Abstract
Aims
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel member of the betacoronaviruses family affecting the lower respiratory tract mainly through binding to angiotensin converting enzyme 2 (ACE2) via its S-protein. Genetic analysis of (ACE2) gene revealed several variants that have been suggested to regulate the interaction with S protein. This study investigates the N720D variant, positioned in the collectrin-like domain (CLD) at proximity to type II transmembrane serine protease (TMPRSS2) cleavage site.
Main methods
The effect of N720D variant on ACE2 structure and thermodynamic stability was studied by DynaMut. HDOCK was utilised to model TMPRSS2 protease binding to ACE2 WT and D720 variant cleavage site. PRODIGY was used to calculate binding affinities and MD simulation tools calculated the at 100 ns for ACE2 apo structure and the ACE2-TMPRSS2 complex.
Key findings
The N720D variant is a more dynamic structure with a free energy change (ΔΔG): −0.470 kcal/mol. As such, introducing a tighter binding affinity of Kd = 3.2 × 10−10 M between TMPRSS2 and N720D variant. RMSD, RMSF calculations showed the N720D variant is less stable, however, RMSF values of the D720-TMPRSS2 complex reflected a slower dynamic motion.
Significance
The hotspot N720D variant in the CLD of ACE2 affected the stability and flexibility of ACE2 by increasing the level of motion in the loop region, resulting in a more favourable site for TMPRSS2 binding and cleavage. Consequently, this would facilitate S-protein binding and can potentially increase viral entry highlighting the importance of variants affecting the ACE2-TMPRSS2 complex.
Keywords: SARS-CoV-2, ACE2, S-protein, TMPRSS2, Thermodynamic stability, Binding, Molecular dynamic simulations
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), similar to the earlier SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), is associated with severe lower respiratory tract disease symptoms such as severe pneumonia and bronchiolitis [36,50]. SARS-CoV-2 is part of the betacoronaviruses family and has a long ORF1ab polyprotein at the 5′ end, followed by structural proteins, glycosylated spike (S), an envelope protein (E), membrane (M), and nucleocapsid (N) proteins [39]. SARS-CoV-2 S-protein binds to angiotensin-converting enzyme 2 (ACE2), facilitating the invasion of the host cells [24,38,43]. Primarily ACE2 is found in the lower respiratory tract [17], which may explain the severe respiratory syndrome associated with SARS-CoV-2 infection [12,48].
ACE2 is an 805 amino acid protease that exists in a monomer, dimer equilibrium [19,42], and is well-known for its role in hypertension. ACE2 exerts its functions through cleaving either Angiotensin I or Angiotensin II into the inactive peptides Angiotensin (1–9) and Angiotensin (1–7), respectively. Angiotensin (1–9) gets further metabolized into Angiotensin (1–7). Angiotensin (1–7) is a vasodilator; hence, ACE2 counteracts the vasoconstrictor effects of ACE-Angiotensin II axis [7,11,33,34,40]. ACE2 consists of an N-terminal peptidase domain (PD), a C-terminal collectrin-like domain (CLD), and a transmembrane helix [9,42,49]. S-protein binding to ACE2 dimer occurs at the receptor-binding domain (RBD) when the S1 subunit interacts with the PD of ACE2 [13,42]. The S-protein binding affinity of SARS-CoV-2 to ACE2 is 10–20-fold higher than SARS-CoV [24], which may explain the high virulence and the increased rate of human to human transmission. Active ACE2 is cleaved on the N-terminus by either metalloprotease ADAM17 (residues 652 to 659) or type II transmembrane serine protease TMPRSS2 (residues 697 to 716) [14]. TMPRSS2 cleavage is required for ACE2 to interact with the S-protein [14]. This was further corroborated on wild type (WT), and knockout (KO) TMPRSS2 in mice infected with SARS-CoV since TMPRSS2 cleavage resulted in an enhanced viral entry [16].
A recent large-scale investigation of ACE2 gene coding sequences variants (1700 variants) and allele frequencies (AF) between different populations reported 32 different variants, including seven hotspot variants [5]. These missense allelic variants included K26R, I468V, A627V, N638S, S692P, N720D, and L731I/F [5,22]. The most prominent mutations on the ACE2 gene were K26R and N720D, where both mutations presented a higher frequency in the European population in comparison to the Chinese and Middle Eastern populations [2,5,22,27]. Cao et al. suggested that these genetic variants were not significantly different between Caucasian and Asian populations but can potentially alter viral binding and entry to host cells [5]. Whereas, Al-Mulla et al. showed that the prevalence of the most ACE2 activating variant N720D was much higher among Europeans (2.5%), when compared to Iranians (0.6%), Kuwaitis (0.3%), Qataris (0.2%) and other global populations (0.4%) and MAF of these variants associated with higher death rate from SARS-CoV-2 infection in the European population compared to the Middle Eastern population [2].
In addition to its high allelic frequency, the significance of the N720D variant emerges from its location in the CLD domain on a dynamic loop region 4 amino acids away from the TMPRSS2 cleavage site [2,4,27]. Since TMPRSS2 cleavage of ACE2 is an essential step in SARS-CoV-2 S-protein binding, any structural changes caused by the N720D variant might affect this process. The aim of this study is to elucidate the impact of structural changes caused by ACE2 N720D variant on TMPRSS2 cleavage and, in turn, S-protein binding. To achieve these objectives, we used computational structural biology tools to predict the effect of the N720D variant on the stability and flexibility of ACE2 structure. In addition, we modelled the binding affinities of TMPRSS2 to ACE2 WT and variant.
2. Results and discussion
2.1. ACE2 N720D mutation
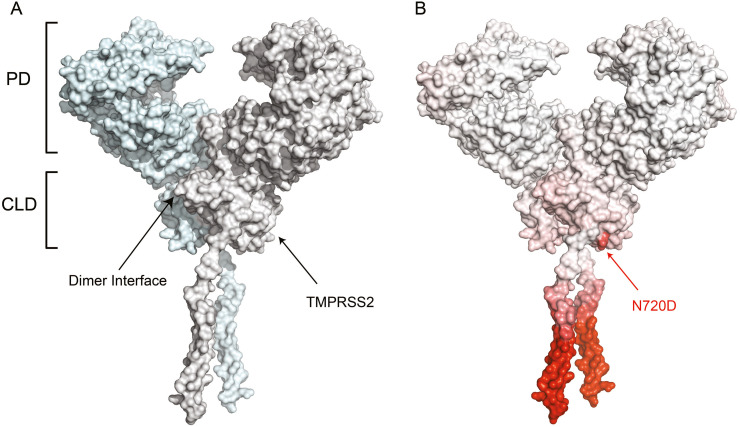
Both K26R on the PD and N720D on the CLD domains have the highest frequency of mutations on ACE2 gene in the European population in comparison to the Chinese and Middle Eastern populations. The PD of ACE2 binds to S-protein with a high affinity of ~15 nM Kd [38]; therefore, a properly functioning ACE2 is essential for viral cell entry. As such, studies have observed the effect K26R and other variants on the structure of the PD that might affect the S-protein binding, and subsequently, the SARS-CoV-2 entry into cells [15,22,27]. Whereas, the N720D variant is located on the same interface of the loop region in close vicinity to the TMPRSS2 cleavage site. Having the mutation in the loop region close to the cleavage site may result in conformational dynamics that affect the binding affinity of TMPRSS2 to ACE2. Here we analysed the structure of the recently published Cryo-EM ACE2 dimer with a 2.9 Å resolution bound to neutral amino acid transporter B0AT1 (PDB ID:6M18) (Fig. 1A) [42]. In our thermodynamic and binding analysis, we modelled out the B0AT1 since, Yan et al. stated that B0AT1 is only found in the kidneys and intestine and not the lungs, in addition, it might hinder access of TMPRSS2 to the cleavage site [42].
Fig. 1.
The effect of N720D mutation on the ACE2 protein structure. A) ACE2 dimer complex promoters 1 and 2 (blue and white, respectively) showing the PD site where the SARS-CoV-2 S protein RBD binds. The CLD region is also shown where ACE2 is cleaved by TMRSS2. B) The single domain of the ACE2 mutation N720D where the red region of the protein depicts the more flexible region of the protein due to the N720D mutation with a (ΔΔG): −0.470 kcal/mol and ΔΔS: 0.070 kcal·mol−1·K−1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Conformational dynamics of N720D
Stability analysis of ACE2 showed that the variant N720D is more dynamic in structure with a free energy change (ΔΔG): −0.470 kcal/mol and entropy of (ΔΔSVib: 0.070 kcal·mol−1·K−1) (Fig. 1B). Hussain et al. measured the free energy change for a series of notable mutations on the PD of ACE2. Most mutations studied on the PD, predicted a less stable ACE2 structure, which in turn may increase the susceptibility of the individual to SARS-CoV-2 [4,22]. The variant K26R, which presented the highest allelic frequency, resulted in a less stable ACE2 structure at 25 °C (ΔΔG −0.34 kcal/mol) and 35 °C (ΔΔG −0.30 kcal/mol) [15]. However, the N720D mutation is positioned in the loop region preceded by the TMPRSS2 cleavage site between residues 697 and 716, which are the third and fourth helices in the CLD of ACE2 (Fig. 1A). Protein loop regions have higher motional dynamics (femtoseconds to seconds) due to the limited covalent interactions in the region [18]. As a result of the mutation, a decrease in ΔΔG and an increase in entropy in the system triggers local unfolding, hence exposing cleavage sites to the surrounding moiety [30] to be readily attacked by proteases such as TMPRSS2.
2.3. ACE2 TMPRSS2 binding
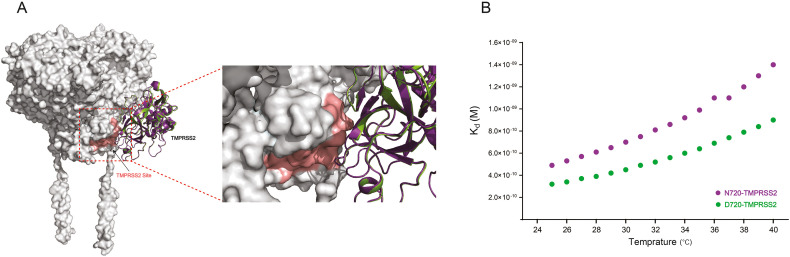
The loop region close to the cleavage site of ACE2 D720 variant displayed a less stable and more dynamic structure. Therefore, we modelled the binding of TMPRSS2 to the cleavage site of ACE2 N720 and D720 variant. The arginine and lysine residues within ACE2 TMPRSS2 cleavage site are essential for cleavage and required for augmentation of SARS-S-driven entry [14]. The ACE2 residues R697 (70.025 Å), K702 (108.684 Å) and R705 (106.398 Å) on the cleavage site to TMPRSS2 HDOCK analysis (Fig. 2A) [44,45]. Whereas, the binding affinities were predicted by PRODIGY using the ACE2-TMPRSS2 binding model (Fig. 2B).
Fig. 2.
A) Modelled TMPRSS2 bound to ACE2 TMPRSS2 cleavage site. B) The dissociation constant Kd for TMPRSS2 bound to ACE2 N720 (purple) and D720 (green). A tighter binding affinity of Kd = 3.2 × 10−10 M between TMPRSS2 and D720 variant than N720 WT. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The binding energy scores (Kd) of ACE2-TMPRSS2 complex (WT and D720 variant) were predicted as a function of temperature [41]. As the temperature increase from 25 °C to 40 °C, there was a 2-fold increase in Kd score, indicating a decrease in binding affinity for both ACE2 WT and D720 variant TMPRSS2 complexes (Fig. 2B). At 25 °C the D720-TMPRSS2 complex showed a tighter binding affinity (3.2 × 10−10 M) in comparison to the WT complex (4.9 × 10−10 M), which is indicative of a more favourable binding between TMPRSS2 and ACE2 D720 variant. Furthermore, the predicted Kd for D720 was less affected as the temperature increased from 25 °C to 40 °C with a ΔKd 9.0 × 10−10 M, in contrast to WT with a ΔKd 1.4 × 10−9 M. Such an increase in thermal energy disrupts the noncovalent interactions between ACE2 and TMPRSS2 resulting in a lower binding affinity [10]. Despite that, the decrease in binding affinity across the temperatures was less apparent for the ACE2 D720 variant complex with TMPRSS2. As such, this tight binding supports the increased susceptibility [21] of ACE2 N720D variant to TMPRSS2 cleavage.
2.4. Molecular dynamic simulations of ACE2 bound to TMPRSS2
To understand the conformational and dynamic features of N720D variant on the stability of ACE2 and the effect on binding to TMPRSS2, molecular dynamics (MD) simulation is an imperative method to explore the behavior of each system in real-time. In this study, 100 ns simulations were run to compare the dynamic behavior of all the systems, including the WT N720-Apo, D720-Apo, N720-TMPRSS2, and D720-TMPRSS2 where the RMSD, RMSF using AMBER package.
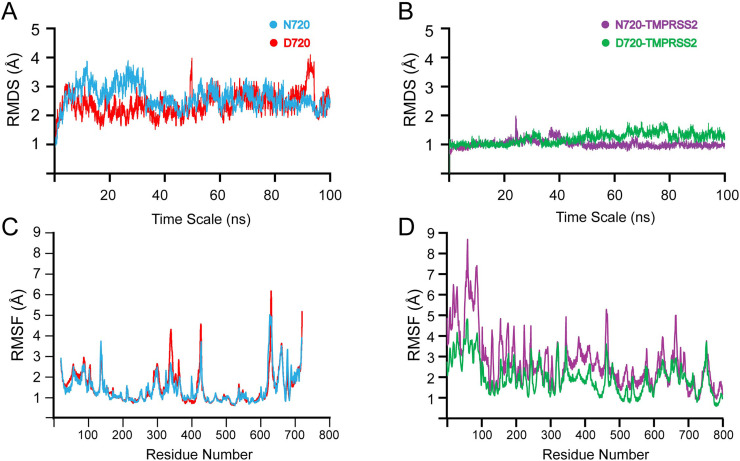
The stability of all the four systems were calculated by Root Mean Square Deviation (RMSD). Fig. 3A depicts the RMSD of the apo structures, with the average RMSD for both WT N720 (blue) and D720 (red) variant was 3.0 Å during the 100 ns simulation. From 0 to 38 ns the WT N720 showed a more rigid structure with a higher RMSD than D720. However, after 40 ns the RMSD value decreased to be 2.5 Å till the end of simulation with no significant convergence. Thus, stability remains uniform for the rest of the time observed during the 100 ns simulation indicating the protein is relatively stable [1,3]. On the other hand, the D720 variant system (Red) appeared relatively lower than N720 WT for the first 38 ns. However, onwards the systems gained equilibrium [25], and higher fluctuations were observed at 55 ns and 90-97 ns indicating significant changes in the conformation of D720 variant. Such major convergence at these two-time points is the results of the D720 being introduced in the system as such, causing instability. Therefore, the variation in conformation may be one of the factors contributing to the destabilization of the protein, making them close to the TMPRSS2 site more accessible cleavage.
Fig. 3.
Molecular dynamic simulations RMSD plots at 100 ns simulations of ACE2 N720 and D720 (A) and ACE2 N720-TMPRSSS2 and D720-TMPRSS2 complex (B). RMSF plot of ACE2 N720 and D720 (C) and ACE2 N720-TMPRSSS2 and D720-TMPRSS2 complex (D).
As for the effect of N720D mutation on stability and dynamics of ACE2-TMPRSS2 complex, RMSD values are shown in Fig. 3B. The WT N720-TMPRSS2 complex remained stable during the simulation with an average RMSD of 1 Å, with the system showing slight convergence at 22 ns and from 35 to 38 ns. Whereas, the D720-TMPRSS2 complex showed a very dynamic behavior, with average RMSD being higher than the WT. The system starts to converge at 30–35 ns; then a major convergence is observed at 60–80 ns with the RMSD continually increasing. Thus, these results signify that the mutation has produced a direct effect on the stability and binding of ACE2. As the RMSD of the mutant complex is still increasing with major convergence at intervals. This is probably due to the integrated mutation at position 720 which causes changes in the conformational dynamics of the proteins and ultimately affecting the interaction with the TMPRSS2. Longer simulation is required for the system to enter the production phase and to better understand the impact of the substitution.
To demonstrate residual flexibility, the root mean square fluctuations (RMSF) were calculated at 100 ns simulations for both apo and complex structures Fig. 3C and D. The N720 WT (blue) shows relatively lower residual flexibility in comparison to D720 variant. The D720 variant (red) shows higher fluctuation, specifically in the region between 320 and 350, 410–420, 605–618, and 695–710. This indicates the mutation has affected the internal dynamics of the protein and causes the residues to be more flexible compared to the N720 WT [1]. However, in the complex system, the N720-TMPRSS2 (purple) showed higher molecular mobility than the variant D720-TMRSS2 complex. The lower mobility observed close to the loop region between residues 695–720 could be the result of the higher binding affinity of TMPRSS2 to the D720 variant.
Thus, the RMSF displayed a higher degree of motion in loop regions and the TMPRSS2 binding site for mutant D720 (Fig. 3C). Loop regions located opposite or close to the cleavage site regulate the dynamic and structural changes of enzyme-protein binding. Consequently, any conformational changes occurring opposite the binding site can in turn, influence the enzyme cleavage process [47]. Therefore, higher RMSF exhibited by the N720D mutation caused higher conformational mobility in the loop region close to the cleavage site, which can result in a more favourable TMPRSS2-ACE2 binding interaction through either conformational selection mechanism or induced-fit mechanism [32,47].
3. Conclusion
The recent variants observed on the ACE2 gene in different populations have gained attention as they might affect the SARS-CoV-2 binding. Since the ACE2 PD bound to S-protein of SARS-CoV-2 has been observed by Cryo-EM studies [38,42], most studies have concentrated on variants on the PD [4,15,22]. Whereas, the effect of N720D variant positioned on the CLD of ACE2 structure has been less examined.
In our current study, we demonstrated the potential importance of the N720D mutation on the stability and flexibility of ACE2. Since ACE2-TMPRSS2 complex structure is not experimentally determined, we modelled the effect of N720D on TMPRSS2-ACE2 interaction. The D720 variant was characterized by an increase in free energy that affected protein stability and flexibility. This was reflected by higher RMSD and RMSF indicating an increased instability due to conformational changes, which elevates the level conformational mobility in the loop region and TMPRSS2 binding site. With the variant D720, ACE2-TMPRSS2 complex depicted a tighter binding affinity, reflected by lower mobility close to the loop region. The effect of the variant D720 on the structural dynamics of ACE2 was emphasized by the calculated Kd for the ACE2-TMPRSS2 complex. The D720 ACE2 variant showed a tighter binding affinity in comparison to ACE2 WT. The more favourable TMPRSS2-ACE2 binding interaction supports the increased susceptibility of ACE2 N720D variant to TMPRSS2 cleavage.
Our report is based on modelling analysis of ACE2 N720D variant to develop an understanding of its interaction with TMPRSS2 and the consequent of SARS-CoV-2 binding. Our finding gives an insight into the potential reason for which N720D was the most frequent variant linked to the higher death rates in Europe. Further computational and functional studies are required to elucidate the clinical importance of ACE2 N720D variant.
In conclusion, based on our modelling analysis of ACE2 variant N720D, we postulate that this variant impacted the stability and flexibility of ACE2, resulting in a more favourable site for TMPRSS2 binding and cleavage. As a result, carriers of this variant have an increased S-protein binding and can potentially have increased viral entry caused by enhanced interaction between ACE2 and TMPRSS2.
4. Methods
The structures of ACE2-B0AT1 (PDB ID:6M18) [38] was used for thermodynamic and structural analysis in this report. The structure of TMPRSS2 was modelled from SWISS-Model [37] and I-TASSER [46]. DynaMut [28] webserver was used to predict the effect of genetic variants on the stability and flexibility on N720D on the ACE2 protein.
Protein-protein docking of ACE2 (PDB ID:6M18) WT(N720) and variant (D720) and TMPRSS2 was done by HDOCK server [44,45], which is based on a hybrid algorithm of template-based modelling and ab initio free docking. The ACE2 residues R697 (70.025 Å), K702 (108.684 Å), and R705 (106.398 Å) on the cleave site to TMPRSS2 residues H296, D345, S441, and D435. Model 1 with the lowest docking energy score and the highest ligand root-mean-square deviation (RMSD) was selected to analyse the binding energy scores (Kd) using PRODIGY server [41]. PRODIGY is a robust predictive system that utilises structural properties of protein-protein interactions, the number of interfacial contacts and non-interacting surfaces to calculate proteins binding affinity [35].
To understand the dynamics and interacting behavior, both the apo N720 and D720 ACE2 and ACE2-TMPRSS2 complexes were subjected to molecular dynamics simulation. Amber 18 package with AMBER ff14SB force field was used to execute the simulations [6,26]. Systems were solvated with TIP3P water box with 18.0 Å distance each side and were neutralized by adding Na+ ions. We used 300 K temperature and 1.0 bar pressure of 1.0 bar using Langevin thermostat and Berendsen Barostat controllers [8,23]. For hydrogen long-range interaction SHAKE algorithm and particle mesh Ewald summation (PME) algorithm was used [20,31]. For the non-bonded interactions 10.0 Å cut-off was fixed. A two-steps gentle minimization followed by heating and equilibration was performed. A 100 ns simulation was carried out at the NPT ensemble. The Cartesian coordinates were stored at every 10 ps. We obtained 5000 frames from each simulation. Systems stability and residual flexibility was also calculated using CPPTRAJ and PTRAJ [29]. For stability RMSD while for flexibility RMSF was calculated.
Acknowledgments
Acknowledgements
This study was supported by the Kuwait Foundation for the Advancement of Sciences.
Ethical approval
Not required.
Declaration of competing interest
All authors have no conflict of interest to declare.
Author contributions
FA-M conceptualized the study. AM collected data and analysed and proposed protein related predictions. SKM and EM calculated the protein kinetics and generated the figures. AM, SKM EM, MA, JA and FA-M wrote and edited the manuscript. All authors discussed the hypothesis, critically read, and revised the manuscript, and gave final approval for publication.
References
- 1.Aier I., Varadwaj P.K., Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016;6:34984. doi: 10.1038/srep34984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Mulla F., Mohammad A., Al Madhoun A., Haddad D., Ali H., Eaaswarkhanth M., et al. 2020. A Comprehensive Germline Variant and Expression Analyses of ACE2, TMPRSS2 and SARS-CoV-2 Activator FURIN Genes From the Middle East: Combating SARS-CoV-2 with Precision Medicine. bioRxiv. [Google Scholar]
- 3.Anwar M.A., Choi S. Structure-activity relationship in TLR4 mutations: atomistic molecular dynamics simulations and residue interaction network analysis. Sci. Rep. 2017;7 doi: 10.1038/srep43807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., et al. 2020. ACE2 Gene Variants May Underlie Interindividual Variability and Susceptibility to COVID-19 in the Italian Population. (medRxiv) (2020.04.03.20047977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr., et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke N.E., Turner A.J. Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012;2012:307315. doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidchack R.L., Handel R., Tretyakov M. Langevin thermostat for rigid body dynamics. J. Chem. Phys. 2009;130:234101. doi: 10.1063/1.3149788. [DOI] [PubMed] [Google Scholar]
- 9.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 10.Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., et al. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gemmati D., Bramanti B., Serino M.L., Secchiero P., Zauli G., Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Y., Kral P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the Airways of Murine Models after coronavirus infection. J. Virol. 2019;93 doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamerzell T.J., Middaugh C.R. The complex inter-relationships between protein flexibility and stability. J. Pharm. Sci. 2008;97:3494–3517. doi: 10.1002/jps.21269. [DOI] [PubMed] [Google Scholar]
- 19.Kohlstedt K., Gershome C., Friedrich M., Muller-Esterl W., Alhenc-Gelas F., Busse R., et al. Angiotensin-converting enzyme (ACE) dimerization is the initial step in the ACE inhibitor-induced ACE signaling cascade in endothelial cells. Mol. Pharmacol. 2006;69:1725–1732. doi: 10.1124/mol.105.020636. [DOI] [PubMed] [Google Scholar]
- 20.Kräutler V., Van Gunsteren W.F., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 21.La D., Kong M., Hoffman W., Choi Y.I., Kihara D. Predicting permanent and transient protein-protein interfaces. Proteins. 2013;81:805–818. doi: 10.1002/prot.24235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q., Cao Z., Rahman P. Genetic variability of human angiotensin-converting enzyme 2 (hACE2) among various ethnic populations. BioRxiv. 2020 doi: 10.1002/mgg3.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y., Pan D., Li J., Zhang L., Shao X. Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. J. Chem. Phys. 2017;146:124108. doi: 10.1063/1.4978807. [DOI] [PubMed] [Google Scholar]
- 24.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;526:165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maximova T., Moffatt R., Ma B., Nussinov R., Shehu A. Principles and overview of sampling methods for modeling macromolecular structure and dynamics. PLoS Comput. Biol. 2016;12:e1004619. doi: 10.1371/journal.pcbi.1004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., III, DeBolt S., et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 27.Renieri A., Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., et al. 2020. ACE2 Variants Underlie Interindividual Variability and Susceptibility to COVID-19 in Italian Population. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky300. (W350-W5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 30.Schrank T.P., Bolen D.W., Hilser V.J. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toukmaji A., Sagui C., Board J., Darden T. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J. Chem. Phys. 2000;113:10913–10927. [Google Scholar]
- 32.Tsai C.J., Ma B., Sham Y.Y., Kumar S., Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44:418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- 33.Uddin M., Mustafa F., Rizvi T.A., Loney T., Suwaidi H.A., Al-Marzouqi A.H.H., et al. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12 doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vangone A., Bonvin A.M. Contacts-based prediction of binding affinity in protein-protein complexes. Elife. 2015;4:e07454. doi: 10.7554/eLife.07454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao L., Sakagami H., Miwa N. ACE2: the key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: demon or angel? Viruses. 2020;12 doi: 10.3390/v12050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 42.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan T., Xiao R., Lin G. Angiotensin-converting enzyme 2 in severe acute respiratory syndrome coronavirus and SARS-CoV-2: a double-edged sword? FASEB J. 2020;34:6017–6026. doi: 10.1096/fj.202000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan Y., Tao H., He J., Huang S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 45.Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx407. (W365-W73) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L.Q., Sang P., Tao Y., Fu Y.X., Zhang K.Q., Xie Y.H., et al. Protein dynamics and motions in relation to their functions: several case studies and the underlying mechanisms. J. Biomol. Struct. Dyn. 2014;32:372–393. doi: 10.1080/07391102.2013.770372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Wada J., Hida K., Tsuchiyama Y., Hiragushi K., Shikata K., et al. Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J. Biol. Chem. 2001;276:17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- 50.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]