Abstract
Cerebral dopamine neurotrophic factor (CDNF) protects dopaminergic neurons against toxic damage in the rodent brain and is in clinical trials to treat Parkinson's disease patients. Yet the underlying mechanism is poorly understood. To examine its significance for neural circuits and behavior, we examined the development of neurotransmitter systems from larval to male adult mutant zebrafish lacking cdnf. Although a lack of cdnf did not affect overall brain dopamine levels, dopaminergic neuronal clusters showed significant abnormalities. The number of histamine neurons that surround the dopaminergic neurons was significantly reduced. Expression of tyrosine hydroxylase 2 in the brain was elevated in cdnf mutants throughout their lifespan. There were abnormally few GABA neurons in the hypothalamus in the mutant larvae, and expression of glutamate decarboxylase was reduced throughout the brain. cdnf mutant adults showed a range of behavioral phenotypes, including increased sensitivity to pentylenetetrazole-induced seizures. Shoaling behavior of mutant adults was abnormal, and they did not display social attraction to conspecifics. CDNF plays a profound role in shaping the neurotransmitter circuit structure, seizure susceptibility, and complex behaviors in zebrafish. These findings are informative for dissecting the diverse functions of this poorly understood factor in human conditions related to Parkinson's disease and complex behaviors.
SIGNIFICANCE STATEMENT A zebrafish lacking cdnf grows normally and shows no overt morphologic phenotype throughout the life span. Remarkably, impaired social cohesion and increased seizure susceptibility were found in adult cdnf KO fish conceivably associated with significant changes of dopaminergic, GABAergic, and histaminergic systems in selective brain areas. These findings suggest that cdnf has broad effects on regulating neurogenesis and maturation of transmitter-specific neuronal types during development and throughout adulthood, rather than ones restricted to the dopaminergic systems.
Keywords: dopamine, GABA, growth factors, histamine, neurotransmitters, Parkinson's disease
Introduction
Neurotrophic factors (NTFs), such as neurotrophins, glial cell line-derived NTF family of ligands, and neurokines, are crucial regulators of neurogenesis and regeneration. These secretory proteins and their signaling receptors are responsible for the survival, maintenance, and synaptic plasticity of nervous systems (Chao, 2003). Several neurodegenerative disorders, such as Parkinson's disease and Alzheimer's disease, are associated with dysregulation of trophic factors (Chao et al., 2006; Mitre et al., 2017).
An unconventional NTF family, which has a distinct two-domain protein structure and trophic effects on dopaminergic neurons, has recently been identified (Parkash et al., 2009; Latge et al., 2015). This evolutionarily conserved NTF family contains two proteins: mesencephalic astrocyte-derived NTF (MANF) and cerebral dopamine NTF (CDNF) (Lindahl et al., 2017). Their protein structure contains two main functional motifs. One is the N-terminus, which is similar to the saposin-like domain that has the capacity of lipid/cell membrane binding. The other is the C-terminus, composed of the unfolded Cys-X-X-Cys (CXXC) motif, the SAP domain of Ku70, and a putative endoplasmic reticulum (ER) retention signal (KDEL/RTDL) at the end of the C-terminal, which may protect cells from ER stress-induced apoptosis (Parkash et al., 2009; Latge et al., 2013, 2015). Both CDNF and MANF are detectable mainly in neurons in adult mouse and human brain, whereas the expression level of CDNF is generally lower than that of MANF (Lindholm et al., 2007; Lindahl et al., 2017). CDNF and MANF both protect dopaminergic neurons against oxidative stress, neurotoxins, cerebral ischemia, and neuroinflammation-induced neuronal death (Liu et al., 2015; Lindahl et al., 2017; Sousa-Victor et al., 2018). As such, CDNF has become recognized as a promising candidate for clinical treatment of Parkinson's disease because of its potent neuroprotective and neurorestorative effects on midbrain dopamine neurons (Lindholm et al., 2016; Lindahl et al., 2017; Nasrolahi et al., 2018).
CDNF protects cultured mesencephalic neurons against α-synuclein-oligomer-induced toxicity (Latge et al., 2015). In 6-OHDA and MPTP-induced Parkinsonian animal models, the application of CDNF and MANF protects and rescues midbrain dopamine neurons (Lindholm et al., 2007; Voutilainen et al., 2011; Airavaara et al., 2012; Ren et al., 2013). In addition to its neuroprotective effects in Parkinson's disease animal models, CDNF improves long-term memory in an APP/PS1 mouse model of Alzheimer's disease (Kemppainen et al., 2015), and reduces Aß25-35-induced ER stress and synaptotoxicity in cultured hippocampus neurons (W. Zhou et al., 2016). Growing evidence suggests that MANF and CDNF are ER stress response proteins involved in the unfolded protein response (UPR) through interactions with glucose-regulated protein 78 (BiP/GRP78) (Arancibia et al., 2018; Yan et al., 2019). Nevertheless, how CDNF responds to ER stress and its functions under healthy conditions remains largely unknown (Lindahl et al., 2017).
The role of CDNF in the development of dopaminergic or other neurons has not been addressed. The role and mechanisms of action of CDNF have remained elusive. To examine the biological function of cdnf and its role in neural systems and behavior, we first generated zebrafish null mutants by CRISPR/Cas9 genome editing. We investigated the role of cdnf on major neurotransmitter systems in the CNS, including dopaminergic, histaminergic, serotonergic, and GABAergic circuits using qRT-PCR, ISH, immunohistochemistry, and high-performance liquid chromatography (HPLC) analysis, as well as locomotor behavioral analysis, from development throughout their life span. We found that adult cdnf mutant zebrafish show abnormal social behaviors and seizure susceptibility phenotypes that are conceivably associated with the multiple impairments of major neurotransmitter networks and deficient neural progenitors during embryonic neurogenesis. These results may provide new evidence that CDNF has a considerable impact on neurogenesis and formation of normal neurotransmitter systems in CNS, which may have important implications for human neurologic disorders.
Materials and Methods
Zebrafish maintenance
Zebrafish were obtained from our WT (Turku) line that has been maintained in the laboratory for more than a decade (Kaslin and Panula, 2001; Sundvik et al., 2011; Chen et al., 2016). Larvae were raised on 14:10 (light/dark, lights on at 8:00 A.M.) cycles at 28°C and fed daily once with flake food and two times with live artemia. Adult fish were raised in continuously cycling Aquatic Habitats Systems with complete exchange of water in each tank every 6-10 min. Circulating water was UV-sterilized, and filtered with foam filters and activated charcoal. Water quality, including temperature (28 ± 0.5°C), pH value (7.4 ± 0.2), and conductivity (450 ± 10 µS), was monitored continuously. Embryos were obtained by natural spawning, collected from the breeding tanks, and staged in hours post-fertilization (hpf), days post-fertilization (dpf), or months post-fertilization (mpf) as previously described (Kimmel et al., 1995). The permits for all experiments were obtained from the Office of the Regional Government of Southern Finland, in agreement with the ethical guidelines of the European convention.
RNA isolation and cDNA synthesis
Total RNA was extracted from 10 pooled larval fish or one dissected adult brain per sample (RNeasy mini Kit; QIAGEN) for qRT-PCR. To synthesize cDNA, 2 μg of total RNA was reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen) with random hexamer primers (Roche Diagnostics) according to the manufacturer's instructions.
ISH
Whole-mount ISH was performed on 4% PFA-fixed embryos and dissected brains as described previously (Chen et al., 2009). Antisense and sense digoxigenin-labeled RNA probes were generated using the digoxigenin RNA labeling kit (Roche Diagnostics) following the manufacturer's instructions. The whole-mount ISH procedure was followed according to the protocol described by Thisse and Thisse (2008), with slight modifications. Briefly, the prehybridization and hybridization steps were conducted at 60°C for all riboprobes. The specificity of the antisense riboprobe hdc and tyrosine hydroxylase 2 (th2) has been described previously (Chen et al., 2009, 2016). The cloning primers for the open-reading frames of cdnf and vGAT cDNA are listed in Table 1. ISH signals were detected with sheep anti-digoxigenin-AP Fab fragments (1:10,000; Roche Diagnostics). The color staining was conducted with chromogen substrates (NBT/BCIP) and incubated in the dark at room temperature. Stained samples were embedded in 80% glycerol and visualized under brightfield optics using a DM IRB inverted microscope (Leica Microsystems) with a DFC 480 charge-coupled device camera. z stacks were processed with Application Suit software (Leica Microsystems), with the multifocal algorithm to identify the gene expression patterns (Chen et al., 2012).
Table 1.
List of primers used in this study
| Gene | Forward primer | Reverse primer | Note | Accession |
|---|---|---|---|---|
| actb1 | CGAGCAGGAGATGGGAACC | CAACGGAAACGCTCATTGC | qRT-PCR | NM_131031.1 |
| cdnf | TGAAGTTCCCTTGGAAGTGCG | TGTGCAGATATTGCACATGGC | RT-PCR cloning | NM_001123281.1 |
| cdnf | TTCTGCAGCCAAAGTGACCG | TCTGTGCTCCAGTCAAGAACC | qRT-PCR | NM_001123281.1 |
| cdnf | CACGAAATCAGCCCTCCAGT | GCTGAAGCCTCTGGCAGATT | crispr_cloning | BX901962.8 |
| cdnf | TGTGTGTGGGGTTTTTGGGA | GCCGATTCTCTTTCCCAGTAGTCTC | HRM | BX901962.8 |
| cdnf | TAGGCCCTCCTCCACCAGCTCT | AAACAGAGCTGGTGGAGGAGGG | sgRNA cloning | NM_001123281.1 |
| gad1b/67 | GGCCAAGGGCACGATTGGGT | GCATGCCACGCAGACTCGGT | qRT-PCR | NM_194419.1 |
| gad2/65 | GCGGAGGCATCGGCTCCAAA | GCCGCAGCTCTCGGCTGTAG | qRT-PCR | NM_001017708.2 |
| gfap | GAAGCAGGAGGCCAATGACTATC | GGACTCATTAGACCCACGGAGAG | qRT-PCR | NM_131373.2 |
| hdc | TTCATGCGTCCTCTCCTGC | CCCCAGGCATGATGATGTTC | qRT-PCR | NM_001102593.1 |
| manf | ACCATGTGCCAGTGGAAAAGA | TCGACGGAGCTCAAGTCAAC | qRT-PCR | NM_001076629.1 |
| rpl13a | AGAGAAAGCGCATGGTTGTCC | GCCTGGTACTTCCAGCCAACTT | qRT-PCR | NM_212784.1 |
| sox2a | CCTATTCGCAGCAAAGCACG | GGAATGAGACGACGACGTGA | qRT-PCR | NM_213118.1 |
| slc17a6a/vGlut2 | CATCCTGTCTACAACTACGGTT | CCAACACCAGAAATGAAATAGCCA | qRT-PCR | NM_001009982.1 |
| slc32a1/vGAT | AATACGCGTCACCACGAGAG | GAGCTCGATGATCTGTGCCA | Cloning | NM_001080701.1 |
| slc32a1/vGAT | CGGACAAGCCCAGAATCACT | CGACGGCGGCGAATATAATG | qRT-PCR | NM_001080701.1 |
| th | GACGGAAGATGATCGGAGACA | CCGCCATGTTCCGATTTCT | qRT-PCR | NM_131149.1 |
| th2 | CTCCAGAAGAGAATGCCACATG | ACGTTCACTCTCCAGCTGAGTG | qRT-PCR | NM_001001829.1 |
qRT-PCR
qRT-PCR was performed with a LightCycler 480 instrument (Roche Diagnostics) using the Lightcycler480 SYBR Green I master mix (Roche Diagnostics) according to the manufacturer's instructions. Primers for amplification were designed by Primer-BLAST (NCBI) and are listed in Table 1. Two housekeeping genes, β-actin and ribosomal protein L13a (rpl13a), were used as reference controls. All primer sets were confirmed to amplify only a single product of the correct size. Cycling parameters were as follows: 95°C for 5 min, followed by 45 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 20 s. Fluorescence changes were monitored with SYBR Green after every cycle. Dissociation curve analysis was performed at the end of the cycles (0.1°C per s increase from 60°C to 95°C with continuous fluorescence readings) to ensure that only a single amplicon (single melting peak) was obtained. All reactions were performed in duplicates, and at least three individual biological replicates were used (sample numbers indicated in the figure legends). Duplicate quantification values were analyzed with the LightCycler 480 software. The data were calculated by the comparative method, using Ct values of β-actin and rpl13a as a reference control (Livak and Schmittgen, 2001). Since the changes of relative gene expression showed the same trend when normalized to the different housekeeping genes (data not shown), only the results from rpl13a are presented.
Establishing zebrafish mutants
CRISPR/Cas9-genome edited fish were generated in our Turku WT strain, based on the description of Hwang et al. (2013). To avoid off-target genomic mutagenesis effects, targeting sites were selected with a minimum of three mismatches in the genome as predicted by the CHOPCHOP software (http://chopchop.cbu.uib.no). The sequence-specific sgRNA template was generated in a pDR274 vector (Addgene, plasmid #42250; oligo sequence listed in Table 1). The sequences of the modified plasmids were verified by Sanger sequencing. sgRNAs were transcribed from linearized template plasmids (Ambion MEGAscritp), and Cas9 mRNA was transcribed in vitro from linearized template plasmid pMLM 3613 (Addgene, plasmid #42251). A mixture containing ∼300 ng/µl Cas9 mRNA and 20 ng/µl sgRNA was injected into fertilized eggs at the one-cell stage. To verify the mutation efficiency of Cas9-sgRNA genome editing, the injected eggs were collected at 24 hpf. DNA was extracted from individual embryos and noninjected controls. PCR amplicons encompassing the targeted sites were amplified and analyzed via Sanger sequencing and high-resolution melting (HRM) analysis (Roche Diagnostics). Mutations were recognized as multiple sequencing peaks at the sgRNA target site. When the mutation efficiency was >50%, the remaining Cas9/sgRNA-injected embryos were raised to adulthood and outcrossed to the WT fish to generate F1 progeny. To collect the tail biopsies, 3 dpf embryos were anesthetized with 0.02% tricaine. The tip of the caudal fin within the pigment gap was removed using a microscalpel, and each larva was placed in an individual well of a 24-well plate with fresh embryonic medium until 5 dpf. We have consistently achieved 100% survival rate from the beginning of this procedure to adulthood. F1 genotyping was done by HRM assays and DNA sequencing. To identify mutated alleles from single embryos, each target locus was PCR-amplified from individual genomic DNA with gene-specific primers (Table 1). PCR products were then cloned and sequenced. Mutated alleles were identified by comparison with the WT sequence. Heterozygous (HET) F1 siblings carrying the same mutations were pooled in one tank and raised to adulthood. Because of the sex imbalance in the F1 generation of cdnf HET fish, F1 male HET fish were outcrossed to Turku female WT fish; female mutants were obtained from the resulting F2 progeny. Genotyping of the F2s was done as described for the F1 generation.
Fin clipping and genomic DNA extraction of adult zebrafish and 3 dpf larvae
To lyse genomic DNA, individual tail clippings were incubated in 50 µl lysis buffer (10 mm Tris-HCl, pH 8.3, 50 mm KCl, 0.3% Tween-20, and 0.3% NP-40) at 98°C for 10 min, followed by incubation on ice for 2 min; 1 µl of Proteinase K (20 mg/ml) was added to remove protein, and the mixture was incubated at 55°C for at least 4 h. To inactivate Proteinase K activity, samples were incubated at 98°C for 10 min and quenched on ice. To detect indel mutations, HRM curve acquisition and analysis were performed. Primers flanking the mutation site were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast; sequences are listed in Table 1). The HRM analysis was performed on a LightCycler 480 instrument (Roche Diagnostics) using the following reaction mixtures: 1× LightCycler 480 HRM master mix (Roche Diagnostics), 2 mm MgCl2, and 0.15 μm primer mixtures. The PCR cycling protocol was as follows: one cycle of 95°C for 10 min; 45 cycles of 95°C for 10 s, 60°C for 15 s, 72°C for 20 s, and melting curve acquisition; one cycle of 95°C for 60 s, and 40°C for 60 s. PCR products were denatured at 95°C for 60 s, renatured at 40°C for 60 s, and melted at 60°C to 95°C with 25 signal acquisitions per degree. Melting curves were generated over a 65°C-95°C range. Curves were analyzed using the LightCycler 480 gene-scanning software (version 1.5) according to the manufacturer's instructions (Roche Diagnostics). To identify deviations of the curves indicative of sequence mutations, a three-step analysis was performed using the Gene Scanning program (Roche Diagnostics) as follows: (1) normalizing the raw melting-curve data by setting the initial fluorescence uniformly to a relative value of 100% and the final fluorescence to a relative value of 0%; (2) determining the temperature threshold at which the entire double-stranded DNA was completely denatured; and (3) further analyzing the differences in melting-curve shapes (threshold setup 0) to cluster the melting curves with similar shapes into the same groups. Those with analogous melting curves were characterized as the same genotype.
Analysis of catecholamines and histamine by HPLC
For each sample, ten 8 dpf larvae were pooled into a group. The dissected brains of 8 mpf or 18 males were flash-frozen in liquid nitrogen and individually homogenized with sonication in 150 µl of 2% perchloric acid. After centrifugation, 10 µl of supernatant was assessed for monoamine concentration measurement. The detection details are described by Sallinen et al. (2009). The results were normalized to the total protein concentration of each sample, which was measured using the Pierce BCA Protein Assay Kit. The HPLC analysis was conducted as a blinded experiment.
Immunocytochemistry
Immunostaining was performed on zebrafish fixed in 2% PFA or 4% 1-ethyl-3 (3-dimethylaminopropyl)-carbodiimide (Carbosynth). For larvae older than 5 dpf, fixed brains were dissected to enhance antigen presentation and improve image quality. Antibody incubations were conducted with 4% normal goat serum and 1% DMSO in 0.3% Triton X-100/PBS for 16 h at 4°C with gentle agitation. Primary antibodies were mouse monoclonal anti-tubulin, acetylated antibody (1:1000; T6793, Sigma Millipore), rabbit anti-histamine 19C (1:5000) (Panula et al., 1990; Sundvik et al., 2011), rabbit anti-TH2 antibody (1:2000) (Semenova et al., 2014), rabbit anti-serotonin antibody (1:1000; S5545, Sigma Millipore), anti-TH (TH1) monoclonal mouse antibody (1:1000; product #22941, Immunostar), anti-GABA 1H (1:1000) (Karhunen et al., 1993; Kukko-Lukjanov and Panula, 2003), rabbit anti‐orexin A (1:1000; Millipore/Chemicon), and mouse anti-zrf1 (Gfap; 1:1000, Zebrafish International Resource Center). The specificities of the anti-GABA (Karhunen et al., 1993) and antihistamine, commercial anti-mouse monoclonal TH, anti-rabbit-TH2, and anti-serotonin antibodies, have been verified previously (Kaslin and Panula, 2001). The following secondary antibodies were applied: AlexaFluor-488 and -568 anti-mouse or anti-rabbit IgG (1:1000; Invitrogen).
Immunocytochemistry following EdU proliferation labeling
To detect the proliferating S-phase dividing cells, the Click-iTEdU AlexaFluor-488 imaging kit (Invitrogen) was used following the manufacturer's instructions, with minor modifications. Briefly, 5 dpf larvae were incubated in 0.5 mm EdU/EIII buffer (zebrafish embryonic medium; 5 mm NaCl, 0.44 mm CaCl2, 0.33 mm MgSO4, and 0.17 mm KCl) with 1% DMSO for 24 h at 28°C. Labeled samples were transferred back to EIII medium for 30 min and fixed in 4% 1-ethyl-3 (3-dimethylaminopropyl)-carbodiimide/PB buffer, pH 7.0, overnight at 4°C with gentle agitation. The skin and lower jaw of the fixed specimens were removed to enhance sample penetration. Dissected brains were incubated with rabbit antihistamine 19C antibody (1:5000) and mouse anti-HuC antibody (1:1000). The secondary antibody AlexaFluor-568 anti-rabbit IgG and AlexaFluor-633 anti-mouse IgG (1:1000; Invitrogen) were applied. After immunostaining, labeled specimens were fixed in 4% PFA/PB for 20 min at room temperature, and then incubated in 1× Click-iT EdU cocktail with the green fluorescent AlexaFluor-488 azide dye for 1 h in the dark at room temperature. After removing the reaction cocktail and rinsing in 1×PBST (PBS and 0.25% Triton X-100) 3 times for 10 min, samples were mounted in 80% glycerol/PBS for confocal microscopy imaging.
Imaging
Brightfield images were taken with a DM IRB inverted microscope (Leica Microsystems) with a DFC 480 charge-coupled device camera. z stacks were processed with Leica Application Suite software and Corel DRAW 2018 software (Chen et al., 2009). Immunofluorescence samples were examined using a TCS SP2 AOBS confocal microscope (Leica Microsystems). For excitation, an Argon laser (488 nm), green diode laser (561 nm), and red HeNe laser (633 nm) were used. Emission was detected at 500-550, 560-620, and 630-680 nm, respectively. Cross-talk between the channels and background noise were eliminated with sequential scanning and frame averaging as previously described (Sallinen et al., 2009). Stacks of images taken at 0.2-1.0 μm intervals were compiled, and the maximum intensity projection algorithm was used to produce final images with Leica Confocal software and Imaris imaging software (version 6.0; Bitplane). Cell numbers were counted in each 1.0 μm optical slice using ImageJ 1.52b software (National Institutes of Health). Fluorescence intensity was quantified in each 1.0 μm optical slice across entire z slices with a standardized-selected ROI under the same parameters using ImageJ 1.52b software. All cell counts and fluorescence intensity measurements were performed by an investigator blinded to the sample type.
Estimation of the posterior swim bladder volume (V3)
The volume of the posterior chamber V3 region (geometric figure-like frustum of a con) was estimated according to the equation (V = (1/3) × π × h × (r12 + r22 + (r1 × r2)) as described in detail previously (Lindsey et al., 2010). Measurements were done on 6-month-old male zebrafish (n = 4 of both genotypes) using Fiji (version 2.0) and were calculated using the lateral-aspect images of the 4% PFA-fixed posterior chamber.
Estimation of the vascular length density of the posterior swim bladder
The vascular length density was estimated using the vessel analysis plugin with Fiji (version 2.0) based on the user manual. The calculation of the vascular length density was presented as the ratio of skeletonized vasculature area to the total area (% area) (Elfarnawany, 2015). Measurements were done on the lateral-aspect images of the 4% PFA-fixed posterior chamber of 6-month-old male zebrafish (n = 4 of both genotypes).
Dark-light flash and sleep behavior test for larval zebrafish
Behavioral trials were done between 11:00 and 16:00. For larval locomotion tracking, 6 dpf zebrafish larvae were individually placed in a 24-well culture dish well containing ∼1.5 ml of EIII medium. The light level was set to ∼330 lux based on the setting of Puttonen et al. (2018). Before each trial, the larvae were habituated in the observation chamber for 10 min, followed by a 30 min locomotion tracking period with the lights on. A dark-light flash response was induced by switching off the lights for 2 min, then turning them back on for 2 min. One experiment consisted of three subsequent periods of white lights on and white lights off. Locomotor activity was monitored for 1 d with continuous illumination by infrared lights while white light remained on from 12:00 to 22:00 on the first day and from 8:00 to 12:00 on the next day. Locomotion response was monitored at room temperature using the Daniovision system (Noldus). Video tracking was analyzed by EthoVision XT 8.5 locomotion tracking software (Noldus).
Social interaction assay
The visually mediated social preference test was based on the setup of Baronio et al. (2018). Briefly, an acrylic apparatus (29 cm length × 19 cm height × 29 cm width) was divided into three arenas by two acrylic partitions. A rectangular compartment in the middle was the testing arena, referred to as the “distal” zone; to one side, the conspecific compartment housed a group of 6 fish, referred to as the “stimulus” zone; the other adjacent compartment was filled with stones and plant mockups, referred to as the “object” zone. A single 6 mpf male adult was placed in the testing arena to allow exploration and analysis of place preference. All experimental fish were raised in a social environment. All behavioral tests were performed between 11:00 and 16:00, and video-recorded from above the tank for 6 min. To quantify social preference, the videos were analyzed with the EthoVision XT 8.5 locomotion tracking software (Noldus), and the amount of time each test fish spent in the proximity of each compartment was quantified.
Novel diving tank assay
The novel tank assay was performed based on Cachat et al. (2010). One day before the experiment, 6 mpf male adult fish with home tanks (19 cm × 34 cm × 21 cm) were habituated in the behavior testing room. In each trial, one fish was placed in a transparent tank (24 cm × 14.5 cm × 5 cm) with 1 L of fish system water. All behavioral tests were performed between 11:00 and 16:00, and video-recorded from the side of the tank for 6 min, using a Basler acA1300-60gm industrial CCD video camera. We performed a three-compartment novel tank test, with digitized divisions between top, bottom, and middle virtual zones. The time spent in each zone was quantified using EthoVision XT 8.5 software. Fish were returned to their home tanks immediately after the test.
Shoaling assay
Five 6 mpf or five 18 mpf male fish per cohort were placed in a round white polyethlene plastic flat-bottomed container (23 cm height, 23 cm diameter) with 2 L of fish system water (5.0 cm depth) based on the description of Green et al. (2012). Before testing, fish were habituated for 15 min followed by video recording for 10 min with a camera at a fixed height (60 cm) from the top of the container. All videos were analyzed with EthoVision XT 8.5 software, using the default setting (the center-point detection of the unmarked animals). The mean of the interfish distance (defined as distance between the body center of every member of the shoal) (Green et al., 2012) was quantified from the average data from all trials (n = 4 trials for the 6 mpf fish, and n = 3 trials for the 18 mpf fish). The proximity duration (in seconds) was defined as the average duration of time a fish stayed close to the shoal fish (i.e., within 2 cm for the 6 mpf fish or 2.5 cm for the 18 mpf fish). The misdetection rate of the video-tracking software was <1%. All behavioral trials were done between 11:00 and 16:00.
Seizures induced by pentylenetetrazole (PTZ) in adult zebrafish
Epileptic seizure stage scores (seizure behavior scores from 0 to 6) were assigned according to Mussulini et al. (2013); 6 mpf male fish were individually exposed to 10 mm PTZ in a 1 L tank (24 cm length × 5 cm width × 14.5 cm height) for 5 min on 3 consecutive days, to induce experimental seizures based on the description of Duy et al. (2017). After PTZ administration and seizure analysis, treated fish were transferred to a clean tank for 1 d until they recovered (i.e., seizure score = 0). They were then killed by decapitation after cold-shock, and brains were dissected for RNA extraction. The PTZ concentration and the exposure period were selected and optimized based on our pilot study, which aimed to determine the shortest time of PTZ exposure that induces a seizure of score V (including fish falling to the bottom of the tank and loss of the body posture for 1-2 s), but allowing full recovery after three daily exposures (Duy et al., 2017). Control fish were subjected to the same procedure but exposed to only clean system water.
Experimental design and statistical analysis
The number of zebrafish for all experiments is indicated in the figure legends. Data analysis was performed by GraphPad Prism software (version 7). Two-tailed p values were generated by one-way ANOVA for multiple comparisons using Tukey's multiple comparison test, two-way ANOVA for multiple comparisons, and Student's unpaired t test for comparison of two groups. Data were presented as mean ± SEM. Statistical significance was considered at p value <0.05.
Results
Zebrafish cdnf 3D structure and mRNA distribution during embryogenesis and in adult tissues
Currently, there is one human homologous cdnf (NM_001123281.1) documented in the latest zebrafish database (GRCz11). The zebrafish cdnf gene is located on chromosome 4 and contains four exons with the exon-intron splice sites conserved in mammalian CDNF and MANF. The open reading frame encodes a protein of 182 amino-acid residues with a 25 amino-acid signal peptide and shares 67.4% and 65.1% of amino-acid sequence similarity with zebrafish manf (NP_001070097) and human CDNF (NP_001025125), respectively. The predicted secondary structure analyzed by the protein homology/analogy recognition engine Phyre2 (Kelley et al., 2015) indicated that zebrafish cdnf contained seven α-helices (Fig. 1A) and eight positioned-conserved cysteine residues. The N-terminus domain is highly similar to human saposin D, and the C-terminus is analogous to the SAP domain (Fig. 1B), suggesting that zebrafish cdnf is structurally conserved with human CDNF (Latge et al., 2013). To determine the spatiotemporal expression of cdnf, whole-mount ISH and qRT-PCR were performed in embryos and adult tissues. The cdnf mRNA was widely expressed at 1 dpf in the brain, eyes, and muscles. In 2 and 3 dpf larvae, cdnf transcripts were detected in the midbrain-hindbrain boundary, hindbrain, otic vesicles, and heart (Fig. 1C). qRT-PCR detected cdnf transcripts as early as 2 hpf (Fig. 1D), and expression gradually increased after 1 dpf. In adult organs, the highest expression level was found in the eyes compared with the brain, kidney, and liver; no sex differences were detected (Fig. 1E).
Figure 1.
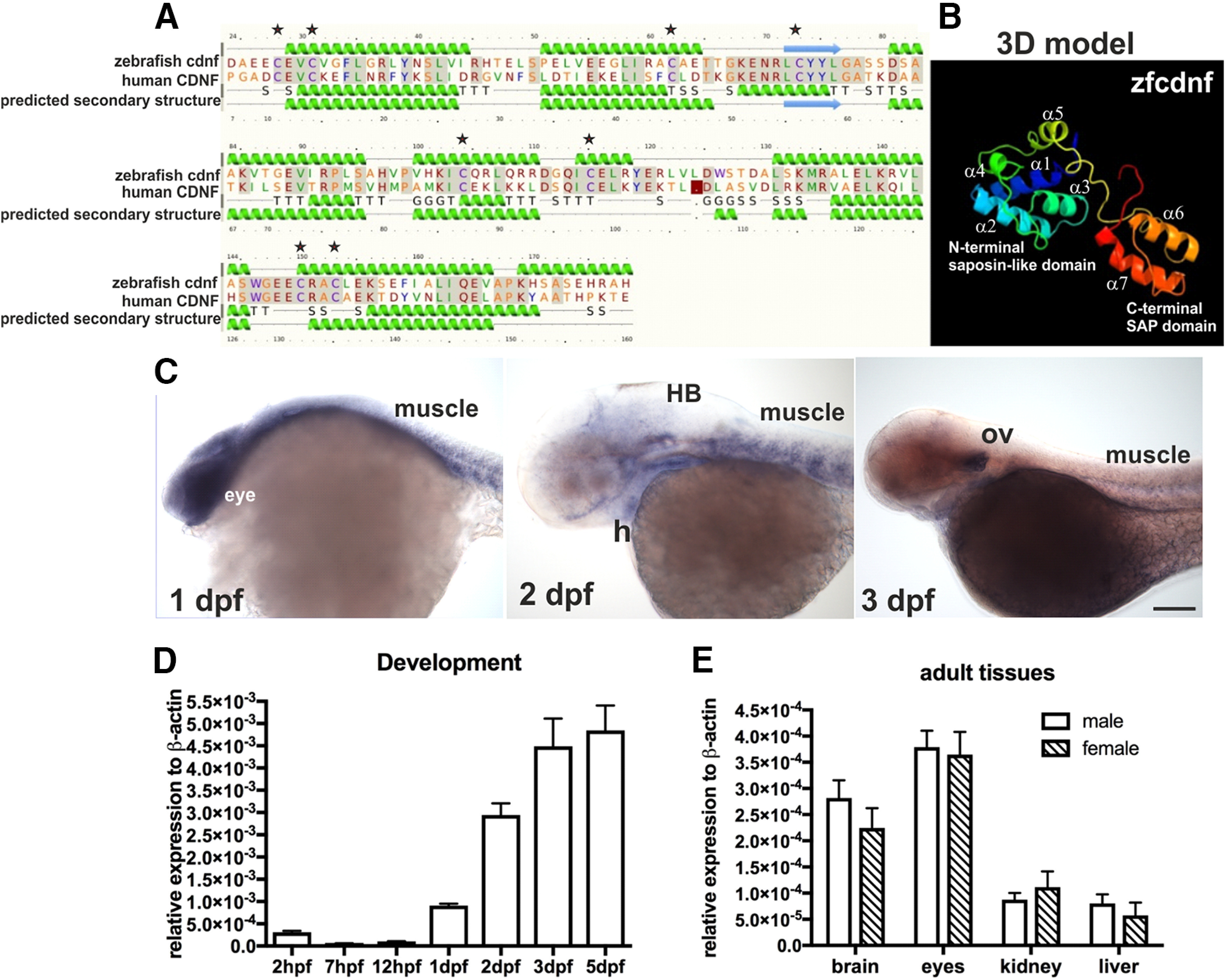
Predicted domain structures and spatiotemporal expression of zebrafish cdnf. A, Secondary structure and (B) predictive 3D model of zebrafish cdnf by Phyre2 modeling with human CDNF. C, Whole-mount ISH highlighting cdnf expression in the head, eyes, heart, muscles, and optic vesicles during development. D, qRT-PCR showing the zygotic cdnf mRNA expression at various developmental stages (one-way ANOVA, F(6,14) = 40.31, p < 0.0001, n = 3), and (E) in various male and female adult organs (two-way ANOVA, F(1,32) = 3.885, p = 0.0574, n = 4/group). Green helices represent α-helices. Blue arrows indicate β-strands. Gray lines indicate the conserved amino-acid residues. G, 3-trun helix; T, hydrogen bonded turn; S, bend; H, heart; HB, hindbrain; ov, optic vesicle. Data are mean ± SEM. One-way or two-way ANOVA was used for statistics. Single asterisk shows the positioned-conserved cysteine residue.
Characterization of the CRISPR/Cas9-generated zebrafish cdnf mutant allele
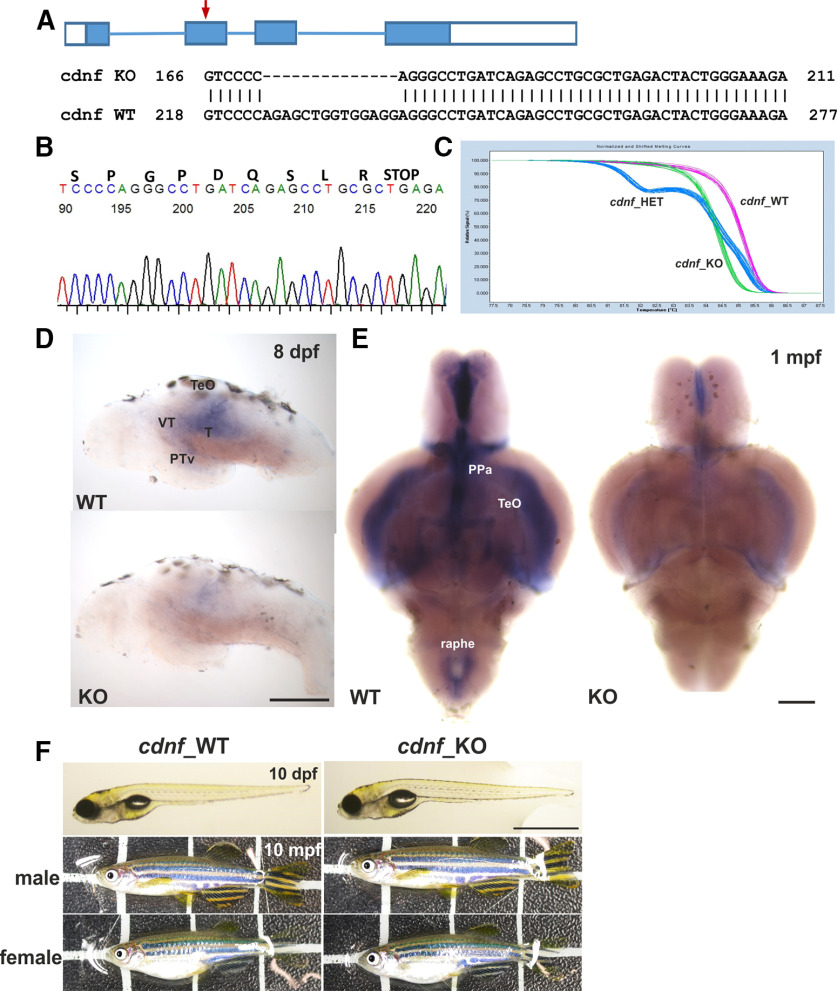
To study the biological function of cdnf, we generated mutations using the CRISPR/Cas9 system. A sequence-specific guide RNA was designed to target exon2 of cdnf (Fig. 2A). We identified two reading frame-shift mutant alleles: one with a 14 bp deletion (Fig. 2A,B) and another with a 1 bp insertion (data not shown). Both lesions are located in exon2, causing premature termination of the protein after amino-acid 49 (NP_001116753.1; Fig. 2B). We also cloned and sequenced the full-length reading frame in cdnf transcripts isolated from the mutant brain. The deletion sequence was the same as the genomic deletion sequence. In this study, the cdnf 14 bp deletion mutant allele was used for subsequent experiments. HET cdnf mutants (F3 and later generations) were mated to generate WT, HET, and homozygous mutant embryos. The ratio of genotyped sibling offspring matched the normal Mendelian ratio (1:2:1). All progenies were tail-clipped at 3 dpf and genotyped using HRM analysis, according to the distinct melting curves of each genotype. The embryos with the same HRM curve were grouped together before 5 dpf (Fig. 2C). ISH results revealed that cdnf expression was mostly abolished in the caudal raphe, ventral part of the posterior tuberculum (PTv), tectum opticum, and ventricular recess of the hypothalamus of cdnf mutant brains (Fig. 2D,E). The faint signal in the cdnf mutant brain may be because of the remaining truncated mRNA. The cdnf mRNA quantification is shown in Figure 3A–C. cdnf homozygous mutants were viable, swam normally, were fertile, and had no gross morphologic phenotype (Fig. 2F).
Figure 2.
CRISPR/Cas9-generated cdnf mutant zebrafish. A, Scheme of CRISPR/Cas9-generated 14-base pair deletion in exon2 of cdnf. B, Sequence chromatogram showing a premature stop codon of a homozygous cdnf mutant zebrafish. C, Results of the HRM analysis showing the distinctive melting curves of each genotype. D, Reduction in cdnf mRNA expression shown in the brains of 8 dpf and E, 1 mpf cdnf mutant fish. F, No obvious gross phenotype in larvae and adult cdnf mutant fish. PPa, anterior parvocellular preoptic nucleus; T, midbrain tegmentum; TeO, tectum opticum. Scale bars: D, 200 µm; F, 10 dpf, 1 mm; 10 mpf, 1 cm.
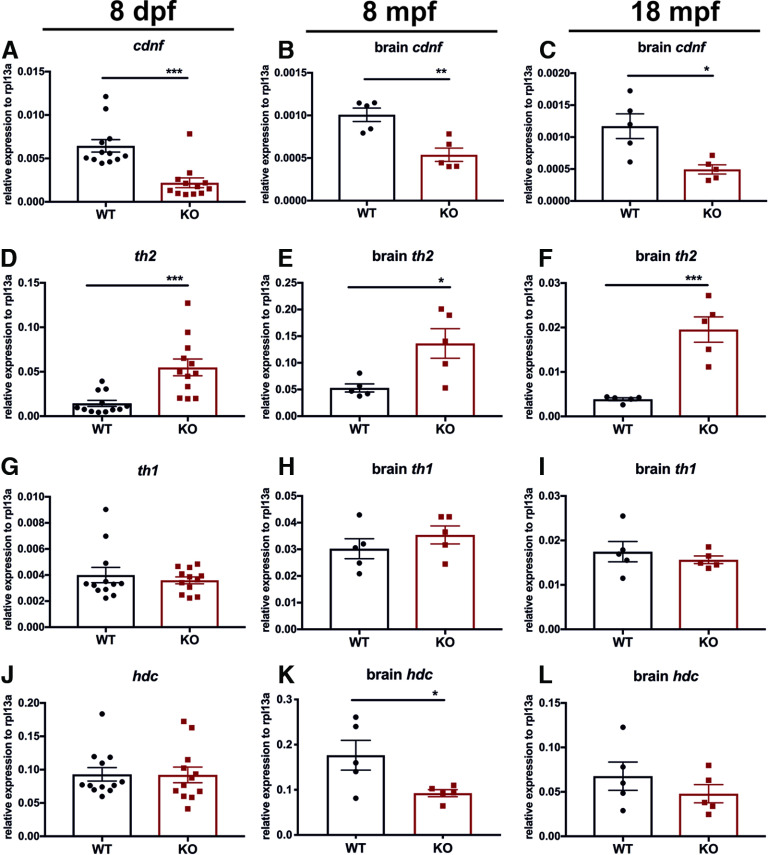
Figure 3.
qRT-PCR analysis of cdnf and neurotransmitter synthesis enzymes at larval, adulthood, and aging stages. A–C, Significant reduction of cdnf mRNA in cdnf-deficient (KO) fish (unpaired t test, t = 4.675, df = 22, two-tailed p = 0.0001; t = 4.245, df = 8, p = 0.0028; t = 3.280, df = 8, p = 0.0112). D–F, Significant increase of th2 transcripts in cdnf-deficient fish (unpaired t test, t = 4.043, df = 22, p = 0.0005; t = 2.901, df = 8, p = 0.0198; t = 5.450, df = 8, p = 0.0006). G–I, Nonsignificant differences in th1 mRNA expression between groups (unpaired t test, t = 0.6318, df = 22, p = 0.5340; t = 1.024, df = 8, p = 0.3358; t = 0.7477, df = 8, p = 0.4760). J–L, Significant reduction of hdc mRNA expression in adult brains (unpaired t test, t = 0.052, df = 22, p = 0.9590; t = 2.473, df = 8, p = 0.0385; t = 1.042, df = 8, p = 0.3278). qRT-PCR analysis relative to expression of the housekeeping gene rpl13a. Values are mean ± SEM; n = 12/group for 8 dpf fish, n = 5/group for 8 mpf brains, n = 5/group for 18 mpf brains in each cdnf KO and WT group. Data are mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; Student's t test.
Upregulation of th2 expression in cdnf mutant fish
We addressed the question of whether cdnf is required for development or maintenance of the dopaminergic neurons in the zebrafish brain. qRT-PCR analysis was performed on 8 dpf larvae, 8 mpf brains, and 18 mpf brains to examine transcript levels of relevant marker genes of the dopaminergic and histaminergic systems (known to depend on dopaminergic system). manf was also analyzed to reveal whether this closely related growth factor is upregulated as a consequence of genetic compensation in the cdnf mutant fish. In zebrafish, gene duplication has led to two nonallelic forms of human orthologous tyrosine hydroxylase (th) that are expressed in the brain in a largely complementary manner (Chen et al., 2009). We thus analyzed the mRNA levels of both tyrosine hydroxylases, th1 and th2. Remarkably, a significant increase of th2 was observed in cdnf mutants compared with their WT siblings (Fig. 3D–F), whereas the expression level of th1 mRNA showed no statistically significant difference (Fig. 3G–I). Additionally, a significant downregulation of histidine decarboxylase (hdc, a histaminergic marker) transcript was observed in 8 mpf mutant brains (Fig. 3J–L). Expression of hdc has been shown to be regulated by dopamine produced by th2 (Chen et al., 2016). We also confirmed that a significant reduction of cdnf transcript remained in cdnf mutants throughout their lifespan (Fig. 3A–C), which agreed with the ISH results (Fig. 2D,E) that most of the truncated cdnf mRNA went through nonsense-mediated mRNA decay pathway in cdnf-deficient mutant fish. The mRNA level of manf, a closely related trophic factor, was not significantly altered (data not shown). The qRT-PCR results revealed that cdnf has an impact on the regulation of dopaminergic and histaminergic gene expression.
A dynamic change of dopaminergic neuron numbers in cdnf mutant fish
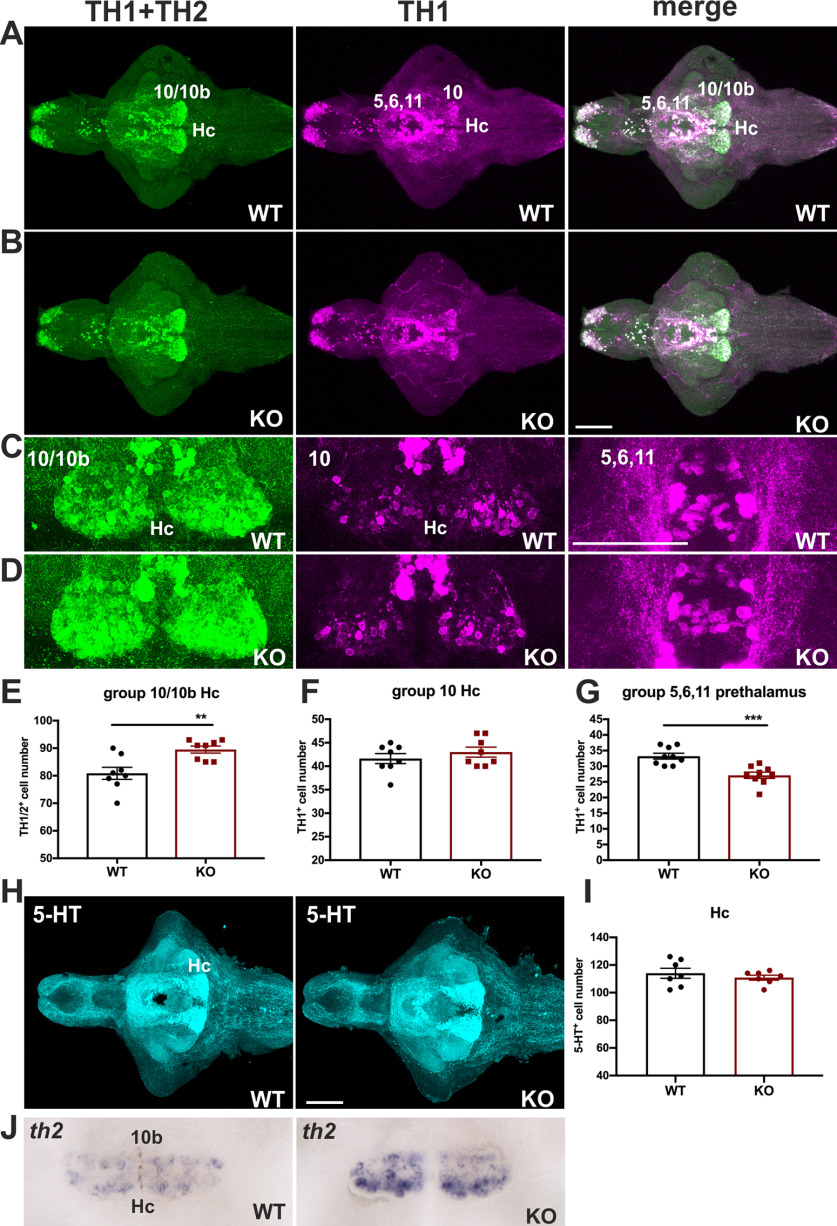
It is evident that dopaminergic signaling regulates the developing hypothalamic neurotransmitter identity (Chen et al., 2016). Because of the prominent upregulation of th2 transcripts in cdnf mutant fish, we then quantified the number of cells of the dopaminergic populations in the prethalamus and caudal hypothalamus (Hc) by immunohistochemistry to assess whether the TH1- and TH2-containing cell numbers were altered. The 8 dpf dissected brains were costained with two antibodies that recognize dopaminergic neurons; one recognizes both TH1 and TH2 (Semenova et al., 2014), and one recognizes only TH1 in zebrafish. A significant increase in TH1- and TH2-positive cell numbers was observed in TH1/TH2 group 10/10b in the HC area of KO mutants compared with their WT siblings (Fig. 4A–E). However, the number of TH1-positive cells was unaffected in this region (Fig. 4F), suggesting that the increase in TH1- and TH2-positive cell numbers in the Hc area of cdnf mutant brains is because of the increase in TH2-containing neurons, consistent with the qRT-PCR results (Fig. 3D–F) and the ISH result (Fig. 4J). Nevertheless, a significant decrease in TH1-positive cell number was found in the prethalamus region (TH1 group 5,6,11) in cdnf mutants (Fig. 4G). On the other hand, the serotonergic population in this location remained intact in KO mutants (Fig. 4H,I). A dynamic change in dopaminergic neuron populations found in cdnf mutant fish suggests that cdnf may function distinctly on dopaminergic populations in different brain areas.
Figure 4.
Selective alteration in dopaminergic systems in cdnf mutant fish. A, Colabeling of zebrafish th1 and th2 of 8 dpf cdnf WT brains. B, Colabeling of zebrafish th1 and th2 of 8 dpf cdnf KO brains. C, Higher-magnification images of TH2 10/10b (Hc) group, TH1 10 group, and TH1 5,6,11 group of cdnf WT brains. D, Higher-magnification images of TH2 10/10b (Hc) group, TH1 10 group, and TH1 5,6,11 group of cdnf KO brains. E, Significant increase in TH1/TH2-immunoreactive cell number in the Hc area (unpaired t test, t = 3.398, df = 14, p = 0.0043, n = 8/group; Hc, 10/10b th population) in the cdnf KO group. F, No significant change in TH1-immunoreactive cell number in the Hc area (unpaired t test, t = 0.9245, df = 14, p = 0.3709, n = 8/group; Hc, 10 th population) in the cdnf KO group. G, Significant decrease in TH1-positive cell number in the prethalamus (unpaired t test, t = 4.472, df = 16, p = 0.0004; n = 8/group, th1 group 5,6,11) in the cdnf KO group. H, 5-HT immunoreactivity in 8 dpf WT larval brains and cdnf KO larval brains (n = 7). I, No significant difference in 5HT-immunoreactive cell numbers in the HC area (unpaired t test, t = 0.7817, df = 12, p = 0.4495, n = 7/group). J, ISH results showing a higher intensity of th2 signals in th2 10b group (Hc) in 8 dpf cdnf KO fish (n = 8). In particular, TH1+TH2 (rabbit anti-th1 and th2 antibody) recognized both zebrafish th1 and th2 dopaminergic neurons. TH1 (mouse anti-tyrosine hydroxylase antibody) specifically recognized zebrafish th1. 5-HT, rabbit anti-serotonin antibody. Data are mean ± SEM. **p < 0.01; ***p < 0.001; Student's t test. Scale bar, 100 µm.
Decreased number of proliferating cells and histaminergic neurons in cdnf mutant fish
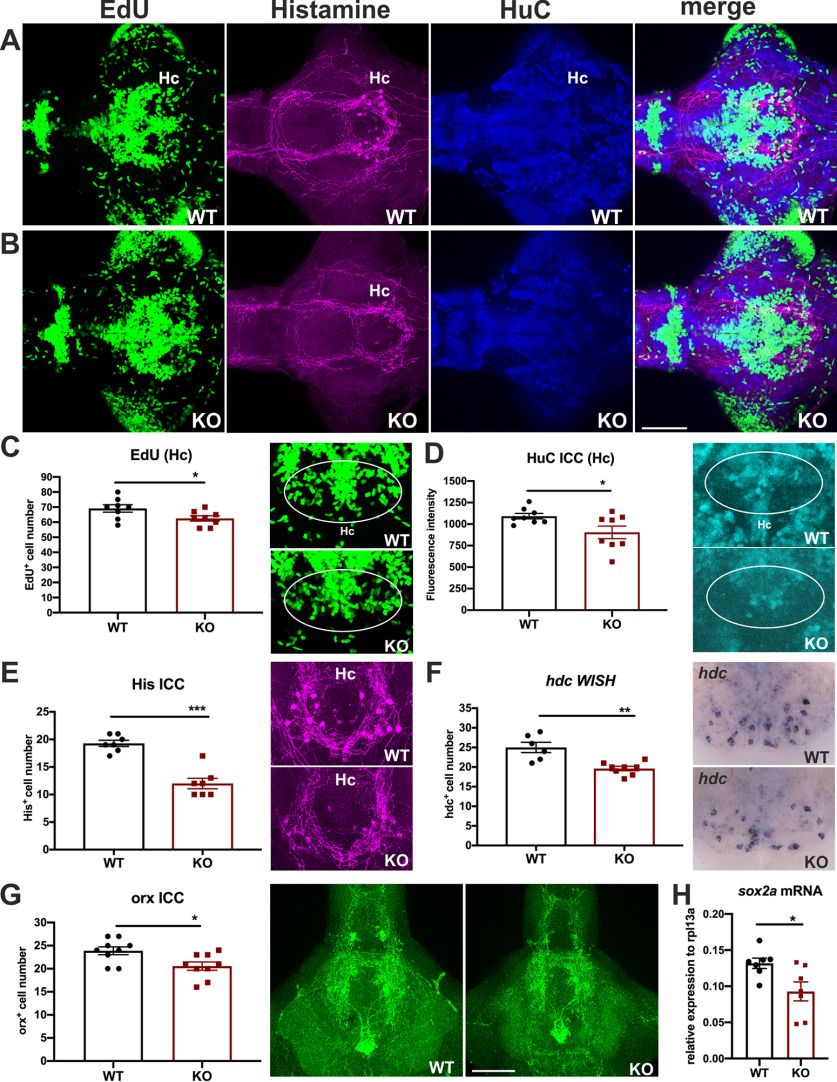
There are no reports about cdnf effects on embryonic neurogenesis. We previously reported that the dopaminergic populations in the Hc play a role in the regulation of histaminergic neuron development in zebrafish (Chen et al., 2016). To investigate whether the loss of functional cdnf affects neurogenesis and whether the upregulated TH2 population in this area disturbs the development of histaminergic neurons in larval cdnf mutants, we studied 8 dpf dissected brains costained with antibodies recognizing (1) histamine (a histamine neuron marker) and (2) HuC (a panneuronal marker) following EdU staining (a proliferation marker) (Fig. 5A,B). A significant reduction of EdU-positive cells (Fig. 5C) as well as lower fluorescence intensity of HuC immunoreactivity (Fig. 5D) was found in the Hc of cdnf mutant fish brains. Moreover, sox2a (neural progenitor marker) mRNA expression level was significantly downregulated in the cdnf mutant larvae (Fig. 5H). Together, these findings suggest that lack of cdnf significantly affects embryonic neurogenesis in the Hc. Similarly, a significant decrease of histaminergic neurons was found in cdnf mutants (Fig. 5E). Consistently, ISH of hdc (an enzyme converting L-histidine to histamine) showed a significant decrease in hdc-positive cell numbers in cdnf KO fish (Fig. 5F). A significant decrease in the number of orexin-positive cells was also found in cdnf KO fish (Fig. 5G). This result was in agreement with reports that the histaminergic system regulates orexin neuron development in zebrafish (Sundvik et al., 2011).
Figure 5.
Deficient proliferating cells and histaminergic neurons in cdnf mutant fish. A, Colabeling of proliferation marker (EdU), antihistamine immunoreactivity, anti-HuC immunoreactivity, and a merged image of triple labeled stacks of an 8 dpf cdnf WT brain. B, Triple labeling of an 8 dpf cdnf KO brain. C, Quantification of EdU-positive cell numbers, and higher-magnification images of EdU-labeled cells in the Hc (unpaired t test, t = 2.193, df = 14, p = 0.0457, n = 8 cdnf WT, and n = 8 cdnf KO fish). D, Quantification of fluorescence intensity of HuC-immunoreactive cells, and higher-magnification images of the Hc (unpaired test, t = 2.362, df = 14, p = 0.0332, n = 8 cdnf WT, and n = 8 cdnf KO fish). E, Quantification of histamine-positive cell numbers, and higher-magnification images of histamine immunostaining images (unpaired t test, t = 6.584, df = 12, p < 0.0001, n = 7 cdnf WT, and n = 7 cdnf KO fish). F, Quantification of histidine decarboxylase (hdc) mRNA-containing cell numbers (unpaired t test, t = 4.124, df = 12, p = 0.0014, n = 6 cdnf WT, and n = 8 cdnf KO fish). G, Quantification of orexin-positive cell numbers, and higher-magnification images of orexin immunostaining images (unpaired t test, t = 2.683, df = 16, p = 0.0163, n = 9 cdnf WT, and n = 9 cdnf KO fish). H, Significant decrease of sox2a transcripts in cdnf-deficient fish (unpaired t test, t = 2.614, df = 12, p = 0.0226). Marked areas were used as ROIs for measurement of fluorescence intensity. Data are mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; Student's t test. Scale bar, 100 µm.
Neurotransmitter profile by HPLC analysis
Because of the alterations of dopaminergic and histaminergic neurons in cdnf mutant fish, HPLC analysis was performed to measure the concentration of dopamine, norepinephrine, serotonin, histamine, and their metabolites throughout the zebrafish lifespan. No statistically significant difference was found in brain dopamine, norepinephrine, and serotonin levels, but a decrease in dopamine metabolites, DOPAC, and homovanillic acid and serotonin metabolite 5-hydroxyindoleaetic acid was found in adult cdnf mutant fish compared with the WT fish brains (Table 2). A reduction in histamine level was found in 8 mpf cdnf mutants, consistent with a reduced expression of histaminergic marker hdc mRNA in 8 mpf cdnf KO brains (Fig. 3K).
Table 2.
HPLC analysis of monoamine concentrations of larvae, adult brains, and aging brainsa
| 19-month-old brain (pmol/mg protein) |
8-month-old brain (pmol/mg protein) |
8 dpf (8 larvae per tube) (pmol/mg protein) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| WT (n = 5) | KO (n = 5) | p | WT (n = 9) | KO (n = 9) | p | WT (n = 3) | KO (n = 3) | p | |
| Dopamine | 22.39 ± 0.3935 | 21.14 ± 0.6298 | 0.13 | 16.52 ± 0.542 | 15.97 ± 0.433 | 0.4451 | 2.834 ± 0.1438 | 3.21 ± 0.2845 | 0.3041 |
| Norepinephrine | 58.79 ± 3.076 | 56.32 ± 2.85 | 0.5707 | 46.29 ± 1.956 | 43.3 ± 1.243 | 0.216 | 7.2 ± 0.1838 | 8.239 ± 0.56 | 0.1528 |
| DOPAC | 4.699 ± 0.3663 | 3.506 ± 0.2053b | 0.0218 | 2.764 ± 0.1298 | 2.452 ± 0.2412 | 0.2713 | 1.721 ± 0.1544 | 1.605 ± 0.08641 | 0.549 |
| Homovanillic acid | 12.48 ± 1.131 | 8.847 ± 0.1623b | 0.013 | 17.54 ± 1.768 | 8.494 ± 1.607b | 0.0016 | 2.235 ± 0.007714 | 4.195 ± 1.205 | 0.1794 |
| 3-Methoxytyramine | 1.539 ± 0.1343 | 1.343 ± 0.2165 | 0.4639 | 5.42 ± 0.6011 | 5.795 ± 0.438 | 0.6205 | 2.854 ± 0.1544 | 3.351 ± 0.4868 | 0.3855 |
| Serotonin | 23 ± 1.162 | 24.63 ± 0.6439 | 0.2543 | 16.83 ± 0.6696 | 19.43 ± 0.6398c | 0.0124 | 3.693 ± 0.157 | 3.902 ± 0.1672 | 0.4142 |
| 5-Hydroxyindoleacetic acid | 19.81 ± 0.9596 | 13.7 ± 0.4645b | 0.0004 | 15.99 ± 0.8659 | 15.08 ± 1.173 | 0.5423 | 3.769 ± 0.03767 | 5.02 ± 0.6834 | 0.1416 |
| Histamine | 9.065 ± 0.4468 | 11.33 ± 1.745 | 0.244 | 5.958 ± 0.516 | 4.285 ± 0.3379b | 0.0154 | 7.805 ± 0.854 | 5.899 ± 1.09 | 0.2406 |
aData are mean ± SEM. Student's t test was used for statistical analysis.
bA significant reduction in KO.
cA significant increase in KO.
Impairments of GABAergic system
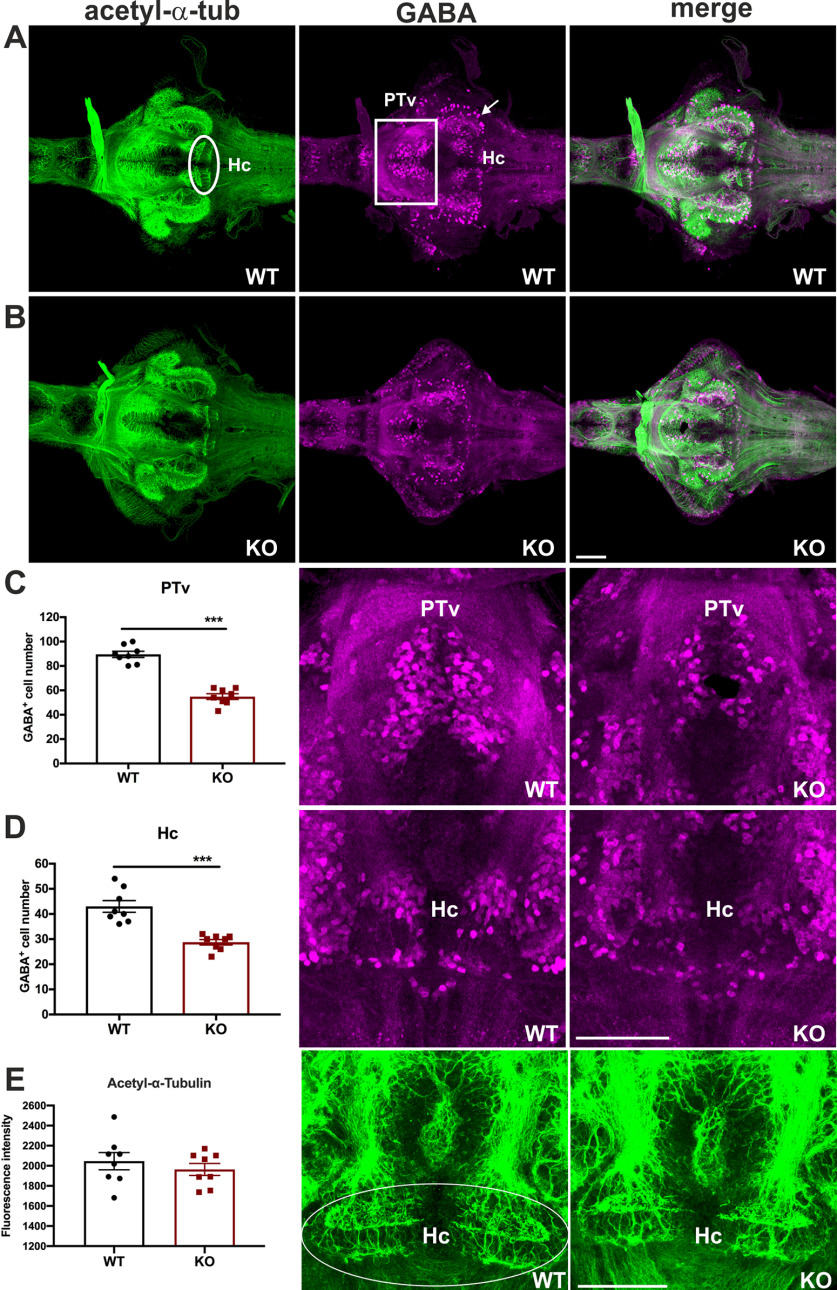
GABA acting through the GABA-A receptor in the adult brain is the major inhibitory neurotransmitter, and GABAergic neurons are widely distributed in the brain to modulate neural activity. It has been reported that MANF can potentiate presynaptic GABAergic inhibition (C. Zhou et al., 2006). Moreover, a dual dopaminergic and GABAergic phenotype is evident in the hypothalamic areas in zebrafish (Filippi et al., 2014). Dopamine signaling deficiency affects the development of GABAergic neurons in zebrafish (Souza et al., 2011). To assess whether the profound change of dopaminergic systems in cdnf mutant fish is associated with abnormalities in the GABAergic system, anti-GABA and antiacetylated-α-tubulin antibodies (axonal α tubulin marker) were used on 8 dpf dissected brains. Significantly fewer GABA-positive cells were observed in the PTv (Fig. 6A–C) and Hc (Fig. 6D) of the mutant fish than in their WT siblings. The axonal tubulin pattern remained intact (Fig. 6A,B,E) in cdnf mutants, suggesting that the loss of functional cdnf causes specific abnormal neurotransmitter systems in the PTv and Hc areas rather than an overall change in neuronal organization.
Figure 6.
Selective impairment of GABAergic system in cdnf KO fish. A, Colabeling of antiacetylated tubulin (showing axons) and GABA with antibody (showing GABAergic cells) of 8 dpf cdnf WT brains. B, Double immunostaining of 8 dpf cdnf KO brains. C, Quantification of GABA-immunoreactive cell numbers in the PTv, and higher-magnification images of GABA-labeled cells (unpaired t test, t = 9.989, df = 14, p < 0.0001, n = 8/group). D, Quantification of GABA-immunoreactive cell numbers in the Hc, and higher-magnification images of GABA-labeled cells (unpaired t test, t = 5.503, df = 14, p < 0.0001, n = 8/group). E, Quantification of fluorescence intensity of acetylated tubulin-immunoreactive projections, and higher-magnification images of acetylated tubulin-immunoreactive projections (unpaired t test, t = 0.7879, df = 14, p = 0.4439, n = 8/group). White rectangles and arrows represent a noteworthy reduction of GABA-staining cells in the PTv and Hc area, respectively. The marked area was used as an ROI for measurement of fluorescence intensity. Data are mean ± SEM. ***p < 0.001 (Student's t test). Scale bar, 100 µm.
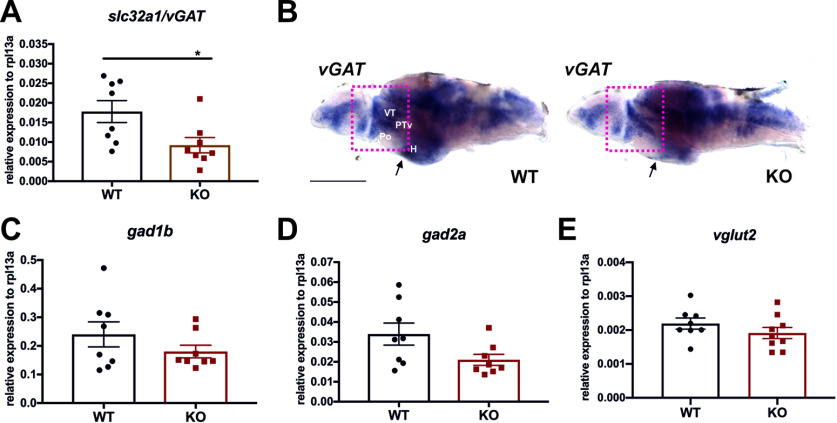
GABA is converted from glutamic acid by glutamic acid decarboxylases (GADs: GAD65/gad2a and GAD67/gad1b), and vesicular GABA transporter (vGAT) is responsible for uptake and storage of GABA in the vesicles in the presynaptic terminals. To study whether the decreased number of GABA-containing cells in cdnf mutants stems from either dysfunction of GABA synthesis or neurotransmission, we performed qRT-PCR to determine the expression level of GABAergic markers slc32a1/vGAT, gad2a/gad65, and gad1b/gda67 in 8 dpf larvae. A significant decrease in slc32a1/vGAT transcripts was detected in cdnf mutant larvae (Fig. 7A), and the ISH results of vGAT in 8 dpf dissected brains showed lower expression in the ventral thalamus, PTv, and hypothalamus (H) areas of cdnf KO mutants than in their WT siblings (Fig. 7B). However, the expression levels of gad1b, gad2a and vGlut2 in whole larvae were not statistically different (Fig. 7C–E). These findings suggest that the decreased number of GABA-positive cells is associated with downregulation of vGAT expression in the cdnf KO larvae.
Figure 7.
slc32a/vGAT downregulation in cdnf KO fish. A, Quantification of slc32a/vGAT mRNA expression relative to the housekeeping gene rpl13a in 8 dpf fish (unpaired t test, t = 2.508, df = 14, p = 0.0251, n = 8/group). B, ISH of vGAT mRNA expression of 8 dpf brains (n = 5). C, Quantification of gad1b mRNA expression relative to rpl13a in 8 dpf fish (unpaired t test, t = 2.077, df = 14, p = 0.0567). D, Quantification of gad2a mRNA expression relative to the rpl13a in 8 dpf fish (unpaired t test, t = 2.508, df = 14, p = 0.0251). E, Quantification of vGlut2 mRNA expression relative to rpl13a in 8 dpf fish (unpaired t test, t = 2.508, df = 14, p = 0.0251). Arrows and red rectangles represent downregulated expression of vGAT in the hypothalamus (H), preoptic region (Po), ventral posterior tuberculum (PTv), and ventral thalamus (VT). Data are mean ± SEM. *p < 0.05 (Student's t test). Scale bar, 100 µm.
Dark-flash and sleep-related behavior
Dopaminergic, histaminergic, and GABAergic circuitries play important roles in the regulation of startle response and sleep-wakefulness behavior (Jones, 2020). cdnf mutant larvae did not show impaired light/dark adaptions by the light-dark flash change (data not shown). Therefore, we examined the sleep-like locomotor behavior of 6 dpf larval fish under a 14:10 (light/dark) regimen (Fig. 8A; n = 16 in each group). Interestingly, during the daylight period, the cdnf mutant larvae were more active than their WT siblings (Fig. 8B), although locomotion during the second daylight period showed no statistically significant differences between groups (Fig. 8C). In contrast, during the dark period, the cdnf mutant larvae moved significantly less time than their WT siblings (Fig. 8D,E). The abnormal locomotor activity during light and dark conditions was thus associated with the dysfunctional neurotransmission in cdnf mutant larvae.
Figure 8.
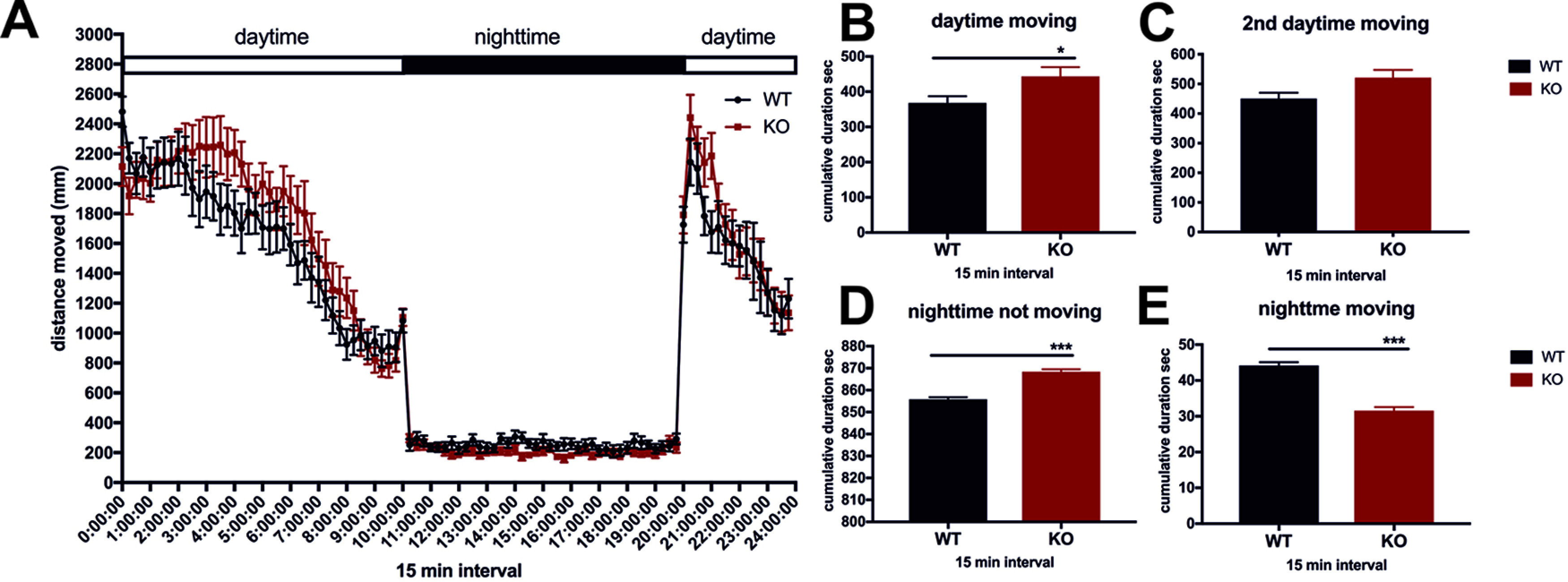
Eight dpf larval sleep-related behavior. A, Locomotor activity under 14:10 (light:dark) illumination conditions (multiple t tests using two-stage step-up method of Benjamini et al., 2006). B, Average cumulative duration (in seconds) in 15 min time bins of movement during light conditions (unpaired t test, t = 2.304, df = 62, p = 0.0246). C, Average cumulative duration of movement during the second treatment of light conditions in 15 min time bins (unpaired t test, t = 2.151, df = 12, p = 0.0525). D, Duration of rest (not moving) during dark conditions in 15 min time bins (unpaired t test, t = 9.159, df = 76, p < 0.0001). E, Average cumulative duration of movement during dark conditions in 15 min time bins (unpaired t test, t = 9.159, df = 76, p < 0.0001). n = 16/group. Data are mean ± SEM. *p < 0.05; ***p < 0.001; Student's t test and one-way ANOVA analysis with multiple comparisons.
Impaired social preference in adult cdnf mutant fish
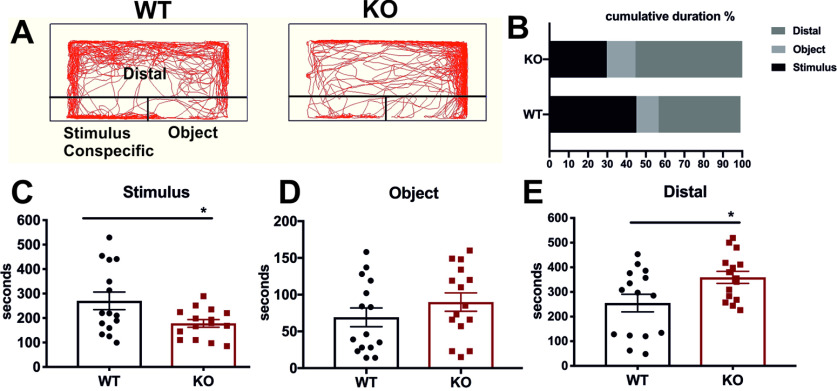
There is growing evidence to suggest that dysregulated neurotransmission causes neuropsychiatric disorders, some of which alter social interactions (Laruelle, 2014; Y. S. Kim and Yoon, 2017; Provensi et al., 2020). Zebrafish are social animals and naturally tend to approach conspecifics by visual choice (Miller and Gerlai, 2011). The social preference test conducted here measures this innate tendency. To investigate the consequences of impaired dopaminergic, histaminergic, and GABAergic systems on adult fish behavior, we performed social preference analysis/visually mediated social preference on 6 mpf male fish by quantifying the amount of time each fish spent in close proximity to conspecifics (“stimulus” arena) compared with the empty “object” arena (Fig. 9A). In comparison with their WT siblings, cdnf mutant fish spent significantly less time in the stimulus/conspecific zone (Fig. 9B,C), but spent more time in the “distal” testing zone (Fig. 9B,E). WT and KO fish spent similar amounts of time in the “object” zone (Fig. 9B,D), indicating that adult cdnf mutant fish show less social preference for conspecifics than the WT fish.
Figure 9.
Social preference deficiency in cdnf KO adult fish. A, Schemes of visually mediated social preference behavior setup and representative movement traces of cdnf WT and KO fish during 10 min recording intervals. B, Ratio of cumulative duration in the distal, object, and stimulus/conspecific zone. C, Cumulative time spent in the stimulus (conspecific) zone (unpaired t test, t = 2.357, df = 28, p = 0.0257). D, Cumulative time spent in the object zone (unpaired t test, t = 1.156, df = 28, p = 0.2575). E, Cumulative time spent in the distal zone (unpaired t test, t = 2.217, df = 28, p = 0.0349). n = 15/group. Data are mean ± SEM. *p < 0.05 (Student's t test).
Anxiolytic behavior appeared in adult cdnf mutant fish
Zebrafish have a natural tendency to spend more time at the bottom of the tank when placed in a novel environment, before gradually migrating to the surface (Kysil et al., 2017). We used a novel tank diving assay to study anxiety-related risk-taking behavior. The novel tank diving area was digitally divided into three zones, and representative swimming tracks are shown in Figure 10A. Compared with their WT siblings, cdnf mutants spent significantly more time exploring the top zone (Fig. 10B,C) and less time in the bottom (Fig. 10E); there was no difference in the time spent in the middle zone between WT and KO siblings (Fig. 10D). There was no significant difference for movement speed between WT and KO siblings, suggesting that impaired bottom-dwelling behavior was not caused by motor defects (Fig. 10F). Moreover, buoyancy is regulated by the swim bladder and its innervation (Finney et al., 2006). We examined the size of the swim bladder (Fig. 10G,H), vasculature (Fig. 10G,I), and TH1 innervation (Fig. 10G,J) of its posterior chamber, and no overt defect was found in the swim bladder of cdnf KO fish (n = 4 in each group).
Figure 10.
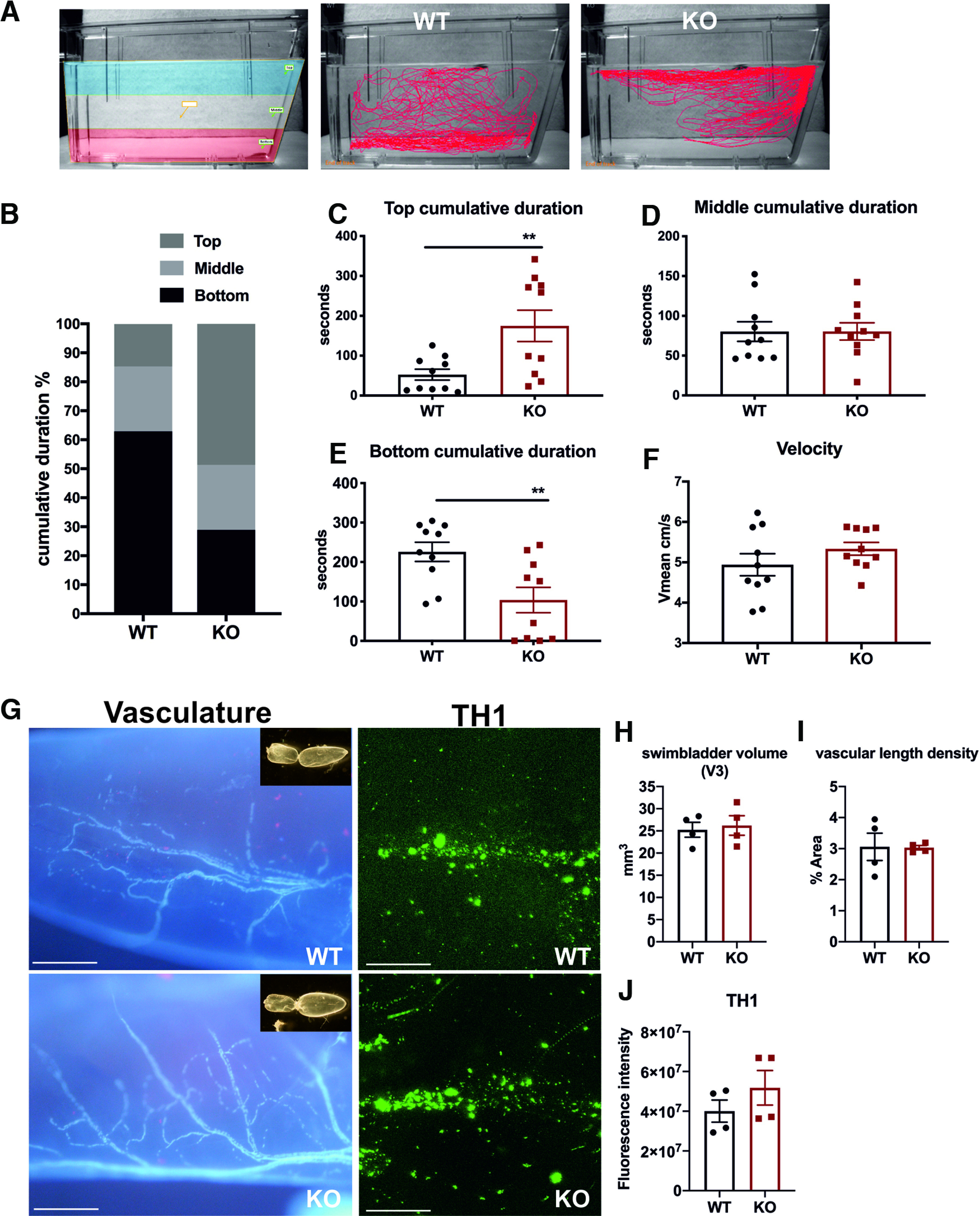
Impaired bottom-dwelling behavior in cdnf KO adult fish. A, Schemes of the novel tank test with three digitized zones and representative movement traces of cdnf WT and KO siblings during a 6 min recording period. B, Ratio of cumulative duration in the top, middle, and bottom zones. C, Amount of time spent in the top zone (unpaired t test, t = 2.942, df = 18, p = 0.0087). D, Amount of time spent in the middle zone (unpaired t test, t = 0.01053, df = 18, p = 0.9917). E, Amount of time spent in the bottom zone (unpaired t test, t = 3.029, df = 18, p = 0.0072). F, Average velocity during the 6 min video tracking period (unpaired t test, t = 1.253, df = 18, p = 0.2261). G, Vasculature by autofluorescence and TH1 immunostaining of the posterior chamber of the swim bladder in cdnf WT and cdnf KO fish. Insets, Dissected swim bladders. H, Quantification of V3 volume of the posterior chamber (unpaired t test, t = 0.3488, df = 6, p = 0.7392). I, Quantification of vascular length density of the posterior chamber (unpaired t test, t = 0.0646, df = 6, p = 0.9506). J, Quantification of TH fluorescence intensity of the posterior chamber (unpaired t test, t = 1.138, df = 6, p = 0.2983). n = 10 for WT and cdnf KO fish in the novel tank analysis. n = 4 for WT and cdnf KO in the swim bladder analysis. Data are mean ± SEM. **p < 0.01; Student's t test. Scale bar, 100 µm.
Decreased shoal cohesion in adult cdnf mutant fish
Zebrafish swim naturally in shoals (Miller and Gerlai, 2011). To test whether cdnf deficiency affects fish shoaling behavior, five 6 mpf fish (male young adult group) or five 18 mpf fish (male adult group) per trial were placed in a plastic cylindrical container (23 cm diameter, and monitored by video tracking for 10 min after a 15 min habituation period (n = 4 trials for young adults, and n = 3 trials for adults). The movement speed, average distance between the test fish and the other four shoal members, and duration of stays in proximity (the nearest interindividual distance defined as <2 cm for young adults and 2.5 cm for adults) were analyzed. cdnf mutant fish showed higher swimming speed during 10 min locomotion activity in both age groups (Fig. 11A,B,E,F). In the young adult group, the interindividual distance was significantly greater in the cdnf mutant group compared with their WT sibling group (Fig. 11C). Furthermore, the time spent in proximity with shoal members was significantly shorter in cdnf mutant groups than in their WT siblings (Fig. 11D). Similar results were obtained in the adult group. Collectively, the cdnf mutant fish were more hyperactive and kept at a greater distance to their neighbors (Fig. 11G), although no significant difference in time spent in proximity was observed in the adult group (Fig. 11H), suggesting that lack of functional cdnf causes social defect phenotypes in adult zebrafish.
Figure 11.
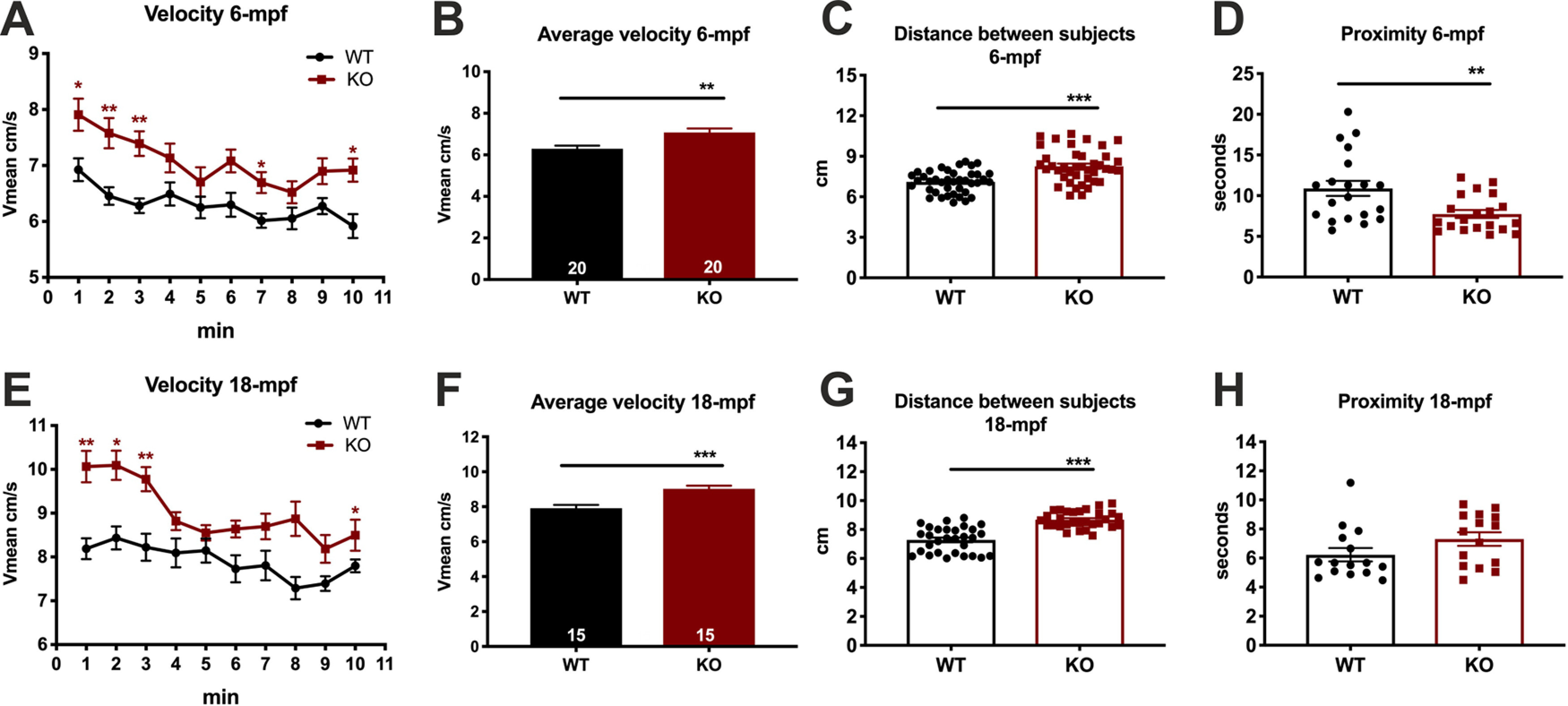
Decreased shoal cohesion in cdnf KO adult fish. Top, Shoaling behavior test on 6 mpf male cdnf WT and KO fish (n = 5 in each trial) during a 10 min video tracking period; four trials were analyzed. A, Average velocity, in 1 min time bins (multiple t tests, 1 min, t = 2.794, df = 38.0, p = 0.0477; 2 min, t = 3.576, df = 38, p = 0.0087; 3 min, t = 4.344, df = 38, p = 0.001; 7 min, t = 2.991, df = 38, p = 0.033; 10 min, t = 3.361, df = 38.00, p = 0.014). B, Average velocity during the 10 min video tracking period (unpaired t test, t = 3.300, df = 38, p = 0.0021). C, Average interindividual distance of total trials (unpaired t test, t = 4.964, df = 78, p < 0.0001). D, Average time spent in the proximity (i.e., <2 cm) of the nearest neighbor. Bottom, Shoaling behavior test on 18 mpf male cdnf WT and KO fish (n = 5 in each trial); three trials were analyzed (unpaired t test, t = 2.973, df = 38, two-tailed p = 0.0051). E, Average velocity, in 1 min time bins (multiple t tests, 1 min, t = 4.351, df = 28.0, p = 0.0016; 2 min, t = 3.898, df = 28, p = 0.0050; 3 min, t = 3.726, df = 28, p = 0.0070; 8 min, t = 3.392, df = 28, p = 0.0145). F, Average velocity during the 10 min video tracking period (unpaired t test, t = 4.122, df = 28, p = 0.0003). G, Average interindividual distance of total trials (unpaired t test, t = 7.264, df = 58, p < 0.0001). H, Average time spent in the proximity of closest neighbor (unpaired t test, t = 1.649, df = 28, p = 0.1103). Data are mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; Student's t test and one-way ANOVA analysis with multiple comparisons.
Increased seizure susceptibility in adult cdnf mutant fish
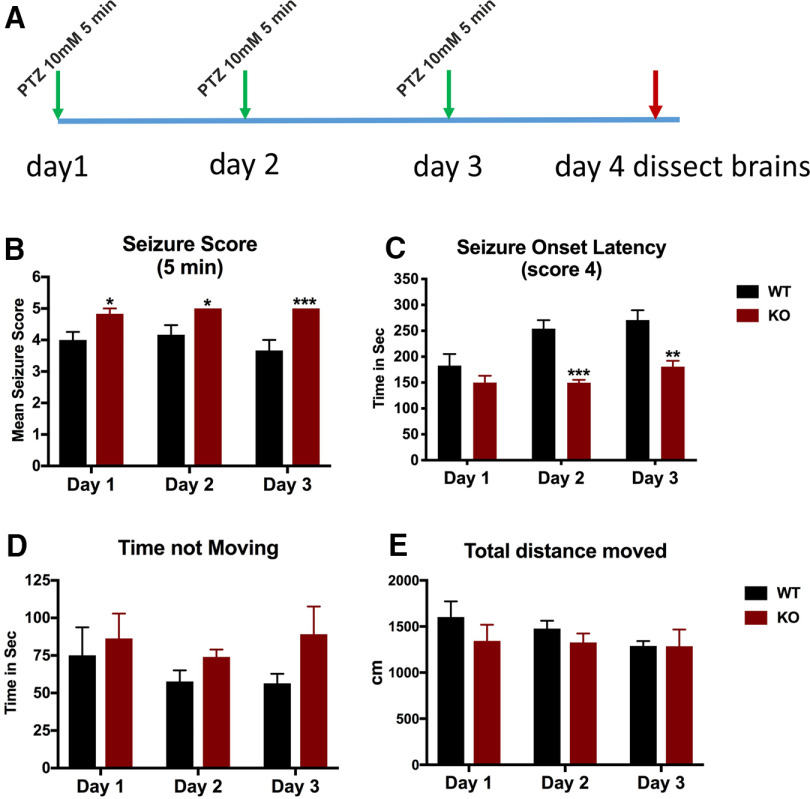
We hypothesized that the impaired GABAergic phenotype found in the cdnf mutant fish may render the mutants more susceptible to drug-induced epileptic seizures. PTZ, a chemoconvulsant drug, is commonly used to induce seizures in rodents and zebrafish by inhibiting GABA-A receptor subunits (Huang et al., 2001; Mussulini et al., 2013). Six-month-old male fish were exposed to 10 mm PTZ (for 5 min periods, over 3 consecutive days) to allow analysis of the molecular consequences of PTZ-induced seizures (Fig. 12A). Seizures were scored based on the description of Mussulini et al. (2013). Briefly, score 3 was recorded when fish showed repetitive circular movements, score 4 included abnormal whole-body rhythmic muscular contractions, and score 5 was characterized by rigidity, loss of body posture, and sinking to the bottom of the tank. None of the tested fish died during the PTZ administration procedure. Throughout the 5 min PTZ administration, a significantly higher percentage of cdnf mutant fish reached score 5 compared with their WT siblings (Fig. 12B), revealing that cdnf mutant fish are more susceptible to PTZ-induced seizures. Moreover, the cdnf mutant fish showed a shorter onset latency to reach score 4 than their WT siblings, particularly on the second and third days of exposure (Fig. 12C). The cdnf mutant fish demonstrated the longest periods of immobility (Fig. 12D), but there were no significant differences between the genotypes in the total distance moved (Fig. 12E).
Figure 12.
Increased seizure susceptibility in cdnf KO adult fish. A, Scheme of the PTZ administration procedure. B, Seizure scores after 5 min of PTZ exposure (two-way ANOVA, F(2,15) = 0.8333, p = 0.4538; multiple t test; day 1, t = 2.712, df = 10, p = 0.0433; day 2, t = 2.712, df = 10, p = 0.0433; day 3, t = 4.000, df = 10, p = 0.0075). C, Seizure onset latency to score of 4 (two-way ANOVA, F(2,30) = 2.973, p = 0.0664; multiple t tests; day 1, t = 1.265, df = 10, p = 0.2344; day 2, t = 6.078, df = 10, p = 0.0004; day 3, t = 4.065, df = 10, p = 0.0045). D, Duration of immobility during 5 min of PTZ exposure (two-way ANOVA, F(2,30) = 0.3478, p = 0.7091; multiple t tests; day 1, t = 0.4509, df = 10, p = 0.6616; day 2, t = 1.853, df = 10, p = 0.2554; day 3, t = 1.674, df = 10, p = 0.2554). E, Total distance traveled during 5 min of PTZ exposure (two-way ANOVA, F(2,30) = 0.4349, p = 0.6513; multiple t tests; day 1, t = 1.053, df = 10, p = 0.6168; day 2, t = 1.158, df = 10, p = 0.6168; day 3, t = 0.0195, df = 10, p = 0.9848). n = 6 in each group. Data are mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; two-way ANOVA analysis with multiple comparisons.
Gene expression in PTZ-treated cdnf mutant and WT fish brains
To investigate the molecular alterations in the zebrafish brain caused by PTZ treatment, qRT-PCR was used to quantify the gene expression of manf (a closely related growth factor), glial fibrillary acidic protein (gfap; an astrocyte marker), and GABAergic and glutamatergic markers. We first confirmed that 32% (Fig. 13A) of the remaining cdnf truncated transcript was detected in untreated cdnf mutant fish brains compared with untreated adult cdnf WT fish brains, which agrees with the qRT-PCR results on larvae (34%; Fig. 3A). The manf expression level did not alter significantly in untreated cdnf KO fish brains (Fig. 13B). A significant downregulation of slc17a6a/vGlut2, slc32a1/vGAT, and gad2a/gad65 mRNA expression was observed in untreated adult cdnf KO fish brains (Fig. 13C–E), but not the expression level of gad1b/gad67 (Fig. 13F). PTZ administration did not significantly alter the mRNA expression of cdnf, manf, GABAergic, and glutamatergic markers in either WT or cdnf mutant fish (Fig. 13A–F), although there was a tendency toward a higher level of mRNA expression of manf and GABAergic markers in PTZ-treated cdnf KO brains than in the untreated cdnf KO fish group (Fig. 13B–F). Remarkably, mRNA expression of gfap was significantly lower in untreated cdnf mutant fish brains than in WT fish brains (Fig. 13G). After PTZ treatment, a significant increase in gfap expression was seen in cdnf mutant fish brains, but not in their WT siblings (Fig. 13G). The immunostaining result of zrf-1-labeled gfap showed that cdnf mutant fish revealed significantly less extensive radial glial fibers in the lateral region of hindbrain than their WT siblings (Fig. 13H,I).
Figure 13.
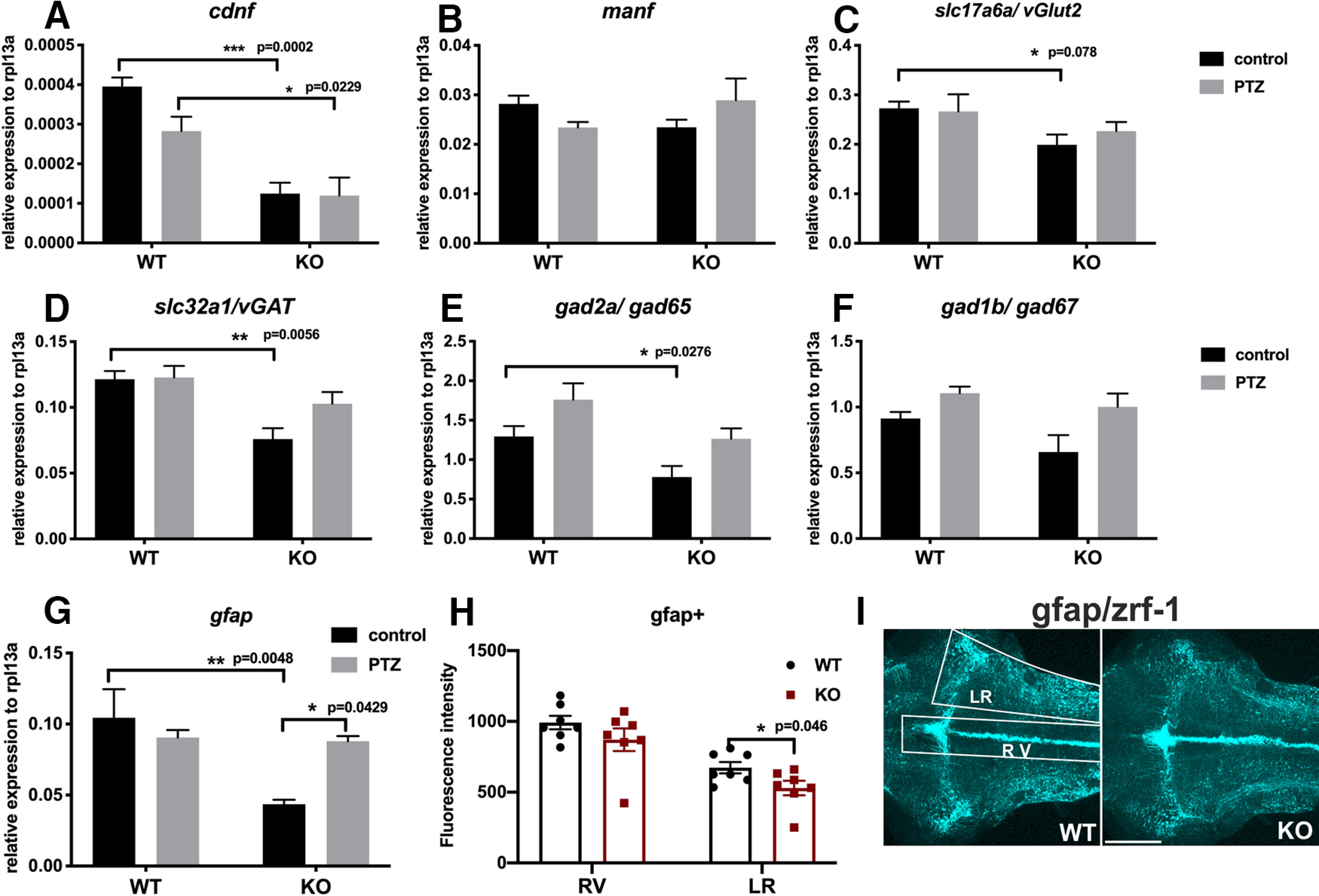
Results of qRT-PCR analysis in PTZ-treated cdnf KO fish. Quantification of relative expression of (A) cdnf (two-way ANOVA, F(1,16) = 40.14, p < 0.0001), (B) manf (two-way ANOVA, F(1,16) = 0.02213, p = 0.8836), (C) slc17a6a/vGlut2 (two-way ANOVA, F(1,16) = 16.16, p = 0.0010), (D) slc32a1/vGAT (two-way ANOVA, F(1,16) = 40.14, p < 0.0001), (E) gad2a/gad65 (two-way ANOVA, F(1,16) = 10.68, p = 0.0048), (F) gad1b/gad67 (two-way ANOVA, F(1,16) = 4.052, p = 0.0613), and (G) gfap (two-way ANOVA, F(1,16) = 8.813, p = 0.0091) in 6 mpf brains with or without PTZ treatment. n = 5 in each group. H, Quantification of fluorescence intensity of zrf-1 immunostaining signals of 10 dpf cdnf WT and KO fish brains (two-way ANOVA, F(1,24) = 33.8, p < 0.0001). I, Quantification of fluorescence intensity of gfap-positive radial glial cells (shown in H) was done in the ROIs shown in white boxes. RV, Rhombencephalic ventricle; LR, lateral region of raphe. Data are mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; two-way ANOVA analysis with Tukey's multiple comparisons test. Scale bar, 100 µm.
Discussion
Despite the apparently typical general development and superficially normal behavior, zebrafish lacking CDNF displayed hyperactivity and impairments in anxiety-related behavior, social preference, and shoal cohesion. Reduction of proliferating cells, neural progenitor markers, and radial glial cells in cdnf mutant larvae brain may cause abnormal neurogenesis, consequently leading to brain dysfunction at later stages. Decreased sociability and increased seizure susceptibility were associated with deficiencies in several neurotransmitter systems, including dopaminergic, GABAergic, and histaminergic neurons. Notably, there was no overall difference in whole-brain dopamine levels, but a detailed examination of the two major dopaminergic systems showed significant abnormalities in cdnf KO fish. A recent study on cdnf−/− mice reports normal levels of brain dopamine and number of nigral dopamine neurons (Lindahl et al., 2020). Our findings lend support to the hypothesis that CDNF acts as a general modulator that regulates neurogenesis and maturation of transmitter-specific neuronal types during development and throughout adulthood, rather than a regulator of only dopaminergic systems. This concept is supported by the expression pattern of cdnf mRNA during development and in the mature brain: it is detected in the anteroposterior axis of the ventricular zones where neurogenesis actively happens from embryonal to adult zebrafish brain (Zupanc et al., 2005; Kizil and Brand, 2011; Schmidt et al., 2013).
There is evidence that the UPR, which is essential in the mechanisms of MANF and CDNF, is associated with the generation and maturation of CNS neurons and circuits. MANF/Armet is upregulated in various forms of ER stress (Apostolou et al., 2008). ATF6 is a transcription factor activated as one component of the ER stress cascade, and its conditional activation induces MANF/Armet in cardiomyocytes (Tadimalla et al., 2008). In addition to ATF6 activation, the UPR cascade consists of activation of inositol-requiring enzyme 6 and protein kinase-like ER kinase and is essential in nervous system development. Additionally, it supports the generation, maturation, and maintenance of CNS neurons (Godin et al., 2016; Hetz and Saxena, 2017). For example, lack of functional BiP/GRP78, an essential component of the UPR, disturbs development of thalamocortical connections in mice (Favero et al., 2013). Moreover, downregulation of the UPR alters generation of progenitor cells and cell fate acquisition in the developing cerebral cortex (Laguesse et al., 2015). In agreement with the structural similarities of MANF and CDNF (Lindahl et al., 2017), CDNF is also activated in the UPR. Expression of CDNF in HEK293-T cells and hippocampal neurons activates the UPR during thapsigargin-induced ER stress in both cell types, and attenuates expression of ER stress activated by the apoptotic proteins CHOP and cleaved caspase 3 (Arancibia et al., 2018). There is clear evidence that exogenously administered CDNF is also neuroprotective (Lindholm et al., 2007; Voutilainen et al., 2015). Indeed, intrastriatally infused CDNF is taken up by neurons and transported retrogradely to, for example, the cortex and substantia nigra in rats, and localized in endosomes rather than the ER (Matlik et al., 2017). Although a cell surface receptor-mediated mechanism for secreted CDNF could be expected, no such mechanism has yet been identified. Moreover, all observed abnormalities are not necessarily direct consequences of a lack of CDNF, but may be secondary to the primary effects of the KO (e.g., alterations in the GABAergic or dopaminergic systems), which are known to regulate neurogenesis and differentiation (Saito et al., 2010; Y. S. Kim and Yoon, 2017).
Using the cdnf null mutant fish generated in this study, we first provide evidence that CDNF plays an important role in the regulation of developing neurotransmitter circuits, including dopaminergic, GABAergic, and histaminergic systems. Moreover, the increased seizure susceptibility revealed by PTZ administration in adult cdnf KO fish may be associated with the deficiency of the GABAergic and glutamatergic systems in cdnf KO fish. As a consequence of this lack of functional cdnf, the dysregulated homeostasis of the neurotransmitter connectivity leads to the impairment of social behaviors.
Loss of cdnf causes a dynamic alteration of dopaminergic systems in the brain
Because of the genome duplication in teleost fish (Postlethwait et al., 2004), two TH genes (Candy and Collet, 2005) complimentarily expressed in the brain (Chen et al., 2009; Filippi et al., 2010; Yamamoto et al., 2010) are found in the zebrafish. In our cdnf KO mutant, the number of TH1 neurons was reduced in the prethalamus area, whereas an increased number of TH2-containing cells (teleost specific paralogous th) appeared in the hypothalamic region. The zebrafish TH1 cell population in the prethalamus is homologous to the mammalian DA population A13 in the zona incerta of the thalamus (Tay et al., 2011), which is more susceptible to neurotoxic MPTP and MPP+ injury in the zebrafish brain (Sallinen et al., 2009). The hypothalamic TH1 groups correspond to A12 and A14 DA groups in the arcuate and periventricular nucleus of the hypothalamus (Tay et al., 2011), respectively. In the L1CAM (neural cell adhesion molecule L1) null mice, abnormal distribution of dopaminergic neurons was evident in A12, A13, and A14 DA groups, but not the A9 group, in the substantia nigra (Demyanenko et al., 2001). It has remained unclear whether cdnf binds to potential signaling receptors to trigger downstream signal pathways. Alternatively, it is possible that, in the zebrafish brain, cdnf serves as a survival-promoting factor affecting the expression of essential regulators that could regulate the number of TH1 dopaminergic neurons but negatively control the TH2-containing cell numbers in a regional-specific fashion. Finally, we cannot exclude the possibility that loss of cdnf per se may induce unpredicted ER stress, oxidative stress, or chronic inflammation, which could result in neurodegeneration in other more vulnerable dopaminergic populations (Sprenkle et al., 2017).
Impaired GABAergic and histaminergic system
It is evident that most of the dopaminergic neurons contain GABA as a cotransmitter in the preoptic area, prethalamus, and hypothalamus regions (Filippi et al., 2014). Histamine neurons also contain GABA in all vertebrates studied thus far (Airaksinen et al., 1992), including zebrafish (Sundvik and Panula, 2012). Moreover, we have reported that dopaminergic signaling plays a crucial role in the specification of hypothalamic neurotransmitter identity (Chen et al., 2016). The number of histaminergic neurons is determined at least in part by dopamine produced by th2-expressing dopaminergic neurons in zebrafish (Chen et al., 2016). The number of histaminergic neurons shows life-long plasticity in adult zebrafish through a Notch1 pathway regulated by presenilin 1 in the γ-secretase complex (Sundvik et al., 2013). Dopamine activation also has a direct impact on GABAergic neuron development in zebrafish larvae (Souza et al., 2011). Consequently, a decreased number of GABA-containing cells and histaminergic neurons found in the hypothalamic area may also be caused by the dynamic alteration of dopaminergic systems, specifically the increased expression of th2 in the hypothalamus, caused by a lack of cdnf. Nevertheless, the wide distribution of abnormal cell populations in cdnf-deficient zebrafish is more likely to derive from a direct effect on early neuronal proliferation, maturation, and transmitter specification.
Neurotransmitter systems associated with abnormal social behaviors
The neurotransmitter phenotype in the developing and mature nervous system is regulated by genetic and environmental cues, to compensate for the changing homeostatic requirements and to maintain the appropriate neuronal circuits during development in nervous system function (Dulcis et al., 2013, 2017). Proper social responses require coordinated neurotransmitter circuits in the CNS. Dysfunction of any main transmitter system is known to cause mental disorders and neurodegenerative diseases (Ng et al., 2015). The behavioral phenotype of the cdnf KO zebrafish is reminiscent of many neurologic and psychiatric conditions, such as attention deficit disorder, autism spectrum disorder, schizophrenia, or epilepsy. The observed sensitivity to PTZ-induced seizures may depend on an abnormal GABAergic system and low expression of vGAT, since PTZ acts via the GABA-A receptors (Saito et al., 2010; Y. S. Kim and Yoon, 2017). The abnormal histaminergic system may be responsible for the low level of anxiety-related behavior observed in our novel tank test, since reducing histamine levels in the adult zebrafish brain by prohibiting the hdc inhibitor α-fluoromethylhistidine has the same effect (Peitsaro et al., 2003). The decrease in hdc expression and number of histamine-containing posterior hypothalamic neurons found in this study may be a direct result of lack of CDNF or a secondary effect of the increased production of dopamine by the th2-expressing neurons in the same cluster of cells (Chen et al., 2016).
In conclusion, this study highlights the novel and broad role of CDNF in shaping the neurotransmitter circuits in the zebrafish brain, and provides evidence that cdnf has an impact on regulation of neural progenitors and maintenance of neurotransmitter properties. Although the cdnf KO fish are superficially normal, the altered transmitter networks produce a range of abnormal behaviors that resemble some human neuropsychiatric conditions, including schizophrenia. Indeed, one study has already shown an association between one SNP/haplotype in the human cdnf gene and schizophrenia characterized by negative symptoms in the Han Chinese population (Yang et al., 2018). Interestingly, ER quality control of protein processing is known to be associated with schizophrenia (P. Kim et al., 2019).
Footnotes
The authors declare no competing financial interests.
This work was supported by the Jane and Aatos Erkko Foundation, Sigrid Juselius Foundation, Magnus Ehrnrooth's Foundation, and Finska Läkaresällskapet. Zebrafish experiments were performed at the Zebrafish Unit of HiLife Infrastructures, funded in part by Biocenter Finland. We thank Henri Koivula, Riikka Pesonen, and Noora Hellen for expert technical help; and Dr. Mart Saarma, Dr. Esa Korpi, and Dr. Marnie Halpern for constructive comments on the manuscript.
References
- Airaksinen MS, Alanen S, Szabat E, Visser TJ, Panula P (1992) Multiple neurotransmitters in the tuberomammillary nucleus: comparison of rat, mouse, and guinea pig. J Comp Neurol 323:103–116. 10.1002/cne.903230109 [DOI] [PubMed] [Google Scholar]
- Airavaara M, Harvey BK, Voutilainen MH, Shen H, Chou J, Lindholm P, Lindahl M, Tuominen RK, Saarma M, Hoffer B, Wang Y (2012) CDNF protects the nigrostriatal dopamine system and promotes recovery after MPTP treatment in mice. Cell Transplant 21:1213–1223. 10.3727/096368911X600948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou A, Shen Y, Liang Y, Luo J, Fang S (2008) Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res 314:2454–2467. 10.1016/j.yexcr.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arancibia D, Zamorano P, Andres ME (2018) CDNF induces the adaptive unfolded protein response and attenuates endoplasmic reticulum stress-induced cell death. Biochim Biophys Acta Mol Cell Res 1865:1579–1589. 10.1016/j.bbamcr.2018.08.012 [DOI] [PubMed] [Google Scholar]
- Baronio D, Puttonen HA, Sundvik M, Semenova S, Lehtonen E, Panula P (2018) Embryonic exposure to valproic acid affects the histaminergic system and the social behaviour of adult zebrafish (Danio rerio). Br J Pharmacol 175:797–809. 10.1111/bph.14124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D (2006) Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93:491–507. 10.1093/biomet/93.3.491 [DOI] [Google Scholar]
- Cachat J, Stewart A, Grossman L, Gaikwad S, Kadri F, Chung KM, Wu N, Wong K, Roy S, Suciu C, Goodspeed J, Elegante M, Bartels B, Elkhayat S, Tien D, Tan J, Denmark A, Gilder T, Kyzar E, Dileo J, et al. (2010) Measuring behavioral and endocrine responses to novelty stress in adult zebrafish. Nat Protoc 5:1786–1799. 10.1038/nprot.2010.140 [DOI] [PubMed] [Google Scholar]
- Candy J, Collet C (2005) Two tyrosine hydroxylase genes in teleosts. Biochim Biophys Acta 1727:35–44. 10.1016/j.bbaexp.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Chao MV. (2003) Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 4:299–309. 10.1038/nrn1078 [DOI] [PubMed] [Google Scholar]
- Chao MV, Rajagopal R, Lee FS (2006) Neurotrophin signalling in health and disease. Clin Sci 110:167–173. 10.1042/CS20050163 [DOI] [PubMed] [Google Scholar]
- Chen YC, Priyadarshini M, Panula P (2009) Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochem Cell Biol 132:375–381. 10.1007/s00418-009-0619-8 [DOI] [PubMed] [Google Scholar]
- Chen YC, Sundvik M, Rozov S, Priyadarshini M, Panula P (2012) MANF regulates dopaminergic neuron development in larval zebrafish. Dev Biol 370:237–249. 10.1016/j.ydbio.2012.07.030 [DOI] [PubMed] [Google Scholar]
- Chen YC, Semenova S, Rozov S, Sundvik M, Bonkowsky JL, Panula P (2016) A novel developmental role for dopaminergic signaling to specify hypothalamic neurotransmitter identity. J Biol Chem 291:21880–21892. 10.1074/jbc.M115.697466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Shibata Y, Maness PF (2001) Altered distribution of dopaminergic neurons in the brain of L1 null mice. Brain Res Dev Brain Res 126:21–30. 10.1016/s0165-3806(00)00129-2 [DOI] [PubMed] [Google Scholar]
- Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC (2013) Neurotransmitter switching in the adult brain regulates behavior. Science 340:449–453. 10.1126/science.1234152 [DOI] [PubMed] [Google Scholar]
- Dulcis D, Lippi G, Stark CJ, Do LH, Berg DK, Spitzer NC (2017) Neurotransmitter switching regulated by miRNAs controls changes in social preference. Neuron 95:1319–1333.e5. 10.1016/j.neuron.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy PQ, Berberoglu MA, Beattie CE, Hall CW (2017) Cellular responses to recurrent pentylenetetrazole-induced seizures in the adult zebrafish brain. Neuroscience 349:118–127. 10.1016/j.neuroscience.2017.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfarnawany MH. (2015) Signal processing methods for quantitative power Doppler microvascular angiography. In: Biomedical Engineering. Ph.D. Electronic Thesis and Dissertation Repository The University of Western Ontario. [Google Scholar]
- Favero CB, Henshaw RN, Grimsley-Myers CM, Shrestha A, Beier DR, Dwyer ND (2013) Mutation of the BiP/GRP78 gene causes axon outgrowth and fasciculation defects in the thalamocortical connections of the mammalian forebrain. J Comp Neurol 521:677–696. 10.1002/cne.23199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi A, Mahler J, Schweitzer J, Driever W (2010) Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J Comp Neurol 518:423–438. 10.1002/cne.22213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi A, Mueller T, Driever W (2014) vglut2 and gad expression reveal distinct patterns of dual GABAergic versus glutamatergic cotransmitter phenotypes of dopaminergic and noradrenergic neurons in the zebrafish brain. J Comp Neurol 522:2019–2037. 10.1002/cne.23524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney JL, Robertson GN, McGee CA, Smith FM, Croll RP (2006) Structure and autonomic innervation of the swim bladder in the zebrafish (Danio rerio). J Comp Neurol 495:587–606. 10.1002/cne.20948 [DOI] [PubMed] [Google Scholar]
- Godin JD, Creppe C, Laguesse S, Nguyen L (2016) Emerging roles for the unfolded protein response in the developing nervous system. Trends Neurosci 39:394–404. 10.1016/j.tins.2016.04.002 [DOI] [PubMed] [Google Scholar]
- Green J, Collins C, Kyzar EJ, Pham M, Roth A, Gaikwad S, Cachat J, Stewart AM, Landsman S, Grieco F, Tegelenbosch R, Noldus LP, Kalueff AV (2012) Automated high-throughput neurophenotyping of zebrafish social behavior. J Neurosci Methods 210:266–271. 10.1016/j.jneumeth.2012.07.017 [DOI] [PubMed] [Google Scholar]
- Hetz C, Saxena S (2017) ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 13:477–491. 10.1038/nrneurol.2017.99 [DOI] [PubMed] [Google Scholar]
- Huang RQ, Bell-Horner CL, Dibas MI, Covey DF, Drewe JA, Dillon GH (2001) Pentylenetetrazole-induced inhibition of recombinant gamma-aminobutyric acid type A (GABA(A)) receptors: mechanism and site of action. J Pharmacol Exp Ther 298:986–995. [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BE. (2020) Arousal and sleep circuits. Neuropsychopharmacology 45:6–20. 10.1038/s41386-019-0444-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhunen T, Airaksinen MS, Tuomisto L, Panula P (1993) Neurotransmitters in the nervous system of Macoma balthica (Bivalvia). J Comp Neurol 334:477–488. 10.1002/cne.903340311 [DOI] [PubMed] [Google Scholar]
- Kaslin J, Panula P (2001) Comparative anatomy of the histaminergic and other aminergic systems in zebrafish (Danio rerio). J Comp Neurol 440:342–377. 10.1002/cne.1390 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen S, Lindholm P, Galli E, Lahtinen HM, Koivisto H, Hamalainen E, Saarma M, Tanila H (2015) Cerebral dopamine neurotrophic factor improves long-term memory in APP/PS1 transgenic mice modeling Alzheimer's disease as well as in wild-type mice. Behav Brain Res 291:1–11. 10.1016/j.bbr.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Kim P, Scott MR, Meador-Woodruff JH (2019) Abnormal ER quality control of neural GPI-anchored proteins via dysfunction in ER export processing in the frontal cortex of elderly subjects with schizophrenia. Transl Psychiatry 9:6. 10.1038/s41398-018-0359-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Yoon BE (2017) Altered GABAergic signaling in brain disease at various stages of life. Exp Neurobiol 26:122–131. 10.5607/en.2017.26.3.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kizil C, Brand M (2011) Cerebroventricular microinjection (CVMI) into adult zebrafish brain is an efficient misexpression method for forebrain ventricular cells. PLoS One 6:e27395. 10.1371/journal.pone.0027395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukko-Lukjanov TK, Panula P (2003) Subcellular distribution of histamine, GABA and galanin in tuberomamillary neurons in vitro. J Chem Neuroanat 25:279–292. 10.1016/s0891-0618(03)00043-7 [DOI] [PubMed] [Google Scholar]
- Kysil EV, Meshalkina DA, Frick EE, Echevarria DJ, Rosemberg DB, Maximino C, Lima MG, Abreu MS, Giacomini AC, Barcellos LJ, Song C, Kalueff AV (2017) Comparative analyses of zebrafish anxiety-like behavior using conflict-based novelty tests. Zebrafish 14:197–208. 10.1089/zeb.2016.1415 [DOI] [PubMed] [Google Scholar]
- Laguesse S, Creppe C, Nedialkova DD, Prevot PP, Borgs L, Huysseune S, Franco B, Duysens G, Krusy N, Lee G, Thelen N, Thiry M, Close P, Chariot A, Malgrange B, Leidel SA, Godin JD, Nguyen L (2015) A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev Cell 35:553–567. 10.1016/j.devcel.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Laruelle M. (2014) Schizophrenia: from dopaminergic to glutamatergic interventions. Curr Opin Pharmacol 14:97–102. 10.1016/j.coph.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Latge C, Cabral KM, Almeida MS, Foguel D (2013) (1)H-, (13)C- and (15)N-NMR assignment of the N-terminal domain of human cerebral dopamine neurotrophic factor (CDNF). Biomol NMR Assign 7:101–103. 10.1007/s12104-012-9388-8 [DOI] [PubMed] [Google Scholar]
- Latge C, Cabral KM, de Oliveira GA, Raymundo DP, Freitas JA, Johanson L, Romao LF, Palhano FL, Herrmann T, Almeida MS, Foguel D (2015) The solution structure and dynamics of full-length human cerebral dopamine neurotrophic factor and its neuroprotective role against alpha-synuclein oligomers. J Biol Chem 290:20527–20540. 10.1074/jbc.M115.662254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Saarma M, Lindholm P (2017) Unconventional neurotrophic factors CDNF and MANF: structure, physiological functions and therapeutic potential. Neurobiol Dis 97:90–102. 10.1016/j.nbd.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Chalazonitis A, Palm E, Pakarinen E, Danilova T, Pham TD, Setlik W, Rao M, Voikar V, Huotari J, Kopra J, Andressoo JO, Piepponen PT, Airavaara M, Panhelainen A, Gershon MD, Saarma M (2020) Cerebral dopamine neurotrophic factor-deficiency leads to degeneration of enteric neurons and altered brain dopamine neuronal function in mice. Neurobiol Dis 134:104696. 10.1016/j.nbd.2019.104696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Makela J, Di Liberto V, Mudo G, Belluardo N, Eriksson O, Saarma M (2016) Current disease modifying approaches to treat Parkinson's disease. Cell Mol Life Sci 73:1365–1379. 10.1007/s00018-015-2101-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm P, Voutilainen MH, Lauren J, Peranen J, Leppanen VM, Andressoo JO, Lindahl M, Janhunen S, Kalkkinen N, Timmusk T, Tuominen RK, Saarma M (2007) Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nature 448:73–77. 10.1038/nature05957 [DOI] [PubMed] [Google Scholar]
- Lindsey BW, Smith FM, Croll RP (2010) From inflation to flotation: contribution of the swim bladder to whole-body density and swimming depth during development of the zebrafish (Danio rerio). Zebrafish 7:85–96. 10.1089/zeb.2009.0616 [DOI] [PubMed] [Google Scholar]
- Liu H, Tang X, Gong L (2015) Mesencephalic astrocyte-derived neurotrophic factor and cerebral dopamine neurotrophic factor: new endoplasmic reticulum stress response proteins. Eur J Pharmacol 750:118–122. 10.1016/j.ejphar.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Matlik K, Vihinen H, Bienemann A, Palgi J, Voutilainen MH, Booms S, Lindahl M, Jokitalo E, Saarma M, Huttunen HJ, Airavaara M, Arumae U (2017) Intrastriatally infused exogenous CDNF is endocytosed and retrogradely transported to substantia nigra. eNeuro 4:ENEURO.0128-16.2017 10.1523/ENEURO.0128-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NY, Gerlai R (2011) Shoaling in zebrafish: what we don't know. Rev Neurosci 22:17–25. [DOI] [PubMed] [Google Scholar]
- Mitre M, Mariga A, Chao MV (2017) Neurotrophin signalling: novel insights into mechanisms and pathophysiology. Clin Sci 131:13–23. 10.1042/CS20160044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussulini BH, Leite CE, Zenki KC, Moro L, Baggio S, Rico EP, Rosemberg DB, Dias RD, Souza TM, Calcagnotto ME, Campos MM, Battastini AM, de Oliveira DL (2013) Seizures induced by pentylenetetrazole in the adult zebrafish: a detailed behavioral characterization. PLoS One 8:e54515. 10.1371/journal.pone.0054515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrolahi A, Mahmoudi J, Akbarzadeh A, Karimipour M, Sadigh-Eteghad S, Salehi R, Farhoudi M (2018) Neurotrophic factors hold promise for the future of Parkinson's disease treatment: is there a light at the end of the tunnel? Rev Neurosci 29:475–489. 10.1515/revneuro-2017-0040 [DOI] [PubMed] [Google Scholar]
- Ng J, Papandreou A, Heales SJ, Kurian MA (2015) Monoamine neurotransmitter disorders: clinical advances and future perspectives. Nat Rev Neurol 11:567–584. 10.1038/nrneurol.2015.172 [DOI] [PubMed] [Google Scholar]
- Panula P, Airaksinen MS, Pirvola U, Kotilainen E (1990) A histamine-containing neuronal system in human brain. Neuroscience 34:127–132. 10.1016/0306-4522(90)90307-p [DOI] [PubMed] [Google Scholar]
- Parkash V, Lindholm P, Peranen J, Kalkkinen N, Oksanen E, Saarma M, Leppanen VM, Goldman A (2009) The structure of the conserved neurotrophic factors MANF and CDNF explains why they are bifunctional. Protein Eng Des Sel 22:233–241. 10.1093/protein/gzn080 [DOI] [PubMed] [Google Scholar]
- Peitsaro N, Kaslin J, Anichtchik OV, Panula P (2003) Modulation of the histaminergic system and behaviour by alpha-fluoromethylhistidine in zebrafish. J Neurochem 86:432–441. 10.1046/j.1471-4159.2003.01850.x [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL (2004) Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet 20:481–490. 10.1016/j.tig.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Provensi G, Costa A, Izquierdo I, Blandina P, Passani MB (2020) Brain histamine modulates recognition memory: possible implications in major cognitive disorders. Br J Pharmacol 177:539–556. 10.1111/bph.14478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttonen HA, Sundvik M, Semenova S, Shirai Y, Chen YC, Panula P (2018) Knockout of histamine receptor H3 alters adaptation to sudden darkness and monoamine levels in the zebrafish. Acta Physiol 222:e12981 10.1111/apha.12981 [DOI] [PubMed] [Google Scholar]
- Ren X, Zhang T, Gong X, Hu G, Ding W, Wang X (2013) AAV2-mediated striatum delivery of human CDNF prevents the deterioration of midbrain dopamine neurons in a 6-hydroxydopamine induced parkinsonian rat model. Exp Neurol 248:148–156. 10.1016/j.expneurol.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Saito K, Kakizaki T, Hayashi R, Nishimaru H, Furukawa T, Nakazato Y, Takamori S, Ebihara S, Uematsu M, Mishina M, Miyazaki J, Yokoyama M, Konishi S, Inoue K, Fukuda A, Fukumoto M, Nakamura K, Obata K, Yanagawa Y (2010) The physiological roles of vesicular GABA transporter during embryonic development: a study using knockout mice. Mol Brain 3:40. 10.1186/1756-6606-3-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallinen V, Torkko V, Sundvik M, Reenila I, Khrustalyov D, Kaslin J, Panula P (2009) MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J Neurochem 108:719–731. 10.1111/j.1471-4159.2008.05793.x [DOI] [PubMed] [Google Scholar]
- Schmidt R, Strahle U, Scholpp S (2013) Neurogenesis in zebrafish: from embryo to adult. Neural Dev 8:3. 10.1186/1749-8104-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova SA, Chen YC, Zhao X, Rauvala H, Panula P (2014) The tyrosine hydroxylase 2 (TH2) system in zebrafish brain and stress activation of hypothalamic cells. Histochem Cell Biol 142:619–633. 10.1007/s00418-014-1240-z [DOI] [PubMed] [Google Scholar]
- Sousa-Victor P, Jasper H, Neves J (2018) Trophic factors in inflammation and regeneration: the role of MANF and CDNF. Front Physiol 9:1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BR, Romano-Silva MA, Tropepe V (2011) Dopamine D2 receptor activity modulates Akt signaling and alters GABAergic neuron development and motor behavior in zebrafish larvae. J Neurosci 31:5512–5525. 10.1523/JNEUROSCI.5548-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenkle NT, Sims SG, Sanchez CL, Meares GP (2017) Endoplasmic reticulum stress and inflammation in the central nervous system. Mol Neurodegener 12:42. 10.1186/s13024-017-0183-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvik M, Panula P (2012) Organization of the histaminergic system in adult zebrafish (Danio rerio) brain: neuron number, location, and cotransmitters. J Comp Neurol 520:3827–3845. 10.1002/cne.23126 [DOI] [PubMed] [Google Scholar]
- Sundvik M, Chen YC, Panula P (2013) Presenilin1 regulates histamine neuron development and behavior in zebrafish, Danio rerio. J Neurosci 33:1589–1597. 10.1523/JNEUROSCI.1802-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvik M, Kudo H, Toivonen P, Rozov S, Chen YC, Panula P (2011) The histaminergic system regulates wakefulness and orexin/hypocretin neuron development via histamine receptor H1 in zebrafish. FASEB J 25:4338–4347. 10.1096/fj.11-188268 [DOI] [PubMed] [Google Scholar]
- Tadimalla A, Belmont PJ, Thuerauf DJ, Glassy MS, Martindale JJ, Gude N, Sussman MA, Glembotski CC (2008) Mesencephalic astrocyte-derived neurotrophic factor is an ischemia-inducible secreted endoplasmic reticulum stress response protein in the heart. Circ Res 103:1249–1258. 10.1161/CIRCRESAHA.108.180679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay TL, Ronneberger O, Ryu S, Nitschke R, Driever W (2011) Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nat Commun 2:171. 10.1038/ncomms1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Voutilainen MH, Back S, Peranen J, Lindholm P, Raasmaja A, Mannisto PT, Saarma M, Tuominen RK (2011) Chronic infusion of CDNF prevents 6-OHDA-induced deficits in a rat model of Parkinson's disease. Exp Neurol 228:99–108. 10.1016/j.expneurol.2010.12.013 [DOI] [PubMed] [Google Scholar]
- Voutilainen MH, Arumae U, Airavaara M, Saarma M (2015) Therapeutic potential of the endoplasmic reticulum located and secreted CDNF/MANF family of neurotrophic factors in Parkinson's disease. FEBS Lett 589:3739–3748. 10.1016/j.febslet.2015.09.031 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ruuskanen JO, Wullimann MF, Vernier P (2010) Two tyrosine hydroxylase genes in vertebrates: new dopaminergic territories revealed in the zebrafish brain. Mol Cell Neurosci 43:394–402. 10.1016/j.mcn.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Yan Y, Rato C, Rohland L, Preissler S, Ron D (2019) MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP. Nat Commun 10:541. 10.1038/s41467-019-08450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu H, Li W, Liu B, Zhang H, Ding S, Lu Y, Jiang T, Lv L (2018) Association between cerebral dopamine neurotrophic factor (CDNF) 2 polymorphisms and schizophrenia susceptibility and symptoms in the Han Chinese population. Behav Brain Funct 14:1. 10.1186/s12993-017-0133-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Xiao C, Commissiong JW, Krnjevic K, Ye JH (2006) Mesencephalic astrocyte-derived neurotrophic factor enhances nigral gamma-aminobutyric acid release. Neuroreport 17:293–297. 10.1097/01.wnr.0000201504.23255.bc [DOI] [PubMed] [Google Scholar]
- Zhou W, Chang L, Fang Y, Du Z, Li Y, Song Y, Hao F, Lv L, Wu Y (2016) Cerebral dopamine neurotrophic factor alleviates Abeta25-35-induced endoplasmic reticulum stress and early synaptotoxicity in rat hippocampal cells. Neurosci Lett 633:40–46. 10.1016/j.neulet.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Zupanc GK, Hinsch K, Gage FH (2005) Proliferation, migration, neuronal differentiation, and long-term survival of new cells in the adult zebrafish brain. J Comp Neurol 488:290–319. 10.1002/cne.20571 [DOI] [PubMed] [Google Scholar]