Abstract
Despite advances in lipid-lowering therapies, people with diabetes continue to experience more limited cardiovascular benefits. In diabetes, hyperglycemia sustains inflammation and preempts vascular repair. We tested the hypothesis that the receptor for advanced glycation end-products (RAGE) contributes to these maladaptive processes. We report that transplantation of aortic arches from diabetic, Western diet–fed Ldlr—/— mice into diabetic Ager—/— (Ager, the gene encoding RAGE) versus WT diabetic recipient mice accelerated regression of atherosclerosis. RNA-sequencing experiments traced RAGE-dependent mechanisms principally to the recipient macrophages and linked RAGE to interferon signaling. Specifically, deletion of Ager in the regressing diabetic plaques downregulated interferon regulatory factor 7 (Irf7) in macrophages. Immunohistochemistry studies colocalized IRF7 and macrophages in both murine and human atherosclerotic plaques. In bone marrow–derived macrophages (BMDMs), RAGE ligands upregulated expression of Irf7, and in BMDMs immersed in a cholesterol-rich environment, knockdown of Irf7 triggered a switch from pro- to antiinflammatory gene expression and regulated a host of genes linked to cholesterol efflux and homeostasis. Collectively, this work adds a new dimension to the immunometabolic sphere of perturbations that impair regression of established diabetic atherosclerosis and suggests that targeting RAGE and IRF7 may facilitate vascular repair in diabetes.
Keywords: Inflammation, Vascular Biology
Keywords: Atherosclerosis, Diabetes, Innate immunity
RAGE impairs regression of diabetic atherosclerosis in mice through IFN signaling and perturbation of macrophage inflammation and cholesterol metabolism.
Introduction
Atherosclerosis, a leading cause of morbidity and mortality, begins in childhood. Fatty streaks are found in early life and may undergo progression, arrest, or regression during their natural history (1). Although pharmacological approaches have yielded success in mediating regression of established atherosclerotic plaques in humans (2), in people with diabetes, lipid- and glucose-lowering therapies are less effective (3). These observations are recapitulated in murine models of diabetic atherosclerosis regression (4–7).
In atherosclerosis regression, prominent roles for macrophage dysfunction have been uncovered through fate mapping, fluorescent bead labeling of monocytes/macrophages, single-cell and bulk RNA-sequencing, and using genetically modified animals and pharmacological treatments (8–18). Roles for impaired emigration of macrophages from established plaques to skewed inflammatory processes, in which overabundance of pro- versus antiinflammatory macrophage gene expression impairs regression, particularly in diabetes, have been uncovered (4, 19).
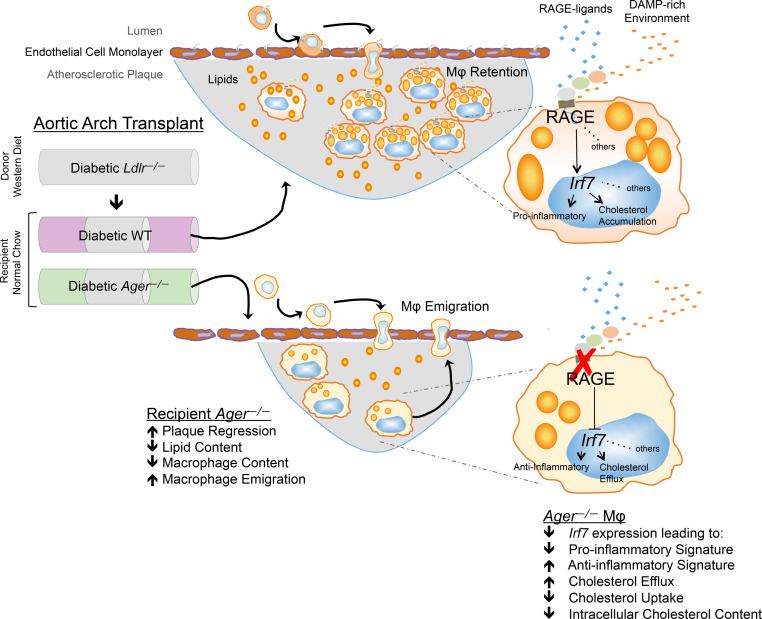
It was in these contexts that we hypothesized a role for the receptor for advanced glycation end-products (RAGE) in impaired regression of atherosclerosis. RAGE, a member of the immunoglobulin superfamily, is expressed on monocytes/macrophages and vascular cells and has been implicated in atherosclerosis progression in mice (20–24). We employed a model of transplantation of donor atherosclerotic aortic arches (25) from mice devoid of the LDL receptor (Ldlr) and fed a Western diet into normolipidemic mice to test the premise that Ager (Ager, the gene encoding RAGE) contributes to macrophage dysfunction and impairs regression of atherosclerosis. We demonstrate that compared with WT recipient mice, deletion of Ager enhanced regression of atherosclerosis. RNA-sequencing of donor versus recipient diabetic macrophages after aortic arch transplantation suggested that Ager deletion downregulated interferon signaling, and, specifically, interferon regulatory factor 7 (IRF7). In macrophages, IRF7 regulated cholesterol metabolism and inflammation. Targeting RAGE and IRF7 might represent an important therapeutic opportunity to accelerate atherosclerosis regression.
Results
RAGE is expressed in regressing atherosclerotic plaques in mice.
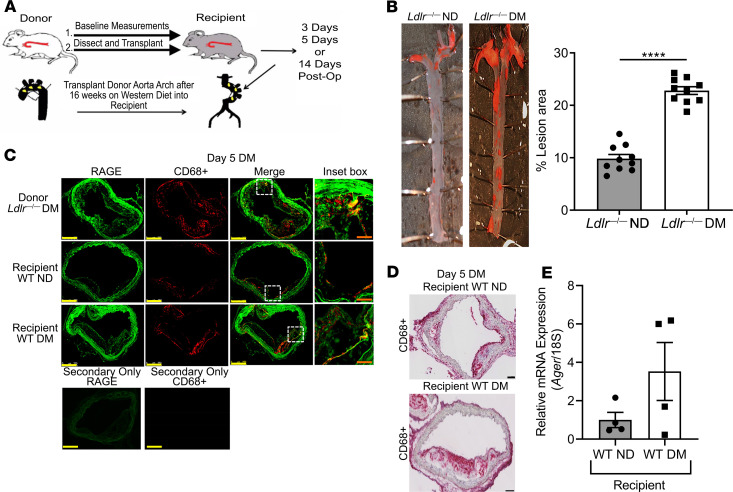
To test the premise that RAGE impairs regression of diabetic atherosclerosis, we rendered male Ldlr—/— mice diabetic with streptozotocin at age 7 weeks. Hyperglycemic and control mice were placed on a Western diet (0.15% cholesterol for 16 weeks). At age 23 weeks, aortic arches from these donor mice were transplanted into male WT or homozygous Ager—/— diabetic or nondiabetic mice (25). The grafted aortic arches were harvested at the indicated times after transplantation (Figure 1A).
Figure 1. RAGE is expressed in regressing atherosclerotic plaques.
(A) Model schematic: atherosclerotic aortic arches from Ldlr—/— mice were transplanted into WT, Ager—/—, diaphanous related formin 1–deficient (Diaph1—/—), or Tg glyoxalase-1 (Glo1) recipient mice and harvested at the indicated times after aortic arch transplantation. (B) Representative images of en face Sudan IV staining of atherosclerotic lesions in nondiabetic Ldlr—/—and diabetic Ldlr—/— mice. Quantification of plaque area as percentage of total aortic surface area is shown; N = 10 mice/group. (C) Immunofluorescence staining for RAGE and CD68 of atherosclerotic plaques 5 days after aortic arch transplantation. Representative images from N = 4 mice/group are shown. Scale bars: 250 μm, and inset, 50 μm. The secondary antibody–alone control is shown. (D) Laser capture microdissection of CD68+ cells 5 days posttransplantation. Representative images from N = 4 mice/group are shown. Scale bar: 100 μm. (E) Ager mRNA expression in macrophages captured from D (N = 4 mice/group). Mean ± SEM. Unpaired t test was performed in B and E. ****P < 0.0001.
We verified that diabetes accelerated atherosclerosis progression in Western diet–fed Ldlr—/— mice, as aortic arches from these mice served as the donors in these experiments. En face analysis of the aorta revealed a significant increase in atherosclerotic lesion area in diabetic versus nondiabetic aortas; P < 0.0001 (Figure 1B). Immunofluorescence microscopy colocalized RAGE to CD68-expressing macrophages in the atherosclerotic plaques of these mice (Figure 1C). When atherosclerotic aortic arches from diabetic Ldlr—/— mice were transplanted into diabetic versus nondiabetic recipient WT mice, laser capture microdissection of lesional CD68-expressing macrophages 5 days posttransplantation revealed trends toward higher Ager mRNA transcripts by about 3-fold (Figure 1, D and E). We tested whether RAGE expression in the recipient mice contributed to impaired regression of atherosclerosis.
RAGE impairs regression of atherosclerosis.
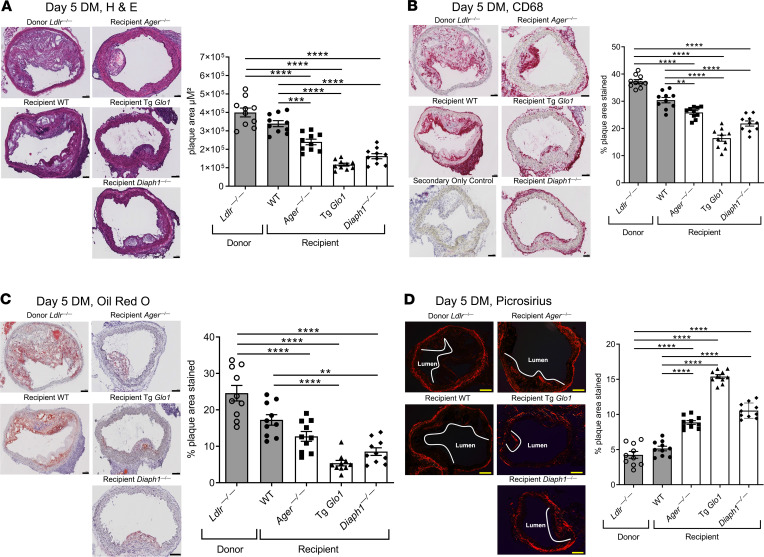
We first tested the role of RAGE in diabetes. Atherosclerotic aortic arches retrieved from diabetic donor Ldlr—/— mice fed a Western diet were transplanted into diabetic WT or Ager—/— mice. Five days later, H&E staining revealed that when the atherosclerotic aortic arches were transplanted into Ager—/— diabetic mice, significantly lower lesion area was observed compared with that in the diabetic donor and diabetic WT recipient mice; P < 0.0001, and P < 0.001, respectively (Figure 2A). There were no significant differences in plaque size when comparing donor aortic arches from diabetic Ldlr—/— mice to the transplanted arches from diabetic WT recipient mice (Figure 2A). With respect to macrophage content, compared with either the diabetic donor aortic arches retrieved from Ldlr—/— mice or the transplanted aortic arches retrieved from WT diabetic recipient mice, the aortic arches retrieved from Ager—/— mice revealed significantly reduced macrophage (CD68) content; P < 0.0001, and P < 0.01, respectively (Figure 2B). There were no significant differences between the diabetic donor Ldlr—/— aortic arches and those retrieved from the WT diabetic recipient mice (Figure 2B). Oil red O staining (neutral lipid) revealed that compared with the donor diabetic Ldlr—/— mice aortic arches, lipid content was significantly reduced in the diabetic recipient mice devoid of Ager but not the WT animals; P < 0.0001 (Figure 2C). Lesional content of collagen, considered a beneficial stabilizing component in human atherosclerotic plaques, was assessed. Whereas there were no significant differences between the diabetic donor Ldlr—/— mice aortic arches and those retrieved from the diabetic WT recipient mice, significantly higher Picrosirius red staining was observed in the diabetic Ager—/— recipient mice; P < 0.0001 (Figure 2D). Significantly higher collagen content was observed in the aortic arches of the diabetic recipient mice devoid of Ager versus the WT diabetic recipient mice; P < 0.0001 (Figure 2D).
Figure 2. RAGE, GLO1, DIAPH1, and regression of atherosclerosis.
Representative images of H&E (A), CD68 (B), Oil red O (C), and Picrosirius red staining (D) of aortic arch donor and transplant sections and their quantification in Ldlr—/— (donor) and WT, Ager—/—, Tg Glo1, and Diaph1—/— diabetic recipient mice 5 days posttransplantation (N = 10 mice/group). Scale bar in A–D: 100 μm. In B, the secondary antibody–alone control is shown. Mean ± SEM; 1-way ANOVA with post hoc Tukey’s test was used. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
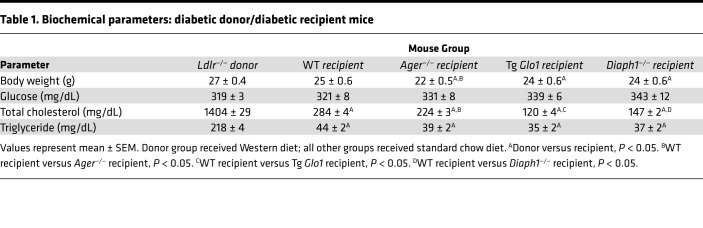
We analyzed metabolic data (Table 1). Ager—/— mice recipients weighed significantly less than the donor Ldlr—/— and the WT recipient mice. There were no significant differences in levels of blood glucose. Levels of total cholesterol were significantly lower in WT and Ager—/— recipient groups versus the donor Ldlr—/— mice; plasma cholesterol was significantly lower in the Ager—/— versus the WT mice. However, only in the donor Ldlr—/— mice, not in the recipient mice (WT and Ager—/—), were significant correlations between atherosclerotic lesion area and plasma cholesterol levels observed (Table 2). The levels of triglyceride were significantly higher in the Ldlr—/— mice versus both recipient groups; no significant differences were observed between the recipient groups.
Table 1. Biochemical parameters: diabetic donor/diabetic recipient mice.
Table 2. Correlation between atherosclerotic lesion area (H&E staining) and total cholesterol levels in diabetic donor and diabetic recipient mice.
As RAGE ligands accumulate in nondiabetic atherosclerosis (20), we transplanted aortic arches from nondiabetic, Western diet–fed Ldlr—/— mice into nondiabetic WT or Ager—/— recipient mice and sacrificed the mice 5 days later. Compared with the nondiabetic donor Ldlr—/— mice aortic arches and the aortic arches that were transplanted into the recipient WT nondiabetic mice, transplantation of atherosclerotic aortic arches into nondiabetic mice devoid of Ager resulted in significantly lower atherosclerotic lesion area (P < 0.0001, and P < 0.01, respectively); lesional macrophage content (CD68) (P < 0.0001); and Oil red O staining (P < 0.0001 and P < 0.001, respectively) and significantly higher collagen content; P < 0.0001) (Supplemental Figure 1, A–D; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137289DS1). Ager—/— recipient mice weighed less than the donor Ldlr—/— mice. Levels of blood glucose were significantly lower in the Ager—/— recipient mice versus the other 2 groups. Levels of total cholesterol were significantly lower in WT and Ager—/— recipient mice versus the donor Ldlr—/— mice but did not differ between the 2 recipient groups. There was no significant correlation between atherosclerotic lesion area and cholesterol levels in the recipient Ager—/— mice, unlike that observed in the WT recipient mice (Supplemental Table 1). Levels of total triglycerides were significantly lower in both recipient groups versus the donor mice (Supplemental Table 1).
To test if the effects of Ager deletion on regression of diabetic atherosclerosis were sustained, recipient diabetic mice that had undergone transplantation of donor aortic arches from diabetic, Western diet–fed Ldlr—/— mice were sacrificed 14 days posttransplantation. Atherosclerotic lesion area, macrophage CD68 content, and Oil red O staining were significantly lower and lesional content of collagen was significantly higher in the diabetic WT and Ager—/— recipient mice compared with the diabetic donor Ldlr—/— mice; P < 0.0001 (Supplemental Figure 2, A–D). All these parameters were significantly improved in the diabetic Ager—/— versus the WT recipient mice; P < 0.05 (Supplemental Figure 2, A–D).
Effects of advanced glycation end-products, oxidative stress, Tg glyoxalase-1 expression, and deletion of Diaph1 on regression of diabetic atherosclerosis.
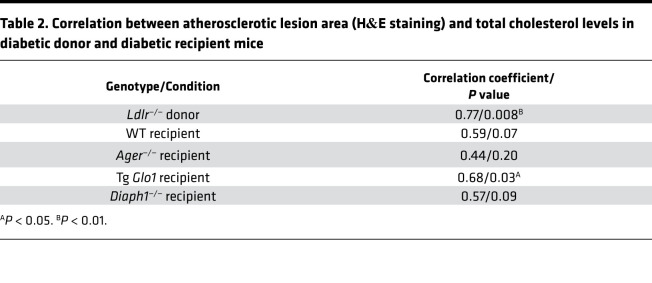
We reasoned that the accumulation of RAGE ligands contributed to the observed differences between Ager-expressing and -deficient mice and probed for RAGE ligands, advanced glycation end-products (AGEs), in plasma and atherosclerotic lesions. AGEs may be generated by hyperglycemia, oxidative stress, and inflammation (20–24, 26, 27). Western blotting revealed that plasma AGE epitopes were significantly higher in diabetic versus nondiabetic WT mice; P < 0.001 (Figure 3A). Aortic arches and atherosclerotic plaques of diabetic Ldlr—/— mice revealed copious AGE immunoreactivity (Figure 3B).
Figure 3. RAGE, AGEs, GLO1, and DIAPH1.
(A) Western blotting of carboxymethyllysine-AGE (CML-AGE) epi-topes or loading control, transferrin, was performed on the plasma from nondiabetic and diabetic WT mice fed normal chow diet. A representative blot is shown from N = 5 mice/group. (B) Staining and quantification of AGEs of aortic arch sections 5 days posttransplantation in diabetic Ldlr—/— (donor), and WT, Tg Glo1 and Diaph1—/— recipient mice. Scale bar: 100 μm. In B, the secondary antibody alone control is shown Representative images are shown from N = 10 mice/group. (C) Staining and quantification of dihydroethidium (DHE) of aortic arch sections 5 days posttransplantation in diabetic Ldlr—/— (donor) and WT, Ager—/—, Tg Glo1, and Diaph1—/— recipient mice. Scale bar: 250 μm. Representative images are shown from N = 10 mice/group. Mean ± SEM. Unpaired t test (A) and 1-way ANOVA with post hoc Tukey’s test (B–C), respectively. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To determine if AGEs impaired regression of diabetic atherosclerosis, we modulated levels of the enzyme GLO1, which detoxifies pre-AGE intermediates and reduces AGE content (28). Tg Glo1 mice, which express murine Glo1, or WT controls were rendered diabetic (29). Five days posttransplantation, significantly lower mean lesion area was observed in the diabetic Tg Glo1 mice versus either the diabetic donor Ldlr—/— mice or the diabetic WT recipient mice; P < 0.0001 (Figure 2A). CD68 staining revealed significant reduction in macrophage content in the Tg Glo1 diabetic mice versus the donor Ldlr—/— mice or the WT recipient animals; P < 0.0001 (Figure 2B). Neutral lipids were significantly lower in the aortas of Tg Glo1 versus the diabetic donor Ldlr—/— mice or the diabetic WT recipient mice; P < 0.0001 (Figure 2C). Content of collagen was significantly higher in the diabetic Tg Glo1 recipient mice versus the diabetic donor Ldlr—/— mice or the diabetic WT recipient mice; P < 0.0001 (Figure 2D). The AGE content in the transplanted atherosclerotic plaques in the diabetic Tg Glo1 recipient mice was significantly lower than that in the aortic arches of the diabetic donor Ldlr—/— or the diabetic WT recipient mice; P < 0.0001 (Figure 3B). Per Table 1, compared with donor Ldlr—/— mice, Tg Glo1 mice weighed significantly less. No differences in levels of blood glucose were observed. Levels of total cholesterol were significantly lower in the Tg Glo1 recipient mice versus the donor Ldlr—/— mice and the WT recipient mice, and the total cholesterol levels in the Tg Glo1 mice correlated with atherosclerotic lesion area (Table 1 and Table 2). The levels of triglycerides were lower in the recipient Tg Glo1 and WT mice versus the donor Ldlr—/— mice (Table 1). Collectively, these data indicate that the reduction of RAGE ligand AGEs accelerated regression in diabetic atherosclerotic plaques.
To identify if RAGE signaling impaired regression of diabetic atherosclerosis, we postulated a role for the cytoplasmic domain binding partner of RAGE, DIAPH1 (30). Deletion of macrophage Diaph1 reduces the upregulation of Egr1 mRNA and activity in hypoxia (31). We rendered mice devoid of Diaph1 and WT mice diabetic and probed regression of atherosclerosis. Five days after transplantation, compared with donor Ldlr—/— mice and WT diabetic recipient mice, the recipient diabetic mice devoid of Diaph1 displayed significantly lower atherosclerotic lesion area, P < 0.0001; significantly lower CD68 macrophage content, P < 0.0001; significantly lower Oil red O staining, P < 0.01; and significantly higher collagen content; P < 0.0001 (Figure 2, A–D). Compared with the diabetic donor Ldlr—/— mice or diabetic WT recipient mice, the aortas of diabetic Diaph1—/— mice displayed significantly lower AGE content; P < 0.0001 (Figure 3B). Dihydroethidium (DHE) staining, a marker for oxidative stress, was significantly lower in the Ager—/— and Diaph1—/— mice compared with donor Ldlr—/— mice; P < 0.0001 (Figure 3C). Compared with the diabetic recipient WT mice, the regressing plaques of Ager—/— or Diaph1—/— recipient diabetic mice displayed significantly lower DHE staining; P < 0.05 and P < 0.001, respectively. Per Table 1, the body weight of the Diaph1—/— mice was significantly lower than that of the donor Ldlr—/— mice but was not different from the WT recipient mice. Levels of blood glucose did not differ. Levels of cholesterol in the recipient Diaph1—/— mice were significantly lower than those in the donor Ldlr—/— mice or the WT recipient mice, but there was no correlation between atherosclerotic lesion area and cholesterol levels in Diaph1—/— mice. Levels of triglyceride were lower than those in the donor mice but did not differ from the levels in WT recipient mice (Table 1 and Table 2). Thus, the RAGE/DIAPH1 signaling axis contributed to impaired regression of diabetic atherosclerosis.
Effects of RAGE on macrophage properties in the regression of diabetic atherosclerosis.
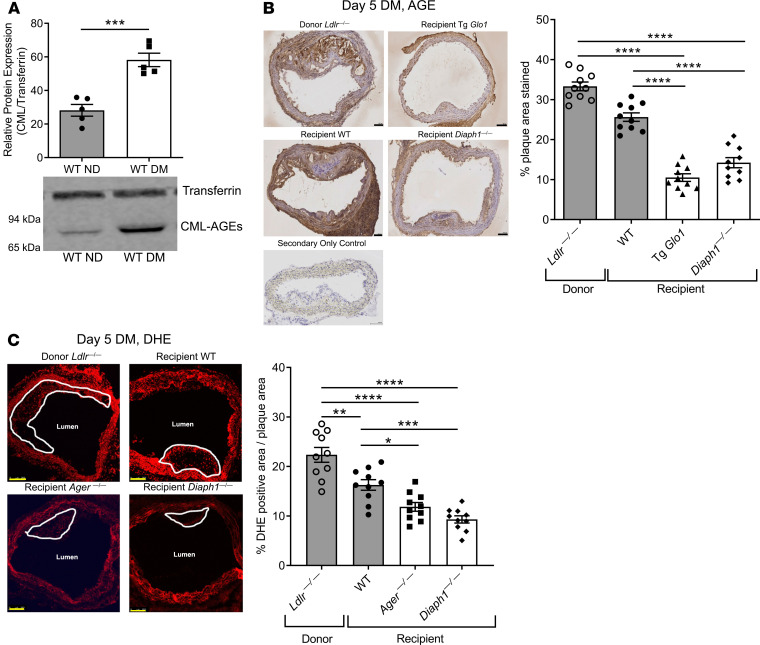
To probe the mechanisms by which RAGE affected macrophage properties, we assessed macrophage proliferation and apoptosis in the regressing diabetic plaques. Compared with diabetic WT recipient mice, Ki67+ (proliferation) immunoreactivity in transplanted aortic arches was significantly lower in the CD68+ macrophages in the diabetic Ager—/— recipient mice; P < 0.01 (Figure 4A). TUNEL staining revealed a significant reduction in TUNEL+/CD68+-expressing macrophages in the aortic arch lesions of the diabetic Ager—/— recipient versus WT mice; P < 0.0001 (Figure 4B).
Figure 4. Effects of RAGE and DIAPH1 on macrophage properties.
(A and B) Ki67 (A) and TUNEL (B) staining was performed on aorta sections from diabetic donor Ldlr—/— mice and WT, Ager—/—, and Diaph1—/— diabetic recipient mice. Scale bar: 250 μm. N = 5 mice/group. (C) Schematic of bead assay for tracking monocyte/macrophages in atherosclerotic plaques. (D) Representative images and quantification of the beads per lesion area from sections from nontransplanted diabetic Ldlr—/— and diabetic WT, Ager—/—, and Diaph1—/— recipient mice 5 days after aortic transplantation. Scale bars: 250 μm, and inset, 50 μm. In D, no-bead control is shown (N = 10 mice/group). (E) Role of RAGE and DIAPH1 in reverse transendothelial migration of macrophages. BMDMs from WT, Ager—/—, and Diaph1—/— diabetic mice were subjected to reverse transendothelial migration assays at 2, 24, and 48 hours. N = 6 mice/group. (F) Gene expression of selected pro- (Nos2 and Tnfa) and antiinflammatory genes (Arg1 and Il10) in BMDMs retrieved from BMDM-MAEC cocultures after transendothelial migration assay at 48 hours. N = 4 mice/group. Mean ± SEM. Unpaired t test or Mann-Whitney U test (A, B, and F) depending on the normality of the data. Kruskal-Wallis test followed by Dunn’s multiple comparisons test (D). One-way ANOVA with post hoc Tukey’s test (E). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To probe in vivo macrophage emigration, (Figure 4C), diabetic Ldlr—/— mice were fed a Western diet for 16 weeks. Eight days before retrieval of aortic arches for transplantation, fluorescently labeled, nondegradable beads were injected into the donor diabetic Ldlr—/— mice via tail vein to label the circulating monocytes (32). The aortic arches of these donor mice were then transplanted into diabetic WT, Ager—/—, or Diaph1—/— mice, and the transplanted arches were harvested 5 days posttransplantation. There were no significant differences in fluorescently labeled bead content in the transplanted aortic arch lesions between the diabetic donor Ldlr—/— mice and the diabetic WT recipient mice. The aortic arch lesional bead content was significantly lower in the diabetic Ager—/— recipient mice and the diabetic Diaph1—/— mice compared with the diabetic donor Ldlr—/— mice (P < 0.001, and P < 0.0001, respectively), when compared with the donor pretransplantation state (Figure 4D). Thus, deletion of Ager or Diaph1 facilitated macrophage emigration from the regressing diabetic plaques.
We tested these concepts in vitro in a reverse transendothelial migration assay in which murine bone marrow–derived macrophage (BMDM) transmigration and reverse transmigration were measured in primary WT murine aortic endothelial cells (MAECs) (Supplemental Figure 3A) (33). WT MAECs were incubated with BMDMs from diabetic WT, Ager—/—, or Diaph1—/— mice. At 2, 24, and 48 hours of incubation, BMDMs devoid of Ager or of Diaph1 displayed significantly higher reverse transmigration compared with WT BMDMs in the coculture with WT MAECs; P < 0.05 (Figure 4E). Thus, deletion of Ager or Diaph1 in BMDMs enhanced reverse endothelial transmigration and supported the conclusions of in vivo bead labeling macrophage studies (Figure 4D).
To test markers of inflammation in the reverse-transmigrated macrophages, we used accutase to remove the adhered reverse-transmigrated macrophages (Supplemental Figure 3B) (34) and examined expression of M1-like macrophage markers, Nos2 and Tnfa, and M2-like macrophage markers, Arg1 and Il10. Compared with WT BMDMs from diabetic mice, reverse-transmigrated BMDMs from Ager—/— diabetic mice displayed significant downregulation of Tnfa (P < 0.05), with trends toward reduced Nos2; P = 0.05 (Figure 4F). In the case of M2 markers, BMDMs from diabetic Ager—/— mice displayed significantly higher expression of Arg1 and Il10 when compared with diabetic WT reverse-transmigrated macrophages; P < 0.05 (Figure 4F).
Effect of RAGE on donor versus recipient-derived plaque macrophage content posttransplantation.
We distinguished the source of the transplanted aortic arch atherosclerotic lesional macrophages, donor versus recipient, and probed their transcriptomes (Supplemental Figure 4A). Donor Ldlr—/— mice (CD45.2) were rendered diabetic and fed a Western diet, and the aortic arches were transplanted into diabetic WT or Ager—/— recipient mice (CD45.1) and were retrieved 3 days posttransplantation.
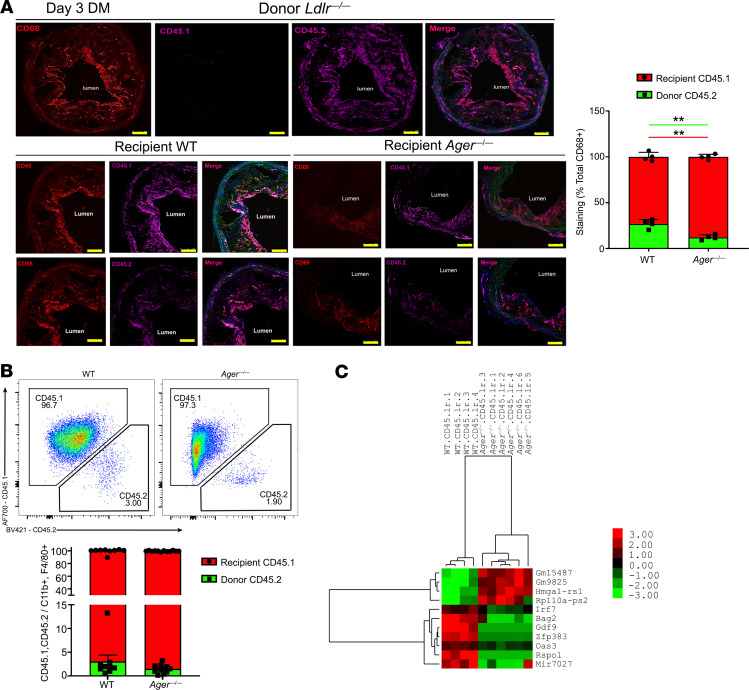
We determined the relative contribution of donor versus recipient macrophages in the transplanted aortic arches with respect to recipient genotype. By immunohistochemistry, 3 days after transplantation, in the WT diabetic recipient mice, the mean recipient CD45.1 macrophage lesional content was 73.1% ± 4.8%, and the mean donor CD45.2 macrophage lesional content was 26.9% ± 4.8%. In the diabetic Ager—/— recipient mice, the mean CD45.1 macrophage lesional content was 87.7% ± 2.8%, and the mean CD45.2 macrophage lesional content was 12.3% ± 1.8%. These data indicated a significantly higher content of recipient CD45.1/CD68+ macrophages and a small but significantly lower content of donor CD45.2/CD68+ macrophages in the diabetic Ager—/— versus WT diabetic recipient lesions; P < 0.01 (Figure 5A). By flow cytometry, although there were no significant differences in the percent recipient CD45.1 versus donor CD45.2 of the CD11B+/F4/80+ macrophage lesional content by recipient genotype, there were trends to higher recipient CD45.1/CD68+ macrophage content in the aortic arch lesions transplanted into the Ager—/— versus the WT diabetic recipient mice (Figure 5B). These findings, consistent with previous reports, indicated that the majority of the aortic arch lesional macrophages in regression are accounted for by newly recruited cells from the recipient mice (18).
Figure 5. Effect of RAGE on the transcriptomes of donor versus recipient macrophages after aortic arch transplantation.
(A) Staining and quantification of CD45.1 and CD45.2 macrophages in aortic grafts. Aortic arches from diabetic Ldlr—/— (CD45.2) donor mice fed Western diet for 16 weeks were transplanted into the diabetic WT or Ager—/— (CD45.1) recipients. Lesions were examined at day 3 posttransplantation (N = 4 mice/group). Scale bar: 250 μm. (B) Flow cytometry plots and quantification of diabetic WT and Ager—/— CD11B+/F4/80+ macrophages from recipient CD45.1 or donor CD45.2 mice (N = 9 WT mice/group; N = 12 Ager—/— mice/group). (C) Hierarchical clustering of differentially expressed genes in recipient macrophages from Ager—/— versus WT diabetic recipient mice (N = 4 WT mice/group and N = 6 Ager—/— mice/group). Mean ± SEM. Unpaired t test and Mann-Whitney U test were performed, respectively, in A and B. **P < 0.01.
RNA-sequencing implicates RAGE-dependent interferon signaling and IRF7 in impaired regression of diabetic atherosclerosis.
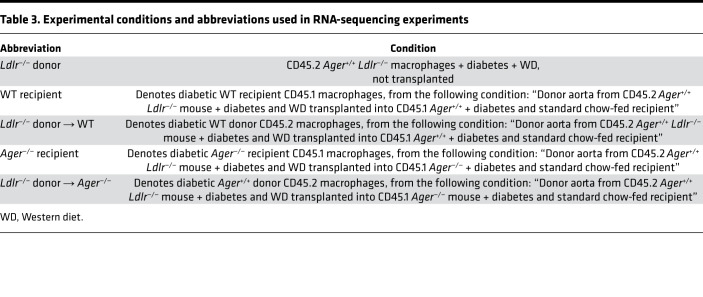
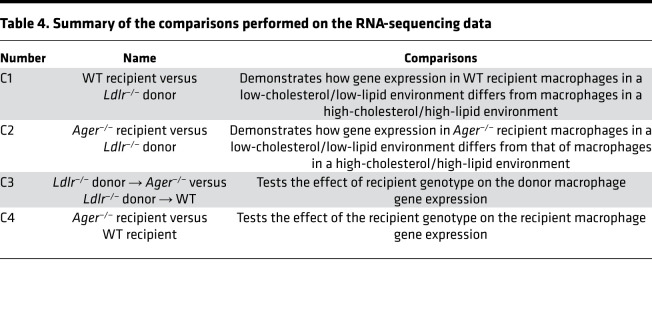
We thus reasoned that the deletion of Ager affected key properties of macrophages that facilitated lesion regression. We performed a transcriptomic analysis of donor versus recipient lesional macrophages sorted from the aortas of recipient mice 3 days after transplantation. Our sorting strategy (CD11B+, F4/80+ CD45.1 or CD45.2) allowed us to examine the transcriptome of donor- versus recipient-derived aortic arch lesional macrophages (Supplemental Figure 4, A–C). Table 3 and Table 4 detail the experimental conditions and the nomenclature used in our RNA-sequencing studies and summarize the 4 major comparisons performed on the RNA-sequencing data in which we consider the transcriptomes of the donor versus the recipient-derived macrophages.
Table 3. Experimental conditions and abbreviations used in RNA-sequencing experiments.
Table 4. Summary of the comparisons performed on the RNA-sequencing data.
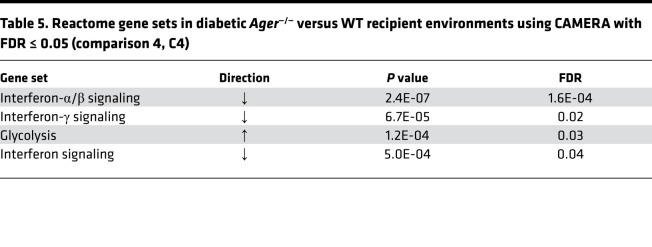
We used Correlation Adjusted Mean Rank Gene Set Test (CAMERA) (35) to compare the number of differentially expressed Reactome gene sets in each comparison (Supplemental Table 2) using FDR 0.05. Table 5 displays the effect of Ager deletion versus the WT recipient genotype on the recipient CD45.1+ macrophages populating the transplanted aortic arch lesions (comparison 4, Supplemental Table 2). Four gene sets were identified, 3 of which related to the downregulation of the interferon signaling pathway (α, β, and γ), and the fourth gene set reflected an increase in glycolysis when comparing Ager—/— versus the WT.
Table 5. Reactome gene sets in diabetic Ager—/— versus WT recipient environments using CAMERA with FDR ≤ 0.05 (comparison 4, C4).
Subsequently, we analyzed differential gene expression. Supplemental Table 6 indicates the number of differentially expressed genes for each of the 4 comparisons using 2 criteria: first, a stringent cutoff with FDR ≤ 0.05, and second, a lenient cutoff with P ≤ 0.001 and FDR ≤ 0.2. No genes were differentially expressed in the donor macrophages (Supplemental Table 6, comparison 3); however, in the recipient macrophages 4 genes were differentially expressed by genotype using the stringent criterion, but 11 genes were differentially expressed by recipient genotype by the lenient cutoff (Figure 5C and Supplemental Table 9). Consistent with the downregulation of the interferon signaling pathways, Irf7 was downregulated in the Ager—/— versus WT diabetic recipient CD45.1 macrophages populating the lesions after transplantation of the atherosclerotic aortic arches.
We focused our attention to the interferon signaling pathway and to Irf7 (Figure 5C), which has not been previously linked to RAGE signaling to our knowledge.
IRF7 is expressed in mouse and human atherosclerosis, is regulated by RAGE ligands, and affects macrophage inflammation and cholesterol metabolism.
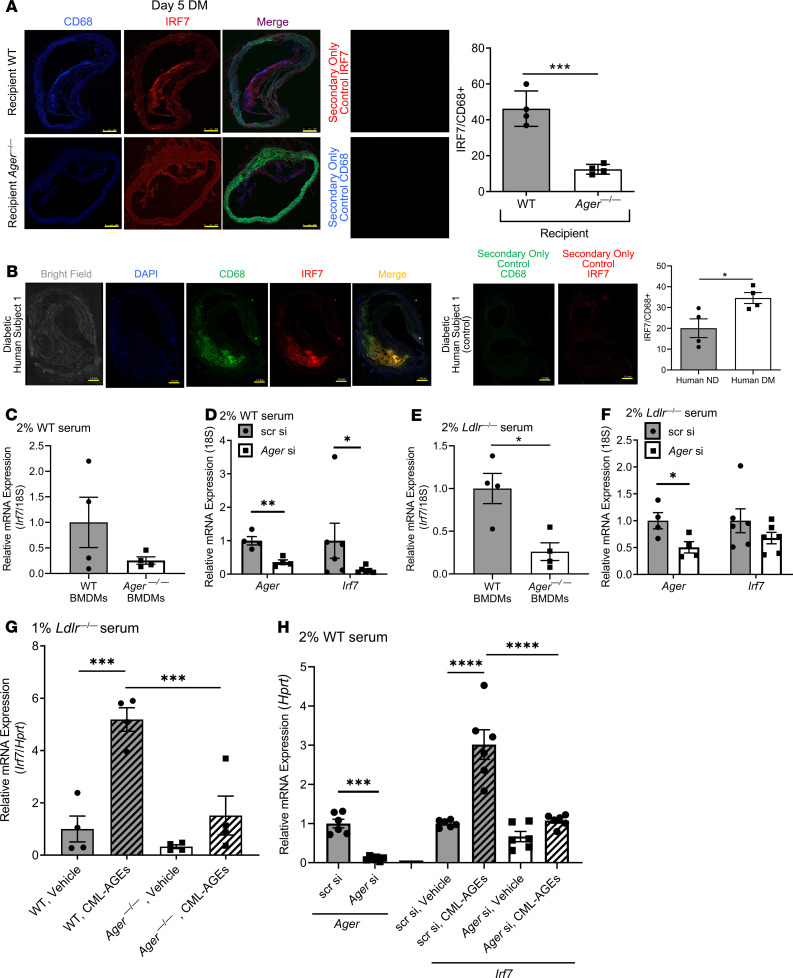
IRF7 colocalized with CD68 in the WT and Ager—/— recipient mice lesions, but the percentage of IRF7 and CD68+ (i.e., double positive) macrophages per lesion area was significantly lower in the Ager—/— versus WT diabetic lesions; P < 0.001 (Figure 6A). IRF7 colocalized with CD68 in human diabetic coronary artery atherosclerotic lesions (Figure 6B, Supplemental Figure 5). Although IRF7 colocalizes with CD68 in nondiabetic atherosclerotic plaques (Supplemental Figure 6), this was significantly lower compared with that in the diabetic subjects; P < 0.05 (Figure 6B).
Figure 6. RAGE regulates the expression of IRF7 in macrophages.
(A) IRF7 and CD68 colocalization in diabetic Ager—/— and WT recipient mice atherosclerotic plaques at day 5 after aortic arch transplantation from Ldlr—/— diabetic mice (N = 4 mice/group). Scale bar: 250 μm. (B) IRF7 colocalizes with CD68 in diabetic human atherosclerotic plaques. Representative image of human coronary atherosclerotic lesions is shown with additional human subject data shown in Supplemental Figure 5 (diabetic subjects) and Supplemental Figure 6 (nondiabetic subjects). Scale bar: 1 mm. Quantification of immunohistochemical staining of human plaque area colocalization of CD68/IRF7 area over total CD68+ staining area calculated as a percentage. (C) Irf7 gene expression in Ager—/— and WT BMDMs exposed to a normolipidemic environment (2% serum from C57BL/6 mice fed a normal chow diet) for 48 hours (2% serum of C57BL/6 mice fed a normal chow diet). N = 4 independent mice/group. (D) Irf7 gene expression in WT BMDMs after 48 hours of Ager or scrambled control knockdown (75 nM) in a normolipidemic environment. Knockdown of Ager by siRNA is shown. N = 6 independent mice/group. (E) Irf7 gene expression in Ager—/— and WT BMDMs exposed to a hyperlipidemic environment for 48 hours (2% serum of Ldlr—/— mice fed a Western diet). N = 4 mice/group. (F) Irf7 gene expression levels in WT BMDMs after 48 hours of Ager or scrambled (75 nM) in a hyperlipidemic environment. Knockdown of Ager by siRNA is shown. N = 6 mice/group. (G) Irf7 gene expression in WT and Ager—/— BMDMs grown in the presence of 1% serum of Ldlr—/— mice fed Western diet for 48 hours, followed by an additional 16 hours in CML-AGE (100 μg/mL) versus vehicle. N = 4 mice/group. (H) Irf7 gene expression in WT BMDMs after 48 hours of Ager or scrambled (75 nM) in a normolipidemic environment in cells treated with CML-AGE (200 μg/mL) or vehicle for 16 hours. Knockdown of Ager is shown. N = 6 mice/group. Secondary antibody alone controls are shown in A and B. Mean ± SEM. Unpaired t test or Mann-Whitney U test was performed to assess the differences in A–F and H (for Ager endpoint) depending on the normality of data. One-way ANOVA with post hoc Holm-Šídák multiple comparisons test was used in G and H (for Irf7 endpoints). *P < 0.05, ***P < 0.001, and ****P < 0.0001.
We tested the relationship between RAGE and IRF7 in vitro. BMDMs isolated from Ager—/— versus WT mice and exposed to serum (2%) from WT C57BL/6 mice displayed trends to lower Irf7 mRNA transcripts (Figure 6C). Ager siRNA knockdown in WT murine BMDMs resulted in significant reduction in Irf7 mRNA transcripts compared with BMDMs exposed to scrambled siRNA; P < 0.05 (Figure 6D).
To mimic the conditions relevant in atherosclerosis, we retrieved serum from Western diet–fed Ldlr—/— mice (36), which is enriched in multiple damage-associated molecular patterns, many of which are RAGE ligands (37, 38). BMDMs retrieved from Ager—/— mice grown in this serum displayed significantly lower levels of Irf7 mRNA versus WT BMDMs; P < 0.05 (Figure 6E). Upon Ager siRNA knockdown in BMDMs, modest reductions in Irf7 mRNA transcripts were observed compared with scrambled siRNA (Figure 6F).
We assessed the effects of a prototypic AGE ligand of RAGE, CML-AGE (39) on the regulation of Irf7 using CML-AGE prepared from human serum albumin (39). WT BMDMs treated with CML-AGE (100 μg/mL) versus vehicle demonstrated a 5-fold increase in Irf7 mRNA; P < 0.001 (Figure 6G). However, in BMDMs from Ager—/— mice exposed to CML-AGE, there was no significant increase in Irf7 versus vehicle (Figure 6G). Compared with WT BMDMs exposed to CML-AGE, the increase in Irf7 mRNA transcripts was significantly higher than that observed in CML-AGE–treated Ager—/— BMDMs; P < 0.001 (Figure 6G). Thus, RAGE ligands directly increase Irf7 mRNA.
We tested if Ager knockdown would prevent CML-AGE–stimulated upregulation of Irf7 mRNA. In the presence of scrambled siRNA and CML-AGE, a 3-fold increase in Irf7 mRNA was observed compared with vehicle; P < 0.0001 (Figure 6H). When BMDMs were treated with CML-AGE in the presence of Ager siRNA, there was no significant increase in Irf7 versus vehicle (Figure 6H). Irf7 mRNA transcripts were significantly higher in CML-AGE–treated BMDMs in the presence of scrambled siRNA versus Ager siRNAs; P < 0.0001 (Figure 6H).
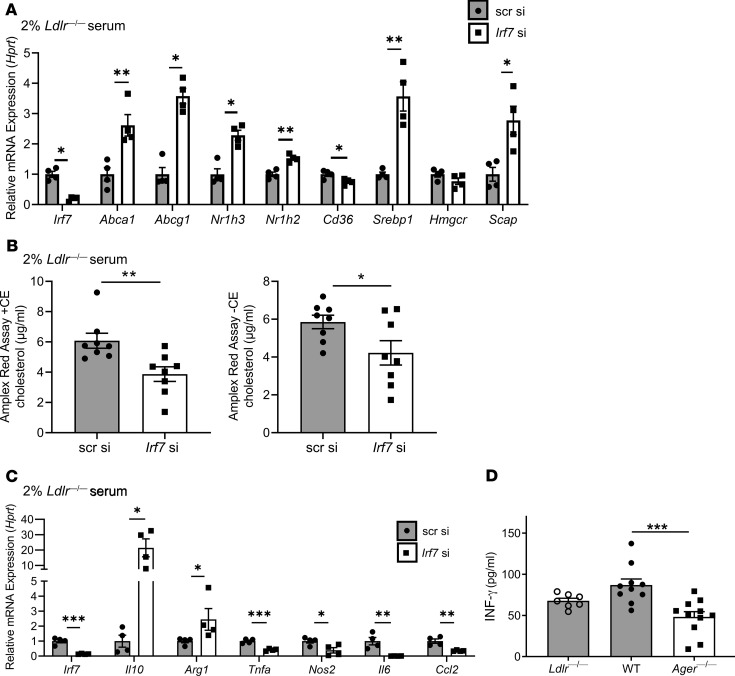
We addressed the effects of IRF7 on cholesterol metabolism and inflammation in BMDMs. Compared with scrambled siRNA, Irf7 knockdown in 2% serum from Western diet–fed Ldlr—/— mice resulted in significant upregulation of genes linked to cholesterol efflux (Abca1 and Abcg1), increased Nr1h2 (LXRβ) and increased Nr1h3 (LXRα), and increased Srebp1 and Scap; P < 0.05 (Figure 7A). Significant reductions in Cd36 (P < 0.05) and no differences in Hmgcr were observed in Irf7-knockdown versus scramble siRNA–treated cells (Figure 7A). In BMDMs exposed to Western diet–fed Ldlr—/— mice serum, Irf7 knockdown resulted in significantly lower levels of cholesterol (Figure 7B). We assessed the effects of Irf7 on BMDM inflammation. Compared with scrambled siRNA, Irf7 knockdown resulted in significant upregulation of Arg1 and Il10 (P < 0.05) and significant reduction in Tnfa, Nos2, Il6, and Ccl2; P < 0.001, P < 0.05, P < 0.01, and P < 0.01, respectively (Figure 7C).
Figure 7. IRF7 regulates macrophage cholesterol content and inflammation.
(A) The expression level of genes involved in cholesterol metabolism after 48 hours of Irf7 or scrambled (75 nM) in WT BMDMs in the presence of a hyperlipidemic environment (2% serum of Ldlr—/— mouse fed a Western diet). N = 4 mice/group. (B) Cellular total cholesterol (left) and free cholesterol (right) content measured using the Amplex Red Cholesterol Assay Kit containing cholesterol esterase or lacking cholesterol esterase (CE), respectively, in WT BMDMs after 48 hours of Irf7 or scrambled (75 nM), in the presence of a hyperlipidemic environment. N = 8 mice/group. (C) The expression level of genes involved in inflammatory processes after 48 hours of Irf7 or scrambled (75 nM) in WT BMDMs in the presence of a hyperlipidemic environment. N = 4 mice/group. (D) Plasma IFN-γ levels were measured by ELISA in diabetic Ldlr—/— donor and WT or Ager—/— diabetic recipient mice 5 days after transplantation. Ldlr—/— diabetic donors (N = 7), WT diabetic recipients (N = 10), and Ager—/— diabetic recipients (N = 11). Mean ± SEM. Unpaired t test or Mann-Whitney U test was performed to assess the difference in A–C depending on the normality of data. One-way ANOVA with post hoc Holm-Šídák multiple comparisons test was used in D. *P < 0.05, **P < 0.01, and ***P < 0.001.
Finally, we tested the levels of IFN-γ in the plasma of diabetic Ldlr—/— mice donor mice and in the diabetic recipient WT or Ager—/— mice. Compared with recipient WT mice, recipient mice devoid of Ager displayed a significant reduction in IFN-γ; P < 0.001 (Figure 7D).
Discussion
Collectively, this work implicates RAGE in impaired regression of atherosclerosis in both diabetic and nondiabetic mice. Although previous studies underscored roles for RAGE in atherosclerosis progression (20-24), here, we demonstrate the maladaptive roles of AGEs, RAGE, and DIAPH1 in impairment of diabetic atherosclerosis regression.
Our studies suggested that RAGE exerted multiple effects on macrophage properties in the regression environment. First, macrophage proliferation, identified as an important contributor to atherosclerosis (40), was significantly reduced in the plaques of Ager—/— versus WT diabetic recipient mice (Figure 4A). Second, TUNEL+ macrophages were significantly lower in the absence of Ager, thereby suggesting that enhanced macrophage survival was associated with more functionally competent macrophages (Figure 4B). Third, macrophage emigration from the regressing plaques was increased in an Ager—/— environment. Furthermore, gene signatures in the reverse-transmigrated macrophages in an in vitro model suggested that Ager deletion reduced proinflammatory markers and upregulated antiinflammatory markers (18).
RNA-sequencing studies revealed that the recipient, not donor-derived, macrophages evoked the major RAGE-dependent pathways in regression (Figure 4C), and RAGE-dependent modulation of Irf7 was pinpointed as a leading candidate gene. In mammals, the 9 IRF family members are transcription factors that regulate type 1 interferon antiviral responses and immune cell differentiation (41, 42). The functions of these molecules are more complex, as IRFs are expressed in myeloid and lymphoid cells and in nonimmune cells, such as vascular cells (43–46) and adipocytes (47, 48).
Although IRF7 has not been tested in atherosclerosis, other members of the IRF family have been studied in murine atherosclerosis progression. Global deletion of Irf1 reduced foam cell formation and atherosclerosis in Apoe—/— mice (49). Global deletion of Irf5 reduced lesion and necrotic core areas in mice devoid of Apoe (50). Dendritic cell deletion of Irf8 significantly reduced atherosclerosis in Ldlr—/— mice, with reduced T cell priming, activation, and differentiation to T helper cells, with consequent marked reduction in B cell activation (51). In Apoe—/— mice, deletion of Irf3 resulted in significant reduction in atherosclerosis; the major effects of IRF3 on atherosclerosis were traced to endothelial cells (52). Neutralization of B cell–activating factor (BAFF) in mice with deficiency of either Apoe or Ldlr increased atherosclerosis, despite depletion of mature B-2 cells. A function of BAFF receptor signaling was the repression of macrophage IRF7-dependent production of proatherogenic CXCL10, which was blocked in the presence of the antagonist (53).
Smooth muscle cell IRF7 (54) plays protective roles through reduction of proliferation consequent to arterial injury (55). Our data indicate that Irf7 knockdown in BMDMs attenuated proinflammatory effects, as shown by the significant reduction of Tnfa, Nos2, Il6, and Ccl2 and upregulated antiinflammatory markers, Il10 and Arg1. In addition, the knockdown of Irf7 in BMDMs exposed to hyperlipidemic conditions resulted in upregulation of cholesterol transporters and downregulation of Cd36, factors favoring efflux of cholesterol. Upregulation of Nr1h3 and Nr1h2 suggests that knockdown of Irf7 in BMDMs imparts an antiatherogenic profile and downregulation of BMDM Irf7 reduced total cellular cholesterol in hyperlipidemic serum. Although global deletion of Irf7 reduced high fat diet–induced obesity, the proximate mechanisms were not elucidated (56). The present body of work adds RAGE-dependent regulation of IRF7 to this network of molecules bridging immune and cholesterol metabolism functions in macrophages, which may contribute to impaired regression of atherosclerosis.
Miao and colleagues exposed human THP1 monocytes to high glucose and showed that this upregulated IRF7 (57), which was analogous to an IRF7-driven inflammatory network that was linked to the risk of type 1 diabetes (58). High glucose upregulates ligands of RAGE. Mediating roles for AGEs were demonstrated in our studies by improved regression of diabetic atherosclerosis in transgenic mice overexpressing Glo1, the enzyme that detoxifies pre-AGE species. It is plausible that non-AGE ligands of RAGE contribute to upregulation of Irf7. Hyperlipidemic serum from Ldlr—/— mice contains TLR ligands, which also interact with RAGE. In vitro, CpG oligodeoxynucleotides and Complement C3a increased production of IFN-α in WT but not Ager—/— BMDMs. In human peripheral blood mononuclear cells, treatment with C3a and CpG oligodeoxynucleotides increased IFN-α, which was reduced by RAGE antagonism (59).
Finally, the regulation and biology of IRF7 bear similarities to those of RAGE. Akin to findings that RAGE ligands upregulate AGER, at least in part through NF-κB, and that RAGE activation perpetuates proinflammatory RAGE ligand generation (60), IRF7-mediated upregulation of IFNs stimulates IFN-mediated upregulation of Irf7 (61, 62). These considerations place RAGE and IRF7 at critical nodes in the mediation of chronic inflammation. Here, we show that RAGE impairs regression of atherosclerosis through perpetuation of inflammation and through impairment of cholesterol metabolism, at least in part through IRF7. Interruption of RAGE/IRF7 may restore reparative mechanisms and accelerate regression of atherosclerosis, particularly in diabetes.
Methods
Animal studies and induction of diabetes.
Male donor mice with atherosclerosis were Ldlr—/—. The recipient male mice, Ager—/— mice (C57BL/6J) (63), Diaph1—/— mice (C57BL/6J) (30), and Tg mice expressing murine Glo1 (C57BL/6J) (29), were bred in-house, and WT C57BL/6J mice were purchased from The Jackson Laboratory. Mice were housed in a specific pathogen–free facility. Certain mice were rendered type 1 diabetic by intraperitoneal injections of streptozotocin (STZ; MilliporeSigma, S0130) at 7 weeks of age. STZ was freshly dissolved in citrate buffer (0.1 mol/L, pH 4.5) and administered at a dose of 55 mg/kg/d, once daily for 5 days. Blood glucose (>250 mg/dL) was measured (Free Style Freedom Lite glucometer) to ensure diabetes was induced. Nondiabetic animals were treated with equal volumes of citrate buffer. Donor Ldlr—/— mice were fed a Western diet (Research Diets, Inc., D01061401Ci; 0.15% cholesterol) for 16 weeks. Recipient mice fed a chow diet were diabetic or control for 3 months before surgery. To test mechanisms of atherosclerosis regression, aortic arch transplantation was performed at 23 weeks of age (8, 9, 18, 25, 64, 65). Buprenorphine was administered by subcutaneous injections every 12 hours for 3 days postsurgery. At sacrifice, whole blood was collected from the aorta. For serum isolation, whole blood was allowed to clot in BD Microtainer SST (365967) and isolated by centrifugation. For plasma isolation, whole blood was collected with EDTA (Thermo Fisher Scientific, BP2482) and isolated by centrifugation.
En face aorta staining.
The full descending aorta was dissected, excised, and pinned (Thermo Fisher Scientific, NC9681411); fixed in 4% formaldehyde for 5 minutes; rinsed in 70% ethanol for 5 minutes; and stained in a working solution of 2.5 g of Sudan IV (Merck Eurolab), 250 mL of 70% ethanol, and 250 mL of acetone filtered through Whatman paper before use. Sudan IV solution was placed on top of the aorta for 10 minutes. This was rinsed twice in 80% ethanol for 3 minutes.
Histological stains.
The pretransplant donor and grafted aortic arches were removed after perfusion of cold PBS, embedded in optimal temperature cutting compound (OCT), and frozen. Murine aortic frozen sections (6 μm thick) were air-dried before the following procedures.
H&E, Oil red O, and Picrosirius red.
Sections were fixed with 10% neutral buffered formalin for 10 minutes at room temperature (RT). H&E staining was performed (66); sections were dehydrated in ethanol, followed by cleaning in xylene and mounting with xylene-based permount (Thermo Fisher Scientific, 15820100). For Oil red O (ORO), sections were incubated with propylene glycol and with preheated ORO solution (American MasterTech, STOROPT) at 60°C. Sections were placed in 85% propylene glycol for 1 minute and counterstained with modified Mayer’s hematoxylin (American MasterTech, HXMMHLT) for 1 minute, rinsed, and mounted with glycerine jelly (Thermo Fisher Scientific, NC0301797). Picrosirius red (PSR) staining was performed using Picrosirius red (Polysciences Inc., 24901-500). Sections were cleaned in xylene and mounted with xylene-based permount (Thermo Fisher Scientific, 15820100).
Dihydroethidium.
Slides were placed into PBS for 10 minutes at RT. In a dark, moist chamber at 37°C, slides were incubated with 10 μM DHE (MilliporeSigma, D7008) for 30 minutes. Hoechst 33258 dye (MilliporeSigma, 23491-45-4) was used to visualize the nuclei. Unstained aortas preincubated with superoxide dismutase–polyethylene glycol from bovine erythrocytes 500 units/mg (MilliporeSigma, S9549) were used for background correction and negative controls.
In situ apoptosis detection (TUNEL assay).
The in situ detection of apoptotic plaque macrophages was performed using the Click-it Plus TUNEL Assay (Invitrogen, Thermo Fisher Scientific, C10619). Atherosclerotic plaque sections were fixed in 4% paraformaldehyde. EdUTP nucleotide was incorporated into dsDNA strand breaks by TdT enzyme and an Alexa Fluor (AF) 647 picolyl azide dye. Sections were counterstained with rat anti-mouse CD68 IgG (Bio-Rad, MCA1957, 1:200 dilution) and secondary donkey anti-rat AF405 IgG (Abcam, ab175670, 1:100 dilution). Sections incubated with labeling solution without terminal transferase were negative controls.
Detection of AGEs.
Sections were fixed in 10% formalin for 15 minutes. Immunohistochemistry was performed using polyclonal rabbit anti-AGE IgG (Abcam, ab23722, 1:200 dilution). AGE epitope was detected with biotinylated horse anti-rabbit (Vector Laboratories, catalog BA-1000, 1:1000 dilution) followed by the application of streptavidin-HRP conjugate. The complex was visualized with 3,3’-Diaminobenzidine and enhanced with copper sulfate. Slides were counterstained with hematoxylin and mounted with permanent medium. Ventana Discovery XT kit (Ventana Medical Systems, 750-701) was used for detection.
Detection of CD68.
Frozen sections were fixed in acetone for 15 minutes. Immunohistochemistry was performed using rat anti-mouse CD68 IgG clone FA-11 (AbD Serotech, MCA1957, 1:1000 dilution). Antibody incubation and detection was performed on a Ventana Discovery XT (Ventana Medical Systems, 750-701) using Ventana’s reagent buffer and detection kits. Slides were counterstained with hematoxylin, air-dried, and heated for 15 minutes at 60°C before mounting with permanent medium.
Detection of RAGE.
Sections were blocked for 1 hour at 20°C with Odyssey Blocking Buffer (LI-COR, 927-40100). Immunofluorescence was performed using goat anti-mouse RAGE IgG (Genetex, GTX27764, 1:150 dilution). RAGE was detected with mouse-adsorbed, biotinylated rabbit anti-rat IgG (Vector Laboratories, Ab150129, 1:1000 dilution). Slides were mounted with gold antifade permanent medium with DAPI.
Detection of IRF7/CD68 in mouse and human sections.
For mouse IRF7, sections were blocked in 10% goat serum. All antibodies were diluted in antibody diluent (Dako, S3022). Rabbit polyclonal anti-mouse IRF7 IgG (Abcam, ab62505) and rat anti-mouse CD68 IgG (Bio-Rad, MCA1957) were diluted 1:200 and incubated overnight at 4°C. Secondary antibodies goat anti-rabbit AF594 IgG (Invitrogen, Thermo Fisher Scientific, A11012) and donkey anti-rat AF405 IgG (Abcam, Ab175670) were diluted 1:100 and incubated 1 hour at RT.
For human IRF7, deidentified human coronary artery atherosclerotic specimens with advanced lesions were obtained from CVPath Institute Sudden Death Registry. The artery segments were fixed in formalin, and 2- to 3-mm segments were embedded in paraffin and cut (5 μm thick). The diagnosis of coronary artery disease and histopathological determination of coronary artery disease were performed by an experienced cardiac pathologist at CVPath Institute. Human sections were deparaffinized using Richard-Allan Scientific 40 Clear-Rite 3 (catalog 6905) and a series of 100%, 90%, 70% ethanol for 5 minutes each, followed by 3 washes. Samples were permeabilized for 10 minutes in 0.2% Triton X-100 in PBS. Sections were blocked for 1 hour in Dako Protein Block, Serum-Free (X0909). All antibodies were diluted in antibody diluent (Dako, S3022). Primary antibodies rabbit polyclonal anti-mouse IRF7 IgG (Abcam, ab62505) and mouse anti-human CD68 IgG (Dako, M0814) were diluted 1:200 and incubated overnight at 4°C. Secondary antibodies goat anti-rabbit AF594 IgG (A11012) and donkey anti-mouse AF488 IgG (A21202) (both Invitrogen, Thermo Fisher Scientific) were diluted 1:100 and incubated 1 hour at RT. Slides were mounted with DAPI.
Detection of CD45.1 versus CD45.2/CD68.
Aortic arches were perfused with PBS, embedded in OCT, and frozen. Sections were stained to detect CD45.1 (SouthernBiotech, 1795-08), CD45.2 (SouthernBiotech, 1800-08), and CD68 (Bio-Rad, MCA1957). Monocytes were stained with anti-CD68 IgG (AF594; red), and anti-CD45.2 IgG (AF647; purple) for donor cells or anti-CD45.1 IgG (AF647; purple) for recipient cells, and slides were mounted with DAPI.
Detection of Ki67/CD68.
Sections were blocked in 10% goat serum and incubated at RT. All antibodies were diluted in antibody diluent (S3022). Primary rabbit anti-mouse Ki67 IgG (Invitrogen, Thermo Fisher Scientific, PA5-19462) and rat anti-mouse CD68 IgG (MCA1957) were diluted 1:200 and incubated overnight at 4°C. Secondary antibodies goat anti-rabbit AF594 IgG (A11012) and donkey anti-rat AF405 IgG (Ab175670) were diluted 1:100 and incubated 1 hour at RT. Slides were mounted with DAPI.
Imaging and quantification.
Digital images were captured using Zeiss LSM 880 laser scanning confocal microscope (for DHE, CD45.1 versus CD45.2/CD68 and Ki67/CD68), Zeiss Axio Observer microscope (mouse and human IRF7/CD68), Zeiss Axioplan wide-field microscope (for PSR and TUNEL), Leica 5500B microscope (for RAGE+), or Leica SCN400 whole-slide scanner up to ×40 original magnification (H&E, ORO, AGEs, CD68+). Using Slidepath Digital Image Hub software (Leica Biosystems), 4 aorta section images/animal were quantified. Image-Pro Plus 7.0 software (Media Cybernetics) was used to determine CD68+, Oil red O+, H&E stain+, AGE+, DHE+, PSR+, RAGE+, and Sudan IV+ areas. Fiji (67) was used to determine CD45.1+ and CD45.2+/ CD68+, IRF7+/ CD68+ areas as well as to identify TUNEL+ and Ki67+/CD68+ in the plaques. All staining and immunohistochemistry and immunofluorescence analyses were performed by an observer blinded to the conditions.
Blood and plasma parameters.
Blood was harvested after a 6-hour fast. Total cholesterol (Infinity, Thermo Fisher Scientific, 948541) and triglyceride (Infinity, Thermo Fisher Scientific, TR22421) concentrations were measured.
Monocyte bead tracking.
Ly6Clo monocytes were labeled in vivo by tail vein injection of 250 μL beads diluted 1:4 in PBS (Fluoresbrite YG microspheres 1 μm, 17154-10, Polysciences Inc.) into Ldlr—/— donor mice before transplantation. (11, 68, 69). Quantification of the number of beads/cross section of the aortic arch at baseline and 5 days after arch transplantation into the diabetic recipients was performed.
Bone marrow–derived macrophages.
Primary BMDMs were isolated from 12-week-old mice (70). Cut-end femora were spun for 6000 g for 1 minute for collection of bone marrow. After addition of medium, single-cell suspensions were plated in bone marrow–conditioned media (71) for 5 days and subjected to experimental conditions.
Laser capture microdissection of CD68-expressing cells.
The aortic arch was cut in half, positioned vertically in OCT, and stored at –80°C for 24 hours under RNase-free conditions. Frozen sections (10 μm) were collected on Arcturus PEN membrane glass slides (Life Technologies, Thermo Fisher Scientific) and fixed with 70% ethanol and partially dehydrated in 70% ethanol followed by washing in 95% ethanol. Eosin Y (VWR Scientific) was used for staining, washed in 95% ethanol, and completely dehydrated in 100% ethanol and xylene (11). After air-drying, foam cells could be identified by light microscopy and were verified by CD68 staining of guiding slides. Atherosclerotic lesion CD68+ cells were laser-captured using Leica LMD6500 laser microdissection system.
Quantitative reverse transcription PCR experiments.
BMDM total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN, 74136). RNA from laser capture–microdissected CD68+ cells was isolated by the Arcturus PicoPure RNA Isolation Kit (Life Technologies, Thermo Fisher Scientific, 12204-01). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad, 1708891) and amplified using TaqMan assays (Supplemental Table 10) using a 7300 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific).
Reverse transendothelial migration assay.
Collagen gels were polymerized with collagen type I (BD 354231) before WT primary MAECs were isolated and seeded (72). Primary BMDMs (4 × 106 cells/mL) were added to confluent endothelial cultures for 2 hours to allow accumulation of macrophages in the subendothelial collagen. Cultures were rinsed to remove nonmigrated cells and incubation was continued. At intervals, cultures were rinsed to remove nonadherent macrophages that had accumulated in the apical compartment by reverse transmigration, and medium was replenished. Macrophages adherent to the apical surface of the endothelium were washed before evaluation of total DNA/well with Yo-Pro-1 (Molecular Probes, Thermo Fisher Scientific, Y3603) At each point, reverse transmigration was calculated as the percentage decrease in the number of macrophages beneath the endothelial monolayer compared with the number that initially accumulated below the endothelium at 2 hours baseline (33). At 48 hours, macrophages were eluted from the coculture with StemPro Accutase Cell Dissociation Reagent (Thermo Fisher Scientific, A1110501) (34). To digest collagen gels containing endothelial cell monolayer and non–reverse-transmigrated cells, 100 μL/well of collagenase D (2 mg/mL in DMEM) was added for 40 minutes at 37°C. The cell suspension was filtered through a cell strainer and subjected to reverse transcription PCR.
Western blot.
Total protein concentrations were quantified using Pierce BSA Protein Assay kit (Thermo Fisher Scientific, 23225). Plasma samples were subjected to electrophoresis (4%–15% SDS-PAGE gels), and specific CML-AGE signals were detected using mouse anti-CML (R&D Systems, Bio-Techne, MAB3247) and anti-transferrin I-20 (1:1000, Santa Cruz Biotechnology, SC-22597) control. Following the primary antibody incubation, blots were exposed to the secondary HRP-conjugated anti-mouse IgG (1:1000, R&D Systems, Bio-Techne, HAF007) and anti-goat IgG (1:1000, GE Healthcare UK Limited). Protein signals were visualized with the Odyssey Imaging System (LI-COR) detection system.
BMDM stimulation.
CML-Human Serum Albumin was synthesized as described (39). BMDMs were stimulated overnight with 100 μg/mL or 200 μg/mL of vehicle or CML-AGEs in normolipidemic (2% serum from C57BL/6J mouse fed chow) or hyperlipidemic conditions (1% or 2% serum from Ldlr—/— mice fed Western diet) conditions.
Ager and Irf7 knockdown.
BMDMs were serum-starved and placed in Opti-MEM Reduced Serum Medium (Gibco, Thermo Fisher Scientific, 31985062) for 2 hours. Cells were transfected using MISSION siRNA Transfection Reagent (MilliporeSigma, S1452) and 75 nM of scrambled siRNA (QIAGEN, 1022076), Ager #4 Flexitube siRNA (QIAGEN, SI001666229), or Irf7 #2 Flexitube siRNA (QIAGEN, SI0077790) for 48 hours in normolipidemic conditions or hyperlipidemic conditions.
Amplex Red assay.
BMDM intracellular cholesterol content was quantified using the Amplex Red Cholesterol Assay Kit (Invitrogen, Thermo Fisher Scientific, A12216) normalized to total protein levels.
ELISA.
Plasma samples were subjected to ELISA for detection of IFN-γ using the Mouse Interferon Gamma sandwich ELISA kit (LifeSpan Biosciences LS-F23028-1).
Flow cytometry.
For CD45.1/CD45.2 validation, whole blood was collected. For macrophage sorting, aortas were perfused with cold PBS, minced, and digested for 1 hour at 37°C with shaking in DMEM containing collagenase I, collagenase XI, hyaluronidase MilliporeSigma, C1639, C9697, H3506), and DNase I enzymes (QIAGEN, 79254) (73, 74), filtered through 100-μM filters (Thermo Fisher Scientific, 22363549) and pelleted by centrifugation. Red blood cells were lysed in RBC Lysis Buffer (eBioscience, Thermo Fisher Scientific, 00-4333-57) at RT, pelleted by centrifugation, and washed in FACS buffer (PBS containing 2 mM EDTA and 0.2% BSA). Single-cell suspensions were blocked with Fc block (1:100, BioLegend, 101302) for 10 minutes at 4°C and labeled with antibodies for 30 minutes at 4°C. For CD45.1/CD45.2 validation, samples were stained with AF700 CD45.1 (1:100, A20, BioLegend, 110724) and BV421 CD45.2 (1:100, 104, BioLegend, 109832). Flow cytometry analyses were performed on FACSCalibur (BD Biosciences). For aortic macrophage sorting, samples were stained with PE-Cy7 F4/80 (1:40, BM8, BioLegend, 123114), APC-Cy7 CD11B (1:100, M1/70, BD Pharmingen, 557657), and the previous AF700 CD45.1 and BV421 CD45.2. Singlets and DAPI– (0.025 g/mL, Invitrogen, Thermo Fisher Scientific, D1306) live cells, F4/80+, CD11B+, and either CD45.1 or CD45.2 were sorted into RLT buffer on a FACSAria II sorter (BD Biosciences) with a 100-μm nozzle. RNA was extracted with RNeasy Plus Micro Kit (QIAGEN, 74004), and only RNA integrity number ≥ 7 were used (RNA 6000 Pico Kit in 2100 Bioanalyzer, Agilent). Data were analyzed using FlowJo software.
RNA-sequencing and bioinformatics.
cDNA samples were prepared using the NuGEN Ovation RNA-Seq system v2 reagents and the Ovation Ultralow DR Multiplex system for the adapter ligation step. Libraries were amplified by 9 cycles. Single-end, 50-nucleotide reads were sequenced on an Illumina 2500 HiSeq using v4 chemistry. FASTQ files were trimmed of low-quality bases and adapters with Trimmomatic (75). RNA-seq reads were aligned to the mouse genome (mm10) using Bowtie2 version 2.1.0 (76) and TopHat2 version 2.0.9 (77). Reads per gene were counted with HTSeq (78). Data were deposited in the Gene Expression Omnibus database (79) (accession GSE142204). Differential expression was analyzed with weighted Limma-Voom (80). Genes were considered expressed if they had at least 1 count per million in at least 2 samples. Genes were considered significantly differentially expressed if the Benjamini-Hochberg (81) FDR ≤ 0.05. An additional, more lenient cutoff of P ≤ 0.001 and FDR ≤ 0.2 was used. All expressed genes were analyzed by Reactome pathway (82) gene lists using CAMERA (35). Hierarchical clustering (83, 84) was performed with Cluster 3.0 (85) using distances based on a modified Pearson’s correlation coefficient and average linkage clustering (83, 84). Genes were mean centered and both genes and samples were clustered. The dendrogram and heatmap were plotted with Java TreeView (86).
Statistics.
Analyses were performed using GraphPad Prism 8.2.0. Data are presented as mean ± SEM. Normality of the data was assessed using the Shapiro-Wilk normality test. A nonparametric test was performed when data did not follow a normal distribution. Independent 2-sample t tests (2 sided) were used to assess the difference between 2 groups of samples (Mann-Whitney U tests were used instead if normality was violated). For over 2 groups, 1-way ANOVA was used, and Tukey’s or Holm-Šídák post hoc test for pairwise comparisons or comparisons of selected groups was performed, respectively. Kruskal-Wallis test with post hoc Dunn’s test was performed instead if the normality test was not passed. Pearson’s correlation coefficient was assessed to evaluate the associations between 2 variables. P < 0.05 denoted statistical significance.
Study approval.
Animal experiments were performed according to the NIH Guide for the Care of Laboratory Animals (National Academies Press, 2011), and protocols were approved by the Institutional Animal Care and Use Committee at NYU Grossman School of Medicine. For human samples, the CVPath Institute was referred autopsy hearts from cases of unexpected sudden death from the Office of the Chief Medical Examiner of the State of Maryland. This study was approved by the CVPath Institutional Review Board as an exempt study (RP0063).
Author contributions
LS, RLD, LEG, EAF, RR, and AMS designed the research and edited the manuscript. LS, RLD, LEG, JL, GD, QL, KR, YV, TB, and MZD performed experiments and edited the manuscript. LS, RLD, LEG, HL, JH, and RAF performed data analyses and edited the manuscript. LG, DF, AVF, and RV provided key materials and edited the manuscript. AMS directed the research and the research team, wrote the original draft of the manuscript, and completed all the edits and final preparation of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the expert assistance of Latoya Woods of the Diabetes Research Program in the preparation of this manuscript. They thank the NYU Histology Core, which is partially supported by NYU Cancer Institute Cancer Center Support Grant 5P30CA016087-31, and the NYU Langone’s Cytometry and Cell Sorting Laboratory, which is partially supported by grant NIH/NCI P30CA016087. These studies were supported by U.S. Public Health Service grants R01HL132516 (to AMS and RR), P01131481 (to EAF, AMS, RR), R01HL141425 (to RV), and R01HL084312 (to EAF).
Version 1. 07/09/2020
Electronic publication
Version 2. 11/11/2024
Gurdip Daffu's email address was updated
Footnotes
Conflict of interest: AMS and RR are inventors on awarded (US 9,353,3078, US 9364472) and pending (US 16/094270, Europe 177864360.0) patent applications assigned to NYU Grossman Medical Center. EAF served as an expert witness on a patent case involving fish oils and received NIH funding. AVF and RV have grant/research/clinical trial support from Leducq Foundation Grant; 480 Biomedical; 4C Medical; 4Tech; Abbott; Accu Medical Inc.; Amgen; Biosensors; Boston Scientific; Cardiac Implants; CeloNova Biosciences; Claret Medical; Concept Medical; Cook; CSI; DuNing, Inc.; Edwards Lifesciences; Emboline; Endotronix; Envision Scientific; Lutonix, BD; Gateway; LifeTech Scientific Corporation; LimFlow; MedAlliance; Medtronic; Mercator; Merill; Microport Medical; Microvention; Mitralign; MitrAssist; NAMSA; Nanova, Inc.; Neovasc; NIPRO; Novogate Medical; Occlutech; OrbusNeich Medical; Phenox; Profusa; Protembis; Qool; ReCor Medical; Senseonics; Shockwave; Sino Medical Sciences Technology Inc.; Spectranetics; Surmodics; Symic; Vesper; W.L. Gore; and Xeltis. AVF has served Abbott Vascular (speaker’s bureau [SB] and consultant/advisory board [CAB]); Amgen (CAB); Biosensors (SB); Boston Scientific (SB and CAB); CeloNova Biosciences (SB and CAB); Cook Medical (SB and CAB); CSI (SB); Lutonix, BD (SB and CAB); Sino Medical Sciences Technology Inc. (SB and CAB); and Terumo Corporation (SB). RV has served Abbott Vascular (SB and CAB); Biosensors (SB); Boston Scientific; CeloNova Biosciences; Cook Medical; Cordis; CSI (SB and CAB for previous 5); Edwards Lifescience (CAB); Lutonix, BD; Medtronic; OrbusNeich Medical; ReCor Medical; Sino Medical Sciences Technology Inc.; Spectranetics (SB and CAB for previous 6); Surmodics (CAB); Terumo Corporation (SB and CAB); W.L. Gore (SB and CAB); and Xeltis (CAB).
Copyright: © 2020, American Society for Clinical Investigation.
Contributor Information
Raquel López-Díez, Email: Raquel.LopezDiez@nyulangone.org.
Lander Egaña-Gorroño, Email: Lander.EganaGorrono@nyulangone.org.
Jianhua Liu, Email: jianhua.liu@mssm.edu.
Jiyuan Hu, Email: Jiyuan.Hu@nyulangone.org.
Gurdip Daffu, Email: Gdaffu@gmail.com.
Qing Li, Email: Qing.Li@nyulangone.org.
Karishma Rahman, Email: karishma.rahman24@gmail.com.
Yuliya Vengrenyuk, Email: Yuliya.Vengrenyuk@mountsinai.org.
M. Zahidunnabi Dewan, Email: Zahidunnabi.Dewan@nyulangone.org.
Liang Guo, Email: lguo@cvpath.org.
Renu Virmani, Email: rvirmani@cvpath.org.
Huilin Li, Email: Huilin.Li@nyulangone.org.
Edward A. Fisher, Email: edward.fisher@nyumc.org.
Ravichandran Ramasamy, Email: ravichandran.ramasamy@nyulangone.org.
Ann Marie Schmidt, Email: AnnMarie.Schmidt@nyulangone.org.
References
- 1.Haust MD. The genesis of atherosclerosis in pediatric age-group. Pediatr Pathol. 1990;10(1-2):253–271. doi: 10.3109/15513819009067112. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls SJ, et al. Effect of two intensive statin regimens on progression of coronary disease. N Engl J Med. 2011;365(22):2078–2087. doi: 10.1056/NEJMoa1110874. [DOI] [PubMed] [Google Scholar]
- 3.Hiro T, et al. Diabetes mellitus is a major negative determinant of coronary plaque regression during statin therapy in patients with acute coronary syndrome--serial intravascular ultrasound observations from the Japan Assessment of Pitavastatin and Atorvastatin in Acute Coronary Syndrome Trial (the JAPAN-ACS Trial) Circ J. 2010;74(6):1165–1174. doi: 10.1253/circj.CJ-09-0766. [DOI] [PubMed] [Google Scholar]
- 4.Parathath S, et al. Diabetes adversely affects macrophages during atherosclerotic plaque regression in mice. Diabetes. 2011;60(6):1759–1769. doi: 10.2337/db10-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Distel E, et al. miR33 inhibition overcomes deleterious effects of diabetes mellitus on atherosclerosis plaque regression in mice. Circ Res. 2014;115(9):759–769. doi: 10.1161/CIRCRESAHA.115.304164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudreault N, Kumar N, Olivas VR, Eberlé D, Stephens K, Raffai RL. Hyperglycemia impairs atherosclerosis regression in mice. Am J Pathol. 2013;183(6):1981–1992. doi: 10.1016/j.ajpath.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagareddy PR, et al. Hyperglycemia promotes myelopoiesis and impairs the resolution of atherosclerosis. Cell Metab. 2013;17(5):695–708. doi: 10.1016/j.cmet.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llodrá J, Angeli V, Liu J, Trogan E, Fisher EA, Randolph GJ. Emigration of monocyte-derived cells from atherosclerotic lesions characterizes regressive, but not progressive, plaques. Proc Natl Acad Sci U S A. 2004;101(32):11779–11784. doi: 10.1073/pnas.0403259101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trogan E, et al. Gene expression changes in foam cells and the role of chemokine receptor CCR7 during atherosclerosis regression in ApoE-deficient mice. Proc Natl Acad Sci U S A. 2006;103(10):3781–3786. doi: 10.1073/pnas.0511043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feig JE, et al. LXR promotes the maximal egress of monocyte-derived cells from mouse aortic plaques during atherosclerosis regression. J Clin Invest. 2010;120(12):4415–4424. doi: 10.1172/JCI38911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feig JE, et al. Reversal of hyperlipidemia with a genetic switch favorably affects the content and inflammatory state of macrophages in atherosclerotic plaques. Circulation. 2011;123(9):989–998. doi: 10.1161/CIRCULATIONAHA.110.984146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121(7):2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pourcet B, et al. LXRα regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ Res. 2011;109(5):492–501. doi: 10.1161/CIRCRESAHA.111.241810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feig JE, et al. Regression of atherosclerosis is characterized by broad changes in the plaque macrophage transcriptome. PLoS One. 2012;7(6):e39790. doi: 10.1371/journal.pone.0039790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanschel A, et al. Neuroimmune guidance cue Semaphorin 3E is expressed in atherosclerotic plaques and regulates macrophage retention. Arterioscler Thromb Vasc Biol. 2013;33(5):886–893. doi: 10.1161/ATVBAHA.112.300941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsey SA, et al. Epigenome-guided analysis of the transcriptome of plaque macrophages during atherosclerosis regression reveals activation of the Wnt signaling pathway. PLoS Genet. 2014;10(12):e1004828. doi: 10.1371/journal.pgen.1004828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrestha E, et al. Poly(ADP-ribose) polymerase 1 represses liver X receptor-mediated ABCA1 expression and cholesterol efflux in macrophages. J Biol Chem. 2016;291(21):11172–11184. doi: 10.1074/jbc.M116.726729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman K, et al. Inflammatory Ly6Chi monocytes and their conversion to M2 macrophages drive atherosclerosis regression. J Clin Invest. 2017;127(8):2904–2915. doi: 10.1172/JCI75005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan C, et al. Human aldose reductase expression prevents atherosclerosis regression in diabetic mice. Diabetes. 2018;67(9):1880–1891. doi: 10.2337/db18-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park L, et al. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat Med. 1998;4(9):1025–1031. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- 21.Bucciarelli LG, et al. RAGE blockade stabilizes established atherosclerosis in diabetic apolipoprotein E-null mice. Circulation. 2002;106(22):2827–2835. doi: 10.1161/01.CIR.0000039325.03698.36. [DOI] [PubMed] [Google Scholar]
- 22.Soro-Paavonen A, et al. Receptor for advanced glycation end products (RAGE) deficiency attenuates the development of atherosclerosis in diabetes. Diabetes. 2008;57(9):2461–2469. doi: 10.2337/db07-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harja E, et al. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE–/– mice. J Clin Invest. 2008;118(1):183–194. doi: 10.1172/JCI32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bu DX, et al. Activation of the ROCK1 branch of the transforming growth factor-β pathway contributes to RAGE-dependent acceleration of atherosclerosis in diabetic ApoE-null mice. Circ Res. 2010;106(6):1040–1051. doi: 10.1161/CIRCRESAHA.109.201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chereshnev I, et al. Mouse model of heterotopic aortic arch transplantation. J Surg Res. 2003;111(2):171–176. doi: 10.1016/S0022-4804(03)00039-8. [DOI] [PubMed] [Google Scholar]
- 26.Anderson MM, Heinecke JW. Production of Nε-(carboxymethyl)lysine is impaired in mice deficient in NADPH oxidase: a role for phagocyte-derived oxidants in the formation of advanced glycation end products during inflammation. Diabetes. 2003;52(8):2137–2143. doi: 10.2337/diabetes.52.8.2137. [DOI] [PubMed] [Google Scholar]
- 27.Anderson MM, Requena JR, Crowley JR, Thorpe SR, Heinecke JW. The myeloperoxidase system of human phagocytes generates Nε-(carboxymethyl)lysine on proteins: a mechanism for producing advanced glycation end products at sites of inflammation. J Clin Invest. 1999;104(1):103–113. doi: 10.1172/JCI3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinohara M, et al. Overexpression of glyoxalase-I in bovine endothelial cells inhibits intracellular advanced glycation endproduct formation and prevents hyperglycemia-induced increases in macromolecular endocytosis. J Clin Invest. 1998;101(5):1142–1147. doi: 10.1172/JCI119885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Distler MG, et al. Glyoxalase 1 increases anxiety by reducing GABAA receptor agonist methylglyoxal. J Clin Invest. 2012;122(6):2306–2315. doi: 10.1172/JCI61319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenmann KM, et al. T cell responses in mammalian diaphanous-related formin mDia1 knock-out mice. J Biol Chem. 2007;282(34):25152–25158. doi: 10.1074/jbc.M703243200. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, et al. Advanced glycation end product (AGE)-receptor for AGE (RAGE) signaling and up-regulation of Egr-1 in hypoxic macrophages. J Biol Chem. 2010;285(30):23233–23240. doi: 10.1074/jbc.M110.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Gils JM, et al. The neuroimmune guidance cue netrin-1 promotes atherosclerosis by inhibiting the emigration of macrophages from plaques. Nat Immunol. 2012;13(2):136–143. doi: 10.1038/ni.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randolph GJ, Luther T, Albrecht S, Magdolen V, Muller WA. Role of tissue factor in adhesion of mononuclear phagocytes to and trafficking through endothelium in vitro. Blood. 1998;92(11):4167–4177. [PubMed] [Google Scholar]
- 34.Kashiyama N, et al. Development of PET imaging to visualize activated macrophages accumulated in the transplanted iPSc-derived cardiac myocytes of allogeneic origin for detecting the immune rejection of allogeneic cell transplants in mice. PLoS One. 2016;11(12):e0165748. doi: 10.1371/journal.pone.0165748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40(17):e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reardon CA, et al. Obesity and insulin resistance promote atherosclerosis through an IFNγ-regulated macrophage protein network. Cell Rep. 2018;23(10):3021–3030. doi: 10.1016/j.celrep.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rai V, Agrawal DK. The role of damage- and pathogen-associated molecular patterns in inflammation-mediated vulnerability of atherosclerotic plaques. Can J Physiol Pharmacol. 2017;95(10):1245–1253. doi: 10.1139/cjpp-2016-0664. [DOI] [PubMed] [Google Scholar]
- 38.Zindel J, Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol. 2020;15:493–518. doi: 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- 39.Kislinger T, et al. N(epsilon)-(carboxymethyl)lysine adducts of proteins are ligands for receptor for advanced glycation end products that activate cell signaling pathways and modulate gene expression. J Biol Chem. 1999;274(44):31740–31749. doi: 10.1074/jbc.274.44.31740. [DOI] [PubMed] [Google Scholar]
- 40.Robbins CS, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med. 2013;19(9):1166–1172. doi: 10.1038/nm.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jefferies CA. Regulating IRFs in IFN driven disease. Front Immunol. 2019;10:325. doi: 10.3389/fimmu.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 43.Simons KH, de Vries MR, de Jong RCM, Peters HAB, Jukema JW, Quax PHA. IRF3 and IRF7 mediate neovascularization via inflammatory cytokines. J Cell Mol Med. 2019;23(6):3888–3896. doi: 10.1111/jcmm.14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan W, et al. PDGFR-β modulates vascular smooth muscle cell phenotype via IRF-9/SIRT-1/NF-κB pathway in subarachnoid hemorrhage rats. J Cereb Blood Flow Metab. 2019;39(7):1369–1380. doi: 10.1177/0271678X18760954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, et al. Interferon regulatory factor-1 together with reactive oxygen species promotes the acceleration of cell cycle progression by up-regulating the cyclin E and CDK2 genes during high glucose-induced proliferation of vascular smooth muscle cells. Cardiovasc Diabetol. 2013;12:147. doi: 10.1186/1475-2840-12-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeVerse JS, et al. Shear stress modulates VCAM-1 expression in response to TNF-α and dietary lipids via interferon regulatory factor-1 in cultured endothelium. Am J Physiol Heart Circ Physiol. 2013;305(8):H1149–H1157. doi: 10.1152/ajpheart.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eguchi J, et al. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7(1):86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumari M, et al. IRF3 promotes adipose inflammation and insulin resistance and represses browning. J Clin Invest. 2016;126(8):2839–2854. doi: 10.1172/JCI86080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du M, et al. Absence of interferon regulatory factor 1 protects against atherosclerosis in apolipoprotein E-deficient mice. Theranostics. 2019;9(16):4688–4703. doi: 10.7150/thno.36862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seneviratne AN, et al. Interferon regulatory factor 5 controls necrotic core formation in atherosclerotic lesions by impairing efferocytosis. Circulation. 2017;136(12):1140–1154. doi: 10.1161/CIRCULATIONAHA.117.027844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clément M, et al. Deletion of IRF8 (interferon regulatory factor 8)-dependent dendritic cells abrogates proatherogenic adaptive immunity. Circ Res. 2018;122(6):813–820. doi: 10.1161/CIRCRESAHA.118.312713. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, et al. Ablation of interferon regulatory factor 3 protects against atherosclerosis in apolipoprotein E-deficient mice. Hypertension. 2017;69(3):510–520. doi: 10.1161/HYPERTENSIONAHA.116.08395. [DOI] [PubMed] [Google Scholar]
- 53.Tsiantoulas D, et al. B cell-activating factor neutralization aggravates atherosclerosis. Circulation. 2018;138(20):2263–2273. doi: 10.1161/CIRCULATIONAHA.117.032790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao GN, Jiang DS, Li H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta. 2015;1852(2):365–378. doi: 10.1016/j.bbadis.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 55.Huang L, et al. Interferon regulatory factor 7 protects against vascular smooth muscle cell proliferation and neointima formation. J Am Heart Assoc. 2014;3(5):e001309. doi: 10.1161/JAHA.114.001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang XA, et al. Interferon regulatory factor 7 deficiency prevents diet-induced obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2013;305(4):E485–E495. doi: 10.1152/ajpendo.00505.2012. [DOI] [PubMed] [Google Scholar]
- 57.Miao F, et al. RNA-sequencing analysis of high glucose-treated monocytes reveals novel transcriptome signatures and associated epigenetic profiles. Physiol Genomics. 2013;45(7):287–299. doi: 10.1152/physiolgenomics.00001.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinig M, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467(7314):460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruan BH, et al. Complement C3a, CpG oligos, and DNA/C3a complex stimulate IFN-α production in a receptor for advanced glycation end product-dependent manner. J Immunol. 2010;185(7):4213–4222. doi: 10.4049/jimmunol.1000863. [DOI] [PubMed] [Google Scholar]
- 60.Li J, Schmidt AM. Characterization and functional analysis of the promoter of RAGE, the receptor for advanced glycation end products. J Biol Chem. 1997;272(26):16498–16506. doi: 10.1074/jbc.272.26.16498. [DOI] [PubMed] [Google Scholar]
- 61.Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17(22):6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441(1):106–110. doi: 10.1016/S0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 63.Liliensiek B, et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest. 2004;113(11):1641–1650. doi: 10.1172/JCI18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siracuse JJ, et al. A20-mediated modulation of inflammatory and immune responses in aortic allografts and development of transplant arteriosclerosis. Transplantation. 2012;93(4):373–382. doi: 10.1097/TP.0b013e3182419829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reis ED, et al. Dramatic remodeling of advanced atherosclerotic plaques of the apolipoprotein E-deficient mouse in a novel transplantation model. J Vasc Surg. 2001;34(3):541–547. doi: 10.1067/mva.2001.115963. [DOI] [PubMed] [Google Scholar]
- 66. Andrés-Manzano MJ, Andrés V, Dorado B. Oil red O and hematoxylin and eosin staining for quantification of atherosclerosis burden in mouse aorta and aortic root. In: Andrés V, Dorado B eds. Methods in Mouse Atherosclerosis. New York, New York, USA: Springer New York; 2015:85–99. [DOI] [PubMed] [Google Scholar]
- 67.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tacke F, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest. 2007;117(1):185–194. doi: 10.1172/JCI28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potteaux S, et al. Suppressed monocyte recruitment drives macrophage removal from atherosclerotic plaques of Apoe–/– mice during disease regression. J Clin Invest. 2011;121(5):2025–2036. doi: 10.1172/JCI43802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;83(1):14.1.1–14.1.14. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daffu G, et al. RAGE suppresses ABCG1-mediated macrophage cholesterol efflux in diabetes. Diabetes. 2015;64(12):4046–4060. doi: 10.2337/db15-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.López-Díez R, et al. Ager deletion enhances ischemic muscle inflammation, angiogenesis, and blood flow recovery in diabetic mice. Arterioscler Thromb Vasc Biol. 2017;37(8):1536–1547. doi: 10.1161/ATVBAHA.117.309714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Epelman S, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40(1):91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nahrendorf M, et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204(12):3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu R, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43(15):e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 82.Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(Database issue):D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Everitt BS, Landau S, Leese M, Stahl D. Cluster Analysis. Hoboken, New Jersey, USA: John Wiley and Sons; 2011. [Google Scholar]
- 85.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 86.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.