Abstract
The G2019S mutation in leucine-rich repeat kinase 2 (LRRK2) is a common cause of Parkinson's disease (PD) and results in age-related dopamine neuron loss and locomotor dysfunction in Drosophila melanogaster through an aberrant increase in bulk neuronal protein synthesis. Under nonpathologic conditions, protein synthesis is tightly controlled by metabolic regulation. Whether nutritional and metabolic influences on protein synthesis can modulate the pathogenic effect of LRRK2 on protein synthesis and thereby impact neuronal loss is a key unresolved question. Here, we show that LRRK2 G2019S-induced neurodegeneration is critically dependent on dietary amino acid content in Drosophila studies with both sexes. Low dietary amino acid concentration prevents aberrant protein synthesis and blocks LRRK2 G2019S-mediated neurodegeneration in Drosophila and rat primary neurons. Unexpectedly, a moderately high-amino acid diet also blocks dopamine neuron loss and motor deficits in Drosophila through a separate mechanism involving stress-responsive activation of 5′-AMP-activated protein kinase (AMPK) and neuroprotective induction of autophagy, implicating the importance of protein homeostasis to neuronal viability. At the highest amino acid diet of the range tested, PD-related neurodegeneration occurs in an age-related manner, but is also observed in control strains, suggesting that it is independent of mutant LRRK2 expression. We propose that dietary influences on protein synthesis and autophagy are critical determinants of LRRK2 neurodegeneration, opening up possibilities for future therapeutic intervention.
SIGNIFICANCE STATEMENT Parkinson's disease (PD) prevalence is projected to rise as populations continue to age, yet there are no current therapeutic approaches that delay or stop disease progression. A broad role for leucine-rich repeat kinase 2 (LRRK2) mutations in familial and idiopathic PD has emerged. Here, we show that dietary amino acids are important determinants of neurodegeneration in a Drosophila model of LRRK2 PD. Restricting all amino acids effectively suppresses dopaminergic neuron loss and locomotor deficits and is associated with reduced protein synthesis, while moderately high amino acids similarly attenuate these PD-related phenotypes through a stress-responsive induction of 5′-AMP-activated protein kinase and autophagy. These studies suggest that diet plays an important role in the development of PD-related phenotypes linked to LRRK2.
Keywords: Drosophila, LRRK2, neurodegeneration
Introduction
Parkinson's disease (PD) pathology is clearly age related, leading to the hypothesis that PD and aging manifest through common mechanisms (Collier et al., 2017). Dietary restriction (i.e., a moderate reduction in food intake) impacts aging in many species including primates (Gems and Partridge, 2013). Evidence in animal models and more recently humans indicates that the beneficial effects of dietary restriction are primarily attributable to reduced protein intake (Kapahi et al., 2004; Grandison et al., 2009; Levine et al., 2014). Dietary protein influences protein synthesis through governing the pool of available amino acids and by signaling to metabolic pathways that couple nutrient sensing to anabolic output. A central amino acid sensor in eukaryotes is the mechanistic target of rapamycin complex 1 (mTORC1) which regulates protein synthesis in response to the supply of specific amino acids (Kim et al., 2002). mTORC1 is evolutionarily conserved in invertebrates such as Drosophila, where the orthologous complex is called TORC1 (Oldham, 2011). Blocking TORC1 signaling in flies by manipulating the pathway components dTsc1, dTsc2, dTOR, or S6K phenocopies the life span-extending effects of dietary restriction (Kapahi et al., 2004). Another important effector of TORC1 is the protein synthesis repressor 4E-BP (eIF4E binding protein), whose activity is potentiated on TORC1 inhibition through dietary restriction. Silencing 4E-BP prevents the maximal life-span extension obtained through dietary restriction, providing direct evidence that enhanced longevity is dependent on TORC1 regulation of protein synthesis (Zid et al., 2009). Despite a link with aging, the role of protein consumption and protein synthesis in age-related neurodegeneration is largely unexplored. Studies in mice and primate toxin models of PD show that restricting calorie intake by reducing the quantity of consumed food (that still contains the same nutrient composition) leads to attenuated motor deficits and nigral dopamine neuron loss (Maswood et al., 2004; Bayliss et al., 2016), but no studies to date have investigated the potential for altering dietary protein levels alone to impact PD-related phenotypes.
A broad role for leucine-rich repeat kinase 2 (LRRK2) mutations in familial and idiopathic PD has been established (Healy et al., 2008; Simón-Sánchez et al., 2009). Expression of the common pathogenic mutation LRRK2 G2019S causes robust age-related and selective loss of dopamine neurons, with accompanying locomotor deficits in Drosophila (Liu et al., 2008). These phenotypes contrast with existing rodent transgenic or knock-in LRRK2 models, which typically display more subtle PD-related phenotypes without overt neuron loss (Herzig et al., 2011), highlighting the utility of Drosophila models as an important tool for understanding LRRK2-induced neurodegeneration. We and others previously showed that expressing LRRK2 G2019S in flies causes elevated neuronal bulk protein synthesis, and that blocking this through protein synthesis inhibitors ameliorates age-related dopamine neuron loss and locomotor dysfunction (Imai et al., 2008; Martin et al., 2014b; Penney et al., 2016). The observation that elevated bulk protein synthesis contributes to PD-related phenotypes in LRRK2 G2019S flies generates a compelling need to understand whether metabolic regulation of protein synthesis through diet can impact dopamine neuron survival with age. Recently, a chemically defined “holidic” Drosophila food medium was described that contains all dietary components necessary to promote optimal fly survival and fecundity (Piper et al., 2014). Holidic food is a completely defined and synthetic medium composed of purified ingredients that contrasts with oligidic medium, which is yeast based and contains natural ingredients but is not amenable to modifying individual nutrient classes.
Here, we used a holidic Drosophila diet approach to investigate the impact of dietary amino acid levels on PD-related phenotypes in LRRK2 G2019S Drosophila. Upon testing a range of amino acid concentrations, we found that substantially reducing amino acid concentration relative to the estimated content of our standard chemically undefined Drosophila food rescues age-related dopamine neuron loss and locomotor dysfunction in LRRK2 G2019S flies. This is associated with a reduction in bulk protein synthesis, but independent of TORC1 activity, autophagy induction, or activation of the sensor of amino acid deficiency GCN2 (general control nonderepressible 2), suggesting that this level of restriction does not constitute amino acid starvation. Elevating dietary amino acid levels within a range unexpectedly suppresses PD-related phenotypes through a distinct mechanism involving stress-responsive induction of 5′-AMP-activated protein kinase (AMPK), with associated enhancement of autophagy and dampening of TORC1 activity in aged flies. We further extend our findings to show that the restriction of amino acids rescues neurite shortening and cell death associated with LRRK2 G2019S expression in rat cortical neurons. Collectively, our results uncover an exquisite sensitivity of LRRK2 G2019S-mediated neurodegeneration to dietary amino acid content in Drosophila and rodent neurons.
Materials and Methods
Drosophila stocks and culture.
The UAS-LRRK2-G2019S and UAS-LRRK2-WT lines were a gift from Wanli Smith (Johns Hopkins University, Baltimore, MD) and were backcrossed to w1118 as described previously (Martin et al., 2014b). Expression of these transgenes was driven by the dopaminergic Ddc-GAL4 driver (BDSC 7010) or the pan-neuronal elav-GAL4 driver (BDSC 8765) as indicated. GAL4 drivers were additionally crossed to w1118 to generate non-LRRK2 transgenic controls. Neurodegenerative phenotypes were assessed using the dopaminergic Ddc-GAL4 driver, while protein synthesis and biochemical markers were assessed in whole heads following pan-neuronal LRRK2 expression (elav-GAL4). Distinct changes in dopamine neurons might not be detected by this approach, but key mechanisms raised through these studies were tested in genetic epistasis experiments for their role in neurodegeneration using the Ddc-GAL4 driver. RNAi fly stocks used were UAS-AMPK-IR 1 (BDSC 25 931); UAS-AMPK-IR 2 (BDSC 32 371) and UAS-Atg1-IR (BDSC 35 177). UAS-Atg1DN (UAS-Atg1KQ#5B) is a dominant-negative form of Atg1 (Scott et al., 2007) and was a gift from Eric Baehrecke (University of Massachusettes, Worcester, MA). For LRRK2 G2019S epistasis experiments, double-transgenic Ddc-GAL4; UAS-LRRK2-G2019S and Ddc-GAL4; UAS-LRRK2-WT lines were generated. All flies were maintained at 25°C and 60% relative humidity under a 12 h light/dark cycle.
Holidic media preparation.
Chemically defined holidic media were prepared following detailed protocols described by Piper et al. (2014) and using the HUNTaa composition of amino acids (Extended Data Fig. 1-1, specific formulations), or an alternative “exome-matched” ratio of dietary amino acids following methods previously described (Piper et al., 2017). All food components were from the same sources as those used in the study by Piper et al. (2014), and, following preparation, holidic media were stored at 4°C until use. The total amino acid-based nitrogen content (millimolar, designated as “N”) of the four diets used were as follows: 25, 50, 200, and 400N. This was achieved by modifying the levels of all essential and nonessential amino acids equally while keeping all other dietary components constant. Sucrose levels were standardized to 50 mM, abbreviated “50S” in all media, as per Piper et al. (2014), to promote optimal fly longevity. Flies were reared on our standard chemically undefined medium and transferred to holidic media as adults to avoid the potential effects of holidic medium or altered amino acid levels on development, as previously reported (Piper et al., 2014). Cohorts (0–3 d old, selecting against flies with visible signs of recent eclosion) were collected under brief anesthesia and transferred to 25N, 50N, 200N or 400N holidic food diets at 25 flies/vial. Flies were housed on a single holidic diet and switched to fresh food every 3 d until testing. For the 25N-phenylalanine (F)/histidine (H)/tryptophan (W) medium used to assess protein synthesis, phenylalanine, histidine, and tryptophan were present at 25N medium levels, while all other amino acids were at 50N levels (Extended Data Fig. 6-1, composition).
Estimation of protein nitrogen molarity in standard Drosophila medium.
The standard chemically undefined medium used in prior studies (Martin et al., 2014b) contained (per liter) the following: yeast 15 g, cornmeal 60 g, agar 5 g, corn syrup 67 ml, propionic acid 4 ml, and tegosept 2.4 ml. Principle sources of protein in this medium are brewer's yeast (15 g/L; ∼40% protein content) and yellow cornmeal (60 g/L; ∼7% protein content). An estimate of protein-derived nitrogen molar content (N) was derived from total protein (10.2 g/L) via multiplying by 0.16 (Jones factor) and then dividing by the molar mass of nitrogen (14 g/mol) = 117 mm N.
Food intake.
Blue dye (erioglaucine disodium salt; catalog #861146, Sigma-Aldrich) was added at 1.5% (w/v) to holidic media during food preparation to assess food intake based on established procedures (Bjordal et al., 2014). Female flies (n = 10 per condition) were transferred to blue dye-labeled holidic medium at the same amino acid level as the medium they had been housed on and kept at 25°C (60% relative humidity) for 2 h. The experiment was terminated by transferring the flies into microtubes and flash freezing in liquid nitrogen. Whole bodies were washed and lysed in ice-cold water. Supernatant was collected after centrifugation at 13,000 rpm for 5 min at 4°C. The amount of ingested blue dye in the supernatant was measured spectrophotometrically at 620 nm (Multiskan FC, Thermo Fisher Scientific). Sample readings were normalized to the number of flies.
Fly brain immunohistochemistry and dopamine neuron counts.
Brains (12–15/condition) from flies aged on holidic media for 6 weeks were harvested, fixed, and permeabilized with 4% PFA in PBS-T, pH 7.4. Fixed brains were blocked in 5% normal donkey serum (catalog #D9663, Sigma-Aldrich) for 1 h at room temperature (RT), followed by incubation with anti-tyrosine hydroxylase (1:1000; catalog #22941, Immunostar) for 72 h at 4°C followed by extensive washing and incubation with Alexa Fluor 488 secondary antibody (1:2000; catalog #A-21202, Thermo Fisher Scientific) for 72 h at 4°C. Brains were washed extensively and whole-mounted in SlowFade Gold Antifade mounting medium (catalog #S36938, Thermo Fisher Scientific). Confocal z-stack images of the stained brains were acquired on Zeiss LSM 780 at 1 µm slice intervals. Projection images of the protocerebral posterior lateral 1 (PPL1) cluster were used for dopamine neuron counts.
Negative geotaxis assay.
Cohorts of 100 female flies (25 flies/vial) aged on holidic media were tested for motor function using negative geotaxis assays performed and analyzed as described previously (Gargano et al., 2005). The performance of flies in a single vial was calculated from the average height climbed by all flies in the vial 4 s after initiating climbing behavior. This generated a single datum (N =1), and the average scores of four vials were tested per line (N = 4).
35S-methionine/cysteine labeling of Drosophila brains.
Ex vivo labeling of protein synthesis in fly brains was conducted based on previously reported methods (Essers et al., 2016). Fly brains (eight per genotype) were harvested in Schneider's medium on ice and transferred to Schneider's medium containing 35S-methionine/cysteine (2 µCi/µl; catalog #NEG709A, PerkinElmer) and 30 μm chloroquine (to inhibit autophagy). Brains were incubated for 20 min at 25°C and washed twice in chilled PBS. On the final wash, all PBS was removed and the brains were flash frozen. Brains were homogenized by pestle and mortar in RIPA extraction buffer on ice. Protein from 50% of the lysate was precipitated by the addition of methanol and heparin [lysate/heparin (100 mg/ml)/methanol volume ratio of 150:1.5:600], centrifuged at 14,000 × g for 2 min, the supernatant was removed, and the pellet air dried. The protein pellet was resuspended in 8 m urea/150 mm Tris, pH 8.5, and 35S-methionine/cysteine incorporation was measured by scintillation counting and normalized to the number of brains. A portion of the remaining RIPA extraction lysate was separated by SDS-PAGE with equal protein loading and transferred to nitrocellulose membrane. The membrane was dried and 35S-methionine/cysteine incorporation was visualized by exposing to a phosphorimaging cassette followed by scanning using a Typhoon scanner.
Western blotting.
Fly heads (25 flies per condition) were harvested at the indicated ages and extracted in a 1% NP-40 lysis buffer, as follow: 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EGTA, 1% (v/v) NP-40, 100 mm NaF, 2 mm β-glycerophosphate, 2% (v/v) phosphatase inhibitor (catalog #P0044, Sigma-Aldrich), and protease inhibitors. Sample protein concentrations were determined by Pierce BCA Protein Assay Kit (catalog #PI3225, Thermo Fisher Scientific), and equal total protein amounts were electrophoretically separated by SDS-PAGE and transferred to nitrocellulose membranes (catalog #PI88018, Thermo Fisher Scientific). Blots were incubated with primary antibody for 18 h at 4°C. Primary antibodies used for Western blotting include the following: phospho (p)-S6K (1:750; catalog #9209, Cell Signaling Technology); p-4EBP (1:750; catalog #2855, Cell Signaling Technology); Non-p-4EBP (1:1000; catalog #4923, Cell Signaling Technology); actin (1:1000; catalog #4967, Cell Signaling Technology); p-LRRK2 (1:750; catalog #ab203181, Abcam); LRRK2 (1:1000; catalog #13046, Cell Signaling Technology); RPS15 (1:500; catalog #WH0006209M1, Sigma-Aldrich); p-AMPK (1:1000; catalog #2535, Cell Signaling Technology); AMPK (1:500; catalog #MABS1232, Millipore), and LAMP1 (1:1000; catalog #ab30687, Abcam). A P-RPS15 antibody used was previously described Martin et al. (2014b). Blots were imaged using a Syngene G:BOX gel doc system. Semiquantitative densitometric analyses were performed using ImageJ software (National Institutes of Health).
Quantitative real-time PCR.
Total RNA was isolated from fly heads (n = 15/condition) using TRIzol reagent (catalog #15596026, Thermo Fisher Scientific) and quantified on NanoDrop 2000c (Thermo Scientific). One hundred nanograms of total RNA was reverse transcribed using Superscript IV first-strand synthesis system (catalog #18091050, Thermo Fisher Scientific), using random hexamers. Quantitative real-time PCR assays were performed in Maxima SYBR Green/ROX qPCR Master Mix (catalog #K0222, Thermo Fisher Scientific) using a Thermo Fisher Scientific QuantStudio Three Real Time PCR system under the following sequence of conditions: 50°C for 2 min, 95°C for 10 min then 40 cycles (c) of 15 s at 95°C and 1 min at variable hybridization temperature. For grim transcript detection, cycle parameters described previously (Zheng et al., 2010) were followed. Primer sequences used in this study are as follows: dSesn: forward, 5′-CTCGACTCGATCCCTCCG-3′; reverse, 5′-CAGGTCATCGAGCTCGTCC-3′; c = 40; hybridization temperature = 58°C; Atg8a: forward, 5′-ACCAGGAACATCACGAGGAG-3′; reverse, 5′-TTAGCTAACTCGCCGTCCAT-3′; c = 40; hybridization temperature = 58°C; Atg9: forward, 5′-TCGTCTGGCTACTTGCCTTT-3′; reverse, 5′-AACCAGGTACGTGACCCAAG-3′; c = 40; hybridization temperature = 58°C; grim: forward, 5′-GTCGTCCTCATCGTTGTTCTGAC-3′; reverse, 5′-CCATCGCCTATTTCATACCCG-3′; c = 40; hybridization temperature = 57.9°C; reaper: forward, 5′-AGTCACAGTGGAGATTCCTGG-3′; reverse, 5′-TGCGATATTTGCCGGACTTTC-3′; c = 40; hybridization temperature = 57°C; actin: forward, 5′-GTGAAATCGTCCGTGACATC-3′; reverse, 5′-GGCAGCTCGTAGGACTTCTC-3′; c = 40; hybridization temperature = 58°C; AMPKα: forward, 5′-GTCAAGGTGGCCGTCAAGAT-3′; reverse, 5′-GGCGTCGATATGACCTGGTA-3′; c = 40; hybridization temperature = 57°C; Atg1: forward, 5′-CCGTGTCGCCAGTTGACAT-3′; reverse, 5′-GGTGATGGTGGCACAAATGG-3′; c = 40; hybridization temperature = 58°C.
Rat cortical neuron culture.
Embryonic day 16 Sprague Dawley rat cortices (BrainBits) were dissociated in trypsin substitute TrypLE enzyme mix (catalog #12-605-010, Thermo Fisher Scientific) for 6 min at 37°C and triturated vigorously with a silanized glass pipette for 1 min. After allowing debris to settle, the supernatant containing dissociated cells was centrifuged at 120 × g for 1 min, resuspended in Neurobasal without phenol red (catalog #12-348-017, Thermo Fisher Scientific)/1% B27 supplement (catalog #17-504-044, Thermo Fisher Scientific)/1% GlutaMAX (catalog #35-050-061, Thermo Fisher Scientific) and plated at a density of 5.0 × 104 cells/well on poly-d-lysine (catalog # P6407, Sigma-Aldrich)-coated 24-well glass-bottom plates (catalog #50-114-9070, Thermo Fisher Scientific). Neurons were cultured at 37°C/5% CO2, and glial growth was inhibited at DIV 4 by the addition of 5-fluoro-2′-deoxyuridine (30 μm; catalog #F0503, Sigma-Aldrich). A 50% fresh culture medium change was performed every 4 d.
Generation of amino acid-diluted rat neuron culture medium.
A Neurobasal-based medium without amino acids was generated using the formulation of standard Neurobasal without phenol red (catalog #12-348-017, Thermo Fisher Scientific) and omitting all amino acids (Extended Data Fig. 7-1). All other standard Neurobasal components were present at standard formula concentrations and using cell culture-grade reagents. This medium was vacuum filtered and combined with standard Neurobasal medium to dilute amino acids to a range of 0.4–0.8× relative to standard Neurobasal medium. All media were then combined with B27 supplement (but no GlutaMAX for l-glutamine as this is an additional source of amino acid) and filtered again.
LRRK2 rat neuron toxicity assays.
Toxicity (loss of neurites and appearance of TUNEL-positive nuclei) in rat cortical neurons overexpressing LRRK2 WT or LRRK2 G2019S was assessed using previously described methods (Martin et al., 2014b) 48 h following plasmid transfection using Lipofectamine 2000 (catalog #11-668-027, Thermo Fisher Scientific). At DIV 10, neurons were switched to Invitrogen Opti-MEM (catalog #115439, Thermo Fisher Scientific) reduced serum medium and transfected with plasmid DNA. Myc-tagged LRRK2 and pEGFP(N3) for tracing neurites at a 10:1 ratio were mixed with Lipofectamine 2000 at a 1:3 ratio in Opti-MEM. Transfection complex was removed after 2 h and replaced with amino acid-modified or standard culture medium (0.4–1.0 dilution, see above for description of amino acid-diluted media) for an additional 48 h of culture. Neurons were washed, fixed (4% PFA in PBS-T at pH 7.4), blocked, permeabilized for 1 h in 10% donkey serum/PBS-T/0.3% Triton-X-100, pH 7.4, and incubated overnight (at 4°C) in mouse anti-GFP (3E6; catalog #A-11 120, Thermo Fisher Scientific) and rabbit anti-LRRK2 (D18E12; catalog #13046, Cell Signaling Technology) primary antibodies, washed extensively, and incubated in anti-mouse Alexa Fluor 488 (1:2000; catalog #A21202, Thermo Fisher Scientific) and anti-rabbit Alexa Fluor 647 (1:2000; catalog #A32733, Thermo Fisher Scientific) secondary antibodies for 1 h at RT. Cell death was assessed by TUNEL assay (with TMR, red-dUTP; catalog #12156792910, Sigma-Aldrich), and nuclei were labeled with Invitrogen DAPI stain (catalog #D1306, Thermo Fisher Scientific) following manufacturer protocols. Neurons were imaged on a Zeiss LSM 780 Confocal Microscope. The vast majority of GFP-positive neurons also overexpressed LRRK2, allowing the effects of LRRK2 expression to be visualized in these neurons. Injured neurons without at least one smooth extension (neurite) twice the length of the cell body were recorded, and the percentage of GFP-positive injured neurons in each experimental group relative to all GFP-positive neurons was derived. The percentage of GFP-positive neurons with TUNEL-positive nuclei was also measured. At least 75 neurons were counted per group per independent experiment.
Protein synthesis surface sensing of translation assay in rat neurons.
The surface sensing of translation (SUnSET) assay permits the nonradioactive monitoring of global protein synthesis in mammalian cells by visualization of low-concentration puromycin incorporation into nascent polypeptide chains (Schmidt et al., 2009). Puromycin incorporation results in nascent chain termination but when administered at low micromolar concentrations does not substantially interfere with global protein synthesis. DIV 10 neurons were cultured in amino acid-modified (see above for description) or standard culture medium for 48 h post-transfection and then exposed to 2 μm puromycin (catalog #540222, Millipore) for 10 min at 37˚C/5% CO2. Following incubation, neurons were immediately fixed (4% PFA in PBS-T at pH 7.4), blocked and permeabilized for 1 h in 10% donkey serum/PBS-T/0.3% Triton X-100, pH 7.4, and incubated for 2 h (at RT) in mouse anti-puromycin 12D10 antibody (catalog #MABE343, Sigma-Aldrich), washed extensively, then incubated in anti-mouse Alexa Fluor 488 secondary antibody (1:2000; catalog #A21202, Thermo Fisher Scientific) for 1 h at RT. Neurons were imaged on a Zeiss LSM 780 Confocal Microscope. Immunofluorescence signal (integrated density counts of cells corrected for background) was quantified using ImageJ where a single n represents the total signal from 20 neurons.
ADP/ATP ratio assay.
The ADP/ATP Ratio Assay Kit (catalog #MAK135, Sigma-Aldrich) was used following all manufacturer instructions. Fly heads were homogenized in ATP reagent and centrifuged for 10 min at 4°C. Luminescence in the supernatant was read using a SpectraMax microplate reader, and then the ADP reagent was added to assess ADP levels and derive ADP/ATP ratio according to manufacturer instructions.
LRRK2 in vitro kinase assay.
LRRK2 kinase assays were performed using purified recombinant GST-tagged LRRK2 (amino acids 970–2527) or its variants Invitrogen D1994A and G2019S (Thermo Fisher Scientific). V5-tagged p70 S6K was affinity purified from HEK293T cells using V5 antibody (V5-10). Proteins were incubated in a kinase assay buffer (20 mm HEPES, pH 7.5, 5 mm EGTA, 20 mm β-glycerophosphate, 20 mm MgCl2, 10 μm ATP and 0.5 µCi of γ-32P-ATP at a combined volume of 30 µl), and the reaction mixture was incubated at 30°C for 30 min. Reactions were quenched by the addition of SDS sample loading buffer and heated to 70°C for 10 min, and then proteins were resolved by SDS-PAGE. Gels were then fixed (50% methanol, 10% acetic acid), stained in Coomassie Brilliant Blue, heat-sealed in hybridization bags, and exposed to a phosphorimaging screen overnight at room temperature for assessing radioactive 32P incorporation. 32P/total protein for LRRK2 and p70 S6K was measured and normalized to that in the D1994A sample.
Experimental design and statistical analyses.
Quantified data presented in all figures are the mean ± SEM, and individual data points are plotted for data with n ≤ 10. Statistical analysis details for each individual experiment are described in figure legends, including the number of flies or number of groups of flies used(n), statistical tests (unpaired Student's t test, ANOVA) with F values and Bonferroni post hoc analysis with associated p values. All statistical analyses were performed using GraphPad Prism software.
Results
Dietary amino acids regulate PD-related phenotypes in Drosophila
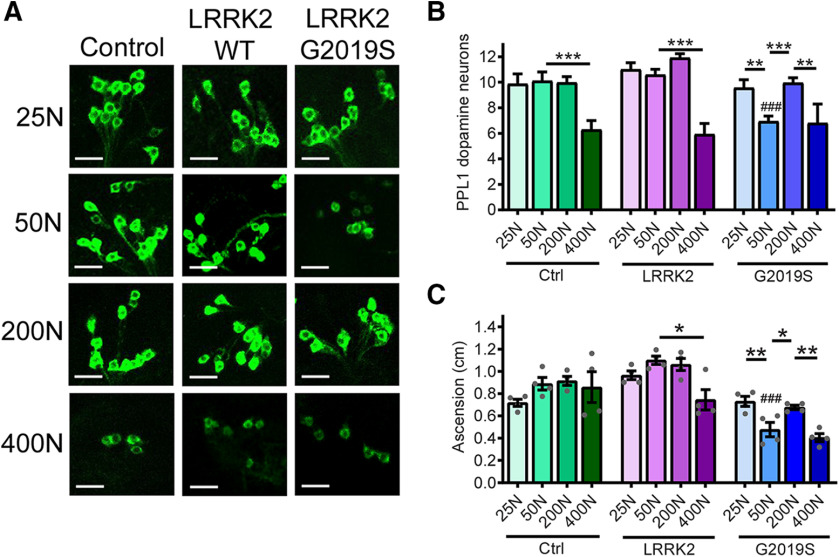
Flies expressing pathogenic human LRRK2 G2019S via the dopaminergic Ddc-GAL4 driver display an age-related loss of dopamine neurons with accompanying locomotor dysfunction at 6 weeks of age (Liu et al., 2008; Martin et al., 2014b). Flies expressing human LRRK2 WT do not exhibit these phenotypes, indicating that they arise from mutation-specific effects. Considering the established contribution of elevated bulk protein synthesis to LRRK2 G2019S-induced neurodegeneration (Imai et al., 2008; Martin et al., 2014b; Penney et al., 2016), we sought to determine whether dietary influences on protein synthesis can impact neurodegeneration. We aged flies expressing human LRRK2 WT or LRRK2 G2019S on a range of holidic diets in which the concentrations of all amino acids isvaried while those of all other food constituents remain constant throughout adulthood (Extended Data Fig. 1-1). In these diets, amino acid content is denoted by the total amino acid-derived millimolar nitrogen concentration (N) of the food (Piper et al., 2014). Amino acid and other nutrient content in the holidic medium is fixed and defined, which contrasts to those of a “standard” Drosophila laboratory diet, which can vary widely between laboratories (Piper et al., 2014) and possibly also between batches of food within the same lab, based on lot-to-lot ingredient variability. Piper et al. (2014) previously demonstrated that aging adult flies on diets with an amino acid concentration range between 0N and 400N causes major effects on median life span, which peaks on 50–100N amino acid diets and dramatically shortens at both extremes of this amino acid range. To assess whether a similar range of dietary amino acids can impact PD-related phenotypes caused by mutant LRRK2, we generated diets varying in amino acid concentrations from 25N to 400N and examined dopamine neuron loss and motor deficits following 6 weeks of aging on these diets. Both dopamine neuron viability and locomotor function are substantially affected by dietary amino acid concentration at this age (Fig. 1). Strikingly, LRRK2 G2019S-dependent dopamine neuron death and locomotor dysfunction are only observed in flies on a 50N amino acid diet. LRRK2 G2019S flies aged on 25N or 200N amino acid diets exhibit a rescue of dopamine neuron viability and motor abilities relative to those on a 50N diet (Fig. 1). The 400N diet elicits a loss of dopamine neurons, which occurs in all fly strains tested, suggesting that it is LRRK2 independent (Fig. 1). There is no significant effect of dietary amino acid levels on locomotor function in young control or mutant LRRK2 flies housed on these diets for 1 week (Fig. 2A), suggesting that the diet effects on neurodegenerative phenotypes seen in aged LRRK2 G2019S flies on a 50N diet are age dependent.
Figure 1.
Dietary amino acids impact PD-related phenotypes in aged flies. A, B, Effect of dietary amino acid levels on dopamine neuron number (immunostained for Tyrosine Hydroxylase) in the PPL1 cluster. Projection images from the right hemisphere of 6-week-old Control (Ctrl), LRRK2 WT, and LRRK2 G2019S fly brains are shown. ANOVAs for diet [F(3,67) = 7.00 (Control); F(3,57) = 23.91 (WT); F(3,57) = 9.57 (G2019S)] and genotype [F(2,53) = 19.32 (50N)] with Bonferroni post-tests; *p < 0.05, **p < 0.01 and ***p < 0.001 for diet effect and ###p < 0.001 for genotype effect, n = 12–15 brains/genotype/diet. Scale bars, 40 µm. C, Negative geotaxis performance of 6-week-old flies [ANOVAs for diet effect, F(3,12) = 1.24 (Control); F(5,18) 26.51 (WT); F(3,12) = 12.28 (G2019S) with Bonferroni post-tests, *p < 0.05, **p < 0.01, and for genotype effect, F(2,9) = 34.32 (50N) with Bonferroni post-test ###p < 0.001, n = 4 replicates of 25 flies/condition]. Genotypes are Control (Ddc-GAL4), LRRK2 WT (Ddc-GAL4/UAS-LRRK2 WT), and G2019S (Ddc-GAL4/UAS-LRRK2 G2019S). Data are the mean ± SEM. Holidic medium formulation is described in Extended Data Figure 1-1.
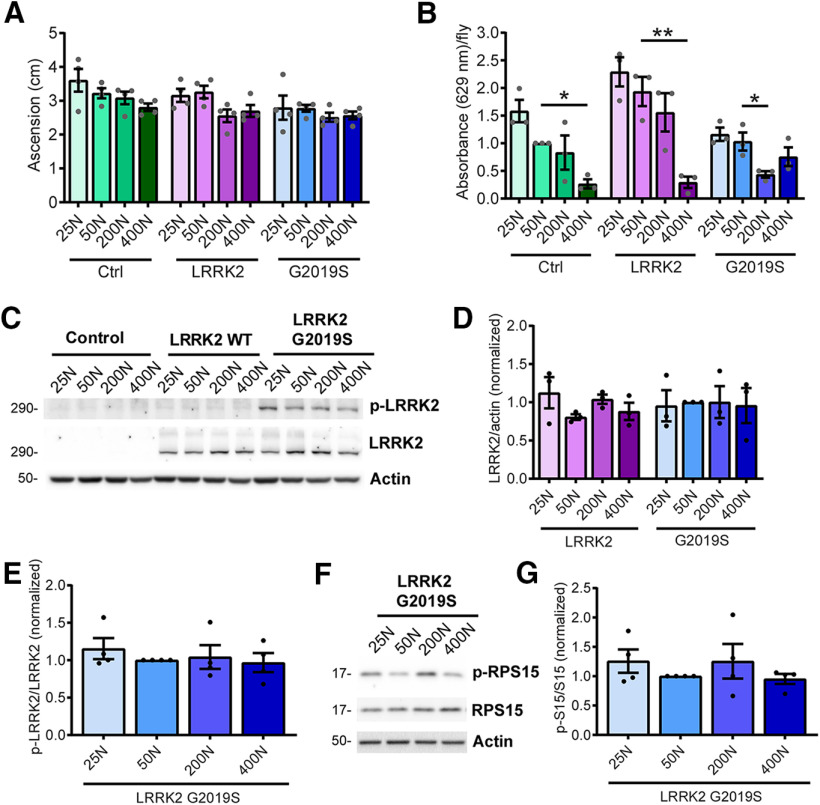
Figure 2.
Characterization of holidic diet effects on young fly locomotion, food intake, and LRRK2 kinase activity. A, No significant effect of diet on locomotor function in 1-week-old Control (Ctrl), LRRK2 WT, or LRRK2 G2019S flies [ANOVAs, F(3,12) = 2.45, p = 0.11 (Control); F(3,12) = 3.41, p = 0.053 (WT); F(3,12) = 0.49, p = 0.49 (G2019S), n = 4 replicates of 25 flies/condition]. Genotypes are Control (Ddc-GAL4), LRRK2 WT (Ddc-GAL4; UAS-LRRK2 WT), and LRRK2 G2019S (Ddc-GAL4; UAS-LRRK2 G2019S). B, Dietary amino acids significantly impact consumption of food [ANOVAs, F(3,8) = 8.18 (Control); F(3,8) = 11.32 (WT); F(3,8) = 5.67 (G2019S), Bonferroni post-test, *p < 0.05, **p < 0.01, n = 3 replicates of 10 flies/condition]. C–E, No effect of diet on human LRRK2 expression or kinase activity assessed by Ser1292 autophosphorylation relative to total LRRK2 in 6-week-old fly heads [ANOVAs, F(3,8) = 1.43, p = 0.31 (LRRK2 WT expression); F(3,8) = 0.02, p =0.99 (LRRK2 G2019S expression); F(3,12) = 0.43 (P-S1292 LRRK2 G2019S), n = 3–4 replicates of 25 heads/condition]. F, G, No effect of diet on phosphorylation of ribosomal protein S15 at T136 (ANOVA, F(3,12) = 0.78, p = 0.52, n = 4 replicates of 25 flies/condition). Genotypes in B–G are Control (elav-GAL4), LRRK2 WT (elav-GAL4; UAS-LRRK2 WT), and G2019S (elav-GAL4; UAS-LRRK2 G2019S). Data are the mean ± SEM.
Composition of holidic diet media. Download Figure 1-1, DOCX file (22KB, docx) .
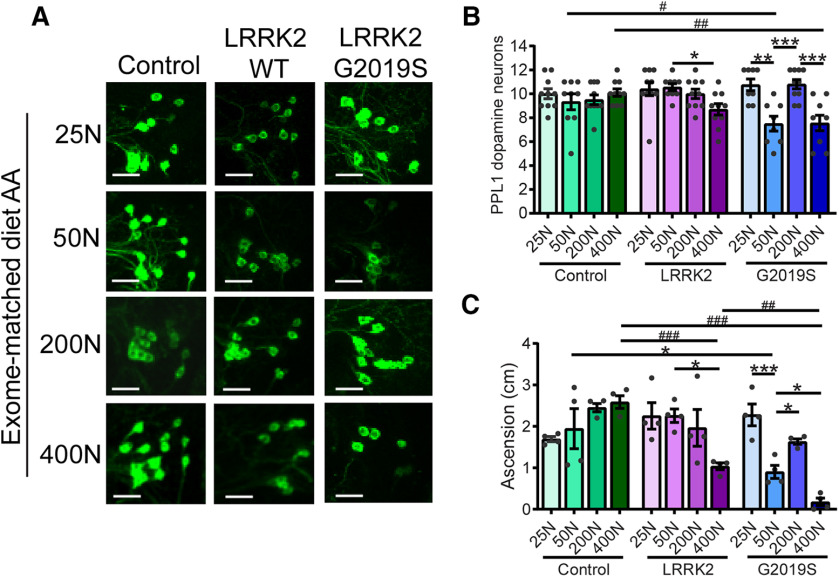
Food intake, measured by the ingestion of blue dye-labeled food, is inversely proportional to dietary amino acid concentration in 6-week-old flies (Fig. 2B), likely because of the rapid satiating effects of high amino acid diets, as has been reported for high-protein diets in Drosophila (Sun et al., 2017), rodents (Solon-Biet et al., 2014), and humans (Westerterp-Plantenga et al., 2009). LRRK2 toxicity is kinase dependent (Greggio et al., 2006), and to probe whether dietary amino acid concentration affects LRRK2 expression or activity in a manner that might modulate PD-related phenotypes, we assessed LRRK2 expression and LRRK2 G2019S autophosphorylation at Ser1292 as a readout of its kinase activity (Sheng et al., 2012). We find no evidence of altered LRRK2 expression or kinase activity in the heads of pan-neuronal LRRK2 transgenic flies (Fig. 2C–E) across dietary amino acid levels. This is further supported by the lack of dietary influence on phosphorylation of the LRRK2 substrate RPS15 (Fig. 2F,G). Following their initial characterization of life span changes with altered dietary amino acid content, Piper et al. (2017) reported a variant holidic diet formulation in which the ratio of individual amino acids within an overall dietary N was modified. This formulation was conceived to better match the dietary amino acid composition of the in silico translated exome, and while the relationship between dietary N and life span was reportedly very similar to the original formulation (Piper et al., 2017), exome-based predictions identified a potential arginine limitation in the original diet. The relationship between dietary amino acid levels (range, 25–400N) and PD-related phenotypes observed in LRRK2 G2019S flies with the original holidic medium are upheld with the modified amino acid formulation (Fig. 3), suggesting that the phenotypic modulation observed via the original formulation is not driven by changes in Arg levels disproportionately to other amino acids. Interestingly, the modified 400N diet did not result in PD-related phenotypes in controls as seen with the original 400N diet, raising the possibility that certain amino acid ratios or the elevation of specific amino acids beyond a threshold point may be important for manifestation of these phenotypes.
Figure 3.
A dietary amino acid formulation designed to match the in silico-translated exome impacts PD-related phenotypes in aged flies. A, B, Effect of exome-matched dietary amino acid levels on dopamine neuron number (immunostained for Tyrosine Hydroxylase) in the PPL1 cluster. Projection images from the right hemisphere of 6-week-old Control, LRRK2 WT, and LRRK2 G2019S fly brains are shown. ANOVAs for diet [F(3,35) = 3.54 (WT); F(3,31) = 12.02 (G2019S)] and genotype [F(2,23) = 7.60 (50N); F(2,26) = 6.79 (400N)]; Bonferroni post-tests: *p < 0.05, **p < 0.01, and ***p < 0.001 for diet; and #p < 0.05, ##p < 0.01 for genotype effect; n = 8–10 brains/genotype/diet. Scale bars, 40 µm. C, Negative geotaxis performance of 6-week-old flies [ANOVAs for diet (F(3,12) = 2.63 (Control); F(3,12) = 4.01 (WT); F(3,12) = 31.07 (G2019S); and genotype (F(2,9) = 5.35 (50N); F(2,9) = 117.3 (G2019S)]; Bonferroni post-test: *p < 0.05 **p < 0.01, ***p < 0.001 for diet; and ##p < 0.01, ###p < 0.001 for genotype effect; n = 4 replicates of 25 flies/condition). Genotypes are Control (Ddc-GAL4), LRRK2 WT (Ddc-GAL4/UAS-LRRK2 WT), and G2019S (Ddc-GAL4/UAS-LRRK2 G2019S). Data are the mean ± SEM.
Before this study, we consistently observed LRRK2 G2019S-induced dopamine neuron loss and locomotor deficits in 6-week-old flies fed standard chemically undefined food using the same Ddc-GAL4 and UAS-LRRK2 G2019S fly strains (Martin et al., 2014b). An estimation of the amino acid nitrogen content of the standard food used in those studies (see Materials and Methods) showed a closest match to the 50N diet used here, where we observe the same PD-related phenotypes. Collectively, these results indicate that the manifestation of LRRK2 G2019S-induced neurodegeneration in Drosophila is highly dependent on dietary amino acid content and, relative to the food used in our previous studies, which is closest to the 50N diet, can be alleviated at both lower (25N) and higher (200N) amino acid diets.
Modulation of LRRK2 neurotoxicity by low-amino acid diets is associated with attenuated protein synthesis
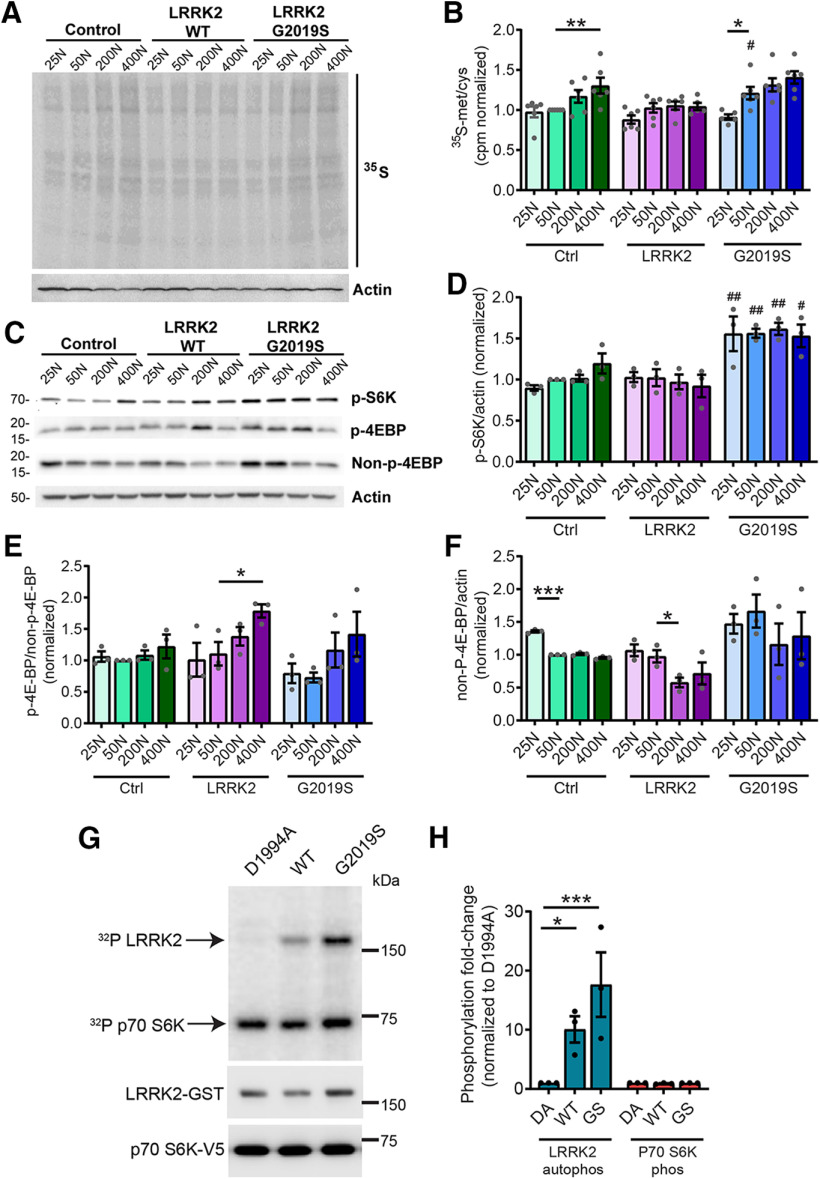
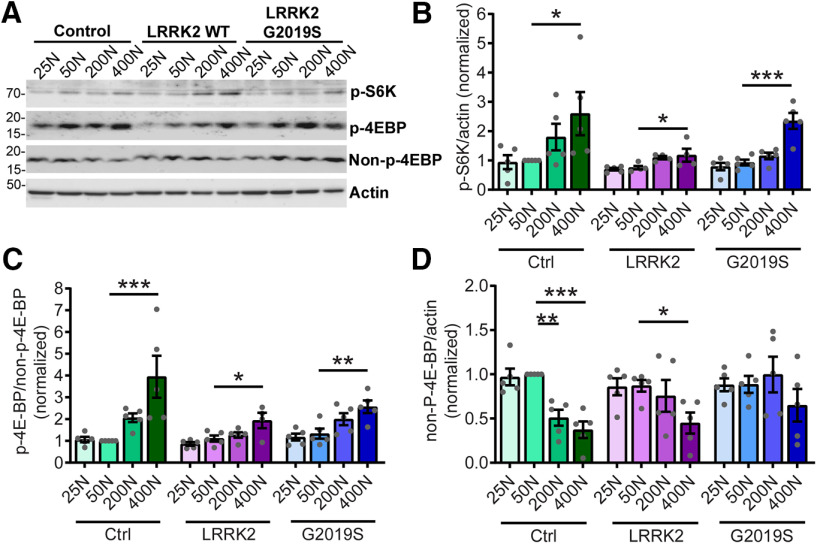
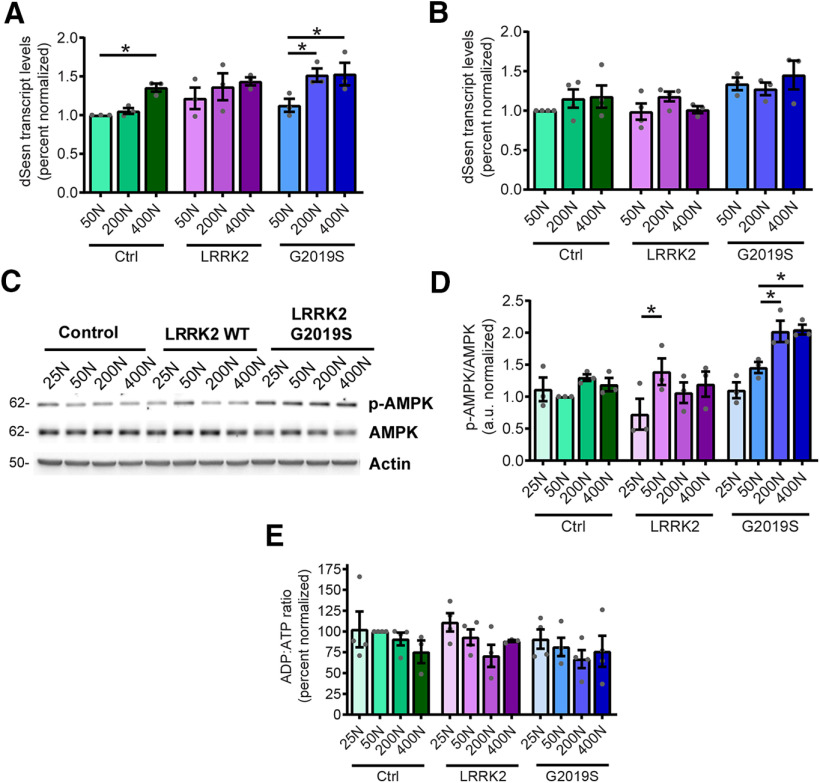
Since aberrant protein synthesis contributes to LRRK2 G2019S-mediated neurodegeneration in multiple models, we hypothesized that dietary amino acid levels modulate neuronal degeneration by directly impacting protein synthesis. To test this, we measured protein synthesis in aged pan-neuronal LRRK2 G2019S-expressing fly brains ex vivo. As expected, there is a trend toward elevated 35S-methionine/cysteine labeling of newly synthesized proteins at higher dietary amino acid levels in the heads of all genotypes (Fig. 4A,B). LRRK2 G2019S flies aged on a 50N diet exhibit higher global protein synthesis relative to control genotype flies aged on the same diet (Fig. 4A,B). This is consistent with a role for elevated protein synthesis in LRRK2 G2019S-induced neurodegeneration on this diet. There is a significant reduction in protein synthesis between 50N and 25N in LRRK2 G2019S flies, hence lowering protein synthesis is associated with the suppression of LRRK2 G2019S neurodegeneration. This is in accordance with prior studies using protein synthesis inhibitors (Martin et al., 2014a,b). Protein synthesis is sensitive to changes in amino acid levels in part via regulation of TORC1 signaling (Kim et al., 2002). TORC1 coordinates nutrient amino acid input with protein synthesis output through multiple channels including direct phosphorylation of downstream targets such as S6K (ribosomal protein S6 kinase) and 4E-BP. Surprisingly, there is no clear effect of dietary amino acid concentration on TORC1 activity assessed via 4E-BP and S6K phosphorylation in 6-week-old fly heads (Fig. 4C–E). While there is a trend for higher 4E-BP phosphorylation with increasing amino acids, consistent with the upregulation of protein synthesis, S6K phosphorylation is fairly constant on all diets. Interestingly, phospho-S6K is significantly higher in LRRK2 G2019S flies relative to control lines at all dietary amino acids, raising the possibility that S6K could be a LRRK2 kinase substrate. However, since human p70 S6K (which shares 73% identity with Drosophila S6K) is not directly phosphorylated by LRRK2 in in vitro kinase assays (Fig. 4G,H), this seems to be an unlikely explanation. These results indicate that the 25N diet suppresses bulk translation in aged mutant LRRK2 flies without any prominent impact on TORC1 activity at 6 weeks.
Figure 4.
Dietary amino acids affect protein synthesis and subtly affect TORC1 activity in aged flies. A, Representative autoradiograph for dietary amino acid effects on protein synthesis measured by 35S-met/cys incorporation in 6-week-old fly brains ex vivo. B, Radioactive counts per minute were divided by the number of brains and normalized to Control 50N values. ANOVAs for diet [F(3,20) = 4.54 (Control); F(3,20) = 2.14 (WT); F(3,19) = 7.92 (G2019S)] with Bonferroni post-tests: *p < 0.05, **p < 0.01; and for genotype [F(2,15) = 4.12 (50N)] with Bonferroni post-test: #p < 0.05, n = 5–6 replicates of 8 brains/genotype/condition. C–E, TORC1 activity was measured via phospho-S6K and phospho-4E-BP levels in 6-week-old fly heads. Effect of genotype but no effect of diet on phospho-S6K [ANOVAs for diet effect, F(3,8) = 3.48, p =0.07 (Control); F(3,8) = 0.24, p =0.87 (WT); F(3,8) = 0.07, p =0.97 (G2019S)] and for genotype [F(2,6) = 7.34 (25N); F(2,6) = 22.32 (50N); F(2,6) = 25.59 (200N); F(2,6) = 5.30 (400N)] with Bonferroni post-tests, ##p < 0.01 relative to Control flies at the same diet; n = 3 replicates of 25 flies/genotype/diet. No significant effect of diet on phospho-4E-BP levels except for LRRK2 WT flies [ANOVAs, F(3,8) = 0.72 (Control); F(3,8) = 3.48 (WT); F(3,8) = 1.79 (G2019S)]; with Bonferroni post-test: *p < 0.05, n = 3 replicates of 25 flies/genotype/diet. F, There is an effect of diet on non-phospho-4E-BP in Control and LRRK2 WT flies but not G2019S [ANOVAs: F(3,8) = 218.7 (Control); F(3,8) = 4.11 (WT); F(3,8) = 0.62 (G2019S)]; with Bonferroni post-test: *p < 0.05, ***p < 0.001; n = 3 replicates of 25 flies/genotype/diet. Genotypes are Control (elav-GAL4), LRRK2 WT (elav-GAL4/UAS-LRRK2 WT), and G2019S (elav-GAL4/UAS-LRRK2 G2019S). G, H, P70 S6K is not directly phosphorylated by LRRK2 in an in vitro kinase assay. Phosphorylation of human P70 S6K (V5 tagged, affinity purified from HEK293T cells) in the presence of purified recombinant LRRK2 (D1994A kinase-dead, wild-type, or G2019S) and 32P-ATP. LRRK2 autophosphorylation (32P-ATP incorporation/protein level, normalized to LRRK2 D1994A) is elevated by LRRK2 and LRRK2 G2019S [two-way ANOVA, F(1,12) = 19.24 (LRRK2 substrate); F(2,12) = 6.00 (LRRK2 variant); Bonferroni post-tests: *p < 0.05, **p < 0.01, n = 3], while P70 S6K phosphorylation is not. Basal P70 S6K phosphorylation is potentially because of copurified kinases from HEK293 cells. Data are the mean ± SEM.
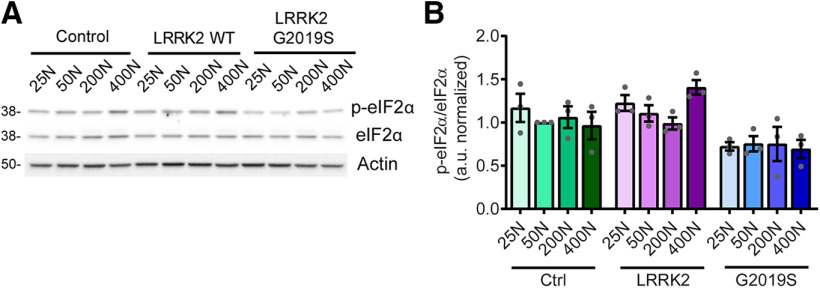
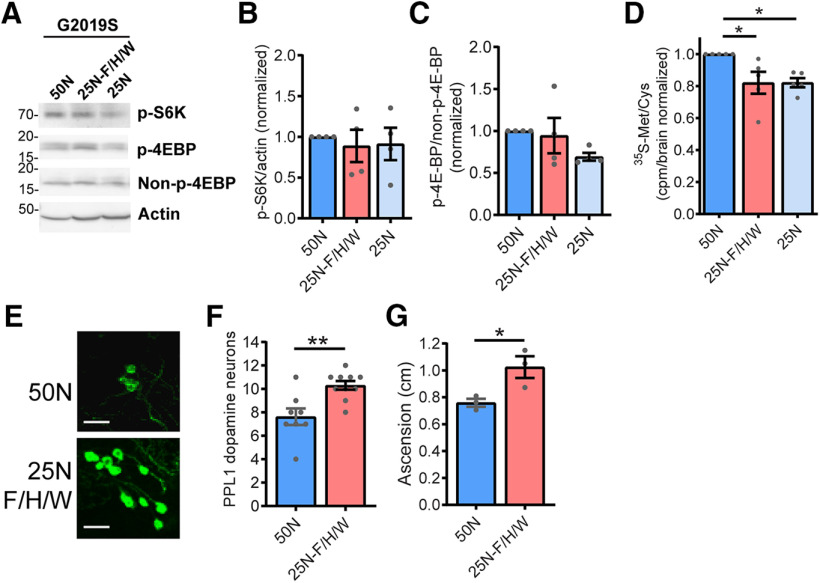
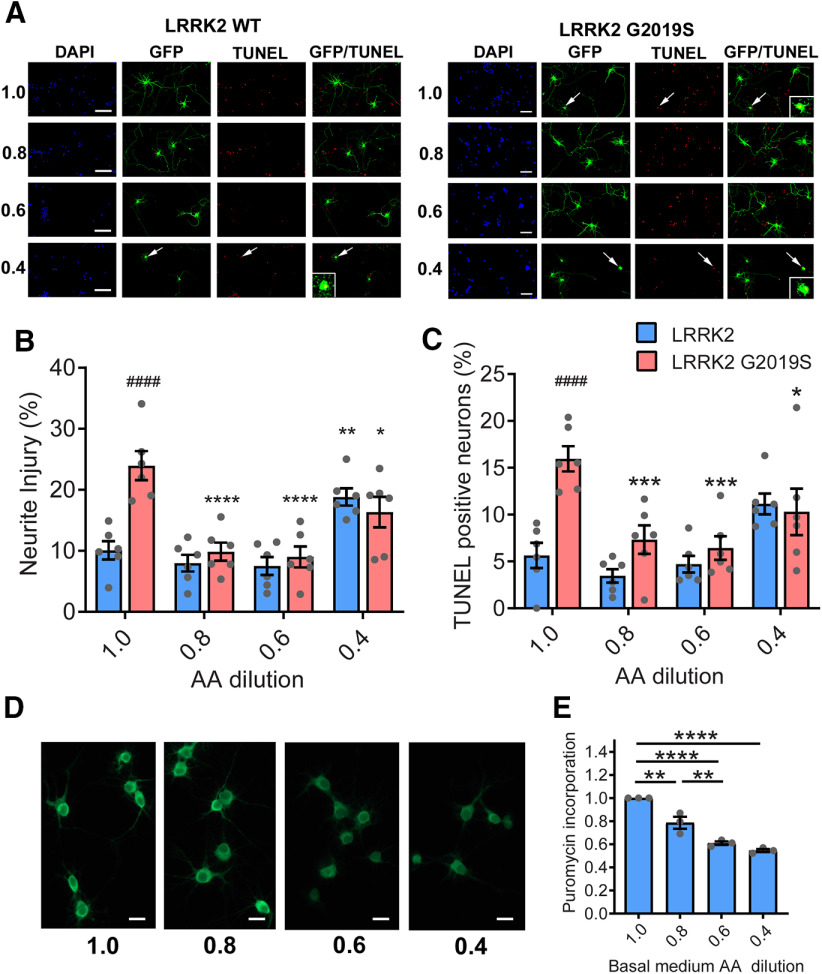
Another cellular mechanism for sensing low amino acid availability is through the activation of the protein kinase GCN2 by uncharged tRNAs, which promote eIF2α phosphorylation and the consequent downregulation of bulk protein synthesis (Soultoukis and Partridge, 2016). However, phospho-eIF2α levels are unaffected by dietary amino acid levels within the range used here (Fig. 5), arguing against this as a possible cause for suppressed protein production. In addition to regulation by nutrient-sensing pathways, protein synthesis is also dependent on the pool of amino acids available for tRNA charging (Elf and Ehrenberg, 2005). This can be readily observed in rapidly dividing organisms such as Escherichia coli, where protein synthesis rates are clearly limited by the rate of supply of amino acids (Elf and Ehrenberg, 2005). To explore whether a reduced pool of amino acids in the 25N diet accounts for the reduction in protein synthesis, we used a modified 50N diet (25N-F/H/W) in which the concentration of just three amino acids, phenylalanine (F), histidine (H) and tryptophan (W), are reduced to 25N levels, while all other amino acids remain at 50N levels (Extended Data Fig. 6-1). These three amino acids are not thought to have a major impact on TORC1 activity, based on experiments in HEK-293T cells (D. Sabatini, personal communication) and are essential amino acids that cannot be synthesized, and therefore must be supplied in food. Indeed, the assessment of phospho-S6K and phospho-4EBP levels demonstrates that the 25N-F/H/W diet does not alter TORC1 signaling relative to the 50N diet in aged flies (Fig. 6A–C). Informatively, reducing just these amino acids to 25N levels lowers protein synthesis in the heads of LRRK2 G2019S flies to virtually the same rates as those seen in flies maintained on the standard 25N diet in an apparently TORC1-independent manner (Fig. 6D). Consistent with a link between protein synthesis and PD-related phenotypes in this model, consuming the 25N-F/H/W diet prevents the loss of dopamine neurons and accompanying locomotor function deficit seen on the 50N diet (Fig. 6E–G). Therefore, restricted amino acid availability in 25N fly diets appears sufficient to prevent manifestation of PD-related phenotypes. To determine whether restricting dietary amino acid content can also modulate LRRK2 toxicity in mammalian systems, we used a rat cortical neuron model of acute LRRK2 toxicity. It is well established that LRRK2 G2019S causes kinase-dependent neurotoxicity in rodent neuron cultures characterized by progressive neurite loss and cell death (Macleod et al., 2006). Strikingly, reducing the concentration of all amino acids in the culture medium by 20–40% during the time of LRRK2 G2019S expression ameliorates neurite loss and cell death caused by mutant LRRK2 (Fig. 7A–C). This is accompanied by the inhibition of global protein synthesis, measured via SUnSET assay (Schmidt et al., 2009; Fig. 7D,E). At a 60% reduction in amino acid concentration, neurite loss and cell death phenotypes become evident in both LRRK2 WT-transfected and LRRK2 G2019S-transfected neurons (Fig. 7A–C), possibly because of a starvation effect at this extent of amino acid restriction. Hence, lowering amino acid levels within a range attenuates LRRK2 G2019S-associated neurotoxicity in both fly brains and mammalian neurons.
Figure 5.
eIF2α phosphorylation is not impacted by dietary amino acids. A, B, No effect of diet on GCN2 pathway activity assessed by eIF2α phosphorylation measurement [ANOVAs: F(2,6) = 0.51 (Control); F(3,8) = 4.30 (WT); F(3,8) = 0.05 (G2019S); n = 3 replicates of 25 flies/condition]. Genotypes are Control (elav-GAL4), LRRK2 WT (elav-GAL4; UAS-LRRK2 WT), and G2019S (elav-GAL4; UAS-LRRK2 G2019S). Data are the mean ± SEM.
Figure 6.
Reducing levels of F, H, and W suppresses bulk translation and PD-related phenotypes in aged LRRK2 G2019S flies without impacting TORC1 activity. A–C, No effect of 25N-F/H/W diet (in which levels of F, H, and W are reduced to 25N levels) on TORC1 activity measured via phospho-S6K and phospho-4E-BP levels in heads from 6-week-old LRRK2 G2019S flies. ANOVAs: F(2,9) = 0.13, p = 0.87 (P-S6K); F(2,9) = 1.73, p = 0.23 (P-4EBP); n = 4 replicates of 25 flies/diet. D, Protein synthesis levels are reduced on the 25N-F/H/W diet measured by 35S-met/cys incorporation in 6-week-old LRRK2 G2019S fly brains ex vivo. ANOVA: F(2,12) = 5.73; Bonferroni post-test: *p < 0.05; n = 5 replicates of 8 flies/condition. E–G, Relative to the 50N diet, the 25N-F/H/W diet improves dopamine neuron viability [unpaired t test: t = 3.57, df =16 (DA neurons); **p < 0.01, n =8-10 brains/genotype; E, F)] and negative geotaxis performance (unpaired t test: t = 3.08, df = 4; *p < 0.05, n = 3 replicates of 25 flies/condition; G) in 6-week-old flies. Genotypes in A–D are elav-GAL4/UAS-LRRK2 G2019S. Genotype in E–G is Ddc-GAL4/UAS-LRRK2 G2019S. Data are the mean ± SEM. Extended Data Figure 6-1 describes the composition of 25N-F/H/W holidic medium.
Figure 7.
Amino acid dilution protects against LRRK2 G2019S toxicity in rat primary neurons. A–C, Effect of culture medium amino acid dilution on LRRK2 G2019S toxicity assessed in rat cortical neurons. Arrows indicate neurons lacking neurites that are TUNEL-positive (see inset magnifications). Amino acid dilutions protect against LRRK2 G2019S-induced neurite injury [two-way ANOVA: F(3,40) = 16.10 (diet effect); F(1,40) = 8.63 (genotype effect); Bonferroni post-tests: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 relative to 1.0 condition for genotype; ####p < 0.0001 relative to LRRK2 control for diet, n = 6] and cell death assessed by TUNEL staining [two-way ANOVA: F(3,40) = 9.12 (diet effect); F(1,40) = 13.89 (genotype effect); Bonferroni post-tests: *p < 0.05, ***p < 0.001 relative to 1.0 condition for genotype; ####p < 0.0001 relative to LRRK2 control for diet, n = 6], while a 0.4-fold dilution increased neurite injury for the LRRK2 WT control (Bonferroni post-test, *p < 0.05). D, E, Protein synthesis, assessed by puromycin incorporation, is progressively reduced by amino acid dilution (ANOVA: F(3,8) = 52.83; Bonferroni post-tests: **p < 0.01, ****p < 0.0001; n = 3 replicates of 20 neurons/condition). Data are the mean ± SEM.
Composition of 25N-F/H/W holidic medium. Download Figure 6-1, DOCX file (22KB, docx) .
Composition of Neurobasal-based medium minus phenol red and amino acids. Download Figure 7-1, DOCX file (22KB, docx) .
AMPK and autophagy activation on high-amino acid diets block PD-related phenotypes in flies
The 200N diet effectively suppresses neurodegenerative phenotypes in aged LRRK2 G2019S flies without reducing protein synthesis (Figs. 1, 3, 4A,B). This indicates that dopamine neuron viability is not always vulnerable to elevated protein synthesis. The data from S6K and 4E-BP phosphorylation suggest that there is no major effect of increasing dietary amino acid levels on TORC1 signaling in aged flies. Previous studies indicate that chronic TORC1 hyperactivity in flies can result in a stress response that triggers feedback inhibition on TORC1 signaling (Lee et al., 2010). To assess whether chronic consumption of a high-amino acid diet triggers TORC1 feedback inhibition in aged flies, we measured TORC1 activity in 1-week-old flies maintained on the same series of holidic diets for the first week of adulthood (Fig. 8). At this age, we observe a more pronounced effect of dietary amino acid levels on TORC1 activity relative to that seen in 6-week-old flies (Fig. 4C–E), particularly at 400N. This is consistent with an age-dependent dampening of dietary effects on TORC1 signaling.
Figure 8.
Impact of dietary amino acids on TORC1 activity in 1-week-old flies. A–C, Diet affects TORC1 activity, measured by phospho-S6K levels and phospho-4E-BP/non-phospho-4E-BP ratio in 1-week-old fly heads [ANOVAs: p-S6K: F(3,16) = 3.04 (Control); F(3,14) = 4.82 (WT); F(3,16) = 18.21 (G2019S); p-4EBP: F(3,16) = 7.70 (Control); F(3,15) = 5.82 (WT); F(3,16) = 6.93 (G2019S); Bonferroni post-tests: *p < 0.05, **p < 0.01, ***p < 0.001 relative to 50N diet for genotype; n = 5 replicates of 25 flies/genotype/diet]. D, Non-phospho-4E-BP levels were decreased at 200N and/or 400N diets in Control and LRRK2 flies [ANOVAs: F(3,16) = 15.72 (Control); F(3,16) = 2.57 (WT); Bonferroni post-tests: *p < 0.05, **p < 0.01, ***p < 0.001, n = 5]. Genotypes are Control (elav-GAL4/+), LRRK2 WT (elav-GAL4/LRRK2 WT), and G2019S (elav-GAL4/LRRK2 G2019S). Data are the mean ± SEM.
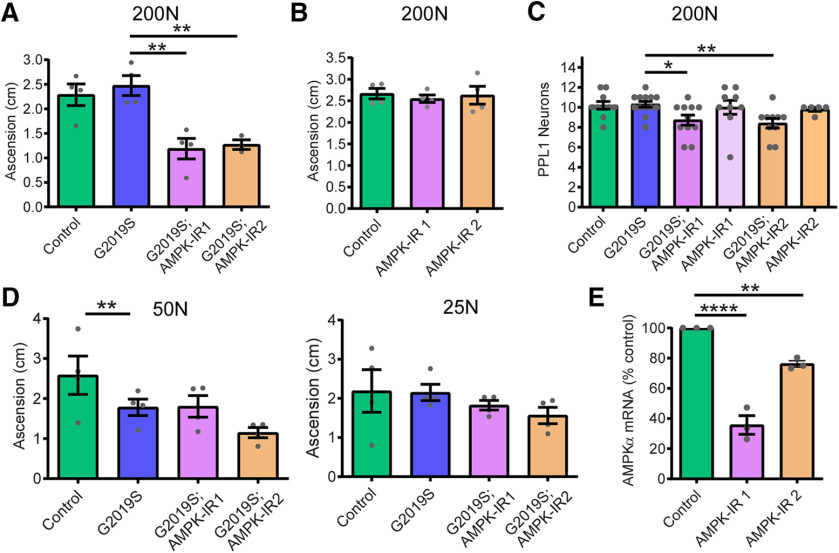
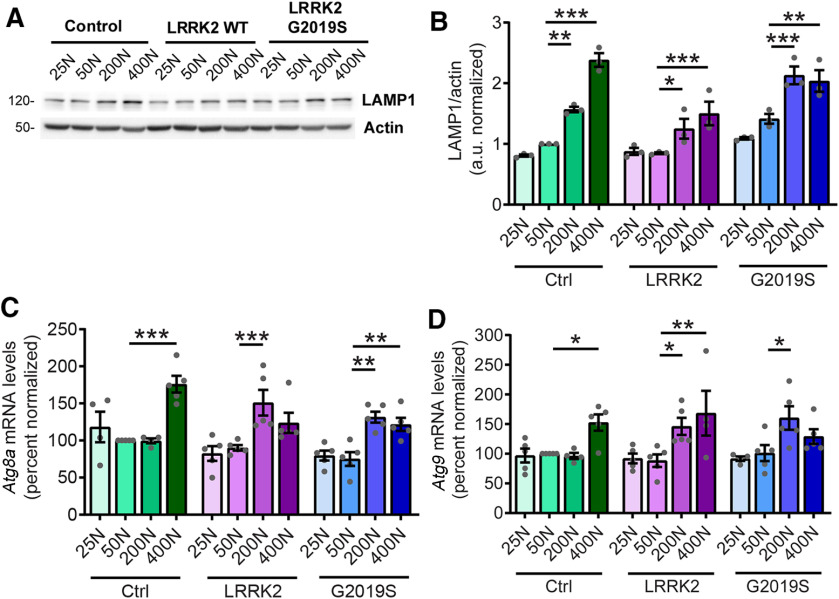
In Drosophila, chronically hyperactive TORC1 has been shown to induce the sestrin dSesn, which exerts feedback inhibition on TORC1 partly through direct activation of AMPK and results in protection against age-related pathologies (Lee et al., 2010). Indeed, we observe that the 200N and 400N diets in LRRK2 G2019S flies and the 400N diet in control flies invoke an age-associated induction of dSesn expression at 5.5 weeks of age (Fig. 9A), which is absent in younger flies (Fig. 9B). The dSesn upregulation by 200N and 400N diets is accompanied by evidence of AMPK activation (increased levels of pAMPK Thr172) in LRRK2 G2019S flies (Fig. 9C,D), which likely dampens the effects of dietary amino acids on TORC1 activity since AMPK is a known inhibitor of TORC1. As we find reduced food intake in flies maintained on higher amino acid diets (Fig. 2B), AMPK could potentially be activated in response to energy stress by sensing elevated AMP/ATP and ADP/ATP ratios. To test this possibility, we assessed ADP/ATP ratios in aged fly heads but found no significant effect of diet in any genotype, suggesting that high-amino acid diets do not cause an overt deficit in ATP (Fig. 9E). To determine whether AMPK is involved in the alleviation of neurodegenerative phenotypes on the 200N diet, we knocked down AMPK and assessed locomotor function in LRRK2 G2019S flies aged on 200N as well as 25N and 50N diets. RNAi-mediated knockdown of AMPK blocks the rescue effect of the 200N diet on locomotor function and dopamine neuron viability (Fig. 10A–C), but has no significant effect on the locomotor deficit seen in LRRK2 G2019S flies aged on a 50N diet or on the locomotor abilities of LRRK2 G2019S flies aged on a 25N diet (Fig. 10D). Additionally, knocking down AMPK alone has no significant effects on negative geotaxis performance or dopamine neuron viability in aged flies (Fig. 10B), consistent with an epistatic effect on locomotor performance in 200N-consuming LRRK2 G2019S flies. Hence, AMPK is necessary for the rescue effect of the 200N diet, but not of the 25N diet. AMPK signaling activates multiple cytoprotective pathways, including autophagy. Accordingly, expression of the autophagosome components Atg8a and Atg9 along with the lysosomal marker LAMP1 are elevated on 200N and 400N diets (Fig. 11A–D), indicating an induction of autophagy on high-amino acid diets. To determine whether autophagy plays a role in suppressing neurodegenerative phenotypes on a 200N diet, we functionally silenced the autophagy-initiating complex gene Atg1 (ortholog of mammalian ULK1). Previous studies have shown that autophagy cannot be induced in flies expressing a kinase-dead Atg1 mutant (Atg1KQ#5B), which functions as a dominant-negative form of this protein (Berry and Baehrecke, 2007; Scott et al., 2007). Expression of dominant-negative Atg1 or RNAi-mediated Atg1 knockdown ablates the rescue effect of the 200N diet on the LRRK2 G2019S locomotor and dopamine neuron phenotypes (Fig. 12A–C), suggesting that augmented autophagy contributes to this rescue. Atg1 knockdown or dominant-negative expression alone does not affect locomotor function or dopamine neuron viability in aged flies (Fig. 12B), and Atg1 loss-of-function does not affect the LRRK2 G2019S locomotor phenotype on the 50N diet or on locomotor function of these flies on a 25N diet (Fig. 12D). Together, these data support a role for AMPK and autophagy activation in the neurodegeneration-suppressing effects of the 200N diet, but not in those of the 25N diet.
Figure 9.
High amino acid diets induce sestrin (dSesn) expression and activate AMPK activity. A, dSesn mRNA levels, measured by quantitative PCR from 5.5-week-old fly heads were significantly increased at 200N and/or 400N diets in Controls and LRRK2 G2019S [ANOVAs: F(2,6) = 27.34 (Control); F(2,6) = 4.42 (G2019S); Bonferroni post-test: *p < 0.05; n = 3 replicates of 15 flies/condition]. B, No significant effect of diet on dSesn levels in Control, LRRK2 WT, or LRRK2 G2019S in 3-week-old fly heads [ANOVAs: F(2,9) = 0.84, p = 0.46 (Control); F(2,8) = 1.83, p = 0.22 (WT); F(2,6) = 0.52, p = 0.62 (G2019S); n = 3-4 replicates of 15 flies/condition]. C, D, AMPK activation, assessed via Thr172 phosphorylation was increased by dietary amino acid elevation in 6-week-old LRRK2 WT and LRRK2 G2019S fly heads [ANOVAs: F(3,8) = 1.89 (WT); F(3,8) = 14.95 (G2019S); Bonferroni post-tests, *p < 0.05; n = 3 replicates of 25 flies/condition]. E, No significant effect of dietary amino acids on ADP/ATP ratio in 6-week-old fly heads [ANOVAs: F(3,11) = 0.75, p = 0.54 (Control); F(3,11) = 2.61, p = 0.1 (WT); F(3,12) = 0.56, p = 0.65 (G2019S)]. Genotypes are Control (elav-GAL4), LRRK2 WT (elav-GAL4; UAS-LRRK2 WT), and G2019S (elav-GAL4; UAS-LRRK2 G2019S). Data are the mean ± SEM.
Figure 10.
AMPKα knockdown blocks the rescue effect of the 200N diet. A, B, AMPKα knockdown via RNAi blocks the rescue effect of 200N diet on locomotor function in 6-week-old LRRK2 G2019S flies (A; ANOVA, F(3,11) = 11.24; Bonferroni post-test, **p < 0.01; n = 4 replicates of 25 flies/condition), but does not affect locomotor function in the absence of LRRK2 G2019S (B; ANOVA: F(2,9) = 0.17, p =0.85; n = 4 replicates of 25 flies/condition). C, Dopamine neuron numbers are significantly lower in LRRK2 G2019S flies following AMPKα knockdown (ANOVA, F(3,37) = 6.12; Bonferroni post-test: *p < 0.05, **p < 0.01; n =8–10 brains/genotype). D, Knockdown of AMPKα has no significant effect on motor performance of 6-week-old G2019S flies on 50 or 25N diets [ANOVAs: F(3,12) = 3.81 (50N); F(3,12) = 0.88 (25N); Bonferroni post-test, **p < 0.01; n = 4 replicates of 25 flies/genotype). E, Pan-neuronal (elav-GAL4) AMPKα RNAi reduces AMPKα transcript levels in whole-head RNA (ANOVA, F(2,6) = 75.27; Bonferroni post-tests: **p < 0.01, ****p < 0.0001; n =3 replicates of 25 heads/genotype). Genotypes in A–E: Control (Ddc-GAL4), G2019S (Ddc-GAL4; UAS-LRRK2 G2019S), and G2019S; AMPK-IR (Ddc-GAL4; UAS-LRRK2 G2019S/UAS-AMPKα-IR) and AMPK-IR (Ddc-GAL4; UAS-AMPK-IR). Data are the mean ± SEM.
Figure 11.
Autophagy markers are elevated with higher-amino acid diets. A, B, Elevated LAMP1 levels across increased dietary amino acids in 6-week-old fly heads [ANOVAs: F(3,8) = 135.4 (Control); F(3,8) = 5.83 (WT); F(3,8) = 16.55 (G2019S); Bonferroni post-tests: *p < 0.05, **p < 0.01, ***p < 0.001; n = 3 replicates of 25 flies/condition]. C, D, Autophagosome components Atg8a (C) and Atg9 (D) mRNA are increased at 200N and/or 400N diets [ANOVAs: for Atg8a: F(3,14) = 11.30 (Control); F(3,16) = 6.74 (WT); F(3,16) = 12.50 (G2019S); for Atg9: F(3,15) = 8.17 (Control); F(3,15) = 4.33 (WT); F(3,15) = 4.54 (G2019S); Bonferroni post-tests: *p < 0.05, **p < 0.01, ***p < 0.001; n = 5 replicates of 15 flies/condition). Genotypes are Control (elav-GAL4), LRRK2 WT (elav-GAL4; UAS-LRRK2 WT), and G2019S (elav-GAL4; UAS-LRRK2 G2019S).
Figure 12.
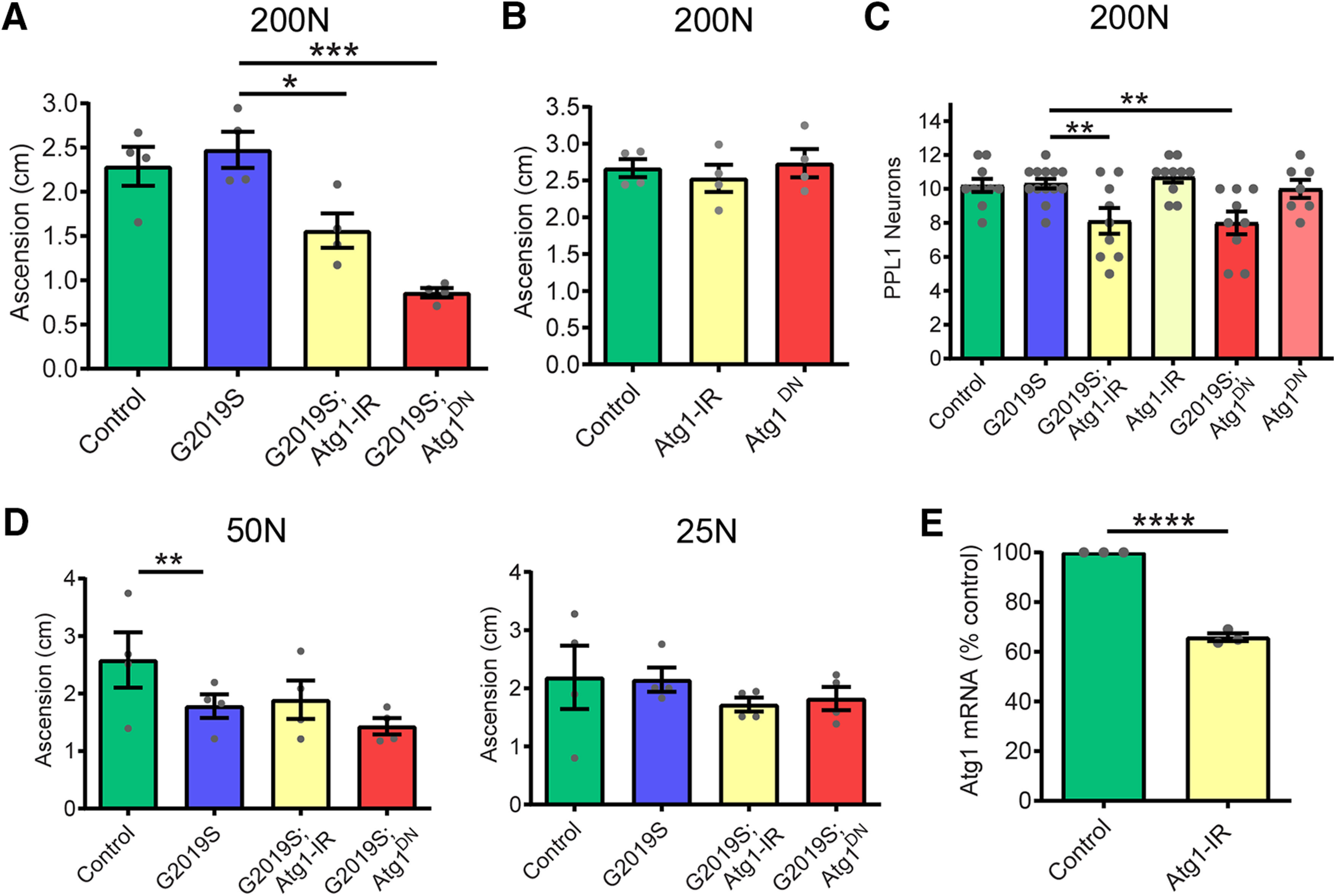
Atg1 loss-of-function blocks the rescue effect of 200N diet. A, Atg1 loss-of-function via RNAi (Atg1-IR) or dominant-negative expression (Atg1DN) blocked the rescue effect of 200N diet on locomotor function in 6-week-old flies (ANOVA, F(3,12) = 16.77; Bonferroni post-tests: *p < 0.05, ***p < 0.001; n = 4 replicates of 25 flies/condition). B, Atg1 loss-of-function alone did not affect negative geotaxis in aged flies (ANOVA, F(2,9) = 0.38, p = 0.69, n = 4 replicates of 25 flies/genotype). C, Atg1 loss-of-function blocks the rescue effect of the 200N diet on dopamine neuron viability (ANOVA, F(3,37) = 6.12; Bonferroni post-test, **p < 0.01; n = 8–10 brains/genotype). D, RNAi-mediated Atg1 knockdown or expression of dominant-negative Atg1DN has no significant effect on motor performance of G2019S flies on 50 or 25N diets for 6 weeks [ANOVAs, F(3,12) = 2.30 (50N); F(3,12) = 0.55 (25N); Bonferroni post-test, **p < 0.01; n = 4 replicates of 25 flies/genotype]. E, Pan-neuronal (elav-GAL4) Atg1 RNAi reduces Atg1 transcript levels in whole-head RNA (Student's unpaired t test: t = 21.11, df = 4; ****p < 0.0001; n = 3 replicates of 25 heads/genotype). Genotypes in A–E are Control (Ddc-GAL4), G2019S (Ddc-GAL4; UAS-LRRK2 G2019S), Atg1-IR (Ddc-GAL4; UAS-Atg1-IR), G2019S; Atg1-IR (Ddc-GAL4; UAS-LRRK2 G2019S/UAS-Atg1-IR), Atg1DN (Ddc-GAL4; UAS- Atg1DN), and G2019S; Atg1DN (Ddc-GAL4; UAS-LRRK2 G2019S/UAS- Atg1DN). Data are the mean ± SEM.
Proapoptotic markers are elevated on a 400N amino acid diet independent of LRRK2
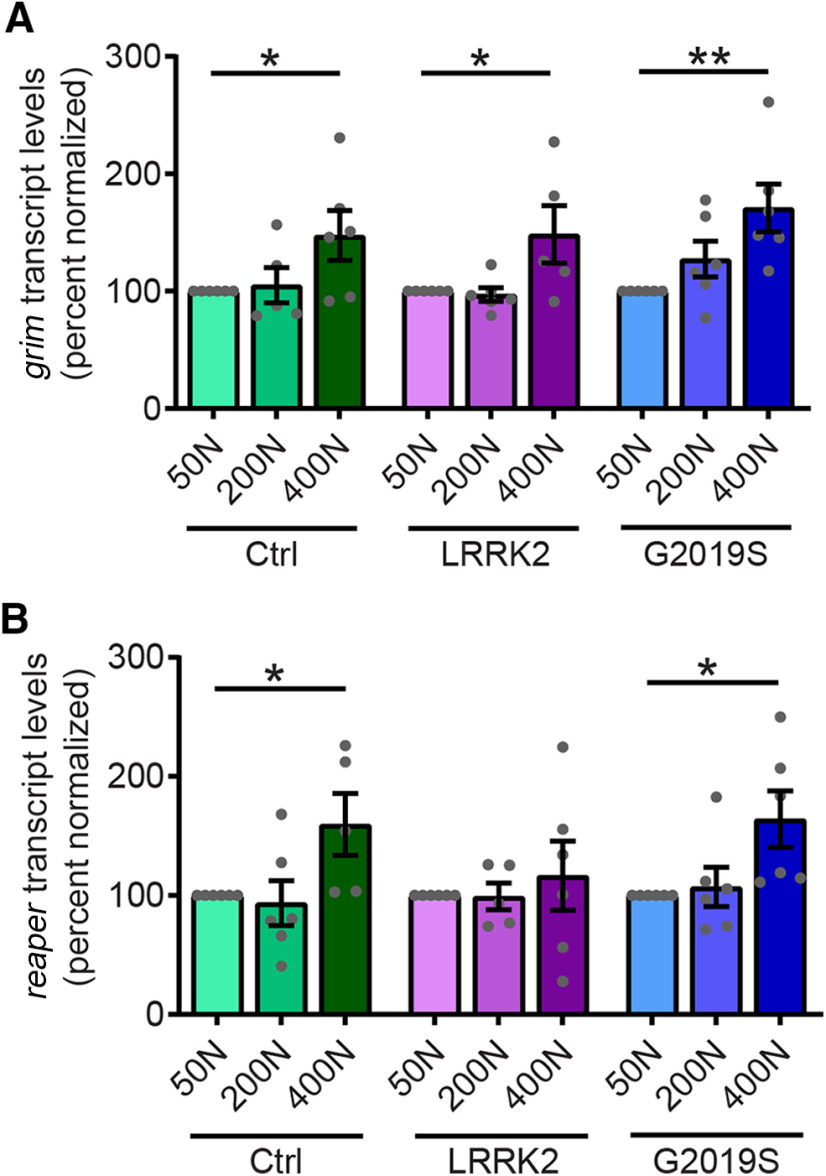
The 400N diet causes PD-related phenotypes in controls as well as LRRK2 G2019S flies (Fig. 1), despite upregulated autophagy markers on this diet (Fig. 11). To probe whether the observed neuronal loss in flies aged on a 400N diet is specific to dopamine neurons or reflects general brain and neuronal demise induced by this diet, we assessed proapoptotic markers across dietary amino acids. In Drosophila, apoptotic stimuli increase the expression of proapoptotic proteins such as Grim, Reaper, and Hid, which bind to DIAP (death-associated inhibitor of apoptosis) and promote caspase release (Zheng et al., 2010). While dopamine neuron loss occurs to a similar extent on both 50N and 400N diets in LRRK2 G2019S flies, expressions of both grim and reaper are elevated exclusively in 400N diet fly heads (Fig. 13). A similar increase in expression of these proapoptotic genes is found in aged control flies on 400N food, suggesting that neuronal apoptosis is promoted by this diet in aged flies independent of mutant LRRK2 expression.
Figure 13.
Chronic consumption of 400N diet leads to elevated proapoptotic gene expression. A, B, The grim (A) and reaper (B) transcript levels assessed by quantitative PCR in 6-week-old fly heads were significantly elevated on the 400N diet [individual ANOVAs: grim: F(2,14) = 3.13 (Control); F(2,14) = 4.69 (WT); F(2,15) = 5.93 (G2019S); reaper: F(2,14) = 4.08 (Control); F(2,14) = 0.28 (WT); F(2,15) = 4.44 (G2019S); Bonferroni post-test: *p < 0.05, **p < 0.01; n = 6 replicates of 15 flies/condition]. Genotypes are Control (elav-GAL4), LRRK2 WT (elav-GAL4; UAS-LRRK2 WT), and G2019S (elav-GAL4; UAS-LRRK2 G2019S). Data are the mean ± SEM.
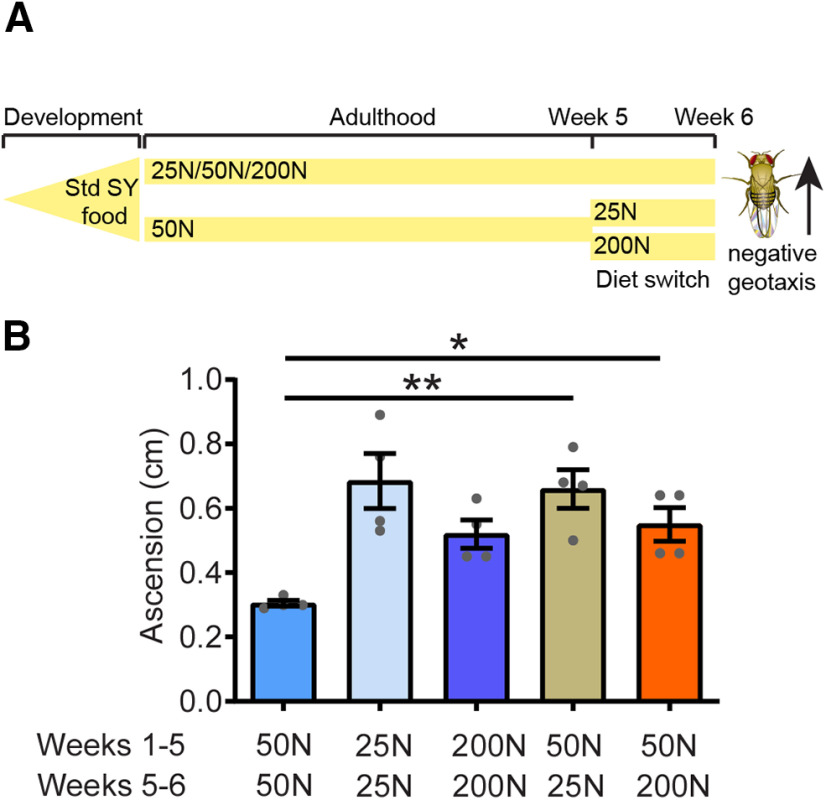
Protective effects of 25 and 200N amino acid diets are upheld in late life diet switch
PD-related phenotypes manifest in LRRK2 G2019S flies at ∼6 weeks of age, suggesting that aging is an important driver of LRRK2-mediated neurodegeneration. It has been proposed that dietary restriction can rapidly affect mortality even when introduced late in life, apparently neutralizing the effects of diet history and decreasing the short-term risk of mortality (Mair et al., 2003). To interrogate whether the beneficial effects of 25N and 200N diets on LRRK2 neurodegeneration require feeding throughout adulthood or whether initiating these diets later in life but before the onset of neurodegenerative phenotypes can still be protective, we performed a diet switch in 5-week-old flies. LRRK2 G2019S flies were maintained on a 50N diet until 5 weeks of age and then switched to either a 25N or 200N diet, or were maintained on 50N food for 1 additional week (Fig. 14). Relative to flies kept on the 50N diet for the whole 6 weeks, a switch to either a 25N or 200N diet for 1 week is sufficient to prevent onset of locomotor deficits at 6 weeks (Fig. 14). These results are consistent with the existence of a time window in organismal aging in which LRRK2 neurotoxicitycan be prevented through amino acid manipulation.
Figure 14.
Rescue effects of dietary amino acids are retained via late life diet switch. A, Schematic representation of the diet switch study. A set of LRRK2 G2019S (Ddc-GAL4; UAS-LRRK2 G2019S) flies were aged on 25, 50, or 200N diets for 6 weeks and tested for locomotor function. Another set of mutant LRRK2 flies were fed 50N diet for 5 weeks and then switched to either 25 or 200N amino acid diet for an additional 1 week. B, All flies were reared on standard sugar/yeast food. Switching flies from a 50N diet to either a 25 or 200N diet significantly improved motor function (ANOVA, F(4,15) = 7.33; Bonferroni post-tests: *p < 0.05, **p < 0.01; n = 4 replicates of 25 flies/condition). Data are the mean ± SEM.
Discussion
The major finding from this study is that neurodegeneration caused by the disease-associated LRRK2 G2019S mutation is highly influenced by dietary amino acid levels in adult Drosophila and in mammalian neuron cultures. Age-related dopamine neuron loss and motor impairment previously observed on standard chemically undefined Drosophila food are recapitulated at similar amino acid levels in a holidic 50N diet fed to flies throughout adulthood. Strikingly, these phenotypes are suppressed by both restriction and moderate elevation of all dietary amino acids relative to 50N through apparently distinct mechanisms.
In prior studies, we demonstrated that administering protein synthesis inhibitors throughout adulthood protected against elevated bulk protein synthesis and associated neurodegeneration in Drosophila (Martin et al., 2014a,b). Here, restricting dietary amino acids from 50N, which is closest in amino acid content to our previous chemically undefined food, to 25N levels similarly lowers protein synthesis and ameliorates neurodegeneration in LRRK2 G2019S flies (Figs. 1, 4A,B). Unexpectedly, this occurs in an apparently TORC1-independent and GCN2-independent manner (Figs. 4C–E, 5). As the control strains exhibit modest or no change in protein synthesis between 25N and 50N diets, a difference in dietary amino acids appears to have minimal effect on protein synthesis in control strains, while 25N levels of amino acids appear to block the LRRK2 G2019S-mediated increase in bulk protein synthesis observed at 50N and previously on chemically undefined diets. Hence, when amino acids are more restricted such as on the 25N diet, it is possible that their levels are limiting to the point where upregulated protein synthesis (e.g., through modulation of ribosomal function as proposed for LRRK2 G2019S; Martin et al., 2014a,b) becomes inhibited. This could conceivably account for a greater impact of the 25N diet on protein synthesis in this strain. Informatively, lowered protein synthesis relative to the 50N diet can be achieved via restricted availability of just phenylalanine, histidine, and tryptophan (Fig. 6D), which are not recognized TOR activity modulators and did not alter p-S6K or p-4EBP levels observed in flies consuming a diet restricted in just these three amino acids relative to the 50N diet (Fig. 6A–C). This suggests that protein synthesis lowering is mediated through a depletion of the amino acid pool available for protein synthesis. Similarly, lowering amino acid levels in the culture medium of primary rat neurons attenuates LRRK2 G2019S-induced neurite loss and cell death with a concomitant reduction in protein synthesis (Fig. 7). Our preliminary evidence supporting a protective effect of amino acid restriction on mammalian neurons warrants future efforts to understand whether disease-related phenotypes in mammalian LRRK2 in vivo models can similarly be prevented.
While amino acid restriction is likely neuroprotective through lowering translation, elevating dietary amino acid concentration from 50N to 200N also blocks dopamine neuron loss and motor deficits in LRRK2 G2019S flies without lowering protein synthesis (Figs. 1, 4). Assessment of 4E-BP and S6K phosphorylation revealed that TORC1 activity generally increases with higher dietary amino acid levels in young adult flies but that this effect becomes more modest in aged flies (Figs. 4C–E, 8A–C). With increasing levels of 4E-BP phosphorylation on higher-amino acid diets, nonphosphorylated 4E-BP levels trend downward as would be expected with a higher proportion of phosphorylated protein (Figs. 4E,F, 8C,D). Sestrins, which are evolutionarily conserved stress-inducible proteins, are negative-feedback regulators of TORC1 signaling and accumulate upon chronic TORC1 activation in Drosophila to protect against age-related pathologies (Lee et al., 2010). Here, we observe a transcriptional induction of the Drosophila sestrin dSesn, in aged flies housed on high -amino acid diets (200N and/or 400N), but no effect of diet on dSesn expression in younger flies, consistent with an age-related dampening of TORC1 responsiveness to dietary amino acids (Fig. 9). As in mammals, dSesn in flies promotes activity of AMPK, which in turn inhibits TORC1 signaling (Lee et al., 2010; Kim and Lee, 2015). The strongest dSesn induction across diet occurs in LRRK2 G2019S flies, where dSesn is upregulated at both 200N and 400N diets relative to 50N, while it only appears to be elevated at 400N in controls. Interestingly, we also observe a more pronounced upregulation of AMPK activity and the autophagy marker LAMP1 in LRRK2 G2019S flies on the 200N diet than in control strains (Figs. 9C,D, 11A,B). This raises the possibility that additional stressors may be present to induce dSesn expression specifically in LRRK2 G2019S flies but not controls on the 200N diet. Prior studies indicate that chronic TORC1 activity induces dSesn expression in a mechanism involving reactive oxygen species (ROS), which are also established activators of mammalian sestrins (Lee et al., 2010), raising the possibility that basal ROS levels might be higher in LRRK2 G2019S flies, as might be suggested by their heightened sensitivity to ROS-generating compounds such as rotenone (Ng et al., 2009).
An established relationship between autophagy and amino acid levels has been described at the level of amino acid starvation, which triggers autophagy induction through GCN2 activation and TORC1 silencing. We see no evidence for the GCN2 pathway being affected by the amino acid range tested here (Fig. 5) or evidence that autophagy markers are elevated by low dietary amino acid levels in this study (Fig. 11A–D). This indicates that amino acid levels on the 25N diet are not low enough to elicit a starvation response, which might potentially be invoked at lower amino acid levels. The induction of autophagy on high-amino acid diets associated with elevated dSesn expression and AMPK activation seen here appears to be a distinct mechanism by which amino acids impinge on autophagy. Reducing expression of AMPK or the autophagy-initiating gene Atg1 eliminates the 200N diet protective effect on LRRK2 G2019S flies, but does not impact the protective effect of the 25N diet, where AMPK and autophagy activity appears similar to that in flies on a 50N diet (Figs. 10, 12). An attractive explanation for these findings is that AMPK activation engenders a neuroprotective effect involving autophagy that staves off neurodegeneration. Upon activation, AMPK limits energy-consuming anabolic processes while promoting catabolic processes such as autophagy. A boost in protein turnover through autophagy could compensate for the elevated bulk protein synthesis caused by LRRK2 G2019S, thereby re-establishing protein homeostasis. Dietary promotion of autophagy via AMPK activation may also protect against the reported perturbation of lysosomal function by mutant LRRK2 (Wallings et al., 2019), additionally promoting a restoration of protein homeostasis. The sestrin–AMPK–autophagy axis acts to limit damage in the face of numerous cell stressors and was previously shown to protect rodent dopaminergic cells against rotenone toxicity (Hou et al., 2015). Likewise, expression of constitutively active AMPK was shown to prevent dopamine neuron loss and motor impairments in rats overexpressing α-synuclein (Bobela et al., 2017) and to protect against the same phenotypes in flies expressing LRRK2 G2019S (Ng et al., 2012). Additionally, it is notable that both translation suppression and AMPK activation were shown to protect against age-related pathology in parkin-null flies (Tain et al., 2009; Ng et al., 2012); thus, it is tempting to speculate that the beneficial effects of dietary manipulations we observe could extend beyond mutant LRRK2. Together, these findings support the potential for AMPK activation to protect against the development of PD phenotypes. Since AMPK can also be activated under conditions of energy deficit and we observe an inverse relationship between dietary amino acid levels and food intake, we considered the possibility that AMPK activation on high amino acid diets could be triggered by low ATP levels. However, ADP/ATP ratios are not affected by the diets used in the study (Fig. 9E), and, additionally, egg laying was also shown in previous studies to track positively with amino acid levels (Piper et al., 2014), further arguing against major energy deficits that are known to impede fecundity in Drosophila (Partridge et al., 2005).
The holidic diet was previously validated for use in adult flies to obtain optimal life span and fecundity while fly development was reported to be slightly delayed on this medium (Piper et al., 2014). Hence, in our study, flies were reared on standard sugar, yeast food, and holidic diet exposure, and amino acid variation was limited to adulthood to avoid potential effects on development. We chose to focus on manipulating dietary amino acid levels because anabolic protein synthesis is strongly linked to dietary protein levels and amino acid nutrient status. In contrast to modifying levels of whole proteins, manipulating free amino acid levels avoids the potential variability of digestion and oligopeptide transport-based uptake of individual proteins (Soultoukis and Partridge, 2016) and is therefore more suitable for examining the postabsorptive effects of dietary amino acids on neurodegeneration. Additionally, evidence from animal models consistently indicates that dietary protein or free amino acid content is a major determinant of organismal life span (Grandison et al., 2009), invoking its potential to impact aging and age-related pathologies. Modifying dietary amino acid levels does impact food intake, resulting in an apparent satiating effect of high-amino acid diets on appetite. This makes it challenging to control for the intake of other dietary components across the amino acid diet range, which could possibly influence the phenotypes assessed here. Importantly, we do not find any evidence of energy stress on high-amino acid diet that could promote degenerative phenotypes. Future detailed investigation into the effect of manipulating other dietary components either alone or in combination is warranted, and while we did not directly explore this possibility within the current study, using the holidic diet approach should facilitate such investigations. Manifestation of PD-related phenotypes in control flies fed a 400N (HUNT amino acid formulation) diet (Fig. 1) occurs independently of LRRK2 expression, which is consistent with the possibility that the consumption of high-amino acid diets may predispose to age-related loss of dopamine neurons in the absence of a disease-linked mutation. This initial observation may be considered interesting in the context of human studies showing a link between PD risk and body mass index (as a correlate of food consumption) or with related factors such as type 2 diabetes and cardiovascular disease (Hu et al., 2006; Biosa et al., 2018). Yet whether the neurodegeneration observed on this 400N diet is specific to dopamine neurons and how directly it is related to the effects of diet on these neurons versus its occurrence as a consequence of a broader physiological effects is unknown and awaits further study.
One challenge in interpreting data from the current study is attempting to identify which dietary N recapitulates the oligidic diet used in prior studies where dietary N was not defined or controlled. We define 50N as closest to the estimated N of our prior yeast-based diet because mathematically this is the case, although the difference between this estimated N and the 200N diet is not much greater. One can calculate at best a theoretical dietary N for prior uncontrolled studies where, in reality, the constituents of any single ingredient such as yeast (the primary nitrogen source) might display batch-to-batch variability and be obtained from multiple sources, which could significantly affect dietary N over long-term studies as previously noted (Piper et al., 2014). Additionally, other nutrient classes differ between the two diets and whether these affect the phenotypes under study is currently unknown. These caveats reduce our confidence in being able to identify a “baseline” diet from prior studies or in being able to accurately compare across studies. This also reinforces the rationale for using a holidic diet approach for diet studies to rigorously control for dietary composition both within a laboratory over the course of time and between independent laboratories. This strengthens the ability to compare and interpret data across multiple independent studies.
In summary, we find that manipulating dietary amino acid levels can potently modulate mutant LRRK2 toxicity in both Drosophila and mammalian systems. Our findings underscore the importance of diet in determining the vulnerability of aging organisms to neurodegeneration and corroborate reports from mouse and primate studies that dietary restriction ameliorates PD-related phenotypes (Bayliss et al., 2016). A growing body of evidence implicates loss of translation control as a causative factor in several neurologic diseases including PD, highlighting the necessity for protein homeostasis to maintain neuronal function. Further understanding the impact of dietary influences on translation in the context of these diseases will help evaluate the potential for dietary intervention to halt their onset or delay their progression.
Footnotes
The authors declare no competing financial interests.
This work was supported by National Institutes of Health Grant P30-NS-061800 to the OHSU Advanced Light Microscopy Core, and an American Parkinson Disease Association Post-Doctoral Fellowship to V.G.C-V.; and by National Institutes of Health Grant K01-AG-050718, an American Parkinson Disease Association Research Grant, and OHSU Neurology Foundation Funds to I.M.
References
- Bayliss JA, Lemus MB, Stark R, Santos VV, Thompson A, Rees DJ, Galic S, Elsworth JD, Kemp BE, Davies JS, Andrews ZB (2016) Ghrelin-AMPK signaling mediates the neuroprotective effects of calorie restriction in Parkinson's disease. J Neurosci 36:3049–3063. 10.1523/JNEUROSCI.4373-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH (2007) Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131:1137–1148. 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosa A, Outeiro TF, Bubacco L, Bisaglia M (2018) Diabetes mellitus as a risk factor for Parkinson's disease: a molecular point of view. Mol Neurobiol 55:8754–8763. 10.1007/s12035-018-1025-9 [DOI] [PubMed] [Google Scholar]
- Bjordal M, Arquier N, Kniazeff J, Pin JP, Léopold P (2014) Sensing of amino acids in a dopaminergic circuitry promotes rejection of an incomplete diet in Drosophila. Cell 156:510–521. 10.1016/j.cell.2013.12.024 [DOI] [PubMed] [Google Scholar]
- Bobela W, Nazeeruddin S, Knott G, Aebischer P, Schneider BL (2017) Modulating the catalytic activity of AMPK has neuroprotective effects against α-synuclein toxicity. Mol Neurodegener 12:80. 10.1186/s13024-017-0220-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier TJ, Kanaan NM, Kordower JH (2017) Aging and Parkinson's disease: different sides of the same coin? Mov Disord 32:983–990. 10.1002/mds.27037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Ehrenberg M (2005) Near-critical behavior of aminoacyl-tRNA pools in E. coli at rate-limiting supply of amino acids. Biophys J 88:132–146. 10.1529/biophysj.104.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers P, Tain LS, Nespital T, Goncalves J, Froehlich J, Partridge L (2016) Reduced insulin/insulin-like growth factor signaling decreases translation in Drosophila and mice. Sci Rep 6:30290. 10.1038/srep30290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargano JW, Martin I, Bhandari P, Grotewiel MS (2005) Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol 40:386–395. 10.1016/j.exger.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Gems D, Partridge L (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol 75:621–644. 10.1146/annurev-physiol-030212-183712 [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L (2009) Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462:1061–1064. 10.1038/nature08619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR (2006) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23:329–341. 10.1016/j.nbd.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Healy DG, Falchi M, O'Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, Ferreira JJ, Tolosa E, Kay DM, Klein C, Williams DR, Marras C, Lang AE, Wszolek ZK, Berciano J, Schapira AHV, et al. (2008) Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol 7:583–590. 10.1016/S1474-4422(08)70117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig MC, Kolly C, Persohn E, Theil D, Schweizer T, Hafner T, Stemmelen C, Troxler TJ, Schmid P, Danner S, Schnell CR, Mueller M, Kinzel B, Grevot A, Bolognani F, Stirn M, Kuhn RR, Kaupmann K, van der Putten PH, Rovelli G, et al. (2011) LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum Mol Genet 20:4209–4223. 10.1093/hmg/ddr348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YS, Guan JJ, Xu HD, Wu F, Sheng R, Qin ZH (2015) Sestrin2 protects dopaminergic cells against rotenone toxicity through AMPK-dependent autophagy activation. Mol Cell Biol 35:2740–2751. 10.1128/MCB.00285-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Jousilahti P, Nissinen A, Antikainen R, Kivipelto M, Tuomilehto J (2006) Body mass index and the risk of Parkinson disease. Neurology 67:1955–1959. 10.1212/01.wnl.0000247052.18422.e5 [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B (2008) Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 27:2432–2443. 10.1038/emboj.2008.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S (2004) Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14:885–890. 10.1016/j.cub.2004.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Kim M, Lee JH (2015) Identification of an AMPK phosphorylation site in Drosophila TSC2 (gigas) that regulate cell growth. Int J Mol Sci 16:7015–7026. 10.3390/ijms16047015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, Karin M (2010) Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327:1223–1228. 10.1126/science.1182228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Suarez JA, Brandhorst S, Balasubramanian P, Cheng CW, Madia F, Fontana L, Mirisola MG, Guevara-Aguirre J, Wan J, Passarino G, Kennedy BK, Wei M, Cohen P, Crimmins EM, Longo VD (2014) Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 19:407–417. 10.1016/j.cmet.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, Ren Q, Jiao Y, Sawa A, Moran T, Ross CA, Montell C, Smith WW (2008) A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A 105:2693–2698. 10.1073/pnas.0708452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A (2006) The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52:587–593. 10.1016/j.neuron.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L (2003) Demography of dietary restriction and death in Drosophila. Science 301:1731–1733. 10.1126/science.1086016 [DOI] [PubMed] [Google Scholar]
- Martin I, Abalde-Atristain L, Kim JW, Dawson TM, Dawson VL (2014a) Abberant protein synthesis in G2019S LRRK2 Drosophila Parkinson disease-related phenotypes. Fly (Austin) 8:165–169. 10.4161/19336934.2014.983382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Kim JW, Lee BD, Kang HC, Xu JC, Jia H, Stankowski J, Kim MS, Zhong J, Kumar M, Andrabi SA, Xiong Y, Dickson DW, Wszolek ZK, Pandey A, Dawson TM, Dawson VL (2014b) Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson's disease. Cell 157:472–485. 10.1016/j.cell.2014.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maswood N, Young J, Tilmont E, Zhang Z, Gash DM, Gerhardt GA, Grondin R, Roth GS, Mattison J, Lane MA, Carson RE, Cohen RM, Mouton PR, Quigley C, Mattson MP, Ingram DK (2004) Caloric restriction increases neurotrophic factor levels and attenuates neurochemical and behavioral deficits in a primate model of Parkinson's disease. Proc Natl Acad Sci U S A 101:18171–18176. 10.1073/pnas.0405831102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL (2009) Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J Neurosci 29:11257–11262. 10.1523/JNEUROSCI.2375-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Guan MS, Koh C, Ouyang X, Yu F, Tan EK, O'Neill SP, Zhang X, Chung J, Lim KL (2012) AMP kinase activation mitigates dopaminergic dysfunction and mitochondrial abnormalities in Drosophila models of Parkinson's disease. J Neurosci 32:14311–14317. 10.1523/JNEUROSCI.0499-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S. (2011) Obesity and nutrient sensing TOR pathway in flies and vertebrates: functional conservation of genetic mechanisms. Trends Endocrinol Metab 22:45–52. 10.1016/j.tem.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Piper MD, Mair W (2005) Dietary restriction in Drosophila. Mech Ageing Dev 126:938–950. 10.1016/j.mad.2005.03.023 [DOI] [PubMed] [Google Scholar]
- Penney J, Tsurudome K, Liao EH, Kauwe G, Gray L, Yanagiya A, R Calderon M, Sonenberg N, Haghighi AP (2016) LRRK2 regulates retrograde synaptic compensation at the Drosophila neuromuscular junction. Nat Commun 7:12188. 10.1038/ncomms12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MD, Blanc E, Leitão-Gonçalves R, Yang M, He X, Linford NJ, Hoddinott MP, Hopfen C, Soultoukis GA, Niemeyer C, Kerr F, Pletcher SD, Ribeiro C, Partridge L (2014) A holidic medium for Drosophila melanogaster. Nat Methods 11:100–105. 10.1038/nmeth.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MDW, Soultoukis GA, Blanc E, Mesaros A, Herbert SL, Juricic P, He X, Atanassov I, Salmonowicz H, Yang M, Simpson SJ, Ribeiro C, Partridge L (2017) Matching dietary amino acid balance to the in silico-translated exome optimizes growth and reproduction without cost to lifespan. Cell Metab 25:1206. 10.1016/j.cmet.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhász G, Neufeld TP (2007) Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17:1–11. 10.1016/j.cub.2006.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Zhang S, Bustos D, Kleinheinz T, Le Pichon CE, Dominguez SL, Solanoy HO, Drummond J, Zhang X, Ding X, Cai F, Song Q, Li X, Yue Z, van der Brug MP, Burdick DJ, Gunzner-Toste J, Chen H, Liu X, Estrada AA, et al. (2012) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med 4:164ra161. 10.1126/scitranslmed.3004485 [DOI] [PubMed] [Google Scholar]
- Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Krüger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, et al. (2009) Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 41:1308–1312. 10.1038/ng.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, Gokarn R, Khalil M, Turner N, Cooney GJ, Sinclair DA, Raubenheimer D, Le Couteur DG, Simpson SJ (2014) The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19:418–430. 10.1016/j.cmet.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultoukis GA, Partridge L (2016) Dietary protein, metabolism, and aging. Annu Rev Biochem 85:5–34. 10.1146/annurev-biochem-060815-014422 [DOI] [PubMed] [Google Scholar]
- Sun J, Liu C, Bai X, Li X, Li J, Zhang Z, Zhang Y, Guo J, Li Y (2017) Drosophila FIT is a protein-specific satiety hormone essential for feeding control. Nat Commun 8:14161. 10.1038/ncomms14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ (2009) Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci 12:1129–1135. 10.1038/nn.2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallings R, Connor-Robson N, Wade-Martins R (2019) LRRK2 interacts with the vacuolar-type H+-ATPase pump a1 subunit to regulate lysosomal function. Hum Mol Genet 28:2696–2710. 10.1093/hmg/ddz088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerterp-Plantenga MS, Nieuwenhuizen A, Tomé D, Soenen S, Westerterp KR (2009) Dietary protein, weight loss, and weight maintenance. Annu Rev Nutr 29:21–41. 10.1146/annurev-nutr-080508-141056 [DOI] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, Grünblatt E, Moran LB, Mandel SA, Riederer P, Miller RM, Federoff HJ, Wüllner U, Papapetropoulos S, Youdim MB, Cantuti-Castelvetri I, et al. (2010) PGC-1α, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med 2:52ra73. ra73. 10.1126/scitranslmed.3001059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139:149–160. 10.1016/j.cell.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composition of holidic diet media. Download Figure 1-1, DOCX file (22KB, docx) .
Composition of 25N-F/H/W holidic medium. Download Figure 6-1, DOCX file (22KB, docx) .
Composition of Neurobasal-based medium minus phenol red and amino acids. Download Figure 7-1, DOCX file (22KB, docx) .