Abstract
The mortality of patients suffering from acute myocardial infarction is linearly related to the infarct size. As regeneration of cardiomyocytes from cardiac progenitor cells is minimal in the mammalian adult heart, we have explored a new therapeutic approach, which leverages the capacity of nanomaterials to release chemicals over time to promote myocardial protection and infarct size reduction. Initial screening identified 2 chemicals, FGF1 and CHIR99021 (a Wnt1 agonist/GSK-3β antagonist), which synergistically enhance cardiomyocyte cell cycle in vitro. Poly-lactic-co-glycolic acid nanoparticles (NPs) formulated with CHIR99021 and FGF1 (CHIR + FGF1-NPs) provided an effective slow-release system for up to 4 weeks. Intramyocardial injection of CHIR + FGF1-NPs enabled myocardial protection via reducing infarct size by 20%–30% in mouse or pig models of postinfarction left ventricular (LV) remodeling. This LV structural improvement was accompanied by preservation of cardiac contractile function. Further investigation revealed that CHIR + FGF1-NPs resulted in a reduction of cardiomyocyte apoptosis and increase of angiogenesis. Thus, using a combination of chemicals and an NP-based prolonged-release system that works synergistically, this study demonstrates a potentially novel therapy for LV infarct size reduction in hearts with acute myocardial infarction.
Keywords: Cardiology, Therapeutics
Keywords: Cardiovascular disease, Cell cycle, Nanotechnology
A combination of chemicals and a nanoparticle-based prolonged-release system demonstrates a potential strategy for left ventricular infarct size reduction in acute myocardial infarction.
Introduction
The mortality of patients suffering from acute myocardial infarction (AMI) is linearly related to the infarct size. As cardiomyocyte regeneration from cardiac progenitor cells is minimal in the mammalian adult heart, salvaging the myocytes that are otherwise destined to die remains the most important objective in cardiovascular sciences. In response to a prolonged coronary artery occlusion, the distal myocardial myocytes undergo membrane rupture followed by release of their intracellular enzymes into the extracellular space. If we assume that AMI is a heterogeneously damaged tissue, then to discover the interventions sparing the border zone myocytes that have been less severely damaged could effectively reduce the infarct size and save lives.
Nanoparticles (NPs) can act as sustained-release delivery vehicles for growth factors and small molecules for an enhanced myocardial recovery in ischemic heart diseases (1, 2). FGF1 is one of these factors, and it has been shown that intramyocardial injection of FGF1-loaded poly-lactic-co-glycolic acid (PLGA) microparticles into infarcted areas improves angiogenesis and/or arteriogenesis, as well as ejection fraction (EF) (3). CHIR99021 (CHIR) is an aminopyrimidine derivative that functions as a Wnt signaling activator. CHIR-loaded NPs efficiently direct reprogramming of fibroblasts into functional cardiac myocytes and might thus be endowed with therapeutic potential in cardiovascular regeneration (4).
Through in vitro study of screening of chemicals that enhance cardiomyocyte cell cycle, we have identified the doses and the combination of CHIR and FGF that are most effective in promoting the cell cycle and antiapoptosis. Using left ventricular (LV) postinfarction remodeling models of mouse AMI or pig ischemia/reperfusion (IR) injury, we have investigated whether the combination of NPs with both CHIR and FGF1 can synergistically protect ischemia-threatened cardiomyocytes from apoptosis, accelerate angiogenesis through promoting the proliferation of endothelial and vascular smooth muscle cells, and consequently enhance myocardial protection. The findings demonstrate that a controlled-release formulation of PLGA NPs encapsulated with CHIR + FGF1 exerted substantial myocardial protective effects and represents a potentially novel strategy for improving postischemic myocardial protection.
Results
Identification of chemicals that promote cell cycle activity of human induced pluripotent stem cell–derived cardiomyocytes.
Using the BrdU incorporation assay, we screened several chemicals for their capacity to enhance the cell cycle activity of cultured human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs). We picked hiPSC-CMs because they are easy to handle and represent a practical system for drug testing and screening (5). These chemicals included Ly294002 (PI3K inhibitor) (6), FGF1 (6, 7), SB203580 and VX702 (p38 MAPK inhibitors) (6), KN93 (Ca2+/calmodulin-dependent protein kinase II inhibitor) (8), Su1498 (Flk-1 inhibitor) (8), and CHIR99021 (Wnt activator and GSK3α and 3β inhibitor) (8, 9). We found a combination of 5 μM CHIR99021 and 100 ng/mL FGF1 was the most potent treatment to induce cell cycle in hiPSC-derived cardiomyocytes (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132796DS1).
Characterization of CHIR-FGF1-NPs.
We have previously shown that PLGA NPs can be used as a platform for slow release (up to 4 weeks) of chemicals to injured animal hearts and that they provide cardioprotection (2). To characterize the long-term cardioprotective function of the FGF/CHIR combination in vivo, we formulated the PLGA NPs with these 2 factors. The size of PLGA NPs was measured using scanning electron microscopy, for both CHIR99021- and FGF1-loaded NPs (Supplemental Figure 2, A and B). Quantification of particle diameter for CHIR-NPs (Supplemental Figure 2C) and FGF1-NPs (Supplemental Figure 2D) yielded values of 123.63 ± 44.48 nm and 129.57 ± 45.94 nm, respectively. The size and shape of CHIR-NPs and FGF1-NPs were uniform. The encapsulation efficiency of CHIR-NPs and FGF1-NPs, i.e., (the amount encapsulated/total amount available) × 100%, was 50.41% and 62.8 ± 1.6%, respectively. The concentration of encapsulated CHIR and FGF1 was 8.07 μg/mg and 1.26 ± 0.03 μg/mg, respectively.
Determination of release kinetics of CHIR- and FGF1-loaded NPs as a function of time, using either NanoDrop via UV-Vis spectrophotometer (for CHIR) or ELISA (for FGF1), and the cumulative percentage of CHIR and FGF1 released from NPs, are shown in Supplemental Figure 2, E and F. When 1000 μg of CHIR- and FGF1-loaded NPs were incubated in 1000 μL of Dulbecco’s phosphate-buffered saline (DPBS), pH 7.4 at 37°C, 55% of the encapsulated CHIR was released during the first day and 85% by day 15 (Supplemental Figure 2E). In contrast, 55% of the encapsulated FGF1 was released during the initial 3 days, and 63% was released by day 10. Notably, between day 1 and day 30, the release kinetics strictly followed the Korsmeyer-Peppas model for FGF1-NPs (Supplemental Figure 2F). Fitting this model, Ct/C0 = ktn, where Ct = concentration at time t; C0 = equilibrium concentration; k = drug release rate constant; and n = diffusion exponent, yielded n = 0.123 and k = 0.324. Next, we investigated the cellular uptake and biodistribution of engineered NPs in both human umbilical vein endothelial cells (HUVECs) and human vascular smooth muscle cells (HVSMCs) in vitro by fluorescence microscopy. The endothelial (red, CD31) and smooth muscle protein 22-α (red, SM22α) cells were able to consistently take up and internalize the engineered NPs (Supplemental Figure 2, G and H).
Last, we explored the localization and tracing of engineered NPs in vivo, using a left anterior descending artery (LAD) ligation mouse model. Following LAD ligation, the coumarin-6–loaded NPs (33.3 μg/μL NPs) were injected into the infarcted mouse heart, at 3 sites. After 24 hours, the whole heart was removed, and the LV border zone tissue sections were subjected to dual immunostaining by antibodies targeting a cardiac-specific regulatory protein, cardiac troponin T (red, cTnT), and an endothelium-associated protein, platelet and endothelial cell adhesion molecule 1/CD31 (white, CD31). Fluorescence microscopic analysis revealed localization and distribution of NPs (green, NPs) within the ventricular tissue, mainly located around the infarct border zone (Supplemental Figure 2, I and J). We further studied whether NPs were preferentially taken up by different cells. We mixed HUVECs, smooth muscle cells (SMCs), hiPSC-CMs, and fibroblasts at the ratio of 1:1:1:1 and treated with coumarin-6–loaded NPs (2 μg/mL) for 24 hours. Our data suggested that there was no preferential uptake by different cells (Supplemental Figure 2K).
Cardioprotection of CHIR + FGF1-NPs in a mouse model of MI.
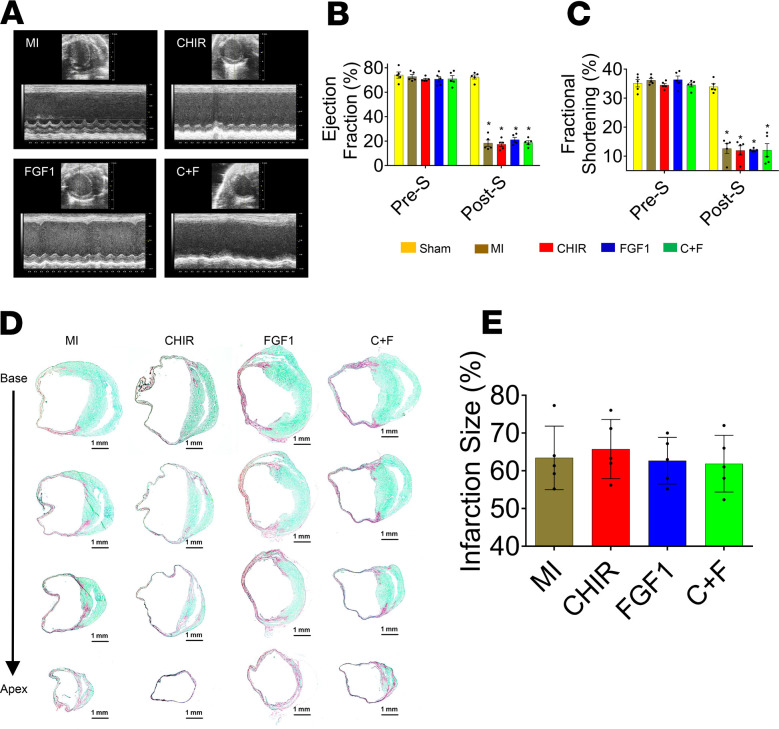
LV function was echocardiographically assessed before MI induction (baseline, pre-S) and 28 days after MI (post-S) (Figure 1A). Parameters of LV function included LV ejection fraction (LVEF) (Figure 1B), fractional shortening (FS) (Figure 1C), end-systolic diameter (ESD), and end-diastolic diameter (EDD) (Figure 1, D and E, respectively). On day 28 post-MI, CHIR + FGF1-NP–treated hearts presented significantly higher contractile indices (EF and FS) and smaller LV dimensions than those of the other treatment groups (Figure 1, B and C). These functional data were corroborated by a significant reduction in infarct size in the CHIR + FGF1-NP treatment group (Figure 1, F and G) compared with the other ones.
Figure 1. CHIR99021 and/or FGF1 NP treatment: assessment of cardiac function and infarct size in a mouse model of MI.
Mice that were treated with intramyocardial injections of different NPs, including CHIR + FGF1-NPs, CHIR-NPs, FGF1-NPs, and empty NPs (nonloaded); MI-only control mice; and sham-operated control mice were subjected to echocardiographic assessments of left ventricular (LV) function (A). Ejection fraction (EF) (B), fractional shortening (FS) (C), end-systolic diameter of the left ventricle (D), and end-diastolic diameter of the left ventricle (E) were assessed before MI induction (pre-S) and on post-MI day 28 (post-S). On day 28 after MI, CHIR + FGF1-NP treatment groups presented significantly greater EF and FS compared with other treatment as well as control groups (B and C) and significantly lower values of systolic/diastolic diameters of the left ventricle. Data are given as means ± SEM. There were 10–12 animals per group. Statistical analysis: 2-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. sham; †P < 0.01 vs. MI; ‡P < 0.05 vs. empty NPs; §P < 0.05 vs. CHIR-NPs, ||P < 0.01 vs. FGF1-NPs. Sirius Red/Fast Green histochemical staining revealing areas of infarcted (red, nonviable) and noninfarcted (green, viable) zones in post-MI day 28 ventricular tissue sections (F). The infarct size was quantified as the ratio of the scar area to the total surface area of the left ventricle and expressed as a percentage, for day 28 samples (G). At day 28, the CHIR + FGF1-NP treatment group showed significant reduction in infarct size compared with other NP treatment groups or the untreated control MI animals. Scale bar: 1 mm (panels in F). Data are given as means ± SEM. There were 10–12 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. MI; †P < 0.01 vs. empty NP; ‡P < 0.01 vs. CHIR-NP; §P < 0.01 vs. FGF1-NP.
We further assessed the compensatory hypertrophy around the periinfarct border zone. Twenty-eight days after the infarct, LV hypertrophy was assessed in this area by quantifying cardiomyocyte cross-sectional area (Supplemental Figure 3A). The MI and NP-treated groups showed significantly higher myocyte cross-sectional areas compared with sham-operated animals. Conversely, the myocyte cross-sectional area was significantly lower in the CHIR + FGF1-NP treatment group compared with the other treatment groups (Supplemental Figure 3B). The heart weight–to–body weight ratio of CHIR + FGF1-NP–treated animals was significantly lower compared with other treatment and MI-untreated animals (Supplemental Figure 3C).
To study whether prolonged release is necessary for the observed cardioprotection, we also tried intramyocardial injection of those chemicals immediately after LAD ligation procedure. All animals were assessed 4 weeks after treatment. Interestingly, we did not observe a similar cardioprotection following a direct intramyocardial injection of free chemicals CHIR with or without FGF1 (Figure 2, A–E); i.e., cardiac function (Figure 2, A–C) and infarct size (Figure 2, D and E) between MI-only and MI plus chemical-treated mice were not significantly different. These data suggest that a prolonged release of these chemicals is important for them to exert their cardioprotective effects.
Figure 2. Direct intramyocardial injection of free CHIR99021 and/or FGF1: assessment of cardiac function and infarct size in a mouse model of MI.
Mice were treated with direct intramyocardial injections of free CHIR, FGF1, and CHIR + FGF1 while MI-only mice served as controls. (A) Echocardiographic assessment of LV function. EF (B) and FS (C) were determined before MI (pre-S) and after 28 days (post-S). On day 28 after MI, no difference was identified among different treatment groups (means ± SEM, 5 animals per group). (D) Fibrosis, as assessed by Sirius Red/Fast Green histochemical staining (scale bar: 1 mm). At day 28, fibrosis, expressed as the ratio of the scar area to the total surface area of the left ventricle, did not differ between groups (E). There were 5 animals per group. Statistical analysis: 2-way ANOVA (B and C) and 1-way ANOVA (E) with Dunn’s multiple comparisons test. *P < 0.01 vs. sham (B and C, post-S).
Assessment of NP-induced cardiomyocyte proliferation and apoptosis in MI mice.
To probe the cellular mechanisms whereby the CHIR-FGF1-NPs mediated cardioprotection, we determined the cardiomyocyte cell cycle in infarcted hearts. To our surprise, cardiomyocyte cell cycle was negligible in either nontreated or CHIR-FGF1-NP–treated hearts (data not shown), which is in contrast to our observation that these NPs promoted cell cycle in cultured hiPSC-CMs (Supplemental Figure 1). These data suggest that cell cycle regulation may be different between humans and lower mammalian hearts.
CHIR is well known for its function in promoting cardiomyocyte differentiation (10). To our knowledge, there is no report about the effects of CHIR on cardiomyocyte apoptosis. However, FGF1 is well known for protecting cardiomyocytes from apoptosis (11). Therefore, we assessed cardiomyocyte apoptosis in the periinfarct border zone. To this end, ventricular sections in this area were subjected to TUNEL assay 3 days after the infarct. At this time point, the combined CHIR + FGF1-NP group was associated with a significantly lower number of apoptotic cells compared with all other (treated and control) groups (Figure 3, A and B), suggesting the synergistic effects of CHIR and FGF1 in cardiac cell sparing.
Figure 3. Detection and quantification of apoptosis by TUNEL assay in the mouse model of MI.
(A) Seventy-two hours posttreatment, the CHIR + FGF1-NP treatment group showed significantly fewer numbers of TUNEL+ cells in the border zone compared with the other treatment groups (scale bar: 50 μm) (B). Cells were also counterstained for nuclei (blue, DAPI). Data are given as means ± SEM. There were 5 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. sham; †P < 0.01 vs. MI; ‡P < 0.01 vs. empty NP; §P < 0.01 vs. CHIR-NPs; ||P < 0.01 vs. FGF1-NPs.
Assessment of NP-mediated neo-angiogenic response in MI mice.
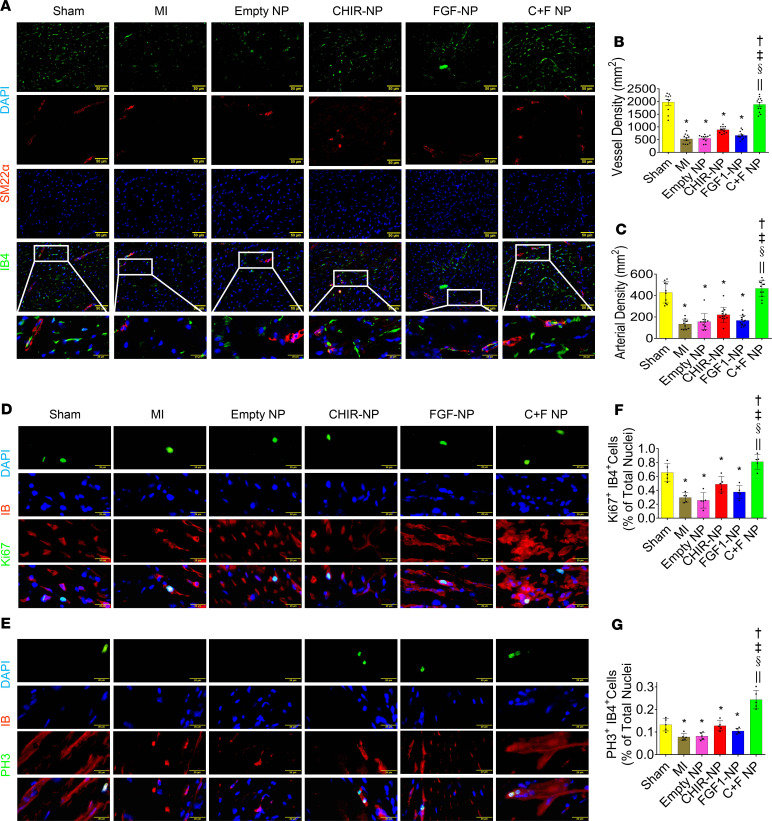
The periinfarct border zones were evaluated for NP-induced neo-angiogenesis by dual immunostaining, using both endothelial and smooth muscle phenotypic markers, i.e., isolectin B4 (red, IB4) and SM22-α (green, SM22α) (Figure 4A). The CHIR + FGF1-NP–treated group showed significantly elevated vessel and arterial density compared with the other NP-treated and untreated MI groups. In this combined group, vascular densities were not significantly different from those of sham-operated mice (Figure 4, B and C). Next, we tried to understand how these CHIR-FGF1-NPs promote angiogenesis in the infarcted mouse hearts. The pro–cell cycle activity effects of these NPs in hiPSC-CMs inspired us to hypothesize that similar effects may exist in vascular cells (ECs). Indeed, we observed increased cell cycle activity of ECs (Figure 4, D–G).
Figure 4. Evaluation of NPs mediated neo-angiogenesis in the mouse model of MI.
(A) Expression of the endothelium-specific lectin, isolectin B4 (red, IB4) and smooth muscle protein, SM22-α (green, SM22α), in the periinfarct border zone sections 28 days after MI (scale bar: 50 μm). (B and C) Graphical expression of angiogenesis data. The CHIR + FGF1-NP–treated group showed significantly elevated vascular densities compared with other NP treatment groups and the untreated MI group, but no significant difference in vascular densities was observed between the CHIR + FGF1-NP treatment group and the sham-operated controls. Data are given as means ± SEM. There were 10–12 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. sham; †P < 0.01 vs. MI; ‡P < 0.01 vs. empty NPs; §P < 0.01 vs. CHIR-NPs; ||P < 0.01 vs. FGF1-NPs. (D–G) Cell cycle activity of endothelial cells assessed by a dual staining for Ki-67 (green) and IB4 (red; D and F) and phospho–histone H3 (PH3) (green) and IB4 (red; E and G). Scale bar: 20 μm. Data are given as means ± SEM. There were 10–12 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. sham; †P < 0.01 vs. MI; ‡P < 0.01 vs. empty NPs; §P < 0.01 vs. CHIR-NPs; ||P < 0.01 vs. FGF1-NPs.
Cardioprotection of CHIR + FGF1-NPs in a pig model of IR injury.
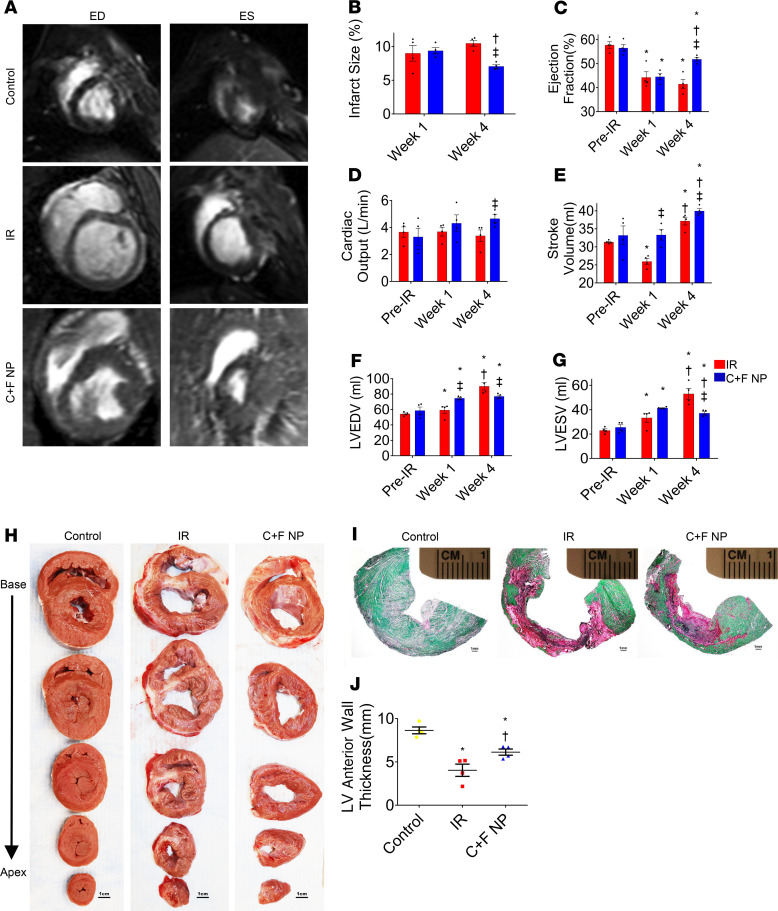
We extended this study to a preclinical large animal, the pig IR model (60-minute ligation of LAD followed by reflow). Cardiac MRI showed that by day 28 after the ischemic insult, hearts that had received NPs loaded with the 2 growth factors (CHIR and FGF1) exhibited greater reductions in infarct size and greater improvements in functional indices than untreated controls only subjected to IR (Figure 5, A–G). At the same time point, the treated group showed a significant reduction in wall thickness compared with healthy controls. However, it was significantly increased when compared with IR animals (Figure 5, H–J).
Figure 5. Assessment of LV morphology and function in a pig model of IR injury.
Cardiac MRI recordings in the experimental groups at end-diastole (ED) and end-systole (ES). At day 28, the CHIR + FGF1-NP-treated group revealed significant reduction in infarct size compared with the untreated IR group (B). On the contrary, EF (C), CO (D), and SV (E) were significantly greater in CHIR + FGF1-NP–treated groups than in untreated IR groups while LV end-diastolic volume and LV end-systolic volume were significantly lower (F and G). Data are given as means ± SEM. There were 4 animals per group. Statistical analysis: 2-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. pre-IR; †P < 0.05 vs. week 1; ‡P < 0.05 vs. IR. Macroscopic areas of infarction/fibrosis/scar (H) at after-IR day 28 in serial transverse sections of fresh (scale bar: 1 cm) representative micrographs of Sirius Red/Fast Green histochemical staining, revealing areas of infarcted (red, nonviable) and noninfarcted (green, viable) zones (I) (scale bar: 1 mm) and quantification of left anterior wall thickness (J). Data are given as means ± SEM. There were 4 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. control; †P < 0.05 vs. IR.
We also assessed the compensatory hypertrophy in the periinfarct border zone. At 28 days after the infarct, the IR and NP-treated groups had significantly greater myocyte cross-sectional areas compared with those of control animals, but this hypertrophy marker was significantly reduced after CHIR + FGF1-NP treatment compared with the untreated IR group (Figure 6, A and B). The LV weight–to–body weight (LVW/BW) ratio was also significantly lower in the treated group (Figure 6C).
Figure 6. Morphometric analysis of LV hypertrophy, cardiomyocyte cell cycle, and apoptosis in a pig model of IR injury.
(A) Expression of a cardiac contractile protein, α-sarcomeric actinin (red, α-SA), and a plasma membrane marker, wheat germ agglutinin (green, WGA), in the periinfarct border zone sections 28 days after MI (scale bar: 50 μm). (B and C) Markers of cardiomyocyte and LV hypertrophy showing a reduction in the treated group. (D–G) Cardiomyocyte cell cycle activity assessed by dual stainings for Ki-67 and cTnT (D and F) and for PH3 and cTnT (E and G). Data are given as means ± SEM. There were 4 animals per group. (H–K) Apoptosis in the infarct, border, and remote zones (IZ, BZ, and RZ, respectively), assessed by staining for TUNEL (red) and the pig cardiac-specific contractile protein (green, α-SA) (scale bar: 50 μm). The number of apoptotic cells did not differ between the control and treated groups in the RZ (K) but was significantly reduced in the treatment group in the IZ and BZ (I and J). Data are given as means ± SEM. There were 4 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test (B, C, and I–K) and t test (F and G). *P < 0.05 vs. control; †P < 0.05 vs. IR (B and C); *P < 0.01 vs. control; †P < 0.05 vs. IR (I–K).
Next, we evaluated cardiomyocyte cell cycle and apoptosis in IR pig hearts. Similar to our observations in the mouse model, no significant pro–cell cycle potential was observed in the IR pig model (Figure 6, D–G). In the infarct zone (IZ) and border zone (BZ), CHIR + FGF1-NP treatment resulted in a significantly smaller number of TUNEL+ cells compared with the IR group, although apoptosis was still significantly higher than in healthy controls (Figure 6, H–J). No significant difference in TUNEL+ cells was observed between the different groups in the remote zone (RZ) (Figure 6K).
We also determined the angiogenesis response following NP treatment in IR injury pigs. In post-IR, day 28 LV sections, immunolocalization of cardiomyocyte, endothelial, and smooth muscle–specific phenotypic markers (Figure 7A) was used to assess angiogenesis in the infarct (Figure 7, B and C), border (Figure 7, D and E), and remote (Figure 7, F and G) zones. In the IZ, both the numbers of IB4+ vessel structures and of IB4+SM22α+ arterial structures were significantly higher than in untreated IR animals but still lower than in healthy controls. In the BZ, the vascular densities were comparable between CHIR + FGF1-NP treatment and control groups and higher than in the untreated IR. In the RZ, no significant difference in vascular densities was observed between groups. Quantification of the percentage of IB4+ cardiac microvascular endothelial cells revealed that the expression levels of both Ki-67 and PH3 were significantly higher in the CHIR + FGF1-NP treatment group compared with the untreated IR and control groups (Figure 7, H–J).
Figure 7. Evaluation of NP-mediated neo-angiogenesis in the pig model of IR injury.
(A) Expression of cardiac-specific regulatory protein (white, cTnT), smooth muscle protein (green, SM22α), and endothelium-specific lectin (red, IB4) in the infarct, border, and remote zones (IZ, BZ, and RZ, respectively) 28 days after MI (scale bar: 20 μm). (B–G) Vasculogenic responses showing that the 2-factor–loaded nanomaterials increased angiogenesis compared with IR-untreated hearts in the infarct (B and C) and border (D and E) zones but not in the remote one (F and G). Data are given as means ± SEM. There were 4 animals per group. (H–J) Proliferation of cardiac microvascular endothelial cells assessed by dual staining for IB4+ cells and both Ki-67 (H and I) and PH3 (H and J) (scale bar: 50 μm). The results suggest that CHIR + FGF1-NPs synergistically promoted pig cardiac microvascular endothelial cell cycle activation and progression. Data are given as means ± SEM. There were 4 animals per group. Statistical analysis: 1-way ANOVA with Dunn’s multiple comparisons test. *P < 0.01 vs. control; †P < 0.05 vs. IR (B–G); *P < 0.05 vs. control; †P < 0.01 vs. IR (I and J).
Treatment with CHIR + FGF1 enhances cell cycle progression of vascular cells in vitro.
To study the mechanisms underlying this chemical-mediated cardioprotection, we used an in vitro cell culture system and treated neonatal cardiomyocytes (NCMs) and the vascular cells (ECs and SMCs) with CHIR-FGF1-NPs. In order to examine the effect of CHIR and/or FGF1 on the activation and proliferation of NCMs, the synchronized in vitro NCM cultures were treated with CHIR (5 μM), FGF1 (100 ng/mL), or CHIR + FGF1 (5 μM, 100 ng/mL, respectively) for 24 hours and evaluated by immunostaining and fluorescence microscopy for the incorporation patterns of BrdU (Supplemental Figure 4A). Quantification of the percentage of MF20+ (red, MF20) NCMs incorporating BrdU (green, BrdU) revealed no significant difference between the various treatment groups, i.e., CHIR, FGF1, and CHIR + FGF1, compared with the control group (Supplemental Figure 4B).
To determine the effect of CHIR and/or FGF1 on the cell cycle progression of HUVECs, the cell cycle–synchronized in vitro HUVEC cultures were treated with CHIR (5 μM), FGF1 (100 ng/mL), or CHIR + FGF1 (5 μM, 100 ng/mL, respectively) for 24 hours and assessed again for various cell cycle–associated markers, such as a proliferation-related nuclear protein, Ki-67 (interphase); PH3 (M phase marker); BrdU (S phase); and Aurora B (cytokinesis). The expression pattern of Ki-67, BrdU, PH3, and Aurora B kinase is shown in Supplemental Figure 5, A–D. Quantification of the percentage of CD31+ (red, CD31) HUVECs demonstrated that the expression levels of all 4 markers were significantly higher in the CHIR + FGF1–treated group compared with either treatment alone (CHIR or FGF1) or control groups (Supplemental Figure 5, E–H). Subsequently, to determine the effect of CHIR and/or FGF1 on the activation and proliferation of HVSMCs, the synchronized in vitro HVSMC cultures were treated with CHIR (5 μM), FGF1 (100 ng/mL), or CHIR + FGF1 (5 μM, 100 ng/mL, respectively) for 24 hours and similarly assessed by immunostaining and fluorescence microscopy for the incorporation of BrdU and the expression patterns of Ki-67 and PH3 (Supplemental Figure 6, A–C). Quantification of the percentage of SM22-α+ (red, SM22α) HVSMCs indicated that the incorporation of BrdU and the expression patterns of Ki-67 and PH3 were significantly greater in the combined treatment group compared with either treatment alone (CHIR or FGF1) or control groups (Supplemental Figure 6, D–F).
To determine the effect of CHIR and FGF1 on the cell cycle progression of fibroblasts, the porcine cardiac fibroblasts were isolated, cultured, cell cycle synchronized, and treated with CHIR + FGF1 (5 μM, 100 ng/mL, respectively) for 24 hours and assessed again for various cell cycle–associated markers, including BrdU (S phase marker) and PH3 (M phase marker). The incorporation of BrdU and the expression pattern of PH3 are shown in Supplemental Figure 7, A and B. Quantification of the percentage of vimentin+ (red, vimentin) fibroblasts demonstrated that the expression levels of both markers were significantly higher in the CHIR + FGF1–treated group compared with control groups (Supplemental Figure 7, A and B).
Global transcriptome profiling of CHIR + FGF1–treated HUVECs by RNA-sequencing.
Total cellular RNA was isolated from CHIR + FGF1–treated (5 μM, 100 ng/mL, respectively) and control HUVECs and subjected to next-generation RNA-sequencing (RNA-Seq). The mRNA profiles indicated that 815 genes out of 18,845 genes (adjusted P < 0.05) showed significant changes in their expression levels. The relative mRNA expression profile of various representative genes related to angiogenesis, cell proliferation, cell senescence, and cell death is shown in Supplemental Figure 8A. Furthermore, quantitative PCR (qPCR) validation of RNA-Seq data showed that cyclin D1, cyclin-dependent kinase 1 (CDK1), CDK4, and c-Myc were significantly upregulated in CHIR + FGF1 treatment groups compared with the other treatment and control groups (Supplemental Figure 8B). Conversely, cyclin D2 was significantly downregulated in all treatment groups compared with the controls (Supplemental Figure 8B).
Similarly, the total cellular protein was extracted from the synchronized HUVEC cultures, i.e., control cells and cells that were exposed to CHIR (5 μM), FGF1 (100 ng/mL), or CHIR + FGF1 (5 μM, 100 ng/mL, respectively) for 24 hours and subjected to Western blot analyses. These assays demonstrated the expression of various cell cycle regulatory proteins, such as cyclins, CDKs, c-Myc, GSK3, and β-catenin, as shown in Supplemental Figure 8C. Semiquantitative Western blot analyses showed that the expression levels of cyclin D1, cyclin H, CDK4, c-Myc, and β-catenin were significantly upregulated in the CHIR + FGF1 treatment groups compared with the other treatment and control groups, as shown in Supplemental Figure 8D. The expression levels of the remaining cyclins (A2, D2, E1, and E2), CDK7, and GSK-3α/β were similarly expressed, as shown in Supplemental Figure 8E. The expression of the receptors for FGF and VEGF indicated that FGF receptor 1 (FGFR1) was significantly upregulated when treated with FGF1 or CHIR + FGF1 (Supplemental Figure 9A) while VEGF receptor 2 was significantly upregulated by CHIR or CHIR + FGF1 compared with the other treatment groups (Supplemental Figure 9B).
Discussion
Mammalian cardiomyocytes lose their cell cycle soon after birth (12, 13). The loss of cardiomyocytes after a coronary occlusive ischemic injury is typically replaced by a fibrotic scar, which is often followed by postinfarction LV remodeling that is characterized by LV dilatation leading to heart failure. In the past few decades, huge efforts have been exerted in reactivating the cell cycle and regeneration capacity of cardiomyocytes in injured hearts. It has been shown that reactivation of cardiomyocyte cell cycle in genetically modified mouse models regenerates the injured myocardium and improves LV function (14, 15). In the present study, we sought to identify small molecules or compounds to stimulate adult cardiomyocyte proliferation and to prevent LV dilatation in hearts with postinfarction LV remodeling. Using the hiPSC-CMs, the initial in vitro screening studies have revealed that a combination of CHIR + FGF1 enhances cardiomyocyte cell cycle. Interestingly, a combination of CHIR + FGF1 indeed conferred cardioprotection when administered in the ischemic hearts, as evidenced by the improved cardiac function (Figure 1, A–E; and Figure 5, A–G) and reduced size of the fibrotic scar (Figure 1, F and G; and Figure 5, H–J) in both mouse and pig models of myocardial infarction. However, in contrast to immature myocytes derived from hiPSCs, these chemicals failed to stimulate cell cycle of mature cardiomyocytes in culture (Supplemental Figure 4) or endogenous cardiomyocytes in both mice and pigs (Figure 6, D–G). This discrepancy may be explained by the difference of the cell cycle regulation network between humans and lower mammalian hearts or by the differences of the immature nature of hiPSC-CMs versus the mature myocytes in vivo. One of these differences, which could be relevant to our findings, is the metabolic switch from glycolysis to oxidative phosphorylation, which seems to be a key driver for arresting cardiomyocyte proliferation (16). Nevertheless, the combination of CHIR + FGF1 robustly stimulated angiogenesis in the left ventricle of hearts in both the mouse model (Figure 4, A–C) and pig model (Figure 7, A–G) of postinfarction LV remodeling. Further studies revealed that the combination of CHIR + FGF1 promotes the cell cycle of ECs and SMCs in vitro (Supplemental Figures 5 and 6) and in vivo (Figure 4, D–G; and Figure 7, H–J). In addition, the combination of CHIR + FGF1 protects cardiomyocytes from undergoing apoptosis in both mice (Figure 3, A and B) and pigs (Figure 6, H–K). Taken together, these data demonstrate for the first time to our knowledge that the combination of CHIR + FGF1 results in cardioprotection in vivo that is evidenced by a substantial reduction of infarct size and by improved LV chamber function.
Previous studies have shown that FGF1-loaded PLGA microparticles result in enhanced recovery of LV function when administered into an infarcted myocardium (3). Similarly, CHIR-loaded NPs efficiently direct reprogramming of fibroblasts into functional cardiomyocytes and have been shown to be an attractive therapeutic option to replenish the lost cardiomyocytes (4). However, the synergetic therapeutic efficacy of the combined use of these 2 factors had not yet been explored. Wnt signaling pathways are expressed in multiple tissues and have pleiotropic effects on cell survival, proliferation, differentiation, migration, and apoptosis (17–19). In addition, the Wnt pathway plays an important role in angiogenesis and vascular remodeling (17, 18, 20), under normal and pathological conditions (21–23). CHIR is an inhibitor of GSK3, which has been shown effective in vitro for driving pluripotent stem cells toward the cardiac mesoderm (24, 25) and promoting their proliferation. Furthermore, inhibition of GSK3β activity has also been shown to induce an apoptotic cascade in human transformed pancreatic epithelial cells but not in nontransformed pancreatic cells by the c-Jun N-terminal kinase signaling pathway (26), thereby suggesting that the regulation of CHIR of apoptosis, whether positive or negative, may depend on both cell type and context (26, 27). In the present study, we used CHIR to activate pharmacologically the canonical Wnt pathway, which synergistically induced robust neo-angiogenesis around periinfarct border zones, reduced the apoptotic rate of vascular cells, and increased the proliferation rate of these cells during ischemia or reperfusion. The results are consistent with the gene expression profile of HUVECs as assessed by RNA-Seq (Supplemental Figure 7). FGF1, also known as acidic FGF, is one of the prototype members of the heparin-binding FGF family (28) and is implicated in cardiogenesis through its synergizing action with bone morphogenetic proteins for inducing mesoderm-to-cardiac differentiation (24). FGF1 is multifunctional and features broad mitogenic and cell survival activities, including cell growth, morphogenesis, tissue repair, tumor growth, and tumor invasion. This protein functions as a modifier of endothelial proliferation and cell migration, as well as an angiogenic factor (29, 30). Nabel and coworkers have demonstrated that recombinant FGF1 could promote intimal hyperplasia and angiogenesis in arteries in vivo (29), and FGF2/6 strongly promotes skeletal myocyte regeneration in conjunction with an increased neo-angiogenic response (31). Additionally, FGF1 has been shown to reduce apoptosis in vascular injury (11, 32–34). Studies using the native form of FGF1 delivered by continuous release into the coronary artery or to the left atrium failed to demonstrate any beneficial effect (35–37). The growth factor did not increase the number of new collateral vessels within a few hours of injection. Thus, alternative mechanisms of FGF-induced reduction of infarct size, i.e., an increase in myocyte viability and resistance to injury (38), restoration of ischemia-impaired endothelium-dependent vasomotion (39), or recruitment of preexisting collaterals, might explain this result (40). Many growth factors are capable of inducing physiologically significant angiogenesis in experimental animals. However, clinical trials, such as the randomized evaluation of VEGF and the angiogenic gene therapy trial, did not produce encouraging results (41–43). It is noteworthy that virtually all clinical trials were carried out as a monotherapy. It is reasonable to hypothesize that administration of a combination of 2 or more growth factors may be superior to single agents (40, 44). In the present study, the combination of CHIR and FGF1 NPs that act on mechanistically distinct signaling pathways resulted in the best outcomes. Future directions may include testing the efficacy and optimizing the dose of these growth factors in combination, which may further improve clinically relevant outcomes (44).
However, another factor that has so far hindered the efficacy of myocardial regenerating strategies is the low retention of biologics (cells or biomolecules) following their direct intramyocardial delivery. Indeed, it makes sense to hypothesize that a single shot of any of these biologics is unlikely to be therapeutically effective because of its expectedly fast clearance from the target tissue. To overcome this hurdle, the ability of HUVECs and HVSMCs to take up PLGA NPs was leveraged to use this biomaterial as a slow-release vehicle intended to extend the exposure time of the myocardium to the particle-loaded agents. PLGA-based nanomaterials have been designed and deployed for the treatment of MI (1, 2) and may promote revascularization of ischemic myocardium, by enabling the sustained release of a therapeutic cocktail of growth factors/cytokines/small molecules. Along with the nature of the delivered compounds, this delivery strategy may have contributed to the superior outcomes yielded by our combined treatment group.
It is noteworthy that apoptosis has been reported to peak during the early postinfarction reperfusion period (45), which may have facilitated the demonstration of the efficacy of the 2 chemicals since they were delivered at the time of the acute injury. Whether similar improvements could be achieved by delaying the treatment to a more chronic phase remains to be determined. We used PLGA NPs to deliver CHIR and FGF1. A testing of nanomaterials and their interaction with these 2 chemicals is necessary in order to optimize the release kinetics and, thus, improve therapeutic effects. PLGA is a US Food and Drug Administration–approved biomaterial, which has been extensively used as a carrier of chemotherapeutic agents (46), and features a long-standing safety record in the clinics, and current efforts at surface functionalization of NPs should further improve the efficiency of their payload delivery (47). On the other hand, from a translational perspective, the issues raised by biomaterials should not be underestimated. These issues include control of degradation kinetics through fine-tuning of their physicochemical properties, assessment of possible inflammatory and immune responses, biodistribution patterns, ultimate fate of the materials once phagocytosed by the mononuclear phagocyte system, scalability, and manufacturing costs.
Methods
Generation and characterization of PLGA NPs.
PLGA NPs were prepared as described previously (2). A double-emulsion (water/oil/water phase) technique for recombinant human FGF1 acidic protein (rhFGF1, aa 16-155 (Novus Biologicals, Inc.) and a single-emulsion (oil/water phase) technique for CHIR99021 were used. FGF1 (water-soluble) and CHIR (water-insoluble) were loaded into separate NPs, and their encapsulation efficiency and release kinetics were determined separately. The NPs were mixed before injection into animal hearts. Briefly, a solution of PLGA (100 mg) in dichloromethane (5 mL) with or without FGF1 (200 μL at 1 mg/mL), CHIR99021 (200 μL at 8 mg/mL), or coumarin-6 (1 mg) was sonicated at 4°C for 2 minutes (40% amplitude, pulse 40 seconds, pause 20 seconds). Next, for FGF1, 20 mL of a 4% (w/v) polyvinyl alcohol–water (PVA-water) solution was added, and for CHIR99021, 20 mL of a 1% (w/v) dimethylamine borane–water solution was added. The mixtures were once again sonicated at 4°C for 2 minutes (40% amplitude, pulse 40 seconds, pause 20 seconds). The resultant mixture was transferred to 10 mL of a 4% (w/v) PVA-water solution and 20 mL of Milli-Q water for FGF1 or 30 mL of Milli-Q water (MilliporeSigma) for CHIR99021, in a 100-mL glass beaker, and stirred for 4 hours until the dichloromethane was evaporated completely. Then, the solution was spun at 1000 g for 10 minutes to remove any aggregate, and the resultant supernatant was centrifuged again at 45,000 g for 20 minutes to collect the NPs. The NPs were washed twice by resuspending them in 50 mL of Milli-Q water. Finally, the suspension was centrifuged at 45,000 g for 20 minutes, frozen at –80°C overnight, lyophilized for 48 hours, and stored at –80°C until further use.
Measurement of the sizes of PLGA NPs was performed using a Quanta scanning electron microscope (Quanta FEG 650). The quantification of particle diameter was performed using NIH ImageJ software. The release kinetics was determined by Slide-A-Lyzer MINI Dialysis Devices, 20K molecular weight cutoffs, 0.5 mL (Thermo Fisher Scientific), suspending the generated CHIR- or FGF1-NPs in the release medium, consisting of DPBS with 0.1% BSA and 0.02% sodium azide. The suspensions were incubated at 37°C under constant shaking, followed by withdrawing and replacing 14.5 mL of the medium at the defined time points. To measure CHIR and FGF1 levels, the collected samples were analyzed using NanoDrop via UV/Vis spectrophotometer (Thermo Fisher Scientific, for CHIR) or an ELISA (Thermo Fisher Scientific) (for FGF1), respectively.
Characterization of cellular uptake and biodistribution of PLGA NPs.
To characterize the cellular uptake of PLGA NPs, HUVECs and HVSMCs were seeded onto a Lab-Tek chamber slide system (Nunc, Thermo Fisher Scientific) at a density of 1.5 × 104 cells/well and expanded with appropriate medium until 70% confluence. Next, the HUVECs/HVSMCs were synchronized by serum deprivation for 24 hours. Afterward, the cells were treated with coumarin-6–loaded NPs (2 μg/mL) and incubated at 37°C with 5% CO2 for 24 hours using appropriate complete medium. Finally, after 24 hours, the cells were washed twice with DPBS, pH 7.4, and were analyzed using a fluorescent microscope for cellular localization and distribution of NPs.
To further study whether NPs were preferentially taken up by different cells, we mixed HUVECs, SMCs, hiPSC-CMs, and fibroblasts with a ratio of 1:1:1:1 and cultured them to 70% confluence. Then, the cells were treated with coumarin-6–loaded NPs (2 μg/mL) and incubated at 37°C with 5% CO2 for 24 hours using appropriate complete medium. Finally, after 24 hours, the cells were washed twice with DPBS, pH 7.4, and were analyzed using a fluorescent microscope (Olympus IX83 Microscope, Olympus America Inc.) for cellular localization and distribution of NPs.
Mouse MI model.
Surgical induction of MI was performed in 8- to 10-week old C57BL/6 mice (The Jackson Laboratory). In brief, mice were intubated and connected to ventilators while breathing 2% isoflurane, USP (Fluriso, VetOne), for maintenance of anesthesia. The heart was exposed through a left thoracotomy, and MI was induced by a permanent ligation of the LAD with an 8-0 nonabsorbable suture (48). NPs were resuspended in DPBS (pH 7.4) containing 0.1% BSA/0.02% sodium azide, and the suspension was sonicated (40% amplitude, ~40-second intervals with 20-second pauses for a total of 2 minutes) on ice. A mixture of 15 μL of NPs loaded with CHIR + FGF1 (20 μg/μL, 13.33 μg/μL, respectively) was immediately delivered into the infarct (1 injection) and border (2 injections) zones by 3 intramyocardial injections (equal volume, 5 μL/site), using a modified Hamilton needle to make the treatment dose of 2400 ng for CHIR (loading efficiency 1 mg CHIR-NPs = 8 μg CHIR) and 200 ng for FGF1 (loading efficiency 1 mg FGF1-NPs = 1 μg FGF1) (n = 12). An equal volume of DPBS (n = 11) or nonloaded NPs (n = 11) was delivered in controls. In the sham group (n = 10), suture was passed around the LAD without ligation. After chest closure, buprenorphine hydrochloride (0.1 mg/kg, Buprenex, Reckitt Benckiser Pharmaceuticals Inc.) and carprofen (5 mg/kg, Rimadyl, Zoetis) were given as intraperitoneal injections for every 12 hours, for 3 and 1 days postsurgery, respectively. Four animals died due to peri-/postoperative complications.
Pig IR injury model.
Surgical induction of myocardial IR injury was performed in 45-day-old female Yorkshire swine (14 kg, Snyder Farms), as described previously (49). Briefly, swine were intubated and connected to ventilators while breathing 2% isoflurane, USP (Fluriso, VetOne), for maintenance of anesthesia. Throughout surgery, the animal body temperature, arterial blood pressure, ECG, and oxygen saturation were continuously monitored. A median sternotomy was performed, and the LAD distal to the second diagonal was identified and completely occluded for 60 minutes to generate AMI, followed by reperfusion.
The NPs were administrated 15 minutes after reperfusion after systemic hemodynamics stabilized. A mixture of 1000 μL of resuspended NPs loaded with CHIR + FGF1 (20 μg/μL, 13.33 μg/μL, respectively) was delivered into the periinfarct zones (equal volume, 200 μL/site × 5), using a modified Hamilton needle to make the treatment dose of 160 μg for CHIR (loading efficiency 1 mg CHIR-NPs = 8 μg CHIR) and 13.3 μg for FGF1 (loading efficiency 1 mg FGF1-NPs = 1 μg FGF1) (n = 4). An equal volume of DPBS (pH 7.4) was delivered in the untreated control group (n = 4). Four size- and weight-matched animals served as additional controls. After chest closure, buprenorphine hydrochloride SR (0.24 mg/kg, Buprenex, Reckitt Benckiser Pharmaceuticals Inc.) was given as subcutaneous injections every 72 hours, up to 3 days postsurgery; and carprofen (4 mg/kg, Rimadyl, Zoetis) was injected intramuscularly for every 24 hours, for 2 days postsurgery. Two animals that were subjected to this IR and protocol (n = 10) died within the first 24 hours after the LAD ligation. One of them died due to operative complications and the other one due to perioperative uncontrolled cardiac arrhythmia.
Echocardiography.
Pre- and post-MI (on day 28), the NP-treated and control untreated mice were subjected to echocardiographic imaging as previously described (48). In brief, animals were maintained under 1.5%–2% isoflurane, USP (Fluriso, VetOne), anesthesia until the heart rate was stabilized at 400 to 500 bpm. The B-mode and 2-dimensional M-mode images were acquired from both long-axis and short-axis views with a high-resolution micro-ultrasound system (Vevo 2100, VisualSonics, Inc.). Finally, the obtained data were analyzed, and LVEF, EF, FS, EDD, and ESD were calculated using Vevo analysis software. The operator was kept blinded to the experimental groups.
Cardiac MRI.
Pre- and post-IR (on day 7 and day 28), the NP-treated and control untreated pigs were subjected to cardiac MRI with a 1.5-Tesla clinical scanner (Siemens Sonta, Siemens Medical System) and a phased-array 4-channel surface coil with ECG gating, as described previously (49, 50). Briefly, animals were anesthetized through inhalation of 2% isoflurane and placed in a supine position within the scanner. LVEF was calculated from a stack of short-axis cine images, using QMASS analytical software program (Medis Medical Imaging Systems). The cine imaging was performed with the following parameters: time to repetition (TR) = 3.1 ms, time to echo (TE) = 1.6 ms, flip angle (FA) = 79°, matrix size = 256 × 120, field of view (FOV) = 340 × 265 mm2, and slice thickness = 6 mm (with a 4-mm gap between slices). Twenty-five phases were acquired across the cardiac cycle. Infarct characterization and quantification were measured by delayed enhancement (DE) cardiac MRI (Magnevist, gadopentetate dimeglumine: 0.20 mmol/kg, i.v. bolus), and the data are presented as the ratio of scar surface area to the total LV surface area. DE-MRI was performed with the following parameters: TR = 16 ms, TE = 4 ms, FA = 30°, matrix size = 256 × 148, FOV = 320 × 185 mm2, and slice thickness = 6 mm (with a 0-mm gap between slices).
Determination of infarct size.
Following the coronary occlusion, at day 28 post-MI, under deep anesthesia, the hearts were excised from groups of 6 mice, and the infarct size was evaluated as described previously (51, 52). Briefly, the excised hearts were fixed in 10% phosphate-buffered formalin for 24 hours. Next, the hearts were embedded in optical cutting temperature compound for cryopreservation. Ten-micron-thick coronal sections of the entire ventricles were cut from base to apex and mounted on glass slides. Next, every 30th section was fixed in Bouin’s solution and stained with 0.04% Sirius Red/0.1% Fast Green collagen staining (all reagents from MilliporeSigma).
In a similar fashion, the LV walls of the pigs were cut transversely into 5 short-axis slices (R1 to R5) from base to apex. Each slice was sequentially cut into 8 samples (S1 to S8) according to coronary perfusion physiology, and the S1 of R2 (i.e., the site of anterior wall for NPs application) was collected and processed for cryosectioning and stained with Sirius Red and Fast Green.
To assess the quantitative changes in infarct size at day 28 after AMI, digital images of the stained sections were captured using an Olympus light microscope. Morphometric analyses were carried out using NIH ImageJ software. The size of the infarct was calculated according to the following formula: Infarct size % = [product of (scar circumferential length × thickness of each of the short axis)/product of (short-axis left ventricle length × thickness of the short axis)] × 100%.
Quantification of angiogenesis.
The angiogenic response in the border zone of infarcted myocardium, i.e., vessel density (capillary/arterial), was evaluated by immunostaining and fluorescence microscopy. Briefly, at 28 days posttreatment (MI or IR), cardiac tissue sections were sequentially stained with cardiac, endothelial, and smooth muscle cell markers, i.e., cTnT, IB4 (Griffonia Simplicifolia Lectin I, GSL I), and SM22α, respectively, and nuclei were counterstained with DAPI (53). We used NE-Cadherin–positive counts/mm2 for all vessel density and SMC22α+ counts/mm2 for arteriolar density.
Screening of chemicals that enhance cardiomyocyte cell cycle.
Using BrdU incorporation assay, we screened several chemicals that were reported to be able to enhance cell cycle activity of cultured cardiomyocytes derived from hiPSCs, including Ly294002 (PI3K inhibitor) (1), FGF1 (1, 2), SB203580 and VX702 (p38 MAPK inhibitors) (1), KN93 (Ca2+/calmodulin-dependent protein kinase II inhibitor) (3), Su1498 (Flk-1 inhibitor) (3), and CHIR99021 (Wnt activator and GSK3α and -3β inhibitor) (3, 4). The maintenance culture of hiPSCs and cardiomyocyte differentiation have been described before (5). In brief, 4 weeks after initiation of cardiomyocyte differentiation, individual chemicals or a combination were used to treat the cells for 24 hours. BrdU (15 μM final concentration) was added to the culture medium 12 hours before cell harvesting. The cells were fixed with ethanol at –20°C for 15 minutes, and BrdU staining was performed with a BrdU Labeling and Detection Kit (Roche Life Science, Inc. 11296736001) as directed by the manufacturer’s instructions. The cells were counterstained for the expression of human cTnT (hcTnT) and human Nkx2.5 to confirm hiPSC-CM identity. The number of nuclei that stained positively were counted, normalized to the total number of hiPSC-CMs (i.e., the number of hcTnT/Nkx2.5–double positive cells), and expressed as a percentage. Each experiment was repeated 3–4 times.
HUVEC culture: expansion and maintenance.
HUVECs were purchased from ATCC (PCS-100-010) and were expanded and subcultured according to the manufacturer’s recommendations. Briefly, cells were thawed, seeded onto a fibronectin-precoated T75 flask (1 μg/cm2 or 0.5 μg/mL), and expanded using a complete endothelial cell growth medium supplemented with 5% fetal bovine serum (FBS), human fibroblast growth factor (hFGF), human epidermal growth factor (hEGF), VEGF, R3-insulin-like growth factor-1, hydrocortisone, ascorbic acid, gentamicin, and amphotericin-B (GA1000) (Clonetics, EGM-2MV BulletKit; Lonza).
HVSMC culture: expansion and maintenance.
HVSMCs (primary pulmonary artery) were procured from ATCC (PCS-100-023) and were expanded and maintained as per the manufacturer’s instructions. Briefly, cells were thawed, plated onto a T75 flask, and expanded using a complete medium 231 (Thermo Fisher Scientific) supplemented with 5% FBS (4.9% v/v), hFGF (2 ng/mL), hEGF (0.5 ng/mL), heparin (5 ng/mL), recombinant human IGF-1 (2 μg/mL), and BSA (0.2 μg/mL) (SMGS, Thermo Fisher Scientific).
NCM culture: isolation and maintenance.
Isolation of NCMs was performed as described in detail previously (6). Briefly, under deep euthanasia, 1-day-old C57BL/6 neonatal mice (The Jackson Laboratory) were subjected to cervical dislocation, and hearts were dissected out from these mice and pooled in ice-cold DPBS, pH 7.4, containing 2 mM 2,3-butanedione monoxime (BDM), and rinsed 3 times. Next, the hearts were minced thoroughly and incubated with 10 mL of isolation medium (20 mM BDM and 0.0125% trypsin in DPBS, pH 7.4, in a 4°C shaking water bath overnight. The tissue fragments were pelleted at 800 g for 8 minutes, the supernatant was discarded, and the obtained pellet was resuspended in 5 mL of digestion medium (1.5 mg/mL collagenase and 20 mM BDM in DMEM) and oxygenated for 1 minute, then incubated at 37°C in a shaking water bath for 20 to 30 minutes. Tissues were dissociated by repeated triturations (at 8-minute intervals). The cells were once again pelleted and resuspended in complete myogenic medium composed of DMEM (4.5 g/l-glucose, l-glutamine, and sodium pyruvate; Cellgro by Mediatech, Inc.) supplemented with 8% horse serum (lot-selected, Gibco, Thermo Fisher Scientific), 5% newborn calf serum (Atlanta Biologicals, Inc.), penicillin (100 U/mL), streptomycin (100 μg/mL), and amphotericin-B (1 μg/mL). Furthermore, to remove the coexisting cardiac fibroblasts and endothelial cells, the above cell suspensions were plated in 100-mm2 cell culture dishes and incubated in a humidified atmosphere of 5% CO2 at 37°C for 30 minutes. Following incubation, the nonadherent myocytes were slightly tapped and were aspirated off immediately from the panning dishes, and the procedure was repeated 2 more times. After panning, the cells were once again pelleted and resuspended in myocyte medium. Finally, the isolated NCMs were quantified using a hemocytometer and plated at a density of approximately 1.5 × 105 cells/cm2 onto cell culture dishes precoated with a monolayer of collagen. Cells were seeded into the wells of a 6-well plate, incubated in a humidified atmosphere of 5% CO2 at 37°C for 48 hours, and observed under a microscope for spontaneous beating and rhythmicity. These NCMs were cultured in complete myocyte medium for an additional 3 days.
Isolation and culture of cardiac fibroblasts.
Cardiac fibroblasts were isolated from heart tissue of postnatal day 3 swine. The tissue was washed twice in DPBS containing antibiotics in sterile conditions and was then cut into small pieces and digested in 10 mL 0.25% trypsin for 45 minutes’ incubation at 37°C in 10-cm dishes. Then the suspension was neutralized with 10 mL of complete culture medium (DMEM, MilliporeSigma) supplemented with 10% FBS, 0.1 mM nonessential amino acid, 1 mM l-glutamine, 0.1 mM 2-mercaptoethanol, 100 U/mL penicillin, 100 μg/mL streptomycin; passed through a 40-μm cell strainer; and pelleted by centrifugation. The pellet was resuspended in 10 mL of complete culture medium. Then the cells were cultured in 10-cm dishes and allowed to adhere for 3 hours. Adherent cells were then gently washed once with prewarmed DPBS supplemented with antibiotics, and the growth medium was replaced. Once they reached 40%–50% confluence, cells were passaged to a gelatin-precoated T75 flask and incubated in humidified 95% air with 5% CO2 at 37°C. Medium was changed every 2–3 days.
HUVECs, HVSMCs, NCMs, and fibroblasts: cell cycle synchronization.
The cell cycle synchronization of HUVEC, HVSMC, NCM, and fibroblast cultures was performed by serum deprivation. In brief, cells were plated onto a T75 flask and expanded to 70% to 80%, using appropriate culture media. Then, the cells were rinsed twice with DPBS (pH 7.4) and incubated with appropriate serum-free culture medium for 24 hours. After 24 hours, they were exposed to CHIR, FGF1, or CHIR + FGF1 for 24 hours and cultured in appropriate complete medium. Finally, the cultures were terminated, and the collected samples were subjected to immunofluorescence staining for cell cycle markers and/or TUNEL assay; for HUVECs, next-generation RNA-Seq was also performed. The data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database and the accession number is GSE138837.
BrdU labeling and detection by immunofluorescence.
In brief, cell cycle–synchronized HUVEC or HVSMC cultures that were treated with CHIR, FGF1, or CHIR + FGF1 were incubated with the S phase marker, BrdU (10 μmol), in a humidified atmosphere of 5% CO2 at 37°C for 12 hours. Then, the cells were fixed with ice-cold 70% ethanol, pH 2.0 (50 mM glycine), for 15 minutes, and BrdU staining was performed using a BrdU Labeling and Detection Kit (Roche Life Science, Inc.) according to the manufacturer’s instructions.
Immunostaining and fluorescence microscopy.
The explanted hearts were collected at day 28 and processed according to previously described protocols (7). In brief, hearts were fixed in ice-cold 4% paraformaldehyde (PFA) for 4 hours followed by overnight immersion in 30% sucrose, at 4°C. Then, 10-μm serial cryosections were generated, and every 30th section was permeabilized with 0.2% Triton X-100, for 10 minutes at room temperature. Next, sections were blocked in 5% donkey serum in DPBS, pH 7.4, for 30 minutes at room temperature. The primary antibodies used are shown in the “Antibodies” section. Primary antibodies were used at 1:10 to 1:10,000 dilution in blocking buffer (1.5% BSA, 100 mM glycine in PBS) for 12 to 16 hours at 4°C. Secondary antibodies (FITC catalog 715-095-150, CY3 catalog 711-165-152, and CY5 catalog 715-157-150 obtained from Jackson ImmunoResearch Laboratory) were used at 1:200 dilution in blocking buffer for 2 hours at room temperature in the dark. Nuclei were stained with DAPI (100 ng/mL; MilliporeSigma). Negative controls for staining included only secondary antibodies. Finally, the sections were analyzed using a fluorescence microscope (Olympus IX81). Similarly, immunostaining was performed on ethanol-fixed cells, i.e., HUVECs, HVSMCs, or NCMs, for various phenotypic as well as cell cycle markers.
Detection of apoptotic cells by TUNEL assay.
The in situ cell death detection kit (ApopTag Fluorescein, Chemicon) was used for the detection and quantification of apoptosis at the single-cell level using a fluorescence microscope. Briefly, samples (cryotissue sections) were collected at defined time points; rinsed twice in DPBS, pH 7.4; and fixed in 4% PFA at room temperature for 20 minutes. The labeling reactions with the TUNEL reaction mixture were carried out as per the manufacturer’s instructions. Finally, the samples were mounted in VECTASHIELD mounting medium with DAPI (Vector Laboratories). A minimum of 8 tissue sections from various zones (ischemic, border, remote) from each heart were evaluated.
Reverse transcription–quantitative real-time polymerase chain reaction.
Total cellular RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) and treated with DNase I to remove genomic DNA contaminants. The reverse transcription reaction was executed using 500 ng of total RNA in a final reaction volume of 20 μL using a SuperScript IV First-Strand Synthesis System (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocols. The cyclin D1 (CCND1), cyclin D2 (CCND2), CDK1, CDK4, catenin-β 1 (CTNNB1), MYC proto-oncogene, BHLH transcription factor (c-Myc), and GAPDH primers were designed using a web-based software, synthesized commercially (Thermo Fisher Scientific), and evaluated for a range of annealing temperature, 58°C to 61°C, for all the primer pairs, as shown in Supplemental Table 1.
All reverse transcription–quantitative real-time PCRs (RT-qPCRs) were carried out using Fast SYBR Green Master Mix (Thermo Fisher Scientific) in a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific), and threshold cycle values were computed using QuantStudio Design and Analysis Software (v1.4.3). The PCR protocol was performed with the primer sequences listed in Supplemental Table 1 and consisted of 40 cycles of denaturation at 94°C for 15 seconds, annealing at 60°C for 15 seconds, and extension at 72°C for 1 minute, followed by a single 7-minute extension period at 72°C. Measurements were normalized to the level of endogenous GAPDH.
Western blotting.
Lysis and extraction of total cellular proteins were carried out using M-PER mammalian protein extraction reagent (Thermo Fisher Scientific), consisting of a cocktail of protease and phosphatase inhibitors (Thermo Fisher Scientific), as per the manufacturer’s instructions. Next, the isolated proteins were quantified using either a Pierce Coomassie plus (Bradford) or a bicinchoninic acid protein assay kit. Proteins were separated by 4% to 20% SDS-PAGE (Precast Gels, Bio-Rad) and electroblotted onto a polyvinylidene difluoride membrane (Trans-Blot Turbo Mini PVDF Transfer Packs, Bio-Rad). Then, the membranes were blocked with 5% BSA, subjected to immunostaining with appropriate dilutions of primary antibodies as shown in “Antibodies,” and detected using HRP-conjugated secondary antibodies. Western blot signals were measured by densitometry and were analyzed using software (AlphaView SA software 3.4, ProteinSimple). The housekeeping protein, GAPDH, was used for Western blot normalization.
Optimization of the concentration of CHIR and FGF1.
To determine the optimal concentration of CHIR and FGF1 for the activation, proliferation, and cell cycle progression of HUVECs, the cell cycle–synchronized in vitro HUVEC cultures were treated with various concentrations of CHIR (0.5 μM, 5 μM, or 50 μM) or FGF1 (10 ng/mL, 100 ng/mL, or 1000 ng/mL) for 24 hours and assessed by immunostaining and fluorescent microscopic analyses for BrdU (S phase marker) incorporation patterns (Supplemental Figure 10, A and B). Quantification of the percentage of CD31+ (red, CD31) HUVECs showed that the incorporation of BrdU was significantly higher in the 5 μM CHIR-treated group than in the 0.5 μM CHIR-treated or control groups (Supplemental Figure 10C). Interestingly, HUVECs treated with a higher concentration of CHIR (50 μM) underwent a significant amount of cellular senescence and death, resulting in complete detachment and floating of cells within 24 hours of treatment (data not shown). Similarly, quantification of the percentage of CD31+ HUVECs revealed that the incorporation of BrdU was significantly greater in the 100 ng/mL FGF1-treated group compared with other treatment (10 ng/mL or 1000 ng/mL) and control groups (Supplemental Figure 10D).
Antibodies.
The following primary antibodies were used in the study: mouse anti–cardiac troponin T monoclonal antibody (1:100, Abcam, catalog ab10214); mouse anti–sarcomeric α-actinin monoclonal antibody (1:100, Abcam, catalog ab9465); goat anti–cardiac troponin I polyclonal antibody (1:100, Abcam, catalog ab188877); mouse anti–myosin 4 monoclonal antibody (1:100, Invitrogen, Thermo Fisher Scientific, catalog 50-6503-82); wheat germ agglutinin (1:1000, Thermo Fisher Scientific, catalog W11261); mouse anti-CD31 monoclonal antibody (1:100, Abcam, catalog ab9498); rabbit anti-CD31 polyclonal antibody (1:100, Abcam, catalog ab28364); goat anti-CD31 polyclonal antibody (1:100, R&D Systems, Bio-Techne, catalog AF3628); rabbit anti-vimentin monoclonal antibody (1:100, Abcam, catalog ab92547); fluorescein-labeled GSL I IB4 (1:10, vector laboratories, catalog FL-1201); rabbit anti–SM22-α polyclonal antibody (1:100, Abcam, catalog ab14106); rabbit anti–cyclin D1 monoclonal antibody (1:1000, Abcam, catalog ab134175); rabbit anti–cyclin D2 polyclonal antibody (1:500, Santa Cruz, catalog sc-593); Cyclin Antibody Sampler Kit (Cyclin A2, E1, E2, and H; CDK4; CDK7) (1:1000, Cell Signaling Technology, catalog 9869T); rabbit anti–c-Myc monoclonal antibody (1:10,000, Abcam, catalog ab32072); mouse anti–histone H3 (phospho S10) monoclonal antibody (1:100, Abcam, catalog ab14955); rabbit anti–phospho–histone H3 (Ser10) polyclonal antibody (1:200, MilliporeSigma, catalog 06-570); rabbit anti–Ki-67 polyclonal antibody (1:100, MilliporeSigma, catalog AB9260); mouse anti–Ki-67 clone Ki-S5 monoclonal antibody (1:100, MilliporeSigma, catalog MAB4190); mouse anti–Aurora B monoclonal antibody (1:100, Abcam, catalog ab3609); rabbit anti–GSK3-β + GSK3-α monoclonal antibody (1:1000, Abcam, catalog ab185141); rabbit anti-GSK3 (α + β) (phospho Y216 + Y279) monoclonal antibody (1:1000, Abcam, catalog ab68476); rabbit anti–β-catenin monoclonal antibody (1:1000, Abcam, catalog ab6302); rabbit anti-FGF receptor 1 polyclonal antibody (1:1000, Abcam, catalog ab10646); and rabbit anti-VEGF receptor 1 + VEGF receptor 2 polyclonal antibody (1:1000, Abcam, catalog ab36844).
Statistics.
All the data are presented as mean ± standard error of the mean (mean ± SEM). For all the experimental data, the differences among the groups were determined by applying 1- or 2-way ANOVA (comparison among more than 2 groups) with adjustment for multiple comparisons by Dunn’s multiple comparisons test and by applying unpaired Student’s t test (2 tailed) when comparing between 2 groups. A P value less than 0.05 was considered statistically significant.
Study approval.
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham and performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals (NIH publication 85-23, National Academies Press, 2011).
Author contributions
CF, WZ, JY, and JZ conceived and designed the project. YO designed and developed the NP formulations. YO and DP generated the NPs. MZ and WZ performed chemical screening on hiPSCs. CF, XL, and YT performed histology, Western blot, and RT-qPCR to test the effects of NPs on hiPSC-CMs, HUVECs, SMCs, and fibroblasts. CF performed mouse surgery, tissue collection, and tissue characterization. WZ and CF performed echocardiography. GPW performed pig surgery and NP injection. CF, YT, and GPW performed pig MRI. CF collected pig tissue and performed histology of these tissue samples. CF, XL, DP, WZ, and JZ performed data analysis and statistical analysis. CF, MTV, PK, PM, WZ, and JZ prepared the manuscript.
Supplementary Material
Acknowledgments
We would also like to thank Mary Flowers Braswell for her donation of the “Echocardiographic Imaging System,” for research purposes. This work was supported by NIH grants (NHLBI R01 grants: HL095077, HL114120, HL131017, HL142627, and U01 HL134764) and by an American Heart Association-National Scientist Development grant (16SDG30410018).
Version 1. 05/26/2020
In-Press Preview
Version 2. 06/18/2020
Electronic publication
Footnotes
WZ’s present address is: Department of Cardiovascular Diseases, Physiology and Biomedical Engineering, Mayo Clinic Arizona, Phoenix, Arizona, USA.
YO’s present address is: College of Dental Medicine, Division of Orthodontics, Columbia University, New York, New York, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e132796.https://doi.org/10.1172/jci.insight.132796.
Contributor Information
Chengming Fan, Email: markfan@uab.edu.
Yasin Oduk, Email: yasinoduk@gmail.com.
Meng Zhao, Email: mengzhao@uab.edu.
Xi Lou, Email: xlou@uab.edu.
Yawen Tang, Email: yawen@uab.edu.
Danielle Pretorius, Email: dpret@uab.edu.
Mani T. Valarmathi, Email: valarmathi64@hotmail.com.
Gregory P. Walcott, Email: gregorywalcott@uabmc.edu.
Philippe Menasche, Email: philippe.menasche@inserm.fr.
Prasanna Krishnamurthy, Email: prasanak@uab.edu.
Wuqiang Zhu, Email: Zhu.Wuqiang@mayo.edu.
Jianyi Zhang, Email: jayzhang@uab.edu.
References
- 1.Binsalamah ZM, Paul A, Khan AA, Prakash S, Shum-Tim D. Intramyocardial sustained delivery of placental growth factor using nanoparticles as a vehicle for delivery in the rat infarct model. Int J Nanomedicine. 2011;6:2667–2678. doi: 10.2147/IJN.S25175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oduk Y, et al. VEGF nanoparticles repair the heart after myocardial infarction. Am J Physiol Heart Circ Physiol. 2018;314(2):H278–H284. doi: 10.1152/ajpheart.00471.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascual-Gil S, Simón-Yarza T, Garbayo E, Prósper F, Blanco-Prieto MJ. Cytokine-loaded PLGA and PEG-PLGA microparticles showed similar heart regeneration in a rat myocardial infarction model. Int J Pharm. 2017;523(2):531–533. doi: 10.1016/j.ijpharm.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira MPA, et al. Dual-drug delivery using dextran-functionalized nanoparticles targeting cardiac fibroblasts for cellular reprogramming. Adv Funct Mater. 2018;28(15):1705134. doi: 10.1002/adfm.201705134. [DOI] [Google Scholar]
- 5.Tzatzalos E, Abilez OJ, Shukla P, Wu JC. Engineered heart tissues and induced pluripotent stem cells: Macro- and microstructures for disease modeling, drug screening, and translational studies. Adv Drug Deliv Rev. 2016;96:234–244. doi: 10.1016/j.addr.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel FB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19(10):1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novoyatleva T, Sajjad A, Pogoryelov D, Patra C, Schermuly RT, Engel FB. FGF1-mediated cardiomyocyte cell cycle reentry depends on the interaction of FGFR-1 and Fn14. FASEB J. 2014;28(6):2492–2503. doi: 10.1096/fj.13-243576. [DOI] [PubMed] [Google Scholar]
- 8.Uosaki H, et al. Identification of chemicals inducing cardiomyocyte proliferation in developmental stage-specific manner with pluripotent stem cells. Circ Cardiovasc Genet. 2013;6(6):624–633. doi: 10.1161/CIRCGENETICS.113.000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Titmarsh DM, et al. Induction of human iPSC-derived cardiomyocyte proliferation revealed by combinatorial screening in high density microbioreactor arrays. Sci Rep. 2016;6:24637. doi: 10.1038/srep24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lian X, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuevas P, et al. Fibroblast growth factor-1 prevents myocardial apoptosis triggered by ischemia reperfusion injury. Eur J Med Res. 1997;2(11):465–468. [PubMed] [Google Scholar]
- 12.Toischer K, et al. Cardiomyocyte proliferation prevents failure in pressure overload but not volume overload. J Clin Invest. 2017;127(12):4285–4296. doi: 10.1172/JCI81870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu W, et al. Regenerative potential of neonatal porcine hearts. Circulation. 2018;138(24):2809–2816. doi: 10.1161/CIRCULATIONAHA.118.034886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu W, Reuter S, Field LJ. Targeted expression of cyclin D2 ameliorates late stage anthracycline cardiotoxicity. Cardiovasc Res. 2019;115(5):960–965. doi: 10.1093/cvr/cvy273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills RJ, et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A. 2017;114(40):E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin AM, D’Amore PA. Wnt signaling in the vasculature. Angiogenesis. 2002;5(1-2):1–9. doi: 10.1023/a:1021563510866. [DOI] [PubMed] [Google Scholar]
- 18.Muley A, et al. Secreted frizzled-related protein 4: an angiogenesis inhibitor. Am J Pathol. 2010;176(3):1505–1516. doi: 10.2353/ajpath.2010.090465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74(4):609–619. doi: 10.1016/0092-8674(93)90509-O. [DOI] [PubMed] [Google Scholar]
- 20.Olsen JJ, et al. The role of Wnt signalling in angiogenesis. Clin Biochem Rev. 2017;38(3):131–142. [PMC free article] [PubMed] [Google Scholar]
- 21.Pate KT, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33(13):1454–1473. doi: 10.15252/embj.201488598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanhollebeke B, et al. Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. Elife. 2015 doi: 10.7554/eLife.06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Nathans J. Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev Cell. 2014;31(2):248–256. doi: 10.1016/j.devcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parikh A, Wu J, Blanton RM, Tzanakakis ES. Signaling pathways and gene regulatory networks in cardiomyocyte differentiation. Tissue Eng Part B Rev. 2015;21(4):377–392. doi: 10.1089/ten.teb.2014.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchand B, Tremblay I, Cagnol S, Boucher MJ. Inhibition of glycogen synthase kinase-3 activity triggers an apoptotic response in pancreatic cancer cells through JNK-dependent mechanisms. Carcinogenesis. 2012;33(3):529–537. doi: 10.1093/carcin/bgr309. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Zhang S, Lu Y, Zhang Y, Wang E, Cui Z. The roles of Notch3 on the cell proliferation and apoptosis induced by CHIR99021 in NSCLC cell lines: a functional link between Wnt and Notch signaling pathways. PLoS One. 2013;8(12):e84659. doi: 10.1371/journal.pone.0084659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess WH, Maciag T. The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem. 1989;58:575–606. doi: 10.1146/annurev.bi.58.070189.003043. [DOI] [PubMed] [Google Scholar]
- 29.Nabel EG, et al. Recombinant fibroblast growth factor-1 promotes intimal hyperplasia and angiogenesis in arteries in vivo. Nature. 1993;362(6423):844–846. doi: 10.1038/362844a0. [DOI] [PubMed] [Google Scholar]
- 30.Murakami M, Simons M. Fibroblast growth factor regulation of neovascularization. Curr Opin Hematol. 2008;15(3):215–220. doi: 10.1097/MOH.0b013e3282f97d98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doukas J, et al. Delivery of FGF genes to wound repair cells enhances arteriogenesis and myogenesis in skeletal muscle. Mol Ther. 2002;5(5 pt 1):517–527. doi: 10.1006/mthe.2002.0579. [DOI] [PubMed] [Google Scholar]
- 32.Uriel S, Brey EM, Greisler HP. Sustained low levels of fibroblast growth factor-1 promote persistent microvascular network formation. Am J Surg. 2006;192(5):604–609. doi: 10.1016/j.amjsurg.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Raju R, et al. A network map of FGF-1/FGFR signaling system. J Signal Transduct. 2014;2014:962962. doi: 10.1155/2014/962962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwakura A, et al. Myocardial ischemia enhances the expression of acidic fibroblast growth factor in human pericardial fluid. Heart Vessels. 2000;15(3):112–116. doi: 10.1007/PL00007264. [DOI] [PubMed] [Google Scholar]
- 35.Unger EF, et al. A model to assess interventions to improve collateral blood flow: continuous administration of agents into the left coronary artery in dogs. Cardiovasc Res. 1993;27(5):785–791. doi: 10.1093/cvr/27.5.785. [DOI] [PubMed] [Google Scholar]
- 36.Unger EF, Shou M, Sheffield CD, Hodge E, Jaye M, Epstein SE. Extracardiac to coronary anastomoses support regional left ventricular function in dogs. Am J Physiol. 1993;264(5 pt 2):H1567–H1574. doi: 10.1152/ajpheart.1993.264.5.H1567. [DOI] [PubMed] [Google Scholar]
- 37.Banai S, et al. Effects of acidic fibroblast growth factor on normal and ischemic myocardium. Circ Res. 1991;69(1):76–85. doi: 10.1161/01.RES.69.1.76. [DOI] [PubMed] [Google Scholar]
- 38.Padua RR, Sethi R, Dhalla NS, Kardami E. Basic fibroblast growth factor is cardioprotective in ischemia-reperfusion injury. Mol Cell Biochem. 1995;143(2):129–135. doi: 10.1007/BF01816946. [DOI] [PubMed] [Google Scholar]
- 39.Sellke FW, et al. Basic FGF enhances endothelium-dependent relaxation of the collateral-perfused coronary microcirculation. Am J Physiol. 1994;267(4 pt 2):H1303–H1311. doi: 10.1152/ajpheart.1994.267.4.H1303. [DOI] [PubMed] [Google Scholar]
- 40.Ware JA, Simons M. Angiogenesis in ischemic heart disease. Nat Med. 1997;3(2):158–164. doi: 10.1038/nm0297-158. [DOI] [PubMed] [Google Scholar]
- 41.Grines CL, et al. A randomized, double-blind, placebo-controlled trial of Ad5FGF-4 gene therapy and its effect on myocardial perfusion in patients with stable angina. J Am Coll Cardiol. 2003;42(8):1339–1347. doi: 10.1016/S0735-1097(03)00988-4. [DOI] [PubMed] [Google Scholar]
- 42.Ripa RS, Wang Y, Jørgensen E, Johnsen HE, Hesse B, Kastrup J. Intramyocardial injection of vascular endothelial growth factor-A165 plasmid followed by granulocyte-colony stimulating factor to induce angiogenesis in patients with severe chronic ischaemic heart disease. Eur Heart J. 2006;27(15):1785–1792. doi: 10.1093/eurheartj/ehl117. [DOI] [PubMed] [Google Scholar]
- 43.Stewart DJ, et al. Angiogenic gene therapy in patients with nonrevascularizable ischemic heart disease: a phase 2 randomized, controlled trial of AdVEGF(121) (AdVEGF121) versus maximum medical treatment. Gene Ther. 2006;13(21):1503–1511. doi: 10.1038/sj.gt.3302802. [DOI] [PubMed] [Google Scholar]
- 44.Johnson T, Zhao L, Manuel G, Taylor H, Liu D. Approaches to therapeutic angiogenesis for ischemic heart disease. J Mol Med. 2019;97(2):141–151. doi: 10.1007/s00109-018-1729-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilahur G, Juan-Babot O, Peña E, Oñate B, Casaní L, Badimon L. Molecular and cellular mechanisms involved in cardiac remodeling after acute myocardial infarction. J Mol Cell Cardiol. 2011;50(3):522–533. doi: 10.1016/j.yjmcc.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 46.Khan I, et al. PLGA Nanoparticles and their versatile role in anticancer drug delivery. Crit Rev Ther Drug Carrier Syst. 2016;33(2):159–193. doi: 10.1615/CritRevTherDrugCarrierSyst.2016015273. [DOI] [PubMed] [Google Scholar]
- 47.Sah H, Thoma LA, Desu HR, Sah E, Wood GC. Concepts and practices used to develop functional PLGA-based nanoparticulate systems. Int J Nanomedicine. 2013;8:747–765. doi: 10.2147/IJN.S40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura Y, et al. Xenotransplantation of long-term-cultured swine bone marrow-derived mesenchymal stem cells. Stem Cells. 2007;25(3):612–620. doi: 10.1634/stemcells.2006-0168. [DOI] [PubMed] [Google Scholar]
- 49.Xiong Q, et al. Bioenergetic and functional consequences of cellular therapy: activation of endogenous cardiovascular progenitor cells. Circ Res. 2012;111(4):455–468. doi: 10.1161/CIRCRESAHA.112.269894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong Q, et al. Functional consequences of human induced pluripotent stem cell therapy: myocardial ATP turnover rate in the in vivo swine heart with postinfarction remodeling. Circulation. 2013;127(9):997–1008. doi: 10.1161/CIRCULATIONAHA.112.000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye L, et al. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell. 2014;15(6):750–761. doi: 10.1016/j.stem.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ong SG, et al. Microfluidic single-cell analysis of transplanted human induced pluripotent stem cell-derived cardiomyocytes after acute myocardial infarction. Circulation. 2015;132(8):762–771. doi: 10.1161/CIRCULATIONAHA.114.015231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao M, et al. Y-27632 preconditioning enhances transplantation of human-induced pluripotent stem cell-derived cardiomyocytes in myocardial infarction mice. Cardiovasc Res. 2019;115(2):343–356. doi: 10.1093/cvr/cvy207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.