Abstract
The amantadine-resistant S31N mutant of the influenza A M2 proton channel has become prevalent in currently circulating viruses. Here, we have solved an X-ray crystal structure of M2(22–46) S31N that contains two distinct conformational states within its asymmetric unit. This structure reveals the mechanism of adamantane resistance in both conformational states of the M2 channel. In the Inwardopen conformation, the mutant Asn31 side chain faces the channel pore and sterically blocks the adamantane binding site. In the Inwardclosed conformation, Asn31 forms hydrogen bonds with carbonyls at the monomer–monomer interface, which twists the monomer helices and constricts the channel pore at the drug binding site. We also examine M2(19–49) WT and S31Nusing solution NMR spectroscopy and show that distribution of the two conformational states is dependent on both detergent choice and experimental pH.
Graphical Abstract
INTRODUCTION
Resistance to influenza antivirals is an ongoing concern. Most currently circulating strains of the virus are resistant to the adamantanes,1,2 which target the influenza A matrix 2 (M2) proton channel.3,4 Some strains are resistant to both neuraminidase (NA) inhibitors and M2 channel blockers.5 The most prevalent mutation in currently circulating influenza viruses is S31N,6–9 which is found in over 98% of adamantine-resistant strains of influenza.1
The M2 channel is homotetrameric and proton selective.10–14 The minimum construct needed for proton conductance is the transmembrane (TM) domain,15,16 which contains gating residues His37 and Trp41.17,18 The TM domain of M2 is well-conserved relative to the rest of the protein,19 with only a small number of mutations resulting in fit viruses. Mutating one residue of the M2 monomer introduces four perturbations into the channel, so pore-lining mutations can have a large effect on conductance. The rate of proton conductance in M2 is finely tuned to allow for the acidification of the viral interior and uncoating of viral RNA before hemagglutinin-mediated membrane fusion occurs.20,21 As a result, only a few pore-lining mutations are observed in transmissible viruses, with the most notable being S31N, V27A, and L26F.2,22,23 These mutations have proton conductance rates and pH activation curves that resemble the wild type (WT) channel.24 Because the S31N mutation is so prevalent, it is an important target for structural characterization and drug discovery.
Two conformational states of the M2 channel have been experimentally characterized: the Inwardclosed and Inwardopen states. Near neutral pH, M2 primarily adopts the Inwardclosed state, in which the C-terminus of the channel is tightly packed and each monomer helix has a kink at residue Gly34. The Inwardclosed state of M2 WT has been characterized using solid-state NMR,25,26 solution NMR,27,28 and X-ray crystallography,29,30 with good agreement between the structures in the TM domain. In the Inwardopen conformation, the C-terminus of the channel opens and the pore-lining residues become exposed to bulk solvent. The Inwardopen conformational state has been characterized primarily using X-ray crystallography.30–32 Bending or straightening of the monomer helices at Gly34 interconverts between these two conformations.33,34In lower pH ranges, conformational equilibrium between the Inwardclosed and Inwardopen states results in peak broadening in NMR spectra.28,33,34
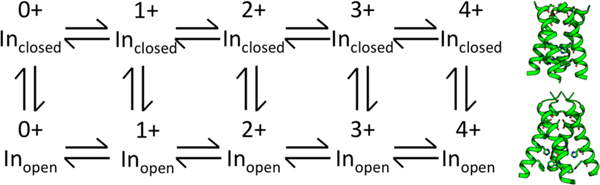
In addition to this conformational equilibrium, simultaneous equilibria exist between the possible charge states of the gatingHis37 tetrad (Figure 1). A number of NMR titration experiments have been carried out to determine the pKa’sassociated with successive protonation of the four His37 gating residues. There exists some variation between the reported values resulting from differences in the hydrophobic environment used for reconstitution.35–39 Nevertheless, in all of these studies, protonation to the +2 charge state occurs slightly above or near neutral pH, with protonation to the +3 state occurring within physiologically relevant low pH ranges (4–5).
Figure 1.
Simultaneous equilibria between the His37 tetrad charge states and the two conformational states, Inwardclosed and Inwardopen.
Though the equilibrium between the two conformational states is influenced by pH (and thus the protonation state of the gating His37 tetrad), the energetic difference between the two conformations is relatively small, and so each conformational state can be isolated in a range of pH conditions. The Inwardopen state is observed in conformational equilibrium with the Inwardclosed state at low pH, as evidenced by NMR and SSNMR studies showing peak broadening27,33,34,40 and increased hydration of the pore at low pH.41,42 However, previous structural studies have also isolated the Inwardopen state at intermediate and high pH.31,32,43 The Inwardclosed state is primarily observed in neutral to high pH ranges but has also been structurally characterized bound to drugs and inhibitors at low pH.30 At very low pH (<5), where all four gating histidines are protonated, the Inwardopen state is the predominant species. Experimental conditions can also influence the conformational landscape of M2, with certain membrane mimetics favoring one conformational state over the other.
Interconversion between the Inwardclosed and Inwardopen conformations is necessary for proton conduction in the M2 channel. In a study using inside-out macro patches of Xenopuslaevis oocytes, IV curves were fitted to a transporter-like model for proton conduction.44 The proton conduction mechanism begins with the channel in the Inwardclosed conformation with the His37 tetrad in the +2 charge state. Protonation to the +3 charge state triggers the channel to convert to the Inwardopen conformation, which opens the Trp41 gate and allows a proton to be released into the viral interior. This is followed by a “recycling” step where the deprotonated (+2) Inwardopen channel converts back to the Inwardclosed conformation. This mechanism is consistent with the channel’s low rate of proton conductance relative to the rate at which protons come on and off the gating His37 residues.45 This is best viewed as a linked equilibrium in which multiple states are explicitly considered as a function of pH.
Here, we have solved a crystal structure of both the Inwardclosed and Inwardopen conformations of M2(22–46) S31N in the same unit cell at low pH (5.0). This motivated us to further examine the pH dependence of the two conformational states using solution NMR. In physiologically relevant pH ranges, we find that the stability of the Inwardclosed conformation reaches a maximum near pH 6 and becomes slightly less favored at higher pH and dramatically less favored at lower pH.
RESULTS AND DISCUSSION
X-ray Crystal Structure of the S31N Mutant in Two Conformational States.
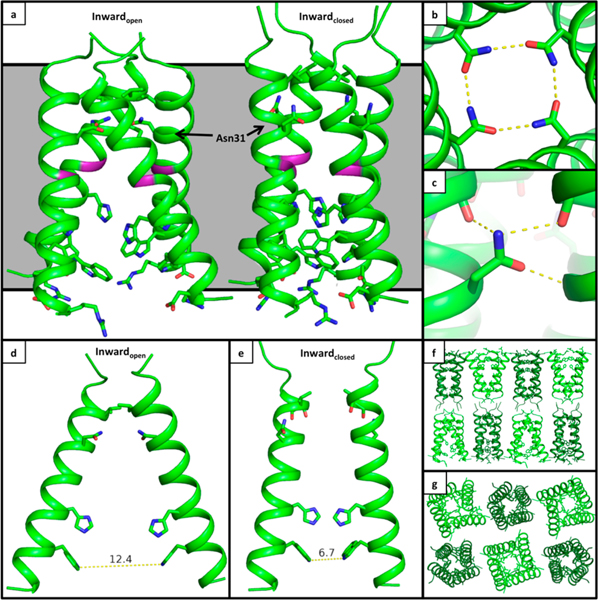
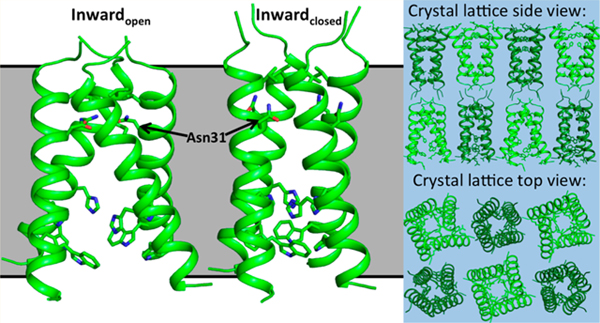
Here, we present an X-ray crystal structure of the drug-resistant M2(22–46) S31N mutant in two distinct conformational states at 2.06 Å resolution (PDB: 6MJH) (Figure 2). The asymmetric unit of these crystals contains two M2 tetramers, with one tetramer adopting the Inwardopen conformation and the second tetramer adopting the Inwardclosed conformation. These crystals have P21 symmetry and lattice dimensions a, b, c (Å) = 36.29, 36.15, 76.45; α, β, γ(deg) = 90, 103.6, 90 (Table S1). The P21 symmetry results in a lattice where the N-terminus of the Inwardopen tetramer packs against the N-terminus of the Inwardclosed tetramer. The two conformations of the channel observed here are consistent with previously solved crystal structures. The Inwardopen tetramer is well-ordered and tightly packed at its N-terminus but more open near the C-terminus.31,32 The N-terminus of the Inwardclosed tetramer has a slightly wider opening relative to that of the Inwardopen conformation, though this region of the pore is narrowed in comparison to previously solved structures of M2 WT. The C-terminus of the Inwardclosed conformation is well packed and consistent with previously solved structures.29,30 We have previously observed a number of crystal structures in a wide range of unit cells and crystal packing environments for both the wild type M2 channel and now also the S31N mutant (Figure S1 and Table S2). The uniformity of the structures in the Inwardopen and Inwardclosed states provides strong evidence that these conformational states are intrinsic to the tetrameric structure rather than being a product of crystal packing.
Figure 2.
X-ray crystal structure of M2(22–46) S31N in two conformational states. (a) The Inwardopen conformation is at left, and the Inwardclosed conformation is at right (PDB: 6MJH). The monomer helices in the Inwardopen conformation are straight, and in the Inwardclosed conformation they are kinked at Gly34 (shown in purple). As previously observed in structure 5C02, Asn31 faces the center of the pore and forms H-bonds with Asn31 side chains from neighboring monomers in the Inwardopen conformation. In the Inwardclosed conformation, Asn31 faces away from the pore and forms H-bonds with carbonyls at the monomer–monomer interface. (b) Top-down view showing hydrogen-bonding networks formed by Asn31 in the Inwardopen conformation. (c) Side view of the hydrogen-bonding network formed by Asn31 in the Inwardclosed conformation. (d) Side view of the Inwardopen conformation, only chains F and H shown. (e) Side view of the Inwardclosed conformation, only chains A and C shown. (f) Side view of crystal lattice from structure 6MJH, with the Inwardopen conformation in light green and the Inwardclosed conformation in dark green. (g) Top-down view of crystal lattice.
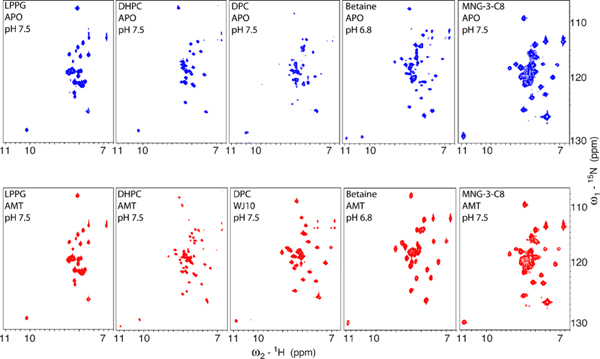
In previous studies, only the Inwardopen conformation of the S31N mutant could be isolated through crystallization. The Inwardopen tetramer seen here is very similar to the previously solved crystal structure of the S31N mutant in this conformational state,46 with an all-atom RMSD of 0.509 Å. Solution NMR screening of M2 in various detergents identified a maltose neopentyl glycol detergent MNG-3-C8 that stabilizes the Inwardclosed conformation (Figure 3). Using this detergentas an additive in crystallization trials allowed us to characterize the M2 S31N mutant in the Inwardclosed conformation for the first time at atomic resolution. The backbone of the Inwardclosed tetramer of M2(22–46) S31N is in good agreement with our previously solved solution NMR structure of M2(19–49) S31N in complex with a small molecule inhibitor of this mutant (2LY0),28 with a CαRMSD of 1.4 Å.
Figure 3.
Conformational equilibrium between the Inwardopen and Inwardclosed conformations for M2(19–49) WT is dependent on detergent choice in 2D NMR experiments. Solution NMR spectra of M2(19–49) WT in multiple detergents, without inhibitors (blue, top), and with inhibitors (red, bottom). The Trp41 indole proton peak (bottom left corner of each spectrum) can be used to distinguish the conformational states; a peak with chemical shift of ∼11.0 ppm corresponds to the Inwardclosed conformation, and a peak with chemical shift of ∼10.2 ppm corresponds to the Inwardopen conformation. From left to right: In LPPG, M2 stays in the Inwardopen conformation in absence and in the presence of amantadine. In DHPC, the presence of amantadine shifts a small portion of the protein in the Inwardopen conformation to the Inwardclosed conformation. In DPC, the inhibitor WJ10 almost fully shifts the Inwardopen to the Inwardclosed conformation. In C14-betaine, both conformations are observed in the absence of any drugs, and addition of amantadine fully shifts the protein to the Inwardclosed conformation. In MNG-3-C8, the protein stays in the Inwardclosed conformation in absence or presence of amantadine.
The networks of ordered waters within the M2 pore resemble those previously observed in crystal structures. In the Inwardopen state, a continuous hydrogen-bonding network connects the waters below Asn31 to the gating His37 residues.The water network within the Inwardclosed state closely resembles that of the X-ray crystal structure of M2 WT (PDB: 3LBW),29 with four ordered waters forming hydrogen bonds to the four His37 δ-nitrogens below and two bridging waters positioned above.
Conformation of the Mutant Asn31 Side Chain.
In the crystal structure of M2(22–46) S31N reported here, the conformation of the resistance-conferring Asn31 side chain differs when the M2 tetramer is in the Inwardopen state vs the Inwardclosed state. In the Inwardopen tetramer, the four Asn31 side chains face toward the aqueous channel pore and form hydrogen bonds with symmetrically repeated Asn31 side chains from neighboring monomer units. The same side chain configuration was observed in our previously solved crystal structure of M2(22–46) S31N in the Inwardopen state (PDB: 5C02).46 However, when M2(22–46) S31N adopts the Inwardclosed conformation in the absence of a drug, these Asn31 side chains face away from the channel pore and instead form hydrogen bonds with carbonyls at the tetramer’s monomer–monomer interface. Interestingly, in a previous solution NMR structure of the M2 S31N mutant in complex with a small molecule inhibitor, the Asn31 side chain carboxamide was stabilized in the inward-facing conformation by interactions with a drug molecule.28 In addition, Chou et al.47 previously reported a solution NMR structure of M2(18–60) S31N in which the Asn31 side chain faces away from the channel’s aqueous pore. However, Chou et al. model the Asn31 side chain as facing into the lipid bilayer. With the greater resolution now available using X-ray crystallography, we can see the Asn31 side chain carboxamide forming a bindentate interaction with a neighboring helix. In summary, it appears that the conformation of Asn31 side chain can be modulated by either the conformational switch between Inwardopen and Inwardclosed state or drug binding. Consequently, understanding this conformational switch is critical for structure-based drug design.
Inwardclosed Asn31 H-Bonding Motif in Other Structures.
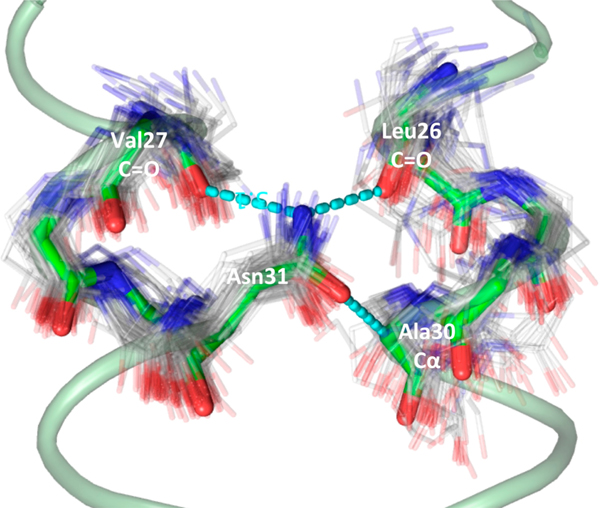
In the M2(22–46) S31N Inwardclosed tetramer, the Asn31 side chain engages in three hydrogen-bonding interactions. The nitrogen of the Asn31 carboxamide forms one intrahelix H-bond with the carbonyl from Val27 and a second H-bond with the Leu26 carbonyl from a neighboring monomer unit. The Asn31 side chain carbonyl oxygen also closely approaches the α-carbon of Ala30 from a neighboring monomer unit, with a distance of 3.3–3.5 Å between the two atoms, which is generally described as a C–H···O bond. The C–H···O hydrogen bonds are commonly observed in α-helical proteins between buried polar side chains and α-carbons, particularly in membrane proteins.48,49
We interrogated the structural abundance of the Asn-to-backbone motif found in the InwardclosedM2 structure (Figure 4). We queried a nonredundant protein structural database of 20,383 structures for the motif, first starting with helical backbone superpositions (C, CA, N, O atoms) with RMSD ≤ 1.5 Å. Backbone residues 26–30 of chain A and 27–31 of chain B were queried with the program MASTER50 for structural matches. All structural matches with an ASN at the position equivalent to 31B of M2 were then filtered by RMSD of the Asn side chain (CB, CG, OD1, ND2 atoms, RMSD ≤ 1 Å), using the program ProDy.51 A total of 114 close matches to the Asn motif were discovered, originating from a diverse set of membrane and soluble proteins (Table S3 contains a list of PDB accession codes). We conclude that this Asn hydrogen bonding motif is commonly observed to link αhelices in a variety of proteins.
Figure 4.
Inwardclosed Asn31 hydrogen bonding motif is commonly observed in the PDB. The Inwardclosed S31N structure backbone from chains A and B is shown with light green ribbon, with green sticks highlighting the search motif and hydrogen bonds shown as dashed cyan lines. Matching PDB structures are shown as gray lines.
Examination of M2 Conformational States Using Solution NMR.
SSNMR studies show that the equilibrium between conformational forms of M2 depends markedly on the phospholipid composition of bilayers in which the protein is reconstituted.33,34,52–54 Similarly, in solution NMR experiments, the conformation of the M2 channel is highly dependent on the micelle or bicelle composition.55 Figure 3 shows NMR spectra for M2(19–49) WT in a variety of detergents before and after addition of an inhibitor, which is known to stabilize the Inwardclosed conformation of M2.33,56 We observe an equilibrium between both conformational states through examination of backbone peaks and also a single peak corresponding to Trp41 in the TM domain. The Inwardclosed conformation has well-dispersed and sharp chemical shifts, while the peaks of the Inwardopen conformation are broader and show less dispersion in the proton dimension. In our previous solution NMR studies,57 we developed a simple, fast way to identify the two conformations through examination of the chemical shift of the Trp41 indole proton. The peak at ~11.0 ppm corresponds to the Inwardclosed conformation, and the peak at ~10.2 ppm corresponds to the Inwardopen conformation. For comparison, the average chemical shift of this Trp indole proton is 10.1 ppm in the BMRB (Biological Magnetic Resonance Data Bank). When M2(19–49) WT is solubilized in LPPG, only the Inwardopen conformation is observed in both the absence and presence of amantadine. When M2(19–49) WT is solubilized in MNG-3-C8 detergent, only the Inwardclosed conformation is observed both with and without amantadine. Using this detergent as an additive in the present study aided crystallization trials of M2(22–46) S31N in the Inwardclosed state. In previous crystallization trials using octyl glucoside as a detergent additive, only the Inwardopen conformation of M2 S31N could be isolated.46 We note that nonionic detergents appear to favor crystallization of M2 and speculate that MNG-3-C8 stabilizes the Inwardclosed conformation because of its additional length relative to OG.
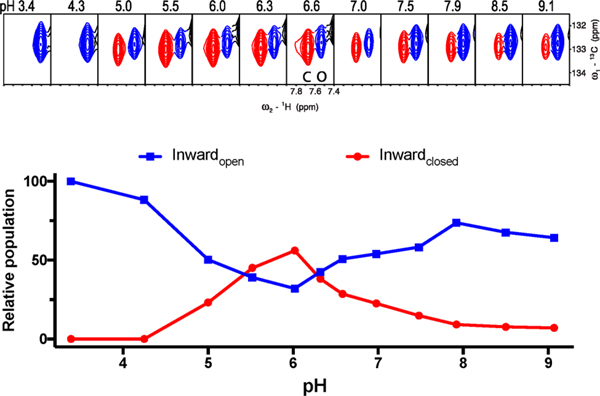
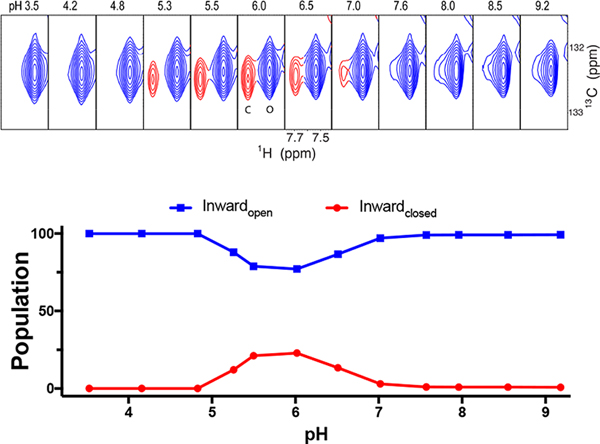
The Inwardopen and Inwardclosed conformational equilibrium of M2 is also highly dependent on pH. After screening a variety of detergents (Figure 3), we observed that C14-betaine allows the two conformations to interexchange within the physiological pH range. In Figures 5 and 6, we selected the aromatic Cδ–Hδ peak of F48 as a probe to perform quantitative analysis of the conformational equilibrium because the F48 peaks fromthe two conformations clearly separate from each other and also do not overlap with any other peaks during the course of titration. In addition, F48 is near the end of TM, so it is sensitive to opening and closing of M2’s C-terminus. The data clearly show that the stability of the Inwardclosed state is relatively low at the highest pH, reaches a maximum as the pH is decreased to near pH 6, and then decreases as the pH is decreased further. The Inwardclosed conformation becomes undetectable at pH 4.3 or lower. Similar behavior is seen for S31N, although the overall stability is somewhat lower in this particular micelle, C14-betaine (Figure 6).
Figure 5.
F48 Cδ–Hδpeak shows conformational equilibration between the Inwardopen(blue) and Inwardclosed (red) states of M2(19–49) WT as a function of pH. Top: Only the Inwardopen state is observed at pH 3.4 and 4.3. In the physiologically relevant pH range (5.0–7.5), both conformational states are observed. The peak of the Inwardclosed conformation (red) gains intensity at pH 5.0, reaches a maximum at pH 6.0, and then decreases at higher pH. The data are normalized to the intensity observed at pH 3.4, and sum to slightly less than 100%, possibly due to a third state that is not detected due to chemical exchange or dynamics. Bottom: Fit of peak intensity vs pH for both conformational states. The intensity profile of the Inwardopen conformation (blue) has a U-like shape with a minimum at pH ∼6.0. The channel primarily adopts the Inwardopen conformation at both very low and very high pH.
Figure 6.
F48 Cδ–Hδpeak of M2(19–49) S31N shows conformational interchange between the Inwardopen and Inwardclosed states of M2(19–49) S31N as a function of pH. Top: Similar to WT as seen in Figure 5, we show the F48 Cδ–Hδ peak from NMR spectra of the M2(19–49) S31N construct in a range of pH conditions. Bottom: Peak intensity vs pH for both conformational states. The intensity profile of the Inwardopen conformation (blue) has a U-like shape with a minimum at pH ∼6.0.
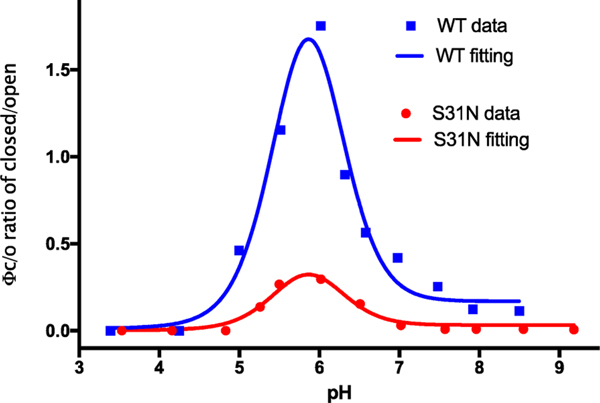
Figure 5 shows that the Inwardclosed state of the WT peptide is somewhat less stable than the Inwardopen state at high pH. Interestingly, the Inwardclosed conformation becomes more stable as the pH is lowered to near 6.0, after which it drops off more precipitously with further decreasing pH, and the Inwardclosed state is not detectable at pH 4.3 or lower. Similar results were seen for the S31N mutant (Figure 6). To explain this behavior, it is important to consider the protonation constants for both states. Thus, the data in the scheme in Figure 1 were used to determine the pKa values for protonation of the Inwardopen and Inwardclosed states (Figure 7, Scheme S1,and Table S4). This analysis indicated that the peak in stability near pH 6 is a direct result of energetic coupling between protonation and conformational change. The computed pKafor the first two protonations of the Inwardclosedstate (6.5 ± 1.1 and 7.4 ± 1.2) are higher than the corresponding pKa for the Inwardopen state (approximately 5.9 ± 0.1), resulting in stabilization of the Inwardclosedstate up to about pH 6 (because here the protons have a higher affinity for the Inwardopen state and, hence, drive the equilibrium toward this state relative to high pH where both His37 residues are neutral). The pKa for the final two protonations of the His37 tetrad are 4.4 ± 0.8 and <4. As a result, decreasing the pH below 6 leads to progressive destabilization, and the Inwardclosed state is entirely destabilized at low pH.
Figure 7.
Fitting of experimental data as a function of pH, giving R square values 0.96 and 0.95 for WT and S31N, respectively, and producing the three pKa’s for the Inwardclosed state (pK1: 6.4 ± 1.1, pK2: 7.4 ± 1.2 and pK3: 4.4 ± 0.8) and one pKa (5.9 ± 0.1) for the Inwardopen state. We set the pK4 as a constant 3, but the fit was insensitive to its value, so long as it was set less than 4.0.
CONCLUSIONS
Proteins in their native environment often have a number of possible conformational states. X-ray crystallography is a powerful technique for structural characterization because of its ability to isolate specific conformations and determine positions of individual atoms within particularly well-ordered crystals. However, conformational heterogeneity exists even within the ordered environment of the crystal lattice, both on the scale of individual side chain conformations and also on the scale of secondary and tertiary structure. Crystallization sometimes results in the isolation of multiple conformational states within a single crystal lattice. Examples of crystal structures containing more than one conformational state include structures of the C. jejuni CmeB multidrug efflux pump,58 an RNA-dependent RNA polymerase from the rabbit hemorrhagic disease virus,17 and an mRNA capping enzyme.59 These conformational states are often important for the function of the protein. In the case of the M2 channel, interconversion between the Inwardclosed and Inwardopen conformational states allows the channel to conduct protons through its narrow transmembrane pore.
Implications for Drug Resistance.
The conformation of the Asn31 side chain holds the key to adamantane resistance in the M2 channel. In the wild-type channel, the Ser31 side chain faces toward the center of the channel pore in both the Inwardopen and Inwardclosed conformational states, with the Ser31 hydroxyl engaging in a single intrahelix hydrogen bond with the carbonyl of Val27.29–32 As a result of this hydrogen bond, Ser31 engages in primarily hydrophobic interactions with the adamantyl cage of amantadine and rimantadine.30,31
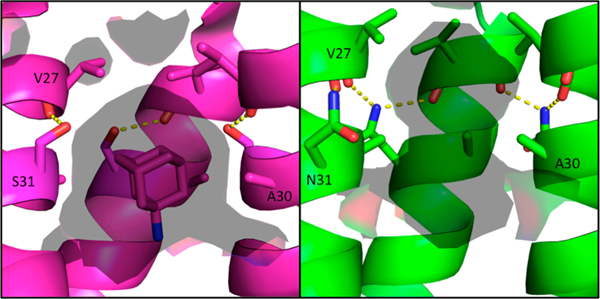
Based on the previously solved structure of S31N in the Inwardopenconformation,46the mechanism of drug resistance in the S31N mutant channel was hypothesized to be a steric block of the binding site as a result of replacing the Ser31 hydroxyl with the larger and more hydrophilic Asn31 carboxamide.46 However, in this newly solved structure of M2(22–46) S31N in the Inwardclosed state, Asn31 faces away from the center of the pore and forms hydrogen bonds at the monomer–monomer interface. The hydrogen bond between the Asn31 side chain and the Leu26 backbone carbonyl from a neighboring monomer twists each monomer helix at the channel’s N-terminus. As a result of this twist, the Ala30 side chain that interacts with the monomer–monomer interface in the WT channel rotates toward the center of the pore in the S31N mutant channel. This constricts the channel at the adamantane binding site and elongates the pore at the N-terminus below the Val27 gate (Figure 8).
Figure 8.
Solvent-accessible surface area inside the M2 pore at the adamantane binding site. Amantadine-bound WT structure in the Inwardclosed conformation 6BKK (left, magenta) vs S31N Inwardclosed conformation (green, right); solvent-accessible surface area (calculated using a 1.4 Å probe) is shown as a transparent surface. In the WT channel, Ser31 forms an intrahelix hydrogen bond and partially faces the pore. In the S31N mutant, Asn31 faces the monomer–monomer interface, twisting the monomer helices such that Ala30 faces the center of the pore. This constricts and elongates the pore at the amantadine binding site for the S31N mutant M2 channel.
The shape of the pore in both conformational states is important for inhibition of the M2 channel. Both amantadine and rimantadine form stable complexes with both conformational states of the M2 WT channel in X-ray crystal structures.30,31 This is consistent with inhibition of proton conduction upon addition of adamantanes at both high and low pH ranges;4,60 although the channel exists in conformational equilibrium at low pH,27,28,34 the adamantanes can bind to both states. The structures presented here will aid the design of new inhibitors to target both conformational states of the adamantane-resistant S31N mutant M2 channel.
Finally, this work provides an in-depth evaluation of the thermodynamically linked protonation and conformational equilibria in M2. The increased stability of the Inwardclosed conformation as the pH is lowered from 8 to 6 directly reflects the well-known stability of partially protonated forms of M2, which have high pKa’s for the first two protonation events on the gating His37 tetrad. In more stabilizing environments (e.g., the detergent MNG-3-C8 or certain lipid compositions),33,34,52–54 the small change in stability between pH 8and 6 does not lead to a large enough change in population of the states to be easily observed. However, as the pH is decreased below pH 6 the decrease in stability is quite pronounced and can be readily observed in many environments. The overall behavior reflects M2’s ability to stabilize protons in the transmembrane pore, with special stabilization of the +2 state relative to either neutral or to +3 and +4 states. This can be understood in a mechanism whereby proton flux depends on the population of channel in the +2 state multiplied by the proton concentration.
METHODS
The M2(22–46) S31N construct was chemically synthesized as previously described61 and crystallized in the lipid cubic phase (LCP) with some modifications to the protocol described by Caffrey and Cherezov.62 Solution NMR was used to screen detergents for the stabilization of the Inwardclosed conformational state; this identified an MNG detergent that was then used as an additive in crystallization trials. Crystals were diffracted at ALS 8.3.1 at a temperature of 100 K. Molecular replacement was carried out using structure 3LBW29 as the starting model for the Inwardclosed tetramer and 5JOO43 as the starting model for the Inwardopen tetramer.
For solution NMR experiments, M2(19–49) WT and M2(19–49) S31N were doubly labeled with 15N and 13C and were prepared as previously described.28 Spectra were recorded at 313 K on a Varian 600 MHz, a Bruker 600 MHz, or a Bruker 900 MHz spectrometer, as described in the Supporting Information.
Supplementary Material
NOTE ADDED IN PROOF.
In the course of reviewing the proofs we noticed that we had failed to reference two key papers concerning solid-state NMR characterization of M2: Mandala et al,63 and Mandala et al.64 These papers provide detailed structural and dynamic characterization of the two states of M2. Moreover they show that interconversion between the states occurs on approximately the same time scale as conductance.
ACKNOWLEDGMENTS
J.L.T., Y.W., N.P., and W.F.D. were supported by NIH Grant Nos. R35-GM122603 and R01-GM117593 and NSF Grant No. 1709506. J.W. was supported by NIH Grant No. R33-AI119187. Use of the LCP crystallization robot was made possible by National Center for Research Resources Grant 1S10RR027234–01. We thank Pil SeokChae (Hanyang University, Seoul, South Korea) for providing MNG detergent for crystallization trials. Data collection was carried out at ALS 8.3.1. Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant No.MR-15–328599 and NIGMS Grant Nos. P30 GM124169 and R01 GM124149. We thank George Meigs and James Holton at ALS 8.3.1 for support during data collection.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b02196.
Supplemental figures and tables and detailed experimental methods (PDF)
Crystallographic data for M2 S31N mutant in two distinct conformational states (PDB code 6MJH) (CIF)
The authors declare no competing financial interest.
REFERENCES
- (1).Bright RA; Medina MJ; Xu XY; Perez-Oronoz G; Wallis TR; Davis XHM; Povinelli L; Cox NJ; Klimov AI Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 2005, 366 (9492), 1175–1181. [DOI] [PubMed] [Google Scholar]
- (2).Furuse Y; Suzuki A; Oshitani H. Large-Scale Sequence Analysis of M Gene of Influenza A Viruses from Different Species: Mechanisms for Emergence and Spread of Amantadine Resistance. Antimicrob. Agents Chemother. 2009, 53 (10), 4457–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hay AJ; Wolstenholme AJ; Skehel JJ; Smith MH The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985, 4 (11), 3021–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wang C; Takeuchi K; Pinto LH; Lamb RA Ion-channel activity of influenza-A virus M(2) protein - characterization of the amantadine block. J. Virol. 1993, 67 (9), 5585–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sheu TG; Fry AM; Garten RJ; Deyde VM; Shwe T; Bullion L; Peebles PJ; Li Y; Klimov AI; Gubareva LV Dual Resistance to Adamantanes and Oseltamivir Among Seasonal Influenza A(H1N1) Viruses: 2008–2010. J. Infect. Dis. 2011, 203 (1), 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Deyde V; Garten R; Sheu T; Smith C; Myrick A; Barnes J; Xu X; Shaw M; Klimov A; Gubareva L. Genomic events underlying the changes in adamantane resistance among influenza A(H3N2) viruses during 2006–2008. Influenza Other Respir. Viruses 2009, 3 (6), 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Deyde VM; Xu X; Bright RA; Shaw M; Smith CB; Zhang Y; Shu Y; Gubareva LV; Cox NJ; Klimov AI Surveillance of Resistance to Adamantanes among Influenza A(H3N2) and A(H1N1) Viruses Isolated Worldwide. J. Infect. Dis. 2007, 196 (2), 249–257. [DOI] [PubMed] [Google Scholar]
- (8).Krumbholz A; Schmidtke M; Bergmann S; Motzke S; Bauer K; Stech J; Dürrwald R; Wutzler P; Zell R. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J. Gen. Virol. 2009, 90 (4), 900–908. [DOI] [PubMed] [Google Scholar]
- (9).Simonsen L; Viboud C; Grenfell BT; Dushoff J; Jennings L; Smit M; Macken C; Hata M; Gog J; Miller MA; Holmes EC The Genesis and Spread of Reassortment Human Influenza A/H3N2 Viruses Conferring Adamantane Resistance. Mol. Biol. Evol. 2007, 24 (8), 1811–1820. [DOI] [PubMed] [Google Scholar]
- (10).Chizhmakov IV; Geraghty FM; Ogden DC; Hayhurst A; Antoniou M; Hay AJ Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996, 494 (2), 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lin TI; Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J. Virol. 2001, 75 (8), 3647–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mould JA; Drury JE; Frings SM; Kaupp UB; Pekosz A; Lamb RA; Pinto LH Permeation and activation of the M-2 ion channel of influenza A virus. J. Biol. Chem. 2000, 275 (40), 31038–31050. [DOI] [PubMed] [Google Scholar]
- (13).Salom D; Hill BR; Lear JD; DeGrado WF pH-Dependent tetramerization and amantadine binding of the trans-membrane helix of M2 from the influenza A virus. Biochemistry 2000, 39 (46), 14160–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Shimbo K; Brassard DL; Lamb RA; Pinto LH Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys. J. 1996, 70 (3), 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Duff KC; Ashley RH The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology 1992, 190 (1), 485–489. [DOI] [PubMed] [Google Scholar]
- (16).Ma CL; Polishchuk AL; Ohigashi Y; Stouffer AL; Schon A; Magavern E; Jing XH; Lear JD; Freire E; Lamb RA; DeGrado WF; Pinto LH Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (30), 12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tang YJ; Zaitseva F; Lamb RA; Pinto LH The gate of the influenza virus M-2 proton channel is formed by a single tryptophan residue. J. Biol. Chem. 2002, 277 (42), 39880–39886. [DOI] [PubMed] [Google Scholar]
- (18).Wang C; Lamb RA; Pinto LH Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys. J. 1995, 69 (4), 1363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dong G; Peng C; Luo J; Wang C; Han L; Wu B; Ji G; He H. Adamantane-resistant influenza A viruses in the world (1902–2013): frequency and distribution of M2 gene mutations. PLoS One 2015, 10 (3), No. e0119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Grambas S; Bennett MS; Hay AJ Influence of amantadine resistance mutations on the pH regulatory function of the M2-protein of influenza-A viruses. Virology 1992, 191 (2), 541–549. [DOI] [PubMed] [Google Scholar]
- (21).Grambas S; Hay AJ Maturation of influenza-A virus hemagglutinin - estimates of the pH encountered during transport and its regulation by the M2 protein. Virology 1992, 190 (1), 11–18. [DOI] [PubMed] [Google Scholar]
- (22).Dong G; Peng C; Luo J; Wang C; Han L; Wu B; Ji G; He H. Adamantane-Resistant Influenza A Viruses in the World (1902–2013): Frequency and Distribution of M2 Gene Mutations. PLoS One 2015, 10 (3), No. e0119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Furuse Y; Suzuki A; Kamigaki T; Oshitani H. Evolution of the M gene of the influenza A virus in different host species: large-scale sequence analysis. Virol. J. 2009, 6, 67–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Balannik V; Carnevale V; Fiorin G; Levine BG; Lamb RA; Klein ML; DeGrado WF; Pinto LH Functional studies and modeling of pore-lining residue mutants of the influenza A virus M2 ion channel. Biochemistry 2010, 49 (4), 696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cady SD; Schmidt-Rohr K; Wang J; Soto CS; DeGrado WF; Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463 (7281), 689–U127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sharma M; Yi M; Dong H; Qin H; Peterson E; Busath DD; Zhou HX; Cross TA Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 2010, 330 (6003), 509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Schnell JR; Chou JJ Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451 (7178), 591–U12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wang J; Wu Y; Ma C; Fiorin G; Wang J; Pinto LH; Lamb RA; Klein ML; Degrado WF Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. U. S. A.2013, 110 (4), 1315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Acharya R; Carnevale V; Fiorin G; Levine BG; Polishchuk AL; Balannik V; Samish I; Lamb RA; Pinto LH; DeGrado WF; Klein ML Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (34), 15075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Thomaston JL; Polizzi NF; Konstantinidi A; Wang J; Kolocouris A; DeGrado WF Inhibitors of the M2 proton channel engage and disrupt transmembrane networks of hydrogen-bonded waters. J. Am. Chem. Soc. 2018, 140 (45), 15219–15226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Stouffer AL; Acharya R; Salom D; Levine AS; Di Costanzo L; Soto CS; Tereshko V; Nanda V; Stayrook S; DeGrado WF Structural basis for the function and inhibition of an influenza virus proton channel. Nature 2008, 451 (7178), 596–U13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Thomaston JL; Alfonso-Prieto M; Woldeyes RA; Fraser JS; Klein ML; Fiorin G; DeGrado WF High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (46), 14260–14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hu FH; Luo WB; Cady SD; Hong M. Conformational plasticity of the influenza A M2 transmembrane helix in lipid bilayers under varying pH, drug binding, and membrane thickness. Biochim. Biophys. Acta, Biomembr. 2011, 1808 (1), 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li C; Qin H; Gao FP; Cross TA Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim. Biophys. Acta, Biomembr. 2007, 1768 (12), 3162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Colvin MT; Andreas LB; Chou JJ; Griffin RG Proton Association Constants of His 37 in the Influenza-A M2(18–60) Dimer-of-Dimers. Biochemistry 2014, 53 (38), 5987–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hu FH; Schmidt-Rohr K; Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J. Am. Chem. Soc. 2012, 134 (8), 3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hu J; Fu R; Nishimura K; Zhang L; Zhou HX; Busath DD; Vijayvergiya V; Cross TA Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (18), 6865–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liao SY; Yang Y; Tietze D; Hong M. The influenza M2 cytoplasmic tail changes the proton-exchange equilibria and the backbone conformation of the transmembrane histidine residue to facilitate proton conduction. J. Am. Chem. Soc. 2015, 137 (18), 6067–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Miao Y; Fu R; Zhou H-X; Cross TA Dynamic Short Hydrogen Bonds in Histidine Tetrad of Full-Length M2 Proton Channel Reveal Tetrameric Structural Heterogeneity and Functional Mechanism. Structure 2015, 23 (12), 2300–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hu FH; Luo WB; Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science 2010, 330 (6003), 505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ghosh A; Qiu J; DeGrado WF; Hochstrasser RM Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc. Natl. Acad. Sci. U. S. A.2011, 108 (15), 6115–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Luo W; Hong M. Conformational changes of an ion channel detected through water-protein interactions using solid-state NMR spectroscopy. J. Am. Chem. Soc.2010, 132 (7), 2378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Thomaston JL; Woldeyes RA; Nakane T; Yamashita A; Tanaka T; Koiwai K; Brewster AS; Barad BA; Chen Y; Lemmin T; Uervirojnangkoorn M; Arima T; Kobayashi J; Masuda T; Suzuki M; Sugahara M; Sauter NK; Tanaka R; Nureki O; Tono K; Joti Y; Nango E; Iwata S; Yumoto F; Fraser JS; DeGrado WF XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (51), 13357–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).DiFrancesco ML; Hansen U-P; Thiel G; Moroni A; Schroeder I. Effect of cytosolic pH on inward currents reveals structural characteristics of the proton transport cycle in the influenza A protein M2 in cell-free membrane patches of xenopus oocytes. PLoS One 2014, 9 (9), No. e107406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Liang R; Swanson JMJ; Madsen JJ; Hong M; DeGrado WF; Voth GA Acid activation mechanism of the influenza A M2 proton channel. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (45), E6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Thomaston JL; DeGrado WF Crystal structure of the drug-resistant S31N influenza M2 proton channel. Protein Sci. 2016, 25 (8), 1551–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Pielak RM; Schnell JR; Chou JJ Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (18), 7379–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Derewenda ZS; Lee L; Derewenda U. The Occurence of C–H···O Hydrogen Bonds in Proteins. J. Mol. Biol. 1995, 252 (2), 248–262. [DOI] [PubMed] [Google Scholar]
- (49).Senes A; Ubarretxena-Belandia I; Engelman DM The Cα—H···O hydrogen bond: A determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (16), 9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhou J; Grigoryan G. Rapid search for tertiary fragments reveals protein sequence-structure relationships. Protein Sci. 2015, 24 (4), 508–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Bakan A; Dutta A; Mao W; Liu Y; Chennubhotla C; Lezon TR; Bahar I. Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics 2014, 30 (18), 2681–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Cady S; Wang T; Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J. Am. Chem. Soc. 2011, 133 (30), 11572–11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Cross TA; Sharma M; Yi M; Zhou HX Influence of solubilizing environments on membrane protein structures. Trends Biochem. Sci. 2011, 36 (2), 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Luo W; Cady SD; Hong M. Immobilization of the influenza A M2 transmembrane peptide in virus envelope-mimetic lipid membranes: a solid-state NMR investigation. Biochemistry 2009, 48 (27), 6361–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Claridge JK; Aittoniemi J; Cooper DM; Schnell JR Isotropic Bicelles Stabilize the Juxtamembrane Region of the Influenza M2 Protein for Solution NMR Studies. Biochemistry 2013, 52 (47), 8420–8429. [DOI] [PubMed] [Google Scholar]
- (56).Yi M; Cross TA; Zhou H-X Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (32), 13311–13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Ma C; Fiorin G; Carnevale V; Wang J; Lamb RA; Klein ML; Wu Y; Pinto LH; DeGrado WF Asp44 Stabilizes the Trp41 Gate of the M2 Proton Channel of Influenza A Virus. Structure 2013, 21 (11), 2033–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Su C-C; Yin L; Kumar N; Dai L; Radhakrishnan A; Bolla JR; Lei H-T; Chou T-H; Delmar JA; Rajashankar KR; Zhang Q; Shin Y-K; Yu EW Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nat. Commun. 2017, 8 (1), 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Håkansson K; Doherty AJ; Shuman S; Wigley DB X-Ray Crystallography Reveals a Large Conformational Change during Guanyl Transfer by mRNA Capping Enzymes. Cell 1997, 89 (4), 545–553. [DOI] [PubMed] [Google Scholar]
- (60).Martin K; Heleniust A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell 1991, 67 (1), 117–130. [DOI] [PubMed] [Google Scholar]
- (61).Cady SD; Wang J; Wu YB; DeGrado WF; Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J. Am. Chem. Soc. 2011, 133 (12), 4274–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Caffrey M; Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009, 4 (5), 706−731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Mandala VS; Liao S-Y; Kwon B; Hong M. Structural Basis for Asymmetric Conductance of the Influenza M2 Proton Channel Investigated by Solid-State NMR Spectroscopy. J. Mol. Biol. 2017, 429 (14), 2192–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Mandala VS; Gelenter MD; Hong M. Transport-Relevant Protein Conformational Dynamics and Water Dynamics on Multiple Time Scales in an Archetypal Proton Channel: Insights from Solid-State NMR. J. Am. Chem. Soc. 2018, 140 (4), 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.