Abstract
ABA INSENSITIVE 5 (ABI5) is a basic leucine zipper (bZIP) transcription factor which acts in the abscisic acid (ABA) network and is activated in response to abiotic stresses. However, the precise role of barley (Hordeum vulgare) ABI5 in ABA signaling and its function under stress remains elusive. Here, we show that HvABI5 is involved in ABA-dependent regulation of barley response to drought stress. We identified barley TILLING mutants carrying different alleles in the HvABI5 gene and we studied in detail the physiological and molecular response to drought and ABA for one of them. The hvabi5.d mutant, carrying G1751A transition, was insensitive to ABA during seed germination, yet it showed the ability to store more water than its parent cv. “Sebastian” (WT) in response to drought stress. The drought-tolerant phenotype of hvabi5.d was associated with better membrane protection, higher flavonoid content, and faster stomatal closure in the mutant under stress compared to the WT. The microarray transcriptome analysis revealed up-regulation of genes associated with cell protection mechanisms in the mutant. Furthermore, HvABI5 target genes: HVA1 and HVA22 showed higher activity after drought, which may imply better adaptation of hvabi5.d to stress. On the other hand, chlorophyll content in hvabi5.d was lower than in WT, which was associated with decreased photosynthesis efficiency observed in the mutant after drought treatment. To verify that HvABI5 acts in the ABA-dependent manner we analyzed expression of selected genes related to ABA pathway in hvabi5.d and its WT parent after drought and ABA treatments. The expression of key genes involved in ABA metabolism and signaling differed in the mutant and the WT under stress. Drought-induced increase of expression of HvNCED1, HvBG8, HvSnRK2.1, and HvPP2C4 genes was 2–20 times higher in hvabi5.d compared to “Sebastian”. We also observed a faster stomatal closure in hvabi5.d and much higher induction of HvNCED1 and HvSnRK2.1 genes after ABA treatment. Together, these findings demonstrate that HvABI5 plays a role in regulation of drought response in barley and suggest that HvABI5 might be engaged in the fine tuning of ABA signaling by a feedback regulation between biosynthetic and signaling events. In addition, they point to different mechanisms of HvABI5 action in regulating drought response and seed germination in barley.
Keywords: abscisic acid, monocots, barley, Hordeum vulgare, water deficit, stress, transcriptomics, ABI5
Introduction
Abscisic acid (ABA) is the crucial regulator of plant responses to abiotic stresses. In the presence of unfavorable conditions, the precise regulation and function of ABA-dependent signaling components ensure the appropriate activity of stress-responsive genes (reviewed by Yoshida et al., 2019), and thus the regulation of physiological processes, such as photosynthesis, stomatal closure (Song et al., 2016; Cai et al., 2017; Saito and Uozumi, 2019), and osmoprotectant biosynthesis (Jones, 2016; Sah et al., 2016; Martignago et al., 2020).
In Arabidopsis thaliana, ABA INSENSITIVE 5 (ABI5) encodes the ABA-dependent, BASIC LEUCINE ZIPPER (bZIP) transcription factor, composed of C1, C2, C3, and bZIP conserved domains (Lopez-Molina et al., 2001; Nakamura et al., 2001). The bZIP domain is responsible for DNA binding, whereas C1, C2, and C3 domains are recognized and phosphorylated by protein kinases (Furihata et al., 2006). C3 is also crucial for ABI5 interaction with another transcription factor, ABI3 (Tezuka et al., 2013). AtABI5 plays a role during early ABA signaling and was shown to be activated shortly after the perception of a stress signal. The formation of ABA-PYRABACTIN RESISTANCE1/PYR LIKE/REGULATORY COMPONENT OF ABA RECEPTOR-PHOSPHATASE 2C complex (ABA-PYR1/PYL/RCAR-PP2C) promotes ABI5 phosphorylation mediated by the SNF1-RELATED PROTEIN KINASE2s (SnRK2s) and thus its activation. Then, AtABI5 binds ABA RESPONSIVE ELEMENTs (ABRE cis-elements) present in the promoters of regulated genes and activates or represses their transcription, often in the interaction with other regulatory proteins (reviewed by Daszkowska-Golec, 2016; Sah et al., 2016; Dejonghe et al., 2018; Yoshida et al., 2019).
ABI5 was described as a regulator of seed germination and early seedling development in the presence of ABA and abiotic stresses (Finkelstein, 1994; Lopez-Molina et al., 2001; reviewed by Skubacz et al., 2016). Several Arabidopsis abi5 alleles have been identified using insertional or physical mutagenesis (Atabi5-1, Atabi5-2, Atabi5-4, Atabi5-5, Atabi5-7, Atabi5-8, and Atabi5-9) (Finkelstein, 1994; Lopez-Molina and Chua, 2000; Carles et al., 2002; Nambara et al., 2002; Zheng et al., 2012; Tezuka et al., 2013). It was demonstrated that Atabi5-2 and Atabi5-5 were insensitive also to salt and osmotic stresses during seed germination (Carles et al., 2002), while Atabi5-1 showed ABA, salt, and osmotic insensitivity both at germination and early seedling stage (Finkelstein et al., 2005; Yuan et al., 2011). Interestingly, the Atabi5-1 showed no differences in other ABA-regulated processes, such as stomata closure during vegetative growth (Finkelstein, 1994; Finkelstein and Lynch, 2000). Although Atabi5-1 was described as “not-wilty” by Finkelstein and Lynch (2000; http://www.arabidopsis.org, AT2G36270) no detailed analysis of the mutant under drought stress has been reported.
AtABI5 expression was observed during a short developmental window, between 48 and 60 h after imbibition, in the presence of drought and salt stress. The increased activity of AtABI5 was related to germination inhibition (Lopez-Molina et al., 2001; Maia et al., 2014). Additionally, AtABI5 function was associated with the repression of primary and lateral root development (Lopez-Molina et al., 2001; Signora et al., 2001). It was proven that AtABI5 downstream target genes were responsible for the inhibition of germination, adaptation to reduced water availability, lower photosynthesis efficiency, and reactive oxygen species (ROS) scavenging (Finkelstein and Lynch, 2000; Kanai et al., 2010; Su et al., 2016; Bi et al., 2017).
Another level of abiotic stress response regulated by AtABI5 involves lipid metabolism where AtABI5 is able to activate the expression of gene encoding triacylglycerol biosynthesis enzyme (Kong et al., 2013). AtABI5 promotes chlorophyll catabolism and inhibits photosynthesis via transcriptional regulation of genes encoding a protein inducing destabilization of LIGHT-HARVESTING COMPLEX FOR PHOTOSYSTEM II (LHCPII) and chlorophyll b reductase (Sakuraba et al., 2014). AtABI5 also regulates expression of gene encoding detoxifying enzyme, CATALASE1 (CAT1) (Bi et al., 2017). Recently, AtABI5 was described as a direct repressor of PHOSPHATE1 (PHO1) gene associated with phosphate homeostasis (Huang et al., 2017).
AtABI5 undergoes complex regulation at the post-translational level by phosphorylation, ubiquitination, sumoylation, and nitrosylation. AtABI5 is phosphorylated by many kinases, such as SnRK2.2, SnRK2.3, CALCIUM-DEPENDENT PROTEIN KINASE 11 (CPK11), and SOS2-LIKE PROTEIN KINASE 5 (PKS5), which results in its activation (Lynch et al., 2012; Zhou et al., 2015). It is noteworthy that all types of AtABI5 modifications take place in the specific amino acid positions of the protein (Miura et al., 2009; Liu and Stone, 2013; Albertos et al., 2015; Yu et al., 2015).
In Arabidopsis AtABI5 is considered as the main regulator of ABA response only during seed germination and early seedling growth, whereas other bZIP transcription factors that recognize the ABRE cis-element, such as ABRE BINDING FACTOR2/ABRE-BINDING PROTEIN1 (AtABF2/AREB1), AtABF4/AREB2, AtABF3, and AtABF1 take part in ABA signaling in vegetative tissues under abiotic stresses (Fujita et al., 2005; Yoshida et al., 2015). It was shown that Atabf4/areb2 and Atabf3 mutants were more sensitive to drought than WT, whereas overexpression of AtABF2/AREB1 ensured better drought tolerance due to increased expression of LEA genes (Fujita et al., 2005; Yoshida et al., 2010). AtABFs/AREBs promote stomatal closure and chlorophyll catabolism (Gao et al., 2016; Qian et al., 2019). Importantly, AtABF2/AREB1, AtABF4/AREB2, AtABF3, and AtABF1 show redundancy during regulation of ABA-mediated drought responses. Atabf2/areb1 Atabf4/areb2 Atabf3 Atabf1 quadruple mutant showed 2.2% survival rate after drought treatment, whereas survival rate of single Atabf/areb mutants was 38.6–57.1% (Yoshida et al., 2010; Yoshida et al., 2015). However, sometimes AtABF genes can act autonomously, e.g. AtABF3 regulates seedling root growth in the presence of ABA (Finkelstein et al., 2005; Yoshida et al., 2010). Moreover, the expression profile of LEA genes in the Atabf2/areb1, Atabf4/areb2, and Atabf3 mutants is not always similar (Yoshida et al., 2010).
The function of barley (Hordeum vulgare) ABI5 homolog as an ABA-dependent transcription factor has been described first in barley seeds, mainly in aleurone layer by Casaretto and Ho (2003). The authors demonstrated that in the presence of ABA, HvABI5 binds ABRE cis-element present in ABA RESPONSE PROMOTER COMPLEX (ABRC). HVA1 and HVA22 encoding LEA proteins have been identified as the HvABI5-activated genes in barley aleurone cells (Casaretto and Ho, 2003). Both proteins ensure the protection from water deprivation in barley seeds. The expression of HVA1 and HVA22 is also dependent on VIVIPAROUS1 (HvVP1), an ortholog of AtABI3 (Casaretto and Ho, 2003). Probably, HvABI5 interaction with HvVP1 is required for HvABI5 activity, similarly to the interaction between ABI5 and ABI3 described in Arabidopsis (Casaretto and Ho, 2003; Bensmihen et al., 2004). Interestingly, HvABI5 expression is auto-activated by HvABI5 protein, as it is observed for AtABI5 in Arabidopsis (Finkelstein and Lynch, 2000; Casaretto and Ho, 2005). It was also found that HvABI5 activity as transcription factor is dependent on serine in 106 position, which probably serves as a target of ABA-dependent phosphorylation by HvSnRK2.1 (HvPKABA1) (Casaretto and Ho, 2005).
HvABI5 shows high similarity to wheat wABI5, rice TRANSCRIPTION FACTOR RESPONSIBLE FOR ABA REGULATION1 (TRAB1), and maize ZmABI5. wABI5, TRAB1, and ZmABI5 were described as ABA-dependent transcription factors regulating abiotic stress responses (Kagaya et al., 2002; Kobayashi et al., 2008; Yan et al., 2012). wABI5 acts as a positive regulator of drought response via expression activation of LEA encoding genes: DEHYDRIN13 (wDHN13), RESPONSIVE TO ABA18 (wRAB18), and wRAB19 (Kobayashi et al., 2008). The increased expression of TRAB1 was observed in rice stress-tolerant cultivar under various abiotic stresses (Paul and Roychoudhury, 2019). On the other side, ZmABI5 negatively regulates abiotic stress responses. ZmABI5 overexpression promoted chlorophyll degradation and reduced activity of detoxifying enzymes, peroxidase (POD), and superoxide dismutase (SOD), under abiotic stresses (Yan et al., 2012). Furthermore, other AtABI5 homologs were identified in rice (OsABI5) and in wheat (TaABI5). Similarly, to ZmABI5, OsABI5 was also described as a negative regulator of stress tolerance (Zou et al., 2008). Wheat TaABI5 was shown to be active only in seeds. TaABI5 overexpression in Arabidopsis seeds caused ABA-hypersensitive germination (Utsugi et al., 2020).
Given the fact that drought stress is considered as the cause of the most severe yield losses worldwide there is a need for extensive development of drought-tolerant cultivars. However, drought tolerance is a very complex trait showing quantitative inheritance and large genotype x environment interactions, therefore developing cultivars better adopted to water deficiency conditions remains a challenge for crop breeding programs (Agarwal et al., 2013; Gilliham et al., 2017). Identification and functional characterization of stress-related genes can help in obtaining plants with a higher tolerance to drought (Joshi et al., 2016; Bailey-Serres et al., 2019). Barley has been suggested as a cereal model for studying mechanisms of abiotic stress adaptation (Dawson et al., 2015), due to its natural ability to cope better with drought stress than other cereals. According to the Food and Agriculture Organization report, barley is the fourth most important cereal crop regarding the harvested acreage (FAO, 2018). These characteristics, together with the assembled genome sequence (IBSC_v2, Ensembl Plants), enhanced the role of barley as a model species for studying genetic and molecular processes underlying drought tolerance in cereals.
In this study we focused on elucidating the role of HvABI5 in ABA signaling during barley response to drought stress. First, we have identified a hvabi5.d mutant using barley TILLING (Targeted Induced Local Lesions IN Genomes) platform created in our laboratory (Szurman-Zubrzycka et al., 2018). The identified TILLING mutant made it possible to initiate studies on the role of HvABI5 in regulation of ABA and drought responses in barley. Taking advantage from Arabidopsis research on abi5 mutants, we analyzed seed germination and early seedling of hvabi5.d in the presence of ABA. Next, we applied drought stress to hvabi5.d seedlings and measured a range of physiological parameters in order to describe the response of hvabi5.d to drought. To gain insight into molecular basis of hvabi5.d drought tolerance we conducted a global transcriptome analysis of the mutant and parent cultivar “Sebastian” exposed to drought treatment. Then, to answer the question whether HvABI5 regulates drought response in the ABA-dependent manner, we analyzed the expression of selected genes related to ABA pathway in hvabi5.d and its wild type parent under drought and ABA treatments. We also studied stomatal conductance of analyzed genotypes after ABA treatment. Our results clearly indicated the role of HvABI5 as the ABA-dependent regulator of response to drought stress in barley. In addition, these findings clearly pointed to different mechanisms of HvABI5 action in regulating drought response and seed germination.
Material and Methods
Plant Material
The hvabi5.d mutant was identified using a TILLING platform (HorTILLUS) developed after chemical mutagenesis of spring barley cultivar “Sebastian” in our laboratory (Szarejko et al., 2017; Szurman-Zubrzycka et al., 2018). The DNA samples from 6,144 M2 plants of the HorTILLUS population were screened with the aim to identify mutations in HvABI5 gene. TILLING of HvABI5 gene was performed based on HvABI5 sequence for “Sebastian” variety present in GenBank (GenBank acc. no. HQ456390.1). Conserved regions of HvABI5 were mapped using CODDLE (Codons Optimised to Discover Deleterious Lesions; http://www.proweb.org/coddle/) and CLUSTAL OMEGA (http://www.ebi.ac.uk/Tools/msa/clustalo/) tools. The analyzed TILLING fragment (1,072 bp) embraced exons encoding most of C1, C2, C3, and bZIP domains, highly conserved and crucial for HvABI5 function. The mutational screening was performed according to the procedure described by Szurman-Zubrzycka et al. (2018). Mutants carrying nucleotide substitutions in homozygous state were identified in M2 or M3 generation and for most of missense mutants, seeds were increased in M4-M5 generation. The type and state of mutation (homo/heterozygous) was analyzed using the CodonCode Aligner software. The hvabi5.d allele was identified in the heterozygous state, therefore homozygous plants were selected in the segregating M3 progeny before using hvabi5.d mutant in the studies. Seven mutants carrying different alleles: alleles (hvabi5.b, hvabi5.d, hvabi5.e, hvabi5.i, hvabi5.o, hvabi5.u, hvabi5.w) were used for preliminary relative water content (RWC) screening after drought treatment in M4-M6 generation. Mutant carrying the hvabi5.d allele was selected for a detailed physiological and molecular studies presented below. Additionally, the homozygous line of hvabi5.d mutant was backcrossed twice with parent variety “Sebastian” to clean the mutant genome from the possible background mutations. The obtained homozygous F4BC2 lines with hvabi5.d allele were further used for basic physiological tests after drought treatment and expression analysis of selected genes.
Prediction of Mutation Significance Using Bioinformatics Software
The conservation of mutated position was checked at the protein level in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) using multi sequence alignment of ABI5 sequences in dicot and monocot species.
Germination Assay in the Presence of ABA
Thirty seeds of hvabi5.d and wild-type (WT) parent cultivar “Sebastian” were sown in a Petri dish (ф=90 mm) containing two layers of Whatman filters with 5 ml distilled water (control) or ABA solution (75 and 300 μM). Four-day stratification at 4°C in darkness was applied to synchronize seed response. Then, the seeds were germinated in a growth chamber (22°C, 16-h-light/8-h darkness, 200 μmol/m−2 s−1 illumination). Germination process was evaluated by a visible appearance of a root on the 1st, 2nd, 3rd, and 4th day after stratification. Germination assay was performed in three biological replications (each Petri dish containing 30 seeds was considered as one biological replicate) and the experiment was repeated three times.
Seedling Development Assay in the Presence of ABA
Four-day-old seedlings of hvabi5.d and WT were used in this experiment. Plants were placed in glass tubes containing 90 ml of Murashige-Skoog (MS) medium or MS supplemented with 50 µM ABA. The treatment was applied for 6 d in a growth chamber (22°C, 16/8 h photoperiod, 200 μmol/m−2 s−1 illumination). The length of the first leaf and the longest seminal root were measured before and after the treatment. At the end of the treatment, chlorophyll a fluorescence was measured and the leaf tissue (0.02–0.05 g) was collected to extract proline. All analyzes were performed in three biological replications, with two plants per replication. The experiment and material collection were repeated three times.
Chlorophyll a Fluorescence
Chlorophyll a fluorescence was measured with the PocketPea fluorimeter (Hansatech Instruments Ltd., England). Before measurement, the leaves were adapted for 30 min in darkness. Next, the leaves were exposed to a pulse of saturating light [3,500 μmol (photon) m−2 s−1] for 1 sec. To analyze the chlorophyll a fluorescence, the JIP-test was applied (Strasser et al., 2004; Kalaji et al., 2016). The OJIP fluorescence transients consist of four phases: O-initial fluorescence level, J-fluorescence at 2 ms, I- fluorescence at 30 ms, and P-maximal fluorescence. The OJIP transients were used to calculate the following parameters: PIABS—performance index for the photochemical activity and φP0—maximum quantum yield of primary photochemistry (Strasser et al., 2000).
Proline Content
Proline concentration (µmol g−1 fresh weight) was determined according to the colorimetric method of Carillo and Gibbon (2011). The absorbance was read at 520 nm using Victor X5 Multilabel Reader (PerkinElmer). The proline content was calculated according to the formula:
where: Absextract is the absorbance of plant extract, blank is the absorbance of clear extraction solution, slope is determined by linear regression of a calibration curve, Volextract is the total extract volume, Volaliquot is the extract volume used for the assay, FW is the weight of the plant material.
Drought Stress Experiment
Drought Stress Treatment
Drought stress was applied as described earlier (Daszkowska-Golec et al., 2017). Briefly, the experiment was carried in boxes (400 x 140 x 175 mm) filled with soil containing a mixture of sandy loam and sand (7:2) with known physicochemical properties. The soil was supplied with nutrient medium (Supplementary Material S1). Based on performed calculations, the water was easily available for plants at 14% of volumetric water content (vwc) in the soil, whereas the severe drought stress was achieved at 1.5%. When plants were grown in 14% vwc, the RWC in leaves was approximately 100%. The soil moisture was measured every day using time-domain reflectometer (TDR) EasyTest (Institute of Agrophysics, Polish Academy of Sciences, Poland). The WT and mutant plants were grown in a greenhouse for 10 d after sowing (DAS) under optimal water conditions (14% vwc), 20/18°C day/night, with a 16/8 h photoperiod, and 420 μE m−2 s−1 light intensity which was provided by fluorescent lamps. Afterward, the soil moisture was decreased by withholding the irrigation under the control of TDR measurements. On 15 DAS, when the soil moisture decreased to 3%, the plants were moved into a growth chamber, where temperature regime was set to 25°C/20°C day/night, with a 16/8 h photoperiod and 420 μEm−2 s−1 light intensity. The severe drought stress (1.5% vwc) lasted 10 d (16–25 DAS). The control plants were grown under the same conditions with optimal water supply (14% vwc) in parallel to the drought treated plants. On 25 DAS the RWC was evaluated for each genotype, according to the procedure described below.
The second leaves were collected from hvabi5.d and WT plants before water withdrawal (10 DAS) and after severe drought (25 DAS) and used for RNA extraction. The anthocyanins content index, flavonols content index, chlorophyll a fluorescence, and chlorophyll content index were analyzed before water withdrawal (10 DAS) and after drought stress (25 DAS). The stomatal conductance was measured under optimal water conditions (10 DAS), on the drought onset (13 and 15 DAS) and after severe drought (25 DAS). On the 25 DAS, RWC, electrolyte leakage (EL), and ABA content were analyzed using plants growing under optimal water supply and under drought stress. The schedule of drought experiment with indicated time-points of all assays are presented in Supplementary Material S2. We conducted all analyses using the second leaf since it was already present when plants entered drought treatment. Each genotype was tested in three biological replicates. One box containing 15 plants per genotype was considered as one replicate.
Relative Water Content
RWC was measured in the second leaf on the last day of drought stress (25 DAS). RWC parameter was calculated according to formula:
where: Fw (fresh weight) is the weight of detached leaf, Tw (turgid weight) is the weight of a leaf after 24 h rehydration in distilled water (leaves were submerged in distilled water in darkness), and Dw (dry weight) is the weight of a leaf dried at 60°C for 48 h. The measurement of RWC was performed in three biological replications (each biological replicate included leaves of three independent seedlings).
Electrolyte Leakage Analysis
The EL was analyzed in drought-treated and control seedlings on 25 DAS (according to the protocol of Bandurska and Gniazdowska-Skoczek, 1995). Briefly, the fragments of the middle part of the second leaf were washed three times in deionized water and kept at 10°C for 24 h. Afterward, the samples were transferred to the room temperature and EL was measured using pH/conductivity meter (CPC-505, Elmetron, Poland). Then, samples were autoclaved for 15 min to damage cells completely and conductivity was measured again at room temperature. Three replications were made for each treatment combination (each biological replicate included leaves of three seedlings). EL was calculated according to formula:
Where: D1 and C1 are the first conductivity measurements for respectively drought and control samples, whereas D2 and C2 are the second (after auctoclaving) conductivity measurements.
Flavonol, Anthocyanin, and Chlorophyll Content Indices Measurement
The measurements of flavonol, anthocyanin, and chlorophyll content indices were performed on the second leave on the 10 and 25 DAS using a Dualex Scientific+TM (Force-A, France). The measurements were performed in three biological replicates, with four seedlings per replicate.
Stomatal Conductance
Stomatal Conductance (mmol m−2 s−1) was measured using AP4 porometer (DELTA-T Devices, Burwell, UK). Measurements were conducted in three biological replicates, each replicate contained three individual plants. The analysis was performed at the exposed, central part of the second leaf adaxial side on 10, 13, 15, and 25 DAS, in three biological replications (each biological replicate included leaves of three independent seedlings).
Endogenous ABA Content
ABA content was measured according to the protocol of Nakurte et al. (2012). A modular HPLC system (Shimadzu, Japan) with SPD-M20A photodiode array detector and a Kinetex™ C18 (4.6 × 250 mm, 5 µm) column (Phenomenex) were used to conduct chromatography. The injection volume was 20 µl and the analysis was performed in the isocratic mode at a flow rate of 1 ml min−1. LabSolutions software was used to evaluate the results (Shimadzu, Japan).
RNA Isolation
RNA was extracted from the second leaves (50–100 mg) of hvabi5.d and WT plants collected on 10 and 25 DAS in three biological replications (each biological replicate represented leaf of one seedling). RNA isolation was conducted using TriPure reagent, according to the modified Chomczynski’s method (Chomczyński and Sacchi, 1987). For the microarray analyses, RNA was additionally purified using precipitation in 1 M lithium chloride and each RNA precipitate was then dissolved in 15 μl of nuclease-free H2O. NanoDrop (ND-1000) spectrophotometer (NanoDrop Technologies, Wilmington, USA) was used for concentration quantification and quality check. RNA integrity was analyzed using Agilent 2100 Bioanalyzer with RNA 6000 Nano chip (Agilent Technologies, Santa Clara, USA).
Microarray Analysis
Microarray Data Analysis
The synthesis, labeling, and hybridization of cDNA and cRNA were performed by the Genomics Core Facility, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany. The microarray data were analyzed using GeneSpring GX 13.0 software (Agilent Technologies) as described earlier (Daszkowska-Golec et al., 2017). A gene was considered to be differentially expressed when the level of its expression differed between the analyzed conditions by at least two times (fold change (FC) ≥ 2; P ≤ 0.05 after FDR correction). The annotation of the Agilent Barley Gene Expression Microarray (Agilent Technologies) was performed against IBSC_v2 of barley genome deposited in Ensembl Plants (v. 45). Functional annotation of differentially regulated genes was carried out using Ensembl Plants tools and the IPK Barley BLAST Server as references (https://plants.ensembl.org/index.html, https://webblast.ipk-gatersleben.de/barley_ibsc/). Three biological replications were used for microarray expression analysis (each biological replicate represented leaf of one seedling).
GO Enrichment Analysis
The GO enrichment analysis was performed using AgriGO 2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/) to identify the functional ontologies for differentially expressed genes (DEGs) and to obtain the categories of biological functions which were overrepresented in the analyzed groups of DEGs. The AgriGO 2.0 enrichment tool reveals the GO terms in the analyzed set of genes in comparison to the GO frequency in the background genome. The statistical significance of identified GO terms was assessed by the hypergeometric distribution followed by the Bonferroni method for multiple testing correction (corrected P ≤ 0.01).
RT-qPCR Analysis of ABA-Related Genes
cDNA was synthetized using 1 µg of total RNA and ReverseAid First Strand cDNA Synthesis Kit (Thermo Scientific). The obtained volume of cDNA (20 µl) was diluted using water (1:5) in order to prepare template for RT-qPCR reaction. Primers were designed using Quant-Prime software (http://www.quantprime.de) and their sequences are given in Supplementary Material S3. The RT-qPCR was performed using SYBR GREEN I on LightCycler 480 Real-Time PCR Instrument (Roche). The program consisted of the initial denaturation step, followed by 45 cycles of 10 s at 95°C, 20 s at 60°C, and 10 s at 72°C. The LinReq software (Ramakers et al., 2003) was used for data analysis. Each sample was normalized using ELONGATION FACTOR 1-α (EF1) and GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH) reference genes (Rapacz et al., 2012). The relative expression level was calculated using expression of reference gene and gene of interest in WT under control conditions, according to delta-delta Ct method (Livak and Schmittgen, 2001). Three biological replications were used for gene expression analysis, each sample was analyzed in two technical replicates.
Prediction of Promoter Transcription Factor Binding Sites
Analysis of promoters of selected ABA pathway genes was conducted using PlanPan 3.0 (http://plantpan.itps.ncku.edu.tw/promoter.php) and PlantRegMap (http://plantregmap.cbi.pku.edu.cn/) platforms. Analyzed promoter sequences included 1,000 bp before the START codon.
ABA Spray Assay
Ten-day-old WT and hvabi5.d seedlings grown in growth chamber (20°C, 16/8 h photoperiod, 200 μmol/m−2 s−1 illumination) under optimal water conditions were sprayed with 25 ml distilled water or 200 μM ABA dissolved in distilled water according to the procedure proposed by Al-Momany and Abu-Romman (2014). The stomatal conductance (mmol m−2 s−1) was measured after 30 min, 3, and 6 h using AP4 porometer (DELTA-T Devices, Burwell, UK). The second leaves were collected from hvabi5.d and WT plants at each time-point and used for RNA extraction. The experiment was performed in five biological replications (each biological replicate represented leaf of one seedling). The method of expression analysis of selected ABA-related genes is described in RT-qPCR Analysis of ABA-Related Genes.
Results
Sequence Analysis of HvABI5 and Its Homologues
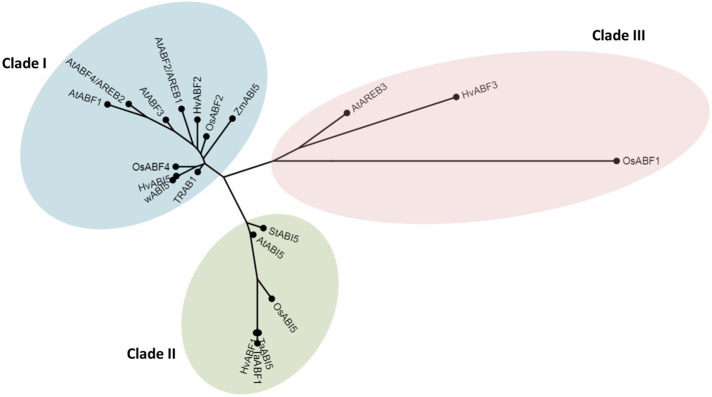
We analyzed the genetic distance between HvABI5 (HORVU5Hr1G068230) and its Arabidopsis, rice (Oryza sativa), wheat (Triticum aestivum), and maize (Zea mays) homologs. Based on phylogenetic analysis, bZIP transcription factors closely related to HvABI5 grouped in three clades (Figure 1). HvABI5 amino acid sequence formed a clade together with most of AtABFs/AtAREBs in Arabidopsis, HvABF2 (HORVU7Hr1G035500) in barley, OsABF2 (Os06t0211200), OsABF4 (Os09g0456200) and TRAB1 (Os08t0472000) in rice, wABI5 (TraesCS5A02G237200) in wheat, and ZmABI5 (Zm00001d018178) in maize. AtABI5 grouped in another clade, together with barley HvABF1 (HORVU3Hr1G084360). HvABI5 showed the highest level of amino acid sequence similarity to wABI5 (94.3%), OsABF4 (73.2%), TRAB1 (62.4%), OsABF2 (56.7%), and ZmABI5 (56.4%) (Supplementary Material S4). Among Arabidopsis genes, the most similar to HvABI5 were AtABF4/AREB2 (52.7%), AtABF2/AREB4 (51.3%), and AtABF3 (50.2%), however AtABI5 also showed a high percentage of similarity to HvABI5 (41.6%). Therefore, we could not clearly indicate the ortholog of HvABI5 in Arabidopsis. Despite the higher similarity of HvABI5 to AtABF/AtAREBs than to AtABI5, we decided not to change its name, since this gene (HORVU5Hr1G068230, Ensembl Plants) was described as HvABI5 in the previous studies of Casaretto and Ho (2003); Seiler et al. (2014) and Ishibashi et al. (2017). Furthermore, our results obtained during physiological analyses of barley hvabi5.d mutant pointed to its clear and obvious ABA-dependent and drought-tolerant phenotype, which was not the case of the single abf mutants in Arabidopsis. Taking these findings together, we decided to use the HvABI5 as a name of an ABA-dependent drought regulator described in this study.
Figure 1.
Relationships between HvABI5 and its homologs. Phylogenetic tree was generated using Phylogeny.fr software (http://www.phylogeny.fr/).
Initial Screening of hvabi5 Mutants for Drought Stress Response
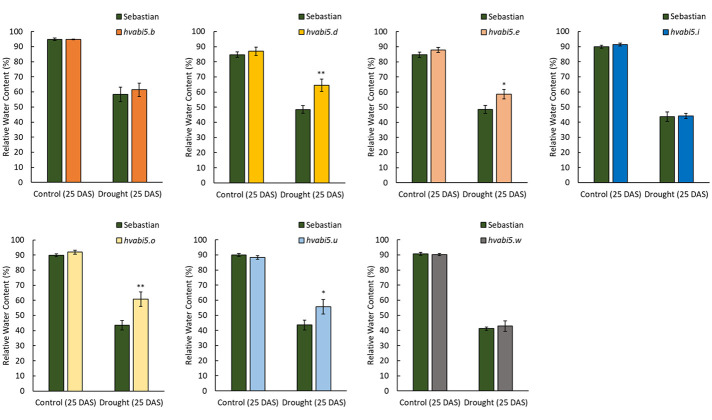
We identified 14 missense mutations in HvABI5 gene (Ensembl Plants, Gene ID HORVU5Hr1G068230; GenBank acc. no. HQ456390.1) after screening of 6,144 M2 plants of HorTILLUS population. All identified mutations were confirmed by sequencing and most of them were G/C to A/T transitions. Seven of 14 identified mutants (Table 1, Supplementary Material S5), after seed increase in M3/M4 generation, were used for RWC screening after drought treatment. Four of the tested mutants (hvabi5.d, hvabi5.e, hvabi5.o, hvabi5.u) exhibited higher RWC than cv. “Sebastian” after exposure to 10-d of severe drought stress (1.5% vwc), while three other (hvabi5.b, hvabi5.i, hvabi5.w) showed no differences in RWC value compared to “Sebastian” (Figure 2). Based on RWC screening after drought treatment, conservation of positions of analyzed mutations in HvABI5 protein (Supplementary Material S5) and seed availability, we selected hvabi5.d mutant for further analysis.
Table 1.
Mutants carrying missense mutations in HvABI5 gene (Ensembl Plants, Gene ID HORVU5Hr1G068230; GenBank acc. no. HQ456390.1) that were analyzed in initial drought stress experiment using Relative Water Content (RWC) test.
| Allele | Mutation position in HvABI5 gene | Position of amino acid substitution in HvABI5 protein | Region in HvABI5 protein | State of mutation in M2 plant |
|---|---|---|---|---|
| hvabi5.b | G1621A | A231T | Non conserved | Homozygous |
| hvabi5.d | G1751A | R274K | Close to bZIP domain | Heterozygous |
| hvabi5.e | G1588A | D220N | Non conserved | Homozygous |
| hvabi5.i | G1229A | G100D | Close to C2 domain | Homozygous |
| hvabi5.o | T1135G | F69V | C1 domain | Homozygous |
| hvabi5.u | G1346A | G139E | Close to C2 domain | Homozygous |
| hvabi5.w | C1445T | P172L | Close to C3 domain | Homozygous |
Figure 2.
The relative water content on 25 DAS under control conditions and 10-d drought treatment in hvabi5.b, hvabi5.d, hvabi5.e, hvabi5.i, hvabi5.o, hvabi5.u, hvabi5.w mutants and parent cv. “Sebastian”. The statistical analysis was performed using the one-way ANOVA followed by Fisher’s least significant difference test (LSD-test) to assess the differences between analyzed genotypes—*P ≤ 0.05, **P ≤ 0.01.
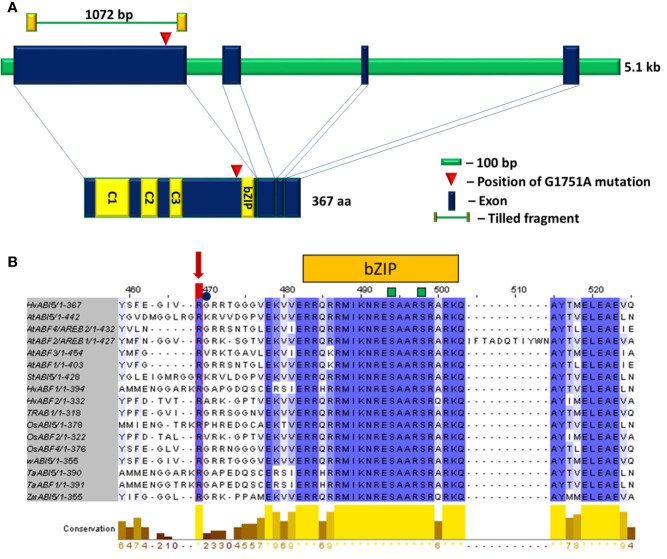
hvabi5.d carries a G1751A transition (position according to Ensembl Plants, Gene ID HORVU5Hr1G068230) which results in the arginine to lysine substitution (R274K) located close to the bZIP domain (Figure 3A), at the highly conserved position across monocot and dicot species (Figure 3B; alignment of the entire ABI5 and ABF protein sequences is presented in Supplementary Material S5). Moreover, the hvabi5.d mutation is located nearby the ubiquitination site (K344) of AtABI5 described in Arabidopsis (Liu and Stone, 2013).
Figure 3.
(A) The structure of HvABI5 gene and HvABI5 protein with the position of hvabi5.d mutation indicated. C1, C2, C3—conserved charged domains, bZIP—basic leucine zipper domain. (B) An alignment of ABI5 and ABF proteins fragment comprising hvabi5.d mutation position. The changed arginine (R274) in hvabi5.d is marked by a red arrow. The square and circle mark phosphorylation and ubiquitination sites, respectively. Blue color indicates conservation of aligned position, yellow bars mark level of conservation. The numbering visible in alignment visualization refers to multi sequence alignment of 17 ABI5 and ABF protein sequences in dicot and monocot species. It does not indicate the position of amino acids substituted in the hvabi5.d mutant given in Tab.1.
Study of hvabi5.d Sensitivity to ABA at Seed Germination and Seedling Development Stages
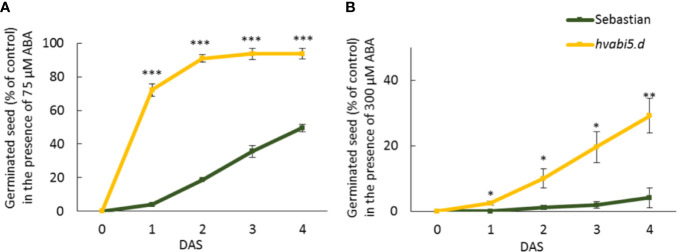
Taking advantage from Atabi5 mutants, we addressed the question of hvabi5.d response to ABA during seed germination. The dynamics of germination of hvabi5.d and its WT “Sebastian” showed no difference under control conditions. However, after ABA treatment, hvabi5.d mutant turned out to be ABA-insensitive. The application of 75 µM ABA reduced germination of the WT to 50% of control, whereas no inhibition was observed in hvabi5.d (Figure 4A). Moreover, in the presence of 300 µM ABA, the rate of hvabi5.d germination reached approximately 30% of control, while germination of the WT seeds was almost completely inhibited (Figure 4B).
Figure 4.
Germination of hvabi5.d and WT “Sebastian” under 75 (A) and 300 µM ABA (B). DAS—day after stratification. The seed germination in the presence of ABA is presented as the percent of the control (untreated seeds) value. The statistical analysis was performed using the T-test to assess the differences between genotypes in the presence of ABA. Statistically significant differences are indicated by asterisks (*P ≤ 0.05. **P ≤ 0.01. ***P ≤ 0.001).
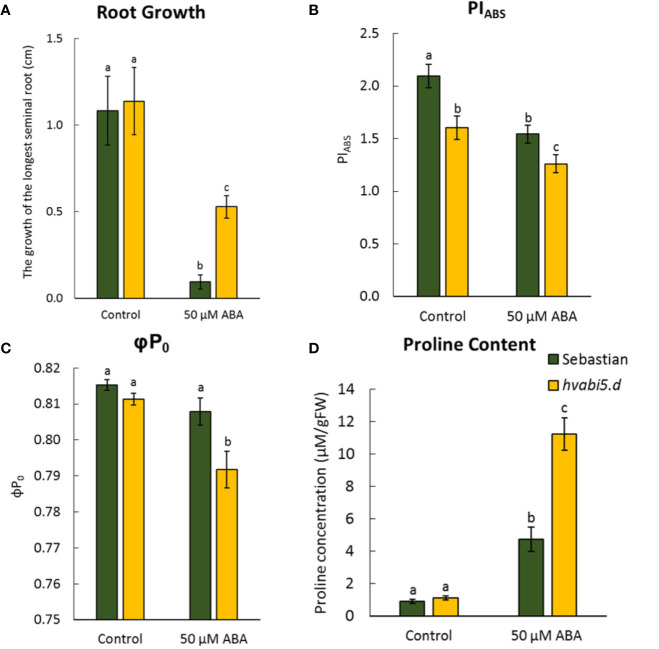
Given the fact that hvabi5.d displayed ABA-insensitivity at germination stage, we analyzed early seedling development of hvabi5.d mutant and the WT parent in the presence of ABA. Although ABA negatively impacted the first leaf growth in both, WT and hvabi5.d, no significant differences in leaf length were observed between these genotypes either in control or exposure to ABA (Supplementary Material S6). However, hvabi5.d showed a much lower ABA-dependent root inhibition, compared to the WT (Figure 5A).
Figure 5.
hvabi5.d response to ABA at seedling stage. (A) The growth of the longest seminal root of seedlings of analyzed genotypes after 6 d of ABA treatment, (B) photosynthesis performance index (PIABS), (C) maximum quantum yield of primary photochemistry (φP0), and (D) proline accumulation in seedlings of analyzed genotypes under control conditions and ABA (50 µM) treatment. The statistical analysis was performed using the two-way ANOVA (P ≤ 0.05) followed by Tukey’s honestly significant difference test (Tukey HSD-test) (P ≤ 0.05) to assess the differences between different growth conditions and genotypes. Statistically significant differences (P ≤ 0.05) are marked by different letters.
Taking into account the regulatory role of ABA in photosynthesis, we analyzed the efficiency of photosynthesis process in hvabi5.d and WT seedlings after ABA treatment. To investigate the photosynthesis performance, we applied the JIP test (Strasser et al., 2004). The obtained OJIP transients were converted into the biophysical parameters. The hvabi5.d seedlings grown under control conditions already showed a lower PIABS value than the WT. ABA treatment decreased the value of PIABS in both genotypes studied, nonetheless, hvabi5.d displayed a significantly more diminished PIABS value, compared to “Sebastian” (Figure 5B). The other analyzed parameter, φP0, was also lower in the mutant than in the parent cultivar after ABA treatment (Figure 5C). The values of φP0 and PIABS indicated the significant decrease of photosynthesis efficiency in hvabi5.d compared to “Sebastian”.
Apart from photosynthesis, we also measured the proline content in both genotypes. Proline is considered as an osmolyte that enables the osmotic adjustment during adaptation to abiotic stress. Although the proline content increased in response to ABA in both genotypes studied, the hvabi5.d accumulated twofold more proline than the WT (Figure 5D).
hvabi5.d Exhibited Drought Tolerance
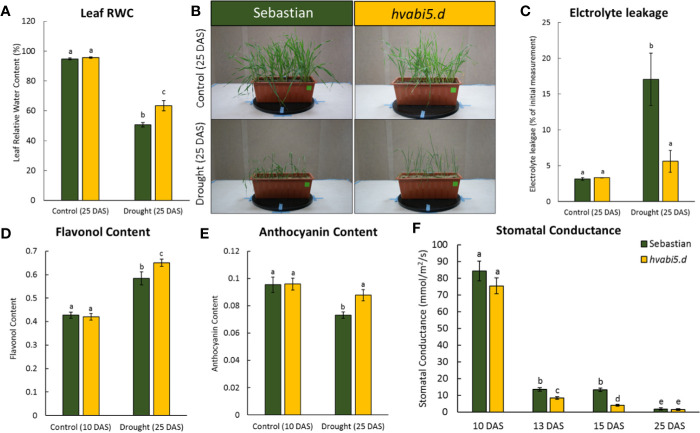
Taking into consideration that hvabi5.d exhibited the ABA-insensitive phenotype at germination and early seedling stages, we investigated its reaction to drought stress in order to determine if HvABI5 regulates drought response in barley. With the aim to characterize the physiological response of hvabi5.d, the leaf RWC, EL, and stomatal conductance were measured. No differences in leaf RWC between hvabi5.d and WT were observed under optimal water supply (control conditions) on 25 DAS. However, after 10-d exposure to drought stress, on the same 25 DAS, the mutant maintained 13% more water in leaf tissues than “Sebastian” according to the RWC value (Figure 6A). Moreover, drought stress induced more severe wilting in “Sebastian” compared to hvabi5.d (Figure 6B). Drought treatment induced a 5.4-fold increase of EL value in “Sebastian”, whereas in hvabi5.d there was no significant difference between control and drought-treated plants on 25 DAS (Figure 6C). We also analyzed flavonol and anthocyanin content index before and after drought treatment in both genotypes. Flavonols and anthocyanins function as antioxidants which scavenge toxic reactive oxygen species (ROS), protecting cells from their negative impact and thus contributing to drought tolerance. Similar levels of both, flavonols and anthocyainins were observed in both genotypes under well-watered conditions (10 DAS), but their content was significantly higher in hvabi5.d than in “Sebastian” under drought (25 DAS) (Figures 6D, E). These results are consistent with leaf RWC measurements. In order to follow the stomata action during the drought stress, we measured the stomatal conductance (gs) that describes the rate of gas exchange and transpiration between leaf and air. We analyzed this parameter before the drought treatment (10 DAS), at the drought onset (13 and 15 DAS), and after the severe drought stress (25 DAS) (Figure 6F). Both genotypes exhibited a vast reduction in stomatal conductance after severe drought. There were no significant differences between mutant and the WT when gs was measured before stress application (10 DAS) and after prolonged drought treatment (25 DAS) (Figure 6F). However, at the drought onset, the hvabi5.d showed respectively 1.5 and threefold lower stomatal conductance on 13 and 15 DAS than the parent variety. The decreased stomatal conductance enables the reduced transpiration via stress-induced stomata closure. It can be assumed that the better response of hvabi5.d to drought may be associated with a faster stomata closure and therefore, the lower water loss.
Figure 6.
hvabi5.d response to drought stress. (A) The relative water content on 25 DAS under control conditions and drought in hvabi5.d and “Sebastian”. (B) The seedling of hvabi5.d and “Sebastian” on 25 DAS under control conditions and drought. (C) The EL on 25 DAS under control conditions and drought in hvabi5.d and “Sebastian” (D) The flavonol content (E) and anthocyanin content on the 10 and 25 DAS in hvabi5.d and “Sebastian”. (F) The stomatal conductance (gs) on the 10, 13, 15, and 25 DAS of analyzed genotypes. The statistical analysis was performed using the two-way ANOVA (P ≤ 0.05) followed by Tukey’s honestly significant difference test (Tukey HSD-test) (P ≤ 0.05) to assess the differences between different growth conditions and genotypes. Statistically significant differences (P ≤ 0.05) are marked by different letters.
Taking into account that hvabi5.d showed the impaired photosynthesis in response to ABA at early seedling growth, we analyzed photosynthesis performance of hvabi5.d and WT when exposed to drought. First, the fluorescence of chlorophyll a was analyzed as the OJIP transient fluorescence (Figure 7A), where the J-I phase is related to reduction of plastoquinone (PQ) and cytochrome (Cyt b6f) and the I-P phase is associated with the reduction of the electron at the acceptor site of photosystem I (PSI) (Kalaji et al., 2016). The drought stress caused decrease of fluorescence only at the I-P phase in the WT, while in the hvabi5.d the decrease of the curve was observed already at the J-I phase and was even much more pronounced at the I-P phase (Figure 7A). These results indicate that photosynthesis is disturbed in hvabi5.d under drought. Moreover, some of the analyzed biophysical photosynthesis parameters were also impaired in hvabi5.d in the presence of drought (25 DAS), among them PIABS and φP0 (Figures 7B, C, Supplementary Material S7). PIABS indicates the amount of absorbed energy by electron acceptors, therefore its lower value denotes the impaired photosynthesis efficiency. The decreased photosynthesis efficiency in the mutant under drought might partially result from a more affected, compared to WT, stomatal conductance observed in the hvabi5.d at the onset of drought. It should be noted that under control conditions no differences in PIABS parameter were noted between the analyzed genotypes. The differences in PIABS value of the mutant observed under control conditions in two experiments (ABA and drought treatment) could arise from the developmental stage of analyzed plants and different experimental conditions, such as temperature and light intensity. Taking into account the impaired photosynthesis of hvabi5.d, we analyzed the content of chlorophyll in both genotypes. We detected no difference in chlorophyll content in genotypes analyzed under control conditions (10 DAS) (Figure 7D). However, we observed a decreased chlorophyll content in the mutant, compared to the WT under drought stress (25 DAS). It could be considered as another, besides the reduced stomatal conductance, cause of decreased photosynthesis efficiency in hvabi5.d.
Figure 7.
The effect of drought stress on photosynthesis efficiency in hvabi5.d and cv. “Sebastian”. (A) Chlorophyll a fluorescence curves during the pulse of light excitation, (B) performance index (PIABS), (C) maximum quantum yield of primary photochemistry (φP0), and (D) chlorophyll content in analyzed genotypes under optimal water supply (10 DAS) and drought conditions (25 DAS). The statistical analysis was performed using the two-way ANOVA (P ≤ 0.05) followed by Tukey’s honestly significant difference test (Tukey HSD-test) (P ≤ 0.05) to assess the differences between different growth conditions and between genotypes. Statistically significant differences (P ≤ 0.05) are marked by different letters.
The Expression of HvABI5-Related Genes Is Up-Regulated in hvabi5.d Under Drought Stress
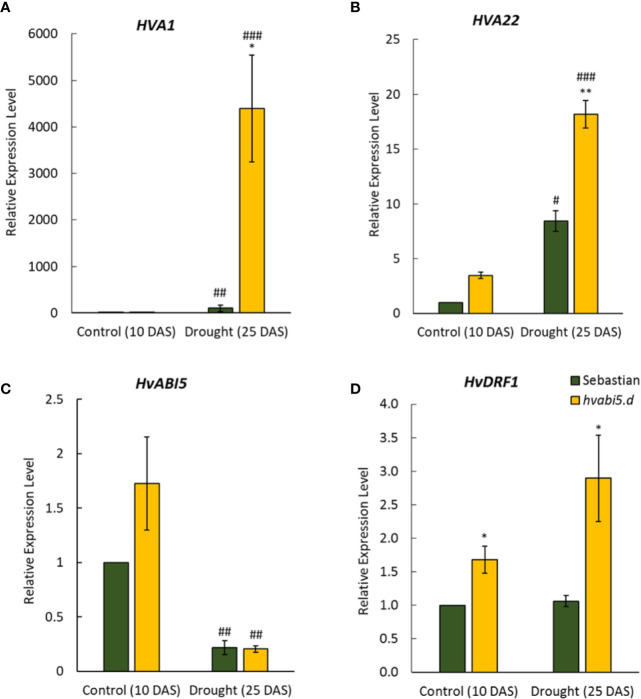
hvabi5.d showed response to ABA and drought tolerant phenotype, therefore we analyzed expression of HvABI5-related genes: HVA1 and HVA22 that are described as HvABI5 target genes (Casaretto and Ho, 2003). HVA1 and HVA22 expression was induced by drought stress in both genotypes, however it was, respectively, 43-fold and 2-fold higher in hvabi5.d than in the WT (Figures 8A, B). On the other side, drought induced a significant reduction of HvABI5 expression, but we did not observed differences between both genotypes analyzed (Figure 8C). Therefore, we investigated expression of the gene encoding another transcription factor, DEHYDRATION-RESPONSIVE FACTOR 1 (HvDRF1), which is known to act synergistically with HvABI5 in activating HVA1 (Xue and Loveridge, 2004). HvDRF1 showed a significantly higher expression in the hvabi5.d mutant already under optimal water conditions compared to its WT (Figure 8D). Drought caused even more pronounced increase of HvDRF1 expression in hvabi5.d, whereas no differences were observed in the WT.
Figure 8.
The expression of HvABI5-related genes in hvabi5.d and WT “Sebastian” under optimal water supply (10 DAS) and drought stress (25 DAS). HvDRF1—DEHYDRATION‐RESPONSIVE FACTOR 1. The statistical significance was estimated by T-test between analyzed genotypes—*P ≤ 0.05, **P ≤ 0.01 and between analyzed points—#P ≤ 0.05, ##P ≤ 0.01, ###P ≤ 0.001.
Transcriptome Analysis of hvabi5.d Under Drought Stress
Taking into account the drought tolerant phenotype of hvabi5.d and the higher expression of HvABI5-related genes, we anticipated that the global transcriptome analysis of this mutant under drought will help to shed light on the molecular mechanisms underlying its response to water deficit. For this purpose, we conducted the transcriptome analysis of hvabi5.d and its WT parent before drought treatment (10 DAS) and after 10-d drought stress application (25 DAS). The reliability of our microarray analysis was validated and confirmed using RT-qPCR analysis of genes which presented a wide range of expression levels in hvabi5.d under drought (Supplementary Material S8).
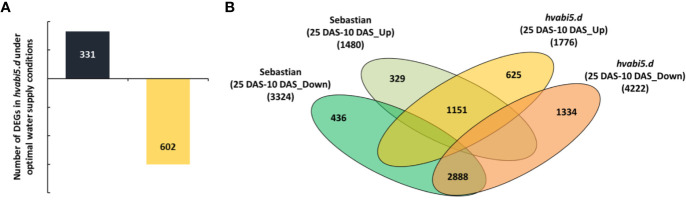
First, we addressed the question whether gene expression levels differed in hvabi5.d and “Sebastian” under optimal water supply, before the drought stress treatment. The comparison of hvabi5.d and WT transcriptomes revealed 933 DEGs (DEGs) in the mutant, among them 331 up- and 602 down-regulated genes (FC ≥ 2; P ≤ 0.05 after FDR correction) (Figure 9A). A relatively high number of DEGs could arise from the low cutoff criterion that was used in the analysis. Taking into account that HvABI5 encodes a transcription factor, we decided to track expression level of a larger number of genes in order to avoid any oversight that might have happened if a stricter criterion was used for analysis. In order to get an insight into biological role of DEGs, the analysis of functional annotation was performed using AgriGO 2.0 (http://http://systemsbiology.cau.edu.cn/agriGOv2/).
Figure 9.
Transcriptome analysis of hvabi5.d and its WT parent “Sebastian” under optimal water conditions (10 DAS) and drought stress (25 DAS). (A) The number of differentially expressed genes (DEGs) in hvabi5.d under optimal water supply (10 DAS) (P ≤ 0.05 after FDR correction; FC ≥ 2). (B) Comparative analysis of the numbers of DEGs after drought in,Sebastian’ and the hvabi5.d mutant. In the Venn diagrams, the subsets of genes that were up-regulated or down-regulated specifically in “Sebastian” and in hvabi5.d after drought stress (25 DAS) compared to control conditions (10 DAS) are shown (P ≤ 0.05 after FDR correction; FC ≥ 2).
Gene ontology (GO) analysis of the sets of genes specifically up- and down-regulated in the mutant indicated several over-represented biological processes (BP) associated with primary metabolism. In the set of up-regulated genes in hvabi5.d we identified processes such as: organonitrogen compound biosynthetic process, translation and protein folding (Supplementary Material S9). Furthermore, genes showing down-regulation specifically in hvabi5.d were categorized, among others, in BPs associated with ribonucleoside triphosphate and nucleoside triphosphate metabolic processes, ATP metabolic process and glycosyl compound metabolic process (Supplementary Material S9). The biological processes were considered as significant when P was less than 0.01 (assessed by the hypergeometric distribution followed by the Bonferroni method for multiple testing correction).
To investigate the transcriptome response to drought of hvabi5.d and its WT, we evaluated the gene expression level after drought stress (25 DAS) in relation to the conditions before drought treatment (10 DAS) for each genotype. Next, the DEGs were filtered out in order to obtain sets of genes specific for mutant and the WT. After drought treatment we found 625 up-regulated and 1,334 down-regulated genes specifically in hvabi5.d, whereas 329 and 436 genes were up- and down-regulated in “Sebastian”, respectively (FC ≥ 2; P ≤ 0.05 after FDR correction) (Figure 9B). To get an insight into the biological relevance of differentially regulated genes, we performed GO enrichment analysis of mutant- and “Sebastian”-specific sets of DEGs (Supplementary Material S10). It indicated that processes, such as: cellular amide metabolic process, translation, and gene expression were overrepresented within the set of genes specifically up-regulated in hvabi5.d under drought (Supplementary Material S10). Therefore, we carried out a more detailed analysis of this set of genes to reveal those, which could be associated with the drought tolerant phenotype of the hvabi5.d mutant (Table 2). Among them were genes directly involved in HvABI5 pathway, such as HVA22 homolog A and Ethylene-responsive transcription factor 4/dehydration responsive element binding protein 3 (ERF4/DREB3). Both these genes are engaged in barley response to drought and showed a significant increase of expression in our experiments (Figure 8). Furthermore, we identified other genes up-regulated specifically in hvabi5.d, which encode proteins associated with abiotic stress tolerance, such as late embryogenesis abundant proteins (LEAs), heat shock proteins (HSPs: Heat shock 70 kDa protein 3, Heat shock 70 kDa protein 15), and heat stress transcription factor C2b (HSFC2B). Some of them were also shown to be involved in detoxification of cytotoxic metabolites, among them genes encoding detoxifying enzymes: PRXs (peroxidase superfamily proteins), SOD ([Cu-Zn]2), and genes associated with flavonoid biosynthesis (Anthocyanidin reductase, Flavanone 3-hydroxylase, Chalcone-flavonone isomerase, and MYB domain protein 30). A closer look at the set of up-regulated genes in hvabi5.d revealed the phytohormonal crosstalk, since genes related to brassinosteroid (Delta(24)-sterol reductase), gibberelin (Gibberellin receptor GID1), and jasmonate (Transcription factor MYC2) signaling were identified in this subset (Table 2).
Table 2.
Differentially expressed genes (DEGs) identified in hvabi5.d after drought treatment associated with adaptation to abiotic stress, flavonoid biosynthesis, detoxification, photosynthesis, and phytohormonal pathways.
| Category | Gene ID | Fold change | Annotation |
|---|---|---|---|
| Adaptation to abiotic stress | HORVU7Hr1G012300 | 8.61 | LEA |
| HORVU6Hr1G081460 | 5.31 | Heat shock 70 kDa protein 3 | |
| HORVU7Hr1G038040 | 3.92 | HVA22 homolog A | |
| HORVU1Hr1G060490 | 3.59 | Ethylene-responsive transcription factor 4 | |
| HORVU7Hr1G088920 | 2.98 | Heat stress transcription factor C-2b | |
| HORVU1Hr1G027420 | 2.92 | Heat shock 70 kDa protein 15 | |
| HORVU5Hr1G120230 | 2.90 | Late embryogenesis abundant protein | |
| HORVU1Hr1G059870 | 2.47 | Late embryogenesis abundant protein D-19 | |
| Flavonoid biosynthesis | HORVU2Hr1G108250 | 33.04 | Anthocyanidin reductase |
| HORVU2Hr1G110130 | 6.19 | Flavanone 3-hydroxylase | |
| HORVU5Hr1G046480 | 3.29 | Chalcone-flavonone isomerase | |
| HORVU5Hr1G123880 | 2.17 | Myb domain protein 30 | |
| Detoxification | HORVU4Hr1G057210 | 2.84 | Peroxidase superfamily protein |
| HORVU4Hr1G057170 | 2.84 | Peroxidase superfamily protein | |
| HORVU2Hr1G021110 | 2.20 | Superoxide dismutase [Cu-Zn] 2 | |
| Photosynthesis | HORVU7Hr1G096250 | −8.18 | High chlorophyll fluorescence phenotype 173 |
| HORVU6Hr1G033160 | −3.53 | Chlorophyll A/B binding protein 3 | |
| HORVU3Hr1G021910 | −3.35 | Tetratricopeptide repeat (TPR)-containing protein | |
| HORVU5Hr1G109940 | −2.22 | Chaperonin-like RbcX protein | |
| HORVU5Hr1G093930 | −2.22 | Chaperonin-like RbcX protein | |
| Auxin pathway | HORVU1Hr1G016700 | −2.59 | Auxin transporter-like protein 3 |
| HORVU7Hr1G084940 | −2.53 | Auxin-responsive protein IAA23 | |
| HORVU3Hr1G032230 | −2.28 | Auxin response factor 1 | |
| HORVU3Hr1G070620 | −2.17 | Auxin-responsive protein IAA6 | |
| HORVU2Hr1G076920 | −2.08 | Auxin response factor 1 | |
| HORVU3Hr1G097200 | −2.05 | Auxin response factor 2 | |
| Gibberellin pathway | HORVU4Hr1G062730 | 13.10 | Gibberellin receptor GID1 |
| HORVU2Hr1G099440 | −2.07 | Terpene synthase 04 | |
| Cytokinin pathway | HORVU3Hr1G027460 | −2.53 | Cytokinin dehydrogenase 2 |
| Brassinosteroid pathway | HORVU7Hr1G120030 | 4.18 | Delta(24)-sterol reductase |
| HORVU5Hr1G114850 | −2.27 | Protein kinase superfamily protein | |
| HORVU3Hr1G068020 | −2.04 | Leucine-rich receptor-like protein kinase family protein | |
| Jasmonic acid pathway | HORVU1Hr1G050560 | 2.71 | Transcription factor MYC2 |
Genes that were down-regulated specifically in hvabi5.d were found to be involved in biological processes related to primary metabolism e.g., translation, peptide metabolic process, organonitrogen compound biosynthetic process, cellular protein metabolic process, and gene expression (Supplementary Material S10). A detailed analysis of the down-regulated DEGs list in the mutant (Table 2) revealed genes associated with chlorophyll (chlorophyll A/B binding protein 3 and high chlorophyll fluorescence phenotype 173), both related to photosynthesis efficiency. Other genes related to photosynthesis, such as genes encoding chaperones involved in RuBisCO assembly (chaperonin-like RbcX proteins) and chaperone associated with photosystem II assembly [tetratricopeptide repeat (TPR)-containing protein] were also identified in the down-regulated group (Table 2). Among the down-regulated set, several genes involved in auxin pathway (auxin-responsive protein IAA6, auxin-responsive protein IAA23, auxin response factors 1, auxin response factor 2, auxin transporter-like protein 3), brassinosteroid signaling (leucine-rich receptor-like protein kinase family protein, Protein kinase superfamily protein), gibberelin synthesis (terpene synthase 04), and cytokinin catabolism (cytokinin dehydrogenase 2) were identified (Table 2).
Based on transcriptome analysis of hvabi5.d response to drought, we assume that a better drought tolerance of the mutant results from of the higher activation of genes associated with the HvABI5 pathway and abiotic stress response mechanisms, together with the changed phytohormonal crosstalk. It should be underlined that we did not observe similar reactions to drought stress in the WT cv. “Sebastian” (Supplementary Material S11).
The Expression of ABA-Related Genes Is Changed in hvabi5.d Under Drought Stress
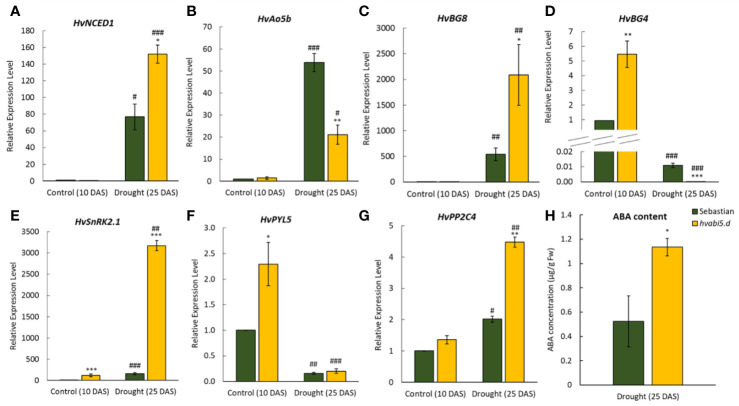
Having shown that hvabi5.d mutant displayed ABA insensitivity during seed germination/early root growth and drought-tolerant phenotype at seedling/tillering stage, we asked next whether HvABI5 regulates barley drought response through the ABA-dependent way. To answer this question, we first analyzed the drought stress response of the genes encoding ABA metabolism and ABA signaling in hvabi5.d and WT. We compared plants grown under optimal water supply before drought (10 DAS) and after drought stress (25 DAS).
The balance between ABA biosynthesis and ABA catabolism is an important factor in activation of stress response (Chan, 2012). 9-cis-epoxycarotenoid dioxygenase-like (HvNCED1) and ABSCISIC ALDEHYDE OXIDASE 5b (HvAO5b) encode key enzymes in ABA biosynthesis. Under optimal water supply (10 DAS) there were no significant differences between the expression level of HvNCED1 and HvAO5b in WT and mutant (Figures 10A, B). Drought stress induced expression of HvNCED1 in both genotypes, however the differences between hvabi5.d mutant and WT were prominent. Drought induced 160-fold increase of expression of HvNCED1 in the mutant and 80-fold-increase in the WT. On the other hand, the expression of HvAO5b gene encoding the last step of ABA biosynthesis raised to the higher level in the WT than in mutant, 55-fold and 20-fold compared to control conditions, respectively.
Figure 10.
The expression of ABA pathway-related genes in hvabi5.d and its WT parent “Sebastian” under optimal water supply (10 DAS) and drought stress (25 DAS). (A) HvNCED1—CAROTENOID CLEAVAGE DIOXYGENASE1, (B) HvAO5b—ABSCISIC ALDEHYDE OXIDASE5b, (C) HvBG8—β-GLUSIDASE8, (D) HvBG4, (E) HvSnRK2.1—SNF1-RELATED PROTEIN KINASE 2.1, (F) HvPYL5—PYRABACTIN RESISTANCE 1-LIKE 5, and (G) HvPP2C4—PROTEIN PHOSPATASE 2C 4. The statistical significance was estimated by T-test between analyzed genotypes—*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and between analyzed points—#P ≤ 0.05, ##P ≤ 0.01, ###P ≤ 0.001. (H) Endogenous ABA content on 25 DAS under drought in hvabi5.d and “Sebastian”. The statistical significance was estimated by T-test between analyzed genotypes—*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
ABA can be also pooled in ABA-glucose esters (Xu et al., 2014). We analyzed the expression of barley β-GLUCOSIDASEs genes, HvBG8 and HvBG4, responsible respectively, for de-conjugation and conjugation of ABA-glucose esters (Figures 10C, D). Under optimal water conditions expression of HvBG8 was very low and did not differ between hvabi5.d and WT. In response to drought stress, a 500-fold increase of HvBG8 expression was observed in the WT and remarkably, a 2,000-fold increase in the mutant. The expression of HvBG4 showed a different pattern of expression than HvBG8, as drought treatment drastically decreased its transcription level in both genotypes. Under optimal water conditions the hvabi5.d mutant showed a 5.5-fold higher expression of HvBG4 compared to “Sebastian”, while after drought treatment HvBG4 was down-regulated to a greater extent in the mutant (−3,300-fold) than in the WT (−91-fold).
The expression analysis of ABA metabolism genes demonstrated that their regulation differed in hvabi5.d and its WT parent. It raised the possibility that HvABI5 might be engaged in the fine tuning of ABA signaling by a feedback regulation between biosynthetic and signaling events. To test this hypothesis the expression of core ABA signaling genes: SNF1-RELATED PROTEIN KINASE2.1 (HvSnRK2.1), PYRABACTIN RESISTANCE 1-LIKE 5 (HvPYL5), and PROTEIN PHOSPATASE2C4 (HvPP2C4) was evaluated in the hvabi5.d and WT before and after drought treatment (Figures 10E–G). HvSnRK2.1 and HvPYL5 were up-regulated in the hvabi5.d compared to the WT already under optimal water conditions. After drought stress the down-regulation of HvPYL5 was observed and no differences between both genotypes were noticed. Interestingly, in response to drought the level of HvSnRK2.1 and HvPP2C4 expression was significantly higher in hvabi5.d than in the WT and the exceptional 3000-fold change of HvSnRK2.1 expression in the mutant, 20 times higher than in “Sebastian” was noted. We discuss these results in the further section of the paper.
Endogenous ABA Content Is Higher in hvabi5.d Than in WT Under Drought
Expression of the key ABA metabolism and signaling genes was induced to much higher level in hvabi5.d than in WT under drought. Therefore, we presumed that endogenous ABA level might differ in the mutant compared to its parent cultivar. To answer this question, we measured ABA content in plants of both genotypes grown under optimal water conditions and exposed to 10-d drought stress. ABA concentration was below detection level under optimal water supply in “Sebastian” and hvabi5.d, whereas in response to drought ABA accumulation increased in both genotypes. Interestingly, the drought stress caused the two-fold higher increase of endogenous ABA content in the hvabi5.d than in the WT (Figure 10H).
Promoters of ABA Biosynthesis and Signaling Genes Contain AtABI5 Binding Motifs
ABI transcription factors (AtABI4 and AtABI5) were shown to be involved in a feedback regulation of ABA biosynthesis and signaling genes through direct binding to their promoters and activating their expression (Shu et al., 2016; Wang et al., 2019). Taking into account the changed expression pattern of ABA biosynthesis and signaling genes in hvabi5.d under drought, we analyzed transcription factor binding sites in their promoters. We found that promoters of HvNCED1, HvSnRK2.1, and HvPP2C4 are enriched in motifs recognized by AtABI5 (Supplementary Material S12). Therefore, HvABI5 might be involved in a feedback regulation of the ABA pathway to ensure the proper amplification of ABA signal in the presence of drought.
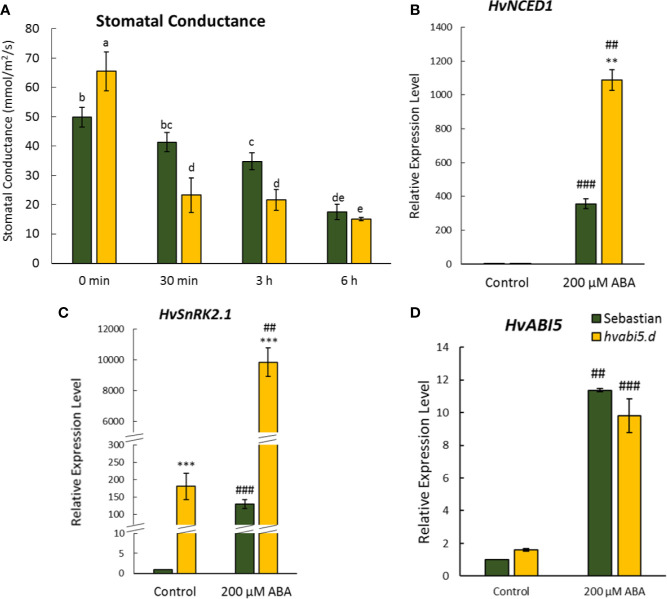
hvabi5.d Showed Faster Stomata Closure and Changed Expression of ABA-Related Genes Under ABA Treatment
To confirm if HvABI5 regulates barley response to drought in the ABA-dependent way, we analyzed stomatal conductance and expression of the selected ABA-related genes in response to exogenously applied ABA. In order to test whether hvabi5.d stomata are sensitive to exogenously applied ABA, we measured stomatal conductance 30 min, 3, and 6 h after ABA spraying (Figure 11A). The reaction of hvabi5.d stomata to ABA was noticed faster than of the WT, already 30 min after treatment. Both genotypes showed the reduction of stomatal conductance 3 and 6 h after ABA treatment, however, 3 h after treatment the stomatal conductance was almost twofold lower in hvabi5.d than in WT. These results indicate that mutant stomata were more sensitive to ABA than stomata of its WT parent. Thus, to check the ABA-dependent response of hvabi5.d and the WT, we analyzed the expression of ABA biosynthesis gene, HvNCED1, and ABA signaling gene, HvSnRK2.1, in leaves of both genotypes 3 h after ABA treatment. It should be underlined that drought treatment activated transcription of both genes to the much higher levels in the hvabi5.d mutant than in “Sebastian”. As previously described, the expression of HvSnRK2.1 was higher in hvabi5.d than in the WT under control conditions. The ABA treatment highly induced HvNCED1 and HvSnRK2.1 expression in both genotypes, nevertheless the expression of HvNCED1 and HvSnRK2.1 was much higher in hvabi5.d compared to WT, 3 and 76 times, respectively (Figures 11B, C). ABA also activated HvABI5 expression, however we observed no differences between hvabi5.d and “Sebastian” (Figure 11D).
Figure 11.
Response of hvabi5.d to ABA spraying. (A) Stomatal conductance of hvabi5.d and its WT parent “Sebastian” after ABA treatment. The statistical analysis was performed using the two-way ANOVA (P ≤ 0.05) followed by Tukey’s honestly significant difference test (Tukey HSD-test) (P ≤ 0.05) to assess the differences between different growth conditions and between genotypes. Statistically significant differences (P ≤ 0.05) are marked by different letters. The expression of (B) HvNCED1—CAROTENOID CLEAVAGE DIOXYGENASE1 (C) HvSnRK2.1—SNF1-RELATED PROTEIN KINASE 2.1, and (D) HvABI5—ABA INSENSITIVE5 in hvabi5.d and its WT 3 h after ABA treatment. The statistical significance was estimated by T-test between analyzed genotypes—**P ≤ 0.01, ***P ≤ 0.001 and between analyzed points—##P ≤ 0.01, ###P ≤ 0.001.
Discussion
Here, we report the first barley mutant (hvabi5.d) carrying a missense mutation in HvABI5 gene (Ensembl Plants, Gene ID HORVU5Hr1G068230). HvABI5 belongs to the ABA-dependent bZIP transcription factors that in response to abiotic stresses regulate directly expression of genes containing ABRE motifs in their promoters (Kagaya et al., 2002; Kobayashi et al., 2008; Hossain et al., 2010a; Yan et al., 2012; Piao et al., 2019). Of the Arabidopsis bZIP genes with ABRE motifs, HvABI5 appears to be more related to AtABFs/AtAREBs than to AtABI5. Both, AtABI5 and AtABFs/AtAREBs regulate plant response to abiotic stresses including drought, although at different developmental stages. AtABI5 takes part in plant response to stress mostly at the germination and seedling stages (Finkelstein and Lynch, 2000; Carles et al., 2002; Bi et al., 2017), while AtABFs/AtAREBs act mostly during vegetative growth and their role seems to be highly redundant (Fujita et al., 2005; Yoshida et al., 2010; Yoshida et al., 2015). In our phylogenetic analysis HvABI5 grouped more closely to AtABFs/AtAREBs than to AtABI5, nevertheless the level of the sequence identity between HvABI5 and AtABFs/AtAREBs and between HvABI5 and AtABI5 were very similar. Although we cannot indicate the functional ortholog of HvABI5 in Arabidopsis, it is possible that barley HvABI5 might share common functions with both, AtABFs/AtAREBs and AtABI5. This presumption is supported by the fact that HvABI5 shows the highest level of similarity to monocot bZIPs such as wheat wABI5, rice TRAB1, OsABF4 and OsABF2, and maize ZmABI5, which act as ABA-dependent transcription factors. wABI5, TRAB1, and OsABF2 were described as positive regulators of response to abiotic stresses, including drought in vegetative tissues of these species (Kagaya et al., 2002; Kobayashi et al., 2008; Hossain et al., 2010a). Close phylogenetic relationship of HvABI5 with wABI5, TRAB1, and OsABF2 enhanced our hypothesis that HvABI5 may act as the ABA-dependent regulator of drought response in barley vegetative tissues.
According to the in silico analysis and alignment with AtABI5, R274K substitution caused by G1751A mutation in the hvabi5.d is located close to the ubiquitination site in the ABI5 protein. The analyzed mutation can generate or diminish the site of post-translational modification in HvABI5, which in turn may change its activity. In this study we confirmed that barley ABI5 regulates ABA response at early developmental stages, as hvabi5.d mutant showed a significantly reduced ABA sensitivity during seed germination and root growth. Similar reaction was reported for Arabidopsis mutants in AtABI5 and AtABFs/AtAREBs genes (Finkelstein, 1994; Carles et al., 2002; Yoshida et al., 2010) and rice mutant in OsABF2 gene (Hossain et al., 2010a). During seedling development in hydroponic conditions in the presence of ABA, hvabi5.d was able to accumulate more proline than its parent cv. “Sebastian” and showed the reduced efficiency of photosynthesis. Moreover, we observed a faster stomatal closure in the mutant compared to WT after ABA spraying of 10-d old seedlings. The results obtained after ABA treatment gave us premises to presume that HvABI5 is involved in the regulation of ABA signaling in barley.
hvabi5.d Showed a Water-Saving Mechanism in Response to Severe Drought Stress
hvabi5.d showed a drought-tolerant phenotype manifested mainly by the ability to store water more efficiently than its WT parent “Sebastian”. Our experiment revealed that the higher RWC in hvabi5.d leaves after severe drought stress might result from a better membrane protection and a faster stomatal closure. The cell membrane stability expressed as percentage of EL is an indicator of the level of tolerance to water stress in plants (Bajji et al., 2002). Our results showed that the cellular membranes of the hvabi5.d were less damaged than the WT after drought treatment.
Better membrane performance in hvabi5.d under drought stress might result from the more efficient activation of protection mechanisms in the mutant, as revealed by transcriptomic analysis. In response to drought, the hvabi5.d showed increased expression of genes encoding late embryogenesis abundant proteins (LEAs) and HSPs (heat shock 70 kDa protein 3, Heat shock 70 kDa protein 15). The LEA and HSP proteins enable adaptation to abiotic stress through protection of proteins and membrane structure (Vinocur and Altman, 2005; Shinozaki and Yamaguchi-Shinozaki, 2007; Bhatnagar-Mathur et al., 2008). AtABI5, AtABFs/AtAREBs, and their monocot homologs were shown to regulate expression of LEA genes (Kobayashi et al., 2008; Yoshida et al., 2010; Yan et al., 2012; Su et al., 2016). In barley, HvABI5 was shown to directly activate HVA1 and HVA22 genes that encode LEA proteins that ensure tolerance to low water availability (Hong et al., 1992; Shen et al., 2001). It was also shown that HVA1 is regulated by other transcription factor, HvDRF1 (Xue and Loveridge, 2004). We observed the increased expression of HVA1, HVA22, and HvDRF1 in hvabi5.d under drought. Furthermore, the global transcriptome analysis also revealed a higher expression of HVA22 homolog A and ethylene-responsive transcription factor 4/dehydration responsive element binding protein 3 (HvDRF1). Therefore, the increased activation of HVA1 and HVA22 in the mutant may result simultaneously from the changed activity of HvABI5 protein and the higher expression of HvDRF1. Taken together, the higher water content and better membrane stability of hvabi5.d under water stress, compared to the WT, can be associated with up-regulation of LEA and HSP genes.
In addition, better membrane stability of hvabi5.d and its drought tolerant phenotype might be related to a higher content of flavonoids: flavonols and anthocyanins. Flavonoids protect plants against ROS under abiotic stress (Di Ferdinando et al., 2012; Davies et al., 2018). The higher expression of genes encoding flavonoid biosynthesis enzymes (anthocyanidin reductase, flavanone 3-hydroxylase, chalcone-flavonone isomerase, and MYB domain protein 30) observed in our study could explain the higher flavonoid content in hvabi5.d under drought. The relationship between AtABI5 and anthocyanins biosynthesis was previously observed in Arabidopsis by Brocard et al. (2002). Another mechanism contributing to increased drought tolerance of hvabi5.d could be related to detoxification enzymes, that similarly to flavonoids protect against ROS under abiotic stress (Anjum et al., 2016; Mishra and Sharma, 2019). The increased expression of genes encoding PRXs (peroxidase superfamily proteins) and SOD ([Cu-Zn] 2) could alleviate drought-induced damages in hvabi5.d. It was shown that ABI5 directly regulated expression of genes encoding detoxification enzymes in Arabidopsis and barley (Bi et al., 2017; Ishibashi et al., 2017). Moreover, over-expression of ZmABI5, a maize homolog of HvABI5, activated expression of ASCORBATE PEROXIDASE (APX) under drought (Yan et al., 2012).
Another physiological trait that ensures a low water loss is stomatal closure in response to water scarcity. We observed a lower stomatal conductance in hvabi5.d mutant than in WT at the onset of drought stress (13 and 15 DAS), but not after application of severe drought (25 DAS). The faster stomata closure can also contribute to the increased drought tolerance of havbi5.d. We discuss the possible mechanism underlying this process in the further part of Discussion.
The altered HvABI5 function in hvabi5.d mutant negatively influenced the photosynthesis efficiency and chlorophyll content under drought stress. We presumed that impaired photosynthesis of hvabi5.d after drought treatment could be the effect of both, the lower stomatal conductance at the onset of drought stress and the lower chlorophyll content. The role of ABA and ABA signaling components in chlorophyll degradation has been well described (Yang et al., 2014; Gao et al., 2016). In Arabidopsis, the involvement of AtABI5 in regulation of photosynthesis was shown in terms of repression of ABA-RESPONSE PROTEIN (ABR) responsible for protection of photosynthesis proteins (Su et al., 2016). Furthermore, AtABI5 directly promoted expression of chlorophyll catabolism-related genes (STAYGREEN 1—SGR1 and NON-YELLOW COLORING 1—NYC1) (Sakuraba et al., 2014). Similarly, AtABFs/AtAREBs were shown to regulate directly genes associated with chlorophyll degradation: SGR1, NYC1 and PHEOPHORBIDE A OXYGENASE (PAO) (Gao et al., 2016). In rice OsABF4 activated directly expression of OsSGR1 and OsNYC1 (Piao et al., 2019). Our transcriptomic data revealed the lower expression level of both, photosynthesis-related genes (chaperonin-like RbcX proteins, TPR-containing protein) and genes associated with chlorophyll function (chlorophyll A/B binding protein 3, high chlorophyll fluorescence phenotype 173) in hvabi5.d under drought.
It should be noted that hvabi5.d is the first ABI5 mutant evaluated in detail in terms of tolerance to drought stress. Arabidopsis Atabi5-1 mutant has not been characterized in this respect, it was only described as “not-wilty” in databases (Finkelstein and Lynch, 2000; http://www.arabidopsis.org, AT2G36270). The same lack of data applies to abi5 mutants of Medicago truncatula and Pisum sativum (Zinsmeister et al., 2016). The only mutants studied in this respect: Atabf4/Atareb2, Atabf3, Osabf1, and Osabf2 showed a lower survival rate after drought application (Hossain et al., 2010a; Hossain et al., 2010b; Yoshida et al., 2010). The changed expression of AtABI5 under drought stress (Brocard et al., 2002), a drought tolerant phenotype of plants over-expressing wABI5 and AtABFs/AtAREBs genes (Kang et al., 2002; Fujita et al., 2005; Kobayashi et al., 2008) and drought sensitive phenotype of lines over-expressing ZmABI5 (Yan et al., 2012) are in agreement with the role of barley ABI5 in drought response presented in this study.
Drought Response of hvabi5.d Is the ABA-Dependent Process
Regarding hvabi5.d response to ABA at early seedling stage and its drought tolerant phenotype we presumed that HvABI5 regulated drought response in the ABA-dependent way. To verify this hypothesis, we analyzed expression of the ABA pathway genes under drought and ABA. Among two genes encoding key enzymes in ABA biosynthesis (HvNCED1 and HvAo5b) analyzed in our study, only HvNCED1, which is a rate-limiting enzyme for ABA biosynthesis, showed a much higher expression in hvabi5.d, compared to WT, in response to drought and ABA. The expression of HvAo5b increased after drought stress in both genotypes. However, the much lower transcription level of HvAo5b in hvabi5.d than in the WT might level out the effect of the high HvNCED1 expression in the mutant. Furthermore, hvabi5.d showed, respectively, higher and lower expression of HvBG8 and HvBG4 after drought. HvBG8 is responsible for ABA de-conjugation, whereas HvBG4 takes part in ABA conjugation. It suggests that ABA is more effectively released from ABA glucosyl esters in hvabi5.d in response to drought. This mechanism might be important for the higher drought tolerance of the mutant.
We investigated also the expression of genes encoding the core ABA signaling, such as HvPYL5, HvPPC4, and HvSnRK2.1. According to the basic model of ABA signal pathway, in the presence of endogenous ABA PYR/PYLs receptors interact with PP2Cs and inhibit phosphatase activity, allowing SnRK2 activation and phosphorylation of target proteins, among them ABI5. In our study, the down-regulation of HvPYL5 and up-regulation of HvPP2C4 was observed in both, hvabi5.d and WT, in response to drought. However, the increase of HvPP2C4 expression was twofold higher in the mutant than in the WT. Moreover, we noticed substantially higher activation of HvSnRK2.1 in hvabi5.d than in WT under drought and ABA. Increased induction of HvSnRK2.1 expression was also observed by Seiler et al. (2014) in drought-tolerant line of barley. It can be presumed that the increased level of HvSnRK2.1 expression in hvabi5.d enables the amplification of ABA signaling and activation of ABA-related genes.
Drought-induced ABA biosynthesis is one of the main processes of plant adaptation to stress (Yoshida et al., 2019). The increased expression of ABA metabolism genes in havbi5.d resulted in the higher ABA content in the mutant compared to the WT. It indicates that HvABI5 takes part in triggering ABA accumulation and subsequent activation of ABA signaling, which in turn ensures response to drought through many mechanisms, among them stomatal closure.
Together, these findings demonstrated the role of HvABI5 in the feedback regulation of components of ABA biosynthesis and signaling under drought stress. We showed a possibility of HvABI5 engagement in a feedback up-regulation of ABA synthesis (via HvNCED1) that may help to amplify the ABA signal under stress and simultaneous up-regulation of PP2C4 that might contribute to avoiding the over-amplification of the stress signal in order to maintain adaptation and performance under stress. Recently AtABF1, AtABF2, AtABF3, AtABF4, and AtABI5 were shown to bind directly to promoters of ABI1 and ABI2, which encode PP2C type phosphatases. This mechanism ensures negative feedback loop in ABA signaling (Wang et al., 2019). Similarly, the up-regulation of HvSnRK2.1 in the hvabi5.d mutant raises the possibility of HvABI5 engagement in the feedback regulation of this kinase activity. It should be underlined that we found ABI5 binding elements in the promoters of HvNCED1, HvPP2C4, and HvSnRK2.1. Thus, it might be possible that HvABI5 directly regulates expression of these genes. Simultaneous upregulation of ABA synthesis via induction of HvNCED1, and ABA signaling via induction of HvPP2C4 and HvSnRK2.1 in hvabi5.d mutant in response to drought stress indicate a fine-tuned control of the amplification of stress signaling and needs further investigations.
Our results strongly support the hypothesis that drought response of hvabi5.d is the ABA-dependent process. Yet, hvabi5.d showed reduced ABA sensitivity at germination stage. It indicates that ABA-dependent function of HvABI5 at seed germination differs from its role in the regulatory pathway involving HvABI5 under drought stress. HvABI5 expression is not restricted to a short developmental window during seed germination and early seedling growth, as shown in Arabidopsis (Maia et al., 2014). It is possible that HvABI5 has acquired new functions during evolution in barley. It has been proved that genes responsible for plant fitness evolve faster than core metabolism genes (Nelissen et al., 2014). Furthermore, many abiotic stress tolerance mechanisms of monocots can differ from those in Arabidopsis (Tester and Bacic, 2005). In Arabidopsis, ABA-mediated response to abiotic stress is regulated by AtABI5 in seeds and AtABFs/AtAREBs in leaves and roots at vegetative stage. However, function of AtABFs/AtAREBs in ABA signaling is highly redundant. Here, we indicate that due to evolution events HvABI5 shares some function with AtABI5 and AtABFs/AtAREBs and acts simultaneously as the ABA-dependent regulator at the seed germination stage and in response to drought in seedling vegetative tissues. Contrary to AtABFs/AtAREBs, the role of HvABI5 under drought seems to be individual since hvabi5.d shows drought tolerant phenotype, while triple Atabf2/areb1 Atabf4/areb2 Atabf3 mutant exhibited significantly disturbed phenotype under drought (Yoshida et al., 2010). On the other side, similarly to AtABFs/AtAREBs, HvABI5 acts in the ABA signaling as a regulator of LEAs expression, chlorophyll catabolism and stomatal closure, and takes part in the feedback regulation of ABA pathway. In monocots, wABI5, OsABF2, TRAB1, and ZmABI5 regulate ABA-mediated drought response by activation of stress responsive genes in seedling vegetative tissues (Kagaya et al., 2002; Kobayashi et al., 2008; Hossain et al., 2010a; Yan et al., 2012). Furthermore, OsABF2 and ZmABI5 were also shown to be active in seeds (Hossain et al., 2010a; Zhang et al., 2019). We revealed that function of HvABI5 as the ABA-dependent regulator of stress response is similar to its close monocot homologs.
Finally, we want to address the issue of the impact of possible background mutations on hvabi5.d phenotype. Taking into account that hvabi5.d was identified in the TILLING population derived from chemical mutagenesis, such a possibility cannot be ruled out. The best way to prove that the observed phenotypic change results from the identified mutation is to complement the mutated allele through genetic transformation with the WT gene. However, in barley the effective transformation is restricted merely to a single cultivar, “Golden Promise”. Cultivar “Sebastian”—the parent of hvabi5.d mutant turn out to be recalcitrant to Agrobacterium transformation in our preliminary experiments (data not shown). However, it should be underlined that hvabi5.d was identified as one of four mutants with different hvabi5 alleles that performed better than WT after drought treatment in our preliminary screening. We examined seven independent TILLING mutants carrying different alleles in the HvABI5 gene for their response to drought stress. Four of these mutants, including hvabi5.d, showed increased water content after drought treatment in the RWC assay. Three other tested alleles showed no significant differences in RWC value compared to WT, which indicates that these missense mutations did not evoke any significant changes in function of HvABI5 protein.
Furthermore, we developed hvabi5.d lines after two back-crosses to the parent cultivar “Sebastian” to clean the mutant from the possible background mutations. The created F4BC2 lines with hvabi5.d allele were then analyzed for the basic physiological traits related to drought response (RWC, flavonol and anthocyanin content, stomatal conductance) after exposure to drought stress. We also examined expression of HvABI5-related genes (HVA1, HVA22, HvDRF1) and genes involved in ABA biosynthesis, metabolism, and signaling (HvNCED1, HvBG8, HvSnRK2, and HvPP2C4) in these lines. All analyzed physiological parameters, as well as expression of all analyzed genes in the hvabi5.d F4BC2 were similar to those observed in the mutant exposed to drought, described in this study (Supplementary Material S13). Taken together our analysis clearly confirmed the association between HvABI5 function and regulation of drought stress response in barley. It should be noted that another approach to linking phenotype and a causative mutation i.e., analysis of their co-segregation in a segregating F2 population cannot be performed for qualitative traits such as drought tolerance. In the case of hvabi5.d it was not possible to distinguish between mutant and WT phenotype in zero-one categories. All tests used in the presented study required biological replications, therefore the analysis of segregation in F2 generation could not be performed on the basis of single plants phenotyping.
Conclusions
The presented results bring the set of new data regarding the function of HvABI5 in the ABA signaling during drought response in barley. We found that HvABI5 takes part in regulation of processes associated with drought stress tolerance, such as membrane protection, flavonoid accumulation, and stomatal closure. HvABI5 activates expression of stress-responsive genes, which ensure plant adaptation to drought stress. We proved that HvABI5 regulates drought response in the ABA-dependent way. Our data indicate that HvABI5 can directly activate ABA biosynthesis and signaling genes and therefore it ensures the proper amplification of ABA signal under drought. We also found that the ABA-dependent regulatory role of HvABI5 during drought response differs from its role at seed germination. Together, these findings increase our understanding of ABI5-dependent modulation of plant response to the abiotic stress. Further analysis is needed to confirm the interaction between HvABI5 and promoters of its putative target genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below:
Author Contributions
IS conceived and supervised the project and contributed to the development of the TILLING platform, IS and AD-G designed the study and contributed to the analysis of results, AC and AD-G performed drought experiment, AC conducted physiological assays and gene expression analysis, and AD-G analyzed photosynthesis and microarray data. MK performed TILLING screen. AC, AD-G, and IS wrote and edited the manuscript.
Funding
This work was supported by the European Regional Development Fund through the Innovative Economy for Poland 2007-2013, project WND-POIG.01.03.01-00-101/08 POLAPGEN-BD “Biotechnological tools for breeding cereals with increased resistance to drought”, task 22 and by the National Science Centre, Poland, project PRELUDIUM 2017/25/N/NZ9/01941 “The role of HvABI5 transcription factor in spring barley (Hordeum vulgare L.) response to drought stress”.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Drs. Vladimir Benes and Paul Collier (Genomics Core Facility at EMBL, Heidelberg) for performing the probe synthesis and microarray hybridizations. Moreover, we would like to thank Dr. Tomasz Płociniczak (Institute of Biology, Biotechnology and Environmental Protection, Faculty of Natural Sciences, University of Silesia in Katowice, Poland) for detection of endogenous ABA content.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01138/full#supplementary-material
References
- Agarwal P. K., Shukla P. S., Gupta K., Jha B. (2013). Bioengineering for salinity tolerance in plants: state of the art. Mol. Biotechnol. 54, 102–123. 10.1007/s12033-012-9538-3 [DOI] [PubMed] [Google Scholar]
- Albertos P., Romero-Puertas M. C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6, 8669. 10.1038/ncomms9669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Momany B., Abu-Romman S. (2014). Cloning and molecular characterization of a flavin-dependent oxidoreductase gene from barley. J. Appl. Genet. 55, 457–468. 10.1007/s13353-014-0227-8 [DOI] [PubMed] [Google Scholar]
- Anjum N. A., Sharma P., Gill S. S., Hasanuzzaman M., Khan E. A., Kachhap K., et al. (2016). Catalase and ascorbate peroxidase—representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. R. 23, 19002–19029. 10.1007/s11356-016-7309-6 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Parker J. E., Ainsworth E. A., Oldroyd G. E., Schroeder J., II (2019). Genetic strategies for improving crop yields. Nature 575, 109–118. 10.1038/s41586-019-1679-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajji M., Kinet J. M., Lutts S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70. 10.1023/A:1014732714549 [DOI] [Google Scholar]
- Bandurska H., Gniazdowska-Skoczek H. (1995). Cell membrane stability in two barley genotypes under water stress conditions. Acta Soc Bot. Pol. 64, 29–32. 10.5586/asbp.1995.005 [DOI] [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561, 127–131. 10.1016/S0014-5793(04)00148-6 [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P., Vadez V., Sharma K. K. (2008). Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 27, 411–424. 10.1007/s00299-007-0474-9 [DOI] [PubMed] [Google Scholar]
- Bi C., Ma Y., Wu Z., Yu Y. T., Liang S., Lu K., et al. (2017). Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol. Biol. 94, 197–213. 10.1007/s11103-017-0603-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard I. M., Lynch T. J., Finkelstein R. R. (2002). Regulation and role of the Arabidopsis ABSCISIC ACID-INSENSITIVE 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129, 1533–1543. 10.1104/pp.005793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Chen G., Wang Y., Huang Y., Marchant D. B., Wang Y. (2017). Evolutionary conservation of ABA signaling for stomatal closure. Plant Physiol. 174, 732–747. 10.1104/pp.16.01848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo P. Y., Gibon and PrometheusWiki contributors (2017). Protocol: extraction and determination of proline. PrometheusWiki 2011, 1–4. [Google Scholar]
- Carles C., Bies-Etheve N., Aspart L., Léon-Kloosterziel K. M., Koornneef M., Echeverria M., et al. (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 30, 373–383. 10.1046/j.1365-313X.2002.01295.x [DOI] [PubMed] [Google Scholar]
- Casaretto J., Ho T. H. D. (2003). The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15, 271–284. 10.1105/tpc.007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaretto J. A., Ho T. H. D. (2005). Transcriptional regulation by abscisic acid in barley (Hordeum vulgare L.) seeds involves autoregulation of the transcription factor HvABI5. Plant Mol. Biol. 57, 21–34. 10.1007/s11103-004-6520-x [DOI] [PubMed] [Google Scholar]
- Chan Z. (2012). Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 100, 110–115. 10.1016/j.ygeno.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Chomczyński P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- Daszkowska-Golec A., Skubacz A., Marzec M., Slota M., Kurowska M., Gajecka M., et al. (2017). Mutation in HvCBP20 (Cap Binding Protein 20) Adapts Barley to Drought Stress at Phenotypic and Transcriptomic Levels. Front. Plant Sci. 8:942. 10.3389/fpls.2017.00942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszkowska-Golec A. (2016). “The role of abscisic acid in drought stress: how aba helps plants to cope with drought stress,” in Drought Stress Tolerance in Plants, vol. 2 . Eds. Hossain M. A., Wani S. H., Bhattachajee S., Burritt D. J., Tran L. S. P. (Cham: Springer International Publishing; ), 123–151. [Google Scholar]
- Davies K. M., Albert N. W., Zhou Y., Schwinn K. E. (2018). Functions of flavonoid and betalain pigments in abiotic stress tolerance in plants. Annu. Plant Rev. Online, 1–41 (1), 21–62. 10.1002/9781119312994.apr0604 [DOI] [Google Scholar]
- Dawson I. K., Russell J., Powell W., Steffenson B., Thomas W. T., Waugh R. (2015). Barley: a translational model for adaptation to climate change. New Phytol. 206, 913–931. 10.1111/nph.13266 [DOI] [PubMed] [Google Scholar]
- Dejonghe W., Okamoto M., Cutler S. R. (2018). Small molecule probes of ABA biosynthesis and signaling. Plant Cell Physiol. 59, 1490–1499. 10.1093/pcp/pcy126 [DOI] [PubMed] [Google Scholar]
- Di Ferdinando M., Brunetti C., Fini A., Tattini M. (2012). “Flavonoids as antioxidants in plants under abiotic stresses,” in Abiotic Stress Responses in Plants. Eds. Ahmad P., Prasad M. N. V. (New York: Springer New York; ), 159–179. [Google Scholar]
- FAO (2018). FAOSTAT Database Collections. Food and Agriculture Organization of the United Nations. Available at: http://faostat3.fao.org/ (Accessed March 19, 2020).
- Finkelstein R. R., Lynch T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S. S., Lynch T. J., Thomas T. L., Rock C. D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI) 5 and ABRE-BINDING FACTOR (ABF) 3. Plant Mol. Biol. 59, 253–267. 10.1007/s11103-005-8767-2 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771. 10.1046/j.1365-313X.1994.5060765.x [DOI] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M. M., Seki M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. 10.1105/tpc.105.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U. S. A. 103, 1988–1993. 10.1073/pnas.0505667103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Gao J., Zhu X., Song Y., Li Z., Ren G., et al. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9, 1272–1285. 10.1016/j.molp.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Gilliham M., Able J. A., Roy S. J. (2017). Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 90, 898–917. 10.1111/tpj.13456 [DOI] [PubMed] [Google Scholar]
- Hong B., Barg R., Ho T. H. D. (1992). Developmental and organ-specific expression of an ABA-and stress-induced protein in barley. Plant Mol. Biol. 18, 663–674. 10.1007/BF00020009 [DOI] [PubMed] [Google Scholar]
- Hossain M. A., Cho J., II, Han M., Ahn C. H., Jeon J. S., An G., et al. (2010. a). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 67, 1512–1520. 10.1016/j.jplph.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Hossain M. A., Lee Y., Cho J., II, Ahn C. H., Lee S. K., Jeon J. S., et al. (2010. b). The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 72, 557–566. 10.1007/s11103-009-9592-9 [DOI] [PubMed] [Google Scholar]
- Huang Y., Sun M. M., Ye Q., Wu X. Q., Wu W. H., Chen Y. F. (2017). Abscisic acid modulates seed germination via ABA INSENSITIVE5-mediated PHOSPHATE1. Plant Physiol. 175, 1661–1668. 10.1104/pp.17.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi Y., Aoki N., Kasa S., Sakamoto M., Kai K., Tomokiyo R., et al. (2017). The interrelationship between abscisic acid and reactive oxygen species plays a key role in barley seed dormancy and germination. Front. Plant Sci. 8:275. 10.3389/fpls.2017.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. M. (2016). A new look at stress: abscisic acid patterns and dynamics at high–resolution. New Phytol. 210, 38–44. 10.1111/nph.13552 [DOI] [PubMed] [Google Scholar]
- Joshi R., Wani S. H., Singh B., Bohra A., Dar Z. A., Lone A. A., et al. (2016). Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 7:1029. 10.3389/fpls.2016.01029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Hobo T., Murata M., Ban A., Hattori T. (2002). Abscisic acid–induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14, 3177–3189. 10.1105/tpc.005272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaji H. M., Jajoo A., Oukarroum A., Brestic M., Zivcak M., Samborska I. A., et al. (2016). Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant 38, 102. 10.1007/s11738-016-2113-y [DOI] [Google Scholar]
- Kanai M., Nishimura M., Hayashi M. (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 62, 936–947. 10.1111/j.1365-313X.2010.04205.x [DOI] [PubMed] [Google Scholar]
- Kang J. Y., Choi H., II, Im M. Y., Kim S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. 10.1105/tpc.010362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi F., Maeta E., Terashima A., Takumi S. (2008). Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol. Plant 134, 74–86. 10.1111/j.1399-3054.2008.01107.x [DOI] [PubMed] [Google Scholar]
- Kong Y., Chen S., Yang Y., An C. (2013). ABA-insensitive (ABI) 948 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett. 587, 3076–3082. 10.1016/j.febslet.2013.07.045 [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S. L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288, 20267–20279. 10.1074/jbc.M113.465369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N. H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. 10.1093/pcp/41.5.541 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 98, 4782–4787. 10.1073/pnas.081594298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T., Erickson B. J., Finkelstein R. R. (2012). Direct interactions of ABA-insensitive (ABI)-clade protein phosphatase (PP) 2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol. Biol. 80, 647–658. 10.1007/s11103-012-9973-3 [DOI] [PubMed] [Google Scholar]
- Maia J., Dekkers B. J., Dolle M. J., Ligterink W., Hilhorst H. W. (2014). Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytol. 203, 81–93. 10.1111/nph.12785 [DOI] [PubMed] [Google Scholar]
- Martignago D., Rico-Medina A., Blasco-Escaméz D., Fontanet-Manzaneque J. B., Caño-Delgado A., II (2020). Drought resistance by engineering plant tissue-specific responses. Front. Plant Sci. 10:1676. 10.3389/fpls.2019.01676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Sharma P. (2019). “Superoxide Dismutases (SODs) and Their Role in Regulating Abiotic Stress induced Oxidative Stress in Plants,” in Reactive Oxygen, Nitrogen and Sulfur Species in Plants. Eds. Hasanuzzaman M., Fotopoulos V., Nahar K., Fujita M. (Hoboken: John Wiley & Sons Ltd; ), 53–88. [Google Scholar]
- Miura K., Lee J., Jin J. B., Yoo C. Y., Miura T., Hasegawa P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U. S. A. 106, 5418–5423. 10.1073/pnas.0811088106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Lynch T. J., Finkelstein R. R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. 10.1046/j.1365-313x.2001.01069.x [DOI] [PubMed] [Google Scholar]
- Nakurte I., Keisa A., Rostoks N. (2012). Development and Validation of a Reversed-Phase Liquid Chromatography Method for the Simultaneous Determination of Indole-3-Acetic Acid, Indole-3-Pyruvic Acid, and Abscisic Acid in Barley (Hordeum vulgare L.). J. Anal. Methods Chem. 2012, 103575. 10.1155/2012/103575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Suzuki M., Abrams S., McCarty D. R., Kamiya Y., McCourt P. (2002). A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., Moloney M., Inzé D. (2014). Translational research: from pot to plot. Plant Biotechnol. J. 12, 277–285. 10.1111/pbi.12176 [DOI] [PubMed] [Google Scholar]
- Paul S., Roychoudhury A. (2019). Transcript analysis of abscisic acid-inducible genes in response to different abiotic disturbances in two indica rice varieties. Theor. Exp. Plant Phys. 31, 249–272. 10.1007/s40626-018-0131-4 [DOI] [Google Scholar]
- Piao W., Kim S. H., Lee B. D., An G., Sakuraba Y., Paek N. C. (2019). Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 70, 2699–2715. 10.1093/jxb/erz095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D., Zhang Z., He J., Zhang P., Ou X., Li T., et al. (2019). Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 70, 435–446. 10.1093/jxb/ery385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Rapacz M., Stępień A., Skorupa K. (2012). Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol. Plant 34, 1723–1733. 10.1007/s11738-012-0967-1 [DOI] [Google Scholar]
- Sah S. K., Reddy K. R., Li J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Uozumi N. (2019). Guard cell membrane anion transport systems and their regulatory components: an elaborate mechanism controlling stress-induced stomatal closure. Plants 8, 9. 10.3390/plants8010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M. Y., Kim J., Paek N. C., Choi G. (2014). Phytochrome interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5, 4636. 10.1038/ncomms5636 [DOI] [PubMed] [Google Scholar]
- Seiler C., Harshavardhan V. T., Reddy P. S., Hensel G., Kumlehn J., Eschen-Lippold L., et al. (2014). Abscisic acid flux alterations result in differential abscisic acid signaling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiol. 164, 1677–1696. 10.1104/pp.113.229062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Chen C. N., Brands A., Pan S. M., Tuan-Hua D. H. (2001). The stress-and abscisic acid-induced barley gene HVA22: developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 45, 327–340. 10.1023/A:1006460231978 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Shu K., Chen Q., Wu Y., Liu R., Zhang H., Wang P., et al. (2016). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. 10.1111/tpj.13109 [DOI] [PubMed] [Google Scholar]
- Signora L., De Smet I., Foyer C. H., Zhang H. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28, 655–662. 10.1046/j.1365-313x.2001.01185.x [DOI] [PubMed] [Google Scholar]
- Skubacz A., Daszkowska-Golec A., Szarejko I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7: 1884. 10.3389/fpls.2016.01884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Xiang F., Zhang G., Miao Y., Miao C., Song C. P. (2016). Abscisic acid as an internal integrator of multiple physiological processes modulates leaf senescence onset in Arabidopsis thaliana. Front. Plant Sci. 7:181. 10.3389/fpls.2016.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R. J., Srivastava A., Tsimilli-Michael M. (2000). “The fluorescence transient as a tool to characterize and screen photosynthetic samples,” in Probing photosynthesis: mechanisms, regulation and adaptation. Eds. Yunus M., Pathre U., Mohanty P. (London: Taylor and Francis; ), 445–483. [Google Scholar]
- Strasser R. J., Tsimilli-Michael M., Srivastava A. (2004). “Analysis of the chlorophyll a fluorescence transient,” in Chlorophyll a Fluorescence. Eds. Papageorgiou G. C., Govindjee (Dordrecht: Springer Netherlands; ), 321–362. [Google Scholar]
- Su M., Huang G., Zhang Q., Wang X., Li C., Tao Y., et al. (2016). The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci. 247, 93–103. 10.1016/j.plantsci.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Szarejko I., Szurman-Zubrzycka M., Nawrot M., Marzec M., Gruszka D., Kurowska M., et al. (2017). “Creation of a TILLING Population in Barley After Chemical Mutagenesis with Sodium Azide and MNU,” in Biotechnologies for Plant Mutation Breeding. Eds. Jankowicz-Cieslak J., Tai T. H., Kumlehn J., Till B. J. (Switzerland: Springer International Publishing; ), 91–111. 10.1007/978-3-319-45021-6 [DOI] [Google Scholar]
- Szurman-Zubrzycka M. E., Zbieszczyk J., Marzec M., Jelonek J., Chmielewska B., Kurowska M. M., et al. (2018). HorTILLUS—A Rich and Renewable Source of Induced Mutations for Forward/Reverse Genetics and Pre-breeding Programs in Barley (Hordeum vulgare L.). Front. Plant Sci. 9 216. 10.3389/fpls.2018.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M., Bacic A. (2005). Abiotic stress tolerance in grasses. From model plants to crop plants. Plant Physiol. 137, 791–793. 10.1104/pp.104.900138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka K., Taji T., Hayashi T., Sakata Y. (2013). A novel abi5 allele reveals the importance of the conserved Ala in the C3 domain for regulation of downstream genes and salt tolerance during germination in Arabidopsis. Plant Signac. Behav. 8, e23455. 10.4161/psb.23455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi S., Ashikawa I., Nakamura S., Shibasaka M. (2020). TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 133, 245–256. 10.1007/s10265-020-01166-3 [DOI] [PubMed] [Google Scholar]
- Vinocur B., Altman A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. 10.1016/j.copbio.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Wang X., Guo C., Peng J., Li C., Wan F., Zhang S., et al. (2019). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221, 341–355. 10.1111/nph.15345 [DOI] [PubMed] [Google Scholar]
- Xu Z. Y., Yoo Y. J., Hwang I. (2014). “ABA Conjugates and Their Physiological Roles in Plant Cells,” in Abscisic acid: Metabolism, Transport and Signaling. Ed. Zhang D. (Dordrecht: Springer Netherlands; ), 77–87. [Google Scholar]
- Xue G. P., Loveridge C. W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37, 326–339. 10.1046/j.1365-313X.2003.01963.x [DOI] [PubMed] [Google Scholar]
- Yan F., Deng W., Wang X., Yang C., Li Z. (2012). Maize (Zea mays L.) homologue of ABA-insensitive (ABI) 5 gene plays a negative regulatory role in abiotic stresses response. Plant Growth Regul. 68, 383–393. 10.1007/s10725-012-9727-x [DOI] [Google Scholar]
- Yang J., Worley E., Udvardi M. (2014). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26, 4862–4874. 10.1105/tpc.114.133769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., et al. (2015). Four A rabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. 10.1111/pce.12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Christmann A., Yamaguchi-Shinozaki K., Grill E., Fernie A. R. (2019). Revisiting the Basal Role of ABA–Roles Outside of Stress. Trends Plant Sci. 24, 625–635. 10.1016/j.tplants.2019.04.008 [DOI] [PubMed] [Google Scholar]
- Yu F., Wu Y., Xie Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 20, 569–575. 10.1016/j.tplants.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Yuan K., Rashotte A. M., Wysocka-Diller J. W. (2011). ABA and GA signaling pathways interact and regulate seed germination and seedling development under salt stress. Acta Physiol. Plant 33, 261–271. 10.1007/s11738-010-0542-6 [DOI] [Google Scholar]
- Zhang Y., Sun Q., Zhang C., Hao G., Wang C., Dirk L. M., et al. (2019). Maize VIVIPAROUS1 interacts with ABA INSENSITIVE5 to regulate GALACTINOL SYNTHASE2 expression controlling seed raffinose accumulation. J. Agric. Food Chem. 67, 4214–4223. 10.1021/acs.jafc.9b00322 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K. S., Guo Y. (2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 109, 12822–12827. 10.1073/pnas.1202630109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Hao H., Zhang Y., Bai Y., Zhu W., Qin Y., et al. (2015). SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 168, 659–676. 10.1104/pp.114.255455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmeister J., Lalanne D., Terrasson E., Chatelain E., Vandecasteele C., Vu B. L., et al. (2016). ABI5 is a regulator of seed maturation and longevity in legumes. Plant Cell 28, 2735–2754. 10.1105/tpc.16.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Guan Y., Ren H., Zhang F., Chen F. (2008). A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66, 675–683. 10.1007/s11103-008-9298-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: