Graphical abstract
Keywords: COVID-19, Clinical sensitivity, In vitro diagnostics, Real-time RT-PCR, Point-of-care, SARS-CoV-2
Abstract
Background
To curb the spread of the COVID-19 (coronavirus disease 2019) pandemic, the world needs diagnostic systems capable of rapid detection and quantification of the novel coronavirus (SARS-CoV-2). Many biomedical companies are rising to the challenge and developing COVID-19 diagnostics. In the last few months, some of these diagnostics have become commercially available for healthcare workers and clinical laboratories. However, the diagnostic technologies have specific limitations and reported several false-positive and false-negative cases, especially during the early stages of infection.
Aim
This article aims to review recent developments in the field of COVID-19 diagnostics based on molecular technologies and analyze their clinical performance data.
Key Concepts
The literature survey and performance-based analysis of the commercial and pre-commercial molecular diagnostics address several questions and issues related to the limitations of current technologies and highlight future research and development challenges to enable timely, rapid, low-cost, and accurate diagnosis of emerging infectious diseases.
Introduction
The last decade has witnessed the rise of epidemics and pandemics such as 2012s coronavirus disease (MERS, middle-eastern respiratory syndrome) in the Middle East [1], 2014s Ebola virus disease (EVD, formerly known as Ebola hemorrhagic fever) in West Africa [2], 2015s zika virus disease in Latin America [3], and 2019s coronavirus disease (COVID-19) in Wuhan, China [4]. These emerging infectious diseases pose a grave threat to human health and the global economy. Lately, the novel coronavirus (SARS-CoV-2, severe acute respiratory syndrome coronavirus 2) aggressively spread throughout the world causing the COVID-19 pandemic [5]. As of 29th July 2020, 185 of 197 UN countries are affected by this pandemic and despite all containment efforts, the number of COVID-19 infected people is rising above 16.5 million with over 655 thousand deaths accounting for the global fatality rate of ~ 3.96% [6], [7]. For COVID-19, the mean incubation period and the mean serial interval are 5.2 days (95% CI: 4.1–7.0 days) and 7.5 days (95% CI: 5.3–19.0 days), respectively; while the basic reproduction number (R0) is reported to be 2.2 (95% CI: 1.4–3.9), i.e. a SARS-CoV-2 carrier can spread it to ~ 2.2 persons on average [8]. Therefore, to limit the spread of SARS-CoV-2 and overcome the COVID-19 crises it is important to identify the suspected individuals and isolate them, which requires indefatigable diagnostic testing.
The dependable diagnostic solutions have been immediately developed and marketed to help early diagnosis of COVID-19 [9], [10], [11]. Thus far, two types of diagnostic tests have been commercialized: one, molecular diagnostics that detect part of the viral genome (i.e. RNA) in the respiratory tract specimens; and second, serological or antibody tests that detect SARS-CoV-2 specific antibodies in serum specimens [10]. Molecular diagnostics are used to identify symptomatic or asymptomatic SARS-CoV-2 carriers, and the basic performance criterion for these tests is high clinical sensitivity to avoid false-negative results. On the other hand, the objective of serological tests is to identify individuals with an active immune response to the SARS-CoV-2 antigen, and the basic performance criterion for these tests is high clinical specificity to rule out false-positive results. Sethuraman et al. [12] published a timeline for the detection of viral RNA (molecular diagnosis) and immune-response antibodies (serological diagnosis), which revealed that molecular diagnostics could identify infected individuals a week before the onset of symptoms while antibodies could only be detected ≥ 8 days after the symptoms appear. Therefore, molecular diagnostics are essentially the only tests for early diagnosis of COVID-19 [13].
This article aims to present a critical performance analysis of commercially available molecular diagnostics and reviews major factors influencing their diagnostic performance. The criteria for the selection of molecular diagnostic tests is two-fold: (a) only those tests are selected that have been approved by major healthcare authorities around the world, and (b) most importantly, those molecular diagnostic kits are shortlisted that have been independently tested by WHO or relevant healthcare authorities for their clinical sensitivity and specificity.
At present, molecular techniques based on the real-time (quantitative) reverse transcriptase-polymerase chain reaction (RT-PCR) are considered the gold standard for COVID-19 diagnosis [14], [15]. Real-time RT-PCR detects amplified SARS-CoV-2 genome in sputum, nasopharyngeal or oropharyngeal swabs, bronchoalveolar lavage fluid, nasal or nasopharyngeal aspirate, and lower respiratory tract aspirates. A typical RT-PCR test can take 4–6 h from sample to result [10]. However, RT-PCR has proven to be extremely handy in clinical settings to consistently perform a large number of tests. As of 29th July 2020, >260 million COVID-19 tests have been performed in the most impacted countries worldwide [16]. Besides, other molecular diagnostic approaches, for instance, loop-mediated isothermal amplification (LAMP) [17], and clustered regularly interspaced short palindromic repeats (CRISPR) [18] are being developed for COVID-19 detection.
Herein, a performance-based review of the commercial and pre-commercial molecular diagnostics is presented. The clinical performance data reported by the manufacturers of commercially available tests are compared with the clinical evaluations performed by independent research labs and healthcare organizations, and the results are reviewed to define upcoming research and development challenges. Since the understanding of molecular diagnostics for COVID-19 is still evolving, their limitations in the current pandemic scenario are deliberated to ask forthcoming research questions that would improve the diagnostic technologies for COVID-19 and future emerging infectious diseases.
Molecular diagnostics
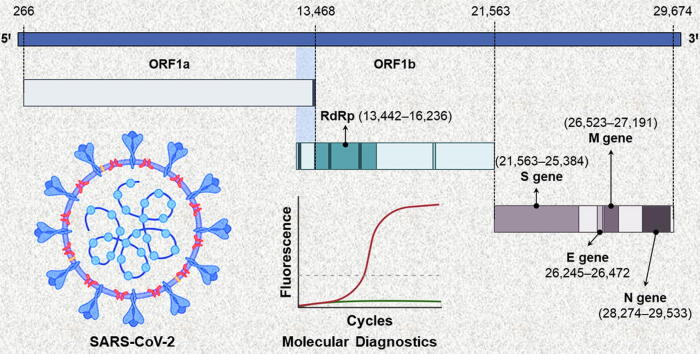
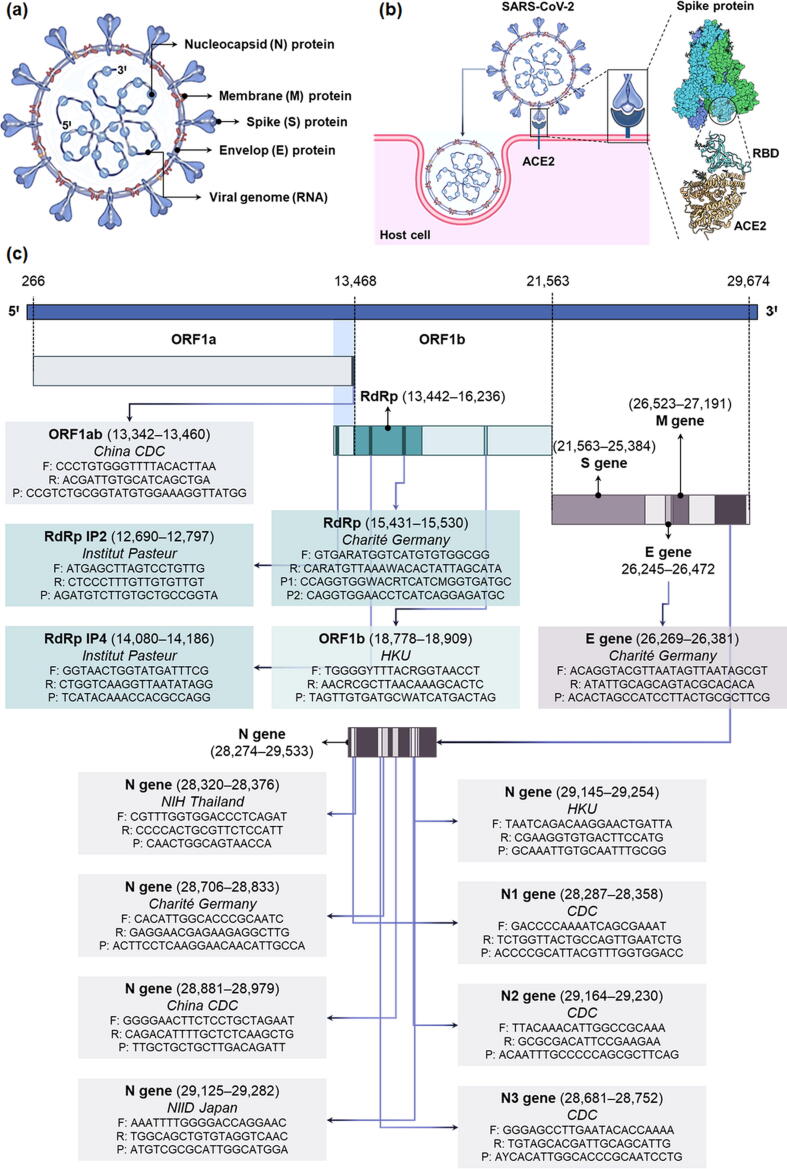
SARS-CoV-2 is a betacoronavirus (β-CoV) that is structurally similar to other coronaviruses (SARS-CoV and MERS-CoV) from the Coronaviridae family [19]. It has a positive-sense, single-stranded RNA (+ssRNA) enclosed in nucleocapsid (N) protein, while envelop (E), membrane (M), and spike (S) proteins form its outer shell or viral envelop (Fig. 1a). The trimeric spike glycoprotein is responsible for interaction with the host cells (Fig. 1b) [20], [21], [22]. Wrapp et al. [23] determined the structure of the spike protein by cryoelectron microscopy. In its predominant conformation, the spike protein exhibits strong binding with host cell’s angiotensin-converting enzyme 2 (ACE2) receptors through one of its receptor-binding domains (RBD) (Fig. 1b) [23], [24]. The viral genome (RNA) is ~ 30,000 nucleotides long comprising a structural gene unit that encodes S, E, M, and N proteins and two large, open reading frame genes (ORF1a and ORF1b) that encrypt sixteen non-structural proteins (NSP) including RNA-dependent RNA polymerase (RdRp) (Fig. 1c) [25], [26].
Fig. 1.
(a) The structure of SARS-CoV-2. (b) A schematic showing the interaction of SARS-CoV-2 with host cells and cellular entry mechanism. The virus first binds to angiotensin-converting enzyme 2 (ACE2) receptors on the host cell membrane through a receptor-binding domain (RBD) on the spike protein. Subsequently, it is endocytosed into the host cell. (c) The genomic constitution of SARS-CoV-2 RNA. The viral genome consists of two large genes: ORF1a, and ORF1b, which encode non-structural proteins (NSP) including RdRp, whereas the smaller structural genomic region hosts S, E, M, and N genes, which encode the structural proteins. CDC: Centers for Disease Control and Prevention; E gene: gene encoding envelop protein of SARS-CoV-2; HKU: Hong Kong University; M gene: gene encoding membrane protein of SARS-CoV-2; N gene: gene encoding nucleocapsid protein of SARS-CoV-2; NIH: National Institute of Health (Thailand); NIID: National Institute of Infectious Diseases (Japan); ORF1a/b: open reading frame 1a and b of SARS-CoV-2; RdRp: RNA-dependent RNA polymerase of SARS-CoV-2; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; S gene: gene encoding spike protein of SARS-CoV-2.
The findings of high-throughput genomic sequencing of SARS-CoV-2 are regularly deposited on the global initiative on sharing all influenza data (GISAID) [27]. The development of oligonucleotides (primers and probes) for molecular diagnosis of COVID-19 was initiated as soon as these findings were made public on 10th Jan 2020 [9], [27]. CDC (USA), China CDC, Charité Germany, Institut Pasteur (France), NIID Japan, Hong Kong University, and NIH Thailand researchers developed forward/reverse primers and probes for real-time RT-PCR-based molecular diagnostics (Fig. 1c). They also published comprehensive protocols for the detection of SARS-CoV-2, which can be found on the WHO’s website [28]. Thanks to these research and development efforts, the first test kits were available for clinical diagnosis of the disease by 4th Feb 2020 [29]. Since then, several molecular diagnostic kits have been commercialized for the detection of the SARS-CoV-2 genome. These diagnostics are approved for emergency use in the current pandemic situation by major healthcare authorities worldwide.
Real-time RT-PCR assays
Real-time RT-PCR is the major workhorse in the field of molecular diagnostics. It has been extensively used for high-throughput screening and early diagnosis of COVID-19, and other infectious diseases in the past. RT-PCR can amplify and detect a single copy of the specific genomic sequence and therefore, it is extremely sensitive [30]. Furthermore, real-time RT-PCT is a quantitative technique as the number of copies of RNA generated in a PCR increases exponentially and is proportional to the amount of starting material, i.e. viral load [31]. At the moment, a vast majority of the commercially available technologies (tests) for early diagnosis of COVID-19 are based on real-time RT-PCR assays [32]. Fig. 2 shows the principle of a real-time RT-PCR diagnostic. In principle, real-time RT-PCR is used to transcribe and amplify the specific SARS-CoV-2 genomic sequence(s). For this purpose, viral RNA is first extracted from the biological specimen collected from the nasal or nasopharyngeal swabs and is purified. Purified RNA template is converted into a cDNA (complementary DNA) by reverse transcriptase (an RNA-dependent DNA polymerase enzyme). The cDNA is subsequently amplified by the PCR (Fig. 2).
Fig. 2.
A molecular representation of the real-time RT-PCR principle. The template (viral RNA) is converted to cDNA (complementary DNA) by reverse transcriptase (RNA dependent DNA polymerase enzyme). Subsequently, cDNA is amplified in a polymerase chain reaction (PCR) in three steps: (1) denaturation of cDNA at 95 °C, (2) annealing of the primers and probe to the respective denatured cDNA strands at 60 °C, and (3) extension or synthesis of RNA copies by DNA polymerase at 72 °C. The amplified products follow the same cycle to generate a large number of RNA copies. TaqMan probe is used to quantify RNA copies by producing fluorescence signal during amplification cycles.
Depending upon the gene target, sequence-specific forward and reverse primers and a dual-labeled fluorogenic probe are designed and utilized in real-time RT-PCR diagnostics, as shown in Fig. 1c. The oligonucleotide probes used in RT-PCR assays are dual-labeled with a fluorophore (fluorescent reporter) and a quencher covalently attached to the 5′ and 3′ ends, respectively; and are often called TaqMan probes. Holland et al. [33] first demonstrated the application of thermostable Thermus aquaticus (Taq DNA polymerase) enzyme’s 5′–3′ exonuclease activity to cleave the fluorogenic probe during PCR and detect the amplified target-specific products. While the TaqMan probe is hybridized to the target sequence, the fluorescence emissions are quenched. However, when the probe is degraded by the DNA polymerase during the PCR, the release of fluorophore results in a fluorescence signal based on the fluorescence resonance energy transfer (FRET) principle [34], [35].
During amplification, the fluorescence signals are detected in real-time and the fluorescence emission data is plotted against the replication cycles (Fig. 2). A fluorescent signal threshold is decided by the standard deviation of the average baseline fluorescence of cycles 3–15 [31], whereas the cycle threshold (Ct) is determined by the number of PCR cycles needed to report a detectable fluorescence, i.e. greater than the fluorescent signal threshold. Therefore, a lower Ct value implies the presence of a greater viral RNA load [12]. In general, for diagnosis of COVID-19 infection, Ct < 40 is considered clinically positive. China CDC recommends a Ct value of 37, i.e. a suspect is judged clinically positive if Ct is < 37 [36]. A Ct value of > 40 is considered clinically negative, while it is recommended to repeat the test if Ct is 37–40 [36].
Commercially available real-time RT-PCR diagnostics for COVID-19 are either one-step or two-step assays. One-step RT-PCR kit utilizes a single tube and buffer for RT and PCR steps, while in a two-step RT-PCR assay RT and PCR are performed separately in different tubes with independently optimized buffers. One-step RT-PCR assays are high-speed, reproducible, and suitable for high-throughput diagnosis of COVID-19 because they require limited sample management and reduce the risk of cross-contamination and human errors [37]. Two-step RT-PCR assays, on the other hand, are generally more flexible and tunable and offer superior sensitivity and low detection limit [38].
To improve the diagnostic efficiency, duplex or multiplex real-time RT-PCR test kits are developed, which enable the simultaneous detection of two or more specific sequences, thereby improving the diagnostic sensitivity and reliability. For instance, FDA-approved Abbott’s RealTime SARS-CoV-2 assay is a dual target RT-PCR assay for the quantitative recognition of RdRp and N genes. It uses an unrelated RNA sequence as an internal control (IC) to validate the PCR, and detects the RdRp, N, and IC target sequences via specific fluorescent-labeled probes. Different fluorophores are used for SARS-CoV-2-specific and IC-specific probes to allow simultaneous detection of these targets. Abbott’s RealTime SARS-CoV-2 assay uses the viral RNA extracted from the nasal or nasopharyngeal swab samples. Rutgers Clinical Genomics Laboratory developed an RT-PCR assay that utilizes self-collected saliva samples for COVID-19 diagnosis. The assay is commercialized as TaqPath™ COVID-19 Combo Kit by Life Technologies (a part of Thermo Fisher Scientific, Inc.), and simultaneously detects ORF1ab, N, and S genes. The test not only offers painless and quick sample collection, high volume testing, and lower exposure risk to healthcare personnel, but higher clinical sensitivity and clinical specificity [39].
Performance of commercial and pre-commercial molecular diagnostics
An overview of the clinical performance in terms of time-to-result, clinical sensitivity (PPA), clinical specificity (NPA), and limit of detection (LoD) of various commercial and pre-commercial molecular diagnostics is presented in Table 1. Thus far, 336 commercially available or pre-commercial (under development) molecular assays for COVID-19 diagnosis are submitted to the Foundation for Innovative New Diagnostics (FIND), which is WHO’s collaborating center for laboratory strengthening and diagnostic technology evaluation [40]. However, a majority of these tests are not evaluated yet and/or are not approved by the WHO for emergency use. This review presents only selected commercial or pre-commercial diagnostics (Table 1), which are already approved by major healthcare authorities around the world and have been independently evaluated [41], [42], [43], [44], [45], [46], [47], [48].
Table 1.
Figures of merit of the modern commercially available (approved) and pre-commercial (research use only) molecular diagnostics for SARS-CoV-2 detection.
| Manufacturer | Test | Target gene | Time to result (hours) | Reported by manufacturers |
Tested independently |
LoD | CLIA Complexity | Regulatory status | Website | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical sensitivity (PPA) | Clinical specificity (NPA) | Clinical sensitivity (PPA) | Clinical specificity (NPA) | Sample size | ||||||||
| Abbott Diagnostics, Inc. | Abbott RealTime SARS-CoV-2 | RdRp, N | ~ 4 | 100% (95% CI: 94.0%–100%) | 100% (95% CI: 88.8%–100%) | 100% (95% CI: 94.0%–100%) | 100% (95% CI: 88.8%–100%) | – | 100 copies/ mL | H | WHO EUL; US FDA EUA; CE-IVD | https://www.molecular.abbott/ |
| Advanced Biological Laboratories SA | UltraGene Combo2Screen SARS-CoV-2 Assay | E, N | ≤ 2 | 100% (95% CI: 84.6%–100%) | 100% (95% CI: 84.6%–100%) | 80% (95% CI: 49.0%–94.3%) | 100% (95% CI: 34.2%–100%) | 12 | 1 × 10–6 TCID50/ mL | H, M | CE-IVD | https://www.ablsa.com/ |
| Altona Diagnostics GmbH | RealStar® SARS-CoV-2 RT-PCR Kits | E, S | ~ 1 | – | – | 92% (95% CI: 81.0%–97.0%) | 100% (95% CI: 96.0%–100%) | – | 10 copies/ PCR | H | US FDA EUA; CE-IVD | https://altona-diagnostics.com/ |
| Anatolia Geneworks | Bosphore Novel Coronavirus (2019-nCoV) Detection Kit | ORF1ab, E | ~ 1.5 | – | 100% | 80% (95% CI: 49.0%–94.3%) | 100% (95% CI: 43.8%–100%) | 13 | < 13 copies/ µL | H | CE-IVD | http://www.anatoliageneworks.com/ |
| Atila BioSystems, Inc. | iAMP COVID-19 Detection Kit (isothermal amplification) | ORF1ab, N | ~ 1 | 100% | 100% | 100% (95% CI: 93.0%–100%) | 99% (95% CI: 95.0%–100%) | – | 4 copies/ µL | H | US FDA EUA | https://atilabiosystems.com/ |
| Becton, Dickinson & Company (BD) | BioGX SARS-CoV-2 Reagents (for BD MAX™ System) | N1 & N2 | ~ 3 | 100% | 100% | 100% (95% CI: 64.6%–100%) | 100% (95% CI: 43.8%–100%) | 10 | 40 copies/ mL | H, M | US FDA EUA; Health Canada; Singapore HSA | https://www.bd.com/ |
| BGI Genomics Co. Ltd. | Real-Time Fluorescent RT-PCR Kit for Detecting SARS-CoV-2 | ORF1ab | ~ 4 | 88.1% (95% CI: 81.2%–92.7%) | 99.6% (95% CI: 97.8%–99.9%) | 100% (95% CI: 95.0%–100%) | 99% (95% CI: 93.0%–100%) | – | 150 copies/ mL | H | US FDA EUA; CE-IVD; NMPA EUA (China); Singapore HSA | https://www.bgi.com/ |
| Cepheid | Xpert® Xpress SARS-CoV-2 | E, N | ≤ 1 | 100% (95% CI: 88.7%–100%) | 100% (95% CI: 90.1%–100%) | 99.5% (95% CI: 97.5%–99.9%) | 95.8% (95% CI: 92.6%–97.6%) | 481 | 0.01 PFU/ mL | H, M | US FDA EUA; Health Canada | https://www.cepheid.com/ |
| 98.3% (95% CI: 90.9%–99.7%) | 100% (95% CI: 92.9%–100%) | 108 | 100 copies/ mL | |||||||||
| 100% (95% CI: 77.2%–100%) | 100% (95% CI: 77.2%–100%) | 26 | – | |||||||||
| Eurobio Scientific | EurobioPlex SARS-CoV-2 Multiplex | RdRp, N | 2–4 | – | – | 100% (95% CI: 64.6%–100%) | 100% (95% CI: 20.6%–100%) | 8 | – | H | CE-IVD | https://www.eurobio-scientific.com/ |
| EUROIMMUN AG (A PerkinElmer Company) | EURORealTime SARS-CoV-2 | ORF1ab, N | ~ 1.5 | 98.2% | 100% | 100% (95% CI: 93.0%–100%) | 98% (95% CI: 93.0%–99.0%) | – | 1 copy/ µL | H | CE-IVD | https://www.euroimmun.com/ |
| GenMark Diagnostics, Inc. | ePlex® SARS-CoV-2 Test | N | ~ 1.5 | – | – | 91.4% (95% CI: 81.4%–96.3%) | 100% (95% CI: 92.9%–100%) | 108 | 1000 copies/mL [45] | H, M | US FDA EUA | https://www.genmarkdx.com/ |
| Hologic, Inc. | Panther Fusion® SARS-CoV-2 Assay | ORF1ab | 2–4 | 100% (95% CI: 94.7%–100%) | 100% (95% CI: 96.6%–100%) | 100% (95% CI: 72.2%–100%) | 90% (95% CI: 59.6%–98.2%) | 20 | 1 × 10–2 TCID50/ mL | H | US FDA EUA | https://www.hologic.com/ |
| Life Technologies (part of Thermo Fisher Scientific, Inc.) | TaqPath™ COVID-19 Combo Kit | ORF1ab, N, S | ≤ 2 | 100% | 100% | 87.5% (95% CI: 52.9%–97.8%) | 100% (95% CI: 20.6%–100%) | 9 | 10 copies/ PCR | H | US FDA EUA; CE-IVD | https://www.thermofisher.com/ |
| OPTI Medical Systems, Inc. | OPTI® SARS-CoV-2 RT PCR Test | N1, N2 | 2–3.5 | – | – | 100% (95% CI: 67.6%–100%) | 100% (95% CI: 20.6%–100%) | 9 | – | H | US FDA EUA | https://www.optimedical.com/ |
| Primerdesign Ltd. (part of Novacyt Group) | COVID-19 genesig® Real-Time PCR assay | RdRp | 2–4 | – | 98.2% | 100% (95% CI: 51.0%–100%) | 100% (95% CI: 93.2%–100%) | 57 | 0.33 copies/ µL | H | WHO EUL; US FDA EUA; CE-IVD | https://www.genesig.com/ |
| Quidel Corporation | Lyra® SARS-CoV-2 Assay | Pp1ab | < 2 | 97% (95% CI: 83.3%–99.4%) | 100% (95% CI: 88.6%–100%) | 75% (95% CI: 40.9%–92.8%) | 0% (95% CI: 0%–79.4%) | 9 | 800 copies/ mL | H | US FDA EUA | https://www.quidel.com/ |
| Roche Molecular Systems, Inc. | cobas® SARS-CoV-2 Test (for cobas® 6800/8800 system | ORF1ab, E | ~ 4 | – | – | 100% (95% CI: 83.9%–100%) | 95% (95% CI: 76.4%–99.1%) | 26 | – | H, M | WHO EUL; US FDA EUA | https://diagnostics.roche.com/ |
| 100% (95% CI: 79.6%–100%) | 100% (95% CI: 56.6%–100%) | 20 | ||||||||||
| Sansure Biotech, Inc. | Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) | ORF1ab, N | ~ 1.5 | 94.3% (95% CI: 84.3%–98.8%) | 99% (95% CI: 96.3%–99.9%) | 93.8% (95% CI: 71.7%–98.9%) | 80% (95% CI: 37.6%–96.4%) | 21 | 200 copies/ mL | H | US FDA EUA; CE-IVD; NMPA (China) | http://www.sansure.com.cn/ |
| SD Biosensor Inc. | STANDARD M nCoV Real-Time Detection kit | ORF1ab, E | 1.5 | 100% (95% CI: 88.6%–100%) | 100% (95% CI: 88.6%–100%) | 100% (95% CI: 93.0%–100%) | 99% (95% CI: 95.0%–100%) | – | 0.5 copies/ µL | H | US FDA EUA; CE-IVD; MFDS EUA (Korea) | http://sdbiosensor.com/ |
| Seegene, Inc. | AllplexTM 2019-nCoV Assay | RdRp, N | < 2 | – | – | 100% (95% CI: 93%–100%) | 100% (95% CI: 96%–100%) | – | – | H | US FDA EUA; CE-IVD; Health Canada; MFDS EUA (Korea); Singapore HSA | http://www.seegene.com/ |
| Shanghai Fosun Long March Medical Science Co., Ltd. | 2019-Novel Coronavirus (2019-nCoV) RT-PCR Detection Kit | ORF1ab, E, N | 2–4 | – | – | 100% (95% CI: 67.6%–100%) | 100% (95% CI: 20.6%–100%) | 9 | 300 copies/ mL | H | RUO | http://www.lm-diagnostics.com.cn/ |
| TIB Molbiol Berlin GmbH/Roche Diagnostics | LightMix® Modular SARS-CoV (COVID19) | E | ~ 1.5 | – | – | 95.2% (95% CI: 85.8–98.8%) | 99.3% (95% CI: 95.8–100%) | 215 | – | H, M | RUO | https://www.tib-molbiol.de/ |
| YouSeq Ltd. | YouSeq Multiplex Covid19 qPCR Kit | RdRp, E, N | 2–4 | 100% (44/44) | 100% (44/44) | 81.2% (95% CI: 57.0%–93.4%) | 100% (95% CI: 56.6%–100%) | 21 | 0.7 copies/ µL | H | RUO | https://youseq.com/ |
Table note: CE-IVD: conformité européenne marked in vitro diagnostic; CLIA: Clinical Laboratory Improvement Amendments of 1988, 42 U.S.C. §263a (USA), while H and M stand for high or medium complexity lab, respectively; CI: confidence interval; CoV: coronavirus; COVID-19: coronavirus disease 2019; E gene: gene encoding envelop protein of SARS-CoV-2; EUA: emergency use authorization; EUL: emergency use listing; FDA: Food and Drug Authority (USA); HSA: Health Sciences Authority (Singapore); MFDS: Ministry of Food and Drug Safety (Korea); N gene: gene encoding nucleocapsid protein of SARS-CoV-2; NMPA: National Medical Products Administration (China); NPA: negative percent agreement; ORF: open reading frame; ORF1ab: open reading frame 1a and b of SARS-CoV-2; PPA: positive percent agreement; Pp1ab: polyprotein 1a and b of SARS-CoV-2; q-PCR: quantitative polymerase chain reaction; RdRp: RNA-dependent RNA polymerase of SARS-CoV-2; RT-PCR: reverse transcriptase-polymerase chain reaction; RUO: research use only; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; S gene: gene encoding spike protein of SARS-CoV-2; US: United States; WHO: World Health Organization.
The most important criterion for a commercial molecular diagnostic is its clinical sensitivity (PPA) and LoD to accurately diagnose COVID-19 at an early stage of infection and to avoid false-negative results. Understandably, during a pandemic of this scale, the standards for authorization of these diagnostics are relaxed to afford the availability of test kits and large-scale screening of the samples. Also, due to the lack of time to pre-screen and test a significant number of samples, it is difficult to accumulate reliable performance data. Thus, 100% clinical sensitivity was reported by several manufacturers, while others directly submitted kits for evaluation without an appropriate number of clinical tests. The tested PPA values for these diagnostics are satisfactory in most cases due to the inherent sensitivity of the real-time RT-PCR method. However, PPA values drop well below 95% in some cases, which may be attributed to the deficiencies in the test kits or sampling and handling errors [49], [50].
Lieberman et al. [41] compared four commercial COVID-19 test kits: Cepheid’s Xpert® Xpress SARS-CoV-2, Hologic’s Panther Fusion® SARS-CoV-2 Assay, DiaSorin’s Simplexa™ COVID-19 Direct RT-PCR Kit, and Roche’s cobas® SARS-CoV-2 Test. They demonstrated that all tests were equally specific, but Cepheid’s Xpert Xpress had higher sensitivity and lower LoD. However, the outcomes were limited by the small sample size (20–26 specimens), they concluded that due to low Ct ranges, the minor sensitivity differences might not have a significant impact on the diagnostic performance during clinical practice [41]. The clinical specificity of these RT-PCR tests was 100%, which meant the design of primers was specific to the genomic sequences of SARS-CoV-2. Moore et al. [51] compared the diagnostic performance of Abbott’s RealTime SARS-CoV-2 and ID NOW™ COVID-19 diagnostics. The later utilizes isothermal amplification and has a fast turnaround time of < 13 min [11]. RealTime SARS-CoV-2 assay takes a longer time, but it is more sensitive and reliable compared to ID NowTM COVID-19 [51].
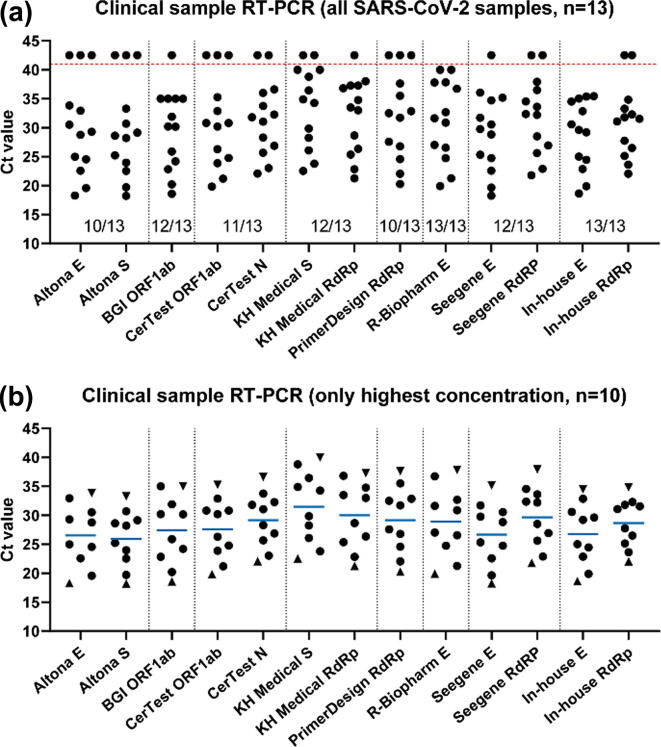
A comparison of seven commercially available RT-PCR diagnostic kits from different manufacturers revealed ≥ 96% efficiency for the detection of low concentrations of SARS-CoV-2 RNA in clinical samples [52]. Compared to the in-house E gene PCR test (Ct ≤ 34.5), the positive samples were successfully identified by these kits with slight variations in Ct values and overall performance (Fig. 3). No crossreactivity to other CoVs was observed and all kits demonstrated 100% specificity [52]. Wang et al. [53] determined the LoD of six RT-PCR kits approved by China NMPA for COVID-19 diagnosis. They observed 100% sensitivity at LoD: 484 copies/mL for BGI Genomics (ORF1ab), Liferiver Biotech (ORF1ab, N & E), DAAN (ORF1ab & N), SanSure (ORF1ab & N), and 968 copies/mL for BioGerm (ORF1ab & N). GeneoDx (ORF1ab & N) demonstrated poor sensitivity and high LoD value (7744 copies/mL), which was attributed to the technical deficiencies in the product’s design that might include reagent instability and inappropriate ratios, irrational oligonucleotide design for primer or probe, and impurities [53].
Fig. 3.
The evaluation of commercially available RT-PCR kits shows variations in the rate of detection and Ct values. (a) Experimental Ct values obtained for commercial RT-PCR assays (n = 13). The data points on top of the horizontal line (red, dotted) are negative, indicated with Ct = 42.5 for plotting purposes. The rate of detection of the RT-PCR kit is mentioned below the data points. (b) The data points for the clinical samples (n = 10) with the highest viral load that were positively identified by all RT-PCR assays. The horizontal lines (blue) indicate the mean Ct value, triangles show the Ct values of the samples with the highest (sample 1) and lowest (sample 10) viral load according to the in-house E gene PCR. E: envelop protein of SARS-CoV-2; RdRp: RNA-dependent RNA polymerase of SARS-CoV-2; N: nucleocapsid protein of SARS-CoV-2; ORF1ab: open reading frame 1a and b of SARS-CoV-2; RT-PCR: reverse transcriptase-polymerase chain reaction; S: spike protein of SARS-CoV-2; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2 (reprinted with permission from [52]; copyright Elsevier, 2020).
Other molecular diagnostic technologies based on LAMP and CRISPR mechanisms are evolving at a rapid pace [54], [55], [56], [57]. However, many of these diagnostics have not been approved by the healthcare authorities worldwide or have not been independently assessed yet. Atila BioSystems’ iAMP COVID-19 Detection Kit uses isothermal amplification (LAMP) is approved for emergency use by FDA and reports 100% sensitivity, low LoD (~4 copies/µL), and 1-h sample-to-result time [58]. Also, the SherlockTM CRISPR SARS-CoV-2 kit is the first and only CRISPR-based diagnostic approved for emergency use by the FDA. It performs quantitative SARS-CoV-2 detection within 1 h with 100% specificity and low LoD (6.75 copies/µL) [59]. Further development and independent assessment of these technologies would provide alternative molecular diagnostic solutions that are faster and suitable for high-throughput screening of suspected individuals.
Limitations and challenges
Molecular diagnostics can be developed rapidly and provide extremely sensitive, specific, and often quantitative detection of the SARS-CoV-2 RNA. However, they are complex, expensive, and slow to deliver. A single RT-PCR test kit may cost over 100 USD, while setting up a diagnostic/processing lab requires more than 15,000 USD, whereas the analysis time is 4–6 h, and sample-to-result turnaround time is more than 24 h [10], [60]. Furthermore, the molecular diagnostics are not deliverable to end-users and are intended only for qualified clinical laboratory personnel and medium- or high-complexity laboratories. Yet, some studies suggest a high false-negative rate of RT-PCR diagnostics for COVID-19 [61], [62], [63]. Erroneous RT-PCR results may be caused by inappropriate sample collection, storage, transfer, purification, and processing. The quality of the RNA extracted from the swabs also affects the results. Other factors such as degradation of purified RNA, the presence of RT-PCR inhibitors, or genomic mutations may cause false-negative results. On the other hand, cross-contamination of samples during collection, pipetting, and processing or technical errors may cause false-positive results. Notwithstanding the probability of these untoward instances, these diagnostics are currently the most accurate and the most sensitive available solutions for the earliest and large-scale detection of SARS-CoV-2.
On the research and development front, molecular assays are still being developed to optimize their clinical sensitivity, LoD, and ease-of-use [64]. The molecular assays may fail to diagnose COVID-19 if they have low sensitivity (and high LoD), while the number of steps involved in handling and processing of biological samples increases the turnaround time and risk of technical errors and cross-contamination. The overall clinical performance of molecular diagnostics is determined by several factors. These factors present continuous research and development challenges for reliable SARS-CoV-2 detection, and are listed below:
COVID-19 viral load in different specimens
For commercial diagnostics, the samples are usually collected from upper and lower respiratory specimens such as nasal, nasopharyngeal, oropharyngeal swabs, sputum, lower respiratory tract aspirates, bronchoalveolar lavage fluid, and nasopharyngeal wash/aspirate or nasal aspirate. However, the biodistribution of virus particles may vary in different specimens [65]. Wang et al. [66] collected 1070 specimens from 205 COVID-19 patients to investigate the biodistribution of SARS-CoV-2 and concluded that the highest positive rates of 93% (14/15) > 69% (72/104) > 62% (5/8) > 32% (126/398) were observed for bronchoalveolar lavage fluid, sputum, nasal swabs, and pharyngeal swabs, respectively. The average Ct was lowest for nasal swabs (Ct = 24.3; 1.4 × 106 copies/mL) that indicated the highest viral loads. In a similar study, the samples collected from 52 suspected COVID-19 patients exhibited a higher positive rate for sputum (77%; 40/52) compared to throat swabs (44%; 23/52) [67]. To et al. [68], [69] suggest the highest positive rates of 92% (11/12) for saliva specimens from deep throat. It is suggested that deep throat saliva sampling may offer early detection of SARS-CoV-2 [70]. However, these studies are not conclusive because the number of samples is not uniform or is very small in certain cases. Furthermore, false-negatives may occur due to inappropriate sample collection time reference to the onset of symptoms or technical deficiencies in sampling methods [12]. Therefore, an RT-PCR result must be validated by the detailed knowledge of the symptoms and epidemiological history, and clinical examination. Moreover, there is a need to perform systematic research and careful analysis with a larger and more diverse sample size to find conclusive evidence of the biodistribution of SARS-CoV-2 in the body.
Diagnostic timing
The knowledge of the detectable SARS-CoV-2 load in the respiratory tract during different stages of infection is still evolving [71]. The viral load kinetics also varies from person to person and depends on several factors including the patients’ epidemiological history, immune response, and treatment or medication effects [72]. Therefore, the timing of the diagnosis is one of the most important limiting factors for molecular diagnostics. Inappropriate diagnostic timing may lead to false-negative results, especially at an early stage of infection [73], [74]. A recent study shows that the highest pharyngeal shedding of SARS-CoV-2 is in the first few days of symptoms and peaks on day 4 with 7.11 × 108 gene equivalents per throat swab, while sputum viral shedding lasted till the sixth week, i.e. end of symptoms [75]. Sethuraman et al. [12] established the diagnostic timeline for RT-PCR tests. It is possible to detect SARS-CoV-2 RNA in nasopharyngeal swab specimens one week before the onset of symptoms as well as in the prodromal phase when the initial symptoms such as malaise, fatigue, fever, and dry cough appear [76], [77]. Therefore, according to the suggested diagnostic timeline [12], nasal or nasopharyngeal swabs specimens stand for early diagnosis of oligosymptomatic or presymptomatic individuals with an epidemiological history, whereas sputum samples should be analyzed to configure complete recovery and avoid false-negative results.
Sensitivity of the oligonucleotide primers and probes
As discussed in the previous section, the clinical sensitivity (PPA) and LoD of an RT-PCR diagnostic play a major role in the early diagnosis of COVID-19. High PPA and very low LoD of the test are desirable to prevent false-negative results. Although multiple factors influence the outcomes of a test, technical deficiencies in the product’s design such as unstable reagents, inappropriate amounts, irrational oligonucleotide design, and impurities should be avoided [53]. Besides, certain primers and probe designs are inherently more sensitive compared to others [78], [79], [80]. For instance, the primer-probe sets for CDC’s N1 and N2 genes have different PPA and LoD with N2 variants showing much lower LoD (356 copies/mL) compared to N1 assay (779 copies/mL). Nalla et al. [81] compared the sensitivity of different RT-PCR primer-probe sets (RdRp/E genes by Charité Germany [82], and N1/N2 genes by CDC, as shown in Fig. 1c). They discovered E-gene by Charité Germany and N2-gene by CDC were the most sensitive assays [81]. The comparison studies reveal high specificity and no cross-sensitivity for different assays as well as comparable sensitivities except RdRp assay by Charité Germany that was least sensitive likely because of a deficiency in the reverse primer [12].
Genomic diversity and mutations
Owing to the rapid pandemic-scale spread of SARS-CoV-2, it shows very high genomic diversity in gene position and nucleotide sequences [83], [84]. The outcomes of the genomic sequencing of SARS-CoV-2 are regularly shared on the GISAID database [27]. Pachetti et al. [85] performed an analysis of 220 SARS-CoV-2 genomic sequences placed on GISAID and found eight novel recurrent genomic mutations, which suggested that SARS-CoV-2 was evolving and different strains of SARS-CoV-2 might coexist. The evolution of SARS-CoV-2 and genetic diversity present a colossal new challenge to molecular diagnostic manufacturers. Osório and Correia-Neves [86] analyzed 1825 high-coverage genomic sequences from the GASAID database. Their analysis revealed that ~ 79% (26/33) primer binding sequences in at least one gene used in RT-PCR were mutated compared to Wuhan-Hu-1 (NC_045512) [86]. Therefore, they concluded that 14% of SARS-CoV-2 variants were not detectable with at least one of the commercialized primers. Consequently, the manufacturers need to regularly optimize the oligonucleotides through frequent analysis of the updated genomic sequences on GASAID to enhance their accuracy and reliability.
Concluding remarks and future perspective
The modern molecular diagnostics are not intended for point-of-care diagnosis of COVID-19 but provide a core diagnostic solution to conduct large numbers of tests in a reasonable timeframe [87]. Li et al. [88] estimated that 80% of the COVID-19 positive cases were initially transmitted by the undetected infections during the early stages of this pandemic. Therefore, accurate diagnosis of the disease is crucial to curb its spread. In the current pandemic situation, the SARS-CoV-2 tests were rapidly developed and approved for emergency use. Rationally, due to the lack of research and development time, the commercialized diagnostics were not validated or optimized with sufficient numbers of clinical samples, which might have led to inaccurate results [53]. Furthermore, false-negative results may have been caused by inadequate diagnostic timing, low sensitivity, or low viral load. Therefore, it is plausible that infected individuals may have been missed by the real-time RT-PCR tests [89], [90], [91]. In a recent study, Liu et al. [92] indicated that RT‐PCR results were potentially erratic, and clinical diagnosis of symptomatic individuals or those with epidemiological history should not rely on RT-PCR only. Ideally, RT-PCR must be combined with clinical examination and computed tomography (CT) to medically judge the suspected individuals [93], [94]. Considering these deficiencies and limitations, continuous optimization of RT-PCR diagnostics is critical to meet the diagnostic standards. In this view, digital RT-PCR technology may enhance the accuracy, sensitivity, and lower detection limits thereby enabling accurate and early diagnosis of COVID-19 [95].
Apart from the aforementioned research and development challenges, the diagnostic efficiency of modern RT-PCR tests is also influenced by the lack of organized knowledge and the novelty of the disease. Thus far, a majority of the research reported in recent literature is rendered with severe limitations. For example, the sample size is not enough to establish convincing conclusions and certain patients have been treated with different medications after different intervals of the onset of symptoms for which the effects on SARS-CoV-2 population, replication, immunity, and efficacy are still unknown [41], [72], [75]. Therefore, on the research front, more structured and systematic pilot studies need to be performed to generate concrete knowledge about the perseverance of SARS-CoV-2 in specific anatomic sites at different times before and after the onset of symptoms. Furthermore, there is a portentous need to monitor, analyze, and regularly update the data about genetic mutations and evolution of SARS-CoV-2 in different communities worldwide. The knowledge progress in these areas will help optimize the molecular diagnostics and relevant protocols.
Also, it is important to keenly follow the emerging knowledge and tangible pieces of evidence in the field to improve current diagnostics as well as to develop more sophisticated tools for rapid and point-of-care detection of SARS-CoV-2. For instance, small-footprint rapid test equipment for cost-effective point-of-care diagnosis of COVID-19 are being developed such as Abbott’s famous ID NOWTM Instrument and ID NOWTM COVID-19 Test Kit and Lumex Instruments’ Microchip RT-PCR COVID-19 Detection System. ID NowTM utilizes isothermal amplification technology and performs rapid tests with 5 min turnaround time for positive and 13 min for negative results. Lumex Microchip utilizes RT-PCR with minimal reagent consumption and diminishes contamination risk and occasional human errors. However, these systems have low sensitivity compared to conventional real-time RT-PCR and exhibit high LoD, i.e. 2 × 104 copies/mL for ID NOWTM test [45] and 9 × 103 copies/mL for Lumex Microchip [96]. Nonetheless, these innovations may help point-of-care diagnosis in remote locations, especially in third-world countries where setting up numerous medium or high-complexity laboratories is not possible. Soon, the development of new techniques may improve the quality of compact and deliverable diagnostic solutions.
Presently, molecular diagnostics combined with a comprehensive clinical examination of suspected asymptomatic or oligosymptomatic individuals may reduce the number of false-negative results. However, an independent and careful assessment of the commercial diagnostic tests should be conducted to identify diagnostic errors and determine the efficiency of approved tests [97]. These approaches would lead to better understand and diagnose COVID-19 and improve the future epidemic readiness for emerging infectious diseases.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
AA extends his appreciation to the Deanship of Scientific Research, University of Hafr Al Batin for funding this work through the research group project number G-107-2020.
Glossary
- ACE2
angiotensin-converting enzyme 2
- CDC
Centers for Disease Control and Prevention
- cDNA
complementary DNA
- CE-IVD
conformité européenne marked in vitro diagnostic
- CLIA
Clinical Laboratory Improvement Amendments of 1988, 42 U.S.C. §263a (USA)
- CI
confidence interval
- CoV
coronavirus
- COVID-19
coronavirus disease 2019
- CRISPR
clustered regularly interspaced short palindromic repeats
- Ct
cycle threshold
- CT
computed tomography
- EUA
emergency use authorization
- EUL
emergency use listing
- EVD
Ebola virus disease
- FDA
Food and Drug Authority (USA)
- FIND
Foundation for Innovative New Diagnostics
- FRET
fluorescence resonance energy transfer
- GISAID
global initiative on sharing all influenza data
- HAS
Health Sciences Authority (Singapore)
- HKU
Hong Kong University
- IC
internal control
- LAMP
loop-mediated isothermal amplification
- LoD
limit of detection
- MERS
Middle-Eastern respiratory syndrome
- MFDS
Ministry of Food and Drug Safety (Korea)
- NIH
National Institute of Health
- NIID
National Institute of Infectious Diseases (Japan)
- NMPA
National Medical Products Administration (China)
- NPA
negative percent agreement
- NSP
non-structural proteins
- ORF
open reading frame
- PPA
positive percent agreement
- RBD
receptor-binding domain
- RdRp
RNA-dependent RNA polymerase
- RT-PCR
reverse transcriptase-polymerase chain reaction
- RUO
research use only
- SARS
severe acute respiratory syndrome
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- UN
United Nations
- WHO
World Health Organization.
Biography

Adeel Afzal (http://aafzal.com/) studied Chemistry at the University of Vienna (Austria) and earned a Ph.D. in 2007. He developed synthetic antibodies as biomimetic coatings for chemical sensors using bulk and surface molecular imprinting techniques. Later, he worked as a Collaborator (Postdoc) at the University of Bari (Italy) and developed nanomaterials-based high-temperature electronic gas sensors. In 2012, he joined King Fahd University of Petroleum and Minerals affiliated colleges in Hafr Al Batin (Saudi Arabia) which is now known as the University of Hafr Al Batin. He is currently an Associate Professor of Chemistry at the College of Science, University of Hafr Al Batin (Saudi Arabia). His research interests include the development of imprinted and functional materials for chemical and biosensors and biomedical diagnostics.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Cotten M., Watson S.J., Kellam P., Al-Rabeeah A.A., Makhdoom H.Q., Assiri A. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. The Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baize S., Pannetier D., Oestereich L., Rieger T., Koivogui L., Magassouba N. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 3.Campos G.S., Bandeira A.C., Sardi S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization W. Coronavirus disease 2019 (COVID-19): situation report, 72; 2020.
- 6.WHO Coronavirus Disease (COVID-19) Dashboard. World Health Organ 2020. https://covid19.who.int (accessed July 29, 2020).
- 7.TrackCorona. TrackCorona - COVID-19 Tracker and Live Map. TrackCorona 2020. https://www.trackcorona.live/ (accessed July 29, 2020).
- 8.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat Biotechnol. 2020;38:382–384. doi: 10.1038/d41587-020-00002-2. [DOI] [PubMed] [Google Scholar]
- 10.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020 doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 11.Satyanarayana M. Companies are racing to develop COVID-19 tests for the US. Will the tests help? CEN Glob Enterp. 2020;98:30–32. doi: 10.1021/cen-09814-cover3. [DOI] [Google Scholar]
- 12.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 13.Abbasi J. The Promise and Peril of Antibody Testing for COVID-19. JAMA. 2020;323:1881–1883. doi: 10.1001/jama.2020.6170. [DOI] [PubMed] [Google Scholar]
- 14.Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) J Mol Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen M, Zhou Y, Ye J, Abdullah AL-maskri AA, Kang Y, Zeng S, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal 2020;10:97–101. https://doi.org/10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed]
- 16.Statista GmbH. COVID-19 tests by country: Number of coronavirus (COVID-19) tests performed in the most impacted countries worldwide as of July 29, 2020. Stat GmbH 2020. https://www.statista.com/statistics/1028731/covid19-tests-select-countries-worldwide/ (accessed July 29, 2020).
- 17.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan A., Kraschel K.L. CRISPR diagnostics: Underappreciated uses in perinatology. Semin Perinatol. 2018;42:525–530. doi: 10.1053/j.semperi.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;180:12. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(281–292) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181(914–921) doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alanagreh L., Alzoughool F., Atoum M. The Human Coronavirus Disease COVID-19: Its Origin, Characteristics, and Insights into Potential Drugs and Its Mechanisms. Pathogens. 2020;9:331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GISAID. Global Initiative on Sharing All Influenza Data. GISAID 2020. https://www.gisaid.org/ (accessed June 2, 2020).
- 28.WHO. Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. World Health Organ 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance (accessed June 5, 2020).
- 29.FDA FDA Emergency Use Authorization of in vitro Diagnostic Products. US Food Drug Adm. 2020 [Google Scholar]
- 30.Artika IM, Wiyatno A, Ma’roef CN. Pathogenic viruses: Molecular detection and characterization. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 2020;81:104215. https://doi.org/10.1016/j.meegid.2020.104215. [DOI] [PMC free article] [PubMed]
- 31.Arya M., Shergill I.S., Williamson M., Gommersall L., Arya N., Patel H.R.H. Basic principles of real-time quantitative PCR. Exp Rev Mol Diagn. 2005;5:209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- 32.Younes N, Al-Sadeq DW, Al-Jighefee H, Younes S, Al-Jamal O, Daas HI, et al. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020;12. https://doi.org/10.3390/v12060582. [DOI] [PMC free article] [PubMed]
- 33.Holland P.M., Abramson R.D., Watson R., Gelfand D.H. Detection of specific polymerase chain reaction product by utilizing the 5’––3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardullo R.A., Agrawal S., Flores C., Zamecnik P.C., Wolf D.E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc Natl Acad Sci. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee L.G., Connell C.R., Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucl Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viral Disease Control Institute. China CDC 2020. http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html (accessed June 9, 2020).
- 37.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Shanti N., Saini A., Stewart C.E. Two-Step Versus One-Step RNA-to-CTTM 2-Step and One-Step RNA-to-CTTM 1-Step: Validity, Sensitivity, and Efficiency. J Biomol Tech JBT. 2009;20:172–179. [PMC free article] [PubMed] [Google Scholar]
- 39.New Rutgers saliva test for coronavirus gets FDA approval 2020. https://www.rutgers.edu/news/new-rutgers-saliva-test-coronavirus-gets-fda-approval (accessed June 10, 2020).
- 40.FIND. SARS-CoV-2 diagnostic pipeline. Found Innov New Diagn FIND 2020. https://www.finddx.org/covid-19/pipeline/ (accessed June 11, 2020).
- 41.Lieberman J.A., Pepper G., Naccache S.N., Huang M.-L., Jerome K.R., Greninger A.L. Comparison of Commercially Available and Laboratory Developed Assays for in vitro Detection of SARS-CoV-2 in Clinical Laboratories. J Clin Microbiol. 2020 doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SARS-CoV-2 Mise à jour le 20/05/20. Société Fr Microbiol 2020. https://www.sfm-microbiologie.org/2020/05/11/covid-19/ (accessed May 22, 2020).
- 43.FIND: Diagnostics resource centre. FIND 2020. https://www.finddx.org/covid-19/ (accessed May 22, 2020).
- 44.Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Test. J Clin Microbiol. 2020 doi: 10.1128/JCM.00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical Evaluation of Three Sample-To-Answer Platforms for the Detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coronavirus (COVID-19) CE IVD: genesig® Real-Time PCR assay. GENESIG Kits Prim Ltd 2020. https://www.genesig.com/products/10039-coronavirus-covid-19-ce-ivd (accessed May 23, 2020).
- 47.National Institute for Communicable Diseases - NICD. NICD 2020. https://www.nicd.ac.za/ (accessed May 22, 2020).
- 48.Poljak M., Korva M., Gašper N.K., Komloš K.F., Sagadin M., Uršič T. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020 doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J Clin Microbiol 2020;58:e00512-20, /jcm/58/6/JCM.00512-20.atom. https://doi.org/10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed]
- 50.Bachelet V.C. Do we know the diagnostic properties of the tests used in COVID-19? A rapid review of recently published literature. Medwave. 2020;20 doi: 10.5867/medwave.2020.03.7891. [DOI] [PubMed] [Google Scholar]
- 51.Moore N.M., Li H., Schejbal D., Lindsley J., Hayden M.K. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV RT-PCR assay for the detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J. Limits of Detection of Six Approved RT–PCR Kits for the Novel SARS-coronavirus-2 (SARS-CoV-2) Clin Chem. 2020 doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan C., Cui J., Huang L., Du B., Chen L., Xue G. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu L, Wu S, Hao X, Li X, Liu X, Ye S, et al. Rapid colorimetric detection of COVID-19 coronavirus using a reverse tran-scriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic plat-form: iLACO. MedRxiv 2020:2020.02.20.20025874. https://doi.org/10.1101/2020.02.20.20025874.
- 56.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020:1–5. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broughton J.P., Deng X., Yu G., Fasching C.L., Singh J., Streithorst J. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. Infect Diseases (except HIV/AIDS) 2020 doi: 10.1101/2020.03.06.20032334. [DOI] [Google Scholar]
- 58.Atila Biosystems: iAMP COVID-19 Detection Kit. Atila Biosyst Inc 2020. https://atilabiosystems.com/our-products/covid-19/ (accessed June 14, 2020).
- 59.Sherlock Biosciences: The SherlockTM CRISPR SARS-CoV-2 kit. Sherlock Biosci 2020. https://sherlock.bio/crispr-sars-cov-2/ (accessed June 14, 2020).
- 60.Ramdas K., Darzi A., Jain S. ‘Test, re-test, re-test’: using inaccurate tests to greatly increase the accuracy of COVID-19 testing. Nat Med. 2020:1–2. doi: 10.1038/s41591-020-0891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet Lond Engl. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet Lond Engl. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta Int J Clin Chem. 2020 doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan JF-W, Yip CC-Y, To KK-W, Tang TH-C, Wong SC-Y, Leung K-H, et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J Clin Microbiol 2020;58:e00310-20, /jcm/58/5/JCM.00310-20.atom. https://doi.org/10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed]
- 65.Jeong J.H., Kim K.H., Jeong S.H., Park J.W., Lee S.M., Seo Y.H. Comparison of sputum and nasopharyngeal swabs for detection of respiratory viruses. J Med Virol. 2014;86:2122–2127. doi: 10.1002/jmv.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin C, Xiang J, Yan M, Li H, Huang S, Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19). Clin Chem Lab Med CCLM 2020;1. https://doi.org/10.1515/cclm-2020-0187. [DOI] [PubMed]
- 68.To K.K.-W., Tsang O.T.-Y., Yip C.C.-Y., Chan K.-H., Wu T.-C., Chan J.M.-C. Novel Coronavirus in Saliva. Clin Infect Dis. 2019;2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. MedRxiv. 2020 [Google Scholar]
- 71.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Exp Rev Mol Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J.Y., Ko J.H., Kim Y., Kim Y.J., Kim J.M., Chung Y.S. Viral Load Kinetics of SARS-CoV-2 Infection in First Two Patients in Korea. J Korean Med Sci. 2020;35 doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I.-P. Novel Coronavirus (2019-nCoV): A Systematic Review. J Clin Med. 2019;2020:9. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 76.Pan Y., Guan H. Imaging changes in patients with 2019-nCov. Eur Radiol. 2020 doi: 10.1007/s00330-020-06713-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Machado C, Gutierrez JV. Anosmia and Ageusia as Initial or Unique Symptoms after SARS-COV-2 Virus Infection 2020. https://doi.org/10.20944/preprints202004.0272.v1.
- 78.Zhen W, Manji R, Smith E, Berry GJ. Comparison of Four Molecular In Vitro Diagnostic Assays for the Detection of SARS-CoV-2 in Nasopharyngeal Specimens. J Clin Microbiol 2020:JCM.00743-20, jcm;JCM.00743-20v1. https://doi.org/10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed]
- 79.Brown JR, O’Sullivan D, Pereira RP, Whale AS, Busby E, Huggett J, et al. Comparison of SARS-CoV2 N gene real-time RT-PCR targets and commercially available mastermixes. BioRxiv 2020:2020.04.17.047118. https://doi.org/10.1101/2020.04.17.047118. [DOI] [PMC free article] [PubMed]
- 80.Okamaoto K., Shirato K., Nao N., Saito S., Kageyama T., Hasegawa H. An assessment of real-time RT-PCR kits for SARS-CoV-2 detection. Jpn J Infect Dis. 2020;advpub. doi: 10.7883/yoken.JJID.2020.108. [DOI] [PubMed] [Google Scholar]
- 81.Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2019;2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Osório N.S., Correia-Neves M. Implication of SARS-CoV-2 evolution in the sensitivity of RT-qPCR diagnostic assays. Lancet Infect Dis. 2020;0. doi: 10.1016/S1473-3099(20)30435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance 2020;25. https://doi.org/10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed]
- 88.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie C., Jiang L., Huang G., Pu H., Gong B., Lin H. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao AT, Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol 2020;n/a. https://doi.org/10.1002/jmv.25855. [DOI] [PMC free article] [PubMed]
- 91.Zhang J, Yan K, Ye H, Lin J, Zheng J, Cai T. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standard for discharge. Int J Infect Dis 2020:S1201971220301260. https://doi.org/10.1016/j.ijid.2020.03.007. [DOI] [PMC free article] [PubMed]
- 92.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;n/a. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imag. 2020;47:1275–1280. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu J, Yu H, Zhang S. The indispensable role of chest CT in the detection of coronavirus disease 2019 (COVID-19). Eur J Nucl Med Mol Imaging 2020. https://doi.org/10.1007/s00259-020-04795-x. [DOI] [PMC free article] [PubMed]
- 95.Lu R, Wang J, Li M, Wang Y, Dong J, Cai W. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge. MedRxiv 2020:2020.03.24.20042689. https://doi.org/10.1101/2020.03.24.20042689.
- 96.Microchip RT-PCR COVID-19 (SARS-CoV-2) Detection System. Lumex Instrum - Lab Anal Equip Manuf 2020. https://www.lumexinstruments.com/applications/covid-19_detection_system.php (accessed June 13, 2020).
- 97.Sharfstein J.M., Becker S.J., Mello M.M. Diagnostic Testing for the Novel Coronavirus. JAMA. 2020;323:1437–1438. doi: 10.1001/jama.2020.3864. [DOI] [PubMed] [Google Scholar]