Abstract
BACKGROUND. Identifying immune correlates of COVID-19 disease severity is an urgent need for clinical management, vaccine evaluation, and drug development. Here, we present a temporal analysis of key immune mediators, cytokines, and chemokines in blood of hospitalized COVID-19 patients from serial sampling and follow-up over 4 weeks.
METHODS. A total of 71 patients with laboratory-confirmed COVID-19 admitted to Beijing You’an Hospital in China with either mild (53 patients) or severe (18 patients) disease were enrolled with 18 healthy volunteers. We measured 34 immune mediators, cytokines, and chemokines in peripheral blood every 4–7 days over 1 month per patient using a bioplex multiplex immunoassay.
RESULTS. We found that the chemokine RANTES (CCL5) was significantly elevated, from an early stage of the infection, in patients with mild but not severe disease. We also found that early production of inhibitory mediators including IL-10 and IL-1RA were significantly associated with disease severity, and a combination of CCL5, IL-1 receptor antagonist (IL-1RA), and IL-10 at week 1 may predict patient outcomes. The majority of cytokines that are known to be associated with the cytokine storm in virus infections such as IL-6 and IFN-γ were only significantly elevated in the late stage of severe COVID-19 illness. TNF-α and GM-CSF showed no significant differences between severe and mild cases.
CONCLUSION. Together, our data suggest that early intervention to increase expression of CCL5 may prevent patients from developing severe illness. Our data also suggest that measurement of levels of CCL5, as well as IL-1RA and IL-10 in blood individually and in combination, might be useful prognostic biomarkers to guide treatment strategies.
Keywords: COVID-19
Keywords: Adaptive immunity
In COVID-19 patients, RANTES elevation associated with mild symptoms early in infection and combined with, IL-1Ra and IL-10 at week 1 of symptom onset may predict patient outcomes.
Introduction
COVID-19 infection has been declared a global pandemic by the WHO, with 1,439,516 confirmed cases over 212 countries by April 10, 2020 (1). COVID-19 is caused by a novel enveloped RNA β coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2, 3). The majority of COVID-19 infections are relatively mild, with clinical features that commonly include fever and cough, with recovery within 2–3 weeks (4, 5). Severe infections are characterized by rapid progression to acute respiratory distress syndrome, septic shock, refractory metabolic acidosis, coagulation disorders, multiorgan failure, and death (6). It is unclear why only a small proportion of patients develop severe illness, but it has been suggested that this relates to both an overreactive adaptive immune response and viral-induced lung pathology (7, 8).
Recent clinical studies found that patients with severe illness had lower levels of CD4+ and CD8+ T cells and higher levels of plasma IL-6 and IL-10 compared with patients with mild illness (7, 9). This combination was associated with reduced patient survival, suggesting that these cytokines may have an important role in viral pathogenesis (10). This has been described as a “cytokine storm,” reflecting an overproduction of immune and inflammatory cells, as well as their cytokines (10, 11). It is thought that a cytokine storm may be an important cause of acute respiratory distress syndrome (9–11). However, a more comprehensive study is required to determine both the potential protective role, as well as pathological role, of immune mediators in disease progression, especially during the early stages of virus infection in severe cases before the development of lung pathology.
In this study, a cohort of 71 patients was followed up with weekly blood tests from hospital admission for up to 4 weeks after onset of symptoms, including mild cases, in nonpandemic circumstances, in which patients do not require hospital admission. Thirty-four immune mediators, cytokines, and chemokines were measured in the blood. Longitudinal analysis was performed to demonstrate the dynamics of cytokine and chemokine production associated with disease progression to severe disease. It is hoped that this will help further clarify the mechanism of immune response to COVID-19 infection, in order to guide more effective interventions for managing patients with severe illness.
Results
Clinical and laboratory characteristics of study subjects.
Infection by SARS-CoV-2 virus in China resulted in a mild disease in the majority of the population. However, a sizeable proportion suffered a more severe and fatal disease. During this phase of the pandemic, the Beijing You’an Infectious Disease Specialist Hospital admitted many patients with mild symptoms; after the exposure to COVID-19 case and/or clinical signs and symptoms reported, nasal and pharyngeal swab specimens were tested by real-time PCR for SARS-CoV-2 virus by the test center. If the result was positive, they were sent to Beijing You’an Hospital. Admission to hospital (first day of blood sampling) is normally 2–6 days after onset of symptoms.
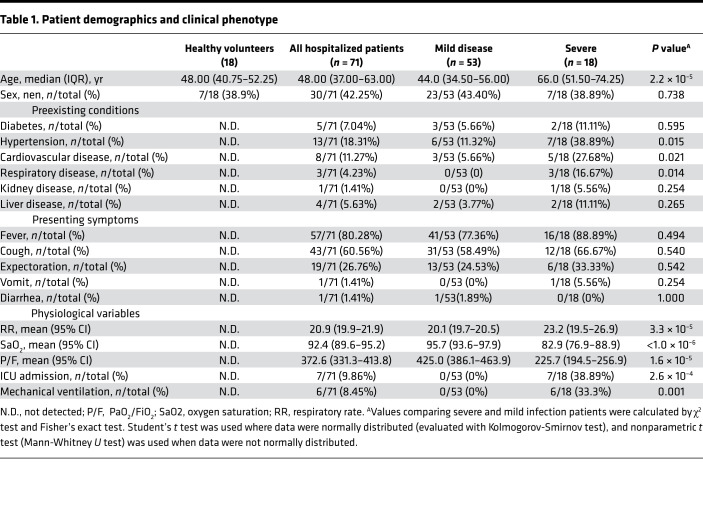
In total, 71 hospitalized patients with laboratory-confirmed COVID-19 and 18 healthy volunteers were recruited to the study. Fifty-three patients were diagnosed with mild disease and 18 patients with severe disease. Age and preexisting hypertension or cardiovascular and respiratory conditions were found to associate with progression of disease (Table 1), which is in agreement with other studies published recently (12, 13). In addition, blood oxygen saturation measured in week 2 in the severe patients was much lower than that in mild patients (P < 1.0 × 10–6). The ratio of arterial oxygen partial pressure (PaO2) to fraction of inspired oxygen (FiO2) in the severe group was also lower than that in mild group (P = 1.6 × 10–5). Demographic and clinical data are shown in Table 1. In addition, the absolute number of WBC, neutrophils, and monocytes in the severe group were much higher than that in the mild group during the first 3 weeks (P < 0.05), while there was no significant difference of lymphocyte between the mild and severe groups (Figure 1).
Table 1. Patient demographics and clinical phenotype.
Figure 1. Dynamic changes of blood cells during COVID-19 infection.
(A–D) The values of white blood cell (A), lymphocyte count (B), neutrophils count (C), and monocyte count (D) in severe and mild infection patients and healthy control were compared with each other. Two-tailed Student’s t test was used to compare parametric continuous data (evaluated with Kolmogorov-Smirnov test), and nonparametric t test (Mann-Whitney U test) was used when data were not normally distributed. The differences between each group were presented with a P value in the table under the diagram. The absolute numbers of white blood cell (A), lymphocyte count (B), neutrophils count (C), and monocyte count (D) in COVID-19 infection patients in weeks 1–4 of onset of symptom and health control were presented in scatter diagrams, in which health control was represented with black circles, mild patients with blue squares, and severe with red triangles. The dynamic changes of WBC, lymphocyte, neutrophils, and monocyte are presented with red lines in severe patients and blue lines in mild patients.
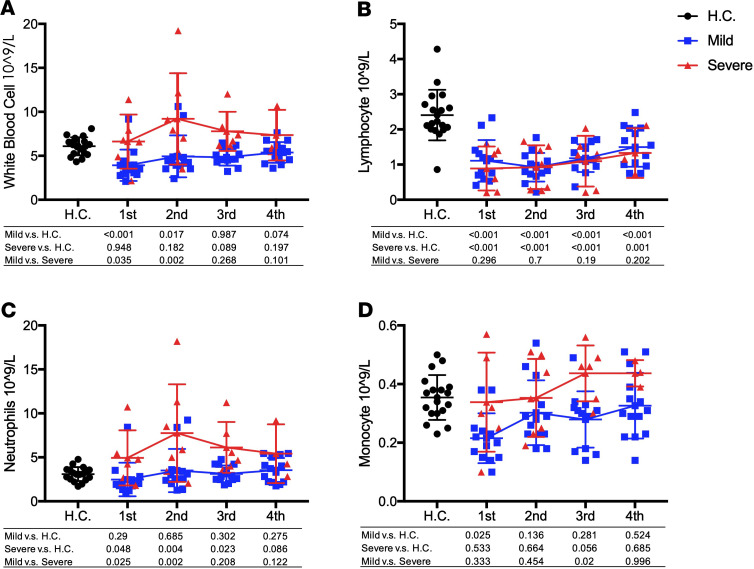
IFN-γ–inducible protein 10 (IP-10) and monocyte chemotactic protein-1 (MCP-1) are significantly associated with disease severity.
IP-10 level was significantly elevated in COVID-19 patients in week 1 of onset of symptoms in both mild and severe groups when compared with healthy volunteer controls (P = 1.36 × 10–8 and 4.39 × 10–8, respectively). Then, in the mild group of patients, IP-10 levels declined from week 2 and returned back to normal on week 4. In severe cases, IP-10 levels remained high level at week 2 and started to decline at week 3 and further by week 4. Significantly higher levels of IP-10 in serum were observed in severe infection compared with mild infection at weeks 2, 3, and 4 (P = 5.31 × 10–3, 0.02, and 1.52 × 10–3, respectively; Figure 1 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.139834DS1).
MCP-1 was also significantly elevated in COVID-19 patients in both mild and severe cases at the beginning of infection, and it remained high at all time intervals when compared with healthy controls (P < 0.05, respectively). Significantly higher levels of MCP-1 in severe cases were observed when compared with mild cases at early an time point of the infection (week 1 and 2; P = 0.047 and 8.62 × 10–5, respectively) but not at later time points (week 3 and 4; P = 0.136 and 0.030, respectively, Supplemental Table 1 and Figure 2).
Figure 2. The dynamic changes of IP-10 and MCP-1 plasma levels between mild and severe COVID-19 infection.
(A and B) The values of IP-10 (A) and MCP-1 (B) in severe and mild infection patients and healthy control were compared with each other. Two-tailed Student’s t test was used to compare parametric continuous data (evaluated with Kolmogorov-Smirnov test), and nonparametric t test (Mann-Whitney U test) was used when data were not normally distributed. The differences between each group were presented with a P value in the table under the diagram. The values of IP-10 (A) and MCP-1 (B) in healthy controls and COVID-19 infection patients in weeks 1–4 of onset of symptom are presented in scatter diagrams, in which health control was represented with black circles, mild patients with blue squares, and severe with red triangles. The dynamic changes of IP-10 and MCP-1 are presented with red lines in severe patients and blue lines in mild patients.
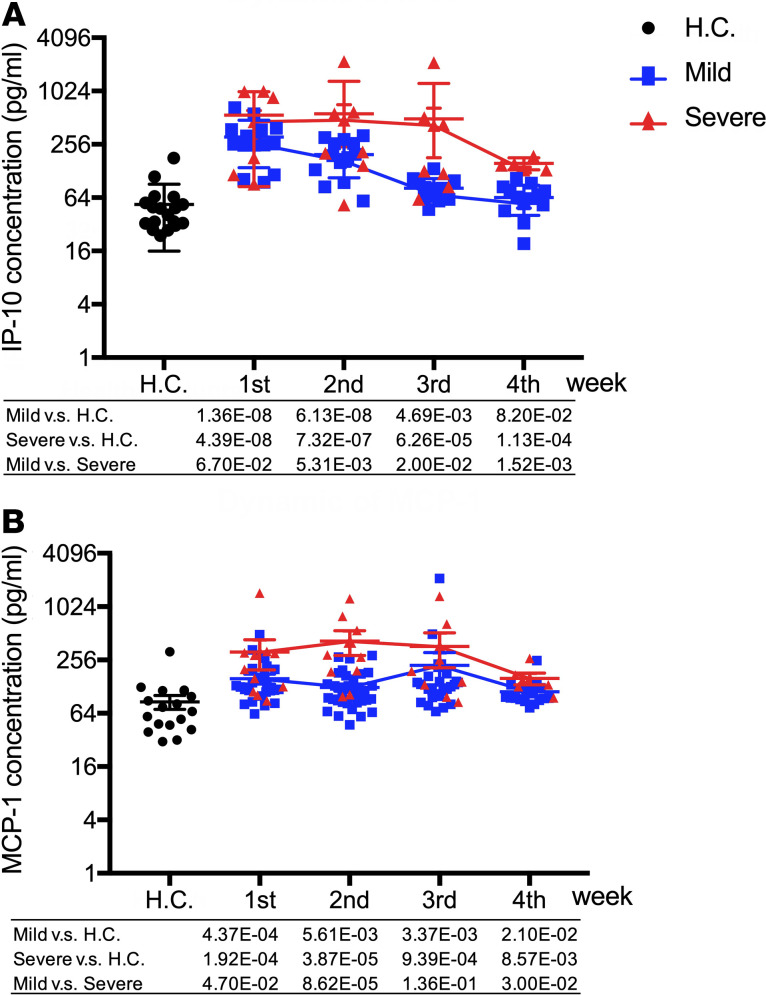
Inhibitory cytokines IL-1RA and IL-10 are significantly elevated in severe cases at an early stage of infection.
We also found that IL-1 receptor antagonist (IL-1RA) levels were elevated in both severe and mild cases and remained at a high level during the 4 weeks of follow-up. A significant difference was only observed in the first 2 weeks after onset of symptoms when we compared severe cases and mild cases (P = 0.037 and 3.78 × 10–3, respectively; Supplemental Table 1 and Figure 2).
IL-10 was only elevated in severe but not mild cases after the virus infection, and similar to IL-1RA, the levels in severe cases were significantly higher than those in mild cases in the first 2 weeks (P = 0.055 and 3.85 × 10–3, respectively; Supplemental Table 1 and Figure 3).
Figure 3. Inhibitory cytokines IL-1RA and IL-10 are significantly elevated in severe cases at early stage of infection.
(A and B) The values of IL-1RA (A) and IL-10 (B) in severe and mild infection patients and healthy control were compared with each other. Two-tailed Student’s t test was used to compare parametric continuous data (evaluated with Kolmogorov-Smirnov test), and nonparametric t test (Mann-Whitney U test) was used when data were not normally distributed. The differences between each group were presented with a P value in the table under the diagram. The values of IL-1RA (A) and IL-10 (B) in healthy controls and COVID-19 infection patients in weeks 1–4 of onset of symptoms are presented in scatter diagrams, in which health control was represented with black circles, mild patients with blue squares, and severe with red triangles. The dynamics of IL-1RA (A) and IL-10 (B) were presented with red lines in severe patients and blue lines in mild patients.
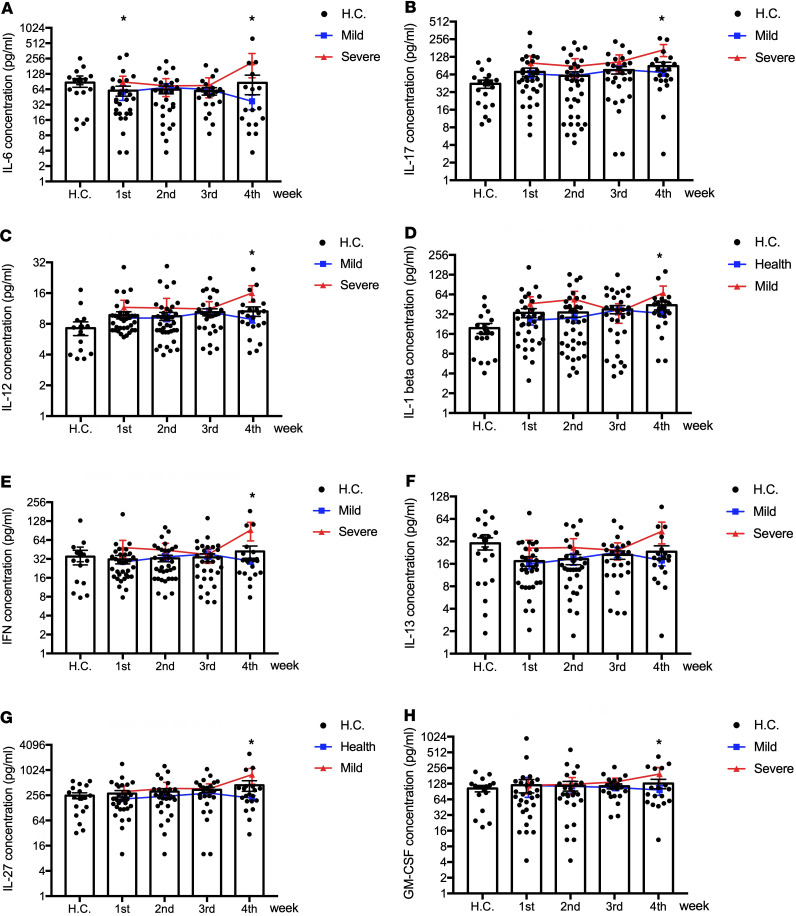
Elevated IL-6, IL-17, IL-12, IL-1β, IFN-γ, and IL-27 were only observed in late-stage severe cases.
Most cytokines observed in previous publications of “cytokine storms” in association with disease severity (9, 10, 14) were observed only in the late stage of severe cases, mostly at 4 weeks after onset of symptom — for example, IL-6, IL-12, IL-1β, IFN-γ, IL-17, and IL-27. No differences were observed in TNF-α or IL-4 between mild and severe cases (Supplemental Table 1 and Figure 4).
Figure 4. Elevated IL-6, IL-17, IL-12, IL-1β, IFN-γ, IL-13, IL-27, and IL-7 in late stages of severe cases.
(A–H) The values of IL-6 (A), IL-17 (B), IL-12 (C), IL-1β (D), IFN-γ (E), IL-13 (F), IL-27 (G) and GM-CSF (H) were compared between severe and mild infection patients by 2-tailed Student’s t test with parametric continuous data (evaluated with Kolmogorov-Smirnov test) and nonparametric t test (Mann-Whitney test) when data were not normally distributed. *P < 0.05. The values of IL-6 (A), IL-17 (B), IL-12 (C), IL-1β (D), IFN-γ (E), IL-13 (F), IL-27 (G) and GM-CSF (H) in healthy controls (H.C.) and COVID-19 infection patients in weeks 1–4 of onset of symptom are presented with black dots in scatter diagrams; the dynamics of cytokines and chemokines are presented with red lines in severe patients and blue lines in mild patients.
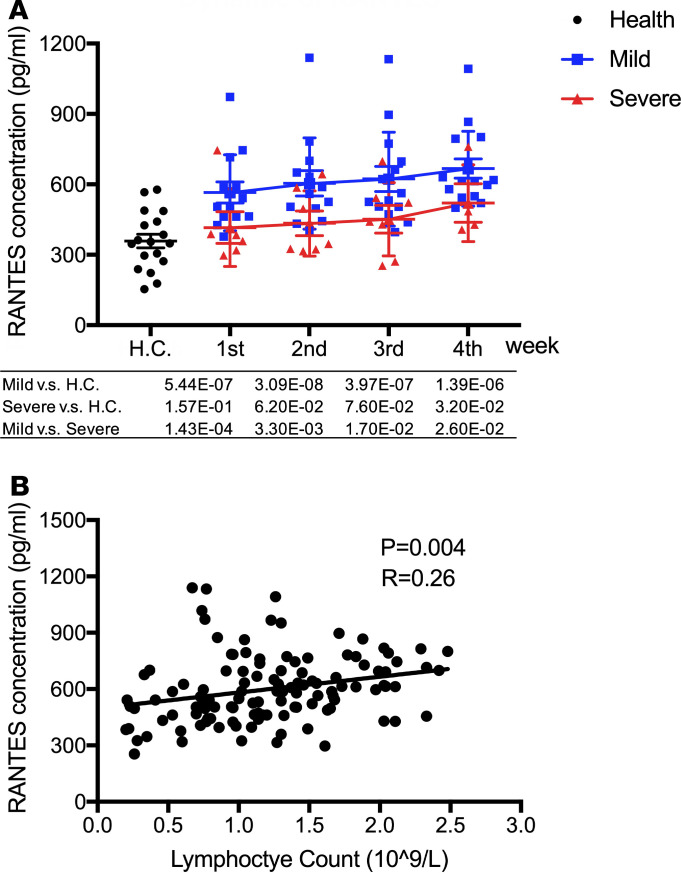
Raised levels of RANTES (CCL5) in sera of mild but not severe COVID-19 patients in the first month of infection.
RANTES, also known as CCL5, was elevated significantly in mild but not severe cases when compared with healthy controls, and it remained high in mild cases after recovery (weeks 3 and 4). In the first week, RANTES in the mild group (638.62 ± 174.81 pg/mL) was much higher than that in healthy controls (358.36 ± 123.44 pg/mL, P = 1.0 × 10–6) and remained high in mild cases during their recovery phase (630.57 ± 171.00 pg/mL in week 3 and 654.14 ± 162.86 pg/mL in week 4). No elevation of RANTES was observed in the severe group during the disease progression, suggesting that RANTES may play an important role in protecting COVID-19 patients from developing severe illness (Supplemental Table 1 and Figure 5A). We also found significant correlation between RANTES level and lymphocyte count (Figure 5B)
Figure 5. High level of RANTES in mild but not severe COVID-19 patients.
(A) The value of RANTES in severe and mild infection patients and healthy control were compared with each other. Two-tailed Student’s t test was used to compare parametric continuous data (evaluated with Kolmogorov-Smirnov test), and nonparametric t test (Mann-Whitney U test) was used when data were not normally distributed. The differences between each group were presented with a P value in the table under the diagram. The values of RANTES in health control and COVID-19 infection patients in weeks 1–4 of onset of symptom was presented in scatter diagrams, in which health control was represented with black circles, mild patients with blue squares, and severe with red triangles. The dynamics of RANTES was presented with red lines in severe patients and blue lines in mild patients. (B) Correlation analysis between RANTES and lymphocyte counts. Correlation analysis between RANTES and lymphocyte counts was performed by linear regression analysis. Each black circle indicates an individual patient; the linear correlation between RANTES and lymphocyte count was presented with a black line.
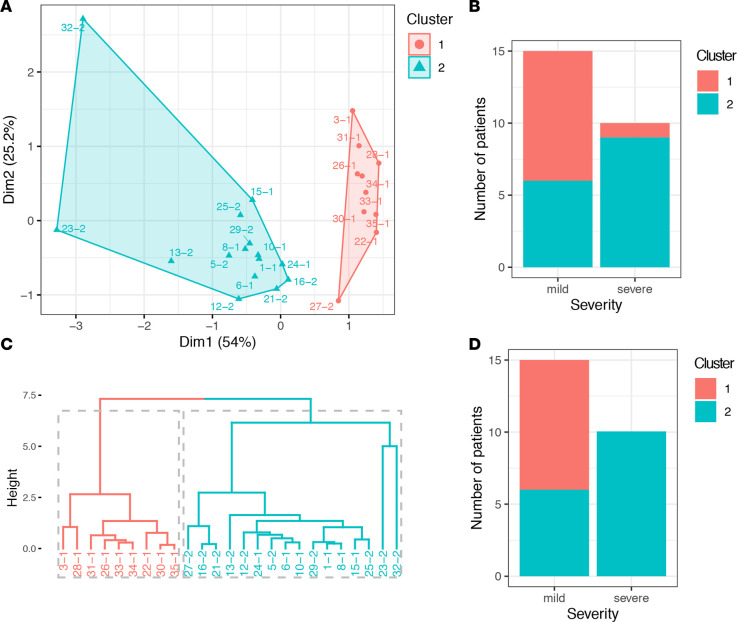
A combination of CCL5, IL-1RA, and IL-10 at week 1 may predict patient outcomes.
To test whether or not the combination of CCL5, IL-1RA, and IL-10 could predict the patient outcomes (mild/severe) at an early stage of the infection, we performed k-means and agglomerative hierarchical clustering analyses using data from the patients in week 1 of onset of symptoms (n = 34) and for whom the CCL5, IL-1RA, and IL-10 levels were assayed (n = 25, Supplemental Table 1). We extracted 2 k-means clusters based on the expression levels of these 3 cytokines across the patients, and we then assigned each cluster to patients with known disease severity and determined whether the cluster assignment could distinguish disease severity. We found 9 of 15 cluster 2 patients (Figure 6A, highlighted in blue) had severe disease compared with only 1 of 10 in cluster 1 (P = 0.018, Fisher’s exact test; Figure 6, A and B). Similarly, when we defined the patient clusters by hierarchical clustering, which is an alternative approach to k-means analysis and does not prespecify the number of clusters, we found 2 distinct clusters (10 of 16 cluster 2 patients had severe disease, relative to 0 of 9 in cluster 1 [P = 0.0028]; Figure 6, C and D). Together, these observations lend further support to a potentially valuable role of CCL5, IL-1RA, and IL-10 measurements in predicting disease severity at an early stage of the COVID-19 infection.
Figure 6. Combination of CCL5, IL-1RA, and IL-10 predict the disease severity.
(A) a cluster plot to visualize k-means clusters with the proportion of variance explained by each component. The 2 distinct clusters are highlighted in red and blue. The patient IDs are shown next to each dots. (B) A bar plot of the number of K-mean cluster 1 and cluster 2 patients with either mild or severe COVID-19 disease. (C) A dendrogram showing agglomerative hierarchical clusters. The height on the y axis represents the distance between 2 clusters. Two major clusters are highlighted in red and blue. The patient IDs are shown at the bottom of the dendrogram. (D) A bar plot of the number of hierarchical cluster 1 and cluster 2 patients with either mild or severe COVID-19 disease.
Discussion
In this study, we have analyzed levels of key immune mediators in the blood of COVID-19 patients with either mild or severe disease followed up for 4 weeks.
We found that the early production of inhibitory mediators such as IL-10 and IL-1RA were significantly associated with severe disease.
Importantly, we also found the chemokine RANTES was significantly elevated in mild but not severe disease from an early stage of the infection and remained significantly higher compared with severe cases during the 4-week follow-up. This suggests a protective role in disease progression, although it could be secondary to other factors such as activated cytotoxic T cells, which produce CCL5 upon antigen stimulation.
Significant elevation of IP-10 and MCP-1 in both mild and severe cases was also observed compared with healthy controls, with significantly higher levels in severe cases when compared with mild cases. This shows similarity to influenza virus infections, where similar associations with disease severity were also observed in both pandemic (pdm2009) H1N1 and avian H5N1 infections (15–18).
The majority of cytokines that are known to be associated with cytokine storm, such as IL-6 and IFN-γ, were only significantly elevated in the late stages of severe COVID-19 illness, while TNF-α and GM-CSF showed no difference between severe and mild cases. Our data imply that a classical cytokine storm may not be the major cause of severe illness in COVID-19 patients (9), highlighting the need for further work in this area.
The IL-1RA is an early inhibitory cytokine that suppresses proinflammatory cytokines and T lymphocyte responses. IL-1RA is a cytokine that controls inflammatory responses during early stages of immune activation (19). IL-1RA competitively binds to the IL-1R (20) and is produced by monocytes, macrophages, or DCs (21, 22). IL-1RA can modulate the production of IL-1 and TNF-α (23) and type I IFN (24). Therefore, early IL-1RA production could affect induction of proinflammatory and antiviral cytokines during the early phase of this coronavirus infection. The role of IL-1RA in the immune response may vary, given the different measured serum concentrations in severe and mild infections. In mild cases, the inhibitory role of elevated IL-1RA may be overridden by the robust adaptive immune responses to the virus. However, in the severe cases, much higher levels of IL-1RA were observed in comparison with mild cases, suggestive of an overactive immune response, which may contribute to the switch from controlled and protective immune environment to inflammation-induced tissue damage. IL-10 was only elevated in severe cases; therefore, the inhibitory role of this cytokine likely contributes to the overall suppression of the immune system, viral control, and disease severity.
RANTES is a chemokine important for T cell homing and migration during acute virus infection, as well as sustaining CD8+ T cell responses during a systemic chronic viral infection (25); RANTES can be expressed by a number of cell types — including T cells, epithelial cells, and platelets — and functions as a chemoattractant for a number of different cells such as monocytes, DCs, T cells, NK cells, and eosinophils. RANTES is produced by CD8+ T cells upon antigen stimulation and is known for its antiviral function in HIV by competing with the virus for the CCR5 receptor (26). Elevated serum levels of the chemokines RANTES observed in mild cases compared with severe cases in early stage of SARS-CoV-2 infection are likely to be produced by virus-specific CD8+ T cells. This is in keeping with the higher percentage of total lymphocyte counts in mild cases at all time points when compared with severe cases. This could imply a protective role for such adaptive T cell responses in mild cases to clear the virus before lung inflammation takes place. This hypothesis is further supported by our data showing significant correlation between RANTES and lymphocyte counts, as well as a recent single cell study showing clonal expansion of CD8+ T cells in broncho-alveolar fluid of mild but not severe COVID-19 patients (27).
We believe our study is timely, and the information should be able to help with the design of planned clinical trials and drug development, as well as the management of individual patients. However, limitations of this study include relatively sample size (n = 73), with a relatively high proportion of female cases with severe disease. Larger cohorts are needed to validate these findings.
In conclusion, our data regarding immune suppression mediated by IL-10 and IL-1RA may be important in promoting progression of infection, which merits further investigation, especially to understand their significance in terms of the pathophysiology occurring at the cellular/tissue level; in addition, our data also suggest increased activity of T cells that produce the chemokine CCL5 may help to prevent patients from developing severe illness. Our data also indicate that measurement of levels of CCL5, as well as IL-1RA and IL-10 in blood individually and/or in combination, could provide useful prognostic biomarkers to guide treatment strategies.
Methods
Study population.
Patients were recruited from Beijing You’an Hospital between January 2020 and March 2020. Inclusion criteria included the following: all available cases presenting consecutively over this period, recruited based on lab confirmation of COVID-19 and independent of severity of illness. Exclusion criteria included the following: HBV and HIV infection, cancer, immmunosupression with high-dose steroids or chemotherapy, or pregnancy. Patient clinical data were collected from Electronic Medical Record System (EMRS), Laboratory Information System (LIS), and Picture Archiving and Communication System (PACS). Plasma was separated from whole blood samples and stored in a –80°C freezer.
Clinical definitions.
COVID-19 was diagnosed according to recommendations by the National Health Commission of China (6). Laboratory-confirmed patients were defined as showing a positive result on high-throughput sequencing or real-time RT-PCR assay of nasal and pharyngeal swab specimens. The degree of severity was identified as mild infection or severe infection. Severe infection was defined as COVID-19–confirmed patients with 1 of the following conditions: respiratory distress with respiratory rate > 30/min; blood oxygen saturation < 93%; arterial PaO2/FiO2 < 300 mmHg; respiratory failure with mechanical ventilation; shock; or other organ failures requiring admission to ICU. Blood tests were taken every 4–7 days over the course of 4 weeks, and the first day of onset of symptom was defined as the first day with clinical signs or symptoms consistent with COVID-19 infection. Patients with severe disease showed mild symptoms during weeks 1 and 2 and developed severe illness in weeks 3 and 4 after the onset of symptoms. Samples from 20 patients were collected at 3–4 time points, and for the remaining patients, samples were collected at 1–2 time points. Healthy volunteers were selected to match the sex and age of patients and to be equal in number to the smallest comparator group (patients with severe disease). The healthy volunteers were recruited in 2017 by You’an bio-bank before COVID-19 emerged and did not have infection with SARS-CoV-2 or diabetes mellitus, hypertension, cardiovascular diseases, respiratory disease, kidney disease, or liver disease.
Measurements of cytokines and chemokines.
We determined the serum cytokine and chemokine levels in healthy volunteers (n = 18) and COVID-19 infection patients in weeks 1 (n = 34), 2 (n = 40), 3 (n = 31), and 4 (n = 22) of onset of symptom. Serum cytokine and chemokine levels were measured using ProcartaPlex Humna Cytokine & Chemokine Panel 1A 34plex (Invitrogen) in Luminex 200 multiplexing instrument (MilliporeSigma). This system allowed us to generate quantitative measurements for 34 different chemokines, cytokines, growth factors, and immune mediators, including IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, Eotaxin, G-CSF, GM-CSF, IFN-α, IFN-γ, IP-10, MCP-1, MIP-1a (CCL3), MIP-1b (CCL4), RANTES, and VEGF in 12.5 μL samples (Supplemental Table 1 and Supplemental Figure 1).
Clustering analysis.
We defined patient clusters by either k-means (Hartigan-Wong algorithm) or agglomerative hierarchical clustering (Ward’s method) based on a similarity measure (Euclidean distance). The analyses were performed using R software (version 3.6.0). We used R package factoextra for visualizing the clusters.
Statistics.
Statistical analysis of the data was performed using the χ2 test for sex analysis. Two-tailed Student’s t test was used to compare parametric continuous data between mild and severe infection groups (evaluated with Kolmogorov-Smirnov test), and nonparametric t test (Mann-Whitney U test) was used when data were not normally distributed. Statistical test differences were considered significant if the P values were less than 0.05. Analyses were performed with SPSS software v25.5 (IBM, New York, USA).
Study approval.
The study was approved by the IRB of Beijing You’an Hospital. Written informed consent was obtained from all patients.
Author contributions
Conception and design were contributed by Y. Zhang and TD. Data analysis was contributed by Y. Zhang, PZ, LQ, Y. Zhao, KL, and YP. Clinical sample and data collection were contributed by RJ, CZ, LQ, KL, BX, LL, YD, YF, AL, JS, XL, ZH, and HX. Writing of the manuscript was contributed by TD and Y. Zhang. Data interpretation and reviewing and editing the manuscript were contributed by TD, Y. Zhang, JCK, AM, GO, and LPH.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2020YFE0202400), Beijing Natural Science Foundation (7191004 and 7202069), Beijing Municipal Science & Technology Commission (Z171100001017078), Beijing municipal administration of hospitals (DFL20181701 and ZYLX201711), Beijing Key Laboratory (BZ0373), and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS), China (2018-I2M-2-002). TD is supported by Medical Research Council (MRC), UK (MR/L018942/1 and MRC Human Immunology Unit Core), and Nuffield Department of Medicine, Oxford University. JCK is supported by a Wellcome Trust Investigator Award (204969/Z/16/Z). JCK, LPH, and GO are, in part, supported by the NIHR Oxford Biomedical Research Centre. We would like acknowledge Richard Newman and Lucy Li for reading through the manuscript and for providing very helpful comments and suggestions.
Version 1. 06/05/2020
In-Press Preview
Version 2. 07/09/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(13):e139834.https://doi.org/10.1172/jci.insight.139834.
Contributor Information
Yan Zhao, Email: 18911380390@163.com.
Ling Qin, Email: 15010068908@163.com.
Ping Zhang, Email: pzhang@well.ox.ac.uk.
Kang Li, Email: bjyahlk@163.com.
Lianchun Liang, Email: llc671215@sohu.com.
Jianping Sun, Email: sun_jp@126.com.
Bin Xu, Email: xubin1016@126.com.
Yanchao Dai, Email: 1484118327@qq.com.
Xuemei Li, Email: 1365737271@qq.com.
Chi Zhang, Email: 18501341600@163.com.
Yanchun Peng, Email: yanchun.peng@imm.ox.ac.uk.
Yingmei Feng, Email: yingmeif13@sina.com.
Ang Li, Email: 13910588941@139.com.
Zhongjie Hu, Email: yfcyt@139.com.
Haiping Xiang, Email: bjxhp@126.com.
Graham Ogg, Email: graham.ogg@ndm.ox.ac.uk.
Ling-Pei Ho, Email: ling-pei.ho@imm.ox.ac.uk.
Ronghua Jin, Email: jin_eagle@yahoo.com.
Julian C. Knight, Email: julian@well.ox.ac.uk.
Tao Dong, Email: tao.dong@imm.ox.ac.uk.
Yonghong Zhang, Email: 13810108505@163.com.
References
- 1. [No authors listed]. Coronavirus disease (COVID-19) pandemic. World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Accessed June 11, 2020.
- 2.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Commission CNH. The new coronavirus pneumonia diagnosis treatment plan (trial version 6). 2020. http://www.gov.cn/zhengce/zhengceku/2020-02/19/content_5480948.htm Accessed June 23, 2020.
- 7.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan WJ, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan WJ, et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55(5):2000547. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. doi: 10.1093/nsr/nwaa037. Liu Y, et al. Elevated plasma level of selective cytokines in COVID-19 patients reflect viral load lung injury [published ahead of print March 9, 2020]. Natl Sci Rev. [DOI] [PMC free article] [PubMed]
- 15.Zhang YH, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayney MS, et al. Serum IFN-γ-induced protein 10 (IP-10) as a biomarker for severity of acute respiratory infection in healthy adults. J Clin Virol. 2017;90:32–37. doi: 10.1016/j.jcv.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 18.Cole SL, et al. M1-like monocytes are a major immunological determinant of severity in previously healthy adults with life-threatening influenza. JCI Insight. 2017;2(7):e91868. doi: 10.1172/jci.insight.91868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14(5):315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arend WP, Malyak M, Guthridge CJ, Gabay C. Interleukin-1 receptor antagonist: role in biology. Annu Rev Immunol. 1998;16:27–55. doi: 10.1146/annurev.immunol.16.1.27. [DOI] [PubMed] [Google Scholar]
- 21.Benedict CA. Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factor Rev. 2003;14(3-4):349–357. doi: 10.1016/S1359-6101(03)00030-3. [DOI] [PubMed] [Google Scholar]
- 22.Dienz O, et al. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 2012;5(3):258–266. doi: 10.1038/mi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oleksowicz L, Dutcher JP. A Review of the New Cytokines: IL-4, IL-6, IL-11, and IL-12. Am J Ther. 1994;1(2):107–115. doi: 10.1097/00045391-199408000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 25.Crawford A, Angelosanto JM, Nadwodny KL, Blackburn SD, Wherry EJ. A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog. 2011;7(7):e1002098. doi: 10.1371/journal.ppat.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270(5243):1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 27. doi: 10.1038/s41591-020-0901-9. Liao MF, et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing [published ahead of print May 12, 2020]. Nat Med . [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.