Abstract
The aim of this work was to develop simultaneous edited MRS of γ-aminobutyric acid (GABA), glutathione (GSH), and ethanol (EtOH) using Hadamard encoding and reconstruction of MEGA-edited spectroscopy (HERMES) at 3T.
Density-matrix simulations of HERMES were carried out and compared with phantom experiments. In vivo experiments were performed in six healthy volunteers about 30 min after alcohol consumption.
Simulations of HERMES showed GABA-, GSH-, and EtOH-edited spectra with low levels of crosstalk and excellent agreement with phantom spectra. In vivo experiments showed well edited GABA signals at 3.0 ppm, GSH at 2.95 ppm, and EtOH at 1.18 ppm in the respective Hadamard combination spectra. Measured integral ratios were 0.082 ± 0.012 for GABA/Cr, 0.037 ± 0.006 for GSH/Cr, and 0.305 ± 0.129 for EtOH/Cr.
Simulated, phantom, and in vivo measurements of HERMES show excellent separation of GABA-, GSH-, and EtOH-edited signals with negligible levels of crosstalk. HERMES allows a threefold acceleration of editing while maintaining spectral quality compared with sequentially acquired MEGA-PRESS measurements.
Keywords: edited MRS, ethanol, GABA, GSH, HERMES
1 |. INTRODUCTION
Proton (1H) MRS is a non-invasive tool for measuring endogenous brain metabolites, such as N-acetylaspartate (NAA), choline (Cho), and creatine (Cr), to investigate both healthy and pathological physiology.1,2 1H MRS also allows in vivo measurement of ethanol (EtOH) in the human brain after alcohol consumption,3 providing insights into the metabolic changes in alcohol-dependent patients, heavy alcohol drinkers, and sober patients.4,5 There is currently little understanding of acute effects of alcohol on levels of the inhibitory neurotransmitter γ-aminobutyric acid (GABA) and the antioxidant glutathione (GSH). Previous MRS studies have demonstrated changes in GABA levels in individuals recovering from alcohol use disorder associated with the duration of withdrawal and the presence of comorbid smoking.6,7 GSH is a key antioxidant in the human brain, critical for protecting the body against oxidative stress caused by reactive oxygen species. Alcohol has been shown to deplete GSH levels, especially in the mitochondria,8 impeding the elimination of reactive oxygen species.
In vivo measurement of GABA and GSH is challenging due to substantial signal overlap and low signal intensity at 3T. Spectral editing reduces signal overlap by selectively targeting spin systems of interest, allowing direct and unambiguous measurements of low-concentration metabolites.9 The most widely used spectral editing technique is Mescher-Garwood Point-Resolved Spectroscopy (MEGA-PRESS10). MEGA-PRESS selectively edits one metabolite per scan, and requires long scan times for adequate signal to noise ratio (SNR), limiting the number of edited experiments that can be performed during a typical research protocol. Therefore, developing a method for measuring GABA, GSH, and EtOH concurrently would substantially reduce scan time, and provide a new tool to investigate neurometabolism in the healthy brain and in alcohol use disorder.
Hadamard Encoding and Reconstruction of Mega-Edited Spectroscopy (HERMES) is a J-difference editing method that selectively detects multiple metabolites simultaneously.11–15 HERMES effectively allows multiple MEGA-PRESS experiments to be conducted at the same time, offering substantial scan time reductions compared with sequentially acquired MEGA-PRESS experiments. HERMES editing of GABA and GSH11 has recently been implemented on all major MR vendor platforms with standardized RF pulse shapes, durations, amplitudes, and timings.16 In this paper, we extend HERMES of GABA and GSH to also edit EtOH. We perform simulation, phantom, and in vivo experiments to establish the feasibility of simultaneous detection of GABA, GSH, and EtOH after alcohol consumption.
2 |. METHODS
MRS experiments were conducted on Siemens MAGNETOM Prisma (Siemens Healthcare, Erlangen, Germany) 3T MRI scanners using 32- and 64-channel head coils for phantom and in vivo experiments, respectively. These experiments were conducted with the universal HERMES sequence16 modified to simultaneously edit in vivo GABA with macromolecules (GABA+), GSH, and EtOH signals.
2.1 |. HERMES of GABA, GSH, and EtOH
The four-step HERMES scheme (labeled A, B, C, D) for simultaneous edited detection of GABA at 3.0 ppm, GSH at 2.95 ppm, and EtOH at 1.18 ppm in the difference spectra is shown in Figure 1. In Experiment A, a dual-lobe editing pulse is applied to GABA at 1.9 ppm and GSH at 4.56 ppm (ONGABA; ONGSH). In Experiment B, a dual-lobe editing pulse is applied to GABA and EtOH at 3.67 ppm (ONGABA; ONEtOH). In Experiment C, a dual-lobe editing pulse is applied to GSH and EtOH (ONGSH; ONEtOH). In Experiment D, no editing pulse is required, and a single-lobe editing pulse was instead applied off resonance at 7.5 ppm (OFFGABA/GSH/EtOH). This editing scheme follows the prior method of editing GABA and GSH, with editing pulse lobes added to invert EtOH spins in Experiments B and C, orthogonally to the scheme for GABA and GSH. The Hadamard combination of A + B − C − D yields the GABA-edited spectrum, A + C − B − D yields the GSH-edited spectrum, and B + C − A − D yields the EtOH-edited spectrum. The editing lobes from Experiment B and Experiment C are not perfectly matched in the vicinity of the Cho signal at 3.2 ppm (Figure 1C), differentially affecting it, and resulting in a residual Cho signal in the GABA- and GSH-edited spectra.
FIGURE 1.
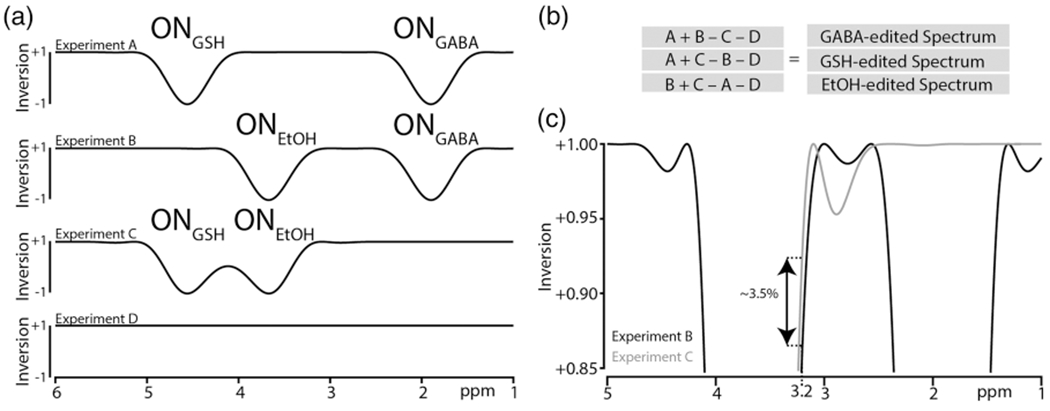
HERMES editing of GABA, GSH, and EtOH: a, inversion profiles for experiments A-D; b, the Hadamard combinations to generate GABA-, GSH-, and EtOH-edited spectra; c, overlap of the inversion profiles from experiments B and C, showing differences in the level of saturation at 3.2 ppm
2.2 |. Acquisition scheme
Since HERMES of GABA, GSH, and EtOH was developed from the multi-vendor standardized HERMES of GABA and GSH,16 the two editing schemes shared the same slice-selective RF pulse durations and amplitudes for voxel localization using PRESS.17 Briefly, the durations of the slice-selective excitation and refocusing pulses were 7.2 ms and 7.0 ms, respectively, at a peak B1 of 13.5 μT. Dual-lobe editing pulses were generated based on a cosine modulation of the universal single-lobe editing pulse.16 When inverting GSH and EtOH, the two editing lobes overlapped substantially, and the cosine splitting of the editing pulse was optimized for inversion using Bloch simulations. The peak B1 values of the single-lobe and dual-lobe editing pulses were 1.07 μT and 2.13 μT, respectively. The durations of the editing pulses were 20 ms and the band-width within which signals are more than 5% inverted is 116 Hz for each lobe.
2.3 |. Simulations, editing efficiency, and phantom experiment
The localized density-matrix simulations of the GABA, GSH, and EtOH spin systems following the HERMES experiment at 3T were performed in FID-A,18 accelerated by the recently published spatial averaging method.19 Ideal excitation and experimental refocusing and editing pulses were used. Simulations were performed on a 101 × 101 two-dimensional spatial array in the dimensions defined by the refocusing pulses spanning 4.5 × 4.5 cm2 (ie the voxel length plus 50% in each dimension), with the following parameters: TE 80 ms; 20 ms editing pulse duration; 8192 data points; 5 kHz spectral width; 2 Hz simulated linewidth; additional line broadening using a 2 Hz exponential filter. Another set of ideal pulse-acquire simulations (simulating “TE 0”) was performed for each metabolite to determine the total available signal (Stotal) in the absence of scalar coupling evolution.
While there is general agreement in the field that editing efficiency expresses a ratio between the yield of an edited experiment and “ideal editing,” there is no standardized definition or agreement over which factors should be included in editing efficiency and which should be omitted. Here, we calculate editing efficiency as the ratio of the integral of the normalized difference spectrum (eg (A − B + C − D)/4) to the “TE 0” integral for a given signal of interest. Since we do not want imperfect localization to be reflected as an editing efficiency loss, and the “TE 0” signal is inherently not localized, we calculate the same ratio for a singlet signal and divide the raw editing efficiency ratio by this “localization efficiency.” With this definition, the editing efficiency includes signal losses due to sub-optimal editing (eg imperfect inversion by editing pulses, TE compromises, triplet center-peak losses) as well as non-uniform evolution of coupling within the localization volume, which is a hybrid localization/editing efficiency loss.20,21 GABA simulations were integrated between 2.9 and 3.1 ppm, GSH between 2.91 and 2.98 ppm, and EtOH between 1.1 and 1.3 ppm.
A single phantom containing GABA, GSH, and EtOH (all 20mM) was prepared. Phantom scan parameters were TE/TR 80/2000 ms; 2048 data points; 2 kHz spectral width; 27 mL voxel; T1- and B1-insensitive (WET) water suppression method for water suppression22; 64 transients (16 per Experiment, A-D). The central peak of the EtOH signal at 1.18 was used to estimate the main magnetic field (B0) drift in the phantom data.
2.4 |. In vivo experiments
2.4.1 |. Participants
Nine adult volunteers (four females, age 35.8 ± 21.1 years (mean ± SD)) were recruited for the study with the approval of the Institutional Review Board of the University of Florida, Gainesville. Participants provided informed consent prior to data collection. All subjects underwent MRS scanning, six after consuming alcohol as described below.
2.4.2 |. Alcohol administration and edited MRS protocol
Prior to participating in the study, participants were instructed to abstain from alcohol and sedating medication for 24 h and fast for 4 h. They were administered a urine drug screen, a pregnancy screen for female participants, and a breath alcohol (BAC) assessment (CMI, Owensboro, KY) to confirm zero blood alcohol concentration. Following confirmation of alcohol administration eligibility, a standard alcohol administration procedure was employed. Six participants were provided with a light snack (220 kcal) approximately 1 h prior to alcohol administration.23 An alcohol dosage targeting a BAC of 0.07 g/dL was calculated for each participant based on a modified version of the Widmark equation (accounting for age, weight, height, and sex).24 Alcohol was prepared for participants by mixing individually calculated doses of 95% EtOH (190 proof, Decon Labs, King of Prussia, PA) at a 1:3 ratio with caffeine free, lemon-lime soda. Participants were given 5 min to consume the beverage. Subjects were brought into the MRI scanner immediately after completion of the beverage and edited MRS data were acquired 31.1 ± 1.4 min (mean ± SD) after beverage consumption. The MRS voxel was prescribed in the midline parietal cortex; outer volume suppression pulses were not used. A T1- and B1-insensitive (WET) water suppression method was used to suppress the water signal,22 and B0 shimming was performed before the start of the scan using the vendor-provided gradient-echo shimming prescan, followed by manual shimming to deliver water linewidths (mean ± SD) of 9.51 ± 1.81 Hz full-width at half-maximum for all subjects. F0 was recalibrated after shimming. The acquisition parameters were the same as the phantom experiments described above, except that 192 transients were acquired. The scan time was about 6.4 min.25 Unsuppressed data were also acquired from each subject. BAC was again measured in each subject after scanning, 69.8 ± 3.4 min after alcohol administration. Data were also acquired with the same scan protocol in the remaining three subjects who had not consumed alcohol.
2.4.3 |. Data processing
In vivo data were analyzed using Gannet,26 modified to accommodate the GABA-, GSH-, and EtOH-edited experiment. Multi-step frequency- and-phase correction was applied to the data to reduce subtraction artifacts,27 followed by a 3 Hz exponential filter and zero padding by a factor of 16. The Cr signal at 3 ppm was used to estimate B0 drift in the in vivo HERMES data before frequency/phase alignment. Finally, the fully processed HERMES sub-spectra were Hadamard-combined to generate the GABA-, GSH-, and EtOH-edited difference spectra. The Hankel singular value decomposition water filtering method was applied to remove the residual water signal.28 To model the three difference-edited signals, weighted nonlinear regression was used, where neighboring co-edited signals were down-weighted to reduce their impact on modeling errors. The GABA+ signal was modeled between 2.60 and 3.55 ppm using a Gaussian function and a linear baseline. The co-edited Cho signal between 3.16 and 3.29 ppm was down-weighted. The GSH-edited spectrum was modeled between 2.25 and 3.5 ppm using a nonlinear baseline, a Gaussian for the GSH signal at 2.95 ppm, and four Gaussians to model the co-edited aspartyl signals. The co-edited Cho signal between 3.13 and 3.3 ppm was down-weighted. The EtOH signal was modeled between 0.6 and 1.8 ppm with two Lorentzians and a linear baseline. The downfield tail of the EtOH signal between 1.29 and 1.51 ppm was down-weighted. The 3.0 ppm Cr signal from Experiment D (OFFGABA/GSH/EtOH) was modeled as a reference signal for calculating GABA+/Cr, GSH/Cr, and EtOH/Cr integral ratios.
Note that the modeling approach of Gannet is simplistic, and more sophisticated linear modeling of these data may prove beneficial in future. Between-subject coefficients of variation (CV) were calculated for all three integral ratios.
3 |. RESULTS
3.1 |. Simulations and phantom experiment
Density-matrix simulations of the GABA, GSH-cysteine, and EtOH spins at the voxel center following the HERMES scheme are shown in Figure 2. As intended, the Hadamard combination of A + B − C − D yields a GABA-edited spectrum, A + C − B − D yields a GSH-edited spectrum, and B + C − A − D yields an EtOH-edited spectrum with low levels of crosstalk. The HERMES experiment conducted in the phantom is shown in the right-hand column, with the GABA signal at 3.0 ppm, GSH signal at 2.95 ppm, and EtOH signal at 1.18 ppm in the respective Hadamard combinations, demonstrating strong agreement with the simulations. The editing efficiencies of GABA, GSH, and EtOH estimated from simulations were 0.37, 0.41, and 0.34, respectively. B0 drift during the phantom experiment was negligible (0.15 ± 0.07 Hz).
FIGURE 2.
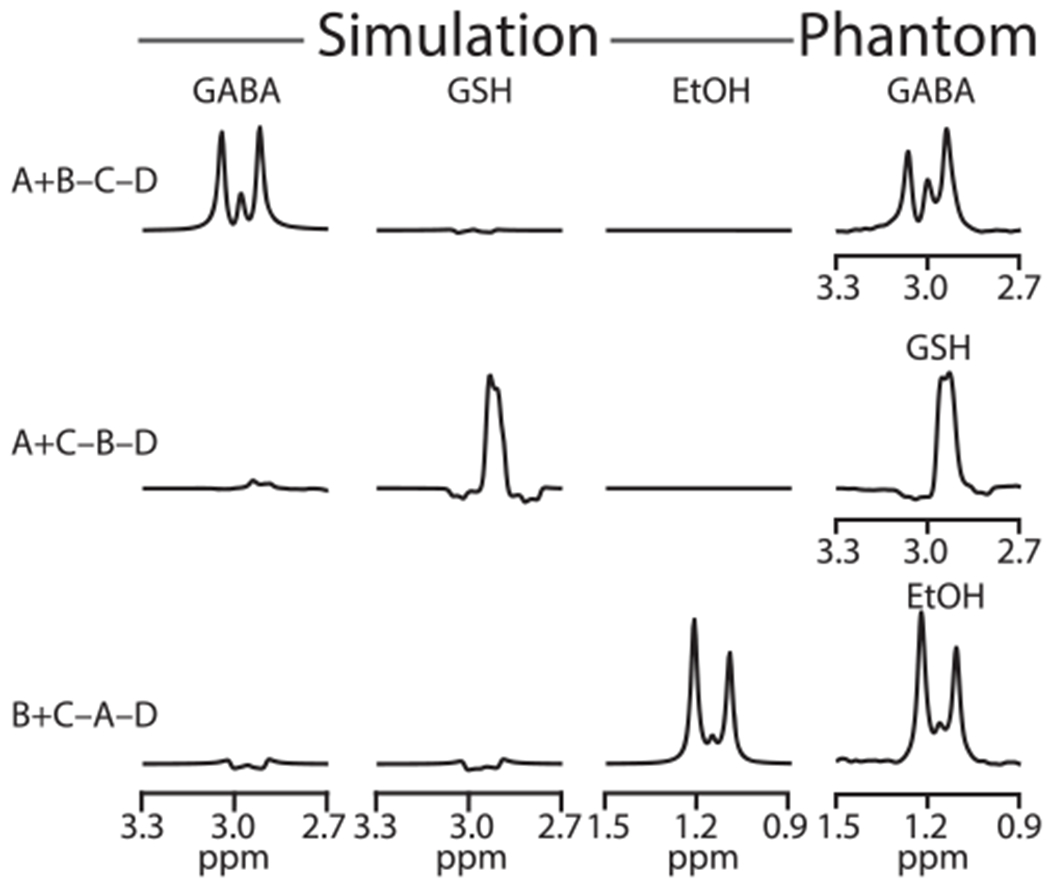
Spatially localized density-matrix simulations and phantom HERMES spectra. A single phantom was used containing GABA, GSH, and EtOH, all at 20mM. Simulated spectra show the GABA-edited spectrum from the Hadamard combination of experiments A + B − C − D, GSH-edited spectrum from A + C − B − D, and EtOH-edited spectrum from B + C − A − D. Phantom experiments also yielded three edited spectra having strong agreement with the simulations
3.2 |. In vivo experiments
Data from one subject are shown in Figure 3, acquired from the midline parietal cortex voxel shown in Figure 3a. The four experiments (A-D) are shown in Figure 3b to demonstrate the location and effect of the editing pulses. The GABA-inverting lobe at 1.9 ppm saturates the NAA signal at 2 ppm, the GSH-inverting lobe saturates the water signal at 4.68 ppm, and the EtOH-inverting lobe saturates several signals in the vicinity of 3.67 ppm. The resulting edited spectra with co-edited signals are shown in Figure 3c, with the GABA, GSH, and EtOH signals in the intended Hadamard combinations. Prominent signals in the spectra are labeled. The B0 offset (mean ± SD) during the in vivo experiment was −1.14 ± 1.95 Hz across all six subjects. in vivo HERMES subspectra and Hadamard-combination spectra, presented as mean ± SD from the six volunteers, are shown in Figure 4a. As intended, the in vivo HERMES experiment resulted in GABA-, GSH-, and EtOH-edited signals at 3 ppm, 2.95 ppm, and 1.18 ppm respectively. The GABA- and GSH-edited spectra have a small negative and a positive Cho signal at 3.2 ppm, respectively. The quantitative analysis of the edited spectra (mean ± SD) yielded integral ratios of 0.082 ± 0.012 for GABA+/Cr, 0.037 ± 0.006 for GSH/Cr, and 0.305 ± 0.129 for EtOH/Cr. The CVs were 15%, 17%, and 42% for GABA+, GSH, and EtOH, respectively. BAC was 0.053 ± 0.004 g/dL about 70 min after alcohol administration. These mean spectra are overlaid in Figure 4b with the mean spectra from three subjects who had not consumed alcohol, indicating that the edited signal at 1.18 ppm is EtOH.
FIGURE 3.
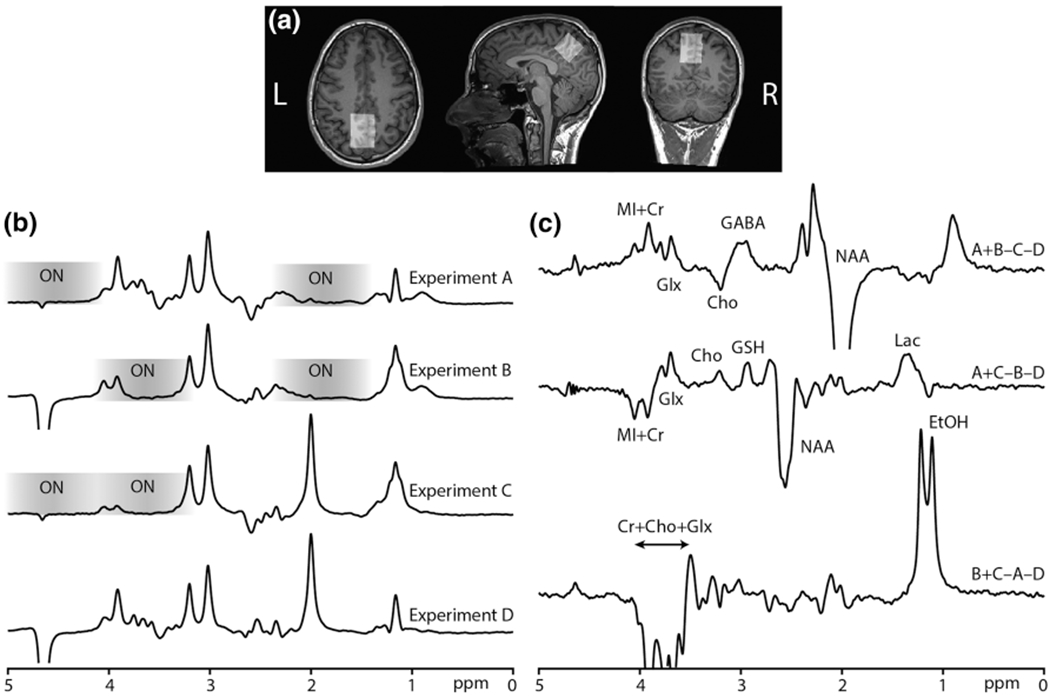
a, MRS voxel placement in the midline parietal cortex. b, experiments A-D are shown separately: Saturated NAA signal in the ONGABA spectra; saturated water signal in the ONGSH spectra; and several signals saturated in the vicinity of 3.67 ppm in the ONETOH spectra. The saturation range of the editing pulses is shown on each spectrum as a grayscale. c, the Hadamard combinations yield GABA-edited (A + B − C − D), GSH-edited (A + C − B − D), and EtOH-edited (B + C − A − D) spectra. The co-edited signals in the edited spectra are shown, specifically NAA, lac (lactate), Glx (glutamate + glutamine), Cho, MI (myo-inositol), and Cr
FIGURE 4.
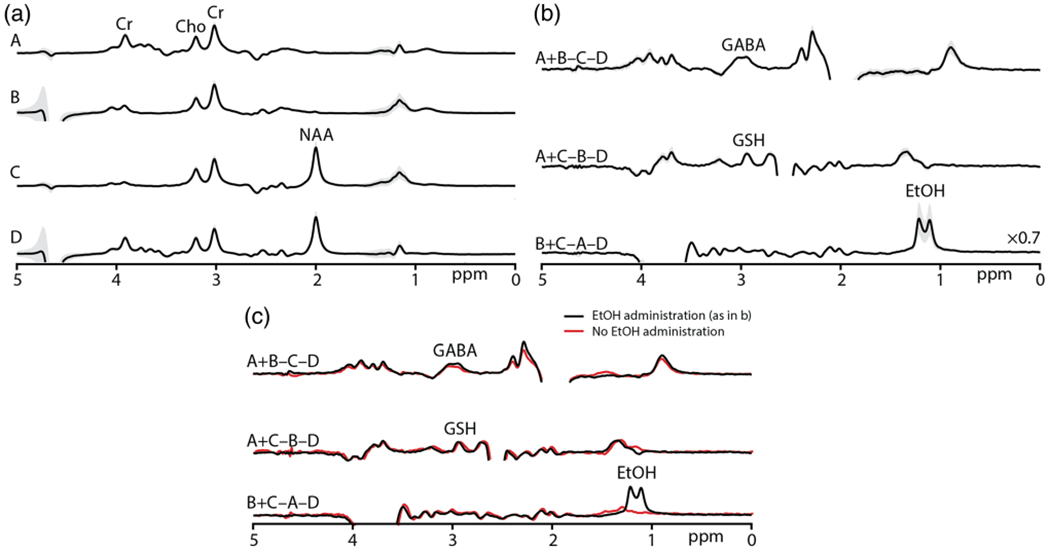
a, average in vivo experiments A, B, C, D and edited spectra from six subjects showing the ±1 SD variability (in gray). b, in vivo experiments yielded the GABA-edited spectra from the Hadamard combination of experiments A + B − C − D, GSH-edited spectra from A + C − B − D, and EtOH-edited spectra from B + C − A − D. c, mean in vivo edited spectra from six subjects are overlaid with the mean HERMES-edited spectra from three subjects who had not consumed alcohol
4 |. DISCUSSION
EtOH is a positive allosteric modulator at GABAA receptors29 that has been suggested to impact levels of GABA and GSH in the brain.30,31 Both these metabolites are present in the human brain at approximately millimolar concentration, requiring long acquisition times (~10 min per metabolite) for adequate SNR in edited spectra. Sequential edited measurements of GABA, GSH, and EtOH would necessitate 30 min acquisitions, limiting the number of brain regions investigated within a typical 1-h MR examination and the time resolution of dynamic studies. In this paper, simultaneous HERMES editing of GABA and GSH11 has been extended to include orthogonal editing of EtOH without an increase in scan time or substantial loss in spectral quality.
Simulations and phantom experiments indicate successful implementation of the new editing sequence, selectively detecting GABA, GSH, and EtOH with excellent separation and low levels of crosstalk. In vivo experiments generated GABA-, GSH-, and EtOH-edited spectra with good spectral quality, allowing quantification of the edited signals. Each in vivo experiment took ~6.4 min per brain region, generating three edited spectra that would otherwise require ~20 min using MEGA-PRESS editing. The time saved from the HERMES acquisition can be allocated for the same acquisition in the other regions of the brain to investigate EtOH-induced regional changes in GABA and GSH, or to allow time-resolved studies of EtOH administration.
DEW-MEGA-PRESS (double editing with MEGA-PRESS)32 has previously been applied to edit both EtOH and GABA30 to investigate EtOH-induced GABA changes in the human brain. DEW allows simultaneous editing in the case where editing target signals and detected signals are all resolved in the spectrum, but cannot separately edit the overlapping GABA and GSH signals. In this HERMES implementation, EtOH was edited orthogonally to the other targets, but it would also be possible to apply EtOH editing pulses in GABA-OFF scans to deliver a GABA/EtOH DEW-MEGA spectrum. The HERMES sequence successfully separates the edited signals from GABA, GSH, and EtOH, implemented within the universal sequence framework,16 permitting future multi-site studies. While editing is required to separate the signals for GABA and GSH from each other and the large Cr signal at 3 ppm, it is not essential for EtOH. However, editing provides the benefit of separating the EtOH signal from lipid contamination in the common event of imperfect signal localization.
This study is intended as a demonstration of methodological feasibility. In vivo measurements were successful in all six subjects, resulting in three edited spectra with good SNR, allowing quantitative analysis. The CV of EtOH was larger than that of GABA+ and GSH, in spite of the larger average EtOH signal, likely reflecting true variation in brain alcohol level, rather than a methodological limitation. Substantial inter-subject differences exist in EtOH absorption, metabolism, and tolerance,33,34 and possibly in MR relaxation effects.35,36 There was also some range among these subjects in the time between the alcohol consumption and the start of edited MRS acquisition.
Although HERMES has the potential to perform multiple MEGA-PRESS acquisitions simultaneously, there is some compromise in terms of experimental freedom comparing one HERMES acquisition with three MEGA-PRESS acquisitions with a total acquisition time three times larger. First, the HERMES measurements all have the same localization, whereas consecutive measurements can probe different brain regions. Second, HERMES measurements all have the same TE, whereas consecutive measurements can vary TE to optimize for a single target. Usually, signals with triplet-like multiplets, such as GABA and EtOH, can be optimally edited at a TE of 1/2 J (~70 ms), while signals with doublet-like multiplets, such as GSH, can be optimally acquired at a TE of 1/J (~140 ms).37 In this current case of HERMES for GABA, GSH, and EtOH, both GABA and EtOH are acquired at a TE longer than is SNR optimal (resulting in editing efficiency losses of under 10%). GSH is acquired substantially below its optimal TE,38 but the editing efficiency losses of this change are greatly mitigated by reduced T2 relaxation losses in vivo. Studies have demonstrated GABA editing at TE values ranging from 68 to 80 ms,39,40 and GSH editing between 68 and 140 ms, without a substantial change in signal intensity.38 Hence, the TE of 80 ms is a reasonable choice for simultaneous editing of GABA, GSH, and EtOH, as demonstrated by simulations and phantom experiments.
The HERMES editing scheme is fundamentally a J-difference editing method, sensitive to B0 drift due to subject motion and scanner instabilities. The HERMES scheme employed in this work spans four TR values compared with two for conventional MEGA-editing, rendering HERMES potentially more susceptible to B0 drift,41 especially for the less rapidly interleaved GABA and EtOH steps (see Figure 1). Incorporation of real-time motion and shim correction would substantially mitigate the effects of B0 drift and increase the efficacy of editing.42–44 Further enhancements can be attained by implementing the other localization methods, such as semi-LASER and LASER (Localization by Adiabatic Selective Refocusing),14,42,45 to reduce chemical shift displacement error and improve editing efficiency. The editing scheme applied suffers to some degree from imperfectly matched treatment of the Cho signal in Experiments B and C, resulting in residual signal in the GABA- and GSH-edited spectra. This makes quantification more challenging, although the Cho signal was successfully incorporated into modeling, so further optimization of editing pulse shapes is warranted.
5 |. CONCLUSION
A four-step HERMES sequence allows simultaneous editing of GABA, GSH, and EtOH in one-third of the scan time of sequentially acquired experiments.
ACKNOWLEDGEMENTS
We would like to thank Brent Foster, Genelle Samson, and Joy Keyanni for their contributions to data collection. Research reported in this publication was supported by the National Institute on Alcohol Abuse and Alcoholism and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award numbers: RAE: R01 EB023963, R01 EB016089, and P41 EB015909; JB and BS: R01AA025337; ECP: K01AA025306. A portion of this work was performed in the McKnight Brain Institute at the National High Magnetic Field Laboratory’s AMRIS Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490* and the State of Florida. This work was supported in part by an NIH award, S10OD021726, for High End Instrumentation.
Funding information
National Institutes of Health, Grant/Award Numbers: R01 EB023963, R01 EB016089, P41 EB015909, R01AA025337, K01AA025306, S10OD021726; State of Florida; National Science Foundation Cooperative Agreement, Grant/Award Number: DMR-1157490*
Abbreviations:
- BAC
breath alcohol
- Cho
choline
- Cr
creatine
- CV
between-subject coefficient of variation
- EtOH
ethanol
- GABA
γ-aminobutyric acid
- GSH
glutathione
- HERMES
Hadamard encoding and reconstruction of MEGA-edited spectroscopy
- LASER
Localization by Adiabatic Selective Refocusing
- MEGA-PRESS
Mescher-Garwood Point-Resolved Spectroscopy
- NAA
N-acetylaspartate
- SNR
signal to noise ratio
- TE
echo time
- TR
repetition time
REFERENCES
- 1.Bonavita S, Di Salle F, Tedeschi G. Proton MRS in neurological disorders. Eur J Radiol. 1999;30:125–131. [DOI] [PubMed] [Google Scholar]
- 2.Rae CD. A guide to the metabolic pathways and function of metabolites observed in human brain 1H magnetic resonance spectra. Neurochem Res. 2014;39:1–36. [DOI] [PubMed] [Google Scholar]
- 3.Hanstock C, Rothman D, Shulman R, Novotny E Jr, Petroff O, Prichard J. Measurement of ethanol in the human brain using NMR spectroscopy. J Stud Alcohol. 1990;51:104–107. [DOI] [PubMed] [Google Scholar]
- 4.Bartsch AJ, Homola G, Biller A, et al. Manifestations of early brain recovery associated with abstinence from alcoholism. Brain. 2007;130:36–47. [DOI] [PubMed] [Google Scholar]
- 5.Meyerhoff DJ, Blumenfeld R, Truran D, et al. Effects of heavy drinking, binge drinking, and family history of alcoholism on regional brain metabolites. Alcohol Clin Exp Res. 2004;28:650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason GF, Petrakis IL, de Graaf RA, et al. Cortical gamma-aminobutyric acid levels and the recovery from ethanol dependence: preliminary evidence of modification by cigarette smoking. Biol Psychiatry. 2006;59:85–93. [DOI] [PubMed] [Google Scholar]
- 7.Behar KL, Rothman DL, Petersen KF, et al. Preliminary evidence of low cortical GABA levels in localized 1H-MR spectra of alcohol-dependent and hepatic encephalopathy patients. Am J Psychiatry. 1999;156:952–954. [DOI] [PubMed] [Google Scholar]
- 8.Wu D, Cederbaum AI. Alcohol, oxidative stress, and free radical damage. Alcohol Res Health. 2003;27:277–284. [PMC free article] [PubMed] [Google Scholar]
- 9.Harris AD, Saleh MG, Edden RA. Edited 1H magnetic resonance spectroscopy in vivo: methods and metabolites. Magn Reson Med. 2017;77:1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11: 266–272. [DOI] [PubMed] [Google Scholar]
- 11.Saleh MG, Oeltzschner G, Chan KL, et al. Simultaneous edited MRS of GABA and glutathione. Neuroimage. 2016;15:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan KL, Puts NA, Schar M, Barker PB, Edden RA. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. 2016;76:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan KL, Saleh MG, Oeltzschner G, Barker PB, Edden RA. Simultaneous measurement of aspartate, NAA, and NAAG using HERMES spectral editing at 3 tesla. Neuroimage. 2017;155:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleh MG, Mikkelsen M, Oeltzschner G, et al. Simultaneous editing of GABA and glutathione at 7T using semi-LASER localization. Magn Reson Med. 2018;80:474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oeltzschner G, Chan KL, Saleh MG, Mikkelsen M, Puts NA, Edden RA. Hadamard editing of glutathione and macromolecule-suppressed GABA. NMR Biomed. 2018;31:e3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleh MG, Rimbault D, Mikkelsen M, et al. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. Neuroimage. 2019;189: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Proc Natl Acad Sci U S A. 1987;508:333–348. [DOI] [PubMed] [Google Scholar]
- 18.Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)—an open source, MATLAB-Based Toolkit. Magn Reson Med. 2017;77:23–33. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, An L, Shen J. Fast computation of full density matrix of multispin systems for spatially localized in vivo magnetic resonance spectroscopy. Med Phys. 2017;44:4169–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edden RA, Barker PB. Spatial effects in the detection of γ-aminobutyric acid: improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58:1276–1282. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser LG, Young K, Matson GB. Numerical simulations of localized high field 1H MR spectroscopy. J Magn Reson. 2008;195:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogg RJ, Kingsley R, Taylor JS. WET, a T1-and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B. 1994;104:1–10. [DOI] [PubMed] [Google Scholar]
- 23.Fillmore MT, Dixon MJ, Schweizer TA. Alcohol affects processing of ignored stimuli in a negative priming paradigm. J Stud Alcohol. 2000;61:571–578. [DOI] [PubMed] [Google Scholar]
- 24.Watson PE, Watson ID, Batt RD. Prediction of blood alcohol concentrations in human subjects. Updating the Widmark equation. J Stud Alcohol. 1981; 42:547–556. [DOI] [PubMed] [Google Scholar]
- 25.Mikkelsen M, Loo RS, Puts NA, Edden RA, Harris AD. Designing GABA-edited magnetic resonance spectroscopy studies: considerations of scan duration, signal-to-noise ratio and sample size. J Neurosci Methods. 2018;303:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: a batch-processing tool for the quantitative analysis of gamma-aminobutyric acid–edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40:1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikkelsen M, Saleh MG, Near J, et al. Frequency and phase correction for multiplexed edited MRS of GABA and glutathione. Magn Reson Med. 2018; 80:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barkhuijsen H, De Beer R, Van Ormondt D. Improved algorithm for noniterative time-domain model fitting to exponentially damped magnetic resonance signals. J Magn Reson. 1987;73:553–557. [Google Scholar]
- 29.Santhakumar V, Wallner M, Otis TS. Ethanol acts directly on extrasynaptic subtypes of GABAA receptors to increase tonic inhibition. Alcohol. 2007; 41:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez R, Behar KL, Watzl J, et al. Intravenous ethanol infusion decreases human cortical γ-aminobutyric acid and N-acetylaspartate as measured with proton magnetic resonance spectroscopy at 4 tesla. Biol Psychiatry. 2012;71:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chitty KM, Lagopoulos J, Hickie IB, Hermens DF. The impact of alcohol and tobacco use on in vivo glutathione in youth with bipolar disorder: an exploratory study. J Psychiatr Res. 2014;55:59–67. [DOI] [PubMed] [Google Scholar]
- 32.Terpstra M, Marjanska M, Henry PG, Tkac I, Gruetter R. Detection of an antioxidant profile in the human brain in vivo via double editing with MEGA-PRESS. Magn Reson Med. 2006;56:1192–1199. [DOI] [PubMed] [Google Scholar]
- 33.Chiu TM, Mendelson JH, Woods BT, Teoh SK, Levisohn L, Mello NK. In vivo proton magnetic resonance spectroscopy detection of human alcohol tolerance. Magn Reson Med. 1994;32:511–516. [DOI] [PubMed] [Google Scholar]
- 34.Mendelson JH, Mello NK. Biologic concomitants of alcoholism. N Engl J Med. 1979;301:912–921. [DOI] [PubMed] [Google Scholar]
- 35.Fein G, Meyerhoff DJ. Ethanol in human brain by magnetic resonance spectroscopy: correlation with blood and breath levels, relaxation, and magnetization transfer. Alcohol Clin Exp Res. 2000;24:1227–1235. [PMC free article] [PubMed] [Google Scholar]
- 36.Sammi MK, Pan JW, Telang FW, et al. Measurements of human brain ethanol T2 by spectroscopic imaging at 4 T. Magn Reson Med. 2000;44:35–40. [DOI] [PubMed] [Google Scholar]
- 37.de Graaf RA, Rothman DL. Detection of gamma-aminobutyric acid (GABA) by longitudinal scalar order difference editing. J Magn Reson. 2001;152: 124–131. [DOI] [PubMed] [Google Scholar]
- 38.Chan KL, Puts NA, Snoussi K, Harris AD, Barker PB, Edden RA. Echo time optimization for J-difference editing of glutathione at 3T. Magn Reson Med. 2017;77:498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothman DL, Petroff O, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90:5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edden RA, Puts NA, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012;68: 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris AD, Glaubitz B, Near J, et al. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72: 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogner W, Gagoski B, Hess AT, et al. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleh MG, Alhamud A, Near J, Kouwe AJ, Meintjes EM. Volumetric navigated MEGA-SPECIAL for real-time motion and shim corrected GABA editing. NMR Biomed. 2016;29:248–255. [DOI] [PubMed] [Google Scholar]
- 44.Edden RA, Oeltzschner G, Harris AD, et al. Prospective frequency correction for macromolecule-suppressed GABA editing at 3T. J Magn Reson Imaging. 2016;44:1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59:1–6. [DOI] [PubMed] [Google Scholar]


