Highlights
-
•
We propose a SAIU compartmental mathematical model that explains the transmission dynamics of COVID-19.
-
•
Perform local and global stability analysis for the infection free and endemic equilibrium point.
-
•
A sensitivity analysis is conducted to identify the most effective parameters with respect to basic reproduction number R0.
Keywords: COVID-19, Mathematical model, Reported and unreported cases, Sensitivity analysis, Isolation
Abstract
The ongoing COVID-19 has precipitated a major global crisis, with 968,117 total confirmed cases, 612,782 total recovered cases and 24,915 deaths in India as of July 15, 2020. In absence of any effective therapeutics or drugs and with an unknown epidemiological life cycle, predictive mathematical models can aid in understanding of both coronavirus disease control and management. In this study, we propose a compartmental mathematical model to predict and control the transmission dynamics of COVID-19 pandemic in India with epidemic data up to April 30, 2020. We compute the basic reproduction number R0, which will be used further to study the model simulations and predictions. We perform local and global stability analysis for the infection free equilibrium point E0 as well as an endemic equilibrium point E* with respect to the basic reproduction number R0. Moreover, we showed the criteria of disease persistence for R0 > 1. We conduct a sensitivity analysis in our coronavirus model to determine the relative importance of model parameters to disease transmission. We compute the sensitivity indices of the reproduction number R0 (which quantifies initial disease transmission) to the estimated parameter values. For the estimated model parameters, we obtained which shows the substantial outbreak of COVID-19 in India. Our model simulation demonstrates that the disease transmission rate βs is more effective to mitigate the basic reproduction number R0. Based on estimated data, our model predict that about 60 days the peak will be higher for COVID-19 in India and after that the curve will plateau but the coronavirus diseases will persist for a long time.
1. Introduction
The continuing novel coronavirus or SARS-CoV-2 epidemic has been declared a pandemic by the World Health Organization (WHO) on March 11, 2020 [1]. According to Worldometer data 3,401,394 total confirmed cases, 239,615 confirmed deaths and 1,083,816 recovered throughout the world as of May 02, 2020 [2]. Novel coronavirus has already exceeded the earlier records of two life-threatening outbreaks, namely Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), posing the major warning to the global public health and economy after the 2nd world war [3]. COVID-19 are a group of enveloped non-segmented with a positive-sense, single-stranded RNA viruses that belongs to the order of Nidovirales, family of Coronaviridae, and subfamily of Orthocoronavirinae and widely spread among the mammals and humans [4]. The novel coronavirus disease is an infectious disease caused by SARS-CoV-2.
A local epidemic of pneumonia was first identified in December 2019 in Wuhan city of Hubei province China, and the mainland China became the epicenter of COVID-19 [5]. Most of the initial stages were usually incorporated to the wholesale Huanan seafood market, which also traded live animals. This communicable coronavirus disease has traits like fever, dry cough, sore throat, breathlessness and fatigue [6]. With human migration through air, the disease has now spread throughout the world as well as the territories of the world, making Europe and USA as new epicenters [7]. It is the third zoonotic human-to-human transmission COVID-19 that has arisen in the present century, after SARS-CoV, which spread around 37 countries and the MERS-CoV, which spread around 27 countries.
In India, the pandemic COVID-19 was first confirmed in the state Kerala on January 30, 2020 when a student returned back from Wuhan, the province of China [8]. The Govt. of India has incorporated social distancing as a precautionary measure to prevent the possibility of a stage-3 human-to-human transmission that can stimulate the spread of the coronavirus diseases. India Govt. also imposed a 14 hours voluntary public curfew (‘Janata Curfiew’) on March 22, 2020 to make aware the people about the peculiar epidemiological traits compared with previous two epidemics of SERS-CoV and MERS-CoV. Moreover, the Govt. of India has declared a 21 days nationwide lockdown from March 25, 2020 to April 14, 2020 to prevent the spread of coronavirus diseases among human, affecting India’s 1.3 billion population. In the second phase, the lockdown has been extended up to May 03, 2020 to combat against COVID-19 pandemic in India. In lack of any specific vaccine or therapeutics, social distancing has been recognized as the most commonly utilized preventive measures to control the novel coronavirus diseases [9]. The main aim of these initiatives are the control of social interactive places, like schools, colleges, theaters, cultural programme and other public spheres, except for essential public services like hospitals, medicine shops, police and fire etc. Indeed the outbreaks of COVID-19 has drastically altered the daily life, health of the publics as well the economy. This is one of the main interests for each and everyone how long this situation will continue and when the disease will be under controlled and the entire world will be returned back to its earlier situation.
As identified by the World Health Organization (WHO), the mathematical models, mainly those formulated in a timely manner, can play a crucial role in allowing public health decision and policy-makers with evidence-based statistics [10], [11]. Mathematical modeling can aid in understanding; (i) how transmissible the disease is, (ii) when does the infectivity become high during the course of epidemic, (iii) how acute the disease is, and (iv) how effectual interventions has been and ought to be. It is not surprising that the researchers throughout the world have been trying to successfully model the coronavirus diseases. The group of modelers throughout the global has accepted the challenge in delineating mathematical models of the transmission dynamics of COVID-19 or SARS-CoV-2 and has rapidly reacted to the ongoing novel coronavirus epidemic. The progression of any outbreak depends on the infectivity of pathogens as well as the available uninfected individuals.
For a novel infection, when the transmission dynamics of an epidemiological disease is unknown, mathematical models play a key role to estimates the number of worst and best case scenarios. It can also aid in estimating the effect of precautionary measures adopted against novel coronavirus. With preventive techniques, the main object is to preserve the basic reproduction number R 0 below 1, to control further development of infection, whereas in a mitigation policy, the main object is to indelicate the effect of outbreak [9]. Recently, some mathematical models have already been investigated to understand the transmission dynamics of peculiar epidemiological traits of COVID-19, and some of these are listed in our references [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. Modeling the dynamics of COVID-19 pandemic is not new and most of the authors focused on the effect of lockdown due to absence of any effective therapeutics or licensed vaccine. In this context A. Atangana [12] studied a mathematical model to study the transmission dynamics of COVID-19 pandemic by using a system of fractional differential equations by using the effect of lockdown. Tang and colleagues [13] proposed a model for COVID-19 by considering symptomatic individuals to get the patients’ epidemiological status and calculated the basic reproduction number 6.47, which is very high for the infectious diseases. Giordano and colleagues [15] developed a new SIDARTHE model for COVID-19 pandemic and predict that restrictive social distancing can reduce the widespread of coronavirus among the human. Sarkar and Khajanchi [14], [17] proposed and analyzed a mathematical model to study the transmission dynamics of COVID-19 or SARS-CoV-2, where they performed the model validation with real data from India and some provinces of India, respectively. Based on the estimated model parameters, the authors performed the short-term prediction as well as long-term prediction. Liu and colleagues [19] developed a mathematical model by considering reported and unreported cases to study the transmission dynamics of novel coronavirus by using data from China. To study the intervention strategies of infectious diseases in an extended version of the classical SEIR (susceptible-exposed-infected-recovered) model that incorporates the fact that asymptomatic and pre-symptomatic infected individuals are believed to play a key role in the transmission dynamics of COVID-19 outbreak [16].
Our aim of this paper is to propose a compartmental mathematical model by introducing reported and unreported symptomatic individuals based on the data from the Republic of India. We address the following important issues regarding the outbreak of COVID-19: how does the outbreak develop in India with respect to the number of reported and unreported cases ? When the epidemic will end from India ? How the basic reproduction number R 0 influence disease outbreak ?
The organization of this manuscript is as follows: in the Section 2 we propose the SAIU mathematical model and its schematic representation. The qualitative properties of the SAIU model is discussed in the Section 3. In the same section, we perform local and global stability analysis of the disease free and endemic equilibrium point in terms of R 0. Also, we establish the criteria for the disease persistence with respect to R 0. In the Section 4, we conduct a sensitivity analysis for the basic reproduction number R 0. Numerical simulations based on the estimated parameter values are presented in the Section 5, and a discussion in the Section 6 concludes our manuscript.
2. Model formulation
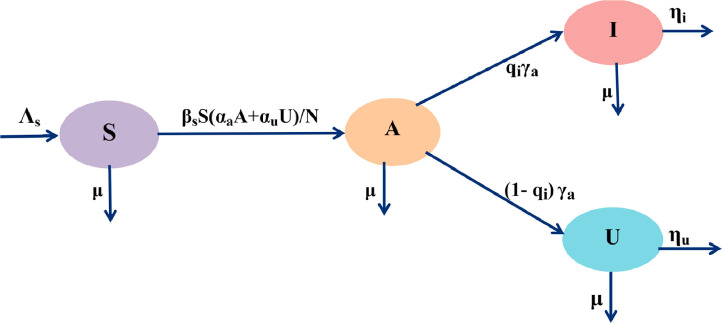
Mathematical modeling of transmission dynamics of infectious diseases are now ubiquitous. A series of mathematical models has been investigated by many researchers to describe the interactive dynamics of infectious diseases [22], [23], [24]. We proposed here a deterministic ordinary differential equation model that can represent the overall dynamics of novel coronavirus or SARS-CoV-2. We stratified the total human population into four compartments, namely susceptible individuals (uninfected), asymptomatic individuals (pauci-symptomatic or clinically undetected), reported symptomatic infected individuals (symptomatic infectious individuals are reported by the public heath service) and unreported symptomatic infected individuals (clinically ill but not reported) to formulate the SAIU (susceptible or uninfected (S) → asymptomatic (A) → reported symptomatic infectious (I) → unreported symptomatic infectious (U)) model. The total size of the population is . We assume that the reported infected individuals will no-longer associate into the infections as they are isolated and move to the hospital or Intensive Care Units (ICU). Thus, only infectious individuals belonging to I(t) or U(t) - spread or transmit the diseases. The COVID-19 transmission is illustrated in the Fig. 1 . The model consists of the following set of nonlinear differential equations:
| (1) |
with the positive initial conditions:
| (2) |
Fig. 1.
The schematic flow diagram represents the susceptible or uninfected (S), asymptomatic (A), reported symptomatic infectious (I) and unreported symptomatic infectious (U) individuals for novel coronavirus disease that persuades the formulation of the SAIU model (1).
Here t ≥ t 0 represents the time in days, t 0 is the starting date for the model system (1) of the epidemic. Some of the parameters may alter in time as control measures are implemented or changed. We performed the theoretical analysis for the model with constant parameters. In our model, we introduce some demographic effects by considering a proportional natural decay rate μ > 0 in each of the four individuals. In addition, our model includes a net inflow of susceptible or uninfected population at a rate Λs per unit time. The uninfected individuals can be decreased following infection, acquired by contact between an uninfected and an infected individual, who may be asymptomatic or unreported infected individuals. The transmission coefficients for these two compartments of infected populations are βsαa and βsαu, respectively. Here, we consider the disease transmission coefficient βs (which models both the contact rates and the infectiousness of novel coronavirus), with adjustment factors for asymptomatic individuals (αa) and unreported symptomatic infected individuals (αu). The effect of the parameters βsαa and βsαu are explicitly associated with the measures like lockdown, social distancing, restriction of movement and shaking hand etc., that actually decrease the number of contacts. Asymptomatic infectious populations develop to reported symptomatic infectious and unreported symptomatic infectious classes at a rate γa with a fraction qi and respectively, that is, 0 < qi < 1. Thus, the average time spent in the asymptomatic infectious individuals is days. The reported symptomatic infected populations (I) and the unreported symptomatic individuals (U) are infectious for an average period of days and days, respectively. In our model formulation, we assume that the reported symptomatic infectious individuals (I) are reported and hospitalized or isolated immediately, and thus no further infections. The asymptomatic infected class (A) can also be considered as having a lower-level symptomatic state.
Albeit, COVID-19 is supposed to be transmitted exclusively by reported symptomatic infectious individuals, a very low rate of transmission by asymptomatic populations cannot yet be ruled out. The SAIU model take into accounts for this probability by utilizing the adjustment parameter αa, where 0 < αa < 1. The adjustment parameter αu > 0 accounts for varying levels of hygiene safeguards during quarantine. Because the quarantine and isolation or hospitalization programmes and hygiene safeguards during quarantine and isolation or hospitalization were implemented and increased continuously after an epidemic. All the infections are obtained from either asymptomatic (A) or unreported symptomatic infectious (U) individuals. The model parameters and their description are provided in the Table 1 and a schematic diagram for the SAIU model is given in the Fig. 1.
Table 1.
Table of biologically relevant parameter values and their interpretation for the SAIU model (1) of COVID-19.
| Symbol | Interpretation | Values (Unit) | Source |
|---|---|---|---|
| net inflow of susceptible individuals | − | − | |
| βs | probability of disease transmission rate | 0.274 | Estimated |
| αa | modification factor for asymptomatic individuals | 0.4775 | Estimated |
| αu | modification factor for reported symptomatic class | 0.695 | Estimated |
| μ | natural death rate for all the individuals | 0.062 | [20] |
| γa | rate of transition from asymptomatic to symptomatic class | 0.29 | Estimated |
| qi | fraction of asymptomatic infectious become reported symptomatic infectious | 0.078 | Assumed |
| average time reported symptomatic individuals have symptoms | 0.009 | Estimated | |
| average time unreported symptomatic individuals have symptoms | 0.05 | Estimated | |
| N | total number of individuals | 1,352,642,280 | [37] |
3. Qualitative properties of the model
3.1. Positive invariance
Here, we shall investigate that all the state variables of the system (1) are non-negative for all time t with initial conditions . In order to prove the positivity, we state the following theorem.
Theorem 3.1
All the solutions (S(t), A(t), I(t), U(t)) of the system (1) with the initial values (2) satisfy S(t) > 0, A(t) > 0, I(t) > 0 and U(t) > 0 for all t > 0, then the system (1) is positively invariant and attracting within .
Proof
The first equation of the system (1), can be written as
where
Thereafter by integration, we obtain the following expression
This shows that S(t) is nonnegative for all t. Further from the second equation of the system (1), we have
which gives
Similarly, from the third equation of the system (1), we get
and this inequality implies
In the similar way, the last equation of the system (1) gives
which leads to
From the above analysis, we can conclude that all the solution trajectories of the system (1) remain positive for all t > 0. Hence the proof. □
3.2. Boundedness
Now we start with the theorem which assure that the solutions of the system (1) is bounded with nonnegative initial values.
Theorem 3.2
The solutions of the system (1) with the initial conditions (2) which initiate in are uniformly bounded in the positively invariant set Φ.
Proof
Here, we will show that all the feasible solutions are uniformly bounded in Φ. From the positivity of solutions, we get
which implies that
Taking we obtain
which gives
Accordingly, we obtain the following positively invariant bounded region
(3) Therefore, all the solution trajectories initiating in will enter Φ with finite time. In the region Φ, the existence, uniqueness and continuity results hold the dynamics of our SAIU model system [25], [26]. Hence the system is well-posed and biologically realistic. □
3.3. Basic reproduction number
The basic reproduction number for the SAIU model can be determined by using the next generation matrix introduced by van den Driessche and Watmough [27]. In order to do this, we consider the nonnegative matrix and the non-singular M-matrix expressing the production of new-infection and transition part, respectively. Our SAIU model system (1) is defined as follows:
Now, F and V can be written as
The basic reproduction number denoted by R 0 is the spectral radius of the next generation matrix:
3.4. Stability analysis of disease-free equilibrium (DFE)
Theorem 3.3
The disease-free equilibrium point E 0(Λs/μ, 0, 0, 0) exists and is locally asymptotically stable for R 0 < 1, otherwise unstable.
Proof
To determine the local stability of E 0(Λs/μ, 0, 0, 0), we compute the Jacobian matrix of the system (1) around the DFE E 0 is given by
The characteristic equation of corresponding to the eigenvalue λ is . From the characteristics equation, two eigenvalues of are real and negative, that is, and (since all parameters are positive) and the other two eigenvalues can be obtained form the following equation
(4) where
Here, we observe that ρ 2 > 0, this implies ρ 1 > 0. Therefore, the quadratic Eq. (4) has two strictly negative real roots or negative real parts if ρ 2 > 0, that is, if R 0 < 1 . Hence, disease-free equilibrium point (DFE) is locally asymptotically stable if R 0 < 1 and unstable for R 0 > 1. □
3.5. Global stability analysis of disease-free equilibrium (DFE)
In this subsection, we study the global stability of the unique disease-free equilibrium point E 0 with the condition R 0 < 1. In order to do this, we use a Lyapunov function similar to those are very classic and used by Korobeinikov & Maini [28], Mcclusky [29], and Khajanchi & Banerjee [30]. Such Lyapunov function take an advantages of all the properties of the function:
| (5) |
which is nonnegative in except at where it become zero. Now, we prove the global stability for E 0 by using the following Theorem.
Theorem 3.4
The disease-free equilibrium E 0 of the SAIU system (1) is globally asymptotically stable if R 0 < 1 and .
Proof
Consider the following Lyapunov function
Here is always nonnegative in the region Φ and attains zero at E 0. We want to show that is negative definite. Differentiate along the solution trajectory is given by
if (i) R 0 < 1 and (ii) with the aid of the relation between the arithmetic means and geometric means, we ensure that and the equality holds only at E 0. Hence, the disease-free equilibrium point E 0 is globally asymptotically stable if R 0 < 1. □
3.6. Persistence of the coronavirus disease
In the Theorem 3.3, we proved that while basic reproduction number R 0 < 1, the coronavirus disease dies out irrespective of the initial size of the epidemic. If R 0 > 1 the disease-free equilibrium E 0 become unstable. Usually it is considered that the infected individuals A(t), I(t) and U(t) will remain persistent for this event. Now, we prove the following theorem to verify the persistence of the coronavirus diseases.
Theorem 3.5
Assume that R 0 > 1. The disease will be uniformly persistent in the sense that there exists an ϱ > 0, such that for every positive solution of the system (1) , holds the following
Also, there exists an endemic equilibrium point in this case.
Proof
According to the theorem by Thieme [31], we prove the uniform persistance. In order to prove this, we consider that
Now, we want to show that the system (1) is uniformly persistent with respect to (G 0, H). Since H contains a unique equilibrium E 0, it is sufficient to show that where Ws(E 0) denotes the stable manifold of the disease-free equilibrium E 0.
Suppose this is not true. Then there is a solution (S(t), A(t), I(t), U(t)) ∈ G 0 of the system (1), such that
Then for any ε > 0, we obtain
for sufficiently large value of t. From the following the system (1), we have
where
and
Note that, is equal to has at least one eigenvalue with positive real part when R 0 > 1. Therefore, ε > 0 is arbitrary, one can make ε small enough so that is positive, where x(B) is the largest real part of the eigenvalue of B. So there exist solutions of the linear system
which can grow exponentially. By comparison, the solutions Q become unbounded as t → ∞. This gives a contradiction to our assumption that the solutions of the model (1) are uniformly bounded. Hence, . By using the Theorem 4.6 in [31], it can be concluded that the system (1) is uniformly persistent with respect to (G 0, H).
Therefore, the system of Eqs. (1) are dissipative (showed in the Theorem 2) and thus by using the Theorem 3.3 in [32] indicates that the model system (1) has an interior equilibrium point (that is, all components are positive). This completes the proof of this theorem. □
3.7. Existence of an endemic equilibrium point
The SAIU model system (1) has an endemic equilibrium point E*(S*, A*, I*, U*) with positive components provided R 0 > 1. Equating the derivatives of the model system (1) to zero and solving the resulting equations. First, we define
| (6) |
By solving the equations in the system (1) at endemic steady state, we obtain that
| (7) |
Plugging the above expression (7) into the Eq. (6), we obtain the nonzero equilibrium of the system (1) satisfying the linear equation, in terms of κ* as follows:
| (8) |
where
Clearly, Υ1 > 0 as > 0, > 0, and > 0, hence we can say that the system (1) has a unique positive endemic equilibrium point whenever R 0 > 1 and no positive equilibrium point whenever R 0 < 1.
Theorem 3.6
The endemic equilibrium E* of the system (1) is locally asymptotically stable if R 0 > 1.
Proof
Introducing and then the system (1) becomes
(9) with and choosing the bifurcation parameter βs.
The Jacobian matrix of the system (9) around the disease free equilibrium E 0 at the threshold point is given by
The eigenvalues of the are and 0. Here 0 is the simple eigenvalue of and the others eigenvalues have negative real parts. Hence, the Center Manifold Theorem can be applied and we get a right eigenvector and a left eigenvector corresponding to the zero-eigenvalue is given by
and
Hence, we have
whose sign determined the local stability criteria of the endemic equilibrium point E*. Substituting the values of all second-order derivatives measured at DFE, E 0 is given by
and
Therefore, a < 0 and b > 0 at a transcritical bifurcation occurs at and unique endemic equilibrium is locally asymptotically stable for R 0 > 1. □
3.8. Global stability of endemic equilibrium point
This subsection is dealing with the global stability of an unique endemic equilibrium point E* with the condition R 0 > 1. In order to show that, we use the Lyapunov functional similar to the Eq. (5) and such Lyapunov functional take advantages of all the properties of the function. Now, we prove the following result.
Theorem 3.7
The endemic equilibrium point E* of the system (1) is exists and globally asymptotically stable if R 0 > 1.
Proof
Let us assume that E* exists and the following function is well defined in . Now, we consider the following Lyapunov functional [33] as
We need to show that is negative definite. Differentiating along the solution trajectories of the system (1), we get
Considering the relation of arithmetic means and geometric means, we ensure that and holds the equality only at E*. Hence, the endemic equilibrium point E* is globally asymptotically stable. □
4. Sensitivity analysis
To determine how best to decrease human impermanence and morbidity due to COVID-19, it is essential to understand the relative importance of the various factors responsible for its transmission. Initial disease transmission is directly associated to the basic reproduction number R 0. We compute the sensitivity indices of the basic reproductive number R 0 to the parameters in the model. These indices allow us how - important each parameter is to disease transmission. Sensitivity analysis is mainly utilized to describe the robustness of model forecasting to the parameter values (since there are usually errors in the collection of data and presumed model parameter values) [34]. Here, we perform a sensitivity analysis of the basic reproductive number R 0 to quantify the fluctuations in the SAIU model parameters. Now from this, we can identify the parameters that have a high impact on the basic reproduction number as well as on the disease transmission. Sensitivity indices permit us to quantify the relative change in a state variable when a parameter alters. The normalized forward sensitivity index of a variable to a parameter is the ratio of the relative change in the variable to the relative change in the parameter. The normalized forward sensitivity index of R 0 with a parameter αu is defined as follows:
Similarly, for other parameter values we can calculate the sensitivity indices of R 0 for the explicit expression for the basic reproduction number. As for example, the sensitivity indices of R 0 with respect to βs is given by
It can be noted that sensitivity indices may depend on several parameters for the SAIU system, but also can be constant, independent of any parameters. As for example, describes that increasing (decreasing) βs by a given percentage increases (decreases) always R 0 by that same percentage. We perform the sensitivity analysis for the parameters with effective care, since a small perturbation in such parameter leads to relevant quantitative changes. On the other hand, the estimation of a parameter with a rather small value for the sensitivity indices does not require much attention to estimate, because a small perturbation in that parameter leads to small changes.
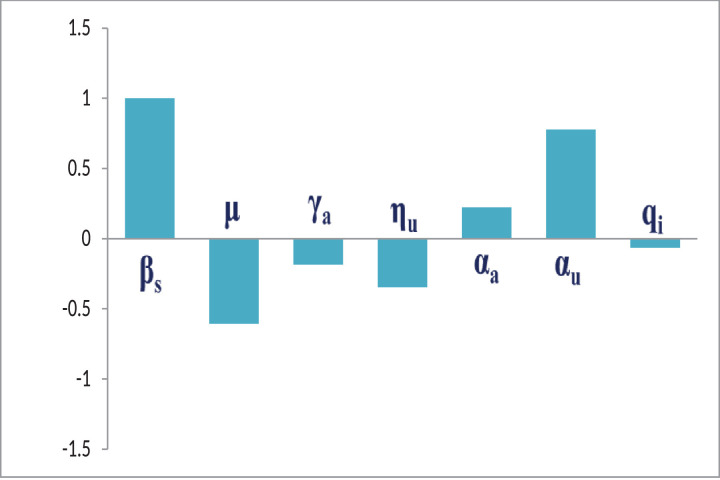
From the Table 2 and the Fig. 5, we can observe that the most sensitive parameters to the basic reproduction number R 0 for the SAIU model system (1) are βs, μ and αu. More precisely, an increase of the value of αu will increase the basic reproduction number R 0 by 77.65% and this happens, in a similar way, for the parameter αa. In contrast, an increase of the value of μ will decrease the basic reproduction number R 0 by 60.60%.
Table 2.
Sensitivity indices of the basic reproduction number R0 to parameters for the SAIU COVID-19 model, evaluated at the baseline parameter values listed in the Table 1.
| Parameters | βs | μ | γa | αa | ηu | αu | qi |
|---|---|---|---|---|---|---|---|
| Values | 1.000 | − 0.6060 | −0.18464 | 0.22348 | −0.34666 | 0.77652 | −0.06474 |
Fig. 5.
The figure shows the sensitivity indices of the basic reproduction number R0 with respect to the each of the system parameters related to R0 for the SAIU model system (1). The baseline parameter values are taken from the Table 1. The simulation exhibits that the most influential parameter is the probability of disease transmission rate (βs), and the least influential parameter is the fraction of asymptomatic infected individuals become reported symptomatic infected individuals at the rate qi. The list of sensitivity indices are given in the Table 2.
5. Numerical simulations
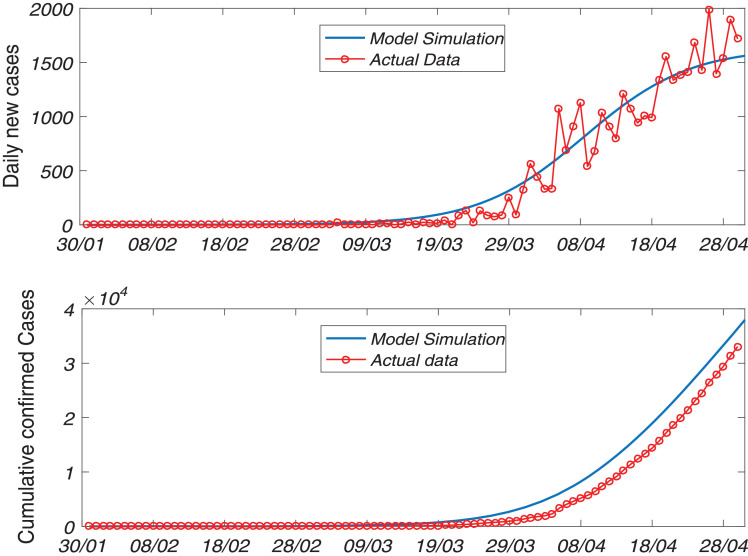
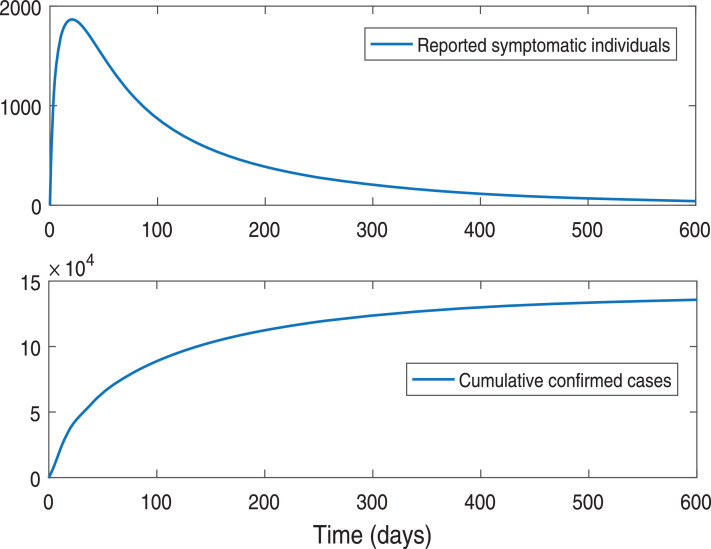
We have calibrated our SAIU model system (1) for the novel coronavirus diseases to the daily new infected cases for the Republic of India. The data are collected from daily new infected cases and confirmed cumulative cases for India for the time period January 30, 2020 to April 30, 2020 from the WHO website [36]. In order to fit the data we use ODE45 in MATLAB and estimate the parameter values that give the best fit for our SAIU model. The proposed SAIU model system (1) has 9 nonnegative parameter values among which, we have estimated 6 parameters, namely βs (probability rate of disease transmission), αa (adjustment factor for asymptomatic individuals), αu (adjustment factor for reported symptomatic infected individuals), γa (transition rate from asymptomatic to symptomatic infected individuals), ηi (average time reported symptomatic infectious have symptoms) and ηu (average time unreported symptomatic infectious have symptoms) based on the sensitivity analysis [35]. To minimize the errors we fit the curve for daily confirmed cases and cumulative confirmed cases, which has been shown in the Fig. 2 and the initial population sizes are given in the figure caption. The parameter values for our SAIU model (1) corresponding to the best fit curve for India are listed in the Table 1, which can be used further to make predictions and simulations of our model.
Fig. 2.
The figures shows the model fitting of daily reported symptomatic infectious individuals (upper panel) and the reported cumulated symptomatic infectious individuals (lower panel) for the SARS-CoV-2 or COVID-19 pandemic in India. The epidemic turning point of the daily reported symptomatic and cumulated cases data from January 30, 2020 to April 30, 2020 (day 1 = January 30, 2020). The observed data points are shown in the red circle and the solid blue line portrays the model simulations. We use the initial size of the population and .
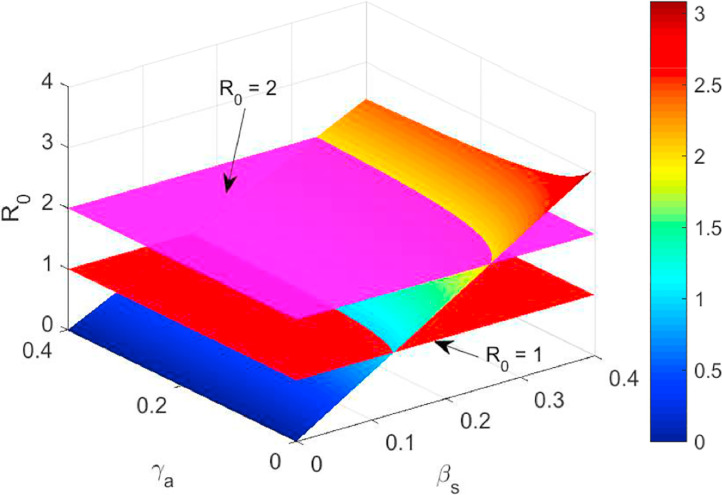
For the set of parameter values listed in the Table 1, we draw a surface plot for the basic reproduction number R 0 with respect to the disease transmission coefficient βs and the transition rate γa from asymptotic individuals to symptomatic individuals. For the basic reproduction number we compute the followings:
From the above expressions, we note that is always positive and becomes negative if . For the listed parameters in the Table 1, we have seen that is negative. As the transmission coefficient βs increases, R 0 will increase and cross the threshold thus leading to the outbreak of the coronavirus diseases. In the Fig. 3 , the red surface indicates the threshold . Moreover, when the transition rate γa of asymptotic individuals to symptomatic individuals increases, R 0 will decrease and if R 0 goes below 1, the coronavirus will die out and the population will be free from COVID-19 or SARS-CoV-2. In the Fig. 3, the magenta surface indicates the threshold . Therefore, our model simulation reveals that the transition rate γa aid in helping to eradicate the coronavirus diseases by reducing the basic reproduction number R 0. Also, we can control the reproduction R 0 by reducing the transmission coefficient βs. The parameter values obtained from data fit for India as are listed in the Table 1, we compute which indicates that the coronavirus diseases spread throughout the India, if we not take preventive measures like social distancing, frequently wash hand by sanitizer etc.
Fig. 3.
The figure shows the basic reproduction number R0 when βs (probability of disease transmission rate) and γa (rate of transition from asymptomatic to symptomatic infectious class) varies. The other parameter values are listed in the Table 1.
For the estimated parameter values in the Table 1, our model predicts that there will be a high peak for the coronavirus diseases around 60 days and after that the peak will be decreased and the curve become plateau (see the Fig. 4 for model prediction). But, the novel coronavirus will persists among the people for a long days. It is extremely difficult to predict the end date of the coronavirus diseases. Thus, we always have to maintain social distancing like lockdown, extension of closing schools and colleges, stop cultural events, bar and shopping mall etc. The SAIU model is concise in framework, and it fortunately captures the course of the COVID-19 or SARS-CoV-2 epidemic, and thus sheds light in understanding the trends of the epidemic.
Fig. 4.
The figures shows the prediction of our SAIU model (1) for the Republic of India. Here, and the initial values are rest of the parameter values are listed in the Table 1. The model simulation demonstrates that about 60 days the peak will be higher for the COVID-19 in India and after that the curve will be flatten but the coronavirus diseases will be continued for a long-time with lesser magnitude.
In order to control the COVID-19, we must have to control the threshold level of R 0. Thus, we plot the sensitivity indices in the Fig. 5 to understand the most sensitive parameters with respect to R 0. From the Fig. 5 and the Table 2, we can see that the parameters βs (disease transmission coefficient), αa (adjustment factor for asymptomatic individuals), and αu (adjustment factor for reported symptomatic infected individuals) have positively correlated sensitivity indices and the parameters μ (natural mortality rate of entire individuals due to COVID-19 deaths), γa (transition rate from asymptomatic individuals to symptomatic individuals), ηu (average time for the unreported individuals have symptoms) and qi (fraction of asymptomatic infected individuals become reported symptomatic individuals) have negatively correlated sensitivity indices. Therefore, the sensitivity graph is very useful to control the basic reproduction number R 0.
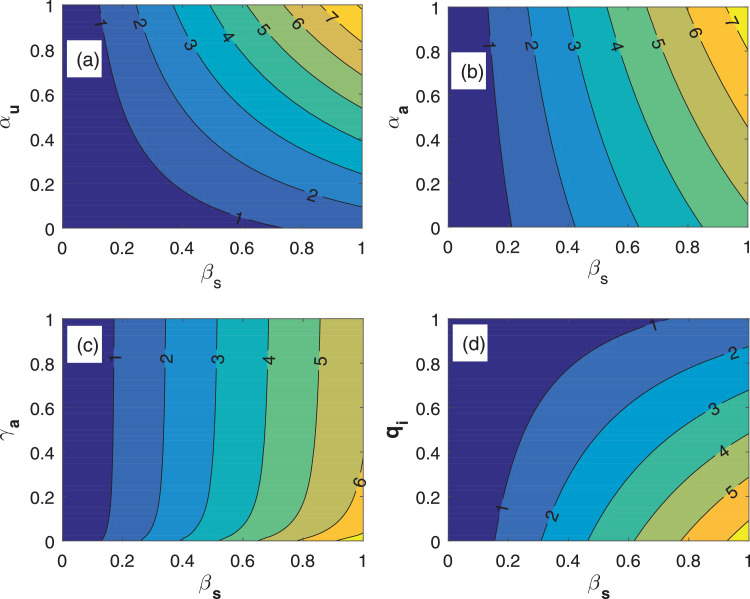
In addition, we draw the contour plots for the basic reproduction number R 0 with respect to the parameters βs versus αu, αa, γa and qi for the SAIU model (1) to study the influence of the control parameters in controlling the reproduction number R 0. From the Fig. 6 ((a), (b)), it can be observed that R 0 increases remarkably for increasing the value of αa, αu and βs. We can notice from the Fig. 6 (c), that R 0 increases for increasing the value of βs and γa but not like Fig. 6 ((a), (b)). Interestingly, the value of R 0 can be controlled by increasing the value of qi (see the Fig. 6 (d)). Thus, we may conclude that the threshold parameter R 0 is highly effective with respect to the parameters who have highly negatively correlated sensitivity indices. For all the sub-plots of the Fig. 6, it can be noticed that the reproduction number R 0 is always increased for increasing values for βs. Thus, to eliminate the coronavirus diseases we must reduce the disease transmission rate and the disease transmission rate can be reduced by controlling βs. Now, we may conclude that the social distancing is the main non-pharmaceutical measure to end the novel coronavirus.
Fig. 6.
Contour plots for the SAIU model system (1). Contour plots of basic reproduction number R0 with respect to the most effective parameters, βs (probability of disease transmission) versus (a) modification factor for the reported symptomatic infected individuals (αu), (b) modification factor for the asymptomatic infected individuals (αa), (c) transition rate (γa) from asymptomatic individuals to symptomatic infected individuals, and (d) a portion of pre-symptomatic infected individuals become symptomatic infected individuals at the rate qi. All the parameter values are listed in the Table 1 except the varied parameters.
6. Discussion
The reported cases of COVID-19 are rising throughout the world and the human-to-human transmission of coronavirus diseases is already established, thus predicting is the highest priority for the control and management the diseases with limited resource. In our study, we proposed and analyzed the SAIU model to study the transmission dynamics of COVID-19 based on the accessible data [36] for India during the time period January 30, 2020 to April 30, 2020. Based on the estimated data our SAIU model predict the outbreak of COVID-19 or SARS-CoV-2 virus. We compute the basic reproduction number R 0, which can be used further for model simulation and predictions.
We studied the SAIU model for COVID-19 assessing the sensitivity indices of the basic reproductive number R 0, as R 0 quantifies the initial disease transmission and the sensitivity indices allow us to describe the relative importance of various parameters in coronavirus transmission. We perform the local and global asymptotic stability analysis for the infection free equilibrium point E 0 in case of R 0 < 1. Furthermore, the SAIU model showed the persistence of diseases for R 0 > 1. The endemic equilibrium point E* is locally asymptotically stable for R 0 > 1. Constructing suitable Lyapunov function followed by Korobeinikov & Maini [28], we showed that the our SAIU model is globally asymptotically stable for R 0 > 1. Theoretically, we showed that at our SAIU model undergoes transcritical bifurcation.
We calibrated our proposed SAIU model to fit with daily and cumulative confirmed cases of India. For the estimated parameter values, we obtained which shows the substantial outbreak of novel coronavirus in India. The reproduction number R 0 can be controlled by reducing the disease transmission rate βs and by increasing the quantity qi also by increasing the adjustment factors αa and αu, which has been shown by contour plot in the Fig. 6. This indicates that the elimination of COVID-19 is possible by maintaining the social distances like contact tracing, lockdown and use precautionary measures. Also, the policymakers as well as the health care agencies should concentrate on successful implementation of control mechanisms to minimize the burden of the coronavirus diseases. Our model simulations nicely capture the increasing trend of the course of the COVID-19 epidemic (see the Fig. 2). Sensitivity indices reveal that the disease transmission rate βs is positively correlated and the proportion rate qi of asymptomatic infected population reported symptomatic infected individuals negatively correlated with respect to the reproduction number R 0. This implies that increasing qi and decreasing the disease transmission rate βs will decrease the reproduction number R 0 and consequently will reduce the disease burden. While investigating the contour plots (see Fig. 6), it can be seen that effective management of disease transmission rate βs is more influential to mitigate the reproduction number R 0 below 1.
Based on the estimated data, our SAIU model predict that there will be a highest peak around 60 days if human-to-human transmission and the personal preventive measures continue with the existing rates. Around 60 days later the peak will be decreased but the CODIV-19 disease will persist for a long time. In absence of any pharmaceutical measures, the public must have to obey the government rules or public health care policies to mitigate the spread of novel coronavirus. It is really difficult to predict the outbreak of COVID-19 in India and throughout the world. In order to get more accurate prediction, we need to get more accurate data. It is worthy to mention that the researchers are working for therapeutics or vaccine to eliminate novel coronavirus and the presence of such pharmaceutical interventions will remarkably change the outcomes.
CRediT authorship contribution statement
Piu Samui: Methodology, Formal analysis, Investigation. Jayanta Mondal: Formal analysis, Methodology, Writing - original draft, Writing - review & editing, Visualization. Subhas Khajanchi: Conceptualization, Software, Validation, Methodology, Funding acquisition, Resources, Writing - original draft, Writing - review & editing, Visualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The study of Subhas Khajanchi is supported by Science and Engineering Research Board (File No. ECR/ 2017/000234), Department of Science & Technology, Government of India. The authors are thankful to the anonymous referees for their careful reading and constructive suggestions/comments which helped in better exposition of the manuscript. The first author is grateful to West Bengal Higher Education Department, Govt. of West Bengal, Bikash Bhavan, India for the financial support (Order No. 52 - Edn(B) / 5 B - 15 / 2017 dated 07. 06. 2017) provided to pursue her Ph.D.
References
- 1.COVID-19 - events as they happen. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-theyhappen.
- 2.COVID-19 report. 2020. https://www.worldometers.info/coronavirus/.
- 3.BBC news. 2020. https://www.bbc.com/news/world-52114829.
- 4.Carlos W.G., Cruz C.S.D., Cao B., Pasnick S., Jamil S. Novel Wuhan (2019-ncov) coronavirus. Am J Respir Crit Care Med. 2020;201(4):7–8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 5.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou L., Ruan F., Huang M. SARS-cov-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.BBC news. https://www.bbc.com/news/world-europe-51876784.
- 8.Government of India, https://www.mygov.in/covid-19.
- 9.Ferguson N., Laydon D., Gilani G.N., Imai N., Ainslie K., Baguelin M., Bhatia S., Boonyasiri A., et al. Report 9: Impact of non-pharmaceutical interventions (NPIS) to reduce covid19 mortality and healthcare demand. 2020.
- 10.Egger M., Johnson L., Althaus C., Schoni A., Salanti G., Low N. Developing WHO guidelines: time to formally include evidence from mathematical modelling studies. F1000Research. 2017;6:1584. doi: 10.12688/f1000research.12367.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khajanchi S., Das D.K., Kar T.K. Dynamics of tuberculosis transmission with exogenous reinfections and endogenous reactivations. Physica A. 2018;497:52–71. [Google Scholar]
- 12.Atangana A. Modelling the spread of COVID-19 with new fractal-fractional operators: can the lockdown save mankind before vaccination? Chaos Soliton Fract. 2020;136:109860. doi: 10.1016/j.chaos.2020.109860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang B., Wang X., Li Q., Bragazzi N.L., Tang S., Xiao Y., Wu J. Estimation of the transmission risk of the 2019-ncov and its implication for public health interventions. J Clin Med. 2020;9(2):462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkar K., Khajanchi S., Nieto J.J. Modeling and forecasting of the COVID-19 pandemic in India. Chaos Soliton Fract. 2020;139:110049. doi: 10.1016/j.chaos.2020.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giordano G., Blanchini F., Bruno R., Colaneri P., Filippo A.D., Matteo A.D., Colaneri M. Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy. Nat Med. 2020;26:855–860. doi: 10.1038/s41591-020-0883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatto M., Bertuzzo E., Mari L., Miccoli S., Carraro L., Casagrandi R. Spread and dynamics of the COVID-19 epidemic in Italy: effects of emergency containment measures. PNAS. 2020;117(19):10484–10491. doi: 10.1073/pnas.2004978117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khajanchi S., Sarkar K. Forecasting the daily and cumulative number of cases for the COVID-19 pandemic in india. Chaos. 2020;30:071101. doi: 10.1063/5.0016240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gumel A.B., Ruan S., Day T., Watmough J., Brauer F., Driessche P.V.d., Gabrielson D., Bowman C., Alexander M.E., Ardal S., Wu J., Sahai B.M. Modelling strategies for controlling SARS outbreaks. Proc R Soc Lond B. 2004;271:2223–2232. doi: 10.1098/rspb.2004.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Magal P., Seydi O., Webb G.B. A COVID-19 epidemic model with latency period. Infect Dis Model. 2020 doi: 10.1016/j.idm.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khajanchi S., Sarkar K., Mondal J., Perc M.. Dynamics of the COVID-19 pandemic in India. 2020. ArXiv:2005.06286.
- 21.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-ncov outbreak originating in Wuhan, china: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson R.M., May R.M. Oxford University Press; London: 1991. Infectious diseases of humans. [Google Scholar]
- 23.Diekmann O., Heesterbeek J.A.P. Wiley; New York: 2000. Mathematical epidemiology of infectious diseases: model building, analysis and interpretation. [Google Scholar]
- 24.Hethcote H.W. The mathematics of infectious diseases. SIAM Rev. 2000;42:599–653. [Google Scholar]
- 25.Khajanchi S. Chaotic dynamics of a delayed tumor immune interaction model. Int J Biomath. 2020;13(2):2050009. [Google Scholar]
- 26.Khajanchi S. Stability analysis of a mathematical model for glioma-immune interaction under optimal therapy. Int J Nonlin Sci Num. 2019;20(3–4):269–285. doi: 10.1515/ijnsns-2017-0206. [DOI] [Google Scholar]
- 27.van den Driessche P., Watmough J. Reproduction numbers and sub-thershold endemic equalibria for compartmental models of disease transmission. Math Biosci. 2002;180:29–48. doi: 10.1016/s0025-5564(02)00108-6. [DOI] [PubMed] [Google Scholar]
- 28.Korobeinikov A., Maini P.K. A Lyapunov function and global properties for SIR and SEIR epidemiological models with nonlinear incidence. Math Biosci Eng. 2004;1(1):57–60. doi: 10.3934/mbe.2004.1.57. [DOI] [PubMed] [Google Scholar]
- 29.Mccluskey C. Global stability for an SIR epidemic model with delay and nonlinear incidence. Nonlinear Anal RWA. 2010;11(4):3106–3109. doi: 10.3934/mbe.2010.7.837. [DOI] [PubMed] [Google Scholar]
- 30.Khajanchi S., Banerjee S. Quantifying the role of immunotherapeutic drug t11 target structure in progression of malignant gliomas: mathematical modeling and dynamical perspective. Math BioSci. 2017;289:69–77. doi: 10.1016/j.mbs.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Thieme H.R. Persistence under relaxed point-dissipativity (with applications to an endemic model) SIAM J Math Anal. 1993;24(2):407–435. [Google Scholar]
- 32.Hale J.K., Waltman P. Persistence in infinite-dimensional systems. SIAM J Math Anal. 1989;20(2):388–395. [Google Scholar]
- 33.Khajanchi S. Uniform persistence and global stability for a brain tumor and immune system interaction. Biophys Rev Lett. 2017;12(4):187–208. [Google Scholar]
- 34.Chitnis N., Hyman J.M., Cushing J.M. Determining important parameters in the spread of malaria through the sensitivity analysis of a mathematical model. Bull Math Biol. 2008;70:1272–1296. doi: 10.1007/s11538-008-9299-0. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee S., Khajanchi S., Chaudhuri S. A mathematical model to elucidate brain tumor abrogation by immunotherapy with t11 target structure. PLoS ONE. 2015;10(5):e0123611. doi: 10.1371/journal.pone.0123611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World health organization (WHO) situation report. (January 30, 2020 - April 30, 2020). http://www.who.int.
- 37.Demographics of India. https://en.wikipedia.org/wiki/Demographics-of-India.