Abstract
BACKGROUND. Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory coronavirus 2 (SARS-CoV-2), has become a pandemic. This study addresses the clinical and immunopathological characteristics of severe COVID-19.
METHODS. Sixty-nine patients with COVID-19 were classified into severe and nonsevere groups to analyze their clinical and laboratory characteristics. A panel of blood cytokines was quantified over time. Biopsy specimens from 2 deceased cases were obtained for immunopathological, ultrastructural, and in situ hybridization examinations.
RESULTS. Circulating cytokines, including IL-8, IL-6, TNF-α, IP10, MCP1, and RANTES, were significantly elevated in patients with severe COVID-19. Dynamic IL-6 and IL-8 were associated with disease progression. SARS-CoV-2 was demonstrated to infect type II and type I pneumocytes and endothelial cells, leading to severe lung damage through cell pyroptosis and apoptosis. In severe cases, lymphopenia, neutrophilia, depletion of CD4+ and CD8+ T lymphocytes, and massive macrophage and neutrophil infiltrates were observed in both blood and lung tissues.
CONCLUSIONS. A panel of circulating cytokines could be used to predict disease deterioration and inform clinical interventions. Severe pulmonary damage was predominantly attributed to both cytopathy caused by SARS-CoV-2 and immunopathologic damage. Strategies that prohibit pulmonary recruitment and overactivation of inflammatory cells by suppressing cytokine storm might improve the outcomes of patients with severe COVID-19.
Keywords: COVID-19, Infectious disease, Pulmonology
Keywords: Apoptosis, Macrophages, Medical statistics
Combined clinical and pathological evaluation characterizes progression in patients with severe COVID-19.
Introduction
Severe acute respiratory coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), an emerging respiratory infectious disease that has become an overwhelming health threat globally. SARS-CoV-2 has affected more than 2 million individuals and caused nearly 120,000 deaths, resulting in an average mortality of 6.33% worldwide as of April 14th, 2020, compared with the influenza mortality rate of less than 1%. The spectrum of SARS-CoV-2 infection ranges from asymptomatic and mild symptoms to pneumonia and life-threatening complications, including acute respiratory distress syndrome (ARDS), septic shock, multiorgan failure, and ultimately death (1, 2). Notably, about 11% of patients with mild symptoms show a sharp deterioration that result in severe manifestations, including respiratory failure, multiorgan failure, or even death (3). The fatality rate of critical patients with COVID-19 was shown to be approximately 61.5% (4). Therefore, early warning of severe COVID-19 and management of individuals with this disease is still a big challenge.
To date, host risk factors have been identified to be associated with critical illness and mortality in patients with COVID-19, but early signs of the risk of disease progression have been reported rarely. Extensive studies have revealed that severe COVID-19 cases are more likely to present lymphopenia and hypoalbuminemia and higher lactate dehydrogenase, C-reactive protein (CRP), ferritin, and D-dimer (2, 3, 5). Both severe acute respiratory syndrome (SARS) and COVID-19 are characterized by an overexuberant inflammatory response that is associated with disease severity. Evidence has shown that CD4+ and CD8+ T lymphocytes were significantly decreased with downregulated IFN-γ expression, while the levels of proinflammatory and antiinflammatory cytokines, including IL-2R, IL-6, IL-10, and TNF-α, were remarkably increased (6, 7). However, whether SARS-CoV-2 per ser or viral infection induced dysregulated immune response contributing to severe lung damage warrants further investigation. In addition, the dynamics of cytokines, pulmonary inflammatory infiltration, and links between abnormal immune response and immunopathology in the lung in COVID-19 remain to be elucidated, and these are vital for clinical management of the disease.
The objective of this study is to fill the gap between clinical and pathological analysis to provide insights into clinical management of patients with severe COVID-19 (severe patients) and to explore the immunopathological features of COVID-19. We hereby assessed the predictors of deterioration by performing detailed comparisons of clinical, radiological, laboratory, and dynamic cytokine data between severe patients and patients with nonsevere COVID-19 (nonsevere patients). The signs of deterioration were further assessed by examination of the immunopathology and ultrastructural pathology of postmortem biopsy specimens from 2 severe patients.
Results
Epidemiology and clinical presentation.
Thirty-six of sixty-nine patients had a definite history of exposure in Hubei. Among the 69 patients, 19 were diagnosed with severe COVID-19 on admission, and 7 patients progressed to severe condition during hospitalization. Three patients were transferred to intensive care units, of whom, 2 patients died of ARDS (8) and septic shock caused by Klebsiella pneumoniae, respectively. Forty-three patients were diagnosed with nonsevere COVID-19.
Fever, cough, headache, diarrhea, and sore throat were common at the onset of illness. Most of the patients had more than 1 sign or symptom. Compared with nonsevere patients, severe patients were significantly older (58 years old [range, 45–75] vs. 39 years old [range, 29–53]; P < 0.001) and more likely to report myalgia or fatigue (14 patients [53.9%] vs. 12 patients [27.9%]; P = 0.031) and dyspnea (7 patients [26.9%] vs. 1 patient [2.3%]; P = 0.004) (Table 1). Severe patients had a higher prevalence of chronic liver disease compared with nonsevere patients (3 patients [11.5%] vs. 0 patients [0%], P = 0.05). The sex distribution, endemic history, and prevalence of underlying comorbidities, such as diabetes, hypertension, cardiovascular disease, and chronic respiratory system disease did not differ between severe and nonsevere patients (P > 0.05) (Table 1). Typical chest radiogram findings include bilateral pulmonary parenchyma ground glass, consolidative pulmonary opacity, and the number of lobes affected by ground glass or combined opacity (Figure 1). Disease severity was associated with higher radiology scores (P < 0.001).
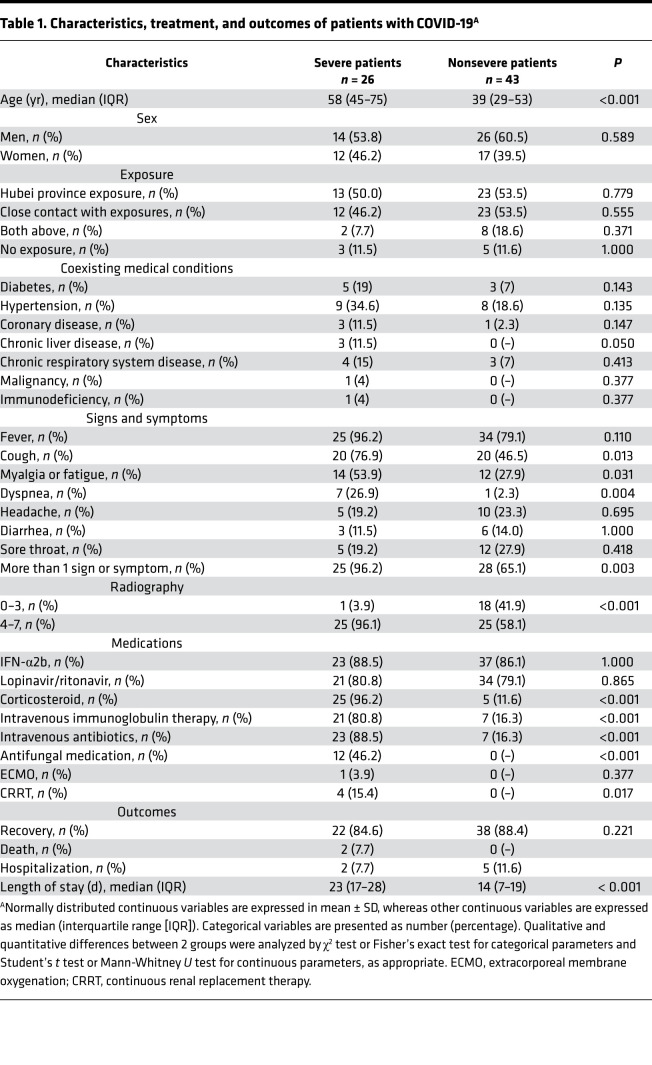
Table 1. Characteristics, treatment, and outcomes of patients with COVID-19A.
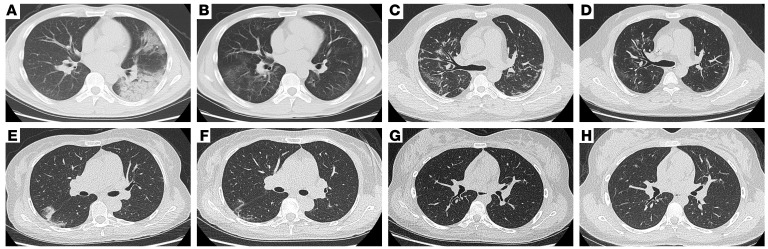
Figure 1. CT images of 2 patients with severe COVID-19 and 2 patients with nonsevere COVID-19.
(A) CT image of a 33-year-old man showing bilateral ground-glass opacities (GGO) partially fused into consolidation at onset of hospitalization and (B) mild GGO in the bilateral lungs. (C) CT image of a 51-year-old man showing bilateral GGO at onset of hospitalization and (D) multiple slightly high dense shadows in the bilateral lungs. (E) CT image of a 53-year-old woman showing bilateral GGO at the onset of hospitalization and (F) absorption after recovery. (G) CT image of a 17-year-old woman showing mild consolidation at the onset of hospitalization and (H) absorption after recovery.
Most patients received antiviral therapy with IFN-α2b by inhalation and lopinavir/ritonavir orally. Systemic corticosteroid therapy (96.2% vs. 11.6%, P < 0.001) and intravenous immunoglobulin therapy (80.8% vs. 16.3%, P < 0.001) were administered more often in severe patients than in nonsevere patients (Table 1). Intravenous antibiotics (88.5% vs. 7%, P < 0.001) and antifungal medication (46.2% vs. N.A., P < 0.001) were administered more frequently in severe patients. In regard to respiratory support, oxygen therapy, especially with a high-flow nasal cannula, was applied more often to severe patients compared with nonsevere patients (P < 0.001). More severe patients received ventilation support (P = 0.006). One patient received extracorporeal membrane oxygenation, and four patients received continuous renal replacement therapy. At follow-up, 60 of 69 patients completely recovered, 7 patients were still hospitalized, and 2 patients died. The length of stay of severe patients was longer than that of nonsevere patients (23 days vs. 14 days, P < 0.001) (Table 1).
Laboratory tests and cytokines in severe and nonsevere patients.
Of the 26 severe patients, 88% had a partial pressure of oxygen/fraction of inspiration oxygen (PaO2/FIO2) value of less than 300; 96% of severe patients had a surplus pulse oxygen (SpO2) value of less than 93%; and 35% of severe patients had a respiratory rate (RR) greater than 30 breaths/min. A SpO2 value of ≤93% at rest was the most useful parameter for defining severity. The pressure of oxygen (68 vs. 85, P < 0.001) and oxygenation index (262 vs. 393, P < 0.001) were much lower in the severe group than in the nonsevere group. The alveolar-arterial oxygen partial pressure difference (A-aDO2, 52 vs. 23, P < 0.001) in severe patients was higher than that in nonsevere patients (Table 2).
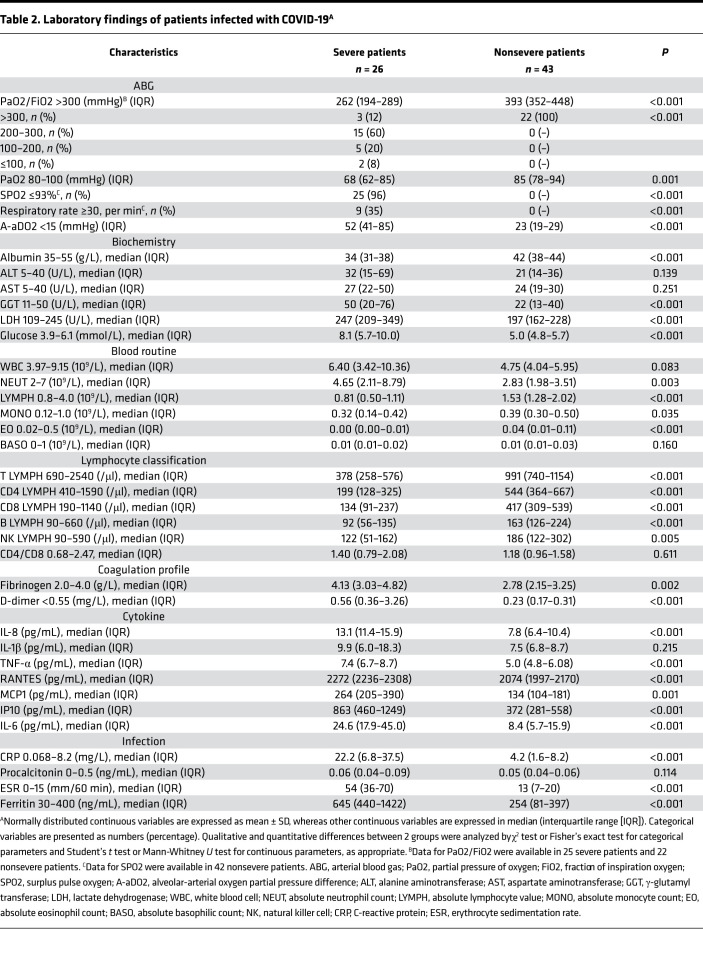
Table 2. Laboratory findings of patients infected with COVID-19A.
There were significant differences in findings from a panel of laboratory tests between severe and nonsevere patients, including cell counts of lymphocytes, neutrophils, and eosinophils (P < 0.05, respectively). We observed a significant decrease in CD4+ and CD8+ T cells in severe patients (P < 0.05, respectively). High levels of blood fibrinogen and D-dimer were present in severe patients (P < 0.05, respectively). In regard to biochemical parameters, albumin (34 vs. 42 g/L, P < 0.001), γ-glutamyl transferase (50 vs. 22 U/L, P < 0.001), and lactate dehydrogenase (247 vs. 197 U/L, P < 0.001) in severe patients were within normal ranges but showed abnormal tendencies and were significantly different from those in nonsevere patients (Table 2). There were significant elevations in blood glucose (8.1 vs. 5.0 mmol/L, P < 0.001), Apo-A1 (0.80 vs. 1.08 g/L, P < 0.001), pro-brain natriuretic peptide (pro-BNP, 217.7 vs. 22.9 ng/mL, P < 0.001), erythrocyte sedimentation rate (54 vs. 13 mm/60 minutes, P < 0.05), ferritin (645 vs. 254 ng/mL, P < 0.05), and CRP (22.2 vs. 4.2 mg/L, P < 0.05) in severe patients (Table 2). Notably, the levels of IL-8 (13.1 vs. 7.8 pg/mL, P < 0.001), TNF-α (7.4 vs. 5.0 pg/mL, P < 0.001), IL-6 (24.6 vs. 8.4 pg/mL, P < 0.001), MCP1 (264 vs. 134 pg/mL, P = 0.001), IP10 (863 vs. 372 pg/mL, P < 0.001), and RANTES (2272 vs. 2074 pg/mL, P < 0.001) were elevated in the severe group compared with those in nonsevere group (Table 2).
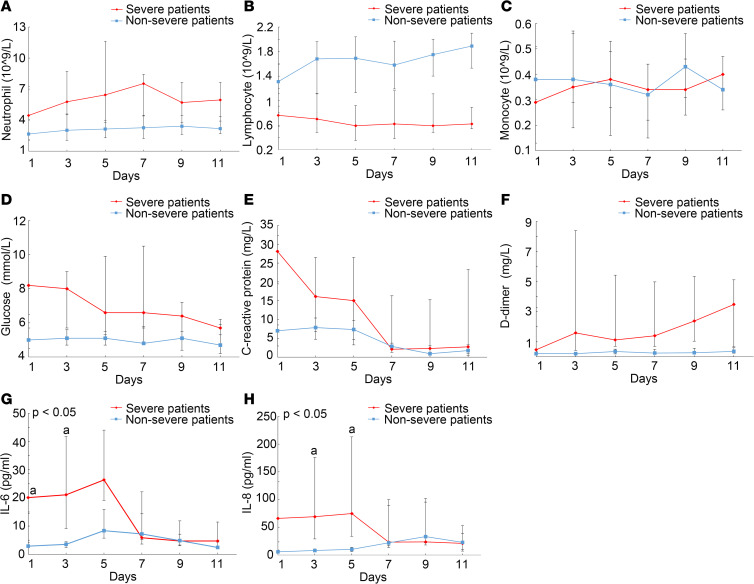
The kinetics of blood neutrophils, lymphocytes, monocytes, IL-6, IL-8, D-dimer, CRP, and glucose based on consecutive samples obtained at 2-day intervals were determined within 11 days from admission day. In the severe group, neutrophilia rose to a peak, while lymphopenia reached its lowest point on day 7 (Figure 2, A and B). Fluctuation of monocyte counts was observed in both the severe and nonsevere group (Figure 2C). Glucose sustained a high level, CPR sustained a decrease, and D-dimer continued to increase during the disease course of COVID-19 in severe patients (Figure 2, D–F). As revealed by Figure 2, G and H, IL-6 and IL-8 levels on admission were remarkably higher in the severe group compared with those in the nonsevere group. Subsequently, the level of IL-6 in the severe group showed a rapid increase, with a peak level on day 5 of hospitalization, on average, whereas serum IL-8 showed a sustained high level until hospital day 5. The dynamics of IL-6 and IL-8 concentrations were highly consistent with disease deterioration.
Figure 2. Dynamic characteristics of laboratory variants between severe patients and nonsevere patients.
Dynamics of blood (A), neutrophils, (B) lymphocytes, (C) monocytes, (D) glucose, (E) C-reactive protein, (F) D-dimer, (G) IL-6, and (H) IL-8 between severe patients (n = 21) and nonsevere patients (n = 25). Data are represented as median (IQR). A generalized linear mixed model was used to compare repeated measures (nonnormal distribution). Data are presented as the mean of triplicate measurements.
Histopathological findings.
In reference to pathology staging of SARS (5), in this study the early phase was defined as a disease course of 7–14 days, with features of acute exudative diffuse alveolar damage (DAD). The late phase was defined as a disease course lasting more than 14 days, or even longer, with features of organizing and fibrotic DAD. Histological examination of lung tissues from 2 cases representative of the acute phase (Figure 3) and late phase (Figure 4) of the disease showed varied pathological features. Case 1 (disease course of 14 days) showed DAD, acute exudative edema, hyaline membrane formation, and desquamation of pneumocytes, which was consistent with the features of early stage (Figure 3A). In case 2 (disease course of 28 days), H&E and Masson’s trichrome staining showed features of interstitial and alveolar fibrosis and pneumocyte hyperplasia, with focal exudative edema and hyaline membrane in alveolar spaces, indicating late-stage pathology of the disease (Figure 4, A and B). In these 2 cases, multinucleated pneumocytes were identified, indicating viral cytopathic-like changes. Viral inclusions were not detected.
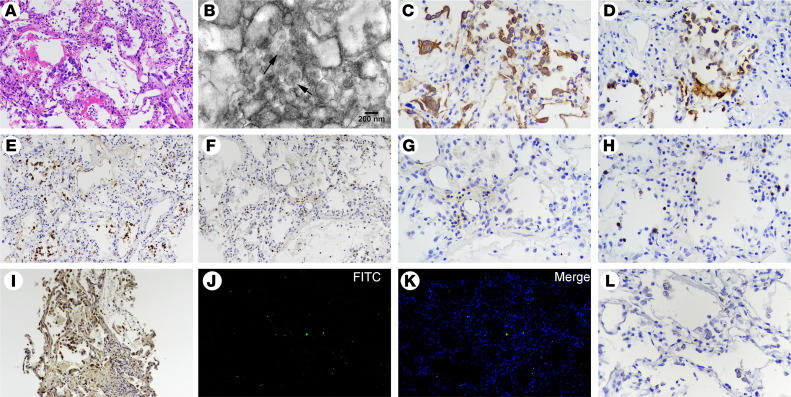
Figure 3. Postmortem lung biopsy specimens from case 1.
(A) Lung tissue showing pulmonary edema with hyaline membranes and desquamation of alveolar epithelial cells (H&E staining; original magnification, ×200). (B) Ultrastructural image showing cytoplasmic viral particles in a type II pneumocyte (arrow), with swollen mitochondria and dilated endoplasmic reticulum (original magnification, ×20,000). (C) CK7+ cells reflecting pneumocytes (original magnification, ×200). (D) SPB+ cells indicating type II pneumocytes with marked vacuolation and mild hyperplasia (original magnification, ×200). (E) Increased CD68+ macrophages with cytomegaly mainly in alveolar spaces (original magnification, ×200). (F) Immunohistochemical staining for MPO, indicating a large number of interstitial infiltrated polymorphonuclear cells (original magnification, ×200). (G) CD4+ and (H) CD8+ T cells distributed in the alveolar septal walls and interstitial areas (original magnification, ×200). (I) Gasdermin D positivity indicating cell pyroptosis (original magnification, ×200). (J and K) TUNEL staining showing apoptotic cells (original magnification, ×200). (L) RNAscope in situ hybridization indicating SARS-CoV-2 nucleic acids, which manifest as spotted brown particles (original magnification, ×200).
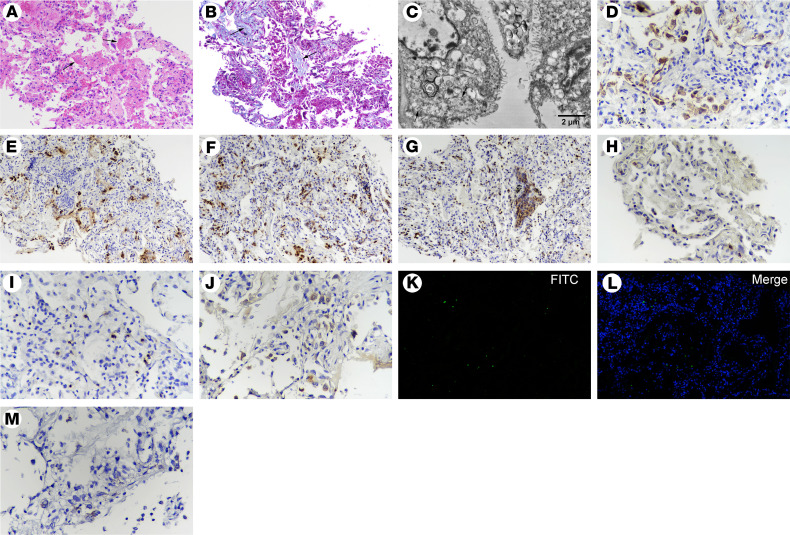
Figure 4. Postmortem lung biopsy from case 2.
(A) H&E and (B) Masson’s trichrome staining showing features of early interstitial and alveolar fibrosis and mild pneumocyte hyperplasia, with focal exudative edema and hyaline membranes (arrows) in alveolar spaces. (C) Ultrastructural image showing cytoplasmic viral particles (arrow) characterized by spherical and spike-like projections in type II pneumocytes, with depleted laminar bodies, swollen mitochondria, and dilated endoplasmic reticulum. (D) CK7+ cells showing pneumocytes (original magnification, ×200). (E) SPB+ cells reflecting type II pneumocytes (original magnification, ×200). (F) Abundantly increased CD68+ macrophages in alveolar spaces. (G) Immunohistochemical staining of MPO, indicating numerous polymorphonuclear cells, aggregated in focal areas of bronchiolitis (original magnification, ×200). (H) A few CD4+ and (I) CD8+ T cells were distributed in the alveolar septal walls and interstitial areas (original magnification, ×200). (J) Gasdermin D+ cells representing cell pyroptosis (original magnification, ×200). (K and L) TUNEL staining showing apoptotic cells (original magnification, ×200). (M) RNAscope in situ hybridization indicating SARS-CoV-2 nucleic acids, which manifest as spotted brown particles (original magnification, ×200).
Ultrastructural findings.
Ultrastructural examination of lung tissues from 2 cases revealed typical viral particles in cytoplasm of pneumocytes, especially in type II pneumocytes, with or without membrane-bound vesicles. The virus particles were spherical and enveloped, with spike-like projections on the surface, most ranged from 60 to 120 nm in diameter, indicating characteristic coronavirus particles (Figure 3B and Figure 4C). Pneumocytes appeared with markedly swollen mitochondria and dilated endoplasmic reticulum. Type II pneumocytes presented hyperplasia, with lager nuclei and nucleus and depleted laminar bodies. Endothelial cells of small blood vessels were swollen and vacuolated.
Immunohistochemical findings.
Immunohistochemical staining for surfactant protein B (SPB) and CK7 showed damaged type II pneumocytes with mild to moderate hyperplasia (Figure 3, C and D, and Figure 4, D and E). The number of CD68+ macrophages with cytomegalic features was substantially increased in alveolar spaces in lung tissues from both deceased cases (Figure 3, E and F). In case 2, some CD68+ cells were foamy or vacuolated (Figure 4F). Numerous neutrophils were observed in the interstitial infiltrates, predominantly in bronchopneumonia in case 2 (Figure 3F and Figure 4G). CD4+ and CD8+ T cells were scattered in lung tissues (Figure 3, G and H, and Figure 4, H and I). Many gasdermin D+ pneumocytes in case 1 and a moderate number of gasdermin D+ pneumocytes in case 2 appeared, which indicated that the main pattern of cell death involved pyroptosis in lung tissues of patients with COVID-19 (Figure 3I and Figure 4J). TUNEL staining indicated an increase of apoptotic cells (40–50 positive cells in case 1 and 15–20 positive cells in case 2; magnification ×200), reflecting an alternative manner of cell death (Figure 3, J and K, and Figure 4, K and L).
RNAscope in situ hybridization findings.
RNAscope in situ hybridization was used to detect SARS-CoV2 nucleic acids in infected cells, which appear with spotted brown particles. Negative controls for in situ hybridization performed with an irrelevant probe showed no positive signal in lung tissues. In situ hybridization results indicated that SARS-CoV-2 RNA was distributed in cytoplasm of many pneumocytes and some macrophages and endothelial cells (Figure 3L and Figure 4M), but no positive signal was detected in infiltrating lymphocytes in lung tissues from both cases.
Discussion
This study addresses the features of severe COVID-19 by comprehensive analysis of clinical, laboratory, radiological, and pathology data. The results showed an overexuberant response of a panel of proinflammatory cytokines, including IL-8, IL-6, TNF-α, IP10, MCP1 and RANTES, in severe patients, and dynamics of serum IL-6 and IL-8 were closely associated with disease progression. As a lung tropic virus, SARS-CoV-2 infection in type II and type I alveolar epithelial cells, as well as endothelial cells in small blood vessels, was illustrated by both TEM and in situ hybridization as leading to lung damage and cell death patterns of pyroptosis and apoptosis. The underlying mechanisms of cytokine storm and lung damage involved the exhaustion of CD4+ and CD8+ T cells, followed by massive infiltration of macrophages and neutrophils into lung tissues, causing the dysregulation of proinflammatory cytokines and chemokines.
Clinically, 3 criteria, the PaO2/FIO2 ratio, SpO2, and RR are often used to define patients with severe COVID-19. In our cohort, because 96% of severe patients had a SpO2 value ≤93% at rest, it seems that SpO2 is the most useful indicator for defining disease severity. Old age and severe symptoms of myalgia, fatigue, and dyspnea were the most common features of severe patients, as described in earlier reports (2, 3, 7). Lymphopenia is a common feature with drastically reduced numbers of CD4+ T cells, CD8+ T cells, and B cells (5, 8, 9). Reduced percentages of monocytes and eosinophils are also common. An increase in the neutrophil-to-lymphocyte ratio usually indicates increased disease severity and a poor clinical outcome. In our cohort, dynamic tests in severe patients found lymphopenia with a remarkable decrease of CD4+ and CD8+ T lymphocytes and neutrophilia as well as a sustained increase in the neutrophil-to-lymphocyte ratio that reached a peak on 7 day after admission, suggesting that physicians should monitor these dynamic characteristics in severe patients. The dramatic reduction in lymphocytes in severe patients might be caused by both overexuberant immune responses and SARS-CoV-2 infection of T lymphocytes (10). Neutrophilia in COVID-19 severity might be ascribed to abnormal elevated IL-8 and granulocyte-macrophage colony-stimulating factor (5). In our study, the substantial neutrophil infiltrate in lung tissues from the 2 deceased cases was assumed to indicate the possible role of neutrophilia in disease severity. In addition, some variables were identified to be associated with COVID-19 severity. An increased level of pro-BNP, as a predictor of heart dysfunction, was associated with disease deterioration. Additionally, increased levels of ferritin and glucose and decreased levels of albumin and Apo-A1 were found to correlate with COVID-19 severity, implying that severe patients were prone to metabolic disorders due to impaired liver function. However, we still lack conclusive evidence to support metabolic dysregulation by SARS-CoV-2 per se.
Previous studies have shown that increased levels of proinflammatory cytokines in blood (e.g., IL-1B, IL-6, IL-12, IL-17, IFN-γ, IP10, MCP1, TNF-α, and IL-15) were associated with pulmonary inflammation and extensive lung damage in patients with SARS and MERS (11–13). Recent evidence suggested that most patients with severe COVID-19 exhibited substantially elevated serum levels of several cytokines, which can even manifest as cytokine storm (14). Consistently, our results showed that patients with severe COVID-19 had an overexuberant response of a panel of proinflammatory cytokines and chemokines, including IL-6, IL-8, TNF-α, IP10, MCP1, and RANTES, compared with nonsevere patients. These inflammatory factors are mainly derived from macrophages and neutrophils, which could be predictors of hypercytokinemia and systemic inflammatory response syndrome. Both IL-6 and IL-8 have been identified to recruit monocytes to drive inflammation by producing a set of chemokines in a mouse model of SARS (15). In this study, we found that the dynamics of serum IL-6 showed a rapid increase, with a peak level on day 5 of hospitalization, on average, whereas serum IL-8 maintained a high level until day 5 of hospitalization in severe patients. The dynamic trends were highly consistent with disease deterioration. Indeed, in this cohort, systemic corticosteroids, which might contribute to the interference of elevated IL-6 and IL-8 levels, were administered to the most severe patients. Based on the pathological findings of massive recruitment of monocytes/macrophages and neutrophils to lung tissues, we speculated that the infiltrating inflammatory cells triggered or exacerbated inflammation and exudation, even ARDS in targeted lung, resulting in a vicious circle of cytokine release that led to cytokine storm.
In the absence of a proven effective therapy for patients with severe COVID-19, currently management consists of supportive care, usually including invasive and noninvasive oxygen support and antibiotic administration. Many patients have received off-label or compassionate-use therapies, including antivirals, antiinflammatory compounds, and convalescent plasma (14, 16, 17). In our cohort, from the early-to-middle period of the SARS-CoV-2 infection outbreak, the treatment strategies were empirically determined according our experiences with the SARS outbreak in 2003. However, the management of COVID-19 has rapidly evolved in the SARS-CoV-2 infection pandemic. In our cohort, 80.8% of severe patients received empirical antiviral therapy, including ritonavir/lopinavir and IFN-α2b, 96.2% of severe patients received systematic corticosteroid therapy, and 80.8% of cases were administered intravenous immunoglobulin. In our cohort, the mortality was 7.7% for severe COVID-19 cases, which is lower than that reported by other centers (5, 18); however, the decrease in mortality could not be ascribed to treatments alone, and other factors such as the relatively young age (58 years old; range, 45–75) of patients in our cohort could not be excluded. Other promising antiviral candidates, including hydroxychloroquine, remdesivir, and favipiravir, are undergoing clinical trials and might provide potential antiviral options (19, 20). A recent study of compassionate treatment with remdesivir for patients with severe COVID-19 reported an improved outcome in oxygen-support status and a decreased mortality of 13% compared with the reported 17%–78% mortality in severe cases (17). Another clinical trial result revealed that favipiravir was independently associated with faster viral clearance of H1N1 flu, SARS-CoV, and Ebola virus infections, encouraging the use of ongoing clinical trials for anti–SARS-CoV-2 infection (ChiCTR2000029600). For severe COVID-19 cases, immediate supplemental oxygen therapy to those with severe acute respiratory infection and respiratory distress, hypoxemia, or shock should be emphasized. The 2 deaths in this study were due to pathological DAD with ARDS and septic shock caused by Klebsiella Pneumoniae, respectively. Thus, prevention of ARDS and sepsis complications in patients with severe COVID-19 is urgent. To suppress inflammatory cytokine storm or ARDS, corticosteroids are not regularly recommended for COVID-19–related lung injury or shock according to WHO guidance. Russell et al. recently reported that no good reason exists to expect patients with COVID-19 to benefit from corticosteroids (21). The development of targeted therapeutics against cytokine storm is still a clinical issue. In our study, a panel of cytokines, IL-6, IL-8, IP10, TNF-α, MCP1, and RANTES, was profoundly elevated in severe patients during disease deterioration. Therefore, cytokine neutralization therapy is likely to be an alternative option. Recently, the preliminary data of a clinical trial of tocilizumab (ChiCTR2000029765), a monoclonal antibody targeting IL-6 receptor, showed improved outcomes in 21 patients with severe COVID-19. Other ongoing clinical trials of monoclonal antibodies targeting IL-17 and TNF-α, such as secukinumab/bimekizumab and infliximab/adalimumab (22) and so forth, will hopefully provide evidence of efficacy against SARS-CoV-2. Additionally, convalescent plasma therapy potentially benefits severe patients with persistent viremia (16).
SARS-CoV-2, a large, positive- and single-stranded RNA coronavirus, is a pneumophila, targeting the epithelial cells of the respiratory tract, resulting in pulmonary DAD. In this study, we demonstrated type II and type I pneumocytes and endothelial cells with conspicuous cytopathy in lung tissues were infected with SARS-CoV-2 by both TEM and in situ hybridization. Reactive hyperplasia of type II pneumocytes in DAD alveolar spaces in turn might facilitate virus infection and spreading due to high expression of angiotensin I converting enzyme 2. SARS-CoV-2 infection in endothelial cells in small vessels might contribute to vasculitis, even thrombosis, which was assumed to be related to increased blood D-dimer and fibrinogen levels in disease exacerbation. Thus, severe pulmonary injury in patients with SARS-CoV-2 was considered to result from both direct viral infection and immunopathological injury. Active cell death, in its many forms, is a fundamental disease process, especially for acute viral infection (23–25). The pathological findings of our study demonstrated that both pyroptosis and apoptosis contributed to lung damage, with pyroptosis as predominant cell death pattern. Pyroptosis is defined as gasdermin-mediated programmed necrosis in response to certain pathogen insults and is critical for immunity (26). A recent study revealed that caspase-cleaved gasdermin D can switch between triggering caspase-3–mediated apoptosis and necrosis induced by TNF (27). In our severe cases, levels of TNF-α and other factors were dramatically upregulated in blood, as were severe inflammatory infiltrates in lung tissues, which could provide possible explanations of pyroptosis as the main pattern as to pulmonary damage.
COVID-19 is an emerging infectious disease, and its immunopathology is highly complex. The possible mechanisms include the dysregulation of cytokines, deficiencies in the innate immune response, direct infection of immune cells, and viral cytopathic effect, among which immunopathological damage to target organs is crucial for the exacerbation of COVID-19. In our study, depletion of CD4+ and CD8+ T cells in both blood and lung tissues from patients with severe COVID-19 suggested the exhaustion of T cells following abnormal activation. A study showed that feline enteric coronavirus infection induced the production of IL-10 and skewed the immune response away from a protective Th1 cell response toward a nonprotective Th2 cell response, thereby diminishing the ability of immune cells to clear the virus (28). Similarly, in severe COVID-19 cases, impaired cellular immunity is not likely only to delay viral clearance, but also to attack lung tissues directly or indirectly. Even more troubling, viral infection might interfere with the normal feedback mechanisms that control inflammation, and proinflammatory chemokines or other cytokines might be produced in large amounts or for an excessive period. Elevated levels of IL-8, IL-6, TNF-α, and IP10 might mediate extensive pulmonary pathology, leading to massive infiltration of neutrophils and macrophages, severe DAD, and even ARDS, which is indicative of immune-mediated damage in severe patients. Collectively, impaired adaptive immune responses and uncontrolled inflammatory innate responses to virus in patients with severe COVID-19 might lead to damage to targeted organs, both locally and systemically, which may be characterized by the unchecked influx of inflammatory cells into targeted sites and cytokine storm. However, uncertainty about the underlying immunopathogenesis, exact cascades of hypercytokinemia, and the timing of the fluctuations of some cytokines during the disease course require further investigation.
This study has some limitations. First, this is a relatively small cohort study; therefore, some results might not be representative of the severe characteristics of COVID-19. Second, our results might not be generalizable to all patients; biopsy and autopsy of deceased cases were not readily available, and only 2 cases of pathological analysis were performed in this cohort. Third, management of severe COVID-19 is rapidly evolving over time, and some treatment regimens for patients with COVID-19 used in early- and middle-endemic periods of the SARS-CoV-2 outbreak were empirical.
In conclusion, this study addressed the characteristics and underlying immunopathology of severe COVID-19 through a comprehensive analysis of clinical and pathological features. Clinically, a panel of laboratory parameters, including dynamic cytokines, could be predictors of disease deterioration, and targeting cytokine intervention, such as with tocilizumab, might be an alternative option for severe COVID-19 cases. Pathologically, severe pulmonary damage was predominantly attributed to both cytopathy caused by SARS-CoV-2 and immunopathologic damage. Therefore, strategies targeting pulmonary recruitment and abnormal activation of mononuclear cells/macrophages and neutrophils through suppression of the inflammatory storm might improve the outcomes of patients with severe COVID-19.
Methods
Patients.
Sixty-nine patients with confirmed COVID-19 included in the present study were admitted to designated centers in China from January 20, 2020, to March 20, 2020. All patients with COVID-19 enrolled in this study were diagnosed and classified by severity according to the Guidelines for the Diagnosis and Treatment of New Coronavirus Pneumonia (version 5) published by the National Health Commission of China (http://www.nhc.gov.cn/xcs/zhengcwj/202002/3b09b894ac9b4204a79db5b8912d4440.shtml). Laboratory confirmation of COVID-19 was performed at the CDC with RT-PCR detection reagents. Severe patients met at least 1 of the following criteria: (a) shortness of breath with a RR ≥30 times/min, (b) oxygen saturation (resting state) ≤93%, or (c) PaO2/FiO2 ≤300 mmHg.
Data collection.
Epidemiological, clinical, laboratory, and radiological data were obtained from electronic medical records. The data were reviewed by a trained team of physicians. The date of the diagnosis of a severe case was determined as the day the patient first met the severity criteria according to medical records. The final date of follow-up was March 29th, 2020.
Radiology.
Digital chest radiographs and CT were performed at admission and during hospitalization. The images were assessed by using a picture archiving and communication system viewer. All chest radiographs and CT images were reviewed independently by 3 trained radiologists with 10 years of experience. Final assessments were reached by consensus. Scoring was performed according to the following 8-stage criteria: 0, bilateral lungs showing clear texture; 1, bilateral lungs showing mild shadows; 2, single small patch shadow; 3, unilateral lung showing multiple patch shadows; 4, bilateral lungs showing multiple patch shadows; 5, unilateral lung showing consolidation; 6, bilateral lungs showing consolidation (less than 50%); and 7, bilateral lungs showing consolidation (more than 50%).
Clinical specimen collection.
Clinical specimens for COVID-19 diagnostic testing were obtained in accordance with China CDC guidelines. Throat swabs and blood specimens were obtained and maintained in viral transport medium. On the admission day, blood samples were collected before any medications were administered. During hospitalization, all blood samples were collected during fasting. Serum samples were collected in serum separator tubes and then centrifuged in accordance with China CDC guidelines. Postmortem lung tissues were biopsied from 2 patients who died of severe COVID-19 as described in Epidemiology and clinical presentation after providing informed consent.
Circulating cytokine and chemokine measurement.
The concentrations of inflammatory cytokines, including IL-6 (YZB/UK 4438-2014, Siemens), IL-8 (YZB/UK 4439-2014, Siemens), IL-1β (YZB/UK 4797-2014, Siemens), and TNF-α (YZB/UK 2641-2012, Siemens) in serum specimens from patients with COVID-19, were measured by chemiluminescence immunoassay according to the manufacturer’s instructions. IP10 (ab100579, abcam), MCP1 (ab179886, abcam), and RANTES (ab174446, abcam) were quantified by enzyme-linked immunosorbent assay according to the manufacturer’s instruction. Triplicate tests of each independent sample were conducted.
Histopathology.
Postmortem lung tissues obtained from patients with COVID-19 were fixed in 10% neutral buffered formalin, paraffin embedded, sectioned at 4 μm, dewaxed, and rehydrated by standard procedures. The slides were stained with H&E. Masson’s trichrome staining was used to assess collagen fibers in lung tissues according to standard methods. The tissues were stained with the trichrome stain kit (HT15-1KT, MilliporeSigma), followed by Biebrich scarlet-acid fuchsin (MilliporeSigma), and differentiated in phosphomolybdic-phosphotungstic acid solution for 10–15 minutes. The sections were stained with aniline blue solution for 5 minutes, differentiated in glacial acetic solution for 5 minutes, dehydrated with 95% ethanol, cleared with xylene, and mounted. The stained sections were observed with a light microscope.
Transmission electron microscope.
Transmission electron microscope (TEM) was performed to observe the ultrastructural changes and SARS-CoV-2 viral particles in lung tissues. In detail, the specimens were fixed with 2.5% glutaraldehyde, postfixed with 1% osmium tetraoxide, rehydrated in a graded series of ethanol concentrations, and embedded in SPIPON812 resin (02660-AB, SPI Supplies). The ultrathin slides were sectioned with a microtome (Leica EM UC6) at approximately 70 nm, collected on copper grids, and stained with uranyl acetate and lead citrate. Images were obtained with TEM (JEM-1011 120, JEOL).
Immunohistochemistry.
Immunohistochemical examination of tissue samples was performed based on a standard protocol. Paraffin-embedded tissue sections were deparaffinized and rehydrated in a graded series of ethanol concentrations. Antigen retrieval was performed by heating the sections at 96°C in 0.01 mol/L citrate buffer (pH 6.0) for 30 minutes. Sections were immersed in 3% hydrogen peroxide to eliminate endogenous peroxidase activity. Slides were then incubated with primary antibodies overnight at 4°C at optimal dilutions. Primary antibodies included anti-human CD4 (ZM-0418, ORIGENE, 1:100), CD8 (ZM-0508, ORIGENE, 1:50), CD68 (Kit-0026, Thermo Fisher Scientific, 1:200), MPO (ZA-0197, MilliporeSigma, 1:150), SPB (ZM-0226, Zeta Corporation, 1:100), CK7(Kit-0021, Thermo Fisher Scientific, 1:100), and gasdermin D (96458 Cell Signaling Technology, 1:200).
TUNEL.
To assess cell apoptosis, a TUNEL assay was performed on tissue slides according to the manufacturer’s instructions. The sections were deparaffinized, rehydrated, and pretreated with proteinase K (DNase free) (ST532, Beyotime Biotechnology). The sections were then washed with PBS and stained with a mixture of TdT and dUTP at 37°C for 60 minutes (C1086, Beyotime Biotechnology). The stained apoptotic cells were observed with a fluorescence microscope.
RNAscope in situ hybridization.
In situ hybridization assay was performed using the RNAscope 2·5 HD-Brown assay (322310, Advanced Cell Diagnostics) to measure SARS-CoV-2 RNA in FFPE tissue sections. RNAscope Probe V-nCoV2019-S for SARS-CoV-2 (848561, target region: 21631–23303 of NC_045512.2), RNAscope Positive Control Probe-Hs-PPIB (313901, target region: 139–989 of NM_000942.4), and RNAscope Negative Control Probe-DapB (310043, target region: 414–862 of EF191515) were performed on serial sections. The assay was performed according to the standard manufacturer’s protocol. Probes were hybridized to a cascade of signal amplification molecules, culminating in the binding of HRP-labeled probes. The assay enhanced the signal further with additional amplification (AMP) steps. Hybridize amplification rounds 1–6 (AMP1–AMP6) were performed. In AMP 1, each slide was taken at a time from the Tissue-Tek Slide Rack, and the excess liquid was removed from the slides. The slides in HybEZ Humidity Control Tray were inserted into oven for 30 minutes at 40°C for AMP1 (15 minutes at 40°C for AMP2, 30 minutes at 40°C for AMP3, 15 minutes at 40°C for AMP4, 30 minutes at room temperature for AMP5, and 15 minutes at room temperature for AMP6). Excess liquid was quickly removed, and the slide was placed in a Tissue-Tek Slide Rack submerged in the Tissue-Tek Staining Dish filled with wash buffer. Slides were washed for 2 minutes at room temperature with occasional agitation. The slides were then stained with DAB substrate per section, and the signals were detected. Slide counterstain was performed in hematoxylin staining solution for 2 minutes at room temperature. The hybridization sections were dehydrated in 70% ethyl alcohol, 95% ethyl alcohol, and xylene and mounted. The hybridization signals were captured using an Aperio AT2 digital slide scanner equipped with a ×40 objective (Leica Biosystems Inc.).
Statistics.
Continuous variables are provided as mean ± SD or median (interquartile range) as appropriate, whereas categorical variables are presented as number (percentage). To compare the continuous variables between 2 groups, 2-tailed Student’s t test or Mann-Whitney U test was performed, as appropriate. A generalized linear mixed model was used to compare repeated measures (nonnormal distribution). The χ2 test and Fisher’s exact test were used for comparing categorical variables. All statistical tests were 2 sided, and a P value of less than 0.05 was defined as statistically significant. Data were analyzed using SAS 9.4 (SAS Institute).
Study approval.
Sample collection and analysis of cases were performed strictly in accordance with the regulations issued by the National Health Commission of China and the ethical standards formulated in the Declaration of Helsinki. This study was approved by the Institutional Review Board of the Fifth Medical Center of PLA General Hospital.
Author contributions
JZ contributed to study concept and design, clinical and pathological analysis, study supervision, and critical revision of the manuscript. JJ contributed to study supervision, clinical data collection, and analysis. S. Li, LJ, XL, and YW contributed to analysis and interpretation of data, creation of tables and figures, literature search, and writing of the manuscript. FL contributed to clinical data collection, analysis, and interpretation and writing of the manuscript. BL contributed to laboratory specimen collection and measurement. TJ contributed to clinical data collection and analysis. WA contributed to radiology data collection and analysis. S. Liu contributed to immunohistochemistry staining and pathological analysis. HL contributed to statistical analysis. PX and L. Zhang contributed to pathological experiments. L. Zhao contributed to creation of figures. HW, JK, and JL contributed to electron microscope examination and analysis. YL contributed to clinical data collection. JM, LH, CZ, and SZ contributed to clinical data and pathological specimen collection and analysis.
Acknowledgments
We thank all the patients and their families involved in this study as well as the numerous doctors, nurses, and civilians working together to fight against COVID-19.
Version 1. 05/19/2020
In-Press Preview
Version 2. 06/18/2020
Electronic publication
Funding Statement
to J. Zhao
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e138070.https://doi.org/10.1172/jci.insight.138070.
Contributor Information
Shaohua Li, Email: shhli55@163.com.
Lina Jiang, Email: jln302@163.com.
Xi Li, Email: 1307025518@qq.com.
Fang Lin, Email: linfang1029@sina.com.
Yijin Wang, Email: yijinwang927015@163.com.
Boan Li, Email: Liboan302@163.com.
Tianjun Jiang, Email: aba302@163.com.
Weimin An, Email: anweimin3021966@163.com.
Shuhong Liu, Email: 18511862409@163.com.
Hongyang Liu, Email: hyliu2356@163.com.
Pengfei Xu, Email: xpf13269339573@163.com.
Lihua Zhao, Email: zhaolihua2008@foxmail.com.
Lixin Zhang, Email: 1058860218@qq.com.
Jinsong Mu, Email: jinsongmu302@163.com.
Hongwei Wang, Email: hongwei304@163.com.
Jiarui Kang, Email: kangjz@126.com.
Yan Li, Email: 17864816533@163.com.
Lei Huang, Email: huangleiwa@sina.com.
Caizhong Zhu, Email: zhucaizhong302@163.com.
Shousong Zhao, Email: 1260345655@qq.com.
Jiangyang Lu, Email: lujy304@163.com.
Junsheng Ji, Email: 740411914@qq.com.
Jingmin Zhao, Email: jmzhao302@163.com.
References
- 1. doi: 10.1001/jama.2020.1097. Phelan AL, Katz R, Gostin LO. The novel coronavirus originating in Wuhan, China: Challenges for Global Health Governance [published ahead of print January 30, 2020]. JAMA . [DOI] [PubMed]
- 2.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507-13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ai T, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin C, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020:ciaa248. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li JY, et al. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22(2):80–85. doi: 10.1016/j.micinf.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong CK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang WK, et al. Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;39(7):1071–1075. doi: 10.1086/423808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72(8):4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Channappanavar R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20(4):398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. doi: 10.1056/NEJMoa2007016. Grein J, et al. Compassionate use of remdesivir for patients with severe Covid-19 [published ahead of print April 10, 2020]. N Engl J Med. https://dx.doi.org/10.1056%2FNEJMoa2007016. [DOI] [PMC free article] [PubMed]
- 18.Cao B, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. doi: 10.2174/0929867327666200416131117. Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus [published ahead of print April 16, 2020]. Curr Med Chem . [DOI] [PubMed]
- 20.Tu YF, et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 2020;21(7):E2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldmann M, et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395(10234):1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green DR. The coming decade of cell death research: five riddles. Cell. 2019;177(5):1094–1107. doi: 10.1016/j.cell.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277(1):61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergsbaken T, Fink SL, den Hartigh AB, Loomis WP, Cookson BT. Coordinated host responses during pyroptosis: caspase-1-dependent lysosome exocytosis and inflammatory cytokine maturation. J Immunol. 2011;187(5):2748–2754. doi: 10.4049/jimmunol.1100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42(4):245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579(7799):415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dean GA, Olivry T, Stanton C, Pedersen NC. In vivo cytokine response to experimental feline infectious peritonitis virus infection. Vet Microbiol. 2003;97(1-2):1–12. doi: 10.1016/j.vetmic.2003.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]