Abstract
BACKGROUND
Prediction of adverse outcomes in cerebral malaria (CM) is difficult. We hypothesized that cell-free DNA (cfDNA) levels would facilitate identification of severe and potentially fatal CM cases.
METHODS
In this retrospective study, plasma from Malawian children with CM (n = 134), uncomplicated malaria (UM, n = 77), and healthy controls (HC, n = 60) was assayed for cfDNA using a fluorescence assay. Host and parasite cfDNA was measured by quantitative PCR. Immune markers were determined by ELISA, Luminex, or cytometric bead array.
RESULTS
Total cfDNA increased with malaria severity (HC versus UM, P < 0.001; HC versus CM, P < 0.0001; UM versus CM, P < 0.0001), was elevated in retinopathy-positive (Ret+) CM relative to Ret– CM (7.66 versus 5.47 ng/μL, P = 0.027), and differentiated Ret+ fatal cases from survivors (AUC 0.779; P < 0.001). cfDNA levels in patients with non–malarial febrile illness (NMF, P = 0.25) and non–malarial coma (NMC, P = 0.99) were comparable with UM. Host DNA, rather than parasite DNA, was the major cfDNA contributor (UM, 268 versus 67 pg/μL; CM, 2824 versus 463 pg/μL). Host and parasite cfDNA distinguished CM by retinopathy (host, AUC 0.715, P = 0.0001; parasite, AUC 0.745, P = 0.0001), but only host cfDNA distinguished fatal cases (AUC 0.715, P = 0.0001). Total cfDNA correlated with neutrophil markers IL-8 (rs = 0.433, P < 0.0001) and myeloperoxidase (rs = 0.683, P < 0.0001).
CONCLUSION
Quantifying plasma cfDNA is a simple assay useful in identifying children at risk for fatal outcome and has promise as a point-of-care assay. Elevated cfDNA suggests a link with host inflammatory pathways in fatal CM.
FUNDING
NIH NCATS (AK), Burroughs-Wellcome (AK), and National Health and Medical Research Council of Australia (SJR).
Keywords: Infectious disease, Inflammation
Keywords: Malaria
Plasma cell-free DNA in children with cerebral malaria is primarily derived from the host and provides information on patient prognosis and potential for fatal outcomes.
Introduction
Malaria, an infectious disease caused by the protozoan parasite Plasmodium, has a broad clinical presentation ranging from asymptomatic or uncomplicated febrile illness to severe and fatal manifestations such as cerebral malaria (CM). CM is a life-threatening condition that disproportionately affects young children living in sub-Saharan Africa. The World Health Organization (WHO) defines CM as an unrousable coma accompanied by Plasmodium falciparum parasitemia and no other cause of coma (1). A large controlled postmortem histopathological study of WHO-defined CM showed that about 25% of clinical CM fatalities were not associated with parasite sequestration in the brain microvasculature (2), the “gold standard” pathological finding in fatal CM. Thus, the WHO CM case definition likely encompasses children with other pathological processes. Our inability to precisely diagnose CM in life has slowed progress in understanding of disease pathogenesis and hindered development of effective adjunctive treatments for CM (3).
Many research groups are working to develop tools to identify and triage patients with CM according to their risk of an adverse outcome. Plasma biomarkers, such as P. falciparum histidine rich protein 2 (PfHRP2) that measures total parasite biomass (4–7) or markers of endothelial cell activation such as Angiopoeitin-2 (8–10), correlate with malaria severity and adverse outcome. Ocular funduscopic abnormalities, known collectively as malarial retinopathy, increase the specificity of CM clinical case definition and correlate with the sequestration of infected erythrocytes in the brain (2, 11–14). MRI of the brain revealed that increased brain volume is a risk factor for death in children with CM (15, 16). However, malaria-endemic regions are resource poor and economically strained. Routine implementation of these tests at clinical sites would require capital-intensive infrastructure for imaging and laboratory assays or extensive ophthalmological training for clinical staff. There is an urgent need to identify point-of-care tools for CM clinical staging that are simple and rapid.
Cell-free nucleic acids were first described in 1948, and it is now appreciated that nucleic acids, released from a variety of cell types, can be detected in plasma and other biofluids (17). Circulating cell-free DNA (cfDNA) may result from cell necrosis, cell apoptosis, or regulated release of exosomes or microvesicles from cells under pathological and inflammatory conditions. Released nucleic acids or vesicles, in turn, can modulate host immune response (18). cfDNA is currently under investigation as a biomarker in various conditions, including cancer, autoimmune disease, sepsis, and parasitic diseases (19–22). For example, recent work identified parasite cfDNA concentration in the plasma as a correlate of disease severity in African children and Asian adults with malaria (23).
Although CM pathogenesis is not fully understood, host inflammation is implicated in the pathogenesis. Total leukocyte counts correlate with both brain swelling and fatal outcome in CM (16), and the proinflammatory cytokine TNF-α also correlates with CM outcome (24). Neutrophils are activated in malaria infection, and host cfDNA is released from neutrophils or found within circulating neutrophil extracellular traps (NETS) composed of host DNA (25–27). Given this rationale, we hypothesized that total plasma cfDNA (consisting of host and parasite DNA) would be elevated in severe disease relative to uncomplicated malaria (UM) and could serve as a biomarker of CM, correlating with CM severity and outcome. We employed a streamlined methodology to measure total cfDNA levels in plasma of Malawian children with malaria and appropriate control children. We found that both total and host-derived cfDNA levels predict clinical severity and CM outcome and correlate with known markers of malaria infection.
Results
Patient characteristics.
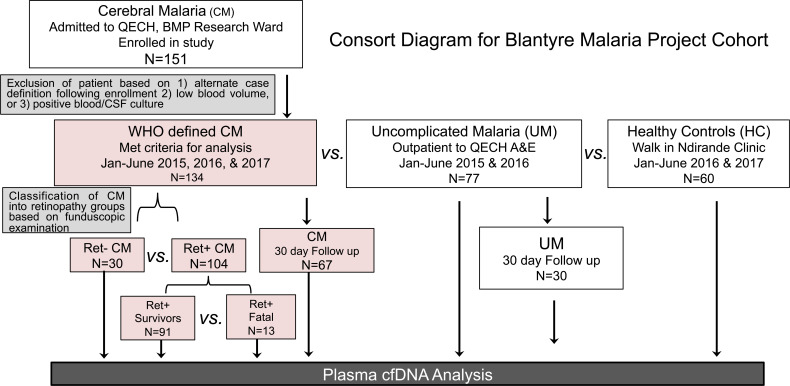
Of the 151 CM cases recruited, 9 cases were later classified as not CM, due to rapid resolution of coma; 5 cases had low blood-draw volumes, excluding them from the study; and 3 cases had revised final diagnoses due to positive cultures in blood or cerebral spinal fluid. A total of 134 CM cases was further classified as retinopathy-positive (Ret+) CM (n = 104) or Ret– CM (n = 30) based upon funduscopic exam (Figure 1 and ref. 13). Demographic and clinical characteristics comparing Ret+ CM with the other clinical groups, Ret– CM, UM, and healthy controls (HC), are presented in Table 1. Relative to Ret+ CM, the proportion of male children and median age for the 3 other clinical groups were comparable. Children with Ret+ CM had significantly lower median levels of packed cell volume (PCV; 24 versus 34%, P < 0.0001), hemoglobin (Hgb, 7.8 versus 9.8 g/dL, P < 0.0001), and platelet count (61 versus 134 × 103/μL, P < 0.0001) and higher PfHRP2 median levels (1343 versus 190 ng/mL, P < 0.0001) relative to children with UM. Ret+ CM cases, compared with Ret– CM had elevated PfHRP2 (1343 versus 469 ng/mL, P = 0.001) and decreased platelet count (61 versus 148x103/μL, P < 0.0001) (Table 1).
Figure 1. Consort diagram of cohort recruitment for BMP study.
Flow chart of Blantyre Malaria Project (BMP) study patient recruitment between 2015 and 2017. CM and UM patients were recruited from Queen Elizabeth Central Hospital (QECH) in Blantyre, Malawi. HC subjects were recruited from Ndirande Health Centre, attending for routine vaccine and health check-up. CM patients were excluded due to failure to meet inclusion criteria (including quick resolution of coma following enrollment), positive blood or CSF cultures, or an alternative diagnosis after enrollment. Plasma samples were collected and analyzed for cfDNA analysis.
Table 1. Clinical parameters.
Total plasma cfDNA levels in malaria patients are elevated with increased disease severity.
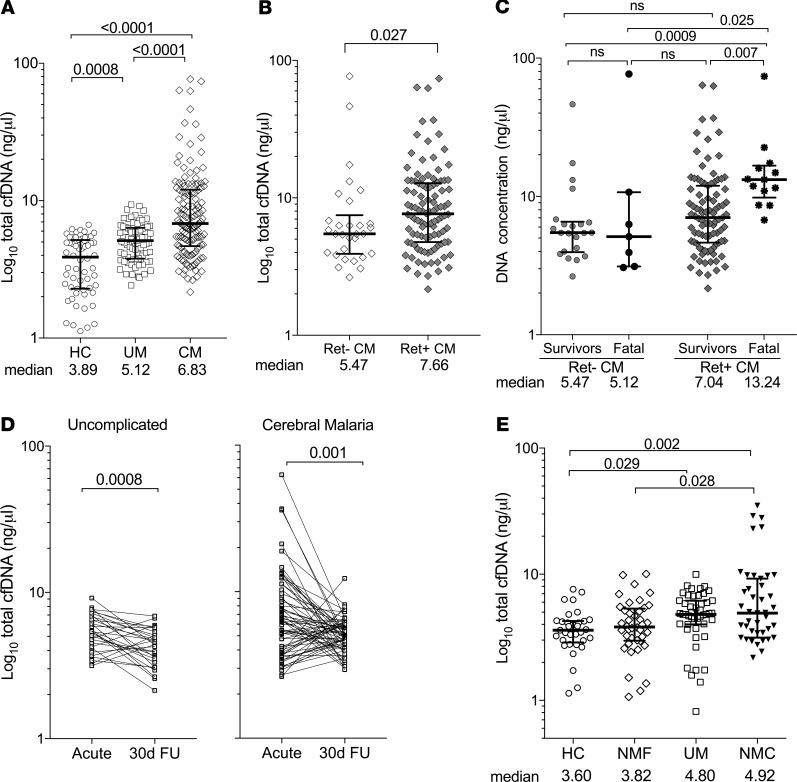
We compared total cfDNA levels between disease states (UM and CM) and healthy individuals (HC) using a fluorescence-based assay. Median levels of circulating total cfDNA were elevated in the 2 clinical malaria groups, UM (5.12 ng/μL; IQR, 3.8–6.4; P = 0.0007) and CM (6.83 ng/μL; IQR, 4.7–12.1; P < 0.0001) relative to HC (3.89 ng/μL; IQR, 2.3–5.2) (Figure 2A). Total plasma cfDNA levels increased with clinical malaria severity (UM versus CM; P < 0.0001) (Figure 2A). We then compared cfDNA levels within the CM group based on retinopathy classification. Ret+ CM cfDNA median levels were moderately but significantly increased (7.66 ng/μL; IQR, 4.8–12.8) relative to Ret– CM (5.47 ng/μL; IQR, 3.9–7.5; P = 0.027) (Figure 2B). When we consider the fatal cases within 2 CM classifications (Ret– versus Ret+), median cfDNA concentration is elevated in Ret+ CM fatal cases (13.24 ng/μL; IQR, 9.8–16.7) relative to Ret+ CM survivors (7.04 ng/μL; IQR, 4.6–11.9; P < 0.001) (Figure 2C). No significant differences in median cfDNA levels were observed between Ret– CM survivors (5.47 ng/μL; IQR, 4.0–6.6) and Ret– CM fatal cases (5.12 ng/μL; IQR, 3.1–10.7; P ≥ 0.99) (Figure 2C). Plasma levels of total cfDNA declined in both UM (P = 0.001) and CM (P = 0.001) between acute presentation and convalescence at 30-day follow-up (30d FU) (Figure 2D).
Figure 2. Total plasma cfDNA levels are elevated with malaria severity.
(A–C) Total plasma cfDNA levels (ng/μL) for clinical and control groups as measured by fluorometry. (A) Healthy controls (HC, n = 60) versus uncomplicated malaria (UM, n = 77) versus cerebral malaria (CM, n = 134). (B) CM classified as Ret– CM (n = 30) versus Ret+ CM (n = 104). (C) Survivors (Ret– CM, n = 22; Ret+ CM, n = 91) versus fatal cases (Ret– CM, n = 7; Ret+ CM, n = 13). (D) Total cfDNA levels in UM (n = 30) or CM (n = 67) convalescence from acute infection to 30-day follow-up (30d FU). (E) Total serum cfDNA levels of healthy controls (HC, n = 33), non–malaria febrile (NMF, n = 40), uncomplicated malaria (UM, n = 47), and non–malaria coma (NMC, n = 49) clinical groups. Shown are median levels ± IQR; statistical significance was determined by Mann-Whitney U test (B), Kruskal-Wallis test for multiple comparisons (A, C, and E), or Wilcoxon signed rank test for paired comparisons (D). P < 0.05 was considered significant.
To determine if elevated total cfDNA concentration is a specific characteristic of P. falciparum infection, we analyzed cfDNA concentrations in sick Malawian children who tested negative for Plasmodium parasites by smear or rapid diagnostic test. These children presented with either nonmalarial, noncoma febrile illness (NMF, n = 40) or non–malarial febrile coma (NMC, n = 49) (Supplemental Table 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136279DS1). Diagnoses for NMF and NMC groups included other infectious diseases such as Salmonella bacteremia or pneumonia for NMF and bacterial or viral meningitis for NMC. In some cases, the etiology of fever or coma could not be determined. Corresponding control groups were also analyzed for comparison: a HC group (n = 33) and a UM group (n = 47). Only serum samples were available for these clinical samples; we, therefore, first ensured that total cfDNA levels were comparable between plasma versus serum (Supplemental Figure 2).
Total cfDNA levels in serum of children with NMF (3.82 ng/μL; IQR, 3.0–5.3) were not significantly different to the corresponding UM control (4.8 ng/μL; IQR, 4.0–6.2; P = 0.25) or the corresponding HC group (3.6 ng/μL; IQR, 2.9–4.3; P ≥ 0.99) (Figure 2E). The median levels of cfDNA in NMC (4.92 ng/μL; IQR, 3.14–9.24) were also comparable with the UM group (P > 0.99) but minimally elevated compared with the HC group (P = 0.019) (Figure 2E).
Host cfDNA is a major contributor to absolute total circulating cfDNA.
We quantified the relative contributions of cfDNA from the human host and the P. falciparum parasite via quantitative PCR (qPCR). Plasma cfDNA median levels were elevated in CM versus UM for both the parasite (P. falciparum 18s rRNA, UM versus CM; 67 versus 268 pg/μL; IQR, 18–346; P < 0.0001) and for the host (Homo sapiens RPP30, UM versus CM; 463 [IQR, 140–1344] versus 2824 [IQR, 670–7303] pg/μL, P < 0.0001) (Figure 3A). Likewise, when the CM group was stratified based on retinopathy, plasma cfDNA for both parasite (Ret– versus Ret+; 48.5 [IQR, 6–391] versus 335 [IQR, 129–895] pg/μL, P < 0.001) and host (Ret– versus Ret+; 650 [IQR, 46–3433] versus 3369 [IQR, 1039–8175] pg/μL, P < 0.001) was elevated in Ret+ CM relative to Ret– CM (Figure 3B).
Figure 3. Host is a major contributor to cfDNA levels in malaria.
(A–C) cfDNA levels (pg/μL) as measured by qPCR to detect parasite (Pf18s gene target, black data points) or host (HsRPP30 gene target, blue data points) from various clinical and control groups. (A) Pf18s, UM (n = 65) versus CM (n = 132); HsRPP30, UM (n = 65) versus CM (n = 103). (B) Ret– CM (n = 29) versus Ret+ CM (n = 103) for Pf18s and HsRPP30. (C) Ret+ survivors versus Ret+ fatal CM for Pf18s and HsRPP30 (survivors, n = 90; fatal, n = 13). Horizontal bar represents median ± IQR. (D) Host/parasite ratio of cfDNA levels presented in a box-and-whisker plot with min-to-max range. Horizontal dotted line across plot demarcates a ratio value equal to 1. Shown are median levels ± IQR. Statistical significance determined by Mann-Whitney U test (A–C) or Kruskal-Wallis test for multiple comparisons (D). P < 0.05 was considered significant.
Among children with Ret+ CM, only the host cfDNA median levels were significantly different and elevated in the fatal cases (8175 pg/μL; IQR, 3838–13,304) versus those that survived (2930 pg/μL; IQR, 862–6901; P = 0.009). Median levels of parasite cfDNA were not significantly different between Ret+ survivors (335 pg/μL) and fatal cases (269 pg/μL, P = 0.719) (Figure 3C). When we consider the median ratio of host cfDNA to parasite cfDNA, we find that host cfDNA is the major contributor by total mass in the 3 clinical malaria groups with ratios above a value of 1 (Figure 3D).
In the Ret+ CM group, the median host/parasite DNA mass ratio (Ret+ CM = 8.34) was higher than in UM (ratio 5.55) or Ret– CM (ratio 4.04), but this difference was not statistically significant. Correlation analysis showed that host cfDNA has a higher correlation coefficient (rs = 0.805, P < 0.0001) to total cfDNA compared with parasite cfDNA (rs = 0.412, P < 0.0001) (Supplemental Figure 3). The host haploid genome (~3234 Mb) is 143 times larger than the P. falciparum genome (22.9 Mb) (28, 29); therefore, we reanalyzed our data to compare genomic equivalents (GE) between Pf18s and HsRPP30. The parasite contribution to cfDNA, when normalized for genome size, was higher than that of the host for both UM (Pf18s versus HsRPP30, 2560 versus 142 GE/μL; P < 0.001) and CM (Pf18s versus HsRPP30, 10,254 versus 916 GE/μL; P < 0.0001) (Supplemental Figure 4).
cfDNA levels correlate with biomarkers of malaria disease severity and neutrophil function.
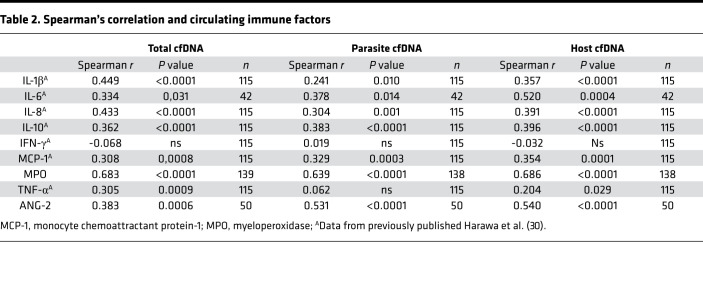
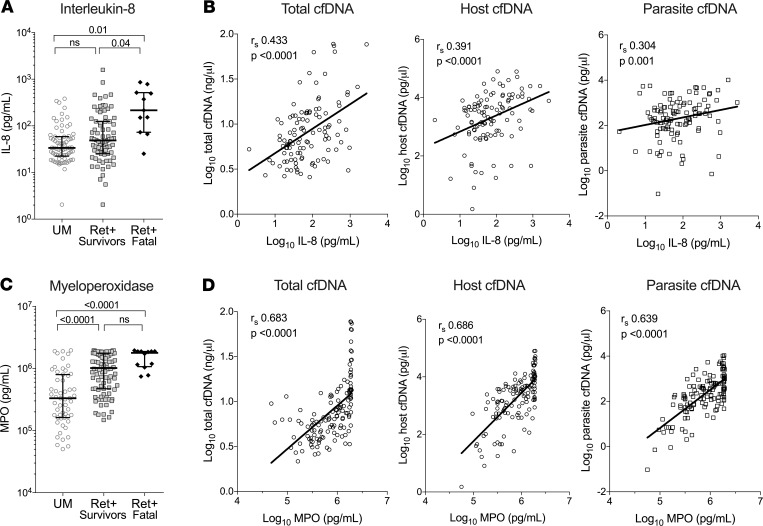
We recently published an analysis of plasma concentrations of inflammatory cytokines in the cohort of CM patients (30). We compared these cytokine data with our total cfDNA results. Spearman’s correlation analysis showed a positive and significant correlation with known inflammatory immune factors, namely IL-1β (rs = 0.449, P < 0.0001), IL-6 (rs = 0.334, P = 0.031), IL-10 (rs = 0.362, P < 0.0001), TNF-α (rs = 0.305, P = 0.001), monocyte chemoattractant protein 1 (MCP-1; rs = 0.308, P = 0.001), and IL-8 (rs = 0.433, P < 0.0001) (Table 2). Of note, IL-8, a neutrophil chemokine, is elevated in fatal cases (216 pg/mL; IQR, 73–515) relative to Ret+ CM survivors (49 pg/mL; P < 0.005) (Figure 4, A and B). We also measured levels of the endothelial cell activation marker, angiopoietin-2 (ANG-2) and found a positive correlation with all 3 measurements of cfDNA (total, rs = 0.383, P = 0.0006; host, rs = 0.540, P < 0.0001; parasite, rs = 0.531, P < 0.0001). The only cytokine that did not correlate with any of the cfDNA measurements was IFN-γ ( P = 0.47) (Table 2).
Table 2. Spearman’s correlation and circulating immune factors.
Figure 4. Plasma cfDNA is associated with markers of neutrophil function.
(A and C) Plasma levels of IL-8 (A) and MPO (C) in UM (n = 73), Ret+ CM survivors (n = 90), and Ret+ CM fatal cases (n = 12). Shown are median levels ± IQR. Statistical significance determined by Kruskal-Wallis test for multiple comparisons. (B and D) IL-8 (B) or MPO (D) were plotted against total, host, or parasite cfDNA and show a positive and significant correlation. Values were log transformed before plotting. Spearman coefficient (rs) and P value of correlation are denoted within graph. P < 0.05 was considered significant. Solid line is a linear regression fit model of data. In B and D, values were log transformed before plotting.
We measured levels of the neutrophil effector, myeloperoxidase (MPO), in the same plasma samples used for cfDNA measurements. MPO levels were elevated in Ret+ CM versus UM cases (1.0 × 106 versus 3.3 × 105 pg/mL, P < 0.0001), but MPO levels were not significantly different between Ret+ CM survivors and Ret+ CM fatal cases (P = 0.118) (Figure 4C). Spearman’s correlation analysis of MPO plasma levels in the CM group correlate with total cfDNA (rs = 0.683, P < 0.0001), host cfDNA (rs = 0.686, P < 0.0001), and parasite cfDNA (rs = 0.639, P < 0.0001) (Figure 4D and Table 2).
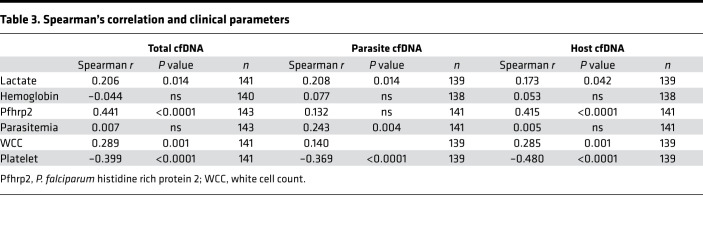
Clinical and biochemical parameters associated with parasite (parasitemia and PfHRP2) or host features (Hgb, lactate, white cell count [WCC], platelet count, and PCV) were analyzed in relation to cfDNA levels in the CM samples. PfHRP2 correlates with total (rs = 0.441, P < 0.0001) and host cfDNA (rs = 0.415, P < 0.0001) in CM patients. PfHRP2 levels did not correlate with parasite cfDNA. Platelet count negatively correlated with all 3 measurements of cfDNA (total, rs = –0.399; host, rs = –0.468; parasite, rs = –0.369; all P < 0.0001). WCC had a significant but modest correlation only with total (rs = 0.289, P = 0.001) and host cfDNA (rs = 0.272, P = 0.001) but not with parasite cfDNA. Parasitemia correlated with Pf cfDNA levels (rs = 0.243, P < 0.001) but did not correlate with host (P = 0.86) or total cfDNA (P = 0.93). Lactate correlated modestly with total (rs = 0.206, P = 0.014) and parasite cfDNA (rs = 0.208, P = 0.014) but not with host cfDNA (P = 0.06) (Table 3).
Table 3. Spearman’s correlation and clinical parameters.
Potential for cfDNA levels as a predictive biomarker for CM outcome.
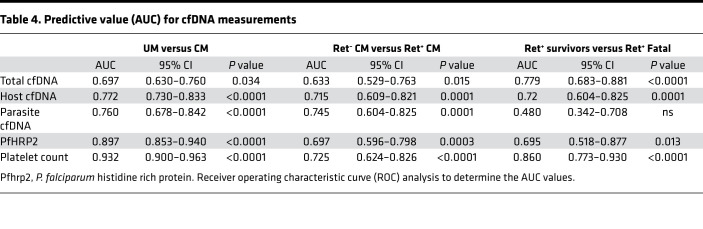
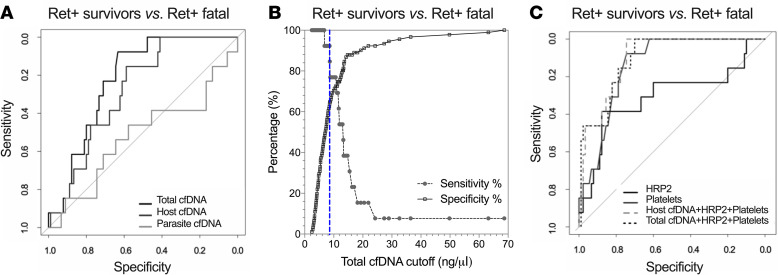
To assess the predictive value of cfDNA measurement as a clinical biomarker, we carried out receiver operating characteristic (ROC) analysis between the various clinical malaria groups. All 3 cfDNA measurements distinguished between UM and CM (AUC, P value: total 0.697, P = 0.034; host 0.772, P < 0.0001; parasite 0.760, P < 0.0001) and between CM retinopathy groups, Ret– CM versus Ret+ CM (AUC, P value: total 0.633, P = 0.015; host 0.715, P = 0.0001; parasite 0.745, P = 0.0001). Ret+ CM fatal cases were distinguished from Ret+ CM survivor cases by total cfDNA (AUC, 0.779; P = 0.001) and host cfDNA (AUC, 0.720; P < 0.0001) but not by parasite cfDNA (AUC, 0.480; P = 0.770) (Table 4 and Figure 5A). A cutoff of > 8.55 ng/μL for total cfDNA (Figure 5B) to discriminate Ret+ CM survivors from Ret+ CM fatal cases gave a sensitivity of 92% (95%CI, 67–100), a specificity of 64% (95%CI, 54–73), and an OR of 21.1 (Table 5).
Table 4. Predictive value (AUC) for cfDNA measurements.
Figure 5. Predictive value of cfDNA as a biomarker of CM fatal outcome.
(A) ROC curves for total cfDNA alone (dark gray curve; AUC, 0.779; P = 0.0006), host cfDNA alone (medium gray curve; AUC, 0.720; P = 0.001), parasite cfDNA (light gray curve; AUC, 0.480; P = ns). (B) Decision plot of sensitivity and specificity plotted against total cfDNA cutoff concentrations for Ret+ CM survivors versus Ret+ CM fatal cases generated from ROC analysis. Vertical blue dashed line demarcates the cutoff point (>8.55 ng/μL) that gives the best combination of sensitivity (92%) to specificity (64%) as determined by Youden Index analysis. (C) ROC curves for HRP2 alone (black solid curve; AUC, 0.691; P = 0.013), platelets alone (gray solid curve; AUC, 0.860; P ≤ 0.0001), and combined variables of total cfDNA + HRP + platelets (dark gray dashed curve; AUC, 0.899; P ≤ 0.0001) and host cfDNA + HRP + platelets (light gray dashed curve; AUC, 0.892; P ≤ 0.0001). ROC, receiver operator curve. P < 0.05 was considered significant.
Table 5. Contingency analysis of cfDNA levels in survivors versus fatal cases.
We carried out logistic regression models combining total or host cfDNA with HRP2 and platelet counts, 2 predictors of malaria severity previously described by us and others (4, 6, 15, 31, 32). The combined models generated an improved case fatality prediction score for total cfDNA + HRP2 + platelets, with an AUC of 0.899 (P < 0.0001), and for host cfDNA + HRP2 + platelets, an AUC of 0.894 (P < 0.0001) (Figure 5C).
Discussion
It is now recognized that increased levels of cfDNA in the circulation are a reflection of pathological processes in human tissues (33–36). Detection of parasite-derived circulating cell-free nucleic acid using PCR methods in biofluids such as saliva, urine, and plasma has previously been described in malaria (23, 37–40). Building upon this, direct PCR amplification of parasite cfDNA from plasma was shown to serve as a measure of disease severity, distinguishing UM from severe forms of malaria in East African children and Asian adults (23).
Here, we describe the utility of measuring total plasma cfDNA as a biomarker of CM outcome and present a streamlined, technically easy and rapid method to quantify plasma cfDNA using the Qubit fluorometer platform. We first analyzed total cfDNA between disease states and show that levels were elevated in clinical malaria (UM and CM) compared with HC. Total cfDNA levels in plasma of patients at hospital presentation also informed about severity, with higher median levels of total cfDNA in CM patients relative to UM. Levels of cfDNA were elevated during acute infection (UM and CM), as levels decreased in convalescence.
Within the CM group, the etiology of Ret– CM is controversial, since it is a heterogeneous clinical group that could represent a progression toward the Ret+ CM spectrum or an alternate severe encephalopathy with incidental P. falciparum parasitemia (41–43). In our analysis, median levels of plasma cfDNA in children with Ret– CM were lower in concentration compared with Ret+ CM and were similar to those of children with UM, supporting evidence that, in terms of cfDNA levels, Ret– CM is more closely related to UM and different from Ret+ CM. However, our study was not sufficiently powered for us to make an adequate evaluation regarding the relevance of cfDNA in Ret– CM.
Total cfDNA levels in patients with other causes of fever (NMF) or coma (NMC), not associated with P. falciparum infection, were similar to cfDNA levels in UM, suggesting that elevated cfDNA levels may be a particular feature of Ret+ CM. Within the NMC group, there were 5 outlier samples with total cfDNA levels > 10 ng/μL. Diagnoses of these patients were confirmed to be sepsis, typhoid fever, or systemic viremia (varicella zoster). Indeed, other such systemic infections that include bacteremia or viremia with inflammation are known to be associated with elevated cfDNA levels in circulation (44). We did not observe a difference between plasma and serum, although some studies have reported higher levels of cfDNA using serum, likely due to lysis of leukocytes (45). Overall, the utility of cfDNA analysis would serve in clinically diagnosed CM cases as a tool to triage and predict those with a potentially severe outcome.
The prognostic value of total cfDNA in children with CM was confirmed within the Ret+ CM group, as total cfDNA levels measured at hospital admission were elevated in those who went on to die of CM. With a cutoff of 8.55 ng/μL, the specificity was 64% but the sensitivity was 92%, suggesting cfDNA could be useful to identify those CM patients with low risk for a fatal outcome, much in the way that low procalcitonin is predictive of nonbacterial causes of pneumonia and sepsis (46).
Measurement of total cfDNA levels in CM patients could also assist clinicians in identifying individuals who require more intensive clinical intervention, including experimental or higher-risk therapies. When combined with PfHRP2 and platelet count, host or total cfDNA had an additive effect in predicting fatal outcome, as determined by ROC analysis. All 3 measurements together (cfDNA, platelets, and PfHRP2) performed better than each variable alone. Although low platelet counts or total cfDNA concentrations are not malaria-specific biomarkers (21, 47), combining these variables with a parasite-specific biomarker such as HRP2 may be useful in generating a clinical decision support tool that incorporates multiple variables.
In addition to platelet counts and PfHRP2, other immune and host markers such as ANG-2 (10), C-X-C motif chemokine 10 (IP-10) (48), chitinase-3–like protein 1 (49), and C-C motif chemokine ligand 5 (RANTES) (50) have previously been described to have prognostic value in distinguishing a fatal outcome in severe malaria and could also be considered to generate clinically informed models. Combined diagnostic and prognostic scoring methods that incorporate multiple biomarkers are in current clinical use for disease states such as sepsis, cancer, and disseminated intravascular coagulation (51–53). Such a scoring method could also be applied to CM to improve case definition and assist in appropriate decision-making for clinical intervention, as well as clinical study interpretations.
Analysis of the cellular origin of plasma cfDNA in malaria shows that the major contributor, by total mass, is from the host. This observation was made previously for P. vivax infection with a positive correlation between increased morbidity and host cfDNA (54). Gillrie et al. (55) first amplified host cfDNA from plasma in an attempt to establish the origins of circulating histones during malaria, but its utility as a biomarker was not explored. Here, we show that total and host cfDNA measurements are elevated in fatal cases. Parasite cfDNA levels, however, did not differ between fatal and nonfatal cases, confirming previous observations (23). Although both HRP2 and parasite cfDNA are derived from ruptured and/or egressed parasites, their levels did not correlate; perhaps cfDNA has a shorter half-life or is more susceptible to degradation or renal, splenic, or liver clearance (56). Our data provide evidence that measures that incorporate global cfDNA, and specifically host cfDNA, are more closely associated with CM fatal outcome than parasite cfDNA, implicating host or global cfDNA in the pathogenic processes of CM. Further studies are needed to evaluate if cfDNA is a biomarker of pathogenic processes or plays a physiological role in CM pathogenesis.
One major source of host cfDNA could be NETs, mesh-like DNA decorated with antimicrobial proteins extruded from activated neutrophils in a process called NETosis that serves as a defense mechanism to trap invading pathogens (57). In malaria, several studies indicate that activated neutrophils and effector functions contribute to pathogenesis and are associated with disease severity (25–27). cfDNA, as a NET remnant, is considered a surrogate marker of circulating NETs and correlates well with other NET markers such as citrullinated histones, elastase, or MPO (57). In support of a role for NETs in CM, levels of cfDNA concentration (total and host) correlate with concentrations of MPO, an effector component of NETs, and with IL-8, a neutrophil chemokine (57), suggesting that the cfDNA detected in patients with malaria may be derived from NETs. Uncontrolled neutrophil effector functions (NETosis, cytokine production) coupled with impaired clearance of neutrophil-derived components (e.g., DNA and histones) has been shown to promote severe systemic inflammation, contributing to pathogenesis (27, 57). Naked cfDNA is also considered a damage-associated molecular pattern, serving as an antigen or auto-antigen to activate innate immune cell pathways (58).
In our analysis, parasite cfDNA levels also correlated with neutrophil markers. This may be explained by the antigenic-like properties present in parasite histones and parasite DNA, which are both recognized as non–self antigens (55, 59–61) that exacerbate innate immune activation and inflammation. Corroborating these presumptions, we further demonstrate that inflammatory markers (IL-1β, IL-6, TNF-α, MCP-1, and IL-10) also positively correlate with all cfDNA measurements, as well as the marker for endothelial activation ANG-2. Therefore, other leukocytes could contribute cfDNA, as the cytokines analyzed are predominantly of monocyte/macrophage origin and recent work suggests that these cell types also undergo a process similar to NETosis (62). Endothelial cells could also be another source for cfDNA in malaria patients.
The simplicity in methodology, high assay sensitivity, and wide detection range of cfDNA concentrations detected among the different clinical malaria groups suggest that plasma DNA quantification may be a useful bedside tool in identifying potentially fatal or severe CM cases in field settings where resources and trained staff are limited. An advantage of the Qubit fluorometry assay is the very low volume (0.5 μL) of serum or plasma required. Access to blood samples is often limited, as phlebotomy in children is considered an invasive procedure. With this assay, a simple finger prick in a field setting would suffice. The ease of the assay and limited need for extensive reagents and expertise may also make it more cost-effective relative to imaging, funduscopy, hematology, or immunology laboratory testing. Further comparative assessment is needed. Given that this work is a retrospective case-control study, the potential of using total cfDNA as a prognostic biomarker for predicting fatal outcome will need to be confirmed and evaluated in larger prospective studies. Nonetheless, the data presented here suggest that cfDNA may be indicative of a unique pathophysiology occurring in severe or potentially fatal CM and that cfDNA warrants further consideration as a biomarker useful for assessing risk for severe and fatal outcomes in CM.
Methods
Study design and cohort recruitment.
This was a case-control study, and children were enrolled from several ongoing malaria studies conducted in Malawi. CM (n = 151) patients were admitted to the Pediatric Research Ward within Queen Elizabeth Central Hospital (QECH) in Blantyre, Malawi, during seasons 2015–2017 from January to June (Figure 1). For UM (n = 77), children presenting to the Accident and Emergency Pediatric Department at QECH were enrolled during seasons 2015–2016 from January to June. HC (n = 60) were children presenting to the Ndirande Health Centre for routine check-up at the vaccination clinic. Inclusion criteria include the following: (a) the CM group included children between the ages of 6 months and 12 years who met the WHO classification of CM+ for P. falciparum parasitemia, a Blantyre coma score (BCS) ≤ 2 that remained following treatment of hypoglycemia and/or seizures, and exclusion of other identifiable causes of coma; (b) the UM group included children 1–12 years old, with a history of fever (>37.8°C), normal mental status as measured by the BCS of 5, peripheral P. falciparum parasitemia, and no overt signs of compromised health, malnutrition, or symptoms of severe malaria (UM cases received treatment per national guidelines); and (c) the HC group included children who were between 1 and 12 years of age, confirmed to be medically well, fully alert, nonfebrile, and negative for malaria Rapid Diagnostic Test (RDT) blood smear and who had no history of malaria within the previous 3 months.
For all cases, 2–4 mL of blood was drawn into citrate anticoagulant tubes, and 0.5 mL of blood was drawn into EDTA tubes at the time of study enrollment. All blood samples were collected by trained clinicians and processed immediately following blood draw by laboratory staff. Within 4 hours of admission, all CM patients underwent thorough clinical examination, and an experienced ophthalmologist carried out direct and indirect ophthalmological funduscopic examinations through dilated pupils. At day 30 after hospital presentation, UM and CM patients returned for a follow-up appointment. Patients underwent a clinical examination, resolution of malaria was confirmed (thick Giemsa smear or RDT), and blood sample processed as above. Cytokine analyses and cfDNA measurements were carried out retrospectively.
Nonmalarial patients with febrile illness and respective control groups (UM and HC) were recruited under the Malaria and Bacterial Bloodstream Infections (MABSI) study. Children aged 6–60 months from Zingwangwa Health Centre in Blantyre, Malawi, were recruited between December 2017 and April 2018. Three groups of children were recruited: MABSI UM (n = 47); nonmalarial, noncomatose febrile (NMF, n = 40, with axillary temperature > 37.8°C at hospital/clinic presentation and malaria RDT test and PCR negative); and HC (n = 33) (Supplemental Figure 1). A 3-mL venous blood sample from each participant at enrollment was collected in serum collection tubes and processed. Recruitment of NMC patients under an ongoing Childhood Aetiologies of Severe Encephalopathy (CHASE) study were as follows: between February 2018 and April 2019, children between the ages of 3 months and 14 years admitted to the Pediatric Research Ward with a suspected central nervous system infection, fever, and deep coma (BCS ≤ 2) met inclusion criteria. Sample collection procedures were identical to the CM cases above with collection of 0.5 mL of serum in serum collection tubes. Children with clinically diagnosed NMC defined as the absence of malaria parasites on admission (n = 49) were included. Of these 49, 6 NMC were excluded from analysis due to a positive detection of P. falciparum DNA in plasma by qPCR, resulting in a total of n = 43 that were analyzed. Consort diagram describing recruitment of MABSI and CHASE cohorts is described in Supplemental Figure 1, and demographic and clinical characteristics are presented in Supplemental Table 1.
Quantification of total (parasite plus host) cfDNA in plasma.
Total cfDNA was measured directly from acute patient plasma using a Qubit 2.0 fluorometer (Invitrogen) (63, 44) according to the manufacturer’s instructions for the Qubit high-sensitivity dsDNA quantification (dsDNA HS assay kit, Invitrogen). All plasma samples were first diluted 1:20. For each measurement, the equivalent of 0.5 μL of plasma diluted in a total volume of 10 μL was added to 190 μL of Qubit working solution. The assay was optimized for plasma dilutions sufficient for a range of detection (0–100 ng/μL) suitable for measurement of baseline levels in HC to elevated levels seen in malaria patients. Quality controls run with experimental assays included standards of known DNA concentration and plasma from healthy donors (data not shown).
Direct quantification of parasite and host DNA in plasma.
We performed amplification of host or parasite cfDNA directly from plasma, with no DNA extraction, using a previously reported method (23) with a primer-probe set targeting Plasmodium 18S rRNA (parasite, PF3D7_1148600) or H. sapiens RPP30 gene (host, NM_001104546.1) (37). Sequences of primers and probes are as follows: Pf18s forward, 5′-GCTCTTTCTTGATTTCTTGGATG-3′, reverse, 5′-AGCAGGTTAAGATCTCGTTCG-3′, and probe: 5′-ATGGCCGTTTTTAGTTCGTG-3′; HsRPP30 forward, 5′-CTGGAAGACCTTCTGGTTATGG-3′, reverse, 5′-CTAGCACCAAGAGACCCATTT-3′, and probe: 5′-ACACTCTCCCAGATAGTTAGAGGCA-3′. Probes were synthesized by Integrated DNA Technologies (IDT) with the following modifications: 5′ FAM/Zen reporter and a 3′ IBFQ quencher. Amplifications were performed in duplicate on Applied Biosystems Step One Plus or Quantstudio 6 PCR systems. To quantify the cfDNA in our experimental samples, we generated standard curves from harvested synchronized parasite cultures at ring stage (P. falciparum 3D7, ATCC) of known parasitemia and concentrations (series of six 5-fold dilutions starting at 1.2 ng/μL in a 20 μL reaction, 5 × 104 genome copies). A standard curve for host gene amplification was generated from gDNA extracted from human foreskin fibroblasts (Boothroyd laboratory, Stanford University, Stanford, California, USA) and was also generated from known concentration (series of six 5-fold dilutions starting at 165 ng/μL in a 20 μL reaction, 5 × 104 genome copies). Standard curves were linear, with an average R2 of 0.98 ± 0.02 for both Pf18s and HsRPP30 target amplification. Nontemplate controls were included on each plate for quality control.
Plasma PfHRP2, MPO, ANG-2, and cytokine quantification.
Plasma samples were analyzed for PfHRP2 by ELISA according to validated methods (4). Samples were diluted 1:100 or 1:500 in PBS and plated in duplicate onto plates coated with anti-HRP2 antibody (Cellabs, KM8BP). PfHRP2 from cultured parasites was plated at multiple dilutions to generate a standard curve for accurate quantification of PfHRP2 levels in the experimental samples. Minor modifications were made to the manufacturer’s protocol, including all incubations carried out at 37°C in a humidified chamber. Upon optical density (OD) measurement, samples that were not in the linear range of the generated standard curve were rediluted (1:10 or 1:1000) and reanalyzed.
MPO concentrations in patient plasma were measured by ELISA (Human MPO ELISA kit, R&D Systems), according to the manufacturer’s protocol, with a modification in the suggested sample dilution. After testing 1:10 versus 1:50 dilutions, the 1:50 dilution of plasma allowed readings within the linear range for both UM and CM samples. OD measurements at 450 nm were made on either a Molecular Devices SpectraMax M2 (University of South Florida) or Biotek ELx800 (University of Malawi) plate reader, and values were converted to pg/mL based on a standard curve using a non–linear curve fit model.
ANG-2 in plasma was measured by custom magnetic bead based Luminex assays (R&D Systems Luminex Assays) specific to detect human ANG-2. As per manufacturer’s protocol, plasma was preclarified to remove particulates at 16,000 g at 4°C for 5 minutes. Clarified plasma was diluted 1:2 with diluent provided and incubated with Luminex beads overnight (18 hours) at 4°C. Washing steps and biotin-conjugated antibody and streptavidin incubations were as stipulated by manufacturer’s protocol. Beads were analyzed with a MagPix analyzer (Luminex Corp.), and concentrations calculated with the provided standard curve using xPONENT software.
Concentrations of plasma cytokines and chemokines including IL-1, IL-6, IL-8, IL-10, IFN-γ, TNF-α, and MCP1 were measured with the Cytokine Bead Arrays (BD Biosciences) (30). Some data from CM patients were previously published (30). Patient plasma was incubated with capture beads and processed according to the manufacturer’s protocol. A CyAn ADP flow cytometer (Beckman Coulter) was used for data acquisition, and data were analyzed with BD FCAP software v3.0 (BD Biosciences).
Statistics.
Descriptive statistics and univariate analyses were performed using Stata 12.1 or Prism 7.0 for Macintosh. For continuous variables, statistical differences were determined by Mann-Whitney U nonparametric analysis, Wilcoxon signed rank (paired data), Kruskal–Wallis with Dunn’s post hoc test for multiple comparisons, or Spearman’s correlation analysis. The data are presented as median ± IQR or Spearman rho (rs). Differences in categorical variables were determined using χ2 test, and the data are presented as n (% of group). ROC analysis was carried out for all cfDNA variables and other clinical variables (platelets and HRP2) with Prism 7.0. The cutoff point for total cfDNA was calculated using Youden Index (J statistic). Multivariate logistic regression models combining cfDNA variables, HRP2, and platelet counts were generated in R software using the glm function with the family and error set to binomial and logit, respectively. Specifically, we regressed survival against every pair-wise combination of the previously listed variables (a total of 5 variables and 10 combinations). The ROC values were calculated using the R verification package, which calculates the area under an ROC curve (AUC) and the associated P value (64). All statistics were conducted with P < 0.05 considered significant.
Study approval.
The study was approved by the IRBs at the University of Malawi, College of Medicine, the Albert Einstein College of Medicine, Michigan State University, and University of South Florida. Ethical approval for the MABSI and CHASE study was obtained from the Malawi College of Medicine Research Ethics Committee. Informed consent was obtained from parents or guardians of all study participants before enrollment. Participants were treated according to Malawi Government guidelines.
Author contributions
KK, AK, and IMV conceptualized the study. IMV, MS, MM, DA, LMT, AK, MN, and VH performed experiments. Data were analyzed by IMV with KK providing oversight/supervision. TK performed multiregression analyses. AK and VH collected and processed patient samples for BMP. SJR and WLM supervised recruitment of UM cases. AA processed patient samples for CHASE. MS and IK processed patient samples for MASBI. KBS and TET recruited patients, provided clinical data for CM patients, and provided intellectual input. SR recruited patients and provided clinical data for CHASE. TSN recruited patients and provided clinical data for MABSI. IMV, AK, and KK wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
First and foremost, we acknowledge the patients and their families for participation in our study and the Blantyre Malaria Project laboratory staff for sharing facilities and aiding with sample collection/processing. This work was supported by a National Health and Medical Research Council of Australia project grant APP1061993 (SJR), NIH National Center for Advancing Translational Science (NCATS) Einstein-Montefiore CTSA TL1TR001072 (AK), and the Burroughs Wellcome Fund (AK).
Version 1. 06/18/2020
Electronic publication
Footnotes
Role of funding: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e136279.https://doi.org/10.1172/jci.insight.136279.
Contributor Information
Iset Medina Vera, Email: isetvera@usf.edu.
Anne Kessler, Email: anne.kessler@nyu.edu.
Li-Min Ting, Email: liminting@usf.edu.
Visopo Harawa, Email: vharawa@gmail.com.
Dylan Allen, Email: dma13d@gmail.com.
Madi Njie, Email: madi.njie@yahoo.com.
McKenze Moss, Email: mckenze.moss@yahoo.com.
Monica Soko, Email: sokomonica2012@gmail.com.
Ajisa Ahmadu, Email: ajisa.ah@gmail.com.
Innocent Kadwala, Email: ikadwala@mlw.mw.
Tonney S. Nyirenda, Email: tnyirenda@medcol.mw.
Kami Kim, Email: kamikim@usf.edu.
References
- 1.Ashley EA, Pyae Phyo A, Woodrow CJ. Malaria. Lancet. 2018;391(10130):1608–1621. doi: 10.1016/S0140-6736(18)30324-6. [DOI] [PubMed] [Google Scholar]
- 2.Taylor TE, et al. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med. 2004;10(2):143–145. doi: 10.1038/nm986. [DOI] [PubMed] [Google Scholar]
- 3.Riggle BA, Miller LH, Pierce SK. Do we know enough to find an adjunctive therapy for cerebral malaria in African children? F1000Res. 2017;6:2039. doi: 10.12688/f1000research.12401.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox LL, et al. Histidine-rich protein 2 plasma levels predict progression to cerebral malaria in Malawian children with Plasmodium falciparum infection. J Infect Dis. 2013;208(3):500–503. doi: 10.1093/infdis/jit176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dondorp AM, et al. Estimation of the total parasite biomass in acute falciparum malaria from plasma PfHRP2. PLoS Med. 2005;2(8):e204. doi: 10.1371/journal.pmed.0020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendriksen IC, et al. Diagnosing severe falciparum malaria in parasitaemic African children: a prospective evaluation of plasma PfHRP2 measurement. PLoS Med. 2012;9(8):e1001297. doi: 10.1371/journal.pmed.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seydel KB, et al. Plasma concentrations of parasite histidine-rich protein 2 distinguish between retinopathy-positive and retinopathy-negative cerebral malaria in Malawian children. J Infect Dis. 2012;206(3):309–318. doi: 10.1093/infdis/jis371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conroy AL, et al. Whole blood angiopoietin-1 and -2 levels discriminate cerebral and severe (non-cerebral) malaria from uncomplicated malaria. Malar J. 2009;8:295. doi: 10.1186/1475-2875-8-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conroy AL, et al. Endothelium-based biomarkers are associated with cerebral malaria in Malawian children: a retrospective case-control study. PLoS ONE. 2010;5(12):e15291. doi: 10.1371/journal.pone.0015291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conroy AL, et al. Angiopoietin-2 levels are associated with retinopathy and predict mortality in Malawian children with cerebral malaria: a retrospective case-control study*. Crit Care Med. 2012;40(3):952–959. doi: 10.1097/CCM.0b013e3182373157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrera V, et al. Severity of retinopathy parallels the degree of parasite sequestration in the eyes and brains of malawian children with fatal cerebral malaria. J Infect Dis. 2015;211(12):1977–1986. doi: 10.1093/infdis/jiu592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birbeck GL, et al. Identification of malaria retinopathy improves the specificity of the clinical diagnosis of cerebral malaria: findings from a prospective cohort study. Am J Trop Med Hyg. 2010;82(2):231–234. doi: 10.4269/ajtmh.2010.09-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewallen S, Taylor TE, Molyneux ME, Wills BA, Courtright P. Ocular fundus findings in Malawian children with cerebral malaria. Ophthalmology. 1993;100(6):857–861. doi: 10.1016/S0161-6420(93)31563-0. [DOI] [PubMed] [Google Scholar]
- 14.Lewallen S, et al. Clinical-histopathological correlation of the abnormal retinal vessels in cerebral malaria. Arch Ophthalmol. 2000;118(7):924–928. [PubMed] [Google Scholar]
- 15.Kessler A, et al. Linking EPCR-Binding PfEMP1 to Brain Swelling in Pediatric Cerebral Malaria. Cell Host Microbe. 2017;22(5):601–614.e5. doi: 10.1016/j.chom.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seydel KB, et al. Brain swelling and death in children with cerebral malaria. N Engl J Med. 2015;372(12):1126–1137. doi: 10.1056/NEJMoa1400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pös O, Biró O, Szemes T, Nagy B. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 2018;26(7):937–945. doi: 10.1038/s41431-018-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahr S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 20.Duvvuri B, Lood C. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseases. Front Immunol. 2019;10:502. doi: 10.3389/fimmu.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weerakoon KG, McManus DP. Cell-Free DNA as a Diagnostic Tool for Human Parasitic Infections. Trends Parasitol. 2016;32(5):378–391. doi: 10.1016/j.pt.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Blauwkamp TA, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 23.Imwong M, et al. Plasma concentration of parasite DNA as a measure of disease severity in falciparum malaria. J Infect Dis. 2015;211(7):1128–1133. doi: 10.1093/infdis/jiu590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski D, et al. TNF concentration in fatal cerebral, non-fatal cerebral, and uncomplicated Plasmodium falciparum malaria. Lancet. 1990;336(8725):1201–1204. doi: 10.1016/0140-6736(90)92827-5. [DOI] [PubMed] [Google Scholar]
- 25.Feintuch CM, et al. Activated Neutrophils Are Associated with Pediatric Cerebral Malaria Vasculopathy in Malawian Children. mBio. 2016;7(1):e01300–e01315. doi: 10.1128/mBio.01300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kho S, et al. Circulating Neutrophil Extracellular Traps and Neutrophil Activation Are Increased in Proportion to Disease Severity in Human Malaria. J Infect Dis. 2019;219(12):1994–2004. doi: 10.1093/infdis/jiy661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knackstedt SL, et al. Neutrophil extracellular traps drive inflammatory pathogenesis in malaria. Sci Immunol. 2019;4(40):eaaw0336. doi: 10.1126/sciimmunol.aaw0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.Harawa V, et al. Brain swelling is independent of peripheral plasma cytokine levels in Malawian children with cerebral malaria. Malar J. 2018;17(1):435. doi: 10.1186/s12936-018-2590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gérardin P, Rogier C, Ka AS, Jouvencel P, Brousse V, Imbert P. Prognostic value of thrombocytopenia in African children with falciparum malaria. Am J Trop Med Hyg. 2002;66(6):686–691. doi: 10.4269/ajtmh.2002.66.686. [DOI] [PubMed] [Google Scholar]
- 32.Hochman SE, et al. Fatal Pediatric Cerebral Malaria Is Associated with Intravascular Monocytes and Platelets That Are Increased with HIV Coinfection. mBio. 2015;6(5):e01390–e01315. doi: 10.1128/mBio.01390-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank MO. Circulating Cell-Free DNA Differentiates Severity of Inflammation. Biol Res Nurs. 2016;18(5):477–488. doi: 10.1177/1099800416642571. [DOI] [PubMed] [Google Scholar]
- 34.Gould TJ, Lysov Z, Liaw PC. Extracellular DNA and histones: double-edged swords in immunothrombosis. J Thromb Haemost. 2015;13 Suppl 1:S82–S91. doi: 10.1111/jth.12977. [DOI] [PubMed] [Google Scholar]
- 35.Paunel-Görgülü A, et al. cfDNA correlates with endothelial damage after cardiac surgery with prolonged cardiopulmonary bypass and amplifies NETosis in an intracellular TLR9-independent manner. Sci Rep. 2017;7(1):17421. doi: 10.1038/s41598-017-17561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Meer AJ, et al. Systemic inflammation induces release of cell-free DNA from hematopoietic and parenchymal cells in mice and humans. Blood Adv. 2019;3(5):724–728. doi: 10.1182/bloodadvances.2018018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamau E, Alemayehu S, Feghali KC, Saunders D, Ockenhouse CF. Multiplex qPCR for detection and absolute quantification of malaria. PLoS ONE. 2013;8(8):e71539. doi: 10.1371/journal.pone.0071539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gal S, Fidler C, Turner S, Lo YM, Roberts DJ, Wainscoat JS. Detection of Plasmodium falciparum DNA in plasma. Ann N Y Acad Sci. 2001;945:234–238. doi: 10.1111/j.1749-6632.2001.tb03891.x. [DOI] [PubMed] [Google Scholar]
- 39.Mharakurwa S, Simoloka C, Thuma PE, Shiff CJ, Sullivan DJ. PCR detection of Plasmodium falciparum in human urine and saliva samples. Malar J. 2006;5:103. doi: 10.1186/1475-2875-5-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nwakanma DC, et al. Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine. J Infect Dis. 2009;199(11):1567–1574. doi: 10.1086/598856. [DOI] [PubMed] [Google Scholar]
- 41.Park GS, Opoka RO, Shabani E, Wypyszynski A, Hanisch B, John CC. Plasmodium falciparum Histidine-Rich Protein-2 Plasma Concentrations Are Higher in Retinopathy-Negative Cerebral Malaria Than in Severe Malarial Anemia. Open Forum Infect Dis. 2017;4(3):ofx151. doi: 10.1093/ofid/ofx151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villaverde C, Namazzi R, Shabani E, Opoka RO, John CC. Clinical Comparison of Retinopathy-Positive and Retinopathy-Negative Cerebral Malaria. Am J Trop Med Hyg. 2017;96(5):1176–1184. doi: 10.4269/ajtmh.16-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small DS, et al. Evidence from a natural experiment that malaria parasitemia is pathogenic in retinopathy-negative cerebral malaria. Elife. 2017;6:e23699. doi: 10.7554/eLife.23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huttunen R, et al. Fatal outcome in bacteremia is characterized by high plasma cell free DNA concentration and apoptotic DNA fragmentation: a prospective cohort study. PLoS ONE. 2011;6(7):e21700. doi: 10.1371/journal.pone.0021700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trigg RM, Martinson LJ, Parpart-Li S, Shaw JA. Factors that influence quality and yield of circulating-free DNA: A systematic review of the methodology literature. Heliyon. 2018;4(7):e00699. doi: 10.1016/j.heliyon.2018.e00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sager R, Kutz A, Mueller B, Schuetz P. Procalcitonin-guided diagnosis and antibiotic stewardship revisited. BMC Med. 2017;15(1):15. doi: 10.1186/s12916-017-0795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franchini M, Veneri D, Lippi G. Thrombocytopenia and infections. Expert Rev Hematol. 2017;10(1):99–106. doi: 10.1080/17474086.2017.1271319. [DOI] [PubMed] [Google Scholar]
- 48.Jain V, et al. Plasma IP-10, apoptotic and angiogenic factors associated with fatal cerebral malaria in India. Malar J. 2008;7:83. doi: 10.1186/1475-2875-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erdman LK, et al. Chitinase 3-like 1 is induced by Plasmodium falciparum malaria and predicts outcome of cerebral malaria and severe malarial anaemia in a case-control study of African children. Malar J. 2014;13:279. doi: 10.1186/1475-2875-13-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John CC, Opika-Opoka R, Byarugaba J, Idro R, Boivin MJ. Low levels of RANTES are associated with mortality in children with cerebral malaria. J Infect Dis. 2006;194(6):837–845. doi: 10.1086/506623. [DOI] [PubMed] [Google Scholar]
- 51.Moxon CA, et al. Laboratory evidence of disseminated intravascular coagulation is associated with a fatal outcome in children with cerebral malaria despite an absence of clinically evident thrombosis or bleeding. J Thromb Haemost. 2015;13(9):1653–1664. doi: 10.1111/jth.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salluh JI, Soares M. ICU severity of illness scores: APACHE, SAPS and MPM. Curr Opin Crit Care. 2014;20(5):557–565. doi: 10.1097/MCC.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 53.den Boer S, de Keizer NF, de Jonge E. Performance of prognostic models in critically ill cancer patients - a review. Crit Care. 2005;9(4):R458–R463. doi: 10.1186/cc3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franklin BS, et al. Plasma circulating nucleic acids levels increase according to the morbidity of Plasmodium vivax malaria. PLoS ONE. 2011;6(5):e19842. doi: 10.1371/journal.pone.0019842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gillrie MR, et al. Plasmodium falciparum histones induce endothelial proinflammatory response and barrier dysfunction. Am J Pathol. 2012;180(3):1028–1039. doi: 10.1016/j.ajpath.2011.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20(8):1057–1067. doi: 10.1080/15384047.2019.1598759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 58.Fischer S. Pattern Recognition Receptors and Control of Innate Immunity: Role of Nucleic Acids. Curr Pharm Biotechnol. 2018;19(15):1203–1209. doi: 10.2174/138920112804583087. [DOI] [PubMed] [Google Scholar]
- 59.Gazzinelli RT, Kalantari P, Fitzgerald KA, Golenbock DT. Innate sensing of malaria parasites. Nat Rev Immunol. 2014;14(11):744–757. doi: 10.1038/nri3742. [DOI] [PubMed] [Google Scholar]
- 60.Rivera-Correa J, et al. Plasmodium DNA-mediated TLR9 activation of T-bet+ B cells contributes to autoimmune anaemia during malaria. Nat Commun. 2017;8(1):1282. doi: 10.1038/s41467-017-01476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sisquella X, et al. Malaria parasite DNA-harbouring vesicles activate cytosolic immune sensors. Nat Commun. 2017;8(1):1985. doi: 10.1038/s41467-017-02083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granger V, et al. Human blood monocytes are able to form extracellular traps. J Leukoc Biol. 2017;102(3):775–781. doi: 10.1189/jlb.3MA0916-411R. [DOI] [PubMed] [Google Scholar]
- 63.Mardis E, McCombie WR. Library Quantification: Fluorometric Quantitation of Double-Stranded or Single-Stranded DNA Samples Using the Qubit System. Cold Spring Harb Protoc. 2017;2017(6):pdb.prot094730. doi: 10.1101/pdb.prot094730. [DOI] [PubMed] [Google Scholar]
- 64.Mason SJ, Graham NE. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: Statistical significance and interpretation. Q J R Meteorol Soc. 2002;128:2145–2166. doi: 10.1256/003590002320603584. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.