Yeast cells lower their intracellular methionine level in response to glucose restriction to regulate metabolism and life span.
Abstract
Caloric restriction (CR) is known to extend life span across species; however, the molecular mechanisms are not well understood. We investigate the mechanism by which glucose restriction (GR) extends yeast replicative life span, by combining ribosome profiling and RNA-seq with microfluidic-based single-cell analysis. We discovered a cross-talk between glucose sensing and the regulation of intracellular methionine: GR down-regulated the transcription and translation of methionine biosynthetic enzymes and transporters, leading to a decreased intracellular methionine concentration; external supplementation of methionine cancels the life span extension by GR. Furthermore, genetic perturbations that decrease methionine synthesis/uptake extend life span. These observations suggest that intracellular methionine mediates the life span effects of various nutrient and genetic perturbations, and that the glucose-methionine cross-talk is a general mechanism for coordinating the nutrient status and the translation/growth of a cell. Our work also implicates proteasome as a downstream effector of the life span extension by GR.
INTRODUCTION
Since the discovery in the early 1930s that reduced food intake extends the life span of rats (1), caloric restriction (CR), defined as a reduction in calorie intake without causing malnutrition, has been shown to extend the life span of a range of species, including yeast, worm, fly, fish, and mouse (2). While the effect on life span for humans remains to be determined, studies in nonhuman primates indicate that CR confers health benefits and possibly extends life span in rhesus monkeys (3, 4), and short-term CR studies in humans evoke metabolic health benefits (5). Thus, the benefits of CR appear to be largely conserved across species, and it is likely that its mechanisms of action may also be conserved. Understanding the molecular mechanisms by which CR extends life span will therefore have important implications for the prevention and treatment of age-associated diseases and for extending human health span.
While the life span phenotype of CR was first observed in laboratory rats, much of the insight into molecular mechanisms has derived from simpler model organisms including the budding yeast Saccharomyces cerevisiae. Budding yeast has been a canonical model for aging research due to its short replicative life span (defined as the number of daughter cells produced by a mother cell prior to senescence) and ease of genetic manipulation (6). In addition, yeast cells can grow on synthetic media of precisely controlled composition, making it possible to isolate the effect of an individual nutrient on life span. For example, CR has been implemented by simply reducing the glucose concentration of the media without affecting other nutrients. A number of experiments have been done by the traditional microdissection on solid plates with different glucose concentrations (7–9), and the aggregated data showed unambiguous life span extension when the glucose concentration is lowered from the standard 2% YPD (yeast extract peptone dextrose) plate to 0.5% or even lower (0.05%), arguing that glucose restriction (GR) in yeast is a robust model for mechanistic studies of CR.
One approach to understand how CR can extend life span involves testing candidate mechanisms, in which specific pathways/regulators known to be important for aging are interrogated. This approach has implicated a number of pathways (7–11). Another approach systematically searches for molecular differences that may account for the life span phenotype, for instance, by characterizing gene expression profiles in the CR and the control samples using microarrays or RNA sequencing (RNA-seq) (12, 13). These studies have revealed some consistent patterns of gene expression changes induced by CR. However, it has been quite challenging to delineate the molecular changes causal to the life span extension, since CR typically induces global gene expression reprogramming involving hundreds or even thousands of genes as cells adjust their gene expression programs in metabolism, stress response, maintenance and repair, and general homeostasis.
With the development of microfluidic devices for yeast replicative aging studies (14–17), it is now possible to monitor cell division and various molecular markers in single mother cells throughout their life span by time-lapse microscopy. In addition, it is easy to maintain a constant environment, for instance, a very low glucose concentration. The constant flow in the microfluidic device removes any secreted molecules from the cells, making it suitable for analyzing cell autonomous effects [a recent study indicated that CR can function non–cell autonomously; (18)]. Furthermore, in a microfluidic device, media can be switched with accuracy and speed, allowing the monitoring of the dynamic response of single cells to CR.
In this work, we investigate the molecular mechanism of life span extension by GR (reducing glucose in the media without changing other nutrients) in yeast, using an approach that combines global gene expression profiling, microfluidics-based single-cell analysis, and candidate-based genetic manipulations. Using ribosome profiling and RNA-seq, we systematically compared the translational and transcriptional profiles of cells grown in GR and normal media, uncovering groups of functionally related genes that are up- or down-regulated. We observed a cross-talk from glucose sensing to the regulation of intracellular methionine: Methionine biosynthetic enzymes and transporters were significantly down-regulated by GR, leading to the decreased intracellular methionine level, and external supplementation of methionine cancels the life span extension by GR without affecting the life span in the normal media. With additional evidence from systematic manipulations of methionine pathway genes and bioinformatic analyses of other long-lived mutants, we were able to place intracellular methionine at a central position for life span regulation. Through analysis of up-regulated gene groups, we found that the translational efficiency of Rpn4, a major transcriptional activator of the proteasome, and the transcription of its targets were up-regulated by GR, leading to increased proteasome activity. Furthermore, we found that Rpn4 is required for the life span extension of GR.
Our study provides strong links between GR and methionine restriction (MR), suggesting that the life span extension caused by GR may be, at least partially, mediated by decreased intracellular methionine, possibly due to overlapping downstream signaling and metabolic pathways. Given the importance of methionine in regulating translation and the metabolic state of the cell, the cross-talk between glucose and methionine might be a general mechanism for coordinating the nutrient status and the translation/growth of a cell with implications beyond the regulation of life span.
RESULTS
GR extends yeast life span in the microfluidic environment
We first set out to independently test whether GR extends yeast replicative life span in microfluidic devices. This is of interest since one recent report indicated no consistent life span extension by GR using a microfluidic device (19), while another report observed life span extension by GR using a different microfluidic device (17). Using an independent microfluidic system described previously (16, 20), we measured the life span in a range of glucose concentrations and found that 0.05% glucose leads to a robust life span extension of 18% in the BY4741 strain, compared with the normal 2% glucose media (2% glucose, mean life span = 20.3 generations, 453 cells; 0.05% glucose, mean life span = 24.0 generations, 436 cells; P = 3.4 × 10−13; see Supplementary File 1 for the raw life span data). The result is from 15 aggregated experiments performed on different dates. This level of life span extension is similar to that observed in standard microdissection studies using the same strain and same level of GR (9).
Ribosomal profiling and RNA-seq revealed a broad spectrum of transcriptional and translational changes induced by GR
To find downstream mechanism(s) for the life span extension caused by GR, we performed global gene expression profiling, comparing cells growing in normal versus GR conditions. We analyzed both the transcriptional and the translational profiles by RNA-seq and ribosomal profiling; the latter technique quantifies the amount of ribosome-protected RNA footprints for all the genes, which directly measures the level of translation (21). We observed a broad spectrum of gene expression changes in response to limited glucose, both at the transcriptional and the translational levels (Fig. 1A and Supplementary File 2). For the majority of the genes with changed expression, the regulation is at the transcriptional level, without a change of translational efficiency (defined as the ratio of the ribosomal footprints to the total RNA), as the fold change of ribosome footprints is the same as the fold change of the transcripts (Fig. 1A; genes close to the fitted line). Overall, there are 229/197 genes whose footprints are up/down by more than twofold in experiments performed using two different protocols (quick dilution of glucose concentration versus spin down and resuspension in low glucose media; see Materials and Method, fig. S1, and Supplementary File 2). However, there is a subset of genes whose translational efficiency is increased or decreased (Fig. 1A, points above or below the fitted line, color coded), indicating that they are under translational regulation. There are 71/17 genes that increased/decreased their translational efficiency by more than twofold.
Fig. 1. RNA-seq and ribosome profiling revealed global transcriptional and translational regulation by GR.
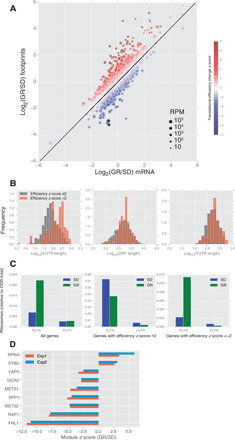
(A) Log2 of the fold change (GR versus SD) of ribosome-protected RNA is plotted against log2 fold change of the total RNA, for all the genes. The size of the dots represents the number of reads (per 1 million total reads) for the gene. Translational efficiency is measured by the deviation from the linear fit (the black line), quantified by a z score (indicated by the color; see Materials and Methods). (B) Length distributions of 5′UTR (left), coding sequence (middle), and 3′UTR (right) for genes with increased translational efficiency (z score >2) and other genes (z score ≤2). (C) Ribosome occupancy of 5′UTR and 3′UTR relative to CDS for all genes (left), genes with increased translational efficiency (z score >2, middle), and genes with decreased translational efficiency (z score <−2, right), under SD and GR conditions. (D) Transcriptional changes between SD and GR. Genes are organized into TF modules. Change of expression of a TF module is quantified by a z score, which measures the collective change of the genes in the module relative to other genes in the genome (see Materials and Methods). Top TF modules with the absolute value of z scores passing 2.5 in both experiments are shown.
We found that genes with increased expression (footprint_fold_change >2) are enriched for cellular processes including monocarboxylic acid metabolism, fatty acid metabolism, and the oxidative stress response, and those with decreased expression (footprint_fold_change <1/2) are enriched for protein synthesis (ribosomal genes) and methionine biosynthesis (Supplementary File 3).
We then analyzed the subset of genes that were subjected to translational regulation by GR. Genes that increased their translational efficiency more than twofold in two independent experiments were highly enriched in processes including carbohydrate metabolism (14 of 71 genes are in this category, P = 5.0 × 10−7; Supplementary File 3). Many genes in this category encode enzymes involved in the breakdown of polysaccharides, consistent with the notion that GR induces gene expression changes to produce sugar from stored carbohydrates. Genes with decreased translational efficiency are enriched in the process of methionine metabolism (4 of 17, P = 7.0 × 10−3; Supplementary File 3). Although ribosomal genes are strongly suppressed at the ribosome footprint level, they are not enriched for in the list with decreased translational efficiency, suggesting that ribosomal genes are mainly regulated at the transcriptional level.
We found that genes with increased translational efficiency tend to have longer 5′ untranslated regions (5′UTRs) compared with other genes; the 3′UTRs and coding sequences (CDSs) of these genes have length distributions similar to those of the other genes (Fig. 1B). This suggests that these genes might be subjected to special translational regulation through their 5′UTR sequences. Consistent with this idea, we found that genes with increased translational efficiency have distinct patterns of ribosome occupancy in their 5′UTR relative to their CDSs. While GR leads to increased ribosome occupancy in the 5′UTR relative to the CDS for most of the genes, this trend is reversed for genes with increased translational efficiency, i.e., for genes with increased translational efficiency, ribosome occupancy of 5′UTR relative to CDS decreased upon GR (Fig. 1C). This trend could result from increased translation initiation at the canonical start site of the CDS, or decreased falloff (before the start site) of ribosomes initiated at the upstream; both scenarios lead to increased translational efficiency. A previously known example of translational regulation with a similar ribosome occupancy pattern is the regulation of Gcn4 through its inhibitory 5′ upstream open reading frames (uORFs) (21–23). We found that the translational efficiency of Gcn4 is significantly increased under GR, consistent with both a previous report (24) and with the finding that Gcn4 is partially required for life span extension by CR (25).
Transcription module analysis identified potential downstream effectors of GR life span extension
Since GR induces a broad spectrum of gene expression changes, it is quite challenging to identify potential downstream effectors of life span extension by analyzing individual genes. To narrow down the potential targets, we analyzed the transcriptional changes by organizing genes into functional groups with shared regulators. We grouped the genes into transcription modules, genes coregulated by the same transcription factor (TF), using the TF-target relationships previously identified by a genome-wide chromatin immunoprecipitation (ChIP)–chip analysis (26); thereafter, we will use the capitalized name of the TF to denote the module. We then analyzed the expression changes of genes in the TF modules collectively by calculating a z score for the whole module (see Materials and Methods). This approach allows for a simpler functional organization of the transcriptome and improves the statistical power when the targets of a TF have small but consistent fold changes. The analysis revealed that a number of TF modules are significantly up- or down-regulated (seven TF modules with z score <−2.5 and two TF modules with z score >2.5 in both experiments; Fig. 1D), with clear functional themes. Examples include decreased expression of ribosomal genes (RAP1, FHL1, and SFP1) and methionine pathway genes (MET31 and MET32), as well as increased expression of oxidative stress response genes (STB5) and genes encoding components of the proteasome (RPN4). The MET31/32 modules have the second highest z score (besides ribosomal gene modules) among those suppressed by GR. The RPN4 module has the highest z score among those induced by GR, but the individual genes in this module changed by ~2-fold or less, indicating the power of the TF module–based approach. This TF module analysis yielded important clues, which led to the identification of methionine and the proteasome as key downstream effectors of GR life span extension, as we will describe below.
The cross-talk from glucose to methionine: GR suppresses methionine transporters and biosynthetic enzymes, leading to decreased intracellular methionine concentration
We first turned our attention to methionine biosynthesis genes and transporters, as the TF module and the Gene Ontology (GO) analyses both suggest that they are strongly repressed by GR, and previous studies indicate that MR itself can extend life span (27, 28). We found that the repression is specific for genes related to sulfur-containing amino acids, as opposed to general amino acids, and occurs at both the transcriptional and the translational levels (Fig. 2A). Both the high-affinity methionine permease Mup1 and the low-affinity methionine permease Mup3 showed down-regulation in the ribosome profiling data. While Mup3 is expressed at a low level, Mup1 is expressed at a level that can be tracked by a fluorescent reporter. Using the microfluidic system, we found that Mup1 is down-regulated in response to GR (Fig. 2B), changing in levels approximately ~30 min after the media switch from 2 to 0.05% glucose and reaching a steady-state level ~30% lower after 2 hours. The mRNA level and translational efficiency of ARO10, a gene encoding the first enzyme for methionine, aromatic, and branched chain amino acid catabolism were strongly up-regulated in 0.05% glucose (footprint increased by 6.7-fold, and translational efficiency increased by 5.0-fold). These gene expression changes operate in the same direction to decrease intracellular methionine levels under GR conditions.
Fig. 2. GR represses the expression of methionine biosynthetic enzymes and transporters and leads to decreased intracellular methionine level.
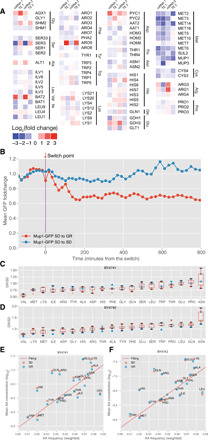
(A) Fold changes of mRNA and translational efficiency for amino acid biosynthetic enzymes and transporters in two independent experiments using different GR protocols (see Materials and Methods). (B) Change of the protein expression of the major methionine transporter Mup1, measured by following the Mup1-GFP reporter in single cells in the microfluidic device. Shown is the average over 23 cells; each was normalized by the mean fluorescence before the switch. The arrow indicates the time point at which the media were switched from SD to GR. (C to F) Change of the intracellular concentration of amino acids after switching from SD to GR. (C and D) Ratios of intracellular amino acid concentrations between GR and SD in the BY4741 strain and BY4742 strain, respectively. Different colors represent different independent experiments. Box plot shows maximum, minimum, median, and top and bottom 20 percentile. (E and F) Amino acid concentration plotted against the usage in the proteome (genome frequency weighted by protein abundance). The red line is a linear fit excluding five charged amino acids (Arg, Lys, His, Glu, and Asp) and one polar amino acid (Gln). There is a log-linear correlation between the intracellular concentration and the usage.
Direct measurement of intracellular amino acid concentration confirmed that methionine levels dropped significantly under the GR condition (Fig. 2, C and D) in both the MATa (BY4741) and the MATα (BY4742) strains; methionine was the second and third most decreased amino acid under GR condition in the two strains, respectively. The other two top ranked amino acids (in terms of fold decrease) are valine and lysine. The decreases in amino acid concentration are selective, as the concentration of some other amino acids increased under GR, e.g., the concentration of asparagine increased significantly under GR in both strains.
To gauge the potential impact of decreased amino acids on translation, we compared the intracellular concentration of amino acids under both the GR and the normal conditions with amino acid usage in the proteome (Fig. 2, E and F). This comparison revealed several interesting features. First, the log of the concentration (thus the chemical potential) is linearly proportional to the usage for most of the amino acids, except for the charged amino acids (Arg, Lys, His, Glu, and Asp) and one polar amino acid (Gln). This suggests that the observed intracellular amino acid concentration might be a result of optimization for protein synthesis. Second, methionine and tryptophan are the two most rarely used amino acids and have the lowest intracellular concentrations. Restriction of each of these two amino acids has also been shown to be sufficient to extend life span in yeast and in other organisms, including mammals (27, 29). In addition, GR further lowers the concentration of methionine to the lowest among all the amino acids, arguing that it might be rate-limiting for protein synthesis. These features have been observed in both the BY4741 (methionine auxotrophic) and the BY4742 (methionine prototrophic) strains, arguing that the cross-talk from glucose to methionine is not an artifact due to a defective methionine biosynthetic pathway.
Methionine supplementation abrogates the life span extension by GR
Since MR was found to extend life span in yeast and several other species, we hypothesized that the life span extension by GR is at least partially mediated through reduced methionine synthesis/uptake. To test this hypothesis, we analyzed the interaction between GR and methionine. We added extra methionine (10 times the level in the normal media, or 10xMet) to both the GR and normal media and observed that life span extension by GR (0.05% glucose) was abrogated. This does not appear to be caused by a deleterious effect of extra methionine, as the same addition did not alter the life span in the normal 2% glucose media (Fig. 3). The same phenomenon was observed in two different strains, BY4741 (Fig. 3A) and BY4741-MET (BY4741 with MET15 knocked in; Fig. 3B), with and without an intact methionine biosynthesis pathway (see Materials and Methods), arguing that this is not a phenomenon peculiar to a methionine auxotrophic strain. We found that the external supplementation of methionine only increases the intracellular concentration of methionine, not any other amino acids (fig. S2), indicating that the life span extension is blocked by increased intracellular methionine. Furthermore, we showed that the life span extension from GR is specifically blocked by methionine supplementation, as supplementation of the other two most decreased amino acids valine and lysine does not shorten the life span under the GR condition (Fig. 3, C and D).
Fig. 3. Adding back extra methionine (10 times the level in SD) to the GR media specifically cancels the life span extension by GR without affecting the life span in the SD media.
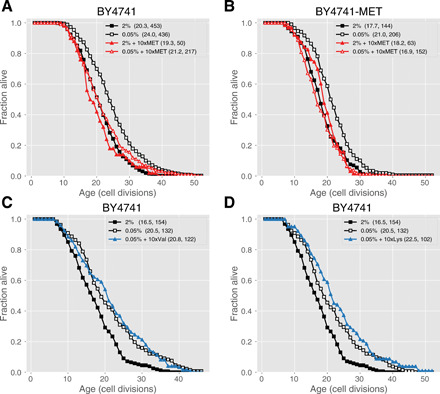
(A) Survival curves of the BY4741 strain under four different conditions: GR (0.05% glucose), GR + 10xMet, SD (2% glucose), and SD + 10xMet. Numbers in parentheses indicate the average life span and the number of cells measured. The life span under GR + 10xMet and SD + 10 × Met is not significantly different from that under SD (P = 0.55 and 0.17, respectively). (B) The same survival curves under the four conditions for a different strain BY4741-MET (BY4741 with MET15 knocked in) with an intact methionine biosynthesis pathway. The life span under GR + 10xMet and SD + 10xMet is not significantly different from that under SD (P = 0.22 and 0.25, respectively). (C) Adding back valine does not abrogate the life span extension by GR. Shown are the survival curves of the BY4741 strain under three different conditions: GR (0.05% glucose), GR + 10xVal, and SD (2% glucose). Life span under GR + 10xVal is not significantly different from that under GR (P = 0.75). (D) Adding back lysine does not abrogate the life span extension by GR. Shown are the survival curves of the BY4741 strain under three different conditions: GR (0.05% glucose), GR + 10xLys, and SD (2% glucose). Life span under GR + 10xLys is not significantly different from that under GR (P = 0.1).
Methionine pathway down-regulation extends life span
Genomic analysis of deletion mutants indicates that the methionine pathway is a central regulator of life span in contexts other than GR. In a systematic search for genes whose expression level may influence life span, we compared the gene expression profiles of ~260 TF deletion mutants (30) in conjunction with their life span data from a recent genome-wide screening (31). We found that methionine and cysteine (which contain sulfur and can interconvert) pathway genes are significantly repressed in the long-lived mutants relative to other TFs, which do not extend life span (see Materials and Methods), suggesting that the life span extension of these mutants may be mediated by the down-regulation of methionine/cysteine pathway genes. We also found that single gene deletions of seven of the eight genes encoding enzymes responsible for methionine biosynthesis lead to life span extension (Fig. 4); this includes all the enzymes catalyzing the reactions upstream of methionine, suggesting that the effect is mediated by methionine or the metabolites downstream of methionine.
Fig. 4. Deletion of methionine biosynthesis genes extends life span.
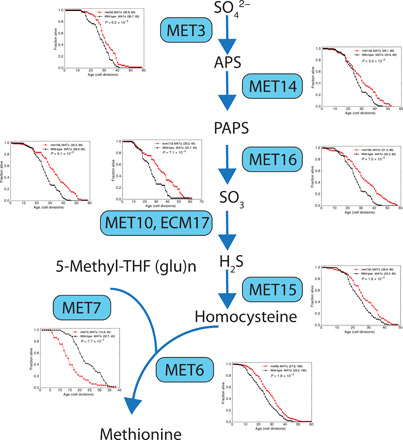
The methionine biosynthetic pathway is shown with enzymes in the boxes and arrows connecting metabolites. Survival curves for the single gene deletion mutants and the wide-type controls are plotted next to the enzymes. Deletion mutants were derived from the BY4742 strain.
Furthermore, we detected an epistatic interaction between GR and genetic perturbations that down-regulated methionine synthesis/uptake. For example, deletion of MET31-a transcriptional regulator for the methionine pathway is sufficient to confer life span extension, and GR does not further extend the life span of this strain (fig. S3). Together, our findings indicate that genetic perturbations blocking methionine synthesis/uptake consistently extend life span, arguing that methionine is a key control point of the life span both in glucose replete and restricted conditions.
Downstream effectors of life span extension: GR up-regulates proteasome expression and activity
We have shown that the transcription module analysis of gene expression changes caused by GR can lead to important clues about the downstream effectors. By following the down-regulated pathways, we found that decreased intracellular methionine may mediate GR life span extension. We next turned our attention to up-regulated TF modules.
One of the most significantly up-regulated TF modules is the RPN4 module (Fig. 1D). Rpn4 is a master transcription regulator of proteasome genes. It is known that proteasome activity declines with aging, and previous gene expression profiling experiments showed that CR induces proteasome expression in mice (13). In addition, deletion of UBR2, which encodes the upstream ubiquitin-protein ligase required for the ubiquitination and, hence, the degradation of Rpn4, leads to increased Rpn4 levels, increased proteasome activity, and extended life span (32, 33). Together, these observations suggest that proteasome may be a key downstream effector of GR’s life span extension.
We found that Rpn4 itself is translationally regulated by GR: While RPN4 transcript level is slightly decreased under GR, the translational efficiency of Rpn4 increased by ~2-fold. This fold change matched quantitatively the fold changes of the transcript level of Rpn4 targets: The majority of Rpn4 targets are up-regulated by ~2-fold under GR (Fig. 5A), suggesting that Rpn4 is mainly regulated at the translational level and that Rpn4 targets are, in turn, transcriptionally regulated. Examination of the ribosome footprint patterns suggests that RPN4 is translationally regulated through its 5′ uORFs: The ribosome occupancy at its 5′UTR increased significantly under GR (Fig. 5B), indicating an increased translation initiation at the upstream uORFs.
Fig. 5. GR up-regulates the translation of RPN4, leading to elevated proteasome expression and activity, and RPN4 is required for the life span extension.
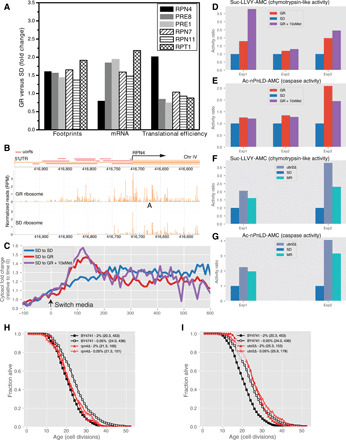
(A) GR induced fold changes of footprint (left), total mRNA (middle), and translational efficiency (right) for Rpn4 and its representative targets. (B) Ribosome footprints in the coding region and the 5′UTR of RPN4 under SD (bottom track) and GR (middle track) conditions. RPN4 coding sequence and 5′uORFs are indicated on the top track. (C) Rpn11-GFP reporter level as a function of time before and after switching from SD to GR media (red curve) or SD to GR + 10xMet (purple curve). The blue curve is the control without media switch. The red, purple, and blue curves are the average over 94, 69, and 123 cells, respectively, where each single-cell curve is first normalized by the mean fluorescence intensity before switch. (D and E) Proteasome activity (chymotrypsin-like and caspase-like) in GR and GR + 10xMet versus SD in three independent experiments. (F and G) Proteasome activity in UBR2 deletion mutant and under the MR condition. (H) RPN4 deletion abolishes the life span extension by GR without affecting the life span in SD. Life spans of RPN4∆ in both SD and GR conditions are not significantly different from WT in SD (P = 0.15 and 0.21, respectively). (I) Deletion of UBR2, a ubiquitin-protein ligase that targets Rpn4 for degradation, extends life span in SD (P = 7.1 × 10−16). GR does not extend the life span of UBR2 deletion mutant further (P = 0.64). Deletion mutants were derived from BY4741.
Using the microfluidic device, we tracked the dynamic expression of the proteasome in single cells before and after switching to the GR media, using a strain in which Rpn11 (one component of the 26S proteasome lid) is tagged by green fluorescent protein (GFP). We observed a transient increase in proteasome expression after switching from synthetic dextrose (SD) to GR media. This transient increase reached a peak about 40% higher than the level seen in the SD media after about 2 hours and then returned back to the level seen in SD media after 4 hours, even while these cells were continuously grown in the GR media (Fig. 5C). Thus, the dynamic expression of proteasome shows adaptation typical of a system with negative feedback. One potential source of this negative feedback is the degradation of Rpn4 by proteasome, regulated by the upstream ubiquitin ligase Ubr2.
To verify that increased proteasome subunit expression leads to increased proteasome activity under GR condition, we tested two distinct proteasomal peptidase activities with proteasome-specific fluorogenic peptide substrates from lysates of cells grown under SD and GR (1 hour after switching from SD to GR) conditions. GR induces both the chymotrypsin-like activity and caspase-like activity of the proteasome (Fig. 5, D and E). We also observed that MR increases proteasome activity, which is consistent with the previous findings (34). Similarly, UBR2 deletion also increases proteasome activity (Fig. 5, F and G), reproducing a previously published result (32).
We found that Rpn4 is required for the life span extension by GR, as RPN4 deletion abolishes the life span extension by GR but does not affect the life span under normal conditions (Fig. 5H). Moreover, we found that GR failed to extend the long life span of the UBR2 deletion mutant further (Fig. 5I), consistent with the hypothesis that both GR and UBR2 deletion exert their effect on life span at least partially through increased proteasome activity.
Since GR leads to decreased intracellular methionine and increased proteasome activity, and MR can induce proteasome activity, we asked whether the increased proteasome activity under GR is caused by decreased intracellular methionine. We measured proteasome activities under GR with external supplementation of methionine and compared the results with those under GR and normal conditions. We found that methionine supplementation does not abrogate the increase in proteasome activity under GR (Fig. 5, D and E). Using the Rpn11-GFP reporter, we also measured proteasome protein level by single-cell tracking using microfluidic devices and found that it has similar induction dynamics under GR with methionine supplementation compared with GR (Fig. 5C). We thus concluded that the increase in proteasome expression/activity under GR does not require a decrease in intracellular methionine, indicating that GR may increase proteasome activity through redundant mechanisms.
DISCUSSION
A comprehensive survey of transcriptional and translational changes between GR and normal conditions
We have found that GR, as implemented by 0.05% glucose in the growth media compared with the normal 2% glucose, robustly extends the replicative life span of yeast in a microfluidic environment. This finding is consistent with the aggregated analysis of traditional life span assays based on microdissection, as well as a recent study based on a different microfluidic design (17). In another study using microfluidic device, Huberts et al. reported no life span extension (19) under GR. We suspect that the different outcomes might be caused by differences in devices and protocols used.
To identify downstream effectors for the life span extension caused by GR, we have performed a comprehensive survey of gene expression differences in cells growing under GR and normal conditions, using ribosomal profiling and RNA-seq. This analysis revealed a broad spectrum of transcriptional and translational changes induced by GR; many features of the gene expression changes are consistent with previous transcriptional and polysome profiling of CR in different species (12) and overlap with the previous ribosome profiling of amino acid or glucose starvation conditions (21, 34, 35). However, our study also revealed many GR-specific transcriptional and translation changes that are relevant to the life span extension phenotype.
The longevity effects of GR may be mediated by the depletion of intracellular methionine—Methionine serves as a key control point for regulating life span in general
CR in animals is typically implemented by reducing food intake, which leads to reduction in proteins (all amino acids), lipids, carbohydrates, and other nutrients simultaneously. It has been suggested that CR may exert its effects through MR, a common dietary intervention shown to have conserved life span effects across species (27, 28). This idea is based on the observations that (i) MR in rodents specifically leads to responses that are similar to those found under CR, and (ii) similar effects have generally not been observed when other amino acids were restricted.
We showed that GR alone, without changing the amino acid composition of the media, triggers a coordinated response in methionine biosynthesis and uptake, suggesting cross-talk between glucose sensing/metabolism and methionine-mediated regulatory control. Methionine biosynthetic enzymes and transporters are repressed by GR at the transcriptional and the translational levels, leading to a decreased intracellular methionine level, something that might already be rate-limiting under SD conditions (Fig. 2, C to F). Although GR also decreases the intracellular concentration of other amino acids, only adding methionine back to the media abrogates life span extension by GR. These observations suggest that life span extension caused by GR may be mediated, at least in part, by the depletion of intracellular methionine, further highlighting the importance of methionine in regulating life span.
Our analysis of the gene expression profiles of longevity mutants adds further support to the general importance of the methionine pathway in regulating life span. In TF deletion mutants with extended life span, we observed that methionine pathway genes were repressed. Moreover, deletion of many of the genes in the methionine biosynthetic pathway leads directly to life span extension.
Together, our study suggests a model in which the intracellular methionine level serves as a key control point, which mediates life span extension caused by various nutrient and genetic perturbations, including GR (Fig. 6). Future work is needed to place other players known to be important for the life span extension by CR in this framework to construct a global picture of how life span is regulated under both normal and CR conditions.
Fig. 6. A graphical model summarizing the findings in this work and some of the previous studies.
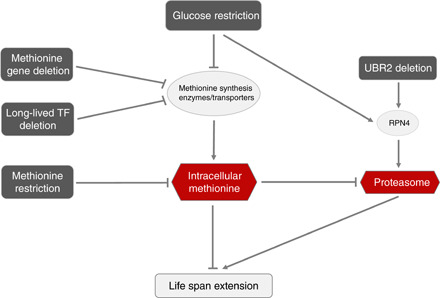
The life span extension by GR is mediated by reduced intracellular methionine (via the repression of methionine biosynthetic enzymes and transporters) and by elevated proteasome activity. MR is known to extend life span. Reducing intracellular methionine extends life span in a more general context beyond GR and MR, such as genetic perturbation of methionine pathway genes or in long-lived TF deletion mutants. Proteasome activity may be regulated by redundant pathways under GR—by intracellular methionine and by an independent pathway, via the translational up-regulation of Rpn4. Elevated proteasome activity is required for the life span extension of GR and the UBR2 deletion mutant.
Cross-talk between glucose and the methionine pathways may be a general strategy to coordinate the cell’s metabolic state, translation, and growth
The cross-talk from methionine to carbon metabolism has been observed previously. For example, Walvekar et al. found that adding methionine to minimum media induces pentose phosphate pathway (PPP) and nucleotide synthesis, coordinating a global anabolic program to promote cell proliferation (36). This finding is consistent with the earlier genetic analysis that found a PPP mutant to be methionine auxotrophic (37). This cross-talk from methionine to PPP is required for the effect of methionine on oxidative stress resistance (38), and MR improves cancer therapy outcome through change of one carbon metabolism (39).
Here, we have observed a cross-talk from glucose sensing to the regulation of intracellular methionine. We found that GR elicits specific transcriptional and translational programs to down-regulate methionine synthesis/uptake. It may be a general phenomenon that altered glucose concentration in the media triggers specific methionine pathway responses. We found that methionine genes were repressed under glucose starvation conditions (35), and induction of methionine genes was observed when glucose was added back to glucose-restricted media (40). We also performed a systematic search for yeast gene expression profiling experiments in which the methionine TF module is significantly repressed; among the top hits, we found experiments measuring the expression change when cells were switched from anaerobic to aerobic growth (aeration of anaerobically grown cells; data from the Gene Expression Omnibus, ID GSE7140) (41). Together, these observations indicate that intracellular methionine is regulated in response to glucose availability, and this response might be mediated through mitochondrial respiration. The phenomenon is conserved across different yeast strains. In our study, we found that intracellular methionine drops in response to GR in two different strains, BY4741 (methionine auxotrophic, with MET15 deleted) and BY4742 (methionine prototrophic), although ribosome profiling and RNA-seq were performed using the BY4741 strain. Glucose starvation represses methionine genes in BY4741 (35), and the glucose addback experiments were performed using the CEN.PK 113-7D strain, which is a prototroph widely used for metabolic engineering (40). Furthermore, gene expression profiling of the anaerobic-to-aerobic switch used a diploid strain, BY4743 (generated from a cross between BY4741 and BY4742), with one copy of the wild-type MET15.
The cross-talk between glucose sensing/metabolism and methionine regulation may be a general mechanism to coordinate the cell’s metabolic state with translation and cell growth. Glucose is the preferred carbon source for yeast cells, and sensing and responding to glucose availability is one of the most important regulatory tasks. Although the majority of gene expression changes under GR are at the transcriptional level, our study also revealed ~90 genes whose translational efficiency was significantly changed, suggesting that translational regulation is an important component of the response. Methionine is a special amino acid coded by the translation initiation codon, and methionyl tRNA (Met-tRNAi) is required for the assembly of the translation initiation complex. It has been proposed that under sulfate or methionine starvation, the signal for G1 arrest is at or beyond the formation of methionyl tRNA, suggesting that methionine controls cell division through translational regulation (42). In addition, methionine can regulate the translational capacity and metabolic homeostasis by modulating tRNA thiolation (34, 43, 44). Thus, methionine may serve as an important control point for translation, and changes in the methionine pathway genes in response to glucose availability may be a general strategy to coordinate the metabolic state of the cell with translation and cell growth.
How does methionine control life span?
It is possible that methionine may exert its influence on life span by directly controlling the translation of some of the key downstream regulators. Although restricting methionine leads to a decrease in general translation, it can also lead to increased translation of some special genes. One example is Rpn4, which plays an important role in GR’s life span extension phenotype.
Besides the direct control of translation, methionine likely influences life span by altering the metabolic program of the cell. As mentioned above, methionine can cross-talk with carbon metabolism by inducing the PPP and a hierarchically organized anabolic program to support cell growth and proliferation (36) and consequently affect life span. One of the derivatives of methionine, S-adenosyl methionine (SAM), is a cofactor that serves as a methyl donor. In a genetic model of CR for Caenorhabditis elegans (eat-2 mutant), it was observed that the enzyme SAMS-1 (SAM synthase) was down-regulated and that RNA interference knockdown of SAM-1 extends the life span of wild-type worms but does not affect the long-lived eat-2 mutants, suggesting that the life span effect of CR is at least in part due to the reduction in SAM (45). In the fly, it has been observed that enhancing SAM catabolism by overexpressing the gene GNMT, which leads to reduced methionine and SAM, extends the life span (46). SAM may mediate the life span effect of methionine by regulating the TORC1 (target of rapamycin complex 1) in yeast and mTORC1 (mammalian target of rapamycin complex 1) in mammals (47, 48). In yeast, methionine represses autophagy through SAM-responsive methylation of the protein phosphatase PPA2 (47), which regulates TORC1 and autophagy through the Iml1p complex, and increased autophagy is required for the life span extension of CR. In mammals, SAM directly binds to SAMTOR to disrupt the SAMTOR-GATOR1 complex (48). SAM may also affect life span through DNA methylation that changes gene expression (49).
Methionine is also coupled to the trans-sulfuration pathway (TSP), and it has been proposed that CR leads to increased expression of the TSP enzyme cystathionine gamma-lyase (CGL), resulting in increased hydrogen sulfide (H2S) production and mediating the benefits of CR, including increased chronological life span of yeast (50). We observed in yeast that both CGL (CYS3) and cystathionine beta synthase (CYS4), two enzymes that can produce H2S in the TSP, are significantly down-regulated by GR and that deletion of methionine biosynthesis genes both up- and downstream of H2S leads to life span extension (Fig. 4). These data suggest that in the yeast-replicative aging model and for the strains we studied, the effector is methionine instead of H2S.
The importance of the proteasome in the life span extension by CR
Our study highlighted the importance of increased proteasome activity for the life span extension by GR. Previous studies showed that induced autophagy is required for the life span phenotype of CR (51), demonstrating the importance of degradation and recycling of damaged cellular components. Targeted degradation by the proteasome is known to be important for controlling cell cycle, gene expression, and response to stress. Our study links this important cellular activity to aging as well. Previous work indicated that deletion of UBR2, a ubiquitin ligase that targets Rpn4 for degradation, increases Rpn4 activity and extends life span (32). Increased proteasome expression has been observed in CR experiments across several species, including worm and mouse (13, 52), but previous studies have not shown that the increase is required for life span extension. Moreover, proteasome activity declines with age in a number of species (53), and genetic approaches to restore proteasome function with age lead to life span extension in C. elegans (52). Together, this collection of evidence and our study suggest that proteasome function is intimately linked to aging. In the context of GR in yeast, we found that the translational up-regulation of Rpn4 leads to the increase in proteasome expression; this increase is possibly caused by pathways redundant to the methionine pathway (Fig. 6).
We observed a dynamic behavior of proteasome that is typical of a system with negative feedback: The proteasome level is transiently increased upon switching from normal to GR media and relaxes back to the baseline levels before switch by a few hours (Fig. 5C). Given that the elevated proteasome level is required for life span extension, this may suggest that an intermittent GR (where the media is switched back and forth between normal and GR) with a time interval comparable to the adaptation time may have even a bigger effect on life span.
The downstream targets of the proteasome relevant to aging in yeast remain to be fully elucidated. While there is evidence for enhanced protein turnover under conditions that lead to life span extension (32), a recent study also identified Mig1 as a specific target of the proteasome relevant for aging (33). Mig1 acts to repress transcription of genes important for respiration while yeast is undergoing fermentation. Thus, the proteasome may have an additional role in coordinating metabolic changes that occur under GR conditions. Whether there are other specific proteasome targets important for longevity in yeast remains to be determined.
Targeting the proteasome has been a useful therapeutic strategy for treating diseases, and much attention has been devoted to proteasome inhibitors; e.g., small-molecule inhibitors have been used for treating cancer (54). In contrast, proteasome activators are much less studied. However, proteasome activators have been shown to have beneficial effects on Huntington’s disease neuronal model cells and to delay the senescence of human fibroblast cells (55), raising the possibility that proteasome activators can be developed into antiaging drugs. Our study suggests that increased proteasome activity may mimic CR and thus confer the life span benefit. In addition to those that directly target the proteasome structure to increase the accessibility of the proteolytic core, small molecules that target the upstream regulators resulting in a coordinated up-regulation of proteasome components may be an effective method to slow aging and mitigate age-associated chronic diseases.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains used for the ribosomal profiling experiments and life span measurements were BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), BY4741-MET (MATa his3Δ1 leu2Δ0 ura3Δ0), and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), and deletion mutants derived from BY4741 and BY4742. Mup1-GFP reporter was taken from the yeast GFP-tagging library (56).
In the replicative life span and ribosome profiling experiments, SD medium was used. The SD medium contained 2% (w/v) glucose, yeast nitrogen base (6.7 g/liter) without amino acid, adenine (20 mg/liter), l-arginine HCL (20 mg/liter), l-aspartic acid (100 mg/liter), l-histidine HCL (20 mg/liter), l-leucine (100 mg/liter), l-isoleucine (30 mg/liter), l-lysine HCL (30 mg/liter), l-methionine (20 mg/liter), l-phenylalanine (50 mg/liter), l-threonine (200 mg/liter), l-tryptophan (20 mg/liter), l-tyrosine (30 mg/liter), uracil (20 mg/liter), l-valine (150 mg/liter), glutamic acid (100 mg/liter), and serine (4 g/liter). The GR media have the similar ingredients as the SD medium except for 0.05% (w/v) glucose concentration. The SD + 10xMet media have the similar ingredients as SD medium except for l-methionine (200 mg/liter). The GR + 10xMet media have the similar ingredients as GR medium except for l-methionine (200 mg/liter). The MR media have the similar ingredients as GR medium except for l-methionine (2 mg/liter). Media were freshly made before the experiment. All nutrients were purchased from Sigma-Aldrich Corporation.
Single-cell observations using microfluidic devices
We used the microfluidic chips previously developed in our laboratory (16). TS-1B autocontrolled syringe pumps were used to inject the medium and the cells into the microfluidic chips at a flow rate of 700 μl/hour for cell loading and 400 μl/hour for life span measurement. Cells were cultured overnight in SD media (2% d-glucose) at 30°C to OD600 (optical density at 600 nm) ~1.0 and then diluted by 10-fold and cultured in the 30°C shaker for another 4 to 6 hours before loading into the microfluidic device.
Image acquisition and analysis
For life span measurement using the microfluidic device, images were taken once every 15 min by a Nikon TE2000 microscope or a Zeiss Axio Observer Z1 with 40× objective. For the Mup1 fluorescent reporter analysis, images were taken once every 15 min with a Zeiss Axio Observer Z1 with Spinning Disc using 63× oil objective. We used the bright-field images for cell segmentation and tracking and measured fluorescence intensity by the total fluorescent signal normalized by the cell area. Image data were analyzed by the customized software Cellseg as previously described (57). For Rpn11 fluorescent reporter analysis, images were taken once every 10 min with a Zeiss Axio Observer Z1 with Spinning Disc using 63× oil objective. Image data were analyzed by a customized python script. Briefly, for each cell, the GFP signal was fitted to a Gaussian mixture model with three components (as the visual inspection of the image reveals distinct intensity in three subcellular localizations: nucleus, cytoplasm, and vacuole). The average signal in the cytoplasm in each cell is used to plot Fig. 5C, which is much stronger than signal in vacuole and less sensitive to the random drifting of focal plane.
Ribosome profiling of cells growing in SD versus GR and other media
We used two different protocols to grow cell culture in SD versus GR conditions.
1) Quick dilution from SD (2% glucose) to GR (0.05% glucose): The initial cell culture was incubated in SD medium overnight to OD600 of 0.8 to 1.0 and then diluted by fivefold using SD media and incubated for another 4 hours. The sample was then divided equally into two aliquots. One was diluted 40 times by prewarmed SD medium with 0% glucose to reach a final 0.05% glucose concentration (the sample), and the other was diluted 40 times by prewarmed SD medium with 2% glucose (the control). Both samples were incubated for another hour before harvesting. All the steps were carried out at 30°C.
2) Spin down and resuspension from SD to GR: The initial cell culture was incubated in SD medium overnight to an OD600 of 0.8 to 1.0 and then diluted by fivefold using SD medium and incubated for another 4 hours. The sample was then divided equally into two aliquots. Cells were separated from the media by spin down at 2000 rcf for 5 min and resuspended into SD and GR media, respectively. All samples were incubated for another hour before harvesting. All the steps were carried out at 30°C.
Ribosomal profiling experiments were carried out using the protocol developed by Ingolia et al. (58). Raw sequences were obtained from Illumina Hiseq 2000.
Amino acid analysis
Cells were cultured overnight in SD medium (2% d-glucose) at 30°C to OD600 ~1.0 and then diluted by fivefold with fresh SD medium and cultured in the 30°C shaker until they reach an OD600 ~0.6 to 0.8. The 400-ml sample was then divided equally into four aliquots. Cells were separated from the media by spin down and resuspended into 100 ml of SD, 100 ml of GR, 100 ml of SD + 10xMet, and 100 ml of GR +10xMet media, respectively. All samples were incubated for another hour before harvesting. After quenching with sodium azide (at 0.1%) and cycloheximide (at 50 μg/ml), the cells were collected by centrifugation and washed with 5 ml of autoclaved water for three times and resuspended in 100 μl of autoclaved water. Then, the cell suspension was boiled for 5 min and centrifuged to collect the supernatant, representing the metabolite extract. This extract was then analyzed by standard PTH (phenylthiohydantoin) derivatization and high-performance liquid chromatography analysis (59) at the Texas A&M University Protein Chemistry Facility, to quantify the concentration of each amino acid present in the extract. These values were then normalized by cell number and size. Note that unmodified cysteine cannot be detected by this method (59).
Proteasome activity assay
Cells were cultured to OD600 ~0.8 to 1.0 and then separated from the original media by spin down and resuspended into SD, GR, or MR media, respectively. All samples were incubated for another hour before harvesting and resuspending in lysis buffer [50 mM tris-HCl (pH 7.5), 0.5 mM EDTA, 5 mM MgCl2, and protease inhibitor cocktail tablet; Roche], and p1000 was use to squirt cell mixture slowly into a 50-ml conical tube with liquid nitrogen, creating frozen droplets floating in the tube. Then, cell breakage was performed by mixer milling. The supernatant was collected at 13,000 rpm for 3 min at 4°C. Protein concentration was determined by a Bradford assay (Bio-Rad) using bovine serum albumin as standard. Proteasomal chymotrypsin-like activity was measured by using Suc-LLVY-AMC (Bachem), and proteasomal caspase-like activity was measured by using Ac-nLPnL-AMC (Bachem). Each reaction volume was 200 μl, including 50 μg of total lysate protein, 100 μM peptide substrate, and lysis buffer. The slope of fluorescence was recorded after incubation for 15 min at 30°C in a SpectraMax M5 with an excitation wavelength of 380 nm and an emission wavelength of 460 nm. The control reactions with a specific proteasome inhibitor MG-132 were performed in each experiment.
Sequence analysis and quantification of differential gene expression
Sequence reads were aligned to the most recent S. cerevisiae genome using SOAPaligner/SOAP2 (2.21) with default setting (60). After trimming off the adapters, reads aligned to ribosomal RNA and tRNA sequences were filtered out. The rest of the reads were then aligned to the genome sequence. Last, reads that did not align to the genome were aligned to all the CDSs to retain those covering the splicing junctions. After the alignment, we counted the number of reads starting at each position across the whole genome; for ribosomal footprinting data, the starting position of each read was shifted by 15 bps toward the 3′ end, to adjust for the offset due to ribosome protection. To get the abundance of reads covering each gene, we sum all reads with the starting position from the start to the stop codon, excluding the first 50 bps from the transcription start site (TSS), to alleviate the effect of the biased distribution of reads around the TSS (21, 58).
For a reliable gene expression comparison between two conditions, we excluded genes with less than 128 total raw reads (combining the reads from the two conditions) (21). We computed the fold change of mRNA or footprint as the ratio of the corresponding reads from the two conditions, with total reads normalized to adjust the median fold change to 1.
Quantification of translational efficiency changes
To measure the change of translational efficiency between two conditions, we first fitted a linear model using the log fold changes of the mRNA and the corresponding log fold changes of the footprint: yi = αxi + β, where xi and yi are the log fold changes of the mRNA and footprint for the i-th gene. After getting the values of α and β, we compute the expected value of the log fold change of the footprint, given the log fold change of mRNA, ri = αxi + β. The deviation between the real fold change and the expected fold change for the footprint is calculated as di = yi − ri, which represents the additional change of the footprint not explained by the mRNA change. Z score is defined as di normalized by the standard deviation: .
Identifying genes associated with a specific GO process that are differentially expressed in long-lived TF deletion mutants
To identifying genes associated with a specific GO process that are differentially expressed in long-lived TF deletion mutants, we used our previously described set of long-lived yeast deletion strains (31) and a set of mRNA expression profiles of 263 TF deletion strains cultured in rich medium (30) as follows. Considering each of the 409 GO processes comprising at least 10 genes, we tabulated a matrix whose rows were transcripts annotated in the process and whose columns were the 12 TF deletion strains that exhibited significant longevity. We calculated the median expression across all the entries of this matrix, mtrue. We next resampled, from the 261 transcriptional profiles of TF deletion strains for which life spans were available, 12 random such strains and used them to tabulate a matrix of expression measurements for all the genes of the GO process, calculating the median expression level mresample across the entries of this matrix. We repeated the latter resampling calculation 10,000 times. We then tabulated plarger, the proportion of the resampled datasets in which mresample was greater than or equal to mtrue, and psmaller, the proportion of resampled datasets in which mresample was less than or equal to mtrue (test on both sides). The final estimated p was taken as 2*min(plarger, psmaller). The GO process “cysteine biosynthetic process” (GO:0019344) had P = 0.0078 in this test, and “methionine biosynthetic process” (GO:0009086) had P = 0.0412.
Calculation of the TF module z scores
We used the TF targets from the analysis by MacIsaac et al. based on the systematic ChIP-chip data, using 0.001 as the P value cutoff and the strongest conservation between species (26). We computed the rank sum test z scores comparing the fold changes of the target genes versus nontarget genes for each TF. The sign of the z score reflects the overall direction of the gene expression change in the modules; positive z score indicates overall induction, and negative z score indicates overall repression. We used TF modules with at least 15 targets for this analysis.
Supplementary Material
Acknowledgments
Funding: This work was funded by the NIH (R01AG058742, R01AG043080, R21AG060129, P30AG013280, GM123139, and P20GM121176). Q.O. acknowledges support from the National Science Foundation of China (NSFC 11434001 and 11774011). M.A.M. acknowledges support from an American Federation for Aging Research Junior Faculty Grant. J.S.W. is an HHMI investigator. Author contributions: K.Z., J.Z., Q.O., and H.L. conceived and designed the experiments. K.Z. and S.R. performed the ribosome profiling and RNA-seq experiments with supervision from J.S.W. K.Z. and Z.C. performed microfluidic-based single-cell life span and reporter analyses. K.Z. and M.P. measured intracellular amino acid concentrations. M.A.M., M.K., and B.K.K. contributed the life span data for the methionine pathway deletion mutants. K.Z. and K.D. performed the proteasome activity assays. C.D. assisted in strain constructions and various biochemical assays. A.S. and R.B.B. performed bioinformatic analysis of long-lived TF deletion mutants. K.Z., J.Z., and H.L. analyzed the data. K.Z., B.K.K., J.S.W., J.Z., and H.L. discussed and interpreted the results. K.Z., J.Z., and H.L. wrote the manuscript with inputs from the other authors. All authors read and edited the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Ribosome profiling and RNA-seq data have also been deposited at GEO (accession number GSE134152). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eaba1306/DC1
REFERENCES AND NOTES
- 1.McCay C. M., Crowell M. F., Maynard L. A., The effect of retarded growth upon the length of life span and upon the ultimate body size: One figure. J. Nutr. 10, 63–79 (1935). [PubMed] [Google Scholar]
- 2.R. Weindruch, R. L. Walford, Retardation of Aging and Disease by Dietary Restriction (Charles C Thomas, Springfield, IL, 1988). [Google Scholar]
- 3.Colman R. J., Anderson R. M., Johnson S. C., Kastman E. K., Kosmatka K. J., Beasley T. M., Allison D. B., Cruzen C., Simmons H. A., Kemnitz J. W., Weindruch R., Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattison J. A., Roth G. S., Beasley T. M., Tilmont E. M., Handy A. M., Herbert R. L., Longo D. L., Allison D. B., Young J. E., Bryant M., Barnard D., Ward W. F., Qi W., Ingram D. K., de Cabo R., Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 489, 318–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus W. E., Bhapkar M., Huffman K. M., Pieper C. F., Das S. K., Redman L. M., Villareal D. T., Rochon J., Roberts S. B., Ravussin E., Holloszy J. O., Fontana L.; CALERIE Investigators , 2 years of calorie restriction and cardiometabolic risk (CALERIE): exploratory outcomes of a multicentre, phase 2, randomised controlled trial. Lancet Diabetes Endocrinol. 7, 673–683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longo V. D., Shadel G. S., Kaeberlein M., Kennedy B., Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 16, 18–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S.-J., Defossez P.-A., Guarente L., Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Lin S.-J., Kaeberlein M., Andalis A. A., Sturtz L. A., Defossez P.-A., Culotta V. C., Fink G. R., Guarente L., Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418, 344–348 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Kaeberlein M., Kirkland K. T., Fields S., Kennedy B. K., Sir2-independent life span extension by calorie restriction in yeast. PLOS Biol. 2, E296 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapahi P., Kaeberlein M., Hansen M., Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res. Rev. 39, 3–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaeberlein M., Powers R. W. III, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K., Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 (2005). [DOI] [PubMed] [Google Scholar]
- 12.S. K. Kim, in Molecular Biology of Aging (Cold Spring Harbor Laboratory Press, 2008), Genome-wide Views of Aging Gene Networks, vol. 51, chap. 9. [Google Scholar]
- 13.Lee C. K., Klopp R. G., Weindruch R., Prolla T. A., Gene expression profile of aging and its retardation by caloric restriction. Science 285, 1390–1393 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Lee S. S., Avalos Vizcarra I., Huberts D. H. E. W., Lee L. P., Heinemann M., Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc. Natl. Acad. Sci. U.S.A. 109, 4916–4920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z., Zhang Y., Zou K., Brandman O., Luo C., Ouyang Q., Li H., Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell 11, 599–606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Luo C., Zou K., Xie Z., Brandman O., Ouyang Q., Li H., Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLOS ONE 7, e48275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jo M. C., Liu W., Gu L., Dang W., Qin L., High-throughput analysis of yeast replicative aging using a microfluidic system. Proc. Natl. Acad. Sci. U.S.A. 112, 9364–9369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei S.-C., Brenner C., Calorie restriction-mediated replicative lifespan extension in yeast is non-cell autonomous. PLOS Biol. 13, e1002048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huberts D. H. E. W., González J., Lee S. S., Litsios A., Hubmann G., Wit E. C., Heinemann M., Calorie restriction does not elicit a robust extension of replicative lifespan in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 111, 11727–11731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou K., Ren D. S., Ou-Yang Q., Li H., Zheng J., Using microfluidic devices to measure lifespan and cellular phenotypes in single budding yeast cells.J. Vis. Exp., 55412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonenberg N., Hinnebusch A. G., Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rolfes R. J., Hinnebusch A. G., Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: Implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13, 5099–5111 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang R., Wek S. A., Wek R. C., Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20, 2706–2717 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., Fox L. A., Dang N., Johnston E. D., Oakes J. A., Tchao B. N., Pak D. N., Fields S., Kennedy B. K., Kaeberlein M., Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacIsaac K. D., Wang T., Gordon D. B., Gifford D. K., Stormo G. D., Fraenkel E., An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7, 113 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee B. C., Kaya A., Gladyshev V. N., Methionine restriction and life-span control. Ann. N. Y. Acad. Sci. 1363, 116–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ables G. P., Johnson J. E., Pleiotropic responses to methionine restriction. Exp. Gerontol. 94, 83–88 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Brown-Borg H. M., Buffenstein R., Cutting back on the essentials: Can manipulating intake of specific amino acids modulate health and lifespan? Ageing Res. Rev. 39, 87–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Z., Killion P. J., Iyer V. R., Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 39, 683–687 (2007). [DOI] [PubMed] [Google Scholar]
- 31.McCormick M. A., Delaney J. R., Tsuchiya M., Tsuchiyama S., Shemorry A., Sim S., Chou A. C.-Z., Ahmed U., Carr D., Murakami C. J., Schleit J., Sutphin G. L., Wasko B. M., Bennett C. F., Wang A. M., Olsen B., Beyer R. P., Bammler T. K., Prunkard D., Johnson S. C., Pennypacker J. K., An E., Anies A., Castanza A. S., Choi E., Dang N., Enerio S., Fletcher M., Fox L., Goswami S., Higgins S. A., Holmberg M. A., Hu D., Hui J., Jelic M., Jeong K.-S., Johnston E., Kerr E. O., Kim J., Kim D., Kirkland K., Klum S., Kotireddy S., Liao E., Lim M., Lin M. S., Lo W. C., Lockshon D., Miller H. A., Moller R. M., Muller B., Oakes J., Pak D. N., Peng Z. J., Pham K. M., Pollard T. G., Pradeep P., Pruett D., Rai D., Robison B., Rodriguez A. A., Ros B., Sage M., Singh M. K., Smith E. D., Snead K., Solanky A., Spector B. L., Steffen K. K., Tchao B. N., Ting M. K., Wende H. V., Wang D., Welton K. L., Westman E. A., Brem R. B., Liu X.-g., Suh Y., Zhou Z., Kaeberlein M., Kennedy B. K., A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 22, 895–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruegel U., Robison B., Dange T., Kahlert G., Delaney J. R., Kotireddy S., Tsuchiya M., Tsuchiyama S., Murakami C. J., Schleit J., Sutphin G., Carr D., Tar K., Dittmar G., Kaeberlein M., Kennedy B. K., Schmidt M., Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLOS Genet. 7, e1002253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao Y., Tsuchiyama S., Yang C., Bulteau A. L., He C., Robison B., Tsuchiya M., Miller D., Briones V., Tar K., Potrero A., Friguet B., Kennedy B. K., Schmidt M., Proteasomes, Sir2, and Hxk2 form an interconnected aging network that impinges on the AMPK/Snf1-regulated transcriptional repressor Mig1. PLOS Genet. 11, e1004968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou K., Ouyang Q., Li H., Zheng J., A global characterization of the translational and transcriptional programs induced by methionine restriction through ribosome profiling and RNA-seq. BMC Genomics 18, 189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zid B. M., O'Shea E. K., Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514, 117–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walvekar A. S., Srinivasan R., Gupta R., Laxman S., Methionine coordinates a hierarchically organized anabolic program enabling proliferation. Mol. Biol. Cell 29, 3183–3200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas D., Cherest H., Surdin-Kerjan Y., Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J. 10, 547–553 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell K., Vowinckel J., Keller M. A., Ralser M., Methionine metabolism alters oxidative stress resistance via the pentose phosphate pathway. Antioxid. Redox Signal. 24, 543–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao X., Sanderson S. M., Dai Z., Reid M. A., Cooper D. E., Lu M., Richie J. P. Jr., Ciccarella A., Calcagnotto A., Mikhael P. G., Mentch S. J., Liu J., Ables G., Kirsch D. G., Hsu D. S., Nichenametla S. N., Locasale J. W., Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kresnowati M. T. A. P., van Winden W. A., Almering M. J. H., ten Pierick A., Ras C., Knijnenburg T. A., Daran-Lapujade P., Pronk J. T., Heijnen J. J., Daran J. M., When transcriptome meets metabolome: Fast cellular responses of yeast to sudden relief of glucose limitation. Mol. Syst. Biol. 2, 49 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. G. Beckhouse, Ph. D, UNSW (2006).
- 42.Unger M. W., Hartwell L. H., Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 73, 1664–1668 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laxman S., Sutter B. M., Wu X., Kumar S., Guo X., Trudgian D. C., Mirzaei H., Tu B. P., Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154, 416–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta R., Walvekar A. S., Liang S., Rashida Z., Shah P., Laxman S., A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. eLife 8, e44795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen M., Hsu A.-L., Dillin A., Kenyon C., New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLOS Genet. 1, 119–128 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Obata F., Miura M., Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat. Commun. 6, 8332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutter B. M., Wu X., Laxman S., Tu B. P., Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 154, 403–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu X., Orozco J. M., Saxton R. A., Condon K. J., Liu G. Y., Krawczyk P. A., Scaria S. M., Harper J. W., Gygi S. P., Sabatini D. M., SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulrey C. L., Liu L., Andrews L. G., Tollefsbol T. O., The impact of metabolism on DNA methylation. Hum. Mol. Genet. 14( suppl. 1), R139–R147 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Hine C., Harputlugil E., Zhang Y., Ruckenstuhl C., Lee B. C., Brace L., Longchamp A., Treviño-Villarreal J. H., Mejia P., Ozaki C. K., Wang R., Gladyshev V. N., Madeo F., Mair W. B., Mitchell J. R., Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 160, 132–144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madeo F., Zimmermann A., Maiuri M. C., Kroemer G., Essential role for autophagy in life span extension. J. Clin. Invest. 125, 85–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vilchez D., Morantte I., Liu Z., Douglas P. M., Merkwirth C., Rodrigues A. P. C., Manning G., Dillin A., RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Vilchez D., Saez I., Dillin A., The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 5, 5659 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Huang L., Chen C. H., Proteasome regulators: Activators and inhibitors. Curr. Med. Chem. 16, 931–939 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katsiki M., Chondrogianni N., Chinou I., Rivett A. J., Gonos E. S., The olive constituent oleuropein exhibits proteasome stimulatory properties in vitro and confers life span extension of human embryonic fibroblasts. Rejuvenation Res. 10, 157–172 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Huh W.-K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K., Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Yang X., Lau K.-Y., Sevim V., Tang C., Design principles of the yeast G1/S switch. PLOS Biol. 11, e1001673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., Weissman J. S., The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinrikson R. L., Meredith S. C., Amino acid analysis by reverse-phase high-performance liquid chromatography: Precolumn derivatization with phenylisothiocyanate. Anal. Biochem. 136, 65–74 (1984). [DOI] [PubMed] [Google Scholar]
- 60.Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J., SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eaba1306/DC1


