Abstract
The adult skin is a typical example of a highly regenerative tissue. Terminally differentiated keratinocytes are shed from the external layers of the epidermis or extruded from the skin as part of the growing hair shaft on a daily basis. These are effectively replenished through the activity of skin-resident stem cells. Precise regulation of stem cell activity is critical for normal skin homeostasis or wound healing and irregular stem cell proliferation or differentiation can lead to skin disease. The scarcity and dynamic nature of stem cells presents a major challenge for elucidating their mechanism of action. To address this, we have recently established a system for visualizing stem cell activity, in real-time or long term, in the intact skin of live mice using 2-photon microscopy. The purpose of this review is to provide essential information to researchers who wish to incorporate 2-photon microscopy and live imaging into their experimental toolbox for studying aspects of skin and stem biology in the mouse model. We discuss fundamental principles of the method, instrumentation and basic experimental approaches to interrogate stem cell activity in the interfollicular epidermis and hair follicle.
Introduction
The replenishment of lost cells to maintain homeostasis and the repair of the skin after wounding rely on the activity of resident stem cells (1–3). Due to their unique properties of self-renewal and multipotency stem cells can be indispensable for normal skin function or extremely detrimental when they deviate from their standard activity (4–8). One of the major impediments on studying adult stem cells is that they usually constitute a relatively small fraction of the total population. While significant progress has been made in this front, with the identification of genes that are preferentially expressed in restricted populations that display stem cell properties in the skin, questions remain regarding the level of heterogeneity within these populations. Another significant challenge is that stem cell activity is by definition a highly dynamic process. While the microscopic analysis of frozen or paraffin embedded skin sections - processed by conventional histology and immunohistochemistry - has been the workhorse of dermatological research, this method is not always sufficient to overcome the unique challenges of studying stem cell activity in vivo. Live imaging modalities such as 2-photon microscopy – especially when combined with powerful mouse genetic models - can provide critical insight into the mechanisms that govern stem cell activity, by enabling the visualization of stem cells within their native environment in the intact living skin (8). In contrast to other tissues, the skin being the most external organ in the adult body offers direct access for observation and requires minimal, non-invasive preparation for imaging (9). This review discusses recent methodological advances in the use of 2-photon microscopy for intra-cutaneous imaging of stem cell activity.
2-Photon microscopy: Principle and advantages
Two-photon laser scanning fluorescent microscopy (abbreviated here as 2-photon microscopy) was introduced in 1990 by Webb and colleagues to mark a turning point for the advancement of intravital imaging (10). 2-photon microscopy is an optical modality which relies on the use of a tunable, pulsed laser that emits light in the infrared spectrum (11). Similar to a confocal microscope, the laser raster-scans the sample and induces the excitation of fluorescent molecules. The major difference between confocal and 2-photon microscopy is defined by the mechanism of fluorophore excitation and the physical properties of infrared light (12). While a single photon in the visible excitation spectrum is sufficient to induce fluorescence, infrared light is less energetic; thus, to induce the transition of the fluorophore to its excited state two photons need to combine their energy by being absorbed at the same time. Such an event is naturally very rare but can be effectively achieved with the use of a high peak-power pulsed laser, which compresses light into intense packets, crowding the photons to increase the probability of two-photons being absorbed simultaneously by the fluorophore. This process provides a key advantage in that 2-photon excitation only occurs at the focal point defined by the objective lens. In contrast, in wide field and confocal microscopy single photons excite – albeit less efficiently – fluorescence continuously as they travel through the sample before reaching the focal plane; therefore, increasing background noise and inducing higher levels of photo-bleaching, photo-damage and photo-toxicity in the sample.
In practical terms, 2-photon microscopy offers four major advantages over other optical fluorescent modalities: 1) The single-plane excitation enables precise control of the contour of the visualized area and true optical sectioning, which together offer high three-dimensional resolution of the observed tissue. 2) Fluorescent proteins and probes have broad excitation spectra in the infrared space (13,14) 2-photon microscopy induces substantially less photo-damage, compared to confocal, which makes it feasible to visualize live tissues for extended periods of time with negligible impact to their normal physiology (15,16). 4) Infrared light scatters significantly less as it travels through the living matter, enabling imaging of cells and structures at much higher depths within the tissue (17). 5) The short-pulse lasers used in 2-photon microscopy are capable of generating contrast from internal tissue structures without the use of exogenous fluorescent probes. For example, in a process known as second harmonic generation (SHG), two photons interacting with non-centrosymmetric structures - such as collagen fibers and elastin, which are abundant in the skin - generate new photons with twice the energy and therefore half the wavelength (18,19). This can be collected as specific signal with the use of appropriate emission filters. These properties make 2-photon microscopy an ideal optical method for dissecting the mechanisms of cellular activity in the skin, given its unique anatomy and stratified organization.
Intra-cutaneous imaging in the live mouse
2-photon microscopy has successfully been implemented in dermatological clinical research (20–25). Efforts to use the technology as a diagnostic tool by visualizing human skin are certainly exciting but mouse models are still invaluable for studying basic biological mechanisms of skin function and disease, especially when trying to understand processes as dynamic and complex as tissue regeneration. We have recently established a method for visualizing stem cell activity in the intact skin of living mice using 2-photon microscopy (8,26). A detailed description of our experimental protocol is available elsewhere (27); however, here we discuss important aspects of the method that will be useful to readers who wish to utilize a live imaging approach to address specific questions in basic skin and stem cell biology. These include instrumentation (Supplement), animal and tissue preparation and mouse genetic tools.
Animal and skin preparation for live imaging
In a typical live imaging experiment involving internal organs this step can be rather complicated and time consuming but this is not the case when imaging the skin (9). A number of different skin mounting apparatuses have been proposed to date and most of them share some common features and requirements (27–30). Anesthesia is one of those requirements, in order to keep the animal and the mounted skin stable for the duration of the experiment. Anesthesia can be easily achieved with injectable (ketamine/xylazine mix) or inhalable (isoflurane) anesthetic. To sustain and monitor the health of the mouse during the course of the experiment a temperature controlled heating plate and sensor are also required. In addition, a pump should also be considered to provide fluids and electrolytes intraperitoneally in a controlled manner, especially if the mouse will be imaged for extended periods of time.
Mice used for live imaging experiments should be bred into an albino background because pigment absorbs intensely in the infrared spectrum. This, may be detrimental to the health of the tissue following prolonged exposure but also produces strong autofluorescence which interferes with signal detection (28,31,32) The mouse skin requires very little preparation before mounting and we typically only remove the external hair shafts using a minimal amount of depilatory cream (Nair), which if carefully applied does not affect the normal activity of the keratinocytes and stem cells in the skin in any appreciable manner. Various skin areas including the ear, back, paw or tail can be easily mounted for imaging (Fig. 1). The ear skin can be particularly thin and tissue structures including the hair follicles are closer to the surface, which gives superior resolution compared to other areas of the skin. However, many hair follicles in the ear especially those closer to the tip do not display the same cycling characteristics as other sites. Conversely, the paw is devoid of hair follicles but is the only site where eccrine sweat glands can be found in the mouse skin. The back skin is routinely used for conventional histology and immunohistochemistry but the tissue is thicker compared to the ear and the hair follicles are difficult to be resolved in their entirety when in full growth phase. The primary concern regardless the site of choice is to maintain stability throughout the experiment and keep the mounted area free of movements that can be caused by breathing or heartbeat - especially when imaging at sub-micron resolution - without compromising the normal physiology of the tissue.
Figure 1.
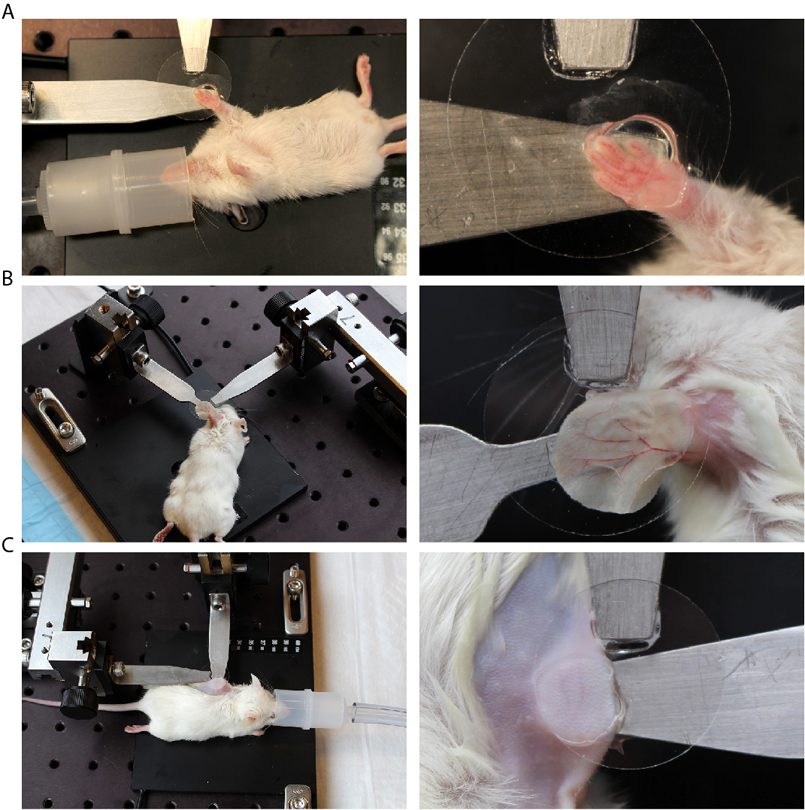
Example of a mounting apparatus for immobilizing (A) the paw, (B) ear and (C) back skin of an anesthetized mouse for intracutaneous imaging.
For mounting, a depilated area of the skin is placed between a flat stable surface and a coverslip. We frequently use the ear because it offers a stable platform for mounting and manipulation. It should be noted that skin in the ear pinna is particularly thin and has hair follicles that display unique cycling characteristics; however, the area connecting the ear to the head more closely resembles the back skin and can be used instead. The use of a coverslip is preferred for two main reasons. First, the coverslip applies downward pressure to the mounted area of the skin and contributes to maintaining overall stability (however, too much pressure can inhibit normal blood flow, so caution should be exercised). Second, the coverslip flattens the surface of the skin, creating a single, starting focal plane from which optical sectioning can commence towards the deeper layers of the tissue. Most objective lenses have a correction collar to minimize spherical aberration caused by the different refractive index of the coverslip.
Visualization of stem cell activity in the interfollicular epidermis
The interfollicular epidermis is particularly attractive for live imaging studies, due to its stratified organization and thickness, which is less than 50 μm in the mouse skin and well within the range of what can be effectively resolved by 2-photon microscopy. To capture the activity of the basal epidermal progenitors that maintain the epidermis the first step is to choose the appropriate signal by which these cells can be visualized and distinguished among the general population. In recent years, a great number of transgenic mouse lines expressing exogenous fluorescent proteins that are suitable for skin research have been reported and many of them are available through commercial repositories. The keratinocytes in the interfollicular epidermis express different genes depending on their differentiation state. For example, keratins 5 and 14 are primarily expressed in the basal layer while involucrin and keratin 10 are expressed in committed keratinocytes undergoing differentiation (33–36). Fluorescent proteins expressed under the control of the respective gene promoters can serve as markers to visualize the individual cell types (37,38).
The use of constitutively expressed markers is not sufficient to study the activity and fate of individual stem cells in the epidermis. Instead a lineage tracing strategy can be implemented which typically involves the use of an inducible Cre-recombinase allele and a Cre reporter allele combined in the same mouse (39–42). Limited induction of the Cre-recombinase activity enables the stable expression of a fluorescent marker in few randomly distributed cells as well as their progeny. By monitoring labeled cells over time one can directly assess their self-renewal and differentiation capacity in vivo. Furthermore, the unique morphology and stratified tissue organization allows the unambiguous determination of the cell fate since the differentiation state of individual keratinocytes can be easily assessed based solely on their morphology and location within the different layers of the epidermis.
For these experiments, we have chosen a Cre reporter mouse strain which expresses a membrane-bound red fluorescent protein in all cells including the epidermal keratinocytes (43; Fig. 2A). Optical sectioning by 2-photon microscopy can easily resolve all the different layers of the epidermis. We combined this Cre reporter with an inducible Cre-recombinase allele that is expressed under the control of the keratin 14 promoter (44). Limited induction removes the floxed sequence of the red fluorescent tag along with the stop codon, which in turn induces the expression of a membrane-bound green fluorescent protein. Two days after induction the epidermis of the treated mouse is imaged and serial optical sections are acquired at 1–2 μm steps to capture the entire volume of the epidermis; starting from the surface until the bottom of the basal layer is reached. This is marked by the appearance of collagen-induced SHG signal. On this first time-point in the series single green-labeled basal keratinocytes that have undergone recombination can be visualized. By identifying and revisiting the exact same areas of the epidermis at regular intervals over the next few days, the same process can be repeated in order to directly evaluate the fate of the traced cells (26,27). At the end of the time-course the epidermal 3-D volumes can be reconstituted from the acquired image stacks of each time point and the activity of individual stem cells can be retrospectively evaluated.
Figure 2.
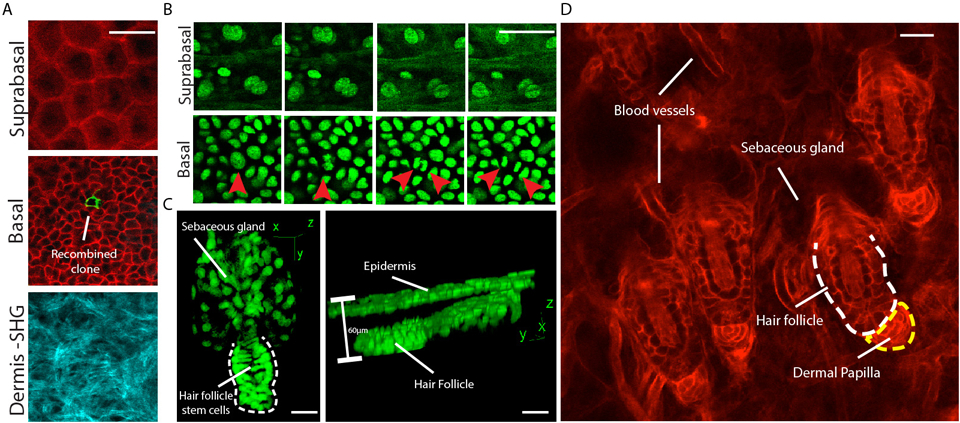
(A) Single optical planes depicting different layers of the interfollicular epidermis. A Cre reporter mouse strain was used which ubiquitously expresses a membrane-bound RFP that converts to membrane-bound GFP (mTmG) after recombination from an inducible Keratin 14-driven Cre recombinase. (B) Time-sequence of keratinocyte activity in the corresponding basal and suprabasal layers of the interfollicular epidermis. A mouse strain was used that expresses GFP fused to histone H2B under the control of the Keratin 14 promoter. Timepoints are 20 min apart and red arrowheads point to a stem cell division in the basal layer. (C) 3D renderings of Telogen hair follicles. (D) A single optical plane from within the dermis showing a group of hair follicles and other cellular elements of the skin. Scale bars: 20 μm.
Visualization of stem cell activity in the hair follicle
Hair follicles are self-contained mini-organs of the mammalian skin with a resident stem cell pool that is sufficient to periodically and stereotypically regenerate new hair appendages throughout life (45). In this process, which is known as the hair cycle and comprises of quiescent, growth and destruction phases (46); a small pool of hair follicle stem cells (47) generate a large number of progenitors which in turn differentiate into the seven lineages that make up the mature and growing hair shaft (38,48). A key question is how stem cells in the hair follicle contribute to the hair regeneration process. Using 2-photon microscopy we previously established the ability to visualize the process of hair regeneration in the intact skin of live mice, in real-time but also long term (8,26).
Dynamic stem cell activity taking place during hair regeneration can be visualized in the mouse, in real time by 2-photon time-lapse imaging. Since time-lapse imaging involves excitation of the sample at frequent intervals, a strong fluorescent reporter resistant to photo-bleaching is generally preferred. Furthermore a reporter that specifically localizes to the cell nucleus is also beneficial since it allows neighboring cells to be spatially resolved. Mice are chosen based on their respective age, since hair follicles are synchronized during the first 6–7 weeks of age and the time of each phase of the hair cycle can be predicted with relative high accuracy. Serial optical sections are typically acquired in 2–3 μm steps at a range that encompasses the entire volume of the hair follicles within the field of view; typically between 50–150 μm depending on the growth phase. The process is repeated at regular intervals for a desired total duration, which can extend to several hours depending on the imaging conditions, the state of anesthesia and the overall health of the mouse during the course of the experiment. Acquired data can be edited after the imaging session is concluded. Corresponding frames from each time point at the desired z-plane are arranged sequentially in a time series to form a movie, which can be qualitatively or quantitatively analyzed (Fig. 2B).
To elucidate the long-term contribution of hair follicle stem cells to hair growth a lineage tracing approach can be implemented and combined with frequent re-visits of the same hair follicles during the regeneration process (26,27). In such experiments and due to the unique physiology of the hair follicle, it’s periodic regeneration process and the major morphogenetic changes that occur at different stages of the hair cycle, the timing of induction is most critical. For this we take advantage of the synchronized cycling of hair follicles in juvenile mice and treat with Tamoxifen at post-natal day 20, when the first hair cycle is concluded and the hair follicles are suspended for a brief time in their most minimal state during the first Telogen phase (Fig. 2C,D). A number of different promoters can be used at this stage to drive the expression of the Cre recombinase and label individual hair follicle stem cells, including Keratin 15, Keratin 19 and Lgr5 (48–50). The contribution of these stem cells can be evaluated by re-visiting the same follicles once they have entered the Anagen phase and by visualizing the localization of their progeny. Since a growing hair follicle is organized in concentric layers of basal undifferentiated and suprabasal differentiated lineages, each with a unique morphology, the differentiation state of each stem cell lineage and therefore their particular contribution to the regeneration process can be accurately assessed. Such initial studies showed that the Telogen hair follicle consists of a heterogeneous pool of progenitor cells, each with a unique activity and contribution during the hair cycle (26).
Considerations and future perspectives
2-photon microscopy is a powerful but relatively new tool in the arsenal of skin and stem cell biologists and as such there are certain considerations and limitations to be taken into account as well as tremendous opportunities for further development of the technique. Even though the method does not require any invasive procedures to visualize elements of the live mouse skin, the tissue as well as the animal are subject to preparatory treatments, such as chemical depilation, mechanical pressure and anesthesia, to name a few, which may have the potential to alter the normal physiology of the tissue. Appropriate controls should always be considered to minimize such concerns. Furthermore, as in most types of optical microscopy a source of signal, usually in the form of an exogenously expressed fluorescent protein, is required for visualization. This can limit the cell types within the skin that can be distinguished or introduce biases towards particular subpopulations, especially when inducible reporter systems are used. Even thought there is currently a limited list of transgenic mouse lines that can be used for live imaging of the skin the list is destined to expand rapidly, especially with the introduction of new genome editing tools such as CRISPR/Cas9 (51). Last, since the method is currently optimized for the murine system due to its genetic amenability questions will always persist regarding relevance to the human skin biology. Live imaging of intact human skin is indeed possible but relies exclusively in indiscriminate contrast mechanisms with low to no cell specificity. A possible adaptation of the technique would employ organotypic cultures established from human primary cells or organ-cultured explants from adult human skin that can easily be genetically manipulated to introduce a variety of fluorescent reporters (52–55). It is certain that broader adaptation of 2-photon microscopy and live imaging approaches will greatly augment dermatological research in years to come.
Supplementary Material
Acknowledgements
P.R. is supported by a Research Scholar Award from the American Skin Association. This work was partially supported by a Pilot and Feasibility grant from the University of Pennsylvania Skin Biology and Diseases Resource-based Center (NIAMS; P30 AR069589). We would like to thank Elaine Fuchs (Rockefeller University) for providing the K14-H2BGFP mice.
Footnotes
Conflicts of Interests
The Authors declare no relevant conflicts of interest.
References
- 1.Hsu Y-C, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat. Med 2014. August6;20(8):847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rompolas P, Greco V. Stem cell dynamics in the hair follicle niche. Semin. Cell Dev. Biol 2013. December17;:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solanas G, Benitah SA. Regenerating the skin: a task for the heterogeneous stem cell pool and surrounding niche. Nat. Rev. Mol. Cell Biol 2013. November;14(11):737–48. [DOI] [PubMed] [Google Scholar]
- 4.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012. March;12(3):170–80. [DOI] [PubMed] [Google Scholar]
- 5.Garza LA, Yang CC, Zhao T, Blatt HB, Lee M, He H, Stanton DC, Carrasco L, Spiegel JH, Tobias JW, Cotsarelis G.Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest. 2011. February;121(2):613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plikus MV, Gay DL, Treffeisen E, Wang A, Supapannachart RJ, Cotsarelis G. Epithelial stem cells and implications for wound repair. Semin. Cell Dev. Biol 2012. December;23(9):946–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonart T, Heenen M, Lejeune O. Epidermal kinetic alterations required to generate the psoriatic phenotype: a reappraisal. Cell Prolif. 2010. June;43(3):321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012. July26;487(7408):496–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi M, Kwok SJJ, Yun SH. In vivo fluorescence microscopy: lessons from observing cell behavior in their native environment. Physiology (Bethesda). 2015. January;30(1):40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denk W, Strickler J, Webb W. Two-photon laser scanning fluorescence microscopy. Science. 1990. April6;248(4951):73–6. [DOI] [PubMed] [Google Scholar]
- 11.Curley PF, Ferguson AI. Actively mode-locked Ti:sapphire laser producing transform-limited pulses of 150-fs duration. Opt Lett. 1991. July1;16(13):1016–8. [DOI] [PubMed] [Google Scholar]
- 12.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol 2003. November;21(11):1369–77. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: new spectral windows for biological nonlinear microscopy. Proc. Natl. Acad. Sci. U.S.A 1996. October1;93(20):10763–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickinson ME, Simbuerger E, Zimmermann B, Waters CW, Fraser SE. Multiphoton excitation spectra in biological samples. J Biomed Opt. 2003. July;8(3):329–38. [DOI] [PubMed] [Google Scholar]
- 15.Koester HJ, Baur D, Uhl R, Hell SW. Ca2+ fluorescence imaging with pico- and femtosecond two-photon excitation: signal and photodamage. Biophys. J 1999. October;77(4):2226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.König K, So PT, Mantulin WW, Tromberg BJ, Gratton E. Two-photon excited lifetime imaging of autofluorescence in cells during UVA and NIR photostress. J Microsc. 1996. September;183(Pt 3):197–204. [PubMed] [Google Scholar]
- 17.Sabino CP, Deana AM, Yoshimura TM, da Silva DFT, França CM, Hamblin MR, et al. The optical properties of mouse skin in the visible and near infrared spectral regions. J. Photochem. Photobiol. B, Biol 2016. April3;160:72–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth S, Freund I. Optical second-harmonic scattering in rat-tail tendon. Biopolymers. 1981. June;20(6):1271–90. [DOI] [PubMed] [Google Scholar]
- 19.Zoumi A, Yeh A, Tromberg BJ. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc. Natl. Acad. Sci. U.S.A 2002. August20;99(17):11014–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masters B, So P, Gratton E. Multiphoton excitation fluorescence microscopy and spectroscopy of in vivo human skin. Biophysical journal. 1997. June1;72(6):2405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S-J, Jee S-H, Dong C-Y. Multiphoton microscopy: a new paradigm in dermatological imaging. European journal of dermatology : EJD 2007. September1;17(5):361–6. [DOI] [PubMed] [Google Scholar]
- 22.Paoli J, Smedh M, Ericson MB. Multiphoton laser scanning microscopy--a novel diagnostic method for superficial skin cancers. Semin Cutan Med Surg. 2009. September;28(3):190–5. [DOI] [PubMed] [Google Scholar]
- 23.Cicchi R, Kapsokalyvas D, Pavone FS. Clinical nonlinear laser imaging of human skin: a review. Biomed Res Int. 2014;2014:903589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yew E, Rowlands C, So PTC. Application of Multiphoton Microscopy in Dermatological Studies: a Mini-Review. J Innov Opt Health Sci. 2014. January3;7(5):1330010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huck V, Gorzelanny C, Thomas K, Getova V, Niemeyer V, Zens K, et al. From morphology to biochemical state - intravital multiphoton fluorescence lifetime imaging of inflamed human skin. Sci Rep. 2016;6:22789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013. October24;502(7472):513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pineda CM, Park S, Mesa KR, Wolfel M, Gonzalez DG, Haberman AM, et al. Intravital imaging of hair follicle regeneration in the mouse. Nat Protoc. 2015. July;10(7):1116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JL, Goh CC, Keeble JL, Qin JS, Roediger B, Jain R, et al. Intravital multiphoton imaging of immune responses in the mouse ear skin. Nat Protoc. 2012Feb;7(2):221–34. [DOI] [PubMed] [Google Scholar]
- 29.Hiratsuka T, Fujita Y, Naoki H, Aoki K, Kamioka Y, Matsuda M. Intercellular propagation of extracellular signal-regulated kinase activation revealed by in vivo imaging of mouse skin. Elife. 2015. January1;4:e05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinselmeyer BH, Lynch JN, Zhang X, Aoshi T, Miller MJ. Video-rate two-photon imaging of mouse footpad - a promising model for studying leukocyte recruitment dynamics during inflammation. Inflamm Res. 2008. March;57(3):93–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lammermann T, Afonso PV, Angermann BR, Wang JM, Kastenmuller W, Parent CA, et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013. June;498(7454):371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng LG, Qin JS, Roediger B, Wang Y, Jain R, Cavanagh LL, et al. Visualizing the neutrophil response to sterile tissue injury in mouse dermis reveals a three-phase cascade of events. J. Invest. Dermatol 2011. October;131(10):2058–68. [DOI] [PubMed] [Google Scholar]
- 33.Coulombe PA, Kopan R, Fuchs E. Expression of keratin K14 in the epidermis and hair follicle: insights into complex programs of differentiation. J. Cell Biol 1989. November;109(5):2295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrne C, Fuchs E. Probing keratinocyte and differentiation specificity of the human K5 promoter in vitro and in transgenic mice. Mol. Cell. Biol 1993. June;13(6):3176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs E, Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980. April;19(4):1033–42. [DOI] [PubMed] [Google Scholar]
- 36.Watt FM. Involucrin and other markers of keratinocyte terminal differentiation. J. Invest. Dermatol 1983. July;81(1 Suppl):100s–3s. [DOI] [PubMed] [Google Scholar]
- 37.Kasparek P, Krenek P, Buryova H, Suchanova S, Beck IM, Sedlacek R. Transgenic mouse model expressing tdTomato under involucrin promoter as a tool for analysis of epidermal differentiation and wound healing. Transgenic Res. 2012. June;21(3):683–9. [DOI] [PubMed] [Google Scholar]
- 38.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science. 2004. January16;303(5656):359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol 2001;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001. January1;97(1):324–6. [DOI] [PubMed] [Google Scholar]
- 41.Van Keymeulen A, Blanpain C. Tracing epithelial stem cells during development, homeostasis, and repair. J. Cell Biol 2012. May 28;197(5):575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kretzschmar K, Watt FM. Lineage tracing. Cell 2012. January20;148(1–2):33–45. [DOI] [PubMed] [Google Scholar]
- 43.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007. September;45(9):593–605. [DOI] [PubMed] [Google Scholar]
- 44.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U.S.A 1999. July20;96(15):8551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paus R, Cotsarelis G. The biology of hair follicles. N. Engl. J. Med 1999. August12;341(7):491–7. [DOI] [PubMed] [Google Scholar]
- 46.Müller-Röver S, Handjiski B, van der Veen C, Eichmüller S, Foitzik K, McKay IA, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol 2001. July;117(1):3–15. [DOI] [PubMed] [Google Scholar]
- 47.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990. June29;61(7):1329–37. [DOI] [PubMed] [Google Scholar]
- 48.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol 2004. April;22(4):411–7. [DOI] [PubMed] [Google Scholar]
- 49.Means AL, Xu Y, Zhao A, Ray KC, Gu G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008. June;46(6):318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet 2008. November;40(11):1291–9. [DOI] [PubMed] [Google Scholar]
- 51.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013. February15;339(6121):819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiede S, Koop N, Kloepper JE, Fässler R, Paus R. Nonviral in situ green fluorescent protein labeling and culture of primary, adult human hair follicle epithelial progenitor cells. Stem Cells. 2009. November;27(11):2793–803. [DOI] [PubMed] [Google Scholar]
- 53.González-González E, Kim Y-C, Speaker TJ, Hickerson RP, Spitler R, Birchall JC, et al. Visualization of plasmid delivery to keratinocytes in mouse and human epidermis. Sci Rep. 2011;1:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simpson CL, Kojima S-I, Getsios S. RNA interference in keratinocytes and an organotypic model of human epidermis. Methods Mol. Biol 2010;585:127–46. [DOI] [PubMed] [Google Scholar]
- 55.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med 2010. December;16(12):1450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


