Abstract
Coronavirus disease 2019 (COVID-19) is a rapidly emerging disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The disease begins as an infection of lungs, which is self-limiting in the majority of infections; however, some develop severe respiratory distress and organ failures. Lung microbiome, though neglected previously have received interest recently because of its association with several respiratory diseases and immunity. Lung microbiome can modify the risk and consequences of COVID-19 disease by activating an innate and adaptive immune response. In this review, we examine the current evidence on COVID-19 disease and lung microbiome, and how lung microbiome can affect SARS-CoV-2 infection and the outcomes of this disease. To date there is no direct evidence from human or animal studies on the role of lung microbiome in modifying COVID-19 disease; however, related studies support that microbiome can play an essential role in developing immunity against viral infections. Future studies need to be undertaken to find the relationship between lung microbiome and COVID-19 disease.
Keywords: Coronavirus disease (COVID-19), Immunity, Infection, Lung microbiome, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
Abbreviations: ACE2, Angiotensin converting enzyme; ARDS, Acute respiratory distress syndrome; ARF, Acute respiratory failure; COPD, Chronic obstructive pulmonary disease; COVID-19, Coronavirus disease 2019; CRP, C reactive protein; IFN, Interferon; PAMP, Pathogen associated molecular patterns; PRR, Pattern recognition receptor; rRNA, Ribosomal RNA; RSV, Respiratory syncytial virus; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; TLR, Toll like receptor; TNF, Tumour necrosis factor
1. Introduction
The rapid outbreak of the coronavirus disease (COVID-19) has crippled global health and economy [1]. As of 21st July 2020, it has affected 188 countries and infected 14,727,753 people causing death of 610,560 patients [2]. The rapid increase in the number of severely ill patients after COVID-19 infection across the globe has resulted in heavy investment in health care infrastructures and medical supplies. To stop the spread of the virus, many countries have imposed a strict international travel ban and put the population into home restriction [3]. The COVID-19 disease is caused by a virus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which emerged in Wuhan, China at the end of 2019 [4]. The disease is highly transmissible among persons through aerosols, and evidence shows the virus can remain viable for up to 3 days on plastic and stainless steel, all of which facilitates the easy propagation of this virus [5].
The usual symptoms of COVID-19 include fever, dry cough, and tiredness. Some patients may have aches and pains, nasal congestion, anosmia, sore throat or diarrhea [6]. A large majority of patients may not develop any symptoms after the viral infection, thereby acting as a hidden carrier for this devastating disease [7]. The common complications of COVID-19 that lead to death include acute respiratory failure (ARF), pneumonia, acute respiratory distress syndrome (ARDS), acute liver injury, acute cardiac injury, acute kidney injury, septic shock, disseminated intravascular coagulation, blood clots and rhabdomyolysis [8], [9], [10], [11]. The development of pneumonia or acute respiratory illness is consistently observed after COVID-19 infection with the majority of the patient showing lesions involving bilateral lungs, which is often the common cause of COVID-19 death [10], [12], [13].
Microbiome refers to the collective genomes of all the microorganisms. It includes every organism (not only bacteria, but also archaea, fungi and viruses), DNA and RNA species, and proteins that can affect health and development of diseases [14]. Microbiota refers to the community of microorganisms, including bacteria, viruses, fungi, and protozoans, that live in a host body [15]. The COVID-19 disease begins with the invasion of lungs by SARS-CoV-2 virus, and the major complications that develop subsequently are related to lung infection and immune response generation, therefore, lung microbiome might play an important role from initiation to the progression of this disease [16].
This review focuses on the potential relationship between COVID-19 disease and lung microbiome. We summarized the most important findings on lung microbiome and its role in immunity and lung disease including COVID-19. We searched for the keywords (SARS-CoV-2) OR (coronavirus disease 2019) OR (COVID-19) AND (microbiome) in PubMed and Scopus for the articles until 20 July 2020. All the original research articles reporting microbiome in COVID-19 were included. We highlight the role of the lung microbiome, and how the microbiome can modulate SARS-CoV-2 attack along with the consequences of viral attack. Finally, we seek the potential role of lung microbiome and immunity in severity of COVID-19 disease. We also discuss the relevant studies supporting the role of microbiome against viral infection and disease complications.
2. Lung microbiome in health and disease
2.1. Microbiome in healthy lung
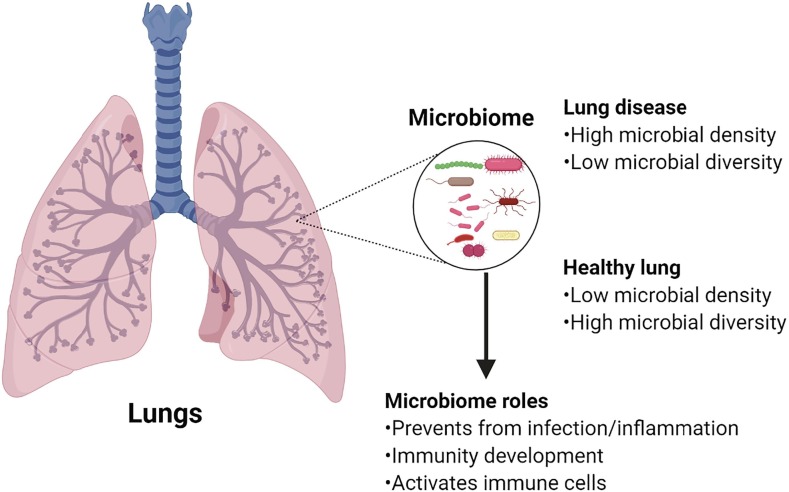
Until very recently, the lung was considered to be a sterile site free from bacteria [17]. Being an environment with favourable temperature, moisture and mucus, and a large surface area that is in regular contact with the external environment, the lung is indeed a microbiome rich site [18]. Lung microbiome is more dynamic and transient than that of the gastrointestinal tract because of bidirectional movement of air and mucus [19], [20]. In a healthy human, the lung microbiota has a low density but harbours a prominent diversity of interacting microbiota. At the phylum level, Firmicutes, Bacteroidetes and Proteobacteria are the most common phyla. At the genus level, Prevotella, Veillonella and Streptococcus are the predominant microorganisms [17], [21]. The composition of lung microbiome is mainly determined by microbial immigration, elimination, and relative growth rates of its members. In lung diseases, these factors can change, and therefore overgrowth of one species with reduction of microbial diversity occurs (shown in Fig. 1 ) [22].
Fig. 1.
Lung microbiome in healthy and diseased lungs and its role.
2.2. Microbiome in lung diseases
During lung pathology, drastic changes in the local environment occur, creating a favourable environment for a specific microbial growth. In one of the common lung disease, asthma, bronchoscopy samples have shown an increased abundance of Haemophilus, Neisseria, Fusobacterium, and Porphyromonas [23], [24]. Recent evidence shows microbiota administration through the oral or intranasal route as probiotics can lower allergic inflammation seen in asthma, highlighting the intricate relationship between microbiota and asthma [25]. In another common lung disease, chronic obstructive pulmonary disease (COPD), Lactobacillus, Fusobacteria, Leptotrichia and Fusobacterium were observed in abundant numbers [26], [27]. The most abundant bacteria seen in cystic fibrosis were Pseudomonas, Staphylococcus, Stenotrophomonas, Achromobacter, and Streptococcus [28], [29]. Recent evidence shows that lung microbiome is altered in lung cancer, and the microbiome can influence cancer development and progression [30], [31].
Emerging evidence shows the relationship between lung microbiota and susceptibility to pneumonia [32]. Pneumonia is a widespread lung disease characterised by infection of lung alveoli leading to an infiltration of inflammatory cells and fluids into the alveoli. This disease can be caused by bacteria, virus, fungi or protozoans [33]. Streptococcus pneumoniae and Haemophilus influenzae type b are the most common bacterial causative agents, whereas, respiratory syncytial virus (RSV) is the most common viral agent for pneumonia [34], [35]. The endotracheal aspirates of patients with ventilator associated pneumonia had a higher bacterial load but a lower abundance of bacterial class, Bacilli, than matched controls [36]. Similarly, the microbiota profile of endotracheal aspirates was different between the mechanically ventilated pneumonia group and the non-pneumonia group. In the intubated patients with pneumonia, Pseudomonas, Corynebacterium, and Rothia were more abundant, while Streptococcus and Prevotella were less abundant as compared to those without pneumonia [37]. The abundance of Firmicutes phyla was reduced in the patients with interstitial pneumonia including the overall phyla richness as compared to healthy group. The abundance of Streptococcus was significantly reduced in interstitial pneumonia patients, while Prevotella and Veillonella were enriched in comparison to healthy volunteers [38]. In addition, the microbial diversity and composition tended to differ according to the causative agent of pneumonia, which suggests that lung microbiome can vary according to the type of pneumonia [39]. Overall, pneumonia is characterized by a shift in lung microbiome, with domination by pneumonia etiologic agent, high microbial biomass, and lowering of microbial diversity [33].
The composition and diversity of upper respiratory tract and lung microbiome have been found to shift in response to respiratory viral infections in humans and animals [40], [41], [42]. Some of the common viruses invading respiratory tract and lungs include rhinovirus, influenza virus, adenovirus, parainfluenza virus and RSV. These respiratory viral infections are quite common during childhood, and they often increase susceptibility to secondary bacterial infection of lungs [41]. The upper respiratory tract microbiome of rhinovirus and RSV infected infants was found to be largely dominated by Moraxella, Streptococcus, Corynebacterium, Haemophilus, and Dolosigranulum [40]. In another study, rhinovirus infection was found to increase the relative abundance of Haemophilus and Neisseria, both of which are associated with secondary lung infections [43]. In mice, influenza virus infection of lungs caused a shift in the composition and diversity of microbial community. The viral infection changed the dominant bacterial class from Alphaproteobacteria to Gammaproteobacteria and Actinobacteria, and this was followed by an increase in the relative abundance of Streptococcus and Staphylococcus [44]. In summary, the lung microbiome is highly dynamic, and infections (bacterial, viral) can lead to rapid alterations in the lung microbiome, and the microbial communities can play a critical role in those infections (shown in Fig. 1).
3. Lung immunity
The alveolar surface is the largest surface area in direct contact to the external environment, so it is constantly exposed to invading microorganisms. Therefore, it harbours several lines of defence against potential infections. A continuous layer of pulmonary epithelial cells constitutes a physical and biological barrier for inhaled substances and pathogens. The mucus coating of the pulmonary epithelium, proteolytic enzymes, defence proteins (immunoglobulins, lactoferrin, defensins) and lysozymes present on the surface and in the fluids around alveoli protect against infection [18]. Pulmonary epithelial cells are in close contact with cells of the immune system, and they also secrete a wide range of cytokines and chemokines. They express cell surface receptors called pattern-recognition receptors (PRRs), such as the toll-like receptors (TLRs), which enable them to recognize pathogen-associated molecular patterns (PAMPs) from viruses, bacteria, fungi, protozoa, and multicellular parasites [45].
Apart from the above mentioned immune mechanisms, lung contains immune cells such as dendritic cells, macrophages (alveolar and interstitial), lymphocytes (T-cells, γδ T cells, NK cells, B-cells), innate lymphoid cells and neutrophils that support innate and adaptive immune mechanism [46]. All these immune cells are involved in eliminating antigens and pathogens from the respiratory tract through phagocytosis or activation of effectors molecules that remove antigens. Regulatory T cells resident within the lungs are vital for the maintenance of immune tolerance to airborne particles, and they are directly influenced by local microbiota [45]. Another type of T cells, called lung resident memory γδ T cells provide a rapid immune response at barrier surfaces to recall antigens previously encountered through lung mucosa, which is apart from its role in cytokine and interferon production to stop viral and bacterial infection [47]. Despite the existing immunity mechanisms, lung infection is unavoidable. In the next section, we will discuss how SARS-CoV-2 initiates lung infection and escape the immune response.
4. SARS-CoV-2 infection and virus immuno-evasion mechanism
In the respiratory tract, SARS-CoV-2 bind to the specific host receptors called angiotensin-converting enzyme 2 (ACE2). This virus has an envelope-anchored spike protein that facilitates virus entry into host cells by first binding to ACE2. The virus then enters the host cell through endocytosis [48]. Inside the cells, the viral genome replicates and makes necessary proteins, which assemble to form new virions. The newly released virions can invade other lung cells [4], [49]. The stress of viral production will lead to cell death, which in turn will trigger a series of immune responses and initiate inflammation reflected as lymphopenia, elevated C reactive protein (CRP) and erythrocyte sedimentation rate [13].
The viral RNAs and PAMPs are detected by the PRRs, such as TLRs inside the cells [50]. The recognition will lead to the activation of the transcription factor nuclear factor-κB (NF-κB) and interferon regulatory factor 3 (IRF3) to induce proinflammatory cytokines such as interferon-α and tumour necrosis factor-beta (TNF-β). The production of type I interferons (IFN) will cause suppression of viral multiplication and prevent the severity of disease [51]. Increase in interleukin-6 (IL-6), CRP, and chemokines such as interferon-γ-inducible protein 10 (IP-10), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1-alpha (MIP-1A), and tumour necrosis factor-alpha (TNF-α) were observed in COVID-19 patients [10], [52].
If the body immune response cannot stop the SARS-CoV-2 multiplication, then a massive degree of lung injury occurs. This degree of damage can generate a substantial immune response from immune effector cells which is characterized by a large amount of cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, and TNF-α) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, and CXCL10) in the body [53]. This condition of hyperinflammation manifested as ‘cytokine storm’ is called as COVID-19 ARDS. This is one of the most dangerous and potentially life-threatening events related to COVID-19 disease [54]. Besides, SARS-CoV-2 infection damages the pulmonary lining altering the composition of normal microbiota, and can promote the growth of specific bacteria to initiate secondary pneumonia [41].
SARS-CoV-2 deploys several evasion mechanisms for defeating host cells. One of the ways is the formation of double-membrane vesicles that lack PRRs, where the virus replicates, leading to failure of recognition by immune cells and therefore avoid host detection [53]. Another feature of this virus, eight surface proteins, can block the action of interferon I, causing the cells to fail to stop viral multiplication [55]. The virus has also developed a strategy to stop host cell recognition by mimicking host cell capping machinery [51]. The SARS-CoV-2 viral infection occurs amid the local environment of diverse microbiota; therefore, it is apparent that lung microbiota can have an impact on the initiation, development, and progression of the COVID-19 disease.
5. Lung microbiome and COVID-19 disease
5.1. Roles of the lung microbiome
Lungs are at the frontline of immunity as they are constantly exposed to a wide variety of external environment. The microbiome has a principal role in shaping pulmonary immunity, and healthy lung has a vast array of microbiota. Microbes boost innate and adaptative immunity (site-specific in lungs and systemic), release factors which assist respiratory functions, and prevent an invasion of lungs from pathogens [56]. The normal microbiota restricts the growth of harmful pathogens that may make their way into the lungs. This growth restriction may occur by several mechanisms, such as, limiting nutrients access and secreting growth inhibitors [57].
Activation of lung immune cells that initiates innate and adaptive immunity requires microbe exposure [58]. The function of one of the vital effector and regulator cells of the lung, γδ T cells, depends on pathogen invasion [47], [59]. Evidence shows that exposure to certain bacterial strains in neonate protects against excessive airway inflammation through the alteration of immune cells [60]. Furthermore, microbial exposure is required for developing innate immunity, as children exposed to the microbial rich environment had a lower rate of asthma and allergic sensitization, and stronger immunity [61]. One of the products of lung’s normal flora showed an allergo-protective effect in the animal model of airway inflammation [62]. In summary, previous studies highlight that lung immunity development requires exposure to diverse microbes (shown in Fig. 1).
5.2. Lung microbiome in COVID-19 disease
To date, only very few studies have explored the microbiome of COVID-19 patients. Emerging evidence from these studies shows that microbial communities in the gut and lung are altered in COVID-19 patients, and the alterations may have a critical impact on the immunity and severity of COVID-19 disease [42], [63], [64], [65], [66]. Though primarily a respiratory virus, the SARS-CoV-2 virus has also been found in the faecal samples from COVID-19 patients. The gut microbiome of COVID-19 patients had a significant reduction of bacterial diversity, and a significantly higher relative abundance of opportunistic pathogens such as Streptococcus, Rothia, Veillonella, and Actinomyces, and a lower abundance of beneficial symbionts as compared to control group. Thus, it appears that gut microbial communities interact with SARS-CoV-2 virus [42]. In another pilot study of 15 COVID-19 patients, the gut microbiome was characterized by enrichment of opportunistic pathogens (Clostridium hathewayi, Actinomyces viscosus and Bacteroides nordii) and depletion of beneficial commensals (Eubacterium ventriosum, Faecalibacterium prausnitzii, Roseburia, Lachnospiraceae taxa) [66]. Moreover, this study showed the severity of COVID-19 disease is directly correlated with the predominance of opportunistic pathogens (Clostridium ramosum and Clostridium hathewayi) and inversely with the predominance of beneficial commensals (Alistipes onderdonkii and Bacteroides ovatus) [66]. Similarly, COVID-19 patients had increased proportions of opportunistic fungal pathogens, Candida albicans, Candida auris, and Aspergillus flavus as compared to controls [65]. In summary, the role of gut microbiome against SARS-CoV-2 infection and disease severity is rapidly emerging, and therefore gut microbiota may be developed as therapeutics for the management of COVID-19 disease in future.
Till date, only two studies have analysed the lung microbiome of COVID-19 patients [63], [64]. In one of the study, bronchoalveolar lavage fluid from COVID-19 patients (n = 8), community-acquired pneumonia patients (n = 25) and healthy controls (n = 20) was investigated, and a significant difference in microbiota composition was observed [64]. Both COVID-19 and community-acquired pneumonia patients had enrichment of pathogenic and commensal bacteria, indicating a degree of microbial dysbiosis in both diseased states [64]. In the other study, the lung post-mortem biopsies from 20 deceased COVID-19 patients were used to study lung microbiome. The most common bacterial genera were Acinetobacter, Chryseobacterium, Burkholderia, Brevundimonas, Sphingobium, and Enterobacteriaceae. The most common fungal genera were Cutaneotrichosporon, followed by Issatchenkia, Wallemia, Cladosporium, Alternaria, Dipodascus, Mortierella, Aspergillus, Naganishia, Diutina, and Candida [63]. Therefore, a mix of bacterial and fungal infection was seen in the COVID-19 patients of this study. Overall, there is very little information about the composition of lung microbiome in COVID-19 patient. Further studies with bigger sample size are required to know more precisely the composition and the role of lung microbiome in the severity of SARS-CoV-2 infections.
5.3. Role of the lung microbiome in COVID-19 disease and COVID-19 ARDS
Although there is no direct evidence if lung microbiome can affect potential SARS-CoV-2 infections and the outcomes of this disease, yet several lines of evidence support that bacteria in lungs have important roles in this disease. Firstly, microbiota dwelling on the respiratory surface can acts as a barrier, thereby preventing viral attachment to host cells. Secondly, microbiota prime the lung immunity, which will fight against viral infection, and exposure to a diverse range of microbiota may build a wider immunity. Such phenomenon is particularly common in the gut, where gut microbiota can protect against potential flu viral infections [67]. In mice, the nasal application of a respiratory normal flora, Corynebacterium pseudodiphtheriticum, increased the TLR3 antiviral response against RSV and enhanced the production of TNFα, IL-6, IFNγ, and IFNβ [68]. Another respiratory tract flora, Staphylococcus aureus, protected against influenza induced lung injury [69]. Recent evidence of the association between COVID-19 disease severity with the gut microbiota seems highly promising [66]. Thus, it appears that lung microbiome may modify propensity to SARS-CoV viral infection.
Till date, it is unknown if lung microbiota can modify the risk of developing severe respiratory complications such as ARDS after SARS-CoV-2 infection. The dysbiosis of lung microbiome may contribute to COVID-19 ARDS because microbial dysbiosis is found to provoke a dysregulated immune response leading to inflammation [22], [54], [70]. In the ARDS unrelated to COVID-19 disease, lung microbiome is found to be enriched with gut-associated bacteria, which demonstrates the relationship between lung microbiome and ARDS [71]. Recently, the altered microbiome (increase in bacterial burden and a decrease in alpha diversity) was shown to be associated with inflammation and mortality in ARDS patients. The change in microbiota population (a reduction in Betaproteobacteria and an increase in Staphylococcus, Streptococcus and Enterobacteriaceae) was significantly associated with serum cytokine IL-6 [72]. Lung microbiome seems to have a critical role in severely ill patients because a recent study found that lung microbiota predicts the clinical outcome and death in those patients [73]. This study showed that increased lung bacterial burden and lung enrichment with gut-associated bacteria were predictive of adverse consequences of ARDS. Thus, in COVID-19 ARDS, the microbiome may have a significant role in determining the severity of the disease, and the outcomes of ARDS. In the future, studies will find if the presence of a specific bacterial population is associated with low risk for SARS-CoV-2 infection and ARDS development.
6. Immunity, COVID-19, and future implications
Based on the current evidence, it appears that immunity is at the forefront for protection against COVID-19 disease. Immunity development is a continuous process and requires exposure to a range of microbiota and environments [74]. Exposure of human population to diverse air microbes that may occur in unhygienic environment is believed to develop more robust immunity, and this may partly account for the variation in the COVID-19 death rate among countries [75]. Several other factors such as the method of delivery at birth, air microbiota of the immediate environment, prenatal and childhood exposures to microbes, respiratory viral illness, indoor and outdoor pollution can have a critical influence in the microbiome composition and immunity development in children [45]. Exposure to air with high air microbial load is found to reduce asthma risk in children [76]. In many developed countries, over the years, there has been a change in diet, sanitary conditions, antibiotics use, exposure to environmental chemicals and the change in the living environment, all of which may have ultimately reduced the exposure to diverse microbes. This factor is believed to be one of the principal factors for the current rise in the number of autoimmune lung diseases, and weak immunity [32], [74].
Till date little is known about the role of lung microbiome in COVID-19 disease. Future studies are highly warranted to solve the complex relationship between lung immunity, lung microbiome and SARS-CoV-2 infection along with this disease complications. Because of the therapeutic benefits of microbiota as probiotics, future studies should also focus on the potential benefits and applicability of microbiota transplantation for the COVID-19 disease. We also emphasize collecting bronchoalveolar samples from the COVID-19 infected patients and perform 16S rRNA gene sequencing in a large sample size.
7. Conclusions
We conclude that lung microbiome can have a profound impact on the susceptibility to SARS-CoV-2 infection and severity of the disease. Moreover, lung microbiome has important contribution for the generation of immune responses against viral attack, and microbiome may affect the outcome of COVID-19 ARDS. Future studies should investigate lung microbiome and immune response in a large cohort of COVID-19 patients, and correlate with the severity of this disease. Future studies should also assess if probiotics treatment can improve COVID-19 disease.
8. Ethics approval and consent to participate
Not required
9. Consent for publication
Not applicable
10. Availability of data and material
Not applicable
Funding
None
Author contributions
Both SK and AS involved in conceptualization, literature review, writing the original draft and review of the draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The figure was created using BioRender.com.
References
- 1.WHO. Coronavirus disease (COVID-2019) situation reports; 2020 [Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
- 2.CSSE JHU. Wuhan coronavirus (2019-nCoV) global cases; 2020.
- 3.Nicola M., Alsafi Z., Sohrabi C., Kerwan A., Al-Jabir A., Iosifidis C., et al. The socio-economic implications of the coronavirus and COVID-19 Pandemic: a review. Int J Surg. 2020 doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Q&A on coronaviruses (COVID-19); 2020 [Available from: https://www.who.int/news-room/q-a-detail/q-a-coronaviruses#:~:text=symptoms.
- 7.Day M. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ. 2020;369 doi: 10.1136/bmj.m1375. [DOI] [PubMed] [Google Scholar]
- 8.Practice B.B. Coronavirus disease 2019 (COVID-19) Complications. 2020 [Google Scholar]
- 9.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368 doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siordia J.A. Epidemiology and clinical features of COVID-19: A review of current literature. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis; 2019. [DOI] [PMC free article] [PubMed]
- 13.Zhu J, Ji P, Pang J, Zhong Z, Li H, He C, et al. Clinical characteristics of 3,062 COVID-19 patients: a meta-analysis. J Med Virol. n/a(n/a). [DOI] [PMC free article] [PubMed]
- 14.Moffatt M.F., Cookson W.O. The lung microbiome in health and disease. Clin Med. 2017;17(6):525–529. doi: 10.7861/clinmedicine.17-6-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumas A., Bernard L., Poquet Y., Lugo-Villarino G., Neyrolles O. The role of the lung microbiota and the gut–lung axis in respiratory infectious diseases. Cell Microbiol. 2018;20(12) doi: 10.1111/cmi.12966. [DOI] [PubMed] [Google Scholar]
- 16.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol 2020;215:108427-. [DOI] [PMC free article] [PubMed]
- 17.Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Invernizzi R., Lloyd C.M., Molyneaux P.L. Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology. 2020;n/a(n/a). doi: 10.1111/imm.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10(2):299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson R.P., Huffnagle G.B. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11(7) doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Beck J.M., Huffnagle G.B., et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson R.P., Martinez F.J., Huffnagle G.B. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384(9944):691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durack J., Lynch S.V., Nariya S., Bhakta N.R., Beigelman A., Castro M., et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140(1):63–75. doi: 10.1016/j.jaci.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilty M., Burke C., Pedro H., Cardenas P., Bush A., Bossley C., et al. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spacova I, Ceuppens JL, Seys SF, Petrova MI, Lebeer S. Probiotics against airway allergy: host factors to consider. Dis Model Mech 2018;11(7):dmm034314. [DOI] [PMC free article] [PubMed]
- 26.Pragman A.A., Kim H.B., Reilly C.S., Wendt C., Isaacson R.E. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sze M.A., Dimitriu P.A., Hayashi S., Elliott W.M., McDonough J.E., Gosselink J.V., et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185(10):1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feigelman R., Kahlert C.R., Baty F., Rassouli F., Kleiner R.L., Kohler P., et al. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome. 2017;5(1):20. doi: 10.1186/s40168-017-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frayman K.B., Armstrong D.S., Carzino R., Ferkol T.W., Grimwood K., Storch G.A., et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax. 2017;72(12):1104–1112. doi: 10.1136/thoraxjnl-2016-209279. [DOI] [PubMed] [Google Scholar]
- 30.Greathouse K.L., White J.R., Vargas A.J., Bliskovsky V.V., Beck J.A., von Muhlinen N., et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19(1):123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S.H., Sung J.Y., Yong D., Chun J., Kim S.Y., Song J.H., et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Wu B.G., Segal L.N. The lung microbiome and its role in pneumonia. Clin Chest Med. 2018;39(4):677–689. doi: 10.1016/j.ccm.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinton L.J., Walkey A.J., Mizgerd J.P. Integrative physiology of pneumonia. Physiol Rev. 2018;98(3):1417–1464. doi: 10.1152/physrev.00032.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO. Pneumonia 2019 [Available from: https://www.who.int/news-room/fact-sheets/detail/pneumonia.
- 35.Gadsby N.J., Russell C.D., McHugh M.P., Mark H., Conway Morris A., Laurenson I.F., et al. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis. 2016;62(7):817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emonet S., Lazarevic V., Leemann Refondini C., Gaïa N., Leo S., Girard M., et al. Identification of respiratory microbiota markers in ventilator-associated pneumonia. Intensive Care Med. 2019;45(8):1082–1092. doi: 10.1007/s00134-019-05660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo S., Park S.Y., Kim Y., Jeon J.P., Lee J.J., Hong J.Y. The dynamics of respiratory microbiota during mechanical ventilation in patients with pneumonia. J Clin Med. 2020;9(3) doi: 10.3390/jcm9030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vázquez-Pérez J.A., Carrillo C.O., Iñiguez-García M.A., Romero-Espinoza I., Márquez-García J.E., Falcón L.I., et al. Alveolar microbiota profile in patients with human pulmonary tuberculosis and interstitial pneumonia. Microb Pathog. 2020;139 doi: 10.1016/j.micpath.2019.103851. [DOI] [PubMed] [Google Scholar]
- 39.Wang H., Dai W., Qiu C., Li S., Wang W., Xu J., et al. Mycoplasma pneumoniae and Streptococcus pneumoniae caused different microbial structure and correlation network in lung microbiota. J Thorac Dis. 2016;8(6):1316–1322. doi: 10.21037/jtd.2016.04.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosas-Salazar C., Shilts M.H., Tovchigrechko A., Schobel S., Chappell J.D., Larkin E.K., et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214(12):1924–1928. doi: 10.1093/infdis/jiw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanada S, Pirzadeh M, Carver KY, Deng JC. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front Immunol 2018;9:2640-. [DOI] [PMC free article] [PubMed]
- 42.Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., et al. Alterations of the Gut microbiota in patients with coronavirus disease 2019 or H1N1 Influenza. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofstra JJ, Matamoros S, van de Pol MA, de Wever B, Tanck MW, Wendt-Knol H, et al. Changes in microbiota during experimental human Rhinovirus infection. BMC Infect Dis 2015;15:336-. [DOI] [PMC free article] [PubMed]
- 44.Gu L., Deng H., Ren Z., Zhao Y., Yu S., Guo Y., et al. Dynamic Changes in the Microbiome and Mucosal Immune Microenvironment of the Lower Respiratory Tract by Influenza Virus Infection. Front Microbiol. 2019;10:2491. doi: 10.3389/fmicb.2019.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lloyd C.M., Marsland B.J. Lung homeostasis: influence of age, microbes, and the immune system. Immunity. 2017;46(4):549–561. doi: 10.1016/j.immuni.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Nicod L.P. Lung defences: an overview. Eur Respir Rev. 2005;14(95):45–50. [Google Scholar]
- 47.Cheng M., Hu S. Lung-resident gammadelta T cells and their roles in lung diseases. Immunology. 2017;151(4):375–384. doi: 10.1111/imm.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boopathi S., Poma A.B., Novel K.P. coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2019;2020:1–10. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Astuti I., Ysrafil. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndrome: Clin Res Rev. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The Cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2(3):264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramírez-Labrada A.G., Isla D., Artal A., Arias M., Rezusta A., Pardo J., et al. The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy. Trends Cancer. 2020;6(2):86–97. doi: 10.1016/j.trecan.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Martin T.R., Frevert C.W. Innate immunity in the lungs. Proc Am Thorac Soc. 2005;2(5):403–411. doi: 10.1513/pats.200508-090JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sommariva M., Le Noci V., Bianchi F., Camelliti S., Balsari A., Tagliabue E., et al. The lung microbiota: role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell Mol Life Sci. 2020 doi: 10.1007/s00018-020-03452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nanno M., Shiohara T., Yamamoto H., Kawakami K., Ishikawa H. gammadelta T cells: firefighters or fire boosters in the front lines of inflammatory responses. Immunol Rev. 2007;215:103–113. doi: 10.1111/j.1600-065X.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- 60.Gollwitzer E.S., Saglani S., Trompette A., Yadava K., Sherburn R., McCoy K.D., et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20(6):642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- 61.Stein M.M., Hrusch C.L., Gozdz J., Igartua C., Pivniouk V., Murray S.E., et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N Engl J Med. 2016;375(5):411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagner S., Harb H., Zhao M., Stein K., Holst O., Ege M.J., et al. Farm-derived Gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy. 2013;68(3):322–329. doi: 10.1111/all.12094. [DOI] [PubMed] [Google Scholar]
- 63.Fan J., Li X., Gao Y., Zhou J., Wang S., Huang B., et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. J Infect. 2020;S0163–4453(20):30429–30431. doi: 10.1016/j.jinf.2020.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuo T., Zhan H., Zhang F., Liu Q., Tso E.Y.K., Lui G.C.Y., et al. Alterations in Fecal Fungal Microbiome of Patients With COVID-19 During Time of Hospitalization until Discharge. Gastroenterology. 2020;S0016–5085(20):34852–34856. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., et al. Alterations in Gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, et al. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep 2019;28(1):245–56.e4. [DOI] [PubMed]
- 68.Kanmani P., Clua P., Vizoso-Pinto M.G., Rodriguez C., Alvarez S., Melnikov V., et al. Respiratory commensal bacteria corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and streptococcus pneumoniae superinfection. Front Microbiol. 2017;8:1613. doi: 10.3389/fmicb.2017.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Li F., Sun R., Gao X., Wei H., Li L.J., et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang D., Xing Y., Song X., Qian Y. The impact of lung microbiota dysbiosis on inflammation. Immunology. 2020;159(2):156–166. doi: 10.1111/imm.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dickson R.P., Singer B.H., Newstead M.W., Falkowski N.R., Erb-Downward J.R., Standiford T.J., et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1(10):16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kyo M., Nishioka K., Nakaya T., Kida Y., Tanabe Y., Ohshimo S., et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir Res. 2019;20(1):246. doi: 10.1186/s12931-019-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickson R.P., Schultz M.J., van der Poll T., Schouten L.R., Falkowski N.R., Luth J.E., et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med. 2020;201(5):555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strachan D.P. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ege M.J., Mayer M., Normand A.-C., Genuneit J., Cookson W.O.C.M., Braun-Fahrländer C., et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable