Abstract
Alopecia areata (AA) is a common autoimmune condition, presenting initially with loss of hair without other overt skin changes. The unremarkable appearance of the skin surface contrasts with the complex immune activity occurring at the hair follicle. AA pathogenesis is due to the loss of immune privilege of the hair follicle, leading to autoimmune attack. Although the literature has focused on CD8+ T cells, vital roles for CD4+ T cells and antigen-presenting cells have been suggested. Here, we use single-cell sequencing to reveal distinct expression profiles of immune cells in murine AA. We found clonal expansions of both CD4+ and CD8+ T cells, with shared clonotypes across varied transcriptional states. The murine AA data were used to generate highly predictive models of human AA disease. Finally, single-cell sequencing of T cells in human AA recapitulated the clonotypic findings and the gene expression of the predictive models.
Keywords: Autoimmunity, Immunology
Keywords: Dendritic cells, Macrophages, T cells
Comprehensive transcriptional investigation identifies immune cell populations in the skin and lymph nodes of murine and human alopecia areata.
Introduction
Alopecia areata (AA) is a common autoimmune condition, affecting an estimated 6 million people in the United States (1), and has a total lifetime risk of approximately 2% (2). Hair loss in AA is due to aberrant immune-mediated attack of hair follicles, resulting in well-demarcated, nonscarring patches of hair loss without overt epidermal changes (3). A subset of patients develops widespread disease that can involve the entire body surface area. Historically, broad-acting immunosuppressants have been used with unreliable outcomes and minimal efficacy, especially in those with severe presentations. Recent genomic investigations (4) and gene expression profiling studies (5) have led to clinical trials involving JAK molecule–targeted therapies (6–8). These emerging clinical trials have shown efficacy in the treatment of AA, although an increasing spectrum of worrisome side effects associated with these inhibitors is being recognized (9). Despite the overall prevalence and well-defined clinical presentation, the underlying pathogenesis of AA is not fully understood and has limited further innovation for AA therapeutics.
T cells are considered the major pathogenic cell type in AA, forming dense, peribulbar lymphocytic infiltrates centered around anagen phase hair follicles in patients and in the C3H/HeJ AA mouse model (10). Adoptive transfer of CD8+ T cells from AA-affected C3H/HeJ mice to syngeneic recipients is sufficient to induce murine AA (11, 12). More recent investigations have linked AA to gene signatures associated with cytotoxicity and CD8+ T cells (4, 5) and have helped further define NKG2D-expressing CD8+ T cells as pathogenic effectors of AA in the murine model. Supporting these observations, genome-wide association studies have found increased overall risk of AA with single-nucleotide variants in NKG2D ligands (13, 14).
However, these approaches likely belie the complex ecosystem of skin immunity and the role of the microenvironment and other cellular participants in AA disease pathogenesis. Mounting evidence implicates a role for CD4+ T cells, including correlation between an increased CD4+/CD8+ T cell ratio and more active disease (15) and the emergence of AA after the transfer of conventional CD4+ T cells in mouse models (12). Furthermore, the disruption of the antiinflammatory, immune-privileged status of the normal anagen hair follicle (16) and the establishment of a proinflammatory environment observed in AA (17) suggest myeloid cells and other antigen-presenting cells (APCs) may play a pivotal role in disease pathogenesis. These populations, as well as other skin-infiltrating immune cell populations, have not been extensively examined in AA pathogenesis.
In this study, we used single-cell mRNA and T cell receptor (TCR) sequencing to investigate the immune cell populations in murine AA in relation to unaffected (UA) control skin. This work provides what we believe to be the first single-cell data set of 18,231 immune cells from the skin and lymph nodes of AA mice and UA mice. Within murine APCs, we found a skewing toward CD11b+ CCR2+ myeloid DCs (mDCs), with enrichment of JAK/STAT signaling and increased expression of both MHC class I and MHC class II genes. Within the T cell populations, we identified CD8+ T cell cluster profiles that implicated and supported their likely pathogenic role in murine AA. AA skin also harbored a shared cluster of mixed CD4+ and CD8+ T cells exhibiting potent IFN-γ expression activity, known to be required for disease induction (4, 18, 19). Furthermore, our data demonstrated extensive sharing of TCR sequences in murine AA but not in control tissues, supporting an antigen-specific immune response in AA. Surprisingly, murine skin AA CD4+ T cell signatures could be used as a classifier for human AA and exhibited the same performance characteristics as CD8+ T cell signatures in deducing disease states in nearly 90% of human whole-skin microarray samples tested. Finally, we conducted single-cell RNA sequencing on skin-infiltrating T cells from lesional and normal human skin, and our results mirrored the findings from the murine model, with distinct clusters enriched for AA-related gene sets and clonal expansion in both CD4+ and CD8+ cells. Taken together, we have demonstrated the use of single-cell expression data in autoimmunity to provide transcriptomic insights into disease process and clinical correlates. In addition, this work offers data on comprehensive skin and lymph node immune cells in murine AA and human skin AA T cells for the field of autoimmune skin pathologies.
Results
Single-cell expression profiling of AA and unaffected immune cells.
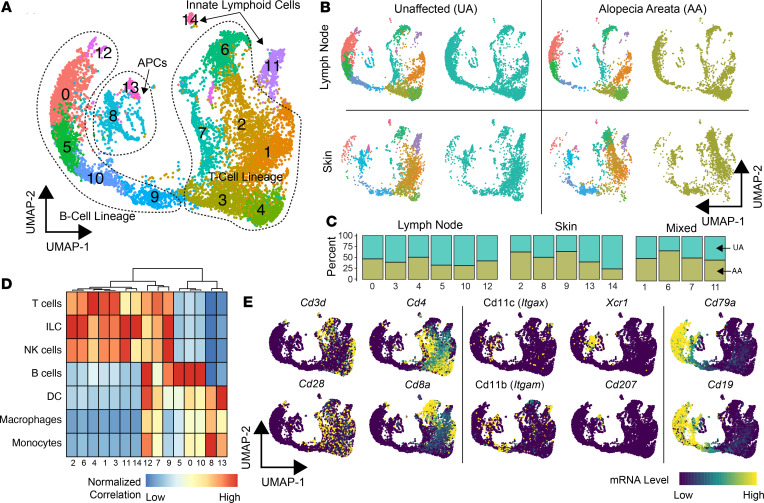
To elucidate the immune cell composition and transcriptomic heterogeneity of murine AA in an unbiased manner, a total of 10,505 immune cells from the skin and lymph nodes of UA (n = 6332) and AA (n = 4173) samples were isolated and sequenced. The multiple sequencing runs were combined into a single uniform manifold approximation and projection (UMAP) and identified 15 immune cell clusters (Figure 1A). Across the UMAP plot, we found a distinct distribution of clusters in which a majority of cells consisted of lymph node (clusters 0, 3, 4, 5, 10, and 12), skin (clusters 2, 8, 9, 13, and 14), or mixed (cluster 1, 6, 7, 11) cells (Figure 1, B and C). Clusters could also be separated by the relative percentage of murine AA versus UA cells, with clusters 2, 6, and 9 possessing enrichment of AA cells (Figure 1C). Using the median gene expression for each cluster, each cluster was assigned to a cell lineage using 2 methods: (a) the correlation of murine pure-cell gene signatures derived from the Immunological Genome Project (20) (Figure 1D) and (b) the analysis of expression patterns of canonical markers (Figure 1E) for T cells (Cd3d, Cd28, Cd4, Cd8), APCs (Itgax [CD11c], Itgam [CD11b], Xcr1, Cd207 [Langerin]), and B cells (Cd79a, Cd19). Using a similar approach, we identified similar distributions of skin and lymph node populations in a second murine AA cohort (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137424DS1), with the exception of isolating a small neutrophil population (0.29% of total sequenced cells, cluster 11). At this global level and in an unbiased manner, differential patterns in cellular composition were apparent between skin and lymph nodes, in which APC clusters 8 and 13 were predominantly found in the skin. We also observed increased T cell clusters 2 and 6 predominantly found in murine AA. Although clusters 2 and 6 were enriched for innate lymphoid cell gene expression profiles (Figure 1D), the T cell classification was given because we recovered TCR sequences from the majority of the cells.
Figure 1. Single-cell immune profiling of m urine skin and lymph node composition in UA controls versus AA.
(A) UMAP plot of the flow-sorted CD45+ murine immune cells from UA (n = 6332) and AA (n = 4173). (B and C) UMAP plots demonstrating the relative distribution of UA and AA, as well as skin and lymph node cells along the UMAP plot (B) and by the breakdown in clusters (C). (D) Normalized correlation values for predicted immune cell phenotypes based on the SingleR R package for each cluster. Cluster of columns based on Euclidean distance between normalized correlation values across all pure immune cell populations in the Immgen database (20). (E) Lineage markers for T cells (Cd3d, Cd4, Cd8a, Cd28), APCs (Itgax, Itgam, Xcr1, Cd207 [Langerin]), and B cells (Cd79a, Cd19).
Consistent with the inability of B cells to recapitulate murine AA upon transfer (11), our analysis revealed minimal evidence that they may be contributing to AA pathogenesis. Among the 5 B cell clusters, only cluster 9 was found to be skin and murine AA predominant. Total B cells in the skin comprised 16.1% of all skin cells compared with 44.1% of lymph node cells. Even this minimal contribution may be aberrant, in that additional murine replicates showed a larger 11.1-fold difference in the secondary murine AA skin to lymph node B cell population (Supplemental Figure 1E).
APCs in murine AA show polarization to CD11b+ DCs with active proinflammatory signaling.
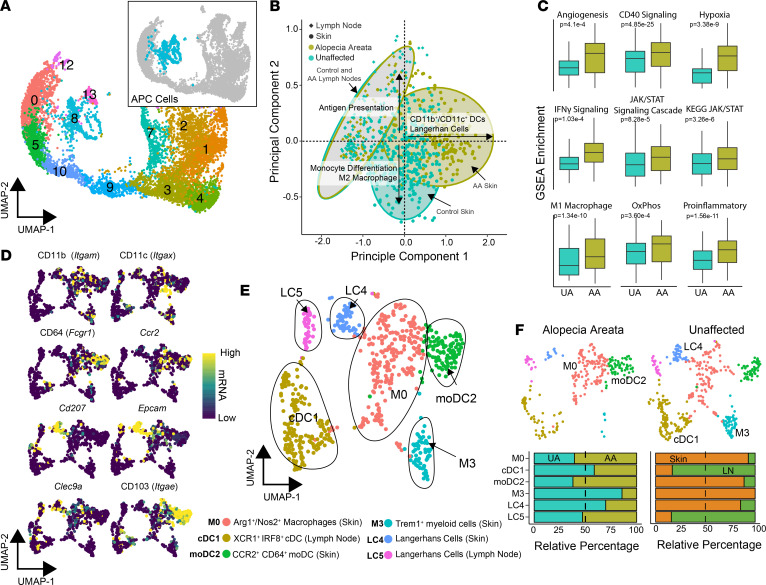
In total, clusters 8 and 13 were composed of 714 APCs, including 276 AA and 438 UA cells (Figure 2A). In order to evaluate the potential functional differences between APCs in murine AA and UA controls, we performed single-sample gene set enrichment analysis (ssGSEA) using gene signatures derived from MSigDB libraries (21) and previously derived myeloid signatures (22). Principal components analysis (PCA) based on the enrichment scores was performed (Figure 2B) and showed striking overlap of APCs from AA and UA lymph nodes (Figure 2B, gray ovals). In contrast, the PCA showed a divergence of ssGSEA enrichment in murine AA skin APCs toward CD11b+ DCs and Langerhans cells (Figure 2B, right x axis) and, in contrast, toward monocytic differentiation and M2 macrophage polarization for UA APCs (Figure 2B, lower y axis). Beyond cell type differentiation, the ssGSEA showed significant increases in angiogenic, CD40, IFN-γ, JAK/STAT, and hypoxic signaling in murine AA APCs (Figure 2C), supporting a proinflammatory signature of this population in AA. In addition, we observed increases in gene sets associated with oxidative phosphorylation and M1 macrophage polarization (Figure 2C) in murine AA.
Figure 2. Murine AA compared with UA skin displays distinct composition and gene expression of APCs.
(A) UMAP plot of the flow-sorted CD45+ murine immune cells focusing on APC clusters: cluster 8 (n = 605) and cluster 13 (n = 109). (B) Unsupervised PCA of ssGSEA APC and Langerhans cell signatures and pathways. (C) ssGSEA enrichment scores for selected signaling pathways comparing UA with AA samples. (D) mRNA expression superimposed on the UMAP plot with canonical markers for APC lineages. (E) UMAP plot for APC cells after scaling mRNA for cell cycle difference. Cluster ID based on gene expression of markers. (F) UMAP plots for AA and UA cells across the new APC clusters with relative contribution of each cluster by UA versus AA sample and skin versus lymph node cells across all single cells, χ2 test; P value less than 0.05 for both comparisons.
In the previous analysis, human APC signatures were used because of the current lack of readily available mouse APC data. We therefore reanalyzed the data in order to label distinct clusters based on characteristic gene expression signatures for the distinct clusters. After correcting for cell cycle states between clusters, the APCs were reclustered and canonical markers for APC were examined (Figure 2, D–F). The numbers per cluster and top markers are summarized in Supplemental Figure 2, A and B. Using the canonical markers, the 6 new murine APC clusters were labeled as follows: M0: Arg1+/Nos2+ macrophages, cDC1: XCR1+ IRF8+ conventional DCs (cDCs), moDC2: CCR2+ CD64+ monocyte-derived DCs (moDCs), M3: Trem1+ macrophages, LC4: Langerhans cells of the skin, and LC5: Langerhans cells of the lymph node (Figure 2E). As detailed by others, moDCs and CD11b+, IRF4-dependent conventional DC2 cells exhibit significant overlap with regard to phenotype and gene expression (23); the moDC2 population labeled here may also be composed of these 2 populations, although the moDC label was favored given the UMAP proximity to tissue macrophages and expression of CD64 (24).
Significant differences in APC composition were identified among disease states and tissue sites. Clusters M0, moDC2, M3, and LC4 were found predominantly in the skin, whereas clusters cDC1 and LC5 were found predominantly in the lymph nodes (Figure 2F). Within AA, there was a greater proportion of the moDCs, whereas UA mouse skin APCs had a predominance of the Trem1+ macrophages (Figure 2F). Interestingly, UA samples had a greater proportion of the LC4 skin-resident Langerhans cells (Figure 2F), which was unexpected based on the PCA of pathway enrichment (Figure 2B). However, expression of canonical Langerhans cell markers, Cd207 (log2 fold change = 6.02, q value = 0.009) and Epcam (log2 fold change 3.39, q value = 0.036), were upregulated in LC4 cells derived from murine AA compared with UA skin cells (Supplemental Figure 2C). With the hypothesis that APCs participate in pathogenicity of AA via the stimulation of T cells, we next examined the expression of MHC genes within the APCs. In general, MHC expression was most consistently detected at heightened levels in clusters M1, moDC2, LC4, and LC5 (Supplemental Figure 1D). We found increases in both MHC class I and MHC class II genes in AA, supporting increased immune reactivity and loss of immune privilege, and implicating a heightened ability to present to T cells. These data indicate that APCs in AA exhibit more proinflammatory signatures and are more highly composed of Arg1+/Nos2+ macrophages and moDCs.
Defining T cell differentiation for the skin and lymph node cells in murine AA.
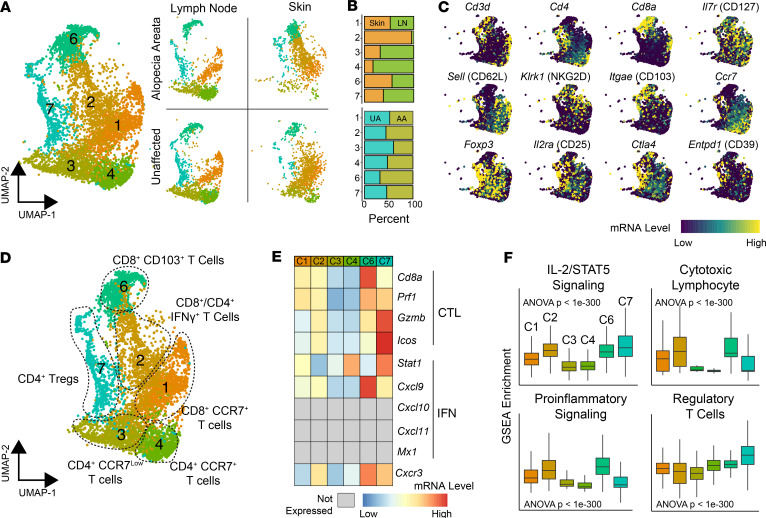
We next focused on gene expression profiling of T cells from murine AA versus UA samples. T cells were defined by the expression of pan-T cell markers and the corresponding TCR sequencing information (Figure 3A). Of the 15 clusters originally identified in the UMAP plot, clusters 1, 2, 3, 4, 6, and 7 were found to correspond to T cells (Figure 3A). Within these clusters, a subset showed a clear distinction between skin (cluster 2) and lymph node (clusters 3 and 4); other clusters had a mixed composition (Figure 3B). Similar to defining APCs, canonical and differential T cell markers were used to assign cluster identification (Figure 3C). Based on these results, clusters 3 and 4 were defined as CD4+CCR7lo and CD4+CCR7+ cells, respectively (Figure 3, C and D). We found similar correlates of the lymph node clusters in an additional biological replicate of murine AA cells (Supplemental Figure 3, cluster 4). Cluster 7 was defined as a Treg population with high levels of Foxp3, CD25 (Il2ra), Helios (Ikzf2), neuropilin (Nrp1), GITR (Tnfrsf18), Tigit, and Ctla4 (Figure 3, C and D). Interestingly, this population also expressed relatively high levels of cytotoxic genes, including Prf1 and Gzmb. For CD8+ T cells, cluster 1 was defined as CD8+CCR7+ T cells with high levels of Ccr7 and CD62L (Sell) expression, indicative of a population with lymph node–trafficking potential, representing most likely naive or central memory T cells. Cluster 2 was composed of CD8+ T cells with admixed CD4+ T cells and expressed high levels of numerous activation markers, including Ifng, Gzmb, Gzma, Fasl, Icam1, Cd44, and CD122 (Il2rb), relative to the other T cell clusters. Cluster 2 was notably found nearly exclusively in the skin, distinguishing this cluster from the other T cell clusters. In contrast, the CD8+ T cell–predominant cluster 6 was found more evenly split between the lymph nodes and the skin, despite expressing high levels of the skin-infiltrating marker CD103. Cluster 6 also demonstrated relatively high levels of genes associated with activation, including Cd44, Cxcr3, Gzmb, and Prf1. NKG2D (Klrk1), a marker found on CD8+ T cell populations in AA with the capability to transfer disease (4), was found most highly expressed in this cluster, as were other genes more commonly associated with NK cells, such as NKG2A (Klrc1), NKG2C (Klrc2), CD94 (Klrd1), and Nkg7. In a secondary AA murine single-cell sequencing run, clusters 2 and 6 appeared to consolidate into a single UMAP cluster (Supplemental Figure 3, cluster 2). Clusters 6 and 7 showed increased expression of the cytotoxic (CTL) and IFN genes used in the ALADIN (5), a gene signature derived from active AA skin tissue (Figure 3E). When ssGSEA was performed, clusters 2, 6, and 7 showed increased IL-2/STAT5 signaling (Figure 3F), and clusters 1, 2, and 6 showed increased CTL and proinflammatory enrichment.
Figure 3. Identification of murine T cell populations in skin and lymph nodes.
(A) Total cell UMAP plot and UMAP separated by tissue and condition for flow-sorted CD45+ murine immune cells focusing on T cell cluster: cluster 1 (n = 1301), cluster 2 (n = 1159), cluster 3 (n = 960), cluster 4 (n = 897), cluster 6 (n = 869), and cluster 7 (n = 681). (B) Relative contribution of cells to each cluster from lymph node versus skin (upper panel) and from UA versus AA samples for all single cells. (C) mRNA expression superimposed on the UMAP plot for general T cell markers (upper and middle rows) and Tregs (bottom row). (D) UMAP plot of the T cell phenotype labels based on gene expression and pathway analysis. (E) Genes from the predictive ALADIN (4) for CTL and IFN. Genes not expressed in T cells are in gray. (F) ssGSEA enrichment scores for selected pathways by T cell cluster; 1-way ANOVA used for comparison across all clusters.
Clonotypic analysis of AA shows clonotypic sharing between clusters and lymph nodes.
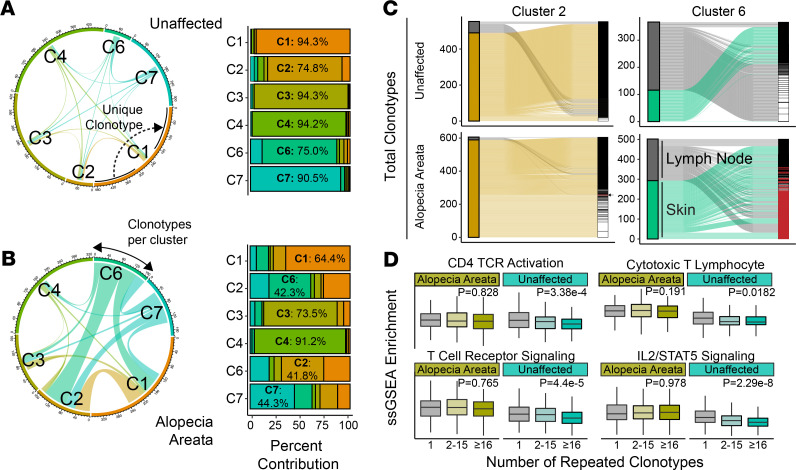
With the clusters defined and differential gene set enrichment identifying the possible role for clusters 2 and 6 in the pathology of murine AA, we next examined TCR diversity in AA versus UA T cells. A full list of clonotypes recovered from the AA and UA T cells is available in Supplemental Table 1. To evaluate differences in clonality and TCR diversity, we analyzed barcode-linked TCR sequences from murine UA and AA samples (Figure 4, A and B). UA sample T cells showed minimal overlap of clonotypes between the 6 T cell clusters: each cluster possessed unshared/unique sequences ranging from 74.8% to 95.3% (Figure 4A). In contrast, AA T cells had clear increases in shared TCRs between clusters: the majority of clonotypes for clusters 2, 6, and 7 had the inverse trend from UA T cells with greater than 55% of nonunique, shared clonotypes (Figure 4B and Supplemental Figure 4). Across UA and AA T cells, lymph node clusters 3 and 4 had the greatest proportion of unique clonotypes (Figure 4B), suggesting a more localized expansion of T cells retained within the skin. In addition to sharing between clusters, focusing on the individual clusters showed an increase in shared clonotypes between skin and lymph nodes in AA cluster 2 IFN-expressing T cells and cluster 6 tissue-infiltrating T cells compared with UA clusters 2 and 6 (Figure 4C). Defining cells by the number of repetitive clonotypes, we formed 3 categories: single clonotypes, clonotypes shared by between 2 and 15 cells, and clonotypes shared by 16 or greater T cells. Across all skin T lymphocytes, ssGSEA was performed to examine activation and signaling pathways (Figure 4D). Interestingly, although murine UA samples showed a stepwise decrease of CD4+ TCR activation and TCR signaling across clonotype categories, there was a notable maintenance of these signatures by clonotype category in murine AA (Figure 4D). This result was mirrored in the analysis of CTL and IL2/STAT5 signaling (Figure 4D).
Figure 4. Murine AA T lymphocytes show greater shared repertoire and sustained CD4 activation.
(A) Shared and unique clonotypes of T cells by cluster for UA samples as a chord diagram (left) and relative proportion bar chart. In the chord diagram, the larger the connection between the cluster, the greater the number of shared clonotypes. Dominant percentage overlap for each cluster is annotated in the bar graph. (B) Shared and unique clonotypes of T cells by cluster for AA samples as a chord diagram (left) and relative proportion bar chart. (C) Alluvial diagrams for the shared clonotypes between skin and lymph node T lymphocytes in skin-predominant T cell clusters. Cluster 2 (overlap coefficient = 7.7%) and cluster 6 (overlap coefficient = 45.5%) in AA had a greater shared repertoire than the UA samples, with overlap coefficients of 0% and 0%, respectively. Clonotypes with 2 or more copies highlighted in white; overlapping clonotypes between skin and lymph nodes highlighted in red. (D) ssGSEA enrichment skin-derived T lymphocytes by grouping the number of repeated clonotypes: 1 (unique clonotypes, AA n = 1056, UA n = 1830), 2–15 (AA n = 385, UA n = 219), and 16 or greater (AA n = 573, UA n = 303); 1-way ANOVA used for comparison across clonotype groups.
Human AA discriminatory performance of the gene signature for CD4+ T cells is equal to that for CD8+ T cells.
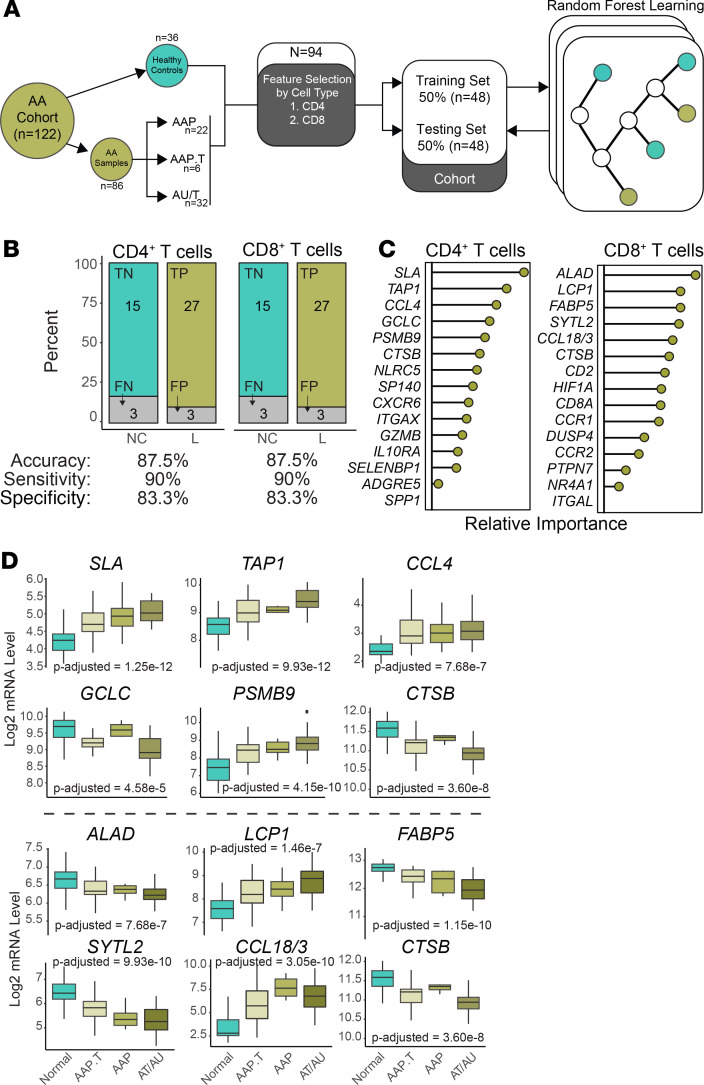
In order to translate our single-cell RNA sequencing findings to human data, we asked whether signatures developed from our mouse model data could predict human AA skin versus control skin in human patient samples (Figure 5A). The pipeline we developed made use of a previously published microarray cohort (5) of 122 skin samples derived from 36 healthy controls and 50 patients with patchy AA (AAP), transient patchy AA (AAP.T), alopecia totalis (AT), or alopecia universalis (AU). For computational feasibility, the list of differentially regulated genes in AA versus control skin CD4+ and CD8+ T cells (Supplemental Table 2) underwent feature selection to identify genes increased in AA with superior discrimination efficiency (Figure 5A). Separating the human normal, healthy controls and AA lesional skin samples, we split the sample groups in half to generate a training cohort and testing cohort with a sample size equal to 48 for each cohort. Random forest learning was used to generate models of the training cohort with the goal of binary classification of healthy, normal controls (NC) and lesion (L) without the inclusion of the paired skin samples (Figure 5A). Models were then applied to the testing cohort and predictions were compared with the disease classification.
Figure 5. Similar performance of CD4+ T cell and CD8+ T cell gene signatures in the discrimination of AA.
(A) Schematic of signature development using feature selection from 1) 180 CD4+ T cell genes and 2) 669 CD8+ T cell genes. Genes were selected by relative importance and trained using the 48 samples of the training cohort. The random forest models were then applied to the training cohort. (B) Discrimination performance of each model by category (upper bar charts) and measures of accuracy, sensitivity, and specificity. P value less than 0.0001 for both model predictions based on Fisher exact test. (C) Composition of the 15-gene cell type signatures displayed in a ranked variable importance plot. (D) Log2 mRNA expression levels for the top 6 genes by relative importance in the CD4+ (upper panel) and CD8+ gene signatures by disease state: normal, AA transient patchy (AAP.T), patchy AA (AAP), or alopecia totalis and universalis (AT/AU). P values based on 1-way ANOVA with correction based on multiple comparisons.
We found the discrimination abilities were equal, using a 15-gene signature based on CD4+ and CD8+ T cell differentially upregulated genes in murine AA. Both signatures had an overall accuracy of 87.5%, sensitivity of 90%, and a specificity of 83.3% (Figure 5B). We next examined the composition of the 15-gene signatures for CD4+ and CD8+ T cell gene signatures (Figure 5C). Several of the top genes in terms of relative performance in the CD4+ T cell model are reported to be associated with human AA, like TAP1 (25), CCL4 (26), PSMB9 (27), or other skin autoimmune pathologies, like SLA (28). Similarly, several of the top genes for the CD8+ T cell model have been associated with AA, like LCP1 (4), SYTL2 (4, 7), and CCL18 (29), or atopic dermatitis, like FABP5 (30). Interestingly, the only shared gene between the CD4+ and CD8+ T cell signatures was cathepsin B (CTSB), a lysosomal cysteine protease, which has been reported to play a role in postdegranulation activities of T cells (31). Given that random forest modeling is an ensemble machine learning approach that uses a modified tree learning algorithm, genes are selected based on the ability to discriminate between human AA versus normal skin. As such, genes display 2 general trends, increasing by severity of clinically evaluated hair loss or conversely, decreasing by severity of hair loss (Figure 5D). For the CD4+ T cell signatures (Figure 5D, upper panel), SLA, TAP1, and PSMB9 had increased expression by hair loss type, with AT and AU having the highest levels of expression. Interestingly, the level of CCL4 was generally increased for all disease states compared with normal. Within the CD8+ T cell signature, 4 of the 6 top genes by model importance were decreased over the disease states, and CCL18/CCL3 and LCP1 were increased in a state-dependent manner (Figure 5D, lower panel). A complete table of all 29 genes for both signatures along with comparisons between disease states is available in Supplemental Table 3.
Single-cell sequencing of human AA skin T lymphocytes demonstrates similar gene expression dynamics and pathway enrichments.
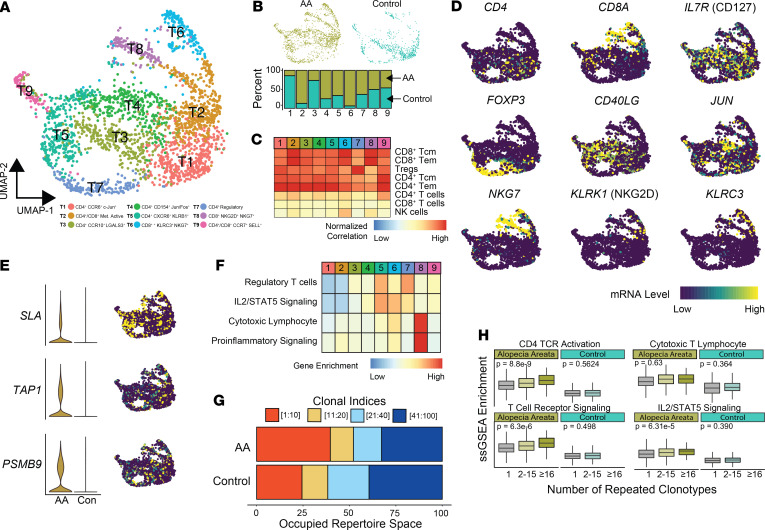
We paired the use of modeling to translate our murine AA findings with performance of single-cell RNA sequencing on T lymphocytes isolated from the skin of a healthy control and AA patient. After processing, a total of 2416 cells (AA n = 1664 and control n = 752) were recovered and formed 9 distinct clusters: T1 (n = 434), T2 (n = 421), T3 (n = 343), T4 (N = 326), T5 (N = 323), T6 (N = 168), T7 (N = 169), T8 (N = 139), and T9 (N = 93) (Figure 6A). Of the clusters identified, T2, T4, T5, T6, and T7 were AA-predominant clusters exhibiting greater than 50% relative contribution from the AA sample (Figure 6B). Using the aforementioned dual approach of correlational analysis of pure immune cell populations (Figure 6C) and canonical/functional T cell markers (Figure 6D), we identified CD4+ (T1, T3, T4, T5, and T7), CD8+ (T6 and T8), and mixed CD4+/CD8+ (T2 and T9) T cell clusters. Distinct from the other CD4+ clusters, T7 correlated with a regulatory T cell signature and had high expression of FOXP3 (Figure 6, C and D), supporting a Treg phenotype. Both the T6 and T8 CD8+ T cell populations expressed KLRK1 (NKG2D): 13.7% and 25.6% of cells in each cluster had detectable expression (Figure 6D). However, the T6 population, composed of 93.6% of AA cells, expressed EOMES, GZMK, and GZMH, as well as NKG7, with greater similarity to the NKG2D-expressing CD8+ T cell population in the murine single-cell analysis (cluster 6 in Figure 3D). Although there was a relatively small difference in expression of NKG2D between these clusters, there was a relative absence of KLRK1 detection in normal T cells in T6 and in less than 5% of normal T cells in T8, supporting the established literature of a specific marker of pathogenic CD8+ T cells in AA. We next examined the expression of the genes derived from the CD4+ and CD8+ signatures. We found the genes downregulated in AA relative to normal were difficult to visualize because of the relative absence of expression or less than 10% of T cells expressing the mRNA species. However, among the most prominent upregulated predictors in our models, we found diffuse distribution of TAP1 and PSMB9 within all clusters (Figure 6E). In contrast, SLA was highly localized to the CD4+ T4 cluster and the distal portions of both T6 and T8 (Figure 6E).
Figure 6. Human single-cell immune profiling of AA T cells recapitulates the findings of the murine analysis.
(A) UMAP plot of the T cell isolated from human AA skin (n = 1664) and control skin (n = 752). (B) Relative contribution of cells to each cluster from normal versus AA samples for all single cells. (C) Normalized correlation values for predicted immune cell phenotypes based on the SingleR R package for each cluster. (D) mRNA expression superimposed on the UMAP plots for lineage and functional markers for T cell populations. (E) Expression comparison and distribution of selected signature markers superimposed on the UMAP plot. (F) Z score–transformed ssGSEA enrichment scores for selected pathways by T cell cluster. (G) The percentage of the top 100 clonotypes that is occupied by the top 1–10, 11–20, 21–40, and 41–100 clones for human AA and control skin. (H) ssGSEA enrichment skin-derived T lymphocytes by grouping the number of repeated clonotypes: 1 (unique clonotypes, AA n = 907, control n = 389), 2–15 (AA n = 262, control n = 122), and 16 or greater (AA n = 51, control n = 0); 1-way ANOVA used for comparison across all clonotype groups.
We next performed ssGSEA in order to identify possible functional differences in the clustering (Figure 6F). We found an increase in IL2/STAT5 signaling associated with a subset of CD4+ (T5) and CD8+ (T6), as well as the regulatory T cell (T7), mirroring our findings in the mouse data set. Interestingly, T8 with 50% AA and control cells had the highest enrichment for both cytotoxic lymphocytes and proinflammatory gene set enrichment (Figure 6F). Using the TCR sequence information, we also examined the clonotype dynamics within the single-cell cohorts, finding the top 10 repeated clonotypes within AA for 39.7% of the repertoire space of the top 100 clones compared with 24.4% in control T cells (Figure 6G). The top 5 most abundant clonotypes in the AA T cells are shown in Supplemental Figure 5. Using the same definition for repetitive clonotypes as the murine AA analysis, we next formed 3 categories: single clonotypes, clonotypes shared by between 2 and 15 cells, and clonotypes shared by 16 or greater T cells. Across all skin T lymphocytes, ssGSEA was performed to examine activation and signaling pathways by clonotype group (Figure 6H). Unlike the dynamics of maintained signaling across murine AA clonotype groupings (Figure 4D), we observed an increase in CD4+ TCR activation, IL2/STAT5 signaling, and general TCR signaling gene sets by increasing clonotype (Figure 6H). Although the decrease across normal skin clonotype groups was not seen, the normal skin gene set enrichment was notably decreased compared with human AA enrichment (Figure 6H). In total, the human AA skin single-cell data largely supported our findings from murine models.
Discussion
AA is a common autoimmune condition globally. Although management guidelines are lacking, current treatments for those with severe disease involve variable-term immune suppression via steroids or other immunosuppressive agents with pronounced risk exposure to side effects. Our growing understanding of AA pathogenesis conceptually includes the breakdown of hair follicle immune privilege (15, 16). What the critical events are in the spatiotemporal development of AA is unclear, but they likely coordinately involve the participation of resident and infiltrating immune architecture. In order to unravel the participation of immune cell populations, we performed single-cell mRNA sequencing on the immune landscape of the murine AA model, analyzing both T cells and APCs, as well as on human skin-infiltrating T cells.
The role of APCs in AA pathogenesis is largely unknown. Within the murine AA APC population, we observed transcriptomic enrichment of profiles associated with Langerhans cells and CD11b+/CD11c+ DCs (Figure 2A), with increased levels of inflammatory and IFN signaling. Despite the transcriptomic enrichment of Langerhans cell profiles, further stratification of the APCs revealed decreased relative representation of Langerhans cells in skin-infiltrating cells in AA (Figure 2E, LC4). The discrepancy is potentially due to a decreased representation of APCs isolated from AA skin and lymph nodes, but also a product of the skin microenvironment, leading to differential enrichment of transcriptomic profiles. Both the LC4 skin Langerhans cells and the moDC2 skin CD11b+ CCR2+ mDCs, which demonstrated increased relative contributions in AA, had the highest level of expression of MHC-I and MHC-II molecules (Supplemental Figure 2D). Single-cell sequencing in atopic dermatitis has grouped these 2 APC populations and reported a variable increase in lesional versus nonlesional skin (32). Further analysis of subpopulations on DCs and Langerhans cells may reveal a greater role in autoimmune skin conditions.
Although evidence suggests CD8+ T cells may be the proximal effector cells in murine and human AA (4, 5, 11, 12), other work has found the transfer of CD4+ T cells to be sufficient to induce AA in mice (12). Additionally, prior studies indicate that CD4+ T cells are the most abundant cell type in the peribulbar immune infiltrate found in human AA (15). In concert with upregulation of both MHC class I and MHC class II molecules, our data suggest AA CD4+ and CD8+ T cells are clonotypically expanded, with extensive overlap of TCR sequences between different single-cell clusters (Figure 4). Notably, in the murine AA T cells, clonotypic overlap was seen predominantly in the C1 CD8+CCR7+, C2 IFN-γ–expressing, and C6 CD8+CD103+ CTL clusters, in line with previous reports (33). The clonally expanded C6 cluster had the highest level of NKG2D (Klrk1) and demonstrated elevated JAK2/STAT5 and cytolytic gene sets (Figure 3), paralleling evidence in prior murine and human AA of these putatively pathogenic pathways (4, 5). Interestingly, the C6 CD8+CD103+ CTL cluster was composed of an even split of skin and lymph node cells, suggesting this population may traffic between these sites.
Interestingly, clonal expansions were also seen in murine C7 Tregs, with 68.1% of clonotypes shared between clusters. The possible expansion of suppressive T cells in AA is counterintuitive for an autoimmune process and does not fit into the framework of recent reports of Tregs in AA, which have demonstrated a reduction in overall number and clonotypic restriction (34, 35). However, our data indicate that immunoregulatory processes are running concurrently, albeit ineffectively, at counteracting the pathogenic autoreactivity. How Tregs contribute to the AA microenvironment and pathogenesis is an open question, although GWAS findings, identifying associations with the pivotal Treg genes CTLA4, ICOS, and IKZF4, further support that changes in this population may affect susceptibility to AA (13, 14, 28). Upregulation of these same genes also seems to be a common feature during infiltration of human and murine Tregs into the skin (36).
An approach to identifying potentially novel markers and drivers of pathogenesis for a disease is the development of signatures. Using genes derived from our mouse single-cell experiment, we developed CD4+ and CD8+ signatures that had equivalent performance, including an accuracy of 87.5%, in discriminating normal and pathological skin, lending support to AA murine modeling for the human disease process (Figure 5, A and B). The use of the random forest approach allowed for the identification of genes that were decreased or increased in disease states. Not only did this approach corroborate other incidentally reported genes in human AA, but the generation of predictive signatures also identified potentially novel genes that may play a role in human AA pathogenesis, like ALAD, SLA, FABP5, and CTSB. In order to further corroborate our findings in mice and signature development, we performed single-cell mRNA sequencing on human T cells derived from the skin of an AA patient. Human single-cell analysis of AA T cells found cytotoxic and IL2/STAT5 signaling enrichment in both CD4+ and CD8+ T cells (Figure 6D). Unlike the mouse clustering analysis, human CD8+ T cells clustered to 2 distinct identities, T6 and T8 (Figure 6). Although gene set enrichment suggests T8 to be highly cytotoxic effector T cells, T6 has a slightly greater expression of KLRK1, highlighting potential functional heterogeneity within CD8+ T cells in AA.
Taken together, our work offers what we believe to be the first single-cell–level snapshot into the complex immune landscape in AA. The resolution offered by single-cell mRNA sequencing allowed us to identify differences in APC and T cells between unaffected and AA skin, which were predictive of human disease state. How these findings are collectively translated into human disease and patient care is the next key step in using these techniques to reveal potentially novel biology. In addition, this work offers data on comprehensive skin and lymph node immune cells in murine AA (n = 18,232) and human skin T cells (n = 2416) for the field of autoimmune skin pathologies.
Methods
Animals.
Mice were kept in pathogen-free conditions at the University of Iowa Animal Facility. C3H/HeJ mice were purchased from The Jackson Laboratory. Lesional skin from AA-affected C3H/HeJ mice was grafted onto C3H/HeJ mice to induce disease as previously described (37).
Single-cell RNA sequencing.
Lymph nodes and skin were collected from AA-affected and UA C3H/HeJ mice. Skin was digested with collagenase type III (MilliporeSigma) and passed through a metal mesh. CD45+ cells (as detected by an anti-CD45 antibody, clone 30-F11, from BioLegend, Inc.) were flow-sorted to isolate skin-infiltrating immune cells. Single-cell suspensions of lymph node cells were made by passing lymph nodes through a metal screen, and CD45+ cells were flow-sorted in parallel with skin immune cells. For sequencing, 5000 individual cells per condition and tissue type were targeted. Sequencing for 5′ gene expression and TCR was performed using the Chromium (10x Genomics) and Illumina sequencing technologies. Amplified cDNA was used to construct both 5′ expression and TCR enrichment libraries. Libraries were pooled and run on separate lanes on an Illumina HiSeq 4000. Each lane consisted of 150–base pair, paired-end reads. Basecalls were converted into FASTQs using the Illumina bcl2fastq software by the University of Iowa Genomics Division. FASTQ files were aligned to the murine genome (mm10) using the CellRanger 3.0.1 pipeline as described by the manufacturer. Across aligned cells, the mean number of reads per cell was 116,563, with an average of 93.1% of reads mapped to the mm10 genome. Single-cell immune profiling of the clonotypes of the CD4+ T cells was performed in conjunction with the single-cell RNA sequencing following the protocols described above using the vdj_GRCm38_alts_ensembl genome build provided by the manufacturer. Details on the secondary murine data set are available in Supplemental Methods.
Human skin collected from the affected scalp of the AA and control patients using a punch tool was minced and digested with Liberase (MilliporeSigma), and CD45+ cells (as detected by an anti-CD45 antibody, clone 2D1, from BioLegend, Inc.) were flow sorted. Library preparation and sequencing were performed as described above. Gene expression FASTQ files were aligned to the human genome (GRCh38) using the CellRanger 3.0.1 pipeline, while clonotype sequencing was aligned to the vdj_GRCh38_alts_ensembl genome build provided by the manufacturer. Human AA and control skin CD45+ cells were sequenced with a mean reads per cell of 171,827 and 95.9% of reads mapping to the GRCh38 genome.
Single-cell data processing and analysis.
Initial processing of immune cells from murine UA skin (n = 2206), murine UA lymph nodes (n = 4160), murine AA skin (n = 2041), and murine AA lymph nodes (n = 2170) was performed using the Seurat R Package (version 3.0.2). Samples were combined into a single data set using canonical correlational analysis and mutual nearest neighbors (MNN) to better harmonize shared populations across sample types (38, 39). Dimensional reduction to form the UMAP plots used the top 30 calculated dimensions and a resolution of 0.6. Cluster markers and differential gene expression analyses were performed using the Wilcoxon rank sum test with an unsupervised approach that involved no filtering of genes. For the purpose of skin-derived and lymph node–derived differential gene expression comparison, CD8+ T cells were defined as T cells with the presence of Cd8a or Cd8b1 expression; CD4+ T cells were defined as the inverse. Single-cell immune phenotyping and single-sample ssGSEA used the SingleR (version 0.2.0) R package (40). For the cell type–based correlations, single-cell expression values were compared with transcriptional profiles from pure cell populations in the Immgen data set (20) or ENCODE project (41). Human AA skin (n = 1720) and control skin (n = 1027) samples were processed in a similar pipeline, with UMAP reduction using 35 dimensions and a resolution of 0.5.
Clonotype analysis was performed using the scRepertoire (version 1.0.0) R package (42) with clonotypes defined as the combination of the genes of the TCR A and B chains and nucleotide sequences. Individual gene sets were derived from the Gene Ontology Consortium (43), the Kyoto Encyclopedia of Genes and Genomes (44), and previously reported pathways (45). Enrichment scores for individual cells were then compiled and underwent PCA using the prcomp() function in the stats (version 3.5.1) R package. Expression data were visualized using the ggplot2 (version 3.1.1) and pheatmap (version 1.0.12) R packages. In addition to the previously mentioned R packages, clonotype visualization used the circlize (version 0.4.6) and ggalluvial (version 0.9.1) R packages. Raw and processed gene expression and TCR data are available at GSE145095.
Patient cohort training and analysis.
Gene expression data from the Affymetrix Human Genome U133 2.0 microarray for 122 samples with disease classifications were downloaded from the Gene Expression Omnibus under the accession code GSE68801 (5). Data were processed using AnnotationDbi (version 1.44.0) and hgu133plus2.db (version 3.2.3) R packages. Gene lists were generated from murine single-cell mRNA sequencing cohort, comparing CD4+ T cells and CD8+ T cells from the skin of AA versus UA controls. Filtered for genes was set at greater than 0.5 log2-fold change, raw P value less than 0.005, and a difference between the percentage of cells expressing the gene greater than 5%. The gene lists were then converted to mouse gene symbols using the biomaRt R package (version 2.38.0) and using the Ensembl genome annotations, and feature selection was performed for CD4+ T cells (n = 266) and CD8+ T cells (n = 822). Of the 202 genes differentially regulated in the CD4+ AA T cells, 180 were converted into human gene symbols. Likewise, the 669 of 703 genes differentially regulated in CD8+ AA T cells were converted into human gene symbols. Ensembl genome annotations converted the mouse Ccl3 to human CCL3 and CCL18, leading to the selection of CCL18 into the models.
Alopecia and normal control skin samples were isolated from the GSE68801 and randomly partitioned into a 50% training cohort (n = 48) and 50% testing cohort (n = 48). For each set of feature-selected genes, random forest models were trained to discriminate lesion versus normal skin samples with the training cohort using the caret (version 6.0-84) R package. The training and feature selection used the repeatedcv method and the twoClassSummary function, splitting the cohort into 10 groups and taking the mean of the error terms across 25 repetitions. The mtry parameter was auto-selected based on the performance across the total number of genes trained. Importance was graphed using the base R functions, with relative importance.
Statistics.
Statistical analyses were performed in R (version 3.5.1). Two-sample significance testing used Welch’s t test; significance testing for more than 3 samples used 1-way ANOVA with Tukey’s honest significance determination for correcting multiple comparisons. Overlap coefficients were calculated using the size of the intersection and dividing by the size of the smaller of the 2 groups. P values of less than 0.05 were considered significant.
Study approval.
All animal procedures were conducted according to the NIH Guide for the Care and Use of Laboratory Animals and under protocols approved by the University of Iowa Institutional Animal Care and Use Committee. Human studies were conducted according to protocols approved by the University of Iowa Institutional Review Board and under the Declaration of Helsinki Principles. The control and AA patients were recruited from the University of Iowa hair disorders clinic in the Department of Dermatology. Informed written consent was received from the participants before inclusion in the study.
Author contributions
AJ and NB were responsible for the initial conception, design, method development, analysis, and writing/review of the manuscript. AJ was the principal study supervisor. AJ, NH, LO, and SC performed experiments and isolated cells. NLB assisted in the development of informatic analyses.
Supplementary Material
Acknowledgments
Funding for this project was provided from the NIH under the K08 award AR069111 (principal investigator, AJ) and from the F30 fellowship CA206255 (principal investigator, NB). This work was supported in part by Merit Review Award I01BX004907 from the United States Department of Veterans Affairs, Biomedical Laboratory Research and Development Service. Research reported in this publication was supported by the National Cancer Institute (NCI) of the NIH under award P30CA086862. We thank Julie McKillip for excellent nursing assistance and Nitin Karandikar, Scott Lieberman, and Ashutosh Mangalam for helpful discussions of the work. The data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility at the University of Iowa, funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veterans Affairs Medical Center, as well as at the Genomics Division of the Iowa Institute of Human Genetics, which is supported, in part, by the University of Iowa Carver College of Medicine and the Holden Comprehensive Cancer Center (NCI of the NIH under award P30CA086862).
Version 1. 05/26/2020
In-Press Preview
Version 2. 07/09/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(13):e137424.https://doi.org/10.1172/jci.insight.137424.
Contributor Information
Nicholas Borcherding, Email: nicholas-borcherding@uiowa.edu.
Sydney B. Crotts, Email: Crotts.Sydney@mayo.edu.
Luana S. Ortolan, Email: Luana.DosSantosOrtolan@seattlechildrens.org.
Nicholas Henderson, Email: nicholas-henderson@uiowa.edu.
Nicholas L. Bormann, Email: nicholas-bormann@uiowa.edu.
Ali Jabbari, Email: ali-jabbari@uiowa.edu.
References
- 1.McMichael AJ, et al. Alopecia in the United States: outpatient utilization and common prescribing patterns. J Am Acad Dermatol. 2007;57(2 suppl):S49–S51. doi: 10.1016/j.jaad.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 2.Safavi K. Prevalence of alopecia areata in the First National Health and Nutrition Examination Survey. Arch Dermatol. 1992;128(5):702. doi: 10.1001/archderm.1992.01680150136027. [DOI] [PubMed] [Google Scholar]
- 3.Alkhalifah A, Alsantali A, Wang E, McElwee KJ, Shapiro J. Alopecia areata update: part I. Clinical picture, histopathology, and pathogenesis. J Am Acad Dermatol. 2010;62(2):177–88. doi: 10.1016/j.jaad.2009.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Xing L, et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med. 2014;20(9):1043–1049. doi: 10.1038/nm.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jabbari A, et al. Molecular signatures define alopecia areata subtypes and transcriptional biomarkers. EBioMedicine. 2016;7:240–247. doi: 10.1016/j.ebiom.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy Crispin M, et al. Safety and efficacy of the JAK inhibitor tofacitinib citrate in patients with alopecia areata. JCI Insight. 2016;1(15):e89776. doi: 10.1172/jci.insight.89776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay-Wiggan J, et al. Oral ruxolitinib induces hair regrowth in patients with moderate-to-severe alopecia areata. JCI Insight. 2016;1(15):e89790. doi: 10.1172/jci.insight.89790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jabbari A, et al. An open-label pilot study to evaluate the efficacy of tofacitinib in moderate to severe patch-type alopecia areata, totalis, and universalis. J Invest Dermatol. 2018;138(7):1539–1545. doi: 10.1016/j.jid.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.[No authors listed] In brief: Risk of pulmonary thromboembolism and death with tofacitinib. Med Lett Drugs Ther. 2019;61(1579):136. [PubMed] [Google Scholar]
- 10.Pratt CH, King LE, Messenger AG, Christiano AM, Sundberg JP. Alopecia areata. Nat Rev Dis Primers. 2017;3:17011. doi: 10.1038/nrdp.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilhar A, Ullmann Y, Berkutzki T, Assy B, Kalish RS. Autoimmune hair loss (alopecia areata) transferred by T lymphocytes to human scalp explants on SCID mice. J Clin Invest. 1998;101(1):62–67. doi: 10.1172/JCI551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McElwee KJ, et al. Transfer of CD8(+) cells induces localized hair loss whereas CD4(+)/CD25(-) cells promote systemicalopecia areata and CD4(+)/CD25(+) cells blockade disease onset in the C3H/HeJ mouse model. J Invest Dermatol. 2005;124(5):947–957. doi: 10.1111/j.0022-202X.2005.23692.x. [DOI] [PubMed] [Google Scholar]
- 13.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betz RC, et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun. 2015;6:5966. doi: 10.1038/ncomms6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todes-Taylor N, Turner R, Wood GS, Stratte PT, Morhenn VB. T cell subpopulations in alopecia areata. J Am Acad Dermatol. 1984;11(2 pt 1):216–223. doi: 10.1016/s0190-9622(84)70152-6. [DOI] [PubMed] [Google Scholar]
- 16.Paus R, Nickoloff BJ, Ito T. A ‘hairy’ privilege. Trends Immunol. 2005;26(1):32–40. doi: 10.1016/j.it.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Tabara K, Kozłowska M, Jędrowiak A, Bienias W, Kaszuba A. Serum concentrations of selected proinflammatory cytokines in children with alopecia areata. Postepy Dermatol Alergol. 2019;36(1):63–69. doi: 10.5114/ada.2019.82826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freyschmidt-Paul P, et al. Interferon-gamma-deficient mice are resistant to the development of alopecia areata. Br J Dermatol. 2006;155(3):515–521. doi: 10.1111/j.1365-2133.2006.07377.x. [DOI] [PubMed] [Google Scholar]
- 19.Gilhar A, Kam Y, Assy B, Kalish RS. Alopecia areata induced in C3H/HeJ mice by interferon-gamma: evidence for loss of immune privilege. J Invest Dermatol. 2005;124(1):288–289. doi: 10.1111/j.0022-202X.2004.23580.x. [DOI] [PubMed] [Google Scholar]
- 20.Heng TS, Painter MW, Immunological Genome Project Consortium The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335):eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashem SW, Haniffa M, Kaplan DH. Antigen-presenting cells in the skin. Annu Rev Immunol. 2017;35:469–499. doi: 10.1146/annurev-immunol-051116-052215. [DOI] [PubMed] [Google Scholar]
- 24.Tamoutounour S, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39(5):925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Kim HK, et al. Association between TAP1 gene polymorphisms and alopecia areata in a Korean population. Genet Mol Res. 2015;14(4):18820–18827. doi: 10.4238/2015.December.28.31. [DOI] [PubMed] [Google Scholar]
- 26.Song T, et al. An integrated model of alopecia areata biomarkers highlights both TH1 and TH2 upregulation. J Allergy Clin Immunol. 2018;142(5):1631–1634.e13. doi: 10.1016/j.jaci.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Carroll JM, McElwee KJ, E King L, Byrne MC, Sundberg JP. Gene array profiling and immunomodulation studies define a cell-mediated immune response underlying the pathogenesis of alopecia areata in a mouse model and humans. J Invest Dermatol. 2002;119(2):392–402. doi: 10.1046/j.1523-1747.2002.01811.x. [DOI] [PubMed] [Google Scholar]
- 28.Jin Y, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44(6):676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuentes-Duculan J, et al. Biomarkers of alopecia areata disease activity and response to corticosteroid treatment. Exp Dermatol. 2016;25(4):282–286. doi: 10.1111/exd.12918. [DOI] [PubMed] [Google Scholar]
- 30.Yamane Y, et al. New horny layer marker proteins for evaluating skin condition in atopic dermatitis. Int Arch Allergy Immunol. 2009;150(1):89–101. doi: 10.1159/000210385. [DOI] [PubMed] [Google Scholar]
- 31.Balaji KN, Schaschke N, Machleidt W, Catalfamo M, Henkart PA. Surface cathepsin B protects cytotoxic lymphocytes from self-destruction after degranulation. J Exp Med. 2002;196(4):493–503. doi: 10.1084/jem.20011836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He H, et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol. 2020;145(6):1615–1628. doi: 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- 33.de Jong A, et al. High-throughput T cell receptor sequencing identifies clonally expanded CD8+ T cell populations in alopecia areata. JCI Insight. 2018;3(19):121949. doi: 10.1172/jci.insight.121949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamed FN, et al. Alopecia areata patients show deficiency of FOXP3+CD39+ T regulatory cells and clonotypic restriction of Treg TCRβ-chain, which highlights the immunopathological aspect of the disease. PLoS One. 2019;14(7):e0210308. doi: 10.1371/journal.pone.0210308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speiser JJ, Mondo D, Mehta V, Marcial SA, Kini A, Hutchens KA. Regulatory T-cells in alopecia areata. J Cutan Pathol. 2019;46(9):653–658. doi: 10.1111/cup.13479. [DOI] [PubMed] [Google Scholar]
- 36.Miragaia RJ, et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity. 2019;50(2):493–504.e7. doi: 10.1016/j.immuni.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McElwee KJ, Boggess D, King LE, Sundberg JP. Experimental induction of alopecia areata-like hair loss in C3H/HeJ mice using full-thickness skin grafts. J Invest Dermatol. 1998;111(5):797–803. doi: 10.1046/j.1523-1747.1998.00380.x. [DOI] [PubMed] [Google Scholar]
- 38.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borcherding N, Bormann NL. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Research. 2020;9(47) doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.