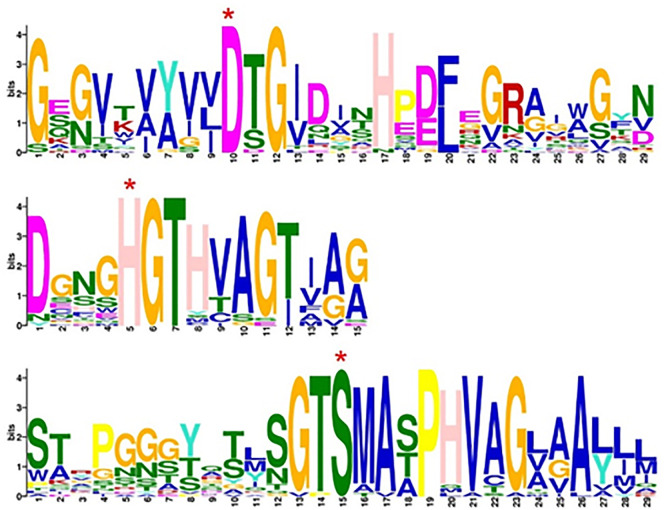Abstract
Keratin is an insoluble and protein-rich epidermal material found in e.g. feather, wool, hair. It is produced in substantial amounts as co-product from poultry processing plants and pig slaughterhouses. Keratin is packed by disulfide bonds and hydrogen bonds. Based on the secondary structure, keratin can be classified into α-keratin and β-keratin. Keratinases (EC 3.4.-.- peptide hydrolases) have major potential to degrade keratin for sustainable recycling of the protein and amino acids. Currently, the known keratinolytic enzymes belong to at least 14 different protease families: S1, S8, S9, S10, S16, M3, M4, M14, M16, M28, M32, M36, M38, M55 (MEROPS database). The various keratinolytic enzymes act via endo-attack (proteases in families S1, S8, S16, M4, M16, M36), exo-attack (proteases in families S9, S10, M14, M28, M38, M55) or by action only on oligopeptides (proteases in families M3, M32), respectively. Other enzymes, particularly disulfide reductases, also play a key role in keratin degradation as they catalyze the breakage of disulfide bonds for better keratinase catalysis. This review aims to contribute an overview of keratin biomass as an enzyme substrate and a systematic analysis of currently sequenced keratinolytic enzymes and their classification and reaction mechanisms. We also summarize and discuss keratinase assays, available keratinase structures and finally examine the available data on uses of keratinases in practical biorefinery protein upcycling applications.
Keywords: α-Keratin, β-Keratin, Keratinases, Keratinase assay, Keratinase classification, Keratinolytic mechanisms, Keratinase crystal structures, Biocatalysis, Protein recycling
1. Introduction
Livestock production is increasing rapidly throughout the world as a result of population growth, increasing incomes, changes in lifestyles and dietary habits. According to the UN Food and Agriculture Organization (FAO), annual global poultry production is projected to reach more than 24.8 billion animals in 2030 and 37.0 billion in 2050 (1.7 kg carcass weight/animal), and bovine and pork meat production also continues to increase (Alexandratos and Bruinsma, 2012). Although the global coronavirus (COVID-19) pandemic in 2020 has caused the United States Department of Agriculture to decrease their global animal protein prognosis in the spring of 2020, the forecast for 2020 for global chicken meat production is still 100.5 million tons, and the global pork production is projected to be 94.3 million tons (United States Department of Agriculture, 2020). The waste or co-products from such animal meat production consists of keratinous materials such as chicken feathers, pig bristles, wool and horns and millions of tons of these co-products are produced each year. For instance, it has been estimated that in 2019 alone more than 4.7 million tons of chicken feathers were produced from poultry processing (Li et al., 2020). Chicken feathers and other keratinous co-products from poultry processing plants and abattoirs are classified in category 3 animal byproducts which entails that they are low-risk materials for animals, the public, and the environment (Verma et al., 2017). They can therefore be considered as an abundant protein or amino acid source for new upcycling processes targeting potential use in e.g. feed, fertilizers, cosmetics, and other applications (Callegaro et al., 2019). Keratinous materials are unique because they are rich in certain amino acids, including in particular the sulfur-containing amino acid, cysteine, other amino acids like glycine, proline, arginine, and the essential amino acids valine, leucine, and threonine. The compact conformation and high stability of keratin is indeed mainly due to disulfide bridges formed among cysteine residues within and between keratin polypeptides (Callegaro et al., 2019). Since keratin is an insoluble protein, its reutilization will have to include partial or complete degradation, without destroying the amino acids, to provide useful biorefinery options for the constituent protein and amino acids.
Keratin is classified as α-keratin and β-keratin according to its secondary structure. The α-keratin is primarily present in so-called mammalian epidermal materials, such as hair, wool and horn, whilst β-keratin is mainly found in birds and reptiles, i.e. in chicken feathers and reptile scales. Dissolution and extraction of keratin is a difficult process compared with extraction of other natural polymers (e.g. chitosan, starch, and collagen). Large-scale use of keratin therefore depends on employing relatively fast and cost-effective extraction-decomposition methods. Present extraction strategies include physical, chemical, physical-chemical and biological methods (Shavandi et al., 2017). However, production of keratin hydrolysates by chemical or physical treatment usually involves use of elevated temperatures and therefore results in degradation of heat sensitive amino acids, such as methionine, lysine and tryptophan, which leads to reduction in the nutritional value of the hydrolysates and lowers the value (Martinez et al., 2020). An alternative approach is to exploit keratinolytic microbes or – maybe better - specific microbial keratinases that are able to catalyze the biodegradation of keratin.
Keratinases (EC 3.4.-.-) are a group of hydrolytic enzymes that can catalyze the degradation of keratin. These keratinolytic enzymes are secreted by different types of microorganisms found in soil, water and on various keratin-rich sources. Keratinases come from bacteria such as Bacillus licheniformis, B. subtilis and Stenotrophomonas maltophilia. Moreover, the actinobacteria Streptomyces albidoflavus and Streptomyces fradiae also secrete keratinases. Fungal keratinases are mainly from Trichophyton rubrum and Microsporum canis. Many expert reviews give details of the sources of keratinolytic organisms, optimal fermentation conditions for producing keratinases, keratinase assays and also keratinase characteristics (Intagun and Kanoksilapatham, 2017; Verma et al., 2017; Martinez et al., 2020). According to the classification of known keratinases in the protease family database (MEROPS), keratinases belong to the serine- and metalloprotease families. The major keratinase family is the S8 family, and the subtilisin subfamily members in particular have keratinolytic activity. Recently, an increasing number of other protease families are also reported to harbor enzymes having keratinolytic activity. For example, family M28 and M3 proteases secreted from the non-pathogenic fungus Onygena corvina have been reported to catalyze degradation of pig bristle keratin, and moreover to act synergistically with enzymes from the S8 family (Huang et al., 2015). In addition, and as discussed later, several other fungal proteases from the M36, S9 and S10 families have been reported to exhibit keratin degrading action (Mercer and Stewart, 2019). Thus, the efficiency of enzymatic keratin degradation may be improved by designing specific blends of keratinases originating from different protease families. Additionally, other proteases, such as disulfide reductase (e.g. cysteine dioxygenase (Kasperova et al., 2013), thioredoxin-disulfide reductase), and other types of enzymes, e.g. lytic polysaccharide monooxygenases (LPMOs) (Lange et al., 2016) and certain enzymes involved in lipoprotein signaling or fatty acid degradation, have also been suggested as possible candidates for contributing to biocatalytic keratin degradation (Lee et al., 2015b). Although microbial and enzymatic keratin degradation has been a subject of research interest for some time, currently available overviews summarize keratinase data, but rarely discuss the enzymatic action details in relation to substrate structure and assessment method. Also, an updated analysis of the protease family classification of keratin-degrading enzymes, possible activity synergies, and a discussion of other enzymes that may play a role in keratin degradation is not available.
The present review aims to provide an overview of the current knowledge of keratin as a substrate for enzymatic attack in relation to enzyme-assisted upcycling of keratin. In addition to summarizing the essential knowledge of keratin, the review provides a systematic investigation of the currently known (sequenced) keratinolytic enzymes and their classification, an analysis of the available keratinase structures, and insight into the structural-functional traits of these enzymes. Also included is a critical discussion of keratinase enzyme assays and a summary of other enzymes that have been suggested to contribute to keratin degradation. Our goal is to provide a better understanding of the roles and functions of keratinases in nature, and help generate a better foundation for using keratin-degrading enzymes in keratin recycling processes.
2. Essentials of keratin biomass as substrate for keratinolytic enzymes
Keratin biomass is among the toughest biological materials known and constitutes the bulk of epidermal appendages such as hair, nails, claws, turtle scutes, horns, beaks and feathers (Wang et al., 2016). As mentioned above, slaughterhouses produce millions of tons of keratin-containing biomass as co-product every year (Sharma and Gupta, 2016; Li et al., 2020). Keratin biomass is extremely rich in protein; for example, ~90% of feather dry matter is protein (Ben Hamad Bouhamed and Kechaou, 2017), while wool contains up to 95% by weight of pure keratin (Eslahi et al., 2013).
Beyond the high cysteine levels (more than 5%), that contribute disulfide bonds which cross-link the protein chains, keratin biomass generally contains high amounts of arginine, serine, proline, valine, leucine, threonine, glutamate, glycine and aspartate, but quite low levels of histidine, lysine, and methionine (Table 1 ). So-called stiff keratinous materials (mainly β-keratin), e.g. claws, beaks, turtle scutes (or shells), notably contains extremely high levels of glycine (>28%) (Table 1). Glycine imparts hydrophobicity, stiffness, and resistance to degradation. Mainly because of the high cysteine and glycine levels, keratin is extremely tough and recalcitrant to biological degradation and has low solubility in water compared with most other proteins. In particular, cysteine at the N- and C-terminals allows intermolecular bonding for hardening, e.g. as is the case in corneous horn material (Fraser and Parry, 2011, Fraser and Parry, 2014).
Table 1.
Amino acid (AA) composition (%) of various keratin sources.
| Amino acids | Mammalian keratin (α-keratin) |
Avian and reptilian keratin (β-keratin) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wool | Human hair | Human nails | Bull horns | Hog hair | Chicken feathers | Penguin feathers (Rachis)a | Penguin feathers (Barbs)a | Lizard claws | Avian beaksb | Avian clawsc | Turtle scutese | |
| Non-polar amino acid | ||||||||||||
| Ala | 3.3 | 5.2 | 6.5 | 7.1 | 4.2 | 8.7 | 5.6 | 4.5 | 4.8 | 6.3 | 6.1 | 3.0 |
| Gly | 4.6 | 6.6 | 6.6 | 11.6 | 4.5 | 13.7 | 10.4 | 8.2 | 28.2 | 31.3 | 30.8 | 30.3 |
| Ile | 3.2 | 3.1 | 3.9 | 4.6 | 3.8 | 3.2 | 5.1 | 5.7 | 2.4 | 1.7 | 1.7 | 1.8 |
| Leu | 7.2 | 7.1 | 9.9 | 9.3 | 8.2 | 8.3 | 12.1 | 9.6 | 3.6 | 7.3 | 5.8 | 8.1 |
| Pro | 5.7 | 9.6 | 4.1 | 2.9 | 5.9 | 9.8 | 11.3 | 12.4 | 9.1 | 9.6 | 9.4 | 10.1 |
| Val | 5.1 | 6.2 | 5.9 | 5.7 | 5.9 | 7.8 | 8.6 | 10.1 | 4.9 | 4.5 | 4.7 | 6.7 |
| Phe | 3.6 | 1.9 | 2.2 | 3.4 | 3.1 | 3.1 | 3.4 | 2.9 | 2.3 | 3.5 | 3.6 | 2.3 |
| Tyr | 5.9 | 1.2 | 2.8 | 2.0 | 3.4 | 1.4 | 4.6 | 4.7 | 4.1 | 8.6 | 9.0 | 12.7 |
| Total | 38.6 | 40.9 | 41.9 | 46.6 | 39.0 | 56.0 | 61.1 | 58.1 | 59.4 | 72.8 | 71.1 | 75.0 |
| Acidic/basic and polar amino acid | ||||||||||||
| Asp | 7.0 | 7.6 | 8.9 | 9.8 | 7.2 | 5.6 | 6.6 | 8.4 | 4.8 | 4.2 | 4.1 | 2.3 |
| Glu | 13.8 | 11.6 | 15.2 | 15.8 | 15.5 | 6.9 | 8.2 | 10.4 | 3.4 | 4.2 | 3.8 | 4.0 |
| Arg | 10.2 | 4.9 | 6.7 | 4.5 | 9.6 | 3.8 | 5.6 | 5.8 | 3.2 | 3.5 | 3.4 | 4.5 |
| His | 1.3 | 1.1 | 0.9 | 1.6 | 1.3 | 0.2 | 1.1 | 0.5 | 2.3 | 1.1 | 1.1 | 1.3 |
| Lys | 3.0 | 3.0 | 4.5 | 3.7 | 3.9 | 0.6 | 0.7 | 1.2 | 1.1 | 0.2 | 0.3 | 0.3 |
| Total | 35.3 | 28.2 | 36.2 | 35.4 | 37.5 | 17.1 | 22.2 | 26.3 | 14.8 | 13.2 | 12.7 | 12.4 |
| Neutral and polar amino acid | ||||||||||||
| Ser | 8.5 | 11.7 | 8.7 | 7.2 | 4.9 | 14.1 | 8.3 | 7.5 | 7.5 | 6.7 | 7.1 | 5.6 |
| Thr | 6.0 | 6.9 | 5.0 | 5.4 | 4.8 | 4.1 | 4.9 | 5.4 | 3.4 | 3.6 | 3.8 | 1.6 |
| Cys | 10.8 | 10.0 | 7.4 | 5.9 | 8.7 | 7.8 | 8.6 | 10.6 | 13.0 | 3.8 | 4.9d | 5.5d |
| Met | 0.6 | 2.5 | 0.8 | 0.5 | 0.6 | 0.1 | 1.0 | 1.7 | 0.8 | 0.8 | 0.5 | 0.1 |
| Total | 25.9 | 31.1 | 21.9 | 19.0 | 19.0 | 26.1 | 22.8 | 25.2 | 24.7 | 14.9 | 16.3 | 12.8 |
| Ref. | (Zoccola et al., 2009) | (Hill et al., 2010) | (Edwards et al., 1998) | (Edwards et al., 1998) | (Esteban et al., 2010) | (Harrap and Woods, 1964) | (Murphy et al., 1990) | (Murphy et al., 1990) | (Gillespie et al., 1982) | (Frenkel and Gillespie, 1976) | (Frenkel and Gillespie, 1976) | (Frenkel and Gillespie, 1976) |
Data from penguin Pygoscelis antarctica, and calculated as μmol/g to composition (%) (w/w) according to the molar mass of amino acids.
Kookaburra beak keratin.
Zinc-precipitable protein isolated from Avian claw (Kookaburra claw).
Content of cysteine, determined as S-carboxymethyl cysteine.
Zinc-precipitable protein isolated from turtle scute (Chelonia mydas).
Based on sulfide content, or cysteine content, a classification of keratin-rich biomass into soft and hard keratin has been proposed. Current reports are conflicting with regard to the criteria for this classification, especially regarding classification of skin. Some authors classify skin as hard keratinous material (Irwin McLean and Moore, 2011; Korniłłowicz-Kowalska and Bohacz, 2011), while others classify skin as soft keratin based on that skin keratin only contains 2–3% cysteine (Safranek and Goos, 1981; Shavandi et al., 2017). However, there is no doubt that feathers, claws, beaks, wool, and hair are all considered as hard keratinous biomass.
Keratin is also classified into α-keratin and β-keratin based on the secondary protein structure. In α-keratin, the α-helical-coils type I (acidic) and type II (basic/neutral) protein chains are coiled together to form elongated α-helix filaments that form fibrils by interchain bonding (Fig. 1, Fig. 2 ), while β-keratin is mainly built of β-sheets (Fig. 3 ). As indicated in Table 1, α-keratin mainly occurs in mammals, while β-keratin is the major component of avian and reptilian tissues. In fact, β-keratin is found only in avian and reptilian epidermis (though one mammal, the pangolin, a unique scaly anteater, has been reported to have both α- and β-keratin in the keratin scales that cover its skin (Wang et al., 2016)), whereas α-keratin is found in mammalian as well as in avian and reptilian keratin (Alibardi, 2003; Greenwold et al., 2014; Ng and Li, 2018). The α-keratin found in the epidermis located between the scales in reptiles and in feathers is responsible for the mechanical strength of the epithelial cells, their adhesiveness, and the changes in shape when stretched (Alibardi, 2007; Skieresz-Szewczyk et al., 2017). By contrast, the β-keratin in reptile scales exhibits limited extensibility yet significant microbiological resistance and hydrophobicity, and serves a protective function in nature (Calvaresi et al., 2016). In addition, some believe that α- and β-keratins are completely unrelated evolutionarily, and it has even been proposed recently that β-keratins should be renamed as corneous β-proteins (Ng and Li, 2018).
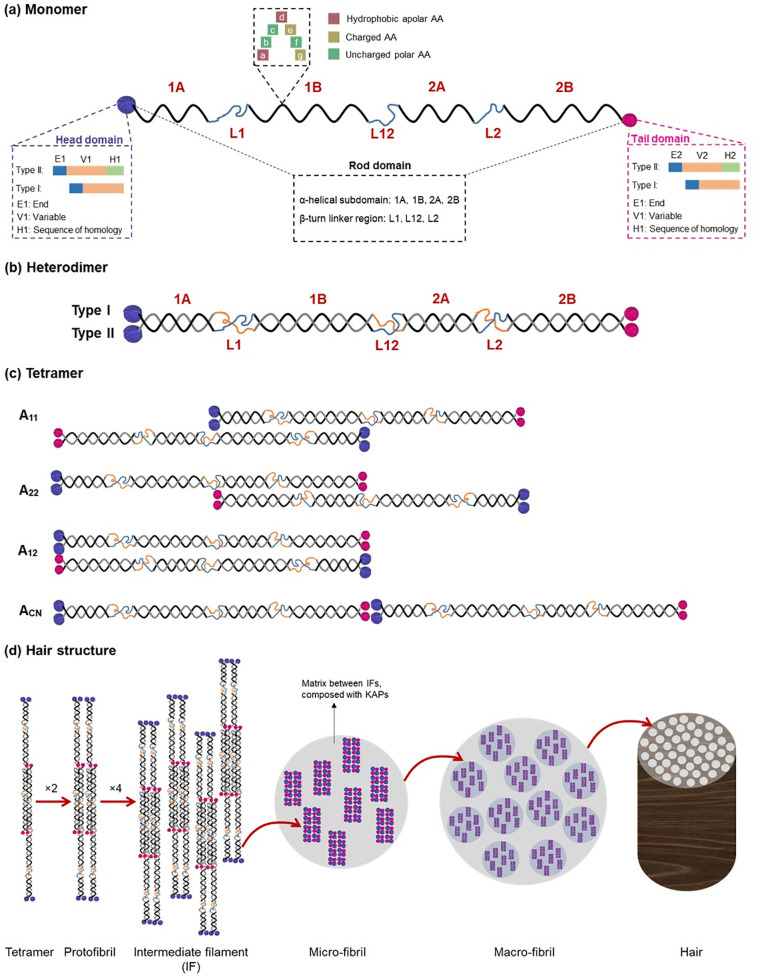
Fig. 1.
α-keratin secondary structure. (a) Monomer structure made up of three parts: head domain (N-terminal domain), rod domain (α-helical domain) and tail domain (C-terminal domain). In addition, the repeating heptad pattern in α-helix is labeled from “a-g” shown in different colors; (b) Heterodimer structure formed from two monomers (one type I keratin monomer and one type II keratin monomer) in parallel alignment; (c) Two heterodimers assembled into a tetramer in four different ways: A11, A22, A12, ACN (Chou and Buehler, 2012); (d) The composition of hair: two tetramers associate into a protofibril, four protofibrils combine into intermediate filament (IF). The IFs are surrounded by sulfide-rich keratin-associated proteins (KAPs).
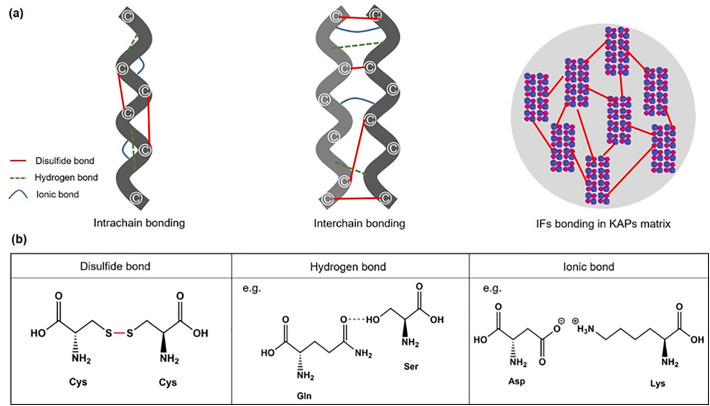
Fig. 2.
Bonds in keratin structures. (a) Intrachain bonds, interchain bonds and intermediate filaments (IFs) bonding in keratin-associated proteins (KAPs) matrix; (b) The formation of disulfide bonds (cysteine-cysteine), hydrogen bonds (e.g. glutamine-serine) and ionic bonds (e.g. aspartic acid-lysine) in protein.
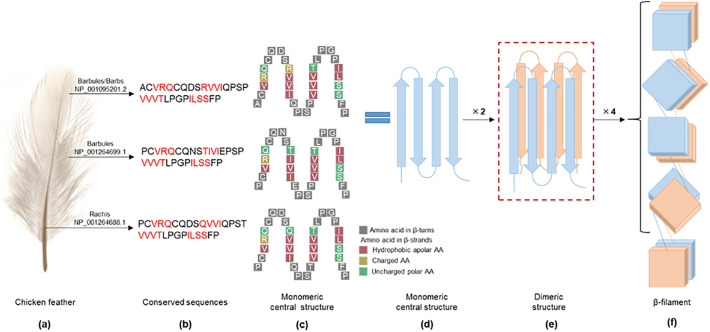
Fig. 3.
Feather structure and secondary β-keratin structure. (a) Feather structure is shown as barbules/barbs, barbules, rachis; (b) Conserved amino acids of the barbules/barbs (NCBI accession number: NP_001095201.2), barbules (NP_001264699.1), feather rachis (NP_001264688.1) (Jin et al., 2017) with the β-strands in each sequence indicated in red; (c) The monomeric central structure of β-keratin in feathers. The structure is based on the amino compositions of the filament framework segments in avian feather keratin (Fraser and Parry, 2008); (d) The central monomeric structure of β-keratin, where the four strands are usually antiparallel; (e) Dimeric structure (β-sandwich) of β-keratin; (f) The basic repeating structure of the β-filaments, with the four dimers rotated by approx. 45° to form a helix structure, with the fifth dimer at the bottom assuming an inverted sandwich position compared to the first dimer (Calvaresi et al., 2016). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.1. α-keratin
As regards α-keratin, the monomer molecular structure contains three parts – a head domain (N-terminal domain), a rod domain and a tail domain (C-terminal domain) – presented in Fig. 1(a). Although the central rod domain is the main driver that sustains self-assembly into filaments, the variable globular nonhelical head and tail domain at the end of intermediate filament proteins exert key roles in the assembly, organization and regulation of intermediate filaments (Lee et al., 2012; Rafik et al., 2004). The rod domain consists of four right-handed α-helical subdomains, 1A, 1B, 2A and 2B, which are separated from one another by non-helical β-turns called ‘linker’ regions (L1, L12 and L2) (Bragulla and Homberger, 2009). As displayed in Fig. 1(a), the α-helical subdomains 1A, 1B, 2A and 2B are composed of repeats of seven amino acids in positions labeled a-g (a heptad pattern) (Smith et al., 2004). Specific hydrophobic apolar amino acids in positions a and d, and charged amino acid residues in positions e and g, stabilize the α-helix (Steinmetz et al., 1998) (Fig. 1a). Two monomers (Type I and Type II monomers) form a left-handed coiled-coil, a so-called heterodimer, as shown in Fig. 1(b) (Bragulla and Homberger, 2009). Two heterodimers can aggregate to form a tetramer in four ways (Fig. 1c) - A11, A22, A12 and ACN - via disulfide bonds and hydrogen bonds (Chou and Buehler, 2012). Then, as illustrated in Fig. 1(d), two such tetramers associate into a protofibril while four protofibrils combine into so-called α-keratin intermediate filaments (IFs). An IF is thus a structure comprised of these tetrameric α-helix structures in a packed assembly. The micro-fibrils contain several of these IFs that bind to each other and cross-link via disulfide bonds in a matrix of sulfide-rich keratin-associated proteins (KAPs) (Marshall et al., 1991). The KAPs include high-glycine tyrosine, high-sulfur, and ultrahigh-sulfur proteins (Rogers, 2004). In hard keratin such as hair, claws and horns, sulfur-rich and ‘ultra’ sulfur-rich KAPs are believed to determine the hardness of these corneous tissues (Alibardi and Segalla, 2011). The macro-fibrils thus appear to consist of larger assemblies of micro-fibril filaments with the KAPs matrix surrounding them (Fig. 2). On the basis of the structural analysis presented above, keratin is clearly a recalcitrant biomass mainly due to the bonding in the structure and the assembly of keratin monomers, dimers, tetramers, and IFs in the KAP matrix. Many intrachain and interchain disulfide bonds, hydrogen bonds and ionic bonds occur to reinforce the structural stability (Fig. 2a, b). In particular, filaments are cross-linked with each other by disulfide bonds in a sulfur-rich KAP matrix (Fig. 2a, b) to form a highly resistant macromolecular assembly (Gong et al., 2012).
Beyond the primary structure post-translational modifications, such as phosphorylation or glycosylation also occur on keratins (Bragulla and Homberger, 2009). Phosphorylation may enable or prevent the interaction of keratins with other molecules, such as signaling molecules, receptor molecules, etc. (Kirfel et al., 2003). Specifically, the phosphorylation of amino acids in the head domain changes the overall net charge of this region to block any interactions with the rod domains of adjacent keratins, and thus prevents the assembly of keratin filaments (Wöll et al., 2007). Glycosylation is another post-translational modification of proteins, which results in an alteration of the binding or signaling functions of keratin (Chou and Omary, 1993). The significance of glycosylation and phosphorylation in relation to enzymatic degradation of keratin appears unexplored.
2.2. β-keratin
Chicken feather is a representative type of β-keratin which contains ordered α-helix as well as β-sheet structures and some disordered structures (Wang et al., 2016). Feathers are made up of barbules, barbs and rachis, as shown in Fig. 3(a), the barbs branch out from the central stiff rachis, and each barb is linked with adjacent barbs by barbules (Reddy and Yang, 2007; Tesfaye et al., 2017b). The structural differences present in different parts of a feather may be related to the variety and structure of the proteins. On average, whole feathers contain 32.2% of α-helix, 53.6% of β-sheets and random coils, while turns make up 14.2% (Ma et al., 2016). However, these average proportions shield the fact that the barb and barbules (“hairs”) fraction has slightly more α-helix than β-sheet structure, whereas the rachis (“stalk”) has more β-sheet than α-helix structure (Ng et al., 2012). The available data for the other β-keratin rich sources, e.g. for turtle scute, confirm that β-keratin has higher glycine content and therefore more non-polar amino acids than α-keratin (Table 1). Some interesting and curious details of the amino acid composition should be noted: penguin feather rachis appears for example to have more non-polar amino acids, especially glycine, than penguin feather barbs (Table 1). This fact may explain why the feather rachis are more hydrophobic and more difficult to degrade than the feather barbs.
The keratin amino acid sequences of rachis, barbules, barbules/barbs in chicken feathers have been recently reported (Jin et al., 2017). Such data allow comparison of conserved keratin sequences of feather rachis (accession number: NP_001264688.1), barbules (NP_001264699.1) and barbules/barbs (NP_001095201.2) (Fig. 3a, b). As many publications show, these three parts of feather keratin contain a 32-residue segment corresponding to the filament framework (Fraser and Parry, 2009, Fraser and Parry, 2008). Based on the amino acid composition of the filament framework segments in such keratin (Fraser and Parry, 2008), each β-sheet includes four β-strands as illustrated in Fig. 3(c) for 31 amino acid residues assigned a β-sheet structure. These four β-strands can be parallel or antiparallel, and the chains are held together by hydrophobic interactions (Wang et al., 2016). From Fig. 3(c), the apolar residues (mainly valine and some isoleucine) are also seen to be on the inner face of the sheet fold, while the charged and cysteine residues are on the sides or turns. The occurrence of a charged residue (arginine) in the outer β-strand in all three keratin structures (Fig. 3c) prevents the exposed edge of one sheet inappropriately bonding to the exposed edge of another sheet (Fraser and Parry, 2009). Proline is found in the turn area in chicken feather keratin (see Fig. 3c) and also in other avian keratin (Fraser and Parry, 2008); proline plays the role of breaking the β-sheet which induces the formation of turns in the conserved β-rich central domain (Calvaresi et al., 2016). This four-strand β-sheet is the socalled monomeric structure of β-keratin (Fig. 3d). Two such monomeric structures pack together via reverse stacking to form a dimer by hydrophobic face-to-face interactions (Fig. 3e) (Calvaresi et al., 2016). In the dimeric structure, salt bridges, hydrogen bonds and disulfide bonds are formed between specific amino acids within the two monomers (Calvaresi et al., 2016). In addition, a head-to-tail assembly of four dimers forms a helix (Fig. 3f). This helix is created through hydrogen bonding and disulfide bonding to produce a left-handed helical ruled surface with four repeating units per turn (where the progressive rotation of the piled dimers is approx. 45°); this is the β-keratin filament (Calvaresi et al., 2016; Wang et al., 2016) (Fig. 3f).
3. Keratinolytic enzymes and keratinase assays
Keratinases are principally type EC 3.4.-.- peptidases (or peptide hydrolases) because their function is to catalyze the hydrolytic cleavage of peptide bonds. “Keratinase” was formerly classified as EC 3.4.4.25, but this particular classification was deleted in 1972, and there is currently neither a 3 or 4-digit EC number for keratinases. The lack of a detailed EC classification is a reflection of the peptidase function not being considered specific because the keratinases in fact merely catalyze keratin degradation by catalyzing the hydrolysis of peptide bonds in keratinous materials. However, a classification is obviously required in order to understand the enzymatic function on this unique substrate. We suggest classifying the keratinolytic enzymes according to their MEROPS protease family relationship, which to some degree corresponds to their substrate attack preference. To our knowledge, all reported keratinolytic enzymes are serine proteases or metallo proteases, but they belong to different protease families (Brandelli et al., 2010). In addition to categorizing the keratinases according to their protease family type, it is highly useful to distinguish their mode of action according to whether they work via endo-attack, exo-attack or exert enzymatic action on peptide-oligomers (oligopeptide-acting). This attack preference differentiation will be discussed in Section 4.
The presence of multiple disulfide bonds in keratin is a key reason for the difficulty of degrading keratin enzymatically. Thus, most keratinases can catalyze keratin degradation only after the disulfide bonds have been broken (reductively cleaved) (Gupta and Ramnani, 2006; Wang et al., 2015). This means that the keratinolytic process may involve two steps: sulfitolysis (S—S bond breakage) and proteolysis (Lange et al., 2016; Peng et al., 2019). Hence in practice, many reports of “keratinase” activity, in fact refer to keratinases that act synergistically with disulfide reductases or reducing agents (e.g. DTT) to break down the complex structure of keratin (Lange et al., 2016). However, what defines a true keratinase (or a superior keratinase) is that it does not rely on the presence of reducing agents or any accompanying disulfide reductase activity (Navone and Speight, 2018). So far, only a few known proteases fit this criterion: for instance, an alkaline protease from Bacillus sp. AH-101, and an S8 serine protease from O. corvina, have been reported to exert high proteolytic activity on keratin biomass in the absence of any added reducing agents (Huang et al., 2015; Takami et al., 1990). In any case, it is necessary to accommodate the fact that keratinases can actually catalyze hydrolysis of peptide bonds in a broad spectrum of protein substrates, including many soluble and insoluble proteins, such as casein, albumin, heme protein, collagen, gelatin, bovine serum albumin, globulin in addition to keratin biomass substrates such as wool, feathers, hair, and porcine bristles (Brandelli et al., 2010). Hence, a ratio of keratinolytic activity to caseinolytic activity of more than 0.5 has been suggested as specifying the keratinase (Gupta et al., 2013b). This ratio gives a relative reference of protease specificity, and works fine as a screening measurement to spot keratinolytic ability e.g. in a crude protease mixture, but the assay may not suffice to compare the rate or the specific keratinolytic activity between different keratinases. For purified proteases, this ratio mainly identifies the endo-keratinolytic proteases but not exo- nor oligo-attacking keratinolytic proteases.
3.1. Keratinolytic enzymes
To date, by using different assays (see Section 3.2), keratinolytic enzymes have been shown to be produced by both bacteria and fungi. Some of the prominent producers are bacterial species, such as B. licheniformis, B. subtilis, and B. pumilus (Fellahi et al., 2016; Gupta and Singh, 2013; Ramnani and Gupta, 2004), as well as actinobacteria, such as Streptomyces fradiae, Nocardiopsis sp. (Li et al., 2007; Mitsuiki et al., 2004). As regards fungi, most keratinolytic enzymes have been described in dermatophytes like T. rubrum and M. canis (Descamps et al., 2002; Zaugg et al., 2008). Furthermore, Onygena spp. such as the non-pathogenic O. equina and O. corvina have also been found to possess keratinolytic ability because they grow on putrefying horns or hooves in nature and proteases from O. corvina have been reported to degrade pig bristles (Huang et al., 2015).
The currently reported keratinolytic enzymes with sequences deposited in the NCBI database are listed in Table 2 . The keratinases were classified into different families according to their amino acid sequence similarities (homology), and validating the categorizations by submitting their sequences to Pfam. This categorization therefore distinguishes the enzyme families according to (a) their functional active sites, and (b) their conserved domains in the MEROPS database. This family classification distributes the keratinolytic enzymes into at least 14 protein families across the serine and metallo proteases: the serine proteases include members of families S1, S9, S8, S10, and S16, and the metallo proteases include members of families M3, M4, M14, M16, M28, M32, M36, M38 and M55 (Table 2). It should be noted that proteases in family M35 have been reported to have the capacity to degrade keratin (Li and Zhang, 2014), but no available sequences have been confirmed to have keratinolytic activity.
Table 2.
Protease family, accession number, source organism, degrading substrate, and some physical characteristics of sequenced keratinolytic enzymes.
| NCBI accession number | Source organism | Original sources | Substrate | No. of AA | Mw (kDa) | Optimum conditions (pH, temp.) | Reference |
|---|---|---|---|---|---|---|---|
| S1 family | |||||||
| BAM67011 | Paenarthrobacter nicotinovorans | Bivalve | Keratin azure | 330 | 23 | – | (Sone et al., 2015) |
| AAO06113 | Nocardiopsis sp. TOA-1 | Tile joint | Keratin | 384 | – | 12.5, 60 °C | (Mitsuiki et al., 2004) |
| AQX39246 | Streptomyces albidoflavus | Poultry soil | Feather | 360 | 36 | 10.0, 50 °C | (Ma et al., 2017) |
| CAH05008 | Streptomyces fradiae | – | Keratin azure | 307 | 26 | 9.0, 55 °C | (Li et al., 2007) |
| S8 family | |||||||
| 1205229A | Parengyodontium album | Soil | Keratin | 277 | 28.9 | 7.5, 37 °C | (Ebeling et al., 1974; Jany et al., 1986) |
| AGK12420 | Stenotrophomonas maltophilia | Poultry farm | Feather | 634 | 48 | 8.0, 60 °C | (Fang et al., 2014) |
| AGK29593 | Stenotrophomonas maltophilia | Poultry farm | Feather | 580 | 40 | 8.0, 60 °C | (Fang et al., 2014) |
| AAK61552 | Fervidobacterium pennivorans | Hot spring | Feather | 699 | 130 | 10.0, 80 °C | (Friedrich and Antranikian, 1996; Kim et al., 2004) |
| ADD51544 | Thermoactinomyces sp. CDF | Campus soil | Feather | 384 | 30 | 11.0, 80 °C | (Wang et al., 2015) |
| AAS86761 | Bacillus licheniformis | – | Feather | 379 | – | – | (Ramnani and Gupta, 2004) |
| AIY62812 | Bacillus subtilis | Poultry soil | Feather | 362 | – | – | (Gupta and Singh, 2013) |
| AKR05134 | Bacillus amyloliquefaciens | – | Feather | 382 | 27 | 11.0, 50 °C | (Yang et al., 2016) |
| ANQ68333 | Bacillus pumilus | Compost | Feather | 383 | 38 | – | (Fellahi et al., 2016) |
| ANQ68334 | Bacillus pumilus | Compost | Feather | 383 | 38 | – | (Fellahi et al., 2016) |
| ACC94305 | Bacillus cereus | Soil | Feather | 917 | 80 | 8.5, 50 °C | (Ghosh et al., 2009) |
| ADD64465 | Bacillus halodurans JB99 | Sugarcane molasses | Feather | 361 | 29 | 11.0, 70 °C | (Shrinivas and Naik, 2011) |
| ARH33809 | Meiothermus taiwanensis | Hot spring | Feather | 400 | 41.3 | 10.0, 65 °C | (Wu et al., 2017) |
| BAQ36632 | Stenotrophomonas maltophilia | – | Keratin azure | 589 | 42 | 10.5, 50 °C | (Jankiewicz et al., 2016) |
| APY18977 | Thermoactinomyces sp. YT06 | Poultry compost | Feather | 389 | 35 | 9.0, 65 °C | (Wang et al., 2017) |
| AAS45672 | Trichophyton benhamiae | Humans | Keratin azure | 399 | 39.7 | – | (Jousson et al., 2004) |
| AAS45666 | Trichophyton benhamiae | Humans | Keratin azure | 400 | 28.2 | – | (Jousson et al., 2004) |
| AAR02423 | Trichophyton rubrum | Humans | Keratin azure | 399 | 28.9 | – | (Jousson et al., 2004) |
| AAR11461 | Trichophyton rubrum | Humans | Keratin azure | 424 | 32.6 | – | (Jousson et al., 2004) |
| AAR02424 | Trichophyton rubrum | Humans | Keratin azure | 396 | 29.0 | – | (Jousson et al., 2004) |
| AJD23187 | Onygena corvina | Horn | Pig bristle | 393 | – | – | (Huang et al., 2015) |
| AJD23193 | Onygena corvina | Horn | Pig bristle | 369 | – | – | (Huang et al., 2015) |
| CAD24008 | Microsporum canis | – | Cat keratin | 485 | 51.3 | – | (Descamps et al., 2002) |
| CAD24009 | Microsporum canis | – | Cat keratin | 427 | 46.1 | – | (Descamps et al., 2002) |
| CAD24010 | Microsporum canis | – | Keratin azure | 397 | 31.5 | – | (Descamps et al., 2002) |
| AAA32703 | Aspergillus niger | – | Keratin azure | 416 | 43.8 | 8.0, 70 °C | (Chen et al., 2015; (Jarai et al., 1994) |
| AHY02992 | Trichophyton mentagrophytes | – | Keratin azure | 412 | – | – | (Yohko et al., 2014) |
| S9 family | |||||||
| AAN03632 | Trichophyton rubrum | Clinical isolate | Keratin | 726 | 78 | 7.0–9.0, − | (Monod et al., 2005) |
| AAS76665 | Trichophyton rubrum | Clinical isolate | Keratin | 775 | 84 | 7.0–9.0, − | (Monod et al., 2005) |
| S10 family | |||||||
| AAS76667 | Trichophyton rubrum | Clinical isolate | Keratin | 652 | 90 | – | (Zaugg et al., 2008) |
| AAS76666 | Trichophyton rubrum | Clinical isolate | Keratin | 662 | 85 | – | (Zaugg et al., 2008) |
| S16 family | |||||||
| AMW33508 | Fervidobacterium islandicum | Hot spring | Feather | 631 | 88 | – | (Kang et al., 2020) |
| M3 family | |||||||
| AJD23200 | Onygena corvina | Horn | Pig bristle | 783 | – | – | (Huang et al., 2015) |
| M4 family | |||||||
| ADP00718 | Pseudomonas aeruginosa | Garden soil | Feather | 475 | – | – | (Sharma and Gupta, 2010) |
| AJD77429 | Geobacillus stearothermophilus | – | Keratin azure | 546 | 57 | 9.0, 60 °C | (Gegeckas et al., 2015) |
| M14 family | |||||||
| ABG67896 | Trichophyton rubrum | Clinical isolate | Keratin | 422 | 42 | – | (Zaugg et al., 2009, Zaugg et al., 2008) |
| M16 family | |||||||
| AMW32060 | Fervidobacterium islandicum | Hot spring | Feather | 406 | 44 | – | (Kang et al., 2020) |
| M28 family | |||||||
| AAS76670 | Trichophyton rubrum | Clinical isolate | Keratin | 373 | 58 | 7.0, 50 °C | (Monod et al., 2005) |
| AJD23165 | Onygena corvina | Horn | Pig bristle | 374 | – | – | (Huang et al., 2015) |
| CAH03796 | Streptomyces fradiae | – | Feather | 461 | 36 | 8.0, 60 °C | (Wu et al., 2010) |
| AJD23207 | Onygena corvina | Horn | Pig bristle | 493 | – | – | (Huang et al., 2015) |
| AAS76669 | Trichophyton rubrum | Clinical isolate | Keratin | 495 | 33 | 7.0, 50 °C | (Monod et al., 2005) |
| M32 family | |||||||
| AMW32563 | Fervidobacterium islandicum | Hot spring | Feather | 489 | 107 | 7.0, 80 °C | (Lee et al., 2015a) |
| M36 family | |||||||
| BAM84176 | Fusarium oxysporum | Soil | Wool cuticle | 632 | 46.8 | 7.0, 50 °C | (Chaya et al., 2014) |
| CAD35288 | Microsporum canis | – | Keratin azure | 633 | – | – | (Brouta et al., 2002) |
| AJD23141 | Onygena corvina | Horn | Pig bristle | 634 | – | – | (Huang et al., 2015) |
| M38 family | |||||||
| AMW33776 | Fervidobacterium islandicum | Hot spring | Feather | 345 | 42 | – | (Kang et al., 2020) |
| M55 family | |||||||
| AMW33601 | Fervidobacterium islandicum | Hot spring | Feather | 279 | 31 | – | (Kang et al., 2020) |
The keratinolytic enzymes in the different families may play different roles in the keratin degrading process, due to differences in their active sites, the preferred substrate cleavage sites and recognition of amino acid length. As is evident from Table 2, the source organisms of these keratinolytic enzymes come from a broad range of natural environments, such as keratin-rich environmental soil, compost, fungus-infected horns, human and animal dermatophytes, and other environments such as bathroom tile-joints, hot springs and even sugarcane molasses (Table 2). The number of amino acids in each enzyme (including N-, C- and available signal peptides) range from 307 to 917 which gives the enzymes molecular weight range of between 26 and 130 kDa (Table 2). Strikingly, the two largest enzymes (with molecular weights of 107 and 130 kDa, respectively) are both from Fervidobacterium species and both were isolated from hot springs. This wide range of habitats indicates the versatility of keratin-degrading organisms, and suggests that microbial keratinolytic enzymes may be quite diverse with regard to their mode of action, biochemical and biophysical properties.
In order to qualify as being keratin degrading, the ability of keratinolytic proteases to degrade keratin has been validated on substrates such as keratin azure (a commercial, colored wool-based keratin), feathers, pig bristles, cat keratin, keratinized tissues and wool cuticles (Table 2) (assays are discussed in detail in Section 3.2). The available data on the optimum reaction conditions of keratinolytic enzymes show that most proteases work at neutral to high alkaline pH of 7.0 to 12.5 and at temperatures of 50–80 °C (Table 2). This wide range of reaction conditions qualifies the keratinolytic enzymes for use in different industrial-scale waste processing facilities. However, in designing keratinolytic enzyme blends, the similarity of optimum conditions of the candidate enzymes also needs to be considered to ensure that all the proteases work efficiently together.
3.2. Keratinase enzyme assays
Many of the keratinolytic enzyme activities have been verified using natural keratin-rich substrates such as feather, pig bristles, wool cuticles etc. (Table 2), where detailed enzyme kinetics studies are difficult. Some more detailed keratinolytic enzyme activity studies using purified or recombinantly produced enzymes have employed colored, modified keratin-derivatives as substrates, e.g. azo-keratin (Gonzalo et al., 2020) and keratin azure, but natural, insoluble keratin substrates such as feather (in fact ball-milled feather powder) (Fang et al., 2013), pig bristles, human hair (Mukhopadhyay and Chandra, 1990), cow's horn (Dozie et al., 1994) have also been used (Table 3 ).
Table 3.
Keratinolytic activity testing methods.
| Substrate and unit definition | Reference |
|---|---|
| Azo-keratin | |
| The amount of enzyme yielding 0.001 (415 nm) absorbance units/h | (Gonzalo et al., 2020) |
| The amount of enzyme that increases the absorbance (440 nm) of the solution by 0.1per mL (U/mL) at the assay conditions | (e Silva et al., 2014) |
| Keratin azure | |
| The increase in absorbance at 450 nm of 0.01 after 20 min in the test reaction compared with the control reaction | (Tork et al., 2016) |
| The amount of enzyme causing an increase of 0.1 in absorbance at 440 nm in 1 min under the assay conditions | (Habbeche et al., 2014) |
| The amount of enzyme required to increase absorbance at 595 nm by 0.01 in 1 h | (Nahar et al., 2016) |
| Human hair | |
| 1 keratinase unit = 0.1 corrected absorbance at 280 nm | (Mukhopadhyay and Chandra, 1990) |
| Cow horn | |
| An increase of 0.01 absorbance unit at 280 nm ml−1 h−1 | (Dozie et al., 1994) |
| Feather powder | |
| 1 mmol tyrosine liberated per min | (Fang et al., 2013) |
| Wool top | |
| The amount of enzyme which liberates 1 μmol tyrosine/min under the assay conditions | (Iglesias et al., 2017) |
| Soluble recombinant chicken feather | |
| The amount of enzyme that produced 1 nmole of free amino groups (equivalent to arginine) as products per min at the assay conditions | (Jin et al., 2017) |
Azo-keratin is a deep red-orange compound with an absorption maximum at 440–450 nm (Riffel et al., 2003). Azo-keratin is not commercially available, but it is possible to prepare this keratinase assay substrate by coupling keratin (derived from various keratin sources) with a diazotized aryl amine to produce a chromophoric derivative, sulfanilic acid-azokeratin (Riffel et al., 2003). Another synthetic colored keratin substrate is keratin azure (Habbeche et al., 2014; Nahar et al., 2016). This substrate is available commercially from Sigma–Aldrich (St. Louis, MO, USA) and is presumably prepared from sheep wool (Scott and Untereiner, 2004). Hence, the keratin azure from sheep wool is particularly useful for α-keratin-degrading enzymes, but β-keratinases are also detected. For deep kinetics analysis and comparisons of keratinases, it seems useful to combine measurements using the keratin azure substrate with parallel assays using azo-keratin prepared from β-rich keratin such as feather. Such comparisons allow assessment of any differentiation in substrate type preference among keratinases, an area that appears very little investigated. An additional point however, is that azo-keratin used as keratinase assay substrate appears to be more sensitive and robust than keratin azure (3-fold) (Gonzalo et al., 2020). The assessment of enzyme action on “genuine” keratin substrates, e.g. pulverized feather, will obviously be a better reflection of the degradation function of a specific keratinase. However, when real keratin substrates are used, more operations and various analytical methodologies need to be employed. Hence, azo dying of genuine keratin biomass appears to be a good choice to develop more selective keratinase assays, and we envisage that more assay combinations may gradually emerge with more focus on keratinase enzyme discovery for keratin biomass refining.
Recently, a particularly elegant type of assay has been developed, involving the use of soluble recombinant chicken feather keratin substrates (Jin et al., 2017). With this assay, it is possible to obtain information about the substrate-enzyme interaction, such as which amino acid residues in the substrate are preferentially bound in the P1/P1’ sites of the enzyme (Jin et al., 2017). Despite these developments, no standard keratinase enzyme assay currently exists and the enzyme unit definitions for keratinase activity vary (Table 3), which makes comparisons of enzyme activities and kinetics across different studies challenging.
4. Classification of enzymes involved in keratin degradation
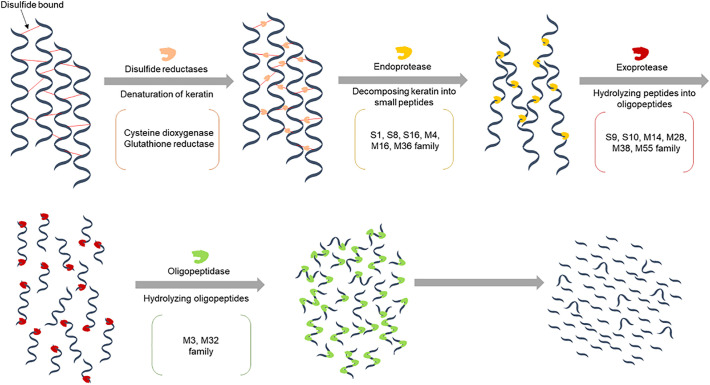
The14 protease families involved in keratin degradation (Table 2) may be further grouped into endo-protease, exo-protease and oligopeptidase families. This additional sub-division of the enzymes is valuable for understanding their roles and potential function in keratin degradation. The complete degradation of keratin to single amino acids requires (at least) three kinds of protease, namely endo-, exo- and oligo-protease, as well as an S—S-bond breaking strategy, which may be done either chemically (such as DTT) or enzymatically, e.g. by disulfide reductase (Lange et al., 2016; Mercer and Stewart, 2019).
Endoproteases catalyze the cleavage of peptide bonds internally within a polypeptide, and keratinolytic endoproteases are classified in the S1, S8, S16, M4, M16, M36 families (Table 2). In contrast, the keratinolytic proteases in the S9, S10, M14, M28, M38 and M55 families are exoproteases, which means that they attack the polypeptide chain at the terminal end (Lange et al., 2016; Mercer and Stewart, 2019). The exo-acting keratinolytic enzyme members in the different families attack from either the N-terminal (S9, M38 and M55 family members) or the C-terminal (S10, M14 and M28 family members); furthermore, synergistic action may take place as exo-acting proteases act on the free peptides released by the action of the keratinolytic endo-proteases. The keratinases acting on the peptide bonds in peptide-oligomers (oligopeptide-acting or oligopeptidases) are categorized in M3 and M32 families. These enzymes can act on shorter peptides and catalyze hydrolysis of small peptides to result in dimeric or trimeric peptides or even individual amino acids. Some other enzymes may have a positive facilitation effect on keratin digestion. For example, LPMOs have been suggested to potentially contribute to keratin degradation in nature (Lange et al., 2016).
4.1. Endoproteases catalyzing keratin hydrolysis
All hitherto identified endo-keratinolytic serine proteases belong to family S1, S8 or S16, whereas the members of family M4, M16 and M36 are metallo proteases.
4.1.1. S1-family keratinolytic endoproteases
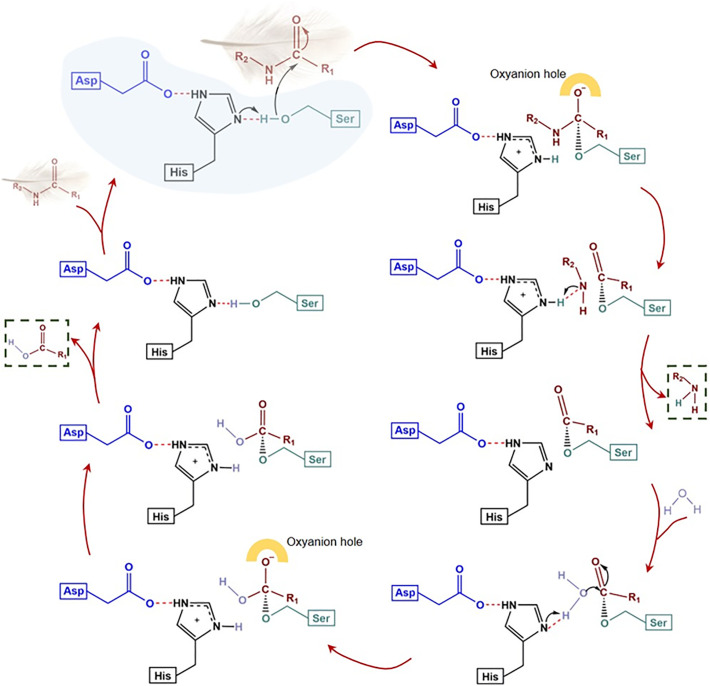
Family S1 is the largest protease family in terms of the number of proteins available in the MEROPS database. All the peptidases in family S1 are endopeptidases and work via a “classic” catalytic triad involving a nucleophilic attack followed by an acid hydrolysis mechanism provided by His, Asp and Ser (presented according to their order in the amino acid sequence) (Rawlings and Barrett, 2013a). The four described keratinolytic S1 family endoproteases (NCBI ID: BAM67011, AAO06113, AQX39246, CAH05008, Table 2) were isolated from very different habitats and organisms. They were tested for their keratinolytic function using different types of keratin assay substrates, yet all four enzymes work optimally at alkaline pH of 9–12.5 (Table 2).
The keratinolytic activity of the enzyme NCBI ID: BAM67011, also called “ANISEP”, derived from Paenarthrobacter nicotinovorans was about 10-fold higher than that of trypsin (Sone et al., 2015). Protein AAO06113 from Nocardiopsis sp. TOA-1 had very high keratinolytic activity, especially at a high alkaline pH of 12.5, where the activity was 5 to 10-fold higher than that of proteinase K and subtilisin (Mitsuiki et al., 2004). The substrate specificity of protein AAO06113 was investigated using synthetic peptides. The K m results show preference for Leu, Ala and Phe residues at the P1 position (Mitsuiki et al., 2004). The keratinolytic enzyme (CAH05008) from Streptomyces fradiae var. k11 gave a keratin/casein ratio of 0.87 (Li et al., 2007). This results indicates that CAH05008 is indeed “keratinolytic” because this high ratio is similar to that of proteinase K, one of the most well described keratinases, considered by some as a benchmark keratinase, based on the same keratinase and caseinase assay. Moreover, the keratinase activity of CAH05008 was much higher than that of proteinase K (Li et al., 2007). From the activity data, the S1 keratinases have comparable ability with S8 keratinases in catalyzing keratin biomass degradation. On this basis, the family S1 proteases deserve further investigation.
4.1.2. S8-family keratinolytic proteases
There are plenty of keratinolytic proteases classified in family S8. After sorting out doublets/identical sequences (using CD-hit protein sequence clustering with a 90% sequence identity threshold), we identified a total of 27 unique keratinolytic proteases in the S8 protease family (Table 2). Like the S1 proteases, the S8 protease family members work via a catalytic triad mechanism, but the active sites with respect to the order of Asp, His, Ser in the sequence (Laskar et al., 2012). Among the 27 S8-family members, the AKR05134 enzyme (also known as KerK) derived from B. amyloliquefaciens has been reported to have the highest keratin specificity with a keratinolytic:caseinolytic activity ratio of 4.76 in the presence of 2 mM DTT and a purified recombinant KerK dosage of 18.75 U/mL (Yang et al., 2016). Other bacterial S8 keratinases are produced by species of Fervidobacterium, Meiothermus, and Stenotrophomonas. Notably the enzyme from Fervidobacterium pennivorans has been reported to be highly thermostable and to act at high pH levels (Table 2); the structure of this enzyme has already been determined (Kim et al., 2004), although it is not clear which structural features are responsible for the alkalinity and thermostability of the enzyme.
Dermatophyte fungi of the class Eurotiomycetes, such as T. rubrum, T. benhamiae and M. canis also produce keratinolytic S8 endoproteases. For example, M. canis produce three different keratinolytic subtilisin-like proteases, Sub1 (CAD24008), Sub2 (CAD24009) and Sub3 (CAD24010). The etiology of the pathogenesis strongly suggests that these proteases are produced by M. canis during invasion of keratinized structures (Descamps et al., 2002). Similarly, Sub3 (AAR11462) and Sub4 (AAR02423) from T. rubrum show appreciable activity on keratin azure is comparable to that of subtilisin Carlsberg and proteinase K (Jousson et al., 2004). In comparison to subtilisin Carlsberg and proteinase K, Sub3 and Sub4 were less active on other protein sources, especially elastin, indicative of the specificity of Sub3 and Sub4 towards keratinous substrates (Jousson et al., 2004). Sub3 (AAR11462) from T. rubrum has 94.5% sequence identity with Sub3 (CAD24010) from M. canis (therefore AAR11462 is not listed in Table 2). The non-pathogenic O. corvina is known to grow on hooves and horns. Huang et al. reported that a recombinantly produced S8 protease from O. corvina appears to be a true keratinase because it is able to catalyze degradation of pig bristles and hooves without addition of reducing agent to break the cysteine bridges in the keratin (Huang et al., 2015).
More detailed examination of S8 protease sequences has shown that the active site Ser in the S8 family members is found within a Gly-Thr-Ser-Xaa-Xaa-Xbb-Pro motif (where Xaa is an aliphatic amino acid and Xbb is a small amino acid) (Rawlings and Barrett, 2013a). In order to reveal the specific amino acid motif in S8 keratinolytic proteases (listed in Table 2), MEME SUITE (Bailey et al., 2009) was used for motif finding. The results (given in Fig. 4 ) indicates that the residues around active sites are highly conserved, with Asp in the Asp-Thr/Ser-Gly motif, His in the His-Gly-Thr-His-Val/Thr-Ala/Ser-Gly/Ser-Thr/Ile motif and Ser in the Gly-Thr-Ser-Met/Ala-Ala-Ala/Ser/Thr-Pro-His-Val motif. These specific motifs may attribute specificity to keratinases compared with other proteases.
Fig. 4.
Sequence logo (from MEME SUITE) of active site areas in S8 keratinolytic enzymes. The active sites Asp (D), His (H), Ser (S) are labeled with an asterisk.
4.1.3. S16-family keratinolytic proteases
Unlike the peptidases in the S1 and S8 family, the active site of members in family S16 is a Ser-Lys catalytic dyad (Rawlings and Barrett, 2013a). One protease in the S16 family (derived from Fervidobacterium islandicum, accession number: AMW33508) has recently been confirmed to be a keratinolytic protease (Kang et al., 2020). The enzymatic rates free amino acids release from chicken feather with addition of purified S16 protease is around 1.3-fold higher than those of a crude F. islandicum extract. The comparison information available for S16 keratinolytic proteases is unclear.
4.1.4. M4-family keratinolytic proteases
There are only two identified and sequenced keratinolytic endoproteases in the M4 family. One of them, the AJD77429 enzyme is referred to as GEOker, originates from the thermophilic bacterium Geobacillus stearothermophilus AD-11 (Table 2). A recombinant version of this enzyme, RecGEOker (expressed in E. coli), has been found to have a consensus zinc-binding HELTH motif which indicates that the enzyme belongs to the zinc metalloproteinases of family M4. Based on studies of the recombinant enzyme, there are strong indications that the His372, Glu373 and His376 catalytic triad is essential for the GEOker enzymatic reaction and the enzyme activity on different protein substrates was ranked as follows: wool keratin > collagen > sodium caseinate > gelatin > BSA (Gegeckas et al., 2015). The enzyme from Pseudomonas aeruginosa (ADP00718) exerts keratin degrading ability (Sharma and Gupta, 2010), however, the specific function has not been investigated.
4.1.5. M16-family keratinolytic proteases
In addition to an S16 keratinolytic protease (see Section 4.1.3), the F. islandicum produce one M16 protease (accession number: AMW32060). Addition of this enzyme on top of a F. islandicum extract also enhanced the feather degradation rate in vitro (more than 1.5 fold) compared with the crude F. islandicum extract only (Kang et al., 2020) but the keratinolytic activity of this protease alone is unknown. Furthermore, it is worth noting that not all family M16 proteases act by endo-attack, for instance, subfamily M16A contains oligopeptidases denoted as insulysin and nardilysin (Rawlings and Barrett, 2013b). The detailed attack mode of family M16 keratinolytic proteases warrants further investigation.
4.1.6. M36-family keratinolytic proteases
Members of the M36 (fungalysins) metalloendopeptidase family have been shown to be involved in degrading different keratinous substrates, and this enzyme family includes proteases secreted by O. corvina (Huang et al., 2015), M. canis (Brouta et al., 2002) and F. oxysporum (Chaya et al., 2014) (Table 2). Family M36 is a metalloprotease family containing a His-Glu-Xaa-Xaa-His (HEXXH) motif of metallopeptidases from clan MA (Rawlings and Barrett, 2013b). The motif serve to coordinate the zinc ion (Xu et al., 2006). Mep4 (accession number: AJD23141) is induced when O. corvina grows on chicken feathers, pig bristle, or dog wool (Huang et al., 2015). The Fusarium-derived enzyme KrtC (accession number: BAM84176) can hydrolyze keratin and has been shown to have a preference for cleaving the amino side of hydrophobic residues with bulky side-chains (Chaya et al., 2014). MEP3 (accession number: CAD35288) was found to be a gene encoding an isolated M. canis 43.5-kDa keratinolytic metalloprotease, and was successfully expressed in Pichia pastoris (Brouta et al., 2002). It is noteworthy that these enzymes can overcome the limited proteolysis on the surface of insoluble keratin particles, which restricts enzyme-substrate interaction (Huang et al., 2015).
4.2. Exoproteases involved in keratin hydrolysis
Current exo-keratinolytic enzymes are classified in the serine protease families S9 and S10 and in the metallo protease families M14, M28, M38 and M55.
4.2.1. S9-family keratinolytic exoproteases
Proteases belonging to the S9 family exerts their catalytic function via a catalytic triad in the order Ser, Asp, His. The representative keratinolytic proteases in the S9 family are AAN03632 (dipeptidyl peptidase V, DPPV) and AAS76665 (dipeptidyl peptidase IV, DPPIV) (Table 2). Both enzymes are from T. rubrum (Monod et al., 2005). Recombinant ruDPPIV is able to catalyze the hydrolysis of synthetic dipeptides Gly-Pro-AMC and Lys-Ala-AMC, while recombinant ruDPPV hydrolyze only Lys-Ala-AMC but not Gly-Pro-AMC (Monod et al., 2005). Another source of DPPIV is the fungus T. tonsurans (Preuett et al., 2010). In addition, secretion of DPPV peptidase activity has been detected from the fungal pathogen T. mentagrophytes (Kaufman et al., 2005) in skin infections. Although the in vitro activity of DPPV and DPPIV in degrading other keratin substrates than skin keratin is unknown, the Trir4 allergen (AAD52012) from T. rubrum (Woodfolk et al., 1998), which has the same amino acid sequence as DPPV (AAN03632), has shown keratinolytic activity on keratin azure. These results demonstrate that Trir4 exerts weak activity against keratin azure (447 units/mg). This keratinolytic activity is 15 times lower than the activity of the more well-known endo-protease proteinase K (7490 units/mg) (Woodfolk et al., 1998). The family S9 exo-keratinases may play a key role in attacking products from endo-protease hydrolyzed keratin protein (Lange et al., 2016; Mercer and Stewart, 2019), but is not highly efficient in directly degrading keratin. Endo- and exo-keratinolytic proteases may degrade keratin synergistically. Similarly, the available research concerning synergism between endo- and exopeptidases is likely to be an essential prerequisite for potent dermatophyte virulence, e.g. when dermatophytes infect keratinized tissues (Monod et al., 2005).
4.2.2. S10-family keratinolytic exoproteases
Family S10 contains only carboxypeptidases (Rawlings and Barrett, 2013a) which hydrolyze the peptides from C-terminal. The proteases AAS76667 (TruScpA) and AAS76666 (TruScpB) are secreted by T. rubrum when degrading compact keratinized tissues (Zaugg et al., 2008). The catalytic triad of TruScpA (Ser238, Asp458, and His516) and TruScpB (Ser240, Asp459, and His517) are in the order of Ser, Asp, His, and belong to the S10 family. In addition, TruScpA and TruScpB enzymes are not secreted into the environment, but are membrane-associated with a glycosylphosphatidylinositol (GPI) anchor. During infection, GPI-anchored carboxypeptidases secreted by T. rubrum may contribute to fungal virulence by cooperating with previously characterized endoproteases and aminopeptidases in the degradation of compact keratinized tissues into assimilable amino acids and short peptides (Zaugg et al., 2008). Therefore, like the S9 keratinolytic proteases, the currently known S10 keratinolytic proteases also promote keratin tissue degradation in concert with keratinolytic endoproteases.
4.2.3. M14-family keratinolytic exoproteases
In a medium containing keratin-soy as sole nitrogen and carbon source, T. rubrum secretes the protease ABG67896 (McpA), which is classified in the M14 protease family (Zaugg et al., 2009, Zaugg et al., 2008). All members of family M14 contain the motif His-Xaa-Xaa-Glu. Most of the peptidases in the M14 family are carboxypeptidases that catalyze the hydrolytic removal of single C-terminal amino acids from polypeptide chains. These enzymes all have a recognition site for the free C-terminal carboxyl group, which is a key determinant of specificity (Riffel et al., 2007). In the McpA enzyme from T. rubrum, the residues His179, Glu182 and His309 are ligands for the catalytic zinc, while other amino acids, e.g. Arg237, Arg255, Tyr311, Tyr362 and Glu385, are important for substrate binding and catalysis in the M14A family (Zaugg et al., 2008). The McpA from T. benhamiae is also known to be highly expressed during in vitro growth on keratin (Tran et al., 2016). Thus McpA in the M14 family may assist in degrading keratin. Another purified M14 keratinolytic metalloprotease, Q1 (no available protein sequence), originating from Chryseobacterium sp. kr6 has also been examined, and reported to have specific keratinolytic activity of 967 U/mg protein with keratin azure as substrate (Riffel et al., 2007).
4.2.4. M28-family keratinolytic exoproteases
The M28 family is the most researched exo-keratinolytic protein family. As listed in Table 2, five proteases in the M28 family have been tested for activity on pig bristle, feathers and keratin. The aminopeptidase CAH03796 in family M28, produced by the keratin-degrading bacterium Streptomyces fradiae var. k11, is known to play a role in the processing of proenzymes (Wu et al., 2010). The conserved motifs H147, D159, E194, D222, and H309, which are all implicated in the coordination of two zinc atoms, are found in this protease (CAH03796). Two T. rubrum derived aminopeptidases AAS76669 (ruLap1) and AAS76670 (ruLap2) have also been reported (Monod et al., 2005). Substrate specificity analysis using different fluorogenic aminoacyl-4-methylcoumaryl-7-amide derivatives has shown that ruLap1 is the most selective for Leu-AMC, while Ser-AMC and Pro-AMC are more efficiently cleaved by ruLap2 (Monod et al., 2005). The tested Laps were not capable of cleaving the Gly-Pro-AMC substrate, which indicates that the presence of a Pro residue in position P1’ affects the action of these enzymes (Monod et al., 2005). Thus, the aminopeptidases Lap1 and Lap2 in family M28 family hydrolyze peptides from the N-terminus until they reach X-pro or X-Ala motifs, which act as a stop point (Mercer and Stewart, 2019). Two exopeptidases (M28 with accession numbers AJD23165 and AJD23207) together with two endopeptidases (S8) from the non-pathogenic fungus, O. corvina, displayed a higher degree of pig bristle degradation compared with degradation by S8 proteases only (Huang et al., 2015). To our knowledge, this is the first record of in vitro synergies of S8 and M28 keratinolytic proteases in degrading keratin, and these data support the view that a combination of exo- and endo-acting proteases can efficiently degrade keratin.
4.2.5. M38- and M55-family keratinolytic exoproteases
The M38 and M55 keratinolytic proteases, with accession number AMW33776 and AMW33601, respectively, are also derived from F. islandicum. Both of the purified M38 and M55 proteases improved the free amino acid release rate by comparison of the rate of F. islandicum extract only (Kang et al., 2020). During keratin degradation by F. islandicum, the M38 protease is presumably responsible for regulating proteolysis, while M55 protease might be activated during starvation conditions and involved in controlling the cell's flow of peptides as degradation products (Kang et al., 2020).
4.3. Oligopeptidases involved in keratin hydrolysis
An oligopeptidase is an enzyme that catalyze cleavage of peptides but not proteins. This property is due to its structure, namely that the active site of this enzyme is located at the end of a narrow cavity that can be reached only by peptides. In the keratin degradation process, oligopeptidase plays an important role in degrading the peptides generated by endo- and exo-protease digestion of compact keratinized tissues. Among the keratinolytic enzymes listed in Table 3, proteins in family M3 and M32 function as oligopeptidases.
4.3.1. M3 and M32-family keratinolytic oligopeptidases
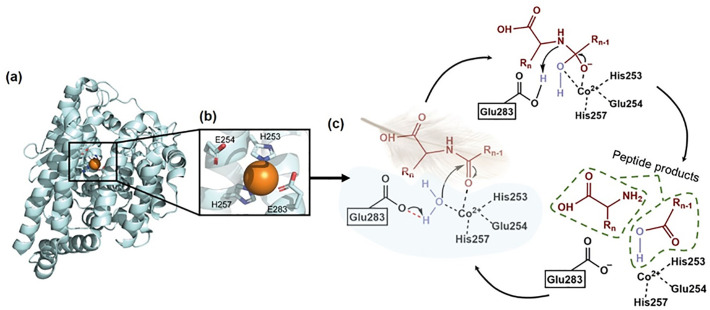
The M3 and M32 family are in the MA (E) clan which contains zinc-dependent metallopeptidases. The two zinc ligands are the histidines in the motif His-Glu-Xaa-Xaa-His (Rawlings and Barrett, 2013b). Protease family M3 includes intracellular oligopeptidases from mammals, fungi and bacteria, which only degrade peptides of certain lengths (between 4 and 17 residues, in the case of thimet oligopeptidase) (Rawlings and Barrett, 2013b). Thus the M3 keratinolytic enzymes can only catalyze hydrolysis of small keratin peptides, i.e. those that are produced from endo- and exo-keratinolytic action on keratin. The first and currently the only described M3 keratinolytic protease, AJD23200, is from the non-pathogenic fungus O. corvina. Previous research has revealed a synergistic function in keratin degradation between three O. corvina proteases, a family M3 keratinase (oligopeptidase), an S8 protease (endo-keratinase), and an M28 protease (exo-keratinolytic enzyme) (Huang et al., 2015). However, the substrate specificity and the oligopeptide size preferences of this M3 protein are not known at present.
Metallo-carboxypeptidases (MCP) of the M32 family are found in many organisms. These enzymes catalyze hydrolysis of the peptide bond at the C-terminus of peptides and have been widely studied (Lee et al., 1994). The protease AMW32563 (FisCP) plays a pivotal role in the decomposition of keratin when added to reaction mixtures containing whole cell extractions derived from F. islandicum AW-1 cells (Lee et al., 2015a). The results show that amino acid production was 2-fold higher in FisCP-supplemented whole cell extractions than in whole cell extractions alone. This feature supports the notion that FisCP together with numerous other highly up-regulated proteases on keratin are mainly involved in the degradation of native chicken feathers (Lee et al., 2015a).
4.4. Other enzymes involved in keratin hydrolysis
As mentioned earlier, the first step in enzymatic keratin degradation is the reductive breakage of the disulfide bonds in keratin (Peng et al., 2019). The enzymes that have been suggested as being able to catalyze the breaking of disulfide bonds include cysteine dioxygenase (EC 1.13.11.20), glutathione reductase (EC 1.8.1.7), alkyl hydroperoxide reductase (EC 1.11.1.15), thioredoxin reductase (EC 1.8.1.9), dihydrolipoyl dehydrogenase (EC 1.8.1.4), peptide methionine sulfoxide reductase (EC 1.8.4.11), phospho-adenosine phosphosulfate reductase (EC 1.8.4.8), ribonucleoside-diphosphate reductase (EC 1.17.4.1) (Peng et al., 2019). Among these, disulfide reductase, cysteine dioxygenase and glutathione reductase have indeed been shown to play an important role in keratinolysis (Grumbt et al., 2013; Łaba et al., 2013). However, research data is scant in this area, so the efficient action of other types of reductases in keratin breakdown cannot be ruled out. Cysteine dioxygenase is a key enzyme in homeostatic regulation of the cysteine level in eukaryotic cells. Because this enzyme is involved in the production of important oxidized metabolites of cysteine, such as pyruvate, sulfite, sulfate, hypotaurine, and taurine in all eukaryotic cells (Kasperova et al., 2013). The relevant research has indicated that when dermatophytes grow in keratin tissues, sulfite formation from cysteine relies on cysteine dioxygenase Cdo1 and sulfite secretion is supported by the sulfite efflux pump Ssu1 (Grumbt et al., 2013).The presence of the reducing agent sulfite may break the S—S bonds in keratin biomass and in this way facilitate keratin degradation. Furthermore, γ-glutamyl transpeptidase (GGT) is a periplasmic enzyme that catalyzes the hydrolysis of glutathione (GSH) to produce cysteinyl-glycine, which is a strong reductant that further reduces disulfide bonds. It has been hypothesized that a GGT-GSH mediated redox might be one of the pathways of sulfitolysis for feather degradation and that cysteinyl glycine might be the active redox moiety (Sharma and Gupta, 2012). The lyase cystathionine gamma-synthase (EC 4.4.1.1) secreted by B. subtilis 8, which catalyzes the breakdown of carbon‑sulfur bonds, has also been confirmed to be involved in and able to promote decomposition of feather keratin (He et al., 2018).
Many disulfide reductases derived from keratinolytic organisms have not yet been identified to a specific type of reductase, but their disulfide reduction activity has been identified using DTNB (5,5′-Dithiobis-(2-Nitrobenzoic Acid)) as substrate. Some researchers have reported efficient synergies between keratinolytic enzymes and disulfide reductases. For instance, Bacillus sp. MTS produces an extracellular alkaline keratinolytic protease and a disulfide reductase while growing in media containing feathers (Rahayu et al., 2012). When these two enzymes are combined, the keratinolytic activity of the mixture on feather substrates (27.5 U/mg) is greatly increased compared to the activity of purified alkaline protease alone (4.5 U/mg) or purified alkaline protease in the presence of reducing agents DTT (12.8 U/mg) (Rahayu et al., 2012). Stenotrophomonas sp. also produce a serine protease and a disulfide bond-reducing protein and is thus capable of degrading native keratin (human hair) (Yamamura et al., 2002). Interestingly, the keratinolytic activity of the serine protease alone was only 83 KU/mg, however, when the serine protease was mixed with the disulfide reductase, the keratinolytic activity was 4450 KU/mg, which was more than 2-fold higher than that of the disulfide reductase combined with proteinase K (2280 KU/mg) (Yamamura et al., 2002). In addition, the disulfide reductase mixed with the serine protease resulted in higher keratinolytic activity compared with the serine protease in presence of the reducing agent DTT (Rahayu et al., 2012; Yamamura et al., 2002). This illustrates that some disulfide reductases are better choices for use in catalyzing the breakage of S—S bonds in keratin. An interesting research focus is the investigation of the keratinolytic efficiency of different disulfide reductases in combination with keratinolytic proteases.
Other enzymes such as LPMOs may exert a possible auxiliary function in degrading keratin (Lange et al., 2016). LPMOs are copper-dependent and utilize molecular oxygen and an electron donor (e.g. ascorbic acid) to catalyze cleavage of glycosidic bonds as has been demonstrated by the action of LPMOs on cellulose, hemicellulose, chitin, and starch. Interestingly, LPMO genes occur consistently in dermato-phytic and keratin-degrading fungi (Busk and Lange, 2015) as well as in keratinolytic non-pathogenic fungi, e.g. O. corvina (Huang et al., 2015). The putative function of (AA11) LPMOs in keratin-degrading fungi is presumed to be the catalytic breakage of glycosyl-bonds between N-acetyl-glucosamine and serine and threonine in the non-coiled head structure of the keratin filaments. Such changes have been shown to loosen the keratin structure or even to de-assemble the keratin filaments (Lange et al., 2016). Besides, enzymes involved in fatty acid degradation may contribute to keratin digestion (Lee et al., 2015b). The surface of wool fiber is rich in lipids, with an outer lipid layer consisting of 18-methyleicosanoic acid along with other fatty acids, which are bound largely through thioester linkages to the cysteine-rich underlying proteins (Ghosh et al., 2014). Enzymes relevant for degradation of fatty acids may be envisaged to assist in breaking the outer lipid layer of keratin so that keratinolytic endoproteases can gain easier access to the internal keratin structure. However, in vitro testing of LPMOs and fatty acid degradation enzymes in hydrolyzing keratin substrates still needs further research.
5. Theories of keratin degradation by enzymes
Although the detailed mechanism of keratin degradation by enzymes is not fully understood, several hypotheses have been suggested to explain the enzymatic degradation events. As outlined above, keratin is packed with disulfide bonds and hydrogen bonds, and both inter- and intra-chain disulfide bonds exist (Fig. 2). To obtain amino acids from keratin-rich materials using keratinolytic enzymes, a preliminary degradation of disulfide bonds is necessary to loosen the keratin structure and make the amino acid chains available for keratinase attack (Gupta et al., 2013b; Peng et al., 2019). The more enzyme attack sites are exposed, the better for enzyme hydrolysis. Based on present research on biodegradation of keratin, an efficient biocatalytic process invariably involves two main processes: reduction of disulfide bonds and hydrolysis of the keratin-polypeptide chain by keratinolytic proteases (Fig. 5 ). If the disulfide bonds are not broken, most keratinases cannot degrade native keratin effectively (Okoroma et al., 2012).For instance, unless reducing agents are present, the WF146-protease from Bacillus sp. cannot catalyze the degradation of feathers (Liang et al., 2010). Analogously, the enzyme Cibenza DP100 did not produce detectable levels of soluble peptides during degradation of hair unless a reducing agent was added to the reaction (Navone and Speight, 2018).
Fig. 5.
Sequential enzymatic keratin degradation process by disulfide reductases and endo-keratinases, exo-keratinases and oligo-keratinases.
As regards the keratin biomass degradation by purified keratinases, the disulfide bonds can be broken either by adding chemical reducing agents or, as mentioned above, by means of enzymes such as cysteine dioxygenase or glutathione reductase (the latter requires the presence of glutathione to accomplish the reaction i.e. the redox half reaction) (Burmester et al., 2011; Descamps et al., 2002). Regarding the process of keratin degradation by keratinolytic organisms, the reducing agent sulfite or disulfide reductase may be secreted from the organism and may be involved in cleavage of disulfide bonds in the keratinous biomass (Grumbt et al., 2013; Kang et al., 2020). Some organisms cannot secrete disulfide reductase or sulfite (e.g. Streptomyces pactum), in this case, the reduction of disulfide bonds must depend on the presence of metabolically active cells and the membrane potential may play a key role (Böckle and Müller, 1997).
Once the disulfide bonds in keratin have been broken, the keratinolytic endoproteases (members of families S1, S8, M4, and M36) act to accomplish the keratinolytic substrate degradation. Keratinolytic exo-proteases (members of the families S9, S10, M14, and M28) cleave peptide chains from both ends, while oligopeptides (members of family M3 and M32) work on oligo-peptides to release individual amino acids or shorter peptides. This sequential degradation, outlined in Fig. 5, has been suggested to take place during for example infectious keratin tissue degradation (Mercer and Stewart, 2019) and during fungal keratin decomposition (Lange et al., 2016).
Apparently, efficient and complete decomposition of keratin to free amino acids cannot be achieved by one keratinolytic protease alone, but seem to require a combination of enzymes, plus presence of a reducing agent or a reducing enzyme to help cleave the S—S bonds. In dermatophytes infecting keratinized tissues, such as Aspergillus spp. and Lactobacillus spp., the Laps (in family M28) and DppIV (in family S9) were shown to synergistically digest the large peptides generated by the endoproteases to give free amino acids and X-Pro dipeptides (Byun et al., 2001). Laps degrade peptides from their N-terminus, while X-Pro acts as a stop sequence. In a complementary manner, these X-Pro sequences can be removed by DppIV and thus allow the Laps access to the next residues (Giddey et al., 2007). Investigations of the keratinases of the keratinolytic non-pathogenic fungus O. corvina suggest that a blend of fungal keratinases – namely an endoprotease (S8), exoprotease (M28), and an oligopeptidase (M3) – act synergistically to break down pig bristle keratin (Huang et al., 2015). Moreover, the enzymology of feather degradation by F. islandicum AW-1, which has recently been reported in detail (Kang et al., 2020), points out that synergies among different types of proteases help the degradation. Hence, after sulfitolysis of feathers, the CPBP family (M48 protease family) intramembrane metalloprotease and five additional membrane proteases (of family S41, S54, S8, M24, A24, respectively) act as key keratinases to catalyze the feather degradation. Subsequently, the released amino acid and peptides are transported into F. islandicum AW-1 for further degradation (involved proteases include proteases belonging to family M16, M38, M55, S16 and T01) (Kang et al., 2020). In addition, the high degradation rates achieved on recalcitrant keratinous material by bacterial consortia also indicates synergies between keratinolytic and other enzymes (Nasipuri et al., 2020).
However, counteracting effects between these different enzymes may also happen during enzyme catalyzed degradation of keratin biomass, mainly because the proteases may attack the other enzymes and/or self-digest. For instance, disulfide bonds in keratinolytic enzymes function to stabilize the enzyme structure. The disulfide reductases or reducing agent added in the hydrolysis reaction may in fact break both keratin and keratinase disulfide bonds; the latter will cause lower enzyme activity and reduce the enzyme stability. Moreover, keratinolytic enzymes themselves can exhibit “autolysis”, especially in the presence of reducing agents (Khan and Ahmad, 2011; Liang et al., 2010). In order to prevent autolysis, the introduction of prolines at the autolytic sites was demonstrated to increase the autolysis resistance of the enzyme under reducing conditions, though sometimes the mutation also changed enzyme specificities (Liang et al., 2010). Synergies and counter-acting effects between the enzymes should be considered when designing keratinase blends for keratin refining. The interactions between the enzymes are complicated and partly unknown, however, and further research is required to identify optimum enzyme combinations for efficient keratinolytic enzyme blends.
6. Three-dimensional structure of keratinolytic enzymes
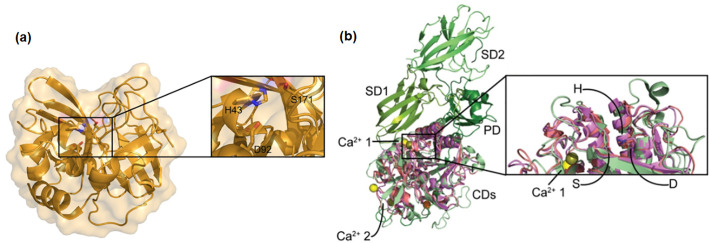
The structures of five keratinolytic enzymes have been X-ray structure analyzed. In addition, the structures of some keratinases in the S8 protease family have been homology modeled based on the crystal structures of subtilisin-like proteases (e.g. PDB identifier of 1MEE, 1SCJ, 1SCN, 3WHI, 3LPA, 3LPC, 3LPD, 3TI9, 3TI7, 1DBI, 1THM, 3AFG) (Fang et al., 2015). The five keratinases for which a crystal structure is known are ANISEP from P. nicotinovorans (Sone et al., 2015), proteinase K from P. album (Betzel et al., 2001; Ebeling et al., 1974; Jany et al., 1986), fervidolysin from F. pennivorans (Friedrich and Antranikian, 1996; Kim et al., 2004), MtaKer from M. taiwanenisis WR-220 (Wu et al., 2017), and FisCP from F. islandicum (Lee et al., 2015a) (Table 4 ). According to the active sites, ANISEP belongs to family S1. Proteinase K, Fervidolysin, MtaKer and subtilisin all belong to family S8, while FisCP is in the metallo-protease family M32.
Table 4.
Keratinolytic enzymes for which the structure is known.
| Protease | Family | NCBI accession number | PDB ID | Active sites | Mw (kDa) | Reference |
|---|---|---|---|---|---|---|
| ANISEP | S1 | BAM67011 | 3WY8 | His43-Asp92-Ser171 | 23 | (Sone et al., 2015) |
| Proteinase K | S8 | 1205229A | 1IC6 | Asp39-His69-Ser224 | 28.9 | (Betzel et al., 2001) |
| Fervidolysin | S8 | AAK61552 | 1R6V | Asp170-His⁎208 -Ser389 | 130 | (Kim et al., 2004) |
| MtaKer | S8 | ARH33809 | 5WSL | Asp39-His72-Ser224 | 41.3 | (Wu et al., 2017) |
| FisCP | M32 | AMW32563 | 5E3X | His253-Glu254-His257-Glu283 (Co2+) | 107 | (Lee et al., 2015a) |
The active site His208 of Fervidolysin is mutated to Ala208 and also show as Ala208 in the structure (PDB ID AAK61552).
6.1. Keratinolytic protease structure in family S1
ANISEP (Fig. 6a) from P. nicotinovorans is significantly more active on keratin azure than on trypsin (Sone et al., 2015). The conserved catalytic triad of ANISEP consists of His43, Asp92, and Ser171 (Fig. 6a), which indicates that ANISEP belongs to Clan PA(S) of the serine protease family (Rawlings and Barrett, 2013a; Sone et al., 2015). The catalytic theory is discussed below (Section 6.2). The active site Ser171 is located in a GGSSG motif (residues 168–172), corresponding to the conserved motif of serine proteases GXSYG (X and Y for any amino acids) (Sone et al., 2015). In clan PA(S) the tertiary structure consists mainly of β-sheets that form a double β-barrel at the core of the enzymes (Rawlings and Barrett, 2013a), which is also the case with ANISEP (Fig. 6a). The active site is located in a shallow cleft that spans the entire enzyme situated between the two β-barrels (Fig. 6a) (Sone et al., 2015).
Fig. 6.
Crystal structures of keratinolytic enzymes of family S1 and S8. (a) Catalytic domain structure of ANISEP, a family S1 keratinase (accession number: BAM67011, PDB ID: 3WY8). The active site catalytic triad amino acids His43-Asp92- Ser171, situated in the upper part of the shallow cleft in the center, are magnified and displayed as sticks. (b) Structures of Fervidolysin (green), Proteinase K (violet), and MtaKer (salmon) family S8 keratinases with the active sites magnified and catalytic triad amino acids, His (H); Asp (D); and Ser (S) displayed as sticks. The CD domain corresponds to the catalytic domain; the PD domain is the propeptide domain; and SD1 and SD2 are β-sandwich domains. The CD domain of Fervidolysin is aligned with the structure of Proteinase K (accession number: 1205229A, PDB ID: 5WSL) and MtaKer (accession number: ARH33809, PDB ID: 5WSL). Proteinase K: Ca2+ ion = yellow, Fervidolysin: Ca2+ ion = orange, MtaKer Ca2+ ion = olive. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
6.2. Keratinolytic protease structure in family S8
More than half of all characterized keratinolytic enzymes belong to the S8 family (Table 2), but the crystal 3D structure has been determined for only three of them, namely Proteinase K (PDB ID 1IC6), Fervidolysin (PDB ID 1R6V) and MtaKer (PDB ID 5WSL) (Table 4). The catalytic domain (CD) of each of these structures consists of seven parallel β-sheets flanked by six (Proteinase K, MtaKer) or nine (Fervidolysin) α-helices (Fig. 6b, Violet: Proteinase K, Green: Fervidolysin, Salmon: MtaKer). Two nearly parallel α-helices are also found in all three structures; these parallel α-helices form the main sites of interaction with the propeptide domain (PD) (Kim et al., 2004).
Though the overall folding of various S8 keratinolytic proteases may differ, they all follow the same mechanism of action through an identical stereochemistry of the catalytic triad (Fig. 7 ) (Betzel et al., 2001). The catalytic triad of the S8 keratinolytic enzymes constitutes Asp, His and Ser in that order in the wild type enzymes (Table 4 and Fig. 6b) (Betzel et al., 2001; Kim et al., 2004; Wu et al., 2017). The His plays a dual role as proton acceptor and donor at different steps in the reaction, while the Asp is thought to bring the His residue into the correct orientation to facilitate nucleophilic attack by Ser (Fig. 7) (Betzel et al., 2001). The whole reaction theory (Hedstrom, 2002) (Fig. 7) is as follows: The keratin substrate binds to the surface of the keratinolytic protease. The nucleophilic Ser acts first as the nucleophile to attack the carbonyl carbon of keratin protein. A tetrahedral intermediate is generated because a pair of electrons from the double bond of the carbonyl oxygen moves to the oxygen (Fig. 7). Next, the electrons move back from the negative oxygen to recreate the bond, generating an acyl-enzyme intermediate and releasing the N-terminus of the peptide. Thus, a larger space is formed for water coming into the reaction to facilitate a nucleophilic attack on the carbonyl carbon of the substrate. The bond between the oxygen of water and the carbon in the substrate is formed to generate another tetrahedral intermediate. In the final stage of the reaction, the carbonyl carbon reforms the double bond with the oxygen. As a result, the C-terminus of the peptide is released. In addition to the catalytic triad, another characteristic of the reaction mechanism is the presence of the oxyanion hole (Fig. 7). The oxyanion hole plays a key role in stabilizing the catalytic tetrahedral intermediate anion and protecting the substrate's negatively charged oxygen from water molecules. In Proteinase K, the negatively charged oxygen ion is stabilized by Ser224 and Asn161 to form an oxyanion hole. As regards Fervidolysin, two peptide nitrogen atoms of Thr388 and Ser389 form the oxyanion hole, where a water molecule mimics the carbonyl oxygen of the scissile peptide bond (Kim et al., 2004). The crystal structure of MtaKer shows that the tetrahedral acyl enzyme intermediate can be stabilized by the oxyanion hole of Asn159 and Thr223 (Wu et al., 2017).
Fig. 7.
Catalytic triad and catalytic mechanism of keratinolytic serine protease enzymes. The nucleophilic Ser first acts as a nucleophile to attack the carbonyl carbon of the keratin protein. A tetrahedral intermediate is generated because a pair of electrons from the double bond of the carbonyl oxygen moves to the oxygen. Next, the electrons move back from the negative oxygen to recreate the bond, generating an acyl-enzyme intermediate and releasing the N-terminus of the peptide. Thus, a larger space is formed to allow water coming into the reaction to facilitate a nucleophilic attack on the carbonyl carbon of the substrate. A bond between the oxygen of water and the carbon in the substrate is formed generating another tetrahedral intermediate. In the final stage of the reaction, the carbonyl carbon reforms the double bond with the oxygen. As a result, the C-terminus of the peptide is released.
One of the major determinants of the substrate specificity of the S8 enzymes is hydrophobic surface pockets (de Kreij et al., 2001; Wu et al., 2017). According to the nomenclature (Schechter and Berger, 1967), the active site residues in the protease are composed of contiguous pockets termed subsites. Amino acid residues in the substrate sequence are consecutively numbered outward from the cleavage sites as Pn…-P2-P1-P1′-P2′-…Pn′, where the scissile bond is located between the P1 and P1′ positions. The subsites in the enzyme are correspondingly labeled as Sn…-S2-S1-S1′-S2′-…Sn′. The S1 subsites in Proteinase K are Ser132-Leu133-Gly134-Gly135 and residues Ala158-Ala159-Gly160-Asn161 at the bottom (Liu et al., 2011). The S1′ subsites are formed by the residues Ser207, His208, Tyr385 and Glu368 in Fervidolysin defined enzyme specificity (Kim et al., 2004). In MtaKer, the S1 binding subsites consists of Ser130-Leu131-Gly132 and Ala156-Ala157-Gly158 (Wu et al., 2017). Overall, the S8 keratinolytic enzyme contains a substrate-binding pocket that is relatively abundant in the nonpolar and smaller-side-chain amino acid residues Ala and Gly. The S1 subsites structure may be associated with specificity for keratin. S8 keratinolytic enzymes prefer to cleave the Phe, Tyr, and Arg at the P1 site of a synthetic pNa substrate (Brandelli et al., 2010).
Many proteases in the S8 family contain one or more calcium ion-binding sites. Two Ca2+ ions are found in Proteinase K (Fig. 6b, yellow Ca2+): the first Ca site contributes to the stabilization of surrounding regions, especially the long connecting loops. The second Ca site stabilizes to some extent the N- and C-terminal regions of the molecular structure (Liu et al., 2011). In MtaKer, two Ca2+ ions are also observed in the structure (Fig. 6b, Ca2+ ion colored in olive). The first Ca2+ (Ca2+ 1) is conserved with the first Ca2+ (Ca2+ 1) of Proteinase K (Fig. 6b, yellow Ca2+ ion), and, similarly, with the first Ca2+ of MtaKer involved in stabilizing the surface loop. The second Ca2+ is not conserved and this Ca2+ comes into contact with the main-chain carbonyl oxygen atoms of the surrounding amino acids residues and two water molecules (Wu et al., 2017). By comparison, only one Ca2+ ion is found in Fervidolysin (Fig. 6b, orange Ca2+) (Kim et al., 2004). The second calcium is not conserved.
6.3. Keratinolytic protease structure in family M32
FisCP from F. islandicum is an M32 keratinolytic protease. Unlike ANISEP (S1family), which consists primarily of β-sheets and S8 keratinolytic enzymes whose structures are a mix of α-helices and β-sheets, FisCP primarily consists of α-helices with a three-stranded β-sheet near the active site (Fig. 8a). The active site of FisCP is located in the HEXXH motif (Table 4) - two His (His253 and His257) and a solvent water molecule coordinated with Co2+ bound to the third active site with Glu283 (Fig. 8a and b) (Lee et al., 2015a). Coordination of the substrate through the scissile carbonyl group is assisted by the presence of the positively charged Co2+ (Fig. 8c). After deprotonation of the water molecule, the resulting hydroxide nucleophilically attacks the scissile carbonyl carbon; this results in protonation of the scissile amide group and cleavage of the peptide bonds to form the peptide products (Fig. 8c) (Szeto et al., 2009).
Fig. 8.
(a) Crystal structure of FisCP (accession number: AMW32563, PDB ID: 5E3X); (b) The active sites of FisCP (His253-Glu254-His257-Glu283 (Co2+)) are displayed as sticks; (c) The mechanism of catalyzing the hydrolysis of protein by FisCP: deprotonation of the nucleophilic water molecule; nucleophilic attack of the resulting hydroxide on the scissile carbonyl carbon; protonation of the scissile amide group, followed by cleavage of the peptide bond (Szeto et al., 2009).
Most M32 proteins are oligopeptidases and the length of their substrate is governed by the length of the active site groove, which closes upon substrate binding (Sharma et al., 2017). The Arg (Arg92 in PfuCP), located at the back of the substrate groove, is 100% conserved in over 600 sequences of M32CPs (Lee et al., 2009). The region surrounding this conserved arginine is re-adjusted during gate closure. The “gate” was found in M32 carboxypeptidases subfamily I (e.g. Thermus aquaticus (TaqCP), Pyrococcus furiosus (PfuCP), Leishmania major (LmaCP)). Hence, M32 subfamily I should be able to cleave only those substrates that are small enough to fit into the narrow substrate groove and allow the active site gate to be closed (Sharma et al., 2017). Furthermore, TaqCP, PfuCP and LmaCP in subfamily I have broad substrate specificity with a C-terminal amino acid preference in the order basic > aliphatic > aromatic >> acidic and the substrate is limited to 7–15 residues (Lee et al., 2009). However, not all M32 proteins have length restriction. For example B. subtilis (BsuCP) and Thermus thermophilus (TthCP) lack conserved Arg, which is why they have no substrate length limitations, although the rate of catalysis may vary depending on the substrate length (Lee et al., 2009). Additionally, unlike the broad substrate specificity in subfamily I, BsuCP does not cleave C-terminal aliphatic and polar amino acids when tested with a series of benzyloxycarbonyl-Ala-X (ZAX) substrates (where X is various amino acids) (Lee et al., 2009). Alignment of FisCP with other M32 proteins demonstrates that FisCP lacks a key Arg residue of substrate length restriction, whereas it has a lysine residue, as also found in BsuCP, which indicates that the substrate length restriction of FisCP differs from CPs in the M32 subfamily I (Lee et al., 2015a). The substrate specificity of FisCP may also be narrower than that of proteases in subfamily I. This substrate discrimination is attributed to the keratin structure. Nevertheless, the exact substrate restriction length and specificity of FisCP still need to be clarified.
7. Applications of keratinolytic enzymes
Keratinolytic enzymes are already applied industrially, for example in detergents, in production of leather, and in bioremediation. Keratinases are attractive detergent agents because they have broad specificity for both soluble and insoluble proteins as substrates. A bio-detergent formulated with alkaline keratinase (from Paenibacillus woosongensis TKB2) can remove blood and egg yolk stains efficiently (Paul et al., 2014). Keratinolytic enzymes produced by Paecilomyces lilacinus (Cavello et al., 2012), Gibberella intermedia (Zhang et al., 2016) and B. pumilus (Gong et al., 2015) have also shown potential in detergent applications. Additionally, keratinolytic enzymes may find use in detergents for cleaning drains and clogged pipes caused for example by hair (Gupta et al., 2013a). In the leather industry, de-hairing by (keratinolytic) enzymes is also considered environmentally friendly (Fang et al., 2017). Keratinolytic enzymes are ideal for this purpose because they exhibit high dehairing activity but no or only very weak collagenolytic activity and elastinolytic activity. For example, a keratinolyic enzyme from Brevibacillus sp. AS-S10-II with no collagen-degrading activity was demonstrated to work as a dehairing agent when tested on goat skin (Rai and Mukherjee, 2011). The enzymes may also reduce the toxicity of wastewater effluents from the leather industry. In addition, industries involved in bio-remediation of soil and wastewater also offer some important applications of keratinolytic enzymes (Brandelli et al., 2010; Gupta et al., 2013a; Sharma and Devi, 2018; Tesfaye et al., 2017a; Verma et al., 2017).
In the agroindustry, one of the most important and potentially significant high-volume applications of keratinases is in keratin co-product management. Keratin harbors at least 17 amino acids (see Table 1). These amino acids may be recycled for use as fertilizer or in animal and aquaculture feed in processes offering high-impact applications of enzymatically degraded keratin such as feathers and pig bristles. Keratinolytic enzymes derived from Bacillus sp. SLII-1 have been shown to be able to degrade chicken feathers and the hydrolysate could in turn substitute about 5% of soybean meal protein in broiler feed. The performance of broiler chickens on this feed was better than with conventional soybean meal protein (Larasati et al., 2017). Similarly, addition of “Versazyme” (a keratinase feed additive produced by B. licheniformis PWD-1) or other keratinases to feed mixtures may improve growth performance, breast meat yield, and gut villus structure of broilers fed diets based on corn and soybean meal (Huang et al., 2018; Wang et al., 2008, Wang et al., 2006). Chicken feather hydrolysates could also serve as a cheap source of liquid organic fertilizer. B. licheniformis AS-S24-I keratinase coupled to iron-oxide magnetic nanoparticles hydrolyzed chicken feathers, and the filtered sterile hydrolysate produced a significant increase in the length and growth of Bengal gram seedlings and increased the soil microbial population (Rai and Mukherjee, 2015). Furthermore, keratinolytic enzymes have been used to suppress nematodes and action of entomopathogenic microorganisms (Brandelli et al., 2010; Gupta et al., 2013a; Verma et al., 2017). A keratinase from Bacillus sp. 50–3 is able to kill Meloidogyne incognita (a root-knot nematode) may be used as biological pesticide (Yue et al., 2011). Overall, keratinolytic enzymes have potential in the agroindustry, and the most relevant enzymes are obtained from Bacillus sp. mostly belonging to the protease S8 family. However, the present review shows that other efficient and specific keratinolytic enzymes may be worth further investigation as well for agroindustrial applications.
In biomedicine, keratinolytic enzymes are applied in treatment of nails, calluses, acne, scars, prions, and skin, and as a cosmetics supplement. For instance, Pure100 keratinase is marketed for treating nail disorders, calluses and for prion decontamination (Gupta et al., 2013a). Keratinolytic enzymes can even contribute to improving drug delivery through the skin (epidermis) (Gupta et al., 2013a). Indeed, keratinolytic enzymes have been added as supplements to cosmetics for use in skin whitening and in exfoliation and for assisting drug permeation (Anandharaj et al., 2016; Gupta et al., 2013a). Keratinolytic enzymes formulated in dehairing cream have also been reported to be particularly effective in removing hair compared to conventional marketed formulations (Sanghvi et al., 2016). Use in prion decontamination is another function of keratinolytic enzymes because the enzymes cleave β-plated protein that is particularly prevalent in prion proteins. Several keratinolytic enzymes showing ability to digest prion protein are produced by B. licheniformis PWD-1 (Langeveld et al., 2003), Streptomyces sp. (Tsiroulnikov et al., 2004), Nocardiopsis sp. TOA-1 (Mitsuiki et al., 2006), Thermoanaerobacter, Thermosipho, and Thermococcus sp. (Suzuki et al., 2006). Furthermore, microbial keratin-rich material hydrolysis can generate bioactive peptides, and this constitutes a point of utmost interest for development of functional ingredients with elevated value (Callegaro et al., 2019), for example, for use in pharmaceuticals and cosmetics (Jin et al., 2018; Yeo et al., 2018). Keratinolytic enzymes may also be used in bioenergy applications by assisting the production of biofuel and biogas. This function is mainly related to keratin hydrolysates serving as nitrogen rich sources for microbial production of methane, fuel pellets and bio‑hydrogen. The contribution of keratinolytic organisms during biohydrogen production has been investigated. Keratinolytic Bacillus sp. degrades keratin first, and then the hyperthermophilic archaeon produces H2 highly efficiently from keratin-derived breakdown products (Bálint et al., 2005). Chicken feathers hydrolyzed by keratinolytic enzymes during pretreatment with Bacillus sp. yielded 124% more CH4 during biogas fermentation than non-pretreated feather (Patinvoh et al., 2016). However, research on keratin-based biogas production is still limited, and further investigations are needed into such fermentation processes.
8. Conclusions and future perspectives
In conclusion, certain microbially derived proteolytic enzymes exhibit significant keratinolytic activity, and thus readily catalyze the degradation of keratin. Keratinolytic enzymes are therefore highly useful for various applications in biomedicine, industry, agroindustry, and bioenergy. Compared with chemical treatment of keratin, enzymes act more gently and do not denature the amino acids. Applications of keratinolytic enzymes are therefore being increasingly explored, and some keratinolytic enzyme-based products have already been commercialized (Gupta et al., 2013a).
Despite such developments, the current knowledge on how keratinolytic enzymes function is limited, especially with regard to the structure-function aspects, substrate specificity, kinetic traits (rate, substrate affinity), biological diversity of the enzymes and their reaction robustness. Hence, further investigations are warranted. In particular, the mechanisms, kinetics, and free energy of catalytic conversion and the significance of supplying reductases for keratinases deserve investigation. Such research will help understand both natural bioconversion of keratin (and its significance in larger natural C- and N-cycles), and the practical use and selection of efficient keratin-converting biocatalysts for new technical applications. A standardized set of assays with proper keratin substrates, i.e. able to distinguish any differences in kinetic rates on α- and β-keratin, and more stable and accurate measurement methods are recommended. Besides, wider use of bioinformatics tools may help further discovery and understanding of keratinases, in particular when it comes to rationally identifying efficient keratinases for use in new upcycling processes of keratinous biomass. In this review, keratinases are classified into different protease families that may relate to different functional theories and products. Therefore, for application purposes, it is important to study the design and optimization of efficient enzyme blends towards different keratin-rich substrate or special products.
Acknowledgements
This work is supported by the Technical University of Denmark and the China Scholarship Council (CSC) grant #201806910049.
References
- Alexandratos N., Bruinsma J. Food Agric. Organ; Rome: 2012. World Agriculture Towards 2030/2050: The 2012 Revision. ESA Work. Pap. No. 12–03. [Google Scholar]
- Alibardi L. Ultrastructural autoradiographic and immunocytochemical analysis of setae formation and keratinization in the digital pads of the gecko Hemidactylus turcicus (Gekkonidae, Reptilia) Tissue Cell. 2003;35:288–296. doi: 10.1016/s0040-8166(03)00050-8. [DOI] [PubMed] [Google Scholar]
- Alibardi L. Keratinization of sheath and Calamus cells in developing and regenerating feathers. Ann. Anat. 2007;189:583–595. doi: 10.1016/j.aanat.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Alibardi L., Segalla A. The process of cornification in the horny teeth of the lamprey involves proteins in the keratin range and other keratin-associated proteins. Zool. Stud. 2011;50:416–425. [Google Scholar]
- Anandharaj M., Sivasankari B., Siddharthan N., Rani R.P., Sivakumar S. Production, purification, and biochemical characterization of thermostable metallo-protease from novel Bacillus alkalitelluris TWI3 isolated from tannery waste. Appl. Biochem. Biotechnol. 2016;178:1666–1686. doi: 10.1007/s12010-015-1974-7. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bálint B., Bagi Z., Tóth A., Rákhely G., Perei K., Kovács K.L. Utilization of keratin-containing biowaste to produce biohydrogen. Appl. Microbiol. Biotechnol. 2005;69:404–410. doi: 10.1007/s00253-005-1993-3. [DOI] [PubMed] [Google Scholar]
- Ben Hamad Bouhamed S., Kechaou N. Kinetic study of sulphuric acid hydrolysis of protein feathers. Bioprocess Biosyst. Eng. 2017;40:715–721. doi: 10.1007/s00449-017-1737-7. [DOI] [PubMed] [Google Scholar]
- Betzel C., Gourinath S., Kumar P., Kaur P., Perbandt M., Eschenburg S., Singh T.P. Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 Å resolution. Biochemistry. 2001;40:3080–3088. doi: 10.1021/bi002538n. [DOI] [PubMed] [Google Scholar]
- Böckle B., Müller R. Reduction of disulfide bonds by Streptomyces pactum during growth on chicken feathers. Appl. Environ. Microbiol. 1997;63:790–792. doi: 10.1128/aem.63.2.790-792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragulla H.H., Homberger D.G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009;214:516–559. doi: 10.1111/j.1469-7580.2009.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandelli A., Daroit D.J., Riffel A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010;85:1735–1750. doi: 10.1007/s00253-009-2398-5. [DOI] [PubMed] [Google Scholar]
- Brouta F., Descamps F., Monod M., Vermout S., Losson B., Mignon B. Secreted metalloprotease gene family of Microsporum canis. Infect. Immun. 2002;70:5676–5683. doi: 10.1128/iai.70.10.5676-5683.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester A., Shelest E., Glöckner G., Heddergott C., Schindler S., Staib P., Heidel A., Felder M., Petzold A., Szafranski K., Feuermann M., Pedruzzi I., Priebe S., Groth M., Winkler R., Li W., Kniemeyer O., Schroeckh V., Hertweck C., Hube B., White T.C., Platzer M., Guthke R., Heitman J., Wöstemeyer J., Zipfel P.F., Monod M., Brakhage A.A. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 2011;12:1–16. doi: 10.1186/gb-2011-12-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk P.K., Lange L. Classification of fungal and bacterial lytic polysaccharide monooxygenases. BMC Genomics. 2015;16:368–381. doi: 10.1186/s12864-015-1601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T., Kofod L., Blinkovsky A. Synergistic action of an X-prolyl dipeptidyl aminopeptidase and a non-specific aminopeptidase in protein hydrolysis. J. Agric. Food Chem. 2001;49:2061–2063. doi: 10.1021/jf001091m. [DOI] [PubMed] [Google Scholar]
- Callegaro K., Brandelli A., Daroit D.J. Beyond plucking: feathers bioprocessing into valuable protein hydrolysates. Waste Manag. 2019;95:399–415. doi: 10.1016/j.wasman.2019.06.040. [DOI] [PubMed] [Google Scholar]
- Calvaresi M., Eckhart L., Alibardi L. The molecular organization of the beta-sheet region in corneous beta- proteins (beta-keratins) of sauropsids explains its stability and polymerization into filaments. J. Struct. Biol. 2016;194:282–291. doi: 10.1016/j.jsb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Cavello I.A., Hours R.A., Cavalitto S.F. Bioprocessing of “hair waste” by Paecilomyces lilacinus as a source of a bleach-stable, alkaline, and thermostable keratinase with potential application as a laundry detergent additive: characterization and wash performance analysis. Biotechnol. Res. Int. 2012;2012:1–12. doi: 10.1155/2012/369308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya E., Suzuki T., Karita S., Hanya A., Yoshino-Yasuda S., Kitamoto N. Sequence analysis and heterologous expression of the wool cuticle-degrading enzyme encoding genes in Fusarium oxysporum 26-1. J. Biosci. Bioeng. 2014;117:711–714. doi: 10.1016/j.jbiosc.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhou B., Xu M., Huang Z., Jia G., Zhao H., Liu G. Prokaryotic expression and characterization of a keratinolytic protease from Aspergillus Niger. Biol. 2015;70:157–164. doi: 10.1515/biolog-2015-0031. [DOI] [Google Scholar]
- Chou C.-C., Buehler M.J. Structure and mechanical properties of human trichocyte keratin intermediate filament protein. Biomacromolecules. 2012;13:3522–3532. doi: 10.1021/bm301254u. [DOI] [PubMed] [Google Scholar]
- Chou C.F., Omary M.B. Mitotic arrest-associated enhancement of O-linked glycosylation and phosphorylation of human keratins 8 and 18. J. Biol. Chem. 1993;268:4465–4472. [PubMed] [Google Scholar]
- Jarai G., Daniel K., Frank P.B. Cloning and characterization of the pepD gene of Aspergillus niger which codes for a subtilisin-like protease. Gene. 1994;139:51–57. doi: 10.1016/0378-1119(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Descamps F., Brouta F., Baar D., Losson B., Mignon B. Isolation of a Microsporum canis gene family encoding three subtilisin-like proteases expressed in vivo. J. Invest. Dermatol. 2002;119:830–835. doi: 10.1046/j.1523-1747.2002.01784.x. [DOI] [PubMed] [Google Scholar]
- Dozie I.N.S., Okeke C.N., Unaeze N.C. A thermostable, alkaline-active, keratinolytic proteinase from Chrysosporium keratinophilum. World J. Microbiol. Biotechnol. 1994;10:563–567. doi: 10.1007/bf00367668. [DOI] [PubMed] [Google Scholar]
- e Silva L.A.D., Macedo A.J., Termignoni C. Production of keratinase by Bacillus subtilis S14. Ann. Microbiol. 2014;64:1725–1733. doi: 10.1007/s13213-014-0816-0. [DOI] [Google Scholar]
- Ebeling W., Hennrich N., Klockow M., Metz H., Orth H.D., Lang H. Proteinase K from Tritirachium album limber. Eur. J. Biochem. 1974;47:91–97. doi: 10.1111/j.1432-1033.1974.tb03671.x. [DOI] [PubMed] [Google Scholar]
- Edwards H.G.M., Hunt D.E., Sibley M.G. FT-Raman spectroscopic study of keratotic materials: horn, hoof and tortoiseshell. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 1998;54:745–757. doi: 10.1016/S1386-1425(98)00013-4. [DOI] [PubMed] [Google Scholar]
- Eslahi N., Dadashian F., Nejad N.H. An investigation on keratin extraction from wool and feather waste by enzymatic hydrolysis. Prep. Biochem. Biotechnol. 2013:624–648. doi: 10.1080/10826068.2013.763826. [DOI] [PubMed] [Google Scholar]
- Esteban M.B., García A.J., Ramos P., Márquez M.C. Sub-critical water hydrolysis of hog hair for amino acid production. Bioresour. Technol. 2010;101:2472–2476. doi: 10.1016/j.biortech.2009.11.054. [DOI] [PubMed] [Google Scholar]
- Fang Z., Zhang J., Liu B., Du G., Chen J. Biochemical characterization of three keratinolytic enzymes from Stenotrophomonas maltophilia BBE11-1 for biodegrading keratin wastes. Int. Biodeterior. Biodegrad. 2013;82:166–172. doi: 10.1016/j.ibiod.2013.03.008. [DOI] [Google Scholar]
- Fang Z., Zhang J., Liu B., Jiang L., Du G., Chen J. Cloning, heterologous expression and characterization of two keratinases from Stenotrophomonas maltophilia BBE11-1. Process Biochem. 2014;49:647–654. doi: 10.1016/j.procbio.2014.01.009. [DOI] [Google Scholar]
- Fang Z., Zhang J., Liu B., Du G., Chen J. Insight into the substrate specificity of keratinase KerSMD from Stenotrophomonas maltophilia by site-directed mutagenesis studies in the S1 pocket. RSC Adv. 2015;5:74953–74960. doi: 10.1039/c5ra12598g. [DOI] [Google Scholar]
- Fang Z., Yong Y.C., Zhang J., Du G., Chen J. Keratinolytic protease: a green biocatalyst for leather industry. Appl. Microbiol. Biotechnol. 2017;101:7771–7779. doi: 10.1007/s00253-017-8484-1. [DOI] [PubMed] [Google Scholar]
- Fellahi S., Chibani A., Feuk-Lagerstedt E., Taherzadeh M.J. Identification of two new keratinolytic proteases from a Bacillus pumilus strain using protein analysis and gene sequencing. AMB Express. 2016;6:42–48. doi: 10.1186/s13568-016-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser R.D.B., Parry D.A.D. Molecular packing in the feather keratin filament. J. Struct. Biol. 2008;162:1–13. doi: 10.1016/j.jsb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Fraser R.D.B., Parry D.A.D. The role of β-sheets in the structure and assembly of keratins. Biophys. Rev. 2009;1:27–35. doi: 10.1007/s12551-008-0005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser R.D.B., Parry D.A.D. The structural basis of the filament-matrix texture in the avian/reptilian group of hard β-keratins. J. Struct. Biol. 2011;173:391–405. doi: 10.1016/j.jsb.2010.09.020. [DOI] [PubMed] [Google Scholar]
- Fraser R.D.B., Parry D.A.D. Amino acid sequence homologies in the hard keratins of birds and reptiles, and their implications for molecular structure and physical properties. J. Struct. Biol. 2014;188:213–224. doi: 10.1016/j.jsb.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Frenkel M.J., Gillespie J.M. The proteins of the keratin component of bird’s beaks. Aust. J. Biol. Sci. 1976;29:467–480. doi: 10.1071/bi9760467. [DOI] [PubMed] [Google Scholar]
- Friedrich A.B., Antranikian G. Keratin degradation by Fervidobacterium pennavorans, a novel thermophilic anaerobic species of the order Thermotogales. Appl. Environ. Microbiol. 1996;62:2875–2882. doi: 10.1128/aem.62.8.2875-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegeckas A., Gudiukaite R., Debski J., Citavicius D. Keratinous waste decomposition and peptide production by keratinase from Geobacillus stearothermophilus AD-11. Int. J. Biol. Macromol. 2015;75:158–165. doi: 10.1016/j.ijbiomac.2015.01.031. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Chakrabarti K., Chattopadhyay D. Cloning of feather-degrading minor extracellular protease from Bacillus cereus DCUW: dissection of the structural domains. Microbiology. 2009;155:2049–2057. doi: 10.1099/mic.0.027573-0. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Clerens S., Deb-Choudhury S., Dyer J.M. Thermal effects of ionic liquid dissolution on the structures and properties of regenerated wool keratin. Polym. Degrad. Stab. 2014;108:108–115. [Google Scholar]
- Giddey K., Monod M., Barblan J., Potts A., Waridel P., Zaugg C., Quadroni M. Comprehensive analysis of proteins secreted by Trichophyton rubrum and Trichophyton violaceum under in vitro conditions. J. Proteome Res. 2007;6:3081–3092. doi: 10.1021/pr070153m. [DOI] [PubMed] [Google Scholar]
- Gillespie J.M., Marshall R.C., Woods E.F. A comparison of lizard claw keratin proteins with those of avian beak and claw. J. Mol. Evol. 1982;18:121–129. doi: 10.1007/BF01810831. [DOI] [PubMed] [Google Scholar]
- Gong H., Zhou H., McKenzie G.W., Yu Z., Clerens S., Dyer J.M., Plowman J.E., Wright M.W., Arora R., Bawden C.S., Chen Y., Li J., Hickford J.G.H. An updated nomenclature for keratin-associated proteins (KAPs) Int. J. Biol. Sci. 2012;8:258–264. doi: 10.7150/ijbs.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Wang Y., Zhang D., Li H., Zhang X., Zhang R., Lu Z., Xu Z., Shi J. A surfactant-stable Bacillus pumilus K9 α-keratinase and its potential application in detergent industry. Chem. Res. Chin. Univ. 2015;31:91–97. doi: 10.1007/s40242-015-4351-8. [DOI] [Google Scholar]
- Gonzalo M., Espersen R., Al-Soud W.A., Cristiano Falco F., Hägglund P., Sørensen S.J., Svensson B., Jacquiod S. Azo dying of α-keratin material improves microbial keratinase screening and standardization. Microb. Biotechnol. 2020 doi: 10.1111/1751-7915.13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwold M.J., Bao W., Jarvis E.D., Hu H., Li C., Gilbert M.T.P., Zhang G., Sawyer R.H. Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles. BMC Evol. Biol. 2014;14:1–16. doi: 10.1186/s12862-014-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt M., Monod M., Yamada T., Hertweck C., Kunert J., Staib P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J. Invest. Dermatol. 2013;133:1550–1555. doi: 10.1038/jid.2013.41. [DOI] [PubMed] [Google Scholar]
- Gupta R., Ramnani P. Microbial keratinases and their prospective applications: an overview. Appl. Microbiol. Biotechnol. 2006;70:21–33. doi: 10.1007/s00253-005-0239-8. [DOI] [PubMed] [Google Scholar]
- Gupta S., Singh R. Statistical modeling and optimization of keratinase production from newly isolated Bacillus subtilis RSE163. Int. J. Adv. Biotechnol. Res. 2013;4:1030–1037. [Google Scholar]
- Gupta R., Rajput R., Sharma R., Gupta N. Biotechnological applications and prospective market of microbial keratinases. Appl. Microbiol. Biotechnol. 2013;97:9931–9940. doi: 10.1007/s00253-013-5292-0. [DOI] [PubMed] [Google Scholar]
- Gupta R., Sharma R., Beg Q.K. Revisiting microbial keratinases: next generation proteases for sustainable biotechnology. Crit. Rev. Biotechnol. 2013;33:216–228. doi: 10.3109/07388551.2012.685051. [DOI] [PubMed] [Google Scholar]
- Habbeche A., Saoudi B., Jaouadi B., Haberra S., Kerouaz B., Boudelaa M., Badis A., Ladjama A. Purification and biochemical characterization of a detergent-stable keratinase from a newly thermophilic actinomycete Actinomadura keratinilytica strain Cpt29 isolated from poultry compost. J. Biosci. Bioeng. 2014;117:413–421. doi: 10.1016/j.jbiosc.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Harrap B.S. t, Woods E.F. Soluble derivatives of feather keratin. 1. Isolation, fractionation and amino acid composition. Biochem. J. 1964;92:8–18. doi: 10.1042/bj0920008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Sun R., Tang Z., Bu T., Wu Q., Li C., Chen H. Biodegradation of feather waste keratin by the keratin-degrading strain Bacillus subtilis 8. J. Microbiol. Biotechnol. 2018;28:314–322. doi: 10.4014/jmb.1708.08077. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Hill P., Brantley H., Van Dyke M. Some properties of keratin biomaterials: kerateines. Biomaterials. 2010;31:585–593. doi: 10.1016/j.biomaterials.2009.09.076. [DOI] [PubMed] [Google Scholar]
- Huang Y., Busk P.K., Herbst F.A., Lange L. Genome and secretome analyses provide insights into keratin decomposition by novel proteases from the non-pathogenic fungus Onygena corvina. Appl. Microbiol. Biotechnol. 2015;99:9635–9649. doi: 10.1007/s00253-015-6805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ma D., Zang J., Zhang B., Sun B., Liu L., Zhang S. Effect of keratinase on ileal amino acid digestibility in five feedstuffs fed to growing pigs. Asian-Australas. J. Anim. Sci. 2018;31:1946–1955. doi: 10.5713/ajas.17.0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M.S., Sequeiros C., García S., Olivera N.L. Newly isolated Bacillus sp. G51 from Patagonian wool produces an enzyme combination suitable for felt-resist treatments of organic wool. Bioprocess Biosyst. Eng. 2017;40:833–842. doi: 10.1007/s00449-017-1748-4. [DOI] [PubMed] [Google Scholar]
- Intagun W., Kanoksilapatham W. A review: biodegradation and applications of keratin degrading microorganisms and keratinolytic enzymes, focusing on thermophiles and thermostable serine proteases. Am. J. Appl. Sci. 2017;14:1016–1023. doi: 10.3844/ajassp.2017.1016.1023. [DOI] [Google Scholar]
- Irwin McLean W.H., Moore C.B. Keratin disorders: from gene to therapy. Hum. Mol. Genet. 2011;20:189–197. doi: 10.1093/hmg/ddr379. [DOI] [PubMed] [Google Scholar]
- Jankiewicz U., Larkowska E., Brzezinska M.S. Production, characterization, gene cloning, and nematocidal activity of the extracellular protease from Stenotrophomonas maltophilia N4. J. Biosci. Bioeng. 2016;121:614–618. doi: 10.1016/j.jbiosc.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Jany K.-D., Lederer G., Mayer B. Amino acid sequence of proteinase K from the mold Tritirachium album limber: proteinase K—A subtilisin-related enzyme with disulfide bonds. FEBS Lett. 1986;199:139–144. doi: 10.1016/0014-5793(86)80467-7. [DOI] [Google Scholar]
- Jin H.S., Park S.Y., Kim K., Lee Y.J., Nam G.W., Kang N.J., Lee D.W. Development of a keratinase activity assay using recombinant chicken feather keratin substrates. PLoS ONE. 2017;12:1–18. doi: 10.1371/journal.pone.0172712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H.S., Song K., Baek J.H., Lee J.E., Kim D.J., Nam G.W., Kang N.J., Lee D.W. Identification of matrix metalloproteinase-1-suppressive peptides in feather keratin hydrolysate. J. Agric. Food Chem. 2018;66:12719–12729. doi: 10.1021/acs.jafc.8b05213. [DOI] [PubMed] [Google Scholar]
- Jousson O., Léchenne B., Bontems O., Mignon B., Reichard U., Barblan J., Quadroni M., Monod M. Secreted subtilisin gene family in Trichophyton rubrum. Gene. 2004;339:79–88. doi: 10.1016/j.gene.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Kang E., Jin H.S., La J.W., Sung J.Y., Park S.Y., Kim W.C., Lee D.W. Identification of keratinases from Fervidobacterium islandicum AW-1 using dynamic gene expression profiling. Microb. Biotechnol. 2020;13:442–457. doi: 10.1111/1751-7915.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperova A., Kunert J., Raska M. The possible role of dermatophyte cysteine dioxygenase in keratin degradation. Med. Mycol. 2013;51:449–454. doi: 10.3109/13693786.2013.794310. [DOI] [PubMed] [Google Scholar]
- Kaufman G., Berdicevsky I., Woodfolk J.A., Horwitz B.A. Markers for host-induced gene expression in Trichophyton dermatophytosis. Infect. Immun. 2005;73:6584–6590. doi: 10.1128/IAI.73.10.6584-6590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.S.A., Ahmad I. In vitro antifungal, anti-elastase and anti-keratinase activity of essential oils of Cinnamomum-, Syzygium- and Cymbopogon-species against Aspergillus fumigatus and Trichophyton rubrum. Phytomedicine. 2011;19:48–55. doi: 10.1016/j.phymed.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Kluskens L.D., De Vos W.M., Huber R., Van Der Oost J. Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin. Acta Sci. Pol. Biotechnol. 2004;335:787–797. doi: 10.1016/j.jmb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Kirfel J., Magin T.M., Reichelt J. Keratins: a structural scaffold with emerging functions. Cell. Mol. Life Sci. 2003;60:56–71. doi: 10.1007/s000180300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korniłłowicz-Kowalska T., Bohacz J. Biodegradation of keratin waste: theory and practical aspects. Waste Manag. 2011;31:1689–1701. doi: 10.1016/j.wasman.2011.03.024. [DOI] [PubMed] [Google Scholar]
- de Kreij A., van den Burg B., Veltman O.R., Vriend G., Venema G., Eijsink V.G.H. The effect of changing the hydrophobic S1’ subsite of thermolysin-like proteases on substrate specificity. Eur. J. Biochem. 2001;268:4985–4991. doi: 10.1046/j.0014-2956.2001.02434.x. [DOI] [PubMed] [Google Scholar]
- Łaba W., Choińska A., Rodziewicz A. The release of sulfur compounds during degradation of feather keratin by two Bacillus strains. Acta Sci. Pol. Biotechnol. 2013;12:29–40. [Google Scholar]
- Lange L., Huang Y., Busk P.K. Microbial decomposition of keratin in nature—a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016;100:2083–2096. doi: 10.1007/s00253-015-7262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld J.P.M., Wang J.-J., Van de Wiel D.F.M., Shih G.C., Garssen G.J., Bossers A., Shih J.C.H. Enzymatic degradation of prion protein in brain stem from infected cattle and sheep. J. Infect. Dis. 2003;188:1782–1789. doi: 10.1086/379664. [DOI] [PubMed] [Google Scholar]
- Larasati D., Tsurayya N., Koentjoro M.P., Prasetyo E.N. Keratinase from newly isolated strain of thermophilic Bacillus for chicken feed modification. AIP Conf. Proc. 2017;1854 doi: 10.1063/1.4985413. 020022-1-020022-10. [DOI] [Google Scholar]
- Laskar A., Rodger E.J., Chatterjee A., Mandal C. Modeling and structural analysis of PA clan serine proteases. BMC. Res. Notes. 2012;5:239–245. doi: 10.1186/1756-0500-5-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Taguchi H., Yoshimura E., Minagawa E., Kaminogawa S., Ohta T., Matsuzawa H. Carboxypeptidase taq, a thermostable zinc enzyme, from Thermus aquaticus yt-1: molecular cloning, sequencing, and expression of the encoding gene in Escherichia coli. Biosci. Biotechnol. Biochem. 1994;58:1490–1495. doi: 10.1080/bbb.58.1490. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Isaza C.E., White J.D., Chen R.P.Y., Liang G.F.C., He H.T.F., Chan S.I., Chan M.K. Insight into the substrate length restriction of M32 carboxypeptidases: characterization of two distinct subfamilies. Proteins. 2009;77:647–657. doi: 10.1002/prot.22478. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Kim M.S., Chung B.M., Leahy D.J., Coulombe P.A. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat. Struct. Mol. Biol. 2012;19:707–715. doi: 10.1038/nsmb.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Dhanasingh I., Ahn J.S., Jin H.S., Choi J.M., Lee S.H., Lee D.W. Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1. Biochem. Biophys. Res. Commun. 2015;468:927–933. doi: 10.1016/j.bbrc.2015.11.058. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Jeong H., Park G.S., Kwak Y., Lee Sang Jae, Lee Sang Jun, Park M.K., Kim J.Y., Kang H.K., Shin J.H., Lee D.W. Genome sequence of a native-feather degrading extremely thermophilic Eubacterium, Fervidobacterium islandicum AW-1. Stand. Genomic Sci. 2015;10:1–9. doi: 10.1186/s40793-015-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang K.-Q. Independent expansion of zincin metalloproteinases in Onygenales fungi may be associated with their pathogenicity. PLoS ONE. 2014;9:e90225. doi: 10.1371/journal.pone.0090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Shi P.J., Han X.Y., Meng K., Yang P.L., Wang Y.R., Luo H.Y., Wu N.F., Yao B., Fan Y.L. Functional expression of the keratinolytic serine protease gene sfp2 from Streptomyces fradiae var. k11 in Pichia pastoris. Protein Expr. Purif. 2007;54:79–86. doi: 10.1016/j.pep.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Li Z., Reimer C., Picard M., Mohanty A.K., Misra M. Characterization of chicken feather biocarbon for use in sustainable biocomposites. Front. Mater. 2020;7:1–12. doi: 10.3389/fmats.2020.00003. [DOI] [Google Scholar]
- Liang X., Bian Y., Tang X.F., Xiao G., Tang B. Enhancement of keratinolytic activity of a thermophilic subtilase by improving its autolysis resistance and thermostability under reducing conditions. Appl. Microbiol. Biotechnol. 2010;87:999–1006. doi: 10.1007/s00253-010-2534-2. [DOI] [PubMed] [Google Scholar]
- Liu S.Q., Liang L.M., Tao Y., Yang L.Q., Ji X.L., Yang J.K., Fu Y.X., Zhang K.Q. In: Pesticides in the Modern World - Pests Control and Pesticides Exposure and Toxicity Assessment. Stoytcheva M., editor. E-Publishing; 2011. Structural and dynamic basis of serine proteases from nematophagous fungi for cuticle degradation; pp. 333–376. [DOI] [Google Scholar]
- Ma B., Qiao X., Hou X., Yang Y. Pure keratin membrane and fibers from chicken feather. Int. J. Biol. Macromol. 2016;89:614–621. doi: 10.1016/j.ijbiomac.2016.04.039. [DOI] [PubMed] [Google Scholar]
- Ma Y., Ke X., Li X., Shu W., Yang W., Liu Y., Yan X., Jia L., Yan H. Expression and characterization of a keratinase encoding gene gm2886 in Streptomyces pactum ACT12 strain. Sheng wu gong cheng xue bao (Chinese J. Biotechnol.) 2017;33:1968–1978. doi: 10.13345/j.cjb.170081. [DOI] [PubMed] [Google Scholar]
- Marshall R.C., Orwin D.F.G., Gillespie J.M. Structure and biochemistry of mammalian hard keratin. Electron Microsc. Rev. 1991;4:47–83. doi: 10.1016/0892-0354(91)90016-6. [DOI] [PubMed] [Google Scholar]
- Martinez J.P.D.O., Cai G., Nachtschatt M., Navone L., Zhang Z., Robins K., Speight R. Challenges and opportunities in identifying and characterising keratinases for value-added peptide production. Catalysts. 2020;10 doi: 10.3390/catal10020184. [DOI] [Google Scholar]
- Mercer D.K., Stewart C.S. Keratin hydrolysis by dermatophytes. Med. Mycol. 2019;57:13–22. doi: 10.1093/mmy/myx160. [DOI] [PubMed] [Google Scholar]
- Mitsuiki S., Ichikawa M., Oka T., Sakai M., Moriyama Y., Sameshima Y., Goto M., Furukawa K. Molecular characterization of a keratinolytic enzyme from an alkaliphilic Nocardiopsis sp. TOA-1. Enzym. Microb. Technol. 2004;34:482–489. doi: 10.1016/j.enzmictec.2003.12.011. [DOI] [Google Scholar]
- Mitsuiki S., Hui Z., Matsumoto D., Sakai M., Moriyama Y., Furukawa K., Kanouchi H., Oka T. Degradation of PrPSc by keratinolytic protease from Nocardiopsis sp. TOA-1. Biosci. Biotechnol. Biochem. 2006;70:1246–1248. doi: 10.1271/bbb.70.1246. [DOI] [PubMed] [Google Scholar]
- Monod M., Léchenne B., Jousson O., Grand D., Zaugg C., Stöcklin R., Grouzmann E. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology. 2005;151:145–155. doi: 10.1099/mic.0.27484-0. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R.P., Chandra A.L. Keratinase of a streptomycete. Indian J. Exp. Biol. 1990;28:575–577. [PubMed] [Google Scholar]
- Murphy M.E., King J.R., Taruscio T.G., Geupel G.R. Amino acid composition of feather barbs and rachises in three species of pygoscelid penguins: nutritional implications. Condor. 1990;92:913–921. doi: 10.2307/1368727. [DOI] [Google Scholar]
- Nahar M., Shishir M.A., Waliullah S., Haque M.S., Ilias M., Karim M.M., Khan S.N., Mozammel Hoq M. Cloning, expression and structure simulation of keratinase from Bacillus licheniformis strain MZK05. Malays. J. Microbiol. 2016;12:182–190. doi: 10.21161/mjm.78515. [DOI] [Google Scholar]
- Nasipuri P., Herschend J., Brejnrod A.D., Madsen J.S., Espersen R., Svensson B., Burmølle M., Jacquiod S., Sørensen S.J. Community-intrinsic properties enhance keratin degradation from bacterial consortia. PLoS ONE. 2020;15:1–26. doi: 10.1371/journal.pone.0228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navone L., Speight R. Understanding the dynamics of keratin weakening and hydrolysis by proteases. PLoS ONE. 2018;13:1–21. doi: 10.1371/journal.pone.0202608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C.S., Li W.-H. Genetic and molecular basis of feather diversity in birds. Genome Biol. Evol. 2018;10:2572–2586. doi: 10.1093/gbe/evy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C.S., Wu P., Foley J., Foley A., McDonald M.-L., Juan W.-T., Huang C.-J., Lai Y.-T., Lo W.-S., Chen C.-F. The chicken frizzle feather is due to an α-keratin (KRT75) mutation that causes a defective rachis. PLoS Genet. 2012;8:e1002748. doi: 10.1371/journal.pgen.1002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoroma E.A., Garelick H., Abiola O.O., Purchase D. Identification and characterisation of a Bacillus licheniformis strain with profound keratinase activity for degradation of melanised feather. Int. Biodeterior. Biodegrad. 2012;74:54–60. doi: 10.1016/j.ibiod.2012.07.013. [DOI] [Google Scholar]
- Patinvoh R.J., Feuk-Lagerstedt E., Lundin M., Sárvári Horváth I., Taherzadeh M.J. Biological pretreatment of chicken feather and biogas production from total broth. Appl. Biochem. Biotechnol. 2016;180:1401–1415. doi: 10.1007/s12010-016-2175-8. [DOI] [PubMed] [Google Scholar]
- Paul T., Das A., Mandal A., Halder S.K., Jana A., Maity C., Dasmohapatra P.K., Pati B.R., Mondal K.C. An efficient cloth cleaning properties of a crude keratinase combined with detergent: towards industrial viewpoint. J. Clean. Prod. 2014;66:672–684. doi: 10.1016/j.jclepro.2013.10.054. [DOI] [Google Scholar]
- Peng Z., Zhang J., Du G., Chen J. Keratin waste recycling based on microbial degradation: mechanisms and prospects. ACS Sustain. Chem. Eng. 2019;7:9727–9736. doi: 10.1021/acssuschemeng.9b01527. [DOI] [Google Scholar]
- Preuett B.L., Schuenemann E., Brown J.T., Kovac M.E., Krishnan S.K., Abdel-Rahman S.M. Comparative analysis of secreted enzymes between the anthropophilic-zoophilic sister species Trichophyton tonsurans and Trichophyton equinum. Fungal Biol. 2010;114:429–437. doi: 10.1016/j.funbio.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Rafik M.E., Doucet J., Briki F. The intermediate filament architecture as determined by X-ray diffraction modeling of hard α-keratin. Biophys. J. 2004;86:3893–3904. doi: 10.1529/biophysj.103.034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahayu S., Syah D., Thenawidjaja Suhartono M. Degradation of keratin by keratinase and disulfide reductase from Bacillus sp. MTS of Indonesian origin. Biocatal. Agric. Biotechnol. 2012;1:152–158. doi: 10.1016/j.bcab.2012.02.001. [DOI] [Google Scholar]
- Rai S.K., Mukherjee A.K. Optimization of production of an oxidant and detergent-stable alkaline β-keratinase from Brevibacillus sp. strain AS-S10-II: application of enzyme in laundry detergent formulations and in leather industry. Biochem. Eng. J. 2011;54:47–56. doi: 10.1016/j.bej.2011.01.007. [DOI] [Google Scholar]
- Rai S.K., Mukherjee A.K. Optimization for production of liquid nitrogen fertilizer from the degradation of chicken feather by iron-oxide (Fe3O4) magnetic nanoparticles coupled β-keratinase. Biocatal. Agric. Biotechnol. 2015;4:632–644. doi: 10.1016/j.bcab.2015.07.002. [DOI] [Google Scholar]
- Ramnani P., Gupta R. Optimization of medium composition for keratinase production on feather by Bacillus licheniformis RG1 using statistical methods involving response surface methodology. Biotechnol. Appl. Biochem. 2004;40:191–196. doi: 10.1042/ba20030228. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J. In: Handbook of Proteolytic Enzyme. Rawlings N.D., Barrett A.J., editors. E-Publishing; 2013. Introduction: serine peptidases and their clans; pp. 2491–2523. [DOI] [Google Scholar]
- Rawlings N.D., Barrett A.J. In: Handbook of Proteolytic Enzymes. Rawlings N.D., Barrett A.J., editors. E-Publishing; 2013. Introduction: metallopeptidases and their clans; pp. 325–370. [DOI] [Google Scholar]
- Reddy N., Yang Y. Structure and properties of chicken feather barbs as natural protein fibers. J. Polym. Environ. 2007;15:81–87. doi: 10.1007/s10924-007-0054-7. [DOI] [Google Scholar]
- Riffel A., Lucas F., Heeb P., Brandelli A. Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch. Microbiol. 2003;179:258–265. doi: 10.1007/s00203-003-0525-8. [DOI] [PubMed] [Google Scholar]
- Riffel A., Brandelli A., Bellato C. de M., Souza G.H.M.F., Eberlin M.N., Tavares F.C.A. Purification and characterization of a keratinolytic metalloprotease from Chryseobacterium sp. kr6. J. Biotechnol. 2007;128:693–703. doi: 10.1016/j.jbiotec.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Rogers G.E. Hair follicle differentiation and regulation. Int. J. Dev. Biol. 2004;48:163–170. doi: 10.1387/ijdb.15272381. [DOI] [PubMed] [Google Scholar]
- Safranek W.W., Goos R.D. Degradation of wool by saprotrohic fungi. Can. J. Microbiol. 1981;28:137–140. doi: 10.1139/m82-015. [DOI] [Google Scholar]
- Sanghvi G., Patel H., Vaishnav D., Oza T., Dave G., Kunjadia P., Sheth N. A novel alkaline keratinase from Bacillus subtilis DP1 with potential utility in cosmetic formulation. Int. J. Biol. Macromol. 2016;87:256–262. doi: 10.1016/j.ijbiomac.2016.02.067. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the acitive site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Scott J.A., Untereiner W.A. Determination of keratin degradation by fungi using keratin azure. Med. Mycol. 2004;42:239–246. doi: 10.1080/13693780310001644680. [DOI] [PubMed] [Google Scholar]
- Sharma R., Devi S. Versatility and commercial status of microbial keratinases: a review. Rev. Environ. Sci. Biotechnol. 2018;17:19–45. doi: 10.1007/s11157-017-9454-x. [DOI] [Google Scholar]
- Sharma R., Gupta R. Extracellular expression of keratinase Ker P from Pseudomonas aeruginosa in E. coli. Biotechnol. Lett. 2010;32:1863–1868. doi: 10.1007/s10529-010-0361-2. [DOI] [PubMed] [Google Scholar]
- Sharma R., Gupta R. Coupled action of c-glutamyl transpeptidase-glutathione and keratinase effectively degrades feather keratin and surrogate prion protein, Sup 35NM. Bioresour. Technol. 2012;120:314–317. doi: 10.1016/j.biortech.2012.06.038. [DOI] [PubMed] [Google Scholar]
- Sharma S., Gupta A. Sustainable management of keratin waste biomass: applications and future perspectives. Braz. Arch. Biol. Technol. 2016;59:1–14. doi: 10.1590/1678-4324-2016150684. [DOI] [Google Scholar]
- Sharma B., Jamdar S.N., Ghosh B., Yadav P., Kumar A., Kundu S., Goyal V.D., Makde R.D. Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations. Biochim. Biophys. Acta, Proteins Proteomics. 2017;1865:1406–1415. doi: 10.1016/j.bbapap.2017.07.023. [DOI] [PubMed] [Google Scholar]
- Shavandi A., Silva T.H., Bekhit A.A., Bekhit A.E.D.A. Keratin: dissolution, extraction and biomedical application. Biomater. Sci. 2017;5:1699–1735. doi: 10.1039/c7bm00411g. [DOI] [PubMed] [Google Scholar]
- Shrinivas D., Naik G.R. Characterization of alkaline thermostable keratinolytic protease from thermoalkalophilic Bacillus halodurans JB 99 exhibiting dehairing activity. Int. Biodeterior. Biodegradation. 2011;65:29–35. doi: 10.1016/j.ibiod.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Skieresz-Szewczyk K., Jackowiak H., Buchwald T., Szybowicz M. Localization of alpha-keratin and beta-keratin (corneous beta protein) in the epithelium on the ventral surface of the lingual apex and its lingual nail in the domestic goose (Anser anser f. domestica) by using immunohistochemistry and raman microspectros. Anat. Rec. 2017;300:1361–1368. doi: 10.1002/ar.23591. [DOI] [PubMed] [Google Scholar]
- Smith T.A., Steinert P.M., Parry D.A.D. Modeling effects of mutations in coiled-coil structures: case study using epidermolysis bullosa simplex mutations in segment 1a of k5/k14 intermediate filaments. Proteins. 2004;55:1043–1052. doi: 10.1002/prot.20089. [DOI] [PubMed] [Google Scholar]
- Sone T., Haraguchi Y., Kuwahara A., Ose T., Takano M., Abe A., Tanaka M., Tanaka I., Asano K. Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease. Protein Pept. Lett. 2015;22:63–72. doi: 10.2174/0929866521666140919100851. [DOI] [PubMed] [Google Scholar]
- Steinmetz M.O., Stock A., Schulthess T., Landwehr R., Lustig A., Faix J., Gerisch G., Aebi U., Kammerer R.A. A distinct 14 residue site triggers coiled-coil formation in cortexillin I. EMBO J. 1998;17:1883–1891. doi: 10.1093/emboj/17.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Tsujimoto Y., Matsui H., Watanabe K. Decomposition of extremely hard-to-degrade animal proteins by thermophilic bacteria. J. Biosci. Bioeng. 2006;102:73–81. doi: 10.1263/jbb.102.73. [DOI] [PubMed] [Google Scholar]
- Szeto M.W.Y., Mujika J.I., Zurek J., Mulholland A.J., Harvey J.N. QM/MM study on the mechanism of peptide hydrolysis by carboxypeptidase A. J. Mol. Struct. THEOCHEM. 2009;898:106–114. doi: 10.1016/j.theochem.2008.06.033. [DOI] [Google Scholar]
- Takami H., Akiba T., Horikoshi K. Characterization of an alkaline protease from Bacillus sp. no. AH-101. Appl. Microbiol. Biotechnol. 1990;33:519–523. doi: 10.1007/bf00172544. [DOI] [PubMed] [Google Scholar]
- Tesfaye T., Sithole B., Ramjugernath D. Valorisation of chicken feathers: a review on recycling and recovery route—current status and future prospects. Clean Techn. Environ. Policy. 2017;19:2363–2378. doi: 10.1007/s10098-017-1443-9. [DOI] [Google Scholar]
- Tesfaye T., Sithole B., Ramjugernath D., Chunilall V. Valorisation of chicken feathers: characterisation of physical properties and morphological structure. J. Clean. Prod. 2017;149:349–365. doi: 10.1016/j.jclepro.2017.02.112. [DOI] [PubMed] [Google Scholar]
- Tork S.E., Shahein Y.E., El-Hakim A.E., Abdel-Aty A.M., Aly M.M. Purification and partial characterization of serine-metallokeratinase from a newly isolated Bacillus pumilus NRC21. Int. J. Biol. Macromol. 2016;86:189–196. doi: 10.1016/j.ijbiomac.2016.01.060. [DOI] [PubMed] [Google Scholar]
- Tran V.D.T., De Coi N., Feuermann M., Schmid-Siegert E., Băguţ E.-T., Mignon B., Waridel P., Peter C., Pradervand S., Pagni M., Monod M. RNA sequencing-based genome reannotation of the dermatophyte arthroderma benhamiae and characterization of its secretome and whole gene expression profile during infection. mSystems. 2016;1:1–18. doi: 10.1128/msystems.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiroulnikov K., Rezai H., Bonch-Osmolovskaya E., Nedkov P., Gousterova A., Cueff V., Godfroy A., Barbier G., Métro F., Chobert J.-M. Hydrolysis of the amyloid prion protein and nonpathogenic meat and bone meal by anaerobic thermophilic prokaryotes and Streptomyces subspecies. J. Agric. Food Chem. 2004;52:6353–6360. doi: 10.1021/jf0493324. [DOI] [PubMed] [Google Scholar]
- United States Department of Agriculuture . 2020. Livestock and Poultry (2020): World Markets and Trade.https://apps.fas.usda.gov/psdonline/circulars/livestock_poultry.pdf Available online at. [Google Scholar]
- Verma A., Singh H., Anwar S., Chattopadhyay A., Tiwari K.K., Kaur S., Dhilon G.S. Microbial keratinases: industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 2017;37:476–491. doi: 10.1080/07388551.2016.1185388. [DOI] [PubMed] [Google Scholar]
- Wang J.J., Garlich J.D., Shih J.C.H. Beneficial effects of versazyme, a keratinase feed additive, on body weight, feed conversion, and breast yield of broiler chickens. J. Appl. Poult. Res. 2006;15:544–550. doi: 10.1093/japr/15.4.544. [DOI] [Google Scholar]
- Wang H., Guo Y., Shih J.C.H. Effects of dietary supplementation of keratinase on growth performance, nitrogen retention and intestinal morphology of broiler chickens fed diets with soybean and cottonseed meals. Anim. Feed Sci. Technol. 2008;140:376–384. doi: 10.1016/j.anifeedsci.2007.04.003. [DOI] [Google Scholar]
- Wang L., Cheng G., Ren Y., Dai Z., Zhao Z.-S., Liu F., Li S., Wei Y., Xiong J., Tang X.-F. Degradation of intact chicken feathers by Thermoactinomyces sp. CDF and characterization of its keratinolytic protease. Appl. Microbiol. Biotechnol. 2015;99:3949–3959. doi: 10.1007/s00253-014-6207-4. [DOI] [PubMed] [Google Scholar]
- Wang B., Yang W., McKittrick J., Meyers M.A. Keratin: structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog. Mater. Sci. 2016;76:229–318. doi: 10.1016/j.pmatsci.2015.06.001. [DOI] [Google Scholar]
- Wang L., Qian Y., Cao Y., Huang Y., Chang Z., Huang H. Production and characterization of keratinolytic proteases by a chicken feather-degrading thermophilic strain, Thermoactinomyces sp. YT06. J. Microbiol. Biotechnol. 2017;27:2190–2198. doi: 10.4014/jmb.1705.05082. [DOI] [PubMed] [Google Scholar]
- Wöll S., Windoffer R., Leube R.E. p38 MAPK-dependent shaping of the keratin cytoskeleton in cultured cells. J. Cell Biol. 2007;177:795–807. doi: 10.1083/jcb.200703174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfolk J.A., Wheatley L.M., Piyasena R.V., Benjamin D.C., Platts-Mills T.A.E. Trichophyton antigens associated with IgE antibodies and delayed type hypersensitivity. J. Biol. Chem. 1998;273:29489–29496. doi: 10.1074/jbc.273.45.29489. [DOI] [PubMed] [Google Scholar]
- Wu B., Shi P., Li J., Wang Y., Meng K., Bai Y., Luo H., Yang P., Zhou Z., Yao B. A new aminopeptidase from the keratin-degrading strain Streptomyces fradiae var. k11. Appl. Biochem. Biotechnol. 2010;160:730–739. doi: 10.1007/s12010-009-8537-8. [DOI] [PubMed] [Google Scholar]
- Wu W.L., Chen M.Y., Tu I.F., Lin Y.C., Eswarkumar N., Chen M.Y., Ho M.C., Wu S.H. The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-04723-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Baldwin D., Kindrachuk C., Hegedus D.D. Serine proteases and metalloproteases associated with pathogenesis but not host specificity in the Entomophthoralean fungus Zoophthora radicans. Can. J. Microbiol. 2006;52:550–559. doi: 10.1139/W06-004. [DOI] [PubMed] [Google Scholar]
- Yamamura S., Morita Y., Hasan Q., Yokoyama K., Tamiya E. Keratin degradation: a cooperative action of two enzymes from Stenotrophomonas sp. Biochem. Biophys. Res. Commun. 2002;294:1138–1143. doi: 10.1016/s0006-291x(02)00580-6. [DOI] [PubMed] [Google Scholar]
- Yang L., Wang H., Lv Y., Bai Y., Luo H., Shi P., Huang H., Yao B. Construction of a rapid feather-degrading bacterium by overexpression of a highly efficient alkaline keratinase in its parent strain Bacillus amyloliquefaciens K11. J. Agric. Food Chem. 2016;64:78–84. doi: 10.1021/acs.jafc.5b04747. [DOI] [PubMed] [Google Scholar]
- Yeo I., Lee Y.J., Song K., Jin H.S., Lee J.E., Kim D., Lee D.W., Kang N.J. Low-molecular weight keratins with anti-skin aging activity produced by anaerobic digestion of poultry feathers with Fervidobacterium islandicum AW-1. J. Biotechnol. 2018;271:17–25. doi: 10.1016/j.jbiotec.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Yohko Y., Mari M., Mohamed Mahdi A., Michel M., Peter S., Tsuyoshi Y. Flippase (FLP) recombinase-mediated marker recycling in the dermatophyte Arthroderma vanbreuseghemii. Microbiology. 2014;160:2122–2135. doi: 10.1099/mic.0.076562-0. [DOI] [PubMed] [Google Scholar]
- Yue X.Y., Zhang B., Jiang D.D., Liu Y.J., Niu T.G. Separation and purification of a keratinase as pesticide against root-knot nematodes. World J. Microbiol. Biotechnol. 2011;27:2147–2153. doi: 10.1007/s11274-011-0680-z. [DOI] [Google Scholar]
- Zaugg C., Jousson O., Léchenne B., Staib P., Monod M. Trichophyton rubrum secreted and membrane-associated carboxypeptidases. Int. J. Med. Microbiol. 2008;298:669–682. doi: 10.1016/j.ijmm.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Zaugg C., Monod M., Weber J., Harshman K., Pradervand S., Thomas J., Bueno M., Giddey K., Staib P. Gene expression profiling in the human pathogenic dermatophyte Trichophyton rubrum during growth on proteins. Eukaryot. Cell. 2009;8:241–250. doi: 10.1128/EC.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.X., Gong J.S., Dou W.F., Zhang D.D., Zhang Y.X., Li H., Lu Z.M., Shi J.S., Xu Z.H. Production and characterization of surfactant-stable fungal keratinase from Gibberella intermedia CA3-1 with application potential in detergent industry. Chem. Pap. 2016;70:1460–1470. doi: 10.1515/chempap-2016-0086. [DOI] [Google Scholar]
- Zoccola M., Aluigi A., Tonin C. Characterisation of keratin biomass from butchery and wool industry wastes. J. Mol. Struct. 2009;938:35–40. doi: 10.1016/j.molstruc.2009.08.036. [DOI] [Google Scholar]