Abstract
Exposure to arsenic, a class I carcinogen, affects 200 million people globally. Skin is the major target organ but the molecular etiology of arsenic-induced skin carcinogenesis remains unclear. As3+-induced disruption of alternative splicing could be involved, but the mechanism is unknown. Zinc finger proteins play key roles in alternative splicing. Arsenite (As3+) can displace zinc (Zn2+) from C3H1 and C4 zinc finger motifs (zfms), affecting protein function. ZRANB2, an alternative splicing regulator with two C4 zfms integral to its structure and splicing function was chosen as a candidate for this study. We hypothesized that As3+ could displace Zn2+from ZRANB2 altering its structure, expression and splicing function. As3+/Zn2+ binding and mutual displacement experiments were performed with synthetic apo-peptides corresponding to each ZRANB2 zfm, employing a combination of intrinsic fluorescence, UV spectrophotometry, zinc colorimetric assay and liquid chromatography-tandem mass spectrometry. ZRANB2 expression in HaCaT cells acutely exposed to As3+ (0 or 5 μM; 0-72 h, or 0-5 μM; 6 h) was examined by RT-qPCR and immunoblotting. ZRANB2-dependent splicing of TRA2B mRNA, a known ZRANB2 target, was monitored by RT-PCR. As3+ bound to, as well as displaced Zn2+ from, each zfm. Also, Zn2+ displaced As3+ from As3+-bound zfms acutely, albeit transiently. As3+ exposure induced ZRANB2 protein expression between 3-24 h and at all exposures tested, but not ZRANB2 mRNA expression. ZRANB2-directed TRA2B splicing was impaired between 3-24 h post-exposure. Furthermore, ZRANB2 splicing function was also compromised at all As3+ exposures, starting at 100 nm. We conclude that As3+ exposure displaces Zn2+ from ZRANB2 zfms, changing its structure and compromising splicing of its targets, and increases ZRANB2 protein expression as a homeostatic response both at environmental/toxicological exposures and therapeutically relevant doses.
Keywords: Alternative splicing, arsenic, zinc finger, ZRANB2
Graphical Abstract
INTRODUCTION
About 200 million individuals in over 70 countries around the globe are chronically exposed to arsenic 1. Most exposure is via contaminated groundwater, although, exposure from food, occupational and industrial sources are also known 2. Acute iatrogenic exposure, as with anti-leukemia chemotherapy 3 or in traditional medicines 4 are also well documented. Chronic exposure results in the development of long term adverse health effects, including multi-organ cancers, non-cancerous skin lesions, as well as higher risk of diabetes, peripheral neuropathy, cardiovascular disorders, etc. 1, 2. Skin cancers, including Bowen’s disease, basal cell carcinoma (BCC) and cutaneous squamous cell carcinoma (cSCC) are the most common forms of arsenic-induced cancer. Squamous cell carcinoma is an insidious form of malignant skin cancer with high rates of recurrence 5. While ultraviolet (UV) radiation is the most well-known and characterized cause of cSCC, arsenic is the second most common. Interestingly, arsenic-induced cSCC appears almost exclusively on covered areas of the body, indicating a distinct molecular etiology compared to UV-induced cSCC. cSCC also can result from sub-chronic iatrogenic arsenic exposures 6.
How arsenic causes skin cancer at the molecular level has been fiercely debated for a considerable period of time. Several hypotheses have been propounded and questioned based on ambiguous experimental design and data that vary with wide disparity in dosage, period of exposure and model cell line/organism used 7-9. Most common theories include induction of DNA damage, clastogenesis, aneuploidy, induction of reactive oxygen/nitrogen species, impairment of DNA damage response and repair, and miRNA dysregulation 10-13. Another mechanism that has come to the forefront of cancer research in the last decade is alternative mRNA splicing. Alternative splicing is a highly regulated process that generates transcriptomic and proteomic diversity by producing multiple mRNA isoforms from a single gene. The process is tightly regulated and in association with nonsense mediated mRNA decay, maintains the cellular profile of canonical protein isoforms under normal conditions. However, dysregulation of alternative splicing can result in adverse health outcomes 14-16. Mounting evidence suggests that dysregulated alternative splicing is a hallmark of carcinogenesis, as well as a potential therapeutic target 15-20. There is also evidence suggesting that individual alternative splicing events are actually components of genome-wide coordinated change of multiple, co-regulated alternative splicing events rather than being isolated incidents 21. Although the mechanisms of such coordination are not well understood, it does put the spotlight onto regulators of alternative splicing as putative targets in carcinogenesis 22.
Many of the proteins involved in splicing contain zinc finger motifs (zfms) 23. These zfms are absolutely critical for RNA binding and consequently splicing 24-26. Recent research suggests As3+ exposure leads to dysregulation of critical DNA repair proteins which often contain zfms 27, 28-30. As3+ binds to C3H1 (3 cysteine and 1 histidine residues) and C4 (four cysteine residues) zfms causing Zn2+ loss from these sites 28, 31, 32. Such binding and consequent Zn2+ loss leads to experimentally demonstrable loss/alteration of protein function 32-34. In fact, abrogation of the function of zfms in DNA repair proteins have been postulated to be a key mechanism in carcinogenesis 35. It is thus reasonable to hypothesize that any splice factor containing C3H1 or C4 zfms is a potential target for As3+ mediated Zn2+ displacement, leading to functional abrogation. However, no study has ever looked at As3+ mediated Zn2+ displacement in any splicing factor. One study documented 104 differential alternative splicing events upon As3+ exposure in the BEAS-2B cell line 36. This observation further suggests that exposure to As3+ could be altering the mechanism of splicing by hitherto unknown mechanisms. This gap in understanding prompted us to determine if As3+ is capable of displacing Zn2+ from a splicing regulatory protein, altering its structure and/or function.
In the present study, we chose ZRANB2 as a zfm containing splice regulator protein of interest. ZRANB2 is an evolutionarily conserved zfm protein with a well-validated role in the regulation of alternative splicing. The protein contains two C4 zfms, each of which recognizes an AGGUAA site and binds to single stranded RNA with high specificity 37. ZRANB2 is responsible for splice site choice that drives alternative splicing and isoform specific gene expression regulation 37-40. Identified splice targets of ZRANB2 include mRNAs encoding TRA2B, DRD2, WDR78 and ACAP1 among others 39-41. Furthermore, ZRANB2 physically interacts with core spliceosomal components U2AF1 and SNRNP70 and this interaction is hypothesized to stabilize the splicing complex 38, 40. Given its established role as a splice regulator, coupled with the presence of two C4 zfms, ZRANB2 is an ideal candidate for the present study.
Here we demonstrate that As3+ binds to each of the two ZRANB2 zfms displacing Zn2+ and altering the structure of these motifs. In addition, we demonstrate that acute As3+ treatment of HaCaT cells leads to changes in ZRANB2 expression as well as differential splicing of ZRANB2 target TRA2B (both at environmentally relevant and therapeutic doses), itself an alternative splicing factor 42, 43. The present study provides a mechanistic understanding of how As3+ exposure can modulate the process of alternative splicing and protein isoform expression profiles.
MATERIALS AND METHODS
Caution: Sodium arsenite is a potent carcinogen and should be handled in accordance with NIH Guidelines for the Use of Chemical Carcinogens.
Chemicals.
Synthetic apo-peptides corresponding to the two ZRANB2 zfms (For sequences, see Table 1) were obtained from GenScript Corp. (Piscataway, NJ, USA) at a purity >95%. Zinc chloride (ZnCl2; CAS 7646-85-7) was obtained from Millipore-Sigma (St. Louis, MO, USA). Fetal Bovine Serum (characterized) was obtained from Hyclone (Logan, UT, USA). Sodium arsenite (NaAsO2; CAS 7784-0698), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP HCl; CAS 5180545-9) and UltraPure™ DNase/RNase-Free Distilled Water, MEM alpha modification media, trypsin, ethylene diamine tetraacetic acid, penicillin/ streptomycin and L-glutamine were obtained from Thermo Fisher Scientific Inc. (Waltham, MA, USA), as were all other chemicals unless mentioned specifically.
Table 1.
Sequence of ZRANB2 Zfm Apo-peptides
| Name | Peptide sequence | Length | Predicted MW (Da) |
Predicted MW - Fully Reduced (Da) |
Location in ZRANB2 Protein |
|---|---|---|---|---|---|
| ZNF1 | MSTKNFRVSDGDWICPDKKCG NVNFARRTSCNRCGREKTT | 40 a.a. | 4583.19 | 4581.15 | 1-40 |
| ZNF2 | AEKSRGLFSANDWQCKTCSN VNWARRSECNMCNTPKYA | 38 a.a | 4370.89 | 4368.96 | 57-94 |
Preparation of Peptide and Metal Solutions.
Each lyophilized apo-peptide was dissolved to a final concentration of 1.25 mM in 0.25 mM TCEP prepared in ultrapure water. Peptide solutions were stored at −80 °C in single thaw aliquots until use. Stock solutions of As3+ and Zn2+ were prepared freshly in ultrapure water immediately before use and serially diluted using ultrapure water, as required.
Metal Binding/Displacement Assays.
In order to determine if one or both of the ZRANB2 zfms bind As3+ and/or Zn2+ and whether these metals can mutually displace each other from either or both of these zfms, we employed a combination of several biophysical techniques, as well as mass-spectrometry as described in the next sections. All these assays were performed using synthetic apo-peptides in cell free systems.
UV-Vis Spectrophotometry.
UV-Vis spectrophotometric experiments were performed to characterize the interaction of As3+ binding and mutual displacement of As3+/Zn2+ from ZRANB2 zfm apo-peptides as described elsewhere with minor modifications 32, 44. Briefly, 100 μM of each ZRANB2 apo-peptide was incubated with increasing molar equivalents (Eq) of As3+ (0–2 Eq at 0.2 Eq intervals; 0-200 μM at 20 μM intervals) for 30 minutes at room temperature. For Zn2+ displacement by As3+, 100 μM of each ZRANB2 apo-peptide was initially incubated with 0.5 Eq Zn2+ (50 μM) for 30 minutes at room temperature, followed by titration with increasing molar equivalents of As3+ (0-2 Eq at 0.25 Eq intervals; 0-200 μM at 25 μM intervals) for 30 minutes at room temperature. At As3+ concentrations higher than 1 Eq following the addition of 1 Eq Zn2+ to the apo-peptides, particulates were seen to form in the reaction mixture, hindering the spectrophotometric detection. Consequently, for Zn2+ displacement reactions for UV-Vis spectrophotometric assay, 0.5 Eq of Zn2+ was used. For As3+ displacement by Zn2+, 100 μM of each ZRANB2 apo-peptide was initially incubated with 1 Eq As3+ (100 μM) for 30 minutes at room temperature, followed by titration with increasing molar equivalents of Zn2+ (0-2 Eq at 0.25 Eq intervals; 0-200 μM at 25 μM intervals) for 30 minutes at room temperature. All the reactions were performed in 20 mM Tris-HCI, pH 7.8 with 250 μM TCEP in a volume of 10 μL. Spectrophotometric measurements were performed using a NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific Inc.) in the wavelength range 230-400 nm 44, 45. A buffer solution containing 20 mM Tris-HCI, pH 7.8 and 250 μM TCEP was used as the blank to set the baseline for the entire wavelength range. Absorbance values at 285 nm (A285) were recorded in each case. Means of duplicate readings were taken for each sample. In another set of As3+ displacement by Zn2+ reactions, the experiments were performed with equimolar amounts of apo-peptide, As3+ and Zn2+ as described earlier in this section, but in a 20 μL reaction volume. Half of the reaction mixture (10 μL) was used for spectrophotometric measurements as described earlier. The remaining half was frozen at −20°C and spectrophotometric measurements were recorded 24 h post-freezing. Each experiment was performed independently three times.
Free Zn2+ Measurement:
Spectrophotometric experiments were performed to characterize the interaction of Zn2+ binding and mutual displacement of As3+/Zn2+ from ZRANB2 zfm apo-peptides as described previously with minor modifications31. Briefly, increasing amounts of each ZRANB2 apo-peptide (0-5 μM) was incubated with 10 μM Zn2+ for 30 minutes at room temperature. For Zn2+ displacement by As3+, 5 μM of each ZRANB2 apo-peptide was initially incubated with 10 μM Zn2+ for 30 minutes at room temperature, followed by titration with increasing concentration of As3+ (0-5 μM) for 30 minutes at room temperature. For As3+ displacement by Zn2+, 5 μM of each ZRANB2 apo-peptide was initially incubated with 5 μM As3+ for 30 minutes at room temperature, followed by titration with increasing concentration of Zn2+ (0-10 μM) for 30 minutes at room temperature. All the reactions were performed in 20 mM Tris-HCI, pH 7.0 with 25 μM TCEP. All the reactions were performed in a volume of 10 μL apart from the As3+ displacement by Zn2+ experiments, which were performed in 20 μL. After the incubations were over, 1 μL of 1 mM 4-(2-pyridylazoresorcinol) was added to each reaction mixture and spectrophotometric measurements were performed using a NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific Inc.) in the wavelength range 300-550 nm 46. The absorbance values at 493 nm were recorded in each case as a measure of free Zn2+ signal in the samples. Means of duplicate readings were taken for each sample. Absorbance readings were converted to free Zn2+ concentration by comparison against a Zn2+ standard curve generated using A493 values. The free Zn2+ concentration was subsequently subtracted from the input Zn2+ concentration to determine the concentration of apo-peptide bound Zn2+. For stability of Zn2+ binding (5 μM apo-peptide incubated with 10 μM Zn2+) and As3+ displacement by Zn2+ experiments, half of the reaction mixture (10 μL) was used for spectrophotometric measurements immediately, while the remaining half was frozen at −20°C and spectrophotometric measurements were recorded 24 h post-freezing. Each experiment was performed independently three times.
Mass Spectrometric Analyses.
Mass spectroscopy analyses were carried out for further characterization of As3+ binding and mutual displacement of As3+/Zn2+ from ZRANB2 zfm apo-peptides. For binding experiments, 1 mM of each ZRANB2 apo-peptide was incubated with As3+ (1 Eq; 1 mM) or Zn2+ (0.5 or 1 Eq; 0.5 or 1 mM) for 30 minutes at room temperature. For Zn2+ displacement by As3+, 1 mM of each ZRANB2 apo-peptide was initially incubated with 0.5 Eq Zn2+ (0.5 mM) for 30 minutes at room temperature, followed by titration with 2 Eq of As3+ (2 mM) for 30 minutes at room temperature. For As3+ displacement by Zn2+, 1 mM of each ZRANB2 apo-peptide was initially incubated with 1Eq As3+ (1 mM) for 30 minutes at room temperature, followed by titration with 1 Eq of Zn2+ (1 mM) for 30 minutes at room temperature. All the reactions were performed in 20 mM Tris-HCI, pH 7.8 with 2.5 mM TCEP in a volume of 10 μL. Samples were diluted with 2% v/v acetonitrile / 0.1% v/v formic acid to a final concentration of 1 pmol peptide/μL. Sample aliquots were dispensed into capped autosampler vials and loaded into a Waters M-Class Acquity UPLC system (Waters Corp., Milford, MA, USA) fitted with a Waters CSH C18 130 Å 1.7 μm 300 μm x 150 mm column, heated at 55 °C, at a flow rate of 10 μL/min. Data were acquired on the Synapt G2-Si Q-Tof (Waters Corp.) using the scan range 400-1500m/z with an MSe method. The .raw data were loaded into MassLynx4.1 (Waters Corp., Milford, MA, USA) and processed with MaxEnt3 (Waters Corp., Milford, MA, USA) for deconvolution and deisotoping to produce precursor and fragment ion mass lists for Skyline template generation. The .raw data were imported into Skyline v4.2.0.19072 using MS1 extracted ion chromatograms 47. For ZNF1 (+5, +6, +7, and +8 charge states) and ZNF2 (+4, +5, +6, and +7 charge states) peptide data, precursor area was extracted for the top three theoretical isotopes of each monitored peptide using a TOF resolving power of 10,000. The area extracted for the fully and partially reduced forms was summed.
Intrinsic Fluorescence Assay.
Intrinsic fluorescence experiments were performed to characterize changes in the folding of the ZRANB2 zfm apo-peptides upon As3+/Zn2+ binding and mutual displacement of As3+/Zn2+, following previously described methods with minor modifications 46, 48. All the reactions were performed in 20 mM Tris-HCI, pH 7.0 with 2.5 μM TCEP in a volume of 2.5 mL. Intrinsic fluorescence spectra were recorded using an excitation wavelength of 280 nm and an emission wavelength range of 290-500 nm (1 nm interval; 1 s integration time; 1 nm excitation bandpass; 5 nm emission bandpass) using the FluoroMax®-3 spectrofluorometer (Jobin Yvon Inc. Edson, NJ, USA). For metal binding experiments, 1 μM of each ZRANB2 apo-peptide was incubated with equimolar amounts of As3+ or Zn2+ for 30 minutes at room temperature followed by collection of intrinsic fluorescence spectra as described earlier. For Zn2+ displacement by As3+, 1 μM of As3+ was added to Zn2+-bound ZRANB2 apo-peptide and incubated for an additional 30 minutes at room temperature, followed by collection of intrinsic fluorescence spectra as described earlier. For As3+ displacement by Zn2+, 1 μM of Zn2+ was added to As3+-bound ZRANB2 apo-peptide and incubated for an additional 30 minutes at room temperature, followed by collection of intrinsic fluorescence spectra as described earlier. Total intrinsic fluorescence over the wavelength range was determined in each case by calculating the area under the curve (AUC). Each experiment was performed independently six times.
Cytotoxicity Assay.
Cytotoxicity in HaCaT cells due to As3+ exposure was quantitated using alamar blue assay. Alamar blue was obtained from Bio-Rad (Hercules, CA, USA) and the assay was performed using the manufacturer’s protocol with minor modifications49. Briefly, 104 cells were seeded per well in a 96 well plate in a volume of 100 μL. Cells were subsequently treated with increasing concentration of As3+ (final concentration 0-10 μM) 24 h post seeding in quadruplicates. Alamar blue was added at different time points (24, 48, 72 h post-treatment), allowed to incubate at 37 °C for 4 h and spectrophotometric readings were taken at 570 and 600 nm using Gen5™ microplate reader (Winooski, VT, USA). The readings were subsequently converted to percentage of live cells compared to untreated control using the equation and the molar extinction coefficient values provided by the manufacturer49.
Cell Culture and As3+ Treatment.
HaCaT cells were the kind gift of Dr. Tai Hao Quan, University of Michigan. The identity of the cells as HaCaT was confirmed by STR mapping (Genetica DNA Laboratories/LabCorp, Burlington, NC, USA). The cells were cultured as described previously50 with minor modifications. Briefly, cells were cultured in MEM alpha modification media supplemented with 10% fetal bovine serum, 100 U/mL penicillin/100 mg/mL streptomycin) and 2 mM L-glutamine. Cultures were maintained at 37°C in a humidified 5% CO2 atmosphere. Twenty-four hours post-seeding, the cells were treated with NaAsO2 by addition of 1000X stock to final [As3+] = 5 μM (or equal volume of ultrapure water as vehicle control). Cells were harvested for RNA and protein extraction 0, 1, 3, 6, 12, 24, 48 and 72 h post-treatment. For the concentration-response experiments, the HaCaT cells were treated with 0, 0.1, 1 and 5 μM As3+ for 6 h. Three independent replicates were obtained per condition per time point/concentration.
RNA Extraction and cDNA Generation.
Total RNA was purified from the cells employing the mirVana RNA Isolation Kit (Thermo Fisher Scientific Inc.) as described previously50. RNA quality was determined using the Agilent RNA 6000 Pico Kit, Eukaryote, version 2.6 and the Agilent 2100 Bioanalyzer instrument (Agilent Technologies Inc., Santa Clara, CA, USA). All samples had RIN (RNA integrity number) > 9 50. cDNA was generated from total RNA using the PrimeScript RT Reagent Kit (TaKaRa Bio USA Inc., Mountain View, CA, USA) as per the manufacturer’s instructions. Briefly, 1 μg of total RNA was reverse transcribed into cDNA using Oligo (dT)20 primer.
Quantitative PCR.
Quantitative PCR of cDNAs was performed using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) following the manufacturer’s recommended protocol. Briefly, 5 ng of cDNA was amplified using highly specific primers (200 nM each): ZRANB2 assay Hs.PT.58.40950918 (Integrated DNA Technologies, Coralville, IA, USA), and GAPDH forward 5’ ACCACAGTCCATGCCATCAC 3’ and reverse 5’ TCCACCACCCTGTTGCTGTA 3’ (Integrated DNA Technologies) in a final volume of 20 μL. The Viia 7 Real-Time PCR System (Thermo Fisher Scientific) was used for amplification and relative quantitation. All samples were run at least in triplicate, using GAPDH as the housekeeping gene. To determine the effect of arsenic exposure on the expression of ZRANB2 mRNA, the threshold cycle (Ct) values for GAPDH from each sample was subtracted from the Ct values for ZRANB2 from the respective sample to obtain a ΔCt value. An average for the ΔCt values was taken for the replicates. ΔCt values for controls at each time point were averaged and this average was subtracted from each ΔCt value for each sample at that time point to obtain a ΔΔCt value. The relative expression of ZRANB2 was calculated as fold change = 2^(−ΔΔCt). For each time point, the mean of control samples was taken as 1 fold and the As3+ exposed samples were expressed as fold change over control.
Analysis of Alternative Splicing.
RT-PCR was performed using cDNA generated from control or As3+ treated HaCaT cells, as described in the preceding sections. Primers were designed against flanking exons 1 (Forward; 5’- AAGGAAGGTGCAAGAGGTTG-3’) and 4 (Reverse; 5’- CGGCAATGGGACCATATTTA-3’) of TRA2B gene, a known splice target of ZRANB2, and obtained from Integrated DNA Technologies. The primers were designed to co-amplify β1 (502 bp) and β3 (368 bp) TRA2B mRNA isoforms 40 in a single reaction. Each amplification reaction was performed using 400 ng cDNA for 36 cycles using 54 °C as the annealing temperature. The amplification products were resolved on 1.5% agarose gels pre-stained with RedSafe™ dye (Bulldog Bio, Portsmouth, NH, USA). Images were acquired with FOTO/Analyst FX (Thermo Fisher Scientific Inc.) and densitometric analysis by employing Image J software 51. Raw data from densitometric analyses were divided by the isoform size (fragment length in bp) to determine the number of events for the β1 and β3 isoforms. Percentage contribution of β3 isoform was determined in each sample under each condition using the following formula: β3 isoform (%) = β3*100/(β1+β3).
Immunoblotting.
Cell lysis and immunoblotting were performed as described previously52 with minor modifications. Briefly, cells were lysed with a solution of 10 mM Tris–HCI pH 7.4, 1 mM EDTA, 0.1% SDS, 180 μg/mL PMSF and 1X protease inhibitor cocktail (Thermo Fisher Scientific Inc.). Lysates were sonicated and centrifuged at 4° C for 15 min to remove insoluble debris. Protein concentrations were determined with BCA assay (Thermo Fisher Scientific Inc.). Proteins were resolved by electrophoresis in SDS polyacrylamide gels of appropriate percentage. The resolved proteins were electro-blotted onto PVDF membranes (Thermo Fisher Scientific Inc.). After staining with Coomassie Brilliant Blue R250 (Thermo Fisher Scientific Inc.) to ensure equal loading and transfer53, membranes were blocked in 5% milk in TBST (10 mM Tris–HCI pH 7.4, 150 mM NaCI, 0.1% Tween 20) at room temperature for 1 h. Blots were subsequently probed with antibodies against Vinculin (Clone E19V; Cat No. 13901; Cell Signaling, Danvers, MA, USA; 1:1000 in 1% skimmed milk in TBST) and ZRANB2 (Clone B-5; Cat No. sc-514200; Santa Cruz Biotechnology, Dallas, TX, USA; 1:100 in 1% skim milk in TBST). Blots were incubated with HRP conjugated secondary antibody as required (in 1% skim milk in TBST), and subsequently with enhanced chemi-luminescent substrate (Thermo Fisher Scientific Inc.) or with SuperSignal™ West Atto Ultimate Sensitivity Substrate (Thermo Fisher Scientific Inc.), and images were acquired using FOTO/Analyst FX. Densitometric analyses were performed using Image J software51.
Statistical Analyses.
For UV-Vis spectrophotometric experiments to characterize As3+ binding to ZRANB2 zfm apo-peptides, blank subtracted A285 values at each As3+ concentration were subtracted from the corresponding blank subtracted A285 value of the native apo-peptide to calculate absorbance difference at 285 nm (ΔA285) ΔA285 values were plotted against As3+ concentration and fitted using one-site specific binding non-linear curve fitting function using GraphPad Prism 5. Kd values as well as r2 values were determined for each curve fit. For mutual displacement of As3+/Zn2+ from ZRANB2 zfm apo-peptides experiments, blank subtracted A285 values at each concentration was subtracted from the blank subtracted A285 value of the native apo-peptide to calculate ΔA285. ΔA285 values were plotted against Zn2+/As3+ concentration and fitted using log(inhibitor) versus response non-linear curve fitting function using GraphPad Prism 5. For free Zn2+ measurements, the relationship between peptide bound Zn2+ and peptide/metal concentrations were fitted using linear regression using GraphPad Prism 5. For intrinsic fluorescence assays, the AUC values were log transformed, tested for normality by Shapiro-Wilk test, and analyzed initially by ANOVA with five levels followed by pairwise comparisons by Fisher’s least significant difference test (as the data were normally distributed). The displacement data for each peptide were subsequently added (ZNF1 Zn2++As3+ Group: ZNF1+Zn+As combined with ZNF1+As+Zn; ZNF2 Zn2++As3+ Group: ZNF2+Zn+As combined with ZNF2+As+Zn) and the four groups (No metal; Zn2+; As3+; Zn2++As3+) were analyzed by two-way ANOVA. Densitometric data for time-course of ZRANB2 protein expression as well as percent of β3 isoform of TRA2B were analyzed using two-way ANOVA with Bonferroni multiple comparisons post-hoc tests (no normality or homoscedasticity assumption). Densitometric data for dose-response of ZRANB2 protein expression as well as percent of β3 isoform of TRA2B were analyzed using one-way ANOVA with Tukey’s multiple comparisons post-hoc tests. p<0.05 was considered to be significant.
RESULTS
UV-Vis Spectrophotometric Analysis of As3+/Zn2+ Interactions with ZRANB2 Zfm Apo-peptides.
Binding of As3+ to sulfhydryl groups of cysteine residues can be monitored by a change in the UV-Vis spectrum between 230-400 nm due to the formation of charge transfer electronic transition 44, 45. We initially characterized the spectrum of each native ZRANB2 apo-peptide in this wavelength range (Supplementary Figure 1). Analysis of the spectrum showed that for each apo-peptide, the absorption maximum was at 285 nm (A285), (Supplementary Figure 1). ΔA285 values were used to quantify As3+ binding as well as mutual displacement of As3+/Zn2+ from ZRANB2 zfm apo-peptides in all subsequent UV-Vis experiments. As3+ binding experiments showed As3+ concentration-dependent increase in ΔA285 (Figure 1A-B and Supplementary Figure 2A-B; Supplementary Table 1). For As3+ mediated Zn2+ displacement, we also observed a concentration-dependent increase in the ΔA285, as expected, suggesting more and more As3+ was binding to each of the apo-peptides at the expense of Zn2+ (Figure 1C-D and Supplementary Figure 2C-D). Conversely, forZn2+ mediated As3+ displacement, we found a concentration-dependent decrease in the ΔA285, suggesting reduction in As-S bonds as As3+ was progressively displaced by Zn2+ (Figure 1E-F and Supplementary Figure 2E-F).
Figure 1.
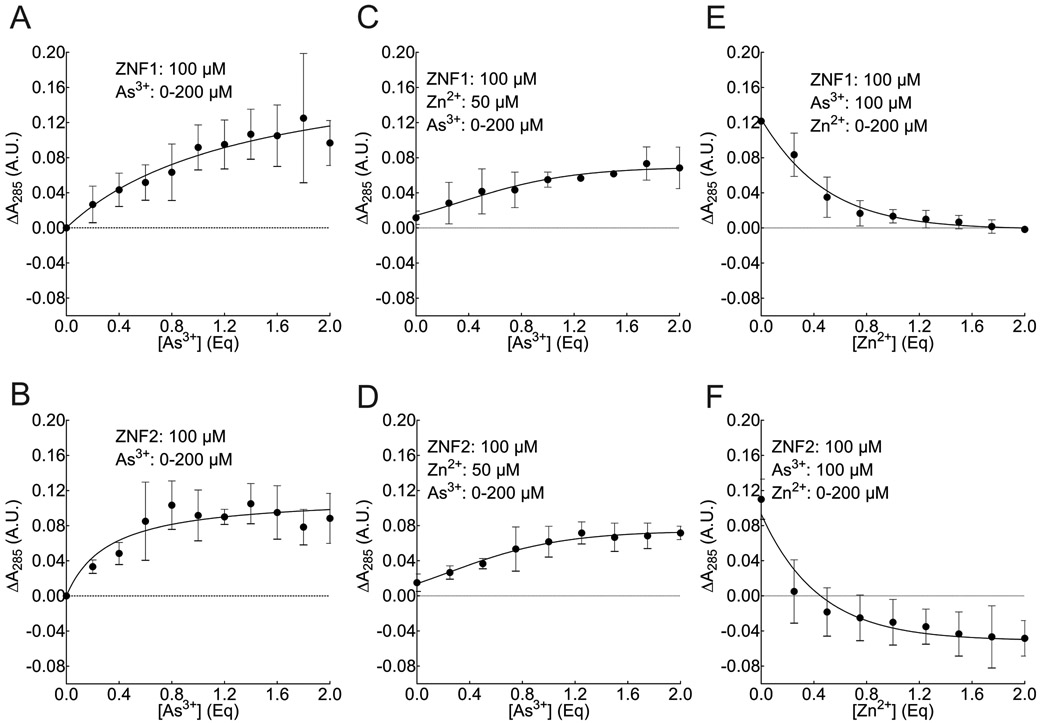
As3+ binding and mutual displacement of Zn2+/As3+ from ZRANB2 zfm apo-peptides using UV-Vis spectrophotometry. A, As3+ binding to ZNF1 (one-site specific binding non-linear curve fit). B, As3+ binding to ZNF2 (one-site specific binding non-linear curve fit). C, Zn2+ displacement by As3+ from ZNF1 (log(inhibitor) versus response non-linear curve fit). D, Zn2+ displacement by As3+ from ZNF2 (log(inhibitor) versus response non-linear curve fit). E, As3+ displacement by Zn2+ from ZNF1 (log(inhibitor) versus response non-linear curve fit). F, As3+ displacement by Zn2+ from ZNF2 (log(inhibitor) versus response non-linear curve fit). In each panel, the closed circle represents ΔA285 values. Each point in each curve represents the Mean ± SD from three independent experiments.
Zinc Colorimetric Analysis of As3+/Zn2+ Interactions with ZRANB2 Zfm Apo-peptides.
While we could demonstrate As3+ binding and mutual displacement of As3+/Zn2+ from ZRANB2 zfm apo-peptides with UV-Vis assay, we could not quantify Zn2+ binding directly by that assay. In order to demonstrate that Zn2+ can bind to one or both ZRANB2 zfm apo-peptides, we performed a Zn2+ colorimetric assay. For Zn2+ binding experiments, the concentration of apo-peptide bound Zn2+ increased linearly with apo-peptide concentration (Figure 2A). Conversely, for As3+ mediated Zn2+ displacement experiments, the concentration of apo-peptide bound Zn2+ decreased linearly with As3+ concentration (Figure 2B). For both these experiments, a tangible difference was observed between the two apo-peptides with respect to how Zn2+ interacts with each of them. For Zn2+ mediated As3+ displacement, the concentration of apo-peptide bound Zn2+ was found to increase linearly with increase in Zn2+ concentration (Figure 2C), demonstrating Zn2+ was capable of displacing As3+ from each apo-peptide.
Figure 2.
Zn2+ Binding and displacement of Zn2+/As3+ from ZRANB2 zfm apo-peptides using spectrophotometric quantification of free Zn2+. A, Increasing amounts of each ZRANB2 apo-peptide (0-5 μM) was incubated with 10 μM Zn2+ for 30 minutes at room temperature. Increase in peptide bound Zn2+ with increase in peptide concentration shows Zn2+ binding to the peptides. B, 5 μM of each ZRANB2 apo-peptide was initially incubated with 10 μM Zn2+ for 30 minutes at room temperature, followed by titration with increasing concentration of As3+ (0-5 μM) for 30 minutes at room temperature. Decrease in peptide bound Zn2+ with increase in As3+ concentration shows displacement of Zn2+ by As3+ from each peptide. C, 5 μM of each ZRANB2 apo-peptide was initially incubated with 5 μM As3+ for 30 minutes at room temperature, followed by titration with increasing concentration of Zn2+ (0-10 μM) for 30 minutes at room temperature. Increase in peptide bound Zn2+ with increase in Zn2+ concentration shows displacement of As3+ by Zn2+ from each peptide. Open circles represent ZNF1 while open squares represent ZNF2 for all the panels. Each point in each curve represents the Mean ± SD from three independent experiments.
Mass Spectrometric Characterization of As3+/Zn2+ Interactions with ZRANB2 Zfm Apo-peptides.
The MS2 spectrum of each apo-peptide (+6 charge state) is shown in Figure 3 A-B. The experimentally determined mass at +6 charge state (764 for ZNF1 and 729 for ZNF2) closely matches the theoretical mass of the fully reduced apo-peptides at the same m/z state (763.8 for ZNF1 and 728.3 for ZNF2). Upon incubation with arsenic alone, the mass shift for each apo-peptide corresponds to As3+ bound form. Each peptide had a characteristic +12 m/z shift (776 for ZNF1 and 741 for ZNF2; Figure 3C-D) at +6 charge state. Similarly, upon incubation with Zn2+ followed by As3+, the same +12 m/z shift (Figure 3E-F) was seen for each peptide (at +6 charge state), characteristic of As3+-bound apo-peptides, suggesting that As3+ displaces Zn2+ from Zn2+-bound ZRANB2 apo-peptides. However, we could not show any mass shift for apo-peptides when incubated with Zn2+ alone (Supplementary Figure 3A-B). This is not entirely unexpected as LC-MS analysis of zfm peptides is known to extract Zn2+ from the peptides, leading to a significant reduction in the mass shift signal 54. Interestingly, upon incubation with As3+ followed by Zn2+, +12 m/z shift was seen (at +6 charge state) for each peptide (Supplementary Figure 3C-D), characteristic of As3+-bound peptides. This was surprising, because, we expected to see mass signature of the native apo-peptides, but not of the As3+-bound peptides, had it been an outcome of Zn2+ extraction from the peptides during ESI-MS/MS analysis.
Figure 3.
Mass spectrometric (ESI-MS/MS) analysis of As3+ binding and displacement of Zn2+ by As3+ from ZRANB2 Zfm Apo-peptides. A, Native ZNF1 apo-peptide mass signature was detected at m/z = 764. B, Native ZNF2 apo-peptide mass signature was detected at m/z = 729. C, As3+ binding to ZNF1 was detected at m/z = 776, which represents a +12 m/z shift compared with the native ZNF1 apo-peptide (m/z = 764). D, As3+ binding to ZNF2 was detected at m/z = 741, which represents a +12 m/z shift against the native ZNF2 apo-peptide (m/z = 729). E, Zn2+ displacement by As3+ from ZNF1 was detected at m/z = 776, which represents a +12 m/z shift against the native ZNF1 apo-peptide (m/z = 764). F, Zn2+ displacement by As3+ from ZNF2 was detected at m/z = 741, which represents a +12 m/z shift against the native ZNF2 apo-peptide (m/z = 729). All the figures represent +6 charge state. Insets represent maginified view of the highest peak in each case.
Intrinsic Fluorescence Analysis of Structural Changes in ZRANB2 Apo-peptides upon Interaction with As3+/Zn2+.
Having demonstrated that each ZRANB2 zfm apo-peptide can bind both As3+ and Zn2+, and that each can displace the other, we wanted to examine if such interactions could affect the tertiary structure of these apo-peptides. For this purpose, change in intrinsic fluorescence of the apo-peptides was monitored 48. The data indicate that Zn2+ binding expectedly promotes folding in each peptide as evidenced by an increase in fluorescence intensity (Figure 4A-B), supporting the notion that the Zn2+-bound form represents the natural folded conformation under physiological conditions 46 that is necessary for RNA binding. As3+ addition, interestingly, promoted folding for ZNF1 but unfolding for ZNF2 (Figure 4A-B). Zn2+ also promoted folding of each peptide under Zn2+ mediated As3+ displacement experimental conditions (Fig 4A-B). Interestingly, there was no difference in intrinsic fluorescence (and hence in folding), compared to Zn2+-bound peptides, when As3+ was added to the Zn2+-bound peptides for the As3+ mediated Zn2+ displacement reactions (Figure 4A-B). These data suggest that interaction with As3+ alters the tertiary structure of either apo-peptide to a lesser extent than interaction with Zn2+ whether in the unfolded state (as compared to the unfolded apo-peptide for the As3+ binding experiment) or the folded state (as compared to the Zn2+ bound peptide for the As3+ mediated Zn2+ displacement experiment). Since the order of addition of As3+ or Zn2+ to the apo-peptides did not make a difference to the intrinsic fluorescence/folding (ZNF1+Zn+As vs. ZNF1+As+Zn and ZNF2+Zn+As vs. ZNF2+As+Zn), these two groups were merged (ZNF1 Zn2++As3+ Group: ZNF1+Zn+As combined with ZNF1+As+Zn; ZNF2 Zn2++As3+ Group: ZNF2+Zn+As combined with ZNF2+As+Zn) and a two way ANOVA analysis with interaction was carried out (No metal; Zn2+; As3+; Zn2++As3+) to examine the effect of each metal individually as well as their potential interaction in terms of ZRANB2 apo-peptide folding (Table 2, Supplementary Fig 4). p Values indicate that addition of either As3+ or Zn2+ individually to each apo-peptide induces tangible structural changes compared to the native apo-peptide. A significant interaction between the two metals was found for ZNF1, but not for ZNF2 (Table 2; Supplementary Figure 4).
Figure 4.

Zn2+/As3+ binding and mutual displacement induces changes in intrinsic fluorescence of ZRANB2 apo-peptides. A, Intrinsic fluorescence of the native ZNF1 apo-peptide changes significantly with both As3+ and Zn2+ binding. Intrinsic fluorescence of Zn2+-bound ZNF1 does not change after displacement of Zn2+ by As3+; whereas displacement of As3+ by Zn2+ significantly alters the intrinsic fluorescence of As3+-bound ZNF1. However, there is no difference in intrinsic fluorescence when both As3+ and Zn2+ are added to ZNF1, irrespective of the order of addition (ZNF1+Zn+As vs. ZNF1+As+Zn). ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05 for pairwise comparison by Fisher’s least significant difference test, following one-way ANOVA with five levels. B, Intrinsic fluorescence of the native ZNF2 apo-peptide changes significantly with both As3+ and Zn2+ binding. Intrinsic fluorescence of Zn2+-bound ZNF2 does not change after displacement of Zn2+ by As3+; whereas displacement of As3+ by Zn2+ significantly changes the intrinsic fluorescence of As3+-bound ZNF2. However, there is no difference in intrinsic fluorescence when both As3+ and Zn2+ are added to ZNF2, irrespective of the order of addition (ZNF2+Zn+As vs. ZNF2+As+Zn). ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05 for pairwise comparison by Fisher’s least significant difference test, following one-way ANOVA with five levels. Please see Table 2 for two-way ANOVA analyses of the data.
Table 2:
Two-way ANOVA p values for As3+/Zn2+ interaction with ZRANB2 zfm apo-peptides
| Parameter | ZNF1 | ZNF2 |
|---|---|---|
| pZn | <.0001 | <.0001 |
| pAs | 0.0058 | 0.0004 |
| pInteraction | 0.0006 | 0.3418 |
Zn2+ Mediated As3+ Displacement from ZRANB2 Zfm Apo-peptides is Transient.
The MS data suggested that Zn2+ was not able to bind to or displace As3+ from either peptide, in contrast to the results obtained using spectrophotometry or colorimetric free Zn2+ analysis. While the apparent lack of Zn2+ binding could be explained by possible Zn2+ extraction from the apo-peptides during LC-ESI-MS analysis 54, we expected to see the mass signature of the native apo-peptides rather than the As3+-bound peptides in the As3+ displacement experiments. We were intrigued by the appearance of the mass signature of As3+ bound peptides in these experiments. Given the samples were stored overnight at −20°C before being analyzed by mass spectrometry, we addressed the hypothesis that As3+ could re-displace Zn2+ in the intervening time period. Consequently, we performed several experiments using different biophysical techniques to address this question. First, we wanted to examine if Zn2+ binding to the apo-peptides was stable. There was no difference in the amount of peptide bound Zn2+ determined by measuring remaining free Zn2+, whether the readings were taken immediately or following storage at −20°C for 24 h (Figure 5A). Having demonstrated that Zn2+ does not come off from the apo-peptides because of storage, we performed experiments to quantify if the Zn2+ was displaced by As3+ during storage. We demonstrate that most of the apo-peptide bound Zn2+ was lost from each apo-peptide after storage for 24 h at −20 °C in the presence of As3+ (Fig 5B). Next, we wanted to explore whether Zn2+ was simply being displaced from the apo-peptides, or As3+ was re-binding to the apo-peptides. To test this, we performed UV-Vis analysis of Zn2+ mediated As3+ displacement experiments immediately upon the completion of the displacement reaction and after 24 h storage at −20 °C. We found that there was no change in ΔA285 for As3+ bound apo-peptide compared to native apo-peptide between the two time points, indicating that As3+ was bound to each of the apo-peptides similarly at both time points (Figures 5B-C). Interestingly, for the Zn2+ mediated As3+ displacement condition, there was a significant decrease in ΔA285 compared to As3+ bound apo-peptide, immediately after the completion of the displacement reaction (Figure 5C-D), indicating a decrease in As-S bond formation owing to As3+ displacement. However, the ΔA285 values were similar to As3+ bound apo-peptide following 24 h storage at −20°C. These results suggest, during 24 h storage, the As-S bond has reformed, replacing the Zn2+ that was bound to the peptide, even at −20°C. Taken together, the data demonstrate that Zn2+ mediated As3+ displacement from ZRANB2 zfm apo-peptides was transient in nature.
Figure 5.

As3+ displacement from ZRANB2 apo-peptides by Zn2+ is transient. A, Zn2+ binding to ZRANB2 apo-peptides is not affected by storage at −20°C for 24 h (colorimetric quantification of free Zn2+). B, Zn2+ can displace As3+ from As3+-bound ZRANB2 apo-peptides when readings are taken immediately following incubation (ZNF1 is represented by open circles, ZNF2 by open squares). Please note that these two graphs (ZNF1-Immediate and ZNF2-Immediate) represent the same dataset as in Figure 2C; added for clarity). However, most of that peptide-bound Zn2+ is lost if the same samples are stored for 24 h (legend: overnight) at −20°C before the readings are taken (ZNF1 is represented by closed circles, ZNF2 by closed squares; colorimetric quantification of free Zn2+). C, UV-Vis spectrophotometric evidence that As3+ displaces Zn2+ from the ZNF1 apo-peptide if the samples are stored for 24 h (legend: overnight) at −20°C before the reading is taken (legend: overnight), but not if the readings are taken immediately following incubation (legend: immediate). When the readings are taken immediately, addition of Zn2+ to the As3+-bound apo-peptide causes a significant decrease in absorbance at 285 nm (absorption maximum for ZNF1). However, if the readings are taken after the same samples have been stored for 24 h at −20°C, there is no difference in absorbance of the As3+ bound peptides with and without Zn2+ addition. ***p<0.001 by Tukey’s multiple comparisons post-hoc test following one-way ANOVA. D, UV-Vis spectrophotometric evidence that As3+ displaces Zn2+ from the ZNF2 apo-peptide if the samples are stored for 24 h (legend: overnight) at −20°C before reading is taken (legend: overnight), but not if the readings are taken immediately following incubation (legend: immediate). When the readings are taken immediately, addition of Zn2+ to the As3+-bound apo-peptide causes a significant decrease in absorbance at 285 nm (absorption maximum for ZNF2). However, if the readings are taken after the same samples have been stored for 24 h at −20°C, there is no difference in absorbance of the As3+ bound peptides with and without Zn2+ addition. ***p<0.001 by Tukey’s multiple comparisons post-hoc test following one-way ANOVA.
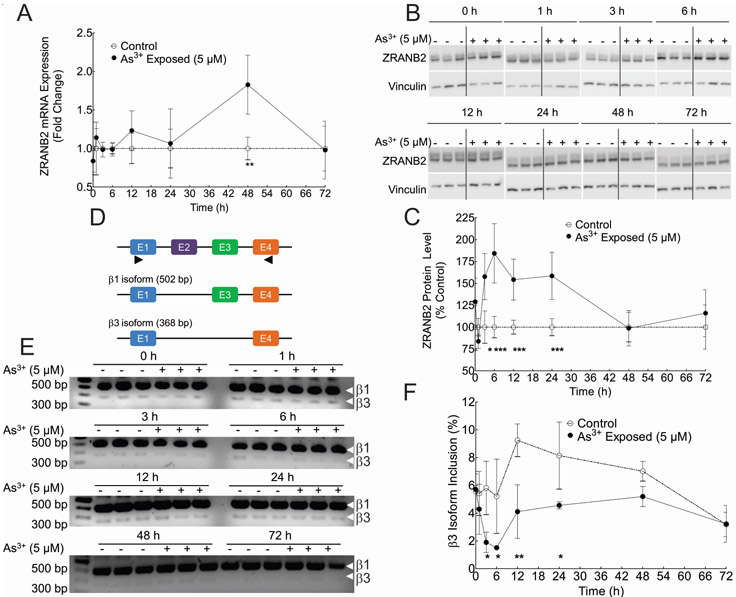
Acute As3+ Treatment Induced ZRANB2 Protein Expression and Altered Splicing Profile in ZRANB2 Target.
Given that ZRANB2 has two C4 zfms, we hypothesized that binding of As3+ to these sites will lead to a decreased function of ZRANB2, leading to an increase in its expression at post-exposure time points, to compensate for the reduced activity. In order to determine a suitable acute As3+ treatment dosage, we carried out cell viability assay by alamar blue. The data demonstrated that 5 μM was the highest dosage that did not induced any toxicity at 72 h time point (Supplementary Figure 5) and hence was chosen for these proof of principle experiments. RT-qPCR data showed that the steady state levels of ZRANB2 mRNA between control and As3+ exposed cells were similar except at the 48 h time point (Figure 6A, Table 3). Immunoblot experiments, however, clearly demonstrate a significant increase in the expression of ZRANB2 protein starting at 3 h post-exposure and continuing until 24 h following which, it returns to basal level (Fig 6B- C, Table 3).
Figure 6.
Time course of ZRANB2 expression and splicing function in HaCaT cells exposed to 5 μM As3+ for 0 – 72 h. A, Quantification of ZRANB2 mRNA n HaCaT cells by RT-qPCR at different time points. The data are represented as fold change compared to unexposed control at each time point. **p<0.01 by Bonferroni post-hoc test following two-way ANOVA. B, ZRANB2 immunoblot from HaCaT cell lysates at different times. C, Densitometric analysis of ZRANB2 protein expression in B. Data are represented as % unexposed control expression at each time point. ***p<0.001, *p<0.05 by Bonferroni post-hoc test following two-way ANOVA. For two-way ANOVA analyses results, please see Table 4. D, Schematic representation of all the TRA2B exons in the region bound by the primers (represented by closed triangles) and the composition of ß1 and ß3 splice isoforms. Please note that the size of the exons/introns is not to scale. E, RT-PCR analysis of ß1 and ß3 splice isoforms at different time points. ß1 is the predominant isoform represented by the 502 bp amplification product, while the ZRANB2-dependent ß3 isoform is represented by the 368 bp product. F, Densitometric analysis of % ZRANB2 dependent ß3 isoform at different times. **p<0.01, *p<0.05 by Bonferroni post-hoc test following two-way ANOVA. For two-way ANOVA analyses results, please see Table 3.
Table 3:
Two-way ANOVA p values for ZRANB2 expression and splice function upon acute As3+ exposure
| Parameter | ZRANB2 mRNA Expression |
ZRANB2 Protein Expression |
ZRANB2 Splice Function |
|---|---|---|---|
| PAs | 0.08 | <0.0001 | <0.001 |
| pTime | 0.03 | <0.0001 | <0.001 |
| pInteraction | 0.03 | <0.0001 | 0.03 |
Zn2+ binding is critical for maintaining both properly folded tertiary structure as well as biological function of zfm proteins 31,46. Having demonstrated that As3+ can bind to each ZRANB2 zfm by displacing Zn2+ under cell free conditions, we wanted to examine if arsenic exposure could modulate the function of ZRANB2 protein under cellular conditions. For this purpose, we looked at the proficiency of ZRANB2 to alternatively splice its well characterized target TRA2B mRNA (Figure 6D). ZRANB2 has been shown to be responsible for the exclusion of exon 3 of TRA2B leading to the generation of β3 isoform (consisting of exons 1 and 4) at the expense of β1 isoform (consisting of exons 1,3 & 4). RT-PCR data unequivocally demonstrate significant reduction of ZRANB2-dependent β3 isoform at 3 h post exposure and continuing until 24 h compared to unexposed control (Figure 6D-E, Table 3). This result indicates that ZRANB2 splicing function is compromised upon acute arsenic exposure.
Environmentally Relevant As3+Treatment Induced ZRANB2 Protein Expression and Altered Splicing Profile in ZRANB2 Target.
Having demonstrated that acute As3+ exposure at therapeutic levels of 5 μM alters both the expression as well as splicing function of ZRANB2, we wanted to examine if lower (and environmentally/toxicologically relevant) exposure to As3+ is capable of eliciting similar effects. The data demonstrate that all the As3+ exposures used in this study (0.1, 1 and 5 μM) significantly induced ZRANB2 protein expression at 6 h (Figure 7A-B), although, there was no significant change in the level of expression within the different exposures. TRA2B splicing data shows a clear exposure response, with the percentage of ZRANB2 dependent β3 isoform significantly reduced at each successive increasing As3+ exposure (Figure 7C-D).
Figure 7.

ZRANB2 expression and splicing function As3+ concentration-response. A, ZRANB2 immunoblot from lysates of HaCaT cells exposed to 0-5 μM As3+ for 6 h. B, Densitometric analysis of ZRANB2 protein expression in panel A. Data are represented as % unexposed control expression at each time point. **p<0.01, *p<0.05 by Tukey’s multiple comparisons post-hoc test following one-way ANOVA. C, RT-PCR analysis of ß1 and ß3 splice isoforms in RNA isolated from HaCaT cells exposed to 0-5 μM As3+ for 6 h. ß1 is the predominant isoform represented by the 502 bp amplification product, while the ZRANB2-dependent ß3 isoform is represented by the 368 bp product. D, Densitometric analysis of ZRANB2 mRNA isoform RT-qPCR in panel C expressed as % ZRANB2 dependent ß3 isoform. ***p<0.001, **p<0.01, *p<0.05 by Tukey’s multiple comparisons post-hoc test following one-way ANOVA.
DISCUSSION
Molecular etiology of arsenic-induced skin cancer is complex and probably consists of multiple cellular mechanisms taking place simultaneously as well as longitudinally. This complexity makes it difficult to gain a clear insight into the global mechanisms that might be operative in the process. The absence of a suitable in vivo model system further hampers understanding. It has been impossible to induce skin cancer in any animal model with arsenic exposure alone, irrespective of the dosage or time of exposure used (reviewed in 55). While several different mechanisms have been suggested for arsenic carcinogenicity, significant gaps still exist in our understanding of how these mechanisms act together to bring about cancer. Furthermore, “interaction with protein sulfhydryls” has long been suggested as a possible mode of arsenic carcinogenicity 56-58; however, an understanding of how this interaction could lead to cancerous outcomes was lacking until recently31, 46, 59.
Zfm proteins are widely distributed in prokaryotes and eukaryotes, including 3% of all human gene products 60. Zfm proteins include a diverse group of members capable of interacting with DNA, RNA and other proteins 61. They can carry out a spectacular range of biological operations including, but not limited to DNA repair, signal transduction, splicing and transcriptional regulation 61, and are widely implicated in cancers 62, 63. Zfms, which are central to the functional flexibility of this class of proteins, are targets for arsenic toxicity. As3+ can bind to C3H1 and C4 zfms, 31, 46 both in vitro and in vivo. Such binding results in the abrogation of the structure as well as function of the concerned protein 33, 44, 64. An overwhelming majority of the studies on As3+-mediated disruption of zfm proteins pertains to DNA repair proteins 34. This is surprising considering that zfm proteins play key roles in almost every major cellular process 61.
Here, we demonstrate for the first time that ZRANB2, a key protein involved in alternative splicing, is targeted by As3+. Biophysical data from our experiments unequivocally demonstrate that As3+ binds to each ZRANB2 zfm stably. We showed As3+ binding directly through mass spectrometry-based mass shift signatures. Furthermore, mass-spectrometry data are corroborated by a dose-dependent increase in As-S bond formation as evidenced by UV-Vis studies. This is the first report of arsenic binding to a zfm protein involved in alternative splicing, although, it is not surprising, given the propensity of arsenic to interact with other zfm proteins 30, 31, 34, 46, 65. Interestingly, the data strongly suggest that the characteristics of As3+ binding and As3+ mediated Zn2+ displacement for two similar C4 zfms of the same protein vary considerably. Such differences could mean that As3+ could be modulating each of the zfms differently, resulting in differential structural and functional outcomes in the physiological milieu. While the Kd values for As3+ binding is quite dissimilar for the two zfms, we would like to point out that UV-Vis spectrophotometry is not the most sensitive technique when it comes to quantitative characterization of binding affinity, especially given the small differences in absorbance between the different As3+ concentrations used. This outcome could be a consequence of the different amino acid residues surrounding the two zfms (Table 1). While structures of each of the zfms of ZRANB2 have been resolved 26,37, it still remains to be seen how the binding of As3+ affects the tertiary structures of these motifs. Clarification of the structures of both the zfms in the As3+ bound state could potentially shed light on why there are differences in As3+-zfm interaction parameters, and what it means in terms of RNA binding/splicing capabilities.
In addition, the data indicate that As3+ is capable of displacing Zn2+ from the Zn2+-bound apo-peptides. This effect is particularly important, as ZRANB2 zfms are Zn2+-bound under physiological conditions. This Zn2+ binding is of utmost importance in maintaining the properly folded state. This folding, in turn is critical to bind RNA molecules and mediate splicing functions 26,37,38. Previous studies, using in vitro assay systems, showed that such As3+ mediated Zn2+ displacement from zfm DNA repair proteins leads to altered DNA binding capacity 32,65. It is thus reasonable to hypothesize that similar Zn2+ displacement in ZRANB2 would lead to structural changes and sub-optimal/faulty RNA binding, adversely affecting its splicing capabilities, or even, altering its usual splicing profile.
Interestingly, we demonstrate that Zn2+ is also capable of displacing As3+ from ZRANB2 zfm peptides using a combination of biophysical techniques. One study showed that 5 μM Zn2+ treatment can restore the Zn2+ content of XPA and PARP-1 to basal levels or higher in HEKn cells treated with 2 μM As3+ 64. However, the study did not examine if comparable changes in Zn2+ content of the proteins could occur in Zn2+ treated cells in the absence of As3+ treatment. The study thus does not refute the possibility that As3+ did not bind all the possible Zn2+ binding sites in the first place. It is possible that increased Zn2+ content upon Zn2+ treatment is an outcome of increased occupancy of the free Zn2+ binding sites, as opposed to occupancy of As3+-bound Zn2+ binding sites. Our data clearly demonstrate a dose-dependent reduction in As-S bond formation upon incubation of As3+-bound apo-peptide with increasing concentrations of Zn2+. This evidence is in agreement with the only previous study available to our knowledge that has studied Zn2+ mediated As3+ displacement using similar As-S bond formation directly 64.
Maintenance of properly folded tertiary structure is important for each of the two ZRANB2 zfms for RNA binding 26,37. Intrinsic fluorescence data show that addition of Zn2+ to the native apo-peptide promotes characteristic folding for each of the two zfms. As3+ addition affects the folding of the two apo-peptides in opposing fashion, promoting folding for ZNF1, while promoting unfolding for ZNF2. This might partially explain the differences in As3+ binding parameters between them. It is interesting to note that As3+ addition has relatively smaller effects on the tertiary structure of each apo-peptide, compared to Zn2+ addition. Moreover, addition of As3+ to the Zn2+-bound apo-peptides does not bring about any significant changes in the tertiary structure, compared to the Zn2+-bound folded conformation (Figure 4). This result can be explained by the fact that Zn2+ coordinates with all 4 cysteine residues, and the resultant Zn2+-bound motif is expected to have a rigid structure. As3+ on the other hand, binds to only three of the four cysteine residues. It is possible that As3+ binds alternately to any three of the four cysteines available, making the conformation less rigid than the Zn2+-bound structure. However, from the two-way ANOVA data (Table 2 and Supplementary Figure 4), it is evident that ZNF1 and ZNF2 interact slightly differently when both As3+ and Zn2+ are present in the system. This result indicates that the two zfms could be modulated in somewhat different manner, consistent with the data from UV-Vis spectrophotometric experiments.
Furthermore, we elucidate that Zn2+ mediated As3+ displacement is transient in nature. This result is a novel finding of the current study. The same reaction mixture that showed Zn2+ mediated As3+ displacement lost most of the bound Zn2+ when stored for 24 h at −20 °C. The observation that Zn2+ binding to apo-peptides in the absence of As3+ is not altered by storage conditions (Figure 5A) argues against the possibility that the Zn2+ loss seen in the presence of As3+ (Figure 5B) is the result of Zn2+ chelation by TCEP, a weak chelator of Zn2+ 71, and a component of the buffer used in these studies. However, it is still possible that weak chelation of Zn2+ may be sufficient to shift the equilibrium in favor of As3+ binding when both are present for an extended period of time. These results also provide one possible explanation as to how we could be seeing the mass signature of As3+-bound ZRANB2 zfm apo-peptides upon Zn2+ mediated As3+ displacement.
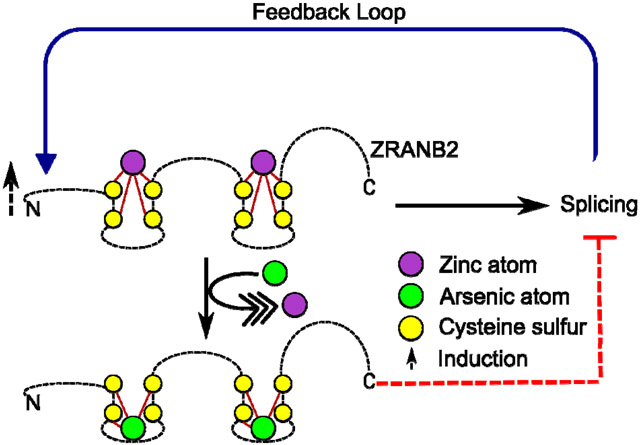
An important question to address was if As3+ mediated Zn2+ displacement from ZRANB2 results in any physiologically relevant outcome, given most of our metal interaction studies were performed with synthetic apo-peptides in cell-free systems. We chose to treat HaCaT cells initially with 5 μM of As3+ for up to 72 h, as this was the highest exposure that did not induce any significant cell mortality in our cell line at the longest time point (Supplementary Figure 5). While the concentration (5 μM) and the time of exposure (up to 72 h) do not emulate a chronic exposure scenario, it does correspond to the plasma As3+ level in subjects following a therapeutic regimen 74. We also chose this treatment regimen as a cost and time effective proof of concept experiment to investigate if it will be worthwhile to pursue future experiments using long term, low dose exposures that accurately simulate in vivo chronic exposures. We show that acute exposure of HaCaT cells to As3+ induces the expression of ZRANB2 protein between 3-24 h post-exposure, although, there is no increase in the steady state mRNA levels of ZRANB2 at these (or earlier) time points. This result could mean that the increase in ZRANB2 expression is perhaps due to stabilization of the ZRANB2 protein, rather than an increase in its steady state mRNA levels. As3+ exposure has previously been demonstrated to inhibit protein degradation by inhibiting autophagosome-lysosome fusion 75. Furthermore, the induction of ZRANB2 protein expression decreased by 48 h. This transient protein induction could signify that most of the arsenic has been metabolized by methylation, glutathionylation, thiolation and possibly effluxed from the cell by 48 h 73, and is consequently, no longer available to bind to ZRANB2. Interestingly, we do see a significant induction in ZRANB2 steady state mRNA levels in the As3+ exposed cells at 48 h post-exposure, when the ZRANB2 protein expression is down to basal level. We are not entirely sure what causes this spike. Furthermore, we demonstrate that acute As3+ exposure in the HaCaT cells results in significant alteration of the functional profile of ZRANB2 leading to aberrant splicing of its downstream target TRA2B. ZRANB2 is known to promote the exclusion of exon 3 of TRA2B, resulting in higher expression of the β3 isoform 40. In our experimental system, the expression of ZRANB2 dependent β3 isoform is significantly suppressed between 3-24 h post-exposure. Interestingly, the ZRANB2 expression is significantly induced at exactly those time points. We speculate that As3+ mediated Zn2+ displacement from ZRANB2 is leading to reduced splicing capability, thereby activating feedback systems to express more and more ZRANB2 to compensate for the observed functional impairment (Figure 6). The cycle of altered function and heightened expression continues possibly till most of the free As3+ capable of binding to ZRANB2 has been removed from the system, possibly by a combination of metabolism, binding and efflux.
Having demonstrated that acute As3+ exposure can alter both expression and function of ZRANB2, we asked if such effects can also occur at much lower environmentally and toxicologically relevant exposures. Our data demonstrate this is true (Figure 7). While both ZRANB2 protein expression and TRA2B splicing function were altered at each As3+ concentration tested, it is noteworthy, that the lowest concentration used (0.1 μM) corresponds to the blood serum level of As3+ in chronically exposed human populations 76. These data suggest that chronic As3+ exposure can alter the splicing profile in chronically exposed populations. This result is particularly interesting, given that the ß1 isoform corresponds to the canonical TRAB2B-205 isoform (288 amino acids) of this gene, while the ß3 corresponds to the TRA2B-203 isoform (188 amino acids) and lacks the first 100 amino acid residues. Although, the functions of the β3 isoform have not been studied, it is interesting to note that this isoform misses a big portion of the RA1 domain which resides within the residues 31-113. Thus, it is fair to speculate that β3 and β1 isoforms might have different RNA binding capacity and possibly different splice targets. As3+ exposure at exposures relevant to both chronic low dose exposures and acute therapeutic doses seems to favor an increase of canonical TRA2B-203 isoform.
TRA2B mRNA is one of the most well-known splice targets of ZRANB2 40. TRA2B mRNA codes for the protein Tra2β which is closely related to the SR family of splice regulator proteins and is a central player in splicing regulation itself. Tra2β is known to be upregulated in several cancers 42. Furthermore, several Tra2β target exons have been experimentally demonstrated to have important pro-oncogenic roles, prime examples being CD44, HIPK3 and NASP 42. TRA2B knockdown leads to widespread altered splicing events in the genome, including many novel and unannotated isoforms 77. It is possible that by altering the expression and function of ZRANB2, As3+ exposure could give rise to genome-wide aberrant alternative splicing and set cells on the path of carcinogenesis via altering the splice patterns of a few target genes like TRA2B.
Although the present work examines the interaction of one alternative splicing zfm protein with one metalloid, these results open up new lines of research in several directions. Several metal containing chemotherapeutic agents (platinum, gold, ruthenium, cobalt, antimony and selenium), as well as toxic and/or carcinogenic metals/metalloids (arsenic, cadmium, chromium, cobalt, nickel, lead) are capable of binding to and displacing Zn2+ from zfm proteins85. Surprisingly, little or no work has been done on zinc displacement from RNA binding proteins including splice regulator proteins, which form a major class of zfm proteins. We speculate that differential alternative splicing as a result of structural and functional abrogation of splice regulating zfm proteins by heavy metals/toxicants/pollutants/therapeutic molecules could be playing an important role both in health and disease.
Supplementary Material
ACKNOWLEDGEMENTS
The authors express their gratitude to Sabine Waigel, Ashley Mitchell Wise and Vennila Arumugam, Genomics Facility, University of Louisville, for technical help and access to the Genomics Facility instruments. We also thank Dr. Robert D. Gray, Brown Cancer Center, University of Louisville, for technical expertise and access to the FluoroMax®-3.
FUNDING
This work was supported in part by NIH grant R01ES027778 and by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant no. P20GM113226-6176. Part of this work was performed with assistance of the University of Louisville Genomics Facility, which is supported by NIH grants P20GM103436 (KY IDeA Networks of Biomedical Research Excellence) and P30GM106396 (UofL J. G. Brown Cancer Center Phase III CoBRE), NIH P20GM113226 (UofL Hepatobiology and Toxicology CoBRE), the J. G. Brown Foundation, and user fees.
Footnotes
DECLARATION OF CONFLICTING INTERESTS
The authors declare no conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFRENCES
- (1).Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, and Suk WA (2013) The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 121, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Banerjee M, Ghosh P, Giri AK (2011) Arsenic-induced Cancers: A Review with Special Reference to Gene, Environment and their Interaction. Genes and Environment 33, 128–140. [Google Scholar]
- (3).Lo-Coco F, Cicconi L, and Breccia M (2016) Current standard treatment of adult acute promyelocytic leukaemia. Br J Haematol 172, 841–854. [DOI] [PubMed] [Google Scholar]
- (4).Lynch E, and Braithwaite R (2005) A review of the clinical and toxicological aspects of 'traditional' (herbal) medicines adulterated with heavy metals. Expert Opin Drug Saf 4, 769–778. [DOI] [PubMed] [Google Scholar]
- (5).Waldman A, and Schmults C (2019) Cutaneous Squamous Cell Carcinoma. Hematol Oncol Clin North Am 33, 1–12. [DOI] [PubMed] [Google Scholar]
- (6).Siefring ML, Lu D, States JC, and Van Hoang M (2018) Rapid onset of multiple concurrent squamous cell carcinomas associated with the use of an arsenic-containing traditional medicine for chronic plaque psoriasis. BMJ Case Rep 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Snow ET, Sykora P, Durham TR, and Klein CB (2005) Arsenic, mode of action at biologically plausible low doses: what are the implications for low dose cancer risk? Toxicol Appl Pharmacol 207, 557–564. [DOI] [PubMed] [Google Scholar]
- (8).Yu HS, Liao WT, and Chai CY (2006) Arsenic carcinogenesis in the skin. J Biomed Sci 13, 657–666. [DOI] [PubMed] [Google Scholar]
- (9).Ghosh P, Banerjee M, Giri AK, and Ray K (2008) Toxicogenomics of arsenic: classical ideas and recent advances. Mutat Res 659, 293–301. [DOI] [PubMed] [Google Scholar]
- (10).Hunt KM, Srivastava RK, Elmets CA, and Athar M (2014) The mechanistic basis of arsenicosis: pathogenesis of skin cancer. Cancer Lett 354, 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cardoso APF, Al-Eryani L, and States JC (2018) Arsenic-Induced Carcinogenesis: The Impact of miRNA Dysregulation. Toxicol Sci 165, 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lee CH, and Yu HS (2016) Role of mitochondria, ROS, and DNA damage in arsenic induced carcinogenesis. Front Biosci (Schol Ed) 8, 312–320. [DOI] [PubMed] [Google Scholar]
- (13).Rossman TG (2003) Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res 533, 37–65. [DOI] [PubMed] [Google Scholar]
- (14).Kim HK, Pham MHC, Ko KS, Rhee BD, and Han J (2018) Alternative splicing isoforms in health and disease. Pflugers Arch 470, 995–1016. [DOI] [PubMed] [Google Scholar]
- (15).Urbanski LM, Leclair N, and Anczukow O (2018) Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 9, e1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Shuai S, Suzuki H, Diaz-Navarro A, Nadeu F, Kumar SA, Gutierrez-Fernandez A, Delgado J, Pinyol M, Lopez-Otin C, Puente XS, Taylor MD, Campo E, and Stein LD (2019) The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature. [DOI] [PubMed] [Google Scholar]
- (17).Song X, Zeng Z, Wei H, and Wang Z (2018) Alternative splicing in cancers: From aberrant regulation to new therapeutics. Semin Cell Dev Biol 75, 13–22. [DOI] [PubMed] [Google Scholar]
- (18).Zhang J, and Manley JL (2013) Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov 3, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Oltean S, and Bates DO (2014) Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318. [DOI] [PubMed] [Google Scholar]
- (20).Chen J, and Weiss WA (2015) Alternative splicing in cancer: implications for biology and therapy. Oncogene 34, 1–14. [DOI] [PubMed] [Google Scholar]
- (21).Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, and Stamm S (2013) Function of alternative splicing. Gene 514, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Radhakrishnan A, Nanjappa V, Raja R, Sathe G, Chavan S, Nirujogi RS, Patil AH, Solanki H, Renuse S, Sahasrabuddhe NA, Mathur PP, Prasad TS, Kumar P, Califano JA, Sidransky D, Pandey A, Gowda H, and Chatterjee A (2016) Dysregulation of splicing proteins in head and neck squamous cell carcinoma. Cancer Biol Ther 17, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fu M, and Blackshear PJ (2017) RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 17, 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hall TM (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15, 367–373. [DOI] [PubMed] [Google Scholar]
- (25).Brown RS (2005) Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 15, 94–98. [DOI] [PubMed] [Google Scholar]
- (26).Plambeck CA, Kwan AH, Adams DJ, Westman BJ, van der Weyden L, Medcalf RL, Morris BJ, and Mackay JP (2003) The structure of the zinc finger domain from human splicing factor ZNF265 fold. J Biol Chem 278, 22805–22811. [DOI] [PubMed] [Google Scholar]
- (27).Asmuss M, Mullenders LH, Eker A, and Hartwig A (2000) Differential effects of toxic metal compounds on the activities of Fpg and XPA, two zinc finger proteins involved in DNA repair. Carcinogenesis 21, 2097–2104. [DOI] [PubMed] [Google Scholar]
- (28).Huestis J, Zhou X, Chen L, Feng C, Hudson LG, and Liu KJ (2016) Kinetics and thermodynamics of zinc(II) and arsenic(NI) binding to XPA and PARP-1 zinc finger peptides. J Inorg Biochem 163, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zhang F, Paramasivam M, Cai Q, Dai X, Wang P, Lin K, Song J, Seidman MM, and Wang Y (2014) Arsenite binds to the RING finger domains of RNF20-RNF40 histone E3 ubiquitin ligase and inhibits DNA double-strand break repair. J Am Chem Soc 136, 12884–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Jiang J, Bellani M, Li L, Wang P, Seidman MM, and Wang Y (2017) Arsenite Binds to the RING Finger Domain of FANCL E3 Ubiquitin Ligase and Inhibits DNA Interstrand Crosslink Repair. ACS Chem Biol 12, 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, and Hudson LG (2011) Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem 286, 22855–22863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sun X, Zhou X, Du L, Liu W, Liu Y, Hudson LG, and Liu KJ (2014) Arsenite binding-induced zinc loss from PARP-1 is equivalent to zinc deficiency in reducing PARP-1 activity, leading to inhibition of DNA repair. Toxicol Appl Pharmacol 274, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, and Liu KJ (2009) Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J Biol Chem 284, 6809–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Shen S, Li XF, Cullen WR, Weinfeld M, and Le XC (2013) Arsenic binding to proteins. Chem Rev 113, 7769–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Witkiewicz-Kucharczyk A, and Bal W (2006) Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol Lett 162, 29–42. [DOI] [PubMed] [Google Scholar]
- (36).Riedmann C, Ma Y, Melikishvili M, Godfrey SG, Zhang Z, Chen KC, Rouchka EC, and Fondufe-Mittendorf YN (2015) Inorganic Arsenic-induced cellular transformation is coupled with genome wide changes in chromatin structure, transcriptome and splicing patterns. BMC Genomics 16, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Loughlin FE, Mansfield RE, Vaz PM, McGrath AP, Setiyaputra S, Gamsjaeger R, Chen ES, Morris BJ, Guss JM, and Mackay JP (2009) The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences. Proc Natl Acad Sci U S A 106, 5581–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Mangs AH, and Morris BJ (2008) ZRANB2: structural and functional insights into a novel splicing protein. Int J Biochem Cell Biol 40, 2353–2357. [DOI] [PubMed] [Google Scholar]
- (39).Yang YH, Markus MA, Mangs AH, Raitskin O, Sperling R, and Morris BJ (2013) ZRANB2 localizes to supraspliceosomes and influences the alternative splicing of multiple genes in the transcriptome. Mol Biol Rep 40, 5381–5395. [DOI] [PubMed] [Google Scholar]
- (40).Adams DJ, van der Weyden L, Mayeda A, Stamm S, Morris BJ, and Rasko JE (2001) ZNF265--a novel spliceosomal protein able to induce alternative splicing. J Cell Biol 154, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cohen OS, Weickert TW, Hess JL, Paish LM, McCoy SY, Rothmond DA, Galletly C, Liu D, Weinberg DD, Huang XF, Xu Q, Shen Y, Zhang D, Yue W, Yan J, Wang L, Lu T, He L, Shi Y, Xu M, Che R, Tang W, Chen CH, Chang WH, Hwu HG, Liu CM, Liu YL, Wen CC, Fann CS, Chang CC, Kanazawa T, Middleton FA, Duncan TM, Faraone SV, Weickert CS, Tsuang MT, and Glatt SJ (2016) A splicing-regulatory polymorphism in DRD2 disrupts ZRANB2 binding, impairs cognitive functioning and increases risk for schizophrenia in six Han Chinese samples. Mol Psychiatry 21, 975–982. [DOI] [PubMed] [Google Scholar]
- (42).Best A, Dagliesh C, Ehrmann I, Kheirollahi-Kouhestani M, Tyson-Capper A, and Elliott DJ (2013) Expression of Tra2 beta in Cancer Cells as a Potential Contributory Factor to Neoplasia and Metastasis. Int J Cell Biol 2013, 843781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Best A, James K, Dalgliesh C, Hong E, Kheirolahi-Kouhestani M, Curk T, Xu Y, Danilenko M, Hussain R, Keavney B, Wipat A, Klinck R, Cowell IG, Cheong Lee K, Austin CA, Venables JP, Chabot B, Santibanez Koref M, Tyson-Capper A, and Elliott DJ (2014) Human Tra2 proteins jointly control a CHEK1 splicing switch among alternative and constitutive target exons. Nat Commun 5, 4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Jiang J, Tam LM, Wang P, and Wang Y (2018) Arsenite Targets the RING Finger Domain of Rbx1 E3 Ubiquitin Ligase to Inhibit Proteasome-Mediated Degradation of Nrf2. Chem Res Toxicol 31, 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Spuches AM, Kruszyna HG, Rich AM, and Wilcox DE (2005) Thermodynamics of the As(NI)-thiol interaction: arsenite and monomethylarsenite complexes with glutathione, dihydrolipoic acid, and other thiol ligands. Inorg Chem 44, 2964–2972. [DOI] [PubMed] [Google Scholar]
- (46).Zhou X, Sun X, Mobarak C, Gandolfi AJ, Burchiel SW, Hudson LG, and Liu KJ (2014) Differential binding of monomethylarsonous acid compared to arsenite and arsenic trioxide with zinc finger peptides and proteins. Chem Res Toxicol 27, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, and Gibson BW (2012) Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics 11, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Royer CA (2006) Probing protein folding and conformational transitions with fluorescence. Chem Rev 106, 1769–1784. [DOI] [PubMed] [Google Scholar]
- (49).Bio-Rad. Method for Measuring Cytotoxicity or Proliferation Using alamarBlue by Spectrophotometry.
- (50).Al-Eryani L, Waigel S, Tyagi A, Peremarti J, Jenkins SF, Damodaran C, and States JC (2018) Differentially Expressed mRNA Targets of Differentially Expressed miRNAs Predict Changes in the TP53 Axis and Carcinogenesis-Related Pathways in Human Keratinocytes Chronically Exposed to Arsenic. Toxicol Sci 162, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Schneider CA, Rasband WS, and Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).McNeely SC, Taylor BF, and States JC (2008) Mitotic arrest-associated apoptosis induced by sodium arsenite in A375 melanoma cells is BUBR1-dependent. Toxicol Appl Pharmacol 231, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Goldman A, Harper S, and Speicher DW (2016) Detection of Proteins on Blot Membranes. Curr Protoc Protein Sci 86, 10 18 11–10 18 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Mattapalli H, Monteith WB, Burns CS, and Danell AS (2009) Zinc deposition during ESI-MS analysis of peptide-zinc complexes. J Am Soc Mass Spectrom 20, 2199–2205. [DOI] [PubMed] [Google Scholar]
- (55).States JC, Barchowsky A, Cartwright IL, Reichard JF, Futscher BW, and Lantz RC (2011) Arsenic toxicology: translating between experimental models and human pathology. Environ Health Perspect 119, 1356–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Kitchin KT, and Wallace K (2006) Arsenite binding to synthetic peptides: the effect of increasing length between two cysteines. J Biochem Mol Toxicol 20, 35–38. [DOI] [PubMed] [Google Scholar]
- (57).Kitchin KT, and Wallace K (2005) Arsenite binding to synthetic peptides based on the Zn finger region and the estrogen binding region of the human estrogen receptor-alpha. Toxicol Appl Pharmacol 206, 66–72. [DOI] [PubMed] [Google Scholar]
- (58).Kitchin KT, and Wallace K (2008) The role of protein binding of trivalent arsenicals in arsenic carcinogenesis and toxicity. J Inorg Biochem 102, 532–539. [DOI] [PubMed] [Google Scholar]
- (59).Zhou X, Cooper KL, Sun X, Liu KJ, and Hudson LG (2015) Selective Sensitization of Zinc Finger Protein Oxidation by Reactive Oxygen Species through Arsenic Binding. J Biol Chem 290, 18361–18369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Klug A (2010) The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem 79, 213–231. [DOI] [PubMed] [Google Scholar]
- (61).Cassandri M, Smirnov A, Novelli F, Pitolli C, Agostini M, Malewicz M, Melino G, and Raschella G (2017) Zinc-finger proteins in health and disease. Cell Death Discov 3, 17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Jen J, and Wang YC (2016) Zinc finger proteins in cancer progression. J Biomed Sci 23, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Munro D, Ghersi D, and Singh M (2018) Two critical positions in zinc finger domains are heavily mutated in three human cancer types. PLoS Comput Biol 14, e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Ding X, Zhou X, Cooper KL, Huestis J, Hudson LG, and Liu KJ (2017) Differential sensitivities of cellular XPA and PARP-1 to arsenite inhibition and zinc rescue. Toxicol Appl Pharmacol 331, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Tam LM, Jiang J, Wang P, Li L, Miao W, Dong X, and Wang Y (2017) Arsenite Binds to the Zinc Finger Motif of TIP60 Histone Acetyltransferase and Induces Its Degradation via the 26S Proteasome. Chem Res Toxicol 30, 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Baglivo I, Russo L, Esposito S, Malgieri G, Renda M, Salluzzo A, Di Blasio B, Isernia C, Fattorusso R, and Pedone PV (2009) The structural role of the zinc ion can be dispensable in prokaryotic zinc-finger domains. Proc Natl Acad Sci U S A 106, 6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).D'Abrosca G, Russo L, Palmieri M, Baglivo I, Netti F, de Paola I, Zaccaro L, Farina B, Iacovino R, Pedone PV, Isernia C, Fattorusso R, and Malgieri G (2016) The (unusual) aspartic acid in the metal coordination sphere of the prokaryotic zinc finger domain. J Inorg Biochem 161, 91–98. [DOI] [PubMed] [Google Scholar]
- (68).Malgieri G, Palmieri M, Russo L, Fattorusso R, Pedone PV, and Isernia C (2015) The prokaryotic zinc-finger: structure, function and comparison with the eukaryotic counterpart. FEBS J 282, 4480–4496. [DOI] [PubMed] [Google Scholar]
- (69).Simpson RJ, Cram ED, Czolij R, Matthews JM, Crossley M, and Mackay JP (2003) CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions. J Biol Chem 278, 28011–28018. [DOI] [PubMed] [Google Scholar]
- (70).Wei D, and Sun Y (2010) Small RING Finger Proteins RBX1 and RBX2 of SCF E3 Ubiquitin Ligases: The Role in Cancer and as Cancer Targets. Genes Cancer 1, 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Krezel A, Latajka R, Bujacz GD, and Bal W (2003) Coordination properties of tris(2-carboxyethyl)phosphine, a newly introduced thiol reductant, and its oxide. Inorg Chem 42, 1994–2003. [DOI] [PubMed] [Google Scholar]
- (72).Maret W (2017) Zinc in Cellular Regulation: The Nature and Significance of "Zinc Signals". Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Roggenbeck BA, Banerjee M, and Leslie EM (2016) Cellular arsenic transport pathways in mammals. J Environ Sci (China) 49, 38–58. [DOI] [PubMed] [Google Scholar]
- (74).Shen ZX, Chen GQ, Ni JH, Li XS, Xiong SM, Qiu QY, Zhu J, Tang W, Sun GL, Yang KQ, Chen Y, Zhou L, Fang ZW, Wang YT, Ma J, Zhang P, Zhang TD, Chen SJ, Chen Z, and Wang ZY (1997) Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 89, 3354–3360. [PubMed] [Google Scholar]
- (75).Wu X, Sun R, Wang H, Yang B, Wang F, Xu H, Chen S, Zhao R, Pi J, and Xu Y (2019) Enhanced p62-NRF2 Feedback Loop due to Impaired Autophagic Flux Contributes to Arsenic-Induced Malignant Transformation of Human Keratinocytes. Oxid Med Cell Longev 2019, 1038932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Pi J, Kumagai Y, Sun G, Yamauchi H, Yoshida T, Iso H, Endo A, Yu L, Yuki K, Miyauchi T, and Shimojo N (2000) Decreased serum concentrations of nitric oxide metabolites among Chinese in an endemic area of chronic arsenic poisoning in inner Mongolia. Free Radic Biol Med 28, 1137–1142. [DOI] [PubMed] [Google Scholar]
- (77).Dichmann DS, Walentek P, and Harland RM (2015) The alternative splicing regulator Tra2b is required for somitogenesis and regulates splicing of an inhibitory Wnt1b isoform. Cell Rep 10, 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Tolins M, Ruchirawat M, and Landrigan P (2014) The developmental neurotoxicity of arsenic: cognitive and behavioral consequences of early life exposure. Ann Glob Health 80, 303–314. [DOI] [PubMed] [Google Scholar]
- (79).Wasserman GA, Liu X, Parvez F, Chen Y, Factor-Litvak P, LoIacono NJ, Levy D, Shahriar H, Uddin MN, Islam T, Lomax A, Saxena R, Gibson EA, Kioumourtzoglou MA, Balac O, Sanchez T, Kline JK, Santiago D, Ellis T, van Geen A, and Graziano JH (2018) A cross-sectional study of water arsenic exposure and intellectual function in adolescence in Araihazar, Bangladesh. Environ Int 118, 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Wasserman GA, Liu X, Loiacono NJ, Kline J, Factor-Litvak P, van Geen A, Mey JL, Levy D, Abramson R, Schwartz A, and Graziano JH (2014) A cross-sectional study of well water arsenic and child IQ in Maine schoolchildren. Environ Health 13, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Kuhn M, Sammartin K, Nabergoj M, and Vianello F (2016) Severe Acute Axonal Neuropathy following Treatment with Arsenic Trioxide for Acute Promyelocytic Leukemia: a Case Report. Mediterr J Hematol Infect Dis 8, e2016023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Saidak Z, Pascual C, Bouaoud J, Galmiche L, Clatot F, Dakpe S, Page C, and Galmiche A (2019) A three-gene expression signature associated with positive surgical margins in tongue squamous cell carcinomas: Predicting surgical resectability from tumour biology? Oral Oncol 94, 115–120. [DOI] [PubMed] [Google Scholar]
- (83).Yuan S, Ding X, Cui Y, Wei K, Zheng Y, and Liu Y (2017) Cisplatin Preferentially Binds to Zinc Finger Proteins Containing C3H1 or C4 Motifs. Eur J Inorg Chem 2017, 1778–1784. [Google Scholar]
- (84).Mendes F, Groessl M, Nazarov AA, Tsybin YO, Sava G, Santos I, Dyson PJ, and Casini A (2011) Metal-based inhibition of poly(ADP-ribose) polymerase--the guardian angel of DNA. J Med Chem 54, 2196–2206. [DOI] [PubMed] [Google Scholar]
- (85).Abbehausen C (2019) Zinc finger domains as therapeutic targets for metal-based compounds – an update. Metallomics 11, 15–28. [DOI] [PubMed] [Google Scholar]
- (86).Sancineto L, Iraci N, Tabarrini O, and Santi C (2018) NCp7: targeting a multitasking protein for next-generation anti-HIV drug development part 1: covalent inhibitors. Drug Discov Today 23, 260–271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.