Abstract
Purpose
The purpose of this study was to establish and analyze a cell model of Leber congenital amaurosis type 16 (LCA16), which is caused by mutations in the KCNJ13 gene encoding Kir7.1, an inward-rectifying potassium ion channel.
Methods
The two guide RNAs specific to the target sites in the KCNJ13 gene were designed and KCNJ13 knock-out (KO) human-induced pluripotent stem cells (hiPSCs) were generated using the CRISPR/Cas9 system. The KCNJ13-KO hiPSCs were differentiated into retinal pigment epithelial cells (hiPSC-RPEs). The KCNJ13-KO in hiPSC-RPEs was confirmed by immunostaining. Phagocytic activity of hiPSC-RPEs was assessed using the uptake of fluorescently labeled porcine photoreceptor outer segments (POSs). Phagocytosis-related genes in RPE cells were assessed by quantitative polymerase chain reaction.
Results
Most of the translated region of the KCNJ13 gene was deleted in the KCNJ13-KO hiPSCs by the CRISPR/Cas9 system, and this confirmed that the Kir7.1 protein was not present in RPE cells induced from the hiPSCs. Expression of RPE marker genes such as BEST1 and CRALBP was retained in the wild-type (WT) and in the KCNJ13-KO hiPSC-RPE cells. However, phagocytic activity and expression of phagocytosis-related genes in the KCNJ13-null hiPSC-RPE cells were significantly reduced compared to those of WT.
Conclusions
We succeeded in generating an RPE model of LCA16 using hiPSCs. We suggest that Kir7.1 is required for phagocytosis of POSs by RPE cells and that impaired phagocytosis in the absence of Kir7.1 would be involved in the retinal degeneration found in LCA16.
Keywords: Kir7.1, KCNJ13, human-induced pluripotent cells, retinal pigment epithelium, phagocytosis
Retinal pigment epithelium (RPE) has multiple functions, such as ion exchange in the subretinal space, maintenance of the visual cycle, secretion of trophic factors (e.g., vascular endothelial growth factor), and formation of the blood–retinal barrier. The RPE also has an important role in the continuous renewal of photoreceptor outer segments (POSs) through phagocytosis.1,2 Leber congenital amaurosis (LCA) is a genetic disease with progressive severe visual impairment, retinal structural abnormalities, and abnormal electroretinograms.3–5 In LCA, there have been numerous known genetic alterations, one of which, LCA16, was found to be associated with KCNJ13 gene mutations.3–5 The KCNJ13 gene encodes the Kir7.1 protein, an inward-rectifying potassium ion channel, which is expressed by the apical microvilli of RPE cells in ocular tissues.6,7 Kir7.1 regulates potassium ions in the subretinal space8 and is known to be involved in photoreceptor maintenance.9–12
There are many questions regarding the pathophysiologic mechanism of LCA16. Previous studies using KCNJ13 conditional knock-out (KO),13 genetic mosaic KO,9 and knock-down8 mice revealed that deletion or decrease of the Kir7.1 protein induces photoreceptor loss and abnormal changes in the electroretinogram. These findings suggest that Kir7.1 dysfunction in RPE cells in the subretinal space may cause structural and functional abnormalities in the retina; however, how the dysfunction of Kir7.1 in the RPE causes LCA symptoms remains unclear.
Here, we aimed to establish an RPE cell model of LCA16 by deleting the KCNJ13 gene using the CRISPR/Cas9 system in human-induced pluripotent stem cells (hiPSCs). KCNJ13-deficient hiPSCs were generated and induced to differentiate into hiPSC-RPE cells, where the functions of KCNJ13 were analyzed.
Methods
Culture of Human iPS Cells
The hiPSC line 454E2,14 generated from healthy human dental pulp cells, was obtained from the RIKEN BioResource Center (Ibaraki, Japan). The hiPSCs were maintained and differentiated as previously described15 (Supplementary Methods).
Gene Editing of hiPSCs Via CRISPR/Cas9 System
By utilizing CRISPRdirect (https://crispr.dbcls.jp),16 more than two candidate target sequences for guide RNA (gRNA) were found in the human KCNJ13 gene (NM_001172416) obtained from the Ensemble database (http://asia.ensembl.org/index.html). To ensure specificity, the hit sequence showing one (“1”) in the column “20 mer + PAM,” which indicates a perfect match with the intended target site, was selected. Only RFX1 was listed as a candidate off-target gene. To delete most of the KCNJ13 gene, two target sequences were determined; one was localized to approximately 100 bases downstream of the start codon, and the other was in the 3′UTR (Fig. 1A). Two CRISPR RNA (crRNA) complementary to the target sequences and the trans-activating CRISPR RNA (tracrRNA) were obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA). The crRNA and tracrRNA were each annealed to construct gRNA according to the manufacturer's instructions. We performed gene editing of hiPSCs according to previous studies17,18 (see Supplementary Methods).
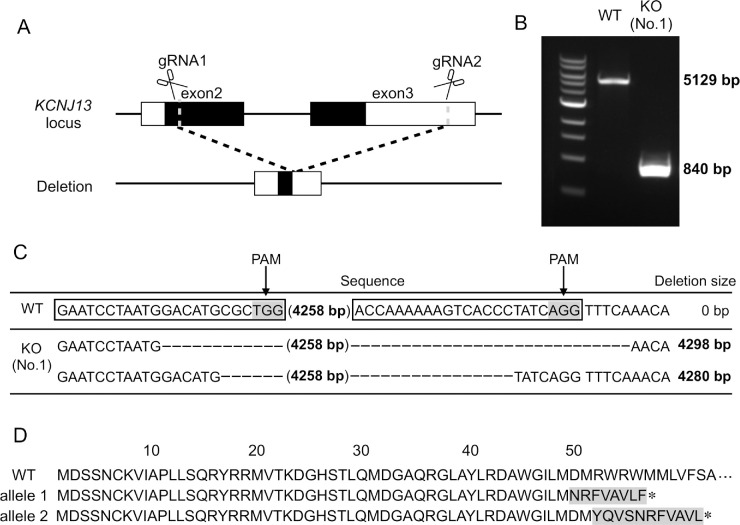
Figure 1.
Establishment of KCNJ13-KO hiPSC. (A) KCNJ13 gene locus and the target sites of designed gRNAs. Schematic diagram of gRNA targeting the human KCNJ13 locus. The black rectangle represents the protein coding region of the KCNJ13 gene. The white rectangle represents the untranslated region of the KCNJ13 gene. The two gRNA sequences are designed to delete exons 2 and 3 from the KCNJ13 gene. (B) PCR products of induced pluripotent stem cell line (iPSC) clones whose KCNJ13 genes were edited by the CRISPR/Cas9 system. PCR was performed using primers to confirm that the intended gene editing occurred in the KCNJ13 locus. The 5129-bp PCR product was observed in the WT iPSC, and the approximately 840-bp PCR fragment was produced in the mutant iPSC. The bi-allelic gene KO was present in clone no. 1. (C) KCNJ13 DNA sequence obtained from WT and KCNJ13-KO iPSCs. KCNJ13-KO was confirmed in clone no. 1, and different mutations occurred in each allele. (D) The amino acid sequences deduced from the nucleotide sequences of the WT and KO alleles are shown. The WT sequence encodes for 360 amino acids. The sequence of allele 1 encodes for 57 amino acids, and that of allele 2 encodes for 62 amino acids.
Immunofluorescence
Immunostaining was performed according to previous reports19,20 with minor modifications (see Supplementary Methods for details). We confirmed expression of undifferentiated marker genes in proliferating hiPSCs and localization of the Kir7.1 protein in the RPE differentiated from intact hiPSCs (hiPSC-RPE). To visualize the cellular polarity and nuclei, hiPSC-RPE cells were co-stained with rhodamine (tetramethylrhodamine)–phalloidin, anti-RPE65, and 4′,6-diamidino-2-phenylindole (DAPI). In the phagocytosis assay, hiPSC-RPE cells were stained by anti-ZO-1 or anti-ezrin to identify the location of incorporated POSs. Immunocytochemical staining was analyzed and images were taken on an LSM 780 confocal microscope equipped with ZEN 2009 software (Carl Zeiss Microscopy GmbH, Jena, Germany).
Western Blot Analysis
Western blot analysis was performed according to previous studies.19,21 Primary antibodies used were as follows: anti-BEST1 (1:1000; Abcam, Cambridge, UK), anti-CRALBP (1:1000; Abcam), and anti-cyclophilin B (1:1000; Cell Signaling Technology, Danvers, MA, USA).
Counting of Protruded Cells Per Unit Area
The hiPSC-RPE cells were seeded onto Transwells (Corning, Inc., Corning, NY, USA) and observed under a microscope at 2 and 4 weeks after seeding. The number of protruded cells above the level of the hiPSC-RPE monolayer was counted in five visual fields; the mean value per unit area (9.63 mm2) was calculated and regarded as the number of protruded cells in the well.
Transmission Electron Microscopy
Samples for electron microscopy were prepared with the modified method reported by Shahi et al.22 Briefly, hiPSC-RPE on Transwells were fixed with 2.5% glutaraldehyde and 2.0% paraformaldehyde in 0.1-M cacodylate buffer (CB) (pH 7.4), for 1 hour at 4°C. The samples were rinsed in 0.1-M CB and then post-fixed in 2% osmium tetroxide (OsO4) in CB for 1.5 hours. After post-fixation, samples were rinsed in CB. The samples were dehydrated via an ethanol series and then infiltrated and polymerized in epoxy resin, Epon 812 or Spurr (Sigma-Aldrich, St. Louis, MO, USA). Ultrathin sections, about 80 nm thick, were obtained with a microtome. The sections were stained with uranyl acetate and lead citrate. The hiPSC-RPE cells were visualized with a transmission electron microscope (H-7650; Hitachi, Tokyo, Japan).
Preparation of Porcine POS
Phagocytic activity of hiPSC-RPE was assayed by using in-house-prepared POSs. Porcine POSs were isolated according to Parinot's procedure.23 Briefly, retina was dissected from freshly sacrificed porcine eyeballs and transferred to a sucrose buffer (20% sucrose; 20-mM tris-acetate, pH 7.2; 2-mM MgCl2; 10-mM glucose; and 5-mM taurine). Suspended retinal tissues were gently layered onto a sucrose gradient and centrifuged (106,000×g) at 4°C for 50 minutes. After centrifugation, only the POS-containing layer was transferred to another tube. The crude POSs were subsequently treated with solutions of Wash 1 (20-mM tris-acetate, pH 7.2, and 5-mM taurine), Wash 2 (10% sucrose; 20-mM tris-acetate, pH 7.2; and 5-mM taurine), and Wash 3 (10% sucrose; 20-mM sodium phosphate, pH 7.2; and 5-mM taurine). Isolated POSs were labeled with fluorescein isothiocyanate (FITC, 2.5 mg/mL; Dojindo Laboratories, Kumamoto, Japan), dissolved in an alkaline solution (0.1-M NaHCO3, pH 8.4, and 0.1-M Na2CO3, pH 11.5), and stored at –80°C until further use. The FITC-labeled POSs were used for phagocytosis assay immediately after they were thawed.
Phagocytosis Assay
Phagocytosis assay was performed according to Davis et al.24 The hiPSC-RPE cells were seeded in 12-well Transwells at 5 × 105 cells/well. After 2 and 4 weeks, the hiPSC-RPE cells were supplemented with 2.5 µg/ml human MFG-E8 (R&D Systems, Minneapolis, MN, USA) and 2 µg/ml human Protein S (Aniara Diagnostica, West Chester, OH, USA) and challenged for 5 hours with 10 POSs per cell. Subsequently, the Transwell membrane on which the hiPSC-RPE cells were cultured was removed, and the cells were processed for immunostaining with anti-ZO-1 or anti-ezrin and nuclear staining with DAPI. The number of POSs below ZO-1 or ezrin was counted as internalized POSs. The number of internalized POSs per cell and the number of POSs per area (µm2) were measured. The mean number of internalized POSs in the KCNJ13-null hiPSC-RPE was normalized to wild-type (WT).
Quantitative PCR Analysis
Quantitative PCR (qPCR) was performed according to Fujita et al.25 with minor modifications. Messenger RNA was isolated from hiPSC-RPE cells, cDNA was synthesized by standard procedures, and quantitative real-time PCR was performed in LightCycler 8-Tube Strips using the LightCycler Nano Instrument and FastStart Essential DNA Green Master (Roche, Basel, Switzerland). The primers used to amplify the phagocytosis marker genes and internal standard, 18s rRNA, and the primers used for qPCR are shown in the Table.
Table.
Sequences of Primers Used in This Study.
| Gene | Forward | Reverse |
|---|---|---|
| KCNJ13 (genotyping) | 5′-ATTTGGTCAAATCAATAAATGCTTG-3′ | 5′-GAATGTCTAAGATTTTCAAACAGCA-3′ |
| ITGAV | 5′-CCTGTGCCTGTGTGGGTGAT-3′ | 5′-GGTGGCGGACCCGTTTA-3′ |
| ITGBV | 5′-TTGGCAGAGAACAACATCAACC-3′ | 5′-TCCTCAGGCTGATCCCAGAC-3′ |
| CD81 | 5′-TCTGGAGCTGGGAGACAAGC-3′ | 5′-GGATGACCAGGCAGGTGAAG-3′ |
| FAK | 5′-GAAGCATTGGGTCGGGAACTA-3′ | 5′-CTCAATGCAGTTTGGAGGTGC-3′ |
| MerTK | 5′-GTGAGGCAGCGTGCATGAAAG-3′ | 5′-GGGCTTTGGGATGCCTTGAG-3′ |
| TMEM175 | 5′-CTTTGCAGCGGCCATCTTC-3′ | 5′-GATGACAGTCACCATCAGCAGGTAA-3′ |
| RNA18SN5 | 5′-GTAACCCGTTGAACCCCATT-3′ | 5′-CCATCCAATCGGTAGTAGCG-3′ |
Statistics
Statistical analysis was performed using SPSS Statistics (IBM, Armonk, NY, USA). The quantitative data of cell protrusion, phagocytosis assay, and expression of phagocytosis-related genes were analyzed by unpaired Student's t-test; P < 0.05 indicated significance.
Results
By use of the Cas9 protein and two gRNAs targeted to KCNJ13 (Fig. 1A), gene editing of hiPSCs was performed and the cells were processed for single cell cloning. First, we verified whether precise gene editing occurred in the manipulated hiPSCs. Consequently, we obtained a KCNJ13-CRISPR cell line (clone no. 1) which had a desired deletion mutation in the KCNJ13 gene (Fig. 1B). The human KCNJ13 gene consists of three exons that encode a Kir7.1 protein of 360 amino acids. Most of exons 2 and 3 were deleted in this cell line, resulting in an N-terminal-only protein of about 60 amino acids, if the mRNA was not degraded (Figs. 1C, 1D, Supplementary Fig. S1). We also examined whether off-target gene editing occurred; however, we did not find any mutations in the only candidate off-target gene, RFX1 (Supplementary Fig. S2). We found that gene-edited hiPSCs expressed undifferentiated marker genes, NANOG, SSEA-4, OCT4, and TRA-1-81 (Supplementary Fig. S3), indicating that stemness or pluripotency was maintained in the hiPSCs after electroporation of CRISPR materials.
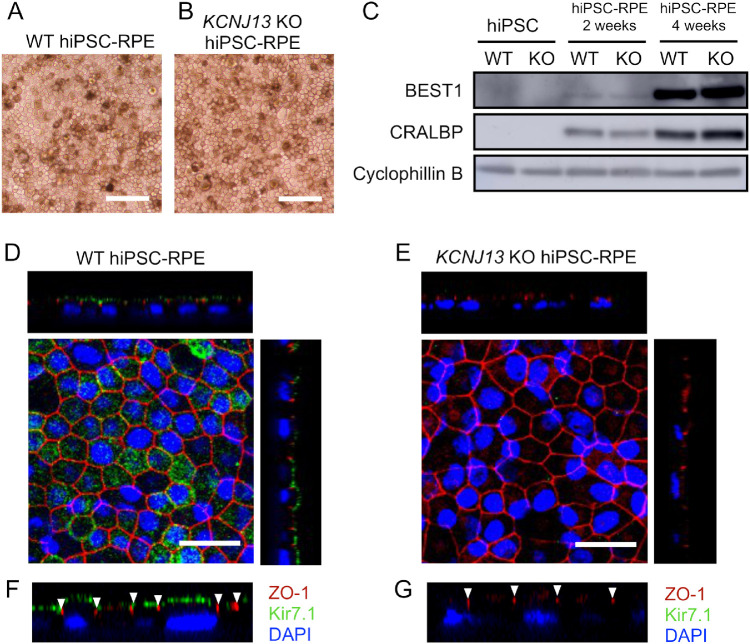
We then induced differentiation of the gene-edited, KCNJ13-deleted iPSCs to RPE cells. When the iPSCs were cultured according to RPE-differentiation protocols,14 WT and KCNJ13-KO cells appeared similarly pigmented and exhibited a cobblestone-like appearance after passage 4 (Figs. 2A, 2B). To verify their RPE differentiation status, we examined expression of RPE marker genes in WT and KCNJ13-KO hiPSC-RPEs. At 2 and 4 weeks after seeding, western blot analysis showed that BEST1 and CRALBP were expressed in WT and KCNJ13-KO hiPSC-RPEs (Fig. 2C). In RPE cells, we verified whether the Kir7.1 protein encoded by the KCNJ13 gene was absent in the KCNJ13-KO cell line. In the case of WT cells, immunostaining showed that Kir7.1 was localized to the apical cell membrane, above ZO-1, which is a phosphoprotein present at tight junctions, and a representative apical structure of epithelial cells (Figs. 2D, 2F). By contrast, Kir7.1 was not observed at all in the KCNJ13-KO hiPSC-RPE (Figs. 2E, 2G). These results indicate that Kir7.1 was lacking in the KCNJ13-KO hiPSC-RPE, and that Kir7.1 is not required for differentiation of iPSCs to RPE cells.
Figure 2.
KCNJ13-KO iPSCs differentiate to RPE, and the expression and localization of Kir7.1 in WT and KCNJ13-KO iPSCs is shown. Cellular morphology of RPE derived from WT (A) and KCNJ13-KO (B) hiPSCs. Both hiPSC-RPE cells show pigmentation and exhibit a cobblestone-like appearance, indicating a normal RPE phenotype. (C) Protein expression of RPE-specific markers in WT and KCNJ13-KO hiPSC-RPE cells. WT hiPSC-RPE and KCNJ13-KO hiPSC-RPE cells were immunostained and observed with a confocal microscope. In WT hiPSC-RPE, Kir7.1 (green) is expressed on the apical side of the cells—that is, on the upper side of ZO-1 (red, arrowhead) (D, F). Kir7.1 is not present in the KCNJ13-KO hiPSC-RPE. Disappearance of Kir7.1 is confirmed in Z-stack images (E, G). Scale bar: 100 µm (A, B); 25 µm (D, E).
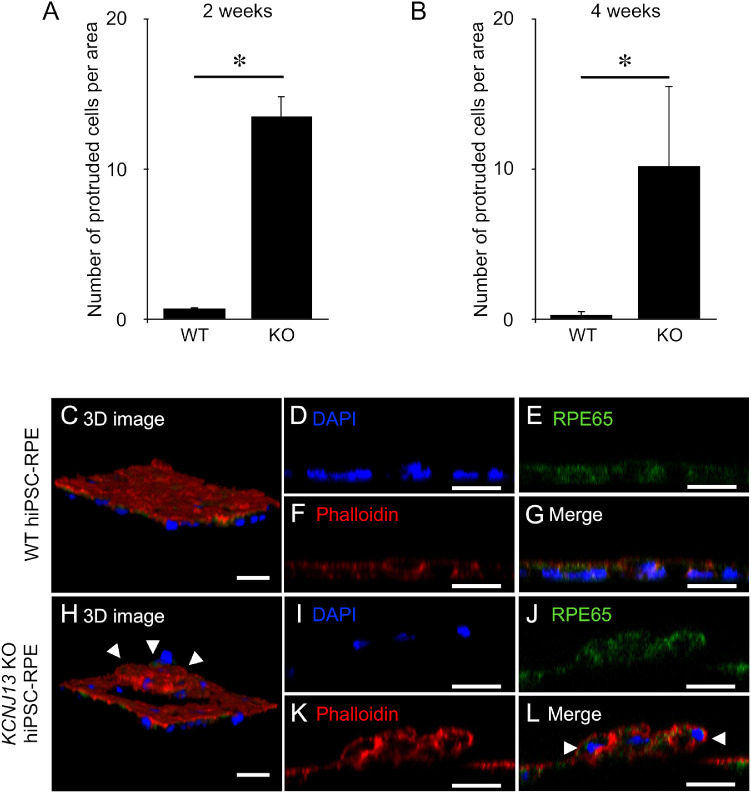
In culturing KCNJ13-KO hiPSC-RPE cells at 2 and 4 weeks after seeding, we found that there were more protruded cells from the bottom of the culture dish compared with WT hiPSC-RPE cells. We counted the number of protruded cells per area and found that there was a significant increase in protruded cells in KCNJ13-KO hiPSC-RPE cells (at 2 weeks, WT: 0.76 ± 0.12, KO: 13.56 ± 1.30, P < 0.001, n = 5; at 4 weeks, WT: 0.32 ± 0.11, KO: 10.24 ± 2.36, P < 0.001, n = 5) (Figs. 3A, 3B). We observed cultured WT and KCNJ13-KO hiPSC-RPE cells by confocal laser microscopy to determine the structure of the cells. We used rhodamine–phalloidin staining to visualize actin filaments and immunostaining with anti-RPE65 antibodies to identify the cytoplasm of RPE cells. The WT hiPSC-RPE cells formed a single epithelial cell layer (Figs. 3C–3G). By contrast, in KCNJ13-KO hiPSC-RPE cells, a subpopulation of cells formed a round elevated layer from a surrounding RPE monolayer (Figs. 3H–3L). Because the protruded cell group was immunoreactive for RPE65 (Figs. 3J, 3L), we concluded that they were differentiated RPE cells. To further see the structure of protruded RPE cells, we observed cultured hiPSC-RPE cells of WT and KCNJ13-KO by transmission electron microscopy. As previously reported,22 the WT hiPSC-RPE cells had elongated apical microvilli, apically distributed melanosomes, and basal membrane infoldings (Supplementary Fig. S4A and not shown). In contrast, although similar microvilli, melanosomes, and basal structures were observed in the single-layered KCNJ13-KO hiPSC-RPE cells (Supplementary Fig. S4B), there were some portions of double-layered cells and cell debris clumping laterally and beneath the RPE cells (Supplementary Fig. S4C).
Figure 3.
Protruded RPE cells were observed in the KCNJ13-KO hiPSC-RPE. (A, B) The number of protruded cells per area is shown (WT and KO, n = 5 for each). Triple staining was performed with anti-RPE65, rhodamine–phalloidin, and DAPI. (C, H) The three-dimensional structures of these cells were analyzed using confocal microscopy. (C–G) Z-stack images of the WT hiPSC-RPE monolayer. (H–L) Z-stack images of the KCNJ13-KO hiPSC-RPE layer, showing protruded RPE cells (arrowheads in H, L) above the basal RPE layer. Data are shown as mean ± SE. *P < 0.05 (Student's t-test). Scale bar: 25 µm (C–L).
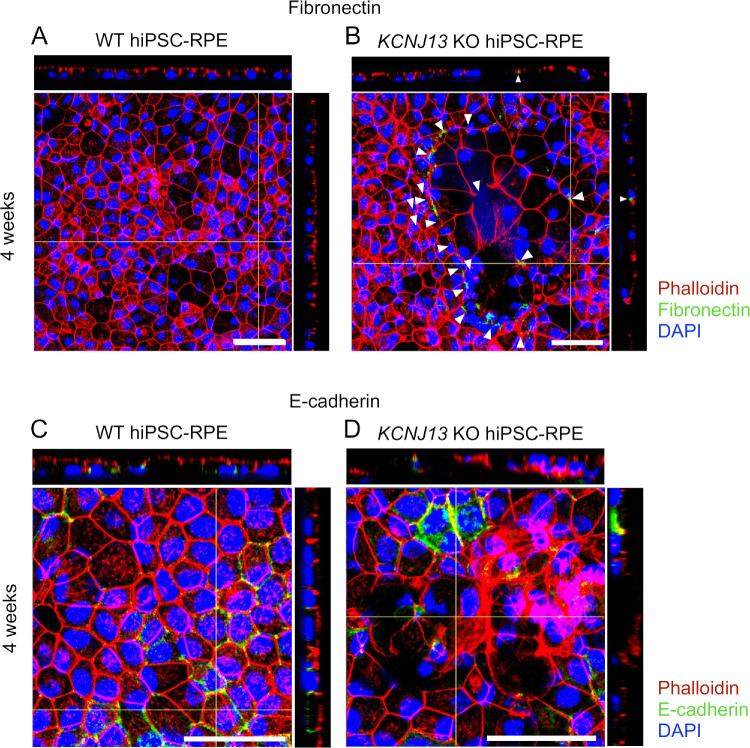
To determine whether the epithelial-to-mesenchymal transition (EMT) is involved in protrusion of RPE cells in the KCNJ13-KO cell populations, we examined expression of EMT markers such as epithelial E-cadherin and mesenchymal fibronectin. Immunofluorescent histochemistry showed that, in the WT hiPSC-RPE, expression of fibronectin was not detected, whereas it was focally detected in the protruded RPE cells of the KCNJ13-KO (Fig. 4A arrowheads in Fig. 4B, Supplementary Fig. S5). In contrast, expression of E-cadherin was detected in the WT hiPSC-RPE, but it was focally lost in the protruded RPE cells of the KCNJ13-KO (Figs. 4C, 4D, Supplementary Fig. S6).
Figure 4.
Expression of EMT markers in WT and KCNJ13-KO hiPSC-RPE. (A) Expression of fibronectin in WT hiPSC-RPE cells at 4 weeks after seeding. (B) Expression of fibronectin in KCNJ13-KO hiPSC-RPE cells at 4 weeks after seeding. (C) Expression of E-cadherin in WT hiPSC-RPE cells at 4 weeks after seeding. (D) Expression of E-cadherin in KCNJ13-KO hiPSC-RPE cells at 4 weeks after seeding. Scale bar: 50 µm (A–D).
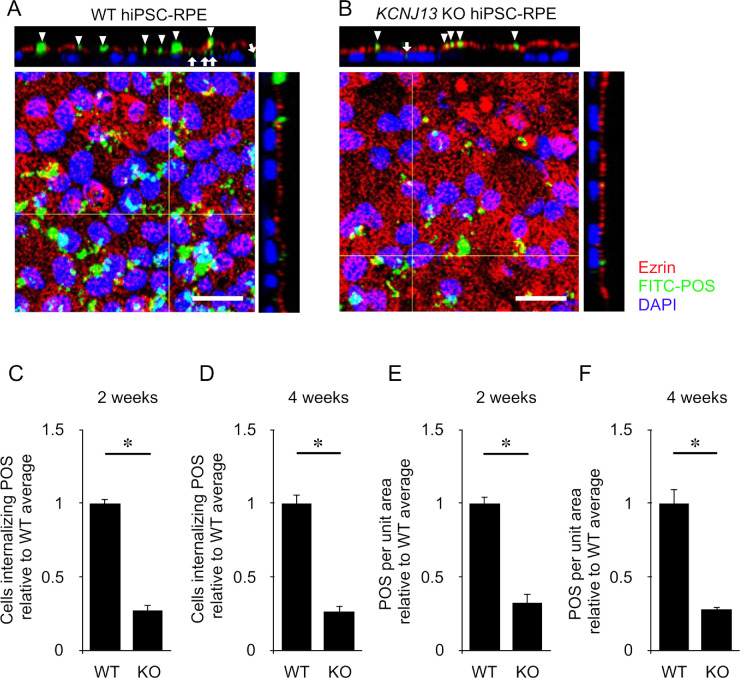
It was reported that phagocytosis of POSs is affected in LCA16 patient-derived hiPSC-RPE cells.22 Therefore, we examined the phagocytic activity of KCNJ13-KO hiPSC-RPE cells and compared it to that of WT (Figs. 5A, 5B). At 2 and 4 weeks after seeding, a phagocytosis assay using fluorescently (FITC)-labeled porcine POSs prepared in-house was performed on WT and KCNJ13-KO (clone no. 1) hiPSC-RPE cells. We counted the number of FITC-labeled POSs per cell and found it significantly decreased in KCNJ13-KO cells (2 weeks, WT: 1.0 ± 0.026, KO: 0.27 ± 0.043, P < 0.001, n = 4; 4 weeks, WT: 1.0 ± 0.043, KO: 0.32 ± 0.057, P < 0.001, n = 4) (Figs. 5C, 5D). The number of internalized POSs per area was also significantly reduced in the KCNJ13-KO cells (2 weeks, WT: 1.0 ± 0.059, KO: 0.26 ± 0.039, P < 0.001, n = 4; 4 weeks, WT: 1.0 ± 0.097, KO: 0.28 ± 0.014, P = 0.005, n = 4) (Figs. 5E, 5F).
Figure 5.
Phagocytic activity of WT and KCNJ13-KO hiPSC-RPE cells. (A) Confocal microscope image of WT hiPSC-RPE 4 weeks after seeding. Z-stack image shows that FITC-POSs (green, arrowheads) are internalized (white arrow) below ezrin (red). (B) Confocal microscope image of KCNJ13-KO hiPSC-RPE 4 weeks after seeding. Z-stack image shows that FITC-POSs (green, arrowheads) are less taken up (white arrow) into cells below ezrin than WT hiPSC-RPE. (C, D) The ratio of cells phagocytosing FITC-POSs relative to WT cells in 2 (C) or 4 (D) weeks (WT and KO, n = 4 each). (E, F) The number of FITC-POSs per area relative to WT cells in 2 (E) or 4 (F) weeks (WT and KO, n = 4 each). Data are shown as mean ± SE. *P < 0.05 (Student's t-test). Scale bar: 20 µm.
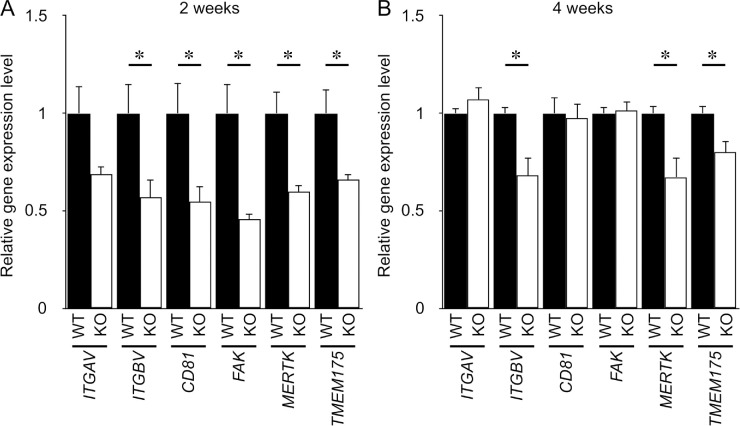
To ensure that phagocytic activity was decreased in KCNJ13-KO hiPSC-RPE cells, we examined the expression level of phagocytosis-related genes to determine the molecular mechanisms underlying the defect (Fig. 6). We examined the expression of integrin αv (ITGAV), integrin β5 (ITGBV), CD81, FAK, and MerTK, known to be involved in the phagocytosis of POSs.26 We also focused on TMEM175, which is an intracellular lysosomal potassium channel regulating lysosomal function.27 qPCR analysis showed that, at 2 weeks after seeding, the relative expression levels of integrin β5, CD81, FAK, MerTK, and TMEM175 were significantly reduced in KCNJ13-KO hiPSC-RPE cells compared with WT hiPSC-RPE cells (Fig. 6A). At 4 weeks after seeding, expression of integrin β5, MerTK, and TMEM175 was significantly decreased in KCNJ13-KO hiPSC-RPE cells compared with WT hiPSC-RPE cells (Fig. 6B). These data indicate that, in the absence of Kir7.1, the phagocytic activity of RPE is disrupted in line with downregulation of these phagocytosis-related genes and a lysosomal gene.
Figure 6.
KCNJ13 deletion decreases expression of genes required for phagocytosis of POSs in hiPSC-RPE. Relative expression levels of mRNA for phagocytosis-related genes at 2 weeks (A) and 4 weeks (B) after seeding. 18s rRNA was used as an internal control. The mean expression levels of each gene in WT hiPSC-RPE were set as 1. Data are shown as mean ± SE. *P < 0.05 (Student's t-test). Each P value is shown as follows: At 2 weeks, ITGAV, P = 0.090; ITGBV, P = 0.038; CD81, P = 0.030; FAK, P = 0.021; MerTK, P = 0.019; and TMEM175, P = 0.047. At 4 weeks, ITGAV, P = 0.334; ITGBV, P = 0.024; CD81, P = 0.817; FAK, P = 0.866; MerTK, P = 0.036; and TMEM175, P = 0.021.
Discussion
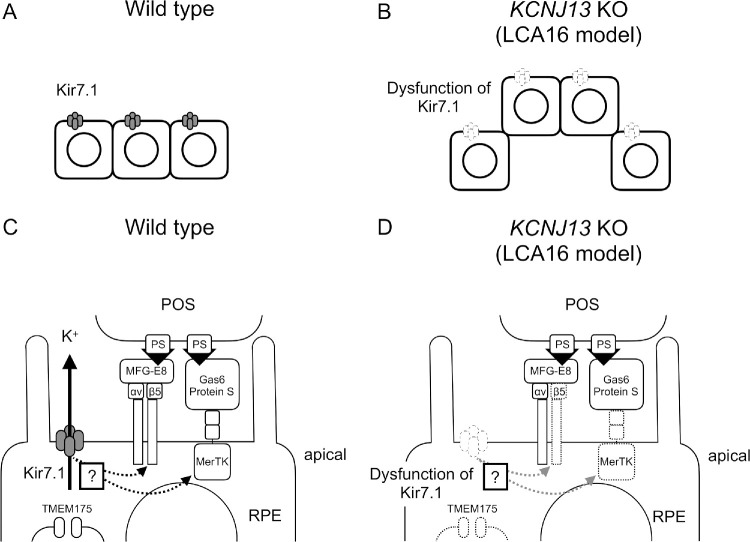
The mechanisms that underlie the symptoms of LCA16 are mostly unknown, and the pathophysiology of structural abnormalities surrounding the affected RPE cells are still puzzling. Here, we have established a disease model of RPE cells for LCA16, derived from human iPSCs by use of a gene editing technique that deletes most of KCNJ13. In this LCA16 model system, the KCNJ13-KO hiPSC-RPE has defects in its own cell alignment and exhibits reduced phagocytic activity of POSs along with decreased expression of phagocytosis-related genes. Until now, there have been no reports showing a direct relationship among the gene function of KCNJ13, the structure of RPE, and the expression of phagocytosis-related genes. This study suggests that structural abnormalities in the microenvironment of RPE cells in LCA16 are attributable to defects in cell alignment of the RPE itself and to insufficient clearance of POSs by the affected RPE (Fig. 7).
Figure 7.
Schematic diagram showing possible mechanisms for POS uptake in normal and LCA16 RPE cells. (A) Normal RPE cells exhibit a monolayer alignment. (B) KCNJ13-KO RPE cells are abnormally aligned and partially protruded from surrounding RPE cells. (C) Normal RPE cells can regulate K+ efflux and phagocytose aged POSs. (D) Uptake of POS is impaired in KCNJ13-KO RPE cells, and POS accumulates in the subretinal space.
Given that disease model animals do not always exhibit phenotypes identical to those found in humans, basic studies using human materials derived from patients have tremendous significance for elucidating the pathophysiological mechanisms underlying diseases. However, as LCA16 is so rare, it is not easy to collect ocular tissues from patients for research. In this study, we succeeded in establishing an LCA16 model of human RPE cells by deleting most of the KCNJ13 gene utilizing the CRISPR/Cas9 system. We employed a modified Hotta's method18 and caused a 4.3-kb deletion in the target gene, KCNJ13, using two gRNAs with the intent to avoid mutation skipping by aberrant splicing. By electroporating the Cas9 protein–gRNA complex into hiPSCs, we obtained cells with intended mutations in both alleles at a rate of 1 per 41 lines (2.4%) and without off-target mutations in the edited cells. This validates that the technique can be applied to gene editing in hiPSCs to generate rare, ophthalmic, genetic disease models precisely and safely.
There are case studies on LCA16,3–5 reporting nonsense or missense mutations in the KCNJ13 gene, in which retinal structures and visual functions were variably disrupted. Despite the wide range of homozygous mutations and compound heterozygosity, the mutant proteins are truncated at the C-terminal intracellular domains, which are critical for Kir channel subunit assembly.28 In these patients, shared fundus findings include nummular pigmentation or mottling of the RPE over the macular region. A spectral domain ocular coherence tomography (OCT) scan has revealed reflective dots at the RPE and sub-RPE focal deposits in addition to loss of outer retinal structures indicating photoreceptor degeneration.3–5 In LCA16 model mice, such as a CRISPR-engineered mosaic mutant and conditional knock-out for Kcnj13, it has been reported that loss of Kir7.1 in the RPE induces photoreceptor degeneration and impairs visual functions.9,13 This demonstrates that mutations in KCNJ13 lead to abnormalities in structures and functions of the RPE and photoreceptors. Additionally, we have shown that KCNJ13-KO hiPSC-RPE exhibits aberrant cell alignment.
Our study clearly shows that what are commonly thought to be “deposits” above the RPE layer as revealed by OCT are misaligned, elevated layers or cell clumps of KCNJ13-deficient RPE cells. Abutting the protruded cells, expression of fibronectin was detected. Because expression of RPE65 and pigment granules was observed even in these protruded cells (Fig. 3J, Supplementary Fig. S4C), it can be concluded that undifferentiated cells or complete mesenchymal cells do not constitute the protruded cell populations among the KCNJ13-KO hiPSC-RPE cells. Rather, it is most likely that partial or incomplete EMT occurs in the protruded KCNJ13-KO hiPSC-RPE cells. This might be related to impaired cell alignment found in the KCNJ13-KO hiPSC-RPE cells and in those of LCA16 patients. This study has also shown that the loss of KCNJ13 in human RPE cells impairs their own phagocytic activity. In LCA16, reduced phagocytosis of RPE likely results in insufficient clearance of POSs and eventual functional disruption and degeneration of outer retinal structures (Figs. 7A–7D).
Many molecules are known to regulate the phagocytic activity of RPE cells: αvβ5 integrins and MerTK membrane receptors are involved in the phagocytosis of POSs by RPE cells.26 In integrin β5-deficient RPE cells, the phagocytic activity is impaired, and age-related accumulation of inclusion bodies was observed.29 In recognizing POSs, MerTK binds to phosphatidylserine via Gas6 and protein S.30 The causal gene in the retinal degeneration model rat (RCS) is MerTK, and phagocytosis of POSs does not occur in RCS rats. RPE cells from mutants in the Gas6 or protein S gene are incapable of engulfment of POSs, resulting in progressive retinal degeneration.30–33 Furthermore, in humans, mutations in the MerTK gene have been found in patients with retinitis pigmentosa.34–36 This study shows that integrin β5 and MerTK are significantly reduced in RPE cells lacking the Kir7.1 potassium channel. Thus, defects in binding and incorporation of POSs would underlie impaired phagocytosis in RPE cells.
TMEM175 is the lysosomal potassium channel,27 and its deficiency is known to impair lysosomal function and increase protein deposits in the cell.37 This study shows that TMEM175 mRNA was decreased in the KCNJ13-KO hiPSC-RPE cells compared with WT hiPSC-RPE cells (Fig. 6). Therefore, it is tempting to speculate that a similar mechanism (that is, reduced autophagosome clearance) might underlie the decreased phagocytosis of POSs in the absence of KCNJ13. The precise molecular link among the Kir7.1 potassium channel, integrin β5, MerTK, and TMEM175 requires further study.
In summary, the loss of KCNJ13 decreases the phagocytic activity of RPE cells, and the resultant defects in renewal of POSs would be involved in degeneration of the outer retina in patients with LCA16. Therefore, as a novel therapeutic intervention for LCA16, reactivation of the phagocytic activity of impaired RPE cells might be promising.
Supplementary Material
Acknowledgments
The authors thank Kumiko Kikuchi and Shiori Ikeda for technical laboratory assistance and Megumi Tsukano (Central Research Laboratory, Okayama University Medical School) for electron microscopic analysis.
Supported by grants from the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (15K10475 18K09410, and 19K18878).
Disclosure: Y. Kanzaki, None; H. Fujita, None; K. Sato, None; M. Hosokawa, None; H. Matsumae, None; F. Shiraga, None; Y. Morizane, None; H. Ohuchi, None
References
- 1. Young RW. The daily rhythm of shedding and degradation of rod and cone outer segment membranes in the chick retina. Invest Ophthalmol Vis Sci. 1978; 17: 105–116. [PubMed] [Google Scholar]
- 2. Feng W, Yasumura D, Matthes MT, LaVail MM, Vollrath D. Mertk triggers uptake of photoreceptor outer segments during phagocytosis by cultured retinal pigment epithelial cells. J Biol Chem. 2002; 277: 17016–17022. [DOI] [PubMed] [Google Scholar]
- 3. Sergouniotis PI, Davidson AE, Mackay DS, et al.. Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause Leber congenital amaurosis. Am J Hum Genet. 2011; 89: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pattnaik BR, Shahi PK, Marino MJ, et al.. A novel KCNJ13 nonsense mutation and loss of Kir7.1 channel function causes Leber congenital amaurosis (LCA16). Hum Mutat. 2015; 36: 720–727. [DOI] [PubMed] [Google Scholar]
- 5. Perez-Roustit S, Marquette V, Bocquet B, et al.. LEBER congenital amaurosis with large retinal pigment clumps caused by compound heterozygous mutations in KCNJ13. Retin Cases Brief Rep. 2017; 11: 221–226. [DOI] [PubMed] [Google Scholar]
- 6. Kumar M, Pattnaik BR. Focus on Kir7.1: physiology and channelopathy. Channels (Austin). 2014; 8: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang D, Zhang X, Hughes BA. Expression of inwardly rectifying potassium channel subunits in native human retinal pigment epithelium. Exp Eye Res. 2008; 87: 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shahi PK, Liu X, Aul B, et al.. Abnormal electroretinogram after Kir7.1 channel suppression suggests role in retinal electrophysiology. Sci Rep. 2017; 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhong H, Chen Y, Li Y, Chen R, Mardon G. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep. 2015; 5: 8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang D, Pan A, Swaminathan A, Kumar G, Hughes BA. Expression and localization of the inwardly rectifying potassium channel Kir7.1 in native bovine retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2003; 44: 3178–3185. [DOI] [PubMed] [Google Scholar]
- 11. Yasuda K, Shimura M, Nakazawa T, et al.. Expression and functional properties of unique inward rectifier K+ channel Kir7.1 in the porcine iris and retinal pigment epithelium. Curr Eye Res. 2003; 27: 279–287. [DOI] [PubMed] [Google Scholar]
- 12. Kusaka S, Inanobe A, Fujita A, et al.. Functional Kir7.1 channels localized at the root of apical processes in rat retinal pigment epithelium. J Physiol. 2001; 531: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roman D, Zhong H, Yaklichkin S, Chen R, Mardon G. Conditional loss of Kcnj13 in the retinal pigment epithelium causes photoreceptor degeneration. Exp Eye Res. 2018; 176: 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kosmidou C, Efstathiou NE, Hoang MV, et al.. Issues with the specificity of immunological reagents for NLRP3: implications for age-related macular degeneration. Sci Rep. 2018; 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamao H, Mandai M, Okamoto S, et al.. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports. 2014; 2: 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015; 31: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park C-Y, Sung JJ, Choi S-H, Lee DR, Park I-H, Kim D-W. Modeling and correction of structural variations in patient-derived iPSCs using CRISPR/Cas9. Nat Protoc. 2016; 11: 2154–2169. [DOI] [PubMed] [Google Scholar]
- 18. Li HL, Fujimoto N, Sasakawa N, et al.. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015; 4: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matoba R, Morizane Y, Shiode Y, et al.. Suppressive effect of AMP-activated protein kinase on the epithelial-mesenchymal transition in retinal pigment epithelial cells. PLoS One. 2017; 12: e0181481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiode Y, Morizane Y, Matoba R, et al.. The role of inverted internal limiting membrane flap in macular hole closure. Invest Ophthalmol Vis Sci. 2017; 58: 4847–4855. [DOI] [PubMed] [Google Scholar]
- 21. Morizane Y, Thanos A, Takeuchi K, et al.. AMP-activated protein kinase suppresses matrix metalloproteinase-9 expression in mouse embryonic fibroblasts. J Biol Chem. 2011; 286: 16030–16038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shahi PK, Hermans D, Sinha D, et al.. Gene augmentation and readthrough rescue channelopathy in an iPSC-RPE model of congenital blindness. Am J Hum Genet. 2019; 104: 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parinot C, Rieu Q, Chatagnon J, Finnemann SC, Nandrot EF. Large-scale purification of porcine or bovine photoreceptor outer segments for phagocytosis assays on retinal pigment epithelial cells. J Vis Exp. 2014; 94: 52100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davis RJ, Alam NM, Zhao C, et al.. The developmental stage of adult human stem cell-derived retinal pigment epithelium Cells influences transplant efficacy for vision rescue. Stem Cell Reports. 2017; 9: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujita H, Ochi M, Ono M, et al.. Glutathione accelerates osteoclast differentiation and inflammatory bone destruction. Free Radic Res. 2019; 53: 226–236. [DOI] [PubMed] [Google Scholar]
- 26. Mazzoni F, Safa H, Finnemann SC. Understanding photoreceptor outer segment phagocytosis: use and utility of RPE cells in culture. Exp Eye Res. 2014; 126: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cang C, Aranda K, Seo Y, Gasnier B, Ren D. TMEM175 is an organelle K(+) channel regulating lysosomal function. Cell. 2015; 162: 1101–1112. [DOI] [PubMed] [Google Scholar]
- 28. Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev. 2010; 90: 291–366. [DOI] [PubMed] [Google Scholar]
- 29. Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J Exp Med. 2004; 200: 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lew ED, Oh J, Burrola PG, et al.. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. Elife. 2014; 3: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burstyn-Cohen T, Lew ED, Través PG, Burrola PG, Hash JC, Lemke G. Genetic dissection of TAM receptor-ligand interaction in retinal pigment epithelial cell phagocytosis. Neuron. 2012; 76: 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bok D, Hall MO. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J Cell Biol. 1971; 49: 664–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duncan JL, LaVail MM, Yasumura D, et al.. An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest Ophthalmol Vis Sci. 2003; 44: 826–838. [DOI] [PubMed] [Google Scholar]
- 34. Ostergaard E, Duno M, Batbayli M, Vilhelmsen K, Rosenberg T. A novel MERTK deletion is a common founder mutation in the faroe islands and is responsible for a high proportion of retinitis pigmentosa cases. Mol Vis. 2011; 17: 1485–1492. [PMC free article] [PubMed] [Google Scholar]
- 35. Mackay DS, Henderson RH, Sergouniotis PI, et al.. Novel mutations in MERTK associated with childhood onset rodcone dystrophy. Mol Vis. 2010; 16: 369–377. [PMC free article] [PubMed] [Google Scholar]
- 36. Shahzadi A, Riazuddin SA, Ali S, et al.. Nonsense mutation in MERTK causes autosomal recessive retinitis pigmentosa in a consanguineous Pakistani family. Br J Ophthalmol. 2010; 94: 1094–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jinn S, Drolet RE, Cramer PE, et al.. TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc Natl Acad Sci USA. 2017; 114: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.