretory proteins or calcium balance perturbation.[11,12] Overload of ER protein folding capacity can lead to an accumulation of misfolded proteins, ER stress and subsequent cell death. Protein disulfide isomerases (PDIs) are resident transmembrane ER proteins that catalyze thiol-disulfide interchanges, which are critical for proper protein folding.[13] There are 21 known genes encoding PDIs, including three members of the Thioredoxin-related transmembrane (TMX) family, TMX-1 (MIM:610527), −2 (MIM:616715), and −3 (MIM:616102). Each TMX family member encodes an N-terminal signal peptide, single-pass transmembrane domain, C-terminal thioredoxin (Trx)-like domain, and a Di-lysine ER retention signal domain, but how they maintain protein homeostasis in the ER remains unknown.[14]
METHODS
Patient recruitment
The procedures followed for recruitment and data collection were in accordance with the ethical standards of the responsible committee on human experimentation at the University of California, San Diego.
DNA extraction and whole exome sequencing
This study was approved by the Institutional Review Board at the respective host institution. All study participants and/or their guardians signed informed consents, and the study was performed in accordance with Health Insurance Portability and Accountability Act (HIPAA) Privacy Rules. DNA was extracted from peripheral blood leukocytes with salt extraction. DNA from the probands was subjected to Agilent Sure-Select Human All Exon v2.0 (44Mb target) and Illumina Rapid Capture Enrichment (37Mb target) library preparation and sequenced on Illumina HiSeq 2000 or 4000 instruments.[15]
Computational analysis
Variant calling and filtering were performed according to a previously described whole exome sequencing pipeline.[15] Variants were filtered if not present in all affected individuals in the family, with zygosity based upon the presumed mode of inheritance. For all families, variants were filtered for minor allele frequency (MAF) >1:1000, PolyPhen-2 scores of <0.9 or GERP score <4.5. Variants were assessed with MutationTaster, and runs of homozygosity were defined with HomozygosityMapper.[16–18]
TMX2 mRNA assessment
Total RNA was extracted from fresh frozen blood on available parents and probands using TRIzol™, quantified by spectrophotometry, and reverse-transcribed using the Superscript III First-Strand cDNA Kit (Invitrogen). PCR analysis of cDNA was performed using dHPLC-purified primers (see supplementary table S2), visualized using the BioRad GelDoc™ and quantified by Azure™ software. hTMX2_ex5–7 primers amplified the region from exon five through exon seven, hTMX2_ex6–7 primers amplify the region between exon six and seven, and hTMX2_ex6–8 primers amplify the region from exon six through exon eight.
RESULTS
Identification of four consanguineous families segregating with microlissencephaly
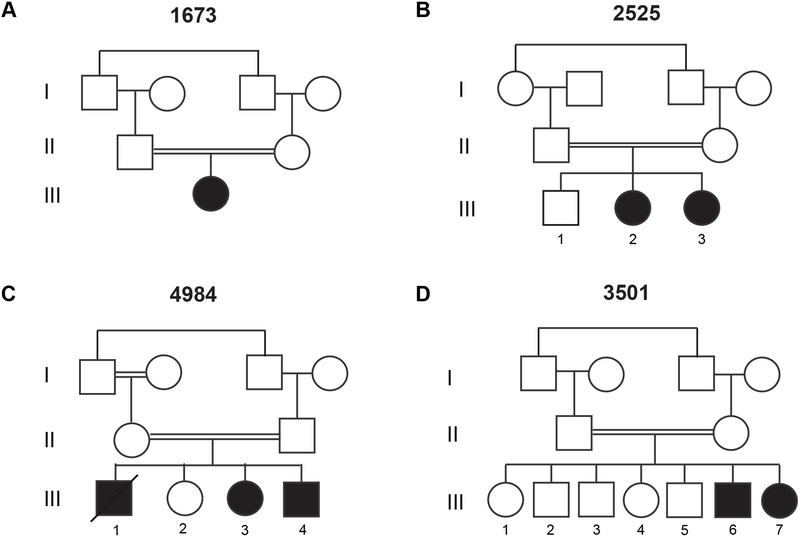
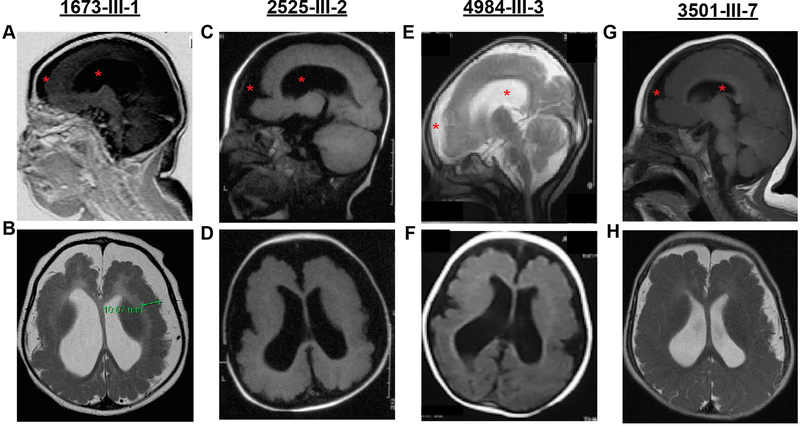
We studied eight affected individuals with neurological phenotypes consistent with microlissencephaly from four unrelated consanguineous families. Family 1673 presented with one affected child (1673-III-1) from a first-cousin marriage. The individual was born at full term with a significantly reduced head circumference (−2SD) and exhibited generalized tonic-clonic (GTC) seizures from birth (figure 1A, table 1). At her last evaluation, she exhibited severe intellectual disability, hypertonia, absence of motor skills, and increased deep tendon reflexes. Brain MRI revealed a thickened cerebral cortical mantle, with diminished gyral folds, with an appearance of pachygyria/lissencephaly, cortical atrophy, corpus callosum hypogenesis, and ventriculomegaly (figure 2A–B, supplementary figure S1A–C).
Figure 1. Consanguineous families with a homozygous recessive mutation in TMX2.
(A-D) Pedigrees of all four families showing first-cousin consanguineous marriages (double bar) with a total of eight affected children. All unfilled members are without neurological disease.
Table 1. Clinical Table.
Clinical representation for affected subjects from all four families. Y:Yes. N:No. N/A: no data available. SD: standard deviations (calculated using SimulConsult’s measurement resources). HC: head circumference. EEG: Electroencephalogram. EMG: electromyography
| Family 1673 | Family 2525 | Family 4984 | Family 3501 | |||||
|---|---|---|---|---|---|---|---|---|
| Individual | III-1 | III-2 | III-3 | III-1 | III-3 | III-4 | III-6 | III-7 |
| Gender | F | F | F | M | F | M | M | M |
| Country of Origin | Saudi Arabia | Pakistan | Egypt | Kuwait | ||||
| Parental Consanguinity | Y | Y | Y | Y | ||||
| Current age (if alive) | 11y | N/A | 6y | N/A | 3y | 2y | 8y | 6y |
| Age of death | N/A | 4y | N/A | 5y | N/A | N/A | N/A | N/A |
| Circumstances of death | N/A | Seizures | N/A | Pneumonia | N/A | N/A | N/A | N/A |
| Mutation | ||||||||
| Genomic (hg38) | Chr11:g.57739039G>A | Chr11:g.57739039G>A | Chr11:g.57739039G>A | Chr11:g.57739039G>A | ||||
| cDNA | c.500G>A | c.500G>A | c.500G>A | c.500G>A | ||||
| Protein | p.Arg205Gln | p.Arg205Gln | p.Arg205Gln | p.Arg205Gln | ||||
| Zygosity | homozygous | homozygous | homozygous | homozygous | ||||
| Perinatal History | ||||||||
| Pregnancy duration (weeks) | full term | full term | full term | full term | 36w | 36w | full term | full term |
| Weight at birth (g) | 2990g | 3300g | 3200g | 3100g | 2800g | 2900g | 3100g | 3100g |
| HC at birth (SD) | −2SD | −3SD | −2SD | −1SD | NA | −2SD | −2SD | −2SD |
| Age at last examination | 11y | 7y | 5y | 4y | 2y | 1y | 8y | 6y |
| Weight at last examination (kg) | 20kg | 17kg | 10kg | 11kg | 8kg | 7kg | 22kg | 12kg |
| Height at last examination (cm) | 125cm | 112cm | 90 cm | 92cm | 75cm | 65cm | 110cm | 85cm |
| HC at last examination (SD) | −3SD | −3SD | −4SD | −4SD | −3SD | −4SD | −4SD | −3SD |
| Psychomotor development | ||||||||
| Gross motor (normal/delayed/absent) | Absent | Delayed | Delayed | Delayed | Delayed | Absent | Delayed | Absent |
| Fine motor (normal/delayed/absent) | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| Language (normal/delayed/absent) | Absent | Absent | Absent | Delayed (babbles) | Absent | Delayed (babbles) | Absent | Delayed |
| Social (normal/delayed/absent) | Absent | Delayed | Absent | Absent | Absent | Absent | Absent | Delayed |
| Seizures | ||||||||
| Age of Onset | Neonatal | 2w | 3w | 1m | 1m | 6w | 2m | 2m |
| Type | GTC | GTC | GTC | GTC | GTC | GTC | GTC | GTC |
| Frequency | 2/w | 2/w | 1/w | 1/m | 1/m | 2/m | 4/m | 2/m |
| Controlled/Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory | Refractory |
| EEG | Multifocal spike/wave | Multifocal spike/wave | Multifocal spike/wave | Multifocal spike/wave | Multifocal spike/wave | Multifocal spike/wave | Multifocal spike/wave | Multifocal spike/wave |
| Neuroimaging | ||||||||
| Lissencephaly spectrum | Y | Y | Y | N/A | Y | N/A | Y | Y |
| Cerebral mantle thickening | Y | N | N | N/A | N | N/A | Y | Y |
| Subcortical band heterotopia | N | N | N | N/A | N | N/A | N | N |
| Corpus callosum hypogenesis | Y | Y | Y | N/A | Y | N/A | Y | Y |
| Cerebellar atrophy | N | N | N | N/A | Y | N/A | Y | Y |
| Brainstem hypoplasia | N | N | N | N/A | Y | N/A | Y | Y |
| Ventriculomegly | Y | Y | Y | N/A | Y | N/A | Y | Y |
| Reduced white matter | Y | Y | Y | N/A | Y | N/A | Y | Y |
| Neurological Examination | ||||||||
| Intellectual disability (mild/moderate/severe) | Severe | Severe | Severe | Severe | Severe | Severe | Severe | Severe |
| Hypertonia | Y | Y | Y | Y | Y | Y | Y | Y |
| Hypotonia | N | N | N | N | N | N | N | N |
| Deep tendon reflexes | Increased | Increased | Increased | Increased | Increased | Increased | Increased | Increased |
| Spastic tetraplegia | Y | Y | Y | Y | Y | Y | Y | Y |
| Ataxia | N | N | N | N | N | N | N | N |
| EMG/Biopsy | N/A | N/A | N/A | N/A | EMG normal | EMG normal | N/A | N/A |
| Sensory | ||||||||
| Vision | Fixes/Follows | Fixes/Follows | Fixes/Follows | Fixes/Follows | Fixes/Follows | Fixes/Follows | Fixes/Follows | Fixes/Follows |
| Hearing | Responds to noise | Responds to noise | Responds to noise | Responds to noise | Responds to noise | Responds to noise | Responds to noise | Responds to noise |
| Other Clinical Features | ||||||||
| Dysmorphism | N | N | N | N | N | N | N | N |
Figure 2. MRIs from affected children display lissencephaly, microcephaly, and brain atrophy.
Top row: Midline sagittal MRI. Bottom row: Axial MRI. Subject ID provided at top. All images are T1-weighted except (B, E, H) which are T2 weighted, selected to best demonstrate the main imaging defects. The lack of cortical folding is apparent in both frontal and occipital regions, consistent with lissencephaly. Red asterisk in top row highlights ventriculomegaly and increased extra-axial fluid accumulation, consistent with brain atrophy and microcephaly. Bottom row shows paucity of cortical gyri and thickened gray matter, which was measured at 10.07 mm (green bracket) in (B), which is roughly twice the width of normal.
Family 2525 had two affected children (2525-III-2, and 2525-III-3) from a consanguineous marriage (figure 1B). Both children were born full term with significantly reduced head circumference (−3SD and −2SD, respectively). They also exhibited GTC seizures in the neonatal period with an onset of 2 weeks and 3 weeks, respectively. At their last evaluation, they were diagnosed with severe intellectual disability, increased deep tendon reflexes, delayed gross motor development, and absence of fine motor and language development (table 1). Brain MRI assessment of the older sibling revealed lissencephaly, reduced white matter volume, cortical atrophy, corpus callosum hypogenesis, and ventriculomegaly. (figure 2C–D, supplementary figure S1D–F).
Family 4984 had three affected children (4984-III-1, 4984-III-3, and 4984-III-4) and one healthy sibling from a consanguineous marriage (figure 1C). The oldest affected child was born at full term with a reduced head circumference (−1SD). He was also diagnosed with severe intellectual disability, delayed gross motor and language development, absent fine motor development, and epilepsy. He died at 5 years of age due to pneumonia. His younger two affected siblings (4984-III-3, and 4984-III-4) were both born at 36 weeks and showed reduced head circumference at last evaluation (−4SD and −3SD, respectively). They exhibited similar symptoms as their older sibling. Brain MRIs were obtained from one child (4984-III-3), and showed lissencephaly, corpus callosum hypoplasia, reduced white matter volume, ventriculomegaly, and cerebellar atrophy (figure 2E–F, supplementary figure S1G–I).
Family 3501 had two affected children (3501-III-6 and 3501-III-7) and five healthy siblings from a consanguineous marriage (figure 1D). Both affected individuals were born at full term with significantly reduced head circumferences (−2SD). They also exhibited neonatal epilepsy, which predated their diagnoses of severe intellectual disability, delayed/absent psychomotor development, and increased tendon reflexes at last evaluation. Brain MRIs for both children showed lissencephaly, cerebral mantle thickening, ventriculomegaly, reduced white matter volume, cerebellar atrophy, and corpus callosum hypoplasia (figure 2G–H, supplementary figure S1J–O).
Identification of a homozygous TMX2 c.500G>A variant in families with microlissencephaly
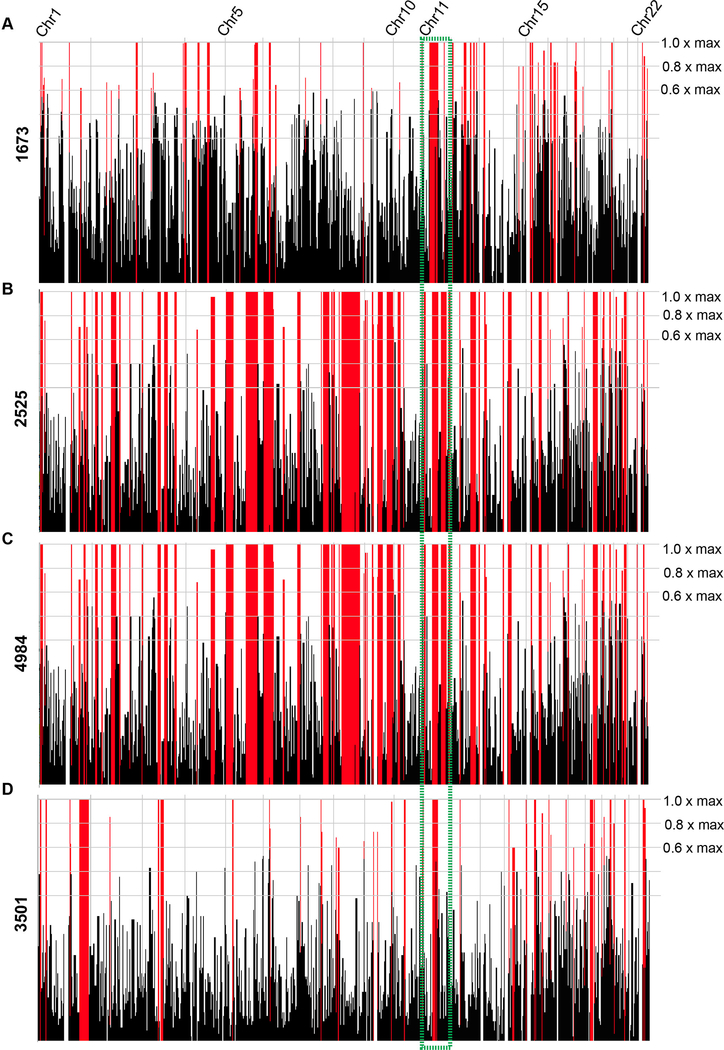
Whole exome sequencing (WES) was performed on blood-derived DNA from the affected child in Family 1673. For variant identification Genome Analysis Toolkit (GATK) workflow identified variants that were intersected with identity-by-descent blocks from HomozygosityMapper.[18,19] Rare potentially deleterious variants were prioritized against an in-house exome database consisting of over 4,000 ethnically matched individuals, in addition to publicly available exome datasets, cumulatively numbering over 10,000 individuals. From this analysis, we identified three variants that passed these filter paramters in the genes RAB3GAP2 (MIM:609275), TMX2, and MOCOS (MIM:613274) (see supplementary table S1). Patient phenotypes did not match the phenotypes reported for RAB3GAP2 and MOCOS, leaving a single, homozygous hg38:Chr11:g.57739039G>A (GenBank: NM_001347898.1) variant in TMX2, located at the last base of exon six of eight (c.500G>A), as the sole candidate. We then performed WES on one affected child each from Families 2525 and 4984, and both affected children from Family 3501, based on matching phenotypic features between the families. Interestingly, we identified the same TMX2 variant recurring in all three families (supplementary table S1). Further, each of the four consanguineous families showed a run of homozygosity that contained TMX2, consistent with the parental consanguinity (figure 3), although the haplotype blocks showed no commonality of shared SNPs of any size (supplementary figure S2). This suggests that the variant arose independently in each family on separate haplotypes, or was so distantly derived that a common haplotype could not be identified. This variant was confirmed by direct Sanger sequencing analysis, and testing of all available family members demonstrated segregation with the phenotype according to a recessive mode of inheritance (supplementary figure S3). We found no additional families in our own cohort of over 5000 individuals, or in the GeneMatcher website, of patients displaying likely causative variants in this gene. The variant was also not identified in the Greater Middle Eastern (GME) Variome (consisting of 2,497 individuals) or any other public database.[20] These analyses made TMX2 the top candidate for pathogenesis.
Figure 3. Homozygosity plots for all four families display homozygosity surrounding the TMX2 gene.
(A-D) Graphical representation of regions of homozygosity, generated by HomozygosityMapper for all 22 autosomes for the affected individual in Family 1673 (III-1, A), one affected individual from Family 2525 (III-2, B), one affected individual from Family 4984 (III-3, C), and both affected individuals from Family 3501 (III-6 and III-7, D). Red peaks indicate homozygosity scores above the given cutoff at 0.8x of the maximum. All plots show a region of homozygosity containing the TMX2 gene (green box).
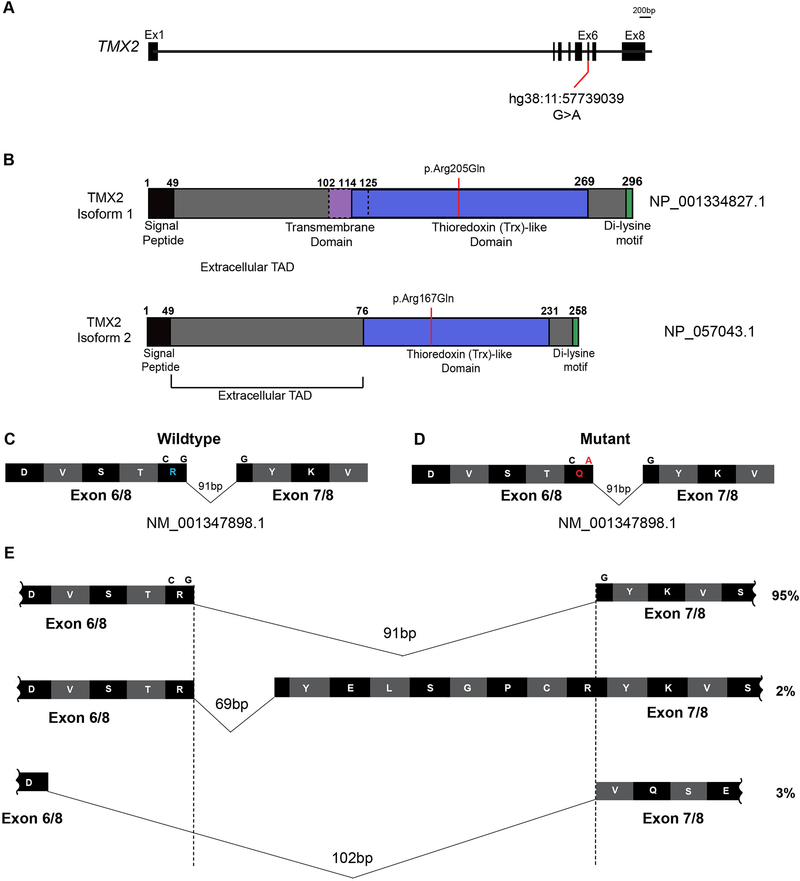
TMX2 encodes for Thioredoxin-related transmembrane protein 2, and the variant introduced an amino acid substitution in the Trx-like domain of the protein (p.Arg205Gln, figure 4A–B). The p.Arg205Gln substitution was also predicted to be disease-causing by MutationTaster.[17] TMX2 has two protein-coding isoforms, isoform 1 and 2 (NP_001334827.1 and NP_057043.1), which differ in the length of the Extracellular Topological Domain (TAD) and absence of the transmembrane domain in isoform 2 (figure 4B), but functional differences between these isoforms are not reported.
Figure 4. TMX2 gene and predicted protein isoforms.
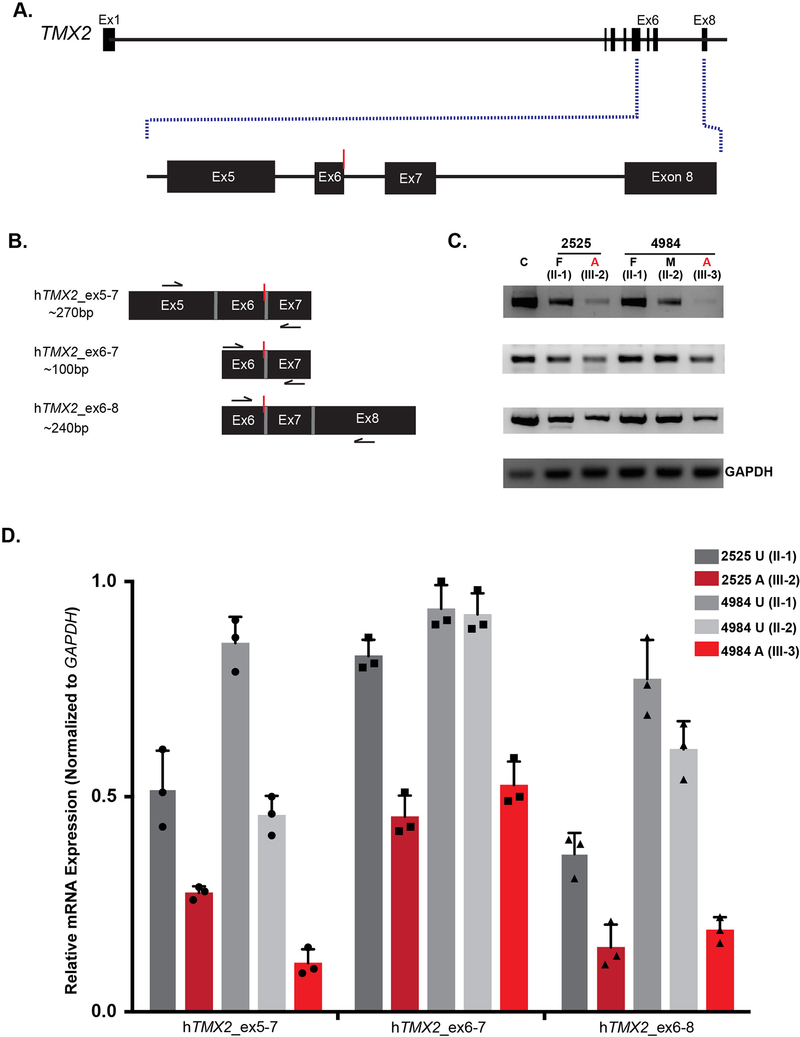
(A) Schematic of TMX2 depicting the genomic map spanning eight exons. Red line indicates the position of the identified variant in exon six in all four families and coordinates within the cDNA map (RefSeq NM_001347898.1). (B) Schematic of the two protein-coding isoforms of Thioredoxin-related transmembrane protein 2 (TMX2) depicting the location of the ‘signal peptide’ (black), extracellular topological associated domain (TAD), transmembrane domain (purple), Thioredoxin (Trx)-like domain (blue), and Di-lysine motif (green). Isoform 2 is 38 amino acids shorter than isoform 1 due to absence of the transmembrane domain and part of the extracellular TAD. Red line indicates the position and coordinates of the amino acid change in affected individuals from all four families within the Trx-like domain. (C-D) Illustration of partial wildtype (C) and mutant (D) residues from exons six and seven surrounding the variant shown in blue and red in wildtype and mutant, respectively. (E) Schematic of the additional splice isoforms of the exon6–7 junction of TMX2, based on the UCSC Genome Browser with predicted percent existence of each isoform in human tissues from GenBank. The percentage of each isoform was calculated based on the average number of Expressed Sequence Tags (ESTs) available, displaying that particular isoform (e.g. BI764082, BG170274, and BI561678, respectively).
As this homozygous variant occurred in the last nucleotide of exon 6 (figure 4D), we considered a potential impact on TMX2 splicing or mRNA stability. In addition to the two canonical protein-coding isoforms, TMX2 is also predicted to undergo alternative splicing at the exon 6–7 junction, which is a particularly small intron. Approximately 95% of observed transcripts from NCBI conform to a 91 bp intron, with the amino acid sequence reading …DVSTRYKVS…. One minor isoform, representing approximately 2% of transcripts, contains a 69 bp intron, with the sequence reading …DVSTRYELSGPCRYKVS…, due to the use of an alternative 3’ splice acceptor site. The other minor isoform, representing approximately 3% of transcripts, conforms to a 102 bp intron, with the sequence reading …DVQSE… (figure 4E), yet contains a premature stop codon approximately 26 amino acids downstream, suggesting a non-functional isoform. Whether or not these isoforms are found in nature or encode for protein has yet to be elucidated.
The TMX2 c.500G>A mutation results in reduced mRNA levels in affected individuals
TMX2 is ubiquitously expressed in adult human tissues (GTEx data, supplementary figure S4). Additionally, TMX2 is expressed in human brain during development at all assessed stages (supplementary figure S5), consistent with a role in this phenotype. Because the variant alters the last base of an exon, we considered whether splicing or RNA stability could be impacted. Thus RNA was extracted from frozen whole blood obtained from the father and one affected individual from Family 2525 as well as both parents and one affected from Family 4984. mRNA transcript levels were assessed using RT-PCR across various regions flanking the variant (figure 5A) and compared to an unrelated control sample. Three primer pairs were designed: 1] the region between exons five and seven, 2] the region between exon six and seven, 3] the region between exons six and eight (figure 5B, supplementary table S2). We did not observe evidence of any novel splice isoforms, such as might result from exon skipping or intron inclusion; however, TMX2 mRNA levels were reduced to 15–50% of control in both samples from affecteds compared with the healthy control, whereas unaffected obligate carriers showed levels between 50–90% of normal (figure 5C–D). The reduced mRNA levels in patients and carriers is consistent with an effect of TMX2 c.500G>A on splicing or RNA stability, and the absence of novel bands in the affected and carrier individuals suggests nonsense mediated decay of aberrantly spliced mRNA.
Figure 5. TMX2 c.500G>A allele associates with reduced mRNA levels in carriers and affected individuals.
(A) Illustration of TMX2 with an expanded view of exons five through eight. Red line indicates the location of the variant. (B) Schematic of primer pairs designed for RT-PCR analysis and expected base pair sizes for the corresponding products. Black arrows indicate the approximate location of the primers. Red line indicates the location of the variant. (C) RT-PCR analysis of RNA extracted from whole blood derived from an unrelated control (C), the unaffected father (II-1) and one affected individual (III-2) from Family 2525, and both parents (II-1 and II-2) and one affected individual (III-3) from Family 4984. There was reduction of TMX2 mRNA levels in affected individuals (red) compared to the normalized expression of 1.0 in the unrelated control (n=3). TMX2 levels of parent carriers (grey) that were intermediate compared with unrelated control. GAPDH was used as a loading control. (D) Quantification of RT-PCR results displaying relative mRNA expression normalized to GAPDH. n=3 independent experiments. Bars show mean ± standard deviation (SD) and individual data points.
Lack of C9ORF72 repeat expansion in TMX2 mutated samples
C9ORF72 gene mutations are the most common cause of Amyotrophic Lateral Sclerosis (ALS, MIM:105400) and Frontotemporal Dementia (FTD, MIM:600274).[20,21] The damaging mutation in C9ORF72 associated with these diseases is an expanded noncoding hexanucleotide repeat (GGGGCC), leading to repeat-associated non-ATG (RAN) translation of a dipeptide repeat protein (DPR).[21,22] Previous transcriptomic analysis using C9ORF72-mutant ALS brain tissue revealed upregulation of genes involved in ER stress,[23] suggesting failed ER stress response in C9ORF72 pathogenesis. Following a CRISPR-Cas9 KO screen for genetic modifiers of DPR toxicity, TMX2 emerged as one of the two strongest protective modifier genes identified.[24] In these experiments, loss of TMX2 suppressed the toxic effect of the C9ORF72 DPR, suggesting that TMX2 is an activator of DPR toxicity. Therefore, we wanted to test the possibility that the TMX2 mutation occurred in the genetic background of C9ORF72 expansion.
To test this, we performed repeat-primed PCR using a previously validated detection method to screen all available DNA samples from three families for the presence of the GGGGCC hexanucleotide repeat expansion (Family 3501 failed analysis due to insufficient amounts of DNA available).[25] All remaining samples were absent for the expanded C9ORF72 repeat, as compared to the negative control and an unrelated positive control sample (figure S6).
DISCUSSION
Here we demonstrate a recurrent variant in TMX2 associated with a severe, recessive neurological disease characterized by microcephaly with lissencephaly. All affected individuals in four families carried an identical homozygous c.500G>A variant in TMX2 and displayed microlissencephaly, global developmental delay, intellectual disability, and epilepsy. The variant led to decreased mRNA steady-state levels, and was not associated with expansion in the C9ORF72 hexanucleotide repeat. Our results implicate a relatively common single allele as a cause for microlissencephaly in humans.
TMX2 is arguably the least studied member of the PDI family. TMX2 was cloned in 2003, and encodes a multidomain transmembrane protein that is enriched on the mitochondria-associated membrane (MAM) of the ER via palmitoylation of two of its cystolically-exposed cysteines.[13,26] The MAM is a domain of the ER that mediates the exchange of ions, lipids and metabolites between the ER and mitochondria, suggesting a potential function in either mitochondria, ER, or both. TMX2 shows ubiquitous expression, with highest levels detected in heart, brain, liver, kidney and pancreas.[12,13,27] Whether TMX2 displays catalytic activity remains unclear, given that the canonical CXXC motif required for oxioreductase activity is replaced by an SXXC motif.[28]
There was one prior mention of TMX2 in human disease, where a cohort of patients with malformations of cortical development was sequenced to identify causes.[29] One family was identified with two children affected with lissencephaly where two candidate genes emerged, one being TMX2.[29] In this family, TMX2 showed compound heterozygous mutations [(c.326A>G; p.Asp109Gly) and (c.691C>T; p.Arg231Trp)], but brain MRI was not available to allow comparisons with our cases. Our results, linking TMX2 to lissencephaly, suggest that, in the previous family, TMX2 was likely the relevant gene.
Mutations in genes that encode ER stress response proteins have been implicated in a range of neuroprogressive disorders among other human diseases, including Parkinson’s disease, Huntington’s disease, and ALS, the latter of which has been studied in connection with ER stress response pathways and apoptosis.[30–33] TMX2 knockdown or CRISPR targeting was able to rescue the cell death and ER stress response toxicity induced by application of the DPR encoded by the C9ORF72 expansion. CRISPR targeting of TMX2 in DPR-expressing human cells showed upregulation of prosurvival unfolded-protein-response-pathway genes including Atf3, and downregulation of calcium-binding and apoptotic proteins.[24] We found no evidence of expansion of the C9ORF72 repeat in patient cells, suggesting that loss of TMX2 was not secondary to effects of C9ORF72, but it is possible that the mutated TMX2 allele is under positive selection, as it appears to have arisen independently on four separate haplotypes. It would be interesting as a next step to understand mechanism by generating an animal model of disease; however, initial attempts indicate that homozygous Tmx2 null mutations in mouse are lethal prior to weaning, with complete penetrance.[34]
The presence of a Trx-like domain in TMX2 suggests a role regulating redox pathways, specifically in relation to the ER stress response. Partial or complete loss of TMX2 may thus predispose to cell death. While we have not linked the phenotypes from our cases to a molecular function in this study, increased ER stress during neurodevelopment has been linked to aberrant neuronal maturation and the subsequent development of neurodevelopmental disorders caused by abnormal neuronal differentiation and maturation.[35–36] We hypothesize that loss of TMX2 in humans may lead to cell-death mediated depletion of neural progenitor cells in the developing cerebral cortex during embryogenesis, producing microlissencephaly. Whether correct, or whether there are secondary effects of TMX2 in neurogenesis or neuronal migration, remain to be explored.
Supplementary Material
Acknowledgments
The authors thank the patients and their families for participation in this study. We would like to thank Grazia Mancini for communicating unpublished results. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 03/03/2018.
Funding: S.G. was sponsored by the Ruth L. Kirschstein Institutional National Research Service Award (T32 GM008666) from the National Institute on Deafness and Other Communication Disorders and by award F31HD095602 from the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank the Broad Institute (U54HG003067 to E. Lander and UM1HG008900 to D. MacArthur) and the Yale Center for Mendelian Disorders (U54HG006504 to M. Gunel). This work was supported by NIH grants R01NS048453, R01NS052455, the Simons Foundation Autism Research Initiative and Howard Hughes Medical Institute (to J.G.G.). This work was supported in part by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02 to B.J. Traynor).
Footnotes
Competing Interests: The authors declare no conflict of interest.
Data availability: Data are available in a public, open access repository. The exome sequencing data from individuals from the University of California, San Diego, study site have been deposited in the Database of Genotypes and Phenotypes under accession number dbGaP:phs000382.v1.p1.
WEB RESOURCES
RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
HomozygosityMapper, http://www.homozygositymapper.org/
Mutation Taster, http://www.mutationtaster.org/
Exome aggregation Consortium, ExAc Browser http://exac.broadinstitute.org/
1000 Genomes, http://browser.1000genomes.org
GME Variome, http://igm.ucsd.edu/gme
The Genotype-Tissue Expression (GTEx) Project, https://www.gtexportal.org/home/
BrainSpan – Atlas of the Developing Human Brain, http://www.brainspan.org/
Human Splicing Finder, http://www.umd.be/HSF/
UCSC Genome Browser, https://genome.ucsc.edu/
SimulConsult – Measurement resources, http://www.simulconsult.com/resources/measurement.html
Python Software Foundation, https://www.python.org/
NCBI database of Genotypes and Phenotypes (dbGaP, http://www.ncbi.nlm.nih.gov/gap)
UniProt, https://www.uniprot.org/
REFERENCES
- 1.Fry AE, Cushion TD, Pilz DT. The genetics of lissencephaly. Am J Med Genet C Semin Med Genet 2014;166C:198–210. [DOI] [PubMed] [Google Scholar]
- 2.Zaki M, Shehab M, El-Aleem AA, Abdel-Salam G, Koeller HB, Ilkin Y, Ross ME, Dobyns WB, Gleeson JG. Identification of a novel recessive RELN mutation using a homozygous balanced reciprocal translocation. Am J Med Genet 2007;143A:939–944. [DOI] [PubMed] [Google Scholar]
- 3.Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, Kentab A, Jan M, Shaheen R, Feng Y, Walsh CA. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am J Hum Genet 2011;88:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radmanesh F, Caglayan AO, Silhavy JL, Yilmaz C, Cantagrel V, Omar T, Rosti B, Kaymakcalan H, Gabriel S, Li M, Sestan N, Bilguvar K, Dobyns WB, Zaki MS, Gunel M, Gleeson JG. Mutations in LAMB 1 cause cobblestone brain malformation without muscular or ocular abnormalities. Am J Hum Genet 2013;92:468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra-Gorur K, Çağlayan AO, Schaffer AE, Chabu C, Henegariu O, Vonhoff F, Akgümüş GT, Nishimura S, Han W, Tu S, Baran B, Gümüş H, Dilber C, Zaki MS, Hossni HA, Rivière JB, Kayserili H, Spencer EG, Rosti RÖ, Schroth J, Per H, Çağlar C, Çağlar Ç, Dölen D, Baranoski JF, Kumandaş S, Minja FJ, Erson-Omay EZ, Mane SM, Lifton RP, Xu T, Keshishian H, Dobyns WB, Chi NC, Šestan N, Louvi A, Bilgüvar K, Yasuno K, Gleeson JG, Günel M. Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron 2014;84:1226–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magen D, Ofir A, Berger L, Goldsher D, Eran A, Katib N, Nijem Y, Vlodavsky E, Tzur S, Behar DM, Fellig Y, Mandel H. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with a loss-of-function mutation in CDK5. Hum Genet 2015;134: 305–314. [DOI] [PubMed] [Google Scholar]
- 7.Jerber J, Zaki MS, Al-Aama JY, Rosti RO, Ben-Omran T, Dikoglu E, Silhavy JL, Caglar C, Musaev D, Albrecht B, Campbell KP, Willer T, Almuriekhi M, Caglayan AO, Vajsar J, Bilguvar K, Ogur G, Abou Jamra R, Gunel M, Gleeson JG. Biallelic mutations in TMTC3, encoding a transmembrane and TPR-containing protein, lead to cobblestone lissencephaly. Am J Hum Genet 2016;99:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, Pushparaj P, Ahmed F, Algahtani HA, Al-Qahtani MH, Saleh Jamal H. Molecular genetics of human primary microcephaly: an overview. BMC Med Genomics 2015;8:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkovich AJ, Ferriero DM, Barr RM, Gressens P, Dobyns WB, Truwit CL, Evrard P. Microlissencephaly: a heterogeneous malformation of cortical development. Neuropediatrics 1998;29:113–9. [DOI] [PubMed] [Google Scholar]
- 10.Parrini E, Conti V, Dobyns WB, Guerrini R. Genetic Basis of Brain Malformations. Mol Syndromol 2016;7:220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Racape M, Van Huyen JPD, Danger R, Giral M, Bleicher F, Foucher Y, Pallier A, Pilet P, Tafelmeyer P, Ashton-Chess J, Dugast E, Pettre S, Charreau B, Soulillou JP, Brouard S. The involvement of SMILE/TMTC3 in endoplasmic reticulum stress response. PLoS One 2011;6:e19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 2000;403:98–103. [DOI] [PubMed] [Google Scholar]
- 13.Meng X, Zhang C, Chen J, Peng S, Cao Y, Ying K, Xie Y, Mao Y. Cloning and identification of a novel cDNA coding thioredoxin-related transmembrane protein 2. Biochem Genet 2003;41:99–106. [DOI] [PubMed] [Google Scholar]
- 14.Matsusaki M, Kanemura S, Kinoshita M, Lee YH, Inaba K, Okumura M. The Protein Disulfide Isomerase Family: from proteostasis to pathogenesis. Biochim Biophys Acta Gen Subj 2019;19:30080–7. [DOI] [PubMed] [Google Scholar]
- 15.Dixon-Salazar TJ, Silhavy JL, Udpa N, Schroth J, Bielas S, Schaeffer AE, Olvera J, Bafna V, Zaki MS, Abdel-Salam GH, Mansour LA, Selim L, Abdel-Hadi S, Marzouki N, Ben-Omran T, Al-Saana NA, Sonmez FM, Celep F, Azam M, Hill KJ, Collazo A, Fenstermaker AG, Novarino G, Akizu N, Garimella KV, Sougnez C, Russ C, Gabriel SB, Gleeson JG. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med 2012;4:138ra78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seelow D, Schuelke M. HomozygosityMapper 2012--bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res 2012;40:W516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7:575–576. [DOI] [PubMed] [Google Scholar]
- 18.Seelow D, Schuelke M, Hildebrandt F, Nurnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res 2009;37:W593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, Gabriel SB, Belkadi A, Boisson B, Abel L, Clark AG, Greater Middle East Variome Consortium, Alkuraya FS, Casanova JL, Gleeson JG. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 2016;48:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo MA, Banks E, Poplin RE, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Radsemakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011;72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M; ITALSGEN Consortium, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ<a/u>, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, Bieniek KF, DeTure M, Lee WC, Biendarra SM, Davis MD, Baker MC, Perkerson RB, van Blitterswijk M, Stetler CT, Rademakers R, Link CD, Dickson DW, Boylan KB, Li H, Petrucelli L. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci 2015;18:1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer NJ, Haney MS, Morgens DW, Jovičić A, Couthouis J, Li A, Ousey J, Ma R, Bieri G, Tsui CK1, Shi Y, Hertz NT, Tessier-Lavigne M, Ichida JK, Bassik MC, Gitler AD. CRISPR–Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat Genet 2018;60:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M; ITALSGEN Consortium, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucelotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynes EM, Bui M, Yap MC, Benson MD, Schneider B, Ellgaard L, Berthiaume LG, Simmen T. Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J 2012;31:451–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics 2012;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep 2005;6:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zillhaardt JL, Poirier K, Broix L, Lebrun N, Elmorjani A, Martinovic J, Saillour Y, Muraca G, Nectoux J, Bessieres B, Fallet-Biano C, Lyonnet S, Dulac O, Odent S, Rejeb I, Ben Jemaa L, Rivier F, Pinson L, Genevieve D, Musizzano Y, Bigi N, Leboucq N, Giuliano F, Philip N, Vilain C, Van Bogaert P, Maurey H, Beldjord C, Artiguenave F, Boland A, Olaso R, Masson C, Nitschke P, Deleuze JF, Bahi-Buisson N, Chelly J. Mosaic parental germline mutations causing recurrent forms of malformations of cortical development. Eur J Hum Genet 2016;4:611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiser BK, Yim D, Chow IT, Gonzalez S, Dai Z, Mann HH, Strong RK, Groh V, Spies TD. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature 2007;447:482–486. [DOI] [PubMed] [Google Scholar]
- 31.Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins.Nat Chem Biol 2010;6:900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, Nunan J, Rembach A, Nagley P, Beart PM, Cheema SS, Horne MK. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem 2006;281:30152–30165. [DOI] [PubMed] [Google Scholar]
- 33.Benham AM. The Protein Disulfide Isomerase Family: Key Players in Health and Disease. Antioxid Redox Signal 2012;16:781–89. [DOI] [PubMed] [Google Scholar]
- 34.https://www.mousephenotype.org/data/genes/MGI:1914208, month accessed: June 2019.
- 35.Kawada K, Lekumo T, Kaneko M, Nomura Y, Okuma Y. ER stress-induced aberrant neuronal maturation and neurodevelopmental disorders. Yakugaku Zasshi 2016;136:811–5. [DOI] [PubMed] [Google Scholar]
- 36.Breuss MW, Nguyen A, Song Q, Nguyen T, Stanley V, James KN, Musaev D, Chai G, Wirth SA, Anzenberg P, George RD, Johansen A, Ali S, Zia-Ur-Rehman M, Sultan T, Zaki MS, Gleeson JG. Mutations in LNPK, Encoding the Endoplasmic Reticulum Junction Stabilizer Lunapark, Cause a Recessive Neurodevelopmental Syndrome. Am J Hum Genet 2018;103:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.