ABSTRACT
Proper orientation of the mitotic spindle is critical for accurate development and morphogenesis. In human cells, spindle orientation is regulated by the evolutionarily conserved protein NuMA, which interacts with dynein and enriches it at the cell cortex. Pulling forces generated by cortical dynein orient the mitotic spindle. Cdk1-mediated phosphorylation of NuMA at threonine 2055 (T2055) negatively regulates its cortical localization. Thus, only NuMA not phosphorylated at T2055 localizes at the cell cortex. However, the identity and the mechanism of action of the phosphatase complex involved in T2055 dephosphorylation remains elusive. Here, we characterized the PPP2CA-B55γ (PPP2R2C)–PPP2R1B complex that counteracts Cdk1 to orchestrate cortical NuMA for proper spindle orientation. In vitro reconstitution experiments revealed that this complex is sufficient for T2055 dephosphorylation. Importantly, we identified polybasic residues in NuMA that are critical for T2055 dephosphorylation, and for maintaining appropriate cortical NuMA levels for accurate spindle elongation. Furthermore, we found that Cdk1-mediated phosphorylation and PP2A-B55γ-mediated dephosphorylation at T2055 are reversible events. Altogether, this study uncovers a novel mechanism by which Cdk1 and its counteracting PP2A-B55γ complex orchestrate spatiotemporal levels of cortical force generators for flawless mitosis.
KEY WORDS: Cdk1, NuMA, PP2A, Dynein, Mitosis, Spindle orientation
Summary: PP2A-B55γ orchestrates mitotic spindle behavior by dephosphorylating NuMA.
INTRODUCTION
Proper orientation and elongation of the mitotic spindle are critical events for defining the accurate positioning of the cleavage furrow and for faithful segregation of the genetic material (Gönczy, 2008; Siller and Doe, 2009; Morin and Bellaiche, 2011). This process further ensures that cell fate determinants are accurately segregated in the resulting daughter cells during asymmetric cell division, including in stem cells (Knoblich, 2008). In animal cells, one of the critical determinants of the control of spindle orientation and elongation is an evolutionarily conserved cortically anchored protein NuMA (LIN-5 in Caenorhabditis elegans; Mud in Drosophila; Siller and Doe, 2009; di Pietro et al., 2016; Bergstralh et al., 2017). Cortical NuMA serves to anchor the microtubule-dependent minus-end-directed motor protein complex dynein and its associated dynactin complex (Kotak, 2019). Motor activity of such cortically anchored dynein–dynactin is assumed to generate pulling forces on the astral microtubules and thus on the spindle apparatus to control mitotic spindle behavior in time and space (Nguyen-Ngoc et al., 2007; Redemann et al., 2010; Laan et al., 2012; Kotak et al., 2012, 2013; Collins et al., 2012; Schmidt et al., 2017; Okumura et al., 2018; Fielmich et al., 2018).
Other than its function in spindle orientation and elongation, NuMA is required for proper assembly as well as maintenance of the mitotic spindle with the help of dynein–dynactin (Yang and Snyder, 1992; Compton and Cleveland, 1993; Merdes et al., 1996, 2000; Hueschen et al., 2019). NuMA is a large protein of 2115 amino acids, and it comprises two globular domains separated by a coiled-coil domain in the middle (Yang et al., 1992). NuMA interacts with dynein through its N-terminus (Kotak et al., 2012), and its C-terminus contains domains that mediate its interaction with LGN (also known as GPSM2), ERM4.1 (EBP41), microtubules, importin-α, and phosphoinositides (Mattagajasingh et al., 1999; Du et al., 2002; Du and Macara, 2004; Woodard et al., 2010; Seldin et al., 2013, 2016; Kiyomitsu and Cheeseman, 2013; Zheng et al., 2014; Kotak et al., 2014; Gallini et al., 2016; Chang et al., 2017). Interestingly, recent studies have highlighted the presence of a conserved region in the C-terminus of NuMA that is necessary for cluster formation, and possibly for generating strong pulling forces at the cell cortex (Okumura et al., 2018; Pirovano et al., 2019).
Because NuMA acts as an essential adaptor for cortical dynein–dynactin in mitosis, its localization is tightly coupled with the mitotic phases. NuMA cortical levels are low in metaphase and substantially increased during anaphase (Kiyomitsu and Cheeseman, 2013; Kotak et al., 2014; Seldin et al., 2013; Zheng et al., 2014). These localization patterns ensure proper spindle orientation and elongation events in metaphase and anaphase, respectively (Kotak et al., 2012, 2014; Collins et al., 2012; Seldin et al., 2013; Zheng et al., 2014). Notably, this tight regulation is achieved with the help of several mitotic kinases including Cdk1–cyclinB (hereafter referred to as Cdk1), Aurora A and Plk1 (Kiyomitsu and Cheeseman, 2012, 2013; Kotak et al., 2013; Seldin et al., 2013; Zheng et al., 2014; Gallini et al., 2016; Kotak et al., 2016; Connell et al., 2017; Sana et al., 2018). We have shown previously that NuMA is directly phosphorylated at threonine 2055 (T2055) by Cdk1 (Kotak et al., 2013). This phosphorylation negatively regulates cortical accumulation of NuMA in metaphase, where cortical NuMA localization is LGN-dependent, as well as in anaphase, where NuMA relies on membrane phosphoinositides for its cortical accumulation (Kiyomitsu and Cheeseman, 2013; Kotak et al., 2014; Seldin et al., 2013; Zheng et al., 2014). Interestingly, loss of the catalytic subunit of PP2A, PPP2CA, abolishes cortical NuMA and dynein distributions (Kotak et al., 2013). However, whether PPP2CA regulates cortical NuMA by directly acting at the Cdk1-phosphorylated residue T2055 remains unknown. PP2A forms a trimeric holoenzyme complex that consists of a catalytic subunit, a scaffold subunit, and variable regulatory subunits of four different families: B55 (also known as PR55 or B), B56 (PR61 or B′), B72 (PR72 or B″) and striatin (PR93 or B‴) (Barr et al., 2011; Moura and Conde, 2019). It is the regulatory subunit that provides the substrate specificity, and thus even if PPP2CA counteracts Cdk1-mediated phosphorylation of T2055, the identity of the regulatory subunit, and the mechanism of its association with NuMA, remains elusive. In general, in stark contrast to mitotic kinases, our knowledge regarding how mitotic phosphatases regulate spindle orientation and spindle elongation remains limited.
In this study, using a combination of cell biology and biochemical experiments, we uncovered that a PP2A complex containing B55γ (also known as PPP2R2C) is involved in T2055 dephosphorylation, and thus for maintaining the cortical pool of NuMA and dynein–dynactin in mitosis. Because cortical NuMA is critical for proper spindle orientation, loss of B55γ caused spindle orientation defects during metaphase. Moreover, we discovered that conserved polybasic residues in the vicinity of the Cdk1 phosphorylation site T2055 are the key for robust spatiotemporal NuMA localization in mitosis. Importantly, data obtained from chemically induced mitotic exit followed by mitotic entry reveal that phosphorylation and dephosphorylation at T2055 are reversible events. In summary, our work identifies a novel B55γ-containing PP2A complex that counteracts Cdk1 and spatiotemporally regulates cortical levels of NuMA and dynein to ensure unperturbed spindle behavior during mitosis.
RESULTS
B55γ regulates cortical levels of NuMA and dynein by dephosphorylating T2055 of NuMA
Cdk1-mediated phosphorylation of NuMA at T2055 (pT2055), prevents its accumulation at the cell cortex in metaphase (Kotak et al., 2013; Seldin et al., 2013). Because RNAi-mediated depletion of PPP2CA negatively impacts cortical localization of NuMA in metaphase (Kotak et al., 2013), one possibility is that PPP2CA counteracts Cdk1-mediated phosphorylation of T2055. Therefore, we sought to investigate whether PPP2CA dephosphorylates NuMA at T2055. As shown previously, we found that acute treatment with the Cdk1 inhibitor RO-3306 led to robust dephosphorylation of T2055, and thus loss of pT2055 signal at the spindle pole (Fig. S1A,B; Kotak et al., 2013). However, T2055 dephosphorylation upon RO-3306 treatment was significantly reduced in cells that were depleted for PPP2CA (Fig. S1B,C). Importantly, among all phosphoprotein phosphatase (PPP) family members that are generally involved in the dephosphorylation of the mitotic substrates (Barr et al., 2011; Moura and Conde, 2019), PPP2CA appeared the only catalytic subunit that was responsible for T2055 dephosphorylation (Fig. S1D).
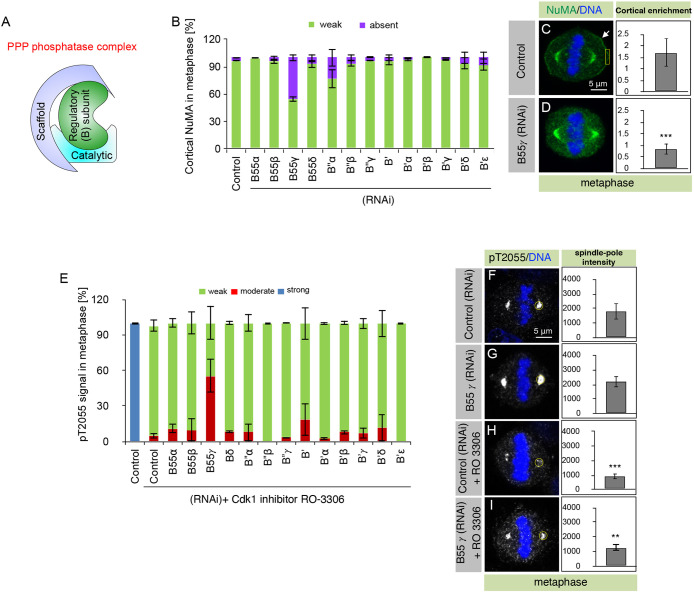
PPP2CA interacts with a regulatory subunit and a scaffold subunit (Fig. 1A), and it is the regulatory subunit that provides substrate specificity to the PPP2CA-containing trimeric complex (Barr et al., 2011; Cundell et al., 2013; Moura and Conde, 2019). To establish which regulatory and scaffold subunits are part of PPP2CA-containing phosphatase holoenzyme involved in pT2055 dephosphorylation, we depleted all 13 regulatory subunits and the two scaffold subunits that could be part of a PPP2CA-based trimeric complex. Interestingly, siRNA-mediated depletion of B55γ and PPP2R1B using multiple siRNAs caused the loss of cortical NuMA and dynein-interacting dynactin subunit p150Glued (also known as DCTN1) (Fig. 1B–D; Fig. S1E,F; data not shown). This impact of B55γ depletion on cortical NuMA was also observed in HeLa Kyoto cells that stably express NuMA with AcGFP (Aequorea coerulescens GFP) and a mono-FLAG epitope (AcGFP–NuMA; Fig. S2A–D). Analogous results were obtained for dynein localization in HeLa cell line that stably expresses GFP fused to the dynein heavy chain DHC1 (also known as DYNC1H1) (Fig. S2E,F). Next, we decided to identify the nature of the regulatory and scaffold subunits whose loss stabilizes pT2055 upon acute Cdk1 inactivation. Remarkably, in this assay as well, only B55γ and PPP2R1B siRNA-treated cells showed significant stabilization of pT2055 (Fig. 1E–I; Fig. S1G–I). Analogous observations were made in non-transformed hTERT-RPE1 cells (data not shown). Overall, these data suggest that PPP2CA–B55γ–PPP2R1B is a bona fide tripartite phosphatase holoenzyme complex involved in NuMA dephosphorylation at T2055, and which thereby orchestrates cortical NuMA levels during mitosis.
Fig. 1.
PP2A-B55γ is required for NuMA dephosphorylation at Cdk1 site T2055. (A) Schematic representation of a PPP phosphatase complex containing a catalytic subunit, a regulatory (B) subunit, and a scaffold subunit. (B) HeLa cells, transfected with control siRNAs or siRNAs against all thirteen different PP2A regulatory subunits, as indicated, were assayed for NuMA localization in metaphase. Cells were fixed 72 h after siRNA transfection and stained for NuMA. Stacked columns show the extent of cortical NuMA that was visually quantified under the epifluorescence microscope, and categorized as either absent (violet bars; example cell is shown in D) or weak (green bars; example cell is shown in C). More than 50 cells were analyzed in each condition, and the experiments were repeated three times. Data are mean±s.d. calculated from the percentage of cells in each category (weak or absent) from three independent experiments. Two or three different siRNAs were tested for each gene (see Table S1). Also, note that there could be potential false negatives in the dataset as we have not assayed the knockdown efficiency for each siRNA. (C,D) Metaphase HeLa cells transfected with control siRNAs (C) or siRNAs against B55γ (D). Cells were fixed 72 h after siRNA transfection and stained for NuMA (green) and DNA (blue). Arrows indicate cortical localization. Quantification of cortical enrichment was performed in an area of size 1.8 μm×4 μm (yellow box; see Materials and Methods for detail). Quantification of mean±s.d. cortical enrichment is shown for ten metaphase cells in each condition. (E) HeLa cells, transfected with control siRNAs or siRNAs against the thirteen regulatory PP2A subunits, and treated with Cdk1 inhibitor RO-3306 (10 μM; Vassilev et al., 2006) for 5 min, were assayed for pT2055 in metaphase. Cells were fixed 72 h after siRNA transfection and stained with an antibody against pT2055. Stacked columns show the extent of pT2055 signal at the spindle poles, as quantified visually under the epifluorescence microscope, and categorized as either weak (comparable to control cells upon RO-3306 treatment, green bars; example cell shown in H), strong (blue bars; example untreated control cell shown in F), or moderate (signal between weak and strong, red bars; example cell shown in I). More than 50 cells were analyzed in each condition, and the experiment was repeated three times. Data are mean±s.d. calculated from the percentage of cells in each category (weak, moderate or strong) from three independent experiments. (F–I) Metaphase HeLa cells transfected with control siRNAs (F), siRNAs against B55γ (G), control cells that were treated with RO-3305 for 5 min (H), or B55γ siRNA-treated cells that were treated with RO-3306 for 5 min (I). pT2055 signal is shown in gray and DNA is shown in blue. Quantification of spindle pole enrichment was performed for ten representative cells using a 16 µm2 region (yellow) and adjusted for background signal (see Materials and Methods for detail). Data are mean ±s.d. P=0.156 between control and B55γ siRNA-treated metaphase cells for the spindle pole, P<0.0001 between control untreated and RO-3306-treated cells, and P=0.0048 between control RO-3306-treated and B55γ siRNA RO-3306-treated cells. **P<0.01, ***P<0.0001 (two-tailed Student's t-test).
The PP2A-B55γ phosphatase complex dephosphorylates pT2055
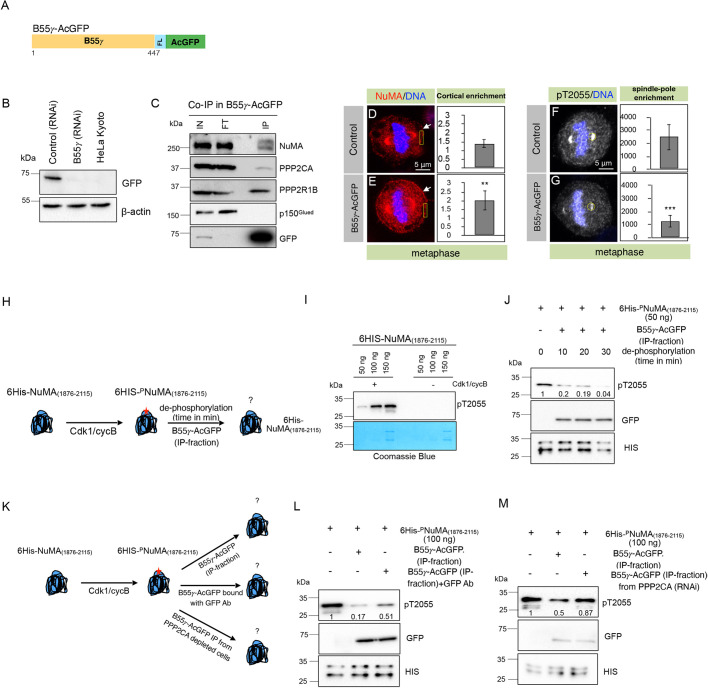
Next, we sought to biochemically determine whether the B55γ-containing PPP2CA-based tripartite complex can dephosphorylate a NuMA peptide sequence that is phosphorylated by Cdk1. To this end, we generated a stable HeLa cell line constitutively expressing low levels of B55γ–AcGFP (Fig. 2A,B). Immunoprecipitates (IP) from nocodazole-synchronized mitotic cell extract made from B55γ–AcGFP-expressing cells, but not those from control cells expressing AcGFP, interacted with endogenous NuMA (Fig. 2C; Fig. S3A). This data suggests that B55γ specifically interacts with NuMA. As expected, the IP fraction from B55γ–AcGFP cells also showed interaction with the catalytic subunit PPP2CA, and the regulatory subunit PPP2R1B, but not with p150Glued (Fig. 2C). Interestingly, ectopic expression of B55γ was sufficient to significantly enrich cortical NuMA and concomitantly decrease the levels of pT2055 (Fig. 2D–G). Next, we attempted to test whether the IP fraction obtained from B55γ–AcGFP cells could dephosphorylate a hexa-His-tagged NuMA(1876–2115) [6His–NuMA(1876–2115)] fragment that had been phosphorylated by recombinant Cdk1 (Fig. 2H). Notably, the B55γ–AcGFP IP fraction could dephosphorylate pT2055 with high specificity, whereas IP fractions that were either made from cells stably expressing AcGFP fused with a non-specific protein (CAAX–FLAG–AcGFP) or another B55 subunit (B55α, also known as PPP2R2A) did not show significant dephosphorylation of pT2055 (Fig. 2I,J; Fig. S3B–E). Furthermore, to ensure the specificity of B55γ in these dephosphorylation experiments, we incubated the B55γ–AcGFP IP fraction with an antibody against GFP to mask the AcGFP-tagged B55γ regulatory subunit prior to the dephosphorylation reaction (Fig. 2K). Interestingly, the IP-fraction incubated with the GFP antibody had a substantially weakened dephosphorylation capability (Fig. 2L). Moreover, the B55γ–AcGFP IP fraction from cells depleted of the catalytic subunit PPP2CA did not support efficient pT2055 dephosphorylation (Fig. 2M; Fig. S3F). Overall, these data strongly support the notion that the B55γ-containing PP2A complex is sufficient to dephosphorylate NuMA at the evolutionarily conserved Cdk1 residue, T2055.
Fig. 2.
PP2A-B55γ dephosphorylates pT2055. (A) Schematic of B55γ construct with FLAG (FL) and AcGFP tags at the C-terminus (B55γ–AcGFP). (B) Characterization of HeLa Kyoto cell line stably expressing B55γ–AcGFP using western blot analysis. β-actin is shown as a loading control. (C) Co-immunoprecipitation (IP) by GFP-Trap from lysates of nocodazole-arrested mitotic HeLa cells stably expressing B55γ–AcGFP. Resulting blots were probed for NuMA, PPP2CA, PPP2R1B, p150Glued, and GFP, as indicated. IN, input (1% of total); IP, 50% of the total immunoprecipitate; FT, flow-through. For GFP detection in the IP fraction, only 10% of the IP fraction was loaded. Note, B55γ interacts with NuMA, but not with the dynein-interacting dynactin subunit p150Glued. (D,E) Metaphase HeLa Kyoto control cells (D), or cells stably expressing B55γ–AcGFP (E) were fixed and stained for NuMA (red) and DNA (blue). Arrows indicate cortical localization, box indicates example region used for quantification. Note enrichment in cortical NuMA in cells expressing B55γ–AcGFP. Data are mean±s.d. cortical NuMA levels of 12 cells. P=0.0061 between control and B55γ–AcGFP-expressing cells (two-tailed Student's t-test). (F,G) Control metaphase HeLa cells (F), or cells stably expressing B55γ–AcGFP (G) fixed and stained for pT2055 (gray) and DNA (blue). Circle indicates example region used for quantification. Note significantly diminished levels of spindle pole pT2055 in cells expressing B55γ–AcGFP. Data are mean±s.d. spindle pole pT2055 levels of 20 cells. P<0.0001 between control and B55γ–AcGFP-expressing cells (two-tailed Student's t-test). (H) Schematic representation of the in vitro dephosphorylation assay whereby 6His–NuMA(1876–2115) is phosphorylated by Cdk1/cyclinB and detected by anti-pT2055 antibody. This phosphorylated substrate was utilized in the dephosphorylation reaction using IP fraction from B55γ–AcGFP-expressing cells. (I) Detection of phosphorylated 6His–NuMA(1876–2115) with anti-pT2055 antibody. Please note that 6His–NuMA(1876–2115) is unstable, therefore two species are observed in the Coomassie-stained gel and western blots. (J) Dephosphorylation reaction with Cdk1/cyclinB-phosphorylated 6His–NuMA(1876–2115) and the B55γ–AcGFP IP fraction at a different times, as indicated. Note that in this and other dephosphorylation experiments, values below the pT2055 western blot represent the band intensity, normalized to the intensity value from the control sample. HIS, blot using anti-His-tag antibody. (K) Schematic representation of the in vitro dephosphorylation assay as mentioned above, however, the dephosphorylation reaction is performed by either incubating the B55γ–AcGFP IP fraction with anti-GFP antibody (GFP-Ab) to mask AcGFP–B55γ, or with a B55γ–AcGFP IP fraction from cells depleted of PPP2CA catalytic subunit. (L) Dephosphorylation reaction with the B55γ–AcGFP IP that was incubated with GFP-Ab before the dephosphorylation reaction. (M) Dephosphorylation reaction with the B55γ–AcGFP IP fraction from the cells depleted for PPP2CA by RNAi. Please note a substantial decrease in dephosphorylation potential was observed for B55γ–AcGFP IP fractions that were either incubated with GFP-Ab (L) or from cells that were depleted for PPP2CA (M). **P<0.01, ***P<0.0001 (two-tailed Student's t-test).
MASTL and its substrate ENSA are dispensable for dephosphorylation of pT2055
In metazoans, during mitotic entry B55 is inhibited by endosulfine-alpha (ENSA) and cAMP-regulated phosphoprotein-19 (ARPP19), two related proteins activated by the protein kinase microtubule-associated serine/threonine kinase-like enzyme (MASTL) (Gharbi-Ayachi et al., 2010; Mochida et al., 2010). Depletion of MASTL, ENSA, or ARPP19 leads to constitutive B55 activity that causes mitotic catastrophe (Cundell et al., 2013; Manchado et al., 2010; Voets and Wolthuis, 2010). Thus, we scrutinized whether RNAi-mediated loss of MASTL impacts cortical NuMA. As reported previously, we observed that RNAi-mediated depletion of MASTL led to cytokinesis defects as well as chromosome congression defects (Fig. S4A,C; Voets and Wolthuis, 2010). However, MASTL depletion did not affect cortical NuMA in metaphase (Fig. S4D,E). Similarly, we did not observe any change in cortical NuMA localization in cells depleted for ENSA (Fig. S4B,F). Next, we analyzed the impact of MASTL and ENSA depletion on pT2055 levels at the spindle poles in metaphase. Interestingly, despite substantial depletion of MASTL or ENSA, as observed by immunoblotting (Fig. S4A,B), their loss did not affect pT2055 levels (Fig. S4G–I). Although the reason why MASTL and ENSA do not regulate the B55γ-containing PP2A complex will be of interest for future work, at this stage our data suggest that, unlike other B55 regulatory subunits such as B55α (Cundell et al., 2013; 2016), the B55γ-containing PP2A complex is not regulated by the MASTL or its substrate ENSA.
B55γ regulates proper spindle orientation
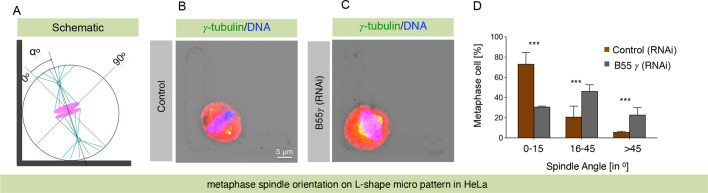
Cortical NuMA-mediated dynein localization is essential for proper spindle orientation in metaphase (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012, 2013). Because B55γ loss markedly reduced cortical NuMA and dynein, we monitored x-y spindle orientation by analyzing fixed cells grown on coverslips with an L-shaped fibronectin micro-pattern (Thery et al., 2005), which were treated with control siRNAs or siRNAs against B55γ. We found that spindle orientation was perturbed in cells depleted of B55γ in contrast to the spindle orientation observed in control cells (Fig. 3A–D), indicating that loss of cortical NuMA and dynein upon B55γ depletion disrupts proper spindle orientation.
Fig. 3.
Loss of B55γ by RNAi misorients the mitotic spindle. (A) Schematic representation of the mitotic spindle on the L-shape micropattern. The mitotic spindle and the centrosomes are shown in cyan, and chromosomes are in magenta. Spindle orientation angles (α°) were determined as depicted, with spindle angle measured relative to the hypotenuse of the micropattern (0°), as shown. (B,C) HeLa cells on an L-shape fibronectin micropattern that were either treated with control siRNAs (B) or RNAi against B55γ (C). Cells were stained for γ-tubulin (green) and DNA (blue). (D) Frequency of angular distribution of spindle positioning from 0°–15°, 16°–45°, and more than 45°. Note the change in the axis of spindle orientation in cells depleted of B55γ. Data are mean±s.d. of n>50 cells. ***P<0.0001 (two-tailed Student's t-test).
PP2A-B55γ controls cortical NuMA levels upon Cdk1 inactivation during anaphase onset
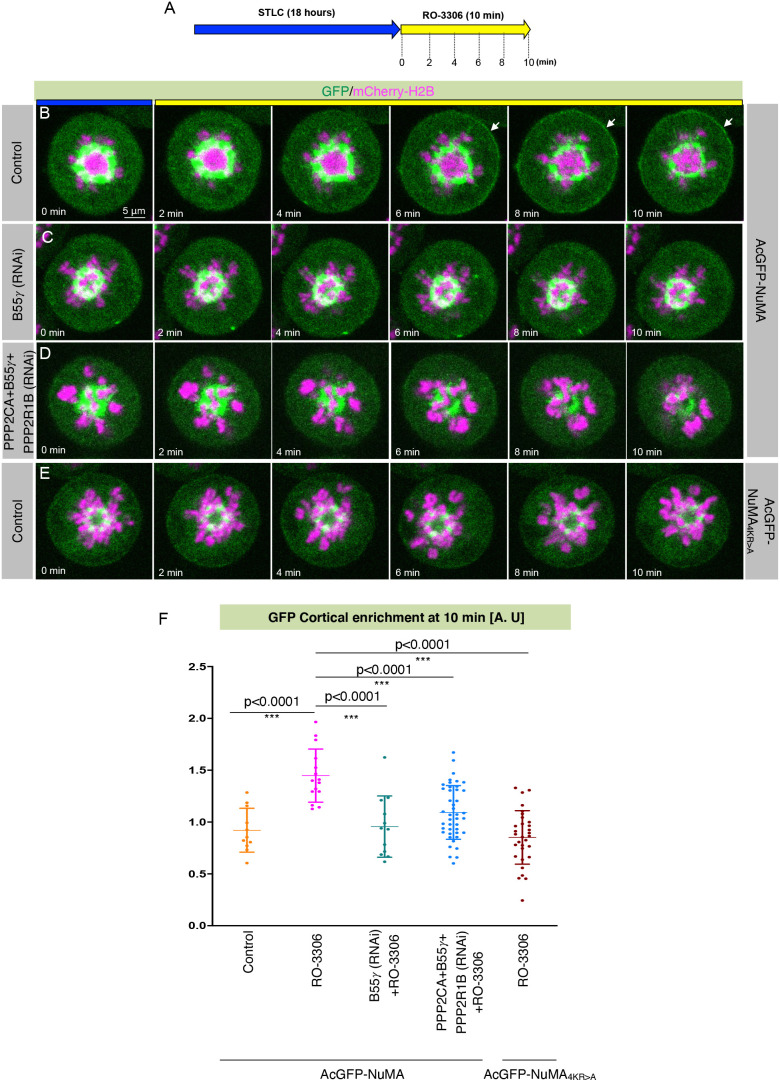
RNAi-mediated loss of B55γ and PPP2CA causes chromosome instability (data not shown; Kotak et al., 2013) and, presumably, this provokes the spindle assembly checkpoint. This observation precluded us from studying the function of the B55γ-based tripartite complex in cortical NuMA and dynein distributions during anaphase. To circumvent this experimental limitation, we sought to investigate the role of B55γ as well as the whole tripartite PPP2CA–B55γ–PPP2R1B complex in anaphase-like conditions. To this end, HeLa Kyoto cells stably co-expressing AcGFP–NuMA and mCherry–H2B were treated with kinesin 5 (KIF11) inhibitor (STLC) to block the cells in prometaphase stage, and subsequently these cells were forced to exit mitosis by the addition of the Cdk1 inhibitor RO-3306 (Fig. 4A). As expected, Cdk1 inactivation for 10 min in STLC-treated cells led to robust cortical NuMA enrichment (Fig. 4B,F). However, this cortical enrichment was highly impaired in cells depleted of either B55γ or PPP2CA–B55γ–PPP2R1B by RNAi (Fig. 4C,D,F). These data support that B55γ, as well as the B55γ-containing tripartite complex, is essential for proper cortical localization of NuMA, not only in metaphase but also in conditions that mimic anaphase.
Fig. 4.
B55γ-PP2A is necessary for NuMA cortical localization in cells mimicking anaphase. (A) Experimental protocol for chemically induced anaphase onset by acute Cdk1 inactivation using RO-3306 treatment in the presence of the kinesin-5 inhibitor STLC for cells stably co-expressing AcGFP–NuMA and mCherry–H2B, or AcGFP–NuMA4KR>A and mCherry–H2B. (B–D) Time-lapse confocal images of HeLa cells stably co-expressing AcGFP–NuMA (green) and mCherry–H2B (magenta) that were either transfected with control siRNAs and treated with STLC and RO-3306 (B, Control), transfected with B55γ siRNAs and treated with STLC and RO-3306 (C), or co-transfected with PPP2CA, B55γ and PPP2R1B siRNAs then treated with STLC and RO-3306 (D). Note the absence of cortical GFP signal in cells that were either transfected with B55γ siRNAs or with PPP2CA, B55γ and PPP2R1B siRNAs, in contrast to control cells. Arrows indicate cortical localization of NuMA. (E) Time-lapse confocal images of HeLa cells stably co-expressing AcGFP–NuMA4KR>A (green) and mCherry–H2B (magenta) and were treated with STLC and RO-3306. Note the substantially weaker cortical GFP signal in these cells upon RO-3306 treatment. (F) Quantification of GFP cortical enrichment in cells as described for B–E. Note significantly reduced cortical signal in cells that were either transfected with B55γ siRNAs, PPP2CA, B55γ and PPP2R1B siRNAs, or expressing AcGFP–NuMA4KR>A upon treatment with RO-3306 for 10 min as indicated. Data show mean±s.d. for more than 10 cells, with individual data points plotted. ***P<0.0001 (two-tailed Student's t-test). A.U., arbitrary units.
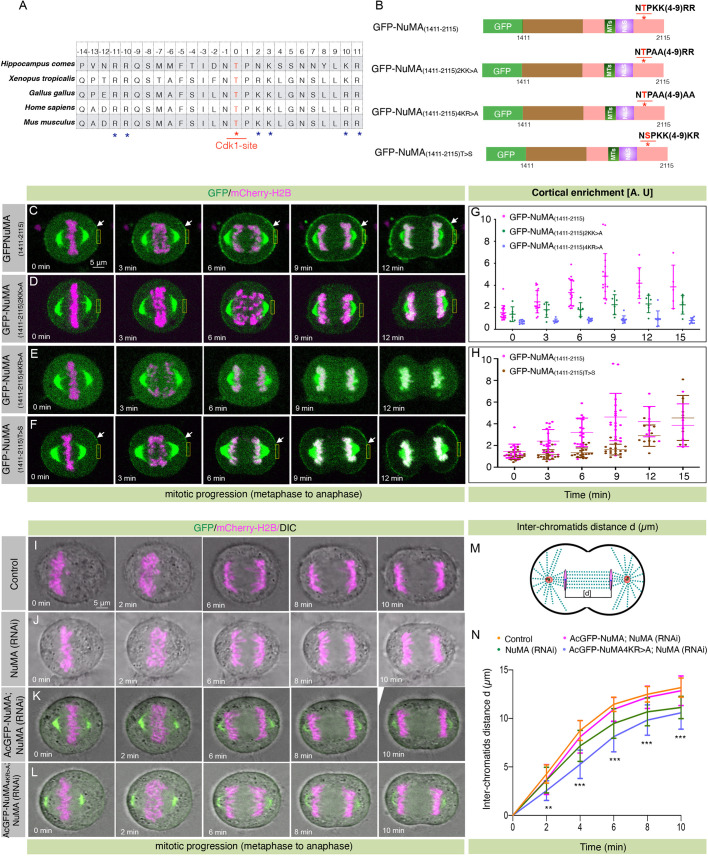
Polybasic amino acid residues in the vicinity of the Cdk1 phosphorylation site T2055 are critical for cortical NuMA localization in mitosis
How does B55γ recognize NuMA phosphorylated by Cdk1 at T2055? B55 substrates were recently shown to possess a bipartite polybasic recognition site flanking a Cdk1 phosphorylation site (Cundell et al., 2016). Importantly, we found that T2055 is surrounded by a few lysine or arginine residues, and these amino acids are evolutionarily conserved (Fig. 5A). To evaluate the function of these polybasic residues for cortical NuMA localization, we mutated either two or four lysine or arginine residues downstream of the Cdk1 phosphorylation site in the GFP-tagged NuMA C-terminal fragment [GFP–NuMA(1411–2115); Fig. 5B; Fig. S5A], which localizes at the cell cortex, similar to the wild-type protein (Kotak et al., 2014; Sana et al., 2018). Mutation of either two lysine, two arginine, or all the four polybasic residues had a significant impact on the cortical localization of GFP–NuMA(1411–2115) (Fig. 5C–E,G; Fig. S5B,D). Notably, we also found that an Escherichia coli-generated hexa-His-tagged mutant of NuMA, where all four basic residues are mutated to alanine was substantially weaker in its interaction with B55γ–AcGFP (Fig. S5E,F). This data indicates that these polybasic residues present in NuMA are critical for efficient interaction with B55γ. The impact on cortical GFP signal in cells expressing polybasic NuMA mutant appeared not because of their inability to localize at the membrane, as the introduction of a phospho-dead mutation at T2055 in GFP–NuMA(1411–2115)2RR>A rescued the cortical signal (Fig. S5B–D). We further established that the impact on cortical NuMA signal in cells expressing GFP–NuMA(1411–2115)4KR>A was not due to impairment of T2055 phosphorylation by Cdk1 at T2055 (Fig. S5G–K). Taken together, these results strengthen our model that B55γ recognizes polybasic amino acids in the vicinity of the Cdk1 phosphorylation site for efficient localization of NuMA at the cell cortex.
Fig. 5.
Polybasic residues in the vicinity of T2055 are necessary for cortical NuMA enrichment. (A) Sequence alignments of NuMA amino acid sequence near T2055 from various NuMA orthologs. A red asterisk marks the Cdk1 phosphorylation site, and blue asterisks show basic amino acids. (B) Schematic representation of wild-type fragment of NuMA(1411–2115) tagged with GFP at the N-terminus, and several mutant forms where either two or four basic residues were mutated to alanine (2KK>A, 4KR>A), or where the conserved threonine residue was mutated to serine (T>S). Asterisks indicate T2055; MTs, region mediating interaction with microtubules; NLS, nuclear localization signal. (C–F) Images from time-lapse microscopy of HeLa cells stably expressing mCherry–H2B and were transfected with GFP–NuMA(1411–2115) (C), GFP–NuMA(1411–2115)2KK>A (D), GFP–NuMA(1411–2115)4KR>A (E), and GFP–NuMA(1411–2115)T>S (F). The GFP signal is shown in green and mCherry signal is in magenta. Note the significant loss of cortical NuMA in cells expressing GFP–NuMA(1411–2115)2KK>A or GFP–NuMA(1411–2115)4KR>A. Also, note the dramatically weaker cortical GFP signal in cells expressing GFP–NuMA(1411–2115)T>S. Arrows indicate cortical GFP localization and boxes indicate examples of regions used for quantification. (G) Quantification of cortical enrichment for cells that underwent metaphase to anaphase transition, as shown in C–E (see Materials and Methods for detail). Data show mean±s.d. for n>5 cells, with individual data points plotted. GFP–NuMA(1411–2115) versus GFP–NuMA(1411–2115)2KK>A; P=0.28, 0.83, 0.032, 0.006, 0.023 and 0.068 for 0, 3, 6, 9, 12 and 15 min, respectively. GFP– NuMA(1411–2115) versus GFP–NuMA(1411–2115)4KR>A; P=0.0003 at 0 min, P<0.0001 at 3–15 min timepoints (two-tailed Student's t-test). (H) Quantification of cortical enrichment for cells that underwent metaphase to anaphase transition, as shown in C and F. Data show mean±s.d. for ≥18 cells, with individual data points plotted. GFP–NuMA(1411–2115) versus GFP–NuMA(1411–2115)T>S; P=0.89, 0.04, 0.002, <0.0001, 0.18 and 0.35 for 0, 3, 6, 9, 12 and15 min, respectively. (I–L) Images from time-lapse microscopy of HeLa Kyoto cells stably expressing mCherry–H2B (I), stably expressing mCherry–H2B and depleted of endogenous NuMA (J), stably co-expressing AcGFP–NuMA and mCherry–H2B and depleted of endogenous NuMA (K), or stably co-expressing AcGFP–NuMA4KR>A and mCherry–H2B and depleted of endogenous NuMA (L). The GFP signal is shown in green, the mCherry signal is in magenta. (M) Schematic representation for the calculation of the distance [d] between inter-chromatids in cells that underwent anaphase progression. (N) Quantification of inter-chromatid distance in cells as shown in I–L. Data are mean±s.d. inter-chromatid distances during anaphase progression for more than 15 cells in each condition (see Materials and Methods). **P=0.0002 at 2 min, ***P<0.0001 for 4–10 min timepoints between AcGFP–NuMA- and AcGFP–NuMA4K>R-expressing cells upon depletion of endogenous NuMA (two-tailed Student's t-test).
Because cortical NuMA promotes proper spindle elongation in anaphase cells (Kotak et al., 2013), and the quadruple mutant of NuMA is unable to localize at the cell cortex, presumably because of the lack of dephosphorylation, we attempted to characterize the impact of this mutant on spindle elongation during anaphase. For this purpose, we utilized a stable HeLa Kyoto cell line co-expressing AcGFP–NuMA4KR>A and mCherry–H2B. This engineered line expresses transgenic AcGFP–NuMA4KR>A, analogous to AcGFP–NuMA, and its endogenous counterpart (Fig. S2A,B). In contrast to the wild-type AcGFP–NuMA-expressing cells, we did not observe significant cortical accumulation of mutated NuMA in cells expressing AcGFP–NuMA4KR>A either in metaphase, anaphase or in cells that mimic anaphase-like conditions (i.e. STLC- and RO-3306-treated cells) (Fig. 4E,F; Fig. S2G–I). Because loss of NuMA impacts bipolar spindle formation (Kotak et al., 2013; Hueschen et al., 2019), we established an experimental regimen where we could study proper spindle elongation in a relatively weak NuMA (RNAi) background without impacting spindle bipolarity and anaphase onset (data not shown). In such a setting, loss of endogenous NuMA significantly affected inter-chromatid distance during anaphase in HeLa cells stably expressing the DNA marker mCherry–H2B alone ( Fig. 5I,J,M,N). However, this phenotype was fully rescued in cells ectopically expressing RNAi-resistant AcGFP–NuMA, but not in cells expressing AcGFP–NuMA4KR>A (Fig. 5K,L,N).
Serine and threonine residues phosphorylated by Cdk1 display differential dephosphorylation kinetics upon mitotic exit (McCloy et al., 2015; Godfrey et al., 2017; Hein et al., 2017). Also, PP2A complexes prefer phospho-threonine over phospho-serine (Pinna et al., 1976; Deana et al., 1982; Deana and Pinna, 1988). Besides, it is further revealed that the catalytic efficiency of PP2A-B55 is almost 20-fold higher for phospho-threonine in comparison with phospho-serine (Hein et al., 2017). Because Cdk1 inactivation robustly dephosphorylates pT2055 through B55γ, and therefore substantially enriches cortical NuMA in metaphase, we wondered whether substitution of the conserved threonine with serine would delay the dephosphorylation of pT2055 upon Cdk1 inactivation in anaphase. To this end, we analyzed the cortical localization of GFP–NuMA(1411–2115)T>S in cells expressing H2B–mCherry during anaphase progression. Remarkably, HeLa cells expressing the serine substitution allele showed slow kinetics of membrane GFP localization in early anaphase compared to cells expressing GFP–NuMA(1411–2115) (Fig. 5C,F,H). Overall, these data suggest that polybasic residues in the vicinity of T2055 are the key for cortical NuMA generation, and therefore for robust spindle elongation. Also, a threonine amino acid residue is a preferred over serine to ensure strong cortical NuMA enrichment upon Cdk1 inactivation during early anaphase.
NuMA phosphorylation at T2055 is a reversible event
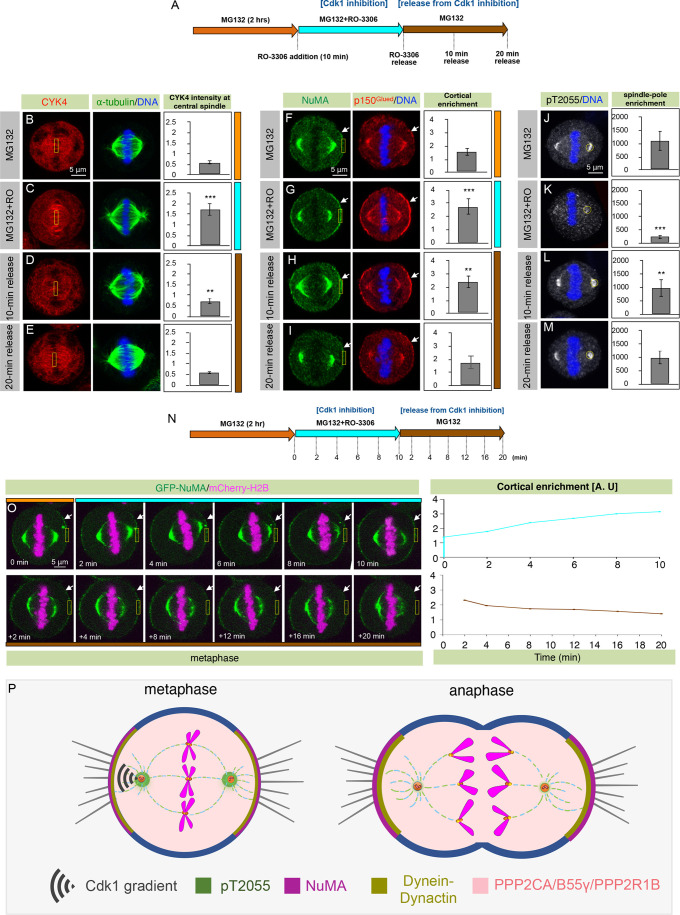
Cdk1 inactivation by RO-3306 promotes cortical NuMA enrichment and loss of pT2055 at the spindle poles. Thus, we wondered whether NuMA dephosphorylation initiated by acute Cdk1 inactivation is reversible. To investigate this, we first incubated mitotically synchronized HeLa cells with the proteasome inhibitor MG132 (to block cyclinB1 degradation), and subsequently we treated these cells with the Cdk1 inhibitor RO-3306 (to promote anaphase entry) (Fig. 6A). MG132- and RO-3306-treated cells showed robust CYK4 (also known as MgcRacGAP or RACGAP1) accumulation at the spindle mid-zone and cortical NuMA accumulation, indicating onset of the anaphase program (Fig. 6B,C,F,G,). Importantly, when the Cdk1 inhibitor was subsequently washed away in the presence of MG132 for 10 and 20 min, we observed time-dependent diminution of CYK4 accumulation at the spindle mid-zone, and reduction in cortical NuMA levels (Fig. 6D,E,H,I). Next, we attempted to study cortical NuMA reversibility by live-recording cells co-expressing AcGFP–NuMA and mCherry–H2B (Fig. 6N). Such MG132-incubated cells, when treated with RO-3306, showed substantial enrichment of AcGFP–NuMA at the cell cortex (Fig. 6O). Importantly, as demonstrated for endogenous NuMA, the excess cortical levels of AcGFP–NuMA were dramatically reduced in a time-dependent manner when the Cdk1 inhibitor was washed away. Next, we tested the reversibility of phosphorylated pT2055 in our experimental conditions. As expected, pT2055 levels were substantially reduced in cells treated with MG132 and RO-3306 (Fig. 6J,K). However, timely activation of Cdk1 upon release from RO-3306-mediated inhibition in the presence of MG132 restored pT2055 levels (Fig. 6K–M). Overall, these data suggest that phosphorylation and dephosphorylation of NuMA at T2055 is dynamic and interconvertible.
Fig. 6.
T2055 phosphorylation is dynamic and reversible. (A) Experimental procedure for chemically induced anaphase onset by acute Cdk1 inactivation using RO-3306 in the presence of proteasome inhibitor MG132, followed by the entry of cells into a metaphase-like state after washing off Cdk1 inhibitor for various time duration as indicated. Please note the color scheme, used throughout this figure, where orange represents cells (fixed or live) that were treated with MG132; cyan represents cells that were treated with MG132 and RO-3306, and brown shows cells that were washed free of RO-3306 using media containing MG132. (B–E) HeLa cells treated with MG132 (B), MG132 and RO-3306 (C), released from Cdk1 inhibition for 10 min (D), or released from Cdk1 inhibition for 20 min (E). Cells were fixed and stained for CYK4 (red), α-tubulin (green) and DNA (blue). Bars show mean±s.d. CYK4 intensity at the central spindle for n>11 cells. Please note the increase in CYK4 intensity in cells treated with RO-3306 and MG132, which was significantly reduced when cells were released from Cdk1 inhibition ( MG132 versus MG132+RO, P<0.0001; MG132 versus 10-min release, P=0.0066; MG132 versus 20-min release from RO-3306, P=0.86; two-tailed Student's t-test). (F–I) HeLa cells treated as described for B–E. Cells were fixed and stained for NuMA (green), p150Glued (red) and DNA (blue). Bars show mean±s.d. cortical NuMA intensity for n>10 cells. Please note the increase in NuMA cortical intensity in cells treated with RO-3306 and MG132, which was significantly reduced when cells were released from Cdk1 inhibition. (MG132 versus MG132+RO, P<0.0001; MG132 versus 20-min release, P=0.3536). (J–M) HeLa cells treated as described in B–E. Cells were fixed and stained for pT2055 (gray) and DNA (blue). Bars show mean±s.d. spindle pole NuMA intensity for n>10 cells. Please note the significant decrease in pT2055 signal at the spindle poles in cells that were treated with RO-3306 and MG132. Also, note that pT2055 intensity was rescued after 10- or 20-min release from Cdk1 inhibition. (MG132 versus MG132+RO, P<0.0001; MG132 versus 10-min release, P=0.24; MG132 versus 20-min release, P=0.13; two-tailed Student's t-test). (N) Experimental protocol for chemically induced anaphase onset followed by mitotic entry, as described above, for live imaging. (O) HeLa cells stably expressing AcGFP–NuMA were treated with RO-3306 in live-imaging conditions for 0–10 min and then released from Cdk1 inhibition for 20 min. Quantification of cortical enrichment is shown on the right for a HeLa cell first treated with the Cdk1 inhibitor RO-3306 for 10 min (top) and then released from this inhibition for the next 20 min in the presence of the proteasome inhibitor MG132 (bottom). More than five videos were recorded, and a representative cell is shown here. A.U, arbitrary units. (P) Model for cortical localization of NuMA and dynein during metaphase and anaphase in human cells. The PPP2CA–B55γ–PPP2R1B phosphatase complex counteracts Cdk1 phosphorylation of T2055 and maintains weak cortical localization of NuMA and the dynein–dynactin complex. This localization is critical for proper spindle orientation in metaphase. However, in anaphase, Cdk1-inactivation causes robust enrichment of cortical NuMA and dynein–dynactin at the cell cortex in a manner dependent on PPP2CA–B55γ–PPP2R1B, and this promotes proper spindle elongation in anaphase. In B–M,O, arrows indicate cortical localization, boxes and circles indicate examples of regions used for quantification. **P<0.01, ***P<0.0001.
DISCUSSION
Protein phosphorylation and dephosphorylation events are critical for regulating several biological processes, such as the cell cycle, proliferation and development, signal transduction pathways, and mitotic progression (Hunter, 1995; Lechward et al., 2001; Virshup, 2000; Bollen et al., 2009; Moura and Conde, 2019). Recently, studies have linked phosphatases with mitotic spindle orientation and elongation (Afshar et al., 2010; Kotak et al., 2013, 2016; Xie et al., 2013). However, the mechanistic understanding relating to these phosphatases and their substrates in orchestrating spindle behavior remained poorly characterized. In this study, we revealed that the existence of a biochemical cross-talk between Cdk1 and a B55γ-based PP2A complex is vital for temporal regulation of cortical NuMA and dynein for proper spindle orientation and elongation (Fig. 6P). Our model suggests that the gradient of Cdk1 activity emanating from the centrosomes that is counteracted by the uniformly weak activity of PP2A-B55γ controls cortical levels of the dynein adaptor NuMA in metaphase. These low levels of cortical NuMA and dynein–dynactin are critical for proper spindle orientation in metaphase. At anaphase onset, Cdk1 inactivation causes robust dephosphorylation of NuMA at T2055, and this leads to substantial cortical enrichment of NuMA and the dynein–dynactin complex in anaphase, which promotes spindle elongation (Fig. 6P).
A B55γ-based PP2A complex involved in NuMA dephosphorylation at T2055
Previous work has uncovered the involvement of PP2A-B55 in several essential processes such as cytokinesis and reassembly of the Golgi apparatus and nuclear envelope during mitotic exit (Schmitz et al., 2010; Cundell et al., 2013). Herein, by conducting a candidate-based chemical genetics screen, we discovered the function of B55γ in cortical accumulation of NuMA during mitosis. B55 (mainly α and δ isoforms) are inhibited in mitosis by ENSA and ARPP19, which are activated by MASTL (also known as Greatwall) (Gharbi-Ayachi et al., 2010; Mochida et al., 2010; Cundell et al., 2013). To our surprise, we did not observe any significant impact upon depletion of MASTL or ENSA on T2055 dephosphorylation in our experimental setting. This data suggests that weak PP2A-B55γ activity independent of MASTL or its substrate ENSA maintains cortical NuMA for proper spindle orientation. Also, it is intriguing that cells with siRNA-mediated depletion of B55γ in anaphase showed cortical NuMA levels comparable to that of control cells. However, we noticed that cells that are presumably strongly depleted of B55γ display chromosomal instability, which triggers the spindle checkpoint, thereby making such cells difficult to follow during anaphase (data not shown). Therefore, the fact that there is no apparent change in the anaphase cortical levels of NuMA could be due to partial depletion of B55γ. In such a scenario, Cdk1 inactivation in anaphase would lead to robust activation of the residual PP2A-B55γ complex. This assumption is supported by the finding that acute inactivation of Cdk1 caused rapid T2055 dephosphorylation kinetics. Therefore, it may well be that Cdk1-mediated phosphorylation on B55γ contributes to its temporal regulation between metaphase and anaphase. This hypothesis aligns with the observation that a phospho-mimicking substitution at the conserved serine (S167) in B55α affects binding of the regulatory (B55α) and catalytic subunits (Schmitz et al., 2010). This regulation could also act at the level of the catalytic subunit PPP2CA, as phosphorylation of T304 (a Cdk1 consensus site) in PPP2CA prevents its association with B55 (Longin et al., 2007). Nonetheless, our data reveal that B55γ and the PPP2CA–B55γ–PPP2R1B complex are necessary to regulate cortical NuMA accumulation upon Cdk1 inactivation in the induced anaphase-like scenario.
Polybasic residues close to the conserved T2055 residue are crucial for spatiotemporal regulation of NuMA localization
Recently, bipartite polybasic amino acid residues upstream and downstream of the Cdk1 phosphorylation site consisting of [S/T]P (with the preference of threonine) were shown to be key to temporal regulation of dephosphorylation events (Cundell et al., 2016). Notably, substrates that have fast dephosphorylation kinetics are more basic than substrates with a slower dephosphorylation rate. It was further postulated that electrostatic interactions between negatively charged residues of PP2A-B55 and positively charged amino acids in the substrates might determine the dephosphorylation rate (Xu et al., 2008; Cundell et al., 2016). Herein, we identified a few polybasic residues in the vicinity of T2055 of NuMA. Notably, the replacement of four evolutionarily conserved positively charged amino acids (K or R) to a neutral amino acid (A) significantly reduced NuMA cortical localization during mitosis. Our data further revealed that this quadruple mutant of NuMA weakly interacted with B55γ. Also, cells expressing this quadruple NuMA mutant were considerably impaired in spindle elongation during anaphase. Thus, our data suggest that a complementary acidic surface on B55γ could recognize these basic residues on NuMA, similar to what has been shown in the context of B55-mediated regulation of Tau and PRC1 dephosphorylation (Xu et al., 2008; Cundell et al., 2016).
Interestingly, our work suggested that the phospho-threonine preference of PP2A-B55γ is critical for cortical NuMA localization in a temporal manner. Previously, it was shown that the PP2A-B55 complex shows an intrinsic choice for phospho-threonine (Cundell et al., 2016; Hein et al., 2017). This preference is not limited to PP2A-B55 complexes; PP2A-B56 complexes also show this bias (Pinna et al., 1976; Deana et al., 1982). However, the molecular details of this preference are unclear. It could well be that the presence of an additional methyl side group present in threonine has a better binding affinity for PP2A-B55 and PP2A-B56.
Overall, our work identifies a novel PP2A-B55γ as an essential trimeric complex involved in a biochemical tug-of-war with Cdk1 to orchestrate cortical levels of NuMA and dynein for proper spindle behavior. Interestingly, reduction in the levels of B55γ is associated with the growth of prostate cancer (Bluemn et al., 2013), and therefore, because of the existing link between spindle orientation and tumorigenesis (Caussinus and Gonzalez, 2005; Quyn et al., 2010; Hehnly et al., 2015; Noatynska et al., 2012), it would be interesting to evaluate whether low levels of expression of B55γ in prostate cancer cells drive cancer progression through spindle misorientation.
MATERIALS AND METHODS
Cell culture, cell synchronization, and transfection
HeLa cells stably expressing GFP–Centrin-1, mCherry–H2B (a kind gift from Arnaud Echard, Institut Pasteur, Paris, France), HeLa Kyoto (generously provided by Daniel Gerlich, IMBA, Vienna, Austria), HeLa Kyoto stably expressing mCherry–H2B, AcGFP–NuMA, AcGFP–NuMA4KR>A, B55γ–AcGFP and GFP–DHC1 (kindly provided by Anthony Hyman, MPI, Dresden, Germany) as well as hTERT1-RPE1 cells, were maintained in high-glucose DMEM with GlutaMAX media supplemented with 10% fetal calf serum (FCS) in a humidified 5% CO2 incubator at 37°C. For monitoring spindle positioning in fixed specimens, HeLa cells were synchronized with 2 mM thymidine (Sigma-Aldrich, T1895) for 20 h, released for 9 h, trypsinized and plated on fibronectin L-shape micropatterns (CYTOO SA), as described previously by Sana et al. (2018). In brief, ∼80,000 cells were placed on a CYTOO chip in a 35 mm culture dish. After 1 h, cells that had not been attached to the micropatterns were removed by gently washing with medium. Cells were then fixed, 9 h after the release, with cold methanol and stained with antibodies against γ-tubulin (GTU88, Sigma-Aldrich).
For siRNA experiments, ∼100,000 cells were seeded in 6-well plates. 20 μM siRNAs in 100 μl RNase-free water and 4 μl of Lipofectamine RNAiMAX (Invitrogen; 13778150) in 100 μl RNase-free water were incubated in parallel for 5 min, mixed for 20 min and then added to 2.5 ml medium per 35 mm dish. siRNA sequences are shown in Table S1.
For transient transfections, cells were transfected at 80% confluency with 4 μg of plasmid DNA in 400 μl serum-free medium either with 6 μl Turbofect (ThermoScientific, R0531) or 4 μl Lipofectamine 2000 (Life Technologies, 11668019) incubated for 15–20 min and added to each well.
For the generation of stable human cells, cells were transfected at 80% confluency with 6 μg of plasmid DNA in 500 μl of JetPRIME buffer with 12 μl of JetPRIME reagent (Polyplus Transfection SA), incubated for 15 min and added to a 10 cm dish.
Drug treatments
Cdk1 inhibition was performed by incubating cells with 10 μM of RO-3306 (Selleckchem, S7747) for either 3, 5, or 10 min as specified in the figure legends. Proteasome inhibitor MG132 (Sigma-Aldrich, M8699) was used at the concentration of 20 μM for 2 h.
To investigate the cortical enrichment of AcGFP-tagged proteins in a monopolar setting, HeLa Kyoto cells stably co-expressing AcGFP–NuMA (wild type or 4KR>A mutant) and mCherry–H2B were synchronized using 7.5 μM Kinesin-5 inhibitor STLC (Sigma-Aldrich, 164739) for 17 h, and thereafter imaged by live recordings for every 2 min, either with or without 10 μM RO-3306.
For the reversal experiment, HeLa cells were synchronized with 2 mM thymidine (Sigma-Aldrich, T1895) for 23 h and then released for 6 h, followed by 5 h of nocodazole (Sigma-Aldrich, M1404) treatment. Cells were washed with the regular medium twice, followed by incubation with 20 μM of MG132 for 2 h to arrest the cells in metaphase. These cells were then treated with 10 μM RO-3306 for 10 min, and subsequently these cells were released in regular medium with MG132 for 0 min, 10 min, and 20 min before fixation and staining. For live-imaging under these conditions, HeLa Kyoto cells co-expressing AcGFP–NuMA and mCherry–H2B were treated with 20 μM of MG132 for 2 h, before treatment with 10 μM RO-3306 for 10 min, followed by RO-3306 release for the next 20 min and live imaging.
Indirect immunofluorescence and time-lapse imaging of HeLa cells
For immunofluorescence, cells were fixed with −20°C methanol for 7 min. Cells were permeabilized in PBS containing 0.05% Triton X-100 (1× PBST) for 1 min. Blocking was performed using 1% bovine serum albumin (BSA; HiMedia, RM3159) in PBST for 1 h, followed by primary antibody incubation for 4 h. Washes were done three times with 1× PBST for 5 min each; cells were then incubated with secondary antibodies for 1 h, counterstained with 1 μg/ml Hoechst 33342 (Sigma-Aldrich, B2261), washed three times for 5 min in PBST and mounted using Fluoromount-G (Southern Biotech, 0100-01). Primary antibodies were used at the following dilutions: 1:200 rabbit anti-NuMA (Santa Cruz, sc-48773), 1:200 mouse anti-p150Glued (Transduction Laboratories, 612709), 1:100 rabbit anti-p2055 NuMA (Kotak et al., 2013), 1:2000 mouse anti-α-tubulin (T6199; Sigma Aldrich) and 1:300 rabbit CYK-4, (Bethyl Laboratories, A302-797A), Secondary antibodies used were 1:500 Alexa Fluor 488 goat anti-mouse (Invitrogen, A11001), 1:500 Alexa Fluor 488 goat anti-rabbit (Invitrogen, A11008), 1:500 Alexa Fluor 568 goat anti-mouse (Invitrogen, A11004) and 1:500 Alexa Fluor 568 goat anti-rabbit (Invitrogen, A11011). Confocal images were acquired on an Olympus FV 3000 confocal laser scanning microscope using a 60× NA 1.4 oil objective, and were processed in ImageJ and Adobe Photoshop, maintaining relative image intensities.
Time-lapse microscopy was conducted on an Olympus FV 3000 confocal laser scanning microscope using a 40× NA 1.3 oil objective (Olympus Corporation, Japan) to image cells in an imaging dish (Eppendorf, 0030740017) at 5% CO2, 37°C, 90% humidity. Precise CO2 and humidity were maintained using Tokai Hit STR stage top incubator (Tokai Hit, Japan). Images were acquired every 3 min or 2 min, capturing 8–10 sections, 3 μm apart, at each time point. Figures containing time-lapse images were made using a single confocal section of the z-stack.
Plasmids and generation of stable cell lines
All NuMA clones were constructed using full-length NuMA as a template with appropriate PCR primer pairs. B55γ was obtained from Transomic technologies (Huntsville, USA) as a glycerol stock (BC032954), and it was amplified from pBluescript (Stratagene, BC032954).
For transient transfection, various NuMA fragments were cloned into pcDNA3-GFP (Merdes et al., 2000; kindly provided by Andreas Merdes, Paul Sabatier, Toulouse). For bacterial expression, NuMA(1876–2115) and NuMA(1876–2115)4KR>A were cloned in the pET28a plasmid and expressed in E. coli as described in Kotak et al. (2013). B55α was amplified from cDNA prepared from HeLa cells and cloned with GFP.
To create an AcGFP- (A. coerulescens GFP; Clonetech) and single FLAG (DYKDDDDK)-tagged version of wild type, mutated NuMA or B55γ in HeLa stable lines, the amplified products were sub-cloned into the pIRES-AcGFP-FLAG plasmid (kindly provided by Mark Patronczki, Boehringer Ingelheim, Vienna, Austria). HeLa Kyoto cells were transfected with AcGFP–NuMA (wild-type or mutant) or B55γ–AcGFP expression constructs. The medium was supplemented with 0.4 μg/ml puromycin (Life Technologies, A1113803) to select for and maintan cell lines expressing pIRESpuro3-based AcGFP-tagged NuMA or B55γ genes. Cells co-expressing mCherry–H2B with AcGFP–NuMA wild-type or mutated protein were selected in a medium containing 500 μg/ml G418 (Life Technologies, 10131035) in addition to puromycin. After 2–3 weeks of antibiotic selection, cells were characterized by immunoblotting and live imaging.
Visual quantification of NuMA and pT2055
For visual quantification of cortical NuMA (Fig. 1B), cells were analyzed under an epifluorescence microscope, and these cells were categorized as either ‘weak’ (as in Fig. 1C) or absent (as in Fig. 1D). For pT2055 quantification at the spindle poles (Fig. 1E; Fig. S1D), metaphase cells were categorized as either ‘strong’ (as in Fig. 1F), ‘moderate’ (as in Fig. 1I), or ‘weak’ (as in Fig. 1H).
Quantification of cortical intensity
Quantification of either cortical NuMA or cortical GFP signal in cells expressing AcGFP or GFP-tagged NuMA protein was determined by calculating the ratio of cortical mean intensity (of an area 1.8 µm ×4 µm, as shown in each figure) divided by the mean intensity value in the cytoplasm (equal area). The raw values for the region of interest (ROI) that were used to measure cortical intensity (ICortex) and cytoplasmic intensity (Icytosol) were subtracted from the background intensity values (Ibg). Ibg values were obtained from a similar area outside the cell. The equation (ICortex−Ibg)/(Icytosol−Ibg) was used for the calculation of cortical intensity. The brightest polar cortical region was used as a selection criterion in control as well as in a given experimental condition. Significance was determined using a two-tailed Student's t-test for each condition in GraphPad Prism 8.
Quantification of spindle pole intensity
Quantification of the spindle pole intensity of phosphorylated NuMA at T2055 was calculated by determining the mean spindle pole intensity from a maximum intensity projection image of an area 16 µm2, as shown in each figure, and correcting for the background signal of a similar area. Significance was determined using a two-tailed Student's t-test for each condition in GraphPad Prism 8.
Spindle elongation and chromosome separation
The distance between the separating chromosomes in cells expressing mCherry–H2B, was quantified every 2 min after anaphase onset using Imaris (Bitplane Inc.). Significance was determined using a two-tailed Student's t-test in GraphPad Prism 8.
Immunoprecipitation and western-blotting
For immunoprecipitation, 3 mg of cell lysates procured from the nocodazole (100 nM)-arrested mitotic HeLa Kyoto cells expressing either AcGFP-tagged or GFP-tagged proteins were incubated with 30 μl GFP-Trap agarose beads (Chromotek, ACT-CM-GFA0050) in lysis buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 25 mM sodium fluoride, 0.1 mM sodium orthovanadate (Sigma-Aldrich, S6508), 0.1 mM PMSF (Calbiochem, 7110), 0.2% Triton-X100, 0.3% NP-40, 100 nM okadaic acid and Complete EDTA-free protease inhibitor (Merck, 539134)] for 4 h at 4°C. The beads were washed three times with wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 25 mM sodium fluoride, 0.1 mM sodium orthovanadate, 0.1 mM PMSF, 0.2% Triton-X100, 0.3% NP-40, 100 nM okadaic acid and complete EDTA-free protease inhibitor) at 4°C. The bead-bound complex was denatured at 99°C in 2× SDS buffer and was analyzed by SDS–PAGE and western blotting.
For pulldown experiments with B55γ, each mg of mitotic cell lysate prepared from cells expressing B55γ–AcGFP was incubated with 25 μg of 6His–NuMA(1876–2115) and 6His–NuMA(1876–2115)4KR>A for 30 min followed by pulldown with GFP-trap, as described above.
For western blotting analysis, HeLa cells synchronized with 100 nM nocadazole for 16–20 h were lysed in lysis buffer (as decribed above) for 2 h on ice. Protein concentration estimation was done using Bradford reagent (Biorad; 500-0001). Cell lysates were denatured at 99°C in 2× SDS buffer and analyzed by SDS–PAGE followed by immunoblotting. For immunoblotting, 1:1000 rabbit anti-NuMA (Santa Cruz, sc-48773), 1:1000 rabbit anti-p2055 NuMA (Kotak et al., 2013), 1:5000 rabbit anti-GFP (Santa Cruz, sc-8334), 1:5000 anti-β-actin (Santa Cruz, 58673), 1:1000 rabbit anti-ENSA (Cell Signaling Technology, 8770S), 1:1000 rabbit anti-MASTL (Cell Signaling Technology, 12069S), 1:2000 rabbit anti-PPP1CA (Bethyl Laboratories, A300-904A), 1:2000 rabbit anti-PPP1CB (Bethyl Laboratories, A300-905A), 1:2000 goat anti-PPP1CC (Santa Cruz, sc6108), 1:2000 rabbit anti-PPP2CA (CST, 2038S), 1:2000 rabbit PPP4C (Bethyl Laboratories, A300-835A), 1:2000 rabbit PPP5C (Bethyl Laboratories, A300-909A), 1:2000 rabbit anti-PPP6C (Bethyl Laboratories,A300-844A), 1:2000 rabbit anti-PPP2R1B (Abcam, EPR10158), 1:1000 mouse anti-p150Glued (Transduction Laboratories, 612709), and 1:5000 rabbit anti-His (Sigma, SAB4301134) were used.
In vitro phosphorylation experiment
1 μg of recombinant 6His–NuMA(1876–2115) was incubated with 0.125 ng of Cdk1/cyclinB (Merck Millipore, 14-450) in 10μl of kinase buffer (50 mM HEPES, pH 7.8, 1 M MgCl2, 1 M KCl, 100 mM EGTA, 200 mg/ml BSA, 0.2 mM ATP) at 30°C for 30 min. The reaction was stopped by adding 20 μM of RO-3306 (Selleckchem, S7747), followed by snap-freezing the reactions in liquid N2. The phosphorylation status of recombinant NuMA was confirmed by western blotting with 1:1000 rabbit anti-p2055 NuMA (Kotak et al., 2013).
In vitro dephosphorylation experiment
HeLa Kyoto cells stably expressing, B55γ–AcGFP or CIBN–CAAX–FLAG–AcGFP or GFP–B55α, and arrested in prometaphase by treatment with 100 nM of nocodazole for 17 h, were lysed in buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.2% Triton-X100, 0.3% NP-40, 0.1 mM PMSF (Calbiochem, 7110), and Complete EDTA-free protease inhibitor (Merck, 539134)] on ice for 2 h. After protein concentration estimation, each mg of cell lysate was incubated with 10 μl of GFP-Trap agarose beads (Chromotek, ACT-CM-GFA0050) for 4 h at 4°C. After extensive washing with wash buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.2% Triton-X100, 0.3% NP-40, 0.1 mM PMSF and Complete EDTA-free protease inhibitor (Merck, 539134)] at 4°C, GFP-bound beads were collected and stored on ice.
For each dephosphorylation reaction, GFP-bound beads equivalent to 1 mg of cell lysate were suspended in 25 μl of dephosphorylation buffer [50 mM Tris-HCl, pH 7.35, 150 mM NaCl, 0.1 mM MnCl2, 1 mM MgCl2, 0.1% NP-40, 0.2 mg/ml BSA, 20μM RO-3306] and was incubated with 50–100 ng of Cdk1/cyclinB-phosphorylated recombinant 6His–NuMA(1876–2115) at 30°C. The reaction was stopped by adding 2× SDS buffer, and the samples were boiled at 99°C. The supernatant was obtained following centrifugation at 13,000 rpm (17,000 g) for 5 min, and was utilized for further analysis by SDS–PAGE and western blotting.
For GFP antibody-mediated inhibition of dephosphorylation, 1 mg equivalent of B55γ–AcGFP-bound beads were incubated with 330 ng of mouse anti-GFP (DSHB-GFP-8H11) at 22°C for 30 min prior to the addition of Cdk1/cyclinB-phosphorylated recombinant 6His–NuMA(1876–2115).
Supplementary Material
Acknowledgements
We thank Andreas Merdes, Daniel Gerlich, Arnaud Echard, Mark Petronczki, and Anthony Hyman for providing us plasmids and cell lines. We are grateful to Jon Pines, Iain Hagan, Phong Tran, Xavier Morin, Helder Maiato and Carlos Conde for providing us critical comments on this manuscript. We thank DST-FIST, UGC Centre for the Advanced Study, DBT-IISc Partnership Program and IISc for the infrastructure support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.K.; Methodology: R.K., S.K.; Validation: R.K., A.R., S.K.; Formal analysis: R.K., A.R., S.K.; Investigation: R.K., S.K.; Resources: S.K.; Data curation: R.K., S.K.; Writing - original draft: S.K.; Writing - review & editing: R.K., A.R., S.K.; Visualization: S.K.; Supervision: S.K.; Project administration: S.K.; Funding acquisition: S.K.
Funding
This work is supported by the Department of Biotechnology, Ministry of Science and Technology, India – Indian Institute of Science Partnership Program and by grants from The Wellcome Trust DBT India Alliance Fellowship (IA/I/15/2/502077 to S.K.). S.K. is a The Wellcome Trust DBT India Alliance Intermediate Fellow. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243857.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243857.reviewer-comments.pdf
References
- Afshar K., Werner M. E., Tse Y. C., Glotzer M. and Gonczy P. (2010). Regulation of cortical contractility and spindle positioning by the protein phosphatase 6 PPH-6 in one-cell stage C. elegans embryos. Development 137, 237-247. 10.1242/dev.042754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A., Elliott P. R. and Gruneberg U. (2011). Protein phosphatases and the regulation of mitosis. J. Cell Sci. 124, 2323-2334. 10.1242/jcs.087106 [DOI] [PubMed] [Google Scholar]
- Bergstralh D. T., Dawney N. S. and St Johnston D. (2017). Spindle orientation: a question of complex positioning. Development 144, 1137-1145. 10.1242/dev.140764 [DOI] [PubMed] [Google Scholar]
- Bluemn E. G., Spencer E. S., Mecham B., Gordon R. R., Coleman I., Lewinshtein D., Mostaghel E., Zhang X., Annis J., Grandori C. et al. (2013). PPP2R2C loss promotes castration-resistance and is associated with increased prostate cancer-specific mortality. Mol. Cancer Res. 11, 568-578. 10.1158/1541-7786.MCR-12-0710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M., Gerlich D. W. and Lesage B. (2009). Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 19, 531-541. 10.1016/j.tcb.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Caussinus E. and Gonzalez C. (2005). Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125-1129. 10.1038/ng1632 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Huang T. L., Shimamoto Y., Tsai S. Y. and Hsia K. C. (2017). Regulation of mitotic spindle assembly factor NuMA by Importin-beta. J. Cell. Biol. 216, 3453-3462. 10.1083/jcb.201705168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E. S., Balchand S. K., Faraci J. L., Wadsworth P. and Lee W.-L. (2012). Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell 23, 3380-3390. 10.1091/mbc.e12-02-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A., and Cleveland D. W. (1993). NuMA is required for the proper completion of mitosis. J. Cell Biol. 120, 947-957. 10.1083/jcb.120.4.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell M. J., Hutter L. H., Bastos Nunes, R., Poser E., Holder J., Mohammed S., Novak B. and Barr F. A. (2016). A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J. Cell Biol. 214, 539-554. 10.1083/jcb.201606033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell M. J., Bastos R. N., Zhang T., Holder J., Gruneberg U., Novak B. and Barr F. A. (2013). The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol. Cell 52, 393-405. 10.1016/j.molcel.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell M., Chen H., Jiang J., Kuan C. W., Fotovati A., Chu T. L., He Z., Lengyell T. C., Li H., Kroll T. et al. (2017). HMMR acts in the PLK1-dependent spindle positioning pathway and supports neural development. Elife 6, e28672 10.7554/eLife.28672.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A. D. and Pinna L. A. (1988). Identification of pseudo ‘phosphothreonyl-specific’ protein phosphatase T with a fraction of polycation-stimulated protein phosphatase 2A. Biochim. Biophys. Acta 968, 179-185. 10.1016/0167-4889(88)90006-7 [DOI] [PubMed] [Google Scholar]
- Deana A. D., Marchiori F., Meggio F. and Pinna L. A. (1982). Dephosphorylation of synthetic phosphopeptides by protein phosphatase-T, a phosphothreonyl protein phosphatase. J. Biol. Chem. 257, 8565-8568. [PubMed] [Google Scholar]
- di Pietro F., Echard A. and Morin X. (2016). Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 17, 1106-1130. 10.15252/embr.201642292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q. and Macara I. G. (2004). Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503-516. 10.1016/j.cell.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Du Q., Taylor L., Compton D. A. and Macara I. G. (2002). LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 12, 1928-1933. 10.1016/S0960-9822(02)01298-8 [DOI] [PubMed] [Google Scholar]
- Fielmich L.-E., Schmidt R., Dickinson D. J., Goldstein B., Akhmanova A. and van den Heuvel S. (2018). Optogenetic dissection of mitotic spindle positioning in vivo. Elife 7, e38198 10.7554/eLife.38198.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallini S., Carminati M., De Mattia F., Pirovano L., Martini E., Oldani A., Asteriti I. A., Guarguaglini G. and Mapelli M. (2016). NuMA phosphorylation by Aurora-A orchestrates spindle orientation. Curr. Biol. 26, 458-469. 10.1016/j.cub.2015.12.051 [DOI] [PubMed] [Google Scholar]
- Gharbi-Ayachi A., Labbe J. C., Burgess A., Vigneron S., Strub J.-M., Brioudes E., Van-Dorsselaer A., Castro A. and Lorca T. (2010). The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330, 1673-1677. 10.1126/science.1197048 [DOI] [PubMed] [Google Scholar]
- Godfrey M., Touati S. A., Kataria M., Jones A., Snijders A. P. and Uhlmann F. (2017). PP2A(Cdc55) phosphatase imposes ordered cell-cycle phosphorylation by opposing threonine phosphorylation. Mol. Cell 65, 393-402.e393. 10.1016/j.molcel.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2008). Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355-366. 10.1038/nrm2388 [DOI] [PubMed] [Google Scholar]
- Hehnly H., Canton D., Bucko P., Langeberg L. K., Ogier L., Gelman I., Santana L. F., Wordeman L. and Scott J. D. (2015). A mitotic kinase scaffold depleted in testicular seminomas impacts spindle orientation in germ line stem cells. Elife 4, e09384 10.7554/eLife.09384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein J. B., Hertz E. P. T., Garvanska D. H., Kruse T. and Nilsson J. (2017). Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat. Cell. Biol. 19, 1433-1440. 10.1038/ncb3634 [DOI] [PubMed] [Google Scholar]
- Hueschen C. L., Galstyan V., Amouzgar M., Phillips R. and Dumont S. (2019). Microtubule end-clustering maintains a steady-state spindle shape. Curr. Biol. 29, 700-708.e705. 10.1016/j.cub.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (1995). Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80, 225-236. 10.1016/0092-8674(95)90405-0 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T. and Cheeseman I. M. (2012). Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14, 311-317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T. and Cheeseman I. M. (2013). Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154, 391-402. 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Kotak S. (2019). Mechanisms of spindle positioning: lessons from worms and mammalian cells. Biomolecules 9, 80 10.3390/biom9020080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C. and Gonczy P. (2014). NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 33, 1815-1830. 10.15252/embj.201488147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C. and Gönczy P. (2012). Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199, 97-110. 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C. and Gönczy P. (2013). NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 32, 2517-2529. 10.1038/emboj.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Afshar K., Busso C. and Gonczy P. (2016). Aurora A kinase regulates proper spindle positioning in C. elegans and in human cells. J. Cell Sci. 129, 3015-3025. 10.1242/jcs.184416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M. P., Vale R. D., Jülicher F., Reck-Peterson S. L. and Dogterom M. (2012). Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 148, 502-514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechward K., Awotunde O. S., Swiatek W. and Muszynska G. (2001). Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim. Pol. 48, 921-933. 10.18388/abp.2001_3858 [DOI] [PubMed] [Google Scholar]
- Longin S., Zwaenepoel K., Louis J. V., Dilworth S., Goris J. and Janssens V. (2007). Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J. Biol. Chem. 282, 26971-26980. 10.1074/jbc.M704059200 [DOI] [PubMed] [Google Scholar]
- Manchado E., Guillamot M., de Cárcer G., Eguren M., Trickey M., García-Higuera I., Moreno S., Yamano H., Canamero M. and Malumbres M. (2010). Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell 18, 641-654. 10.1016/j.ccr.2010.10.028 [DOI] [PubMed] [Google Scholar]
- Mattagajasingh S. N., Huang S.-C., Hartenstein J. S., Snyder M., Marchesi V. T. and Benz E. J. (1999). A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein. J. Cell Biol. 145, 29-43. 10.1083/jcb.145.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy R. A., Parker B. L., Rogers S., Chaudhuri R., Gayevskiy V., Hoffman N. J., Ali N., Watkins D. N., Daly R. J., James D. E. et al. (2015). Global phosphoproteomic mapping of early mitotic exit in human cells identifies novel substrate dephosphorylation Motifs. Mol. Cell. Proteomics 14, 2194-2212. 10.1242/jcs.184416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J. D. and Cleveland D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447-458. 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W. C. and Cleveland D. W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851-862. 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Maslen S. L., Skehel M. and Hunt T. (2010). Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330, 1670-1673. 10.1126/science.1195689 [DOI] [PubMed] [Google Scholar]
- Morin X. and Bellaiche Y. (2011). Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102-119. 10.1016/j.devcel.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Moura M. and Conde C. (2019). Phosphatases in mitosis: roles and regulation. Biomolecules 9, 55 10.3390/biom9020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T., Afshar K. and Gonczy P. (2007). Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9, 1294-1302. 10.1038/ncb1649 [DOI] [PubMed] [Google Scholar]
- Noatynska A., Gotta M. and Meraldi P. (2012). Mitotic spindle (DIS)orientation and DISease: cause or consequence? J. Cell Biol. 199, 1025-1035. 10.1083/jcb.201209015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M., Natsume T., Kanemaki M. T. and Kiyomitsu T. (2018). Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. Elife 7, e36559 10.7554/eLife.36559.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna L. A., Donella A., Clari G. and Moret V. (1976). Preferential dephosphorylation of protein bound phosphorylthreonine and phosphorylserine residues by cytosol and mitochondrial “casein phosphatases”. Biochem. Biophys. Res. Commun. 70, 1308-1315. 10.1016/0006-291X(76)91045-7 [DOI] [PubMed] [Google Scholar]
- Pirovano L., Culurgioni S. and Carminati M. (2019). Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions. Nat. Commun. 10, 2208 10.1038/s41467-019-09999-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quyn A. J., Appleton P. L., Carey F. A., Steele R. J. C., Barker N., Clevers H., Ridgway R. A., Sansom O. J. and Näthke I. S. (2010). Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6, 175-181. 10.1016/j.stem.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Redemann S., Pecreaux J., Goehring N. W., Khairy K., Stelzer E. H., Hyman A. A. and Howard J. (2010). Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE 5, e12301 10.1371/journal.pone.0012301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana S., Keshri R., Rajeevan A., Kapoor S. and Kotak S. (2018). Plk1 regulates spindle orientation by phosphorylating NuMA in human cells. Life Sci Alliance 1, e201800223 10.26508/lsa.201800223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Fielmich L. E., Grigoriev I., Katrukha E. A., Akhmanova A. and van den Heuvel S. (2017). Two populations of cytoplasmic dynein contribute to spindle positioning in C. elegans embryos. J. Cell Biol. 216, 2777-2793. 10.1083/jcb.201607038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M. H. A., Held M., Janssens V., Hutchins J. R. A., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A. I., Poser I. et al. (2010). Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12, 886-893. 10.1038/ncb2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L., Poulson N. D., Foote H. P. and Lechler T. (2013). NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell 24, 3651-3662. 10.1091/mbc.e13-05-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L., Muroyama A. and Lechler T. (2016). NuMA-microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. Elife 5, e12504 10.7554/eLife.12504.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K. H. and Doe C. Q. (2009). Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11, 365-374. 10.1038/ncb0409-365 [DOI] [PubMed] [Google Scholar]
- Thery M., Racine V., Pepin A., Piel M., Chen Y., Sibarita J. B. and Bornens M. (2005). The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7, 947-953. 10.1038/ncb1307 [DOI] [PubMed] [Google Scholar]
- Vassilev L. T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D. C. and Chen L. (2006). Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA 103:10660-10665. 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup D. M. (2000). Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 12, 180-185. 10.1016/S0955-0674(99)00074-5 [DOI] [PubMed] [Google Scholar]
- Voets E. and Wolthuis R. M. (2010). MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle 9, 3591-3601. 10.4161/cc.9.17.12832 [DOI] [PubMed] [Google Scholar]
- Woodard G. E., Huang N.-N., Cho H., Miki T., Tall G. G. and Kehrl J. H. (2010). Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30, 3519-3530. 10.1128/MCB.00394-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Jüschke C., Esk C., Hirotsune S. and Knoblich J. A. (2013). The phosphatase PP4c controls spindle orientation to maintain proliferative symmetric divisions in the developing neocortex. Neuron 79, 254-265. 10.1016/j.neuron.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Chen Y., Zhang P., Jeffrey P. D. and Shi Y. (2008). Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol. Cell 31, 873-885. 10.1016/j.molcel.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Lambie E. J. and Snyder M. (1992). NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. 116, 1303-1317. 10.1083/jcb.116.6.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H. and Snyder M. (1992). The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell 3, 1259-1267. 10.1091/mbc.3.11.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Wan Q., Meixiong G. and Du Q. (2014). Cell cycle-regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol. Biol. Cell 25, 606-619. 10.1091/mbc.e13-08-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.