Abstract
Lupus nephritis, one of the most serious manifestations of systemic lupus erythematosus (SLE), has a heterogeneous clinical and pathological presentation. For example, proliferative nephritis identifies a more aggressive disease class that requires immunosuppression. However, the current classification system relies on the static appearance of histopathological morphology, which does not capture differences in the inflammatory response. Therefore, a biomarker grounded in the disease biology is needed in order to understand the molecular heterogeneity of lupus nephritis and identify immunologic mechanism and pathways. Here, we analyzed the patterns of 1000 urine protein biomarkers in 30 patients with active lupus nephritis. We found that patients stratify over a chemokine gradient inducible by IFN-γ. Higher values identified patients with proliferative lupus nephritis. After integrating the urine proteomics with the single-cell transcriptomics of kidney biopsies, we observed that the urinary chemokines defining the gradient were predominantly produced by infiltrating CD8+ T cells, along with natural killer and myeloid cells. The urine chemokine gradient significantly correlated with the number of kidney-infiltrating CD8+ cells. These findings suggest that urine proteomics can capture the complex biology of the kidney in lupus nephritis. Patient-specific pathways could be noninvasively tracked in the urine in real time, enabling diagnosis and personalized treatment.
Keywords: Autoimmunity, Nephrology
Keywords: Lupus, Proteomics, Th1 response
Urine proteomics identify the IFN-γ response as the major source of variability among lupus nephritis patients that associates with intrarenal CD8+ T cell infiltration.
Introduction
Lupus nephritis is one of the most severe manifestations of systemic lupus erythematosus (SLE) (1). Lupus nephritis affects up to 50% of patients, leading to end-stage renal disease in 10%, and carries an 8-fold increase in mortality compared with the general population (2–8). Since nephritis is usually asymptomatic, patients with SLE are serially screened for the presence of abnormal urine protein (9). When proteinuria is elevated, a renal biopsy is obtained to confirm the diagnosis and guide treatment. Histopathology classifications subgroup lupus nephritis into 6 classes based on the presence, amount, and location of inflammation and fibrosis (10). For example, the presence of intraglomerular immune cell infiltration (classes III and IV) identifies the most aggressive proliferative form of SLE nephritis, which is treated with stronger immunosuppression. Membranous glomerulonephritis (class V) is characterized by thickened glomerular basal membranes, subepithelial immune complex deposition, and absence of intraglomerular infiltration. Endocapillary proliferation and membranous disease frequently overlap in the “mixed” phenotype.
Although the current treatment approach to lupus nephritis is loosely grounded in the morphological classification, findings from recent studies have challenged this paradigm. Kidney biopsies repeated after 1 year showed a transition in class in up to 70% of cases, from nonproliferative to proliferative and vice versa, with the second biopsy having better prognostic value (11, 12). These findings indicate that lupus nephritis is a dynamic process, and therefore disease state inferred from kidney biopsies at one time point may have a limited value. In addition, recent studies have underscored the prognostic importance of interstitial disease, which is inadequately addressed by current histologic classification schemes (13). Finally, routine histopathology of lupus nephritis cannot evaluate the underlying molecular pathways that may inform personalized treatment choices. Recent single-cell RNA-Seq studies have revealed the complex network of immune cells, and their diversity, in the context of lupus nephritis (14–16). These studies offer promise for more personalized strategies for therapeutics and for prevention of renal fibrosis.
As patients with the same histological class may have dramatically different outcomes (17), there is an unmet need for a new biology-based classification of lupus nephritis that can dissect the heterogeneity. Ideally, such immunologic classification needs to be easily queried during the course of the disease to dynamically assess changes in response to intervention over months (rather than year). Several previous studies explored potential biomarkers to assess and predict lupus nephritis by using targeted and systematic proteomics approaches (18–21). However, these have not yet been standardized for clinical practice and do not consider lupus nephritis biological endotypes (18). In this study, we sought to identify patterns in the urine proteome that can identify distinct groups of SLE patients and infer the ongoing renal molecular pathology. We quantified 1000 urine protein biomarkers in active lupus nephritis and identified that patients stratify on an immune activation gradient. Integration with renal single-cell RNA-Seq revealed that the urine signature reflected intrarenal secretion by immune infiltrating cells and that IFN-γ is a main driver of such increased immune activation.
Results
Urine proteomics stratifies lupus nephritis patients on an immune activation gradient.
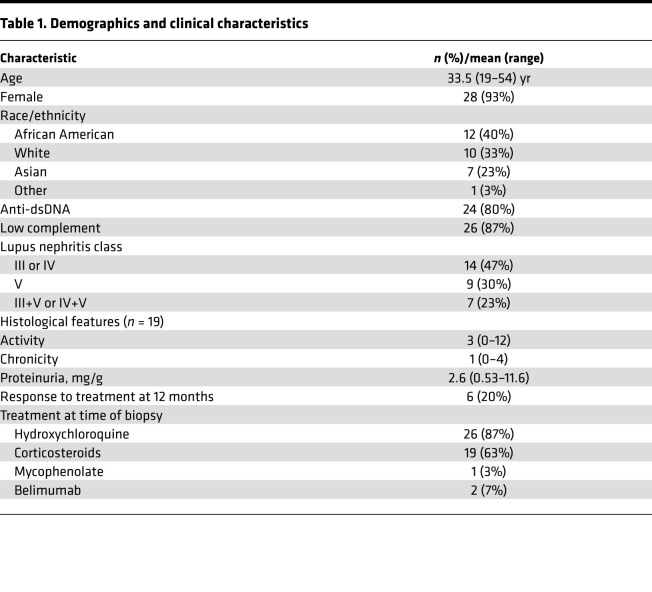
This study was initiated to address whether we could classify lupus nephritis patients based on distinct biological patterns in the urine. We quantified 1000 analytes, including cytokines, growth factors, and other soluble markers, in urine samples from 30 SLE patients with proteinuria and kidney biopsy samples taken on the same day confirming lupus nephritis (class III, IV, or V), as part of phase 1 of the Accelerating Medicines Partnership (AMP; Table 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.138345DS1) (14). To determine patterns of inflammation in an unbiased manner, we used principal component analysis (PCA), a dimension reduction technique that identifies major axes of variation in the urine proteome. For example, the first 2 principal components (PCs) clearly separated healthy donors from SLE patients based on distinct patterns of urinary protein excretion (Supplemental Figure 2). Within lupus nephritis, the first PC (PC1) stratified patients on a gradient rather than distinct clusters (Figure 1A). We noted that patients with higher PC1 values were more likely to have proliferative lupus nephritis, while the patients with pure membranous lupus nephritis had PC1 values that were close to 0 or negative (Figure 1, A and B), suggesting that PC1 detected a biological response shared by many patients with proliferative lupus nephritis. No association was detected between PC1 and demographics or technical confounders such as batch, proteinuria, age, or sex. There was no association with prednisone use or dosage (Supplemental Figure 3). PC2 was associated with site of urine collection (Supplemental Figure 4), and therefore it was not evaluated further.
Table 1. Demographics and clinical characteristics.
Figure 1. Lupus nephritis patients stratify over an immune activation gradient.
(A and B) PCA (n = 30) of the first 2 PCs of the urine proteome (% variance explained is indicated). Patients with higher PC1 value almost exclusively have proliferative lupus nephritis (class III, IV, or mixed). P value calculated by t test. (C) Top and bottom 10 PC1 loading values of the measured urine protein. (D) Top 10 enriched pathways detected by PC1 using Gene Ontology Biological Process, indicating the biological significance of PC1.
Next, we sought to characterize the biological significance of the PC1 gradient. As PCs are determined by the weighted sum of all the analytes, we tested whether a molecular pathway was enriched using the weight (“loading”) of each analyte on PC1 (Figure 1C). Gene set enrichment analysis (GSEA) revealed that PC1 detected chemotaxis pathways (FDR < 0.01) (Figure 1D and Supplemental Figure 5): in particular, a pattern of chemokines secreted in response to IFN-γ, IL-1β, and TNF and directed to attract monocytes, NK cells, and CD8+ T cells.
Together, these findings indicate that urine proteomics analyses identified differential expression of a specific immune activation signature in patients with lupus nephritis and that stronger signals were associated with proliferative lupus nephritis.
Urine cytokines reflect intrarenal production by myeloid and cytotoxic lymphocytes.
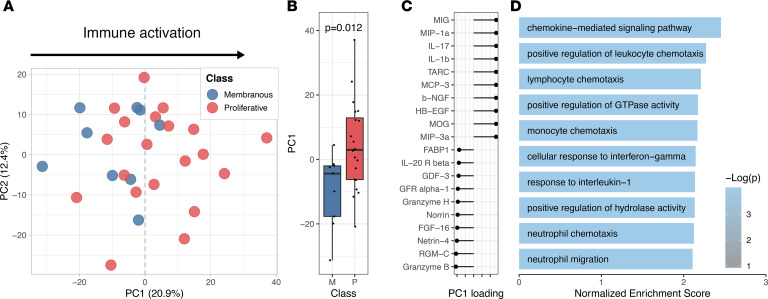
We asked whether the chemokines detected in the urine were indicative of intrarenal production by kidney-infiltrating immune cells. To answer this question, we analyzed single-cell transcriptomics from 24 renal biopsies of patients with lupus nephritis (Figure 2, A and B) (14). We carried out single-cell transcriptomic analysis on genes coding for the proteins found in 2 of the most enriched pathways in the urine PC1 (“chemokine-mediated signaling pathway,” GO:0070098; and “cellular response to interferon-gamma,” GO:0071346) (Supplemental Figure 6) in the renal cells. A chemokine score was defined as the sum of the normalized expression of the aforementioned genes. While these chemokines were expressed by most infiltrating immune cells as well as epithelial cells, dominant expression was observed in myeloid, NK cells, and CD8+ T cells (Figure 2, C–F).
Figure 2. The chemokine pathway detected by urine proteomics reflects intrarenal production by myeloid, CD8+, and NK cells.
(A) Uniform manifold approximation and projection (UMAP) plot of single-cell RNA-Seq of 24 renal biopsies of patients with active lupus nephritis (medium resolution clustering). (B) The identity of the cell clusters was determined by the expression of lineage markers. (C–F) A chemokine and an IFN-γ response score based on the expression of the markers in the top urine PC1 pathways (“chemokine-mediated signaling pathway,” GO:0070098; and “cellular response to interferon-gamma,” GO:0071346)) identified kidney-infiltrating myeloid, CD8+, and NK cells as the main producers of the chemokines in response to IFN-γ detected in the urine. (G) Distribution of IFN-γ–, TNF-, and IL-1β–expressing cells. (H) Distribution of the total unique molecular identifiers (UMIs) mapped to IFN-γ, TNF, and IL-1β as a proxy for the amount of cytokine produced by each cell type.
To evaluate whether the proteins found in the urine derived from serum as a consequence of glomerular or tubular damage, we repeated the analysis after adjustments for albumin or β2-microglobulin, respectively. Results were virtually identical to the unadjusted analysis (Supplemental Figure 7).
These findings suggest that the chemokines identified by urine proteomics derive from intrarenal chemokine production particularly by myeloid, NK, and CD8+ T cells.
IFN-γ is mostly produced by infiltrating CD8+ and NK cells in all patients with lupus nephritis.
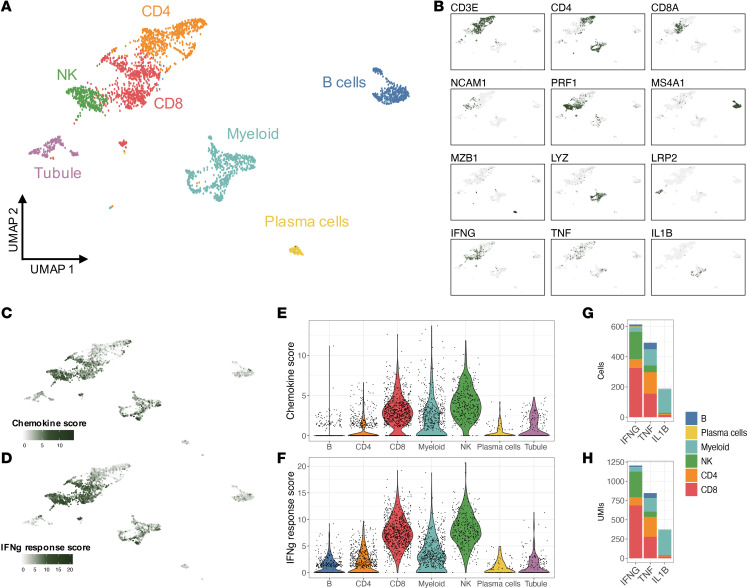
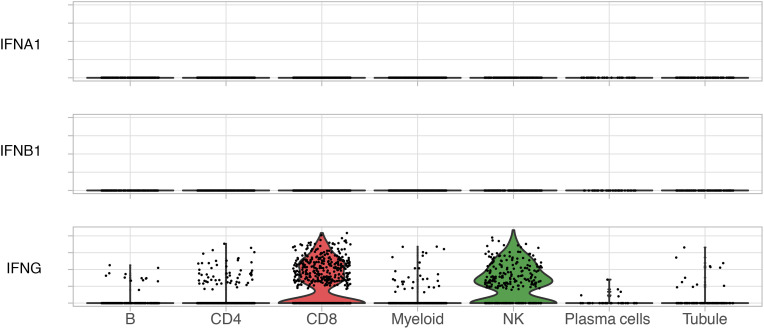
Pathway analysis indicated a chemokine pattern inducible by IFN-γ, IL-1β, or TNF. We quantified kidney-infiltrating cells expressing such cytokines. Of the 3 cytokines, IFN-γ–positive cells were the most abundant, followed by TNF and IL-1β (Figure 2G). IFN-γ was mostly produced by CD8+ T and NK cells; TNF by myeloid, CD8+, and CD4+ T cells; and IL-1β almost exclusively by myeloid cells (Figure 2, B and H). Local transcription of these cytokines suggests that their transcriptional signatures in the kidney and protein levels in the urine are a direct consequence of intrarenal production. Since patients contributed a different number of cells to the data set, we evaluated whether these findings were biased by a small number of patients contributing more cells. IFN-γ+ cells were identified in all patients, ranging between 4% and 45% of the total immune cells (mean 18%); TNF+ cells in 4%–32% (mean 16%); and IL-1β+ in 0%–13% (mean 7%) (Table 2 and Supplemental Figure 8). In contrast, we did not detect any transcription of type 1 IFNs (Figure 3) or type 2 immunity (Th2) cytokines such as IL-4, IL-5, or IL-13 (Supplemental Figure 9), suggesting that a type 1 (Th1) immune response (22) is ubiquitous in patients with lupus nephritis including nonproliferative histological classes and that IFN-γ (type 2 IFN) is the main IFN being produced by immune cells in lupus kidney disease.
Table 2. Prevalence cytokine-positive cells based on single-cell RNA-Seq.
Figure 3. Type 2 IFN (IFN-γ), but not type 1 (IFN-α or IFN-β), is expressed in lupus nephritis kidney.
Violin plots of the expression of type 1 (IFNA1 and IFNB1) and type 2 (IFNG) IFNs by single-cell RNA-Seq. No other type 1 IFN (i.e., IFNA2–21, IFNB3, IFNK, etc.) was detected (data not shown).
Urine proteomics may predict the intrarenal cellular infiltrate.
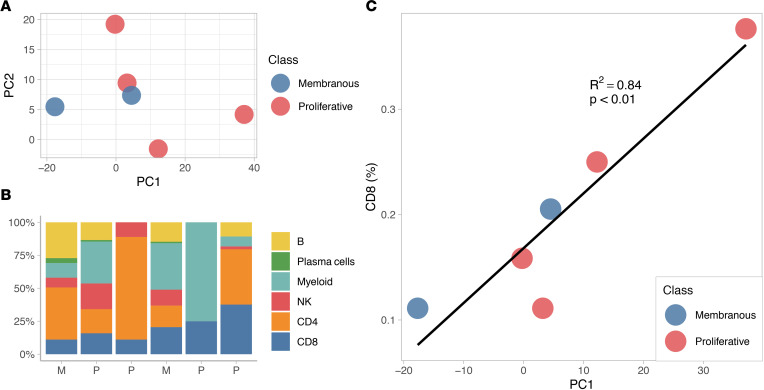
We then hypothesized that urine proteomics could be used to infer the composition of kidney-infiltrating cell types in lupus nephritis. We analyzed 6 patients with matching urine proteomics and single-cell transcriptomics on a renal biopsy sample collected the same day. Of these, 4 patients were diagnosed with proliferative lupus nephritis (including mixed) and 2 with pure membranous lupus nephritis based on histopathology (Figure 4A). Single-cell transcriptomics revealed a heterogenous cellular infiltrate regardless of histopathological class (Figure 4B). We found that urine proteomics PC1 values strongly correlated with the relative abundance of kidney-infiltrating CD8+ T cells (r2 = 0.84, P < 0.01; Figure 4C). There was no statistically significant correlation with the other cell populations. While needing validation in a larger cohort, these findings suggest that urinary protein patterns would predict the cell composition of the renal immune infiltrate in lupus nephritis and that CD8+ T cells, the major producer of IFN-γ, may be the key contributor of the immune activation signature in the urine.
Figure 4. Urine proteomics may predict the intrarenal cellular infiltrate.
Six patients had matching urine proteomic and single-cell RNA-Seq data. Their coordinates on the urine proteomics PCA shown in Figure 1 are plotted in A. (B) Distribution of kidney-infiltrating immune cells determined by single-cell RNA-Seq. Patients are ordered based on their urine proteomics PC1 value as they appear in A. (C) Correlation of the urine PC1 values and the frequency of kidney-infiltrating CD8+ T cells.
Discussion
Urine is an ideal noninvasive specimen for repetitive assessment of lupus nephritis. It contains the by-products of the ongoing renal biological processes in real time, is highly noninvasive, and can be easily monitored over time. Here, we show that urine proteomics in patients with lupus nephritis may detect biologically relevant pathways that are mirrored in the kidney tissue. We found that (i) patients with higher expression of urinary chemokine and immune activation pathways almost exclusively have proliferative lupus nephritis; (ii) these chemokines are renally produced by infiltrating myeloid, NK, and CD8+ T cells; and (iii) urine proteomics may predict the composition of the renal cellular infiltrate.
The current classification of lupus nephritis relies on morphological features that may not fully capture the complex, dynamic, and heterogeneous biology. We showed that systematic, unbiased analysis of the urine proteome may reveal the active immunological pathways in the kidney. Not surprisingly, patients with a stronger chemokine signal in the urine have glomerular immune cell infiltration, as seen in proliferative lupus nephritis. Strikingly, the chemokines detected are part of a specific immune response inducible by IFN-γ, IL-1β, and TNF that matches the inflammatory environment in the kidney. The universal and numerically substantial presence of IFN-γ–positive cells suggests a strong type 1 (Th1) immune response (22). In fact, the analysis of the single-cell transcriptome of kidney-infiltrating immune cells revealed IFN-γ production in all patients, but not type 2 (Th2) response cytokines (IL-4, IL-5, and IL-13). Furthermore, the IFN-γ–inducible urinary PC1 strongly correlated with the predominance of infiltrating CD8+ T cells, the major source of IFN-γ in lupus nephritis. Together, these findings suggest an ongoing IFN-γ–driven response in the kidneys of patients with lupus nephritis (supporting the presence of nonexhausted T cells), which can be detected in real time in the urine by measurement of chemokines. These chemokines are directed to attract mostly macrophages, neutrophils and lymphocytes, and especially other IFN-γ–producing cells via ligation of CXCR3, CXCR4, and CCR5 that may amplify the immune response in a feed-forward loop.
We performed an unsupervised analysis to detect biology-driven differences rather than looking for markers that associated with prespecified outcomes such as histological class. While a stronger immune activation signal in the urine identified patients with proliferative lupus nephritis, a substantial group of patients classified as proliferative were grouped with patients with nonproliferative disease. Compared with those with an elevated immune activation signal, these patients may represent those whose nephritis differs in the molecular pathways involved, or those in a different stage of the inflammatory process. It is also conceivable that investigation of the relevant molecular pathways would provide a better assessment of the disease process in lupus nephritis compared with the current classification system based on glomerular endocapillary infiltration in histology. Since histological class may vary in more than 50% of patients on re-biopsy, the dynamic identification of precise disease-driving pathways may better inform treatment selection, development of new treatments, and patient selection for clinical trials. Urine proteomics might also obviate biopsy sampling error. The next phase of AMP studies is aimed to molecularly dissect lupus nephritis and reclassify it based on disease biology.
Our results highlight the importance of IFN-γ in lupus nephritis as a potential driver and coverable therapeutic target. For example, an increase in IFN-γ activity is the first abnormality detected, years before the diagnosis of SLE, preceding the appearance of autoantibodies and the dysregulation of type 1 IFNs (23). IFN-γ receptor polymorphisms are linked to the risk of developing SLE; and IFN-γ polymorphisms result in increased production of IFN-γ that predisposes SLE patients to developing proliferative rather than membranous lupus nephritis (24, 25). The immune response in proliferative lupus nephritis has in fact been shown to be skewed toward the Th1 axis, which is IFN-γ predominant (24, 26, 27). In mouse models, IFN-γ is a crucial cytokine for the development of lupus nephritis, and its blockage prevents and ameliorates kidney disease as well as reducing mortality (28–30). Strikingly, type 1 IFNs are not required for the generation of germinal centers and the development of lupus nephritis in some mouse models, in contrast to IFN-γ signaling (30). IFN-γ has a very pleiotropic effect involving cellular events implicated in lupus nephritis, including the recruitment and activation of neutrophils, CD8+ and CD4+ T cells, NK cells, and macrophages (31). Importantly, this molecule induces the formation of germinal centers, T follicular helper cell development, and class switch in B cells (32). In the gut, IFN-γ mediates the death of intestinal stem cells, suggesting that a similar mechanism may have implications for irreversible kidney damage and fibrosis (33). In humans, encouragingly positive phase II trials showed the efficacy of blocking the IFN-γ pathway in extrarenal lupus (34–36). In an ustekinumab trial, for example, clinical response was associated with reduction of the IFN-γ signature, rather than type 1 IFN (37). In aggregate, these findings suggest that IFN-γ is a central cytokine in lupus nephritis, and, given the acceptable safety profile of its direct blockage (38), further studies in lupus nephritis should pursue IFN-γ inhibition.
An IFN gene response, especially type 1, is the archetypal signature of SLE (39–41). We and others have previously showed a strong type 1 IFN signature in lupus nephritis (14–16). There is a large overlap between the gene signatures induced by type 1 and type 2 IFNs (42–44), as indicated by shared signaling pathways. Each IFN type can induce the production of the other one (43), eventually leading to stimulation from both sides and thus a mixed signature. Here, we showed the presence of IFN-γ–producing cells in lupus nephritis along a typical chemokine signature in the urine, suggesting that IFN-γ is central to the pathogenesis of lupus nephritis. Type 1 IFN intrarenal transcripts were not detected in our studies, suggesting that, assuming the absence of technical limitations, type 1 IFNs may originate outside the kidney or arise prior to the clinical events leading to a kidney biopsy.
We acknowledge the limitations of our study. We did not detect an association with clinical variables such as renal activity/chronicity or medication use. There are likely other major detectable molecular pathways that could identify distinct patient groups. However, the relatively limited sample size did not allow for further exploration without the risk of a type 1 error. Nevertheless, our findings are confirmed by other smaller and independent studies that assessed the concentration of a fraction of our analytes in the urine of patients with lupus nephritis (45). Further studies, as those ongoing with the phase 2 of the AMP, are needed to extend and validate these results. Second, while our platform did not assay the whole urine proteome, it allowed for interpretability, a broad assessment of the immune response, and limited major confounders from batch effect and unalignable peptides, as commonly seen in classical mass spectrometry experiments. To limit this bias, we used a self-contained enrichment strategy (GSEA). In order to identify protein patterns independently of the amount of proteinuria, we scaled the analyte concentration within and between patients. As this might introduce a bias related to this data set, a larger study will be needed to validate the findings and to develop a convenient assay to apply in clinical practice. However, the high level of infiltrating IFN-γ–producing cells supports the biological plausibility of our results. Finally, the single-cell RNA-Seq did not include certain cell types, such as stromal and epithelial cells. These cells were underrepresented by design in order to enrich for immune cells but are an important source of cytokines and will be evaluated in future studies. Neutrophils were also not detected, likely because they did not survive the freeze-thaw process. However, urine proteomics detected a chemokine pattern implicated in neutrophil recruitment consistent with their known involvement in lupus nephritis (46).
In summary, we demonstrated that the complex molecular biology in kidney biopsies could be captured in urine samples from patients with lupus nephritis. These processes provide insight into patient-specific disease pathogenesis that can address the issue of heterogeneity in SLE. These pathways can be tracked in the urine in real time and may not only improve diagnosis but also guide selection, dosing, and monitoring of treatment, thus paving the way to developing a biologically grounded liquid biopsy.
Methods
Patients and sample collection.
This study enrolled SLE patients with urine protein/creatinine ratio greater than 0.5 undergoing clinically indicated renal biopsy. Only patients with a pathology report confirming lupus nephritis were included in the study. Renal biopsies were scored by a renal pathologist according to the International Society of Nephrology/Renal Pathology Society (ISN/RPS) guidelines and NIH activity and chronicity scales. Urine specimens were acquired at 2 clinical sites in the United States (Johns Hopkins University and New York University). The total urine volume was split into two 50-mL Falcon tubes. Urine cells were pelleted by centrifugation at 200 g for 10 minutes, and the supernatant was aliquoted and stored at –80 °C. Serological features and complement levels were determined at the clinical visit preceding the biopsy. Proteinuria was measured on or near the day of the biopsy.
Urine Quantibody assay.
The Kiloplex Quantibody protein array platform (QAH-CAA-X00, RayBiotech Life) was used for screening urine samples. The array was spotted with 1000 capture antibodies specific for 1000 different proteins in quadruplicate. All urine samples were clarified by centrifugation, and then diluted to yield a total protein concentration within the working range (0.5–1 mg/mL) before application to the arrays. Briefly, protein standards and urine samples were incubated on the array for 2 hours to allow the proteins to bind to the capture antibodies. A biotinylated antibody cocktail comprising 1000 detection antibodies was subsequently added for incubation for 2 hours. Finally, streptavidin-Cy3 was added and left to incubate for 1 hour. Washing was performed between each step to remove the unbound reagents. After a final wash and dry, the slides were read with a fluorescence scanner, and data were extracted from the image using a vendor-provided GAL file with suitable microarray analysis software. Creatinine was measured for each urine sample (KGE005, R&D Systems), and all data were creatinine normalized before analysis. For the complete list of the analytes measured in the urine, see “Kiloplex targets” in the supplemental material.
Renal tissue single-cell RNA-Seq.
Renal tissue was collected, stored, and processed as previously described (14). Briefly, research biopsy cores were collected from consented subjects as an additional biopsy pass or tissue from routine clinical passes. Only biopsies with confirmed lupus nephritis were included. Kidney tissue was frozen on site and shipped to a central processing location, where it was thawed and disaggregated. Individual cells were retrieved and sorted by flow cytometry. For each sample, 10% was allocated to sort CD10+CD45− epithelial cells as single cells, and the remaining 90% was used to sort CD45+ leukocytes as single cells. For each single cell, the whole gene expression profile was sequenced using the CEL-Seq2 method (47). The relative abundance of the cell clusters was calculated as the proportion of cells annotated to one cluster over all the immune cells for each patient. The data reported in this article, including the clinical and serological data of the study participants, have been deposited in the ImmPort repository (accession SDY997). The raw single-cell RNA-Seq data have been deposited in dbGAP (accession phs001457.v1.p1)
Statistics.
PCA: Urine protein concentrations were scaled (Z-normalized) within and across patients after log normalization. Scaling within samples favors the identification of the sample-specific pattern of expression regardless of the total urine protein concentration. PCs were computed using the R Project statistics package version 3.5.2.
Pathway enrichment analysis: GSEA (48) was performed using the genes coding for the protein assessed by the Quantibody assay using the Gene Ontology Biological Process database (49, 50). Pathways with at least 10 (but fewer than 500) genes represented in the Quantibody assay were included in the analysis. Enrichment P values were computed based on 10,000 permutations and adjusted for multiple comparisons using the Benjamini-Hochberg procedure (i.e., FDR) (51). The pathways captured by PC1 were determined based on the loading of each analyte on PC1. Adjusted P values less than 0.05 were considered significant.
Study approval.
Human study protocols were approved by the institutional review boards at Johns Hopkins University and New York University, and written informed consent was received from all participants.
Author contributions
AF, MP, JB, and CM conceptualized the project. AF, MP, DF, JMT, HMB, PI, RC, and BD acquired samples and supervised the processing. TZ and CM generated the proteomic data. AF, MP, JB, DF, JM, HMB, PI, RC, AA, AD, DAR, CCB, NH, DW, and BD contributed to sample collection, processing, analysis, and interpretation of the renal biopsy scRNA-Seq. WA coordinated the network effort. AF devised the analytical plan, interpreted the results, and wrote the manuscript with the guidance of MP, SR, YZ, and LM, and the contribution of JB, DAR, RC, HMB, and PI.
Supplementary Material
Acknowledgments
We thank the participating AMP network clinical sites and participants. We thank Ilya Korsunski, Maria Gutierrez, Fan Zhang, and Felipe Andrade for their critical review of the analytical plan and help with the interpretation of the results. This work was supported by the AMP Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Co., Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi, and Takeda Pharmaceuticals International Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development. See Supplemental Acknowledgments for details on network authors. Funding was provided through grants from the NIH (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2- AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, UM2- AR067678, and AR074096). The Hopkins Lupus Cohort is funded by NIH AR 69572.
Version 1. 05/12/2020
In-Press Preview
Version 2. 06/18/2020
Electronic publication
Funding Statement
AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development Funding was provided through grants from the National Institutes of Health
Hopkins Lupus Cohort
Footnotes
Conflict of interest: JB is a consultant for Bristol-Myers Squibb and received research grant support from Exagen; HMB is a consultant for Exagen and in the speakers bureau for Ra Pharmaceuticals; CM has consulted for Equillium; MP is a consultant for AbbVie, Aleon, Amgen, Annenberg Center for Health Sciences, AstraZeneca, Blackrock, Bristol-Myers Squibb, Decision Resources, Exagen, Glenmark, GSK, INOVA, IQVIA, Janssen, Lilly, Merck EMD Serono, Novartis, Sanofi Japan, Thermo Fisher, and UCB,and received research grant support from AstraZeneca, Exagen, GSK, and Lilly.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e138345.https://doi.org/10.1172/jci.insight.138345.
Contributor Information
Andrea Fava, Email: afava1@jh.edu.
Chandra Mohan, Email: cmohan@uh.edu.
Ting Zhang, Email: tzhang30@Central.UH.EDU.
H. Michael Belmont, Email: Michael.Belmont@nyulangone.org.
Peter Izmirly, Email: peter.izmirly@nyumc.org.
Robert Clancy, Email: Robert.Clancy@nyumc.org.
Jose Monroy Trujillo, Email: jmonroy2@jhmi.edu.
Yuji Zhang, Email: Yuzhang@som.umaryland.edu.
Laurence Magder, Email: LMAGDER@epi.umaryland.edu.
Deepak A. Rao, Email: darao@bwh.harvard.edu.
Arnon Arazi, Email: arnonar@broadinstitute.org.
Celine C. Berthier, Email: celineb@med.umich.edu.
Anne Davidson, Email: adavidson1@nshs.edu.
Betty Diamond, Email: bdiamond@nshs.edu.
Nir Hacohen, Email: NHACOHEN@PARTNERS.ORG.
David Wofsy, Email: david.wofsy@ucsf.edu.
William Apruzzese, Email: WAPRUZZESE@PARTNERS.ORG.
Soumya Raychaudhuri, Email: soumya@broadinstitute.org.
Michelle Petri, Email: mpetri@jhmi.edu.
References
- 1.Fava A, Petri M. Systemic lupus erythematosus: diagnosis and clinical management. J Autoimmun. 2019;96:1–13. doi: 10.1016/j.jaut.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcón GS, et al. Systemic lupus erythematosus in three ethnic groups: II. Features predictive of disease activity early in its course. LUMINA Study Group. Lupus in minority populations, nature versus nurture. Arthritis Rheum. 1998;41(7):1173–80. doi: 10.1002/1529-0131(199807)41:7<1173::AID-ART5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 3.Mok CC, Kwok RCL, Yip PSF. Effect of renal disease on the standardized mortality ratio and life expectancy of patients with systemic lupus erythematosus. Arthritis Rheum. 2013;65(8):2154–2160. doi: 10.1002/art.38006. [DOI] [PubMed] [Google Scholar]
- 4.Lim SS, et al. The incidence and prevalence of systemic lupus erythematosus, 2002-2004: the Georgia lupus registry. Arthritis Rheum. 2014;66(2):357–368. doi: 10.1002/art.38239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferucci ED, et al. Prevalence and incidence of systemic lupus erythematosus in a population-based registry of American Indian and Alaska Native people, 2007-2009. Arthritis Rheum. 2014;66(9):2494–2502. doi: 10.1002/art.38720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dall’Era M, et al. The incidence and prevalence of systemic lupus erythematosus in San Francisco County, California: the California Lupus Surveillance Project. Arthritis Rheum. 2017;69(10):1996–2005. doi: 10.1002/art.40191. [DOI] [PubMed] [Google Scholar]
- 7.Izmirly PM, et al. The incidence and prevalence of systemic lupus erythematosus in New York County (Manhattan), New York: the Manhattan Lupus Surveillance Program. Arthritis Rheum. 2017;69(10):2006–2017. doi: 10.1002/art.40192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somers EC, et al. Population-based incidence and prevalence of systemic lupus erythematosus: the Michigan lupus epidemiology and surveillance program. Arthritis Rheum. 2014;66(2):369–378. doi: 10.1002/art.38238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn BH, et al. American College of Rheumatology guidelines for screening, treatment, and management of lupus nephritis. Arthritis Care Res. 2012;64(6):797–808. doi: 10.1002/acr.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weening JJ, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol. 2004;15(2):241–250. doi: 10.1097/01.ASN.0000108969.21691.5D. [DOI] [PubMed] [Google Scholar]
- 11.Greloni G, et al. Value of repeat biopsy in lupus nephritis flares. Lupus Sci Med. 2014;1(1):1–6. doi: 10.1136/lupus-2013-000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rosa M, et al. A prospective observational cohort study highlights kidney biopsy findings of lupus nephritis patients in remission who flare following withdrawal of maintenance therapy. Kidney Int. 2018;94(4):788–794. doi: 10.1016/j.kint.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Clark MR, Trotter K, Chang A. The pathogenesis and therapeutic implications of tubulointerstitial inflammation in human lupus nephritis. Semin Nephrol. 2015;35(5):455–464. doi: 10.1016/j.semnephrol.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arazi A, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20(7):902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Der E, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. 2019;20(7):915–927. doi: 10.1038/s41590-019-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Der E, et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight. 2017;2(9):e93009. doi: 10.1172/jci.insight.93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginzler EM, et al. Mycophenolate mofetil or intravenous cyclophosphamide for lupus nephritis. N Engl J Med. 2005;353(21):2219–2228. doi: 10.1056/NEJMoa043731. [DOI] [PubMed] [Google Scholar]
- 18.Soliman S, Mohan C. Lupus nephritis biomarkers. Clin Immunol. 2017;185:10–20. doi: 10.1016/j.clim.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Kiani AN, et al. Urinary vascular cell adhesion molecule, but not neutrophil gelatinase-associated lipocalin, is associated with lupus nephritis. J Rheumatol. 2012;39(6):1231–1237. doi: 10.3899/jrheum.111470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunner HI, et al. Association of noninvasively measured renal protein biomarkers with histologic features of lupus nephritis. Arthritis Rheum. 2012;64(8):2687–2697. doi: 10.1002/art.34426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiani AN, et al. Serum osteoprotegrin (OPG) in subclinical atherosclerosis in systemic lupus erythematosus. Lupus. 2017;26(8):865–870. doi: 10.1177/0961203316682101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Annunziato F, Romagnani C, Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol. 2015;135(3):626–635. doi: 10.1016/j.jaci.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Munroe ME, et al. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann Rheum Dis. 2016;75(11):2014–2021. doi: 10.1136/annrheumdis-2015-208140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyake K, et al. Genetically determined interferon-gamma production influences the histological phenotype of lupus nephritis. Rheumatology (Oxford) 2002;41(5):518–524. doi: 10.1093/rheumatology/41.5.518. [DOI] [PubMed] [Google Scholar]
- 25.Nakashima H, et al. The combination of polymorphisms within interferon-γ receptor I and receptor 2 associated with the risk of systemic lupus erythematosus. FEBS Lett. 1999;453(1–2):187–190. doi: 10.1016/s0014-5793(99)00701-2. [DOI] [PubMed] [Google Scholar]
- 26.Uhm WS, et al. Cytokine balance in kidney tissue from lupus nephritis patients. Rheumatology. 2003;42(8):935–938. doi: 10.1093/rheumatology/keg255. [DOI] [PubMed] [Google Scholar]
- 27.Masutani K, et al. Predominance of Th1 immune response in diffuse proliferative lupus nephritis. Arthritis Rheum. 2001;44(9):2097–2106. doi: 10.1002/1529-0131(200109)44:9<2097::AID-ART360>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 28.Schwarting A, Wada T, Kinoshita K, Tesch G, Kelley VR. IFN-gamma receptor signaling is essential for the initiation, acceleration, and destruction of autoimmune kidney disease in MRL-Fas(lpr) mice. J Immunol. 1998;161(1):494–503. [PubMed] [Google Scholar]
- 29.Haas C, Ryffel B, Le Hir M. IFN-γ receptor deletion prevents autoantibody production and glomerulonephritis in lupus-prone (NZB x NZW)F1 mice. J Immunol. 1998;160(8):3713–3718. [PubMed] [Google Scholar]
- 30.Jackson SW, et al. B cell IFN-γ receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J Exp Med. 2016;213(5):733–750. doi: 10.1084/jem.20151724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard KM, Cauvi DM, Toomey CB, Morris KV. Kono DH. Interferon-gamma and systemic autoimmunity. Discov Med. 2013;16(87):123–131. [PMC free article] [PubMed] [Google Scholar]
- 32.et al. IFN-γ receptor and STAT1 signaling in B cells are central to spontaneous germinal center formation and autoimmunity. J Exp Med. 2016;213(5):715–732. doi: 10.1084/jem.20151722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takashima S, et al. T cell-derived interferon-γ programs stem cell death in immune-mediated intestinal damage. Sci Immunol. 2019;4(42):1–15. doi: 10.1126/sciimmunol.aay8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vollenhoven RF, et al. Efficacy and safety of ustekinumab, an IL-12 and IL-23 inhibitor, in patients with active systemic lupus erythematosus: results of a multicentre, double-blind, phase 2, randomised, controlled study. Lancet. 2018;392(10155):1330–1339. doi: 10.1016/S0140-6736(18)32167-6. [DOI] [PubMed] [Google Scholar]
- 35.Wallace DJ, et al. Baricitinib for systemic lupus erythematosus: a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet. 2018;392(10143):222–231. doi: 10.1016/S0140-6736(18)31363-1. [DOI] [PubMed] [Google Scholar]
- 36.Gadina M, et al. Translational and clinical advances in JAK-STAT biology: the present and future of jakinibs. J Leukoc Biol. 2018;104(3):499–514. doi: 10.1002/JLB.5RI0218-084R. [DOI] [PubMed] [Google Scholar]
- 37. doi: 10.1136/lupus-2019-lsm.251. Jordan, J et al. 251 Type II but not type I interferon signifies clinical response to ustekinumab in patients with systemic lupus erythematosus [abstract]. Lupus Sci Med . 2019;6. Accessed May 14, 2020. [DOI]
- 38. DeBenedetti FD, et al. Interferon-gamma (IFN-γ) neutralization with emapalumab and time to response in patients with macrophage activation syndrome (MAS) complicating systemic juvenile idiopathic arthritis (s-JIA) who failed high-dose glucocorticoids [abstract]. Arthritis Rheum. 2019;71L(suppl 10): abstract L06. https://acrabstracts.org/abstract/interferon-gamma-ifn-%ce%b3-neutralization-with-emapalumab-and-time-to-response-in-patients-with-macrophage-activation-syndrome-mas-complicating-systemic-juvenile-idiopathic-arthritis-s-jia-who/ Accessed May 14, 2020.
- 39.Baechler EC, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U SA. 2003;100(5):2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirou KA, et al. Coordinate overexpression of interferon-α-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50(12):3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 41.Bennett L, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197(6):711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall JC, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc Natl Acad Sci U S A. 2012;109(43):17609–17614. doi: 10.1073/pnas.1209724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusinova I, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(Database issue):D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catalina MD, Bachali P, Geraci NS, Grammer AC, Lipsky PE. Gene expression analysis delineates the potential roles of multiple interferons in systemic lupus erythematosus. Commun Biol. 2019;2:140. doi: 10.1038/s42003-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klocke J, et al. Mapping urinary chemokines in human lupus nephritis: potentially redundant pathways recruit CD4+ and CD8+ T cells and macrophages. Eur J Immunol. 2017;47(1):180–192. doi: 10.1002/eji.201646387. [DOI] [PubMed] [Google Scholar]
- 46.Villanueva E, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. 2011;187(1):538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner A, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. doi: 10.1101/060012. Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation [preprint]. bioRxiv 2016;060012. Posted on bioRxiv June 20, 2016. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.