Abstract
Scleraxis is a basic helix-loop-helix transcription factor that plays a central role in promoting tenocyte proliferation and matrix synthesis during embryonic tendon development. However, the role of scleraxis in the growth and adaptation of adult tendons is not known. We hypothesized that scleraxis is required for tendon growth in response to mechanical loading and that scleraxis promotes the specification of progenitor cells into tenocytes. We conditionally deleted scleraxis in adult mice using a tamoxifen-inducible Cre-recombinase expressed from the Rosa26 locus (ScxΔ) and then induced tendon growth in Scx+ and ScxΔ adult mice via plantaris tendon mechanical overload. Compared with the WT Scx+ group, ScxΔ mice demonstrated blunted tendon growth. Transcriptional and proteomic analyses revealed significant reductions in cell proliferation, protein synthesis, and extracellular matrix genes and proteins. Our results indicate that scleraxis is required for mechanically stimulated adult tendon growth by causing the commitment of CD146+ pericytes into the tenogenic lineage and by promoting the initial expansion of newly committed tenocytes and the production of extracellular matrix proteins.
Keywords: Cell Biology, Stem cells
Keywords: Extracellular matrix, Pericytes
Scleraxis is required for adult tendon growth and plays an important role in differentiation of perivascular progenitor cells into tenocytes.
Introduction
Tendons are made up of an extracellular matrix (ECM) containing primarily type I collagen, as well as other collagens, elastin, and various proteoglycans (1). Tendon ECM is arranged in a hierarchical manner, with densely packed collagen fibers wrapped by layers of basement membrane (1). Tenocytes, or tendon fibroblasts, are the main cell type in tendons and are thought to be responsible for the production, organization, and maintenance of the tendon ECM (2). Tendons are surrounded by an outermost basement membrane called the epitenon, which provides blood and nerve supply to the tendon (1, 3). The organization of the ECM allows the tendon to properly transmit forces from muscle to bone and allow for locomotion and to respond to mechanical stimuli (4, 5). Mechanical loading can increase tendon cross-sectional area (CSA) up to 30% (6) and improve tendon mechanical properties (5, 7), but less is known about the cellular and molecular mechanisms behind tendon growth and adaptation in adult animals.
Scleraxis (Scx) is a basic helix-loop-helix (bHLH) transcription factor that is regulated by TGF-β signaling and is required for the proper embryonic development of tendons (8, 9). Scleraxis is the earliest detectible marker for differentiated tendon cells (10) and activates downstream tendon differentiation genes such as type I α1 chain collagen 1 (Col1a1), mohawk (Mkx), and tenomodulin (Tmnd) through its interaction with the bHLH transcription factor E47, among others (11–13). Scleraxis is expressed through birth and the early postnatal period, but beyond 3 months of age in mice, scleraxis expression in tendons is limited to the epitenon (6). Loss of scleraxis results in the loss or severe disruption of long tendons throughout the body (8). In adult animals, mechanical loading increases scleraxis expression and promotes the proliferation of scleraxis-expressing cells in the epitenon, which correlates to increased type I collagen levels and increased CSA of the tendon (6, 14, 15), while immobilization reduces scleraxis levels (16). Although scleraxis is required for proper embryonic tendon development, it is not known whether scleraxis is required for growth of tendons in adult animals.
Pericytes, or perivascular stem cells, make up a population of multipotent stem cells surrounding small blood vessels and are capable of differentiation into many types of mesenchymal cells, including fibroblasts (17). Tendon pericytes express the marker CD146 (Mcam) (15, 18, 19). Most of the tenocytes within tendons of adult animals appear to be terminally withdrawn from the cell cycle, with low rates of cell proliferation observed in homeostatic and mechanically loaded tendons (14). Since pericytes are thought to be a progenitor cell population in adult tendons (15, 18, 19), and scleraxis is required for the proper embryonic development of tendons, the objective of this study was to determine the role of scleraxis in adult tendon growth. We hypothesized that scleraxis is required for proper tendon growth in adult animals, and that scleraxis promotes the commitment of pericytes into the tendon lineage. To test this hypothesis, we conditionally deleted scleraxis in adult animals using Rosa26CreERT2/CreERT2 Scxfl/fl mice and induced tendon growth via mechanical overload of the plantaris tendon. Additionally, we performed a series of in vitro experiments with pericytes and tenocytes to gain further mechanistic insight into the observed structural and transcriptional changes in mechanically stimulated tendons that lack scleraxis.
Results
Overview.
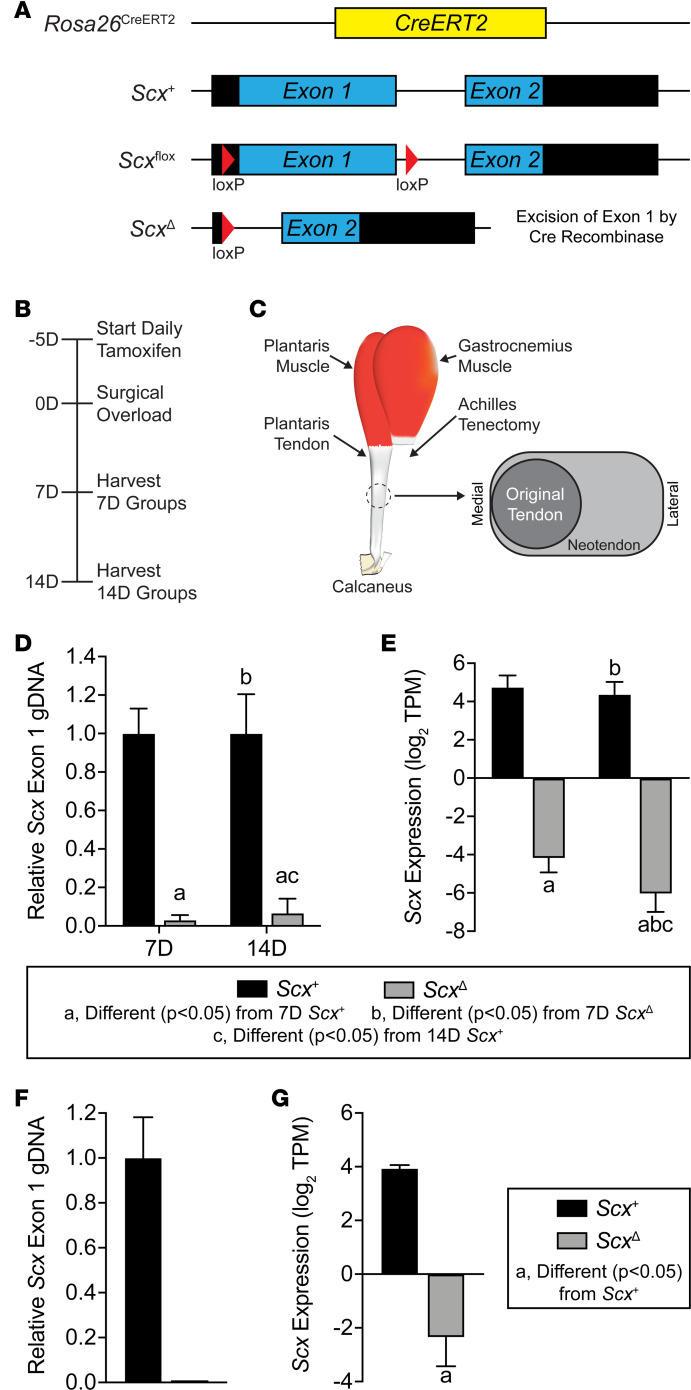
To study the role of scleraxis in adult tendon growth, we generated Rosa26CreERT2/CreERT2 Scxfl/fl mice to allow for the inactivation of scleraxis upon treatment with tamoxifen (referred to as ScxΔ mice), while Rosa26CreERT2/CreERT2 Scx+/+ mice maintain the expression of Scx after tamoxifen treatment (referred to as Scx+ mice). An overview of the Rosa26CreERT2, Scx+, Scxfl, and ScxΔ alleles is provided in Figure 1A. Following tamoxifen treatment, we induced a supraphysiological overload of the plantaris tendons of Scx+ and ScxΔ mice by synergist ablation and then analyzed tendons at either 7 days (7D) or 14D after surgery (Figure 1, B and C). We also isolated cells from Scx+ and ScxΔ mice to further mechanistically study the effects of scleraxis on the behavior of tenocytes in vitro. To verify efficient deletion of scleraxis, we isolated DNA and RNA from tendons of animals treated with tamoxifen, and we cultured cells treated with 4-hydroxytamoxifen (4HT). There was a 97% and 93% reduction in Scx exon 1 abundance in 7D and 14D plantaris tendons, respectively (Figure 1D). Cultured tenocytes of ScxΔ mice had a 99% reduction in exon 1 abundance compared with Scx+ tenocytes (Figure 1F). RNA sequencing (RNAseq) data also revealed a significant downregulation in Scx transcript levels in plantaris tendons and cultured cells (Figure 1, E and G). These data confirm the efficient conditional deletion of scleraxis in the models used in this study.
Figure 1. Experimental overview.
(A) Overview of the alleles used to perform targeted deletion of scleraxis in this study, including the constitutive CreERT2 (Rosa26CreERT2), scleraxis WT (Scx+), scleraxis floxed (Scxfl), and scleraxis loss-of-function (ScxΔ) alleles. (B) Timeline of tamoxifen treatment, synergist ablation/plantaris growth procedure, and tissue harvest. Tamoxifen was delivered on a daily basis beginning 5 days before surgery and continued through tissue harvest. (C) Overview of synergist ablation/plantaris growth procedure. A large segment of the Achilles tendon was carefully removed from the animal, resulting in compensatory hypertrophy of the synergist plantaris muscle and tendon. A neotendon area of new tendon extracellular matrix formed around the original tendon, growing in a lateral direction toward the area where the Achilles was previously located. (D and F) Relative genomic DNA (gDNA) abundance of exon 1 of scleraxis to exon 2, as measured using qPCR, in plantaris tendons of Scx+ and ScxΔ mice either 7D or 14D after synergist ablation (D) or in cultured Scx+ or ScxΔ tenocytes (F). (E and G) Expression of Scx, measured using RNAseq and quantified as transcripts per kilobase million (TPM) reads, in plantaris tendons of Scx+ and ScxΔ mice either 7D or 14D after synergist ablation (E), or in cultured Scx+ or ScxΔ tenocytes (G). Values are mean ± CV (D and F) or mean ± SD (E and G). (D and E) Differences between groups were tested using a 2-way ANOVA; a, significantly different (P < 0.05) from 7D Scx+; b, significantly different (P < 0.05) from 7D ScxΔ; c, significantly different (P < 0.05) from 14D Scx+. (F and G) Differences between groups were tested using a t test; a, significantly different (P < 0.05) from Scx+ tenocytes. N = 4 per group.
Scleraxis deletion impairs tendon growth.
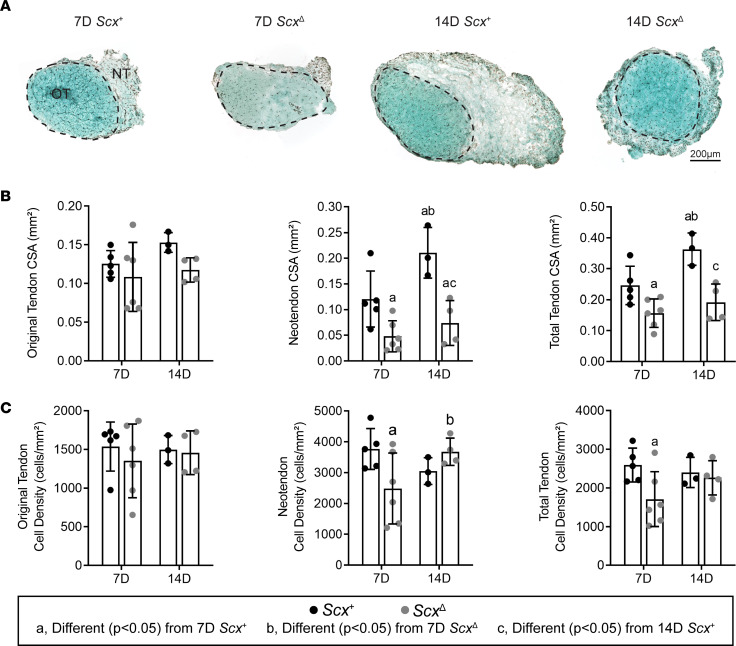
Following synergist ablation, Scx+ mice demonstrated an increase in plantaris tendon CSA through the formation of the neotendon matrix, while this response was blunted in ScxΔ mice (Figure 2A). The original tendon CSA and cell density did not change between groups (Figure 2, B and C). However, at 7D, ScxΔ mice had a 60% reduction in neotendon CSA, which resulted in a 36% smaller total tendon area (Figure 2B). At 14D, ScxΔ mice had a 65% decrease in neotendon CSA and a 47% decrease in overall tendon CSA compared with Scx+ mice (Figure 2B). For cell density, at 7D, there was a 34% decrease in the neotendon and total tendon of ScxΔ mice (Figure 2C). Collagen fibril size distributions were significantly different between the original tendon and neotendon of Scx+ and ScxΔ mice at 7D and 14D, with ScxΔ mice generally demonstrating a greater proportion of larger collagen fibrils (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.138295DS1).
Figure 2. Effect of scleraxis deletion on tendon growth.
(A) Representative Fast Green–stained cross-section histology from Scx+ and ScxΔ mice at either 7D or 14D after synergist ablation/plantaris growth procedure, demonstrating general morphology and cell density. The original tendon (OT) and neotendon (NT) are indicated by the hashed line. Scale bar: 200 μm. (B and C) Quantification of original tendon, neotendon, and total tendon cross-sectional area (CSA) (B) and cell density (C). Values are mean ± SD. Differences between groups were tested using a 2-way ANOVA; a, significantly different (P < 0.05) from 7D Scx+; b, significantly different (P < 0.05) from 7D ScxΔ; c, significantly different (P < 0.05) from 14D Scx+. N ≥ 3 mice per group.
Scleraxis deletion increases pericyte density.
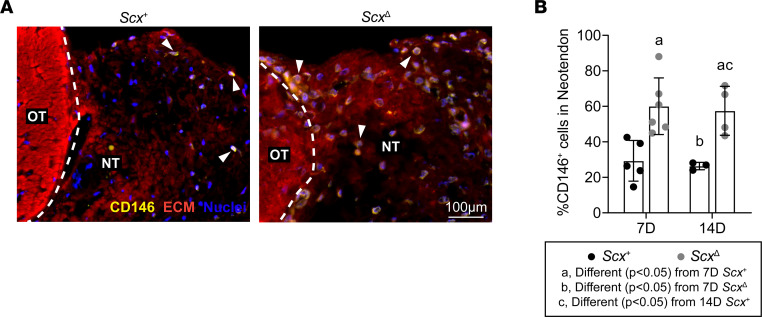
Using IHC, we measured the abundance of CD146+ pericytes within the neotendon of mechanically overloaded plantaris tendons (Figure 3A). Compared with ScxΔ mice, there was an approximate 2-fold increase in the percentage of pericytes in the neotendons of Scx+ mice at each time point (Figure 3B).
Figure 3. Effect of scleraxis deletion on pericyte density.
(A) Representative immunofluorescence histology demonstrating the presence of CD146+ pericytes in tendons of Scx+ and ScxΔ mice 14D after synergist ablation/plantaris growth procedure. The original tendon (OT) and neotendon (NT) are indicated by the hashed line. CD146, yellow; extracellular matrix (ECM), red; nuclei, blue. Scale bar: 100 μm. White arrowheads indicate the presence of CD146+ pericytes. (B) Quantification of the CD146+ pericytes as a percentage of total cells in the neotendon in Scx+ and ScxΔ mice at 7D or 14D. Values are mean ± SD. Differences between groups were tested using a 2-way ANOVA; a, significantly different (P < 0.05) from 7D Scx+; b, significantly different (P < 0.05) from 7D ScxΔ; c, significantly different (P < 0.05) from 14D Scx+. N ≥ 3 mice per group.
Scleraxis deletion impacts the proteome of tendons 14D after synergist ablation.
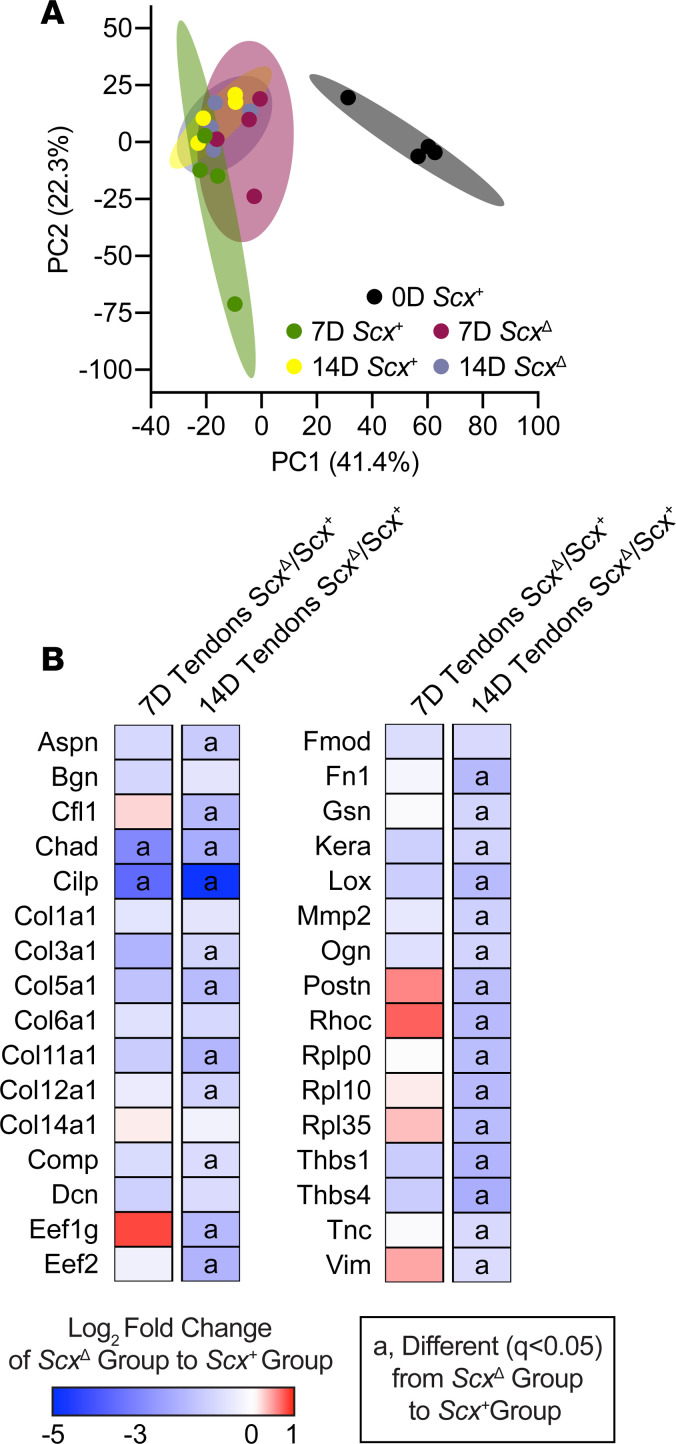
As we observed noticeable differences between the size of tendons from mechanically loaded Scx+ and ScxΔ mice, we performed mass spectrometry–based (MS-based) label-free proteomics to look at changes in tendon matrix composition. Principal component analyses (PCA) demonstrated differences between overloaded groups and the control nonoverloaded group, as well as some nonoverlapping areas between the Scx+ and ScxΔ groups (Figure 4A). In total, 313 proteins out of the 453 that were detected were found to be significantly different between Scx+ and ScxΔ mice for at least 1 time point, with 298 of these proteins being significantly different at 14D. We report select proteins in Figure 4B, with the entire data set available in Supplemental Table 1. At 7D, the matrix proteins chondroadherin (Chad) and cartilage intermediate layer protein (Cilp) were reduced in ScxΔ tendons, which was also noted at 14D. Additionally, at 14D, several collagens, including Col3a1, Col5a1, Col11a1, Col12a1, were reduced in ScxΔ tendons compared with Scx+ tendons. Proteins involved in ECM production, remodeling, and mechanotransduction — such as asporin (Aspn), fibronectin (Fn1), lysyl oxidase (Lox), matrix metalloproteinase 2 (Mmp2), osteoglycin (Ogn), thrombospondins 1 and 4 (Thbs1, Thbs4), and tenascin C (Tnc) — were lower in tendons of ScxΔ mice (Figure 4B). Ribosomal proteins Rplp0, Rpl10, and Rpl35; eukaryotic translation elongation factors Eef1g and Eef2; and mechanotransduction proteins cofilin 1 (Cfl1), gelsolin (Gsn), Ras homolog family member C (Rhoc), and vimentin (Vim) were also less abundant in ScxΔ tendons at 14D (Figure 4B).
Figure 4. Effect of scleraxis deletion on the proteome of tendons.
(A) Principal component analysis (PCA) of mass spectroscopy–based proteomics data of tendons from control nonoverloaded (0D) Scx+ tendons, as well as from Scx+ and ScxΔ mice at 7D or 14D after synergist ablation/plantaris growth procedure. (B) Heatmaps demonstrating the log2 fold change in selected proteins between Scx+ and ScxΔ tendons at 7D and 14D. Differences between groups were tested using FDR-corrected t tests; a, different (q < 0.05) between Scx+ and ScxΔ groups at a given time point. N = 4 mice per group.
Scleraxis deletion impacts the transcriptome of tendons and cultured tenocytes.
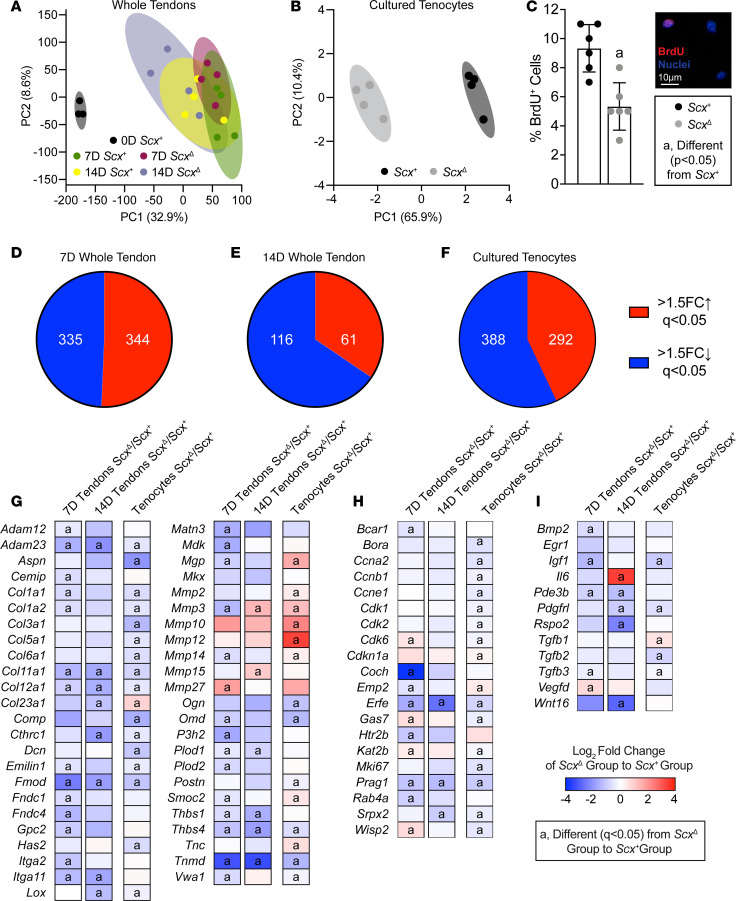
We then performed RNAseq in order to analyze transcriptional changes in overloaded plantaris tendons from Scx+ and ScxΔ mice, and in tenocytes cultured from tail tendons. Tail tendons were selected instead of plantaris tendons, as the size is much larger, allowing us to obtain high numbers of low-passage cells for experiments. PCA demonstrated both unique and overlapping regions for tendons of Scx+ and ScxΔ mice at 7D and 14D, and all 4 groups were different from control, nonoverloaded tendons (Figure 5A). More divergence in genotypes was observed in cultured tenocytes (Figure 5B). Seven days and 14D after synergist ablation, there were 679 and 177 transcripts, respectively, that were significantly different and at least 50% differentially regulated (Figure 5, D and E). In vitro, there were 680 genes that were significantly different between Scx+ and ScxΔ tenocytes (Figure 5F).
Figure 5. Effect of scleraxis deletion on transcriptional changes of whole tendons and cultured tenocytes.
(A and B) PCA of RNA sequencing data of plantaris tendons from control nonoverloaded (0D) Scx+ tendons, as well as from Scx+ and ScxΔ mice at 7D or 14D after synergist ablation/plantaris growth procedure (A), and from cultured Scx+ and ScxΔ tenocytes (B). (C) Proliferating tenocytes, isolated from tail tendons, as quantified by BrdU+ nuclei (red) as a percentage of total nuclei (blue) in Scx+ and ScxΔ tenocytes. Representative BrdU– and BrdU+ cells are shown in the inset. Scale bar: 10μm. (D–F) The number of genes that with > 1.5-fold upregulation (red) or > 1.5-fold downregulation (blue), and with q < 0.05, in ScxΔ tendons compared with Scx+ tendons at 7D (D) or 14D (E) after synergist ablation/plantaris growth procedure, or in cultured ScxΔ tenocytes (F) compared with Scx+ tenocytes. (G–I) Heatmaps demonstrating the log2 fold change in selected genes from RNA sequencing that are components or regulators of the extracellular matrix or tendon differentiation (G), involved in cell migration or proliferation (H), or growth factors, cytokines, and signaling molecules (I). The fold change value is displayed for ScxΔ relative to Scx+ group. (C) Values are mean ± SD. Differences between Scx+ and ScxΔ groups were tested using a t test. (G–I) Differences between groups were tested using DESeq2; a, different (q < 0.05) between Scx+ and ScxΔ groups at a given time point for whole tendons, or for cultured tenocytes. (A, D, E, G–I) N ≥ 3 tendons. (B, C, F–I) N = 6 replicates per tenocyte group.
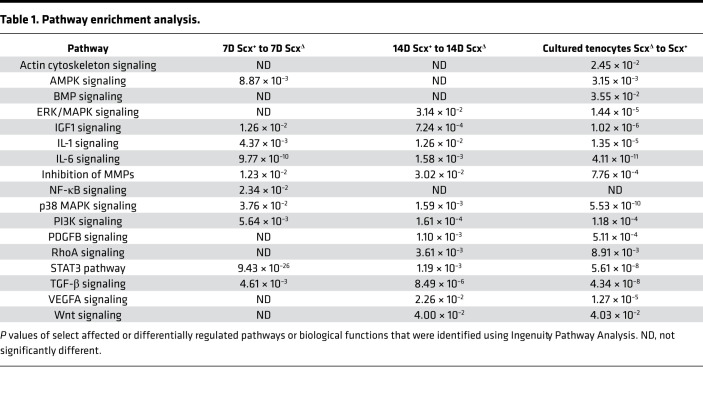
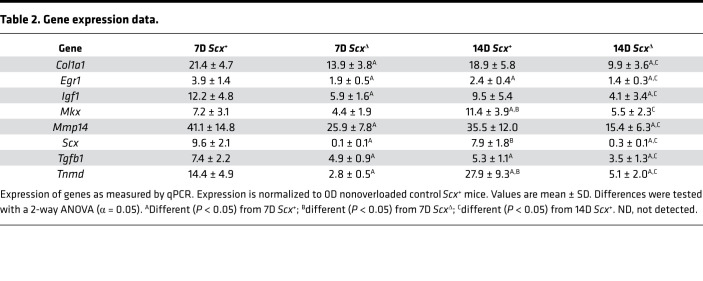
Pathway enrichment analysis was performed for both experiments to determine signaling pathways predicted to be different between groups (Table 1). Several of the pathways identified were involved with growth and differentiation, cytoskeletal signaling, and ECM production. Based on these findings, and from genes that are known to be important for tenocyte proliferation and tenogenesis, we selected several genes for reporting in the manuscript, with the full data set available on NIH Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo). We also performed quantitative PCR (qPCR) validation for a select set of genes of interest identified from RNAseq data, and we generally observed similar patterns of fold-change differences between tendons of Scx+ and ScxΔ mice (Table 2). Many genes in the whole-tendon RNAseq data were also differently expressed in the cultured cells.
Table 1. Pathway enrichment analysis.
Table 2. Gene expression data.
Expression of components or regulators of the ECM — including the collagens Col1a1, Col1a2, Col11a1, and Col12a; small leucine-rich proteoglycans Aspn, Dcn, Fmod, and Omd; and proteases Adam12 and Adam23 — were downregulated in ScxΔ whole tendons and cultured tenocytes, while some matrix degradation genes, such as Mmp10 and Mmp12, were upregulated (Figure 5G). There were also genes like Col23a1, Mgp, Mmp14, and Smoc2 that displayed differential expression patterns between whole tendons and cultured tenocytes (Figure 5G).
Genes involved in cell proliferation and migration were different between genotypes, mostly at the 7D time point (Figure 5H). Many cyclins and cyclin-dependent kinases — such as Ccna2 and Cdk1, 2, and 6, as well as the cell proliferation marker Mki67 — were significantly downregulated in the cultured ScxΔ tenocytes compared with Scx+ tenocytes, with similar patterns in whole-tendon data (Figure 5H). Consistent with this, cell proliferation, as measured with BrdU incorporation, was reduced by 44% in ScxΔ tenocytes compared with Scx+ tenocytes (Figure 5C). Several growth factors and signaling molecules were also reduced in ScxΔ tendons and cells, including Igf1, Pdgfrl, Tgfb2, and Tgfb3 (Figure 5I).
Scleraxis deletion reduces the ability of pericytes to commit to the tenogenic lineage.
As we observed an accumulation of CD146+ pericytes in the neotendon of ScxΔ mice compared with Scx+ mice, we sought to evaluate the effect of scleraxis deletion on the commitment of pericytes to the tenogenic lineage in vitro. We first wanted to identify culture conditions that would result in pericyte differentiation into tenocytes. We found that culturing pericytes from on type I collagen–coated plates, which is similar to the mature tendon matrix, induced morphological changes in cells to make them appear like tenocytes and express Col1a1, Mkx, Scx, and Tnmd (Supplemental Figure 2, A and B). However, culturing pericytes on tissue culture plastic alone, or on basement membrane–derived Matrigel, which is similar to the perivascular matrix of tendon, did not induce markers of tenogenesis (Supplemental Figure 2, A and B).
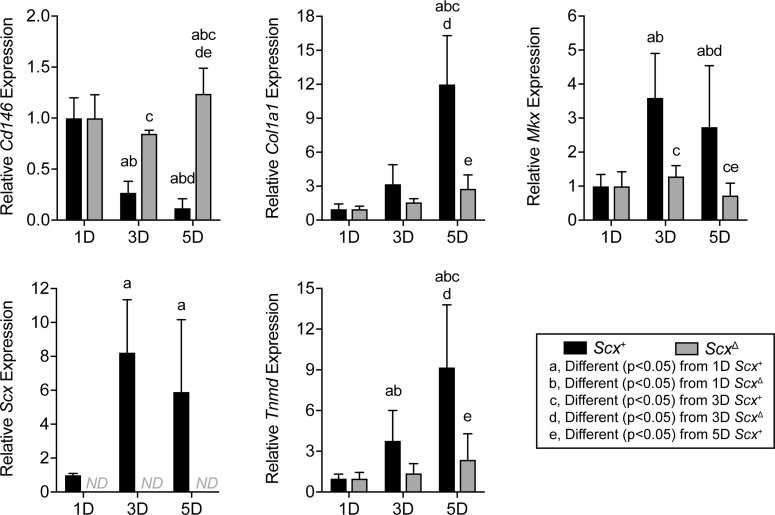
After establishing that pericytes isolated from tendons and placed on type I collagen appear to enter the tenogenic lineage, we sought to determine how the deletion of scleraxis would impact this process. Scx+ and ScxΔ CD146+ pericytes were cultured on type I collagen substrates for 1D, 3D, and 5D, and qPCR was performed (Figure 6). As expected, Scx was undetectable in the ScxΔ cells at all time points, but it increased at 3D and 5D in the Scx+ cells (Figure 6). Relative expression of CD146 decreased over time in the Scx+ group, while it increased in the ScxΔ group (Figure 6). Col1a1, Mkx, and Tnmd expression significantly increased over time in the Scx+ cells and to a much greater extent than in ScxΔ cells (Figure 6).
Figure 6. Effect of scleraxis deletion on gene expression in cultured pericytes.
Gene expression (Cd146, Col1a1, Mkx, Scx, and Tnmd) of CD146+ MACS-sorted pericytes isolated from tail tendons of Scx+ and ScxΔ mice cultured on type I collagen gels for 1D, 3D, or 5D. Values are mean ± CV. Differences between groups were tested using 2-way ANOVA; a, significantly different (P < 0.05) from 1D Scx+; b, significantly different (P < 0.05) from 1D ScxΔ; c, significantly different (P < 0.05) from 3D Scx+; d, significantly different (P < 0.05) from 3D ScxΔ; e, significantly different (P < 0.05) from 5D Scx+. Values are mean ± CV. N = 6 replicates per group.
Discussion
While the cellular components and genetic program that are responsible for the initial embryonic formation and elongation of tendons are well understood (20–23), less is known about growth of tendon tissue in adult animals. Using the synergist ablation model, we previously identified the formation of a neotendon matrix that develops around the original tendon matrix, and we discovered that this new matrix was populated with proliferative, scleraxis-expressing cells (14). The aim of our study was to identify whether scleraxis was required for tendon adaptation to growth stimuli. To study tendon growth, we used the synergist ablation mouse model, in which the Achilles tendon is surgically removed, resulting in a supraphysiological growth stimulus to the plantaris tendon and muscle (15, 24–26). A neotendon matrix consisting of immature collagen and other ECM proteins forms around the plantaris tendon, and over a month, this ECM matures and resembles the original tendon matrix (15). Our results indicate that scleraxis is required for the proper expansion, formation, and remodeling of tendon ECM during mechanical loading and that growth defects in scleraxis-deficient tendons appear to occur in part due to a reduction in the ability of pericytes to differentiate into the tendon cell lineage.
The existing tenocytes within adult tendons appear to be mostly postmitotic cells that do proliferate in response to mechanical loading (14, 27, 28). Increased numbers of tenocytes that occur in response to growth stimuli have been postulated to occur through differentiation of tissue-resident progenitor cells into tenocytes (15, 18, 19, 28, 29). TGF-β signaling plays an important role in the proper embryonic development of tendons by regulating scleraxis and other signaling events (9, 20). Pathway enrichment analysis in the current study demonstrated a potentially important role for TGF-β signaling and other pathways such as IL-1, IL-6, p38 MAPK, and STAT3 in postnatal tendon growth. Several studies have suggested that one of the populations of progenitor cells in adult tendon tissue is CD146+ pericytes, located adjacent to tendon vasculature (15, 18, 19, 30). In a rat model of patellar tendon injury and repair, these cells participate in tenogenic differentiation when stimulated with CTGF (18). In the patellar tendon, approximately 0.8% of cells are CD146+ (18), which is consistent with the relatively low amount of vasculature within homeostatic adult tendon (3). Previous studies in our laboratory using the synergist ablation model in rats showed that these cells appear to migrate from their niche in the vasculature and proliferate within the neotendon (15). In the current study, despite smaller tendon CSAs in ScxΔ mice, the relative abundance of CD146+ pericytes was twice as high in the neotendon of ScxΔ mice compared with Scx+ mice. When pericytes were cultured in tenogenic conditions, deleting scleraxis prevented the ability of these cells to express tenogenic markers. When taken together, these in vivo and in vitro findings indicate that scleraxis is required for the commitment of pericytes into the tenogenic lineage. Furthermore, the higher percentage of CD146+ pericytes in the neotendon of ScxΔ mice likely occurs due to the failure of these cells to properly differentiate into tenocytes. Alternatively, scleraxis could inhibit pericyte proliferation in vivo. While we did not directly measure pericyte proliferation in tendons, the in vitro data suggest that a reduction in the specification or differentiation of pericytes into the tenogenic lineage is more likely to be responsible for the greater abundance of pericytes observed in ScxΔ mice.
During early postnatal development, committed tenocytes (sometimes referred to as tenoblasts) are able to proliferate for a period of time (28), and a similar proliferative capacity likely exists for newly committed tenocytes that arise from CD146+ progenitors. This is further supported by previous work that demonstrated scleraxis-expressing cells in the neotendon of mechanically stimulated tendons also display uptake of the cell proliferation marker EdU (14). In the current study, several genes that are involved in cell proliferation were downregulated in ScxΔ tendons or cells, including numerous cyclins and cyclin-dependent kinases, as well as the cell proliferation marker Mki67. Furthermore, ScxΔ tenocytes displayed a proliferation rate that was nearly half that of Scx+ tenocytes. These results indicate that scleraxis also induces postnatal tenocyte proliferation.
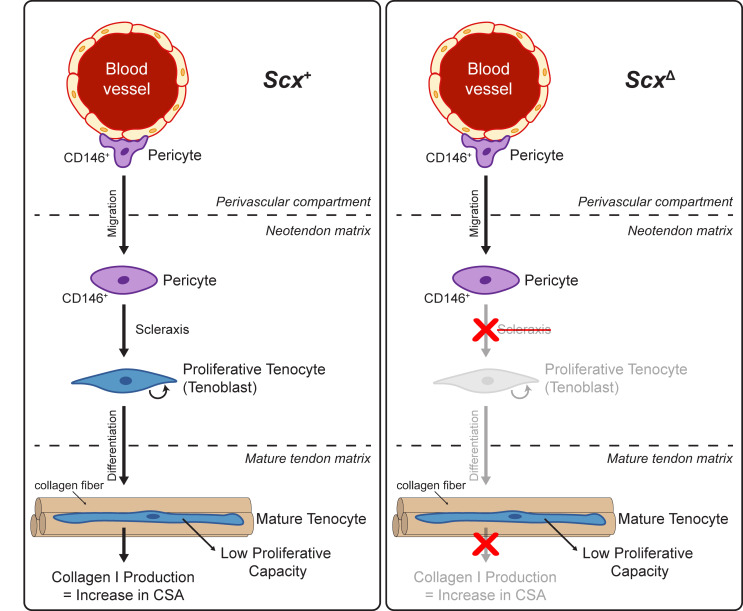
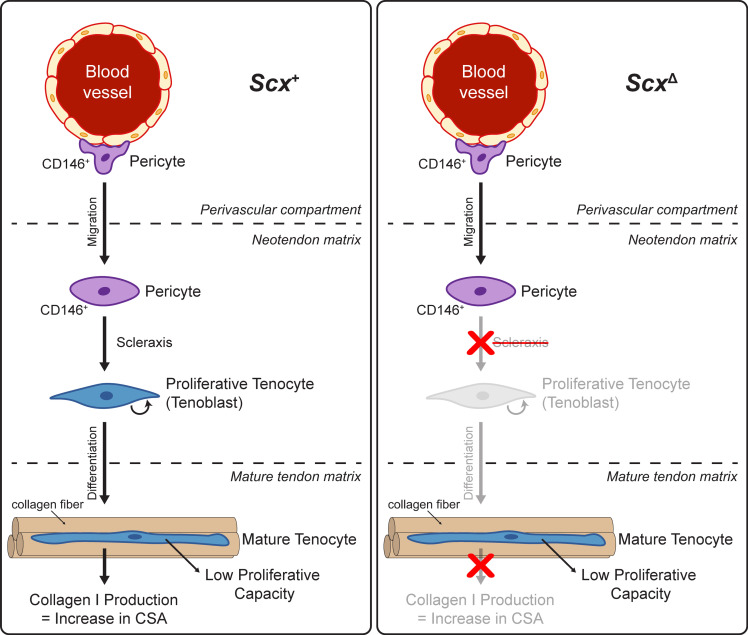
Scleraxis appears to be important in embryonic tendon development, in part, by directing the expression of type I collagens, tenomodulin, and fibronectin (12, 13, 31–33). Mechanical loading increases the expression of scleraxis, type I and III collagen, and tenomodulin in tendons (6, 14, 34, 35), and our transcriptomic analyses demonstrated that expression of components or regulators of the ECM, including Col1a1, Mkx, and Tnmd, were found to be reduced in the absence of scleraxis in whole tendons and in cultured tenocytes. Proteoglycans and glycoproteins that compose the tendon ECM, such as asporin, fibronectin, osteoglycin, thrombospondins 1 and 4, and tenascin C were lower in tendons of ScxΔ mice at 14D, although no change in the expression of many genes involved with heparin sulfate, chondroitin sulfate, or hyaluronic acid synthesis were observed in whole tendons. Differences in MMPs, FACIT collagens, and other factors that regulate type I collagen fibril diameter may also explain the apparent differences in fibril size distributions in electron micrographs. IGF1 signaling can induce protein synthesis in tenocytes (26), and Igf1 expression was downregulated in scleraxis-deficient mice and cultured cells, along with reduced ribosomal proteins and translation elongation factor proteins in tendons of ScxΔ mice. The growth deficiency of ScxΔ tendons was most pronounced at 14D, and proteomics analyses at this time point identified a reduction in several collagens and proteoglycans. These results indicate that, similar to embryonic development, scleraxis plays an important role in adult tendon growth by directing the expression of ECM proteins. When combined with other findings in this manuscript, we propose a model of mechanically stimulated adult tendon growth, whereby scleraxis plays a central role in the commitment of CD146+ pericytes to the tenogenic lineage, the initial expansion of newly committed tenocytes, and the production of ECM proteins (Figure 7).
Figure 7. Proposed model of scleraxis role in adult tendon growth response to mechanical loading.
In WT (left) scleraxis controls the differentiation of CD146+ pericytes into a tenogenic lineage. When scleraxis is deleted (right), CD146+ cells still migrate to the neotendon but are unable to differentiate and do not participate in ECM synthesis.
Despite providing substantial insight into the role of scleraxis in adult tendon adaptation, this study is not without limitations. We only studied male mice, as we previously demonstrated a general lack of differences between the Achilles tendons of male and female mice (36), and we anticipate the results are likely applicable to both sexes. Results from the plantaris tendon may not be applicable to all tendons, as different tendons display extensive divergence in their function and transcriptional profiles (37). Only 2 time points were used following overload to study scleraxis function. These time points were selected due to high levels of scleraxis expression in a previous study using the same tendon growth model (14), but additional insight would likely be gained from studying long-term changes in scleraxis-deficient tendon growth. While we achieved efficient knockdown of scleraxis in whole tendons and cultured tenocytes, there are likely some cells in which scleraxis was not deleted. Although we and others demonstrated that CD146+ pericytes likely serve as progenitor cells for tenocytes, we did not test this with genetic lineage tracing. We also did not directly determine which genes that scleraxis directly regulates by recruitment or repression of transcription machinery to promoter or enhancer sequences. Immune cells likely contribute to tendon growth, and we did not directly quantify the abundance of these cell populations with histology or flow cytometry. However, in the extended RNAseq data set, we did not observe differences in the expression of the pan-hematopoietic cell marker Cd45, macrophage markers (such as Cd11b, Cd68, Cd163, Cd206), or T cell markers (Cd3, Cd4, Cd8) between tendons of Scx+ and ScxΔ mice. Last, while the synergist ablation model is useful in the study of mechanical load–induced tendon growth, the model induces supraphysiological growth that exceeds the loading that is used clinically to induce tendon hypertrophy, such as progressive resistance exercise. There may be a larger inflammatory stimulus in the synergist ablation model compared with resistance exercise, but we think the fundamental mechanisms of tendon growth and ECM synthesis are similar between the 2 conditions.
Tendons of adult animals display an ability to grow and adapt in response to mechanical loading (5–7), and deficiencies in this process may lead to the development of painful and debilitating tendinopathies (38, 39). We found that scleraxis plays an important role in the growth response of adult tendons to mechanical loading by promoting the commitment and expansion of new tenocytes, as well as the production of tendon ECM components. Scleraxis may therefore be a useful biomarker in assessing the response of rehabilitation interventions to treat tendinopathies. This study also provides further support for the potential therapeutic use of pericytes in the treatment of tendon disorders. While scleraxis appears to be a critical transcription factor for tendon growth throughout a lifespan, additional studies that integrate mechanotransduction with molecular genetics and cell biology will provide further insight into the basic mechanisms of tendon growth and contribute to the development of novel biomarkers of early tendinopathic changes and improved therapies for tendinopathies.
Methods
Mice.
WT C57BL/6J (WT, strain 000664) mice and Rosa26CreERT2 mice (strain 008463), in which the tamoxifen-sensitive CreERT2 recombinase enzyme is expressed from the ubiquitous Rosa26 locus (40), were obtained from the Jackson Laboratory. Scxfl/fl mice in which loxP sites flank the coding region of exon 1 of scleraxis (8), and transgenic ScxGFP mice in which GFP is driven by 4kb of the scleraxis promoter (41), were also used (kindly provided by Ronen Schweitzer, Shriners Hospitals for Children, Portland, OR.). Genotype of mice was determined by PCR analysis of DNA obtained from tail tendon biopsies. After performing initial crosses between Rosa26CreERT2/CreERT2 and Scxfl/fl mice, we generated Rosa26CreERT2/CreERT2 Scxfl/fl mice to allow for the inactivation of scleraxis upon treatment with 4HT in vitro or tamoxifen in vivo (referred to as ScxΔ mice), while Rosa26CreERT2/CreERT2 Scx+/+ mice maintain the expression of Scx after 4HT or tamoxifen treatment (referred to as Scx+ mice). An overview of the Rosa26CreERT2, Scx+, Scxfl, and ScxΔ alleles is provided in Figure 1A. Male mice 4–6 months of age were used in this study.
Scleraxis inactivation in mice.
Rosa26CreERT2/CreERT2Scxfl/fl mice were treated daily with an i.p. injection of 2 mg of tamoxifen (T5648, MilliporeSigma) dissolved in 50 μL corn oil to activate CreERT2 recombinase and generate ScxΔ mice (26). Scx+ mice underwent the same tamoxifen treatment regime, but as they did not have loxP sites flanking scleraxis, no inactivation of scleraxis occurred. Tamoxifen treatment began 5D before synergist ablation procedure or isolation of tendons for cell culture, and it continued on a daily basis until tissue was harvested for analysis.
Synergist ablation tendon growth procedure.
Bilateral synergist ablation procedures were performed as described (14, 26, 42). An overview of the time points and surgical procedures are shown in Figure 1, B and C. In this procedure, the Achilles tendon is surgically excised. This prevents the gastrocnemius and soleus muscles from plantarflexing the talocrural joint, resulting in compensatory hypertrophy of the adjacent plantaris muscle and tendon. The tendon growth pattern is eccentric, with new tendon tissue, or neotendon, expanding in a mostly lateral direction (Figure 1C and Figure 2A).
Mice were deeply anesthetized with isoflurane, and the hind limbs were shaved and scrubbed with chlorhexidine solution. A small incision was created in the skin above the posterior hindpaw plantarflexor tendons, and a 3–4 mm portion of the Achilles tendon was isolated and excised, leaving remnant stumps at the myotendinous junction and calcaneus. Care was taken to not injure the plantaris tendon during this procedure. The skin was closed with GLUture (Zoetis), buprenorphine (0.1 mg/kg, s.c.) was administered for postoperative analgesia, and ad libitum weight bearing and cage activity were allowed in the postoperative period.
Animals recovered for a period of 7D or 14D, during which time they received daily treatment with tamoxifen. At the end of 7D or 14D, mice were euthanized by exposure to CO2, followed by cervical dislocation. Plantaris tendons were removed and prepared for either DNA isolation, histology, electron microscopy, or RNA isolation. Up to 20 mice per time point and genotype were used in experiments.
Scleraxis knockdown efficiency.
To determine the efficiency of scleraxis knockdown, DNA was isolated from plantaris tendons or cultured cells using a DNeasy kit (QIAGEN). DNA was quantified with a NanoDrop 2000 system (Thermo Fisher Scientific), and qPCR was conducted in a real-time thermal cycler using iTaq Universal SYBR Green Supermix (Bio-Rad) and primers specific for the region of exon 1 of scleraxis flanked by loxP sites (forward 5′-GACCGCAAGCTCTCCAAGAT-3′; reverse 5′-ACGACCGCTGTGGAAGAAAG-3′), or exon 2 of scleraxis, which is outside of the loxP sites (forward 5′-CGCAGGTCCCCAAGAGCACG-3′; reverse 5′-GGCCTGGGTCAGTGTTCGGC-3′). The abundance of exon 1 of scleraxis was measured relative to exon 2 using the 2–ΔCt method, and it was further normalized to the Scx+ group for whole tendons or to Scx+ tenocytes.
Histology.
Distal portions of plantaris tendons were snap-frozen in liquid nitrogen–cooled isopentane and stored at –80°C until use. Tendons were sectioned and stained with Fast Green and hematoxylin as described previously (MilliporeSigma) (14). Fast Green and hematoxylin sections were imaged on a BX51 microscope (Olympus). For IHC, tendons were sectioned and permeabilized in Triton X-100 (0.2%) and blocked with 5% goat serum. Sections were incubated overnight in primary antibodies against CD146 (ab75769, AbCam) and then incubated in goat secondary antibodies conjugated to Alexa Fluor 555 (A32732, Thermo Fisher Scientific). Samples were also stained with DAPI (MilliporeSigma) to visualize nuclei and wheat germ agglutinin conjugated to Alexa Fluor 488 (WGA-AF488, W11261, Thermo Fisher Scientific) to visualize the ECM. Images were taken on a A1 confocal microscope equipped with a high-resolution camera (Nikon). ImageJ (NIH) was used for quantitative measurements. Cell density was calculated by the number of nuclei per unit area of the tendon. The percentage of CD146+ pericyte cells in the neotendon was measured by dividing the total number of CD146+/DAPI+ cells by the total number of DAPI+ cells.
Transmission electron microscopy.
Proximal tendon portions were fixed in 1% tannic acid and 1% glutaraldehyde in Sorenson’s buffer, followed by postfixation in 2% osmium tetroxide (Electron Microscopy Sciences). Samples were then dehydrated using a graded ethanol series and embedded in EMBed 812 resin (Electron Microscopy Sciences) using a graded resin and propylene oxide series. The 1-μm transverse sections were cut with a diamond knife ultramicrotome and imaged using a 1400-plus transmission electron microscope with a high-resolution digital camera (JEOL). Collagen fibril areas were calculated using ImageJ, as described (43).
Proteomics.
Label-free proteomics was performed as reported in previous studies (36, 44) at the University of Liverpool Centre of Genomic Research. Soluble proteins were extracted by homogenizing plantaris tendons in 25 mM ammonium bicarbonate and 10 mM iodoacetamide, followed by centrifugation and removal of the supernatant (15 minutes at 15000 × g and 4°C). Then, 50 μg of soluble protein was further reduced and alkylated; in solution, trypsin digestion was performed. The remaining pellet was resuspended in 500 μL of 4M Guanidine-HCl extraction buffer (GnHCl), 65 mM dithiothreitol, 50 mM sodium acetate, pH 5.8, for 48 hours at 4°C with shaking (MilliporeSigma). The samples were then centrifuged (15 minutes at 15000 × g and 4°C), and the supernatant was removed and subjected to in solution-trypsin digest on 10 μL of resin (StrataClean, Agilent) followed by reduction and alkylation. The digests (2 μL, corresponding to 2.5 μg of peptides) were loaded onto a spectrometer (Q-Exactive Quadrupole-Orbitrap, Thermo Fisher Scientific) on a 1-hour gradient with an intersample 30-minute blank loaded.
Raw spectra were converted to mgf files using Proteome Discovery (Thermo Fisher Scientific), and resulting files were searched against the UniProt mouse sequence database using a Mascot server (Matrix Science). Search parameters used were: peptide mass tolerance, 10 ppm; fragment mass tolerance, 0.01 Da; +1,+2,+3 ions; unique peptides ≥ 2; missed cleavages, 1; and instrument type ESI-TRAP. Variable modifications included carbamidomethylation of cysteine residues and oxidation of methionine. Label-free quantification was performed using ProgenesisQI software (Waters) as described previously (36, 44). In brief, the MS/MS peak list was searched against the UniProt mouse reviewed database on Mascot using the same search parameters as mentioned above. To account for changes in tendons size, relative protein abundance was normalized to tendon mass, and MetaboAnalyst 4.0 (45) was used to quantify differences between proteins from Scx+ and ScxΔ mice. Proteomics data for this study has been deposited at the ProteomeXchange Consortium via the PRIDE partner repository (accession PXD017944; https://www.ebi.ac.uk/pride).
RNAseq and gene expression.
RNA was isolated from tendons and cultured tenocytes, and it was quantified as previously described (26, 46). Plantaris tendons or cultured cells were homogenized in QIAzol (QIAGEN), and RNA was purified with a miRNeasy Micro Kit (QIAGEN) supplemented with DNase I (QIAGEN). RNA concentration and quality was determined using a NanoDrop 2000 (Thermo Fisher Scientific) and a TapeStation 2200 (Agilent Technologies). RNA integrity numbers for whole-tendon samples were > 7.5 and, for cultured cells, were > 9.5.
To sequence RNA, sample concentrations were normalized and cDNA pools were created for each sample, and they were then subsequently tagged with a barcoded oligo adapter to allow for sample-specific resolution. RNAseq of whole tendons was conducted by the University of Michigan Sequencing Core using HiSeq 2500 system (Illumina) with 50-bp single-end reads. RNAseq of cultured cells was conducted by the Weill Cornell Medical College Genomics Core using a NovaSeq 6000 system (Illumina) with 50-bp paired-end reads. Standard Illumina reagents were used for library preparation and sequencing. Raw RNAseq data was quality checked using FastQC v0.10.0 (Barbraham Bioinformatics). Alignment to the reference genome (mmu10, UCSC) was conducted using the STAR aligner, and differential expression was calculated using DESeq2 (47). A FDR procedure was applied to correct for multiple observations. Sequencing data has been deposited to NIH GEO (accession GSE145864; https://www.ncbi.nlm.nih.gov/geo). Ingenuity Pathway Analysis (IPA) software (QIAGEN) was used to perform gene enrichment analysis of RNAseq data described (48, 49).
For qPCR, RNA was reverse transcribed into cDNA using iScript Reverse Transcription reagents (Bio-Rad) and then amplified in a real-time thermal cycler using iTaq Universal SYBR Green Supermix (Bio-Rad). Primer sequences for specific have been previously published (26, 27). Relative copy number of transcripts in the 7D and 14D Scx+ and ScxΔ mice was normalized to 0D nonoverloaded Scx+ controls using the linear regression of efficiency method (50) and log2 transformed before analysis.
Cell culture.
Tail tendons were isolated from Scx+, ScxΔ, or ScxGFP mice, and cells were cultured as described previously (26, 27). Tendon fascicles were carefully isolated from tail tendons, minced, and placed in DMEM with 0.2% type II collagenase (Thermo Fisher Scientific) for 1 hour at 37°C with agitation. CD146+ pericytes were isolated from digested cell suspensions of Scx+ or ScxGFP mice using anti-CD146 magnetic microbeads (130-092-007, Miltenyi Biotec), and placed into 24-well tissue culture dishes (Corning). Pericytes were expanded in DMEM containing 10% FBS and 1% antibiotic-antimycotic (AbAm; Thermo Fisher Scientific), referred to as growth medium, for a period of up to 5D. When experiments involved the evaluation of scleraxis inactivation, growth medium was supplemented with 10 μM of 4HT to ensure efficient deletion of Scx.
Pericytes from ScxGFP mice were placed either directly onto plastic, a thin gel of growth factor–reduced Matrigel (Corning), or type I collagen reconstituted from rat tail tendons as described (27). General cell morphology and GFP abundance from cells of ScxGFP mice was measured by visualizing cells in an EVOS FL microscope (Thermo Fisher Scientific) and overlaying GFP fluorescence filters with phase contrast images. Pericytes from Scx+ and ScxΔ mice were isolated and cultured on type I collagen gels and were expanded in growth medium containing 10 μM of 4HT. RNA was isolated, and gene expression analysis was performed as described above.
Tenocytes were cultured from digested tail tendons, plated on 100 mm type I collagen–coated dishes (Corning), and expanded in growth medium. Fibroblasts were passaged upon reaching 70% confluence onto 6-well dishes coated with type I collagen (Corning), and media was switched to DMEM containing 2% horse serum and 1% AbAm for a period of 2D. RNA was then isolated, and RNAseq and gene expression analysis was performed as described above.
To measure cell proliferation of Scx+ and ScxΔ tenocytes at 50% confluence, 20 μM of BrdU (MilliporeSigma) was added to growth media for 1 hour. Tenocytes were then fixed in ice-cold methanol and permeabilized with 0.5% Triton X-100, and the BrdU epitope was exposed by denaturing DNA using 2 M HCl. Plates were then incubated with anti-BrdU antibodies (G3G4, Developmental Studies Hybridoma Bank), secondary antibodies conjugated to AF555 (A21127, Thermo Fisher Scientific), and DAPI. The number of BrdU+ nuclei as a percentage of total nuclei was quantified in 5 randomly selected fields of a single experiment from 6 independent experiments performed per group. Plates were imaged in an EVOS FL microscope and quantified using ImageJ (NIH).
Statistics.
Results are presented as mean ± SD or mean ± coefficient of variation (CV). Prism version 8.0 (GraphPad Software) was used to conduct statistical analyses for all data except RNAseq. A 2-way ANOVA followed by Fisher’s post hoc sorting (α = 0.05) evaluated the interaction between time after synergist ablation and Scx knockdown. For cell culture experiments, differences between groups were tested with an unpaired 2-tailed Student’s t test (α = 0.05), or a 1- or 2-way ANOVA followed by Fisher’s post hoc sorting (α = 0.05). For CSA quantification of fibrils, differences between the bins of the 2 groups were tested with a χ2 test (α = 0.05). Replicates for whole tissue experiments are tendons from individual animals, while cell culture experiments represent technical replicates of individual or pooled animals.
Study approval.
Animal studies were approved by the University of Michigan (protocol PRO00003566) and Hospital for Special Surgery/Weill Cornell Medical College/Memorial Sloan Kettering (protocol 2017-0035) IACUCs. Experimental procedures were conducted according to approved protocols and in accordance with the US Public Health Policies on Human Care and Use of Laboratory Animals.
Author contributions
JPG, MMS, and CLM designed research; JPG, MMS, and YAK performed research; JPG, MMS, YAK, EC, and CLM analyzed data; JPG, MMS, and CLM drafted and edited the manuscript; and all authors approved of the final version of the manuscript.
Supplementary Material
Acknowledgments
The authors would like to acknowledge technical assistance from Deborah Simpson at the University of Liverpool. The ScxGFP and Scxfl mice used in this study were provided by Ronen Schweitzer at the Shriners Hospital for Children in Portland, Oregon, USA. This work was supported by grants R01-AR063649 and F31-AR065931 from the NIH.
Version 1. 05/28/2020
In-Press Preview
Version 2. 07/09/2020
Electronic publication
Footnotes
Conflict of interest:
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(13):e138295.https://doi.org/10.1172/jci.insight.138295.
Contributor Information
Jonathan P. Gumucio, Email: jgumucio@umich.edu.
Martin M. Schonk, Email: schonkm@hss.edu.
Yalda A. Kharaz, Email: Y.Ashraf-Kharaz@liverpool.ac.uk.
Eithne Comerford, Email: Eithne.Comerford@liverpool.ac.uk.
Christopher L. Mendias, Email: MendiasC@hss.edu.
References
- 1.Davis ME, Gumucio JP, Sugg KB, Bedi A, Mendias CL. MMP inhibition as a potential method to augment the healing of skeletal muscle and tendon extracellular matrix. J Appl Physiol. 2013;115(6):884–891. doi: 10.1152/japplphysiol.00137.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gumucio JP, Sugg KB, Mendias CL. TGF-β superfamily signaling in muscle and tendon adaptation to resistance exercise. Exerc Sport Sci Rev. 2015;43(2):93–99. doi: 10.1249/JES.0000000000000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noah AC, et al. Adaptive and innate immune cell responses in tendons and lymph nodes after tendon injury and repair. J Appl Physiol. 2020;128(3):473–482. doi: 10.1152/japplphysiol.00682.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AH, Elliott DM. Comparative multi-scale hierarchical structure of the tail, plantaris, and Achilles tendons in the rat. J Anat. 2019;234(2):252–262. doi: 10.1111/joa.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnusson SP, Kjaer M. The impact of loading, unloading, ageing and injury on the human tendon. J Physiol (Lond) 2019;597(5):1283–1298. doi: 10.1113/JP275450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendias CL, Gumucio JP, Bakhurin KI, Lynch EB, Brooks SV. Physiological loading of tendons induces scleraxis expression in epitenon fibroblasts. J Orthop Res. 2012;30(4):606–612. doi: 10.1002/jor.21550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kjaer M, et al. Extracellular matrix adaptation of tendon and skeletal muscle to exercise. J Anat. 2006;208(4):445–450. doi: 10.1111/j.1469-7580.2006.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murchison ND, et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134(14):2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 9.Pryce BA, Watson SS, Murchison ND, Staverosky JA, Dünker N, Schweitzer R. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development. 2009;136(8):1351–1361. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang AH, Lu HH, Schweitzer R. Molecular regulation of tendon cell fate during development. J Orthop Res. 2015;33(6):800–812. doi: 10.1002/jor.22834. [DOI] [PubMed] [Google Scholar]
- 11.Carlberg AL, Tuan RS, Hall DJ. Regulation of scleraxis function by interaction with the bHLH protein E47. Mol Cell Biol Res Commun. 2000;3(2):82–86. doi: 10.1006/mcbr.2000.0195. [DOI] [PubMed] [Google Scholar]
- 12.Léjard V, et al. Scleraxis and NFATc regulate the expression of the pro-alpha1(I) collagen gene in tendon fibroblasts. J Biol Chem. 2007;282(24):17665–17675. doi: 10.1074/jbc.M610113200. [DOI] [PubMed] [Google Scholar]
- 13.Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol. 2006;298(1):234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 14.Gumucio JP, Phan AC, Ruehlmann DG, Noah AC, Mendias CL. Synergist ablation induces rapid tendon growth through the synthesis of a neotendon matrix. J Appl Physiol. 2014;117(11):1287–1291. doi: 10.1152/japplphysiol.00720.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz AJ, Sarver DC, Sugg KB, Dzierzawski JT, Gumucio JP, Mendias CL. p38 MAPK signaling in postnatal tendon growth and remodeling. PLoS One. 2015;10(3):e0120044. doi: 10.1371/journal.pone.0120044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dideriksen K, et al. Tendon collagen synthesis declines with immobilization in elderly humans: no effect of anti-inflammatory medication. J Appl Physiol. 2017;122(2):273–282. doi: 10.1152/japplphysiol.00809.2015. [DOI] [PubMed] [Google Scholar]
- 17.Caplan AI. New MSC: MSCs as pericytes are Sentinels and gatekeepers. J Orthop Res. 2017;35(6):1151–1159. doi: 10.1002/jor.23560. [DOI] [PubMed] [Google Scholar]
- 18.Lee CH, et al. Harnessing endogenous stem/progenitor cells for tendon regeneration. J Clin Invest. 2015;125(7):2690–2701. doi: 10.1172/JCI81589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Q, Lui PP, Lee YW. In vivo identity of tendon stem cells and the roles of stem cells in tendon healing. Stem Cells Dev. 2013;22(23):3128–3140. doi: 10.1089/scd.2013.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan GK, et al. Tgfβ signaling is critical for maintenance of the tendon cell fate. Elife. 2020;9:e52695. doi: 10.7554/eLife.52695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenthal N, et al. Development of migrating tendon-bone attachments involves replacement of progenitor populations. Development. 2018;145(24):dev165381. doi: 10.1242/dev.165381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz AG, Long F, Thomopoulos S. Enthesis fibrocartilage cells originate from a population of Hedgehog-responsive cells modulated by the loading environment. Development. 2015;142(1):196–206. doi: 10.1242/dev.112714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang AH, et al. Requirement for scleraxis in the recruitment of mesenchymal progenitors during embryonic tendon elongation. Development. 2019;146(20):dev182782. doi: 10.1242/dev.182782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olesen JL, et al. Expression of insulin-like growth factor I, insulin-like growth factor binding proteins, and collagen mRNA in mechanically loaded plantaris tendon. J Appl Physiol. 2006;101(1):183–188. doi: 10.1152/japplphysiol.00636.2005. [DOI] [PubMed] [Google Scholar]
- 25.Calve S, Isaac J, Gumucio JP, Mendias CL. Hyaluronic acid, HAS1, and HAS2 are significantly upregulated during muscle hypertrophy. Am J Physiol, Cell Physiol. 2012;303(5):C577–C588. doi: 10.1152/ajpcell.00057.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Disser NP, Sugg KB, Talarek JR, Sarver DC, Rourke BJ, Mendias CL. Insulin-like growth factor 1 signaling in tenocytes is required for adult tendon growth. FASEB J. 2019;33(11):12680–12695. doi: 10.1096/fj.201901503R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugg KB, et al. Postnatal tendon growth and remodeling require platelet-derived growth factor receptor signaling. Am J Physiol, Cell Physiol. 2018;314(4):C389–C403. doi: 10.1152/ajpcell.00258.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinstein M, Dingwall HL, O’Connor LD, Zou K, Capellini TD, Galloway JL. A distinct transition from cell growth to physiological homeostasis in the tendon. Elife. 2019;8:e48689. doi: 10.7554/eLife.48689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi Y, et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13(10):1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- 30.Tarafder S, et al. Tendon stem/progenitor cells regulate inflammation in tendon healing via JNK and STAT3 signaling. FASEB J. 2017;31(9):3991–3998. doi: 10.1096/fj.201700071R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Docheva D, Hunziker EB, Fässler R, Brandau O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol Cell Biol. 2005;25(2):699–705. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimoto Y, Takimoto A, Watanabe H, Hiraki Y, Kondoh G, Shukunami C. Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci Rep. 2017;7:45010. doi: 10.1038/srep45010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagchi RA, Lin J, Wang R, Czubryt MP. Regulation of fibronectin gene expression in cardiac fibroblasts by scleraxis. Cell Tissue Res. 2016;366(2):381–391. doi: 10.1007/s00441-016-2439-1. [DOI] [PubMed] [Google Scholar]
- 34.Mendias CL, Gumucio JP, Lynch EB. Mechanical loading and TGF-β change the expression of multiple miRNAs in tendon fibroblasts. J Appl Physiol. 2012;113(1):56–62. doi: 10.1152/japplphysiol.00301.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinemeier KM, et al. Expression of collagen and related growth factors in rat tendon and skeletal muscle in response to specific contraction types. J Physiol (Lond) 2007;582(Pt 3):1303–1316. doi: 10.1113/jphysiol.2007.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarver DC, Kharaz YA, Sugg KB, Gumucio JP, Comerford E, Mendias CL. Sex differences in tendon structure and function. J Orthop Res. 2017;35(10):2117–2126. doi: 10.1002/jor.23516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Disser NP, et al. Widespread diversity in the transcriptomes of functionally divergent limb tendons. J Physiol (Lond) 2020;598(8):1537–1550. doi: 10.1113/JP279646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell AL, et al. IL-21 receptor expression in human tendinopathy. Mediators Inflamm. 2014;2014:481206. doi: 10.1155/2014/481206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mead MP, Gumucio JP, Awan TM, Mendias CL, Sugg KB. Pathogenesis and Management of Tendinopathies in Sports Medicine. Transl Sports Med. 2018;1(1):5–13. doi: 10.1002/tsm2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 41.Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236(6):1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 42.Sugg KB, Korn MA, Sarver DC, Markworth JF, Mendias CL. Inhibition of platelet-derived growth factor signaling prevents muscle fiber growth during skeletal muscle hypertrophy. FEBS Lett. 2017;591(5):801–809. doi: 10.1002/1873-3468.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starborg T, et al. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat Protoc. 2013;8(7):1433–1448. doi: 10.1038/nprot.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashraf Kharaz Y, Zamboulis D, Sanders K, Comerford E, Clegg P, Peffers M. Comparison between chaotropic and detergent-based sample preparation workflow in tendon for mass spectrometry analysis. Proteomics. 2017;17(13-14):1700018. doi: 10.1002/pmic.201700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gumucio JP, et al. Reduced mitochondrial lipid oxidation leads to fat accumulation in myosteatosis. FASEB J. 2019;33(7):7863–7881. doi: 10.1096/fj.201802457RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudgens JL, Sugg KB, Grekin JA, Gumucio JP, Bedi A, Mendias CL. Platelet-Rich Plasma Activates Proinflammatory Signaling Pathways and Induces Oxidative Stress in Tendon Fibroblasts. Am J Sports Med. 2016;44(8):1931–1940. doi: 10.1177/0363546516637176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendias CL, Schwartz AJ, Grekin JA, Gumucio JP, Sugg KB. Changes in muscle fiber contractility and extracellular matrix production during skeletal muscle hypertrophy. J Appl Physiol. 2017;122(3):571–579. doi: 10.1152/japplphysiol.00719.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutledge RG, Stewart D. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 2008;8:47. doi: 10.1186/1472-6750-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.