Abstract
Purpose
Retinal detachment (RD) disrupts the nutritional support and oxygen delivery to photoreceptors (PRs), ultimately causing cell death. High-mobility group box 1 (HMGB1) can serve as an extracellular alarmin when released from stressed cells. PRs release HMGB1 after RD. The purpose of this study was to investigate the relationship between HMGB1 and PR survival after RD.
Methods
Acute RD was created by injection of hyaluronic acid (1%) into the subretinal space in C57BL/6 mice and mice with a rhodopsin-Cre-mediated conditional knockout (cKO) of HMGB1 in rods (HMGB1ΔRod). Immunofluorescence (IF) in retinal sections was used to localize HMGB1, rhodopsin, and Iba-1 proteins. Optical coherence tomography and electroretinography were used to quantify retinal thickness and function, respectively. The morphology of the retina was assessed by hematoxylin and eosin.
Results
HMGB1 protein was localized to the nuclei of all retinal neurons, including PRs, with cones staining more intensely than rods. HMGB1 protein was also found in the inner and outer segments of cones but not rods. Creation of RD caused a dramatic increase of HMGB1 protein IF in rods. cKO of HMGB1 in rods did not affect retinal structure or function. However, after RD, loss of rods and reduction in the thickness of the outer nuclear layer were significantly increased in the HMGB1ΔRod retinas as compared to the control. Interestingly, depletion of HMGB1 in rods did not affect the activation and mobilization of microglia/macrophages normally seen after RD.
Conclusions
Increased HMGB1 expression in stressed rods may represent an intrinsic mechanism regulating their survival after RD.
Keywords: mouse, retina, photoreceptor, retinal detachment, HMGB1
Retinal detachment (RD) describes the separation of photoreceptors (PRs) from the retinal pigment epithelium (RPE), thus creating a physical barrier to the diffusion of oxygen and nutrients from the choroidal vasculature.1,2 PR cells are highly vulnerable to the insults of hypoxia and nutrient deprivation because of substantial metabolic demands to support visual phototransduction and outer segment production. Our previous studies have shown that PR death after RD is primarily driven by Fas receptor activation.3 The ability of Fas to initiate PR death after RD, however, is attenuated by the expression of IL-6 and activation of intrinsic prosurvival factors and pathways, such as Fas apoptotic inhibitory molecule 2 and autophagy.4–7
High-mobility group box 1 (HMGB1) is a ubiquitous nonhistone DNA-binding protein normally localized in the nuclei of many cell types8,9 that acts as a danger-associated molecular pattern (DAMP) signaling molecule when released by dying or necrotic cells.10 HMGB1 has multiple functions depending on its cellular location. In the nucleus, HMGB1 binds to nucleosomes, alters chromosomal structure, bends DNA, and alters access of DNA-binding proteins and DNA polymerases.11–13 HMGB1 colocalizes with euchromatin, which contains an abundance of actively transcribed DNA, and HMGB1 has been shown to enhance transcriptional activity of a large number of genes.9,14–16 HMGB1 can translocate to the cytoplasm, where it promotes autophagy in response to stress and helps control the transition of autophagy to apoptosis during proinflammatory conditions.17–19 HMGB1 is also actively secreted by stressed cells or passively released by necrotic cells.20–25 In the extracellular space, HMGB1 acts as an alarmin by binding to the receptor for advanced glycation end products (RAGE) or the Toll-like receptors, TLR2 or TLR4.26–29 Extracellular HMGB1 contributes to microglial activation and proinflammatory cytokine production in several models of ocular diseases, including experimental autoimmune uveitis30 and diabetic retinopathy.31–35 Inhibition of HMGB1 suppresses microglial activation, blunts proinflammatory mediator production, and improves neurologic deficits in cerebral ischemic and hemorrhagic brain injuries.36–38
Despite the abundant reports regarding HMGB1 functions in neuronal injury, knowledge is limited regarding the functions of HMGB1 as an endogenous danger signaling molecule in response to PR injury, especially after RD. A potential role for HMGB1 in human RD is strongly suggested by the fact that HMGB1 levels are significantly increased in the vitreous of patients with RD compared to non-RD controls.39 Considering the involvement of HMGB1 in central nervous system inflammatory responses, it is reasonable to hypothesize that RD induces PR release of HMGB1, which is detected by microglia and promotes their response, eventually affecting PR cell survival. In this work, we characterize the expression pattern and subcellular accumulation of HMGB1 in the mouse retina before and after RD. In addition, we generated conditional knockout (cKO) of HMGB1 in rod PRs to investigate the effect that loss of HMGB1 has on rod PR survival, both at baseline and in response to RD. We observed that levels of HMGB1 protein are normally relatively low in rod PRs and increase in response to RD. Surprisingly, we found that cKO of HMGB1 in rods does not affect microglial activation or migration toward the stressed PRs. Instead, cKO of HMGB1 in rods accelerates PR cell death in the detached retinas. In other words, HMGB1 may represent an intrinsic signal contributing to PR survival after RD.
Materials and Methods
Animals
All experiments were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and approved by the University Committee on the Use and Care of Animals of the University of Michigan. Transgenic mice with rhodopsin promoter–driven Cre recombination of the HMGB1 gene in rod PRs were used. HMGB1flox/flox strain40 was generated and provided by Dr. Allan Tsung, University of Pittsburgh Medical Center. Rhodopsin-Cre transgenic mouse41 was generated by Dr. Ching-Kang Chen, University of Utah, and provided by Dr. Thomas A. Ferguson, Washington University School of Medicine. HMGB1flox/flox (Rho)Cre+ mice (denoted as HMGB1ΔRod) were generated by crossing a HMGB1flox/flox mouse with a rhodopsin-Cre mouse. Rho-Cre+ mice served as controls. Nrl−/− mouse42 was generated and provided by Dr. Anand Swaroop, National Eye Institute. All mice were genotyped via PCR analysis of genomic DNA collected from tail tips. Mice were bred and housed in vivarium at the University of Michigan Kellogg Eye Center under the standard 12-hour light/12-hour dark cycle.
Experimental Model of Retinal Detachment
Mice were anesthetized by intraperitoneal injection of ketamine (50 mg/kg bodyweight) and xylazine (5 mg/kg bodyweight) and pupils were dilated with topical application of tropicamide (1%) and phenylephrine (2.5%). RD was created in adult mice (8–12 weeks old) as previously reported.43 Briefly, a penetrating incision was made in the sclera approximately 1 mm posterior to the limbus with a 30-gauge needle. The tip of a 35-gauge needle attached to a microsyringe (NANOFIL-100 injector system; World Precision Instruments, Sarasota, FL, USA) was inserted through the sclerotomy into the vitreous cavity. The needle was then advanced through the retina to allow for the injection of 1 to 2 µL 10 mg/mL sodium hyaluronate (Healon; Abott Medical Optics, Santa Ana, CA, USA) into the subretinal space until one-third to one-half of the neurosensory retina was separated from the underlying RPE. The contralateral eyes did not receive an incision, needle insertion, or retinal detachment.
Intraocular Fluid Collection
Intraocular fluid was collected as previously reported with slight modification.30,44 Briefly, eyeballs were enucleated and the muscles and other accessory tissues were removed. The eyeball was immersed in 200 µL PBS with protease inhibitor (PI) (Complete, cat. 11697498001; Roche, Mannheim, Germany), and the cornea and lens were removed to create an eyecup. The eyecup samples, including choroid and sclera, were minced, and centrifuged at 14,000 rpm for 10 minutes, and supernatant containing aqueous, vitreous, and subretinal fluid was collected. Duplicate samples of 100 µL of each eyecup supernatant were used to measure the levels of soluble HMGB1 using a Mouse HMGB1 ELISA Kit (cat. OKCD04072; Aviva Systems Biology, San Diego, CA, USA) according to the manufacturer's instructions. The pelleted eyecup pieces were homogenized and processed for nuclear and cytoplasmic protein extraction using the NE-PER nuclear and cytoplasmic extraction kit (cat. 78833; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions.
Western Blot Analysis
Extracted nuclear and cytoplasmic proteins from eyecup pieces were solubilized in nuclear extraction reagent with PI and cytoplasmic extraction reagent with PI, respectively, and protein concentrations were measured by DC protein assay (cat. 5000116; Bio-Rad, Hercules, CA). Then, 15 mg of protein from each sample was separated on 4% to 15% SDS-PAGE (Mini-PROTEAN TGX; Bio-Rad) and then transferred to a polyvinylidene fluoride membrane (Bio-Rad). After blocking with 3% BSA (Sigma-Aldrich, St. Louis, MO, USA) in Tris-buffered saline (TBS; Bio-Rad), membrane was incubated with primary antibody to HMGB1 (cat. PA1-10926; Thermo Fisher Scientific) overnight at 4°C under slight agitation. After three washes with 0.1% Tween-20 in TBS, membrane was incubated with peroxidase-conjugated secondary antibody (cat. G-21234; Thermo Fisher Scientific) for 2 hours at room temperature (RT). Signals were visualized by enhanced chemiluminescence (SuperSignal West Dura Substrate; Thermo Fisher Scientific) and detected by cSeries Capture Software (c500; Azure Biosystems, Dublin, CA, USA). Densitometry was quantified by ImageJ software (National Institutes of Health, Bethesda, MD). Probing with antibody to lamin A/C (cat. sc-376248; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as a loading control for nuclear extracts, while probing with antibody to glyceraldehyde 3-phosphate dehydrogenase (cat. AM4300; Applied Biosystems, Foster City, CA, USA) was used as a loading control for cytoplasmic extracts.
Histology and Immunohistochemistry
Following euthanasia, mouse eyes were immediately removed and fixed in 4% formaldehyde overnight at 4°C. For histology, whole eyes were processed in an automatic tissue processor following standard protocols for paraffin embedding. Serial 6-µm-thick sections were obtained with a Shandon AS325 microtome (Thermo Fisher Scientific). Sections were deparaffinized in xylene and rehydrated in a series of descending concentrations of ethyl alcohol. After rinsing in distilled water, sections were stained with hematoxylin and eosin for general histology and photographed with a Leica DM6000 microscope (Leica Microsystems, Wetzlar, Germany). The total number of cells in the outer nuclear layer (ONL) was measured using ImageJ software (National Institutes of Health) as previously reported.7,45 Briefly, three noncontinuous sections that cross the optic nerve head were chosen and continuous serial images in the detached portion of the retina were captured under 20× magnification. The nuclei count in the ONL, the area of ONL, and the retina (from the outer edge of the ONL to the inner limiting membrane) were measured. The ONL area was normalized to the total retinal area (ONL area divided by total retinal area) to account for the possible differences due to the different angles in sectioning to allow for intersample comparison. For each group, measurements were made on three sections of each of three to five eyes.
For immunohistochemistry using paraffin sections, rehydrated sections underwent heat antigen retrieval by boiling the sections in 10 mM citrate buffer (pH 6.0) for 10 minutes before immunostaining procedures. For immunofluorescence (IF) of retinal cryosections, the anterior segments were removed to form an eyecup. Then the eyecups were washed three times in PBS, cryoprotected in increasing concentrations of sucrose in PBS (5% for 1 hour at RT, 10% for 1 hour at RT, and 20% overnight at 4°C), and embedded in a 1:1 mixture of 20% sucrose in PBS and optimal cutting temperature compound (Tissue Tek 4583; Sakura Finetek, Tokyo, Japan). Samples were sectioned on a Leica CM3050S cryostat (Leica Microsystems) at 10-µm thicknesses. Deparaffinated and frozen sections were first treated with 5% goat serum in PBS with 0.1% Triton X-100 (PBST) for 1 hour and then incubated with primary antibody to HMGB1 (cat. PA1-10926; Thermo Fisher Scientific), ionized calcium-binding adapter molecule 1 (Iba-1) (cat. 019-19741; Wako Chemicals USA Corporation, Richmond, VA, USA), rhodopsin (cat. NBP1-48334; Novus Biologicals, Centennial, CO, USA), Cre-recombinase (cat. MAB3120; Millipore, Burlington, MA, USA), or lectin peanut agglutinin (PNA; cat. L32458; Thermo Fisher Scientific) overnight at 4°C. Following three washes in PBST, sections were incubated with secondary antibody (Thermo Fisher Scientific) for 1 hour at RT, then counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Sections were mounted with Gel Mount Aqueous Mounting Media (Biomeda, Foster City, CA, USA) and visualized under a confocal microscope (SP5; Leica Microsystems) with fixed detection gains and fixed laser powers to allow for intersection comparison.
Retinal Flat Mount
Mouse eyes were enucleated and fixed with 4% paraformaldehyde in PBS for 30 minutes at RT. After several rinses with PBS, the anterior segments were removed under a dissecting microscope to create posterior eyecups. Four radial incisions were made from the perimeter of the eyecup halfway to the optic disc to make a flat preparation. Retinas were carefully detached from the eyecups by positioning forceps between the neuroretina and the underlying layers. After three washes with 0.3% PBST, retinas were blocked with blocking buffer containing 10% normal goat serum in PBST at RT for 2 hours, followed by incubation with primary antibody to Iba-1 (cat. 019-19741; Wako Chemicals USA Corporation) in blocking buffer at 4°C over two nights. Retinas were washed with PBST and then incubated with a mixture of secondary antibody and DAPI in blocking buffer at 4°C overnight. After washes with PBST, retinas were flat-mounted on slides with inner retina upward. Based on DAPI staining, the entire thickness of the retina was imaged and confocal slices were Z-stacked together to create merged images of the retina.
Electroretinography
A Celeris Electroretinography (ERG) System (Diagnosys, Lowell, MA, USA) was used to assess retinal function. Mice were dark adapted overnight and then treated with a drop of 0.5% tropicamide to stimulate eye dilation and a drop of 0.5% proparacaine to numb the corneas. The mice were anesthetized as previously stated, and a drop of 0.3% hypromellose was used to cushion the corneal contact with the electrode/stimulators. Scotopic responses were sequentially recorded for white light stimuli of 0.01, 10, and 32 cd•s/m2 intensity. Mice were then light adapted for 10 minutes and photopic responses were recorded at 10, 32, and 100 cd•s/m2 of white light stimuli. Photopic flicker responses were then recorded using 20-cd•s/m2 flashes cycled at 9.9 Hz. During testing, body temperature was maintained at 37°C by the system's built-in heating element.
Spectral Domain Optical Coherence Tomography
A spectral domain optical coherence tomography (OCT) system (Bioptigen, Inc., Durham, NC, USA) was used to generate OCT images. Mice were anesthetized and lubricant eye drops were used to keep the cornea hydrated throughout the procedure. A volume scan of the retina on a 1.6-mm × 1.6-mm square with the optic nerve head in the center or the edge was performed on the attached and detached retinas. For the attached retinas, OCT data were analyzed by a masked reader using InVivoVue Diver 2.4 software (Bioptigen, Inc.). The thicknesses of the total retina and outer nuclear layer were measured at four points (nasal, temporal, superior, and inferior) at a distance of 500 µm from the optic nerve head (as indicated by the Bioptigen software).
Due to the angle created by retinal detachment, a novel method using ImageJ was developed to ensure accurate measurement of distance from the optic nerve head and of retinal layer thicknesses in the nonorthogonal detached portion of the retina. The retinal thickness was measured at 500 pixels away from the optic nerve head. A correction factor was applied at two separate points: first, using ImageJ, the angle of the detachment (θ) formed between the detached retina and the RPE was measured in degrees and converted to radians (θR). Cosine (θR) was calculated to generate a horizontal correction factor. This factor was multiplied by 500 pixels to obtain the distance from the optic nerve head in the orthogonal plane that corresponds to a distance of 500 pixels on the angled retina. Then, the thickness of the ONL and the retinal thickness from the inner limiting membrane (ILM) to the outer limiting membrane (OLM) were measured in the radial direction (perpendicular to the RPE layer), and a cosine (θR) correction factor was again applied to calculate the true layer thicknesses. The ratio of ONL/(ILM-OLM) was then calculated.
Statistical Analyses
The statistical analyses were performed using Prism 6 software (GraphPad Software, Inc., San Diego, CA, USA). Data were presented as the mean ± SD. Experiments with only two treatment or condition groups were analyzed using the unpaired Student's t-test. Paired Student's t-test was used to compare the changes of HMGB1 protein levels in the nuclear and cytoplasmic fractions of the RD and fellow eyes. Experiments with more than two groups were analyzed using one-way ANOVA followed by a Bonferroni post hoc correction. Significance was accepted at P < 0.05.
Results
HMGB1 Expression Pattern in the Mouse Retina
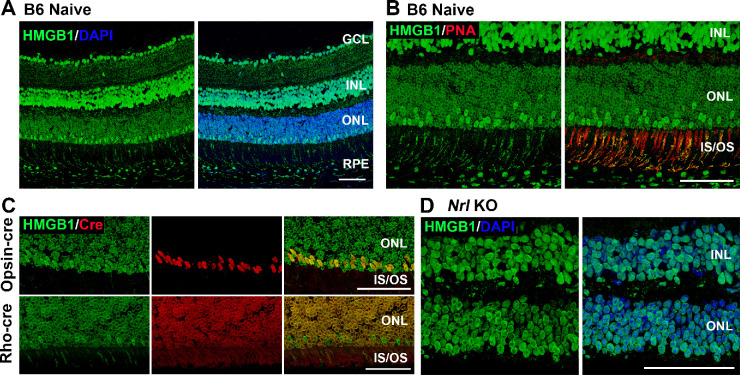
Consistent with the previous study in rat retinas by Hoppe et al.,16 HMGB1 protein was detected in the nuclei of all retinal cell types in C57BL/6 mice (Fig. 1A). HMGB1 protein was also apparent in the inner and outer segments of PRs. Whereas cells in the ganglion cell layer, the inner nuclear layer, and the retinal pigment epithelium demonstrated a relatively homogeneous nuclear HMGB1 IF pattern, the nuclei of the PR cells showed a characteristic ring-like pattern staining of the nuclear periphery (Figs. 1A, 1B). This is also consistent with the observations of Hoppe et al.16 in rat PRs. Based on an inverse colocalization with DAPI staining, transmission electron microscopy, and colocalization with acetylated histone H3, these authors concluded that HMGB1 was localized to low-density and transcriptionally active euchromatin in PR nuclei. Interestingly, we detected a subpopulation of PR cells with more intense immunostaining for HMGB1 situated along the outer edge of the ONL. Moreover, the punctuate HMGB1 IF observed in the inner and outer segments was mainly localized in the regions positive for PNA, which stains cone photoreceptor sheaths (Fig. 1B). These observations prompted us to investigate whether the nuclei with higher HMGB1 represent cone PRs. To test this, HMGB1 protein was colocalized with Cre protein expressed in M-opsin-Cre (M-cone-targeted) and rhodopsin-Cre (rod-targeted) mice. As shown in Figure 1C, the cells with brighter immunopositivity for HMGB1 consistently colocalized with the Cre-positive cells in the M-opsin-Cre mouse retinas and not in the rhodopsin-Cre mouse retina. Notably, the Cre IF staining in PRs colocalized with HMGB1 and replicated the ring-like nuclear pattern in these cells. In addition, the Nrl−/− mouse retina, which has only cone-like PRs,42 displayed homogeneously strong HMGB1 immunostaining throughout the ONL (Fig. 1D). These results suggest that HMGB1 expression is relatively low in rod PRs compared to other retinal neurons, including cone PRs.
Figure 1.
The expression pattern of HMGB1 in the mouse retina. (A) Naive retinas of 8-week-old C57BL/6J mice were probed with anti-HMGB1 antibody (Ab). Note low HMGB1 Ab IF in most of the nuclei in the ONL. (B) Naive retinas of C57BL/6 mice were probed with anti-HMGB1 Ab (green) and PNA (red). Note juxtaposition of PNA binding (indicating cone inner and outer segments) with a minority of photoreceptor nuclei with high HMGB1 Ab binding. (C) Colocalization of HMGB1 (green) and Cre (red) in the naive retinas of opsin-Cre and rhodopsin-Cre (Rho-cre) mice on a C57BL/6J background. Note the colocalization of high HMGB1 IF with opsin-Cre IF but no colocalization with rho-Cre IF. (D) Naive retinas of Nrl−/− mice (cone-like photoreceptors only) were probed with anti-HMGB1 Ab and nuclei were counterstained with DAPI. Note that HMGB1 IF is comparable in the ONL and INL. GCL, ganglion cell layer; INL, inner nuclear layer; IS/OS, inner and outer segments. Scale bars: 50 µm.
Retinal Detachment Increases HMGB1 Protein Expression in Rod PR Cells and Triggers Translocation of HMGB1 From the Nucleus
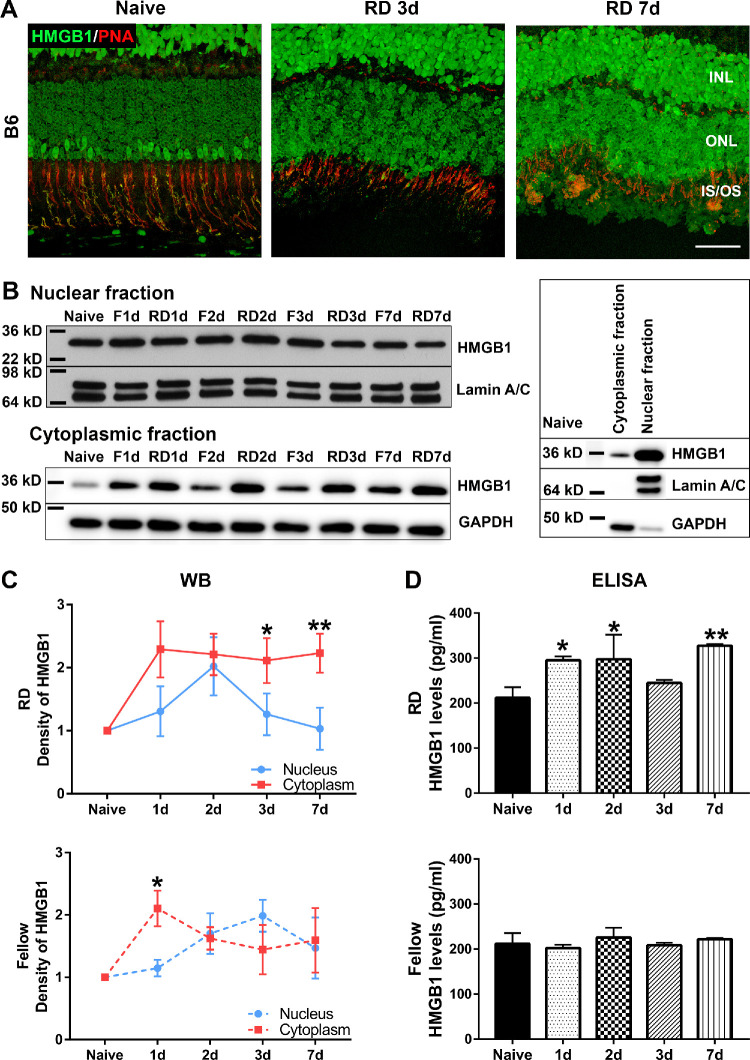
HMGB1 protein was reported to be increased in the vitreous of human eyes with RD.39 To further explore the effects of RD on HMGB1 protein expression, we injected 1% sodium hyaluronate into the subretinal space to create a detachment and assessed HMGB1 expression by IF. RD enhanced the immunopositivity for HMGB1 in rod PRs at 3 days post RD (dprd), as compared to the retinas from naive mice without RD (Fig. 2A). The apparent difference in the intensity of immunostaining between rods and cones was lost in the RD eyes. The increased HMGB1 staining in rods was maintained at 7 dprd (Fig. 2A).
Figure 2.
RD triggers the translocation and release of HMGB1. RD was created by subretinal injection of 1% hyaluronic acid in the left eyes of C57BL/6 mice. The contralateral eyes did not receive any treatment and served as fellow controls. (A) Costaining of HMGB1 Ab (green) and PNA (red) in the naive retina and detached retinas at 3 and 7 dprd. Note the relatively low HMGB1 IF in most PR nuclei and the spatial juxtaposition of high HMGB1 IF in some PR nuclei with PNA staining of cone inner and outer segments. Scale bar: 50 µm. (B–D) Mouse eyes were enucleated at 1, 2, 3, and 7 dprd. Eyecups were minced and supernatants collected for ELISA to measure the levels of soluble HMGB1. (B) Insoluble eyecup pieces underwent extraction of nuclear and cytoplasmic proteins for Western blot. Lamin A/C and glyceraldehyde 3-phosphate dehydrogenase were used as loading controls for the nuclear fraction and cytoplasmic fraction, respectively. Blots on the right side show the relative amounts of HMGB1 protein in cytoplasmic and nuclear fractions in naive retinas and the efficiency of fractionization. (C) Graphs show the quantification of protein levels based on densitometry of the Western blots (n = 3). (D) Graphs show levels of HMGB1 in the eyecup supernatant (n = 4) measured by ELISA. Data were plotted with mean ± SD. One-way ANOVA. *Indicates significant difference in comparison with naive. Statistical significance was set at P < 0.05. F, fellow.
Under nonstress conditions, HMGB1 typically resides in the nucleus. Upon injury, HMGB1 translocates into the cytoplasm and is released into the extracellular space by dying and necrotic cells. To investigate the translocation of HMGB1 after RD, we collected eyecup samples at 1, 2, 3, and 7 dprd and measured HMGB1 expression in different cellular compartments. The protein levels of HMGB1 in the nuclear and cytoplasmic fractions were evaluated by Western blot (WB), while its levels in the eyecup supernatant were determined by ELISA (Figs. 2B–2D). WBs showing the relative levels of HMGB1 protein in the nuclear and cytoplasmic fractions and the efficiency of fractionation of proteins from these two cellular compartments from naive eyecup samples are shown on the right side of Figure 2B. Most of HMGB1 is located in the nucleus, with a relatively small amount of HMGB1 in the cytoplasm. RD induced a gradual increase of HMGB1 protein in the nuclear fraction, with significant elevation detected at 2 dprd (1.75-fold increase compared with naive), followed by reduction back to baseline at 3 and 7 dprd. HMGB1 protein in the cytoplasmic fraction, however, significantly increased at 1 dprd and remained elevated out to 7 days after detachment. Furthermore, HMGB1 levels in the eyecup supernatant were significantly elevated at 1 dprd (1.4-fold), 2 dprd (1.4-fold), and 7 dprd (1.5-fold). Interestingly, HMGB1 protein levels in the fellow, nondetached eyes also showed gradual increases in the nuclear fraction, and significant increase in the cytoplasmic fraction at 1, 2, 3, and 7 dprd. The fellow eyes did not show significant changes in the levels of HMGB1 in the eyecup supernatant as detected by ELISA. Paired Student's t-test was used to compare the changes of HMGB1 levels in the nuclear and cytoplasmic fractions (Fig. 2C). In the detached retinas, RD induced significant translocation of HMGB1 from the nucleus to cytoplasm at both 3 and 7 dprd. In the fellow retinas, RD induced the significant translocation only at 1 dprd. WB analysis also showed significantly increased HMGB1 levels in the cytoplasm of fellow eyes at 1 dprd (Fig. 2C). However, the immunofluorescence analysis did not detect the marked increase in rod HMGB1 content in the fellow eyes that was observed in the detached eyes (Fig. 3A). Taken together, these data demonstrate that RD triggers an increase in HMGB1 staining in the rod PRs, coinciding with general translocation of HMGB1 from nuclei to the cytoplasm and eventual release into the extracellular space. Inexplicably, RD also causes a transient perturbation of HMGB1 expression and localization in the fellow eye.
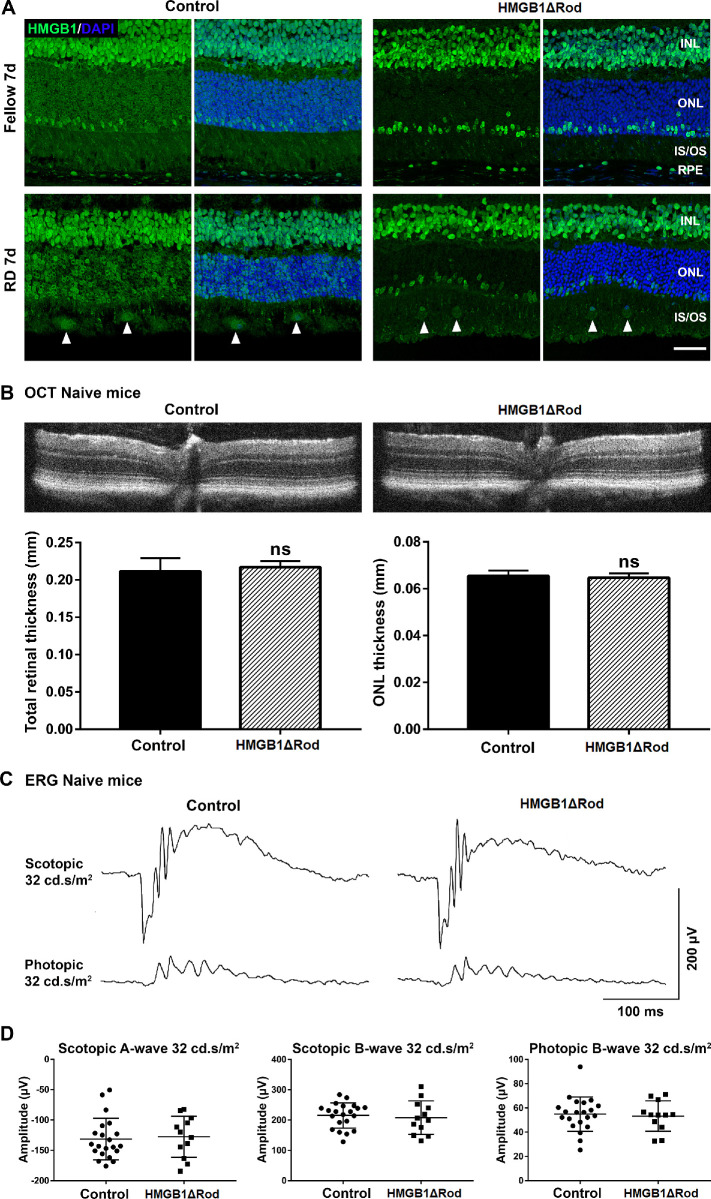
Figure 3.
Conditional knockout of HMGB1 in rod photoreceptors does not affect retinal thickness or ERG response at baseline. (A) Fellow and detached retinas at 7 dprd were probed with HMGB1 Ab to confirm the complete and specific knockout of HMGB1 in rod photoreceptors in HMGB1ΔRod mice. Rho-Cre+ mice served as controls. Nuclei were counterstained with DAPI. Arrowheads show the infiltrated cells positive for HMGB1 IF. Scale bar: 50 µm. (B–D) OCT imaging and ERG analyses were performed in naive HMGB1ΔRod and control mice at 20 weeks of age. Represented OCT images are shown in (B). The thicknesses of the total retina and ONL were measured at four points (nasal, temporal, superior, and inferior) at a distance of 500 µm from the optic nerve head. Represented ERG response traces are shown in (C). The amplitudes of scotopic a-wave and b-wave and photopic b-wave at the intensity of 32 cd•s/m2 are shown in (D). Control mice, n = 11; HMGB1ΔRod mice, n = 6. Data were plotted with mean ± SD. Paired Student's t-test. ns, not significant.
Conditional Knockout of HMGB1 in Rod PRs Does Not Affect Baseline Retinal Thickness or ERG Response
To directly investigate the functions of HMGB1 in rod PRs, the HMGB1 gene was disrupted in rods to generate HMGB1ΔRod mice. In control mice expressing rhodopsin promoter-driven Cre but lacking floxed HMGB1 alleles, HMGB1 IF was again apparent in all PRs and bright in a subset of PRs located in the outer region of the ONL (Fig. 3A). As previously observed, RD increased HMGB1 in most PRs at 7 dprd. In 8-week-old HMGB1ΔRod mice, however, HMGB1 IF was selectively lost in most PRs (rods) while bright HMGB1 staining in the subset of PRs located in the outer region of the ONL (cones) was preserved (Fig. 3A). Notably, the infiltrated cells (arrowheads in Fig. 3A) in the inner and outer segment region of the detached retinas were also immunopositive for HMGB1. These data validate the anti-HMGB1 antibody and IF methods as faithfully detecting HMGB1 protein in PRs and confirm that at baseline, cones contain considerably more HMGB1 protein than do rods.
Given the presence of HMGB1 in rods, albeit at a level lower than in cones, we sought to determine whether the loss of HMGB1 in rods affected normal rod physiology, such as retinal thickness or visual function. Retinal thickness was assessed by the OCT in vivo retinal imaging system, and visual function was determined by measuring the ERG responses under both scotopic and photopic conditions. No significant difference was detected in either the total retinal thickness or ONL thickness of naive retinas in HMGB1ΔRod mice and their controls at 20 weeks of age (Fig. 3B). We tested rod function by performing dark-adapted scotopic single-flash ERG (Figs. 3C, 3D). The average amplitudes of scotopic a-wave and b-wave responses were not significantly different in the naive retinas of the HMGB1ΔRod and the control mice. As expected, no significant difference in the amplitude of photopic b-wave response was detected in these two groups of mice. These results demonstrate that lack of HMGB1 expression in rods does not affect baseline retinal thickness, PR number, or ERG responses.
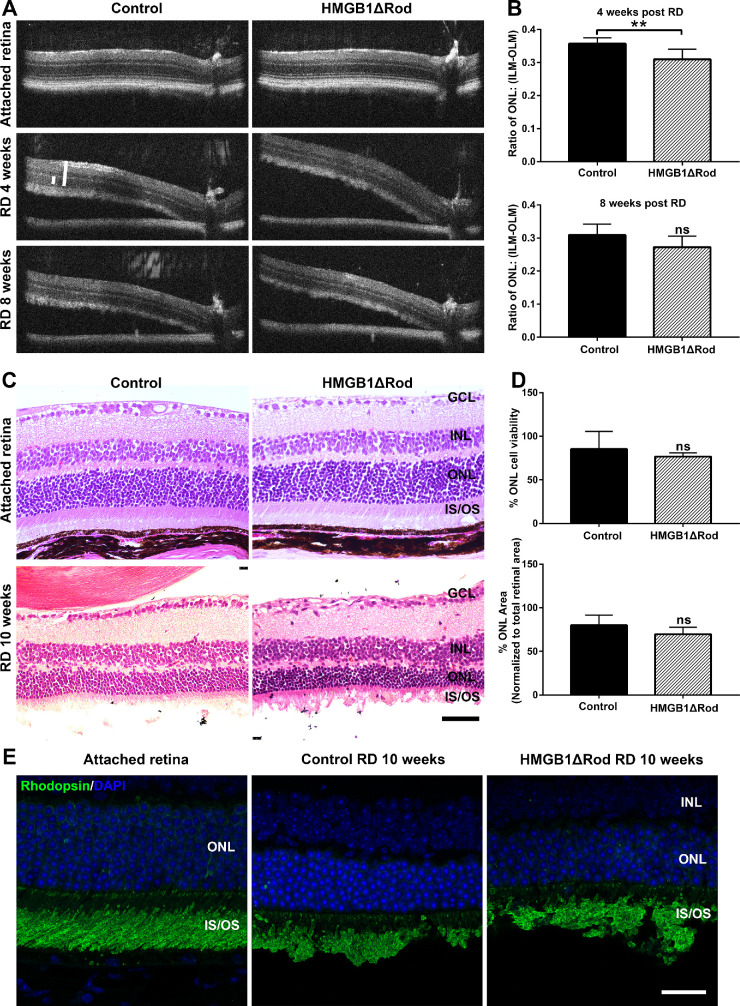
Lack of HMGB1 in Rod PRs Accelerates the Rate of Thinning of the ONL Following RD
Given that HMGB1 protein expression was apparently increased in rod PRs following RD, we hypothesized that HMGB1 plays a role in rod PR survival under this stressed condition. Because the detached region of the retina is no longer orthogonal to the OCT light path, an adjustment based upon the angle of the detached retina to the RPE was used to determine the appropriate distance away from the optic nerve head at which the OCT images should be evaluated to ensure that the retinal thicknesses were measured at equivalent retinal regions in each mouse (Fig. 4). Because the thicknesses of the ONL and the entire retina from the ILM to the OLM were measured along the OCT light path and thus at a nonorthogonal angle to the retina, the correction was also applied to adjust measured thicknesses. The normalized ONL thickness was then calculated as ONL/(ILM-OLM). As shown in Figures 4A and 4B, the normalized ONL thickness at 4 weeks following RD was significantly reduced in HMGB1ΔRod mice compared to the controls. The same set of mice was reimaged by OCT at 8 weeks post-RD. At this time point, the normalized ONL thickness in the detached HMGB1ΔRod retinas was slightly but not significantly thinner than in detached control retinas. Histology performed at 10 weeks post-RD was consistent with the OCT results (Figs. 4C, 4D). Although the HMGB1ΔRod group showed a slightly greater decrease in numbers of nuclei in the ONL and in normalized ONL area after RD than did the controls, the difference was not statistically significant. Rhodopsin IF analysis showed that accumulation of rhodopsin in outer segments was normal in the HMGB1ΔRod group (Fig. 4E), suggesting that the translocation of rhodopsin into outer segments was not dependent upon HMGB1. These results indicate that a lack of HMGB1 protein in rods accelerated retinal thinning in detached retinas and that HMGB1 plays a protective role in the early rod PR response to stress following RD.
Figure 4.
Conditional knockout of HMGB1 from rods accelerates their degeneration during RD. (A) OCT images in attached and detached retinas at 4 and 8 weeks post-RD in HMGB1ΔRod and control mice. Due to the angle created by retinal detachment, a novel method was developed to measure the thickness of the retina to ensure that corresponding points on the retinas were measured (see Materials and Methods). (B) The thickness of ONL and the thickness from the ILM to the OLM were measured at 500 pixels away from the optic nerve head using ImageJ. The ONL/(ILM-OLM) ratio was calculated and graphed. (C) Hematoxylin and eosin staining of retinal sections was performed at 10 weeks post-RD in HMGB1ΔRod and control mice. (D) The nuclei count in the ONL in the detached portion of retinal sections was performed using ImageJ. The ONL area and the area between the ILM and OLM in the detached portion of the retina were also measured. The ONL/(ILM-OLM) area ratio was calculated. Normalized data were graphed with controls set as 100%. (E) Retinal sections of HMGB1ΔRod and control mice at 10 weeks post-RD were probed with antirhodopsin Ab. Nuclei were counterstained with DAPI. Scale bars: 50 µm. **P < 0.01 (n = 5, unpaired Student's t-test). ns, not significant.
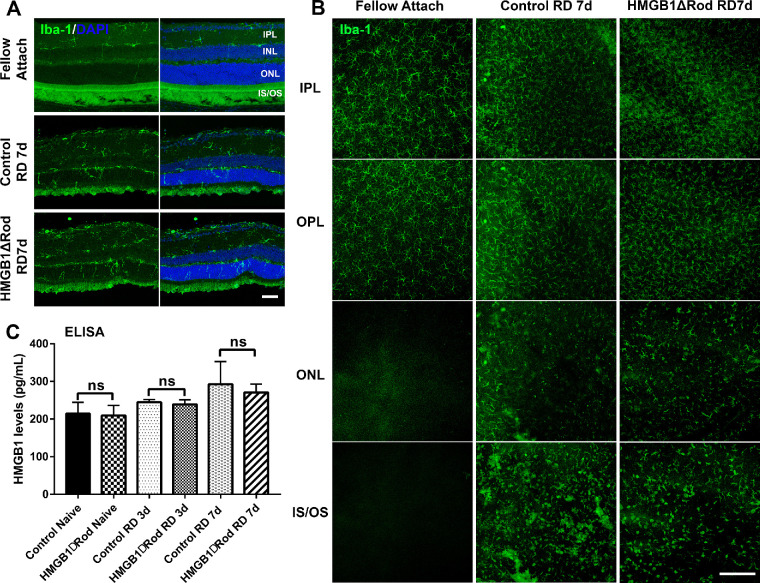
HMGB1 cKO in Rods Does Not Influence the Activation and Mobilization of Iba-1–Positive Cells to Stressed PRs After RD
Once released into the extracellular space, HMGB1 can act as an alarmin and contribute to the initiation of inflammatory response in a number of disease states.30–33 Released HMGB1 is responsible for microglial activation, as well as proinflammatory cytokine and chemokine production in several neuronal diseases.34,35 We and others previously demonstrated that microglia/macrophages are activated, proliferate, and mobilize to the ONL and subretinal space following RD.46,47 The translocation and secretion of HMGB1 into the eyecup supernatant following RD led us to hypothesize that HMGB1 plays a role in the recruitment of microglia/macrophages toward stressed PRs after RD. To test this hypothesis, we examined the effect of HMGB1 cKO in rod PRs on the morphologic response and mobilization of cells positive for Iba-1, a marker for both microglia and macrophages, at 7 dprd. Consistent with our previous results,46 RD triggered the activation, proliferation, and migration of Iba-1+ cells to the outer retina, as evidenced by morphologic changes, an increased number of Iba-1+ cells in the inner and outer plexiform layers, and the presence of Iba-1+ cells in the ONL and in proximity to PR inner and outer segments in detached retinal regions. Retinas of fellow eyes did not exhibit evidence of activation or mobilization of Iba-1+ cells. Importantly, when assessed in retinal cryosections or flat-mounted retinas, no apparent differences in the recruitment of Iba-1+ cells into the stressed PR regions were observed between HMGB1ΔRod and control mice (Figs. 5A, 5B). Given that extracellular HMGB1 is responsible for the mobilization of microglia in other neuronal diseases, we hypothesized that cKO of HMGB1 in rods may not affect the total amount of extracellular HMGB1 released by RD. We measured extracellular HMGB1 in HMGB1ΔRod and control mice following RD and found no significant difference between these two groups of mice (Fig. 5C). These results suggest that lack of HMGB1 in rod PRs did not affect the total levels of extracellular HMGB1 and that HMGB1 release from rods is not necessary for the activation and mobilization of microglia/macrophages toward the stressed PRs after RD.
Figure 5.
Conditional knockout of HMGB1 in rods does not affect microglial activation or mobilization. (A) Retinas of HMGB1ΔRod and control mice were sectioned at a 30-µm thickness and probed with anti–Iba-1 antibody to view activation and mobilization of microglia/macrophage at 7 dprd. Nuclei were counterstained with DAPI. Scale bar: 50 µm. (B) Entire retinas of HMGB1ΔRod and control mice were probed with anti–Iba-1 antibody and flat-mounted for confocal imaging at 7 dprd. Multiple confocal images were Z-stacked to create merged images of different layers of the retina based on the DAPI staining. Scale bar: 200 µm. (C) Supernatants from eyecups were collected and the levels of HMGB1 in HMGB1ΔRod and control mice at 3 and 7 dprd were measured by ELISA. One-way ANOVA (n = 4). IPL, inner plexiform layer; IS/OS, inner and outer segment; ns, not significant; OPL, outer plexiform layer.
Discussion
Retinal detachment causes PR cells to lose direct contact with the RPE, and it exposes them to liquefied vitreous that leaks into the subretinal space and forms a diffusional barrier to the flux of oxygen and nutrients coming from the RPE and the choroidal vasculature.48 The resultant hypoxia and nutrient deprivation eventually lead to PR cell death and loss of vision. Identifying factors that contribute to PR cell survival will provide valuable insights into therapies to prolong cell survival and improve visual outcomes. In this study, we examined the role of HMGB1, a multifunctional protein, in regulating PR survival after RD. We have identified the differential expression of HMGB1 protein in rods versus cones and the unique upregulation of HMGB1 protein in rods after RD. Loss of HMGB1 in rods resulted in a more rapid loss of these cells following RD, demonstrating the importance of HMGB1 in contributing to rod survival of the stress caused by detachment.
The localization of HMGB1 protein in normal and detached rat and human retinas has been previously reported by two separate groups.16,39 The apparent relatively low expression of HMGB1 in rods and high expression in cones was not noted in these studies. However, in the study by Hoppe et al.,16 the image of HMGB1 IF in a human retinal section (see Fig. 1G) shows relatively low intensity in most PRs and a group of PRs in the outer region of the ONL that exhibits higher intensity. The striking similarity to our images in the mouse retina suggests that HMGB1 expression is relatively low in rods and higher in cones of humans as well. The underlying reason for this differential expression of HMGB1 in rods versus cones is unknown but may indicate different functions for HMGB1 in these two PR cell types. The present study also found that HMGB1 IF increased in rod PRs of mice following RD. One of the aforementioned previous studies39 also showed that HMGB1 IF increased in PRs in the rat retina after RD.
In addition to increased HMGB1 IF detected in rod PRs after RD, in the detached retina, HMGB1 protein was promptly translocated into the cytoplasm and was released into the extracellular space. However, the source of HMGB1 that we detected in the supernatant of the mouse eyecups may not be derived exclusively from stressed or dying PRs. As well as being released by stressed and necrotic cells, HMGB1 is actively secreted by immune cells.20–22,24,25,49,50 We observed the presence of cells in the subretinal space that were positive for HMGB1 (Fig. 3A). Arimura et al.39 made a similar observation and declared these subretinal cells to be macrophages. Thus, it is possible that microglia and/or infiltrating leukocytes contribute to extracellular levels of HMGB1 after RD.
Interestingly, we also observed that RD in one eye affects HMGB1 expression in the fellow eye, even though no treatment was performed on the fellow eyes. HMGB1 protein expression increased after RD in the contralateral eyes but not in the extracellular space. One possible explanation is that insults in one eye may induce a systemic acute response that affects the fellow eye and subsequently induces HMGB1 translocation. Another possible explanation is that insults in one eye may propagate signals through the optic nerves and across the optic chiasm to affect the fellow eye. Hence, we recommend that naive animals without manipulation of either eye be used as a baseline control. The implications of the elevated expression and translocation of HMGB1 in the fellow eyes warrant further investigation. It should be noted that no microglial activation or migration was detected in the fellow eyes (Fig. 5).
HMGB1 was originally identified as a nuclear protein that can regulate transcription by altering the structure of DNA and facilitating the interaction of other proteins with DNA.51 Overexpression of HMGB1 is detected in many tumor cells and is necessary for cell cycle progression, presumably through granting access of certain transcriptional factors to DNA.52–54 RD triggers early alterations in retinal gene expression.46,55 We observed enhanced HMGB1 expression in rods in the detached retinas. It is therefore possible that HMGB1 upregulation contributes to the rod PR gene transcription response following RD. By conditionally disrupting hmgb1 in rods, we directly investigated the effects of loss of HMGB1 function in these photoreceptors. While the loss of HMGB1 in rods had no effect on the baseline structure and ERG response of the retina, it did result in a slightly more rapid loss of these cells during RD. Despite this more rapid loss of rod cells, the detachment-induced PR loss in HMGB1ΔRod retinas and controls eventually reached the same level, suggesting that the protective effect of HMGB1 mitigates but does not eliminate the drive to death following RD.
While the mechanism underlying the protective effect of HMGB1 on rod cells during RD remains to be elucidated, one potential mechanism is the effect of cytoplasmic HMGB1 on the autophagy pathway. Cytoplasmic HMGB1 promotes autophagy, a conserved survival pathway adopted by many cells to respond to environmental stress.17,18 HMGB1 does this by protecting autophagy regulatory proteins such as beclin1 and ATG5 from calpain-mediated cleavage.19 Our laboratory has previously demonstrated that RD triggers autophagy in PRs and that this functions to reduce PR death during RD.56 We suggest that loss of HMGB1 in rods impedes autophagy in response to RD, promoting rod PRs to enter apoptosis at a faster rate.
HMGB1 is a DAMP molecule that, when released, triggers and propagates proinflammatory responses to neuronal stress.24,50,57 Extracellular HMGB1 causes microglial activation through binding to RAGE, TLR2, or TLR435,58,59 and contributes to the neuronal deficits in the postischemic or hemorrhagic brain.36–38 We have demonstrated the RD-induced translocation of HMGB1 from the nucleus to the cytoplasm and eventual release to the extracellular space. Arimura et al.39 observed loss of HMGB1 IF in PRs with TUNEL-positive nuclei after RD in the rat retina and concluded that this indicated release of HMGB1 from apoptotic cells. These authors also found that HMGB1 levels were increased in the vitreous of patients with RD and that it correlated with the concentrations of monocyte chemoattractant protein 1, suggesting that HMGB1 plays a role in the inflammatory response to RD. Contrary to our expectation, lack of HMGB1 in rods did not seem to affect microglial activation or mobilization after RD. This may reflect that HMGB1 release is not necessary to the microglial response to RD, possibly because other DAMP signals compensate for the loss of this alarmin from the rods. However, cKO of HMGB1 from rods did not affect the extracellular HMGB1 levels, as measured in the eyecup supernatant, either at baseline or post-RD, such that a firm conclusion regarding the necessity of HMGB1 release cannot be made. Another possibility is that because cone PRs express more HMGB1 than do rods, cones may be responsible for the release of HMGB1 into the extracellular space to attract microglia toward the stressed PR layer. It has been shown that blue light–induced retinal injury in R91W;Nrl−/– mice (all-cone retina) induces a more pronounced microglial/macrophage migration/infiltration and more cell death than in wild-type controls,60 which would be consistent with this explanation.
In conclusion, we demonstrate that HMGB1 protein is expressed in all the neurons of the retina and that there is a differential expression of this factor in PR cells, with cones expressing higher levels than rods in the naive retinas. Separation of the neuroretina from the underlying RPE stimulates rod PRs to express more HMGB1 protein and also triggers the translocation of HMGB1 protein from the nucleus to the cytoplasm and eventually to the extracellular space. Our data suggest that HMGB1 plays a protective role in rod cells and that this effect may be cell autonomous. The molecular mechanisms underlying this effect are yet to be defined.
Acknowledgments
Supported by NIH 4T32 EY013934-15 (BXR), R01 EY020823 (DNZ, SFA), R01-EY029349 (SFA), and P30 EY007003 (Kellogg Eye Center Core Center for Vision Research).
Disclosure: B.X. Ross, None; J. Choi, None; J. Yao, None; H.M. Hager, None; S.F. Abcouwer, None; D.N. Zacks, None
References
- 1. Piccolino FC, de la Longrais RR, Ravera G, et al.. The foveal photoreceptor layer and visual acuity loss in central serous chorioretinopathy. Am J Ophthalmol. 2005; 139: 87–99. [DOI] [PubMed] [Google Scholar]
- 2. Hisatomi T, Sakamoto T, Murata T, et al.. Relocalization of apoptosis-inducing factor in photoreceptor apoptosis induced by retinal detachment in vivo. Am J Pathol. 2001; 158: 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zacks DN, Boehlke C, Richards AL, Zheng QD. Role of the Fas-signaling pathway in photoreceptor neuroprotection. Arch Ophthalmol. 2007; 125: 1389–1395. [DOI] [PubMed] [Google Scholar]
- 4. Besirli CG, Chinskey ND, Zheng QD, Zacks DN. Autophagy activation in the injured photoreceptor inhibits Fas-mediated apoptosis. Invest Ophthalmol Vis Sci. 2011; 52: 4193–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chong DY, Boehlke CS, Zheng QD, Zhang L, Han Y, Zacks DN. Interleukin-6 as a photoreceptor neuroprotectant in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci. 2008; 49: 3193–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chinskey ND, Zheng QD, Zacks DN. Control of photoreceptor autophagy after retinal detachment: the switch from survival to death. Invest Ophthalmol Vis Sci. 2014; 55: 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pawar M, Busov B, Chandrasekhar A, Yao J, Zacks DN, Besirli CG. FAS apoptotic inhibitory molecule 2 is a stress-induced intrinsic neuroprotective factor in the retina. Cell Death Differ. 2017; 24: 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bustin M, Hopkins RB, Isenberg I. Immunological relatedness of high mobility group chromosomal proteins from calf thymus. J Biol Chem. 1978; 253: 1694–1699. [PubMed] [Google Scholar]
- 9. Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME. GR and HMGB1 interact only within chromatin and influence each other's residence time. Mol Cell. 2005; 18: 109–121. [DOI] [PubMed] [Google Scholar]
- 10. Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukocyte Biol. 2007; 81: 1–5. [DOI] [PubMed] [Google Scholar]
- 11. Stros M, Polanska E, Kucirek M, Pospisilova S. Histone H1 differentially inhibits DNA bending by reduced and oxidized HMGB1 protein. PLoS One. 2015; 10: e0138774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blair RH, Horn AE, Pazhani Y, Grado L, Goodrich JA, Kugel JF. The HMGB1 C-terminal tail regulates DNA bending. J Mol Biol. 2016; 428: 4060–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balliano A, Hao F, Njeri C, Balakrishnan L, Hayes JJ. HMGB1 stimulates activity of polymerase beta on nucleosome substrates. Biochemistry. 2017; 56: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mitsouras K, Wong B, Arayata C, Johnson RC, Carey M. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol Cell Biol. 2002; 22: 4390–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci. 2001; 26: 167–174. [DOI] [PubMed] [Google Scholar]
- 16. Hoppe G, Rayborn ME, Sears JE. Diurnal rhythm of the chromatin protein Hmgb1 in rat photoreceptors is under circadian regulation. J Comp Neurol. 2007; 501: 219–230. [DOI] [PubMed] [Google Scholar]
- 17. Tang D, Kang R, Livesey KM, et al.. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010; 190: 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang D, Kang R, Livesey KM, Zeh HJ III, Lotze MT. High mobility group box 1 (HMGB1) activates an autophagic response to oxidative stress. Antioxid Redox Signal. 2011; 15: 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu X, Messer JS, Wang Y, et al.. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J Clin Invest. 2015; 125: 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Bloom O, Zhang M, et al.. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999; 285: 248–251. [DOI] [PubMed] [Google Scholar]
- 21. Bonaldi T, Talamo F, Scaffidi P, et al.. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003; 22: 5551–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu B, Nakamura T, Inouye K, et al.. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012; 488: 670–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu B, Antoine DJ, Kwan K, et al.. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc Natl Acad Sci USA. 2014; 111: 3068–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002; 418: 191–195. [DOI] [PubMed] [Google Scholar]
- 25. Rovere-Querini P, Capobianco A, Scaffidi P, et al.. HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep. 2004; 5: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park JS, Svetkauskaite D, He Q, et al.. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004; 279: 7370–7377. [DOI] [PubMed] [Google Scholar]
- 27. Taguchi A, Blood DC, del Toro G, et al.. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000; 405: 354–360. [DOI] [PubMed] [Google Scholar]
- 28. Stern D, Yan SD, Yan SF, Schmidt AM. Receptor for advanced glycation endproducts: a multiligand receptor magnifying cell stress in diverse pathologic settings. Adv Drug Deliv Rev. 2002; 54: 1615–1625. [DOI] [PubMed] [Google Scholar]
- 29. van Beijnum JR, Buurman WA, Griffioen AW. Convergence and amplification of toll-like receptor (TLR) and receptor for advanced glycation end products (RAGE) signaling pathways via high mobility group B1 (HMGB1). Angiogenesis. 2008; 11: 91–99. [DOI] [PubMed] [Google Scholar]
- 30. Jiang G, Wang Y, Yun J, et al.. HMGB1 release triggered by the interaction of live retinal cells and uveitogenic T cells is Fas/FasL activation-dependent. J Neuroinflammation. 2015; 12: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santos AR, Dvoriantchikova G, Li Y, et al.. Cellular mechanisms of high mobility group 1 (HMGB-1) protein action in the diabetic retinopathy. PLoS One. 2014; 9: e87574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abu El-Asrar AM, Mohammad G, Nawaz MI, Siddiquei MM. High-mobility group box-1 modulates the expression of inflammatory and angiogenic signaling pathways in diabetic retina. Curr Eye Res. 2015; 40: 1141–1152. [DOI] [PubMed] [Google Scholar]
- 33. Abu El-Asrar AM, Siddiquei MM, Nawaz MI, Geboes K, Mohammad G. The proinflammatory cytokine high-mobility group box-1 mediates retinal neuropathy induced by diabetes. Mediators Inflamm. 2014; 2014: 746415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim JB, Sig Choi J, Yu YM, et al.. HMGB1, a novel cytokine-like mediator linking acute neuronal death and delayed neuroinflammation in the postischemic brain. J Neurosci. 2006; 26: 6413–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takizawa T, Shibata M, Kayama Y, et al.. High-mobility group box 1 is an important mediator of microglial activation induced by cortical spreading depression. J Cereb Blood Flow Metab. 2017; 37: 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim SW, Jin Y, Shin JH, et al.. Glycyrrhizic acid affords robust neuroprotection in the postischemic brain via anti-inflammatory effect by inhibiting HMGB1 phosphorylation and secretion. Neurobiol Dis. 2012; 46: 147–156. [DOI] [PubMed] [Google Scholar]
- 37. Haruma J, Teshigawara K, Hishikawa T, et al.. Anti-high mobility group box-1 (HMGB1) antibody attenuates delayed cerebral vasospasm and brain injury after subarachnoid hemorrhage in rats. Sci Rep. 2016; 6: 37755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang D, Liu K, Wake H, Teshigawara K, Mori S, Nishibori M. Anti-high mobility group box-1 (HMGB1) antibody inhibits hemorrhage-induced brain injury and improved neurological deficits in rats. Sci Rep. 2017; 7: 46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arimura N, Ki-i Y, Hashiguchi T, et al.. Intraocular expression and release of high-mobility group box 1 protein in retinal detachment. Lab Invest. 2009; 89: 278–289. [DOI] [PubMed] [Google Scholar]
- 40. Huang H, Nace GW, McDonald KA, et al.. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: a role for intracellular high-mobility group box 1 in cellular protection. Hepatology. 2014; 59: 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li S, Chen D, Sauve Y, McCandless J, Chen YJ, Chen CK. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis. 2005; 41: 73–80. [DOI] [PubMed] [Google Scholar]
- 42. Mears AJ, Kondo M, Swain PK, et al.. Nrl is required for rod photoreceptor development. Nat Genet. 2001; 29: 447–452. [DOI] [PubMed] [Google Scholar]
- 43. Zacks DN, Hanninen V, Pantcheva M, Ezra E, Grosskreutz C, Miller JW. Caspase activation in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci. 2003; 44: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 44. Murakami Y, Matsumoto H, Roh M, et al.. Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsRNA-induced retinal degeneration. Cell Death Differ. 2014; 21: 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Besirli CG, Chinskey ND, Zheng QD, Zacks DN. Inhibition of retinal detachment-induced apoptosis in photoreceptors by a small peptide inhibitor of the fas receptor. Invest Ophthalmol Vis Sci. 2010; 51: 2177–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kiang L, Ross BX, Yao J, et al.. Vitreous cytokine expression and a murine model suggest a key role of microglia in the inflammatory response to retinal detachment. Invest Ophthalmol Vis Sci. 2018; 59: 3767–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakazawa T, Hisatomi T, Nakazawa C, et al.. Monocyte chemoattractant protein 1 mediates retinal detachment-induced photoreceptor apoptosis. Proc Natl Acad Sci U S A. 2007; 104: 2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S, Linsenmeier RA.. Hyperoxia improves oxygen consumption in the detached feline retina. Invest Ophthalmol Vis Sci. 2007; 48: 1335–1341. [DOI] [PubMed] [Google Scholar]
- 49. Gardella S, Andrei C, Ferrera D, et al.. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002; 3: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012; 8: 195–202. [DOI] [PubMed] [Google Scholar]
- 51. Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005; 5: 331–342. [DOI] [PubMed] [Google Scholar]
- 52. Alexandrova EA, Beltchev BG.. Differences between HMG1 proteins isolated from normal and tumour cells. Biochim Biophys Acta. 1987; 915: 399–405. [DOI] [PubMed] [Google Scholar]
- 53. Kohlstaedt LA, Cole RD.. Effect of pH on interactions between DNA and high-mobility group protein HMG1. Biochemistry. 1994; 33: 12702–12707. [DOI] [PubMed] [Google Scholar]
- 54. Kohlstaedt LA, Cole RD. Specific interaction between H1 histone and high mobility protein HMG1. Biochemistry. 1994; 33: 570–575. [DOI] [PubMed] [Google Scholar]
- 55. Hollborn M, Francke M, Iandiev I, et al.. Early activation of inflammation- and immune response-related genes after experimental detachment of the porcine retina. Invest Ophthalmol Vis Sci. 2008; 49: 1262–1273. [DOI] [PubMed] [Google Scholar]
- 56. Shelby SJ, Angadi PS, Zheng QD, Yao J, Jia L, Zacks DN. Hypoxia inducible factor 1alpha contributes to regulation of autophagy in retinal detachment. Exp Eye Res. 2015; 137: 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Abraham E, Arcaroli J, Carmody A, Wang H, Tracey KJ. HMG-1 as a mediator of acute lung inflammation. J Immunol. 2000; 165: 2950–2954. [DOI] [PubMed] [Google Scholar]
- 58. Maroso M, Balosso S, Ravizza T, et al.. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat Med. 2010; 16: 413–419. [DOI] [PubMed] [Google Scholar]
- 59. Mohammad G, Siddiquei MM, Othman A, Al-Shabrawey M, Abu El-Asrar AM. High-mobility group box-1 protein activates inflammatory signaling pathway components and disrupts retinal vascular-barrier in the diabetic retina. Exp Eye Res. 2013; 107: 101–109. [DOI] [PubMed] [Google Scholar]
- 60. Geiger P, Barben M, Grimm C, Samardzija M. Blue light-induced retinal lesions, intraretinal vascular leakage and edema formation in the all-cone mouse retina. Cell Death Dis. 2015;6: e1985. [DOI] [PMC free article] [PubMed] [Google Scholar]