Abstract
In situ hybridization (ISH) and immunohistochemistry (IHC) are essential tools to characterize SARS-CoV-2 infection and tropism in naturally and experimentally infected animals and also for diagnostic purposes. Here, we describe three RNAscope®-based ISH assays targeting the ORF1ab, spike, and nucleocapsid genes and IHC assays targeting the spike and nucleocapsid proteins of SARS-CoV-2.
The world is currently experiencing the devastating effects of the coronavirus disease 2019 (COVID-19) pandemic caused by the newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 has severely challenged the health care systems in most countries around the world, with increased demand for rapid diagnosis and treatment of seriously ill patients. It has been demonstrated that SARS-CoV-2 can infect domestic (i.e., cats and ferrets) and wild animals (e.g., tigers, lions, and mink), causing increasing concerns amongst animal owners [1].
In situ hybridization (ISH) and immunohistochemistry (IHC) techniques allow visualization of viral nucleic acid and protein antigens, respectively, within tissues and cells. These methods offer a semi-quantitative identification of target nucleic acids and proteins, respectively, while conserving topological information of expression within cells and tissues, with respect to specific cellular/tissue structures. This critical information is, in fact, lost with other detection methods, such as western blotting, qPCR/RT-qPCR, or single-cell RNAseq, for which cells and tissues must be dissociated. ISH and IHC are well established and widely used in research and routine laboratory diagnostics [2, 3]. Since the 1970s, RNA ISH has been a valuable tool for investigating molecular mechanisms of cellular and molecular pathology. Currently, multiple approaches exist to carry out RNA ISH [4–9], and among them, the RNAscope® technology excels for robustness, specificity, and sensitivity [6, 7, 10–13]. This technique takes advantage of a variation of the branched DNA or “tree” amplification method. In contrast, IHC performance depends heavily on the existence of a specific antibody with high affinity for its antigen with minimum background staining and good performance in formalin-fixed tissues.
The development of suitable preclinical animal models is paramount for studying COVID-19 pathogenesis and evaluating the efficacy of vaccines and therapeutics (i.e., antivirals). For this purpose, the development of SARS-CoV-2-specific ISH and IHC are both critical for the assessment of viral distribution, cell tropism, and cytopathology within tissues, complementing classical histopathology, various molecular tools, and serological assays. Also, validation of these tools can be of significant utility for postmortem diagnosis of SARS-CoV-2 in animals (and humans) within the context of veterinary diagnostic laboratories using formalin-fixed tissues, which render the virus inactive and safer to test under BSL2 conditions. Thus, the objective of this study was to develop RNAscope® ISH and IHC methods for the detection of SARS-CoV-2-specific antigen and RNA in infected cells that can be utilized for both research (e.g., studies involving experimentally and naturally infected animals) and diagnostic purposes.
For this study, confluent Vero cells (CCL-81™, ATCC, Manassas, VA, USA) were infected with the WA1 strain of SARS-CoV-2 (USA-WA1/2020 strain; BEI Resources, ATCC, Manassas, VA, USA) at a multiplicity of infection (MOI) of 1. Twenty-four hours postinfection, mock-infected and SARS-CoV-2-infected monolayers were fixed in 10% formalin, and cell pellets were embedded in paraffin. Here, we briefly describe the SARS-CoV-2-specific ISH and IHC procedures. The list of reagents, including catalog numbers, as well as detailed protocols for these assay, can be obtained by contacting the authors.
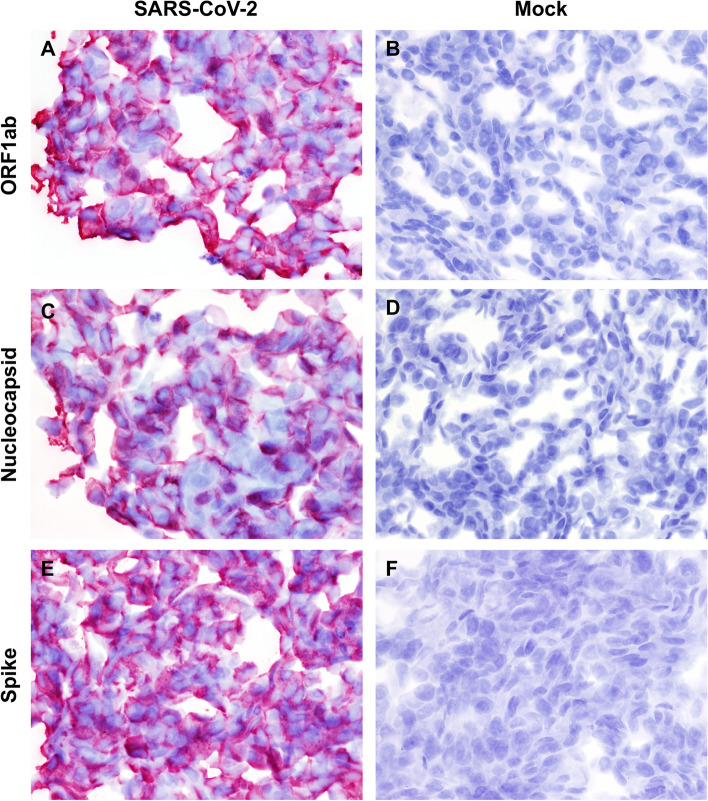
For RNAscope® ISH, a total of three antisense probes targeting the nucleocapsid (N, nucleotide [nt] 28,274-29,533), spike (S, nt 21,563-25,384) and open reading frame 1ab (ORF1ab, nt 266-13,467) of SARS-CoV-2 WA1 strain (GenBank accession number MN985325.1) were designed and manufactured by a commercial company (Advanced Cell Diagnostics [ACD], Newark, CA; Table 1). Four micron sections of formalin-fixed paraffin-embedded mock-infected and SARS-CoV-2-infected Vero cells were mounted on positively charged Superfrost® Plus Slides (VWR, Radnor, PA). The RNAscope® ISH assay was performed using an RNAscope 2.5 HD Red Detection Kit (ACD) as described previously [10, 14, 15]. Briefly, deparaffinized sections were subjected to target retrieval for 15 min at 98-102 °C in 1X Target Retrieval Solution, dehydration in 100% ethanol for 10 min, and Protease Plus treatment for 20 min at 40 °C in a HybEZ™ oven (ACD). Slides were subsequently incubated with a ready-to-use probe mixture for 2 h at 40 °C in the HybEZ™ oven, and the signal was amplified using a specific set of amplifiers (AMP1-6) as recommended by the manufacturer). The signal was detected using a Fast Red solution (Red B: Red A in a 1:60 ratio) for 1-10 minutes at room temperature. Slides were counterstained with 50% Gill hematoxylin I (Sigma Aldrich, St Louis, MO) for 2 min, and bluing was performed using 0.02% ammonium hydroxide in water. Slides were finally mounted with Ecomount® (Biocare, Concord, CA). Probes specific for dihydrodipicolinate reductase B mRNA of Bacillus subtilis (DapB) and peptidylprolyl isomerase B (PPIB) were used as negative and positive controls to assess the assay specificity and RNA integrity, respectively. The antisense probes targeting the N, S, and ORF1ab genes generated equal and very strong intracytoplasmic and membranous signals in SARS-CoV-2-infected Vero cell pellets. In contrast, there was no staining in mock-infected Vero cells or Vero cells hybridized with the DapB probe (Fig. 1).
Table 1.
Antisense probe targets for detection of SARS-CoV-2 strain WA1 (GenBank accession number MN985325.1) by RNAscope® ISH
| Probe name | Target | Nucleotide position | Source |
|---|---|---|---|
| V-nCoV-N | Nucleocapsid (N) | 28,274-29,533 | Advanced Cell Diagnostics |
| V-nCoV2019-S | Spike (S) | 21,563-25,384 | Advanced Cell Diagnostics |
| V-nCoV-orf1ab-O1 | ORF1ab | 266-13,467 | Advanced Cell Diagnostics |
Fig. 1.
Detection of SARS-CoV-2 RNA via RNAscope® ISH in SARS-CoV-2 WA1 strain-infected (A, C, E) and mock-infected (B, D, F) Vero cells. (A and B) ORF1ab, (C and D) nucleocapsid (N) and (E and F) spike (S)-specific RNAscope® ISH. Strong and diffuse intracytoplasmic labeling is evident in infected cells for all of the designed probes. No specific labeling is evident in mock-infected Vero cells incubated with SARS-CoV-2-specific probes. Fast Red, 400X
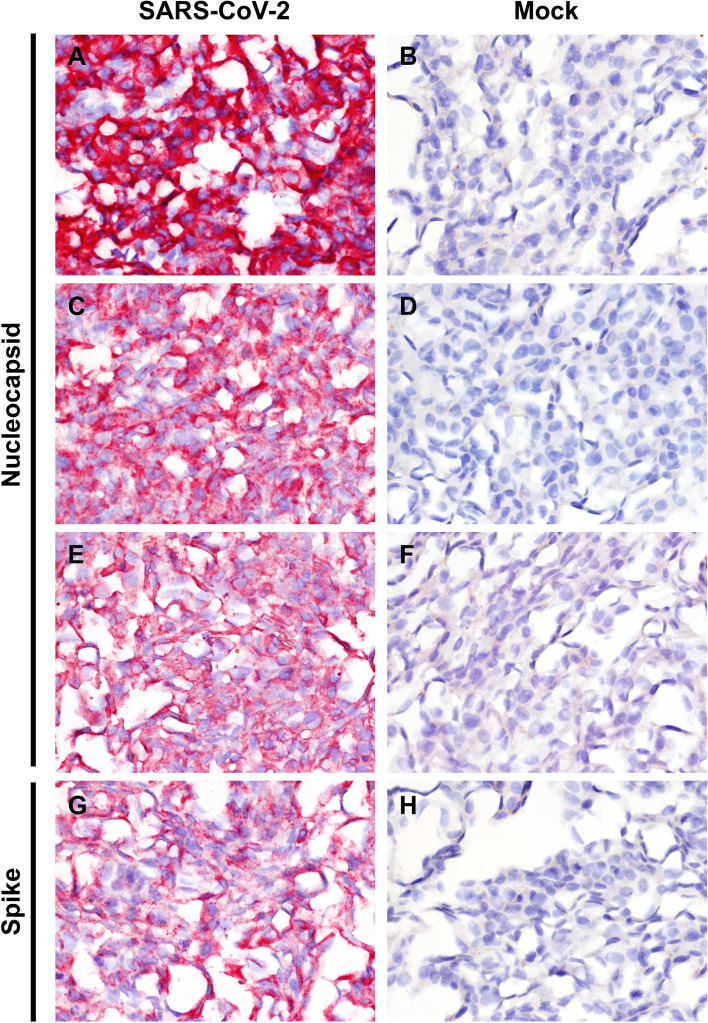
For IHC, 4 μm sections of formalin-fixed paraffin-embedded mock-infected and SARS-CoV-2-infected Vero cells were mounted on positively charged Superfrost® Plus slides and subjected to IHC using three different antibodies directed to the nucleocapsid (N) and one antibody directed to the spike (S) protein (Table 2). IHC was performed using the automated BOND-MAX and a BOND Polymer Refine Red Detection Kit (Leica Biosystems, Buffalo Grove, IL) as described previously [10]. Following automated deparaffinization, heat-induced epitope retrieval (HIER) was performed using a ready-to-use citrate-based solution (pH 6.0; Leica Biosystems) at 100 °C for 20 min. Sections were then incubated with each antibody (Table 2) for 30 min at room temperature, followed by a rabbit anti-mouse IgG (30 minutes) and/or a polymer-labeled goat anti-rabbit IgG coupled with alkaline phosphatase (30 minutes). Fast Red was used as the chromogen (15 minutes), and counterstaining was performed with hematoxylin. Slides were mounted using a permanent mounting medium (Micromount®, Leica Biosystems). Infected and mock-infected sections were incubated without the primary antibodies as controls. The antibodies specific for the N and S proteins showed equivalent cytoplasmic labeling only in SARS-CoV-2-infected Vero cells (Fig. 2). However, among the four antibodies used in this study, clone 6F10, specific for the N protein, showed the most intense staining of the infected cells.
Table 2.
Monoclonal and polyclonal antibodies used for immunocytochemical detection of SARS-CoV-2
| Specificity | Clone | Species, isotype | Working concentration | Source |
|---|---|---|---|---|
| Nucleocapsid (N) | 4B21 | Mouse, IgG | 0.25 μg/ml | Creative Diagnostics |
| Nucleocapsid (N) | 6F10 | Mouse, IgG | 1 μg/ml | BioVision |
| Nucleocapsid (N) | NA* | Rabbit polyclonal | 5 μg/ml | Thermo Fisher |
| Spike (S) | 1A9 | Mouse, IgG1 | 0.25 μg/ml | GeneTex |
*NA, not applicable
Fig. 2.
Detection of SARS-CoV-2 antigen via IHC in SARS-CoV-2 WA1 strain-infected (A, C, E, G) and mock-infected (B, D, F, H) Vero cells. (A and B) anti-nucleocapsid (N) monoclonal antibody 6F10, (C and D) anti-nucleocapsid (N) monoclonal antibody 4B21, (E and F) anti-nucleocapsid (N) polyclonal antibody, and (G and H) anti-spike (S) monoclonal antibody 1A9. Strong and diffuse intracytoplasmic labeling is evident in infected cells with all antibodies. No specific staining is evident in mock-infected Vero cells. Fast Red, 400X
Here, we describe the development of three antisense probes for the detection of three gene targets of SARS-CoV-2 (namely the ORF1ab, S, and N genes) using the highly sensitive and specific RNAscope®-based ISH assay. Concurrently, SARS-CoV-2-specific IHC assays were developed. Of the four commercial antibodies used for IHC, the monoclonal antibody clone 6F10 specific for the N protein provided the best staining of SARS-CoV-2-infected cells. The assays developed in this study are readily adaptable for the detection of SARS-CoV-2 in tissues from humans and animals, including those utilized as preclinical animal models of COVID-19 for studying the efficacy of vaccines and therapeutics. Furthermore, the development of IHC and ISH tools is of utmost significance for understanding the pathogenesis of SARS-CoV-2 by characterizing the viral tissue distribution/cellular tropism in animal models and humans. Most importantly, ISH and IHC will complement other quantitative molecular methods in the assessment of vaccine and therapeutic efficacy studies by effectively analyzing the dynamics of viral distribution and clearance within a tissue/cellular context. The tools developed and reported here can be easily multiplexed using automated systems and substantially contribute to understanding SARS-CoV-2 pathogenesis.
Acknowledgments
The authors would like to acknowledge Ms. Cheryl Johnson and other members of the histopathology and immunohistochemistry section at the Louisiana Animal Disease Diagnostic Laboratory (LADDL), Louisiana State University (LSU) School of Veterinary Medicine (SVM), for assistance in slide preparation for RNAscope® and IHC assays, and Katy Griffin at the United States Department of the Interior, U.S. Geological Survey, National Wildlife Health Center for the SARS-CoV-2 virus culture. The use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Abbreviations
- COVID-19
coronavirus disease 2019
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- ISH
In situ hybridization
- IHC
Immunohistochemistry
- BSL2
Biosafety level 2
- MOI
Multiplicity of infection
- FFPE
Formalin-fixed paraffin-embedded
- S
Spike
- N
Nucleocapsid
- ORF1ab
Open reading frame 1ab
Funding
This study was supported by Louisiana State University, School of Veterinary Medicine start-up fund (PG 002165) to Dr. Udeni B. R. Balasuriya, NIH-CEIRS, and NBAF Transition Funds from the State of Kansas and funds from the NIAID Centers of Excellence for Influenza Research and Surveillance under contract number HHSN 272201400006C to Dr. Juergen A. Richt.
Availability of data and material
For further detail on protocols, please contact the authors directly.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest. The use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Consent for publication
All of the authors have agreed to the submission of this manuscript and to be responsible for its contents.
Ethics approval
This article does not contain any studies with living animals performed by any of the authors.
Footnotes
Handling editor: John Ziebuhr.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McNamara T, Richt JA. Glickman L (2020) A critical needs assessment for research in companion animals and livestock following the pandemic of COVID-19 in humans. Vector Borne Zoonotic Dis. 2020;20(6):393–405. doi: 10.1089/vbz.2020.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNicol AM, Farquharson MA. In situ hybridization and its diagnostic applications in pathology. J Pathol. 1997;182(3):250–261. doi: 10.1002/(SICI)1096-9896(199707)182:3<250::AID-PATH837>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Shi SR, Shi Y, Taylor CR. Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades. J Histochem Cytochem. 2011;59(1):13–32. doi: 10.1369/jhc.2010.957191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin L, Lloyd RV. In situ hybridization: methods and applications. J Clin Lab Anal. 1997;11(1):2–9. doi: 10.1002/(SICI)1098-2825(1997)11:1<2::AID-JCLA2>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenny D, Shen LP, Kolberg JA. Detection of viral infection and gene expression in clinical tissue specimens using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. 2002;50(9):1219–1227. doi: 10.1177/002215540205000909. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Wang MX, Su N, Wang LC, Wu X, Bui S, et al (2014) RNAscope for in situ detection of transcriptionally active human papillomavirus in head and neck squamous cell carcinoma. J Vis Exp 2014;(85):51426. 10.3791/51426. [DOI] [PMC free article] [PubMed]
- 8.Yin VP. In situ detection of microRNA expression with RNAscope probes. Methods Mol Biol. 2018;1649:197–208. doi: 10.1007/978-1-4939-7213-5_13. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen R, Nielsen PS, Jensen TH. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005;11(11):1745–1748. doi: 10.1261/rna.2139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carossino M, Loynachan AT, James MacLachlan N, Drew C, Shuck KM, Timoney PJ, et al. Detection of equine arteritis virus by two chromogenic RNA in situ hybridization assays (conventional and RNAscope(R)) and assessment of their performance in tissues from aborted equine fetuses. Arch Virol. 2016;161(11):3125–3136. doi: 10.1007/s00705-016-3014-5. [DOI] [PubMed] [Google Scholar]
- 11.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Jr, et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun. 2016;1(1):68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roe CJ, Siddiqui MT, Lawson D, Cohen C. RNA in situ hybridization for Epstein-barr virus and cytomegalovirus: comparison with in situ hybridization and immunohistochemistry. Appl Immunohistochem Mol Morphol. 2019;27(2):155–159. doi: 10.1097/PAI.0000000000000568. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Svensson Akusjarvi S, Sonnerborg A, Neogi U. Characterization of inducible transcription and translation-competent HIV-1 using the RNAscope ISH technology at a single-cell resolution. Front Microbiol. 2018;9:2358. doi: 10.3389/fmicb.2018.02358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carossino M, Dini P, Kalbfleisch TS, Loynachan AT, Canisso IF, Shuck KM, et al. Downregulation of microRNA eca-mir-128 in seminal exosomes and enhanced expression of CXCL16 in the stallion reproductive tract are associated with long-term persistence of equine arteritis virus. J Virol. 2018;92(9):e00015–e00018. doi: 10.1128/JVI.00015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carossino M, Dini P, Kalbfleisch T, Loynachan AT, Canisso IF, Cook RF, et al. Equine arteritis virus long-term persistence is orchestrated by CD8+ T lymphocyte transcription factors, inhibitory receptors, and the CXCL16/CXCR6 axis. PLoS Pathog. 2019;15(7):e1007950. doi: 10.1371/journal.ppat.1007950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
For further detail on protocols, please contact the authors directly.