Abstract
Spinal muscular atrophy (SMA) is an inherited neuromuscular disorder caused by reduced expression of the survival motor neuron (SMN) protein. SMN has key functions in multiple RNA pathways, including the biogenesis of small nuclear ribonucleoproteins that are essential components of both major (U2-dependent) and minor (U12-dependent) spliceosomes. Here we investigated the specific contribution of U12 splicing dysfunction to SMA pathology through selective restoration of this RNA pathway in mouse models of varying phenotypic severity. We show that virus-mediated delivery of minor snRNA genes specifically improves select U12 splicing defects induced by SMN deficiency in cultured mammalian cells, as well as in the spinal cord and dorsal root ganglia of SMA mice without increasing SMN expression. This approach resulted in a moderate amelioration of several parameters of the disease phenotype in SMA mice, including survival, weight gain, and motor function. Importantly, minor snRNA gene delivery improved aberrant splicing of the U12 intron–containing gene Stasimon and rescued the severe loss of proprioceptive sensory synapses on SMA motor neurons, which are early signatures of motor circuit dysfunction in mouse models. Taken together, these findings establish the direct contribution of U12 splicing dysfunction to synaptic deafferentation and motor circuit pathology in SMA.
Keywords: Neuroscience
Keywords: Neurodegeneration
Correction of minor U12-dependent splicing dysfunction improves spinal muscular atrophy phenotypes.
Introduction
Removal of introns from precursor mRNAs (pre-mRNAs) is a finely regulated nuclear process carried out by the spliceosome, a highly dynamic RNA protein machine (1). There are 2 functionally distinct spliceosomes comprising different subsets of small nuclear ribonucleoproteins (snRNPs) that are dedicated to the excision of different types of introns based on their specific splicing consensus sequences (2). The majority of introns, referred to as U2-type introns, are excised by the major (U2-dependent) spliceosome that includes U1, U2, U4/U6, and U5 snRNPs. Additionally, a small fraction of introns, referred to as U12-type introns, are processed through the low-abundance minor (U12-dependent) spliceosome formed by U11, U12, U4atac/U6atac, and U5 snRNPs (2). U5 is the only snRNP common to both spliceosomes, and minor snRNPs are about 100-fold less abundant than major snRNPs (3), reflecting the fact that U12 introns account for less than 1% of all introns (4, 5).
The accurate removal of U12 introns is essential for organism development (6–8), and its disruption by mutations in components of the minor spliceosome has recently been associated with human disease (9). Prominent examples are autosomal recessive developmental disorders caused by loss-of-function mutations in the U4atac snRNA gene, including microcephalic osteodysplastic primordial dwarfism type I (10, 11), Roifman syndrome (12) and Lowry Wood syndrome (13), all of which are characterized by postnatal growth retardation and microcephaly, among other clinical features. Interestingly, U12-type introns and their relative positions are evolutionarily conserved and not randomly distributed across the genome (14–17). Rather, they are enriched in information processing genes implicated in DNA and RNA metabolism as well as vesicular transport and voltage-gated ion channels that have especially important functions in neurons. Accordingly, disturbance of U12 splicing has also been associated with neurodevelopmental and neurological deficits (9), including early-onset cerebellar ataxia caused by homozygous mutations in the U12 snRNA gene (18). The first link between U12 splicing dysfunction and neurodegenerative disease came from studies of spinal muscular atrophy (SMA) (19–21), and further evidence implicating this RNA pathway in motor neuron disease has been accumulating from studies of amyotrophic lateral sclerosis (ALS) (22–25). To date, however, conclusive evidence that U12 splicing defects contribute to the etiology of these neurodegenerative diseases has been missing and ideally requires demonstration that selective functional restoration of this RNA pathway leads to phenotypic improvement in animal models.
Here we sought to determine whether U12 splicing dysfunction contributes to the SMA phenotype in mouse models with varying disease severity. SMA is an autosomal recessive neurodegenerative disorder characterized by loss of motor neurons, atrophy of voluntary muscles, and motor impairment (26, 27). SMA is caused by a ubiquitous reduction in the survival motor neuron (SMN) protein due to homozygous deletions or mutations of the Survival motor neuron 1 (SMN1) gene with preservation of a nearly identical gene called SMN2 that produces low levels of functional SMN (28). SMN is essential for the biogenesis of Sm class snRNPs of the major and minor spliceosomes (29, 30). Specifically, SMN mediates the assembly of a heptameric ring of Sm proteins on a conserved sequence of each snRNA to form the protein core required for the biogenesis, stability, and function of snRNPs (31, 32). SMN-dependent impairment of snRNP assembly leads to a preferential decrease of minor snRNPs of the U12 spliceosome in SMA mice (19, 33, 34), and accumulation of U12 splicing defects has been documented in a variety of cellular and animal models of SMN deficiency (20, 21, 35, 36). Importantly, we previously showed that dysregulation of a U12 intron–containing gene we called Stasimon (also known as Tmem41b) is responsible for specific neuronal phenotypes induced by SMN deficiency in Drosophila and zebrafish models of SMA (21). Furthermore, we recently demonstrated the direct contribution of Stasimon to motor circuit pathology in a severe mouse model of SMA (37). These and other studies support the notion that snRNP dysfunction leads to perturbation of RNA splicing of select genes that in turn contribute to SMA (21, 33, 37–41), but a specific pathogenic role of U12 splicing dysfunction in mouse models of the disease has not been directly demonstrated.
Here, we devised a means to functionally enhance the U12 splicing pathway in the context of SMN deficiency through virus-mediated delivery of minor snRNA genes. This approach was effective in selectively correcting U12 splicing defects without increasing SMN levels or influencing other RNA processing events regulated by SMN. Furthermore, enhancing U12 splicing ameliorated disease phenotypes in SMA mice, leading to prolonged survival and improved motor function. Importantly, minor snRNA gene delivery improved missplicing of the Stasimon gene and robustly rescued the loss of proprioceptive sensory synapses on motor neurons, which represent early signatures of motor dysfunction in SMA mice (21, 37, 42, 43). These findings highlight a direct pathogenic role of U12 splicing dysregulation in mouse models of SMA and reveal the functional requirement of minor snRNPs for the maintenance of synaptic integrity in mammalian sensory-motor circuits.
Results
Correction of SMN-dependent U12 splicing defects in mammalian cells by virus-mediated expression of minor snRNAs.
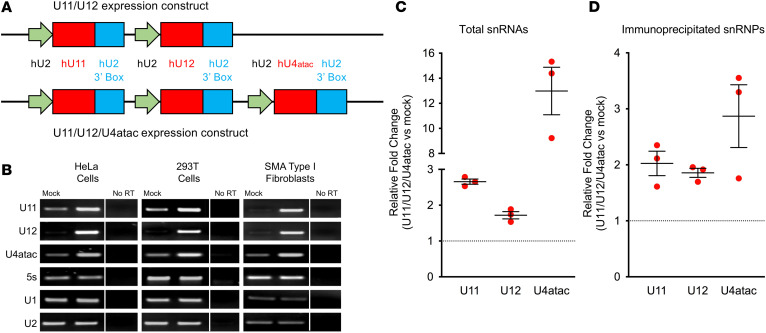
To investigate the role of U12 splicing dysfunction in SMA, we constructed lentiviral and self-complementary adeno-associated virus serotype 9 (AAV9) vectors that contained expression cassettes for human U11, U12, and U4atac snRNAs under the control of the human U2 promoter (44) (Figure 1A). For initial validation of vector-derived minor snRNA overexpression, HeLa and HEK293T cells as well as fibroblasts from a patient with type I SMA (GM03813) (45) were transduced with the U11/U12/U4atac-expressing lentivirus, and RNA was extracted 72 hours later for reverse transcription PCR (RT-PCR) analysis (Figure 1B). The 5S ribosomal RNA (rRNA) as well as U1 and U2 major snRNAs were used as internal controls. We found a robust increase in the expression of human U11, U12, and U4atac snRNAs above the endogenous levels in each of the human cell types examined (Figure 1B). Moreover, the expression of endogenous U1, U2, and 5S rRNA was unchanged, indicating that the overexpression of minor snRNAs was specific and did not alter the levels of major snRNAs. To determine the incorporation of vector-derived minor snRNAs into snRNPs, we transiently transfected HEK293T cells with the U11/U12/U4atac lentiviral construct and performed coimmunoprecipitation experiments with anti-SmB antibodies under stringent conditions followed by RT-PCR analysis (Figure 1, C and D). The increased amounts of immunoprecipitated snRNAs from U11/U12/U4atac-transfected cells relative to mock-transfected controls are consistent with proper formation of the Sm core on the overexpressed minor snRNAs.
Figure 1. Transgenic expression of minor snRNAs in human cell lines.
(A) Schematics of viral constructs harboring cassettes for the expression of human U11, U12, and U4atac snRNAs driven by human U2 promoter with 3′ box at the 3′ end. (B) Semiquantitative RT-PCR analysis of U11, U12, and U4atac snRNA expression 72 hours after transduction of the U11/U12/U4atac lentiviral construct in HeLa, HEK293T, and SMA type I patient fibroblasts (GM03813). U1 and U2 major snRNAs as well as 5S rRNA were used as controls. Lanes that were run on the same gel but were noncontiguous are separated by a vertical line. (C) RT-PCR analysis of the total levels of minor snRNA overexpression in cell extracts from HEK293T cells transiently transfected with the U11/U12/U4atac lentiviral construct for 48 hours relative to mock-transfected cells used as controls. The scatter plot shows the fold change in the relative amounts of U11, U12, and U4atac overexpression over endogenous minor snRNA levels in mock-transfected cells, which were arbitrarily set to 1 (dotted line). Individual data points, mean, and SEM from 3 independent experiments are shown. (D) Equal amounts of cell extract from either U11/U12/U4atac or mock-transfected HEK293T cells as in C were immunoprecipitated with anti-SmB antibodies, followed by RNA purification and RT-PCR analysis. The scatter plot shows the fold change in the amounts of immunoprecipitated minor snRNAs from U11/U12/U4atac-transfected cells relative to the levels from mock-transfected cells, which were arbitrarily set to 1 (dotted line). Individual data points, mean, and SEM from 3 independent experiments are shown.
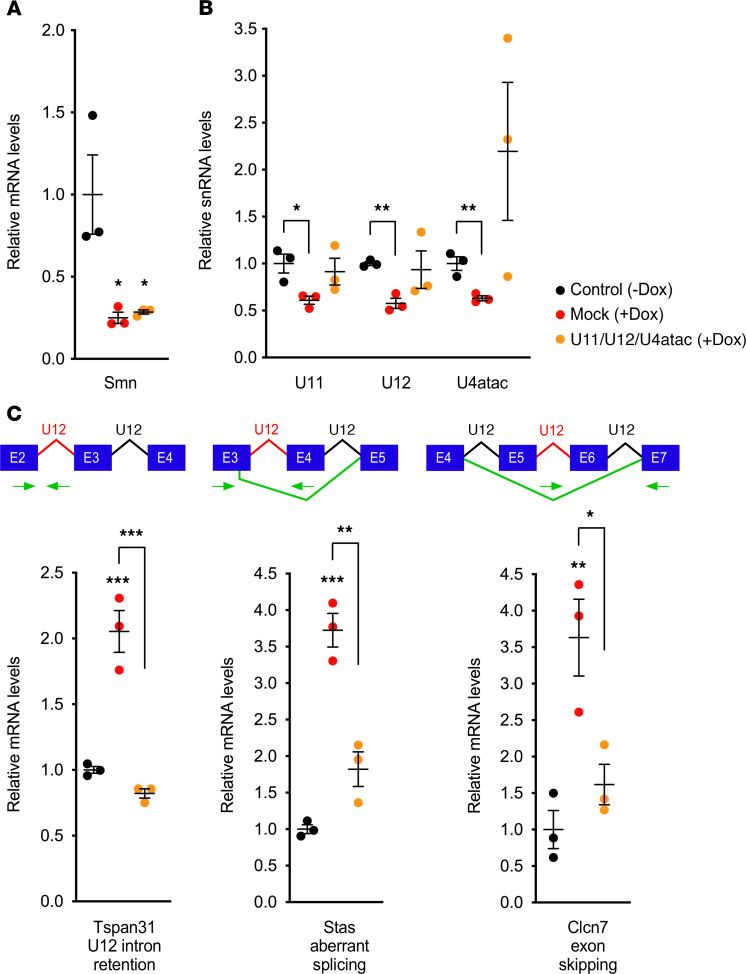
Next, we sought to determine the effects of minor snRNA overexpression on U12 splicing in a setting of SMN deficiency. To do so, we used previously characterized mouse NIH 3T3-SmnRNAi fibroblasts in which doxycycline-inducible (Dox-inducible) RNA interference (RNAi) knockdown of endogenous Smn causes snRNA reduction and U12 splicing impairment (21). Following transduction with the U11/U12/U4atac lentivirus or mock treatment, NIH 3T3-SmnRNAi cells were cultured for 5 days in the presence of Dox to induce knockdown of Smn. RT-PCR analysis showed that the efficient reduction of Smn mRNA expression upon Dox treatment was not altered by transduction with the U11/U12/U4atac lentivirus (Figure 2A). In contrast, the reduction in the steady-state levels of U11, U12, and U4atac snRNAs induced by SMN deficiency in NIH 3T3 was corrected by lentivirus-mediated expression of minor snRNAs, with U11 and U12 being similar to, and U4atac exceeding, the levels found in control cells with normal Smn (Figure 2B).
Figure 2. Lentivirus-mediated expression of minor snRNAs improves U12 splicing defects induced by SMN deficiency in mammalian cells.
(A and B) RT-PCR analysis of the levels of Smn mRNA (A) and the indicated minor snRNAs (B) in Dox-treated, Smn-deficient NIH 3T3-SmnRNAi cells that were either mock transduced or transduced with lentivirus expressing human U11, U12, and U4atac snRNAs relative to untreated NIH 3T3-SmnRNAi cells with normal Smn levels (black, control – Dox; red, mock + Dox; orange, U11/U12/U4atac + Dox). Note that primers were designed to detect both endogenous mouse snRNAs and virally delivered human snRNAs. The scatter plots show individual data points, mean, and SEM from 3 independent experiments normalized to the control group. Statistics were performed using multiple t tests with the Benjamini, Krieger, and Yekutieli correction for multiple comparisons. *P < 0.05; **P < 0.01. (C) RT-PCR analysis in the same experimental groups as in A of U12 intron retention in Tspan31 mRNA, aberrant splicing of Stasimon (Stas) mRNA, and exon skipping in Clcn7 mRNA — all of which are previously characterized SMN-dependent U12 splicing defects in NIH 3T3 cells (21). Schematics of the RNA processing events monitored in the assay are shown at the top. The scatter plots show individual data points, mean, and SEM from 3 independent experiments normalized to the control group. Statistics: 1-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001. All other pairwise comparisons between groups are not statistically significant.
Having established a means to restore minor snRNA levels, we then investigated their potential effects in correcting U12 splicing dysfunction in SMN-deficient NIH 3T3 cells. To do so, we analyzed representative U12-dependent mRNA processing events that we have previously demonstrated to be dysregulated in response to SMN deficiency in NIH 3T3 cells (21). These include increased retention of the U12 intron in the Tetraspanin 31 (Tspan31) mRNA, accumulation of an aberrantly spliced Stasimon mRNA due to activation of a cryptic 5′ splice site, and an alternatively spliced chloride voltage-gated channel 7 (Clcn7) mRNA in which both exons flanking the U12 intron are skipped because of inefficient U12 splicing (Figure 2C). Consistent with our previous study (21), RT-PCR analysis of these target mRNAs confirmed U12 splicing dysfunction in SMN-deficient NIH 3T3 cells relative to control cells (Figure 2C). Importantly, all these splicing defects were strongly corrected by transduction of the U11/U12/U4atac lentivirus in SMN-deficient NIH 3T3 cells (Figure 2C).
Together, these results indicate that virus-mediated gene delivery is an effective means to counteract the reduction in the levels of U11, U12, and U4atac minor snRNAs and improve U12 splicing dysfunction induced by SMN deficiency in mammalian cells.
AAV9-mediated minor snRNA gene delivery improves Stasimon U12 splicing in SMA mice.
We sought to test our virus-based approach of snRNA gene delivery in vivo in order to investigate the contribution of U12 splicing dysfunction in the SMNΔ7 mouse model of severe SMA. These mice are knocked out for the mouse Smn gene and harbor 2 copies of the human SMN2 gene as well as multiple copies of a human SMNΔ7 transgene (46). SMNΔ7 SMA mice display reduced weight gain, severe motor dysfunction, and death by the second postnatal week. We used intracerebroventricular (ICV) injection of ~1 × 1011 viral genome particles (vgp) of either AAV9-U11/U12 or AAV9-U11/U12/U4atac in SMNΔ7 SMA mice at postnatal day 1 (P1) as previously described (47–49). We then isolated spinal cords from AAV9-injected SMA mice as well as untreated SMA mice and unaffected heterozygous mice at P9 and measured the levels of U11, U12, and U4atac snRNAs by RT-PCR. Given that primers that would specifically and selectively detect individual snRNAs from one species but not the other could not be designed; we relied on primers that detect both endogenous (mouse) and virally expressed (human) snRNAs with identical efficiency (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130574DS1). Consistent with previous studies (19, 33, 34), we found a reduction in the levels of U11, U12, and U4atac in the spinal cord of untreated SMA mice relative to unaffected controls. However, we did not find an increase in the levels of these snRNAs in SMA mice injected with AAV9-U11/U12 or AAV9-U11/U12/U4atac (Figure 3A). While seemingly surprising, the lack of an increase at whole-tissue levels likely reflects the tropism of AAV9, which is highly effective in transducing motor neurons but not the vast majority of other spinal cells as indicated by immunostaining experiments of SMA mice injected with AAV9-GFP (Supplemental Figure 1A). Therefore, snRNA overexpression in AAV9-targeted cells, such as motor neurons, would be masked by the lack thereof in the much larger proportion of other spinal cells. Consistent with this interpretation, we found that the levels of minor snRNAs were also not increased in the spinal cord of SMA mice injected with AAV9-SMN (Figure 3A), while these snRNAs were restored in the spinal cord of SMA mice following pharmacological treatment with compounds that induce widespread SMN upregulation (50).
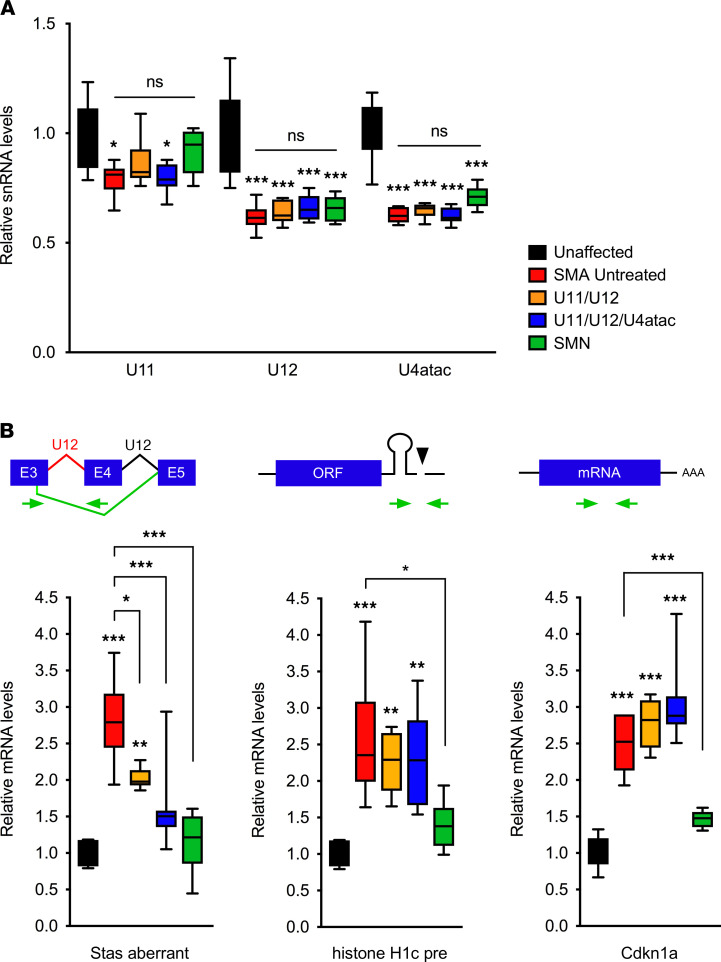
Figure 3. AAV9-mediated delivery of minor snRNAs improves Stasimon U12 splicing defects in the spinal cord of SMA mice.
(A) RT-PCR analysis of minor snRNA levels at P9 in the spinal cord of unaffected control mice (n = 6) and SMA mice that were either untreated (n = 6) or ICV injected with AAV9-U11/U12 (n = 6), AAV9-U11/U12/U4atac (n = 8), and AAV9-SMN (n = 8) as indicated. Note that primers were designed to detect both endogenous mouse snRNAs and virally delivered human snRNAs. The box-and-whiskers graph shows the median (middle line), interquartile range (box), and minimum and maximum (whiskers). Statistics were performed with 1-way ANOVA with Tukey’s post hoc test. *P < 0.05; ***P < 0.001; ns, no significance. (B) RT-PCR analysis of the levels of aberrantly spliced Stasimon mRNA (21), 3′ end–extended histone H1c precursor mRNA (52), and Cdkn1a mRNA (39) in the spinal cord from the same groups as in A at P9. Schematics of the RNA processing events monitored in the assay are shown at the top. The box-and-whiskers graph shows the median, interquartile range, and minimum and maximum. Statistics were performed with 1-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance.
We next investigated whether injection of AAV9-U11/U12 or AAV9-U11/U12/U4atac could correct defective U12 splicing of Stasimon mRNA in the spinal cord of SMA mice. Consistent with a previous study (21), an aberrantly spliced Stasimon mRNA isoform accumulates because of impaired excision of the U12 intron in the spinal cord of untreated SMA mice relative to unaffected control mice (Figure 3B). This splicing defect was significantly corrected in SMA mice injected with AAV9-U11/U12 or AAV9-U11/U12/U4atac as well as upon injection of AAV9-SMN (Figure 3B), which was used as a positive control. To expand our analysis to other U12 splicing events dysregulated in SMA, we investigated 3 genes (Myh9, Myo10, and Rasgrp3) for which increased U12 intron retention was previously validated in the spinal cord of a different mouse model of SMA (36). Interestingly, although we showed defective splicing of the U12 introns of Myh9, Myo10, and Rasgrp3 mRNAs in the spinal cord of SMNΔ7 SMA mice relative to unaffected controls, these defects were not corrected by treatment with either AAV9-U11/U12/U4atac or AAV9-SMN (Supplemental Figure 1B). These unexpected results suggest that these U12 splicing changes occur in spinal cells that are not efficiently transduced by AAV9 and that dysregulation of these genes does not significantly contribute to SMA pathology.
Given that Stasimon missplicing is prominent in SMA proprioceptive neurons that reside in the dorsal root ganglia (DRGs) (21), we analyzed lumbar DRGs isolated from SMA mice treated with AAV9-U11/U12 and AAV9-SMN relative to untreated controls (Supplemental Figure 2). Previous studies have documented highly efficient transduction of DRG neurons, including proprioceptive sensory neurons (43, 51). We found here that the strong accumulation of aberrantly spliced Stasimon mRNA in SMA DRGs was markedly reduced by minor snRNA gene delivery and fully corrected by SMN restoration (Supplemental Figure 2).
To determine the selectivity of the effects, we also monitored well-established mRNA processing events in other SMN-regulated RNA pathways that are disrupted in SMA but unrelated to U12 splicing. Specifically, we analyzed the U7-dependent 3′ end processing of histone H1c mRNA (52) and the expression of cyclin-dependent kinase inhibitor 1A (Cdkn1a) mRNA that is mediated by p53 activation through dysregulation of Mdm2 and Mdm4 alternative splicing (41, 53). The accumulation of both 3′ end–extended H1c mRNA and Cdkn1a mRNA induced by SMN deficiency in the spinal cord of SMA mice was not corrected by AAV9-U11/U12 or AAV9-U11/U12/U4atac, while it was normalized by treatment with AAV9-SMN as expected (Figure 3B), indicating specificity of the effects of minor snRNA gene delivery in improving U12 splicing of Stasimon in SMA mice.
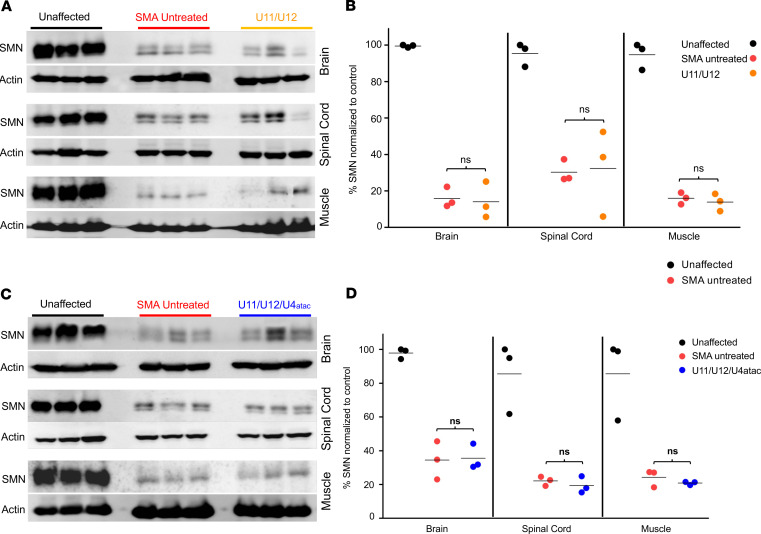
Last, it was important to establish that SMN levels were not elevated following AAV9-mediated minor snRNA gene delivery in SMA mice. Therefore, we harvested disease-relevant tissues from treated mice at P9 and performed Western blot analysis. Compared with untreated SMA samples, SMN protein expression in brain, spinal cord, and skeletal muscle (gastrocnemius) was not increased in AAV9-treated relative to untreated SMA mice (Figure 4). These results demonstrate that treatment with either AAV9-U11/U12 or AAV9-U11/U12/U4atac vectors does not elevate SMN protein levels in SMA mice, indicating that the improvement in U12 splicing is independent of SMN induction.
Figure 4. Minor snRNA gene delivery does not increase SMN protein expression in SMA mice.
Tissues for Western blot analysis were collected from brain, spinal cord, and gastrocnemius muscle. Western blot images showing SMN expression levels in tissues collected at P9 from heterozygous (unaffected) control mice as well as untreated and U11/U12- (A) or U11/U12/U4atac-treated (C) SMA mice. No significant changes in the SMN levels were observed in all tested tissues comparing samples from untreated and U11/U12- (B) or U11/U12/U4atac-treated (D) SMA mice. All SMA samples were predictably lower than unaffected samples. Actin was used as a loading control. Comparisons were analyzed by Student’s t test. *P < 0.05. Data expressed as mean ± SEM. n = 3 animals per group. Data in the scatter plots are presented as percentage of SMN normalized to control.
Minor snRNAs improve motor function and extend survival in severe and intermediate SMA mouse models.
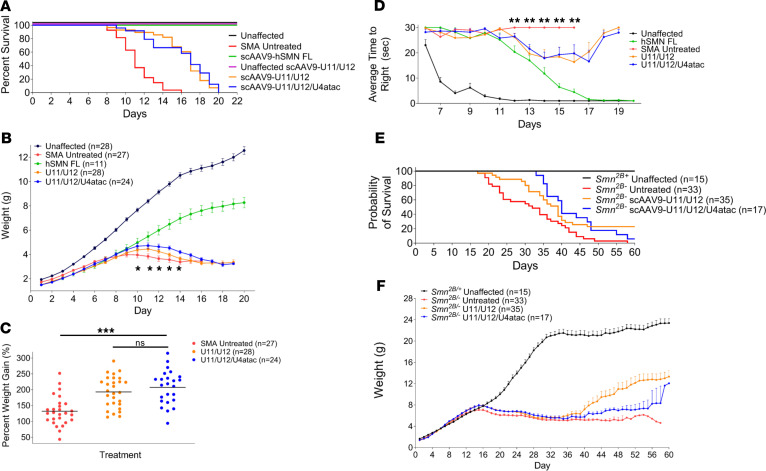
We next investigated the phenotypic effects of AAV9-U11/U12 and AAV9-U11/U12/U4atac in SMNΔ7 SMA mice following a single ICV injection of ~1 × 1011 vgp at P1. To assess any potential toxicity associated with this treatment, similar injections were also performed in unaffected, heterozygous littermates. Moreover, AAV9 expressing full-length human SMN was used as a positive control that induces robust rescue of the SMA phenotype as reported previously (54–56). Compared with the untreated SMA mice with a median survival of 11 days (Figure 5A), the introduction of U11 and U12 snRNAs extended life span to a median of 17 days, with the longest-lived animals reaching 20 days (Figure 5A). Similar results were observed with the AAV9-U11/U12/U4atac vector, where the median survival was extended to 16 days, with a maximal life span of 20 days (Figure 5A). Treatment with each of the vectors also increased total body weight compared with untreated SMA animals over a time course analysis and when measured as the percentage of weight gain relative to birth weight (Figure 5, B and C).
Figure 5. Phenotypic analysis of AAV9-mediated minor snRNA gene delivery in mouse models of SMA.
(A) AAV-mediated gene delivery of human U11/U12 and U11/U12/U4atac snRNAs prolonged survival of severe SMNΔ7 SMA mice. The Kaplan-Meier survival curve indicates significant life extension for SMNΔ7 SMA mice injected with either AAV9-U11/U12 (orange) or AAV9-U11/U12/U4atac (blue) compared with the untreated SMA mice (red). Unaffected animals (black) and SMA mice injected with AAV9 carrying human full-length SMN (green) were used as a positive control. Healthy littermates were also treated with AAV9-U11/U12 (purple) to test for any potential toxicity of the minor snRNAs. Log-rank Mantel-Cox test (P < 0.0001). (B) The average weight per group is plotted across days for the surviving animals in each cohort. Significant weight gain was observed from P10 through P14 in SMNΔ7 mice injected with AAV9-U11/U12 or AAV9-U11/U12/U4atac relative to untreated SMA mice. Statistical analyses were performed by 2-way ANOVA, where an asterisk denotes significant weight gain difference between snRNAs treated and untreated cohorts (P ≤ 0.03). (C) Scatter plot depicting the percentage weight gained from birth to peak. Analysis of percentage weight gain showed significant improvement in peak weight gain in SMNΔ7 SMA mice injected with AAV9-U11/U12 or AAV9-U11/U12/U4atac relative to untreated SMA mice. Statistics: 1-way ANOVA with Tukey’s post hoc test. (D) Righting reflex measurements. Time-to-right (TTR) measurements were initiated on P6 and recorded along animals’ life span. SMNΔ7 SMA mice injected with AAV9-U11/U12 or AAV9-U11/U12/U4atac showed significantly improved TTR from P12 through P16 relative to untreated SMA mice (P = 0.004; 1-way ANOVA; Tukey’s multiple-comparisons test). *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance. (E and F) Phenotypic assessment in the intermediate Smn2B/– mouse model after delivery of AAV9 expressing minor splicing snRNAs showed a significant extension in survival (E) and increased weight gain (F). Log-rank Mantel-Cox test (P < 0.0001) for survival measurements.
Motor behavior was assessed by measuring the righting reflex from a prone position, an assay used in this model as a reliable measurement of motor function (48, 57). This analysis started at P6 and motor performance was recorded daily for each group continuously throughout the life span. As expected, unaffected controls performed much better than untreated SMNΔ7 SMA mice, and the AAV9-SMN–treated SMA animals were able to right as quickly as the unaffected animals by P17 (Figure 5D). Treated SMA mice in the U11/U12 and U11/U12/U4atac cohorts showed improved righting reflexes and were able to right themselves faster starting at P12, at which time the untreated SMA animals were no longer able to stand (Figure 5D). These results demonstrate that delivery of minor snRNAs significantly improves important parameters of disease in a severe mouse model of SMA. Although the overall phenotypic benefit is relatively moderate, a complete rescue would not be expected because there are several other pathways regulated by SMN that contribute to SMA pathology that are not targeted for correction with our approach (37, 39, 41, 52).
To assess the role of minor snRNAs in a milder disease context, we next investigated their effects in Smn2B/– mice, which are a well-described intermediate model of SMA harboring 1 Smn-knockout allele and 1 hypomorphic Smn allele in which exon 7 is mostly skipped due to the mutation of an exonic splicing enhancer (58). These SMA mice have a delayed onset of neuromuscular pathology, and motor dysfunction appears in the third postnatal week (59). First, we analyzed the expression of minor snRNAs, which was not previously described in this mouse model, and found significant reduction in the levels of U11 and U12 but not U4atac in the spinal cord of Smn2B/– mice relative to WT controls at P18 (Supplemental Figure 3A). We then analyzed the same representative SMN-dependent mRNA processing events that are dysregulated in SMNΔ7 SMA mice (Figure 3B) and found that Stasimon’s U12 splicing, histone H1c mRNA 3′ end processing, and Cdkn1a expression are also dysregulated in the spinal cord of Smn2B/– mice at P18 (Supplemental Figure 3B). Increased intron retention in Myh9, Myo10, and Rasgrp3 mRNAs — which is observed in severe SMA mice (Supplemental Figure 1B) (36) — was not detected in Smn2B/– mice (Supplemental Figure 3B). Nevertheless, the disruption of Stasimon splicing is conserved across models and provides evidence for defective U12 splicing in Smn2B/– mice. Next, we injected Smn2B/– mice with AAV9 carrying either U11/U12 or U11/U12/U4atac minor snRNAs (~1 × 1011 vgp) at P2 by ICV delivery. The median survival for untreated Smn2B/– mice was 31 days, whereas the median survival of Smn2B/– mice treated with U11/U12 or U11/U12/U4atac snRNAs was substantially extended to 39 and 41 days, respectively (Figure 5E). Moreover, the introduction of these minor snRNAs improved the average weight gain when compared with the untreated SMA animals from 40 days of age onward (Figure 5F). Together, these results indicated that minor snRNAs contribute to the disease phenotype in a milder SMA mouse model.
Minor snRNAs do not prevent neuromuscular pathology in severe SMA mice.
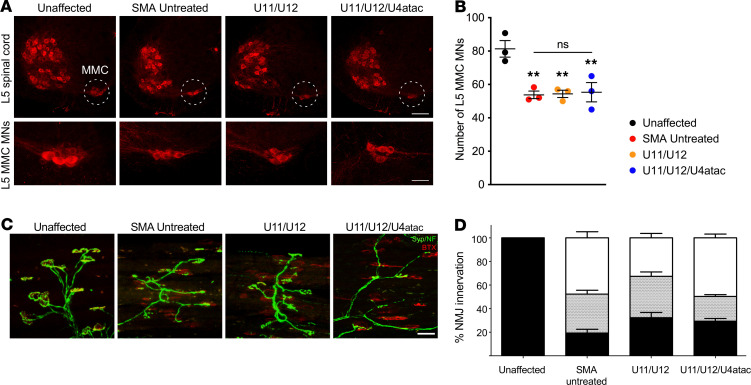
To identify the cellular basis for the improvement of AAV9-mediated delivery of minor snRNAs on motor behavior of SMA mice, we sought to investigate the effects on several morphological parameters of the motor circuit that are strongly affected by SMN deficiency and contribute to motor dysfunction in the SMNΔ7 severe model of SMA (60, 61). First, we focused on the degeneration of vulnerable SMA motor neurons, a hallmark of the disease in both patients and mouse models. To do so, we delivered AAV9-U11/U12 and AAV9-U11/U12/U4atac in SMA mice by ICV injection at P1 and quantified the total number of vulnerable motor neurons that reside in the medial motor column (MMC) of the lumbar L5 segment of the spinal cord at P11 using ChAT immunohistochemistry and confocal microscopy as previously described (41) (Figure 6). Consistent with previous studies (41, 42, 53), we found that approximately 40% of L5 MMC motor neurons are lost in untreated SMA mice relative to unaffected controls at P11 (Figure 6A). Moreover, we found that neither AAV9-U11/U12 nor AAV9-U11/U12/U4atac was able to prevent motor neuron loss in SMA mice (Figure 6B). We recently showed that Stasimon dysfunction contributes to the degeneration of SMA motor neurons by promoting p38 MAPK-mediated phosphorylation of p53 (37). It is possible that a partial degree of correction of Stasimon’s splicing by minor snRNA expression in motor neurons was insufficient to prevent induction of the neurodegenerative process.
Figure 6. AAV9-mediated delivery of minor snRNA genes does not prevent motor neuron degeneration and neuromuscular junction pathology in SMA mice.
(A) ChAT immunostaining of L5 spinal segments at P11 from unaffected control mice and SMA mice that were either untreated or ICV injected with AAV9-U11/U12 and AAV9-U11/U12/U4atac as indicated. In the top panels, dotted circles identify MMC motor neuron pools. Scale bar: 200 μm. Bottom panels display representative images of L5 MMC motor neuron pools from the same treatment groups at higher magnification. Scale bar: 50 μm. (B) AAV9-mediated delivery of minor snRNA genes does not prevent motor neuron degeneration. Total number of L5 MMC motor neurons from the same groups as in A at P11. The scatter plots show individual data points, mean and SEM from 3 mice per group. Statistics were performed with 1-way ANOVA with Tukey’s post hoc test. **P < 0.01; ns, no significance. (C) Images of NMJ innervation in the vulnerable longissimus capitis muscle collected at P12 from unaffected control mice and SMA mice either untreated or injected with AAV9-U11/U12 and AAV9-U11/U12/U4atac. Immunochemistry labeling is as follows: α-bungarotoxin (α-BTX, shown in red), synaptophysin and neurofilament (Syn/NF, shown in green). Scale bar: 40 μm. (D) No improvement in NMJ pathology in SMNΔ7 mice treated with U11, U12, and U4atac minor splicing snRNAs. Bar graph representing the percentage of fully innervated (black), partially innervated (gray stippled), and denervated (white) NMJs from the same treatment groups. NMJ analysis was done by blinded counts for a minimum of 4 fields of view per muscle type from n = 3 animals per treatment. Statistical analysis showed no significant difference between treated and untreated SMA groups using 1-way ANOVA with Tukey’s post hoc test (P = 0.50).
Neuromuscular junction (NMJ) pathology has been well characterized in the SMNΔ7 mouse model of SMA, demonstrating that there are subsets of muscle groups particularly vulnerable to SMN deficiency (62–64). In order to examine the effects of our treatments on NMJ phenotypes, mice of each experimental group were perfused on P12, and the longissimus capitis, a vulnerable muscle implicated in the control of head stability and movement, was isolated and analyzed (Figure 6C). We found that the longissimus capitis from SMA mice injected with AAV9-U11/U12 or AAV9-U11/U12/U4atac showed no significant improvement in the proportion of denervated and partially innervated NMJs when compared with the same muscle from untreated SMA controls (Figure 6D). Furthermore, we examined additional aspects of NMJ development by analyzing end plate morphology in the sternocleidomastoid muscle, which previous studies showed to be less mature in SMA mice than in the healthy unaffected animals (65). During normal neuromuscular development, NMJs of healthy animals become progressively more complex, increase in size, and develop from a plaque-like structure to a perforated, pretzel-like shape, whereas the NMJs in SMA mice remain undersized and plaque-like with reduced numbers of acetylcholine receptors (62, 63). We found that at P12 about 85% of the NMJs in SMA mice treated with AAV9-U11/U12 or AAV9-U11/U12/U4atac were developmentally delayed and displayed a plaque-like morphology (defined by the lack of perforations in the acetylcholine receptor clusters) similar to untreated SMA mice. In contrast, only about 20% of the NMJs in the healthy cohorts lacked perforations (Supplemental Figure 4). Overall, our analysis of NMJ morphology and end plate maturity showed that minor snRNA gene delivery failed to ameliorate neuromuscular pathology in SMA mice.
Minor snRNAs rescue the loss of proprioceptive synapses on motor neurons of SMA mice.
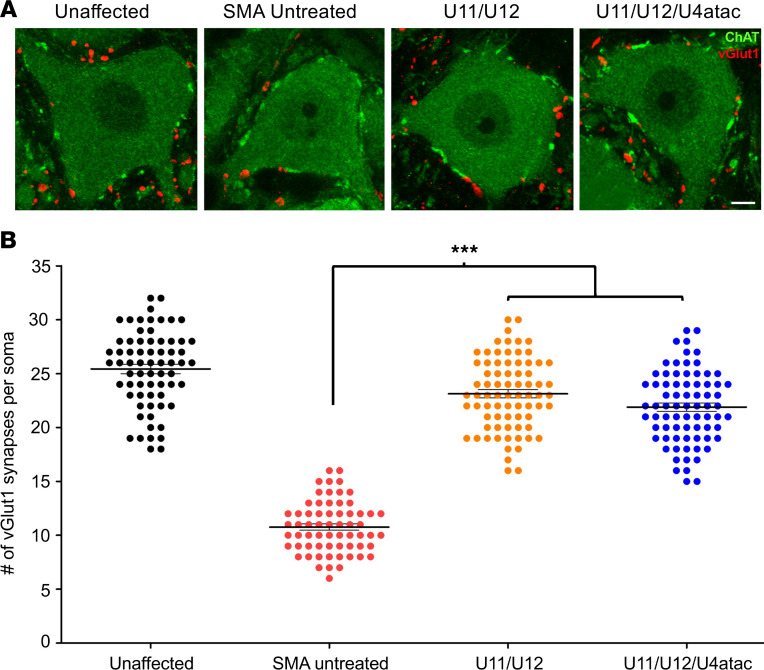
Afferent innervation of motor neurons is critical for motor control, and this element of the motor circuit is severely disrupted in SMA (60, 61). In particular, the loss of VGluT1+ synapses on SMA motor neurons has been well characterized both morphologically and functionally in SMNΔ7 mice (42, 43, 62, 66, 67). Additionally, it is well established that all the VGluT1+ synapses onto somata and dendrites of motor neurons are exclusively derived from proprioceptive afferent neurons (68–70). To determine the effect of minor snRNAs on the loss of these excitatory sensory synapses, we quantified VGluT1+ proprioceptive boutons juxtaposed to the somata of ChAT+ motor neurons located in the L3–L5 lumbar segments of the spinal cord of SMA mice at P12 (Figure 7A). Consistent with previous studies, we found that untreated SMA animals had a strong reduction in the number of VGluT1+ synapses onto motor neuron somata compared with unaffected controls (Figure 7B). Remarkably, we observed a robust rescue in the number of proprioceptive synapses impinging on motor neurons of SMA mice treated with AAV9-U11/U12 or AAV9-U11/U12/U4atac compared with untreated SMA controls (Figure 7, A and B). Furthermore, our results indicated that AAV9-mediated expression of either U11/U12 or U11/U12/U4atac snRNAs was equally effective in preventing the stripping of proprioceptive SMA synapses (Figure 7B). Collectively, these results implicate minor snRNA dysfunction in the process underlying the loss of sensory synapses on motor neurons of SMA mice.
Figure 7. Loss of proprioceptive synapses onto motor neurons is rescued by minor snRNA gene delivery in SMA mice.
(A) Immunostaining showing ChAT+ motor neurons (green) and VGluT1+ proprioceptive synapses (red) from the ventral horn of the lumbar spinal cord (L3–5) of unaffected control mice and SMA mice either untreated or injected with AAV9-U11/U12 and AAV9-U11/U12/U4atac. Age at tissue harvest was P12. Scale bar: 20 μm. (B) The scatter plot diagram shows the number of glutamatergic synapses impinging on motor neuron somata from the same experimental groups as in A. Each individual dot represents the number of VGluT1+ synapses per motor neuron somata. For each treatment group and control groups, n = 4–5 mice and ~15 motor neurons per animal. Both treatment groups show significant preservation of central proprioceptive synapses compared with the untreated SMA mice. One-way ANOVA was applied where significance is represented by ***P ≤ 0.001.
Discussion
It is increasingly evident that SMN has multiple functions in RNA metabolism and plays a central role in gene regulation (71, 72). However, the specific contribution of individual SMN-dependent RNA pathways to SMA pathology is only beginning to emerge. Identifying which RNA pathways and downstream genes — among many potentially dysregulated events — are directly relevant to the disease phenotype is critical to elucidate molecular mechanisms and may help uncover therapeutic approaches distinct from SMN upregulation. This study aimed to address these issues by investigating the contribution of reduced assembly of minor snRNPs and dysregulated U12 splicing to SMA pathology in mouse models of the disease, which provide the best recapitulation of the human condition both genetically and phenotypically (26, 60). We utilized AAV9-mediated gene delivery of minor snRNAs in SMA mice to demonstrate functional improvement in the splicing of U12 introns as well as moderate but significant phenotypic benefit, including extension of survival and enhanced motor function independent of any alteration of SMN levels. Importantly, these effects were associated with a robust rescue of the loss of proprioceptive sensory synapses impinging on SMA motor neurons, an early signature of motor dysfunction in mouse models of the disease (42, 43, 62). Taken together, these findings reveal that U12 splicing dysfunction contributes directly to the loss of synapses and altered sensory-motor connectivity induced by SMN deficiency, providing new insight into the RNA-mediated mechanisms of SMA pathogenesis.
This work was based on the premise that phenotypic benefit from selective restoration of individual RNA pathways regulated by SMN in a mouse model of the disease would provide conclusive evidence of their involvement in the etiology of SMA. Additionally, correction of molecular events that are directly dependent on the biological function(s) of SMN would provide confidence that the most proximal steps in the pathogenic cascade are being targeted. To date, however, none of the RNA pathways implicated in SMA pathology has been evaluated for its specific contribution to the disease phenotype in mouse models, partly because of the lack of adequate tools for uncoupling distinct SMN functions and testing their respective role in disease pathogenesis. Here, we addressed this issue by focusing on the well-established activity of SMN in snRNP assembly and in particular on the biogenesis of minor snRNPs of the U12 spliceosome, which are prominently reduced in SMA mouse models (19, 33, 34). We developed a means to selectively enhance U12 splicing in the setting of SMN deficiency through virus-mediated minor snRNA gene delivery. Our approach is based on the reasoning that overexpression of minor snRNAs, which are about 100-fold less abundant than major snRNAs (2, 3), could tilt the equilibrium in favor of their increased incorporation into functional snRNPs even under conditions in which SMN-mediated assembly is severely reduced. As proof of concept for the validity of this approach, we show specific elevation of minor snRNA expression levels following lentiviral transduction of U11, U12, and U4atac genes in various cultured mammalian cells with either normal or reduced SMN levels (Figure 1 and Figure 2B). Using a previously established cell model system in NIH 3T3 fibroblasts (21), we further show that this approach was effective not only in counteracting the reduction in minor snRNAs but also in correcting U12-dependent splicing deficits induced by SMN deficiency (Figure 2C), thereby validating minor snRNA gene delivery as a suitable approach to selectively improve the functionality of this SMN-dependent RNA pathway.
Building on the in vitro results with mammalian cells, we went on to show that AAV9-mediated delivery of minor snRNA genes in SMA mice improves defective U12 splicing of the Stasimon mRNA in the spinal cord and lumbar DRGs of severe SMA mice (Figure 3B and Supplemental Figure 2), which we previously associated with sensory-motor circuit dysfunction in several animal models of SMA (21, 37). The effects are specific and unrelated to SMN induction because other SMN-regulated RNA events, such as histone mRNA 3′ end processing (52) and p53-mediated Cdkn1a mRNA upregulation (41, 53), were not corrected (Figure 3B), and SMN levels did not change (Figure 4). Interestingly, the AAV9 vector expressing U11 and U12 was as effective as the one harboring U11, U12, and U4atac on Stasimon pre-mRNA splicing in vivo, suggesting that the SMN-dependent reduction of U11 and U12 snRNAs has a greater functional impact than U4atac. U11 and U12 snRNPs associate with U12 introns as a preformed di-snRNP complex that simultaneously binds the 5′ splice site and branch point sequence (73) and contribute to exon definition (15). Defects in these processes because of reduced availability of U11/U12 di-snRNP are consistent with the observed minor splicing abnormalities induced by SMN deficiency resulting from disruption of the initial recognition of U12 introns.
Our results point to a direct contribution of U12 splicing dysfunction to SMA pathology in mouse models of the disease with varying degrees of severity as we found that minor snRNA gene delivery ameliorated disease phenotypes of severe and milder SMA mice (Figure 5). There was a modest but significant extension of survival and weight gain from birth to peak as well as a transient improvement of motor function in severe SMA mice. Increased survival and enhanced weight gain were also found in a milder model of SMA. Consistent with these results, we show that SMN deficiency decreases the levels of U11 and U12 snRNAs as well as disrupts U12 splicing of Stasimon mRNA in the spinal cord of milder SMA mice (Supplemental Figure 3), providing direct evidence for dysregulation of this RNA pathway that was previously investigated only in severe mouse models of SMA (19, 21, 35, 36, 39). Moreover, overall similar phenotypic benefit was observed in SMA mice injected with either AAV9-U11/U12 or AAV9-U11/U12/U4atac, as is the case for Stasimon splicing correction. In agreement with other studies in which specific aspects of SMA pathology have been corrected independent of SMN upregulation (41, 53, 74), it is not surprising that the degree of phenotypic benefit associated with minor snRNA delivery is relatively limited and wears out as disease pathogenesis progresses toward the end stage. This likely reflects the multifactorial nature of SMA pathology (75, 76) and the involvement of other SMN-dependent RNA pathways beyond U12 splicing (71, 72). Additionally, our approach relies on the low levels of functional SMN present in SMA that likely limits the amount of minor snRNPs that can assemble following AAV9 treatment. Therefore, it is also possible that our results underestimate the potential contribution of U12 splicing dysfunction to SMA pathology because of incomplete correction of this RNA pathway as compared with what is accomplished with SMN restoration.
We highlight a functional requirement of minor snRNPs for the maintenance of synaptic integrity in mammalian sensory-motor circuits, the disruption of which contributes to SMA. Previous studies have characterized several defects induced by SMN deficiency in the motor circuit of mouse models that contribute to neuromuscular pathology and are clinically relevant features of SMA (60, 61), including dysfunction and loss of specific synapses as well as death of select motor neuron pools. Here we looked at their potential link with U12 splicing dysfunction and found that it specifically contributed to disrupt sensory-motor synaptic connectivity in severe SMA mice (Figure 6 and Figure 7). The reduction of VGluT1+ proprioceptive synapses on spinal motor neurons is an early pathogenic event in SMA mice (42, 62). Moreover, dysfunction and loss of sensory synapses is a cell-autonomous process caused by SMN deficiency within proprioceptive neurons that reduces the excitatory drive on SMA motor neurons (43), resulting in motor neuron hyperexcitability, altered firing, and compromised muscle contraction. Remarkably, we found that minor snRNA gene delivery in SMA mice robustly rescued the number of VGluT1+ synapses on the somata of lumbar motor neurons relative to untreated SMA mice, thereby linking U12 splicing dysfunction to motor neuron deafferentation (Figure 7). It is plausible that preservation of proprioceptive synapses contributes to benefits in motor function of severe SMA mice and is mediated by the effects of minor snRNAs on correcting splicing of Stasimon — an ER-resident transmembrane protein implicated in the regulation of autophagy (77–80) — in proprioceptive DRG neurons. This interpretation is consistent with early studies in which we identified Stasimon as a U12 intron–containing gene essential for sensory-motor circuit function that contributes to SMN-dependent neuronal phenotypes in a Drosophila model of SMA (21) as well as our recent results with AAV9-mediated Stasimon restoration in SMA mice (37). Additionally, the dysregulation of Stasimon splicing that we report here in milder SMA mice, in which the loss of VGluT1+ synapses onto motor neurons has previously been documented (81), suggests that Stasimon dysfunction may contribute to deafferentation across mouse models of the disease. In contrast to minor snRNA gene delivery, however, AAV9-Stasimon overexpression does not increase survival or weight gain in severe SMA mice (37). It is therefore likely that dysregulation of U12 intron–containing genes other than Stasimon might be involved in distinct aspects of SMA pathology as recently suggested (36). Interestingly, we tested a handful of other U12 intron–containing genes dysregulated in SMA mice but found that, in contrast to Stasimon mRNA, they were corrected neither by minor snRNA gene delivery nor by AAV9-SMN. These results suggest that, while Stasimon is misspliced in disease-relevant neurons (21), these other changes occur in other spinal cells that are not efficiently transduced by AAV9 and are unlikely to contribute to SMA pathology. Future studies are needed to reveal the identity and potential role in disease of other U12 intron–containing genes regulated by SMN.
In summary, beyond mutations genetically linking minor snRNPs to human disease (9), U12 splicing dysfunction has been implicated in the pathogenesis of motor neuron diseases, such as SMA (19–21, 35, 36) and ALS (22–24), but direct evidence for a functional involvement of this pathway in the disease process has been missing. Through selective enhancement of minor snRNA function, our study supports the conclusion that defects in U12 splicing play a role in SMA pathogenesis and expands the range of human disorders associated with dysregulation of this RNA processing pathway.
Methods
Supplemental Methods are available online with this article.
Study approval.
Research animals were housed and treated in accordance with the guidelines of the Animal Care and Use Committees at the University of Missouri and at Columbia University following the regulations established by the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
EYO, PFY, LP, and CLL conceived the study; PFY, EYO, and CLL designed the methodology; PFY, EYO, FL, MVA, ZF, KKYL, CPK, LP, and CLL investigated; EYO, PFY, FL, MVA, and KKYL validated results; EYO and MVA conducted formal analysis; EYO and PFY wrote the original draft; EYO, PFY, MVA, LP, and CLL reviewed and edited the draft; CPK, LP, and CLL provided resources; and CPK, LP, and CLL supervised the study.
Supplementary Material
Acknowledgments
This work was supported by grants from FightSMA (to CLL), the Gwendolyn Strong Foundation (to CLL), and NIH/National Institute of Neurological Disorders and Stroke R21NS106490 (to CLL), R21NS077038 (to LP), and R01NS102451 (to LP). EYO, PFY, and CLL would like to thank John Marston for technical assistance with the mouse colony and animal husbandry.
Version 1. 06/09/2020
In-Press Preview
Version 2. 06/18/2020
Electronic publication
Footnotes
Conflict of interest: CLL is the chief scientific officer of Shift Pharmaceuticals.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e130574.https://doi.org/10.1172/jci.insight.130574.
Contributor Information
Erkan Y. Osman, Email: erikosman@gmail.com.
Meaghan Van Alstyne, Email: mev2138@cumc.columbia.edu.
Pei-Fen Yen, Email: buolia@gmail.com.
Francesco Lotti, Email: fl2219@cumc.columbia.edu.
Zhihua Feng, Email: zfeng@usc.edu.
Karen K.Y. Ling, Email: kling@ionisph.com.
Chien-Ping Ko, Email: cko@usc.edu.
Livio Pellizzoni, Email: lp2284@cumc.columbia.edu.
Christian L. Lorson, Email: lorsonc@missouri.edu.
References
- 1.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4(12):960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 3.Montzka KA, Steitz JA. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci U S A. 1988;85(23):8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34(14):3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35(Database issue):D110–D115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otake LR, Scamborova P, Hashimoto C, Steitz JA. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol Cell. 2002;9(2):439–446. doi: 10.1016/S1097-2765(02)00441-0. [DOI] [PubMed] [Google Scholar]
- 7.Baumgartner M, et al. Minor spliceosome inactivation causes microcephaly, owing to cell cycle defects and death of self-amplifying radial glial cells. Development. 2018;145(17):dev166322. doi: 10.1242/dev.166322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markmiller S, et al. Minor class splicing shapes the zebrafish transcriptome during development. Proc Natl Acad Sci U S A. 2014;111(8):3062–3067. doi: 10.1073/pnas.1305536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma B, Akinyi MV, Norppa AJ, Frilander MJ. Minor spliceosome and disease. Semin Cell Dev Biol. 2018;79:103–112. doi: 10.1016/j.semcdb.2017.09.036. [DOI] [PubMed] [Google Scholar]
- 10.He H, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332(6026):238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edery P, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332(6026):240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 12.Merico D, et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman syndrome by disrupting minor intron splicing. Nat Commun. 2015;6:8718. doi: 10.1038/ncomms9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farach LS, et al. The expanding phenotype of RNU4ATAC pathogenic variants to Lowry Wood syndrome. Am J Med Genet A. 2018;176(2):465–469. doi: 10.1002/ajmg.a.38581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, Krainer AR. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA. 1997;3(6):586–601. [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Krainer AR. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol. 1999;19(5):3225–3236. doi: 10.1128/MCB.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coady TH, Lorson CL. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip Rev RNA. 2011;2(4):546–564. doi: 10.1002/wrna.76. [DOI] [PubMed] [Google Scholar]
- 17.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2(6):773–785. doi: 10.1016/S1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 18.Elsaid MF, et al. Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann Neurol. 2017;81(1):68–78. doi: 10.1002/ana.24826. [DOI] [PubMed] [Google Scholar]
- 19.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2(9):e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonné R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet. 2011;20(4):641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 21.Lotti F, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151(2):440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onodera O, Ishihara T, Shiga A, Ariizumi Y, Yokoseki A, Nishizawa M. Minor splicing pathway is not minor any more: implications for the pathogenesis of motor neuron diseases. Neuropathology. 2014;34(1):99–107. doi: 10.1111/neup.12070. [DOI] [PubMed] [Google Scholar]
- 23.Ishihara T, et al. Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22(20):4136–4147. doi: 10.1093/hmg/ddt262. [DOI] [PubMed] [Google Scholar]
- 24.Reber S, et al. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 2016;35(14):1504–1521. doi: 10.15252/embj.201593791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuiji H, et al. Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol Med. 2013;5(2):221–234. doi: 10.1002/emmm.201202303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10(8):597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tisdale S, Pellizzoni L. Disease mechanisms and therapeutic approaches in spinal muscular atrophy. J Neurosci. 2015;35(23):8691–8700. doi: 10.1523/JNEUROSCI.0417-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 29.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582(14):1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8(4):340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meister G, Eggert C, Bühler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11(24):1990–1994. doi: 10.1016/S0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 32.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298(5599):1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 33.Workman E, et al. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet. 2009;18(12):2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133(4):585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jangi M, et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci U S A. 2017;114(12):E2347–E2356. doi: 10.1073/pnas.1613181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doktor TK, et al. RNA-sequencing of a mouse-model of spinal muscular atrophy reveals tissue-wide changes in splicing of U12-dependent introns. Nucleic Acids Res. 2017;45(1):395–416. doi: 10.1093/nar/gkw731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon CM, et al. Stasimon contributes to the loss of sensory synapses and motor neuron death in a mouse model of spinal muscular atrophy. Cell Rep. 2019;29(12):3885–3901.e5. doi: 10.1016/j.celrep.2019.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler C, et al. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19(19):2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruggiu M, et al. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol Cell Biol. 2012;32(1):126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, et al. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci U S A. 2013;110(48):19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Alstyne M, Simon CM, Sardi SP, Shihabuddin LS, Mentis GZ, Pellizzoni L. Dysregulation of Mdm2 and Mdm4 alternative splicing underlies motor neuron death in spinal muscular atrophy. Genes Dev. 2018;32(15–16):1045–1059. doi: 10.1101/gad.316059.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mentis GZ, et al. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69(3):453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fletcher EV, et al. Reduced sensory synaptic excitation impairs motor neuron function via Kv2.1 in spinal muscular atrophy. Nat Neurosci. 2017;20(7):905–916. doi: 10.1038/nn.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyd DC, Pombo A, Murphy S. Interaction of proteins with promoter elements of the human U2 snRNA genes in vivo. Gene. 2003;315:103–112. doi: 10.1016/S0378-1119(03)00717-0. [DOI] [PubMed] [Google Scholar]
- 45.Stabley DL, et al. Establishing a reference dataset for the authentication of spinal muscular atrophy cell lines using STR profiling and digital PCR. Neuromuscul Disord. 2017;27(5):439–446. doi: 10.1016/j.nmd.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 47.Coady TH, Baughan TD, Shababi M, Passini MA, Lorson CL. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS One. 2008;3(10):e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osman EY, Yen PF, Lorson CL. Bifunctional RNAs targeting the intronic splicing silencer N1 increase SMN levels and reduce disease severity in an animal model of spinal muscular atrophy. Mol Ther. 2012;20(1):119–126. doi: 10.1038/mt.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passini MA, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3(72):72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, et al. Pharmacokinetics, pharmacodynamics, and efficacy of a small-molecule SMN2 splicing modifier in mouse models of spinal muscular atrophy. Hum Mol Genet. 2016;25(10):1885–1899. doi: 10.1093/hmg/ddw062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mende M, et al. Sensory-derived glutamate regulates presynaptic inhibitory terminals in mouse spinal cord. Neuron. 2016;90(6):1189–1202. doi: 10.1016/j.neuron.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tisdale S, et al. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3’-end formation of histone mRNAs. Cell Rep. 2013;5(5):1187–1195. doi: 10.1016/j.celrep.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon CM, et al. Converging mechanisms of p53 activation drive motor neuron degeneration in spinal muscular atrophy. Cell Rep. 2017;21(13):3767–3780. doi: 10.1016/j.celrep.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foust KD, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28(3):271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Robbins KL, Glascock JJ, Osman EY, Miller MR, Lorson CL. Defining the therapeutic window in a severe animal model of spinal muscular atrophy. Hum Mol Genet. 2014;23(17):4559–4568. doi: 10.1093/hmg/ddu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer K, et al. Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Mol Ther. 2015;23(3):477–487. doi: 10.1038/mt.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butchbach ME, Edwards JD, Burghes AH. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis. 2007;27(2):207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowerman M, Murray LM, Beauvais A, Pinheiro B, Kothary R. A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology. Neuromuscul Disord. 2012;22(3):263–276. doi: 10.1016/j.nmd.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 59.Eshraghi M, McFall E, Gibeault S, Kothary R. Effect of genetic background on the phenotype of the Smn2B/- mouse model of spinal muscular atrophy. Hum Mol Genet. 2016;25(20):4494–4506. doi: 10.1093/hmg/ddw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Alstyne M, Pellizzoni L. Advances in modeling and treating spinal muscular atrophy. Curr Opin Neurol. 2016;29(5):549–556. doi: 10.1097/WCO.0000000000000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shorrock HK, Gillingwater TH, Groen EJN. Molecular mechanisms underlying sensory-motor circuit dysfunction in SMA. Front Mol Neurosci. 2019;12:59. doi: 10.3389/fnmol.2019.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One. 2010;5(11):e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ling KKY, Gibbs RM, Feng Z, Ko CP. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2012;21(1):185–195. doi: 10.1093/hmg/ddr453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kline RA, et al. Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases. PLoS Genet. 2017;13(3):e1006680. doi: 10.1371/journal.pgen.1006680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee YI, Mikesh M, Smith I, Rimer M, Thompson W. Muscles in a mouse model of spinal muscular atrophy show profound defects in neuromuscular development even in the absence of failure in neuromuscular transmission or loss of motor neurons. Dev Biol. 2011;356(2):432–444. doi: 10.1016/j.ydbio.2011.05.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thirumalai V, Behrend RM, Birineni S, Liu W, Blivis D, O’Donovan MJ. Preservation of VGLUT1 synapses on ventral calbindin-immunoreactive interneurons and normal locomotor function in a mouse model of spinal muscular atrophy. J Neurophysiol. 2013;109(3):702–710. doi: 10.1152/jn.00601.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schultz AJ, Rotterman TM, Dwarakanath A, Alvarez FJ. VGLUT1 synapses and P-boutons on regenerating motoneurons after nerve crush. J Comp Neurol. 2017;525(13):2876–2889. doi: 10.1002/cne.24244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alvarez FJ, Villalba RM, Zerda R, Schneider SP. Vesicular glutamate transporters in the spinal cord, with special reference to sensory primary afferent synapses. J Comp Neurol. 2004;472(3):257–280. doi: 10.1002/cne.20012. [DOI] [PubMed] [Google Scholar]
- 69.Rotterman TM, Nardelli P, Cope TC, Alvarez FJ. Normal distribution of VGLUT1 synapses on spinal motoneuron dendrites and their reorganization after nerve injury. J Neurosci. 2014;34(10):3475–3492. doi: 10.1523/JNEUROSCI.4768-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mentis GZ, Alvarez FJ, Shneider NA, Siembab VC, O’Donovan MJ. Mechanisms regulating the specificity and strength of muscle afferent inputs in the spinal cord. Ann N Y Acad Sci. 2010;1198:220–230. doi: 10.1111/j.1749-6632.2010.05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li DK, Tisdale S, Lotti F, Pellizzoni L. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol. 2014;32:22–29. doi: 10.1016/j.semcdb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donlin-Asp PG, Bassell GJ, Rossoll W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr Opin Neurobiol. 2016;39:53–61. doi: 10.1016/j.conb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 73.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5’ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13(7):851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ackermann B, et al. Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum Mol Genet. 2013;22(7):1328–1347. doi: 10.1093/hmg/dds540. [DOI] [PubMed] [Google Scholar]
- 75.Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19(1):40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 76.Shababi M, Lorson CL, Rudnik-Schoneborn SS. Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J Anat. 2014;224(1):15–28. doi: 10.1111/joa.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Alstyne M, Lotti F, Dal Mas A, Area-Gomez E, Pellizzoni L. Stasimon/Tmem41b localizes to mitochondria-associated ER membranes and is essential for mouse embryonic development. Biochem Biophys Res Commun. 2018;506(3):463–470. doi: 10.1016/j.bbrc.2018.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morita K, et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol. 2018;217(11):3817–3828. doi: 10.1083/jcb.201804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moretti F, et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 2018;19(9):e45889. doi: 10.15252/embr.201845889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shoemaker CJ, Huang TQ, Weir NR, Polyakov NJ, Schultz SW, Denic V. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019;17(4):e2007044. doi: 10.1371/journal.pbio.2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cerveró C, et al. Glial activation and central synapse loss, but not motoneuron degeneration, are prevented by the Sigma-1 receptor agonist PRE-084 in the Smn2B/- mouse model of spinal muscular atrophy. J Neuropathol Exp Neurol. 2018;77(7):577–597. doi: 10.1093/jnen/nly033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.