Abstract
Interferon regulatory factor 1 (IRF1) regulates diverse biological functions, including modulation of cellular responses involved in tumorigenesis. Genetic mutations and altered IRF1 function are associated with several cancers. Although the function of IRF1 in the immunobiology of cancer is emerging, IRF1-specific mechanisms regulating tumorigenesis and tissue homeostasis in vivo are not clear. Here, we found that mice lacking IRF1 were hypersusceptible to colorectal tumorigenesis. IRF1 functions in both the myeloid and epithelial compartments to confer protection against AOM/DSS-induced colorectal tumorigenesis. We further found that IRF1 also prevents tumorigenesis in a spontaneous mouse model of colorectal cancer. The attenuated cell death in the colons of Irf1–/– mice was due to defective pyroptosis, apoptosis, and necroptosis (PANoptosis). IRF1 does not regulate inflammation and the inflammasome in the colon. Overall, our study identified IRF1 as an upstream regulator of PANoptosis to induce cell death during colitis-associated tumorigenesis.
Keywords: Immunology, Oncology
Keywords: Cancer, Colorectal cancer, Innate immunity
IRF1-deficient mice are hypersusceptible to colorectal tumorigenesis, with attenuated cell death in the colons caused by defective PANoptosis.
Introduction
Interferon regulatory factor 1 (IRF1) is a transcription factor induced in response to interferons (1). IRF1 plays crucial roles in various physiological and pathological processes, including microbial infection, inflammation, innate and adaptive immune responses, and tumor immune surveillance (2, 3). IRF1 controls the gene expression of guanylate-binding proteins, the inducible nitric oxide synthase, and caspase-1 (CASP1) (2, 4), which have been implicated in various inflammatory diseases. While IRF1 is protective in innate immunity-related inflammatory diseases, it is pathogenic in chronic inflammatory ailments. NOD mice deficient in IRF1 are resistant to developing diabetes compared with IRF1-sufficient NOD mice (5). Similarly, IRF1-deficient mice exhibit reduced susceptibility to experimental autoimmune encephalomyelitis (6). We have previously found that IRF1 mediates NLRP3 and AIM2 inflammasome activation during microbial infection (4, 7, 8). These inflammasomes are associated with providing protective responses against colorectal cancer (9, 10).
Chronic inflammation can predispose tissue to cancer development. The tumor suppressor function of IRF1 was first identified in Irf1–/– mouse embryonic fibroblasts (MEFs), which failed to undergo cell death in response to γ irradiation, a phenotype similar to that observed in MEFs lacking the tumor suppressor p53 (11). Numerous other studies have also elucidated the involvement of IRF1 in cell proliferation and apoptosis (12–15). IRF1 mediates CASP8- or CASP9-driven extrinsic or intrinsic apoptosis in different cancer models. IRF1 expression induces apoptosis and inhibits tumor growth in mouse mammary cancer cells both in vitro and in vivo (16). Previous clinical studies have also indicated that loss of IRF1 may affect the development of specific human cancers (17). Specifically, loss of IRF1 has been reported in esophageal and gastric cancers and pulmonary metastasis (18, 19). In addition, a polymorphism in the IRF1 gene is detected at a high frequency in human breast cancer lines (20).
Colorectal cancer is one of the leading causes of adult cancer-related deaths, and it is projected that 147,950 new cases will be diagnosed in the United States in 2020 (21). The abnormal proliferation of colonic epithelial cells as a result of mutations in oncogenes and tumor suppressor genes develops into colorectal cancer (10). Although the ability of apoptotic cell death modulation to prevent colorectal cancer has gained significant attention, most cancer cells develop resistance to apoptosis, underscoring the importance of other modes of cell death, such as pyroptosis and necroptosis, in preventing colorectal cancer (22). Pyroptosis is mediated by gasdermin D (GSDMD) cleavage upon inflammasome activation (23), whereas necroptosis is mediated by mixed lineage kinase domain-like pseudokinase (MLKL) upon activation of its upstream kinases receptor-interacting serine/threonine-protein kinase 1 (RIPK1) and RIPK3 (24). However, the role of GSDMD and MLKL in colorectal cancer is unknown. The 3 programmed cell death pathways, pyroptosis, apoptosis, and necroptosis (PANoptosis), exhibit crosstalk that allows them to switch from one mode to another under specific conditions. For instance, upon CASP8 inhibition, the activity of RIPK3 is increased, and cells undergo necroptosis (25). Likewise, RIPK3 inhibition enhances the apoptotic activity of CASP8 (26). The finding that CASP8 and FADD regulate both the canonical and noncanonical NLRP3 inflammasome demonstrates an association between apoptosis and pyroptosis (27). Furthermore, the apoptotic caspases CASP3 and CASP7 specifically block pyroptosis by cleaving GSDMD at a different site from that of inflammatory caspases (28). Studies have shown that certain molecules, including Z-DNA–binding protein 1 (ZBP1) during influenza A virus infection (7) and TGF-β–activated kinase 1 (TAK1) (29), can act as master regulators of all 3 pathways, leading to the establishment of the concept of PANoptosis (30). Moreover, we recently found that CASP6 is a critical regulator of ZBP1-mediated NLRP3 inflammasome activation and PANoptosis (31). Genetic evidence for the crosstalk among pyroptosis, apoptosis, and necroptosis has also been shown in autoinflammatory disease (32). Additionally, recent studies have shown an association between necroptosis and pyroptosis in colorectal cancer (33, 34). However, the regulation and relevance of cell death executed by this integrated model remain unknown in this disease. Hence, promoting PANoptosis (30) could hold great therapeutic potential for colorectal cancer. Given that IRF1 is a transcription factor, it can induce the expression of upstream death receptors, ligands, caspases, or proteases that subsequently prepare the cell to undergo PANoptosis. Although IRF1 has been implicated in the immunobiology of some cancers, its precise role in the development of colorectal cancer remains unknown.
Here, we found that Irf1–/– mice were hypersusceptible to colitis-associated colorectal cancer (CAC). Although the production of proinflammatory cytokines was similar to that of WT mice, there was decreased cell death in the colons of Irf1–/– mice, which was associated with the impaired activation of apoptotic caspases (CASP7 and CASP3) and pyroptotic GSDMD, and reduced necroptosis. Overall, this study identifies IRF1 as a regulator of PANoptosis to restrict tumorigenesis.
Results
IRF1 inhibits colitis-associated tumorigenesis.
To investigate the role of IRF1 in CAC, we used the established azoxymethane/dextran sodium sulfate (AOM/DSS) model to induce tumorigenesis in the colons of mice. WT and Irf1–/– mice were given a single injection of the DNA-damaging agent AOM and administered 3 cycles of DSS in drinking water (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136720DS1). First, we compared the expression of the gene encoding IRF1 between tumor and nontumor tissue obtained from the colons of WT mice 80 days after AOM injection. There was decreased Irf1 expression in tumor tissue compared with nontumor tissue in WT mice (Supplemental Figure 1B). In line with this, tumor tissue from WT mice exhibited reduced protein levels of IRF1 in comparison with nontumor tissue (Supplemental Figure 1C). Moreover, we observed decreased IRF1 protein expression in both tumor and nontumor tissue at day 80 when compared with the normal colons of WT mice (day 0; Supplemental Figure 1C), suggesting an association between IRF1 and CAC. To further understand the relevance of the association between IRF1 and the development of colorectal cancer, we analyzed publicly available data from The Cancer Genome Atlas database to understand this association in humans. The analyzed data revealed a statistically significant reduction in IRF1 expression in the colons of patients with late-stage (stage IV) colorectal cancer (Supplemental Figure 1D). Furthermore, survival analyses in patients with colorectal cancer revealed shorter survival for patients with lower IRF1 expression (Supplemental Figure 1E), suggesting that decreased IRF1 expression is associated with poor prognosis in human colorectal cancer.
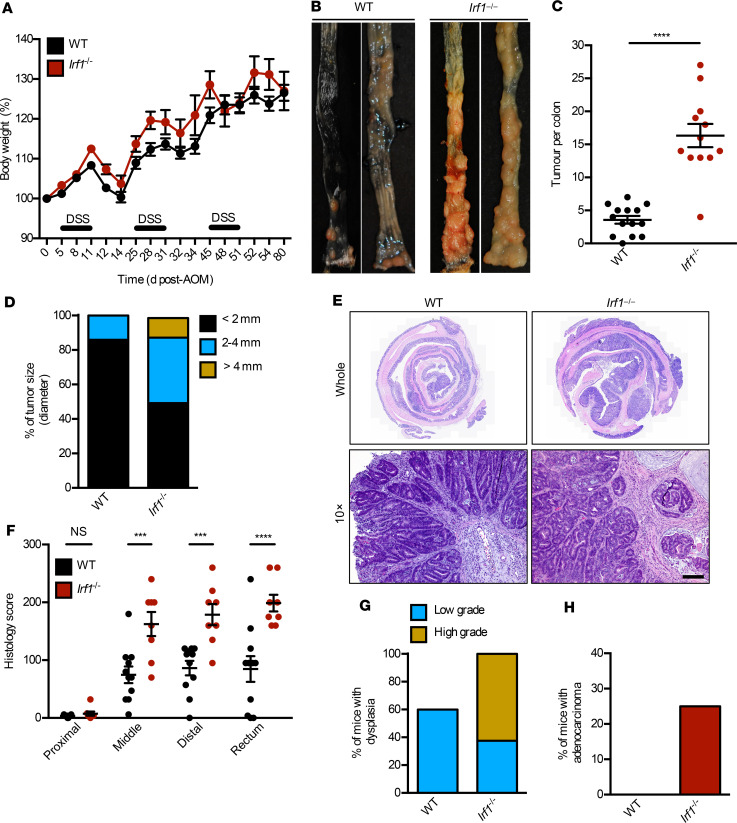
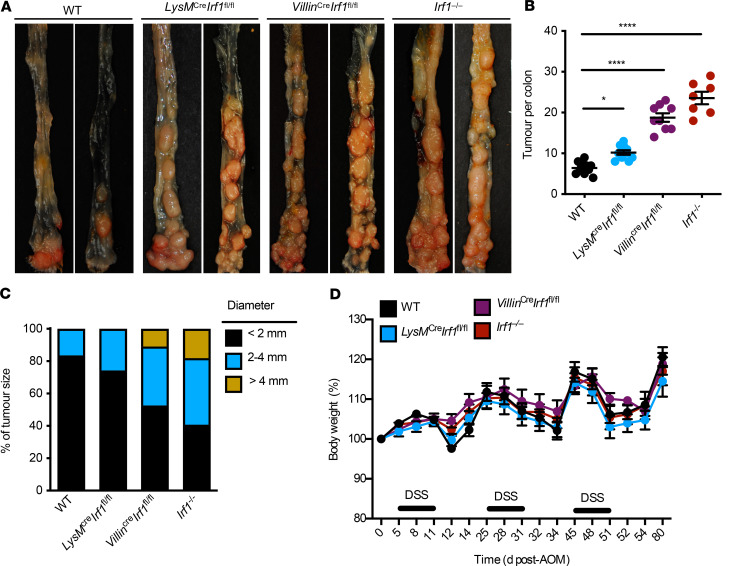
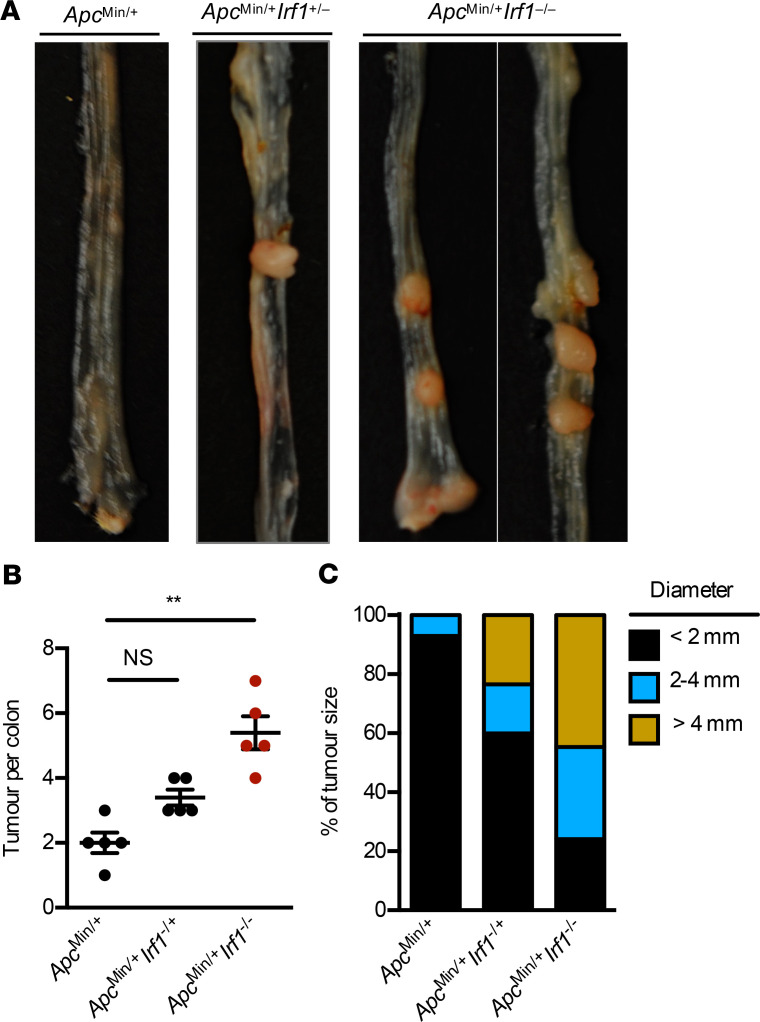
To understand the mechanistic role of IRF1 in colorectal tumorigenesis, we monitored body weight change and prevalence of tumors in WT and Irf1–/– mice 80 days after AOM injection. Both genotypes lost body weight similarly after each cycle of DSS (Figure 1A). However, the colons of Irf1–/– mice had a higher tumor burden in terms of both number of tumors and tumor size compared with WT mice (Figure 1, B–D). Histopathological analysis revealed thickening of the colons in Irf1–/– mice relative to those in WT mice (Figure 1E). Histological hallmarks associated with inflammation, ulceration, hyperplasia, and the extent or severity of damage were more frequently identified in the middle and distal colons and the rectums of Irf1–/– mice compared with the corresponding regions in WT mice (Figure 1F). Irf1–/– mice suffered from a high grade of dysplasia and adenocarcinoma more frequently than WT mice (Figure 1, G and H), suggesting that IRF1 restricts colitis-associated colorectal tumorigenesis. To determine the tissue-specific role of IRF1, we generated mice lacking IRF1 in myeloid cells and epithelial cells and subjected these mice to AOM/DSS. The colons of mice lacking IRF1 in either myeloid cells (LysMCreIrf1fl/fl) or epithelial cells (VillinCreIrf1fl/fl) presented with higher tumor burden in terms of both number and size of tumors compared with WT mice (Figure 2, A–C), despite a lack of significant body weight change among these mice after DSS administration (Figure 2D). Moreover, mice lacking IRF1 globally displayed more tumors in the colons compared with mice lacking IRF1 in myeloid cells or epithelial cells only (Figure 2, A and B), suggesting that IRF1 restricts the development of tumorigenesis by functioning in both myeloid cells and epithelial cells. To further investigate the role of IRF1 in restricting tumorigenesis, we used a spontaneous mouse model of colon cancer. In this model, the mouse line containing a heterozygous mutation in the gene encoding adenomatous polyposis coli (ApcMin/+) was crossed with Irf1−/− mice. We found that ApcMin/+Irf1−/− mice had a higher tumor burden than ApcMin/+ control mice in terms of both the number and size of tumors, suggesting that IRF1 also plays a tumor suppressive role in a sporadic model of colon cancer (Figure 3).
Figure 1. IRF1 prevents colitis-associated colorectal tumorigenesis.
(A) Body weight change of WT (n = 10) and Irf1–/– (n = 10) mice from 1 experiment (representative of 3 independent experiments). (B) Representative images of colon tumors in WT and Irf1–/– mice 80 days after injection of azoxymethane (AOM). (C) Number of colon tumors in WT (n = 14) and Irf1–/– (n = 12) mice. (D) Percentage of tumors of various sizes 80 days after AOM injection. (E) Representative H&E staining of colon tumors. Scale bar: 200 μM. (F) Histological scores 80 days after injection of AOM. (G) Percentage of mice with dysplasia 80 days after AOM injection. (H) Percentage of mice with adenocarcinoma 80 days after AOM injection. Data are from 1 experiment (representative of 3 independent experiments). Each symbol represents 1 individual mouse (C and F). ***P < 0.001; ****P < 0.0001. The 2-tailed t test (C) and 2-way ANOVA (F) were used. Data are represented as mean ± SEM.
Figure 2. IRF1 functions in both the myeloid and epithelial cell to prevent colitis-associated colorectal tumorigenesis.
(A) Representative images of colon tumors in WT, LysMCreIrf1fl/fl, VillinCreIrf1fl/fl, and Irf1–/– mice 80 days after azoxymethane (AOM) injection. (B) Number of colon tumors in WT (n = 10), LysMCreIrf1fl/fl (n = 10), VillinCreIrf1fl/fl (n = 10), and Irf1–/– (n = 7) mice. (C) Percentage of tumors of various sizes in WT, LysMCreIrf1fl/fl, VillinCreIrf1fl/fl, and Irf1–/– mice 80 days after AOM injection. (D) Body weight change in WT, LysMCreIrf1fl/fl, VillinCreIrf1fl/fl, and Irf1–/– mice 80 days after AOM injection. Each symbol represents 1 individual mouse in B. *P < 0.01; ****P < 0.0001. One-way ANOVA (B) was used. Data are representative of 2 independent experiments. Data are represented as mean ± SEM.
Figure 3. IRF1 prevents colorectal cancer in an ApcMin/+ model of tumorigenesis.
(A) Representative images of colon tumors in 120-day-old littermate ApcMin/+, ApcMin/+Irf1+/-, and ApcMin/+Irf1−/− mice. (B) Number of colon tumors in 120-day-old littermate ApcMin/+ (n = 5), ApcMin/+Irf1+/– (n = 5), and ApcMin/+Irf1−/− (n = 5) mice. (C) Percentage of tumors of various sizes in 120-day-old littermate ApcMin/+, ApcMin/+Irf1+/–, and ApcMin/+Irf1−/− mice. Each symbol represents 1 individual mouse in B. **P < 0.01. One-way ANOVA (B) was used. Data are represented as mean ± SEM.
IRF1 does not regulate colonic inflammation.
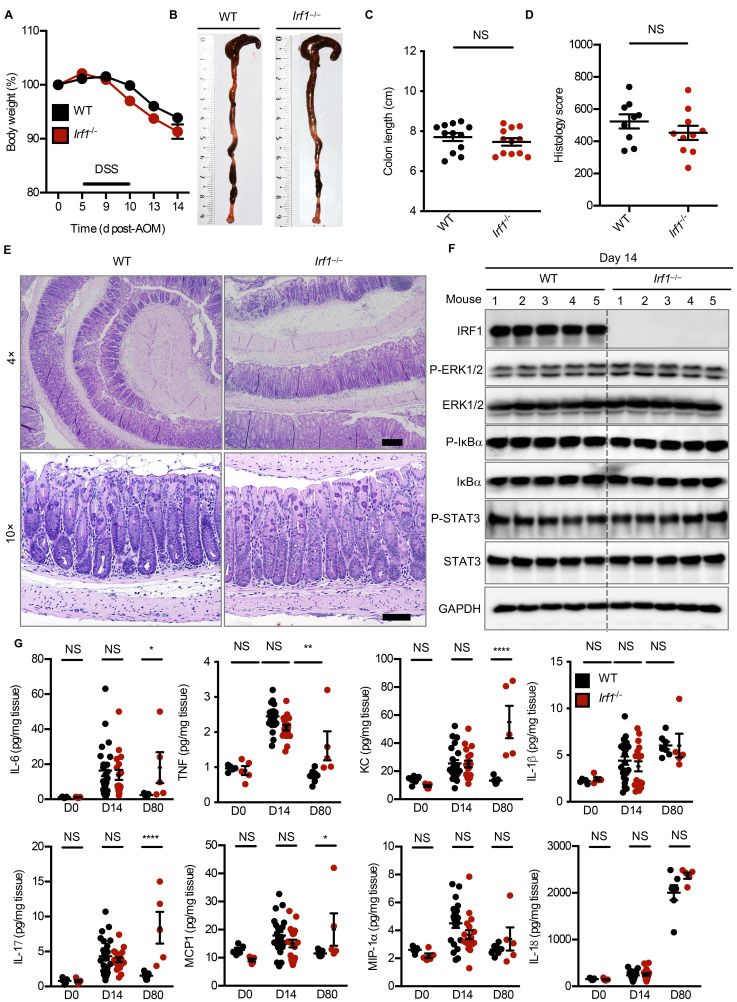
Considerable evidence indicates that inflammation exacerbates the development of CAC (35). Hence, an increased inflammatory response in the colons of Irf1–/– mice after DSS administration may predispose them to tumorigenesis. To investigate the role of IRF1 in colitis, cohorts of WT and Irf1–/– mice were subjected to AOM injection and a single round of DSS. Body weight change and inflammatory responses were monitored in these mice for 14 days after AOM injection. There was no significant difference in weight change, colon length, and overall histological scores associated with inflammation, ulceration, hyperplasia, and the extent or severity of damage between WT and Irf1–/– mice (Figure 4, A–E). Consistent with this observation, major inflammatory signaling pathways were similarly activated in the colons of these mice (Figure 4F). Inflammatory cytokines and chemokines produced during colitis affect tumor development by providing survival cues to tumor cells (10). Despite the known role of IRF1 in inflammasome activation, we found the levels of major inflammatory cytokines and chemokines to be similar in the colons of WT and Irf1–/– mice at day 14 after AOM injection (Figure 4G). However, some of the cytokines, such as IL-6, TNF, KC, IL-17, and MCP-1, were found at higher levels in the colons of Irf1–/– mice than in the colons of WT mice 80 days after AOM injection (Figure 4G). This increase may be a consequence of overt tumor growth in the IRF1-deficient mice. Moreover, we used flow cytometry to profile the immune cell composition in the colons of WT and Irf1–/– mice 14 days after AOM injection. We observed similar frequencies of macrophages, neutrophils, dendritic cells, B cells, and CD4+ T cells (Supplemental Figure 2) in the colons of WT and Irf1–/– mice at day 0 and day 14 after AOM injection. However, the frequency of CD8+ T cells in the colons of Irf1–/– mice was significantly lower than in the colons of WT mice at both day 0 and day 14. This is likely due to the role of IRF1 in T cell selection processes and the differentiation of CD8+ cells (36). Altogether, these data suggest that IRF1 is dispensable for the regulation of inflammatory responses during colitis.
Figure 4. IRF1 does not regulate inflammation in the colon.
(A) Body weight change of WT (n = 10) and Irf1–/– (n = 10) mice from 1 experiment (representative of 3 independent experiments). (B and C) Representative images of colon (B) and length of colon (C) in WT (n = 10) and Irf1–/– (n = 10) mice 14 days after azoxymethane (AOM) injection. (D) Histological scores. (E) Representative H&E staining of colon. Scale bar: 500 μM. (F) Immunoblot analysis of IRF1, phosphorylated and total ERK1 and ERK2 (P-ERK1/2 and ERK1/2, respectively), phosphorylated and total IκBα (P-IκBα and IκBα, respectively), phosphorylated and total STAT3 (P-STAT3 and STAT3, respectively), and GAPDH (loading control) in colons of WT and Irf1–/– mice. Blots represent data from the same biological samples run in parallel. (G) Levels of inflammatory cytokines in the colons of WT and Irf1–/– mice at day 0, 14, and 80 after AOM injection. Each symbol represents 1 individual mouse (C, D, and G). *P < 0.05; **P < 0.01; ****P < 0.0001. The 2-tailed t test (C and D) or 1-way ANOVA (G) were used. Data are from 1 experiment (representative of 3 independent experiments) (A–F) or pooled from 2 independent experiments (G). Data are represented as mean ± SEM.
IRF1 regulates PANoptosis.
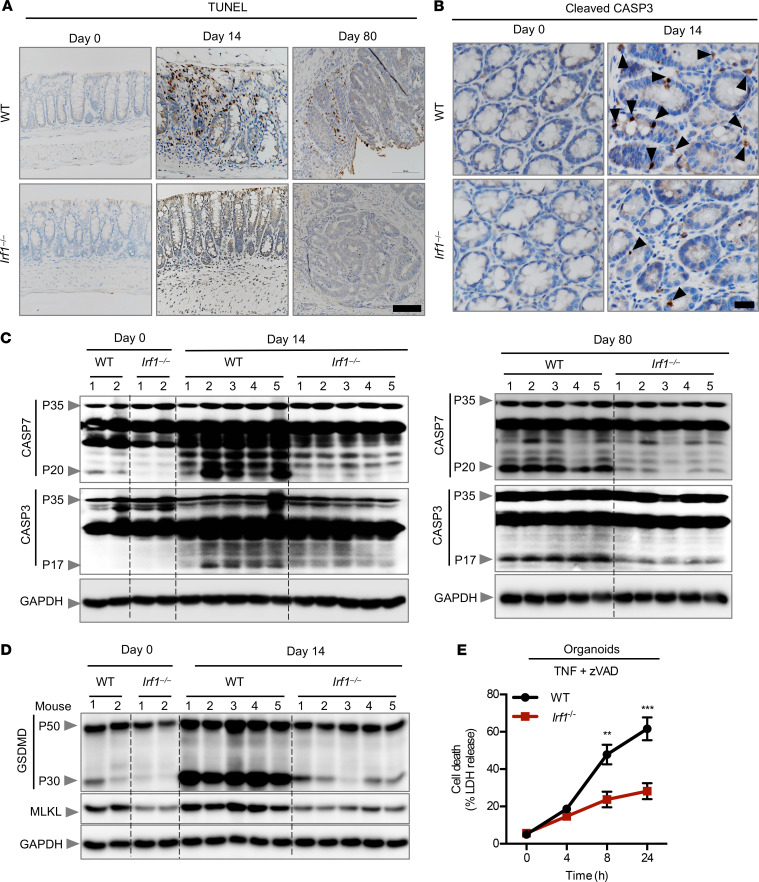
Colonic cells undergo cell death following inflammation and DNA damage, which may inhibit colorectal tumorigenesis (37). Dysregulation of or an acquired resistance to cell death eventually leads to the development of CAC (38–40). Given that the increased susceptibility to tumor development in Irf1–/– mice could not be explained by differences in inflammasome activity or inflammatory mediators, we asked whether IRF1 regulated cell death following exposure to AOM and DSS. We used immunohistochemistry to identify the cells undergoing death in the colons of WT and Irf1–/– mice. We observed a reduced number of TUNEL+ cells in the colons of Irf1–/– mice 14 and 80 days after AOM injection compared with those of WT mice, whereas no difference was observed in untreated (day 0) WT and Irf1–/– mice (Figure 5A). In line with reduced cell death in the colons of Irf1–/– mice, we observed defective CASP3 and CASP7 activation in the colons of Irf1–/– mice 14 and 80 days after AOM injection with respect to WT mice (Figure 5, B and C), suggesting that IRF1 plays a role in apoptosis under these conditions. The roles of the inflammatory cell death pathways pyroptosis and necroptosis in CAC have not been clearly defined to date (9, 10). Pyroptosis is primarily implicated in clearing microbial pathogens during infection. However, recent studies have focused on tumor pyroptosis as an alternative mode of cell death for the treatment of cancer (41). Herein, we found reduced expression and activation of the pyroptosis executioner, GSDMD, in the colons of mice lacking IRF1 (Figure 5D), suggesting that IRF1 regulates pyroptosis. Consistent with the reduced expression of GSDMD observed in Irf1–/– mice, recent studies have found that IRF1 and IRF2 transcriptionally induce GSDMD for pyroptotic cell death (42, 43). An alternative mode of programmed cell death that overcomes resistance to both apoptosis and pyroptosis by triggering and amplifying antitumor immunity in cancer therapy is necroptosis (44). Necroptosis is executed by MLKL, which is phosphorylated and activated to form pores in the membrane (24). Necroptosis has been shown to be involved in the regression of colorectal tumors (34). In our study, colons of mice lacking IRF1 showed reduced expression of MLKL compared with the colons of WT mice (Figure 5D). In addition, we observed reduced cell death in organoids (Figure 5E) and bone marrow–derived macrophages (BMDMs) (Supplemental Figure 3) derived from Irf1–/– mice as compared with those of WT mice in response to the necroptotic trigger TNF + zVAD, further supporting the role of IRF1 in necroptosis. In principle, a multitude of different stimuli can initiate the activation of cell death machinery, which subsequently leads to different modes of cell death (45). To further understand the role of IRF1 in different modes of cell death, we stimulated BMDMs with staurosporine, shikonin, and RSL3, which have been extensively used as classical inducers of apoptosis, necroptosis, and ferroptosis, respectively (46). The dynamics of cell death induced by these triggers were similar between WT and Irf1–/– BMDMs, suggesting that IRF1 in macrophages is dispensable for mediating cell death in response to these classical triggers (Supplemental Figure 4, A–C). Overall, we identified IRF1 as a master regulator of PANoptosis in the colon during colorectal tumorigenesis through its function in both the myeloid and epithelial compartment to prevent colorectal cancer.
Figure 5. IRF1 regulates PANoptosis.
(A) Representative images of TUNEL staining of colons from azoxymethane/dextran sulfate sodium–treated (AOM/DSS-treated) WT and Irf1–/– mice on days 0, 14, and 80. (B) Representative images of cleaved caspase-3 (CASP3) staining of colon tissues from DSS-treated WT and Irf1–/– mice on days 0 and 14 after AOM injection. Scale bar: 100 μM. (C) Immunoblot analysis of the pro- and cleaved forms of CASP3 and caspase-7 (CASP7) in colons of WT and Irf1–/– mice on days 0, 14, and 80 after AOM injection. Blots represent data from the same biological samples at the indicated time point run in parallel. (D) Immunoblot analysis of GSDMD and MLKL in colons of WT and Irf1–/– mice 14 days after AOM injection. Blots represent data from the same biological samples run in parallel. (E) Cell death analysis of intestinal organoids derived from WT and Irf1–/– mice after stimulation with TNF + zVAD. **P < 0.01; ***P < 0.001. The 2-tailed t test (E) was used. Data are representative of 3 independent experiments. Data are represented as mean ± SEM.
Discussion
Our understanding of the tumor-suppressive function of IRF1 is largely based on several in vitro studies where IRF1 has been shown to regulate cellular proliferation and apoptosis (12–15, 47). However, how IRF1 affects tumorigenesis in a physiological setting has remained unexplored. In this study, we used an AOM/DSS-induced model and a sporadic model to investigate the role of IRF1 in colorectal tumorigenesis and showed that mice deficient in IRF1 were more susceptible to the development of CRC. It is known that continuous colonic inflammation predisposes tissue to CRC development. However, inflammasome signaling inhibits the development of CRC, and we have previously identified the role of IRF1 in inflammasome activation (4). The tumor-suppressive function of IRF1 was not associated with the regulation of inflammasome activation and inflammatory cytokines in the colon. Colitis results in the production of inflammatory cytokines and chemokines that drive tumor development and provide survival signals for tumor cells (10). However, these cytokines and chemokines were similarly produced in the colons of WT and Irf1–/– mice, indicating an inflammation-independent role for IRF1 in colon tumorigenesis. This inflammation independence is further supported by increased tumor burden in ApcMin/+ mice deficient in IRF1. Despite the similar cytokine profiles, we observed reduced cell death in the colons of Irf1–/– mice compared with WT mice, suggesting a cell-intrinsic function for IRF1 in inhibiting survival signaling in response to these inflammatory cytokines. The reduced cell death in Irf1–/– BMDMs in response to TNF + zVAD but not in response to staurosporine, shikonin, and RSL3 further supports a cell-intrinsic function for IRF1-mediated inhibition of survival signaling. Indeed, inflammatory cytokines, especially TNF and IFN-γ, are induced during colitis and act synergistically to cause cell death (48). Future studies are warranted to understand the role of IRF1 in cytokine-mediated cell death.
The appearance of an additional lower-molecular-weight product of IRF1 in the tumor tissue may be a cleaved form or a splice variant. Overtly activated executioners CASP3 and CASP7 may further cleave IRF1 in the tumor microenvironment to inactivate it and thus possibly promote tumor progression. These caspases are well known for their ability to cleave various signaling proteins, switching them into functionally inactive forms (49). Other studies have suggested that splicing aberrations in the IRF1 gene also account for the loss of IRF1 expression (50, 51). Although we observed the role of IRF1 in myeloid and epithelial cells in conferring protection against colon tumorigenesis, the function of IRF1 in other cell types cannot be overlooked. For instance, the presence of myofibroblasts around adenomatous colorectal polyps and primary tumor sites has previously been described, and their prevalence correlates with a higher rate of colon cancer recurrence (52, 53). The loss of IRF1 in subepithelial myofibroblasts and fibroblasts could contribute to the increased tumor formation in whole-body knockout mice. Future studies are warranted to understand the role of IRF1 in these fibroblasts and other cell types to prevent tumor formation in the colon.
Although the expression and activation of the pyroptosis executioner, GSDMD, is reduced in the colons of mice lacking IRF1, the production of IL-18 in the colon was similar in both the WT and Irf1–/– mice, suggesting that either a low level of GSDMD cleavage is sufficient or that other compensatory pathways exist for the release of IL-18 in the colon. The similar amount of IL-18 in both genotypes also implies that IRF1 provides protection against CAC independent of IL-18 signaling (54, 55). Consistent with the reduced expression of GSDMD observed in Irf1–/– mice, recent studies have found that IRF1 and IRF2 transcriptionally induce GSDMD for pyroptotic cell death (42, 43).
Because cancer cells can develop resistance to apoptosis induced by currently available chemotherapeutics (22), therapeutic agents that activate multiple forms of cell death in parallel could overcome this limitation in cancer therapy. In addition, molecular switching from one form of cell death to another could be an effective strategy for efficient killing of cancerous cells. Therefore, IRF1 could be an effective target in modulating multiple forms of cell death to enhance the effectiveness of cancer therapy.
Overall, in this study, we identified IRF1 as a master regulator of PANoptosis in the colon to suppress tumorigenesis. Understanding the precise functions of IRF1 in regulating these forms of cell death could open new avenues in the treatment of different forms of cancer.
Methods
Mice.
Irf1–/– mice (56) have been previously described. CMV-Cre mice were used to delete the conditional floxed Irf1 allele to generate the Irf1–/– mice. B6.129P2-Lyztm1(cre)Ifo/J (004781, The Jackson Laboratory) and B6.Cg-Tg(Vil-cre)997Gum/J mice (004586, The Jackson Laboratory) were used to delete the conditional floxed Irf1 allele in a cell-type–specific manner. C57BL/6J-ApcMin/+/J mice (002020, The Jackson Laboratory) were used as an ApcMin model of colorectal tumorigenesis. All mice were bred at St. Jude Children’s Research Hospital.
AOM/DSS model of colorectal tumorigenesis.
Both male and female mice were injected with 10 mg AOM (MilliporeSigma) per kg body weight according to previously established protocols (35). Five days later, 2.5% DSS (9011-18-1, Affymetrix) was given in the drinking water for 6 days, followed by regular drinking water for 2 weeks. This cycle was repeated twice with 2% DSS, and mice were sacrificed on day 80 (Supplemental Figure 1A). For day 14 samples, mice were injected with AOM and, after 5 days, administered 2.5% DSS for 6 days. Mice were then administered regular water for 3 days and sacrificed. Mice were cohoused for 4 weeks before AOM injection and remained separated over the course of the experiment. No randomization or blinding was performed.
ApcMin model of colorectal tumorigenesis.
ApcMin/+ mice were genetically crossed with Irf1–/– mice to generate littermate ApcMin/+, ApcMin/+Irf1+/–, and ApcMin/+Irf1–/– mice. These littermates were harvested to look for the prevalence of tumors in the colon.
Histology and microscopy analysis.
Colons were rolled into a “Swiss roll” and fixed in 10% formalin and then processed and embedded in paraffin by standard techniques. Longitudinal sections of 5-μm thickness were stained with H&E and examined by a pathologist blinded to the experimental groups. Colitis scores were assigned based on inflammation, ulceration, hyperplasia, and the extent or severity of the damage. Severity scores for inflammation were assigned as follows: 0, normal (within normal limits); 2, minimal (mixed inflammation, small, focal, or widely separated, limited to lamina propria); 15, mild (multifocal mixed inflammation, often extending into submucosa); 40, moderate (large multifocal lesions within mixed inflammation involving mucosa and submucosa); 80, marked (extensive mixed inflammation with edema and erosions); and 100, severe (diffuse inflammation with transmural lesions and multiple ulcers). Scores for ulceration were assigned as follows: 0, normal (none); 2, minimal (only 1 small focus of ulceration involving fewer than 5 crypts); 15, mild (a few small ulcers, up to 5 crypts); 40, moderate (multifocal ulcers, up to 10 crypts); 80, marked (multifocal to coalescing ulcers involving more than 10 crypts each); and 100, severe (extensive to diffuse, with multiple ulcers covering more than 20 crypts each). Scores for hyperplasia were assigned as follows: 0, normal; 2, minimal (some areas with crypts elongated and increased mitoses); 15, mild (multifocal areas with crypts elongated up to twice the normal thickness, normal goblet cells present); 40, moderate (extensive areas with crypts up to 2 times normal thickness, reduced goblet cells); 80, marked (mucosa over twice the normal thickness, hyperchromatic epithelium, reduced or rare goblet cells, possibly foci of arborization); and 100, severe (mucosa twice the normal thickness, marked hyperchromasia, crowding/stacking, absence of goblet cells, high mitotic index, and arborization). Damage extent scores were assigned as follows: 0, normal (rare or inconspicuous lesions); 2, minimal (less than 5% involvement); 15, mild (multifocal but conspicuous lesions, 5%–10% involvement); 40, moderate (multifocal, prominent lesions, 10%–50% involvement); 80, marked (coalescing to extensive lesions or areas of inflammation with some loss of structure, 50%–90% involvement); and 100, severe (diffuse lesion with effacement of normal structure, >90% involvement).
TUNEL staining was performed using the In Situ Cell Death Detection Kit (11684817910, MilliporeSigma) according to the manufacturer’s instructions. Cleaved CASP3 staining was performed according to the manufacturer’s instructions (catalog 4704, Essen Biosciences). Tissues were counterstained with hematoxylin.
Cytokine measurement by ELISA.
Cytokines in the colon were measured by ELISA according to manufacturer’s instructions. IL-18 was measured using an ELISA kit (catalog BMS618-3TEN, eBioscience). All other cytokines were measured using a multiplex ELISA (catalog MCYTOMAG-70K, MilliporeSigma).
Western blotting.
Cell lysates and culture supernatants were combined in caspase lysis buffer (containing protease inhibitors, phosphatase inhibitors, 10% NP-40, and 25 mM DTT) and sample loading buffer (containing SDS and 2-mercaptoethanol) for immunoblot analysis of caspases. For immunoblot analysis of signaling components, supernatants were removed, and cells were washed once with PBS, followed by lysis in RIPA buffer and sample loading buffer. Proteins were separated by electrophoresis through 8%–12% polyacrylamide gels (57). Following electrophoretic transfer of proteins onto PVDF membranes (IPVH00010, MilliporeSigma), nonspecific binding was blocked by incubation with 5% skim milk, and then membranes were incubated with primary antibodies against CASP3 (9662, Cell Signaling Technology [CST]), cleaved CASP3 (9661, CST), CASP7 (9492, CST), cleaved CASP7 (9491, CST), CASP8 (AG-20T-0138-C100, AdipoGen), cleaved CASP8 (8592, CST), IRF1 (8478, CST), P-ERK (9101, CST), ERK (9102, CST), P-IκBα (9241, CST), IκBα (9242, CST), P-STAT3 Tyr705 (9131, CST), STAT3 (9139, CST), and GAPDH (5174, CST). Membranes were then washed and incubated with the appropriate HRP-conjugated secondary antibodies (111-035-047, anti-rabbit; 315-035-047, anti-mouse; 705-035-003, anti-goat; Jackson ImmunoResearch Laboratories). Proteins were visualized using Luminata Forte Western HRP Substrate (WBLUF0500, MilliporeSigma).
RT-PCR analysis.
RNA was extracted using TRIzol (15596026, Thermo Fisher Scientific) according to the manufacturer’s instructions. The isolated RNA was reverse-transcribed into cDNA using a First-Strand cDNA Synthesis Kit (4368814, Applied Biosystems). Real-time quantitative PCR was performed on an ABI 7500 RT-PCR instrument using 2× SYBR Green (4368706, Applied Biosystems) and the appropriate primers. Sequences for qRT-PCR primers are listed in Supplemental Table 1.
Cell culture and real-time imaging for cell death.
Primary mouse BMDMs were cultured as described previously (35). BMDMs (5 × 105 cells/well) were seeded in 24-well plates. Cells were stimulated with 50 ng/mL TNF (315-01A, PeproTech), 50 ng/mL LPS (Invivogen), 25 μM zVAD-FMK (Calbiochem), 25 μM Nec1 (MilliporeSigma), 1 μM staurosporine (S5921, MilliporeSigma), 5 μM shikonin (S7576, MilliporeSigma), or 5 μM RSL3 (SML2234, MilliporeSigma) and stained with propidium iodide (P3566, Life Technologies) following the manufacturers’ protocols. The plate was scanned, and fluorescence and phase-contrast images (4 image fields/well) were acquired in real time every hour after stimulation. Resulting images were analyzed using the software package supplied with the IncuCyte imager (Essen Bioscience).
Flow cytometry.
Colons were first dissected and then washed with ice-cold PBS. Washed colons were cut into small pieces, which were incubated in PBS containing 10 mM HEPES, 5 mM EDTA, and 1 mM DTT at 37°C for 30 minutes with gentle shaking to remove the epithelial layer. The colon segments were further digested in RPMI medium containing 0.5 mg/mL collagenase D at 37°C for 1.5 hours. The supernatant from the digested colon segments was passed through a 70-μm cell strainer and enriched using 37.5% percoll to isolate lamina propria cells. The following monoclonal antibodies were used for flow cytometry: Gr1 (RB6-8C5; 108426), F4/80 (BM8; 123116), and CD4 (GK1.5; 100408) from BioLegend; CD19 (1D3; 35-0193-U025), NK1.1 (PK136; 65-5941-U025), TCRb (H57-597; 20-5961-U100), and CD45.2 (104; 60-0454-U025) from Tonbo Biosciences; CD11b (M1/70; 48-0112-82) from Invitrogen; CD8a (53-6.7; 48-0081-82) from eBioscience; and CD11c (HL3; 557401) from BD Pharmingen. Cells were gated on live single-cell populations and hematopoietic cells using the CD45.2 gate followed by separation of each of the specific cell populations using the following cell surface markers: macrophages (CD11b+, F4/80+), dendritic cells (CD11c+ Gr1–), neutrophils (CD11b+, Gr1hi), NK cells (NK1.1+), CD8 T cells (TCRb+, CD8+, CD4–), and CD4 T cells (TCRb+, CD8–, CD4+).
Colon organoid culture.
Mouse colon stem cells were cultured using IntestiCult Organoid Growth Media according to the manufacturer’s instructions (STEMCELL Technologies). The whole colon was removed from untreated WT and Irf1–/– mice and rinsed with cold PBS. The colon was cut into 2-mm segments and washed 20 times with cold PBS. Colonic segments were incubated in Gentle Cell Dissociation Reagent (STEMCELL Technologies) and rotated on a rocking platform for 15 minutes at room temperature, followed by resuspension in 0.1% BSA in PBS. Dissociated colonic crypts were vigorously suspended. Cells were filtered through 70-μm strainers, mixed with Matrigel (356230, Corning), and plated in 24-well culture plates (3738, Corning). IntestiCult Organoid Growth Media was added to the cell culture plates to immerse the Matrigel.
Human cancer patient data analysis.
Data from patients at different stages of disease during CRC were obtained from the publicly available The Cancer Genome Atlas database. Patients were classified into either high or low IRF1 mRNA expression groups using the best expression cutoff criterion described in the Human Protein Atlas (58, 59).
Statistics.
Statistical significance was determined by the 2-tailed Student’s t test or 1- or 2-way ANOVA using GraphPad Prism v7.0. The specific test used is indicated in the respective figure legends. P < 0.05 was considered statistically significant.
Study approval.
All animal studies were conducted under protocols approved by St. Jude Children’s Research Hospital’s committee on the use and care of animals and in accordance with all relevant ethical guidelines.
Author contributions
RK and TDK conceptualized the study. RK, BRS, and EL designed the methodology. RK, BRS, EL, BB, RKSM, PS, ST, and ARB performed the experiments. RK, BRS, EL, BB, RKSM, PS, ST, HM, ARB, and PV conducted the analysis. RK wrote the original draft; all authors reviewed and edited the draft and approved the final version. TDK acquired the funding and provided resources and supervision.
Supplementary Material
Acknowledgments
We thank members of the Kanneganti lab for their comments and suggestions. Several images were acquired at the St. Jude Cell & Tissue Imaging Center, which is supported by St. Jude Children’s Research Hospital and National Cancer Institute grant P30 CA021765-35. We also thank Rebecca Tweedell for scientific editing and writing support. Work from our laboratory is supported by the US NIH (grants AI101935, AI124346, AR056296, and CA163507 to TDK) and the American Lebanese Syrian Associated Charities (to TDK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 06/18/2020
Electronic publication
Version 2. 06/18/2020
Figure 4 was updated.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(12):e136720.https://doi.org/10.1172/jci.insight.136720.
Contributor Information
Rajendra Karki, Email: rajendra.karki@stjude.org.
Ein Lee, Email: ein1201@gmail.com.
Balaji Banoth, Email: balaji.banoth@stjude.org.
Harisankeerth Mummareddy, Email: harimumm@gmail.com.
Peter Vogel, Email: Peter.Vogel@stjude.org.
Thirumala-Devi Kanneganti, Email: Thirumala-Devi.Kanneganti@STJUDE.ORG.
References
- 1.Fujita T, Reis LF, Watanabe N, Kimura Y, Taniguchi T, Vilcek J. Induction of the transcription factor IRF-1 and interferon-beta mRNAs by cytokines and activators of second-messenger pathways. Proc Natl Acad Sci USA. 1989;86(24):9936–9940. doi: 10.1073/pnas.86.24.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamijo R, et al. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263(5153):1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 3.Bouker KB, et al. Interferon regulatory factor-1 (IRF-1) exhibits tumor suppressor activities in breast cancer associated with caspase activation and induction of apoptosis. Carcinogenesis. 2005;26(9):1527–1535. doi: 10.1093/carcin/bgi113. [DOI] [PubMed] [Google Scholar]
- 4.Man SM, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16(5):467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakazawa T, et al. Complete suppression of insulitis and diabetes in NOD mice lacking interferon regulatory factor-1. J Autoimmun. 2001;17(2):119–125. doi: 10.1006/jaut.2001.0531. [DOI] [PubMed] [Google Scholar]
- 6.Buch T, Uthoff-Hachenberg C, Waisman A. Protection from autoimmune brain inflammation in mice lacking IFN-regulatory factor-1 is associated with Th2-type cytokines. Int Immunol. 2003;15(7):855–859. doi: 10.1093/intimm/dxg086. [DOI] [PubMed] [Google Scholar]
- 7.Kuriakose T, et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016;1(2):aag2045. doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briard B, et al. Fungal ligands released by innate immune effectors promote inflammasome activation during Aspergillus fumigatus infection. Nat Microbiol. 2019;4(2):316–327. doi: 10.1038/s41564-018-0298-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karki R, Man SM, Kanneganti TD. Inflammasomes and cancer. Cancer Immunol Res. 2017;5(2):94–99. doi: 10.1158/2326-6066.CIR-16-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19(4):197–214. doi: 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka N, et al. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature. 1996;382(6594):816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- 12.Romeo G, Fiorucci G, Chiantore MV, Percario ZA, Vannucchi S, Affabris E. IRF-1 as a negative regulator of cell proliferation. J Interferon Cytokine Res. 2002;22(1):39–47. doi: 10.1089/107999002753452647. [DOI] [PubMed] [Google Scholar]
- 13.Kim EJ, Lee JM, Namkoong SE, Um SJ, Park JS. Interferon regulatory factor-1 mediates interferon-gamma-induced apoptosis in ovarian carcinoma cells. J Cell Biochem. 2002;85(2):369–380. doi: 10.1002/jcb.10142. [DOI] [PubMed] [Google Scholar]
- 14.Pizzoferrato E, et al. Ectopic expression of interferon regulatory factor-1 promotes human breast cancer cell death and results in reduced expression of survivin. Cancer Res. 2004;64(22):8381–8388. doi: 10.1158/0008-5472.CAN-04-2223. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, et al. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ. 2010;17(4):699–709. doi: 10.1038/cdd.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim PK, et al. IRF-1 expression induces apoptosis and inhibits tumor growth in mouse mammary cancer cells in vitro and in vivo. Oncogene. 2004;23(5):1125–1135. doi: 10.1038/sj.onc.1207023. [DOI] [PubMed] [Google Scholar]
- 17.Chen FF, Jiang G, Xu K, Zheng JN. Function and mechanism by which interferon regulatory factor-1 inhibits oncogenesis. Oncol Lett. 2013;5(2):417–423. doi: 10.3892/ol.2012.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogasawara S, et al. Common deleted region on the long arm of chromosome 5 in esophageal carcinoma. Gastroenterology. 1996;110(1):52–57. doi: 10.1053/gast.1996.v110.pm8536888. [DOI] [PubMed] [Google Scholar]
- 19.Tamura G, et al. Two distinct regions of deletion on the long arm of chromosome 5 in differentiated adenocarcinomas of the stomach. Cancer Res. 1996;56(3):612–615. [PubMed] [Google Scholar]
- 20.Bouker KB, et al. The A4396G polymorphism in interferon regulatory factor 1 is frequently expressed in breast cancer cell lines. Cancer Genet Cytogenet. 2007;175(1):61–64. doi: 10.1016/j.cancergencyto.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 21. American Cancer Society. Cancer facts & figures 2020. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2020/cancer-facts-and-figures-2020.pdf Accessed on June 10, 2020.
- 22.Mohammad RM, et al. Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol. 2015;35 Suppl:S78–S103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24(1):105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11(10):700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 26.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343(6177):1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 27.Gurung P, et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol. 2014;192(4):1835–1846. doi: 10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taabazuing CY, Okondo MC, Bachovchin DA. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem Biol. 2017;24(4):507–514.e4. doi: 10.1016/j.chembiol.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malireddi RKS, et al. TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J Exp Med. 2018;215(4):1023–1034. doi: 10.1084/jem.20171922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malireddi RKS, Kesavardhana S, Kanneganti TD. ZBP1 and TAK1: master regulators of nlrp3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front Cell Infect Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng M, Karki R, Vogel P, Kanneganti TD. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell. 2020;181(3):674–687.e13. doi: 10.1016/j.cell.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukens JR, et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 2014;516(7530):246–249. doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia X, et al. The role of pyroptosis in cancer: pro-cancer or pro-”host”? Cell Death Dis. 2019;10(9):650. doi: 10.1038/s41419-019-1883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He GW, et al. Regression of apoptosis-resistant colorectal tumors by induction of necroptosis in mice. J Exp Med. 2017;214(6):1655–1662. doi: 10.1084/jem.20160442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karki R, et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature. 2016;540(7634):583–587. doi: 10.1038/nature20597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penninger JM, et al. The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity. 1997;7(2):243–254. doi: 10.1016/S1074-7613(00)80527-0. [DOI] [PubMed] [Google Scholar]
- 37.Watson AJ. An overview of apoptosis and the prevention of colorectal cancer. Crit Rev Oncol Hematol. 2006;57(2):107–121. doi: 10.1016/j.critrevonc.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Plati J, Bucur O, Khosravi-Far R. Dysregulation of apoptotic signaling in cancer: molecular mechanisms and therapeutic opportunities. J Cell Biochem. 2008;104(4):1124–1149. doi: 10.1002/jcb.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Safa AR. Resistance to cell death and its modulation in cancer stem cells. Crit Rev Oncog. 2016;21(3-4):203–219. doi: 10.1615/CritRevOncog.2016016976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labi V, Erlacher M. How cell death shapes cancer. Cell Death Dis. 2015;6:e1675. doi: 10.1038/cddis.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagarajan K, Soundarapandian K, Thorne RF, Li D, Li D. Activation of pyroptotic cell death pathways in cancer: an alternative therapeutic approach. Transl Oncol. 2019;12(7):925–931. doi: 10.1016/j.tranon.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thygesen SJ, Stacey KJ. IRF1 and IRF2 regulate the non-canonical inflammasome. EMBO Rep. 2019;20(9):e48891. doi: 10.15252/embr.201948891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kayagaki N, et al. IRF2 transcriptionally induces GSDMD expression for pyroptosis. Sci Signal. 2019;12(582):eaax4917. doi: 10.1126/scisignal.aax4917. [DOI] [PubMed] [Google Scholar]
- 44.Su Z, Yang Z, Xie L, DeWitt JP, Chen Y. Cancer therapy in the necroptosis era. Cell Death Differ. 2016;23(5):748–756. doi: 10.1038/cdd.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281(5381):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 46.Ke B, Tian M, Li J, Liu B, He G. Targeting programmed cell death using small-molecule compounds to improve potential cancer therapy. Med Res Rev. 2016;36(6):983–1035. doi: 10.1002/med.21398. [DOI] [PubMed] [Google Scholar]
- 47.Stang MT, et al. Interferon regulatory factor-1-induced apoptosis mediated by a ligand-independent fas-associated death domain pathway in breast cancer cells. Oncogene. 2007;26(44):6420–6430. doi: 10.1038/sj.onc.1210470. [DOI] [PubMed] [Google Scholar]
- 48.Kalai M, et al. Regulation of the expression and processing of caspase-12. J Cell Biol. 2003;162(3):457–467. doi: 10.1083/jcb.200303157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widmann C, Gibson S, Johnson GL. Caspase-dependent cleavage of signaling proteins during apoptosis. A turn-off mechanism for anti-apoptotic signals. J Biol Chem. 1998;273(12):7141–7147. doi: 10.1074/jbc.273.12.7141. [DOI] [PubMed] [Google Scholar]
- 50.Harada H, et al. Accelerated exon skipping of IRF-1 mRNA in human myelodysplasia/leukemia; a possible mechanism of tumor suppressor inactivation. Oncogene. 1994;9(11):3313–3320. [PubMed] [Google Scholar]
- 51.Tzoanopoulos D, et al. Low expression of interferon regulatory factor-1 and identification of novel exons skipping in patients with chronic myeloid leukaemia. Br J Haematol. 2002;119(1):46–53. doi: 10.1046/j.1365-2141.2002.03829.x. [DOI] [PubMed] [Google Scholar]
- 52.Tsujino T, et al. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin Cancer Res. 2007;13(7):2082–2090. doi: 10.1158/1078-0432.CCR-06-2191. [DOI] [PubMed] [Google Scholar]
- 53.Koliaraki V, Pallangyo CK, Greten FR, Kollias G. Mesenchymal cells in colon cancer. Gastroenterology. 2017;152(5):964–979. doi: 10.1053/j.gastro.2016.11.049. [DOI] [PubMed] [Google Scholar]
- 54.Man SM, et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 2015;162(1):45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185(8):4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer. 2016;16(7):447–462. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 57.Sharma BR, Karki R, Lee E, Zhu Q, Gurung P, Kanneganti TD. Innate immune adaptor MyD88 deficiency prevents skin inflammation in SHARPIN-deficient mice. Cell Death Differ. 2019;26(4):741–750. doi: 10.1038/s41418-018-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhlen M, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 59.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.