Abstract
Honey bee viruses are one of the most important pathogens that have contributed to the decrease in honey bee colony health. To analyze the infection dynamics of honey bee viruses, quantification of viral gene expression by RT-qPCR is necessary. However, suitable reference genes have not been reported from viral and RNAi studies of honey bee. Here, we evaluated the expression of 11 common reference genes (ache2, rps18, β-actin, tbp, tif, rpl32, gadph, ubc, α-tubulin, rpl14, and rpsa) from Apis mellifera (Am) and Apis cerana (Ac) under Israeli acute paralysis virus (IAPV), chronic bee paralysis virus (CBPV), and Chinese sacbrood virus (CSBV) infection as well as dsRNA-PGRP-SA treatment, and we confirmed their validation by evaluating the levels of the defensin 1 and prophenoloxidase (ppo) genes during viral infection. Our results showed that the expression of selected genes varied under different viral infections. ache2, rps18, β-actin, tbp, and tif can be used to normalize expression levels in Apis mellifera under IAPV infection, while the combination of actin and tif is suitable for CBPV-infected experiments. The combination of rpl14, tif, rpsa, ubc, and ache2 as well as more reference genes is suitable for CSBV treatment in Apis cerana. Rpl14, tif, rps18, ubc, and α-tubulin were the most stable reference genes under dsRNA treatment in Apis mellifera. Furthermore, the geNorm and NormFinder algorithms showed that tif was the best suitable reference gene for these four treatments. This study screened and validated suitable reference genes for the quantification of viral levels in honey bee, as well as for RNAi experiments.
Keywords: honey bee, RT-qPCR, reference genes, bee virus, dsRNA treatment
Introduction
Honey bees provide a significant pollination service for many agricultural crops and wild plants. However, a decline in bee populations has been recently observed in the United States and parts of European countries (Brutscher et al., 2015). Although multiple biotic and abiotic factors contribute to colony decline, several epidemiological and temporal monitoring studies have indicated that pathogens play an important role in colony loss (Brutscher et al., 2015). Among these pathogens, RNA viruses are the major impacting factors, including two Apis mellifera viruses, Israeli acute bee paralysis virus (IAPV) (Maori et al., 2007) and chronic bee paralysis virus (CBPV) (Genersch and Aubert, 2010), as well as one Apis cerana virus, namely, Chinese sacbrood virus (CSBV) (Li et al., 2018).
Israeli acute paralysis virus, a positive-sense RNA virus in the family Dicistroviridae, has a widespread impact on honey bee health and has been linked with colony losses (Chen et al., 2014; Galbraith et al., 2015). RNA-seq data indicated that IAPV attacks several core genes in insect immune pathways (Toll, JAK-STAT and RNA interference pathways) (Galbraith et al., 2015). Although CBPV has not been classified, its genome, which is made up of two segments of single-stranded RNA in a non-enveloped anisometric capsid, shows homology to the Nodaviridae and Tombusviridae families (Coulon et al., 2017). Exposure of honey bees to CBPV alone promoted downregulation of immune-related genes, such as dorsal-1a and ppo (Coulon et al., 2017). CSBV, a geographic strain of SBV isolated from A. cerana in China, can cause fatal infections to A. cerana larvae by decreasing antimicrobial peptide levels (Shan et al., 2017). The quantification of viral and immune gene expression levels is one of the most important aspects when studying host and virus interactions (Marcial-Quino et al., 2016). RNA interference (RNAi) technology is frequently used as a research tool to study the function, transcription and regulation of gene including immune genes (Vyas et al., 2017; Zhu et al., 2019). Introducing exogenous double stranded RNA (dsRNA) into the insect cells activates RNAi pathways that normally function to induce antiviral responses (Dietrich et al., 2017). To analyze the expression differences in immune genes during viral infection and RNAi process, RT-qPCR is used to assess gene expression by normalization with reference genes. Most gene expression studies focused on honey bee select actin as an internal control (Chen et al., 2014), though RPS5 (Wheeler et al., 2006) and EIF S8 (Hunt et al., 2007) are also occasionally used. However, the identification of reference genes was not made in advance when these studies assessed viral dynamics and the changes in corresponding target genes.
As summarized by Shakeel et al. (2018), the identification and validation of reference genes for qRT-PCR have been performed in various insect species. For honey bee, the stability of reference genes has been investigated in different developmental stages and tissues (Lourenco et al., 2008; Reim et al., 2013; Moon et al., 2018a), and even after bacterial challenge (Scharlaken et al., 2008). However, suitable candidate reference genes have not been validated during honey bee viral infection. Moreover, most of these genes displayed variable expression levels under different experimental conditions (Thellin et al., 2009).
Thus, we evaluated the expression of 11 reference genes under infection with three different viruses and under experimental RNAi conditions to identify the most suitable reference genes for different conditions. Here, 11 reference genes, including A. mellifera ribosomal protein L32 (Amrpl32), A. mellifera 40S ribosomal protein S18 (Amrps18), A. mellifera TATA-box-binding protein (Amtbp), A. melliferaα-tubulin alpha-1C chain (Amα-tubulin), A. mellifera glyceraldehyde-3-phosphate dehydrogenase 2 (Amgadph), A. mellifera acetylcholinesterase 2 (Amache2), A. mellifera translation initiation factor eIF-2B subunit delta (Amtif), A. mellifera ubiquitin-conjugating enzyme E2-22 kDa (Amubc), A. mellifera actin related protein 1 (Amβ-actin), Apis cerana 60S ribosomal protein L14 (Acrpl14), Apis cerana 40S ribosomal protein SA (Acrpsa), Actbp, Acα-tubulin, Acgadph, Acache2, Actif, Acubc, and Acβ-actin, were analyzed in A. mellifera under IAPV or CBPV infection as well as RNAi experimental conditions. The corresponding genes from A. cerana were also used to determine the most stable reference genes for RT-qPCR under CSBV infection. Five common algorithms, including NormFinder, BestKeeper, Delta-Ct, geNorm, and RefFinder, were used to evaluate reference gene expression. We also tested the reliability of the reference genes by investigating the expression levels of two immune genes, defensin 1 and prophenoloxidase (ppo), under different viral infections. We hope that this report will provide a basis for future studies on gene expression in honey bee under viral infection.
Materials and Methods
Collection of Virus-Infected Honey Bees
Israeli acute paralysis virus-infected A. mellifera adults with obvious paralysis symptom (Maori et al., 2007) were collected from three different apiaries in Beijing, Miyun, China, while CBPV-infected A. mellifera adults with paralysis symptom (Olivier et al., 2008) were collected from three different apiaries in Guangdong Province in China. They were tested by RT-PCR with special primers (Diao et al., 2018). Healthy A. cerana larvae were collected from three different apiaries in Guangdong Province in China. CSBV-infected larvae were taken from naturally infected colonies with obvious cystic phenotypes and symptoms. The samples were immediately transported to the laboratory on dry ice to avoid the degradation of the active substances therein.
We collected seemingly healthy bee samples from the experimental apiary at the Institute of Apicultural Research (IAR), Chinese Academy of Agricultural Sciences, Beijing, China. Newly emerged bees (A. mellifera) were obtained from brood frames taken from the experimental honey bee hives and maintained in an incubator at 30°C and 60% relative humidity (RH) for approximately 12 h with access to a 50% sucrose solution. Honey bee samples were divided into five groups with three repetitions per group with 30 bees. The first, second, and third groups consisted of bees treated with IAPV, CBPV, and CSBV, respectively; the fourth group consisted of bees treated with dsRNA-pgrpsa; and the fifth group was used to test the reliability of the reference genes. Each group contained a blank control, except for the dsRNA experiment, which had dsRNA-GFP as a control.
To gain reliable samples, PCR detection was performed. Samples in IAPV group were collected every day (24, 48, 72, and 96 h) and followed by PCR using IAPV primers (Supplementary Table S1). Similarly, samples in CBPV group were collected every day (0, 24, 48, 72, and 96 h) and followed by PCR using CBPV primers (Supplementary Table S1). The naturally CSBV-infected larvae and the healthy larvae of A. cerana were collected from 2th to 6th-instar and followed by PCR using CSBV primers (Supplementary Table S1). Samples in dsRNA treatment group were collected every day (24, 48, 72, and 96 h) and followed by PCR using PGRP-SA primers (Supplementary Table S1).
Purification and Inoculation of IAPV and CBPV
To purify the viruses, the IAPV (IAPV-BJ, KX421583.1) and CBPV (CBPV-HB, MF175174.1, and MF175173.1) field strains originating from paralyzed bees showing clinical symptoms of viral infection were collected in May 2017 in China. IAPV and CBPV were screened for predominance in the samples, as well as the absence of other common viruses. IAPV was purified as described by Maori et al. (2007). CBPV was purified from the heads of bees by ultracentrifugation in a 10 to 40% (w/v) sucrose gradient as previously described by Olivier et al. (2008). To avoid interference by other viruses, PCR was run to identify whether there was infection by four other common honey bee viruses before virus purification (Supplementary Table S1), such as black queen cell virus (BQCV), acute bee paralysis virus (ABPV), deformed wing virus (DWV) and aphid lethal paralysis virus (ALPV) following the method of Diao et al. (2018) and Yang et al. (2019). The purified virus particles were dissolved in sucrose at −80°C and aliquoted until use.
Purified virus particles (5 μl, 1 μg/μl) with about 4.0 × 104 genome equivalent copies was injected into the abdominal intersegment space of 30 individual newly emerged bees taken from a healthy colony. For the IAPV and CBPV infection groups, 5 μg of IAPV and CBPV, respectively, was injected into newly emerged honey bees (A. mellifera). The naturally infected CSBV and normal control A. cerana larvae were collected from the 2nd to 6th instars. For the dsRNA group, dsRNA-PGRP-SA and dsRNA-GFP (5 μg) were injected into newly emerged honey bees (A. mellifera) at 0 and 48 h, respectively. The honey bee samples were detected for six other viruses before they were used for the infection experiments. The infected bees were maintained in an incubator at 30°C, and their mortality was calculated each day. Bees in control group were injected with PBS buffer.
RNA Extraction and RT-qPCR
Total RNA was extracted with the TRIol Kit (Ambion, Life Technologies, United States) following the manufacturer’s instructions. A NanoDrop 2000 (Thermo Scientific, United States) was used to check the quality of each RNA sample. RNA samples with A260/280 ratios ranging from 1.8 to 2.2 were used for cDNA synthesis with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the manufacturer’s instructions. The cDNAs of all samples were stored at −20°C until use.
The specific primers for the reference genes were designed using Primer-BLAST (Table 1). The RT-qPCR reaction system consisted of 12.5 μL 2 × SYBR Premix Ex TaqTM II (Takara, Dalian, China), 0.5 μL each of the forward and reverse primers (10 mM), 2 μL cDNA template, and 9.5 μL double-distilled H2O for a total volume of 25 μL. The RT-qPCR conditions followed the manufacturer’s instructions. All reactions were performed using SYBR Green Premix and amplified under the following cycling conditions: an initial cycle at 95°C; 40 cycles of 95°C for denaturation, 25 s at 55°C for annealing and 20 s at 72°C for extension; and the generation of a melting curve consisting of a single peak to rule out non-specific products and primer dimers afterward. The RT-qPCR analysis was performed with three biological replicates for each sample and three technical replicates for each biological replicate and was measured as the mean Ct value. The results were analyzed using the 9600 plus Software.
TABLE 1.
Primers used for RT-qPCR of the candidate reference genes.
| Gene name | Gene symbol | Primer sequence (5′–3′) | Amplicon size (bp) | GenBank accession no. |
| Apis mellifera ribosomal protein L32 | Amrpl32 | F:CGTCACCAGAGTGATCGTTACA | 244 | NM_001011587.1 |
| R:CCCCATGAGCAATTTCAGCAC | ||||
| Apis mellifera 40S ribosomal protein S18 | Amrps18 | F:GGTGTTGGTCGTCGTTATGC | 269 | XM_625101.6 |
| R:CGCAAACCTCTATGAGCACG | ||||
| Apis mellifera TATA-box-binding protein | Amtbp | F:GCAACCTCAAACACCGCAAA | 121 | XM_623085.6 |
| R:TGCAGAAGCCGGTGTCATAG | ||||
| Apis mellifera tubulin alpha-1C chain | Amα-tubulin | F:TGAGTCAGACAGTAGTCGGAT | 126 | XM_006558697.3 |
| R:TACCCATTTGGACACCAGCC | ||||
| Apis mellifera glyceraldehyde-3-phosphate dehydrogenase 2 | Amgadph | F:TACCGCTTTCTGCCCTTCAA | 142 | XM_393605.7 |
| R:GCACCGAACTCAATGGAAGC | ||||
| Apis mellifera acetylcholinesterase 2 | Amache2 | F:GACGCGAAGACCATATCCGT | 140 | NM_001040230.1 |
| R:TCTGTGTCCTTGAAGTCCGC | ||||
| Apis mellifera translation initiation factor eIF-2B subunit delta | Amtif | F:TGAAACAAGAGGAACATGTCTAA | 121 | XM_006559190.3 |
| R:CGTTCTGCTTTTATTTCCTCCC | ||||
| Apis mellifera ubiquitin-conjugating enzyme E2-22 kDa | Amubc | F:TCAAAAGGGTGAACACGAGC | 208 | XM_016917677.1 |
| R:AGTTCGCTGAATGCGTCAAC | ||||
| Apis mellifera actin related protein 1 | Amactin | F:TTGTATGCCAACACTGTCCTT | 120 | NM_001185146.1 |
| R:TGGCGCGATGATCTTAATTT | ||||
| Apis cerana 60S ribosomal protein L14 | Acrpl14 | F:TTGCTTCCGGTAAACCATGC | 261 | XM_017051728.2 |
| R:ATGTTTGCAGCTTCCCATGC | ||||
| Apis cerana 40S ribosomal protein SA | Acrpsa | F:TTGGAAGTCTCCTAGTGGCA | 314 | XM_017053220.2 |
| R:GCTCTTTGTCCAGTTTGTCGG | ||||
| Apis cerana TATA-box-binding protein | Actbp | F:AAGTCCGATGACCCCAGCTA | 302 | XM_028665297.1 |
| R:TAGCTGCAAAACCTAACTTCTGAAT | ||||
| Apis cerana tubulin alpha-1 chain | Actubulin | F:GTTCGACTGTGCGTTGTGTG | 211 | XM_017057619.2 |
| R:TGACCATCAGGTTGGATGCC | ||||
| Apis cerana glyceraldehyde-3-phosphate dehydrogenase 2 | Acgadph | F:CTGCACAGACCCGAGTGAAT | 120 | XM_017062468.2 |
| R:GCAACAACCTGAGCACCAAA | ||||
| Apis cerana acetylcholinesterase | Acache2 | F:CTGGTCTTGGTAAAGGGCCA | 303 | XM_017066417.2 |
| R:CCTGGACGAAGCCCTAACAC | ||||
| Apis cerana translation initiation factor eIF-2B subunit delta | Actif | F:TGGAACTGCTCAAGTTGCCT | 241 | XM_028664590.1 |
| R:ATCGGCTGGTGTAACATCGT | ||||
| Apis cerana ubiquitin-conjugating enzyme E2-22 kDa | Acubc | F:GCTGCTGCCATGACTTTACG | 280 | XM_017048857.2 |
| R:TAGCGCGTTCCAAATCCCAA | ||||
| Apis cerana actin | Acacetin | F:CGAGCACGGTATCATCACCA | 329 | JX899419.1 |
| R:CCAAGTCCAGACGGAGGATG | ||||
| Apis mellifera defensin 1 | AmDef1 | F:AAGAACGTGCCGACAGACAT | 121 | NM_001011616.2 |
| R:TCGCAATGACCTCCAGCTTT | ||||
| Apis mellifera phenoloxidase subunit A3 | Amppo | F:AGATGGCATGCATTTGTTGA | 332 | NM_001011627.1 |
| R:CCACGCTCGTCTTCTTTAGG | ||||
| Apis cerana defensin 1 | AcDef1 | F: AATTCGAGCCACTTGAGCATCC | 148 | XM_017050425.2 |
| R: CAATGACCTCCAGCTTTACCCAA |
PCR Detection for Viral and Immune Genes
The primer sequences, orientation and references are provided in Supplementary Table S1. The cDNAs of all samples were gained in the above, followed by the PCR cycles. The PCR reaction consisted of a total 20 μL volume containing 10 μL 2 × GoTaq reaction buffer (Promega, United States), 0.5 μL 10 μM of the sense and antisense primers, 1 μL of cDNA, and 8 μL nuclease-free water. The cycling conditions were as follows: 1 min at 95°C; 33 cycles of 30 s at 94°C, 30 s at 55 and 72°C for 1 min; a final extension of 10 min at 72°C; and cooling to 4°C. The PCR amplification products were separated in a 2% agarose gel stained with GV II (BIOMEC, China) and photographed with a FR-200A luminescent and fluorescent biological image analysis system (Furi, China). The product size was determined using a 100-bp molecular size ladder.
dsRNA Synthesis
The specific primers for PGRP-SA and GFP tagged with T7 promoter sequence were designed using Primer-BLAST to amplify the templates for synthesis of dsRNA (Supplementary Table S2). Amplification was performed under normal PCR conditions as follows: 30 s at 95°C; followed by 35 cycles of 8 s at 95°C, 30 s at 56°C, and 30 s at 72°C; and a final extension incubation at 72°C for 5 min. The PCR templates were purified using a Qiagen purification kit (Qiagen, Germantown, MD, United States). dsRNA-PGRP-SA and dsRNA-GFP were produced using a T7 RNAi Transcription Kit (Vazyme, China) according to the manufacturer’s instructions. The reaction product was subjected to DNase and RNAse digestion, and then incubated at 37°C for 8 h. Subsequently, the samples were dried at 37°C for 10 min and resuspended in 20–40 μL of nuclease-free H2O. The quality of the dsRNA was checked by electrophoresis and quantified with a spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). Then, 5 μg each of PGRPSA dsRNA and GFP dsRNA was injected into newly emerged A. mellifera at 0 and 48 h, respectively.
Stability of Expression of Selected Reference Genes
The stability of expression of the candidate reference genes was evaluated by five statistical algorithms, namely, the delta CT method (Silver et al., 2006), BestKeeper (Pfaffl et al., 2004), geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), and the online platform RefFinder (Xie et al., 2012). geNorm is based on the principle that the expression ratio of two reference genes should be constant in all samples, regardless of the experimental conditions or sampling time (Hellemans et al., 2007). The gene expression stability (M) is defined as the average pairwise variation compared with all other tested candidate genes by geNorm, and the reference genes with the lowest M-value remained (Vandesompele et al., 2002). Pairwise variation (Vn/Vn + 1) analysis between the normalization factors (NFn and NFn + 1) was performed by geNorm to determine the optimal number of reference genes (Vandesompele et al., 2002). NFn was also calculated by the stepwise inclusion of a less stable gene until the (n + 1)th gene had no significant contribution to the newly calculated NFn + 1 (Vandesompele et al., 2002). In particular, if the pairwise variation Vn/n + 1 between the two sequential normalization factors NFn and NFn + 1 was lower than the cut-off value of 0.15, it was considered that NFn + 1 is not required (Peng et al., 2018). NormFinder was used to estimate changes and intra- and intergroup expression variation among the candidate reference genes (Andersen et al., 2004). The most stable gene is the one with the lowest stability value according to the intra- and intergroup variability of each gene (Andersen et al., 2004). BestKeeper determines the most stably expressed genes according to the standard deviation (SD) and coefficient of variation (CV) of all the Ct values for each gene (Pfaffl et al., 2004). When the SD values of genes are <1, they are considered stable (Marcial-Quino et al., 2016). Similarly, the delta-Ct method was used to identify suitable housekeeping genes based on the average SD value of each gene (Silver et al., 2006). To identify the optimal reference genes, RefFinder, a user-friendly web-based comprehensive tool that provides a comprehensive ranking evaluation based on the geometric mean and integrates with other computational algorithms (geNorm, NormFinder, BestKeeper, and the comparative Delta-Ct method), was used to reduce bias or avoid contradictory results caused by using individuals (Peng et al., 2018). Individual genes were assigned an appropriate weight and the geometric mean of their weights was calculated for the overall final ranking based on each program.
Validation of the Selected Reference Genes
The three strategies for expression profile normalization using the reference genes in each treatment are as follows: (1) the optimal reference gene from all the samples, (2) the optimal reference gene from each treatment, and (3) the least stable reference gene from each treatment.
To validate the candidate reference genes, the relative expression levels of two immune genes (defensin1 and prophenoloxidase) were used to evaluate the reference genes by RT-qPCR. The 2–ΔΔCT method was employed to validate and quantify the expression levels of def1 and ppo after viral infection. One-way ANOVA was used to analyze the significance and the means were compared using Tukey’s multiple comparison test (P < 0.05) using the GraphPad Prism software.
Results
Identification of Primer Specificity and the Efficiency of the Candidate Reference Genes
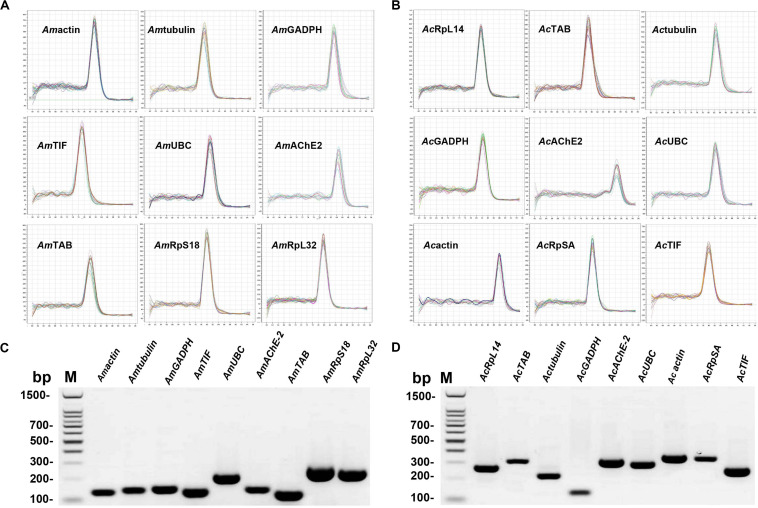
The reference gene candidates in this study were selected based on the following two criteria: (a) genes routinely used in honey bees or insects for transcript normalization and (b) genes described as stably expressed under environmental stress. Eleven candidate reference genes were selected for A. mellifera and A. cerana under different treatments. The amplicon sizes of the target genes were between 100 and 320 bp with primer lengths of 17–25 bp and a 40–60% GC content. Primer specificity for each gene was validated by melting curve analysis during qRT-PCR, and a single peak was observed in each reaction (Figures 1A,B). Primer specificity for all genes was further tested using 2.0% agarose gels. A single PCR amplification band of the expected size was observed, suggesting that primer dimers and non-specific amplified products were not generated (Figures 1C,D). Additionally, the amplification efficiencies ranged from 92.2 to 107% based on the standard curves for the 18 genes. Moreover, correlation coefficients (R2) were greater than 0.99, which fit the RT-qPCR requirements (Supplementary Table S3).
FIGURE 1.
Confirmation of primer specificity and amplicon size. (A) Melting curve analysis of the Apis mellifera candidate reference genes. All RT-qPCR products had a single melting curve indicating the breakdown of only one PCR product. (B) Melting curve analysis of Apis cerana candidate reference genes. All RT-qPCR products had a single melting curve indicating only one PCR product. (C) Amplification results for Apis mellifera candidate genes from cDNA template. (D) Amplification results for Apis cerana candidate genes from cDNA template. M: DL1500 DNA marker.
Characterization of Candidate Reference Genes
To analyze the expression profiles of the candidate reference genes, sample validation was performed by PCR (Supplementary Table S1). As shown in Supplementary Figure S1, the samples from the IAPV-treated group were positive but the control groups were not (Supplementary Figure S1A). Similarly, positive results were also observed for the CBPV and CSBV groups (Supplementary Figures S1B,C). The results for the dsRNA treatment groups showed that the expression of PGRP-SA had been markedly downregulated compared to the control group (Supplementary Figure S1D).
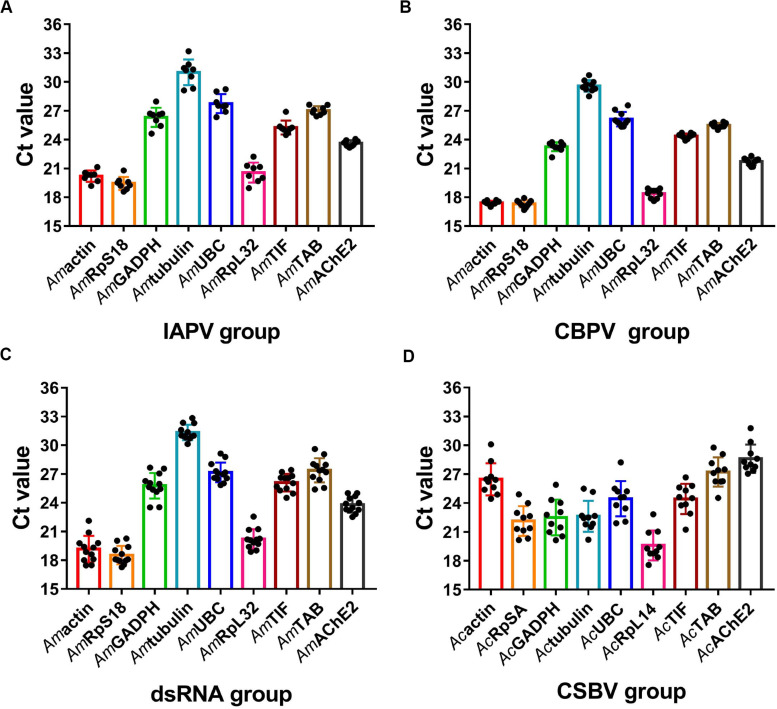
A reliable reference gene should show constant expressions level under different experimental conditions (Galli et al., 2015). We next evaluated the expression levels of the candidate reference genes and found that the average Ct values of the reference genes in different samples ranged from 16 to 32 (Figure 2). In the A. mellifera groups, RPS18 was the most abundant with the lowest mean Ct value at 19.43 ± 0.67 in the IAPV-treated group (Figure 2A), 17.31 ± 0.37 in the CBPV-treated group (Figure 2B) and 18.52 ± 0.96 in the dsRNA-treated group (Figure 2C). In contrast, Amα-tubulin transcription showed the lowest level with the highest mean Ct value at 30.99 ± 1.33 in the IAPV-treated group, 29.58 ± 0.62 in the CBPV-treated and 31.33 ± 0.80 in the dsRNA-treated group (Figure 2). In the A. cerana group, ache2 exhibited the smallest SD value (28.6 ± 1.47) but had the lowest expression level (Figure 2D). In contrast, rpl14 exhibited the smallest Ct value (19.59 ± 1.56) but had a larger SD value. Tif was a moderately stable gene with Ct average values at 25.25 ± 0.73 in the IAPV-treated group, 24.38 ± 0.26 in the CBPV-treated group, 26.10 ± 0.9 in the dsRNA-treated group and 24.43 ± 1.57 in the CSBV-infected A. cerana groups (Figure 2). However, reference genes from the CSBV-infected group exhibited larger SD values, suggesting that studies should carefully consider when only using one gene as an internal reference for the quantification of genes related to CSBV in A. cerana (Figure 2D).
FIGURE 2.
Expression profiles of candidate reference genes in different groups. Scatter plot analysis showing the raw Ct values of the candidate reference genes in (A) IAPV-infected honey bee. (B) CBPV-infected honey bee. (C) honey bee exposed to dsRNA PGRPSA treatment and (D) CSBV-infected A. cerana larvae.
Stability of the Candidate Reference Genes Under Different Viral Infections and dsRNA Treatment
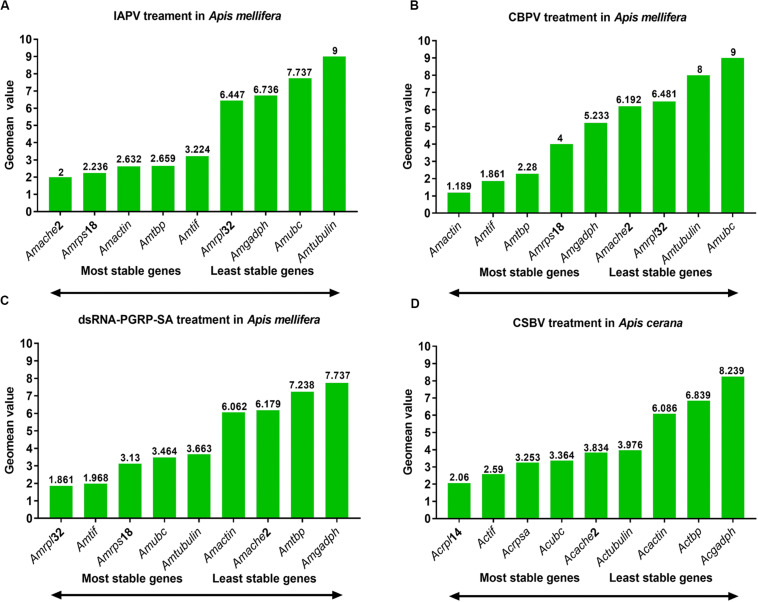
The rankings for the expression stability of these 11 genes in honey bee under treatment with IAPV, CBPV, CSBV or dsRNA was analyzed using different algorithms (Table 1 and Figure 3). In the IAPV treatment group, Amrps18, Amtif, and Amache2 were considered the most stable genes with the lowest M value according to the geNorm and NormFinder analysis (Table 2). However, the ranking obtained with BestKeeper and Delta CT showed that Amache2, Amtbp, and Amβ-actin were the top three stable candidate reference genes (Table 2). To identify the most suitable reference genes, the results of these four programs were integrated with RefFinder (He et al., 2019), which showed that the stability of these genes in A. mellifera treated with IAPV was as follows: Amache2, Amrps18, Amactin, Amtbp, Amtif, Amrpl32, Amgadph, Amubc, and Amα-tubulin (Figure 3A). However, Amubc and Amα-tubulin were ranked as the least stable genes in the IAPV-infected group.
FIGURE 3.
Expression stability of candidate reference genes analyzed by RefFinder. Expression stability of candidate reference genes during (A) IAPV infection, (B) CBPV infection, and (C) CSBV infection in A. cerana larvae as well as (D) honey bees treated with dsRNA-PGRP-SA.
TABLE 2.
Gene expression stability ranked by the geNorm, NormFinder, and BestKeeper algorithms.
| Treatments | Gene |
GeNorm |
NormFinder |
BestKeeper |
delta-CT |
|||||
| M | R | SV | R | CV | SD | R | SD | R | ||
| IAPV | Amβ-actin | 0.749 | 4 | 0.02 | 3 | 2.14 | 0.432 | 3 | 0.588 | 3 |
| Amrps18 | 0.659 | 1 | 0.018 | 2 | 2.41 | 0.468 | 5 | 0.669 | 4 | |
| Amgadph | 0.820 | 5 | 0.077 | 6 | 2.77 | 0.729 | 6 | 0.996 | 7 | |
| Amα-tubulin | 1.373 | 8 | 0.267 | 8 | 3.22 | 0.999 | 9 | 1.332 | 9 | |
| Amubc | 0.839 | 6 | 0.101 | 7 | 2.66 | 0.737 | 7 | 0.985 | 6 | |
| Amrpl32 | 0.871 | 7 | 0.038 | 4 | 4.13 | 0.850 | 8 | 1.041 | 8 | |
| Amtif | 0.697 | 2 | 0.014 | 1 | 1.73 | 0.437 | 4 | 0.726 | 5 | |
| Amtbp | 0.749 | 4 | 0.053 | 5 | 1.34 | 0.363 | 2 | 0.446 | 2 | |
| Amache2 | 0.742 | 3 | 0.018 | 2 | 1.08 | 0.255 | 1 | 0.318 | 1 | |
| CBPV | Amβ-actin | 0.380 | 1 | 0.015 | 2 | 0.96 | 0.166 | 1 | 0.211 | 1 |
| Amrps18 | 0.430 | 3 | 0.015 | 2 | 1.64 | 0.285 | 4 | 0.669 | 8 | |
| Amgadph | 0.464 | 4 | 0.030 | 4 | 1.35 | 0.314 | 6 | 0.463 | 5 | |
| Amα-tubulin | 0.640 | 7 | 0.096 | 6 | 1.59 | 0.470 | 8 | 0.617 | 7 | |
| Amubc | 0.732 | 8 | 0.067 | 5 | 2.28 | 0.595 | 9 | 0.750 | 9 | |
| Amrpl32 | 0.488 | 5 | 0.030 | 4 | 2.07 | 0.381 | 7 | 0.467 | 6 | |
| Amtif | 0.380 | 1 | 0.013 | 1 | 0.81 | 0.197 | 2 | 0.262 | 2 | |
| Amtbp | 0.407 | 2 | 0.030 | 4 | 0.85 | 0.218 | 3 | 0.263 | 3 | |
| Amache2 | 0.507 | 6 | 0.021 | 3 | 1.32 | 0.287 | 5 | 0.376 | 4 | |
| dsRNA | Amβ-actin | 1.023 | 5 | 0.033 | 3 | 5.55 | 1.064 | 8 | 1.384 | 9 |
| Amrps18 | 1.152 | 7 | 0.049 | 4 | 5.55 | 1.064 | 8 | 0.963 | 4 | |
| Amgadph | 1.032 | 6 | 0.063 | 6 | 3.85 | 0.992 | 7 | 1.332 | 8 | |
| Amα-tubulin | 1.255 | 8 | 0.147 | 9 | 2.03 | 0.636 | 1 | 0.617 | 1 | |
| Amubc | 0.895 | 3 | 0.058 | 5 | 2.82 | 0.766 | 4 | 1.024 | 5 | |
| Amrpl32 | 0.856 | 2 | 0.030 | 2 | 3.60 | 0.727 | 3 | 1.030 | 6 | |
| Amtif | 0.806 | 1 | 0.014 | 1 | 2.94 | 0.768 | 5 | 0.914 | 3 | |
| Amtbp | 1.032 | 6 | 0.080 | 7 | 3.45 | 0.946 | 6 | 1.263 | 7 | |
| Amache2 | 0.925 | 4 | 0.082 | 8 | 2.97 | 0.707 | 2 | 0.843 | 2 | |
| CSBV | Acacetin | 1.450 | 4 | 0.124 | 8 | 4.39 | 1.161 | 4 | 1.676 | 7 |
| Acrpsa | 2.077 | 9 | 0.059 | 3 | 5.73 | 1.267 | 7 | 1.550 | 3 | |
| Acgadph | 1.580 | 6 | 0.108 | 7 | 6.73 | 1.514 | 9 | 1.852 | 9 | |
| Acα-tubulin | 1.490 | 5 | 0.077 | 5 | 4.85 | 1.098 | 2 | 1.604 | 6 | |
| Acubc | 1.758 | 7 | 0.062 | 4 | 5.42 | 1.325 | 8 | 1.833 | 8 | |
| Acrpl14 | 1.435 | 3 | 0.037 | 1 | 6.13 | 1.200 | 6 | 1.555 | 4 | |
| Actif | 1.371 | 1 | 0.053 | 2 | 4.86 | 1.187 | 5 | 1.574 | 5 | |
| Actbp | 1.769 | 8 | 0.178 | 9 | 4.22 | 1.149 | 3 | 1.526 | 2 | |
| Acache2 | 1.428 | 2 | 0.099 | 6 | 3.82 | 1.093 | 1 | 1.467 | 1 | |
M, gene expression stability; R, ranking; SV, stability value; SD, standard deviation; CV, coefficient of variation.
As shown in Table 2, Amtif and Amtbp were the top two stable candidate reference genes in A. mellifera infected with CBPV based on the ranking with geNorm, Delta-CT and BestKeeper. However, the ranking obtained with NormFinder was Amtif, Amactin and Amrps18, which was different from that obtained with geNorm. Based on the comprehensive analysis by RefFinder, the stability of these genes in A. mellifera treated with CBPV was as follows: Amβ-actin, Amtif, Amtbp, Amrps18, Amgadph, Amache2, Amrpl32, Amubc, and Amα-tubulin (Figure 3B). Because Amubc and Amα-tubulin were ranked as the least stable genes in IAPV-infected and CBPV-infected A. mellifera, they were not recommended as reference genes for qRT-PCR analyses in honey bee infected with viruses (Figures 3A,B).
For A. cerana larvae infected with CSBV, Actif, Acache2, and Acrpl14 were the top three stable candidate reference genes based on the geNorm algorithms (Table 2). Acrpl14, Actif, and Acrpsa were the top three stable candidate reference genes according to NormFinder. However, the ranking obtained with BestKeeper showed that Acache2, Acα-tubulin and Actbp were the top three stable candidate reference genes. Furthermore, the ranking obtained with the Delta CT method showed that Acache2, Actbp, and Acrpsa were the top three suitable genes (Table 2). The outcome from integration with RefFinder showed that the stability of the genes in A. cerana infected with CSBV was as follows: Acrpl14, Actif, Acrpsa, Acubc, Acache2, Acα-tubulin, Acacetin, Actbp, and Acgadph (Figure 3C). However, according to their larger M values (1.3 to 2.0), none of the reference genes used in CSBV-infected A. cerana was stable (Table 2).
For A. mellifera treated with dsRNA-PGRP-SA and dsRNA-GFP, Amtif, Amrpl32 and Amubc were the top three stable candidate reference genes according to the geNorm algorithms (Table 2). Amtif, Amrpl32 and Amactin were the top three stable candidate reference genes based on NormFinder. However, the ranking obtained with BestKeeper showed that Amα-tubulin, Amache2, and Amrpl32 were the top three stable candidate reference genes. Moreover, the ranking by the Delta-CT method showed that Amα-tubulin, Amache2 and Amtif were the top three reference genes (Table 2). According to the RefFinder results, the stability of the genes in A. mellifera treated with dsRNA was as follows: Amrpl32, Amtif, Amrps18, Amubc, Amα-tubulin, Amactin, Amache2, Amtbp, and Amgadph (Figure 3D).
Determination of the Optimal Number of Reference Genes
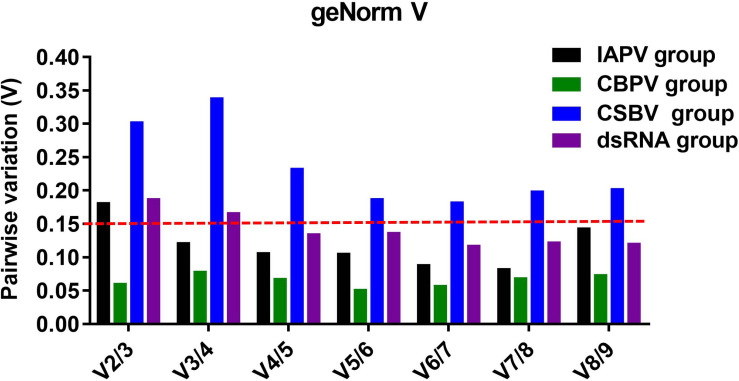
To determine the optimal number of reference genes, we used geNorm to calculate the pairwise variation. In the IAPV treatment group, V3/V4 < 0.15 indicated that three reference genes was sufficient to accurately normalize RT-qPCR data (Figure 4). Therefore, Amache2, Amrps18 and Amactin were considered to be a suitable combination of reference genes for RT-qPCR analyses in honey bee infected with IAPV. In the CBPV treatment group, V2/V3 < 0.15 indicated that two reference genes was sufficient to normalize RT-qPCR data. Thus, Amactin and Amtif were considered to be a suitable combination of reference genes for RT-qPCR. In contrast, in the CSBV treatment group, Vn/Vn + 1 > 0.15 indicated that more reference genes are required and should be combined for RT-qPCR data normalization. In the dsRNA treatment group, V4/V5 < 0.15 suggested that rpl32, tif, rps18, and ubc were a suitable combination of reference genes for RT-qPCR analyses in honey bee treated with dsRNA (Figure 4).
FIGURE 4.
Determination of the optimal number of reference genes for normalization based on the pairwise variation values calculated by geNorm. The value of Vn/Vn + 1 < 0.15 means n should be the optimal number of reference genes selected for RT-qPCR analysis among IAPV-infected (black), CBPV-infected (green), and CSBV-infected (blue) honey bees, as well as honey bees exposed to dsRNA-PGRPSA (purple).
Validation of the Selected Reference Genes
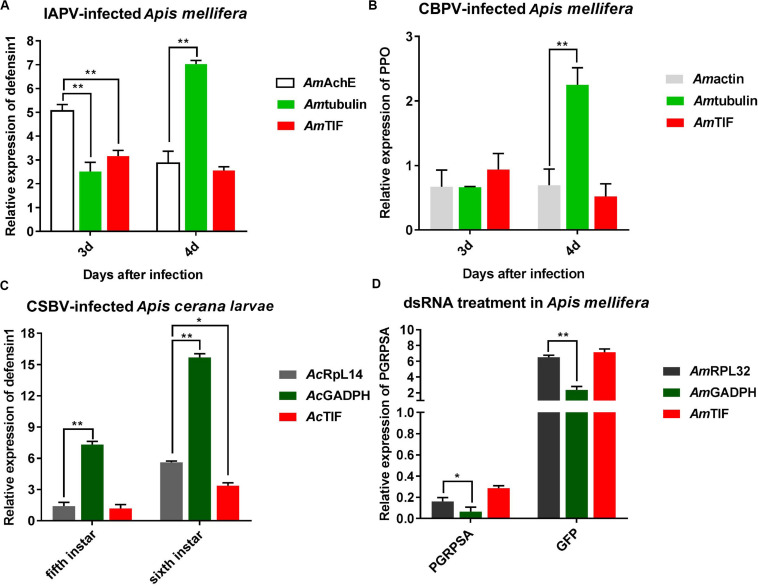
As shown in Figure 5A, the relative expression levels of the defensin 1 and ppo genes varied based on the different reference genes. In the IAPV-infected group, the relative expression levels of defensin 1 were downregulated from 5.1- to 2.9-fold and from 3.2- to 2.6-fold when Amache2 and Amtif were used as reference genes, respectively, while the expression levels of defensin 1 were upregulated from 2.5- to 7-fold if Amα-tubulin was the reference gene. In the CBPV-infected group, the normalization of ppo expression levels to Amactin or Amtif as reference genes exhibited similar downregulated expression trends for both CBPV-infected and uninfected honey bees (Figure 5B). However, the relative expression levels of ppo were upregulated to 2.25-fold at 96 h after CBPV infection when Amα-tubulin was the reference gene (Figure 5B). In the CSBV-infected group, the relative expression levels of defensin 1 were upregulated from 1.4- to 5.6-fold and from 1.2- to 3.4-fold in 5th and 6th CSBV-infected honey bee larvae when we used Acrpl14 and Actif as reference genes, respectively, while the relative expression levels of defensin 1 were upregulated from 7.3- to 15.3-fold when Acgadph used as the reference gene (Figure 5C). In contrast, when Acgadph was used for normalization, the expression levels of defensin 1 exhibited a significant difference from that of Acrpl14 (P < 0.01). In the dsRNA treatment group, the relative expression levels of pgrpsa were downregulated 0. 16-, 0. 06-, and 0.28-fold after 72 h of dsRNA-pgrpsa treatment when Amrpl32 (best), Amgadph (worst) and Amtif (modest) were used as the reference genes (Figure 5D), respectively, while the relative expression levels of pgrpsa were upregulated 6. 5-, 2. 4-, and 7.2-fold at 72 h after dsRNA-gfp treatment if we used Amrpl32, Amgadph and Amtif as the reference genes, respectively (Figure 5D). This result showed that normalization of pgrpsa expression using the best (Amrpl32) and modest (Amtif) reference genes exhibited nearly similar outcomes after dsRNA treatment, though its expression exhibited significant differences (P < 0.05 and P < 0.01) compared to those of Amrpl32 and Amgadph as reference genes (Figure 5D).
FIGURE 5.
Validation of reference gene stability. (A) Relative expression levels of defensin 1 in IAPV-infected honey bee. (B) Relative expression of ppo in CBPV-infected honey bee. (C) Relative expression of defensin 1 in CSBV-infected honey bee larvae. (D) Relative expression of PGRP-SA in honey bees treated with dsRNA-PGRP-SA. Bars represent the means and standard deviations of three biological replicates. Asterisks indicate significant differences. (*P < 0.05 and **P < 0.01).
Discussion
RT-qPCR is an important, simple and practical technique for assessing gene expression when compared to other quantitative methods such as Northern blotting, in situ hybridization and RNA-seq technology (Peng et al., 2018; Adeyinka et al., 2019; He et al., 2019). The application of accurate reference genes is crucial for the quantification of gene expression in A. mellifera and A. cerana. In our study, five algorithms were used to identify the stability of reference genes. Ache2, rps18, actin, tbp, and tif are suitable to normalize the gene expression levels in IAPV-infected A. mellifera. The combination of actin and tif is suitable for CBPV-infected A. mellifera, while the combination of rpl14, tif, rpsa, ubc, and ache2 as well as more reference genes is suitable for CSBV-infected A. cerana larvae. This result suggests that further studies should be performed to select candidate reference genes for CSBV infection. Moreover, rpl14, tif, rps18, ubc, and α-tubulin showed the most stable expression in A. mellifera under dsRNA treatment.
Our results showed that reference genes were different under different viral infection conditions. An increasing number of studies have reported that using a single reference gene to normalize RT-qPCR might be insufficient and inaccurate for the quantification of gene expression (Yang et al., 2018). As shown in our four experiments, the number of suitable reference genes was greater than two (Figure 4). Additionally, the stability of these reference genes was different under different experimental conditions. As determined by RefFinder (Figure 3), tif was a relatively stable reference gene under all of our experimental conditions. Actually, tif have been widely used as housekeeping genes for gene expression analysis during Bombyx mori Cytoplasmic Polyhedrosis Virus (BmCPV) and Bombyx mori Bidensovirus (BmBDV) infection (Guo et al., 2016). Ache2 was the most stable reference gene under IAPV infection, while β-actin was the most stable reference gene under CBPV infection. In contrast, α-tubulin exhibited the worst stability under IAPV and CBPV infection. AcRpl14 and Amrpl32 were the most stable reference genes under CSBV infection and dsRNA treatment, respectively, while Acgadph and Amgadph exhibited the worst stability under CSBV infection and dsRNA treatment, respectively. However, gapdh and rps18 were suggested to be the optimal reference genes for RT-qPCR-based under the labor-specific gene expression, bacterial infected and developmental gene expression in Western honey bee as shown in Table 2 (Scharlaken et al., 2008; Reim et al., 2013; Moon et al., 2018a). Previous studies have shown that some ribosome-associated genes have been used as stable internal reference genes for quantitative analysis (Wang et al., 2019). For instance, Freitas et al. (2019) found that rpl32, rps5, and rps18 were identified as suitable reference genes for the different tissues of stingless bees. Similarly, rpl23 was identified the reliable reference gene in bumblebees challenged by IAPV (Niu et al., 2014; Table 3). Although our study showed that rpl32 displayed stable expression in A. mellifera under dsRNA treatment, it exhibited the worst stability during IAPV and CBPV infection in A. mellifera (Figure 3), while rps18 was the better reference gene in A. mellifera under dsRNA treatment or viral infection (Table 3).
TABLE 3.
Comparison analysis on stability of candidate reference genes in stingless bees, bumblebee, Apis mellifera and Apis cerana.
| Bees | Condition | Most stable reference genes | References |
| Stingless bees (Frieseomelitta | Development | rpl32, rps18 | Freitas et al., 2019 |
| varia, Melipona quadrifasciata, | Sex | rpl32, rps18, rps5, tbp-af | Freitas et al., 2019 |
| and Scaptotrigona bipunctata) | Tissues, organs and body parts | rps18, rps5, tbp-af | Freitas et al., 2019 |
| Bacterial injection | rps18, rpl32, tbp-af | Freitas et al., 2019 | |
| Pesticide exposure | rps5, tbp-af, rpl32, rps18 | Freitas et al., 2019 | |
| Bombus terrestris | IAPV infection | ppai, rpl23, ubi | Niu et al., 2014 |
| Apis mellifera | Abdomens | rpl32, rps18, or gapdh | Moon et al., 2018b |
| Head | rps18, gapdh | Moon et al., 2018a | |
| Bacterial infection | actin, rpS18, gapdh | Scharlaken et al., 2008 | |
| Development | gapdh, rpl32, ef1α-f1 | Reim et al., 2013 | |
| IAPV infection | ache2, rps18, actin, tbp, tif | This study | |
| CBPV infection | actin, tif, tbp, rps18 | This study | |
| dsRNA treatment | rpl32, tif, rps18, ubc, tubulin | This study | |
| Apis cerana | CSBV infection | rpl14, tif, rpsa, ubc, ache2, tubulin | This study |
Elongation factor 1 alpha-f1: ef1α-f1, peptidylprolyl isomerase A: ppia, polyubiquitin:UBI.
Likewise, the stability of the same reference gene varied under different experimental conditions. Although actin is used as an internal control in most gene expression studies during honey bee viral infection, such as IAPV infection (Chen et al., 2014), CSBV infection (Shan et al., 2017), and CBPV infection (Coulon et al., 2017), actin exhibited poorer stability during CSBV infection (Figure 3). Gadph, the other most commonly used reference gene, also showed expression variability from the 2nd to 6th honey bee infected with CSBV infection as well as dsRNA treatment (Figure 3). Similarly, α-tubulin, another commonly used reference gene, exhibited worse stability under IAPV and CBPV infection. This might result due to interactions between α-tubulin and acetylation, as Zhang et al. (2016) found that acetylated α-tubulin will enhance viral titers by promoting fusion with viral inclusion bodies.
This study systematically analyzed 11 reference genes for RT-qPCR under viral infection in A. mellifera and A. cerana. These results will facilitate future studies in honey bee under different viral infection conditions.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
CH and YD conceived the manuscript. YD, HZ, SY, LiZ, and LinZ performed the experiments. YD analyzed the data and wrote the manuscript. CH revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. We thank for the support from the National Natural Science Foundation of China (No. 31572471), the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2019-IAR), the Fundamental Research Funds for CAAS (Y2020PT17), and Young Talent Program of CAAS, GDAS Special Project of Science and Technology Development (2018GDASCX-0107).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01715/full#supplementary-material
Sample collection and validation by RT-PCR. (A) IAPV was injected into newly emerged A. mellifera after 24, 48, 72, and 96 h. (B) CBPV was injected into newly emerged A. mellifera after 0, 24, 48, 72, and 96 h. (C) The larvae naturally infected with CSBV were validated from the 5th to 9th instar. (D) dsRNA-PGRP-SA and dsRNA-GFP were injected into newly emerged A. mellifera and validated at 24, 48, 72, and 96 h.
Primer used for detection common viruses and PGRP-SA.
Primer sequences and amplicon size for dsRNA synthesis of PGRP-SA and GFP.
The standard curve for the quantification RT-qPCR of the reference genes.
References
- Adeyinka O. S., Tabassum B., Nasir I. A., Yousaf I., Husnain T. (2019). Identification and validation of potential reference gene for effective dsrna knockdown analysis in chilo partellus. Sci. Rep. 9:13629. 10.1038/s41598-019-49810-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C. L., Jensen J. L., Ørntof T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Brutscher L. M., Daughenbaugh K. F., Flenniken M. L. (2015). Antiviral defense mechanisms in honey bees. Curr. Opin. Insect. Sci. 7 71–82. 10.1016/j.cois.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. P., Pettis J. S., Corona M., Chen W. P., Li C. J., Spivak M. (2014). Israeli acute paralysis virus: epidemiology, pathogenesis and implications for honey bee health. PLoS Pathog. 10:e1004261. 10.1371/journal.ppat.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon M., Schurr F., Martel A. C., Cougoule N., Dubois E. (2017). Metabolisation of thiamethoxam (a neonicotinoid pesticide) and interaction with the chronic bee paralysis virus in honey bees. Pestic. Biochem. Phys. 144 10–18. 10.1016/j.pestbp.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Diao Q. Y., Li B. B., Zhao H. X., Wu Y. Y., Guo R., Dai P. L., et al. (2018). Enhancement of chronic bee paralysis virus levels in honeybees acute exposed to imidacloprid: a Chinese case study. Sci. Total Environ. 630 487–494. 10.1016/j.scitotenv.2018.02.258 [DOI] [PubMed] [Google Scholar]
- Dietrich I., Shi X. H., McFarlane M., Watson M., Blomström A. L., Skelton J. K., et al. (2017). The antiviral RNAi response in vector and non-vector cells against orthobunyaviruses. PLoS Neglect. Trop. D. 1:5 10.1371/journal pntd.ooo5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas F. C. P., Depintor T. S., Agostini L. T., Luna-Lucena D., Nunes F. M., Bitondi M. M. G., et al. (2019). Evaluation of reference genes for gene expression analysis by real-time quantitative pcr (qpcr) in three stingless bee species (hymenoptera: apidae: meliponini). Sci Rep. 9:17692. 10.1038/s41598-019-53544-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D. A., Yang X., Ni O. E. L., Yi S., Grozinger C., Schneider D. S. (2015). Parallel epigenomic and transcriptomic responses to viral infection in honey bees (apis mellifera). PLoS Pathog 11:e1004713. 10.1371/journal.ppat.1004713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli V., Borowski J. M., Perin E. C., Messias R., da S., Labonde J., et al. (2015). Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 554 205–214. 10.1016/j.gene.2014.10.049 [DOI] [PubMed] [Google Scholar]
- Genersch E., Aubert M. (2010). Emerging and re-emerging viruses of the honey bee (apis mellifera l.). Vet Res. 41:54. 10.1051/vetres/2010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Jiang L., Xia Q. (2016). Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkwormbombyx mori. Mol Genet Genomics. 291 999–1004. 10.1007/s00438-015-1125-4 [DOI] [PubMed] [Google Scholar]
- He S., An T. A. R., Liu S. (2019). Validation of reliable reference genes for RT-qPCR Studies of Target gene expression in Colletotrichum camelliae During spore germination and mycelial growth and interaction with host plants. Front. Microbiol. 10:2055. 10.3389/fmicb.2019.02055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt G. J., Amdam G. V., Schlipalius D., Emore C., Sardesai N., Williams C. E., et al. (2007). Behavioral genomics of honey bee foraging and nest defense. Naturwissenschaften 94 247–267. 10.1007/s00114-006-0183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ming L., Dongliang F., Qingyun D., Jian W., Liqin L., et al. (2018). Preparation and application of egg yolk antibodies against chinese sacbrood virus infection. Front. Microbiol. 9:1814. 10.3389/fmicb.2018.01814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco A. P., Mackert A., Cristino A. D. S., Simoes Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39 372–385. 10.1051/apido:2008015 [DOI] [Google Scholar]
- Maori E., Lavi S., Mozes-Koch R., Gantman Y., Peretz Y., Edelbaum O., et al. (2007). Isolation and characterization of israeli acute paralysis virus, a dicistrovirus affecting honey bees in israel: evidence for diversity due to intra- and inter-species recombination. J. Gen. Virol. 88 3428–3438. 10.1099/vir.0.83284-0 [DOI] [PubMed] [Google Scholar]
- Marcial-Quino J., Fierro F., De la Mora-De la Mora I., Enríquez-Flores S., Gómez-Manzo S., Vanoye-Carlo A., et al. (2016). Validation of housekeeping genes as an internal control for gene expression studies in Giardia lamblia using quantitative real-time PCR. Gene 581 21–30. 10.1016/j.gene.2016.01.018 [DOI] [PubMed] [Google Scholar]
- Moon K., Lee S. H., Kim Y. H. (2018a). Validation of quantitative real-time pcr reference genes for the determination of seasonal and labor-specific gene expression profiles in the head of western honey bee, apis mellifera. PLoS One 13:e0200369. 10.1371/journal.pone.0200369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K., Lee S. H., Kim Y. H. (2018b). Evaluation of reference genes for quantitative real-time pcr to investigate seasonal and labor-specific expression profiles of the honey bee abdomen. J. Asia Pac Entomol. 21 1350–1358. 10.1016/j.aspen.2018.10.014 [DOI] [Google Scholar]
- Niu J. Z., Cappelle K., Miranda J. R., Smagghe G., Ivan M. (2014). Analysis of reference gene stability after Israeli acute paralysis virus infection in bumblebees Bombus terrestris. J. Invertebr. Pathol. 115 76–79. 10.1016/j.jip.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Olivier V., Blanchard P., Chaouch S., Lallemand P., Schurr F., Celle O., et al. (2008). Molecular characterisation and phylogenetic analysis of chronic bee paralysis virus, a honey bee virus. Virus Res. 132 1–68. 10.1016/j.virusres.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Peng S., Liu L., Zhao H., Wang H., Li H. (2018). Selection and validation of reference genes for quantitative real-time PCR normalization under ethanol stress conditions in Oenococcus oeni SD-2a. Front. Microbiol. 9:892. 10.3389/fmicb.2018.00892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper-excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Reim T., Thamm M., Rolke D., Blenau W., Scheiner R. (2013). Suitability of three common reference genes for quantitative real-time pcr in honey bees. Apidologie 44 342–350. 10.1007/s13592-012-0184-3 [DOI] [Google Scholar]
- Scharlaken B., de Graaf D. C., Goossens K., Brunain M., Peelman L. J., Jacobs F. J. (2008). Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honey bee, Apis mellifera, after a bacterial challenge. J. Insect. Sci. 8:33 10.1673/031.008.3301 [DOI] [Google Scholar]
- Shakeel M., Alicia R., Bin T. U., Fengliang J. (2018). Gene expression studies of reference genes for quantitative real-time pcr: an overview in insects. Biotechnol Lett. 40 227–236. 10.1007/s10529-017-2465-4 [DOI] [PubMed] [Google Scholar]
- Shan L., Liuhao W., Jun G., Yujie T., Yanping C., Jie W., et al. (2017). Chinese sacbrood virus infection in asian honey bees, (apis cerana cerana), and host immune responses to the virus infection. J. Invertebr. Pathol. 150 63–69. 10.1016/j.jip.2017.09.006 [DOI] [PubMed] [Google Scholar]
- Silver N., Best S., Jiang J., Thein S. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time pcr. BMC Mol. Biol. 7:33. 10.1186/1471-2199-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O., Elmoualij B., Heinen E., Zorzi W. (2009). A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol. Adv. 27 323–333. 10.1016/j.biotechadv.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas M., Raza A., Yousaf Ali M., Ashraf M. A., Mansoor S., Shahid A. A., et al. (2017). Knock down of whitefly gut gene expression and mortality by orally delivered gut gene-specific dsRNAs. PLoS One 1:e0168921. 10.1371/journal.pone.0168921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Meng Q., Zhu X., Sun S., Gao S., Gou Y., et al. (2019). Evaluation and validation of reference genes for quantitative real-time PCR in Helopeltis theivora Waterhouse (Hemiptera: Miridae). Sci. Rep. 9:13291. 10.1038/s41598-019-49479-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. E., Buck N., Evans J. D. (2006). Expression of insulin pathway genes during the period of caste determination in the honey bee, apis mellifera. Insect. Mol. Biol. 15 597–602. 10.1111/j.1365-2583.2006.00681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., Zhang B. (2012). Mirdeepfinder: a mirna analysis tool for deep sequencing of plant small rnas. Plant Mol. Biol. 80 75–84. 10.1007/s11103-012-9885-2 [DOI] [PubMed] [Google Scholar]
- Yang D. H., Zhao H. X., Shi J. M., Xu X., Wu Y. Y., Guo R., et al. (2019). Discovery of aphid lethal paralysis virus in vespa velutina and apis cerana in china. Insects 10:157. 10.3390/insects10060157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Pan H., Yuan L., Zhou X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. 10.1038/s41598-018-20612-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Jiang Y., Cheng Q., Yi Z., Chen M. (2016). Inclusion body fusion of human parainfluenza virus type 3 regulated by acetylated α-α-tubulin enhances viral replication. J. Virol. 91:e1802-16. 10.1128/JVI.01802-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Shan J., Li R., Liang P., Gao X. (2019). Identification and RNAi-based function analysis of chitinase family genes in diamondback moth, plutella xylostella. Pest Manag. Sci. 75, 1951–1961. 10.1002/ps.5308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample collection and validation by RT-PCR. (A) IAPV was injected into newly emerged A. mellifera after 24, 48, 72, and 96 h. (B) CBPV was injected into newly emerged A. mellifera after 0, 24, 48, 72, and 96 h. (C) The larvae naturally infected with CSBV were validated from the 5th to 9th instar. (D) dsRNA-PGRP-SA and dsRNA-GFP were injected into newly emerged A. mellifera and validated at 24, 48, 72, and 96 h.
Primer used for detection common viruses and PGRP-SA.
Primer sequences and amplicon size for dsRNA synthesis of PGRP-SA and GFP.
The standard curve for the quantification RT-qPCR of the reference genes.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.