Analysis of convergent collateral sensitivity phenotypes may guide the design of antibiotic therapies in P. aeruginosa infections.
Abstract
The analysis of trade-offs, as collateral sensitivity, associated with the acquisition of antibiotic resistance, is mainly based on the use of model strains. However, the possibility of exploiting these trade-offs for fighting already resistant isolates has not been addressed in depth, despite the fact that bacterial pathogens are frequently antibiotic-resistant, forming either homogeneous or heterogeneous populations. Using a set of Pseudomonas aeruginosa-resistant mutants, we found that ceftazidime selects pyomelanogenic tobramycin-hypersusceptible mutants presenting chromosomal deletions in the analyzed genetic backgrounds. Since pyomelanogenic resistant mutants frequently coexist with other morphotypes in patients with cystic fibrosis, we analyzed the exploitation of this trade-off to drive extinction of heterogeneous resistant populations by using tobramycin/ceftazidime alternation. Our work shows that this approach is feasible because phenotypic trade-offs associated with the use of ceftazidime are robust. The identification of conserved collateral sensitivity networks may guide the rational design of evolution-based antibiotic therapies in P. aeruginosa infections.
INTRODUCTION
Antibiotic effectiveness, currently compromised by the spread of antibiotic resistance (AR), requires not only innovation but also conservation, which may allow for improved use of current antibiotics (1). For this conservation, understanding of the trade-offs associated with AR acquisition—such as increased susceptibility to a second drug after use of the first, a phenomenon first described in the 1950s as collateral sensitivity (2)—might be particularly relevant. Various studies have been undertaken trying to exploit the evolutionary constraint imposed by collateral sensitivity patterns (3, 4), such as combinatory therapy (5, 6) or alternating collaterally susceptible drug pairs (7–9).
Despite progress in study of the collateral sensitivity phenomenon, some questions remain to be answered. In particular, there is still only limited information on the evolutionary conservation of collateral sensitivity patterns, not only between different species (10, 11) but also within different members from the same species (12–15). In the case of Pseudomonas aeruginosa, some studies have revealed that isolates of this pathogen obtained from patients with chronic infections and treated with distinct antibiotic classes present convergence in their collateral sensitivity phenotypes (9), while others have described major differences in the collateral sensitivity patterns associated with the acquisition of resistance to one antibiotic between replicates of the same strain of P. aeruginosa evolving in parallel (13). These discrepancies may lie in the degree of reproducibility of the evolutionary pathways leading to AR. In this respect, it is known that the type of resistance mutations present in a given genetic background may be restricted because of epistatic interactions (10, 16–21) and that the cumulative acquisition of AR mutations in different loci reduces the variety of pathways, leading to AR (22, 23). Since contingency may be relevant not only for AR evolution but also for the acquired collateral sensitivity phenotype (24), knowing the degree of conservation of collateral sensitivity patterns associated with the use of a specific drug in different genetic backgrounds, particularly in the case of mutants already presenting a phenotype of AR, is of special interest.
In a previous study, we observed that P. aeruginosa PA14 populations experimentally evolved in the presence of ceftazidime displayed a robust collateral sensitivity to amikacin (25), as a consequence of selection of large chromosomal deletions upon 1 day of adaptive laboratory evolution (ALE) (25). The chromosomal deletions included hmgA, which encodes an enzyme whose lack of activity leads to the hyperproduction of the brown pigment pyomelanin (26); galU, whose inactivation reduces ceftazidime susceptibility (27); and mexXY, which encodes a multidrug resistance (MDR) efflux pump that contributes to P. aeruginosa intrinsic aminoglycosides resistance (28). Up to 13% of patients with cystic fibrosis (CF) are infected by P. aeruginosa pyomelanin-producing mutants (29) that, in agreement with the phenotype of the mutants selected after ALEs, are also hypersusceptible to aminoglycosides (30). Although the fact that pyomelanin increases resistance to oxidative stress and favors bacterial persistence in chronic lung infections (26) has been considered the most likely explanation for in vivo selection of those mutants, this genetic event has also been reported in different strains of P. aeruginosa subjected to ALE in the presence of other β-lactams (31–34). Therefore, it remains to be established whether these deletions are selected by the antibiotic treatment or merely represent an adaptation to the lung environment of patients with CF (35), as was previously proposed (34, 35). Even further, it remains to be answered whether these deletions are selected upon ceftazidime treatment in different genetic backgrounds of P. aeruginosa, in particular, in mutants resistant to other antibiotics; or on the contrary, whether the evolutionary robustness of this genetic event, and of its phenotypic effects, is limited. In this study, by constructing a set of resistant mutants previously identified in different ALE experiments of P. aeruginosa PA14 (16, 36) in the presence of antibiotics, and by submitting them, as well as the wild-type PA14 strain, to ALE in the presence of ceftazidime, we have determined the robustness of an early event of ceftazidime resistance evolution that is associated with collateral sensitivity to tobramycin. Further, by the recreation of heterogeneous pyomelanogenic populations belonging to each genetic background and the alternation of tobramycin with ceftazidime, we found that driving evolution toward hypersusceptibility to the first drug is generally feasible, at least in the genetic backgrounds analyzed. This finding supports the possibility of rationally designing treatments based on collateral sensitivity convergence in P. aeruginosa.
RESULTS
As mentioned above, the conservation of collateral sensitivity to a given drug within different genetic backgrounds of a species may be contingent on the degree of conservation of the evolutionary routes toward resistance to the first drug used for selection. We and others have reported the early selection of large chromosomal deletions that contain genes encoding the intrinsic aminoglycosides resistance efflux pump MexXY (28), when ALE experiments in the presence of different β-lactams, including ceftazidime (31), piperacillin (32), or meropenem (33, 34), were performed. These data suggest that this genetic event could be one of the first evolutionary steps in the evolution of P. aeruginosa toward β-lactam resistance. However, since these studies were limited to a single wild-type genetic background, it remained to be analyzed whether a similar evolutionary pattern could be followed by P. aeruginosa PA14 strains presenting different genetic backgrounds, in particular, different antibiotic-resistant mutants. That being so, collateral sensitivity to aminoglycosides, such as tobramycin, a drug that forms part of usual therapy regimens against P. aeruginosa (37), would also be conserved.
Construction and characterization of P. aeruginosa mutants
Our first objective was to analyze the evolutionary conservation of ceftazidime resistance evolution in different genetic backgrounds, consisting of mutants derived from different P. aeruginosa PA14 ALEs in the presence of antibiotics (see table S1). It has been recently suggested that AR mutations may associate with either robust or variable collateral sensitivity patterns in different genetic backgrounds, depending on whether they lead to “target” or “regulatory alterations,” respectively (10). Taking this hypothesis into account and resorting to previous different in-house ALE experiments (16, 36), we constructed a broad spectrum of strains containing single mutations that affect regulatory proteins (NfxB, ParR, or MexZ), nonregulatory proteins (NuoD or OrfN), or simultaneously containing both types of mutations. The mutant containing mutations in nfxB, phoQ, frr, and pmrB was dubbed MDR6, and the mutant containing mutations in fusA, orfN, pmrB, mexZ, gabP, ptsP, and nuoD was dubbed MDR12 (see table S1 for detailed information). The susceptibility of each mutant to different antibiotics, including those of interest in this study—tobramycin and ceftazidime—is shown in Table 1.
Table 1. MICs (μg/ml) of different antibiotics for the single and multiple P. aeruginosa PA14 mutants used in this work.
MICs ≥2-fold of the MICs for the wild-type PA14 strain are highlighted in bold. TOB, tobramycin; TGC, tigecycline; CAZ, ceftazidime; CIP, ciprofloxacin; IPM, imipenem.
| TOB | TGC | CAZ | CIP | IPM | |
| PA14 | 1 | 6 | 1 | 0.094 | 0.75 |
| nfxB177 | 1 | 32 | 1.5 | 3 | 1 |
| parR87 | 1.5 | 8 | 1 | 0.125 | 2 |
| orfN50 | 3 | 32 | 3 | 0.19 | 2 |
| nuoD184 | 2 | 4 | 1 | 0.047 | 0.75 |
| mexZ43 | 1.5 | 8 | 1 | 0.38 | 1.5 |
| MDR6* | 2 | 48 | 1.5 | 0.19 | 1.5 |
| MDR12† | 32 | 64 | 1 | 0.5 | 1.5 |
*MDR6 mutant presents mutations in nfxB, phoQ, frr, and pmrB.
†MDR12 mutant presents mutations in fusA, orfN, pmrB, mexZ, gabP, ptsP, and nuoD.
Evolutionary robustness of first steps of P. aeruginosa ceftazidime resistance evolution and collateral sensitivity to tobramycin
To determine whether chromosomal deletions containing mexXY would be early selected during P. aeruginosa evolution in presence of ceftazidime in the set of mutants mentioned above, as it was the case in the wild-type strain PA14 (25), four biological replicates of each single (nfxB177, parR87, mexZ43, orfN50, and nuoD184) and multiple mutants (MDR6 and MDR12), and the wild-type PA14 strain, were subjected to ALE in presence or absence of ceftazidime (a total of 64 populations) for 3 days. Upon 1 day of experimental evolution, almost every P. aeruginosa population challenged with antibiotic hyperproduced pyomelanin (27 of 32 populations; Fig. 1A). This result is consistent with the presence of deletions that include hmgA, as those described in our previous study, because pyomelanin accumulation is due to the lack of homogentisate 1,2-dioxygenase activity provided by HmgA (26). Since we had previously determined a cause-effect relationship between the presence of chromosomal deletions containing hmgA and mexXY and the hyperproduction of pyomelanin and hypersusceptibility to aminoglycosides, respectively (25), the susceptibility of each final population to tobramycin was analyzed. All the pyomelanogenic populations obtained after the short-term evolution in presence of ceftazidime were resistant to ceftazidime and hypersusceptible to tobramycin, when compared to the parental strain from which they evolved (Fig. 1B and table S2), even when the mutants were originally less susceptible to tobramycin than the wild-type P. aeruginosa PA14 strain. In particular, tobramycin minimal inhibitory concentration (MIC) was reduced by up to 4-fold in PA14, 2.6-fold in nfxB177, 3-fold in parR87, 7.9-fold in orfN50, 6-fold in mexZ43, 5.3-fold in MDR6, and 10.7-fold in MDR12. Consistent with the linkage between pyomelanin production and deletion of a chromosomal region containing mexXY, nonpyomelanogenic populations (parR87, replicate 1; and nuoD184, all replicates) were not tobramycin hypersusceptible (Fig. 1B and table S2). Further analysis of the results from the experimental evolution study has revealed that six of eight genetic backgrounds presented a significantly (P < 0.01 in all cases) reduced MIC to tobramycin, compared to their parental strains, after the ceftazidime short-term evolution. To further analyze whether chromosomal deletions containing mexXY could be associated with the phenotype of hypersusceptibility, every evolved population, as well as their original parental strain, was genotyped (fig. S1A). A 163-bp polymerase chain reaction (PCR) fragment corresponding to mexXY was detected in every parental strain, as well as in nonpyomelanogenic populations (parR87, replicate 1; and nuoD184, all replicates) and in a pyomelanogenic mixed population (PA14, replicate 2). Consistent with the observed increase in tobramycin susceptibility of most evolved populations (table S2), every pyomelanogenic tobramycin-hypersusceptible population lacked mexXY (fig. S1A). Overall, these results suggest that chromosomal deletions containing mexXY are consistently selected at first steps of ceftazidime resistance evolution in P. aeruginosa, at least in the genetic backgrounds analyzed. To further test whether this observation could be conditioned by the number of replicates used during the assay for each genetic background, 32 replicates of one of the mutants (mexZ43) were subjected to evolution under the same conditions used before, in presence or absence of ceftazidime. We observed that upon 1 day of experimental evolution, a high number of mexZ43 populations challenged with antibiotic hyperproduced pyomelanin (28 of 32 populations; Fig. 1D), just 4 of 32 populations did not become pyomelanogenic (Fig. 1D). Although we are aware that there is a space for unpredictability of ceftazidime resistance evolution in the mutants analyzed, the results here described suggest that alternative genetic evolutionary trajectories in ceftazidime resistance evolution may be limited, and that phenotypic convergence toward collateral sensitivity to tobramycin is robust in P. aeruginosa, even in the case of antibiotic-resistant mutants, as those here analyzed (table S1), after ceftazidime treatment (table S2). However, we are fully aware that all mutants described here derive from P. aeruginosa PA14, and the generalization of our results to other strains will require the analysis of the effect of short-term ceftazidime evolution on a broad and diverse set of clinical strains of P. aeruginosa.
Fig. 1. Analysis of early steps in the evolution of P. aeruginosa wild-type strain and antibiotic-resistant mutants in the presence of CAZ.
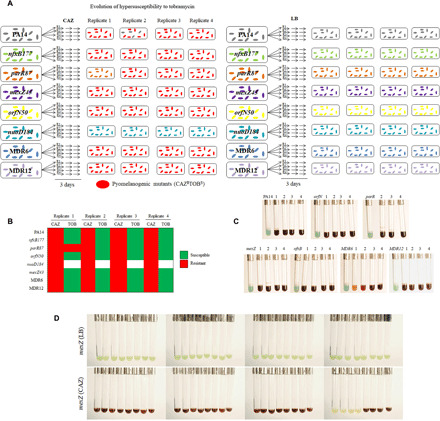
(A) Scheme of the resultant phenotype after the evolution of PA14, single (nfxB177, parR87, mexZ43, orfN50, and nuoD184) and multiple (MDR6 and MDR12) mutants, in the presence of CAZ [left part of (A)] or in the absence of antibiotic [LB; right part of (A)] for 3 days (see Materials and Methods). Pyomelanin hyperproduction was observed in 27 of 32 populations evolved in the presence of CAZ (red-colored cells). (B) Diagram showing convergence toward hypersusceptibility to TOB in the different genetic backgrounds and replicates, analyzed after short-term evolution on CAZ. In all cases, acquisition of a pyomelanogenic phenotype is associated with collateral sensitivity to TOB, irrespective of the genetic background of the evolving strain. MIC values of TOB and CAZ are included in table S2. (C) Isolation of pyomelanogenic clones from each 27 pyomelanogenic population [left part of (A); red-colored cells] obtained in the presence of CAZ. As shown in the figure, the early steps of evolution in presence of CAZ of P. aeruginosa, which lead to collateral sensitivity to TOB and pyomelanin production, are conserved among the different antibiotic-resistant mutants analyzed. (D) Pyomelanogenic phenotype of mexZ43 mutant after 1-day evolution in the presence of CAZ or in the absence of antibiotic (LB). Pyomelanin hyperproduction was observed in 28 of 32 populations evolved on CAZ. Photo credits for (C) and (D): Inés Poveda, Centro Nacional de Biotecnología. Permission for using these images is not required.
Evolution of heterogeneous pyomelanogenic populations of P. aeruginosa subjected to tobramycin/ceftazidime sequential evolution
We have shown that ceftazidime selects, in the short term, pyomelanogenic mutants presenting collateral sensitivity to tobramycin in P. aeruginosa, at least in the set of resistant mutants analyzed here (table S1). Nevertheless, there is an extensive heterogeneity within populations of P. aeruginosa in the CF lungs (38), frequently including mutants already presenting a pyomelanogenic phenotype (29, 30), which is usually associated with a reduced susceptibility to ceftazidime. Since this heterogeneity might impair the chances of exploiting the observed tobramycin collateral sensitivity associated with the use of ceftazidime in these heterogeneous populations, we tested the possibility of alternating the antibiotics, first, by using tobramycin and then second, ceftazidime, for reducing the chances of pyomelanogenic heterogeneous populations to escape from the antibiotic challenge.
The strategy would consist of three stages: A first step on tobramycin may force extinction of the ceftazidime-resistant (tobramycin-hypersusceptible) part of the populations, a second step on ceftazidime may drive evolution toward tobramycin hypersusceptibility, and then the extinction of the tobramycin-hypersusceptible cells after switching back to tobramycin (Fig. 2) would be expected. Although this strategy would be also potentially applicable to populations not resistant to ceftazidime, being reduced to a first step on ceftazidime followed by a second one on tobramycin, we decided to analyze its effectiveness in a more complex situation of clinical relevance, as it is the case of heterogeneous pyomelanogenic populations. In particular, diverse populations derived from the set of mutants were analyzed in this work (see table S1). To that end, we isolated a pyomelanogenic clone from each of the 27 pyomelanogenic populations obtained after ceftazidime short-term evolution (Fig. 1A). To note here that although the original clones from which these populations were derived belonged to a diverse set of genetic backgrounds, consisting of antibiotic-resistant mutants that contain mutations in regulatory, nonregulatory, or both types of proteins (Table 1 and table S1), all the isolated clones that hyperproduced pyomelanin (Fig. 1C) were significantly (P < 0.001 in all cases) more susceptible to tobramycin than their respective parental strain (table S3), and lacked mexXY (fig. S1B). Then, we recreated a total of 27 heterogeneous pyomelanogenic populations by mixing each of the 27 pyomelanogenic clones with its respective parental strain in a 1:1 ratio. The heterogeneous populations were dubbed PA14 +1 to +4, nfxB177 +1 to +4, parR87 +2 to +4, mexZ43 +1 to +4, orfN50 +1 to +4, MDR6 +1 to +4, and MDR12 +1 to +4. These populations were first subjected to tobramycin short-term evolution for 3 days (see Materials and Methods), and the capacity of these populations to either evolve toward tobramycin resistance or to go extinct was analyzed (Fig. 3). Since pyomelanogenic clones are less susceptible to ceftazidime (i.e., >256 μg/ml in all the parR87 clones; see “b” data in table S4) than the parental strain from which they evolved (1 μg/ml in parR87; see “a” data in table S4), ceftazidime MIC values were used to verify the extinction of the pyomelanogenic part of each population. After 3 days of evolution in the presence of tobramycin, the ceftazidime MICs for every population (i.e., 1 μg/ml in parR87 heterogeneous populations; see the “First step (TOB)” data in table S4) were close to the ceftazidime MIC for the parental strain [compare First Step (TOB) to the “a and b” MIC data in table S4]. These data support that the pyomelanogenic part of every population is extinct after the first step of sequential evolution. To further confirm the extinction of the pyomelanogenic clones, the phenotype (green/yellow versus brown color) of 20 clones from each of the 27 populations, a total of 540 clones, were isolated and grown in liquid medium (brown color is poorly appreciated in colonies) to detect any escape of ceftazidime-resistant cells from tobramycin treatment. As shown in Fig. 3B, none of the clones produced pyomelanin, confirming the extinction of the pyomelanogenic part of every population, a fact that was previously hinted by the ceftazidime MIC values of the resultant populations (see table S4). Besides that, each resultant population presented an increased tobramycin MIC, up to 48-fold, depending on the genetic background and replicate (table S4).
Fig. 2. General model illustrating evolution of heterogeneous pyomelanogenic populations of P. aeruginosa subjected to TOB/CAZ sequential evolution.
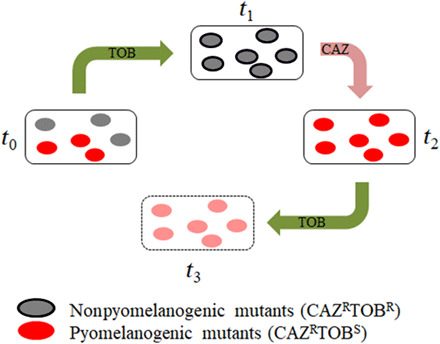
Evolution of a heterogeneous population containing a pyomelanogenic (CAZ resistant) subpopulation starts when TOB is added at time zero (t0). In the presence of TOB, there is an extinction of TOB-hypersusceptible pyomelanogenic mutants (red-colored cells) and TOB becomes ineffective (t1). Then, treatment is switched to CAZ, and TOB-resistant cells (contoured gray-colored cells) become TOB-hypersusceptible (t2). Treatment would be switched back to TOB, resulting in the elimination of TOB-hypersusceptible cells (t3). This strategy would also be potentially applicable to initial populations not resistant to CAZ (t1), being reduced to a first step on CAZ (leading to t2), followed by a second step on TOB (resulting in t3).
Fig. 3. Analysis of TOB/CAZ sequential evolution of heterogeneous pyomelanogenic populations of P. aeruginosa.

(A) Diagram showing the evolution of heterogeneous populations (dubbed +1, +2, +3, and +4) containing each parental strain: PA14, nfxB177, parR87, mexZ43, orfN50, MDR6, or MDR12 and four individual pyomelanogenic clones belonging to the same genetic background, during first step of sequential evolution in the presence of TOB (left) and second step in the presence of CAZ (right), for 6 days (see Materials and Methods). In the case of parR87, only three pyomelanogenic clones from independent CAZ-evolved populations (see Fig. 1, A and C) could be isolated. Hypersusceptibility to TOB (contoured and noncontoured red-colored cells) is observed in 23 of 27 populations (see table S4), and pyomelanin production (noncontoured red-colored cells) is observed in 17 of 27 populations. (B) Analysis of extinction of the pyomelanogenic part of the heterogeneous pyomelanogenic populations after a first step of sequential evolution in the presence of TOB [(A) section, left]. The phenotype (color) of 20 clones isolated from each heterogeneous population, after 3 days of TOB evolution, was observed in liquid medium and compared with the color (brown) of each pyomelanogenic parental strain (upper left corner of each plate). In agreement with data shown in table S4, which points to the extinction of pyomelanogenic populations by comparison of CAZ MIC value of each heterogeneous population with the ones of their parental strains and pyomelanogenic clones, the color of the 540 clones analyzed indicated that pyomelanogenic clones were extinct after first step of sequential evolution on TOB. Photo credits for (B): Fernando Sanz-García, Centro Nacional de Biotecnología.
At this point, we specifically focused on the switch from tobramycin to ceftazidime, the second step of the sequential evolution (Fig. 2). Although we had observed conservation of tobramycin collateral sensitivity after evolution in presence of ceftazidime within the analyzed set of mutants of P. aeruginosa (Fig. 1 and table S2), a critical point would be to determine whether this genetic event would also be selected by ceftazidime in the tobramycin-resistant mutants obtained after the first step of evolution of the populations in the presence of tobramycin. Hence, we switched the selective pressure from tobramycin to ceftazidime (see Materials and Methods). As shown in Fig. 3, 17 of 27 populations hyperproduced pyomelanin. A second critical point emerged here: The degree of sensitization to tobramycin of highly resistant mutants to the said antibiotic was uncertain. To determine whether these populations presented an increased susceptibility to tobramycin, as it was observed in the wild-type PA14 background and in most of mutants analyzed in this work (Fig. 1 and table S2), the tobramycin MIC was determined in all the evolved populations. Twenty-three of 27 populations presented an important increase in their sensitivity to tobramycin after switching the selective pressure from tobramycin to ceftazidime, reducing the MIC by up to 128-fold in PA14, 48-fold in nfxB177, 21-fold in parR87, 96-fold in orfN50, 43-fold in mexZ43, 4-fold in MDR6, and 11-fold in MDR12 (Fig. 4 and table S4). This analysis revealed that five of seven heterogeneous pyomelanogenic populations presented a substantially reduced MIC to tobramycin after the second step of sequential evolution on ceftazidime (table S4). These results suggest that it could be possible to exploit the tobramycin collateral sensitivity associated with the use of ceftazidime by switching back selective pressure to tobramycin, although there may be some limitations depending on the genetic background. This was the case of the multiple resistant mutants MDR6 and MDR12, which did not present a relevant reduction in tobramycin MIC after the switch to ceftazidime.
Fig. 4. MICs (μg/ml) of TOB for heterogeneous pyomelanogenic populations of P. aeruginosa after TOB/CAZ sequential evolution.
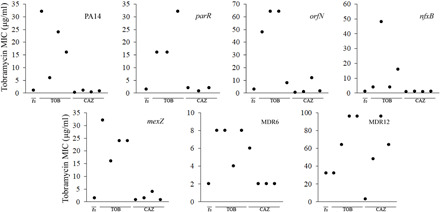
Evolution of TOB MICs (μg/ml) of heterogeneous pyomelanogenic populations after sequential evolution on TOB/CAZ (see Fig. 3). Each plot shows the TOB MIC values for a parental strain (PA14, parR87, orfN50, nfxB177, mexZ43, MDR6, or MDR12), indicated as t0 in the x axis, and for four heterogeneous pyomelanogenic populations (represented as black circles) after first evolution on TOB (indicated as TOB in the x axis), followed by second evolution on CAZ (indicated as CAZ in the x axis). Only three heterogeneous pyomelanogenic populations of parR87 were analyzed. TOB MICs decreased after switching from TOB to CAZ by up to 128-fold in PA14, 48-fold in nfxB177, 21-fold in parR87, 96-fold in orfN50, 43-fold in mexZ43, 4-fold in MDR6, and 11-fold in MDR12. MIC values are shown in table S4.
The fact that some populations were hypersusceptible to tobramycin, even without having suffered chromosomal deletions containing mexXY (6 of 27 populations; Fig. 3 and fig. S1C) or being mixed populations (4 of 27 populations; Fig. 3 and fig. S1C), indicates that reciprocal collateral sensitivity between ceftazidime and tobramycin may occur even in the absence of these deletions, a feature that remains to be explored in detail. Overall, our results indicate that this strategy could potentially be applicable from complex situations similar to the ones explored in the current work (heterogeneous pyomelanogenic ceftazidime-resistant populations) to other ones (populations not resistant to ceftazidime).
Cross-resistance and collateral sensitivity patterns of P. aeruginosa after tobramycin/ceftazidime sequential evolution
We have recently described that P. aeruginosa replicate populations subjected to ribosome-targeting antibiotics may present common changes in the susceptibility to other antibiotics, besides those used along selection (16, 36). However, important differences have been described in the collateral sensitivity phenotype among replicate populations of the same P. aeruginosa strain adapted to one antibiotic (13). We have recently reported that a loss-of-function mutant of P. aeruginosa PA14, differing from its parental strain in the activity of just one regulator, not directly linked to AR, presents different patterns of collateral sensitivity and cross-resistance phenotype when it acquires resistance to ribosome-targeting antibiotics (16). Since historical contingency may restrict the evolution of collateral sensitivity and cross-resistance phenotypic outcomes, we wondered whether the populations obtained after tobramycin and ceftazidime sequential evolution, besides presenting a convergent hypersusceptibility to tobramycin, may converge toward the phenotypes of cross-resistance or collateral sensitivity to antibiotics found in other structural families. To address this question, the MICs of a set of antibiotics were determined for the 27 populations. A general pattern of cross-resistance to aztreonam, imipenem, and chloramphenicol, as well as significant collateral sensitivity to fosfomycin, tobramycin, and tetracycline, was observed (P < 0.0001 in all cases) (Fig. 5 and table S5). Since the combination fosfomycin-tobramycin has been found to be synergistic against biofilms of CF P. aeruginosa strains (39) and against P. aeruginosa PAO1 in anaerobic environments (40), we propose that the switch back to tobramycin (Fig. 2) could also be replaced, if necessary, by the combination fosfomycin-tobramycin. To analyze the relative efficacy of the two possible options, we subjected the resultant populations obtained after tobramycin/ceftazidime sequential evolution to ALE in either tobramycin or the combination fosfomycin-tobramycin, applying twice the MIC of each parental strain. We observed that 8 of 27 populations were able to escape from the switch back to tobramycin (orfN50 +3, mexZ43 +2 and +3, MDR6 +1 and +3, and MDR12 +2 to +4). This result agrees with the fact that although most of the populations (23 of 27) presented an important reduction in the tobramycin MIC after sequential evolution on tobramycin and ceftazidime, by up to 128-fold in PA14, 48-fold in nfxB177, 21-fold in parR87, 96-fold in orfN50, 43-fold in mexZ43, 4-fold in MDR6, and 11-fold in MDR12 (Fig. 4), tobramycin MIC values were close to those of parental strains. None of the populations survived the combination fosfomycin-tobramycin, possibly due to the synergistic effect of these antibiotics (39). We therefore propose that use of the fosfomycin-tobramycin combination is more effective than switching back to tobramycin after that treatment.
Fig. 5. Diagram showing the degree of convergence of cross-resistance and collateral sensitivity in heterogeneous pyomelanogenic populations of P. aeruginosa obtained after TOB/CAZ sequential evolution.
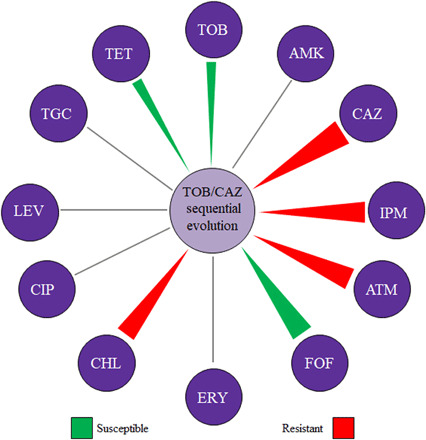
Collateral sensitivity and cross-resistance to antibiotics from different structural families were analyzed in the 27 populations obtained after sequential evolution. A population is classified as “susceptible” or “resistant” when there was an MIC change with respect to the parental strain value. Triangles indicate antibiotics where a predominant change toward resistance (red) or susceptibility (green) with respect to the parental strain was observed. Thickness of the triangle depends on the percentage of conservation of said phenotype. MIC values (μg/ml) are included in table S5. AMK, amikacin; ATM, aztreonam; FOF, fosfomycin; ERY, erythromycin; CHL, chloramphenicol; LEV, levofloxacin; TET, tetracycline.
DISCUSSION
Bacterial evolution is known to be one of the main causes of the current AR problem; but in-depth analysis of this evolution could also help to tackle this issue through the exploitation of the evolutionary trade-offs (as collateral sensitivity) associated with AR acquisition (2, 4). However, the feasibility of this approach requires the collateral sensitivity phenotypes of different resistant mutants to be robust and reproducible (41). In this study, we describe the robustness of collateral sensitivity to tobramycin associated with the short-term use of ceftazidime in an array of P. aeruginosa antibiotic-resistant mutants, chosen on the basis of their differences both in resistance phenotype and in the functions affected by the mutations that they harbor. We propose that the observed evolutionary trade-offs could be exploited for treating both clonal and heterogeneous pyomelanogenic infections. Patients with CF are usually infected by heterogeneous P. aeruginosa populations (38) that include pyomelanogenic mutants (29, 30), which frequently present resistance to β-lactams, a feature that could compromise the use of ceftazidime. However, we have found that it is possible to drive the extinction of the pyomelanogenic mutants, first, by using tobramycin and then second, by driving the evolution of the remaining population toward tobramycin hypersusceptibility, using ceftazidime. A bottleneck for the application of this strategy would be the durability over time of tobramycin hypersusceptibility. However, it is important to highlight that the populations obtained after tobramycin/ceftazidime alternation present also collateral sensitivity to fosfomycin. We observed that it is possible to replace the switch back to tobramycin by a fosfomycin-tobramycin combination, which results in higher efficacy. These results point to the possibility of exploiting specific evolutionary trade-offs for tackling the problem of AR. However, we are aware that a detailed analysis determining the degree of conservation of the short-term ceftazidime resistance evolution in a broad and diverse set of clinical strains of P. aeruginosa would be required. Overall, our results and those of others, previously described (41) suggest that the analysis of phenotypic convergence and, in particular, the aspects that deal with the collateral sensitivity of P. aeruginosa AR mutants, is an important step forward in the rational design of therapeutic approaches capable of reducing the AR burden.
MATERIALS AND METHODS
Growth conditions and antibiotic susceptibility assays
Bacteria were grown in LB at 37°C, with shaking at 250 rpm in glass tubes. MICs of ceftazidime, aztreonam, imipenem, tobramycin, amikacin, tigecycline, tetracycline, ciprofloxacin, levofloxacin, chloramphenicol, fosfomycin, and erythromycin were determined at 37°C in Mueller-Hinton (MH) agar, using E-test strips (MIC Test Strip, Liofilchem).
Mutant construction
Four single mutants of P. aeruginosa (nfxB177, parR87, mexZ43, and nuoD184) were constructed by inserting each mutant allele (table S1) by homologous recombination into the wild-type PA14, while the orfN50 single mutant was previously obtained (16). Mutant alleles were obtained by PCR from previous in-house evolved populations (16, 36), leaving approximately 500 bp upstream and downstream the corresponding single-nucleotide polymorphism, using the oligonucleotides described in table S6. PCR products containing Hind III restriction sites were cloned into the Hind III–digested and dephosphorylated pEX18Ap vector (42) and then introduced by transformation into the conjugative Escherichia coli S17-1 strain. Subsequently, conjugation and mutant selection were performed, as described elsewhere (42), using carbenicillin (350 μg/ml) and 10% sucrose. In all cases, the presence of the mutations was confirmed by Sanger sequencing. To obtain mutants containing multiple mutations, 10 independent resistant clones from end point evolved populations (16) on tobramycin or tigecycline were selected and mutations confirmed by Sanger sequencing. From them, two multiple mutants (table S1), tobramycin or tigecycline resistant, respectively, were chosen. The mutant allele of lasR was replaced by homologous recombination with that of the wild type in the two selected clones, using the above-described strategy and oligonucleotides encompassed in table S6.
Short-term ALE in presence of ceftazidime
Five single mutants, two multiple mutants, and PA14, four replicates of each, were subjected to short-term ALE in presence or absence of ceftazidime, resulting in a total of 64 independent bacterial populations (32 populations grown in presence of ceftazidime and 32 control populations grown without antibiotic). Cultures were grown at 37°C and 250 rpm for 3 days. Every day, the cultures were diluted (1/125), adding 8 μl of bacteria in 1 ml of fresh LB, either containing or lacking ceftazidime at the concentration that hinders the growth of each P. aeruginosa genetic background under these culture conditions (4 μg/ml for PA14, mexZ43, and MDR12; 5 μg/ml for nfxB177, orfN50, and MDR6; 3 μg/ml for parR87; and 2 μg/ml for nuoD184). During the 3 days, the concentration of ceftazidime was maintained. Every replicate population was preserved at −80°C at the end of the experimental evolution. In addition, the MIC of the antibiotic used for selection in populations (ceftazidime) and of the one to which ceftazidime evolution gives rise to collateral sensitivity (tobramycin), was determined at 37°C in MH agar using E-test strips.
Sequential tobramycin/ceftazidime experimental evolution
Pyomelanogenic clones were isolated from every individual pyomelanogenic replicate population of each genetic background previously submitted to short-term evolution in the presence of ceftazidime, resulting in a total of 27 pyomelanogenic clones (see above). Overnight bacterial cultures from each pyomelanogenic clone and its parental strain were normalized to an optical density at 600 nm of 4.0 and then mixed in a 1:1 (pyomelanogenic clone:parental strain) ratio, obtaining 27 heterogeneous populations. Cultures were grown at 37°C and 250 rpm for 6 days. Every day, during the first 3 days, the cultures were diluted (1/125) in fresh LB containing the tobramycin concentration that hinders the growth of each P. aeruginosa genetic background under these culture conditions (1 μg/ml for PA14; 1.5 μg/ml for nfxB177, parR87, mexZ43, and MDR6; 4 μg/ml for orfN50; and 12 μg/ml for MDR12). During the 3 days, the concentration of tobramycin was maintained. At the end of the first step of sequential experimental evolution, every replicate population was preserved at −80°C, and the MIC of ceftazidime and tobramycin was determined at 37°C in MH agar using E-test strips. The 27 populations were grown, from glycerol stocks, and every day, during the last 3 days, the cultures were diluted (1/125) in fresh LB containing ceftazidime, as described in the above-mentioned section of Material and Methods (see the “Short-term ALE in presence of ceftazidime” section). Every final population was preserved at −80°C at the end of the second step of sequential experimental evolution, and the MIC of tobramycin was determined at 37°C in MH agar using E-test strips.
Analysis of the presence/absence of mexXY in the evolved populations
The presence of chromosomal deletions including mexXY in the different genetic backgrounds and their respective evolved populations was analyzed by determining the absence of a 163-bp PCR fragment belonging to mexXY in 2% agarose gel. Primers used for mexXY genotyping are included in table S6.
Statistical analysis
Data were subjected to pre hoc and post hoc analyses to identify relevant differences, using either analysis of variance (ANOVA), Friedman’s, or χ2 tests and Dunnett’s or Fisher’s exact test with Hochberg correction, as implemented in R.
Supplementary Material
Acknowledgments
We thank J. Ramón Valverde, from the Servicio de Computación Científica del CNB, for the helpful discussion and the assistance with the manuscript, and I. Poveda, from the Servicio de Fotografía del CNB, for the photograph support. Funding: This work was supported by the Instituto de Salud Carlos III (grant number RD16/0016/0011), cofinanced by the European Development Regional Fund “A Way to Achieve Europe,” by grant S2017/BMD-3691 InGEMICS-CM, funded by Comunidad de Madrid (Spain) and European Structural and Investment Funds and by the Spanish Ministry of Economy and Competitivity (BIO2017-83128-R). F.S.-G. is the recipient of a FPU fellowship from MINECO. Author contributions: S.H.-A. participated in the design of the study and performed experimental work, and F.S.-G. performed experimental work. J.L.M. participated in the design of the study. All authors participated in writing the manuscript and approved the submitted version. Competing interests: All authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eaba5493/DC1
REFERENCES AND NOTES
- 1.Laxminarayan R., Antibiotic effectiveness: Balancing conservation against innovation. Science 345, 1299–1301 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Szybalski W., Bryson V., Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 64, 489–499 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baym M., Stone L. K., Kishony R., Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pál C., Papp B., Lázár V., Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 23, 401–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munck C., Gumpert H. K., Wallin A. I. N., Wang H. H., Sommer M. O. A., Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 6, 262ra156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa C., Beardmore R., Schulenburg H., Jansen G., Antibiotic combination efficacy (ACE) networks for a Pseudomonas aeruginosa model. PLOS Biol. 16, e2004356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imamovic L., Sommer M. O., Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 5, 204ra132 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Kim S., Lieberman T. D., Kishony R., Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 111, 14494–14499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamovic L., Ellabaan M. M. H., Dantas Machado A. M., Citterio L., Wulff T., Molin S., Krogh Johansen H., Sommer M. O. A., Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 172, 121–134.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knopp M., Andersson D. I., Predictable phenotypes of antibiotic resistance mutations. mBio 9, e00770-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apjok G., Boross G., Nyerges Á., Fekete G., Lázár V., Papp B., Pál C., Csörgő B., Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Mol. Biol. Evol. 36, 1601–1611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podnecky N. L., Fredheim E. G. A., Kloos J., Sørum V., Primicerio R., Roberts A. P., Rozen D. E., Samuelsen Ø., Johnsen P. J., Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat. Commun. 9, 3673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbosa C., Trebosc V., Kemmer C., Rosenstiel P., Beardmore R., Schulenburg H., Jansen G., Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 34, 2229–2244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lázár V., Pal Singh G., Spohn R., Nagy I., Horváth B., Hrtyan M., Busa-Fekete R., Bogos B., Méhi O., Csörgő B., Pósfai G., Fekete G., Szappanos B., Kégl B., Papp B., Pál C., Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 9, 700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lázár V., Nagy I., Spohn R., Csörgő B., Györkei A., Nyerges Á., Horváth B., Vörös A., Busa-Fekete R., Hrtyan M., Bogos B., Méhi O., Fekete G., Szappanos B., Kégl B., Papp B., Pál C., Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 5, 4352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernando-Amado S., Sanz-García F., Martínez J. L., Antibiotic resistance evolution is contingent on the quorum-sensing response in Pseudomonas aeruginosa. Mol. Biol. Evol. 36, 2238–2251 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Vogwill T., Kojadinovic M., MacLean R. C., Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc. Biol. Sci. 283, 20160151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trindade S., Sousa A., Xavier K. B., Dionisio F., Ferreira M. G., Gordo I., Positive epistasis drives the acquisition of multidrug resistance. PLOS Genet. 5, e1000578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward H., Perron G. G., Maclean R. C., The cost of multiple drug resistance in Pseudomonas aeruginosa. J. Evol. Biol. 22, 997–1003 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Salverda M. L., Dellus E., Gorter F. A., Debets A. J., van der Oost J., Hoekstra R. F., Tawfik D. S., de Visser J. A., Initial mutations direct alternative pathways of protein evolution. PLOS Genet. 7, e1001321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kryazhimskiy S., Rice D. P., Jerison E. R., Desai M. M., Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344, 1519–1522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jochumsen N., Marvig R. L., Damkiær S., Jensen R. L., Paulander W., Molin S., Jelsbak L., Folkesson A., The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 7, 13002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novais A., Comas I., Baquero F., Cantón R., Coque T. M., Moya A., González-Candelas F., Galán J.-C., Evolutionary trajectories of beta-lactamase CTX-M-1 cluster enzymes: Predicting antibiotic resistance. PLOS Pathog. 6, e1000735 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichol D., Rutter J., Bryant C., Hujer A. M., Lek S., Adams M. D., Jeavons P., Anderson A. R. A., Bonomo R. A., Scott J. G., Antibiotic collateral sensitivity is contingent on the repeatability of evolution. Nat. Commun. 10, 334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz-García F., Hernando-Amado S., Martínez J. L., Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime-avibactam. Antimicrob. Agents Chemother. 62, e01379-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Rojas A., Mena A., Martín S., Borrell N., Oliver A., Blázquez J., Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology 155, 1050–1057 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R. E., Martínez J. L., Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to β-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4159–4167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda N., Sakagawa E., Ohya S., Gotoh N., Tsujimoto H., Nishino T., Contribution of the MexX-MexY-oprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 2242–2246 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer-Hamblett N., Rosenfeld M., Gibson R. L., Ramsey B. W., Kulasekara H. D., Retsch-Bogart G. Z., Morgan W., Wolter D. J., Pope C. E., Houston L. S., Kulasekara B. R., Khan U., Burns J. L., Miller S. I., Hoffman L. R., Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am. J. Respir. Crit. Care Med. 190, 289–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hocquet D., Petitjean M., Rohmer L., Valot B., Kulasekara H. D., Bedel E., Bertrand X., Plésiat P., Köhler T., Pantel A., Jacobs M. A., Hoffman L. R., Miller S. I., Pyomelanin-producing Pseudomonas aeruginosa selected during chronic infections have a large chromosomal deletion which confers resistance to pyocins. Environ. Microbiol. 18, 3482–3493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabot G., Florit-Mendoza L., Sánchez-Diener I., Zamorano L., Oliver A., Deciphering β-lactamase-independent β-lactam resistance evolution trajectories in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 73, 3322–3331 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Yen P., Papin J. A., History of antibiotic adaptation influences microbial evolutionary dynamics during subsequent treatment. PLOS Biol. 15, e2001586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabot G., Zamorano L., Moyà B., Juan C., Navas A., Blázquez J., Oliver A., Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob. Agents Chemother. 60, 1767–1778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wardell S. J. T., Rehman A., Martin L. W., Winstanley C., Patrick W. M., Lamont I. L., A large-scale whole-genome comparison shows that experimental evolution in response to antibiotics predicts changes in naturally evolved clinical Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63, e0169-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rau M. H., Marvig R. L., Ehrlich G. D., Molin S., Jelsbak L., Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ. Microbiol. 14, 2200–2211 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Sanz-García F., Hernando-Amado S., Martínez J. L., Mutational evolution of Pseudomonas aeruginosa resistance to ribosome-targeting antibiotics. Front. Genet. 9, 451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheer S. M., Waugh J., Noble S., Inhaled tobramycin (TOBI®): A review of its use in the management of Pseudomonas aeruginosa infections in patients with cystic fibrosis. Drugs 63, 2501–2520 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Workentine M. L., Sibley C. D., Glezerson B., Purighalla S., Norgaard-Gron J. C., Parkins M. D., Rabin H. R., Surette M. G., Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLOS ONE 8, e60225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Díez-Aguilar M., Morosini M. I., Köksal E., Oliver A., Ekkelenkamp M., Cantón R., Use of calgary and microfluidic bioflux systems to test the activity of fosfomycin and tobramycin alone and in combination against cystic fibrosis Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 62, e01650-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCaughey G., Gilpin D. F., Schneiders T., Hoffman L. R., McKevitt M., Elborn J. S., Tunney M. M., Fosfomycin and tobramycin in combination downregulate nitrate reductase genes narG and narH, resulting in increased activity against Pseudomonas aeruginosa under anaerobic conditions. Antimicrob. Agents Chemother. 57, 5406–5414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbosa C., Römhild R., Rosenstiel P., Schulenburg H., Evolutionary stability of collateral sensitivity to antibiotics in the model pathogen Pseudomonas aeruginosa. eLife 8, e51481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P., A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/32/eaba5493/DC1


