Abstract
Poly(ADP-ribosyl)ation (PARylation) is a reversible post-translational modification regulating various biological pathways including DNA damage repair (DDR). Rapid turnover of PARylation is critically important for an optimal DNA damage response and maintaining genomic stability. Recent studies show that PARylation is tightly regulated by a group of enzymes that can erase the ADP-ribose (ADPR) groups from target proteins. The aim of this review is to present a comprehensive understanding of dePARylation enzymes, their substrates and roles in DDR. Special attention will be laid on the role of these proteins in the development of cancer and their feasibility in anticancer therapeutics.
Overview
ADP-ribosylation is a dynamically regulated and reversible post-translational modification that plays an important role in numerous biological processes such as DNA damage repair [1] [2] [3]. It is predominantly catalyzed by poly(ADPR) polymerases (PARPs) which are a17 member protein superfamily [4]. These enzymes use nicotinamide adenine dinucleotide (NAD+) as co-substrate and covalently attach ADPR moiety onto their target proteins with nicotinamide as a byproduct during the chemical reaction [5]. Some PARP family members including PARP1, PARP2, PARP5A (aka tankyrase 1) and PARP5B (aka tankyrase 2) can add additional ADPR moieties on the first ADPR, leading to the formation of ADP-ribosyl polymer (PARylation) [6]. In each ADPR unit, both distal ribose and ribose close to adenine can be covalently linked with the distal ribose of another ADPR unit via α(1→2) O-glycosidic bonds. Thus, the ADPR polymers form both linear or branched chains (Figure 1) [7] [8]. In addition, due to lack of key catalytic residues, PARPs including PARP3, PARP4, PARP6, PARP10, PARP12, PARP14, PARP15 and PARP16 can only attach one ADPR moiety onto their target proteins for mono(ADP-ribosyl)ation (MARylation) [1], whereas PARP9 and PARP13 are enzymatically inactive and cannot catalyze ADP-ribosylation [9] [10] [11].
Figure 1.
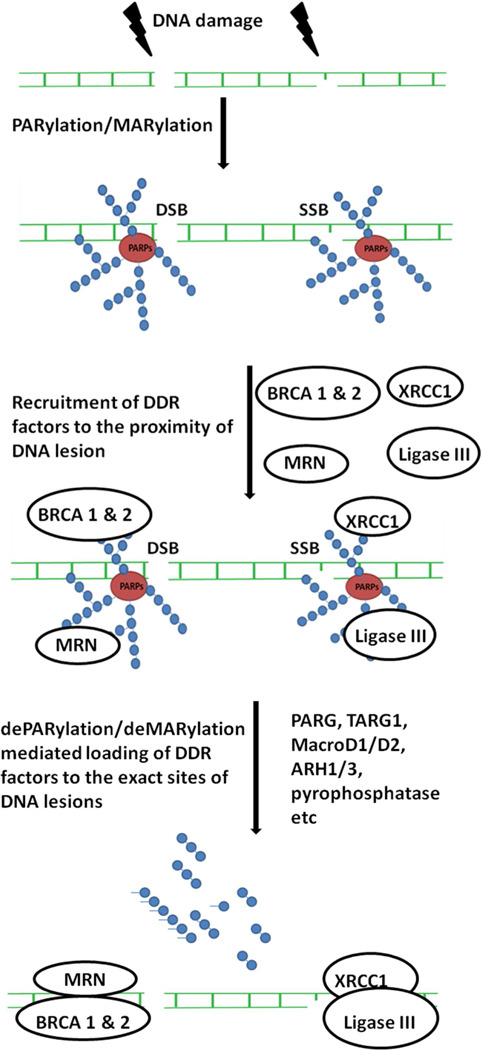
Schematic diagram showing that DNA damage-induced PARylation and dePARylation are sequential steps to mediate the recruitment of DDR response proteins. Both single-stranded (SSB) and double-stranded (DSB) breaks on the target DNA are shown. All the poly(ADP-ribose) polymerases (PARPs) are represented as a single color bubble along with the attached PAR chains. Branched, linear PAR chains and mono-ADPR moieties attached to the PARPs are shown. The DNA damage response (DDR) proteins are recruited to the vicinity of the DNA breaks by the ADP-ribosylation. The ADP-ribosylation signals are removed by dePARylation/deMARylation enzymes as shown leading to the recruitment of DDR proteins to the actual damaged site.
ADP-ribosylation acts as a signal during DDR by recruiting DDR machinery via recognition of ADPR at DNA lesions (Figure 1). Upon DNA damage, PARP1 acts as a DNA damage sensor to recognize single and double-strand break ends, and initiates PARylation quickly at DNA lesions (Figure 1) [12]. Accumulated evidence shows that PARP1 accounts for 80 – 90 % of DNA damage-induced PARylation, and the PAR chains can extend to more than 100 units on the substrates including PARP1 itself, histones and other chromatin associated proteins [4]. In addition to PARP1, others PARPs, such as PARP2 and PARP3, are also recruited to DNA lesions and facilitate PARylation on chromatin remodelers [1]. The amino acid residues modified by ADP-ribosylation include glutamic acid, aspartic acid, lysine, arginine, serine and cysteine (Figure 2) [12] [13] [14] [15]. PARylation is considered as the first wave of signals at DNA lesions to mediate the recruitment of DNA damage repair machinery [12].
Figure 2.
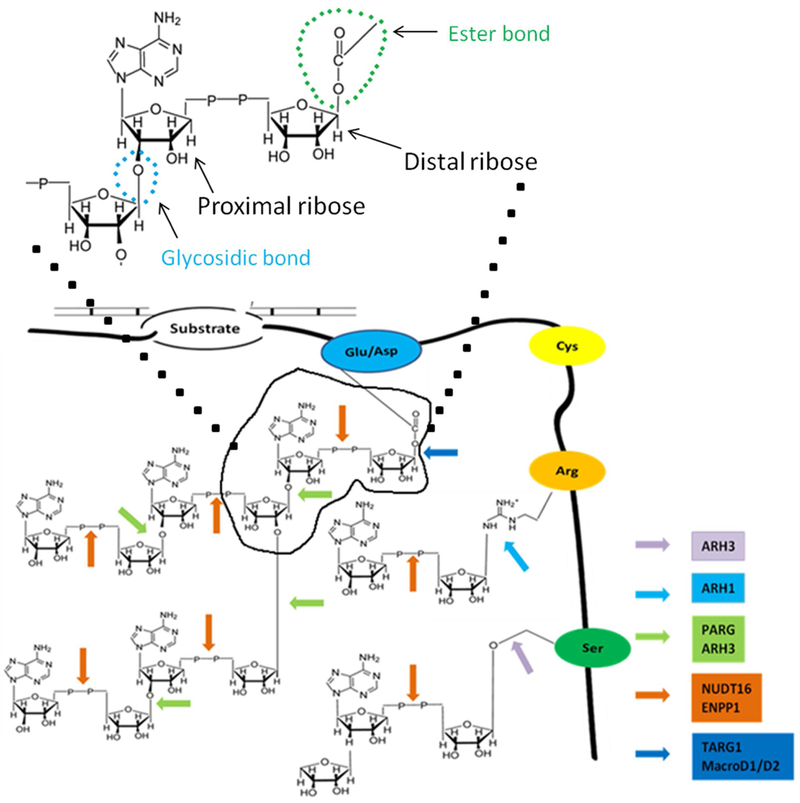
Schematic diagram depicting dePARylation and deMARylationon on a target protein. Ester bond, glycosidic bond, proximal and distal ribose in an ADPR are depicted in the top panel. The enzymes involved in the degradation of the modifications are shown with different color arrows. The arrows point to the bonds digested by the corresponding enzyme. The acceptor protein is shown along with an embedded peptide chain containing different amino acids as beads with their names as depicted. PAR polymer chain with a branch site and MAR sites on the target protein are also depicted. The damaged DNA is depicted on top of the substrate.
However, ADP-ribosylation has to be removed quickly and effectively so that the recruited DDR factors can act on the damaged DNA and genetic homeostasis is maintained. Otherwise, those ADPR-binding DDR factors will be trapped at the vicinity of DNA lesions. The removal of ADP-ribosylation is mediated by dePARylation enzymes (Figure 1) (Table 1). For hydrolysis of ADPR polymer, it is primarily catalyzed by poly(ADPR) glycohydrolase (PARG). PARG digests the glycosidic bonds in the PAR chains and releases ADPR residues [16]. However, the last ADPR moiety attached to the PARylated protein by an ester bond is resistant to PARG [17]. Instead, other ADPR hydrolases including MACROD1/D2, TARG1 and ARH1/3 cut the last ADPR residue or digest MARylation from target proteins (Figure 1) [18] [4]. Here, we will discuss detailed structure and function of these dePARylation enzymes as the downstream effectors of ADP-ribosylation during DNA damage response and cancer therapy.
Table 1.
ADPribosylation degrading proteins, functions and inhibitors
| Name | Function* | Inhibitor/s |
|---|---|---|
| PARG | dePARylation deMARylation, deMARylation from DNA, | Salicylanilides[104] |
| RBPI-3 [103] | ||
| Gallotannin[122] | ||
| PDD00017273[108] | ||
| TARG1 | dePARylation, OAADPr hydrolysis, deMARylation from DNA, | No |
| MacroD1 | deMARylation from Asp and Glu, OAADPr hydrolysis | No |
| MacroD2 | deMARylation from Asp and Glu, OAADPr hydrolysis, deMARylation from DNA, | No |
| ARH1 | deMARylation from Arg | No |
| ARH3 | dePARylation, deMARylation, deMARylation from Ser, deMARylation from DNA, | No |
| ENPP1 | deMARylation from DNA, phosphatase, | No |
| NUDT16 | Phosphatase, release PAR from proteins | No |
| NUDT5 | Phosphatase-acts on free OADPr | TH5427119 |
See text for details
DePARylation in DNA damage repair
PARylation is dynamically regulated at DNA lesions. Following PARylation, dePARylation occurs at DNA lesions to release DNA damage repair factors, so that these DNA damage repair factors can be deposited on the DNA lesions [12]. Otherwise, the DDR machinery will be trapped at the vicinity of DNA lesions and repair will be impaired. Thus, dePARylation occurs very quickly at DNA lesions. It has been shown that the half-life of PARylation at the DNA lesions is only a few minutes due to its turnover by a powerful dePARylation [19]. To date, several dePARylation enzymes have been identified (Figure 2). Based on their hydrolysis targets, we categorized them into PARG, deMARylase and pyrophosphatase.
PARG
The DNA damage-induced PARylation is primarily terminated by PARG [20] [21], the key dePARylase which in humans is a 976 amino acids long protein. PARG superfamily includes 55kDa hPARG55, 60kDa hPARG60, 111kDa (hPARG111), 102 kDa (hPARG102) and 99 kDa (hPARG99) proteins formed due to alternative splicing of the parent mRNA [22] [23]. It is an evolutionarily conserved enzyme whose sequence is highly conserved especially in C terminal catalytic region across all kingdoms from prokaryotes to human [24] [25]. The N terminal regulatory region whose sequence varies between different species is involved in the recruitment of PARG to the sites of DNA damage [26]. The crystal structure of Tetrahymena thermophila PARG catalytic domain was determined recently in association with PAR chain [25]. The first human PARG structure was solved in complex with an ADPR dimer substrate. The crystal structure suggests that the adenine moiety of ADPR interacts with Tyr795 which positions the ribose-ribose glycoside bond near the catalytic site. The catalytic region is contributed by Glu755, Glu756 and Asp737 flanked by three Gly residues [27].
The catalytic domain of PARG is an endo and exo glycosidase and digests 1’ −2’ glycosidic bond linked between two ADPR units in the PAR chain. However, the endo-glycosidase property of PARG is stronger (100 times) than its exo-glycosidase activity. Moreover, linear and larger PAR chains tend to be hydrolyzed with higher affinity (Km < 0.3 μM) than shorter or branched polymers (Km≈10 μM) [28] [29]. Besides PAR length, PARG activity is also regulated by the cellular concentration of PAR. The degradation of PAR by PARG involves a biphasic decay in which the earliest phase small chains and monomeers are generated from the longer PAR chains. The second phase which is proceeds slowly than the first phase involves exo PARG activity. The endo activity of PARG is predominantly observed on long PAR chains and this activity is limited by the length of a PAR chain. The endo activity is completely abolished in a PAR chain containing less than 40 ADPr units [28] [29] [30]
Thus, digestion of AR by PARG generates both ADPR moieties and shorter PAR chains from the terminal and internal PAR residues respectively.
Although PARG was identified in 1971 [31], purification of PARG were unsuccessful until recently because PARG readily undergoes proteolysis during purification. In addition, the cellular level of PARG is very low (2,000 molecules per cell) [28] [32] compared to that of PARP1 (200,000 molecules per cell [33] ; however, unlike PARP1 which switches from active to inactive form depending on its binding to DNA ends [34], PARG is constitutively active [28]. The PARG hydrolysis activity is 50–70 times stronger than the polymerization potential of PARP [19]. The short half-life (few minutes) of PAR polymers is mainly owing to the strong enzymatic activity of PARG [19]. PARG hydrolyzes both linear and branched PAR chains and functions as the major enzyme responsible for rapid PAR turnover [35]. However, PARG cannot break the ester bond holding the first ADPR to the proteins and the removal of the most proximal ADPR moieties functions as the rate-limiting step in PARylation-dePARylation switch [36] [17] [37].
A coordinated PARP-PARG pathway is the hallmark of a productive DNA damage response. PARG is recruited to the DNA damage sites immediately after the formation of PAR chains. The recruitment is mediated via the interaction between the catalytic domain of PARG and PAR chains [17] as well as its association with PCNA [38]. The interaction between PARG and PCNA is guided by the acetylation of K409 on PARG, which assists in the recruitment of PARG to the sites of DNA breaks [39]. Upon recruitment to the damaged site, PARG immediately degrades the PAR chains on PARP1. Loss of PAR on the PARP1 favors its ubiquitination by E3-ligase CHFR and consequent proteasomal degradation [40]. Nevertheless, mouse embryonic fibroblasts (MEFs) with knockdown of PARG are hypersensitive to DNA damage, suggesting that PARG-mediated dePARylation is required for DNA damage repair. PARG knockdown cells show suboptimal recruitment of XRCC1 to the damaged site which in turn severely affects the signature signal of DNA damage i.e. H2AX phosphorylation. Thus, PARG-deficient cells have compromised SSB and DSB repair [41]. In our recent studies on PARG deficient cells, we observed that PARylation is remarkably prolonged at DNA lesions, which traps DNA damage repair machinery on the long PAR chains (unpublished results from X.Y). It could be one of the major reasons explaining the critical role of PARG-mediated erasing of the PARylation for cell survival wherein the depletion of PARG leads to embryonic lethality in mice due to accumulated PAR chains [42].
In addition to directly participating in DNA damage repair, PARG also indirectly regulates DDR via other cellular events. For example, once PAR is digested, PARG is also involved in the conversion of ADPR into ATP in association with ADPR pyrophosphorylase (ARPP) [43]. The ATP generated by these enzymes at the vicinity of a damaged site is required for optimal DNA ligation. Furthermore, due to the negative charge of PAR, PAR polymers drive chromatin relaxation and favor transcription [44]. Thus degradation of PAR by PARG restores chromatin conformation. Moreover, PARG reaction products i.e. oligo PAR and ADPR can function as cell death signaling molecules [45]. These metabolites will be particularly enriched inside the cells in case of PAPR1 hyperactivation. PARP1 hyperactivation can be occurring not only by DNA damage but also by hypoxia, hypoglycemia, oxidative stresses etc [46]. PARP1 mediated PAR chains will be rapidly degraded by PARG producing huge amounts of oligo PAR and ADPR. This leads to cell death in a caspase independent pathway known as parthantos. PARP1 depletion will rapidly exhaust the cellular NAD+ reservoirs depriving the cells of its energy resource [47] [35]. The oligo PAR chains formed translocate from the nucleus into mitochondria. Once, inside the mitochondria, PAR chains release apoptosis inducing factor (AIF) from the inner mitochondrial membrane. The truncated AIF localizes to the nucleus, interacts with H2AX and initiates DNA fragmentation. Moreover, apoptosis is assisted by the ADPR moieties through its cyclic intermediate cADPR. cADP mobilizes Ca2+ within the cells due to the activation of calcium channels like TRPM2, disturbs calcium homeostasis which in turn favors truncation of AIF [48]. The cell death pathway mediated by PAR is majorly controlled by the PAR/PARG ratio which in turn directly controls the exo-endoglycosidase catalysis of PARG. Under normal cellular metabolism, PARG functions mainly as an exo-glycohydrolase. However, irreparable cellular insults lead to formation of huge amounts of PAR which are rapidly degraded by endo-glycohydrolase property of PARG and the endo-activity is itself activated by the PAR lengths with increased PAR chains formed under increased DNA damage. The endo-glycohydrolase activity leads to a further increase in oligo PAR chains favoring cell death [25]. A recent report suggests that ADP-ribose analogs like 2′-deoxy-ADPR formed by the hydrolysis of PAR by PARG can function as more potent activators of TRPM2 channels than ADP-ribose itself [49]. Two research groups have published data suggesting that apart from PAR, PARG can hydrolyze mono-ADP ribosylation (MAR) on terminal phosphate groups on DNA. DNA mono-ADP ribosylation is reportedly catalyzed by PARP1, PARP2 and PARP3 on complex DNA breaks under in-vitro conditions [50] [51] [52]. PARG also interacts with RNA binding protein HuR. A research group recently suggested that PARG mRNA interacts with RNA binding proteins HuR may explain a possible mechanism of resistance to PARP inhibitor treatment. The authors found that olaparib treatment lead to the binding of HuR to the PARG mRNA in 3’ UTR. The binding favors PARG upregulation which in turn will be associated with an increased frequency of DDR due to the digestion of PAR [53] Collectively, PARG plays a key role in regulating DDR.
DeMARylation by macro domain containing enzymes including Terminal ADPR protein glycohydrolase 1 (TARG1/OARD1/C6orf130), MacroD1 and MacroD2.
Macro domain containing proteins are highly conserved and identified in viruses, archaea, bacteria, plants and mammals. In humans, so far eleven macrodomain containing proteins have been reported [54] TARG1, MacroD1 and MacroD2 belong to the family of macrodomain containing proteins which remove MARylation or the last ADPR moiety in PAR chain that is linked to Asp or Glu. TARG1 indeed was initially identified as the enzyme involved in the removal of ADP-ribose from OAADPr [55]. The role of these three enzymes in the removal of mono-ADPR from proteins was simultaneously reported by two independent groups in 2013 [56] [57]. It was observed that PARP10 mediated MARylation was completely reversed by treatment with MacroD1, MacroD2 or TARG1 [56]. Consequently, redundant deMARylation of the terminal ADPR by these enzymes culminates the dePArylation signaling pathway.
TARG1 (177 amino acids) catalysis involves binding of highly conserved Lys84 on TARG1 with ADPR. Lys84 then mediates a nucleophilic attack on the C1’ atom of the distal ribose leading to the formation of the Lys84-ADPR intermediate and regenerates an unmodified protein. This intermediate is then cleaved by another conserved residue Asp125-mediated catalysis. To support this molecular mechanism, mutations of Asp125 or Lys84 on TARG1 abolish the enzymatic activity. Moreover, mutations in Lys84 significantly decreased the recruitment of TARG1 to the site of DNA damage, suggesting that this residue mediates the recognition of ADPR at DNA lesions [18]. Additionally, knockdown of TARG1 increases the sensitivity of 293T cells to genotoxic stress. Homozygous mutations in TARG1 have been linked to the development of severe neurodegenerative diseases in humans. The predictive mutations in these patients involves the formation of truncated proteins lacking catalytic activity [58] TARG1 is recruited to the sites of DNA damage by PAR and is involved in the removal of terminal ADP ribose units attached to Glu and Asp. Moreover, TARG1 can release an entire PAR chain, albeit weakly, from an acceptor protein due to the hydrolysis of the ester bond holding the proximal ADPR with Asp or Glu on an acceptor protein. TARG1 mediated removal of ADPR and PAR chains from PARP1 suppresses the catalytic activity of PARP1 and ensures that the PARP1 protein returns back to the inactive state [58].
TARG1 shuttles between the nucleus and nucleolus, and the movement is governed by DNA damage response [59]. The shuttling is lost upon genotoxic stress by H2O2 wherein it predominantly localizes to the sites of DNA damage due to its binding with PAR. It also interacts with proteins involved in DNA damage response like XRCC1, XRCC2, XRCC5, LIG3, etc [59] [58].
MacroD1 (323 amino acids) and MacroD2 (425 amino acids) catalyze the hydrolysis of ultimate proximal ADPR left after the digestion of PAR by PARG. The catalytic macrodomain fold is almost identical in both proteins and both localize to the sites of DNA damage [18]. The catalytic process involved in the removal of ADPR from acceptor proteins has been elucidated for MacroD2 in conjugation with ADP. The catalytic region shares homologies with PARG catalytic domain (similar to macrodomain) in both binding and orientation of bound ADPR. ADPR is buried in a deep cleft in which the distal ribose bends towards the α-phosphate group. This constrained conformation is flanked by two glycine-rich loops and a water molecule in the region [56] [57]. Originally, it was proposed that the water molecule gets activated due to the ADPR and the activated water molecule carries a nucleophilic attack on the substrate [57] [25]. However, recent reports suggest that the water molecule in the catalytic site is activated by the Asp102, which in turn carries out a nucleophilic attack on the C1′ atom of the distal ribose of ADPR, thus releasing the ADPR [56] [60].
Like MacroD1, MacroD2 also recognizes MARylation and removes ADPR from Asp or Glu [56]. It is recruited to the sites of DNA damage and the recruitment is also governed by ATM-mediated phosphorylation on MacroD2. Phosphorylation occurs on two serine residues within an SQ/TQ motif localized at the C terminal end of the protein. The phosphorylation leads to the export of MacroD2 from the nucleus to the cytoplasm [61]. Thus the sequestration of MacroD2 in the cytoplasm by phosphorylation could be a novel regulatory mechanism controlling ADPR removal. Notability, MacroD1 has been lately associated with the hydrolysis of ADPR on dsDNA from both 5’ and 3’ ends [62].
ADP-ribosyl-acceptor hydrolases (ARHs)
ADP-ribosyl-acceptor hydrolases is a protein family involved in the discharge of mono-ADPR moieties from proteins. ARH proteins play an important role in completely reversing PARylation, as PARG cannot remove the proximal ADPR units attached to the acceptor proteins. The ARH family consists of three members ARH1, ARH2 and ARH3. These proteins are encoded by three different genes each giving rise to an identical size (39kDa) protein sharing similar sequence. However, only ARH1 and ARH3 are known to participate in DNA damage response and the function and target substrates of ARH2 are still unknown [18] [4].
Mammalian ARH1(357 amino acids) is involved in the removal of α-ADP–ribose from the arginine residues of a target protein [63]. The ADP–ribose moiety is attached to the guanidine group of arginine by mono-ADP-ribosyltransferases (ART) using NAD+ as ADPR donor. Thus an N-glycosidic bond is formed between ADPR and arginine. Mammals contain seven ART proteins (ART1–7) and among them, ART1, 2, 5, 6 and 7 can catalyze ADP-ribosylation on arginine. ADPR being negatively charged changes the biological properties of the acceptor proteins. In addition, the ADPR group due to its bulky nature modulates the interaction of acceptor proteins with its cognate targets. The target substrates are mostly DDR proteins and the presence of ADPR increases their interaction with other downstream DDR proteins [64]. This is particularly true when actin polymerization is inhibited due to the attachment of ADP-ribose on arginine 177 in G-actin [65]. A well-studied function of arginine ADP-ribosylation is in the activation ATP gated channel P2X7 purinergic receptor by ADPR. Under normal conditions, the activation of P2X7 channel in murine T lymphocytes requires high extracellular ATP concentration (EC50 ~500μM). However, in presence of high NAD concentration, ART2.2 catalyzes ADP-ribosylation on R125 of P2X7 channel which activates the channel even under low ATP concentration and leads to cell death [66].
ADP-ribosylation cycle of arginine switches on and off the nitrogen fixation enzyme dinitrogen reductase in diazotrophic bacteria and controls nitrogen fixation [67]. Moreover, cholera-toxin from Vibrio Cholera functions like an ART and transfers ADP-ribose on the arginine residues of stimulatory guanine nucleotide binding protein (GS alpha) resulting in the activation of adenylyl cyclase. The activation leads to the formation of secondary messenger cAMP which in turn promotes secretion of fluids and electrolyte from the intestine of infected patients [68].
ARH3 (363 amino acids), also known as ADPRHL2 localizes to all cellular compartments i.e. nucleus, cytoplasm and mitochondria [69] . ARH3 has the highest ADPR hydrolysis activity among the ARH family proteins with majorities of ARH3 localized to the nucleus. The crystal structure of apo form was determined in conjugation with ADP and Mg 2+ ions at the catalytic site [69]. Recently, we and others further determined the structure of ARH3 with ADPR using X-ray crystallography [70] [71]. We observed that ARH3 monomer binds to an ADPR unit along with two Mg 2+ ions. ARH3 has a compact α-helix structure matching with other two ARH isoforms i.e. ARH1 and ARH2. The catalytic site contains two Mg2+ ions which are shifted 0.6 Å towards distal ribose when bound to ADPR. The catalytic site contains Glu41, a water351 molecule and multiple acidic residues functioning together as the predominant catalytic center and holding the ADPR by multiple hydrogen bonds. Yasin et al. described in detail the mechanism used by ARH3 in binding and hydrolysis of its substrate ADPR. Their results suggested that a conformational change in the ARH3-ADPR complex favors an open state structure in which the Glu41 motif region allows substrate binding. Once the ARH3 is present in unliganded conformation, the Glu41 motif region switches to an enzymatically inactive closed conformation [71].
Our studies suggest that ARH3 is recruited to the sites of DNA damage within 30 seconds after the damage. Moreover, we observed that three key residues S148A, Y149A and H182A are required for the damage induced recruitment and ARH3 with mutations in these residues fails in its recruitment. We induced DNA damage in U2OS cell line and observed that knockdown of ARH3 or key reside mutations in ARH3 severely impairs DNA damage repair kinetics [70]. ARH3 plays an important role in DNA damage repair by cleaving the glycosidic bond between ADPR and serine [72]. Serine ADP-ribosylation is one of the dominant forms of ADP-ribosylation deposited by PARP1 and PARP2 on histones and other proteins during DNA damage repair [73]. A huge proportion of human proteins have been reported to be modified by serine ADP-ribosylation [74] [75] [76], which can be removed by ARH3 treatment but not by PARG or MACRD1/D2 hydrolyases [72]. Moreover, endogenous levels of serine-ADP-ribosylation are increased in ARH3 knockout cells, and the normal levels of serine-ADP-ribosylation can be restored by an ectopic introduction of ARH3 [72].
Although cells lacking ARH3 retain DNA damage-induced ADP-ribosylation, ARH3−/− mice are viable [36]. ARH3−/− MEFs are sensitive to H2O2 treatment causing cell death by a caspase-independent pathway. ARH3−/− cells undergo nuclear shrinkage, chromatin condensation and display of phosphotidylserine on the cell surface. The cell viability in ARH3−/− MEFs post H2O2 treatment can be restored by PARP1 inhibitor treatment or siRNA mediated PARP1 knockdown, suggesting its important function downstream of the PARP1 pathway during DDR. The nuclear PAR level after the H2O2 treatment is higher in ARH3−/− MEFs than in wild type MEFs [36].
Pyrophosphatases
In addition to the enzymes that digest glycosidic bonds, recent studies have demonstrated the involvement of NUDIX superfamily and ENPP in PAR metabolism. NUDIX (“X” nucleoside diphosphates linked to variable moiety X) superfamily hydrolases are found across all kingdoms of life and are mostly involved in the hydrolysis of pyrophosphates. These proteins have a unique catalytic motif (Nudix box) structure containing 23 amino acids GX5EX7REUXEEXGU, where U represents an aliphatic or hydrophobic residue [77]. Some members digest the phosphate groups of free ADPR released following the digestion of PAR/MAR. NUDT16 (195 amino acids) is the only known NUDIX protein which can digest both PAR and MAR from an acceptor protein. However, being a pyrophosphatase, NUDT16 digestion product is different from the canonical enzymes involved in dePARylation or deMARylation i.e a ribose-5’-phosphate (R5P) is left on the target amino acid substrate [78]. Interestingly, two decades before the role of NUD16 was observed, a lysosomal storage disease causing progressive neurodegenerative disorder and renal failure was observed in a patient characterized by the accumulation of R5P containing proteins in lysosomes [79]. This suggests that these proteins could be a substrate for a NUDIX based dePARylation/deMARylation enzyme and identification of the corresponding NUDIX may help in understanding the molecular mechanism of this disease.
Another NUDIX, NUDT5 (232 amino acids) interacts with PAR and PARG besides degrading free ADPR generated after the digestion of PAR. The digestion product generates AMP and ribose-5’-phosphate (R5P) similar to NUDT16. A recent report suggests that both AMP, as well as ATP can be generated by NUDT5 catalysis which can be used for chromatin remodeling[43].
ENPP (Ectonucleotide Pyrophosphatase/Phosphodiesterase) (925 amino acids) is a non-NUDIX phosphodiesterase with broad specificity capable of digesting phosphodiester/pyrophosphate bonds. Recent studies suggest that ENPP participates in the digestion of PAR and MAR. ENPP catalysis is similar to NUDT16 and NUDT5 i.e. leaves behind R5P on the acceptor proteins. ENPP is localized to the extracellular membrane and its enzymatic activity may be involved in digestion of extracellular ADRR [80]. ENPP is localized to the extracellular membrane and its enzymatic activity may be involved in digestion of extracellular PAR; which can function in immune response [81]. Moreover, extracellular activities of ENPP1 can generate AMP and inorganic pyrophosphate which inhibits bone formation [82]. Loss of ENPP in Enpp1−/− mice significantly reduced the survival of long-lived plasma cells [83]; however, the relation between ENPP expression, survival and the toxic ADPR degradation is still unknown.
Targeting DePARylation/DeMARylation in cancer treatment
The rationale to identify novel therapeutic targets in PARylation-mediated DDR
DDR is required to maintain chromosome stability in response to DNA damaging insults by recruiting various DDR factors to the proximity of DNA lesions. Thus, suppression of PARylation impairs DNA damage repair and induces apoptosis of tumor cells with DNA damage repair defects. Based on the molecular mechanism of PARylation in DNA damage repair, various therapeutic interventions targeting PARylation are used for the treatment of cancers with DNA damage repair defects [84]. In addition, PAR molecules differ from other biopolymers due to the uniqueness of the bonding nature in the PAR chains. The polymers α(1→2) O-glycosidic bonds is an exclusively nature of PAR polymers and is not present in other bio-molecules. As such absolute specificity can be achieved by the inhibitors targeting proteins that hydrolyze the PAR chains.
Since PARP1 and PARP2 are enzymes that catalyze majorities of PARylation in response to DNA damage, most well-studied PARP inhibitors are targeted against PARP1 and PARP2 for the treatment of cancers with DNA damage repair defects such as BRCA tumors[85]. To date, olaparib, niraparib [86] and rucaparib [87] have been approved by the U.S. FDA for the treatment of stage IV ovarian cancers with BRCA1/2 mutations. In addition, olaparib has been approved for the treatment of triple-negative breast cancers with BRCA1/2 mutations [88] [84] [89].
The therapeutic success of olaparib in BRCA1/2 cancers led to a spike in the use of PARP inhibitors in a variety of other cancer types. Several other PARP inhibitors are currently in clinical trials for other cancer types including breast, prostate, small cell lung, colon cancer, melanoma, and glioblastoma [90] [91] [92] [93]. Unfortunately, like many other cancers, cells treated with PARP inhibitors acquire resistance to the inhibitors over the course of treatment. This is predominantly due to the reversion of BRCA1/2 mutations or by the drug-induced mutations in other components of the repair pathway like hyperactivation of NHEJ pathway [94], loss of 53BP1 [95] [96], mutations in RAD51C/D [97], ribonucleotide mediated PARP trapping [98], loss of PTIB [99] within the MLL3/4 nuclease complex and loss of PARG [100]. Moreover, PARP inhibitors only suppress the enzymatic activities of PARP1 and PARP2. Other PARPs that participate in DDR may have redundant roles to cover the loss of PARP1 and PARP2 during DDR. Nevertheless, chemo-resistance of cancer cells to PARP inhibitors can be circumvented by the development of alternative inhibitors targeting different components of the PARylation. Notably, DePARylation is a following step of PARylation for releasing DDR factors from PAR chains to DNA lesions [12]. The absence of one or more components of the dePARylation pathway may sequester DDR factors and compromise DDR. Thus, targeting dePARylation is a novel therapeutic approach for treating cancers with DDR defects.
PARG as a therapeutic target
PARG carries out the majority of PARylation reversal and is required for proper DDR. Like PARP1-deficient cells, PARG-deficient cells are hypersensitive to genotoxic insults such as ionizing radiation (IR) and alkylating agents[32] [101]. In addition, cellular inhibition of PARG in BRCA-deficient cells phenocopies their PARP inhibitor hypersensitivity [102] [42]. These biochemistry properties suggest that PARG can be an effective chemo-drug target. Moreover, unlike PARP family enzymes, PARG is monogenic and does not share redundant functions with other enzymes during dePARylation [22]. Thus PARG inhibition can be achieved with high specificity when compared to PARP inhibitors [103].
Salicylanilides were developed as the first class of cell-permeable PARG inhibitors with druggable features [104]. These inhibitors probably acting as PAR analogs were effective in squamous cell carcinoma cells and were found to be inhibiting both PARG and PARP1. Yet another PARG inhibitor, RBPI-3 was developed against T. thermophila PARG [105]. However, the inhibition pharmacology of these compounds could not be observed by yet another study which finally led to the identification of the currently available cell-permeable PARG inhibitor “PDD00017273” with high activity at 0.3 μM concentration. When breast cancer cell line “MCF-7” was treated with PDD00017273, there was an increase in γH2AX foci formation, a significant increase in PARylation and reduced cell viability [106]. The inhibitor was particularly effective in MCF-7 cells lacking HR machinery such as BRCA1, BRCA2 [107], BARD1, PALB2, or FAM175A. PDD00017273 caused replication fork stalling, and increased RAD51 foci suggesting of a defect in homologous recombination (HR). Consequently, PDD00017273 treatment in these defective cells led to lethality [108]. This PARG inhibitor also sensitized MCF-7 and triple-negative breast cancer (TNBC) cells to IR, which is primarily due to the accumulation of PARylation. Moreover, PARG inhibition increased γH2AX foci after IR treatment and the foci were reduced to the basal level much quicker than that by olaparib inhibition. In addition, after IR treatment, DNA-PKcs (NHEJ marker) foci were rapidly induced in PARG inhibitor treatment, while RAD51 (HR marker) foci were delayed with respect to control treated cells. These results suggest that IR-induced DSBs show an increased tendency of repair by NHEJ after PARG inhibition. In the presence of PDD00017273, majorities of the cells were arrested in G2/S checkpoint and the cell population in mitosis was significantly decreased, suggesting that PARG inhibition affects the cell cycle progression to mitosis [109].
Recent studies also suggest that combination therapy involving PARG inhibition can be used to improve the clinical activity of antineoplastic drugs in human cancer cells e.g combination therapy involving PARG inhibitor (ethacridine) and epidermal growth factor receptor (EGFR) inhibitor (erlotinib) can be used to induce apoptosis in AML cells resistant to EGFR inhibition [110]. Erlotinib is an EGFP inhibitor used to induce killing of AML cells. However, some cell lines such as TEX and OCI-AML2 are resistant to erlotinib treatment. The resistance can be circumvented by the combination therapy of erlotinib with ethacridine. The two drugs when combined together induce lethal levels of reactive oxygen species in AML cancer cell lines. The mode of synergism involves an erlotinib induced increase in intracellular concentration of ethacridine in these cells which in turn induced the levels of PAR.
The role of ARHs in cancer suppression
Mouse embryonic cell lacking ARH1 are completely devoid of removing the ADPr units attached to Arg-ADPr. It has been shown that mouse embryonic fibroblasts (MEFs) derived from ARH1−/− and ARH1+/− mice proliferated faster and formed colonies on soft agar, signifying that ARH1 may act as a tumor suppressor. This was particularly evidenced by the formation of subcutaneous tumors in nude mice by ARH1−/− cells. Furthermore, ARH1−/− and ARH1+/− mice developed a variety of tumors within 3 months and 6 months respectively [111]. The development of tumors was directly linked to the expression of the ARH1 protein with a decrease in the expression promoting tumorigenesis [112]. Human cancer database shows the presence of recurrent mutations in the catalytic domain of ARH1 and loss of heterozygosity (LOH) in ARH1 gene in cancers from different tissues [112]. Collectively, these studies suggest that loss of ARH1 may induce tumorigenesis.
The role of MacroD1/D2 in cancer suppression
Both MacroD1 and D2 are involved in tumorigenesis. MacroD1 functions as an estrogen response gene and is up-regulated in a majority (~40%) of breast cancers [113]. ER-alpha up-regulates MacroD1 which in turn regulates ER-alpha by a positive feedback signal. Increased level of MacroD1 favors cellular proliferation and siRNA mediated knockdown of MacroD1 reduces the proliferation of estrogen-responsive cancer cells [114].
Recently MacroD2 amplification was observed in a subset of breast cancers which can grow in absence of estrogen pathway. These cancers are resistant to anti-ER-alpha drug tamoxifen, and the resistance can be circumvented by siRNA mediated knockdown of MacroD2 in cancer cell lines. Moreover, MacroD2 overexpression was linked to the poor prognosis in breast cancers [115]. Recent reports suggest that MacroD2 gene may function as a common fragile site and get focally deleted in some human colorectal cancers [116] [117] [118] [119]. The deletions were mapped (microdeletions or whole gene) in 30 % of TCGA analyzed colorectal cancers. Loss of MacroD2 was associated with increased (dosage-dependent manner) auto-inhibitory MARylation on PARP1 which in turn impaired its catalytic activity. The reduced PARP1 activity led to a compromised DNA damage response and chromosome instability. These studies suggest that MacroD2 functions as a tumor suppressor and its deficiency in ApcMin/+ mice were associated with the stimulated growth of intestinal cancers and chromosome abnormalities [120].
The role of pyrophosphatases in cancer suppression
NUDT5 levels are elevated in breast cancer and it functions as a progestin dependent factor in this cancer [43]. As already stated NUDT5 can generate ATP at the vicinity of damaged DNA. The ATP, in turn, modulates gene expression and chromatin remodeling in breast cancer cells in response to progestin signaling. Nevertheless, NUDT5 mediated chromatin remodeling can be suppressed by NUDT5 inhibitor TH5427. TH5427 competes with ADPR for the catalytic pocket in NUDT5 and blocks hormone-dependent growth in breast cancer cells. Thus, NUDT5 can function as a promising chemotherapeutic target for the treatment of breast cancer [121].
Conclusion and future prospects:
The FDA approval on olaparib for the treatment of stage-IV ovarian cancer represents a significant breakthrough and demonstrates the usefulness of inhibitors targeting the ADP-ribosylation writers. However, the emergence of olaparib resistant cancers necessities the development of alternative therapeutics targeting the readers and erasers of PARylation. DePARylation represents a sequential downstream pathway to ADP-ribosylation and attracts significant potential as a therapeutic target for cancer therapy. However, structural and functional properties of ADP-ribosylation degrading enzymes are elusive. The function of different PAR degrading enzymes must be deciphered in the context of DNA damage which will help researchers in the development of most potent and specific therapies. Signals which activate the erasers and the signature sequences left at the target site following the removal of ADP-ribosylation need to be ascertained. Nevertheless, specific and cell permeable potent inhibitors against known PAR/MAR metabolizing enzymes in-vivo need to be developed to exploit the therapeutic potential of these proteins in cancer treatment. Since, the expression of dePARylation enzymes (e.g. PARG 2000 molecules/cell) is much lower than PARylation enzymes (e.g. PARP1 is nearly 2, 000,000 molecules/cell), targeting dePARylation may have greater therapeutic effects on clinical cancer treatment.
Acknowledgements
M.A.K. is a recipient of the award “Supporting Innovation in Cancer and Aging Research” from The City of Hope Center on Cancer and Aging. The work in X.Y. lab is supported by grants from National Institutes of Health (CA130899 and CA187209 to X.Y.). X.Y. is a recipient of Research Scholar Award from Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gupte R, Liu Z, Kraus WL, PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes, Genes Dev 31 (2017) 101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J, High speed of fork progression induces DNA replication stress and genomic instability, Nature 559 (2018) 279–284. doi: 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- [3].Li Z, Li Y, Tang M, Peng B, Lu X, Yang Q, Zhu Q, Hou T, Li M, Liu C, Wang L, Xu X, Zhao Y, Wang H, Yang Y, Zhu W-G, Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair, Cell Res 28 (2018) 756–770. doi: 10.1038/s41422-018-0048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lüscher B, Bütepage M, Eckei L, Krieg S, Verheugd P, Shilton BH, ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease, Chem. Rev 118 (2018) 1092–1136. doi: 10.1021/acs.chemrev.7b00122. [DOI] [PubMed] [Google Scholar]
- [5].Barkauskaite E, Jankevicius G, Ahel I, Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation, Mol. Cell 58 (2015) 935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- [6].Bai P, Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance, Mol. Cell 58 (2015) 947–958. doi: 10.1016/j.molcel.2015.01.034. [DOI] [PubMed] [Google Scholar]
- [7].Leung AKL, Poly(ADP-ribose): An organizer of cellular architecture, J. Cell Biol 205 (2014) 613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen Q, Kassab MA, Dantzer F, Yu X, PARP2 mediates branched poly ADP-ribosylation in response to DNA damage, Nat. Commun 9 (2018) 3233. doi: 10.1038/s41467-018-05588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Palazzo L, Mikoč A, Ahel I, ADP-ribosylation: new facets of an ancient modification, FEBS J 284 (2017) 2932–2946. doi: 10.1111/febs.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Lüscher B, Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation, Mol. Cell 32 (2008) 57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- [11].Karlberg T, Klepsch M, Thorsell A-G, Andersson CD, Linusson A, Schüler H, Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein, J. Biol. Chem 290 (2015) 7336–7344. doi: 10.1074/jbc.M114.630160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu C, Vyas A, Kassab MA, Singh AK, Yu X, The role of poly ADP-ribosylation in the first wave of DNA damage response, Nucleic Acids Res 45 (2017) 8129–8141. doi: 10.1093/nar/gkx565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhen Y, Zhang Y, Yu Y, A Cell-Line-Specific Atlas of PARP-Mediated Protein Asp/Glu-ADP-Ribosylation in Breast Cancer, Cell Rep 21 (2017) 2326–2337. doi: 10.1016/j.celrep.2017.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang Y, Wang J, Ding M, Yu Y, Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome, Nat. Methods 10 (2013) 981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- [15].Leutert M, Menzel S, Braren R, Rissiek B, Hopp A-K, Nowak K, Bisceglie L, Gehrig P, Li H, Zolkiewska A, Koch-Nolte F, Hottiger MO, Proteomic Characterization of the Heart and Skeletal Muscle Reveals Widespread Arginine ADP-Ribosylation by the ARTC1 Ectoenzyme, Cell Rep 24 (2018) 1916–1929.e5. doi: 10.1016/j.celrep.2018.07.048. [DOI] [PubMed] [Google Scholar]
- [16].Bonicalzi M-E, Haince J-F, Droit A, Poirier GG, Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when?, Cell. Mol. Life Sci. CMLS 62 (2005) 739–750. doi: 10.1007/s00018-004-4505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I, The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase, Nature 477 (2011) 616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rack JGM, Perina D, Ahel I, Macrodomains: Structure, Function, Evolution, and Catalytic Activities, Annu. Rev. Biochem 85 (2016) 431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- [19].Alvarez-Gonzalez R, Althaus FR, Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents, Mutat. Res 218 (1989) 67–74. [DOI] [PubMed] [Google Scholar]
- [20].Fisher AEO, Hochegger H, Takeda S, Caldecott KW, Poly(ADP-Ribose) Polymerase 1 Accelerates Single-Strand Break Repair in Concert with Poly(ADP-Ribose) Glycohydrolase, Mol. Cell. Biol 27 (2007) 5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG, Mode of action of poly(ADP-ribose) glycohydrolase, Biochim. Biophys. Acta 1219 (1994) 342–350. [DOI] [PubMed] [Google Scholar]
- [22].Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK, Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments, Exp. Cell Res 297 (2004) 521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- [23].Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK, Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity, Exp. Cell Res 313 (2007) 2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lin W, Amé JC, Aboul-Ela N, Jacobson EL, Jacobson MK, Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase, J. Biol. Chem 272 (1997) 11895–11901. [DOI] [PubMed] [Google Scholar]
- [25].Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, Lafite P, Ahel M, Mitchison TJ, Ahel I, Leys D, Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities, Nat. Commun 4 (2013) 2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK, Human poly(ADP-ribose) glycohydrolase (PARG) gene and the common promoter sequence it shares with inner mitochondrial membrane translocase 23 (TIM23), Gene 314 (2003) 181–190. [DOI] [PubMed] [Google Scholar]
- [27].Tucker JA, Bennett N, Brassington C, Durant ST, Hassall G, Holdgate G, McAlister M, Nissink JWM, Truman C, Watson M, Structures of the human poly (ADP-ribose) glycohydrolase catalytic domain confirm catalytic mechanism and explain inhibition by ADP-HPD derivatives, PloS One 7 (2012) e50889. doi: 10.1371/journal.pone.0050889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O, Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose), J. Biol. Chem 261 (1986) 14902–14911. [PubMed] [Google Scholar]
- [29].Malanga M, Althaus FR, Poly(ADP-ribose) molecules formed during DNA repair in vivo., J. Biol. Chem 269 (1994) 17691–17696. [PubMed] [Google Scholar]
- [30].Braun SA, Panzeter PL, Collinge MA, Althaus FR, Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase, Eur. J. Biochem 220 (1994) 369–375. doi: 10.1111/j.1432-1033.1994.tb18633.x. [DOI] [PubMed] [Google Scholar]
- [31].Miwa M, Sugimura T, Splitting of the ribose-ribose linkage of poly(adenosine diphosphate-robose) by a calf thymus extract, J. Biol. Chem 246 (1971) 6362–6364. [PubMed] [Google Scholar]
- [32].Amé J-C, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V, Radiation-induced mitotic catastrophe in PARG-deficient cells, J Cell Sci 122 (2009) 1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- [33].PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1 | Nucleic Acids Research | Oxford Academic, (n.d.). https://academic.oup.com/nar/article/45/22/12834/4599181 (accessed December 18, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eustermann S, Wu W-F, Langelier M-F, Yang J-C, Easton LE, Riccio AA, Pascal JM, Neuhaus D, Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1, Mol. Cell 60 (2015) 742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G, The recognition and removal of cellular poly(ADP-ribose) signals, FEBS J 280 (2013) 3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- [36].Oka S, Kato J, Moss J, Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase, J. Biol. Chem 281 (2006) 705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- [37].Wielckens K, Schmidt A, George E, Bredehorst R, Hilz H, DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins, J. Biol. Chem 257 (1982) 12872–12877. [PubMed] [Google Scholar]
- [38].Mortusewicz O, Fouquerel E, Amé J-C, Leonhardt H, Schreiber V, PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms, Nucleic Acids Res 39 (2011) 5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kaufmann T, Grishkovskaya I, Polyansky AA, Kostrhon S, Kukolj E, Olek KM, Herbert S, Beltzung E, Mechtler K, Peterbauer T, Gotzmann J, Zhang L, Hartl M, Zagrovic B, Elsayad K, Djinovic-Carugo K, Slade D, A novel non-canonical PIP-box mediates PARG interaction with PCNA, Nucleic Acids Res 45 (2017) 9741–9759. doi: 10.1093/nar/gkx604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu C, Wu J, Paudyal SC, You Z, Yu X, CHFR is important for the first wave of ubiquitination at DNA damage sites, Nucleic Acids Res 41 (2013) 1698–1710. doi: 10.1093/nar/gks1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gao H, Coyle DL, Meyer-Ficca ML, Meyer RG, Jacobson EL, Wang Z-Q, Jacobson MK, Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase, Exp. Cell Res 313 (2007) 984–996. doi: 10.1016/j.yexcr.2006.12.025. [DOI] [PubMed] [Google Scholar]
- [42].Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stöger T, Poirier GG, Dawson VL, Dawson TM, Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality, Proc. Natl. Acad. Sci. U. S. A 101 (2004) 17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wright RHG, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, Nacht AS, Samino S, Font-Mateu J, Vicent GP, Wierer M, Trabado MA, Schelhorn C, Carolis C, Macias MJ, Yanes O, Oliva B, Beato M, ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling, Science 352 (2016) 1221–1225. doi: 10.1126/science.aad9335. [DOI] [PubMed] [Google Scholar]
- [44].Wei H, Yu X, Functions of PARylation in DNA Damage Repair Pathways, Genomics Proteomics Bioinformatics 14 (2016) 131–139. doi: 10.1016/j.gpb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Andrabi SA, Kim NS, Yu S-W, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM, Poly(ADP-ribose) (PAR) polymer is a death signal, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Parthanatos, a messenger of death. - PubMed - NCBI, (n.d.). https://www.ncbi.nlm.nih.gov/pubmed/19273119 (accessed December 18, 2018). [Google Scholar]
- [47].Berger NA, Poly(ADP-Ribose) in the Cellular Response to DNA Damage, Radiat. Res 101 (1985) 4–15. doi: 10.2307/3576299. [DOI] [PubMed] [Google Scholar]
- [48].Virág L, Robaszkiewicz A, Rodriguez-Vargas JM, Oliver FJ, Poly(ADP-ribose) signaling in cell death, Mol. Aspects Med 34 (2013) 1153–1167. doi: 10.1016/j.mam.2013.01.007. [DOI] [PubMed] [Google Scholar]
- [49].Fliegert R, Bauche A, Wolf Pérez A-M, Watt JM, Rozewitz MD, Winzer R, Janus M, Gu F, Rosche A, Harneit A, Flato M, Moreau C, Kirchberger T, Wolters V, Potter BVL, Guse AH, 2’-Deoxyadenosine 5’-diphosphoribose is an endogenous TRPM2 superagonist, Nat. Chem. Biol 13 (2017) 1036–1044. doi: 10.1038/nchembio.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zarkovic G, Belousova EA, Talhaoui I, Saint-Pierre C, Kutuzov MM, Matkarimov BT, Biard D, Gasparutto D, Lavrik OI, Ishchenko AA, Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation, Nucleic Acids Res 46 (2018) 2417–2431. doi: 10.1093/nar/gkx1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Munnur D, Ahel I, Reversible mono-ADP-ribosylation of DNA breaks, FEBS J 284 (2017) 4002–4016. doi: 10.1111/febs.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Talhaoui I, Lebedeva NA, Zarkovic G, Saint-Pierre C, Kutuzov MM, Sukhanova MV, Matkarimov BT, Gasparutto D, Saparbaev MK, Lavrik OI, Ishchenko AA, Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro, Nucleic Acids Res 44 (2016) 9279–9295. doi: 10.1093/nar/gkw675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chand SN, Zarei M, Schiewer MJ, Kamath AR, Romeo C, Lal S, Cozzitorto JA, Nevler A, Scolaro L, Londin E, Jiang W, Meisner-Kober N, Pishvaian MJ, Knudsen KE, Yeo CJ, Pascal JM, Winter JM, Brody JR, Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors, Cancer Res 77 (2017) 5011–5025. doi: 10.1158/0008-5472.CAN-16-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation | Nature Reviews Molecular Cell Biology, (n.d.). https://www.nature.com/articles/nrm3601#macrodomaincontaining-proteins (accessed December 18, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Orphan Macrodomain Protein (Human C6orf130) Is an O-Acyl-ADP-ribose Deacylase, (n.d.). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3195580/(accessed December 18, 2018). [DOI] [PMC free article] [PubMed]
- [56].Rosenthal F, Feijs KLH, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, Lüscher B, Hottiger MO, Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases, Nat. Struct. Mol. Biol 20 (2013) 502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- [57].Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG, A family of macrodomain proteins reverses cellular mono-ADP-ribosylation, Nat. Struct. Mol. Biol 20 (2013) 508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sharifi R, Morra R, Denise Appel C, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I, Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease, EMBO J 32 (2013) 1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bütepage M, Preisinger C, von Kriegsheim A, Scheufen A, Lausberg E, Li J, Kappes F, Feederle R, Ernst S, Eckei L, Krieg S, Müller-Newen G, Rossetti G, Feijs KLH, Verheugd P, Lüscher B, Nucleolar-nucleoplasmic shuttling of TARG1 and its control by DNA damage-induced poly-ADP-ribosylation and by nucleolar transcription, Sci. Rep 8 (2018) 6748. doi: 10.1038/s41598-018-25137-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hirsch BM, Burgos ES, Schramm VL, Transition-state analysis of 2-O-acetyl-ADP-ribose hydrolysis by human macrodomain 1, ACS Chem. Biol 9 (2014) 2255–2262. doi: 10.1021/cb500485w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Golia B, Moeller GK, Jankevicius G, Schmidt A, Hegele A, Preißer J, Tran ML, Imhof A, Timinszky G, ATM induces MacroD2 nuclear export upon DNA damage, Nucleic Acids Res 45 (2017) 244–254. doi: 10.1093/nar/gkw904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Agnew T, Munnur D, Crawford K, Palazzo L, Mikoč A, Ahel I, MacroD1 Is a Promiscuous ADP-Ribosyl Hydrolase Localized to Mitochondria, Front. Microbiol 9 (2018). doi: 10.3389/fmicb.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Maehama T, Nishina H, Katada T, ADP-ribosylarginine glycohydrolase catalyzing the release of ADP-ribose from the cholera toxin-modified alpha-subunits of GTP-binding proteins, J. Biochem. (Tokyo) 116 (1994) 1134–1138. [DOI] [PubMed] [Google Scholar]
- [64].Laing S, Unger M, Koch-Nolte F, Haag F, ADP-ribosylation of arginine, Amino Acids 41 (2011) 257–269. doi: 10.1007/s00726-010-0676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Margarit SM, Davidson W, Frego L, Stebbins CE, A steric antagonism of actin polymerization by a salmonella virulence protein, Struct. Lond. Engl. 1993 14 (2006) 1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- [66].Seman M, Adriouch S, Scheuplein F, Krebs C, Freese D, Glowacki G, Deterre P, Haag F, Koch-Nolte F, NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor, Immunity 19 (2003) 571–582. [DOI] [PubMed] [Google Scholar]
- [67].Ludden PW, Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes, Mol. Cell. Biochem 138 (1994) 123–129. doi: 10.1007/BF00928453. [DOI] [PubMed] [Google Scholar]
- [68].Kato J, Zhu J, Liu C, Moss J, Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice, Mol. Cell. Biol 27 (2007) 5534–5543. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mashimo M, Kato J, Moss J, Structure and function of the ARH family of ADP-ribose-acceptor hydrolases, DNA Repair 0 (2014) 88–94. doi: 10.1016/j.dnarep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang M, Yuan Z, Xie R, Ma Y, Liu X, Yu X, Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation, J. Biol. Chem (2018) jbc.RA118.004284. doi: 10.1074/jbc.RA118.004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pourfarjam Y, Ventura J, Kurinov I, Cho A, Moss J, Kim I-K, Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition, J. Biol. Chem (2018) jbc.RA118.003586. doi: 10.1074/jbc.RA118.003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fontana P, Bonfiglio JJ, Palazzo L, Bartlett E, Matic I, Ahel I, Serine ADP-ribosylation reversal by the hydrolase ARH3, ELife 6 (n.d.). doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, Ahel I, Serine is the major residue for ADP-ribosylation upon DNA damage, ELife (2018). doi: 10.7554/eLife.34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, Palazzo L, Stockum A, Ahel I, Matic I, Serine is a new target residue for endogenous ADP-ribosylation on histones, Nat. Chem. Biol 12 (2016) 998–1000. doi: 10.1038/nchembio.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bonfiglio JJ, Colby T, Matic I, Mass spectrometry for serine ADP-ribosylation? Think o-glycosylation!, Nucleic Acids Res 45 (2017) 6259–6264. doi: 10.1093/nar/gkx446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, Matic I, Serine ADP-Ribosylation Depends on HPF1, Mol. Cell 65 (2017) 932–940.e6. doi: 10.1016/j.molcel.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].McLennan AG, The Nudix hydrolase superfamily, Cell. Mol. Life Sci. CMLS 63 (2006) 123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Palazzo L, Thomas B, Jemth A-S, Colby T, Leidecker O, Feijs K, Zaja R, Loseva O, Puigvert JC, Matic I, Helleday T, Ahel I, Processing of protein ADP-ribosylation by Nudix hydrolases, Biochem. J 468 (2015) 293–301. doi: 10.1042/BJ20141554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Williams JC, Chambers JP, Liehr JG, Glutamyl ribose 5-phosphate storage disease. A hereditary defect in the degradation of poly(ADP-ribosylated) proteins, J. Biol. Chem 259 (1984) 1037–1042. [PubMed] [Google Scholar]
- [80].Palazzo L, Daniels CM, Nettleship JE, Rahman N, McPherson RL, Ong S-E, Kato K, Nureki O, Leung AKL, Ahel I, ENPP1 processes protein ADP-ribosylation in vitro, FEBS J 283 (2016) 3371–3388. doi: 10.1111/febs.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Krukenberg KA, Kim S, Tan ES, Maliga Z, Mitchison TJ, Extracellular poly(ADP-ribose) is a pro-inflammatory signal for macrophages, Chem. Biol 22 (2015) 446–452. doi: 10.1016/j.chembiol.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hessle L, Johnson KA, Anderson HC, Narisawa S, Sali A, Goding JW, Terkeltaub R, Millan JL, Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization, Proc. Natl. Acad. Sci. U. S. A 99 (2002) 9445–9449. doi: 10.1073/pnas.142063399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang H, Gonzalez-Garcia I, Traba J, Jain S, Conteh S, Shin D-M, Qi C, Gao Y, Sun J, Kang S, Abbasi S, Naghashfar Z, Yoon J, DuBois W, Kovalchuk AL, Sack MN, Duffy P, Morse HC, ATP-degrading ENPP1 is required for survival (or persistence) of long-lived plasma cells, Sci. Rep 7 (2017) 17867. doi: 10.1038/s41598-017-18028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lin KY, Kraus WL, PARP Inhibitors for Cancer Therapy, Cell 169 (2017) 183. doi: 10.1016/j.cell.2017.03.034. [DOI] [PubMed] [Google Scholar]
- [85].Ashworth A, Lord CJ, Synthetic lethal therapies for cancer: what’s next after PARP inhibitors?, Nat. Rev. Clin. Oncol 15 (2018) 564–576. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- [86].Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, Fabbro M, Ledermann JA, Lorusso D, Vergote I, Ben-Baruch NE, Marth C, Mądry R, Christensen RD, Berek JS, Dørum A, Tinker AV, du Bois A, González-Martín A, Follana P, Benigno B, Rosenberg P, Gilbert L, Rimel BJ, Buscema J, Balser JP, Agarwal S, Matulonis UA, ENGOT-OV16/NOVA Investigators, Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer, N. Engl. J. Med 375 (2016) 2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- [87].Drew Y, Ledermann J, Hall G, Rea D, Glasspool R, Highley M, Jayson G, Sludden J, Murray J, Jamieson D, Halford S, Acton G, Backholer Z, Mangano R, Boddy A, Curtin N, Plummer R, Phase 2 multicentre trial investigating intermittent and continuous dosing schedules of the poly(ADP-ribose) polymerase inhibitor rucaparib in germline BRCA mutation carriers with advanced ovarian and breast cancer, Br. J. Cancer 114 (2016) 723–730. doi: 10.1038/bjc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, Konecny GE, Coleman RL, Tinker AV, O’Malley DM, Kristeleit RS, Ma L, Bell-McGuinn KM, Brenton JD, Cragun JM, Oaknin A, Ray-Coquard I, Harrell MI, Mann E, Kaufmann SH, Floquet A, Leary A, Harding TC, Goble S, Maloney L, Isaacson J, Allen AR, Rolfe L, Yelensky R, Raponi M, McNeish IA, Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial, Lancet Oncol 18 (2017) 75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- [89].Lord CJ, Ashworth A, PARP inhibitors: Synthetic lethality in the clinic, Science 355 (2017) 1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].O’Sullivan CC, Moon DH, Kohn EC, Lee J-M, Beyond Breast and Ovarian Cancers: PARP Inhibitors for BRCA Mutation-Associated and BRCA-Like Solid Tumors, Front. Oncol 4 (2014). doi: 10.3389/fonc.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brown JS, Sundar R, Lopez J, Combining DNA damaging therapeutics with immunotherapy: more haste, less speed, Br. J. Cancer 118 (2018) 312–324. doi: 10.1038/bjc.2017.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Sun C, Fang Y, Yin J, Chen J, Ju Z, Zhang D, Chen X, Vellano CP, Jeong KJ, Ng PK-S, Eterovic AKB, Bhola NH, Lu Y, Westin SN, Grandis JR, Lin S-Y, Scott KL, Peng G, Brugge J, Mills GB, Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers, Sci. Transl. Med 9 (2017). doi: 10.1126/scitranslmed.aal5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li L, Karanika S, Yang G, Wang J, Park S, Broom BM, Manyam GC, Wu W, Luo Y, Basourakos S, Song JH, Gallick GE, Karantanos T, Korentzelos D, Azad AK, Kim J, Corn PG, Aparicio AM, Logothetis CJ, Troncoso P, Heffernan T, Toniatti C, Lee H-S, Lee J-S, Zuo X, Chang W, Yin J, Thompson TC, Androgen receptor inhibitor–induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer, Sci Signal 10 (2017) eaam7479. doi: 10.1126/scisignal.aam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Montoni A, Robu M, Pouliot É, Shah GM, Resistance to PARP-Inhibitors in Cancer Therapy, Front. Pharmacol 4 (2013). doi: 10.3389/fphar.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, Drost R, Wientjens E, Ji J, Aly A, Doroshow JH, Cranston A, Martin NMB, Lau A, O’Connor MJ, Ganesan S, Borst P, Jonkers J, Rottenberg S, Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors, Cancer Discov 3 (2013) 68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J, 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers, Nat. Struct. Mol. Biol 17 (2010) 688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, Harrell MI, Kuiper MJ, Ho G-Y, Barker H, Jasin M, Prakash R, Kass EM, Sullivan MR, Brunette GJ, Bernstein KA, Coleman RL, Floquet A, Friedlander M, Kichenadasse G, O’Malley DM, Oza A, Sun J, Robillard L, Maloney L, Bowtell D, Giordano H, Wakefield MJ, Kaufmann SH, Simmons AD, Harding TC, Raponi M, McNeish IA, Swisher EM, Lin KK, Scott CL, AOCS Study Group, Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma, Cancer Discov 7 (2017) 984–998. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zimmermann M, Murina O, Reijns MAM, Agathanggelou A, Challis R, Tarnauskaitė Ž, Muir M, Fluteau A, Aregger M, McEwan A, Yuan W, Clarke M, Lambros MB, Paneesha S, Moss P, Chandrashekhar M, Angers S, Moffat J, Brunton VG, Hart T, de Bono J, Stankovic T, Jackson AP, Durocher D, CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions, Nature 559 (2018) 285–289. doi: 10.1038/s41586-018-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Chaudhuri AR, Callen E, Ding X, Gogola E, Duarte AA, Lee J-E, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A, Replication fork stability confers chemoresistance in BRCA-deficient cells, Nature 535 (2016) 382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gogola E, Duarte AA, de Ruiter JR, Wiegant WW, Schmid JA, de Bruijn R, James DI, Llobet SG, Vis DJ, Annunziato S, van den Broek B, Barazas M, Kersbergen A, van de Ven M, Tarsounas M, Ogilvie DJ, van Vugt M, Wessels LFA, Bartkova J, Gromova I, Andújar-Sánchez M, Bartek J, Lopes M, van Attikum H, Borst P, Jonkers J, Rottenberg S, Selective Loss of PARG Restores PARylation and Counteracts PARP Inhibitor-Mediated Synthetic Lethality, Cancer Cell 33 (2018) 1078–1093.e12. doi: 10.1016/j.ccell.2018.05.008. [DOI] [PubMed] [Google Scholar]
- [101].Feng X, Zhou Y, Proctor AM, Hopkins MM, Liu M, Koh DW, Silencing of Apoptosis-Inducing factor and poly (ADP-ribose) glycohydrolase reveals novel roles in breast cancer cell death after chemotherapy, Mol. Cancer 11 (2012) 48. doi: 10.1186/1476-4598-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cortes U, Tong W-M, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang Z-Q, Depletion of the 110-Kilodalton Isoform of Poly(ADP-Ribose) Glycohydrolase Increases Sensitivity to Genotoxic and Endotoxic Stress in Mice, Mol. Cell. Biol 24 (2004) 7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Bitler BG, Watson ZL, Wheeler LJ, Behbakht K, PARP inhibitors: Clinical utility and possibilities of overcoming resistance, Gynecol. Oncol 147 (2017) 695–704. doi: 10.1016/j.ygyno.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Steffen JD, Coyle DL, Damodaran K, Beroza P, Jacobson MK, Discovery and structure-activity relationships of modified salicylanilides as cell permeable inhibitors of poly(ADP-ribose) glycohydrolase (PARG), J. Med. Chem 54 (2011) 5403–5413. doi: 10.1021/jm200325s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Finch KE, Knezevic CE, Nottbohm AC, Partlow KC, Hergenrother PJ, Selective Small Molecule Inhibition of Poly(ADP-Ribose) Glycohydrolase (PARG), ACS Chem. Biol 7 (2012) 563–570. doi: 10.1021/cb200506t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].James DI, Smith KM, Jordan AM, Fairweather EE, Griffiths LA, Hamilton NS, Hitchin JR, Hutton CP, Jones S, Kelly P, McGonagle AE, Small H, Stowell AIJ, Tucker J, Waddell ID, Waszkowycz B, Ogilvie DJ, First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib, ACS Chem. Biol 11 (2016) 3179–3190. doi: 10.1021/acschembio.6b00609. [DOI] [PubMed] [Google Scholar]
- [107].Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells. - PubMed - NCBI, (n.d.). https://www.ncbi.nlm.nih.gov/pubmed/22333589 (accessed September 5, 2018). [DOI] [PubMed] [Google Scholar]
- [108].Gravells P, Grant E, Smith KM, James DI, Bryant HE, Specific killing of DNA damage-response deficient cells with inhibitors of poly(ADP-ribose) glycohydrolase, DNA Repair 52 (2017) 81–91. doi: 10.1016/j.dnarep.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gravells P, Neale J, Grant E, Nathubhai A, Smith KM, James DI, Bryant HE, Radiosensitization with an inhibitor of poly(ADP-ribose) glycohydrolase: A comparison with the PARP1/2/3 inhibitor olaparib, DNA Repair 61 (2018) 25–36. doi: 10.1016/j.dnarep.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Rotin LE, MacLean N, Aman A, Gronda M, Lin F-H, Hurren R, Wang X, Wrana JL, Datti A, Al-Awar R, Minden MD, Schimmer AD, Erlotinib synergizes with the poly(ADP-ribose) glycohydrolase inhibitor ethacridine in acute myeloid leukemia cells, Haematologica 101 (2016) e449–e453. doi: 10.3324/haematol.2016.146894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kato J, Zhu J, Liu C, Stylianou M, Hoffmann V, Lizak MJ, Glasgow CG, Moss J, ADP-ribosylarginine hydrolase ARH1 regulates cell proliferation and tumorigenesis, Cancer Res (2011) canres.100733.2011. doi: 10.1158/0008-5472.CAN-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kato J, Vekhter D, Heath J, Zhu J, Barbieri JT, Moss J, Mutations of the functional ARH1 allele in tumors from ARH1 heterozygous mice and cells affect ARH1 catalytic activity, cell proliferation and tumorigenesis, Oncogenesis 4 (2015) e151. doi: 10.1038/oncsis.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Han W-D, Mu Y-M, Lu X-C, Xu Z-M, Li X-J, Yu L, Song H-J, Li M, Lu J-M, Zhao Y-L, Pan C-Y, Up-regulation of LRP16 mRNA by 17beta-estradiol through activation of estrogen receptor alpha (ERalpha), but not ERbeta, and promotion of human breast cancer MCF-7 cell proliferation: a preliminary report, Endocr. Relat. Cancer 10 (2003) 217–224. [DOI] [PubMed] [Google Scholar]
- [114].Liao D-X, Han W-D, Zhao Y-L, Pu Y-D, Mu Y-M, Luo C-H, Li X-H, [Expression and clinical significance of LRP16 gene in human breast cancer], Ai Zheng Aizheng Chin. J. Cancer 25 (2006) 866–870. [PubMed] [Google Scholar]
- [115].Mohseni M, Cidado J, Croessmann S, Cravero K, Cimino-Mathews A, Wong HY, Scharpf R, Zabransky DJ, Abukhdeir AM, Garay JP, Wang GM, Beaver JA, Cochran RL, Blair BG, Rosen DM, Erlanger B, Argani P, Hurley PJ, Lauring J, Park BH, MACROD2 overexpression mediates estrogen independent growth and tamoxifen resistance in breast cancers, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 17606–17611. doi: 10.1073/pnas.1408650111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Cancer Genome Atlas Network, Comprehensive molecular characterization of human colon and rectal cancer, Nature 487 (2012) 330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Rajaram M, Zhang J, Wang T, Li J, Kuscu C, Qi H, Kato M, Grubor V, Weil RJ, Helland A, Borrenson-Dale A-L, Cho KR, Levine DA, Houghton AN, Wolchok JD, Myeroff L, Markowitz SD, Lowe SW, Zhang M, Krasnitz A, Lucito R, Mu D, Powers RS, Two Distinct Categories of Focal Deletions in Cancer Genomes, PLOS ONE 8 (2013) e66264. doi: 10.1371/journal.pone.0066264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].van den Broek E, Dijkstra MJJ, Krijgsman O, Sie D, Haan JC, Traets JJH, van de Wiel MA, Nagtegaal ID, Punt CJA, Carvalho B, Ylstra B, Abeln S, Meijer GA, Fijneman RJA, High Prevalence and Clinical Relevance of Genes Affected by Chromosomal Breaks in Colorectal Cancer, PloS One 10 (2015) e0138141. doi: 10.1371/journal.pone.0138141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Briffa R, Um I, Faratian D, Zhou Y, Turnbull AK, Langdon SP, Harrison DJ, Multi-Scale Genomic, Transcriptomic and Proteomic Analysis of Colorectal Cancer Cell Lines to Identify Novel Biomarkers, PloS One 10 (2015) e0144708. doi: 10.1371/journal.pone.0144708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sakthianandeswaren A, Parsons M, Mouradov D, MacKinnon RN, Catimel B, Liu S, Palmieri M, Love CG, Jorissen RN, Li S, Whitehead L, Putoczki TL, Preaudet A, Tsui C, Nowell CJ, Ward RL, Hawkins NJ, Desai J, Gibbs P, Ernst M, Street I, Buchert M, Sieber OM, MACROD2 Haploinsufficiency Impairs Catalytic Activity of PARP1 and Promotes Chromosome Instability and Growth of Intestinal Tumors, Cancer Discov (2018) CD-17–0909. doi: 10.1158/2159-8290.CD-17-0909. [DOI] [PubMed] [Google Scholar]
- [121].Page BDG, Valerie NCK, Wright RHG, Wallner O, Isaksson R, Carter M, Rudd SG, Loseva O, Jemth A-S, Almlöf I, Font-Mateu J, Llona-Minguez S, Baranczewski P, Jeppsson F, Homan E, Almqvist H, Axelsson H, Regmi S, Gustavsson A-L, Lundbäck T, Scobie M, Strömberg K, Stenmark P, Beato M, Helleday T, Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells, Nat. Commun 9 (2018) 250. doi: 10.1038/s41467-017-02293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].The poly(ADP-ribose) glycohydrolase inhibitor gallotannin blocks oxidative astrocyte death. - PubMed - NCBI, (n.d.). https://www.ncbi.nlm.nih.gov/pubmed/10841343/(accessed December 21, 2018). [DOI] [PubMed] [Google Scholar]


