Abstract
Type I IFN (IFN-I) production by plasmacytoid DCs (pDCs) occurs during acute HIV-1 infection in response to TLR7 stimulation, but the role of pDC-derived IFN-I in controlling or promoting HIV-1 infection is ambiguous. We report here a sex-biased interferogenic phenotype for a frequent single-nucleotide polymorphism of human TLR7, rs179008, displaying an impact on key parameters of acute HIV-1 infection. We show allele rs179008 T to determine lower TLR7 protein abundance in cells from women, specifically — likely by diminishing TLR7 mRNA translation efficiency through codon usage. The hypomorphic TLR7 phenotype is mirrored by decreased TLR7-driven IFN-I production by female pDCs. Among women from the French ANRS PRIMO cohort of acute HIV-1 patients, carriage of allele rs179008 T associated with lower viremia, cell-associated HIV-1 DNA, and CXCL10 (IP-10) plasma concentrations. RNA viral load was decreased by 0.85 log10 (95% CI, −1.51 to −0.18) among T/T homozygotes, who also exhibited a lower frequency of acute symptoms. TLR7 emerges as an important control locus for acute HIV-1 viremia, and the clinical phenotype for allele rs179008 T, carried by 30%–50% of European women, supports a beneficial effect of toning down TLR7-driven IFN-I production by pDCs during acute HIV-1 infection.
Keywords: AIDS/HIV, Immunology
Keywords: Innate immunity
A frequent TLR7 SNP that affects protein dosage and IFN-I production by plasmacytoid dendritic cells is associated with variation in HIV-1 acute viremia in women.
Introduction
The intracellular TLRs — TLR3, TLR7, TLR8, and TLR9 — recognize autologous and microbial nucleic acids and are critical components of innate antiviral immunity (1). RNA and DNA ligands engaging the endosomal TLRs of plasmacytoid DCs (pDCs) — TLR7 and TLR9, respectively — elicit a vigorous production of the type I IFNs (IFN-I), IFN-α and -β (2). Cell-associated and, less efficiently, cell-free HIV-1 virions signal predominantly through TLR7 (3, 4), and elevated plasma IFN-I marks the acute and late chronic phases of HIV-1 infection (5, 6).
Studies of macaques infected with SIV to model the pathogenesis of human AIDS identified pDCs as the critical source of IFN-I in vivo (7–10). Blocking the IFN-I receptor early during acute SIV infection increased the viral burden and led to distinctly shorter times to death from AIDS (11). Conversely, continued treatment with IFN-I resulted in increased SIV replication and accelerated CD4+ T cell loss, suggesting that the time pattern of IFN-I signaling is determinant in the outcome of SIV infection (11). In line with the macaque model, depleting the pDCs of humanized mice prior to HIV-1 infection boosted viral replication and abolished serum IFN-I elevation and the expression of IFN-stimulated genes (12). These investigations are supportive of a beneficial role for IFN-I during early-stage HIV-1 infection, opposing the spread of the virus and its expansion within lymph nodes. Nonetheless, innate immunity might play a paradoxical role at the mucosal epithelial barrier shortly after HIV-1 entry by facilitating viral dissemination (13, 14). A double-edged effect of IFN-I is revealed also by the course of SIV infection in its natural host, such as African green monkeys, where the IFN-I response is toned down during the acute-to-chronic phase transition and the animals do not progress to AIDS despite patent viremia (15). The nonpathogenicity of SIV in African green monkeys is linked to lower expression of IFN-stimulated genes in the blood and tissues (lymph nodes, gut) of this host during chronic SIV infection — by comparison with macaques and humans infected with a closely related virus, respectively SIVmac and HIV-1 (15, 16). A deleterious role of pDC-derived IFN-I production has been illustrated also by the lymphocytic choriomeningitis virus (LCMV) murine model of persistent viral infection. Here, blockade of IFN-I signaling, induced even before infection, promoted viral clearance (17, 18). Long-term viral control brought about by early IFNAR blockade was associated with downregulation of IL-10 and PD-L1, improved CD4+ T cell function, and restoration of lymphoid architecture (17, 18).
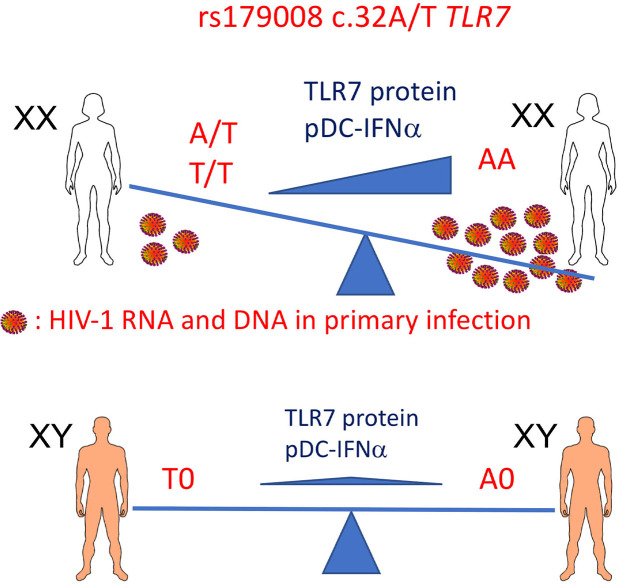
These observations highlight the importance of the timing and duration of IFN-I signaling to the balance of its opposite antiviral and immunosuppressive effects during acute and chronic HIV-1 infection (19). We have exploited natural genetic variation to investigate the direct contribution of the TLR7-driven production of IFN-I by pDCs in the course of acute HIV-1 infection. Variation in the genes for TLR7 and the other nucleic acid–sensing intracellular TLRs is constrained by strong purifying selection against mutations suppressing or modifying the function of the encoded receptor (20). Single-nucleotide polymorphisms (SNPs) of the TLR7 gene, which is located on the X-chromosome, have nevertheless been associated with changes in parameters of HIV-1 infection (21, 22). We chose to study a frequent SNP of the TLR7 coding sequence, rs179008 (NM_016562.3:c.32A>T), which was found to be associated with higher viral load at set point and accelerated decline of CD4+ T cell counts in male patients, and with a diminished cytokine response to a TLR7 agonist in the leukocytes of male allele T carriers (22). This SNP was also, intriguingly, associated with higher prevalence of HIV-1 infection in women (22). The minor allele rs179008 T is common worldwide, except East Asia, and is especially frequent among European populations, where 30%–50% of women are homozygous or heterozygous carriers. Moreover, rs179008 is a missense SNP substituting a leucine for a glutamine (p.Gln11Leu) at protein level, which warrants enquiry into the impact of this variation on TLR7 biology. In this work, we analyzed the functional differences between the rs179008 alleles in the production of IFN-I by pDCs, as well as the mechanism of action of the SNP on TLR7 protein expression. In addition, we describe a pattern of association between this TLR7 polymorphism and key parameters of acute HIV-1 infection in women that supports the notion of a deleterious role for excess TLR7-driven IFN-I production during the early stages of infection.
Results
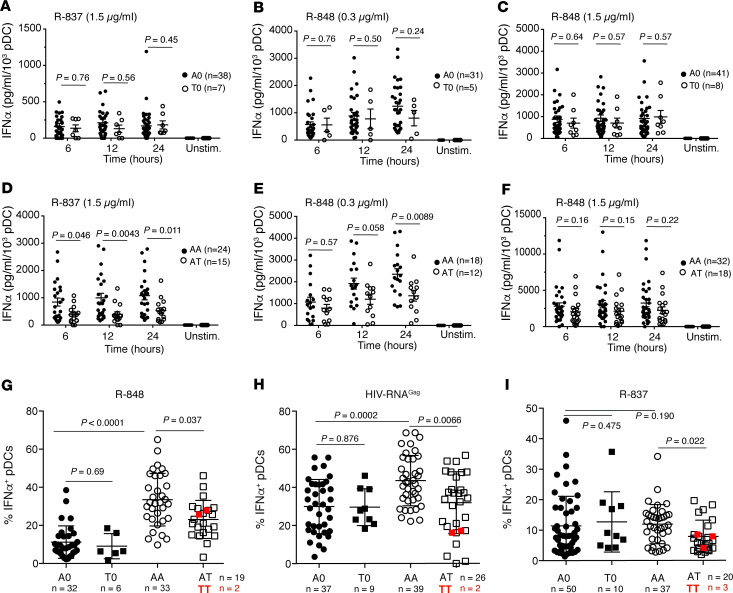
TLR7-dependent IFN-αproduction is impaired in female carriers of rs179008 T. There is a strong sex bias in the TLR7-driven production of IFN-α by human pDCs, with higher frequencies of IFN-α–producing cells in women than in men (23–25). We therefore investigated the impact of rs179008 in either sex separately. We first examined PBMC production of IFN-α in response to TLR7 agonists in male (i.e., hemizygous) carriers of either rs179008 allele and in A/T heterozygous women relative to A/A homozygotes. In this context, the pDC fraction of PBMCs is solely responsible for the early-onset production of IFN-I (25, 26). We accordingly normalized IFN-α measurements to pDC numbers to quantify the effect of rs179008 in time-course experiments where male or female PBMCs were stimulated ex vivo with either R-837 (imiquimod; a TLR7 ligand) or R-848 (resiquimod; a ligand of TLR7 and TLR8). Contrary to the earlier study (22), we observed no significant differences in IFN-α production between A/0 and T/0 males, regardless of agonist dosage (Figure 1, A–C). In female PBMCs, by contrast, heterozygous carriage of allele T was associated with substantially lower production of IFN-α upon stimulation with either 1.5 μg/mL R-837 or 0.3 μg/mL R-848 (Figure 1, D and E). The response to 1.5 μg/mL R-848, possibly a near-saturation dose (26), showed a similar trend but differences by genotype were not statistically significant (Figure 1F). In line with previous reports of female-biased responses (23), PBMCs from A/A women consistently produced much more IFN-α than comparable male cells of A/0 genotype in response to R-848 or R-837, regardless of dosage (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136047DS1). IFN-α production by pDCs was, thus, selectively blunted in female carriers of the rs179008 T allele, and the responses for the genotypes represented in these experiments ranked A/A > A/T > A/0 = T/0.
Figure 1. Sex-specific effect of the rs179008 T allele on the IFN-α expression phenotype.
(A–F) Quantitation of IFN-α in culture supernatants of male (A–C) and female (D–F) PBMCs stimulated ex vivo with TLR7 ligands as shown, sampled 6, 12, and 24 hours after stimulation or 24 hours for nonstimulated (Unstim.) control cells. ELISA IFN-α values (pg/mL) were normalized to flow-cytometrically determined pDC numbers for each donor; see Supplemental Figure 2A for the Lin−CD123+BDCA4+ gating strategy. Each dot represents 1 donor; horizontal bars indicate mean values ± SEM. The number of donors by genotype is indicated in the plot legend. Groups were compared by a 2-way ANOVA followed by Sidak’s multiple comparisons test. N.D., not detectable (i.e., below the 30 pg/mL detection threshold of the assay). (G–I) Frequency of IFN-α–producing pDCs. Freshly isolated male and female PBMCs were stimulated with 0.3 μg/mL R-848 (G), HIV-1–derived GagRNA1166 (H), or 1 μg/mL R-837 (I). The frequency of IFN-α–producing pDCs was determined by flow cytometry, as above. Each dot represents 1 donor; horizontal bars indicate mean values ± SD. Groups were compared by the Kruskal-Wallis test corrected for multiple comparisons by controlling the FDR (Benjamini-Hochberg method). Data for a small number of T/T female donors studied in parallel are plotted (in red) but were not included in the statistical tests.
We also assessed the effect of rs179008 on the frequency of IFN-α– or TNF-α–producing pDCs following stimulation with TLR7 ligands, using the flow cytometric strategy in Supplemental Figure 2. We stimulated PBMCs with GagRNA1166, a HIV-1–derived synthetic RNA ligand of TLR7 and TLR8 (24), in parallel with the imidazoquinoline compounds R-837 and R-848. IFN-α–producing pDCs were boosted most efficiently by GagRNA1166 and R-848, and TNF-α–producing pDCs by R-837 (Supplemental Figures 2 and 3). Consistent with the data in Figure 1, carriage of the T allele was associated with a reduced frequency of IFN-α–producing pDCs in women but not in men (Figure 1, G–I). The most prominent genotypic bias in women occurred with R-848 and GagRNA1166 (Figure 1, G and H), along with the largest male-female differences in the frequency of IFN-α–producing pDCs (Figure 1, G and H). No significant impact of the T allele was observed in the frequency of TNF-α–producing pDCs, and a small sex bias was observed only upon PBMC stimulation with GagRNA1166 (Supplemental Figure 3C). This argues against a general dysfunction of pDCs in allele T carriers and suggests that the rs179008 genotype fine-tunes the TLR7-driven production of IFN-α in female pDCs, specifically.
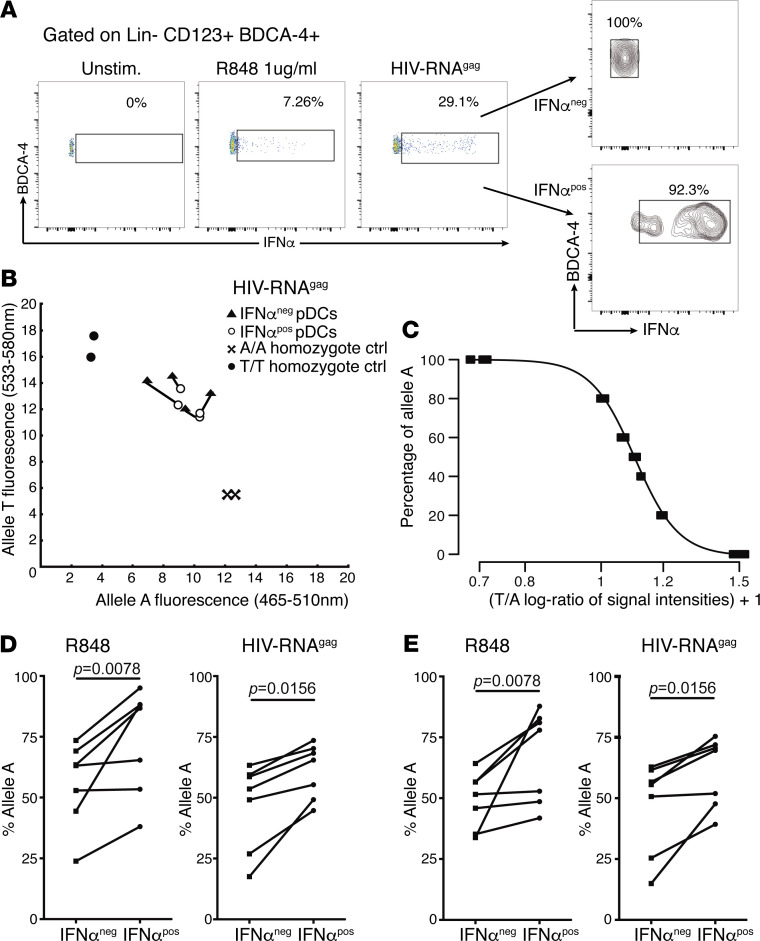
Female pDCs expressing the rs179008 T allele produce less IFN-α. The immune cells of women express either 1 or both alleles of TLR7 in mosaic fashion, as the allele on the inactive X chromosome may evade repression (27). More than half of the cells express monoallelic transcripts, and we developed an assay to determine whether IFN-α production following TLR7 engagement depends, at the cellular level, on the expressed rs179008 allele. PBMCs from A/T heterozygous women were stimulated with either R-848 or GagRNA1166; next, pDCs were labeled for lineage-specific markers, surface-immunostained for self-secreted IFN-α, and sorted into IFN-α+ or IFN-α– pDC pools (Figure 2A) that were analyzed for the proportions of transcripts from either allele (Figure 2, B–D). In a group of healthy women, IFN-α–secreting pDCs displayed enrichment in allele A TLR7 transcripts relative to same-donor IFN-α– cells, as measured by an allele-specific PCR assay (27) (Figure 2D) and corroborated by deep sequencing of the TLR7 amplimers (Figure 2E). These experiments demonstrated a depressed interferogenic response to TLR7 engagement in female allele T–expressing pDCs, relative to the allele A–expressing cells from the same donor.
Figure 2. Allele A enrichment among TLR7 transcripts from IFN-α–producing female pDCs.
(A) Flow cytometric analysis of IFN-α–secreting pDCs. Freshly isolated PBMCs from heterozygous rs179008 A/T women were stimulated with either 1 μg/mL R-848 (n = 7 donors) or GagRNA1166 (HIV-RNAGag) (n = 7 donors); next, pDCs were surface-stained for self-secreted IFN-α and sorted into IFN-α+ and IFN-α– subsets. (B) Within-donor comparisons of TLR7 transcripts from the IFN-α+ and IFN-α– pDC subsets using KASP allele-specific PCR; results from a representative experiment encompassing A/A and T/T homozygous control cells. (C) Standard curve for the KASP assay, giving the relative abundance of allele A in the TLR7 transcript pool as a function of the fluorescence intensities from the A and T allelic probes. (D) Percentages of allele A transcripts expressed in the IFN-α+ and IFN-α– cell populations, determined with the standard curve in C. (E) Corroboration by deep-sequencing of the allelic proportions in the TLR7 cDNA amplimers from each sample in D. Group comparisons in D and E by Wilcoxon’s paired signed-rank test.
The rs179008 A > T substitution determines reduced protein expression in vitro.
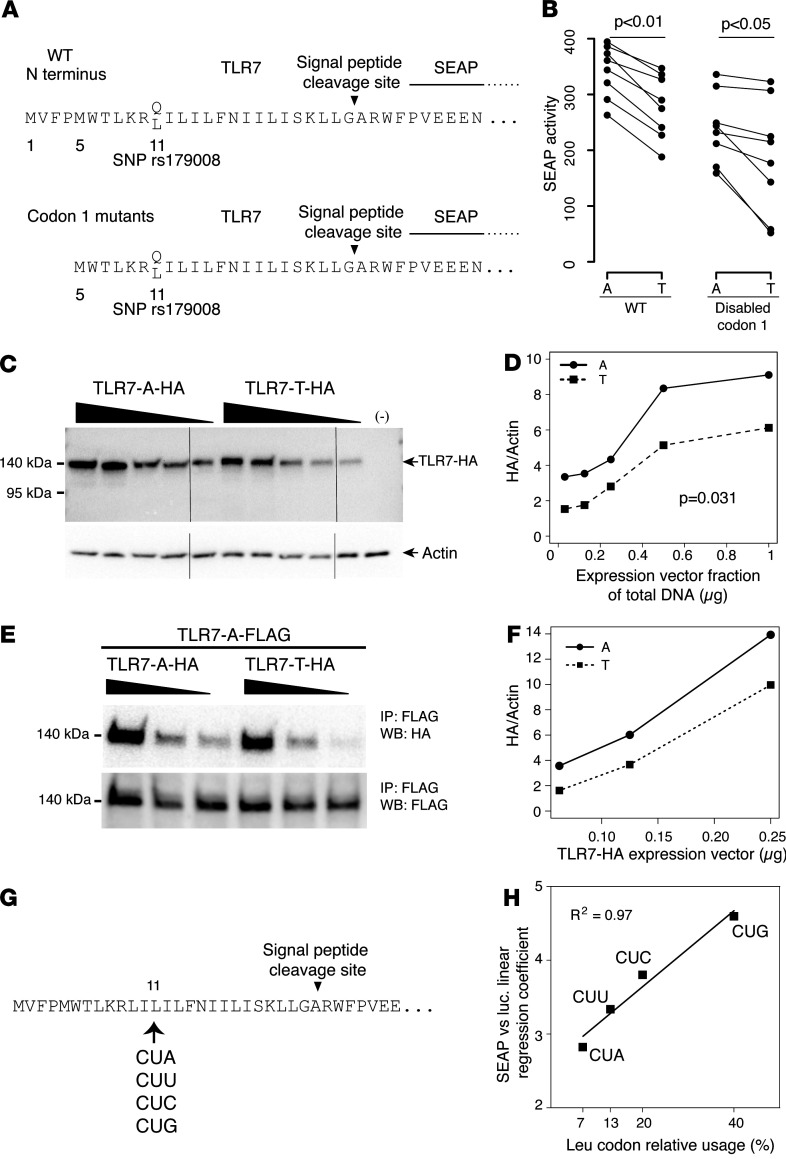
In assessing the mechanism of the allelic bias in interferogenesis, neither in silico predictions nor PCR amplification of the relevant TLR7 cDNA region indicated differences in TLR7 pre-mRNA splicing (Supplemental Figure 4, A and B). Likewise, modeling of mRNA folding failed to map the polymorphic base to regions of RNA secondary structure. Importantly, we ruled out a negative effect of the SNP on TLR7 mRNA abundance (Supplemental Figure 4, C and D). The substitution of a hydrophilic glutamine by a hydrophobic leucine residue (p.Gln11Leu) occurs within the N-terminal signal peptide of TLR7, which determines the cotranslational translocation of the nascent protein to the ER. Combined analysis of the amino acid sequence with the SignalP 4.1 and SignalP 3.0 software (see Methods) predicted a 26–amino acid signal peptide for both alleles, with a more favorable score for the Leu11 (rs179008 T) allele (0.837 versus 0.748 on a 0–1 scale, with a 0.450 threshold). The difference in scores is consistent with the longer span of the hydrophobic h-region of the signal peptide, Leu11– Ile21 in the Leu11 variant protein versus Ile12–Ile21 in the Gln11 allele (Figure 3A), because the central h-region is critical to the interaction of a signal peptide with the signal recognition particle and with the ER pore complex (28, 29). Overall, this argues against a deleterious effect of the Gln to Leu substitution per se on signal peptide function.
Figure 3. Decreased Leu11 (rs179008 T) TLR7 protein expression and secretion reporter activity.
(A) N termini of the TLR7-SEAP fusions encoded by reporter plasmids, with an allelic Gln11 (rs179008 A) or Leu11 (rs179008 T) signal peptide from TLR7 joined to a signal peptideless SEAP moiety. Parallel vector pairs encoded TLR7-SEAP proteins starting at either Met1 or Met5. (B) SEAP reporter activity in transfection supernatants of HEK 293T cells; paired allele A and allele T data from the same experiment. Wilcoxon’s paired signed-rank test. (C and D) Western blot analysis of HEK 293T cells transfected with varying amounts of expression vectors encoding Gln11 and Leu11 full-length TLR7 carrying a C-terminal HA peptide tag. Western blot and reporter enzyme data below are representative of at least 3 independent experiments. (D) Densitometry of TLR7-HA bands, normalized to β-actin. P value from the 2-way ANCOVA test comparing the 2 series of expression values for the A and T vectors. (E and F) Western blot analysis of TLR7 dimers. HEK 293T cells were cotransfected with a fixed amount of FLAG-tagged Gln11 TLR7 expression vector and decreasing amounts of HA-tagged Gln11 or Leu11 TLR7 vector. Proteins were IP with anti-FLAG antibody, and Western blots (WB) successively probed with anti-HA and anti-FLAG antibodies. (F) Densitometry of TLR7-HA bands normalized to TLR7-FLAG. (G) Synonymous-codon variants of the Leu11 TLR7-SEAP reporter vector. (H) SEAP versus control luciferase activity in HEK 293T transfection supernatants for the synonymous vectors, plotted against the respective human codon usage frequencies. R2 value from a Pearson test for correlation analysis.
To experimentally address the functional fitness of the TLR7 signal peptide, we engineered a pair of secretion reporter vectors encoding a secreted alkaline phosphatase (SEAP) with its autologous signal peptide replaced by amino acids 1–30 from TLR7; the vectors were identical except for the single-nucleotide change in codon 11 (Figure 3A). Transfection experiments on human embryonic kidney (HEK) 293T cells showed an approximately 20% decrease in the activity of the secreted reporter enzyme for Leu11 (T) relative to the Gln11 (A) form (Figure 3B). A second vector pair carried a mutated codon 1 to force translation initiation at a second in-frame AUG (Met5). Conservation of Met5 among placental mammals suggested alternative TLR7 translation initiation at this site; in addition, this Met codon opens the main reading frame in a minor alternative splicing isoform of TLR7 mRNA (30). These vectors retained approximately 70% of measured SEAP activity in cell culture supernatants and again showed lower activity for the Leu11 version (Figure 3B). These results evidenced lower activity attributable to the A-to-T change, specifically, but were in contradiction with the hierarchy of scores from the in silico signal peptide predictions.
We transfected HEK 293T cells with expression vectors for each TLR7 variant tagged with a C-terminal HA or FLAG epitope and analyzed recombinant TLR7 expression by Western blot. In line with the SEAP reporter experiments, we observed a 30%–40% decrease in TLR7 synthesis for the Leu11 (rs179008 T) form relative to the Gln11 protein (Figure 3, C and D). Next, we coexpressed FLAG-tagged Gln11 TLR7 with either Gln11 or Leu11 HA-tagged TLR7 to analyze TLR7 intracellular dimers. Immunoprecipitation with anti-FLAG antibodies followed by Western blot analysis with anti-HA antibodies confirmed that the rs179008 A > T substitution was associated with decreased TLR7 protein synthesis and homodimer formation in vitro (Figure 3, E and F).
The effect of rs179008 on translation is related to codon usage bias.
The CTA (CUA) Leu11 codon is the least frequent (7%) of the 6 leucine codons in H. sapiens genes, according to the Codon Usage Database (see Methods database sources) (31). Relative codon usage positively correlates with translation efficiency and the abundance of the corresponding anticodon tRNAs (reviewed in ref. 32). In allele rs179008 T, our analysis of the adaptation of the 5′ end of the TLR7 coding sequence to the human codon usage bias (Supplemental Figure 5) predicted the Leu11 CUA codon to be the first low-adaptiveness codon (33) met by the elongation-competent ribosome under the 5′-to-3′ scanning model for translation (34). As such, this codon could throttle translation efficiency through a longer ribosome residence time (35) relative to the higher-adaptiveness Gln11 CAA codon of rs179008 A. To explore the hypothesis of a codon effect on translation, we introduced single-nucleotide mutations (CTA to CTC, CTT, or CTG) in the Leu11 TLR7-SEAP reporter plasmid to generate a set of 4 expression vectors differentiated by their synonymous CTN Leu11 codon. By encoding the same protein (Figure 3G), codon-specific effects were dissected from signal peptide function in the reporter system. SEAP reporter activity was positively correlated with relative codon usage (Figure 3H), with the most (CUG) and least frequent (CUA) codons at opposite ends of the range of activities. Codon 11 is, thus, a likely control point for TLR7 mRNA translation, and this effect could involve interference with translation initiation, the rate-limiting step in protein synthesis, owing to the proximity of the polymorphic codon to the 5′ end of the TLR7 coding sequence (36).
The T allele lowers TLR7 protein expression in female PBMCs specifically.
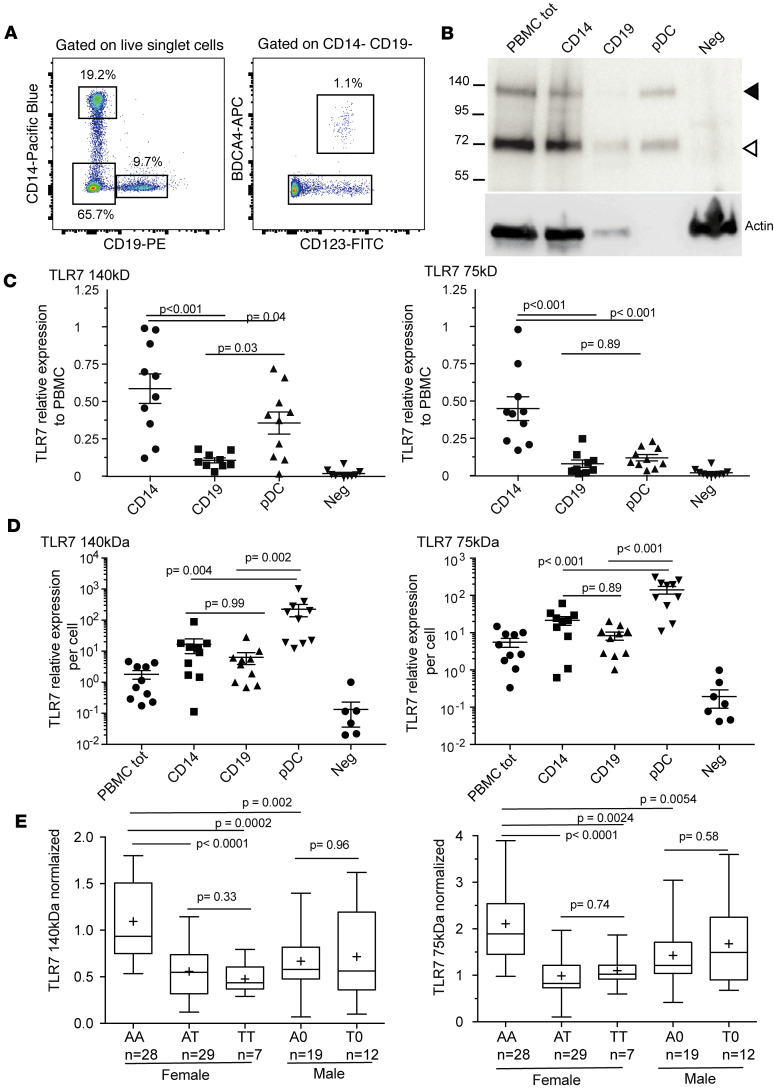
We recently characterized a commercially available monoclonal antibody to the C-terminus of human TLR7 that correctly detects natural and recombinant TLR7 in Western blotting, but not TLR7 modified with a C-terminal epitope tag (Supplemental Figure 6 and ref. 27). By this method, we quantified the full-length (140-kDa) and proteolytically mature (75-kDa) forms of TLR7 in a deconvolution experiment to characterize the principal TLR7-producing cell populations among steady-state PBMCs (Figure 4, A and B). Most of the TLR7 signal came from CD14+ monocytes, followed by pDCs and then by B cells, with regard to both the full-length and mature forms of TLR7 (Figure 4C). A small proportion of TLR7 (<5%) came from the CD14−CD19−BDCA4− cell fraction mainly comprising T lymphocytes and NK cells. When the densitometric data were expressed on a per-cell basis (Figure 4D), pDCs exhibited the highest level of full-length TLR7, with a 100-fold enrichment over unsorted PBMCs, followed by CD14+ monocytes and B cells. A similar hierarchy was found for the mature form of TLR7.
Figure 4. Quantification by Western blotting of TLR7 protein in PBMC subpopulations.
(A) Gating in the flow cytometric isolation from cryopreserved PBMCs of CD14+ monocytes, CD19+ B lymphocytes, CD123+BDCA4+ pDCs, and the CD14−CD19−BDCA4− cell fraction for Western blot analysis. (B) Representative blot for a single human donor. In each lane, the number of cells in the sample was proportional to the relative abundance of the cell type among total PBMCs. Neg, CD14−CD19−BDCA4− cell fraction. (C) Relative contribution from each cell type. Densitometric analysis of TLR7 for 9 different donors. The 140- and 75-kDa forms were quantitated separately and normalized to same-donor signals for total PBMCs. (D) Densitometry normalized to the number of cells represented in each lane, displaying per-cell quantities of TLR7 in the different cell types. (E) TLR7 quantitation in total cryopreserved PBMCs from female or male donors classified by rs179008 genotype. The TLR7 densitometric signal was first normalized to β-actin and then to the corresponding values of a standard PBMC cell lysate as described in Methods. Data presented as a box spanning the 25th to 75th percentiles, with the line in the middle of the box plotted at the median and whiskers going down to the smallest value and up to the largest; “+” denotes the mean. P values from Kruskal-Wallis test corrected for multiple comparisons by controlling the FDR (Benjamini-Hochberg method).
Next, we quantified by Western blotting the impact of the T allele on TLR7 protein expression within PBMCs of different rs179008 genotypes (Supplemental Figure 6). PBMCs from women heterozygous (A/T) and homozygous (T/T) for the T allele expressed significantly less TLR7 protein than A/A women (Figure 4E). We observed no significant differences between A/0 and T/0 male donors (Figure 4E). By contrast, we observed a significant 2-fold excess of full-length TLR7 in A/A women over A/0 men (Figure 4E), as well as a trend in the same direction for the 75-kDa mature form (Figure 4E), in agreement with our earlier work (27) noting greater TLR7 protein levels in female than in male human PBMCs.
The hypomorphic effect of rs179008 T on TLR7 protein expression is maintained in postmenopausal women.
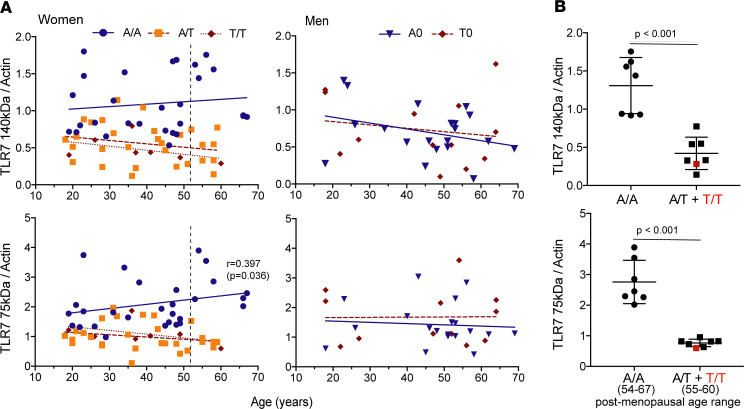
The functional responses of pDCs from women decrease after menopause as they are partly dependent on estrogen-receptor signaling (25, 37). Premenopausal plasma levels of estradiol, which range between 50 and 65 pg/mL in women of various ethnicities (38), decline below 20 pg/mL within 1 year after the final menstrual period in the majority of White women (38). Plotting the TLR7 protein quantitation data in Figure 4E against the age of donors (Figure 5A), however, failed to show a dependency of the quantitative effect of rs179008 on age. Only a modest positive correlation (r = 0.397, P = 0.036) was observed here for mature TLR7 expression in A/A women (Figure 5A). In men, TLR7 expression was noncorrelated with age or with the rs179008 allele (Figure 5A).
Figure 5. The rs179008 T allele lowers TLR7 protein expression in PBMCs from both pre- and postmenopausal women.
(A) Expression data for the 140- and 75-kDa forms of TLR7 presented in Figure 4E, plotted here against donor age. The dashed line represents the median age of the final menstrual period (51.5 years) (38). Spearman’s correlation coefficients and corresponding P values are shown when P < 0.05. Some significant regressions were observed for women (P < 0.0001) but not men. (B) Women above the age of 53 were selected for further comparisons of TLR7 protein expression between A/A homozygotes (n = 7) and T allele carriers, A/T (n = 6) and T/T (n = 1) as indicated. P values from Mann-Whitney U tests, 2-sided.
The median age of women at the final menstrual period is 51.5 years (38). In our data for likely postmenopausal women, older than 53 years of age, A/A women displayed markedly higher levels of the full-length and mature forms of TLR7 than the carriers of allele T (P < 0.001; Figure 5B). These results show that the hypomorphic effect of rs179008 T on TLR7 protein expression is present on either side of menopause, which supports the hypothesis that this effect is not primarily dependent on estrogen signaling.
The T allele is associated with reduced clinical parameters of HIV-1 infection in women.
In studying the effect of rs179008 on HIV-1 infection, we explored the previously reported association (22) with the acquisition of HIV-1 by women in 2 separate case-controlled settings, restricted to White women to minimize genetic diversity regarding rs179008 allele frequency distribution and other potential genetic determinants. The ANRS EP53 X-LIBRIS study observed a cohort of female White patients infected by sexual transmission before menopausal status (n = 90) compared with a control group (n = 90) of healthy women from the same age stratum and geographical origin (Toulouse, France). The second comparison involved adult White females infected by sexual transmission included in the French ANRS PRIMO multicenter cohort (n = 151) upon diagnosis of primary HIV-1 infection, compared with a control group (n = 87) of healthy females from throughout France. Neither comparison supported a significant genetic association of rs179008 with HIV-1 acquisition through the sexual route in women, and a meta-analytical combination of the 2 studies pointed to nonassociation (OR = 1.00; 95% CI, 0.67–1.51; Supplemental Figure 7).
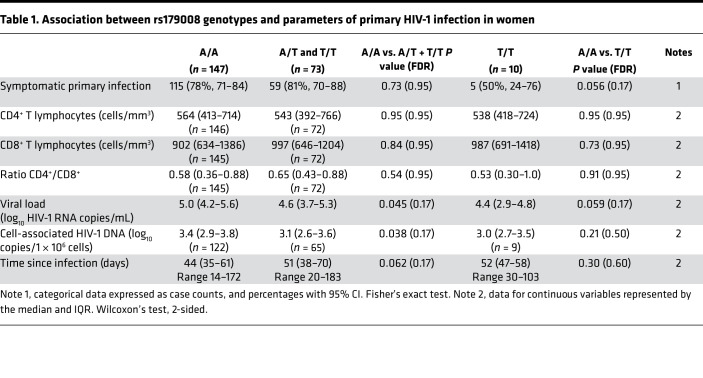
By contrast, we observed an association between rs179008 and clinical parameters of primary infection at the time of cohort inclusion. Women of White and African ethnicity of the ANRS PRIMO cohort were analyzed together to expand the number of cases to n = 220. Carriage of rs179008 T was associated with a lower plasma viral load with 4.6 log10 RNA copies/mL (IQR, 3.7–5.3) for the combined A/T and T/T genotypes versus 5.0 log10 RNA copies/mL (IQR, 4.2–5.6) among A/A homozygotes (P = 0.045, Wilcoxon’s test). Cell-associated HIV-1 DNA was also reduced in women harboring rs179008 T: 3.1 log10 DNA copies/106 PBMCs (IQR, 2.6–3.6) for the combined A/T and T/T genotypes versus 3.4 log10 DNA copies/1 × 106 PBMCs (IQR, 2.9–3.8) among A/A homozygotes (P = 0.038, Wilcoxon’s test). The frequencies of circulating CD4+ and CD8+ T cells were not significantly different between carriers and noncarriers of allele rs179008 T (Table 1). We next looked for potentially greater contrasts by comparing the opposite A/A and T/T homozygous genotypes (Table 1). We controlled for the confounding of RNA viral loads dependent on time since infection by using a fractional polynomial regression (Supplemental Figure 8 and Supplemental Note 2). The regression model showed a difference of −0.85 log10 (95% CI, −1.51 to −0.18), a 7-fold decrease in viral load, for the T/T relative to the A/A genotype (P = 0.013; regression P value). In addition, only 50% of T/T homozygotes (5 of 10) displayed symptomatic infections at inclusion, versus 78.2% (115 of 147) among the A/A patients (P = 0.056; Fisher’s exact test). Among the commonly reported clinical signs and symptoms of primary HIV-1 infection (39) are tonsillitis and pharyngitis, which were absent among T/T patients (Supplemental Table 1).
Table 1. Association between rs179008 genotypes and parameters of primary HIV-1 infection in women.
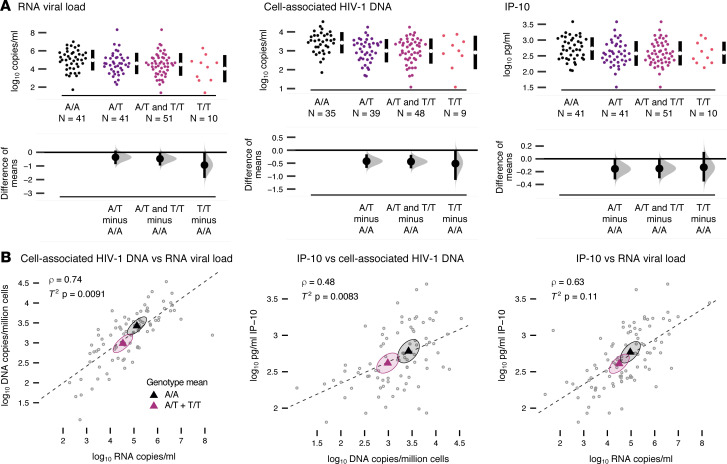
We measured additional plasmatic parameters of HIV-1 infection and the immune response, namely IFN-α and CXCL10 (IP-10), among the 10 T/T PRIMO patients plus randomly drawn subsets of the A/A and A/T groups (n = 41 for each genotype; n = 92 total). IP-10 was used as a surrogate marker enabling more robust detection and quantitation than IFN-I; in the PRIMO subcohort, detectable IFN-α was, in fact, associated with significantly higher IP-10 values (Supplemental Figure 9). Splitting the subcohort based on the symptomatic or asymptomatic presentation of primary HIV-1 infection resulted in 2 groups displaying clear, significant differences with each other in respect to RNA viral load and cell-associated HIV-1 DNA, IP-10 levels, and detectable IFN-α. All these parameters were lower in the asymptomatic fraction (Supplemental Table 2), which, as noted above, was enriched among the rs179008 T/T relative to the A/A patients. This subcohort showed the previously noted association of rs179008 T carriage with lower values for RNA viral load and cell-associated HIV-1 DNA, confirming that the randomly drawn subsets were representative of the source group. We also observed a trend for lower IP-10 values (Figure 6A and Supplemental Table 3). These 3 parameters were in positive correlation with one another (Figure 6B). Thus, the comparisons of the rs179008 T carrier (A/T, T/T) and noncarrier (A/A) groups revealed significant differences for RNA viral load combined with cell-associated HIV-1 DNA, for IP-10 combined with cell-associated HIV-1 DNA (Figure 6B), and for the combination of the 3 parameters (P = 0.023, Hotelling’s T2 test). Lastly, mean RNA viral load among T/T patients was here 0.97 log10 units lower than in rs179008 A/A patients in the data adjusted for time since infection, a 9-fold difference (P = 0.025; 2-tailed Student’s t test).
Figure 6. Effect of the rs179008 genotype on IP-10 and HIV-1 nucleic acid measurements across a PRIMO subcohort.
ELISA quantitation of IP-10 was performed on the 10 PRIMO female patients of rs179008 T/T genotype plus randomly drawn subsets of A/A and A/T women from the same cohort (n = 92 total). (A) Estimation plots comparing rs179008 genotype groups against the A/A baseline with regard to viral load, HIV-1 cell–associated DNA (n = 83 total), and IP-10 values. Top, raw data groups with mean ± SD shown as a gapped line. Effect size is plotted underneath as the difference in means and its 95% CI estimated by bootstrap resampling; the shaded curve denotes the resampling distribution. (B) Pairwise relationships between the 3 HIV-1 infection parameters. Pearson’s ρ correlation coefficient and the regression line are shown on each scatter plot, together with the bivariate mean and its 95% confidence ellipse for A/A homozygotes and for carriers of allele rs179008 T. P values from Hotelling’s T2 test on the difference of mean vectors between the 2 groups.
Genetic association with HIV-1 viral load in the wider context of TLR7 polymorphism.
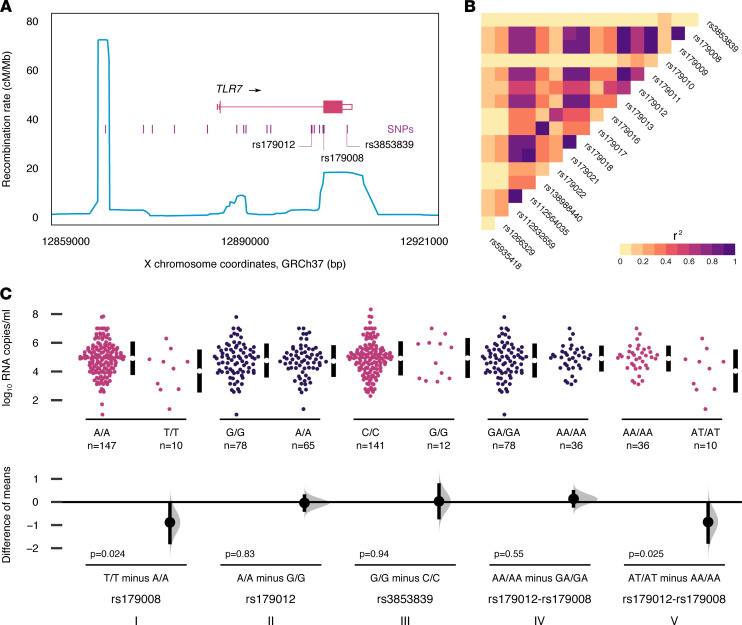
rs179008 is one of a group of frequent TLR7 SNPs within a 40-kb linkage disequilibrium (LD) block on the X chromosome (Figure 7A). Pairwise LD is moderately high (r2 = 0.63) between rs179008 and the frequent intronic tag SNP, rs179012, previously associated with HIV-1 set-point viral load (21), but it is weak (r2 = 0.04) between rs179008 and a frequent functional SNP in the 3′ UTR of TLR7, rs3853839 (Figure 7, A and B), in genetic association with the risk of developing systemic lupus erythematosus (40). We carried out an estimation analysis to compare the effect of each SNP on HIV-1 viral load among the PRIMO female patients (n = 220) (Figure 7C). For rs179008, we again observed a clear difference in mean viral loads between the opposite A/A and T/T homozygous genotypes (Figure 7C), as expected from our earlier comparisons (Table 1). By contrast, we found the difference between opposite homozygous genotypes to be very weak or absent for rs179012 (G/G versus A/A; Figure 7C) and rs3853839 (C/C versus G/G; Figure 7C). To control for confounding due to LD between rs179008 and rs179012, we considered rs179012–rs179008 diplotypes where the genotype for one of the SNPs was the same in the 2 groups of patients compared; in this way, the effect of the second SNP could be assessed while removing the effect of the first (Figure 7C). This fully corroborated the direction and magnitude of the effect for rs179008 with independence from rs179012. Taken together, our biochemical and genetic association studies support the conclusion that rs179008 is a functional polymorphism of TLR7 in its own right.
Figure 7. Effect of TLR7 SNPs on viremia during HIV-1 primary infection.
(A) Map of TLR7 and SNPs with minor allele frequency > 0.1, including the 3 SNPs analyzed for association with viral load. Blue line, recombination rates from the Phase II HapMap project. (B) Heatmap of pairwise linkage disequilibrium (r2) for the frequent SNPs in A ordered by position along TLR7. Allelic frequencies and r2 values were derived from individual genotypes for the European populations in the 1000 Genomes Project Phase 3. (C) Estimation plot comparing viremia values between PRIMO females of opposite homozygous genotypes for rs179008, rs179012, and rs3853839. See Supplemental Note 1 for the parsing of genotype and diplotype notations. To control for linkage disequilibrium between rs179012 and rs179008, sections (IV) and (V) compare homozygous rs179012–rs179008 diplotypes: (IV) compares opposite rs179012 genotypes while keeping the rs179008 genotype uniform and (V) compares opposite rs179008 genotypes given a uniform rs179012 genotype. Data may be duplicated across plot sections. P values from Student’s t tests, 2-sided.
Discussion
We observed here that the rs179008 SNP of TLR7 exerts in female patients a significant effect on key parameters in the clinical course of HIV-1 acute infection. Although recent findings strongly suggest a detrimental effect of pDCs and IFN-I during the chronic phase of HIV-1 infection, the early-stage role of IFN-I produced by pDCs as a result of TLR7 activation is still not fully understood (41). We have addressed this issue firstly through the functional characterization of a frequent TLR7 SNP, rs179008, which we found to exert a quantitative effect on TLR7 protein synthesis and pDC IFN-I production in women specifically, and secondly by analyzing the association of TLR7 polymorphisms with clinical parameters of HIV-1 infection. Our results demonstrate a protein-level and interferogenic cellular phenotype of rs179008. Carriage of the rs179008 T allele lowered ex vivo TLR7-driven IFN-I production by pDCs from heterozygous A/T women relative to A/A homozygotes. Mechanistically, we show this allele to determine lower TLR7 protein expression in female leukocytes, which mirrors the reduced frequency of IFN-α−producing female pDCs. Lastly, we observed a beneficial association between the lower-interferogenesis allele, rs179008 T, and lower levels of informative correlates of acute HIV-1 infection severity in women.
These results may appear counterintuitive in light of previous studies supporting a beneficial function of pDCs and IFN-I signaling through the control of viral replication during acute infection in macaque and humanized mouse models (11, 12). The effects of innate immunity on HIV-1 infection, however, are complex and not consistently positive. The recruitment and activation of pDCs and monocytes in the vaginal mucosa of female macaques early after local SIV inoculation helps attract CD4+ T cells, which can then be infected to boost viral production and dissemination (13, 42). Conversely, prolonged exposure to IFN-I is associated with unfavorable outcomes in chronic HIV and SIV infection (19). A deleterious effect is, thus, conceivable for TLR7-driven activation of pDCs and other innate immune cells, even during acute-phase HIV-1 infection, by recruiting virus target cells at mucosal portals of entry (14) or initiating immunosuppressive pathways mediated by pDC-derived IFN-I (17, 18). Such early mechanisms might be toned down in women carrying the rs179008 T allele of TLR7, blunting the initial spread of the virus. Our results highlight that the dosage of IFN-α during acute HIV infection is critical. The effects of the polymorphism on the outcome of chronic HIV-1 disease deserve further investigations.
We show also that this nonsynonymous polymorphism of the signal peptide controls protein dosage at the translational level through the mRNA sequence itself, rather than by altering signal peptide function. The connection of genotype to phenotype via translation is compatible with the expected loss of the protein’s allelic character when the signal peptide is cotranslationally cleaved off on entering the ER. Overall codon usage in TLR7 is known to be markedly suboptimal (43), and translation of the mRNA relatively inefficient (43, 44), which makes translation a likely rate-limiting step in TLR7 expression. Local translation efficiency depends on the abundance of aminoacyl-tRNAs with compatible anticodons, and this is positively correlated with the relative usage of synonymous codons (45). Given the 5′ to 3′ mRNA scanning direction of ribosomes (34), the Leu11 CUA (rs179008 T) codon could play a translation rate–limiting role as the first rare codon in the path of a translation-competent ribosome (Supplemental Figure 5). In addition, the ribosome at codon 11 may sterically hinder translation initiation, the rate-limiting step in protein synthesis, because the span between codons 1 and 11 is similar to the 20- to 30-nucleotide footprint of a ribosome (46) and less than the 32- to 39-nucleotide pitch of ribosomes in tightly packed polysomes (47). This would be in line with evidence that the effect of nonoptimal codons on translation efficiency is greatest near the 5′ end of the coding sequence (36). Further experimental work addressing translation directly will be necessary to confirm this mechanism. To our knowledge, the notion of a sex bias in regulatory mechanisms of translation has yet to be explored in the context of human biology, and it is uncertain whether the sex dependency we have observed in the TLR7 quantitative protein phenotype for rs179008 (Figure 4) could be mediated by translational regulation. The nondependency of TLR7 quantitation on women’s age (Figure 5) strongly suggests that estrogens are not involved in the sex-biased phenotype because this is observable beyond menopause, despite the expected decline in estrogen levels (38).
In agreement with earlier reports (23–25), we observed a sex bias in the TLR7-driven production of IFN-α by human pDCs, with higher frequencies of IFN-α–producing cells in women than in men. This was most noticeable when comparing women and men of similar A/A and A/0 rs179008 genotypes. The sex bias in the TLR7-mediated response of pDCs arises from mechanisms implicating estrogen signaling (25, 37, 48) and the X-chromosome complement (37). The X-linked TLR7 gene escapes X chromosome inactivation in female immune cells, including pDCs, and was associated with higher TLR7 protein expression in leukocytes from rs179008 A/A women compared with A/0 men (27). In line with the concept of TLR7 dosage–dependent effects determined at the chromosome level, we found here the carriage of allele rs179008 T in females to be associated both with a decrease in TLR7 protein relative to A/A homozygous pDCs and with lower per-cell IFN-I production. Future studies of the functional response of pDCs from women should, therefore, consider the genotype for this polymorphism of TLR7.
Oh et al. (22) identified rs179008 as a determinant of HIV-1 infection prevalence in women. Our observations fail to support an effect of the SNP on HIV-1 acquisition by the sexual route in women. This discrepancy might be explained by the small size of the healthy control group in that study (22), where the frequency of the T allele was clearly lower than in relevant reference populations, leading to overestimation of effect size (49). In that study, the Leu11 (rs179008 T) allele of TLR7 was associated with more severe HIV disease progression in men (22). We show that this allele is not associated with lower constitutive TLR7 protein expression or pDC interferogenesis in men, as initially suggested by Oh et al. (22), but we cannot exclude a sex-specific effect of rs179008 on TLR7 expression in other immune cell types important for the progression of chronic HIV-1 disease. In addition to B lymphocytes, pDCs, and monocytes, TLR7 is expressed at lower levels in human CD4+ T cells (50), where TLR7 activation has been shown to induce anergy and to promote infection by HIV-1 (50). Conceivably, the polymorphism might alter HIV-1 disease progression through mechanisms dependent on sex, the phase of infection, and the immune cell population involved. A limitation of the present study is that data on HIV-1 patient cohorts are observational and circumscribed to women, which does not allow for a comparison with the longitudinal study of the effect of rs179008 on CD4+ T cell counts in men as previously reported by Oh et al. (22).
Our observations led us to propose that increased dosage of TLR7 protein is conducive to a higher viral burden during acute HIV-1 infection in women and correlates with a predictor of the speed of disease progression. The reduced TLR7 protein values and pDC interferogenesis associated with allele rs179008 T were accompanied by significantly lower viremia and cell-associated HIV-1 DNA measurements in women from the ANRS-PRIMO cohort. We also investigated the association between rs179008 T and further clinical parameters of HIV-1 infection, namely plasma IP-10 (CXCL10) and IFN-α levels, among a representative subset of these female patients. Plasma IP-10 is known to display a robust positive correlation with viral load (51–53) and is responsible for the recruitment of CXCR3+ target T cells, fueling viral dissemination and the establishment of a viral reservoir (54). Elevated blood IP-10 during primary HIV-1 infection predicts rapid disease onset more robustly than peak viremia or the CD4+ cell nadir (53, 55). Although pDCs can produce IP-10 through direct TLR7 stimulation, this cytokine can be indirectly induced in cells such as monocytes and mDCs by pDC-derived IFN-I (56, 57). We found here, as expected, that patients with detectable plasma IFN-α exhibited higher levels of IP-10 and that both IFN-α and plasma IP-10 were associated with symptomatic HIV-1 infection. Importantly, rs179008 T was significantly associated with lower levels of a triad of positively correlated parameters: RNA viral load, the cell-associated HIV-1 DNA reservoir, and plasma IP-10. In addition, the higher frequency of asymptomatic clinical presentations at diagnosis for the T/T homozygous genotype implies a trend for a delayed onset of the symptoms of acute infection.
Heritable quantitative variation of TLR7 dosage and function provides here valuable insights into the role of this receptor as a determinant of acute viral load. Our data point to TLR7 as a key effector molecule, with a dominant deleterious action of higher TLR7 dosage during acute HIV-1 infection in women. This, in turn, argues for a beneficial effect of toning down a strong IFN-I response early on, even before the acute-to-chronic phase transition. Corroboration of the genetic association with disease parameters in those populations where the rs179008 T allele is frequent will be important.
Methods
Patients and healthy controls.
The X-LIBRIS patient cohort consisted of 90 nonmenopausal, HIV-1+ White women aged 18 or older, infected by sexual transmission before menopausal status, and with sustained undetectable plasma viral load for a median of 7.5 years (IQR, 4–10) under combined antiretroviral therapy, followed up at the Purpan Hospital in Toulouse, France. The healthy control group for the X-LIBRIS cohort consisted of 90 nonmenopausal White women, approximately age-matched to the patient group and from the same geographic area, and was exclusive of subjects suffering from immune dysfunction or from acute or chronic infection. An additional DNA sample panel representing 87 nonrelated healthy French women was sourced from the multicentric collections of the Centre d’Etude du Polymorphisme Humain (CEPH, Fondation Jean Dausset, Paris). The French ANRS-PRIMO CO6 cohort enrolls patients presenting with primary HIV-1 infection, as previously described (58). We selected female patients with available frozen PBMC or whole blood samples. All patients gave written informed consent. The date of infection was alternatively defined as the date of the incomplete Western blot minus 1 month, the date of diagnosis of symptomatic acute infection minus 15 days, or the midpoint between a negative and a positive HIV antibody test. Further blood samples of anonymous healthy blood donors were sourced from the local blood transfusion center (Etablissement Français du Sang, Toulouse, France) in compliance with French regulations.
Genomic DNA extraction and SNP genotyping.
Genomic DNA was extracted from cryopreserved or freshly isolated PBMCs using a NucleoSpin Tissue kit, or from frozen blood using a NucleoSpin Blood kit (both kits from Macherey-Nagel) and genotyped for TLR7 SNPs by a KASP allele-specific PCR assay (LGC Genomics) run on a LightCycler 480 II instrument (Roche).
Analysis of IFN-αproduction by blood mononuclear cells. PBMCs were freshly isolated from whole blood by Pancoll (PAN-Biotech) gradient separation and resuspended in R10 medium — i.e., RPMI 1640 (Sigma-Aldrich) containing 10% heat-inactivated FBS (Sigma-Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (Life Technologies). The cells were seeded into 96-well plates (3 × 105 cells/well) and stimulated with either resiquimod (R-848; 1.5 μg/mL or 0.3 μg/mL) or imiquimod (R-837; 1.5 μg/mL; both from InvivoGen). IFN-α was quantified by ELISA (PBL Biomedical Laboratories) in culture supernatants sampled 6, 12, and 24 hours after stimulation. Supernatants from 24-hour cell cultures without any TLR7 ligands were used as controls. ELISA values were normalized to the percentage of pDCs in the respective PBMC preparation and expressed as IFN-α produced per 1 × 103 pDCs as described elsewhere (25).
Intracellular cytokine staining.
Freshly isolated PBMCs (2.5 × 106 cells/mL) were alternatively stimulated with R-848 (0.3 μg/mL or 1 μg/mL), R-837 (1 μg/mL), or 30 μg/mL HIV-1–derived GagRNA1166 synthetic oligoribonucleotides (24) (Eurogentec) complexed with 1,2-dioleoyloxy-3-trimethylammonium-propane (DOTAP; Roche), during 5 hours. Nonstimulated cells served as a negative control. Brefeldin A (eBioscience) was added for the last 3 hours of culture. Human PBMCs were surface labeled with anti–BDCA-4-allophycocyanin (REA774, Miltenyi Biotec, 130-111-894), anti–CD123-phycoerythrin-cyanine 5 (9R5, BD Biosciences, 551065) and anti–Lin-FITC antibody conjugates (lin1, BD Biosciences, 340546). Cells were then fixed with 2% paraformaldehyde in PBS, permeabilized with 0.5% saponin, and stained for intracellular cytokine production using anti–IFN-α–phycoerythrin (LT27:295, Miltenyi Biotec, 130-092-601) and anti–TNF-α–Alexa700 (MAb11, BD Biosciences, 557996). Flow cytometry analysis was performed on a BD Biosciences LSRII instrument, and pDCs were defined as the Lin−CD123+BDCA-4+ cell fraction (Supplemental Figure 2). The frequency of cytokine-producing pDCs was determined by subsequent data processing with the FlowJo software.
Intraindividual analysis of the impact of rs179008 on IFN-αproduction by pDCs. Freshly isolated PBMCs (10 × 106 cells/mL) from healthy rs179008 A/T heterozygous women were stimulated for 5 hours with 1 μg/mL R-848 or with GagRNA1166 complexed with DOTAP. Cells were then surface-labeled for IFN-α production using an IFN-α secretion assay from Miltenyi Biotec (catalog 130-094-161), according to the manufacturer’s instructions. Briefly, stimulated cells were coated with IFN-α capture reagent (anti–IFN-α monoclonal antibody conjugated to a cell surface-specific monoclonal antibody) and then incubated for 20 minutes at 37°C to allow IFN-α secretion. Cells were then surface-stained with IFN-α detection antibody (anti–IFN-α monoclonal antibody conjugated to phycoerythrin) and for BDCA-4, CD123, and Lin as above. pDCs were then sorted on a FACSAria II instrument (BD Biosciences) into Lin−CD123+BDCA-4+IFN-α+ and Lin−CD123+BDCA-4+IFN-α− populations. RNA from both purified populations was extracted using the Absolutely RNA Nanoprep kit (Agilent Technologies) and retrotranscribed with the Maxima H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific). The cDNA was subjected to target amplification using TLR7 PCR primers flanking the SNP (oligonucleotides 1F and 2R below) followed by KASP typing as above to determine the rs179008 allele of origin of TLR7 transcripts. The percentage of allele A expression in each cell population was given by the standard curve for the KASP PCR assay (Figure 2C) as a 4-parameter logistic function of the log-ratio of the A and T fluorescence intensities. The curve was generated using genomic TLR7 PCR amplimers from male (hemizygous) A/0 and T/0 subjects in different mix ratios.
Ultra-deep sequencing.
A 273-nucleotide–long TLR7 cDNA fragment encompassing the position of the rs179008 SNP was generated by PCR as above. The PCR products were amplified on capture beads in water-in-oil emulsion microreactors, and pyrosequencing was performed on a Roche 454 GS Junior system. The sequence reads were quantified with the GS amplicon variant analyzer (Roche).
Analysis of pre-mRNA splicing.
RNA from PBMCs was isolated and reverse-transcribed as above. The exon 2–exon 3 splice junction in the TLR7 cDNA was amplified by PCR with forward primer 1F: 5′-CCATGCCATCAAGAAAGTTG-3′ straddling the exon 1–exon 2 junction, and reverse primer 2R: 5′-TCTGTGCAGTCCACGATCA-3′ hybridizing 155 bp downstream from the canonical TLR7 exon 2–exon 3 junction. PCR products were analyzed by agarose gel electrophoresis.
Quantitation of TLR7 transcripts.
B cells were isolated from PBMCs with an immunomagnetic negative selection kit from STEMCELL Technologies. Total RNA was then extracted with the RNeasy Mini kit (Qiagen) and reverse-transcribed with Maxima reverse transcriptase (Thermo Scientific) using random hexamer primers. Quantitative PCR (qPCR) was performed in the presence of SYBR Green (Roche) on a LightCycler 480 II instrument, and TLR7 expression was determined by the ΔCt method relative to the housekeeping gene, GAPDH. The PCR primer pairs were TACCTGGATGGAAACCAGCTACT and CAAGGCTGAGAAGCTGTAAGCTA (TLR7 mRNA), and AGCACCAGGTGGTCTCCTCT and CCAAATTCGTTGTCATACCAG (GAPDH mRNA). Kits and reagents were used according to the manufacturer’s instructions.
Quantification of TLR7 protein expression by Western blotting.
Cell lysates were prepared in Laemmli sample buffer (Invitrogen), sheared through a 31G needle, and total protein was quantified by a bicinchoninic acid protein assay (Pierce). Samples were heated for 10 minutes at 70°C in the presence of a reducing agent (Invitrogen). Protein (20–25 μg per lane) were fractionated by SDS-PAGE on precast 4%–15% gradient gels (Bio-Rad) and transferred to Amersham Hybond 0.45-μm PVDF membranes (GE Healthcare). The membranes were blocked with 5% skim milk and 0.1% Tween-20 in PBS, probed overnight with antibodies to β-actin (mouse monoclonal IgG1, clone AC-15; MilliporeSigma), the human influenza hemagglutinin (HA) epitope tag (rat monoclonal IgG, clone 3F10; Roche), the FLAG epitope tag (mouse monoclonal IgG1, clone M2; MilliporeSigma), or the C-terminus of human TLR7 (rabbit monoclonal IgG, clone EPR2088[2]; Abcam) and finally incubated with suitable peroxydase-conjugated secondary antibodies: anti–mouse IgG 7076, anti–rabbit IgG 7074, or anti–rat IgG 7077 (Cell Signaling Technology). Chemiluminescent detection was carried out with Amersham ECL Select or ECL Prime reagents (GE Healthcare) as necessary, and densitometric analysis was performed with the Image Lab 5.0 software (Bio-Rad).
TLR7 protein quantification in healthy subjects of the different rs179008 genotypes was performed on cryopreserved PBMCs. PBMCs were thawed and cultured at 37°C for 2 hours before counting, cell lysate preparation, and Western blotting as above. The densitometric signals of the TLR7 145-kDa and 75-kDa forms were normalized to β-actin and then to an internal standard PBMC lysate that was loaded in each gel for intergel data normalization as described in Supplemental Figure 6.
TLR7-SEAP reporter vectors.
The signal peptide reporter vectors encoded a SEAP where the autologous signal peptide was replaced by an allelic form of the human TLR7 signal peptide (Figure 3A). The TLR7 and SEAP moieties connected at a proline residue analogous to Pro31 in TLR7 and to Pro20 in SEAP (GenBank U89937.1) (see below). Preparatory analyses with the SignalP 4.1 software (59) predicted in each case a signal peptide with a near-identical profile to that of the parental TLR7 form (not shown). The SEAP gene under the control of the human EF-1α promoter was released from pYSEAP (60) (Addgene plasmid no. 37326) as a 3.5-kb SpeI-SalI fragment and inserted into a modified pBC KS+ vector (Stratagene) to generate pYSEAP-JM562. Next, a 360-bp fragment of pYSEAP-JM562 overlapping the 5′ end of the SEAP coding sequence was excised with EcoRI and NdeI, and it was in-frame replaced by one of a series of synthetic TLR7-SEAP DNA fragments. The hybrid inserts were generated by successive PCR steps with the Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific) and included 69 proximal nucleotides of the TLR7 5′-untranslated region to preserve a predicted RNA stem-loop structure around the start codon (not shown). Initial TLR7 cDNA amplifications were seeded with reverse-transcribed pDC or PBMC RNA from male donors carrying the relevant rs179008 allele. Where necessary, single-base changes in TLR7 codons 1 or 11 (Figure 3G) were introduced in the design of PCR primers; to force translation initiation at Met5, the major ATG initiation codon was made a TTG triplet. In the Leu11 synonymous versions, the natural CTA leucine codon was replaced by a CTC, CTT, or CTG leucine codon. Vectors for leucine codons TTA and TTG were not generated. In all plasmid constructs, any regions derived from synthetic DNA fragments were subjected to complete DNA sequencing to rule out unwanted sequence variation. The sequence of a prototypic TLR7-SEAP reporter vector has been deposited in the GenBank database under accession number KX035095 (see below).
Metridia luciferase expression vector.
Vector pPgk1-M.Luc was assembled by inserting the Metridia luciferase gene from pDonor-hβ-Actin-hMLuc (61) (a gift from Shawn Lupold, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA) downstream from the murine Pgk1 promoter in pJM1950 (62), as a NcoI-XhoI fragment replacing the initial EGFP gene.
Plasmid transfection.
Human embryonic kidney (HEK) 293T/17 cells (from ATCC, CRL-11268) were grown in DMEM supplemented with 5% or 10% heat-inactivated FBS and 20 μg/mL gentamicin, and seeded into 24-well culture plates the day before transfection (1 × 105 cells/well in 1 mL of culture medium). Plasmid DNA was complexed with TurboFect cationic polymer transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, 1 μg DNA in 100 μL serum-free medium was mixed with 2 μL of TurboFect, and 15–30 μL of this transfection medium were added to a culture well. Transfection supernatants and cells were harvested 48 hours later and stored frozen at −20°C until use; for the firefly luciferase assay, the cells were lysed in 1× passive lysis buffer (Promega), and the lysates were stored at −75°C. For transfection normalization, either a Metridia luciferase vector, pPgk1-M.Luc, or a firefly luciferase vector, pGL4.23 (Promega), were mixed with the reporter vector to 10% of total DNA.
Expression of full-length TLR7.
Human TLR7 expression vectors were derived from pUNO1-hTLR7-HA3× (InvivoGen), encoding TLR7 (Gln11) tagged at its C-terminus with 3 tandem copies of the HA epitope. First, the neo gene was inserted into the NotI site of pUNO1-hTLR7-HA3×, and kanamycin selection was used in propagating the resulting plasmid and its derivative plasmids. Similar to the strategy for TLR7-SEAP plasmid assembly, synthetic DNA inserts were substituted for a restriction fragment to generate an allele T (Leu11) version of the HA-tag vector (AgeI, ClaI); A and T versions with the natural (nontagged) C-terminus of TLR7 restored (BspEI, NheI); and an allele A version with the HA3× tag replaced by 3 tandem copies of the FLAG epitope (BamHI, NheI). Transfection of HEK 293T/17 cells was performed as above in 24-well plates, with TLR7 vector dosage adjusted by dilution with carrier plasmid DNA. Cells from replicate transfections were harvested after 48 hours for Western blot analysis
Enzyme assays.
SEAP activity in transfection supernatants was fluorometrically quantitated in the presence of the substrate compound, 4-methylumbelliferyl phosphate (4-MUP; MilliporeSigma), as described by Kolniak and Sullivan (63), with slight modifications. Firefly luciferase activity in cell lysates was assayed with Luciferase Assay System reagents from Promega according to the manufacturer’s instructions. Chemiluminescence was measured in triplicate over a 10-second period, with a 2-second delay, on a Berthold Centro LB960 microplate luminometer. The Metridia luciferase assay involved 20 μL/well of transfection supernatant or a dilution thereof in PBS and 100 μL/well of substrate buffer: 6.3 μm coelenterazine (from Biaffin) in 220 mM sodium phosphate buffer, pH 7.2, with 0.1% BSA, 100 mM NaCl, 2.5 mM EGTA, and 1.3 mM sodium azide; chemiluminescence was measured over 10 seconds, with a 0.1-second delay (all reagents from Sigma). Suitable positive control transfection supernatants were frozen at −20°C in aliquots and used throughout as standards in the SEAP and Metridia luciferase assays.
Quantification of IFN-αand IP-10 in plasma from HIV-infected patients. The concentration of CXCL-10 (IP-10) was measured by a 2-site sandwich ELISA (Quantikine kit, R&D Systems, DIP100). The plasma concentration of IFN-α was determined on a Simoa HD-1 analyzer using the Simoa Human IFN-α kit (Quanterix) following the manufacturer’s instructions. The lower limit of quantification for this assay was 0.014 pg/mL.
Database sources.
Recombination rate data from the Phase II HapMap project (64) were downloaded from ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2011-01_phaseII_B37/. Lists of TLR7 polymorphisms, allelic frequencies, and individual genotype data were downloaded from the Ensembl database at http://www.ensembl.org/ Human codon usage data was from the Codon Usage Database (31) at http://www.kazusa.or.jp/codon/ For DNA sequences, accession numbers are given with reference to the NCBI GenBank database at https://www.ncbi.nlm.nih.gov/nucleotide/
Statistics.
Statistical analyses were performed using Stata 14 (StataCorp), SAS 9.4 (SAS Institute), and Prism 6 (GraphPad Software), including ANOVA tests and adjustment for multiple hypotheses by the Benjamini-Hochberg (FDR) method, Fisher tests, Mann-Whitney tests, and Wilcoxon tests. All statistical tests were 2-tailed and P < 0.05 was considered to be statistically significant. Specialized analyses, and correlation and Hotelling tests, were performed in the R computing environment as described in Supplemental Note 2. Aminoterminal protein sequences were analyzed for the presence of a signal peptide and the location of the signal peptide cleavage site using the SignalP 4.1 software (59) at http://www.cbs.dtu.dk/services/SignalP/, with the setting “Input sequences do not include TM regions” and default D-cutoff values. For a more sensitive prediction of the cleavage site (59), complementary analyses were performed with SignalP version 3.0 (65) at http://www.cbs.dtu.dk/services/SignalP-3.0/ Codon adaptation analysis of the 5′-terminal region of the TLR7 coding sequence was performed with the gcua software at http://gcua.schoedl.de/seqoverall_v2.html
Study approval.
The PRIMO cohort was previously registered with ClinicalTrials.gov under identifier NCT03148964. The X-LIBRIS cohort was previously registered with ClinicalTrials.gov under identifier NCT01952587. This study was conducted in adherence to the ethical principles of the Declaration of Helsinki and approved by the relevant medical ethics board, Comité de Protection des Personnes Sud-Ouest et Outre-Mer I, Toulouse, France. Informed consent was obtained in writing from the patients and control subjects, and experiments were performed in accordance with institutional and regional guidelines.
Author contributions
PA, JEM, JI, MMT, PD, LM, and JCG conceived and designed the study. PA, JEM, CC, AY, SL, AE, and CP performed experiments. PA, JEM, AS, AE, PD, MMT, LM, and JCG analyzed and interpreted the data. JEM and JCG prepared the manuscript with input from their coauthors. All authors read and approved the final version of the manuscript. PA, JEM, and CC contributed equally to this work. PA initiated the study, JEM and CC performed the mechanistic and genetic association studies, CC analyzed TLR7 expression in human PBMCs; this determined the authorship order.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from F. L’Faqihi, A.L. Iscache, and V. Duplan at the flow cytometry facility (CPTP); E. Lhuillier, C. Naylies, A. Emile, and Y. Lippi for DNA sequencing and RNA analyses at the GeT-Purpan and GeT-TRiX facilities; and the technical assistance of M. Requena, M. Cazabat, and R. Carcénac. We also thank Anaïs Bellin-Robert and Guilhem Vazzoler for assistance during plasmid cloning; G. von Heijne and H. Nielsen for comments on SignalP; H. Blanché-Koch for curating the CEPH sample panel; S. Lupold for the Metridia luciferase vector; and W. Chowdhury and S. Chavanas for advice on luciferase assays. This work was supported by grants from the French National Agency for Research on AIDS and Viral Hepatitis (ANRS, EP-53 study), Fondation pour la Recherche Médicale (DEQ20180339187), and Conseil Régional Occitanie-Midi-Pyrénées. PA was supported by a fellowship from SIDACTION. MMT received grants from the ANRS and the Fondation Beytout.
Version 1. 06/18/2020
Electronic publication
Funding Statement
one year PhD fellowship to Pascal Azar
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e136047.https://doi.org/10.1172/jci.insight.136047.
Contributor Information
Pascal Azar, Email: pascal.azar@gmail.com.
Claire Cenac, Email: claire.cenac@inserm.fr.
Arnoo Shaiykova, Email: shaiykova.arnoo@gmail.com.
Ali Youness, Email: ali.youness@inserm.fr.
Sophie Laffont, Email: sophie.laffont-pradines@inserm.fr.
Asma Essat, Email: asma.essat@inserm.fr.
Jacques Izopet, Email: izopet.j@chu-toulouse.fr.
Caroline Passaes, Email: caroline.pereira-bittencourt@pasteur.fr.
Pierre Delobel, Email: pierredelobel@yahoo.fr.
Laurence Meyer, Email: laurence.meyer@inserm.fr.
Jean-Charles Guéry, Email: jean-charles.guery@inserm.fr.
References
- 1.Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. 2014;426(6):1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15(8):471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beignon AS, et al. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest. 2005;115(11):3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lepelley A, et al. Innate sensing of HIV-infected cells. PLoS Pathog. 2011;7(2):e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Sydow M, Sönnerborg A, Gaines H, Strannegård O. Interferon-alpha and tumor necrosis factor-alpha in serum of patients in various stages of HIV-1 infection. AIDS Res Hum Retroviruses. 1991;7(4):375–380. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- 6.Herbeuval JP, et al. Differential expression of IFN-alpha and TRAIL/DR5 in lymphoid tissue of progressor versus nonprogressor HIV-1-infected patients. Proc Natl Acad Sci USA. 2006;103(18):7000–7005. doi: 10.1073/pnas.0600363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malleret B, et al. Primary infection with simian immunodeficiency virus: plasmacytoid dendritic cell homing to lymph nodes, type I interferon, and immune suppression. Blood. 2008;112(12):4598–4608. doi: 10.1182/blood-2008-06-162651. [DOI] [PubMed] [Google Scholar]
- 8.Harris LD, et al. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol. 2010;84(15):7886–7891. doi: 10.1128/JVI.02612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kader M, et al. Blocking TLR7- and TLR9-mediated IFN-α production by plasmacytoid dendritic cells does not diminish immune activation in early SIV infection. PLoS Pathog. 2013;9(7):e1003530. doi: 10.1371/journal.ppat.1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruel T, et al. Plasmacytoid dendritic cell dynamics tune interferon-alfa production in SIV-infected cynomolgus macaques. PLoS Pathog. 2014;10(1):e1003915. doi: 10.1371/journal.ppat.1003915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandler NG, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511(7511):601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, et al. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog. 2014;10(7):e1004291. doi: 10.1371/journal.ppat.1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458(7241):1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haase AT. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med. 2011;62:127–139. doi: 10.1146/annurev-med-080709-124959. [DOI] [PubMed] [Google Scholar]
- 15.Jacquelin B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119(12):3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campillo-Gimenez L, et al. Nonpathogenesis of simian immunodeficiency virus infection is associated with reduced inflammation and recruitment of plasmacytoid dendritic cells to lymph nodes, not to lack of an interferon type I response, during the acute phase. J Virol. 2010;84(4):1838–1846. doi: 10.1128/JVI.01496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340(6129):207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson EB, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340(6129):202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti A. HIV: The mixed blessing of interferon. Nature. 2014;511(7511):537–538. doi: 10.1038/nature13517. [DOI] [PubMed] [Google Scholar]
- 20.Barreiro LB, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5(7):e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackelprang RD, et al. Toll-like receptor polymorphism associations with HIV-1 outcomes among sub-Saharan Africans. J Infect Dis. 2014;209(10):1623–1627. doi: 10.1093/infdis/jit807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh DY, et al. A frequent functional toll-like receptor 7 polymorphism is associated with accelerated HIV-1 disease progression. AIDS. 2009;23(3):297–307. doi: 10.1097/QAD.0b013e32831fb540. [DOI] [PubMed] [Google Scholar]
- 23.Berghöfer B, Frommer T, Haley G, Fink L, Bein G, Hackstein H. TLR7 ligands induce higher IFN-alpha production in females. J Immunol. 2006;177(4):2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- 24.Meier A, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15(8):955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seillet C, et al. The TLR-mediated response of plasmacytoid dendritic cells is positively regulated by estradiol in vivo through cell-intrinsic estrogen receptor α signaling. Blood. 2012;119(2):454–464. doi: 10.1182/blood-2011-08-371831. [DOI] [PubMed] [Google Scholar]
- 26.Gibson SJ, et al. Plasmacytoid dendritic cells produce cytokines and mature in response to the TLR7 agonists, imiquimod and resiquimod. Cell Immunol. 2002;218(1-2):74–86. doi: 10.1016/S0008-8749(02)00517-8. [DOI] [PubMed] [Google Scholar]
- 27.Souyris M, et al. TLR7 escapes X chromosome inactivation in immune cells. Sci Immunol. 2018;3(19):eaap8855. doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- 28.Voorhees RM, Hegde RS. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife. 2015;4:e07975. doi: 10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voorhees RM, Hegde RS. Structure of the Sec61 channel opened by a signal sequence. Science. 2016;351(6268):88–91. doi: 10.1126/science.aad4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philbin VJ, et al. Identification and characterization of a functional, alternatively spliced Toll-like receptor 7 (TLR7) and genomic disruption of TLR8 in chickens. Immunology. 2005;114(4):507–521. doi: 10.1111/j.1365-2567.2005.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28(1):292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quax TE, Claassens NJ, Söll D, van der Oost J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol Cell. 2015;59(2):149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. Measurement of average decoding rates of the 61 sense codons in vivo. Elife. 2014;3:e03735. doi: 10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19(1):20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laffont S, et al. X-Chromosome complement and estrogen receptor signaling independently contribute to the enhanced TLR7-mediated IFN-α production of plasmacytoid dendritic cells from women. J Immunol. 2014;193(11):5444–5452. doi: 10.4049/jimmunol.1303400. [DOI] [PubMed] [Google Scholar]
- 38.Tepper PG, et al. Trajectory clustering of estradiol and follicle-stimulating hormone during the menopausal transition among women in the Study of Women’s Health across the Nation (SWAN) J Clin Endocrinol Metab. 2012;97(8):2872–2880. doi: 10.1210/jc.2012-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melhuish A, Lewthwaite P. Natural history of HIV and AIDS. Medicine. 2018;46(6):P356–361. [Google Scholar]
- 40.Deng Y, et al. MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS Genet. 2013;9(2):e1003336. doi: 10.1371/journal.pgen.1003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrat FJ, Su L. A pathogenic role of plasmacytoid dendritic cells in autoimmunity and chronic viral infection. J Exp Med. 2019;216(9):1974–1985. doi: 10.1084/jem.20181359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang L, et al. Epithelium-innate immune cell axis in mucosal responses to SIV. Mucosal Immunol. 2017;10(2):508–519. doi: 10.1038/mi.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee BL, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman ZR, Young JM, Ingolia NT, Barton GM. Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. Proc Natl Acad Sci USA. 2016;113(10):E1362–E1371. doi: 10.1073/pnas.1518976113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149(1):202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 46.Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myasnikov AG, Afonina ZA, Ménétret JF, Shirokov VA, Spirin AS, Klaholz BP. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat Commun. 2014;5:5294. doi: 10.1038/ncomms6294. [DOI] [PubMed] [Google Scholar]
- 48.Griesbeck M, et al. Sex Differences in Plasmacytoid Dendritic Cell Levels of IRF5 Drive Higher IFN-α Production in Women. J Immunol. 2015;195(11):5327–5336. doi: 10.4049/jimmunol.1501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19(5):640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 50.Dominguez-Villar M, Gautron AS, de Marcken M, Keller MJ, Hafler DA. TLR7 induces anergy in human CD4(+) T cells. Nat Immunol. 2015;16(1):118–128. doi: 10.1038/ni.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keating SM, et al. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of women. AIDS. 2011;25(15):1823–1832. doi: 10.1097/QAD.0b013e3283489d1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts L, et al. Plasma cytokine levels during acute HIV-1 infection predict HIV disease progression. AIDS. 2010;24(6):819–831. doi: 10.1097/QAD.0b013e3283367836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liovat AS, et al. Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS One. 2012;7(10):e46143. doi: 10.1371/journal.pone.0046143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paiardini M, Müller-Trutwin M. HIV-associated chronic immune activation. Immunol Rev. 2013;254(1):78–101. doi: 10.1111/imr.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ploquin MJ, et al. Elevated Basal Pre-infection CXCL10 in Plasma and in the Small Intestine after Infection Are Associated with More Rapid HIV/SIV Disease Onset. PLoS Pathog. 2016;12(8):e1005774. doi: 10.1371/journal.ppat.1005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Decalf J, et al. Plasmacytoid dendritic cells initiate a complex chemokine and cytokine network and are a viable drug target in chronic HCV patients. J Exp Med. 2007;204(10):2423–2437. doi: 10.1084/jem.20070814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmons RP, et al. HIV-1 infection induces strong production of IP-10 through TLR7/9-dependent pathways. AIDS. 2013;27(16):2505–2517. doi: 10.1097/01.aids.0000432455.06476.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novelli S, et al. Long-term Therapeutic Impact of the Timing of Antiretroviral Therapy in Patients Diagnosed With Primary Human Immunodeficiency Virus Type 1 Infection. Clin Infect Dis. 2018;66(10):1519–1527. doi: 10.1093/cid/cix1068. [DOI] [PubMed] [Google Scholar]
- 59.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 60.Pastrana DV, et al. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology. 2004;321(2):205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 61.Lupold SE, Johnson T, Chowdhury WH, Rodriguez R. A real time Metridia luciferase based non-invasive reporter assay of mammalian cell viability and cytotoxicity via the β-actin promoter and enhancer. PLoS One. 2012;7(5):e36535. doi: 10.1371/journal.pone.0036535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Compton SH, et al. Stable integration of large (>100 kb) PAC constructs in HaCaT keratinocytes using an integrin-targeting peptide delivery system. Gene Ther. 2000;7(18):1600–1605. doi: 10.1038/sj.gt.3301280. [DOI] [PubMed] [Google Scholar]
- 63.Kolniak TA, Sullivan JM. Rapid, cell-based toxicity screen of potentially therapeutic post-transcriptional gene silencing agents. Exp Eye Res. 2011;92(5):328–337. doi: 10.1016/j.exer.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.International HapMap Consortium. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.