Abstract
The emergence of COVID-19 has triggered many works aiming at identifying the animal intermediate potentially involved in the transmission of SARS-CoV-2 to humans. The presence of SARS-CoV-2-related viruses in Malayan pangolins, in silico analysis of the ACE2 receptor polymorphism and sequence similarities between the Receptor Binding Domain (RBD) of the spike proteins of pangolin and human Sarbecoviruses led to the proposal of pangolin as intermediary. However, the binding affinity of the pangolin ACE2 receptor for SARS-CoV-2 RBD was later on reported to be low. Here, we provide evidence that the pangolin is not the intermediate animal at the origin of the human pandemic. Moreover, data available do not fit with the spillover model currently proposed for zoonotic emergence which is thus unlikely to account for this outbreak. We propose a different model to explain how SARS-CoV-2 related coronaviruses could have circulated in different species, including humans, before the emergence of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, Pangolin, Zoonosis, Zoonotic model
1. Introduction
Since the earliest times, it has been in the human nature to look for a culprit (human or animal) during epidemics, leading often to irrational behaviors aimed at eliminating the threat. With the emergence of COVID-19, the designated culprits were bats and pangolins. However, what was their actual role in the process leading to the current pandemic?
2. Main
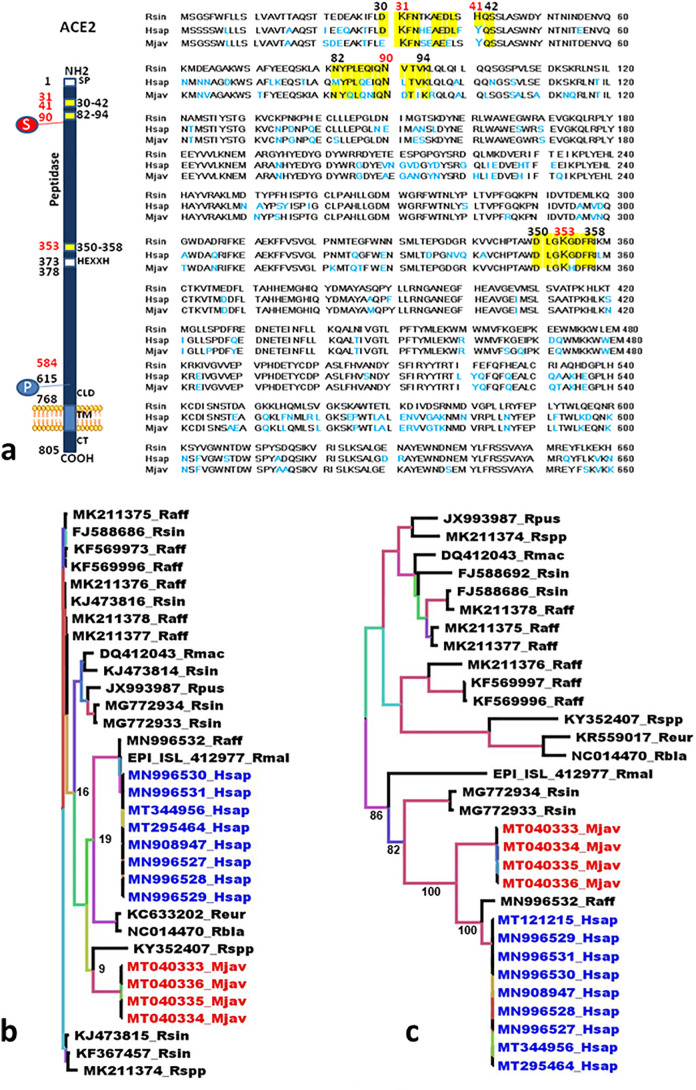
SARS-CoV-2 which recently affected the human population worldwide is suspected to have originated from bats. It was found to be closely related to the Sarbecoviruses MN996532_raTG13 and RmYN02 from the Chinese horseshoe bats Rhinolophus affinis and Rhinolophus malayanus, respectively (Zhou et al., 2020a; Zhou et al., 2020b) There is no evidence of direct transmission of Sarbecoviruses from bats to humans yet (Afelt et al., 2018a). The only direct Sarbecovirus infections of humans have been linked to laboratory accidents during the SARS epidemic (Heymann et al., 2004; Della-Porta, 2008; Normile, 2004a, Normile, 2004b, Normile, 2004c; Normile and Vogel, 2003; Senio, 2003; Watts, 2004: WHO, 2004; Webster, 2020). To date, no higher incidence of Sarbecovirus infections has been reported in anthropized ecosystems even among employees of guano farms who come into direct contact with bat feces (guano) even though about 22% of bats from guano farms release coronaviruses in their feces, among which almost 5% are Sarbecoviruses (Berto et al., 2017; Afelt et al., 2018b). Assuming a lack of direct bat-to-human Sarbecovirus transmission and the need of a reservoir according to the spillover theory of zoonotic emergence, many teams have attempted to identify the animal serving as an intermediate. The Malayan or Javan pangolin (Manis javanica) was suspected of being this intermediate host based on, 1) its ACE2 receptor sequence, 2) the presence of Sarbecoviruses related to SARS-CoV-2 (Liu et al., 2019; Liu et al., 2020a; Andersen et al., 2020; Han, 2020) in animals smuggled from the Indomalayian region, and 3) the presence of pangolins in wet markets in China where they are considered a delicacy and a component of traditional pharmacopeia. Several studies, based on metagenomics, have concluded on a diversity of the pangolin's virome, the close relationship of the ACE2-binding region of the viral spike named “Receptor Binding Domain” (RBD) to that of SARS-CoV-2 and the in silico model prediction of recombinants between pangolins and R. affinis bats Sarbecoviruses (Liu et al., 2019; Lam et al., 2020; Zhang et al., 2020; Lau et al., 2020; Wahba et al., 2020; Wong et al., 2020).. However, this is purely an in silico modeling and bats and pangolins from which Sarbecoviruses have been described are found in very distant regions and display very different biology and ecology, making such a recombination event very questionable. Furthermore, a recent in depth genomic analysis demonstrated that there was no recombination in SARS-CoV-2 and that it has circulating at least in bats for several decades (Boni et al., 2020). Furthermore, the drawback with metagenomic analyses is that there is no evidence that the different parts come from the same virus and the observed recombinants might result from artifactual assembly mosaics. The SARS-CoV-2 S1 spike protein binds the human ACE2 cell surface molecules (Yan et al., 2020; Zhao et al., 2020a, Zhao et al., 2020b; Wang and Cheng, 2020). The analysis of 3-D structures of ACE2 indicated that the amino acids found in the regions 30–41, 82–93 and 353–358 play a major role in interactions with the viral S1 spike (Wrapp et al., 2020; Hoffmann et al., 2020). This opened the way for a whole series of in silico analyses of ACE2 polymorphism intended to predict which putative intermediate host animal might bear the ACE2 receptor best suited to capture a SARS-CoV-2-like Sarbecovirus transmissible to humans (Liu et al., 2020a; Shi et al., 2020; Lambert et al., 2020; Qiu et al., 2020; Luan et al., 2020; Zhao et al., 2020a, Zhao et al., 2020b; Stawiski et al., 2020; Liu et al., 2020b; Damas et al., 2020). Besides pangolin, SARS-CoV-2 was also reported to bind to ACE2 from Chinese horseshoe bats, civet, cat, turtle, ferret, monkey, dog, Chinese hamster, buffalo, cow, sheep, swine and even pigeon (Liu et al., 2020a; Shi et al., 2020; Lambert et al., 2020; Qiu et al., 2020; Luan et al., 2020; Zhao et al., 2020a, Zhao et al., 2020b; Stawiski et al., 2020; Liu et al., 2020b; Damas et al., 2020; Devaux et al., 2020). Nevertheless, the focus was put only on pangolin since it hosts Sarbecoviruses and is sold in wet markets. Furthermore, the binding affinity of the pangolin ACE2 receptor for SARS-Cov-2 RBD was reported to be low (Damas et al., 2020). The ACE2 receptor from Rhinolophus sinicus (Chinese horseshoe bat) is characterized by the SARS-CoV-2 favorable amino acids K31, N82, N90, K353 but lacks the Y amino acid residue at position 41 (Y41H), while Manis javanica (Sunda pangolin) expresses the favorable amino acids K31, Y41, N82, N90, K353 and Homo sapiens (human) expresses K31, Y41, N90, K353 and lacks the N82 residue (N82M) (Fig. 1a). These amino acid substitutions in ACE2 do not impact the global 3-D structure of the molecule even though they can alter the electrostatic potential surface of the molecule thereby affecting the affinity of the viral spike for ACE2 (Devaux et al., 2020). The ACE2 receptors from three species of pangolin, i.e. Manis pentadactyla, M. Janavica and Phataginus tricuspis, display a binding score for SARS-CoV-2 ranging from low to very low (Andersen et al., 2020). The RBD is the most variable region in the spike protein (Andersen et al., 2020). It is not a sequence isolated from the rest of the spike protein and its coding sequence is not independent from the rest of the genome. The active conformation of the RBD largely depends on the specific conformation of the spike protein and interactions with other amino acids within the protein. In silico structural modeling of a short sequence cannot provide evidence of full similarity, only hints. Furthermore, the RBD is under positive selection, i.e. host driven (Zhang et al., 2007; Tang et al., 2009) and converging adaptive evolution might thus occur resulting in a similar structure to recognize a similar active site of a receptor. A virus identical or highly similar to SARS-CoV-2 must be isolated from an animal species in order to formally identify this intermediary, which is not the case with pangolins' Sarbecoviruses. The RdRp (polymerase) sequences from Sarbecoviruses of R. affinis (MN996532_raTG13) and R. malayanus (RmYN02_EPI_ISL_412977), both from Yunnan, China, are closer to SARS-CoV-2 than the RdRp from pangolins' coronaviruses (Fig. 1b). The RdRp from SARS-CoV-2 is also closer to the RdRp from Sarbecoviruses isolated from Rhinolophus euryale (KC633202) and Rhinolophus blasii (NC014470) from Bulgaria than to the RdRp from pangolin. Conversely, the Sarbecovirus from pangolin is, according to the RdRp sequence, closer to a Rhinolophus bat from Kenya (KY352407) (Fig. 1b). Furthermore, SARS-CoV-2 falls into a separate branch than Sarbecoviruses from pangolin (Fig. 1b). This indicates that SARS-CoV-2-like Sarbecoviruses circulate within the genus Rhinolophus worldwide. The S gene coding for the spike protein, provides a similar feature although less sequences are available. SARS-COV-2 and pangolins' Sarbecoviruses belong to different clusters separated by high bootstraps (100), the spike gene from R. affinis Sarbecovirus (MN996532) being closer to SARS-CoV-2 (Fig. 1c). The conclusion coming from these data is, in agreement with a previous study (Liu et al., 2020c), that the pangolin is not the intermediate species in the transmission of SARS-CoV-2 from bats to humans. It appears to only be a parallel host infected by a Sarbecovirus very closely related to SARS-CoV-2 with whom it shares a common ancestor. Furthermore, the COVID-19 epidemic did not start in December 2019 in the Huanan Seafood Wholesale Market (HSWM) where pangolins were supposed to be sold but earlier and outside HSWM (Frutos et al., 2020). Another conclusion is that very closely related Sarbecoviruses circulate in different hosts worldwide raising the question of the dynamic of zoonotic emergence and the role played by wildlife. Two hypothetic models of zoonotic emergence can be envisioned, the well-known spillover model and an alternative model we propose to name the circulation model. The spillover model theorizes that zoonotic emergence starts as a consequence of a zoonotic pressure (Plowright et al., 2017). In this model, the virus is developing into an epizootic stage in an animal population, reaching the threshold needed for interspecies transmission and following contact with humans, develop within the human populations. Socio-economic factors and demographic dynamic trigger the epidemic or pandemic expansion of the disease (Plowright et al., 2017; Frutos et al., 2020). According to the spillover model, the disease already exists as an epizootic and, thus, identifying the animal reservoir is essential to stop viral spread. However, when considering the last three main coronavirus epidemics, i.e. SARS, MERS and COVID-19, several requirements for the spillover model are not fulfilled. First of all, no epizootic necessary to reach the level of zoonotic pressure required for the spillover was ever recorded for either SARS, MERS or COVID-19. Palm civets and dromedaries were identified as intermediate hosts carrying viruses similar to SARS and MERS, respectively (Song et al., 2005; Azhar et al., 2014; Briese et al., 2014). However, they were identified by sampling in the absence of epizootics. Infected masked palm civets were found in markets in Guangdong and Hong Kong (Song et al., 2005). Infected dromedaries were found in the Arabian Peninsula and in Africa (Briese et al., 2014: Müller et al., 2014). Noteworthy, camels used in the Arabian Peninsula are imported from Africa (Younan et al., 2016) and no MERS outbreak or related epizootic were ever reported in Africa. Until now, no animal intermediate was formally identified in the COVID-19 pandemic. Currently, only hypotheses exclusively based on in silico models and predictions have been put forward. Two closely related bat CoVs were identified but both from samples collected before the outbreak and outside Hubei, i.e. Yunnan (Zhou et al., 2020a; Zhou et al., 2o20b). The proposed circulation model for zoonotic emergence complies with the observations done until now. In this model there is no requirement for zoonotic pressure or epizootic episode prior to the emergence of a human disease. No epizootics were reported in civets or camels for SARS or MERS, respectively. Furthermore, there is no animal reservoirs displaying a disproportionate risk of triggering a zoonosis and potentially zoonotic viruses are evenly spread in all animal taxa (Mollentze and Streicker, 2020). Searching for a culprit in the wild is not realistic (Mollentze and Streicker, 2020). According to the circulation model there is a broad circulation of viruses in different species, including humans, upon contact but with no epidemic to follow. This fits with the observation that humans have been a lot more exposed to various viruses than expected and without any related epidemic (Pike et al., 2010). Out of 60 viruses involved in zoonoses, 59 are RNA viruses (Brook and Dobson, 2015). Several RNA viruses, among which the Coronaviruses, including SARS-CoV and SARS-CoV-2, were shown to undergo a quasispecies evolutionary process (Zhang et al., 2007; Tang et al., 2009; Plowright et al., 2017; Song et al., 2005). This process postulates that there is no specific preadaptation of the virus to the host but instead a post-exposure, host-driven selection of viruses displaying the best propensity to evade immune surveillance and replicate. Contact, low affinity receptor interaction and lack of molecular interference during replication are enough to establish productive infection after what the virus will follow in-host selection. This is compatible with the high diversity observed in the spike proteins of Coronaviruses (Andersen et al., 2020) which is under positive selection, i.e. host driven (Zhang e al, 2007; Tang et al., 2009; Briese et al., 2014; Tang et al., 2006; Xu et al., 2004). In line with this model, the RBD from SARS-CoV-2 is not fully optimized for human ACE2 (Andersen et al., 2020). According to the circulation model, what really prepares the ground for the epidemic is simply an accidental event, i.e. a mutation, recombination or reassortment in the virus genome. The virus is already present in an animal population close to humans or even in humans, and this mutation makes it more invasive and/or pathogenic. This was very recently reported in SARS-CoV-2 (Korber et al., 2020) and beyond coronaviruses has been observed in influenza, chikungunya or Zika viruses (Webster et al., 1982: Tsetsarkin and Weaver, 2011; Yuan et al., 2017).
Fig. 1.
Comparative analysis of ACE2 and Sarbecoviruses sequences.
1a. Comparative analysis of ACE2 sequences from bat, human and pangolin. Clustal Omega multiple sequence alignment (EMBL-EBI bioinformatic tool; Copyright © EMBL 2020), was used to compare the extracellular 1 to 660 amino acids portion of the ACE2 protein sequences of bat (Rsin, Rhinolophus sinicus; GenBank: AGZ48803.1), human (Hsap, Homo sapiens; GenBank: BAB40370.1), and pangolin (Mjav, Manis javanica; NCBI Reference Sequence: XP_017505752.1),. Some of the amino acids important for viral tropism are in red (previous studies showed that residues 31 K, 41Y, 90 N and 353 K are important for viral spike binding to human ACE2). Within the regions considered important for the interaction with the spike of SARS-CoV-2 (regions 30–42, 82–94 and 350–358, respectively), the conserved amino acids with respect to the bat ACE2 sequence are highlighted in yellow. Amino acids that differ from the bat ACE2 sequence are in light blue.
b. Phylogenetic analysis of the RdRp gene. The alignment of the full RdRp genes was performed with MUSCLE from the SeaView package (Gouy et al., 2010). The tree was built using the maximum likelihood method under the GTR model with 500 repeats. The tree was rooted using the RdRp sequence of a 229E-related bat coronavirus (KT253278) as outgroup. Blue: RdRp sequences from human SARS-CoV-2. Red. RdRp sequences from pangolins' Sarbecoviruses.
c. Phylogenetic analysis of the S (spike) gene. The alignment of the full S (spike) genes was performed with MUSCLE from the SeaView package (Gouy et al., 2010). The tree was rooted using the spike sequence of a Neoromicia capensis coronavirus (KC869678) as outgroup. The tree was built using the maximum likelihood method under the GTR model with 500 repeats. Blue: RdRp sequences from human SARS-CoV-2. Red. RdRp sequences from pangolins' Sarbecoviruses.
3. Conclusion
The real triggers for epidemic and pandemics are the societal organization and society-driven human/animal contacts and amplification loops provided by the modern human society, i.e. contacts, land conversion, markets, international trades, mobility, etc. (Pike et al., 2010; Frutos et al., 2020). A major positive effect of the circulation model is that the focus is put on these human activities and not on wildlife. We must reconsider the way we interact with Nature. Pangolins, bats and other animals are not responsible for the epidemics or pandemics affecting humans. Blaming wildlife for zoonotic emergence may result in useless and highly damaging culling, mass slaughter and loss of biodiversity.
Ethics declarations
No human samples or clinical data were used.
Authorship
All authors have read the manuscript and agreed on the submitted version. All authors participated to the writing of the article.
Funding
This study was supported by IHU Méditerranée Infection, University of Marseille, CNRS and CIRAD in France, by University of Barcelona in Spain and by Xiamen University in P.R. China. This study was partly supported by the Bill & Melinda Gates Foundation (INV-005834), the Science and Technology Program of Fujian Province (No: 2020Y0002), and the Xiamen New Coronavirus Prevention and Control Emergency Tackling Special Topic Program (No: 3502Z2020YJ03). The funders had no role in study design, data collection and analysis, decision.
Declaration of Competing Interest
None.
References
- Afelt A., Devaux C., Serra-Cobo J., Frutos R. Bats, bat-borne viruses and environnemental changes. In: Mikolla H., editor. Bats. IntechOpen; 2018. pp. 113–131. [Google Scholar]
- Afelt A., Frutos R., Devaux C. Coronaviruses, and deforestation: toward the emergence of novel infectious diseases? Front. Microbiol. 2018;9:702. doi: 10.3389/fmicb.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Berto A., Anh P.H., Carrique-Mas J.J., Simmonds P., Van Cuong N., Tue N.T. Detection of potentially novel paramyxovirus and coronavirus viral RNA in bats and rats in the Mekong Delta region of southern Viet Nam. Zoonoses Public Health. 2017;65:30–42. doi: 10.1111/zph.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.Y., Perry B., Castoe T., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Briese T., Mishra N., Jain K., Zalmout I.S., Jabado O.J., Karesh W.B. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio. 2014;5 doi: 10.1128/mBio.01146-14. (e01146–14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook C.E., Dobson A.P. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. bioRxiv. 2020 doi: 10.1101/2020.04.16.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Porta T. Laboratory accidents and breaches in biosafety–they do occur! Microbiol. Austr. 2008;29:62–65. [Google Scholar]
- Devaux C.A., Pinault L., Osman I.O., Raoult D. Can ACE2 receptor polymorphism predicts species susceptibility to SARS-CoV-2? Sci. Rep. 2020 doi: 10.21203/rs.3.rs-25753/v1. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutos R., Lopez Roig M., Serra-Cobo J., Devaux C.A. COVID-19: the conjunction of events leading to the coronavirus pandemic and lessons to learn for future threats. Front. Med. 2020;7:223. doi: 10.3389/fmed.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Han G.Z. Pangolins Harbor SARS-CoV-2-related coronaviruses. Trends Microbiol. 2020;28:515–517. doi: 10.1016/j.tim.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D.L., Aylward R.B., Wolff C. Dangerous pathogens in the laboratory: from smallpox to today’s SARS setbacks and tomorrow’s polio-free world. Lancet. 2004;363:1566–1568. doi: 10.1016/S0140-6736(04)16234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W., Gnanakaran S.G., Yoon H., Theiler J., Abfalterer W. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.29.069054. [DOI] [Google Scholar]
- Lam T.T.Y., Shum M.H.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. 2020 doi: 10.1101/2020.02.13.945485. No. 2020.02.13.945485. [DOI] [PubMed] [Google Scholar]
- Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I. Tumor necrosis factor- convertase ADAM17 mediates regulated Ectodomain shedding of the severe-acute respiratory syndrome-coronavirus SARS-CoV receptor, angiotensin-converting Enzyme-2 ACE2. J. Biol. Chem. 2020;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Luk H.K.H., Wong A.C.P., Li K.S.M., Zhu L., He Z. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Chen W., Chen J.P. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins Manis javanica. Viruses. 2019;11:979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H. Composition and divergence of coronavirus spike proteins and hostACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Zhao X., Ji F., Ren W. Functional and genetic analysis of viral receptor ACE2 Orthologs reveals broad potential host range of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.22.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jiang J.Z., Wan X.F., Hua Y., Li L., Zhou J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;56:165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollentze N., Streicker D.G. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl. Acad. Sci. 2020;117:9423–9430. doi: 10.1073/pnas.1919176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A. MERS coronavirus neutralizing antibodies in camels, eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014;20:2093. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Second lab accident fuels fears about SARS. Science. 2004;303:26. doi: 10.1126/science.303.5654.26. [DOI] [PubMed] [Google Scholar]
- Normile D. Mounting lab accidents raise SARS fears. Science. 2004;304:659–661. doi: 10.1126/science.304.5671.659. [DOI] [PubMed] [Google Scholar]
- Normile D. Lab accidents prompt calls for new containment program. Science. 2004;304:1223–1225. doi: 10.1126/science.304.5675.1223a. [DOI] [PubMed] [Google Scholar]
- Normile D., Vogel G. Early indications point to lab infection in new SARS case. Science. 2003;301:1642–1643. doi: 10.1126/science.301.5640.1642a. [DOI] [PubMed] [Google Scholar]
- Pike B.L., Saylors K.E., Fair J.N., LeBreton M., Tamoufe U., Djoko C.F. The origin and prevention of pandemics. Clin. Infect. Dis. 2010;50:1636–1640. doi: 10.1086/652860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., Lloyd-Smith J.O. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zhao Y.B., Wang Q., Li J.Y., Zhou Z.J., Liao C.H., Ge X.Y. Predicting the angiotensin converting enzyme 2 ACE2 utilizing capability as the receptor of SARS-CoV-2. Microb. Infect. Pre-proof. 2020 doi: 10.1016/j.micinf.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senio K. Recent Singapore SARS case a laboratory accident. Lancet Infect. Dis. 2003;3:679. doi: 10.1016/S1473-3099(03)00815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti F. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. bioRxiv. 2020 doi: 10.1101/2020.04.07.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Cheung J.L., Chu I.M., Sung J.J., Peiris M., Chan P.K. The large 386-nt deletion in SARS-associated coronavirus: evidence for quasispecies? J. Infect. Dis. 2006;194:808–813. doi: 10.1086/507044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Li G., Vasilakis N., Zhang Y., Shi Z., Zhong Y. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol. Biol. 2009;9:52. doi: 10.1186/1471-2148-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K.A., Weaver S.C. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Jain N., Fire A.Z., Shoura M.J., Artiles K.L., McCoy M.J., Jeong D.E. Identification of a pangolin niche for a 2019nCoV-like coronavirus through an extensive metagenomic search. bioRxiv. 2020 doi: 10.1101/2020.02.08.939660. No. 2020.02.08.939660. [DOI] [Google Scholar]
- Wang P.H., Cheng Y. Increasing host cellular receptor-angiotensin converting enzyme 2 ACE2 expression by coronavirus may facilitate 2019-nCoV infection. bioRxiv. 2020 doi: 10.1101/2020.02.24.963348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. SARS under control, but lab-safety questions remain. Lancet. 2004;363:1780. doi: 10.1016/S0140-6736(04)16344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G. Wet markets—a continuing source of severe acute respiratory syndrome and influenza? Lancet. 2020;363:234–236. doi: 10.1016/S0140-6736(03)15329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Laver W.G., Air G.M., Schild G.C. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296:115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- WHO 2004. https://www.who.int/csr/don/2004_04_30/en/ (accessed July 30, 2020)
- Wong M.C., Cregeen S.J.J., Ajami N.J., Petrosino J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. 2020 doi: 10.1101/2020.02.07.939207. [DOI] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang Z., Wang F.S. SARS-associated coronavirus quasispecies in individual patients. N. Engl. J. Med. 2004;350:1366–1367. doi: 10.1056/NEJMc032421. [DOI] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younan M., Bornstein S., Gluecks I.V. MERS and the dromedary camel trade between Africa and the Middle East. Trop. Anim. Health Prod. 2016;48:1277–1282. doi: 10.1007/s11250-016-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Huang X.Y., Liu Z.Y., Zhang F., Zhu X.L., Yu J.Y. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science. 2017;358:933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- Zhang X., Hasoksuz M., Spiro D., Halpin R., Wang S., Vlasova A. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology. 2007;363:1–10. doi: 10.1016/j.virol.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30 doi: 10.1016/j.cub.2020.03.063. (1346-1351.e2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen D., Szabla R., Zheng M., Li G., Du P. Broad and differential animal ACE2 receptor usage by SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.19.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020 doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]