Abstract
Heterotopic ossification (HO) is defined as abnormal differentiation of local stromal cells of mesenchymal origin, resulting in pathologic cartilage and bone matrix deposition. Cyr61, CTGF, Nov (CCN) family members are matricellular proteins that have diverse regulatory functions on cell proliferation and differentiation, including the regulation of chondrogenesis. However, little is known regarding CCN family member expression or function in HO. Here, a combination of bulk and single-cell RNA sequencing defined the dynamic temporospatial pattern of CCN family member induction within a mouse model of trauma-induced HO. Among CCN family proteins, Wisp1 (also known as Ccn4) was most upregulated during the evolution of HO, and Wisp1 expression corresponded with chondrogenic gene profile. Immunohistochemistry confirmed WISP1 expression across traumatic and genetic HO mouse models as well as in human HO samples. Transgenic Wisp1LacZ/LacZ knockin animals showed an increase in endochondral ossification in HO after trauma. Finally, the transcriptome of Wisp1-null tenocytes revealed enrichment in signaling pathways, such as the STAT3 and PCP signaling pathways, that may explain increased HO in the context of Wisp1 deficiency. In sum, CCN family members, and in particular Wisp1, are spatiotemporally associated with and negatively regulate trauma-induced HO formation.
Keywords: Bone Biology, Stem cells
Keywords: Cartilage
Endogenous CCN family member WISP-1 inhibits trauma-induced heterotopic ossification.
Introduction
Heterotopic ossification (HO) is the abnormal collection of bone and cartilage matrix in soft tissues, incited by traumatic insult (1). In its severe manifestations, HO can be disabling, owing to joint contracture and chronic pain (2). Although HO can form in diverse anatomic sites, including tendon, ligament, and muscle, it consistently derives from tissue-resident connective tissue mesenchymal progenitor cells (1, 3–6). Here, predominantly local stromal/fibroblastic cells of mesenchymal origin within connective tissue (3, 4, 6–8) undergo abnormal osteochondral differentiation producing pathologic extracellular matrix of bone and cartilage (1, 9).
Matricellular proteins are one link between the extracellular matrix and cell fate determination decisions. The Cyr61, CTGF, Nov (CCN) family consists of 6 structurally related matricellular proteins with cysteine-rich regulatory proteins (10) that bind to a plethora of extracellular ligands, in turn regulating cell proliferation and differentiation. Some CCN family members regulate cartilage and bone formation, including, for example, CTGF (also known as CCN2) (11–13) and WISP1 (also known as CCN4) (14, 15). In particular, WISP1 is expressed in perichondral and preosteoblastic cells during mammalian skeletal development, and this finding is recapitulated in mouse fracture repair (16). Although WISP1 exerts clear positive regulation of osteogenic differentiation (15, 17), its role in cartilage differentiation is more context dependent. For example, WISP1 overexpression antagonizes BMP2- or growth differentiation factor 5–induced (GDF5-induced) chondrogenesis (16), while, in other observations, WISP1 enhances TGF-β3–induced chondrogenesis (18). In addition, WISP1 appears to play an aggravating role in cartilage degradation in osteoarthritis, potentially through an increase in expression of matrix metalloproteinases (19, 20). Despite their roles in modulating osteochondral cell proliferation and differentiation, virtually nothing is known regarding CCN family members, including WISP1, in the aberrant cell fate determination seen in HO.
In the present study, we hypothesized that CCN family members would be dynamically regulated during the evolution of HO and would modulate disease progression. We first surveyed CCN family member expression within a mouse model of extremity trauma–induced HO. Bulk and single-cell RNA (scRNA) sequencing demonstrated an enrichment in most CCN family members during HO formation, and, in particular, Wisp1 was most upregulated. scRNA sequencing, in combination with immunohistochemistry in mouse and human tissues, demonstrated that mesenchymal progenitor cells and their osteochondral progeny were the major source of WISP1 during HO formation. Transgenic Wisp1LacZ/LacZ-knockin mice demonstrated more exuberant endochondral ossification during HO, and downstream pathway analysis identified putative signaling changes associated with increased endochondral ossification within Wisp1-deficient cells.
Results
Wisp1 is enriched in traumatic HO in vivo.
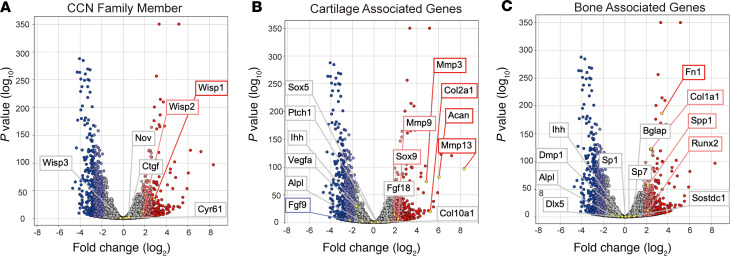
CCN family members were first examined in a model of trauma-induced HO. Complete Achilles tenotomy, coupled with cutaneous dorsal burn injury, results in a well-characterized incidence of HO at the tenotomy site, replicating human disease (21–25). First, bulk RNA sequencing was performed at the tenotomy site and compared with the contralateral tendon site of the same animal (serving as an internal control) at 3 weeks after injury (Figure 1). A 3-week time point was chosen for analysis as most representative of the cartilaginous phase of HO in this model (24). Five of six CCN family members showed an increase in the HO site, as compared with the contralateral uninjured limb (Figure 1A). Of these CCN family members, Wisp1 showed the highest increase (2.8694 fold change [FC], +6 SD). Wisp2 (also known as Ccn5) was also increased (1.8642 FC, +3 SD), while Ctgf (0.5615 FC, +0.5 SD), Cyr61 (0.2931 FC, +0.5 SD), and Nov (0.5231 FC, +0.5 SD) were marginally increased. Wisp3 (also known as Ccn6) showed a modest reduction (–0.2347 FC, –0.5 SD).
Figure 1. Bulk total RNA sequencing identifies Wisp1 enrichment in posttraumatic heterotopic ossification.
Bulk RNA sequencing of Achilles tendon tenotomy site. Tissue was collected from the soft tissue around the injury site from the posterior compartment between the muscular origin and calcaneal insertion of Achilles tendon. Corresponding soft tissue, including intact Achilles tendon, was also harvested from the uninjured contralateral hind limb. In each image, data are presented in volcano plots and logarithmic fold change is shown on the x axis (log2). The y axis depicts P value (log10). Data were derived from n = 3 pooled samples per group, with expression examined at 3 weeks after injury. (A) CCN family members, including Ctgf, Cyr61, Nov, Wisp1, Wisp2, and Wisp3. (B) Selective cartilage-associated genes. (C) Selective bone-associated genes. All analyses were performed on the same data set. Light red and dark red dots indicate genes with a >2 SD increase and a >3 SD increase in comparison with uninjured control, respectively. Light blue and dark blue dots indicate genes with a >2 SD decrease and a >3 SD decrease in comparison with uninjured control, respectively.
So as to confirm aberrant osteochondral differentiation in HO bulk sequencing, characteristic genes markers associated with cartilage and bone cells were assessed (Figure 1, B and C). Cartilage-associated genes, such as SRY-box transcription factor 9 (Sox9) (1.9189 FC, >+3 SD), aggrecan (Acan, 6.0439 FC, +6 SD), and type 2 collagen (Col2a1, 5.2387 FC, +6 SD) all showed an increase in expression within the HO site. Likewise, type 10 collagen (Col10a1, 0.3442 FC, +0.5 SD) showed a moderate increase. Genes associated with osteogenesis also showed an increase at this time point (Figure 1C), including Runt-related transcription factor 2 (Runx2, 2.1093 FC, >+3 SD), osteopontin (Spp1, 2.1479 FC, >+3 SD), type 1 collagen (Col1a1, 2.4458 FC, >+3 SD), osteocalcin (Bglap, 0.8528 FC, +1 SD), and osterix (Sp7, 0.4037 FC, >+0.5 SD).
scRNA sequencing identifies Wisp1 enrichment in chondrocyte-like mesenchymal cells within HO.
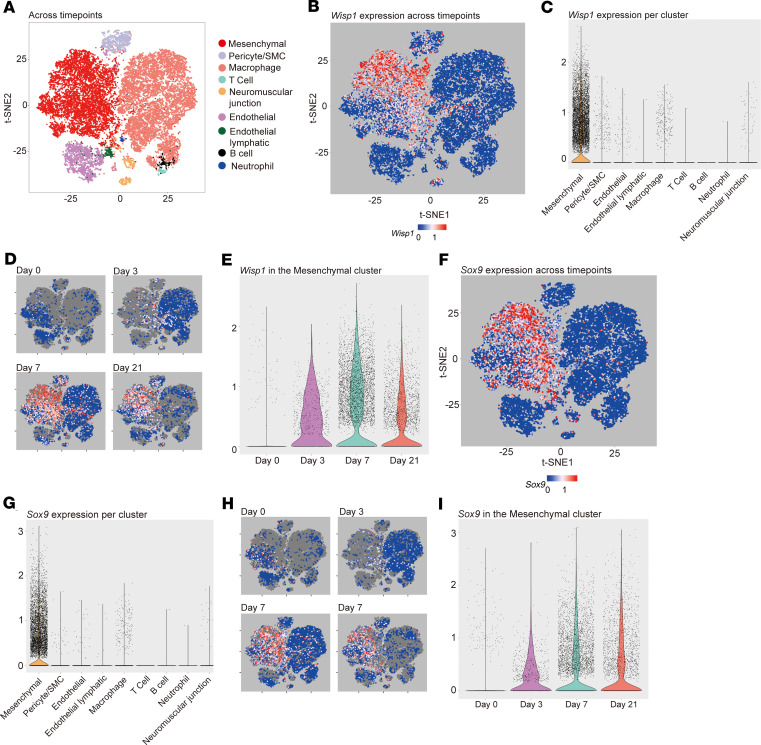
Expression of CCN family members, including Wisp1, was next examined in detail across the dynamic time course of HO formation by scRNA sequencing (Figure 2 and Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.135432DS1). Nine cell clusters were defined (Figure 2A), which broadly included 3 mesodermal populations (mesenchymal, pericyte/vascular smooth muscle, and neuromuscular junction cells), 2 endothelial populations (endothelial and endothelial lymphatic cells), and 4 inflammatory cell populations (macrophage, B cell, T cell, and neutrophil). A list of markers used to define each cluster is shown in Supplemental Table 1. Single-cell sequencing at the site of HO formation was performed and analyzed before injury (day 0) and at the early time points of 3, 7, and 21 days after injury in relation to uninjured control (Figure 2, B and C). Overall, Wisp1 was principally expressed in the mesenchymal cell cluster, typified by expression of platelet-derived growth factor receptor α (Pdgfra), paired related homeobox 1 (Prrx1), and leptin receptor (Lepr) (Figure 2, B and C). Within mesenchymal cells, Wisp1 showed mean normalized expression of 0.46, and it was expressed in 52% of cells. In contrast, in nonmesenchymal cells (8 other cell clusters combined), the average normalized expression of Wisp1 was 0.02, and it was expressed in 3% of cells. Changes in Wisp1 expression over the time course of HO development was next assessed (Figure 2, D and E). As shown by t-distributed stochastic neighbor embedding (t-SNE) plots (Figure 2D) or violin plots (Figure 2E), Wisp1 gene expression was inconspicuous at baseline and overtime increased in the mesenchymal cluster, peaking at day 7, and remaining elevated at day 21 after injury when cartilage is visible histologically.
Figure 2. Single-cell RNA sequencing identifies Wisp1 enrichment in Sox9-expressing mesenchymal cells during posttraumatic heterotopic ossification.
Single-cell RNA sequencing of the heterotopic ossification (HO) induction site was examined at 0, 3, 7, and 21 days after injury. (A) t-SNE plots of pooled cells from all 4 time points after HO induction, identifying 9 cell clusters. Characteristic markers used to define each cluster are listed in Supplemental Table 1. (B and C) Wisp1 expression within 9 cell clusters pooled across all time points, shown by (B) t-SNE plots or (C) violin plots. (D and E) Wisp1 expression across time points of HO induction (days 0, 3, 7, and 21), shown by (D) t-SNE plots or (E) violin plots within the mesenchymal cell cluster only. (F and G) SRY-box transcription factor 9 expression within all 9 cells clusters pooled across all time points, shown by (F) t-SNE plots or (G) violin plots. (H and I) Sox9 expression across time points of HO induction (days 0, 3, 7, and 21), shown by (H) t-SNE plots or (I) violin plots within the mesenchymal cell cluster only. Data represent injury sites from n = 3–4 animals per time point.
For comparison, other CCN family members were also assessed by scRNA sequencing (Supplemental Figure 1). With the exception of Wisp3, all CCN family members were detected within the mesenchymal cluster of the HO site, ranging from 35% (Nov) to 72% (Cyr61) of the mesenchymal cells. Unlike Wisp1, other CCN family members, such as Ctgf and Cyr61, were identified across multiple cell clusters (Supplemental Figure 1).
Next, Wisp1 expression among 9 cells clusters (markers used to identify different clusters are shown in Supplemental Table 1) in HO was compared with cartilage-associated markers, including Sox9 (Figure 2, F–I) and Acan and Col2a1 (Supplemental Figure 2). Sox9, the master chondrogenic transcription factor, showed an expression pattern that closely mirrored that of Wisp1 (Figure 2, F–I). Sox9 showed an overlapping expression within the mesenchymal cluster (compare t-SNE plots in Figure 2, F and B) and a similar induction pattern over time after trauma (Figure 2, H vs. D, or Figure 2, I vs. E). Acan, and to a lesser extent Col2a1, showed a similar expression pattern of expression (Supplemental Figure 2). These findings were replicated when the mesenchymal cluster was isolated and subclustering was performed (Supplemental Figure 3). Four subclusters were identified within the mesenchymal cluster (Supplemental Figure 3A). Overlap between Wisp1 expression and cartilage-associated genes was again observed. Thus, among CCN family members, Wisp1 demonstrated a prominent increase in expression after trauma, particularly in mesenchymal cells with a chondrocyte-like gene profile.
WISP1 immunolocalization in mouse models of HO.
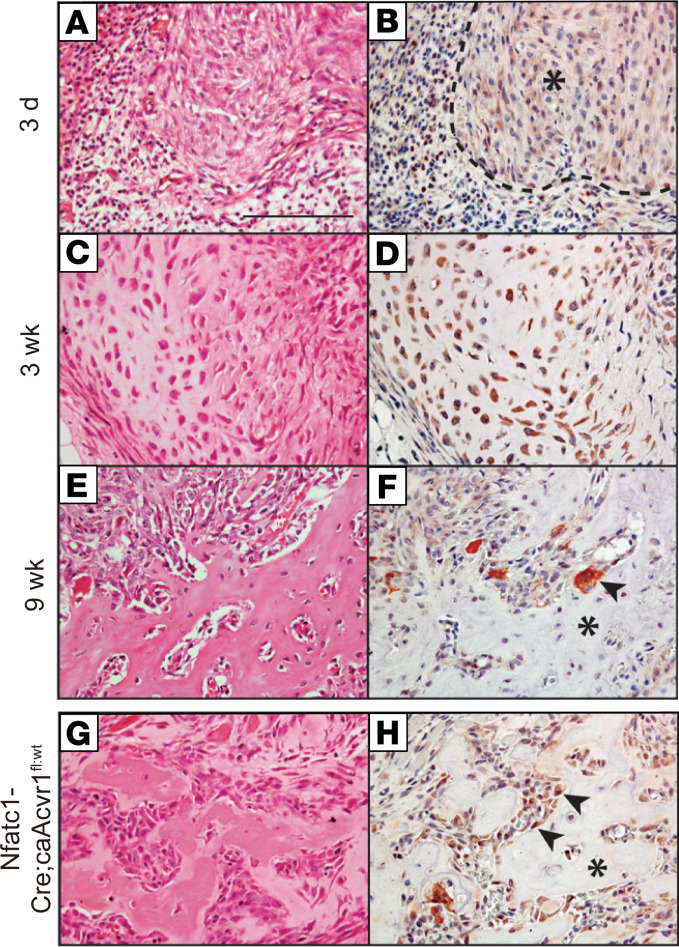
To validate these transcriptional analyses, WISP1 expression was next studied using immunohistochemistry in mouse models of HO (Figure 3). First, WISP1 immunohistochemical staining was performed at sequential time points after trauma, which highlighted the fibroproliferative phase (3 days, Figure 3, A and B), cartilaginous phase (3 weeks, Figure 3, C and D), and osseous phase of HO (9 weeks, Figure 3, E and F). WISP1 immunoreactivity was seen within fibroblastic mesenchymal cells of the injury site at the 3-day time point and was weak to moderate in intensity (asterisk, Figure 3B). In the cartilaginous phase, stronger and more widespread WISP1 immunoreactivity was present within chondrocytes (Figure 3D). In the osseous phase, bone-lining cells and some osteocytes showed some WISP1 immunoreactivity, which was weak to intermediate in intensity (Figure 3F). Multinucleated osteoclasts also showed WISP1 immunostaining. A similar pattern was observed using Wisp1-LacZ reporter, in which X-gal staining was predominantly seen in bone lining of the HO site at 9 weeks after injury (Supplemental Figure 4).
Figure 3. WISP1 immunohistochemistry corresponds with osteochondral cells in heterotopic ossification mouse models.
(A–F) WISP1 immunohistochemical staining in a posttraumatic model of mouse heterotopic ossification (HO). Images of H&E-stained sections and corresponding IHC are shown. (A and B) At days 3 after HO induction, WISP1 immunoreactivity is observed among reactive fibroblastic cells in early HO (asterisk). (C and D) At 3 weeks after HO induction, strong WISP1 immunoreactivity is present in cartilaginous areas in developing HO. (E and F) At 9 weeks after induction, WISP1 immunohistochemical staining is present in bone-lining cells, scattered osteocytes, and multinucleated giant cells (arrowhead). The asterisk indicates woven bone matrix. (G and H) WISP1 immunohistochemical staining in ACVR1-transgenic animals (fibrodysplasia ossificans progressiva [FOP] mouse model, Nfatc1-Cre; ca-Acvr1fl/WT), in which spontaneous bone formation occurs in the early postnatal period. Images are taken from animals at 2 weeks of age, with immunostaining in bone-lining cells (arrowheads), and in multinucleated osteoclasts. The asterisk indicates woven bone matrix. Scale bar: 100 μm.
Finally, WISP1 protein expression was examined within an fibrodysplasia ossificans progressiva–like (FOP-like) mouse model (Figure 3, G and H). In this model, the mutant Acvr1 allele (Q207D mutation) is driven by expression of Nfatc1-Cre (26–29). Resulting pups carrying both transgenes (Nfatc1-Cre; ca-ACVR1fl/WT) developed widespread HO by postnatal days 4–5. On H&E sections, the histologic appearance was similar to that of the osseous phase of trauma-induced HO. Likewise, WISP1 immunoreactivity was similar to traumatic HO and predominantly found in bone-lining cells. In sum, WISP1 was robustly expressed across multiple cells types, giving rise to HO, including chondrocytes and bone-lining osteoblasts.
WISP1 immunolocalization among human nongenetic HO.
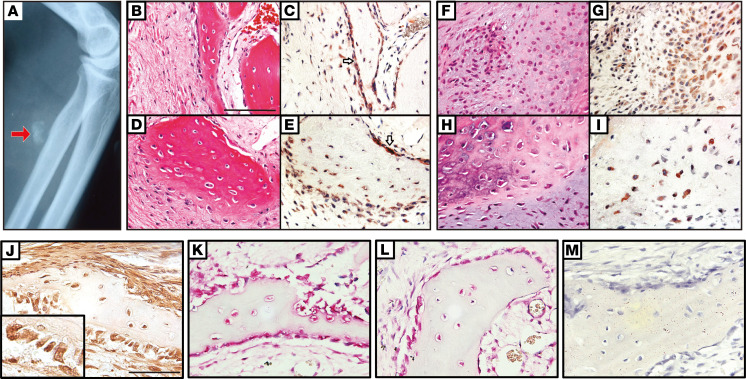
Next, to translate our findings to human HO, the surgical pathology case files of 12 human HO specimens were examined for WISP1 expression. All clinical cases were examples of sporadic (nongenetic) HO. Patient age, sex, and location of HO are summarized in Supplemental Table 2. An example of plain film radiographic appearance of 1 case is shown, demonstrating flocculent radiodensity within the soft tissue (Figure 4A). WISP1 immunoreactivity was noted among bone-lining cells as well as osteocytes within bone matrix (Figure 4, B–E). Cartilaginous areas of HO also showed strong WISP1 immunoreactivity (Figure 4, F–I). Spatial overlap with transcription factors associated with bone and cartilage differentiation were next assessed (Figure 4, J–M). WISP1-immunostained sections again highlighted predominantly bone-lining cells (Figure 4J, brown), which corresponded with spatial distributions of both RUNX2 (Figure 4K, red) and SOX9 (Figure 4L, red). In all cases, immunostaining without primary antibody served as a negative control (Figure 4M). Thus, and similar to mouse HO models, WISP1 protein expression was primarily found in bone- and cartilage-associated cell types in human HO.
Figure 4. WISP1 immunohistochemistry corresponds with osteochondral cells in human heterotopic ossification.
(A) Plain film image of a representative case of heterotopic ossification (HO) in a patient, demonstrating flocculent radiodensity within the soft tissue. The arrow indicates the HO location in the flocculent radiodensity. (B–I) WISP1 immunohistochemical staining among human HO samples, with images from corresponding H&E-stained sections. Supplemental Table 2 indicates patient demographics and location of HO for each sample tested. (B–E) WISP1 immunoreactivity among bone-lining cells (white arrows) and bone-entombed osteocytes. Images of H&E stained sections and corresponding WISP1 IHC are shown. (F–I) WISP1 immunoreactivity among cartilaginous tissues within human HO. (J–M) Correspondence of WISP1 immunoreactivity with SOX9 and RUNX2 immunohistochemical staining in bone-lining cells. (J) WISP1 IHC (brown), with high-magnification inset of bone-lining cells (original magnification, ×20). (K) RUNX2 immunohistochemical staining in adjacent section (red). (L) RUNX2 immunohistochemical staining in adjacent section (red). (M) Negative control IHC in adjacent section, without primary antibody. Scale bar: 100 μm. n = 12 human samples used. Hematoxylin was used for counterstaining in all slides.
Wisp1–/– mice demonstrate increased trauma-induced HO.
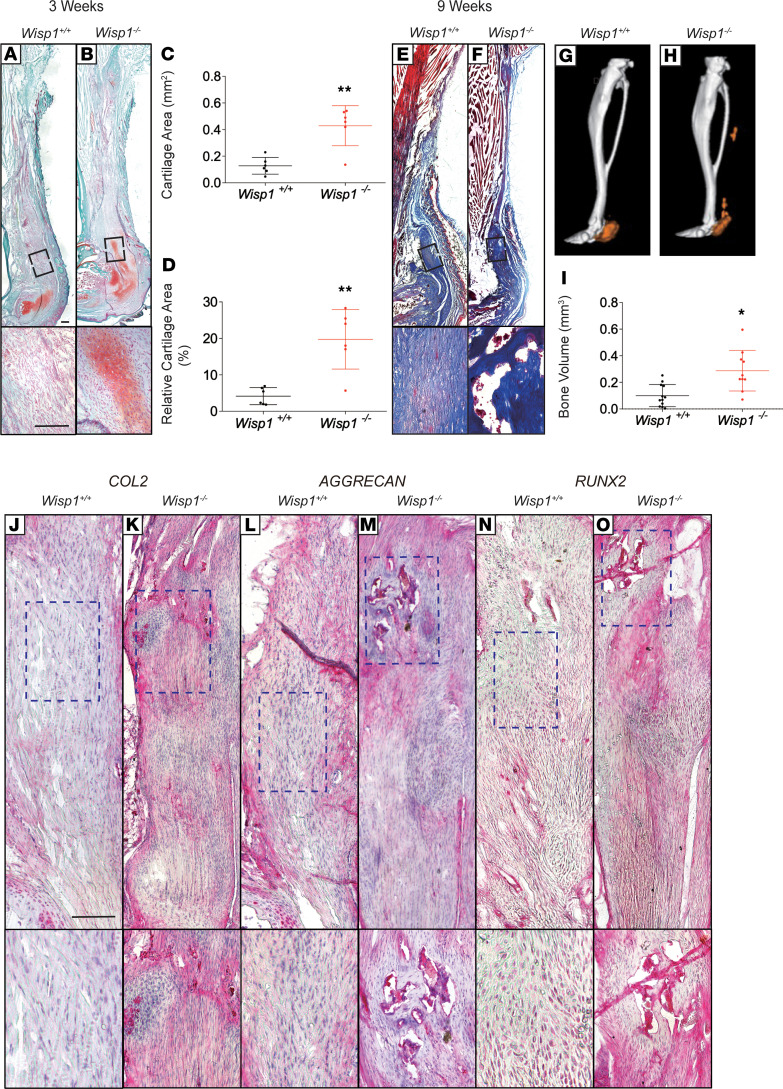
We next returned to the mouse trauma-induced HO model in the context of Wisp1 global knockin animals (30), in which exons 1–5 of the allele are replaced with a LacZ-neo cassette. Wisp1+/+ and Wisp1–/– (Wisp1LacZ/LacZ) littermates were subjected to burn/tenotomy, followed by examination of the resulting HO 3 and 9 weeks later (Figure 5). Safranin O/fast green staining on samples 3 weeks after tenotomy was used to assess cartilage (Figure 5, A and B). Histomorphometric analysis of the mean area of cartilage using histologic cross sections of the injury site showed an increase in cartilage formation among Wisp1–/– animals (Figure 5, C and D). Masson’s trichrome staining on samples 9 weeks after tenotomy was used to highlight bone matrix, which was likewise more conspicuous among Wisp1–/– animals (Figure 5, E and F). μCT was used to analyze new bone formation in HO at 9 weeks after tenotomy, again demonstrating a significant (P < 0.05) increase in bone formation in HO among Wisp1–/– animals, which was notable at the proximal and distal tenotomy sites (Figure 5, G and H). μCT quantification of bone volume at the HO injury site demonstrated a significant increase in bone volume among Wisp1–/– animals (Figure 5I).
Figure 5. Transgenic Wisp1-knockout mice demonstrated increased posttraumatic heterotopic ossification.
Heterotopic ossification (HO) induction was performed in Wisp1–/– (Wisp1LacZ/LacZ) mice, in which in which exons 1–5 of the alleles are replaced by a LacZ-neo cassette. Wisp1+/+ littermates were used as control animals. (A and B) Representative Safranin O red/fast green staining within the injury site, 3 weeks after tenotomy. Cartilage appears orange, while tendon or bone appear green. n = 6 mice per genotype. (C and D) Histomorphometric quantification of relative cartilage among Wisp1+/+ and Wisp1–/– injury sites. Analysis performed on n = 6representative samples per genotype. (E and F) Masson’s trichrome staining within the site 9 weeks after tenotomy. Bone appears dark blue, while tendon appears light blue. (G and H) μCT reconstruction of bone formation within the injured tendon among Wisp1+/+ and Wisp1–/– animals. Reconstruction shown from a sagittal perspective. Heterotopic bone appears orange, while native bone appears gray. (I) μCT quantification of heterotopic bone volume (BV). Each dot indicates an individual mouse, with mean and SD indicated. n = 12 mice per genotype. (J–O) Immunohistochemical staining among Wisp1+/+ and Wisp1–/– animals at the sites 9 weeks after tenotomy, including (J and K) type II collagen (COL2), (L and M) aggrecan (ACAN), (N and O) Runt-related transcription factor 2 (RUNX2). *P < 0.05, **P < 0.01, ***P < 0.001 in comparison with Wisp1+/+ animals. Differences were calculated between groups by Student’s t test. Data represent mean ± SD. Scale bar: 100 μm; original magnification, ×20 (bottom images, J–O).
Immunohistochemical staining among injury sites from Wisp1+/+ and Wisp1–/– animals was next performed. Foci of stronger expression of the cartilage-associated antigens COL2 (Figure 5, J and K) and ACAN (Figure 5, L and M) were more common in Wisp1–/– mouse injury sites, as compared with those in Wisp1+/+ littermates. Likewise, RUNX2 (Figure 5, N and O) was more common in injured areas of Wisp1–/– animals. In aggregate, radiographical, histological, and immunohistochemical analyses demonstrated increased trauma-induced endochondral ossification among transgenic Wisp1–/– mice.
Changes in the transcriptome of Wisp1–/– soft tissue mesenchymal progenitor cells.
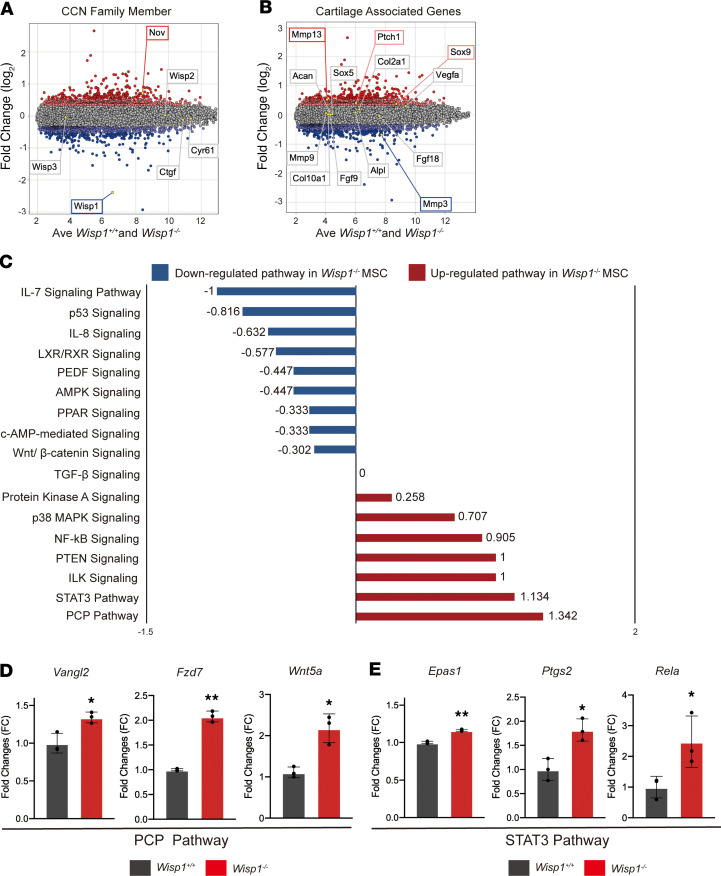
Having observed an increase in HO within Wisp1–/– animals, we sought to better explain this with isolated in vitro analysis of soft tissue–derived stromal cells. For this purpose, adipose tissue derived–stromal cells (ASCs) were first chosen, as ASCs are an easily cultured cell population that can serve as a representative extraskeletal multipotent stromal cell. Wisp1+/+ and Wisp1–/– adipose stromal cells were examined for global changes in gene expression via Clariom D microarray among cells in confluent monolayer culture at equivalent passage. Evaluation of CCN family members was first performed, which confirmed deletion of Wisp1 among Wisp1–/– mice (blue box, Figure 6A). Interestingly, the CCN family member Nov was significantly increased among Wisp1–/– cells (0.7162 FC, >+3 SD) (red box, Figure 6A). No significant change in other CCN family members was seen.
Figure 6. Wisp1-null ASCs demonstrate signaling pathway changes associated with increased chondrogenic differentiation.
Microarray analysis of Wisp1–/– (Wisp1LacZ/LacZ) and Wisp1+/+ mouse ASCs by Clariom D Array GeneChip. (A and B) Microarray gene expression is presented as a MvA (mean versus average) plot. Light red and dark red dots indicate genes with a >2 SD and >3 SD increase in expression among Wisp1–/– vs. Wisp1+/+ cells, respectively. Light blue and dark blue dots indicate genes with a >2 SD and >3 SD decrease in expression among Wisp1–/– vs. Wisp1+/+ cells, respectively. (A) CCN family member gene expression in Wisp1–/– versus Wisp1+/+ cells. (B) Selective cartilage-associated gene expression in Wisp1–/– versus Wisp1+/+ cells. (C) QIAGEN Ingenuity Pathway Analysis (IPA) demonstrating signaling pathway changes among Wisp1–/– ASCs. Positive Z scores indicate pathway enrichment among Wisp1–/– cells. Negative Z scores indicate pathway enrichment among Wisp1+/+ cells. See Supplemental Tables 4–6 for complete pathway analysis. (D) Selected target genes of the planar cell polarity (PCP) signaling pathway and (E) STAT3 signaling pathway among Wisp1–/– and Wisp1+/+ primary tenocytes. *P < 0.05, **P < 0.01 in comparison with Wisp1+/+ cells. Differences were calculated between groups by Student’s t test. Data represent mean ± SD and triplicate experimental replicates in biological duplicate.
Representative genes associated with chondrogenic differentiation were next assessed among Wisp1–/– cells (Figure 6B). Enrichment in several cartilage-associated genes was observed among Wisp1–/– cells, including the transcription factor Sox9 (0.3709 FC, +3 SD) and the Hedgehog signaling-responsive gene Patched1 (Ptch1) (0.4054 FC, +3 SD) (red boxes, Figure 6B). Other cartilage-associated genes were relatively similar among undifferentiated Wisp1–/– versus Wisp+/+ cells, such as markers of chondrocyte differentiation, including Col2a) (0.1811 FC, +2 SD), Acan (0.1156 FC, +1 SD), alkaline phosphatase (Alpl) (0.085 FC, +1 SD), and Col10a1 (0.501 FC, +0.5 SD).
To gain further insight into downstream pathway activation among Wisp1-deficient cells, QIAGEN Ingenuity Pathway Analysis (IPA) was next performed (Figure 6C and Supplemental Tables 4–6). A number of pathways associated with the positive regulation of cartilage or bone formation were upregulated among Wisp1–/– cells. This included signaling pathways promoting in vitro chondrogenesis or cartilage repair, including planar cell polarity (PCP) signaling (Z score = 1.342) (31), STAT3 signaling (Z score = 1.134) (32), phosphatase and tensin homolog (PTEN) signaling (Z score = 1) (33), and PKA signaling (Z score = 0.258) (34) (Supplemental Table 4). p38 MAPK signaling, which promotes chondrocyte proliferation, chondrogenesis, and osteogenesis (35), was also upregulated in Wisp1–/– cells (Z score = 0.707).
In order to confirm the activation of the signaling pathway from IPA analysis, selected genes were validated by qPCR in primary tenocyte cultures from Wisp1–/– animals. Here, gene members of the PCP and STAT3 pathways that showed prominent upregulation in Wisp1–/– ASCs were analyzed. PCP pathway genes, including Wnt family member 5A (Wnt5a), VANGL PCP protein 2 (Vangl2), and frizzled-7 (Fzd7) all showed similar upregulation in Wisp1–/– tenocytes (Figure 6D). Likewise, STAT3 pathway genes, including endothelial PAS domain protein 1 (EPAS1), prostaglandin-endoperoxide synthase 2 (Ptgs2), and REL-associated protein (Rela) were likewise increased among Wisp1–/– tenocytes (Figure 6E).
Conversely, downregulated signaling pathways among Wisp1–/– cells included several pathways that negatively regulate chondrogenesis or promote cartilage matrix degradation, such as AMP-activated protein kinase (AMPK) signaling (Z score = –0.447) (36) and pigment epithelium-derived factor (PEDF) signaling (Z score = –0.447) (Supplemental Table 5) (37). Pathways with no change between Wisp1+/+ and Wisp1–/– cells are shown in Supplemental Table 6. In sum, isolated Wisp1–/– cells have a gene expression profile associated with an increase in osteochondral markers and changes in several pathways that would predispose to a chondrogenic cell fate.
Osteochondrogenic differentiation is skewed among Wisp1–/– mesenchymal progenitor cells.
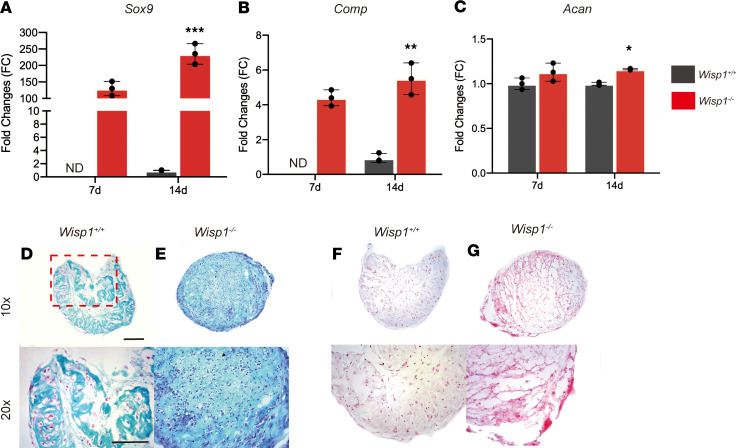
As mentioned, WISP1 has context-dependent effects on chondrogenic differentiation (18). To further investigate, isolated Wisp1+/+ and Wisp1–/– ASCs were exposed to chondrogenic or osteogenic culture conditions (Figure 7 and Supplemental Figure 5). Wisp1–/– cells showed an increase in chondrogenic differentiation in micromass culture (Figure 7). This included an increase in chondrogenic gene expression by qPCR, including Sox9 (Figure 7A), Comp (Figure 7B), and Aggrecan (Figure 7C). Similar findings were observed using sections of micromasses after 21 days in culture, as assessed by Alcian blue (Figure 7, D and E) and Safranin O red staining (Figure 7, F and G).
Figure 7. Chondrogenic differentiation among Wisp1–/– adipose-derived stromal cells.
(A–C) Gene expression for markers of chondrogenic differentiation, assessed by micromass culture at days 14 of differentiation. (A) Sox9, (B) cartilage oligomeric matrix protein (Comp), and (C) Acan. (D–G) Micromass culture of Wisp1+/+ and Wisp1–/– adipose derived–stromal cells in chondrogenic differentiation medium. (D and E) Representative images of Wisp1+/+ micromass culture sections stained with Alcian blue. (F and G) Representative images of Wisp1+/+ and Wisp1–/– micromass culture sections stained with Safranin O red. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar: 100 μm. Original magnification, ×10 (bottom row, D–G); ×20 (bottom row, D–G). Differences were calculated between groups by Student’s t test. Data represent mean ± SD and triplicate experimental replicates in biological duplicate.
Next, osteogenic differentiation was assessed among Wisp1+/+ and Wisp1–/– ASCs (Supplemental Figure 5). Results showed the opposing trend among Wisp1–/– cells, including a reduction in osteogenic gene expression, including Alpl, type 1 collagen (Col1), Bglap, and Runx2 (Supplemental Figure 5, A–D). Likewise, alizarin red staining performed at day 14 of differentiation confirmed a reduction in mineralization of Wisp1–/– cells (Supplemental Figure 5E).
Discussion
Here, we observed that CCN family members, including Wisp1, demonstrated dynamic induction during the process of trauma-induced HO. These findings were observed across mouse and human tissues. Further, transgenic Wisp1-null mice demonstrated a tendency to form more endochondral bone formation after soft tissue trauma. Several potential downstream signaling pathways altered among Wisp1–/–adipose stromal cells were identified that would predispose to a chondrogenic phenotype, including STAT3 (32), PKA (31), and p38 MAPK signaling (34).
Our findings are just one example of how the matricellular protein WISP1 alters cellular fate and differentiation decisions. Past studies have implicated WISP1 in the balance of osteoblast versus adipocyte formation (38, 39) and, more broadly, epithelial cells adopting a mesenchymal cell fate (epithelial-mesenchymal transition) (40–43). As CCN family members, including WISP1, have numerous extracellular binding partners it is understandable that they may have context-dependent effects on cellular differentiation. In terms of chondrogenesis, WISP1 overexpression antagonizes BMP2- or GDF5-induced chondrogenesis (16), while, in other observations, WISP1 enhances TGF-β3–induced chondrogenesis (18). Further, deficiency of Wisp1 appears to be protective against articular cartilage catabolism in mouse models. For example, previous data has shown a significant decrease in cartilage degeneration in collagenase-induced arthritis using another Wisp1-knockout mouse (19). Data in osteoarthritis have also shown a decrease in cartilage degeneration and an increase in cartilage matrix formation in Wisp1-knockout mice, and similar findings in WISP1 stimulated osteoarthritic human chondrocytes (19, 20). In our models, Wisp1 deletion led to a significant increase in aberrant endochondral bone formation after trauma, and isolation of Wisp1-null adipose stromal cells demonstrated signaling pathway changes consistent with a chondrogenic cell fate. Understanding that the effects of WISP1 signaling on cartilage differentiation have been somewhat nonuniform across studies, it is important to recognize that the tissue source, species for cell derivation, and method of Wisp1 inhibition (acute knockdown versus global genetic knockout) all may have contributory roles in any discrepant findings.
Pathway analyses and pathway target genes activation demonstrated that Wisp1-null adipose stromal cells showed some broad-ranging changes in multiple pathways associated with the regulation of chondrogenic differentiation. Interestingly, noncanonical Wnt signaling (PCP signaling) was activated, while Wnt/β-catenin signaling was downregulated among Wisp1-null cells. In agreement with findings in other cell lines (44), Smad-independent TGF-β signaling via PKA signaling and STAT3 was upregulated, while no change in canonical TGF-β signaling was observed among Wisp1-null cells. The STAT3 signaling pathway has shown to positively regulate chondrogenic differentiation in human mesenchymal stromal cells (45). Likewise, and in agreement with findings in other cell lines (46), noncanonical BMP signaling was activated via P38 MAPK signaling among Wisp1-null cells. The differential expression of selected target genes of the chondrogenic differentiation–related signaling pathway were confirmed among primary tenocytes. For instance, Rela has been reported to be involved in chondrogenic differentiation and cell survival and prevent chondrocyte apoptosis (47). The confluence of these pathway changes would be expected to include a predisposition to cartilage differentiation, as we observed in vivo after soft tissue injury.
The chondrogenic and osteogenic induced ASC differentiation also confirmed the observation in mouse HO. Our chondrogenic differentiation results showed a significant increase in chondrogenic associated genes in Wisp1-null cells. On the other hand, osteogenic induced culture showed a decrease in osteogenic differentiation in Wisp1-null ASCs. This suggested that in a context-dependent manner Wisp1 can negatively regulate chondrogenic differentiation but positively regulate osteogenic differentiation. In our in vivo results, Wisp1-null mice showed a conspicuous increase in cartilage after injury, while bone formation was increased to a lesser degree. Further study must be performed to understand the cell- and context-dependent effects of Wisp1 in other models of endochondral ossification.
Important limitations exist in the study. Our in vitro findings were notable for a prominent and possibly compensatory increase in the CCN family member Nov among Wisp1-null cells. In previous studies with genetic deletion of CCN family member proteins, other CCN family members can play a compensatory role (48). For example, overexpression of Ccn3 in murine embryonic skin fibroblast–derived NIH3T3 cells results in a significant decrease in expression of Ccn2 and Ccn4, suggesting the compensatory nature of these CCN family member proteins in organ fibrosis (49, 50). As a second potential limitation, global knockout of Wisp1 not only affects mesenchymal cells, but likely has secondary effects on inflammatory cell types, such as macrophages and T lymphocytes (51). In future studies, use of tissue-specific Wisp1-knockout animals may better elucidate mesenchymal cell intrinsic functions of WISP1 in the regulation of HO. However, based on our scRNA-sequencing data, Wisp1 is predominantly expressed in adipose stromal cells and only expressed at low levels in other cell subpopulations. This suggests that a tissue-specific knockout for mesenchymal cells would likely have the same phenotype as the global knockout used here.
In conclusion, among CCN family members, Wisp1 expression is strongly associated with bone and cartilage across mouse and human HO. Studies in transgenic Wisp1-knockout animals suggest a role for Wisp1 in the negative regulation of trauma-induced HO and implicate pathways associated with enhanced cartilaginous differentiation in this process. To date, there are no clinically approved treatments for trauma-induced HO that use specific molecular targets (1). Improved understanding of CCN family signaling in HO may open the door to more targeted therapies for this common and potentially severe condition.
Methods
Bulk RNA sequencing.
Bulk RNA sequencing of RNA was performed using RNA from the Achilles tendon tenotomy site. Tissue was collected from the soft tissue around the injury site. Tissue was dissected between the muscular origin and calcaneal insertion of Achilles tendon. Corresponding soft tissue, including intact Achilles tendon, was also harvested from the uninjured contralateral hind limb. n = 3 mouse tissues were isolated at 3 weeks after injury. Tissues were immediately stabilized after isolation using RNAlater (Ambion). Total cellular RNA was extracted from the specimen using the TRIZOL reagent kit (Ambion) and was examined per condition with the Affymetrix Gene 2.1 ST.
Mouse ASC and tenocyte isolation, differentiation, and analyses.
For ASC collection, Wisp1+/+ and Wisp1–/– mice were sacrificed and the inguinal fat pad tissues were harvested. Tissues were sequentially washed in PBS. The tissues were placed on a clean Petri dish lid and finely minced. Digestion of fat was achieved by 1 mg/mL collagenase type II. For tenocyte collection, Wisp1+/+ and Wisp1–/– mice were sacrificed and the Achilles tendon was harvested and digested with 3 mg/mL collagenase type I and 4 mg/mL dispase type II (52). Both tissues were incubated in a 37°C shaker (160 g) for 1 hour. Equal amounts of growth media were added (DMEM, 10% FBS, 1% penicillin/streptomycin), and cells were passed through a 40-μm strainer. The filtered cells were centrifuged at 450 g for 10 minutes, and the cell pellet was resuspended in 10 mL growth media. Cells were seeded into a 10-cm dish. Media were changed every 3 days unless otherwise noted.
For chondrogenic differentiation, mouse ASCs at passages 3–5 were used. Cells were cultured in high-density micromass as modified protocol (53) or 1 × 104 cells/well in 24-well plates for monolayer culture. The MesenCult-ACF Chondrogenic Differentiation Kit (StemCell Technology, catalog 05455) was used. Medium was changed every 3 days, and micromasses were analyzed for up to 14 days. Micromasses were fixed in 4% PFA, placed in 30% sucrose and embedded with optimal cutting temperature compound. Serial sections of 10 μm were made, hydrated, and subsequently stained with Alcian blue (MilliporeSigma) or Safranin O (MilliporeSigma) using previously published methods (54).
Osteogenic differentiation medium consisted of 10 mM β-glycerophosphate and 50 μM ascorbic acid in DMEM + 15% FBS. Alkaline phosphatase staining was performed using the Leukocyte Alkaline Phosphatase Kit (MilliporeSigma). Briefly, cells were seeded in 24-well plates at a density of 1 × 104 cells/well. Cells were cultured under osteogenic differentiation conditions for 14 days before staining. Cells were then washed with PBS and fixed with formalin for 10 minutes at room temperature. Following fixation, cells were stained using the Leukocyte Alkaline Phosphatase Kit (MilliporeSigma) according to the manufacturer’s protocol. Cells were incubated in alkaline phosphatase stain for 15 minutes at 37°C and then washed with PBS. Relative staining was quantified using Adobe Photoshop CC2015.
For the detection of mineralization, cells were seeded in growth medium in 24-well plates at a density of 1 × 104 cells/well. Twenty-four hours after seeding cells, basal medium was replaced with ODM in triplicates per each treatment for 12 days. Cells were washed with PBS and fixed with 4% paraformaldehyde. Following fixation, cells were stained with 2% alizarin red (MilliporeSigma) at room temperature for 15 minutes and then washed with deionized water and allowed to dry.
RNA isolation and quantitative real-time PCR.
Gene expression was assayed by quantitative RT-PCR. Primer sequences are shown in Supplemental Table 7. Briefly, total RNA was extracted using Trizol (Invitrogen). 1 μg total RNA from each sample was subjected to first-strand cDNA synthesis using the SuperScript III Reverse-Transcriptase Kit (Life Technologies) to a final volume of 20 μL. The reverse transcription reaction was performed at 65°C for 5 minutes, followed by 50°C for 50 minutes and 85°C for 5 minutes.
The quantitative real-time PCR (qRT-PCR) reaction was performed using SYBR green RT-PCR master mix (Thermo Fisher Scientific) and an ABI PRISM 7300 qRT-PCR system instrument (Applied Biosystems). qRT-PCR was performed using 384-well optical plates at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and at 60°C for 60 seconds. The relative quantification of gene expression was performed using a comparative CT method according to the manufacturer’s protocol and was normalized to the expression levels of Gapdh in each sample.
RNASeq — 10x single-cell genomics.
For scRNA-sequencing analysis, the tenotomy site was harvested by microdissection (days 3, 7, and 21 after injury) and compared with baseline uninjured tissue at the same site. n = 3–4 mice per time point were used. Baseline (t0) and tissue samples harvested on days 3, 7, and 21 were digested for 45 minutes in 0.3% type 1 collagenase and 0.4% Dispase II (Gibco Invitrogen) in RPMI medium at 37°C under constant agitation at 160 g. Digestions were subsequently quenched with 10% FBS RPMI and filtered through 40-μm sterile strainers. Cells were then washed in PBS with 0.04% BSA, counted, and resuspended at a concentration of approximately 1000 cells/μL. Cell viability was assessed with trypan blue exclusion on a Countess II (Thermo Fisher Scientific) automated counter, and only samples with >85% viability were processed for further sequencing.
Single-cell 3′ library generation was performed on the 10× Genomics Chromium Controller following the manufacturer’s protocol for the v2 reagent kit (10× Genomics). Cell suspensions were loaded onto a Chromium Single-Cell A chip along with reverse-transcription (RT) master mix and single-cell 3′ gel beads, aiming for 2000–6000 cells per channel. The cDNA was amplified, and SPRIselect (Beckman Coulter) beads were cleaned and quantified and then enzymatically fragmented and size selected using SPRIselect beads to optimize the cDNA amplicon size before library construction. An additional round of double-sided SPRI bead cleanup was performed after end repair and A-tailing. Another single-sided cleanup was done after adapter ligation. Indexes were added during PCR amplification and a final double-sided SPRI cleanup was performed. Libraries were sequenced on a HiSeq 4000 (Illumina) using a HiSeq 4000 PE Cluster Kit (PN PE-410-1001) with HiSeq 4000 SBS Kit (100 cycles, PN FC-410-1002) reagents, loaded at 200 pM following Illumina’s denaturing and dilution recommendations. Cell Ranger Single Cell Software Suite 1.3 was used to perform sample demultiplexing, barcode processing, and single-cell gene counting (alignment, barcoding, and unique molecular identifiers [UMI] count) at the University of Michigan Biomedical Core Facilities DNA Sequencing Core.
The sequencing data were first preprocessed using the 10× Genomics software Cell Ranger and aligned to mm10 genome. Downstream analysis steps were performed using Seurat 2.3.0. Cells with fewer than 500 genes per cell, with more than 60,000 UMIs, or expressing a fraction of mitochondrial UMIs higher than 0.1 were filtered for quality control. The number of UMIs (UMI fraction related to mitochondrial genes >0.1) and the number of genes, mitochondrial genes, and cell cycle genes (S and G2M scores by CellCycleScoring Seurat function) were regressed before further analysis. The downstream analysis was performed with Seurat 2.3.0. The time point sets were merged with canonical correlation analysis based on the intersection of the variable genes for time point set. Unsupervised clustering (Louvain algorithm) was used to identify cell populations.
Transgenic strains.
Unless otherwise stated, experiments were performed on age-matched 8-week-old, mixed-sex C57BL/6J WT mice or Wisp1-knockin mice. C57BL/6J mice were purchased from The Jackson Laboratory. Wisp1 global knockout mice were generated in the Lyons laboratory and genotyped as per prior reports (30). In the Wisp1-knockin mouse, exons 1–5 are replaced by a LacZ-neo cassette. For these experiments, Wisp1+/+ and Wisp1–/– (i.e., Wisp1LacZ/LacZ) littermates were compared. To generate an FOP-like genetic model, mice carrying the floxed constitutively active allele of ACVR1 (ACVR1 carrying Q207D mutation, ca-ACVR1fl/WT) and Nfatc1-Cre–transgenic mice were used for breeding, as previously described (26–29). Resulting pups carrying both transgenes (Nfatc1-Cre; ca-ACVR1fl/WT) developed spontaneous heterotopic bone by postnatal days 4–5.
Histological and immunohistochemical analyses.
Five-mm-thick paraffin sections of mouse and human tissues were stained with H&E. Additional sections were analyzed by indirect immunohistochemistry, with antibodies summarized in Supplemental Table 3. Briefly, unstained sections were deparaffinized in xylene and a series of graded ethanol solutions and rehydrated using PBS. The slides were treated with Trypsin Enzymatic Antigen Retrieval Solution (ab970, Abcam) for 10 minutes at 37°C and then incubated with 3% hydrogen peroxide (20 minutes at room temperature) and 5% goat serum (1 hour at room temperature). Next, slides were probed with primary antibodies at a dilution of 1:200 overnight. Subsequently, for slides probed with rabbit host primary antibodies, the sections were washed and incubated with biotinylated secondary antibody (BA-1000, Vector Laboratories) and VECTASTAIN Elite ABC HRP reagent (PK-6100, Vector Laboratories) for 1 hour and 30 minutes at room temperature. Color development was achieved by treatment with the chromogen DAB (SK-4100, Vector Laboratories) and was carried out for 5–10 minutes. The slides were then blocked with BLOXALL Blocking Solution (SP-6000, Vector laboratories) and 2.5% normal horse serum (MP-5402, Vector Laboratories) for 10 and 20 minutes at room temperature. For slides probed with mouse host primary antibodies, the sections were washed and incubated with IMPRESS-AP reagent (MP-5402, Vector Laboratories) for 30 minutes at room temperature. Color development was achieved by treatment with the Alkaline Phosphatase Substrate (SK-5100, Vector Laboratories) and was carried out for 5–10 minutes. Finally, the sections were rinsed in distilled water, counterstained with hematoxylin (H-3401, Vector Laboratories), and mounted on glass slides before microscopic evaluation.
X-gal staining.
Cryosections of Wisp1-LacZ tenotomy sites were washed in PBS for 10 minutes. Samples were washed (wash buffer, 200 μL 1 M MgCl2, 10 mg sodium deoxycholate, 0.02% Np-40 [Triton X-100] in 100 mL 0.1 M PBS) for 15 minutes 3 times. Samples were placed in fresh X-gal staining solution (200 μL X-gal stock, 100 μL 0.5 M potassium ferricyanide, 100 μL 0.5 M potassium ferrocyanide, 200 μL 1 M Tris, 9.4 mL wash buffer) for 96 hours at 37°C. Samples were then washed in wash buffer for 15 minutes twice and counterstained with Nuclear Fast Red (N8002, MilliporeSigma) for 5 minutes, placed under running tap water for 1 minutes, dehydrated, and mounted using permanent mounting medium.
Positive immunoreactivity was detected following ABC complex (PK-6100, Vectastain Elite ABC Kit, Vector Laboratories Inc.) incubation and development with AEC chromogen (K346911-2, Dako). Negative controls for each antibody consisted of incubation with secondary antibody in the absence of primary antibody. Sections were counterstained in Modified Mayer’s Hematoxylin (Thermo Fisher Scientific) for 30 seconds and placed under running water for 5 minutes. Slides were mounted using aqueous mounting medium (Dako). Photomicrographs were acquired using Olympus BX51 (UPLanFL).
Identification of human nongenetic HO samples.
Twelve cases of nongenetic HO were identified in our surgical pathology archives (Johns Hopkins University and University of California, Los Angeles). All material was coded so as to protect the confidentiality of personal health information. All cases were reviewed by 2 independent pathologists verifying the diagnostic accuracy. Patient demographics are provided in Supplemental Table 2.
μCT imaging and analysis.
A total of 22 mouse (11 in each group) hind limbs were harvested and imaged 9 weeks after injury (Bruker MicroCT, 35-μm resolution and 357 μA/70kV/beam). Scans were analyzed by blinded operators manually spinning around ectopic tissue and computing volumes at 800 Hounsfield units (HU) (GE Healthcare v2.2, Parallax Innovations rc18).
RNA Clariom D microarray analyses.
RNA from Wisp1+/+ and Wisp1–/– adipose stromal cells was isolated as per prior methods (39). RNA isolation was performed as previously described (55). RNA transcript levels were quantified by microarray analyses on the Clariom D Array GeneChip (Affymetrix; Thermo Fisher Scientific). Raw expression was RNA normalized and underwent analysis of variance using the PARTEC GS 7.0 platform. Microarray analysis was conducted in the Johns Hopkins Transcriptomics and Deep Sequencing Core Facility. Gene expression values for Wisp1–/– to Wisp1+/+ animals are shown as log2 FC.
Data availability.
Expression data that support the findings of this study have been deposited in Gene Expression Omnibus (GEO) (accession GSE 126060, GSE 127956, and GSE 126118).
Statistics.
Statistical analyses were performed in IBM SPSS Statistics 16.0. Quantitative data are expressed at mean ± SD. Two-tailed Student’s t test was used for 2-group comparisons, and 1-way ANOVA test was used for comparisons of 3 or more groups, followed by Tukey’s post hoc test.
Study approval.
All animal procedures were carried out in accordance with the guidelines provided in the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by IACUC of the University of Michigan (PRO0007390). All animals were housed in IACUC-supervised facilities at 18°C–22°C on a 12-hour light/dark cycle, with ad libitum access to food and water. Cases were obtained under approval from the University of California, Los Angeles, IRB with a waiver of informed consent.
Author contributions
GCYH collected and assembled data, provided data analysis and interpretation, and wrote the manuscript. S. Marini collected and assembled data. CAM collected and assembled data. JX collected and assembled data. YW collected and assembled data. SN collected and assembled data. CH collected and assembled data. DS collected and assembled data. S. Miller collected data and wrote the manuscript. CP collected and assembled data. EM provided study materials. KML provided study materials. SN, CH, and DS collected and assembled data. BL provided study materials and financial support and approved the final manuscript. AWJ provided study materials, conception, and design; wrote the manuscript; provided financial support; and approved the final manuscript.
Supplementary Material
Acknowledgments
AWJ was supported by the NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 AR070773 and K08 AR068316), the NIH/National Institute of Dental and Craniofacial Research (R21 DE027922), US Army Medical Research Acquisition Activity (W81XWH-18-1-0121, W81XWH-18-1-0336, and W81XWH-18-1-0613), the American Cancer Society (RSG-18-027-01-CSM), and the Orthopaedic Research and Education Foundation, with funding provided by the Musculoskeletal Transplant Foundation, and the Maryland Stem Cell Research Foundation. BL was funded by the NIH (R01GM123069 and R01AR071379), the American College of Surgeons Clowes Award, and the International Fibrodysplasia Ossificans Progressiva Association Research Award.
Version 1. 06/02/2020
In-Press Preview
Version 2. 07/09/2020
Electronic publication
Footnotes
Conflict of interest: AWJ is on the scientific advisory board for Novadip Biosciences and receives research funding from Novadip Biosciences and MTF Biologics.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(13):e135432.https://doi.org/10.1172/jci.insight.135432.
Contributor Information
Ginny Ching-Yun Hsu, Email: chsu46@jhu.edu.
Simone Marini, Email: smarini@umich.edu.
Stefano Negri, Email: stefano.negri@jhu.edu.
Yiyun Wang, Email: ywang412@jhmi.edu.
Jiajia Xu, Email: jxu76@jhmi.edu.
Charles Hwang, Email: umichuck@umich.edu.
Carolyn A. Meyers, Email: cannmeyers@gmail.com.
Sarah Miller, Email: sarahfmiller12@gmail.com.
Edward McCarthy, Email: mccarthy@jhmi.edu.
Karen M. Lyons, Email: klyons@mednet.ucla.edu.
Benjamin Levi, Email: blevi@umich.edu.
Aaron W. James, Email: aaronwjames1@gmail.com.
References
- 1.Meyers C, et al. Heterotopic ossification: a comprehensive review. JBMR Plus. 2019;3(4):e10172. doi: 10.1002/jbm4.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranganathan K, et al. Heterotopic ossification: basic-science principles and clinical correlates. J Bone Joint Surg Am. 2015;97(13):1101–1111. doi: 10.2106/JBJS.N.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal S, et al. Inhibition of Hif1α prevents both trauma-induced and genetic heterotopic ossification. Proc Natl Acad Sci USA. 2016;113(3):E338–E347. doi: 10.1073/pnas.1515397113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal S, et al. Scleraxis-lineage cells contribute to ectopic bone formation in muscle and tendon. Stem Cells. 2017;35(3):705–710. doi: 10.1002/stem.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loder SJ, et al. Characterizing the circulating cell populations in traumatic heterotopic ossification. Am J Pathol. 2018;188(11):2464–2473. doi: 10.1016/j.ajpath.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey D, et al. Two tissue-resident progenitor lineages drive distinct phenotypes of heterotopic ossification. Sci Transl Med. 2016;8(366):366ra163. doi: 10.1126/scitranslmed.aaf1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal S, et al. BMP signaling mediated by constitutively active activin type 1 receptor (ACVR1) results in ectopic bone formation localized to distal extremity joints. Dev Biol. 2015;400(2):202–209. doi: 10.1016/j.ydbio.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kan L, Peng CY, McGuire TL, Kessler JA. Glast-expressing progenitor cells contribute to heterotopic ossification. Bone. 2013;53(1):194–203. doi: 10.1016/j.bone.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocks M, et al. Vascular patterning in human heterotopic ossification. Hum Pathol. 2017;63:165–170. doi: 10.1016/j.humpath.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perbal A, Perbal B. The CCN family of proteins: a 25th anniversary picture. J Cell Commun Signal. 2016;10(3):177–190. doi: 10.1007/s12079-016-0340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanaan RA, Aldwaik M, Al-Hanbali OA. The role of connective tissue growth factor in skeletal growth and development. Med Sci Monit. 2006;12(12):RA277–RA281. [PubMed] [Google Scholar]
- 12.Kubota S, Takigawa M. Role of CCN2/CTGF/Hcs24 in bone growth. Int Rev Cytol. 2007;257:1–41. doi: 10.1016/S0074-7696(07)57001-4. [DOI] [PubMed] [Google Scholar]
- 13.Kubota S, Takigawa M. The role of CCN2 in cartilage and bone development. J Cell Commun Signal. 2011;5(3):209–217. doi: 10.1007/s12079-011-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalakkal P, N Ferreira A, C Da Costa G, A Aras M. Functional lingual arch with hinge-type lockable dentulous component. Int J Clin Pediatr Dent. 2017;10(3):302–308. doi: 10.5005/jp-journals-10005-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono M, Inkson CA, Kilts TM, Young MF. WISP-1/CCN4 regulates osteogenesis by enhancing BMP-2 activity. J Bone Miner Res. 2011;26(1):193–208. doi: 10.1002/jbmr.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French DM, et al. WISP-1 is an osteoblastic regulator expressed during skeletal development and fracture repair. Am J Pathol. 2004;165(3):855–867. doi: 10.1016/S0002-9440(10)63348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda A, et al. WNT1-induced secreted protein-1 (WISP1), a novel regulator of bone turnover and Wnt signaling. J Biol Chem. 2015;290(22):14004–14018. doi: 10.1074/jbc.M114.628818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshioka Y, et al. CCN4/WISP-1 positively regulates chondrogenesis by controlling TGF-β3 function. Bone. 2016;83:162–170. doi: 10.1016/j.bone.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bosch MH, et al. WISP1/CCN4 aggravates cartilage degeneration in experimental osteoarthritis. Osteoarthr Cartil. 2017;25(11):1900–1911. doi: 10.1016/j.joca.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 20.van den Bosch MHJ, et al. Increased WISP1 expression in human osteoarthritic articular cartilage is epigenetically regulated and decreases cartilage matrix production. Rheumatology (Oxford) 2019;58(6):1065–1074. doi: 10.1093/rheumatology/key426. [DOI] [PubMed] [Google Scholar]
- 21.Peterson JR, et al. Effects of aging on osteogenic response and heterotopic ossification following burn injury in mice. Stem Cells Dev. 2015;24(2):205–213. doi: 10.1089/scd.2014.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson JR, et al. Early detection of burn induced heterotopic ossification using transcutaneous Raman spectroscopy. Bone. 2013;54(1):28–34. doi: 10.1016/j.bone.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson JR, et al. Treatment of heterotopic ossification through remote ATP hydrolysis. Sci Transl Med. 2014;6(255):255ra132. doi: 10.1126/scitranslmed.3008810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson JR, et al. Direct mouse trauma/burn model of heterotopic ossification. J Vis Exp. 2015;(102):e52880. doi: 10.3791/52880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal S, et al. Diminished chondrogenesis and enhanced osteoclastogenesis in leptin-deficient diabetic mice (ob/ob) impair pathologic, trauma-induced heterotopic ossification. Stem Cells Dev. 2015;24(24):2864–2872. doi: 10.1089/scd.2015.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal S, et al. Analysis of bone-cartilage-stromal progenitor populations in trauma induced and genetic models of heterotopic ossification. Stem Cells. 2016;34(6):1692–1701. doi: 10.1002/stem.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB. BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128(22):4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- 28.Kakudo N, Kusumoto K, Wang YB, Iguchi Y, Ogawa Y. Immunolocalization of vascular endothelial growth factor on intramuscular ectopic osteoinduction by bone morphogenetic protein-2. Life Sci. 2006;79(19):1847–1855. doi: 10.1016/j.lfs.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 29.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287(1):134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, Hu Z, Lyons KM. Design and analysis of CCN gene activity using CCN knockout mice containing LacZ reporters. Methods Mol Biol. 2017;1489:325–345. doi: 10.1007/978-1-4939-6430-7_28. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y. Wnt signaling in development and disease. Cell Biosci. 2012;2(1):14. doi: 10.1186/2045-3701-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SY, et al. The therapeutic effect of STAT3 signaling-suppressed MSC on pain and articular cartilage damage in a rat model of monosodium iodoacetate-induced osteoarthritis. Front Immunol. 2018;9:2881. doi: 10.3389/fimmu.2018.02881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J, et al. Sustained Akt signaling in articular chondrocytes causes osteoarthritis via oxidative stress-induced senescence in mice. Bone Res. 2019;7:23. doi: 10.1038/s41413-019-0062-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142(5):817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez-Carballo E, Gámez B, Ventura F. p38 MAPK signaling in osteoblast differentiation. Front Cell Dev Biol. 2016;4:40. doi: 10.3389/fcell.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bandow K, Kusuyama J, Kakimoto K, Ohnishi T, Matsuguchi T. AMP-activated protein kinase (AMPK) activity negatively regulates chondrogenic differentiation. Bone. 2015;74:125–133. doi: 10.1016/j.bone.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Klinger P, et al. PEDF is associated with the termination of chondrocyte phenotype and catabolism of cartilage tissue. Biomed Res Int. 2017;2017:7183516. doi: 10.1155/2017/7183516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrand N, Béreziat V, Moldes M, Zaoui M, Larsen AK, Sabbah M. WISP1/CCN4 inhibits adipocyte differentiation through repression of PPARγ activity. Sci Rep. 2017;7(1):1749. doi: 10.1038/s41598-017-01866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers CA, et al. WISP-1 drives bone formation at the expense of fat formation in human perivascular stem cells. Sci Rep. 2018;8(1):15618. doi: 10.1038/s41598-018-34143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang AC, Lien MY, Tsai MH, Hua CH, Tang CH. WISP-1 promotes epithelial-mesenchymal transition in oral squamous cell carcinoma cells via the miR-153-3p/Snail axis. Cancers (Basel) 2019;11(12):E1903. doi: 10.3390/cancers11121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng W, Fernandez A, McLaughlin SL, Klinke DJ. WNT1-inducible signaling pathway protein 1 (WISP1/CCN4) stimulates melanoma invasion and metastasis by promoting the epithelial-mesenchymal transition. J Biol Chem. 2019;294(14):5261–5280. doi: 10.1074/jbc.RA118.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YZ, et al. WISP1 silencing confers protection against epithelial-mesenchymal transition of renal tubular epithelial cells in rats via inactivation of the wnt/β-catenin signaling pathway in uremia. J Cell Physiol. 2019;234(6):9673–9686. doi: 10.1002/jcp.27654. [DOI] [PubMed] [Google Scholar]
- 43.Heise RL, Stober V, Cheluvaraju C, Hollingsworth JW, Garantziotis S. Mechanical stretch induces epithelial-mesenchymal transition in alveolar epithelia via hyaluronan activation of innate immunity. J Biol Chem. 2011;286(20):17435–17444. doi: 10.1074/jbc.M110.137273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pütter L. WNT1-inducible signaling pathway protein-1 (WISP1), a novel critical protein in hepatotoxic liver injury. Biology. 2017. https://eldorado.tu-dortmund.de/bitstream/2003/36040/1/Dissertation_P%C3%BCtter%20.pdf Accessed June 23, 2020.
- 45.Kondo M, et al. Contribution of the interleukin-6/STAT-3 signaling pathway to chondrogenic differentiation of human mesenchymal stem cells. Arthritis Rheumatol. 2015;67(5):1250–1260. doi: 10.1002/art.39036. [DOI] [PubMed] [Google Scholar]
- 46.Venkatesan B, et al. WNT1-inducible signaling pathway protein-1 activates diverse cell survival pathways and blocks doxorubicin-induced cardiomyocyte death. Cell Signal. 2010;22(5):809–820. doi: 10.1016/j.cellsig.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi H, et al. Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat Commun. 2016;7:13336. doi: 10.1038/ncomms13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubota S, Takigawa M. The CCN family acting throughout the body: recent research developments. Biomol Concepts. 2013;4(5):477–494. doi: 10.1515/bmc-2013-0018. [DOI] [PubMed] [Google Scholar]
- 49.Abd El Kader T, et al. Anti-fibrotic effect of CCN3 accompanied by altered gene expression profile of the CCN family. J Cell Commun Signal. 2013;7(1):11–18. doi: 10.1007/s12079-012-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peidl A, Perbal B, Leask A. Yin/Yang expression of CCN family members: transforming growth factor beta 1, via ALK5/FAK/MEK, induces CCN1 and CCN2, yet suppresses CCN3, expression in human dermal fibroblasts. PLoS One. 2019;14(6):e0218178. doi: 10.1371/journal.pone.0218178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, et al. WISP1-αvβ3 integrin signaling positively regulates TLR-triggered inflammation response in sepsis induced lung injury. Sci Rep. 2016;6:28841. doi: 10.1038/srep28841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song N, Armstrong AD, Li F, Ouyang H, Niyibizi C. Multipotent mesenchymal stem cells from human subacromial bursa: potential for cell based tendon tissue engineering. Tissue Eng Part A. 2014;20(1-2):239–249. doi: 10.1089/ten.tea.2013.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naftali-Shani N, et al. Left ventricular dysfunction switches mesenchymal stromal cells toward an inflammatory phenotype and impairs their reparative properties via toll-like receptor-4. Circulation. 2017;135(23):2271–2287. doi: 10.1161/CIRCULATIONAHA.116.023527. [DOI] [PubMed] [Google Scholar]
- 54.Jang Y, et al. Centrifugal gravity-induced BMP4 induces chondrogenic differentiation of adipose-derived stem cells via SOX9 upregulation. Stem Cell Res Ther. 2016;7(1):184. doi: 10.1186/s13287-016-0445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen W, et al. Nfatc2 is a primary response gene of Nell-1 regulating chondrogenesis in ATDC5 cells. J Bone Miner Res. 2011;26(6):1230–1241. doi: 10.1002/jbmr.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression data that support the findings of this study have been deposited in Gene Expression Omnibus (GEO) (accession GSE 126060, GSE 127956, and GSE 126118).