Abstract
The aim of the present study was to elucidate the genetic features of early-onset colorectal cancer (CRC), particularly the genetic mutations that may be regarded as prognostic and/or predictive markers in CRC and other malignancies. In total, 40 patients with non-polyposis CRC aged 35 or younger were selected. The formalin-fixed, paraffin-embedded tumors acquired were subjected to mismatch repair (MMR) protein immunochemical staining and gene analysis with next-generation sequencing (44 exons, 17 genes; Ion Torrent Sequencing Platform). A total of 11 (27.5%) tumors presented with MMR protein deficiency (dMMR) and 26 (65%) tumors harbored one or more genetic mutations, including K-RAS proto-oncogene (35%), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA; 20%), B-Raf proto-oncogene (5%), erb-b2 receptor tyrosine kinase 2 (5%), discoidin domain receptor tyrosine kinase 2 (5%), N-RAS proto-oncogene (2.5%), KIT proto-oncogene (2.5%), TSC complex subunit 1 (2.5%), DNA methyltransferase 3 alpha (2.5%) and ABL proto-oncogene 1 (2.5%). Of the dMMR tumors, 81.8% (9/11) of cases presented with mutations in the tested genes, while only 58.6% (17/29) of the MMR-proficient (pMMR) tumors presented with these (P=0.158). PI3KCA was frequently mutated in dMMR tumors compared to pMMR tumors (P=0.025). In a subgroup with a family history of CRC, the dMMR status (P<0.001) and PIK3CA genetic mutation status (P=0.01) were more frequently observed compared to the other two groups (with a family history of other cancer types or no malignancy). Almost all patients who had relatives with CRC presented with both dMMR and other genetic mutations, while this was not observed in the patients who had relatives with other types of carcinoma. Certain genetic mutations that are rarely reported in CRC were only identified in those patients with a family history of carcinoma. In conclusion, non-polyposis CRC in young adults presents as a distinct entity with a unique set of genetic features. However, investigation of more cases in further studies is required to verify the present results.
Keywords: early-onset colorectal cancer, biomarker, genetic mutation, mismatch repair, PI3KCA, next-generation sequencing
Introduction
Colorectal cancer (CRC) is the third most prominent type of cancer worldwide, which is commonly diagnosed in patients above the age of 50 years (1–3). The incidence of CRC among adolescents and young adults has exhibited an increase over the past decades (1–3), particularly in patients aged 20–39 years (4,5). Early-onset CRC usually presents at an advanced stage at the time of diagnosis with a more aggressive biological behavior, which is unique to this subset of CRC (6–8). A study indicated that the prevalence of hereditary cancer syndromes in CRC patients aged 35 years or younger was ~35% (7). This group of patients (aged 35 years or younger) has comparatively more concerns in various aspects that require to be addressed (including the impact on fertility) (9), which raises a compelling need for the identification of prognostic markers and optimization of treatment strategies. However, studies focusing on the genetic features of early-onset CRC in patients aged 35 or younger are limited (7).
The carcinogenesis of CRC is described by a genetic model of cancer comprising the sequential accumulation of genetic alterations. There are two distinct genetic pathways: The adenomatous polyposis coli (APC)/β-catenin pathway, which exhibits sequential alterations of genes including APC, K-RAS proto-oncogene (KRAS) and tumor protein 53 (TP53), and the microsatellite instability (MSI) pathway, which comprises a deficiency in DNA mismatch repair (MMR) genes (10,11). In addition, the methylation of CpG islands, an epigenetic alteration, has been recognized as an early event involved in the development of CRC (12). The genetic alterations of non-polyposis CRC in younger patients manifest as Lynch syndrome (LS), which exhibits germline mutations in MMR genes; and sporadic CRC, which presents as more complex and diverse genetic alterations (13).
Certain genetic test panels have been reported to be a useful tool with which to identify multiple genetic mutations (14). However, the genetic alterations that were identified from these pathways or panels have limited clinical utility at present. The major molecular alterations validated as significant markers are high levels of MSI/defective MMR (MSI-H/dmmr; good prognosis, insensitive to 5-fluorouracil chemotherapy), mutation in the B-Raf proto-oncogene (BRAF) [poor prognosis, resistance to anti-epidermal growth factor receptor (EGFR) antibodies] and mutations in KRAS/N-RAS proto-oncogene (NRAS) genes (resistance to anti-EGFR antibodies) (15,16).
More genes, particularly prognosis- and treatment-associated genes, should be identified in order to enhance the current understanding of early-onset CRC. The identification of these genes may aid in the development of more specific and suitable genetic analyses and may also help to improve management strategies. There are certain therapies that were initially used to target specified molecular alterations in a particular tumor type and have been successfully utilized in other cancer types. A successful example of this is the status regarding anaplastic lymphoma kinase (ALK) gene rearrangement, which was initially identified in anaplastic large-cell lymphomas, and is currently an important indicator for ALK inhibitor treatment in lung cancer (17). It would be of interest to determine whether CRC exhibits certain genetic mutations that are already considered as prognostic and/or predictive markers in other cancer types and also to determine what the frequencies of these genetic mutations are. Thus, after reviewing the literature and searching certain databases (COSMIC, https://cancer.sanger.ac.uk/cosmic and My Cancer Genome, http://www.mycancergenome.org), the present study selected 44 exons of 17 genes, which have been reported to be prognostic and/or predictive markers in CRC and other malignancies (Table SI) for analysis. The aim of the present study was to use next-generation sequencing (NGS) to identify these genetic mutations in a cohort of patients with early-onset non-polyposis CRC. Of note, a set of genetic mutations was identified, which was rarely reported previously, as well as other features, which may indicate that this subset of CRC has unique genetic features.
Materials and methods
Patient selection
A total of 40 patients aged 35 years or younger who had undergone surgery for CRC at West China Hospital (Sichuan University, Chengdu, Sichuan, China) between November 2014 and May 2015 were selected for the present study, which was performed according to protocols approved by the West China Hospital Institutional Review Board (Chengdu, China). They primarily presented with solitary tumors without polyposis or any clinical history of inflammatory bowel disease. Information regarding medical history and family history and pathological data were collected.
Immunohistochemistry (IHC) of MMR protein
Tissue sections (4-µm-thick) were cut from the formalin-fixed, paraffin-embedded (FFPE) tumors. The slides had undergone deparaffinization, rehydration and antigen retrieval, and endogenous peroxidase activity was blocked by incubation with 3% H2O2 at room temperature for 25 min. The sections were then mounted on a DAKO Autostainer Link 48 and then exposed to the ready-to-use mutL homolog 1 (MLH1; cat. no. IR07961), mutS homolog 2 (MSH2; cat. no. IR08561), mutS homolog 6 (MSH6; cat. no. IR08661), PMS1 homolog 2 (PMS2) (cat. no. IR08761) rabbit and mouse monoclonal antibodies (Dako; Agilent Technologies, Inc.) at 37°C for 1 h. The antigen-antibody reaction was visualized with the Envision kit (Dako; Agilent Technologies, Inc). Finally, the slides were counterstained with hematoxylin.
The tumors were considered to be devoid of MLH1, MSH2, MSH6 and PMS2 expression if no nuclear staining of tumor cells was detected. However, tumor cells that were strongly and weakly stained were considered as positive. The surrounding stromal cells, lymphocytes served as an internal positive control (18).
Genomic DNA extraction
The FFPE colorectal tumors obtained during surgery were analyzed and for this, biopsy samples were extracted from them. The sample blocks which harbored tumors that covered >50% of the area were selected by a pathologist (DJ). The genomic DNA was extracted from the FFPE samples using the QIAamp DNA FFPE Tissue kit (Qiagen GmbH) as per the manufacturer's protocol. The DNA concentration was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) and normalized to 20–50 ng/µl. The DNA samples were then stored at −20°C until use.
Ion Torrent Personal Genome Machine (PGM) library preparation and sequencing
Libraries were prepared using the CureSeq PAN-Cancer Panel (ACCB Biotech Ltd.) on the Ion Torrent™ System according to the manufacturer's protocol. The genomic regions of selected genes (Table I) were amplified using pooled primer pairs, followed by ligation with adaptors and barcodes. The hot points and exons that were tested in the 17 genes are listed in Table I. Following purification, the libraries were quantified using a Qubit dsDNA HS Assay kit on a Qubit 2.0 fluorimeter (Life Technologies; Thermo Fisher Scientific, Inc.) diluted to a concentration of 3 ng/ml and pooled in equal volumes. The library pool was clonally amplified in an emulsion PCR reaction using Ion Sphere Particles on the OneTouch 2 instrument (Life Technologies; Thermo Fisher Scientific, Inc.). Subsequently, template-positive ion sphere particles were enriched on the Ion OneTouch ES (Life Technologies; Thermo Fisher Scientific, Inc.). Following enrichment, sequencing primers and polymerase were added from the Ion PGM™ Sequencing Supplies 200 v2 kit (Life Technologies; Thermo Fisher Scientific, Inc.). The mentioned procedures were performed according to the manufacturer's protocol. The libraries were loaded onto an Ion 318 chip (Life Technologies; Thermo Fisher Scientific, Inc.) and sequenced on an Ion Torrent PGM (Life Technologies; Thermo Fisher Scientific, Inc.) instrument to generate the sequencing data.
Table I.
Detected gene panel and mutation sites used in the present study.
| Gene name | Detected mutation sites |
|---|---|
| KRAS | Exons 2 and 3 |
| NRAS | Exons 2 and 3 |
| BRAF | Exons 11 and 15 |
| PIK3CA | Exons 9 and 20 |
| KIT | Exons 9, 11, 13, 14 and 17 |
| PDGFRA | Exons 12, 14 and 18 |
| EGFR | Exons 18, 19, 20 and 21 |
| ERBB2 | Exon 20 |
| DDR2 | Exon 18 |
| ALK | Codon 1196, 1202 and 1206 at exon 23, and codon 1269 at exon 25 |
| RET | Codon 634 at exon 11, codon 918 at exon 16 |
| SMO | Codon 473 at exon 8 |
| TSC1 | Exon 15 |
| FLT3 | Codon 835 at exon 20, exon 14 and 15 |
| NPM1 | Exon 11 |
| DNMT3A | Codon 882 at exon 23, exons 15–22 |
| ABL1 | Codons 253–255 at exon 4, codons 299 and 317 at exon 5, and codons 351–359 at exon 6 |
KRAS, KRAS proto-oncogene, GTPase; NRAS, NRAS proto-oncogene; BRAF, B-Raf proto-oncogene, serine/threonine kinase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; KIT, KIT proto-oncogene, receptor tyrosine kinase; PDGFRA, platelet derived growth factor receptor alpha; EGFR, epidermal growth factor receptor; ERBB2, erb-b2 receptor tyrosine kinase 2; DDR2, discoidin domain receptor tyrosine kinase 2; GTPase; ALK, ALK receptor tyrosine kinase; RET, ret proto-oncogene; SMO, smoothened, frizzled class receptor; TSC1, TSC complex subunit 1; FLT3, fms related receptor tyrosine kinase 3; NPM1, nucleophosmin 1; DNMT3A, DNA methyltransferase 3 alpha; ABL1, ABL proto-oncogene 1.
Variant calling
The generated data were initially processed using the Ion Torrent pipeline software Torrent Suite (Life Technologies; Thermo Fisher Scientific, Inc.) to generate sequence reads, trim adapter sequences and filter and remove poor signal-profile reads. Initial variant calling from the sequencing data was generated using Torrent Suite Software v3.4 with the plug-in ‘variant caller’. In order to eliminate erroneous base calling and generate the final variant calling, the variants in amplicon AMPL339432 (PIK3CA, exon13, ch3: 178938822-178938906) were eliminated as previously described (19). The sequence read distribution across the 67 amplicons generated from the 40 FFPE specimens was normalized to 300,000 reads per sample (Fig. S1).
Somatic mutations and bioinformatic validation
The detected mutations were compared with variants in the 1000 Genomes Project (https://www.internationalgenome.org) and 6500 exomes of National Heart, Lung, and Blood Institute Exome Sequencing Project (https://www.nhlbi.nih.gov), COSMIC database (http://cancer.sanger.ac.uk/cosmic), MyCancerGenome database (https://www.mycancergenome.org) and certain pieces of published literature to assess the mutations in CRC in PubMed database using the gene names as keywords.
Statistical analysis
Categorical variables were examined using the Chi-squared or Fisher's exact test. A two-sided P<0.05 was considered to indicate a statistically significant difference. All the statistical analyses were performed using SPSS software (version 24.0 for Windows; IBM Corp.).
Results
Clinicopathological characteristics
The clinicopathological features of the patients of the present study are summarized in Table II. The cohort consisted of 22 males and 18 females with a mean age of 30 years (range, 18–35 years). The rectum was the most common tumor site, followed by the right colon and the left colon. Half of the patients presented with stage III carcinoma at the time of diagnosis, followed by stage II, stage I and stage IV. In total, 6 patients had poorly differentiated tumors, including 4 cases of signet ring cell carcinoma. In addition, 15 of the patients had relatives within three generations who had been diagnosed with carcinoma and 9 patients had relatives who were diagnosed with CRC.
Table II.
Patient demographics and clinicopathological characteristics.
| Case no. | Age (years) | Gender | Family history (number of relatives with cancer) | Tumor site | Differentiationa | Perineural invasion | AJCC stage |
|---|---|---|---|---|---|---|---|
| C1 | 28 | F | 1, FDR with CRC | Right colon | 2 | No | IIIb |
| C2 | 34 | M | 1, FDR with CRC | Right colon | 2 | No | IIa |
| C3 | 31 | F | 1, FDR with CRC | Right colon | 1 | No | IIa |
| C4 | 26 | F | 1, FDR with CRC | Right colon | 3 | No | IIIb |
| C5 | 31 | F | 1, FDR with CRC | Rectum | 2 | No | IIb |
| C6 | 33 | M | 2, FDR with CRC | Right colon | 1 | No | IIa |
| C7 | 33 | M | 1, SDR with CRC | Rectum | 3 | No | IIIc |
| C8 | 34 | F | 1, SDR with CRC | Rectum | 1 | No | I |
| C9 | 34 | M | 2, SDR with CRC; 1, TDR with CRC | Rectum | 2 | No | IIIc |
| C10 | 34 | M | 1, FDR with other cancer | Right colon | 2 | Yes | IIIb |
| C11 | 33 | F | 1, FDR with other cancer | Right colon | 2 | No | IIIb |
| C12 | 33 | M | 1, FDR with other cancer | Right colon | 2 | No | IIa |
| C13 | 31 | M | 1, FDR with other cancer; 1, SDR with other cancer | Rectum | 2 | No | IVa |
| C14 | 31 | M | 1, SDR with other cancer | Right colon | 2 | No | IIIb |
| C15 | 25 | M | 1, SDR with other cancer | Rectum | 2 | No | I |
| C16 | 27 | M | None | Right colon | 1 | No | IIc |
| C17 | 26 | M | None | Left colon | 2 | No | IIb |
| C18 | 27 | F | None | Rectum | 2 | No | IIIb |
| C19 | 31 | M | None | Rectum | 3 | No | IIIb |
| C20 | 31 | M | None | Rectum | 2 | No | IIIa |
| C21 | 27 | M | None | Rectum | 1 | No | I |
| C22 | 18 | M | None | Right colon | 3 | No | IIIc |
| C23 | 23 | M | None | Right colon | 3 | No | IIIb |
| C24 | 30 | F | None | Left colon | 2 | No | IIIb |
| C25 | 27 | F | None | Rectum | 3 | No | IIa |
| C26 | 33 | F | None | Rectum | 2 | No | IIIb |
| C27 | 29 | M | None | Left colon | 2 | No | IIIc |
| C28 | 28 | M | None | Left colon | 1 | No | IIb |
| C29 | 34 | F | None | Right colon | 2 | No | IIIa |
| C30 | 28 | F | None | Right colon | 2 | No | IIIa |
| C31 | 35 | F | None | Right colon | 2 | No | IIIb |
| C32 | 35 | F | None | Right colon | 2 | No | IIb |
| C33 | 30 | F | None | Rectum | 2 | No | IV |
| C34 | 26 | F | None | Rectum | 2 | Yes | IV |
| C35 | 34 | M | None | Rectum | 1 | No | Ib |
| C36 | 33 | M | None | Left colon | 2 | No | IIIb |
| C37 | 35 | F | None | Right colon | 2 | No | IIIc |
| C38 | 27 | F | None | Rectum | 2 | No | IIb |
| C39 | 32 | M | None | Rectum | 2 | No | IIb |
| C40 | 35 | M | None | Rectum | 2 | No | IIa |
Degree of differentiation: 1, well; 2, intermediate; 3, poor. F, female; M, male; CRC, colorectal cancer; FDR, first-degree relative; SDR, second-degree relative; TDR, third-degree relative; AJCC, American Joint Committee on Cancer.
MMR protein expression
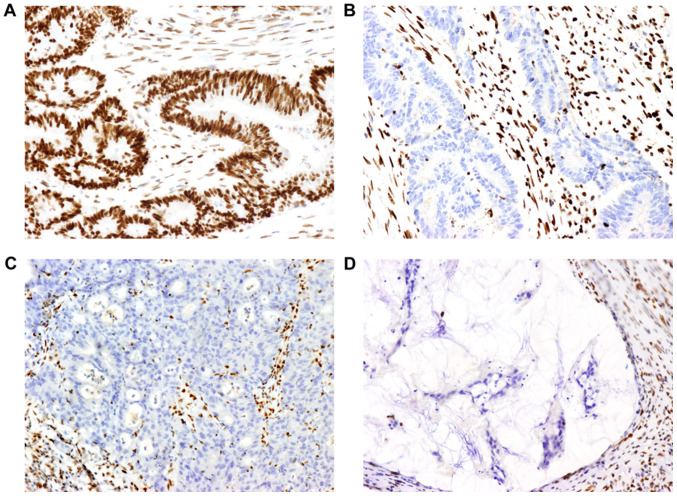
Representative IHC images for MMR are presented in Fig. 1. The results regarding MMR are summarized in Table III. The absence of at least one MMR protein was observed in 11 tumors. No solitary MLH1 absence was observed. Solitary MSH2 or PMS2 absence was observed in three and one of the cases, respectively. Furthermore, five of the tumors presented with both MSH2 and MSH6 absence and two of the tumors exhibited both MLH1 and PMS2 protein absence. In total, 7 out of the 17 cancers that were located in the right colon presented with MMR protein absence and among the 18 rectal cancers, only 4 samples exhibited MMR protein absence. MMR proteins were observed in all of the 5 cases of cancer of the left colon.
Figure 1.
Images of MMR proteins assessed by immunohistochemistry on paraffin-embedded colorectal cancer specimens. (A) Positive expression of MLH1 protein (magnification, ×200). (B, C and D) Lack of expression of (B) MSH2, (C) MSH6 and (D) PMS2 protein (magnification, ×200). Positive expression is defined as the MMR protein being expressed in the tumor cell nucleus. A negative result is defined as the absence of nuclear staining detected in tumor cells, while the surrounding stromal cells and lymphocytes present with staining and serve as an internal positive control. MMR, mismatch repair; MLH1 mutL homolog 1, DNA mismatch repair protein MLH, putative; MSH2, mutS homolog 2; MSH6, mutS homolog 6; PMS2, PMS1 homolog 2, mismatch repair system component.
Table III.
Mismatch repair status and genetic mutations present in this group of early-onset nonpolyposis colorectal cancers.
| A, Family history of CRC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Case no. | MMR expressiona | Gene 1 | Mutation 1 | Gene 2 | Mutation 2 | Gene 3 | Mutation 3 | Gene 4 | Mutation 4 |
| C1 | MSH2(−) | PIK3CA | p.E545G | ||||||
| C2 | MLH1(−), PMS2(−) | KRAS | p.G12D | PIK3CA | p.H1047R | KIT | p.L682fs | DDR2 | p.V797F |
| C3 | MSH2(−), MSH6(−) | KRAS | p.G13D | ||||||
| C4 | MSH2(−), MSH6(−) | ERBB2 | p.V777M | KRAS | p.G12D | ||||
| C5 | MSH2(−), MSH6(−) | PIK3CA | p.H1047R | ||||||
| C6 | MSH2(−) | PIK3CA | p.H1047Y | DNMT3A | p.R882H | DNMT3A | p.A741V | DNMT3A | p.A572G |
| C7 | PMS2(−) | KRAS | p.G13D | BRAF | p.V600E | ||||
| C8 | MSH2(−), MSH6(−) | PIK3CA | p.E545K | ||||||
| C9 | MSH2(−), MSH6(−) | WT | |||||||
| B, Family history of other cancer types | |||||||||
| Case no. | MMR expressiona | Gene 1 | Mutation 1 | ||||||
| C10 | MLH1(−), PMS2(−) | WT | |||||||
| C11 | (+) | WT | |||||||
| C12 | (+) | ERBB2 | p.C805S | ||||||
| C13 | (+) | PIK3CA | p.E545K | ||||||
| C14 | (+) | WT | |||||||
| C15 | (+) | DDR2 | p.V797L | ||||||
| C, No family history of cancer | |||||||||
| Case no. | MMR expressiona | Gene 1 | Mutation 1 | Gene 2 | Mutation 2 | ||||
| C16 | MSH2(−) | KRAS | p.G12D | ||||||
| C17 | (+) | WT | |||||||
| C18 | (+) | WT | |||||||
| C19 | (+) | WT | |||||||
| C20 | (+) | NRAS | p.G12D | ||||||
| C21 | (+) | ABL1 | p.V323F | ||||||
| C22 | (+) | WT | |||||||
| C23 | (+) | WT | |||||||
| C24 | (+) | KRAS | p.G12D | ||||||
| C25 | (+) | KRAS | p.G13D | PIK3CA | p.E545K | ||||
| C26 | (+) | WT | |||||||
| C27 | (+) | BRAF | p.V600E | ||||||
| C28 | (+) | KRAS | p.Q61L | ||||||
| C29 | (+) | KRAS | p.G12V | ||||||
| C30 | (+) | KRAS | p.G12D | ||||||
| C31 | (+) | PIK3CA | p.T1025S | ||||||
| C32 | (+) | WT | |||||||
| C33 | (+) | KRAS | p.G12D | ||||||
| C34 | (+) | WT | |||||||
| C35 | (+) | WT | |||||||
| C36 | (+) | WT | |||||||
| C37 | (+) | KRAS | p.G12V | ||||||
| C38 | (+) | KRAS | p.G12V | ||||||
| C39 | (+) | TSC1 | p.Q654E | ||||||
| C40 | (+) | KRAS | p.G12V | ||||||
(+) indicates that the four tested MMR proteins (MLH1, MSH2, MSH6, PMS2) are positive; protein of negative expression are listed. The order of gene mutation is listed by the read number of each mutation. MMR, mismatch repair; CRC, colorectal cancer; WT, wild-type; KRAS, KRAS proto-oncogene, GTPase; NRAS, NRAS proto-oncogene; BRAF, B-Raf proto-oncogene, serine/threonine kinase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; KIT, KIT proto-oncogene, receptor tyrosine kinase; ERBB2, erb-b2 receptor tyrosine kinase 2; DDR2, discoidin domain receptor tyrosine kinase 2; GTPase; TSC1, TSC complex subunit 1; DNMT3A, DNA methyltransferase 3 alpha; ABL1, ABL proto-oncogene 1.
Genetic mutations detected by NGS
The results regarding genetic mutation are summarized in Table III and Fig. 2. In total, 26 of the tumors (26/40, 65%) carried one or more of the tested genetic mutations. Single mutations were observed in 21 of the tumors, double mutations in 3 tumors and three or more gene mutations in 2 tumors. The most common mutated gene was KRAS, followed by phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), BRAF, erb-b2 receptor tyrosine kinase 2 (ERBB2), discoidin domain receptor tyrosine kinase 2 (DDR2), NRAS, KIT proto-oncogene (KIT), TSC complex subunit 1 (TSC1), DNA methyltransferase 3 alpha (DNMT3A) and ABL proto-oncogene 1 (ABL1). The specific codon data are presented in Table III.
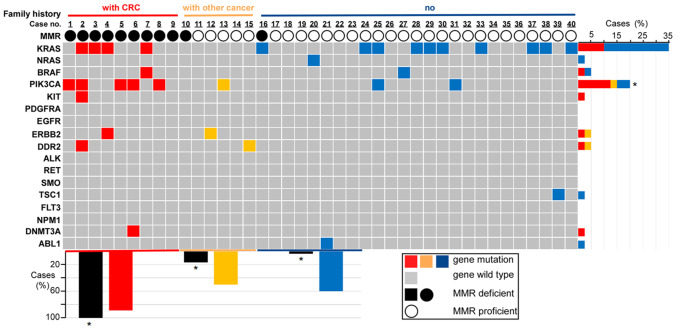
Figure 2.
Family history, MMR status and genetic mutations for each sample. The association between genetic mutation and family history or the MMR status is provided. Family history and MMR status are represented by differently colored bars or circles. For family history (first row in the figure), red represents the cases (cases from 1 to 9) with a family history of CRC, yellow represents the cases (cases from 10 to 15) with a family history of other types of cancer and blue represents the cases (cases from 16 to 40) without any family history of cancer. For the MMR status (third row in the figure), the solid black circle represents the dMMR status (loss of expression of at least one MMR protein), the hollow black circles represent the pMMR status (case exhibits positivity for MLH, MSH2, MSH6 and PMS2 proteins). The grid figure shows the genetic mutations of each sample (grey grids represent the cases without genetic mutations tested in the experiment, while the genetic mutational cases are colored differently according to their family history status) and all the tested genes in the study are listed on the left. The bar chart on the right illustrates the percentages of mutational cases with a different family history (differently colored bars indicate a different family history status) for each gene. For instance, the top bar indicates that 35% of the cases in this group of patients had a KRAS gene mutation, of which 10% of total cases had a family history of CRC (red bar), while the other 25% of total cases had no family history of cancer (blue bar). The lower bar chart illustrates the percentages of dMMR cases (black bars) and the percentages of genetic mutational cases (differently colored bars based on different family history) classified by three different family history statuses (colored differently on the horizontal axis). For instance, for the cases with a family history of CRC (red horizontal axis), 100% showed dMMR (black bar on the left) and 88.9% of these cases (patients with CRC family history) had genetic mutations (red bar). *P<0.05 (patients with CRC family history vs. patients with other cancer history and vs. patients with no family history). CRC, colorectal cancer; MMR, mismatch repair; d/pMMR, MMR deficient/proficient; KRAS, KRAS proto-oncogene, GTPase; NRAS, NRAS proto-oncogene; BRAF, B-Raf proto-oncogene, serine/threonine kinase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; KIT, KIT proto-oncogene, receptor tyrosine kinase; PDGFRA, platelet derived growth factor receptor alpha; EGFR, epidermal growth factor receptor; ERBB2, erb-b2 receptor tyrosine kinase 2; DDR2, discoidin domain receptor tyrosine kinase 2; GTPase; ALK, ALK receptor tyrosine kinase; RET, ret proto-oncogene; SMO, smoothened, frizzled class receptor; TSC1, TSC complex subunit 1; FLT3, fms related receptor tyrosine kinase 3; NPM1, nucleophosmin 1; DNMT3A, DNA methyltransferase 3 alpha; ABL1, ABL proto-oncogene 1.
Correlation between MMR status and genetic mutations
Out of the 11 dMMR tumors, 9 samples (9/11, 81.8%) exhibited genetic mutations in the genes tested in the present study, while only 17 samples of the 29 MMR-proficient (pMMR) tumors exhibited genetic mutations (P=0.158). KRAS/NRAS was mutated in 5 dMMR tumors and in 10 pMMR tumors (P=0.469). PI3KCA was mutated in 5 dMMR tumors and 3 pMMR tumors (P=0.025). KIT and DNMT3A mutations were only detected in dMMR right colon cancers (but not in all dMMR right colon cancers). TSC1 and ABL1 mutations were only observed in pMMR rectal cancers (but not in all pMMR rectal cancers). BRAF, ERBB2 and DDR2 mutations were identified both in dMMR and pMMR cancers (not all cancers). The results are presented in Fig. 2.
MMR status and genetic mutations classified by family history
The results regarding MMR status and genetic mutations classified by family history are illustrated in Fig. 2. All of the 9 patients who had relatives with CRC presented with tumors with dMMR, the prevalence of which differed significantly from the other groups (100 vs. 16.7% of patients with a family history of other cancer types vs. 4% of patients without a family history of cancer; P<0.001). In total, 8 out of the 9 patients with a family history of CRC presented with mutations in genes that were assessed in the present study. The mutation of PIK3CA was more frequently detected in the group with a family history of CRC compared to that in the other two groups (P=0.01), while the other genetic features exhibited no statistically significant differences (Fig. 2).
The coincidence of dMMR and genetic mutations occurring simultaneously was not observed in the 6 patients who had relatives with other types of carcinoma. There were genetic mutations in only 3 samples, while dMMR was observed in another single case separately. ERBB2, DDR2, KIT and DNMT3A mutations were only detected in patients who presented with a family history of carcinoma. For the other 25 patients who did not have any family history of cancer, 15 cases exhibited genetic mutations together with pMMR.
Discussion
CRC is recognized as a highly heterogeneous type of tumor at the molecular level (20). The most important characteristic of young patients with CRC is considered to be the high prevalence of hereditary cancer syndromes (7). However, the results of the present study revealed the genetic features of this subgroup of CRC from a different perspective. The present study focused on the genetic mutations, which are already recognized as prognostic and/or predictive markers in multiple malignancies and various noteworthy results were obtained. These results may aid in the understanding of the genetic features associated with this subset of patients with CRC. Most importantly, these results may serve as a basis to investigate these newly identified mutations in future research.
Early-onset non-polyposis CRC is a distinct entity with unique genetic features. Accumulating data suggest that early-onset CRC may have distinct clinicopathological features, including a more frequent occurrence in the rectum (32-57.7%), manifesting as mucinous (12.6%) or signet ring (10.8%) carcinoma and exhibiting poor differentiation features (20.4%) on histological analysis, and that diagnosis is mostly made at stages III and IV (8,21–23). All of these features were also determined in the present study.
In the cohort of the present study, the frequency of loss of MMR function was 27.5%, which may be close to that of CRC in general (15-20%) (24). The mutation frequencies in the RAS signaling pathway (KRAS, 35%; NRAS, 2.5%; and BRAF, 5%) and the PI3K/AKT pathway (PIK3CA, 20%) were similar to the general CRC series (KRAS, 35%; NRAS, 1–6%; BRAF, 5–15%; and PIK3CA, 10–30%) (25–27). However, certain distinct molecular features were still recognized. First, mutations were identified in certain receptor tyrosine kinases (ERBB, 2.5%; DDR, 2.5%; KIT, 2.5%; and ABL1, 2.5%) which have been rarely studied in CRC. This result indicates that these receptor tyrosine kinases, which participate in several signaling pathways, may be involved in the development of CRC. Furthermore, these tested genes are mutated more frequently in dMMR (81.8%) tumors than in pMMR (58.6%) tumors. This result is in accordance with the observations of a previous study, which suggests that multiple genes, including transforming growth factor beta receptor 2 (TGFBR2; ~90%, TGFB signaling), activin A receptor type 2A (~86%, activin signaling), BAX (~50%, apoptosis) and phosphatase and tensin homolog (~30%, growth regulation) are frequently mutated in MSI-H/dmmr CRC (28). Finally, the association of the MMR status with genetic mutations may be classified based on family history. Patients with a family history of CRC exhibited a higher mutational load in the present study.
Another gene that should be mentioned is DNMT3A, a DNA methyltransferase and an epigenetic modifier. This gene has mutations that may be detected in ~20% of acute myeloid leukemia and is considered to be a marker of poor prognosis (29,30). However, DNMT3A has not been identified in CRC (31,32). In the present study, a tumor sample from a 33-year-old patient exhibited a DNMT3A mutation and the absence of MSH2 protein expression. Of note, both parents of this patient had CRC at the age of 45 and 56 years, respectively. Further research is required on this particular gene, since there may be a noteworthy biological association.
Certain recommendations may be made regarding stratified genetic analysis for early-onset non-polyposis CRC. The genetic features discovered in the present study may aid clinicians in the development of more effective strategies with which to analyse genetic mutations in early-onset CRC. First, this involves testing for more RAS gene mutations. The RAS gene status is crucial for patients who are being considered for anti-EGFR antibody therapy (33). For early-onset CRC, it may be necessary to test for multiple RAS gene mutations, as the mutation may occur at rare locations that render treatment ineffective. This result is consistent with the findings of a previous study, which suggested testing for broad RAS genes (34). Furthermore, it is advisable to provide a more extensive genetic analysis for patients with early-onset CRC with dMMR. It has been observed that dMMR tumors presented with more genetic mutations in comparison to pMMR cancers. In addition, it is advised that more extensive gene testing is considered for patients with a family history of CRC, even in cases in which the MSI-H/dMMR status is fully known. In the present study, almost all patients who were related to individuals with CRC had both dMMR status and genetic mutations. Finally, for patients with no family history of cancer, or those with a family history of other cancer types, it is important to further understand the status of other genes, even though the tumors do not have dMMR status or may not be LS, since 50–60% of the patients presented with other genetic mutations in the present study. On the other hand, this suggests that 40–50% of tumors from these two groups exhibited genetic wild-type mutations based on the testing results from the current 17-gene panel. Thus, a larger number of such cases require to be investigated and other types of genetic alterations, other genes or even epigenetic modifications require to be explored in future research.
There are various challenges and opportunities in treating non-polyposis early-onset CRC. Emerging evidence suggests that early-onset CRC has a more progressive biological behavior and a worse prognosis (6,35). This subset of patients has comparatively more concerns in a number of aspects, including the effects of treatment on fertility (8). These features raise a compelling concern as to the identification of prognostic markers and optimal treatment strategies, which may differ from the older-aged subset of patients.
The present study not only identified certain mutations in various pathways that may be considered as prognostic and predictive markers, but also provided insight into therapy to be identified and developed in future research. Due to the mutations detected in the present study, it is suggested that these receptor tyrosine kinases and certain rarely reported genes should be investigated for non-polyposis early-onset CRC. Furthermore, it is crucial that further research determines whether these targeted agents may also be efficient in early-onset CRC due to the fact that there are established treatment strategies available for these kinases that are normally utilized in other cancer types (36–38). Continued focus on this tumor entity may provide an opportunity to identify more targetable genetic changes that may improve the current understanding and treatment of this disease.
It should be noted that the present study still has certain limitations. One limitation is the relatively small sample size in the present study. Although unique genetic features of patients with CRC aged ≤35 years were identified, only 40 patients were included. However, the incidence of early-onset CRC is relatively low, particularly in patients aged ≤35 years (~1.5% in all CRC patients) (5). Furthermore, only the MMR IHC analysis rather than both MMR and MSI analysis was performed in the present study. However, according to the high consistency (>95%) between MMR IHC and MSI PCR results reported (39), the MMR status may precisely represent the MSI status and vice versa. Another limitation is that a 17-gene panel with testing in hot mutational spots on 44 exons was applied, which do not include intron mutations. Finally, the number of selected genes was limited, while the major goal of the present study was to reveal the genetic mutations that are relevant to treatment and prognosis. The panel used in the present study included genes that are biomarkers for the treatment and prognosis in multiple cancer types and certain genes which have targeted therapeutic agents in clinical practice or other studies. Therefore, further studies using larger sample sizes and investigating more genetic alterations are required in order to clarify the genetic alterations occurring in young patients with CRC.
In conclusion, the present pilot study tested genetic mutations in 40 patients with early-onset non-polyposis CRC using the NGS technique. The results reveal genetic mutations at different frequencies and a possible association between the tested genes with the MMR status or with family history. The present study provides evidence that may aid in the understanding of the genetic features of patients with early-onset CRC and provides insight into potential gene-targeted therapeutic agents for early-onset non-polyposis CRC. However, investigation of a greater number of such cases is required in order to obtain further information and to confirm the present findings.
Supplementary Material
Acknowledgements
The authors would like to thank Dana Philips for her help revising the language of this manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (grant no. 81401990).
Availability of data and materials
All of the analyzed results are included in this article. The original sequencing data were not submitted to a database, as a further study is underway. Datasets used and or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DJ and CS designed the study; CL, YW and LS performed experiments; DJ and CS analyzed and interpreted the data; DJ and CS wrote the manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to participate
This study was approved by the West China Hospital Institutional Review Board, Sichuan University (Chengdu, China; no. K2016031). Informed consent of patients was obtained at the time of tissue collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stigliano V, Sanchez-Mete L, Martayan A, Anti M. Early-onset colorectal cancer: A sporadic or inherited disease? World J Gastroenterol. 2014;20:12420–12430. doi: 10.3748/wjg.v20.i35.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh KE, Taylor TH, Pan CG, Stamos MJ, Zell JA. Colorectal cancer incidence among young adults in California. J Adolesc Young Adult Oncol. 2014;3:176–184. doi: 10.1089/jayao.2014.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey CE, Hu CY, You YN, Bednarski BK, Rodriguez-Bigas MA, Skibber JM, Cantor SB, Chang GJ. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 5.Vuik FE, Nieuwenburg SA, Bardou M, Lansdorp-Vogelaar I, Dinis-Ribeiro M, Bento MJ, Zadnik V, Pellisé M, Esteban L, Kaminski MF, et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut. 2019;68:1820–1826. doi: 10.1136/gutjnl-2018-317592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Barrak J, Gill S. Presentation and outcomes of patients aged 30 years and younger with colorectal cancer: A 20-year retrospective review. Med Oncol. 2011;28:1058–1061. doi: 10.1007/s12032-010-9639-4. [DOI] [PubMed] [Google Scholar]
- 7.Mork ME, You YN, Ying J, Bannon SA, Lynch PM, Rodriguez-Bigas MA, Vilar E. High prevalence of hereditary cancer syndromes in adolescents and young adults with colorectal cancer. J Clin Oncol. 2015;33:3544–3549. doi: 10.1200/JCO.2015.61.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel RL, Jakubowski CD, Fedewa SA, Davis A, Azad NS. Colorectal Cancer in the Young: Epidemiology, prevention, management. Am Soc Clin Oncol Educ Book. 2020;40:1–14. doi: 10.1200/EDBK_279901. [DOI] [PubMed] [Google Scholar]
- 9.Yantiss RK, Goodarzi M, Zhou XK, Rennert H, Pirog EC, Banner BF, Chen YT. Clinical, pathologic, and molecular features of early-onset colorectal carcinoma. Am J Surg Pathol. 2009;33:572–582. doi: 10.1097/PAS.0b013e31818afd6b. [DOI] [PubMed] [Google Scholar]
- 10.Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 12.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–1225 e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peltomaki P, de la Chapelle A. Mutations predisposing to hereditary nonpolyposis colorectal cancer. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/S0065-230X(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 14.Haraldsdottir S, Hampel H, Tomsic J, Frankel WL, Pearlman R, de la Chapelle A, Pritchard CC. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147:1308–1316 e1. doi: 10.1053/j.gastro.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Comprehensive Cancer Network, corp-author. Colon cancer treatment guidelines. https://www.nccn.org/professionals/physician_gls/default.aspx#colon. [Oct 19;2018 ]; [Google Scholar]
- 16.National Comprehensive Cancer Network, corp-author. Rectal cancer treatment guidelines. https://www.nccn.org/professionals/physician_gls/default.aspx#rectal. [Aug 7;2018 ]; [Google Scholar]
- 17.Shaw AT, Engelman JA. ALK in lung cancer: Past, present, and future. J Clin Oncol. 2013;31:1105–1111. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Carbonell L, Ruiz-Ponte C, Guarinos C, Alenda C, Paya A, Brea A, Egoavil CM, Castillejo A, Barbera VM, Bessa X, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012;61:865–872. doi: 10.1136/gutjnl-2011-300041. [DOI] [PubMed] [Google Scholar]
- 19.Xu Z, Huo X, Ye H, Tang C, Nandakumar V, Lou F, Zhang D, Dong H, Sun H, Jiang S, et al. Genetic mutation analysis of human gastric adenocarcinomas using ion torrent sequencing platform. PLoS One. 2014;9:e100442. doi: 10.1371/journal.pone.0100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, Koong AC, Kunz PA, Fisher GA, Ford JM, et al. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: An adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25:1128–1139. doi: 10.1038/modpathol.2012.61. [DOI] [PubMed] [Google Scholar]
- 22.O'Connell JB, Maggard MA, Liu JH, Etzioni DA, Livingston EH, Ko CY. Do young colon cancer patients have worse outcomes? World J Surg. 2004;28:558–562. doi: 10.1007/s00268-004-7306-7. [DOI] [PubMed] [Google Scholar]
- 23.Ballester V, Rashtak S, Boardman L. Clinical and molecular features of young-onset colorectal cancer. World J Gastroenterol. 2016;22:1736–1744. doi: 10.3748/wjg.v22.i5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li SKH, Martin A. Mismatch repair and colon cancer: Mechanisms and therapies explored. Trends Mol Med. 2016;22:274–289. doi: 10.1016/j.molmed.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Wang J, Han X, Yang H, Wang S, Lin D, Shi Y. Effectors of epidermal growth factor receptor pathway: The genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8:e81628. doi: 10.1371/journal.pone.0081628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 28.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 31.Kim MS, Kim YR, Yoo NJ, Lee SH. Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. APMIS. 2013;121:85–94. doi: 10.1111/j.1600-0463.2012.02940.x. [DOI] [PubMed] [Google Scholar]
- 32.Li WL, Xiao MS, Zhang DF, Yu D, Yang RX, Li XY, Yao YG. Mutation and expression analysis of the IDH1, IDH2, DNMT3A, and MYD88 genes in colorectal cancer. Gene. 2014;546:263–270. doi: 10.1016/j.gene.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 33.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 34.Al-Shamsi HO, Alhazzani W, Wolff RA. Extended RAS testing in metastatic colorectal cancer-Refining the predictive molecular biomarkers. J Gastrointest Oncol. 2015;6:314–321. doi: 10.3978/j.issn.2078-6891.2015.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou CL, Chang SC, Lin TC, Chen WS, Jiang JK, Wang HS, Yang SH, Liang WY, Lin JK. Differences in clinicopathological characteristics of colorectal cancer between younger and elderly patients: An analysis of 322 patients from a single institution. Am J Surg. 2011;202:574–582. doi: 10.1016/j.amjsurg.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Bai Y, Kim JY, Watters JM, Fang B, Kinose F, Song L, Koomen JM, Teer JK, Fisher K, Chen YA, et al. Adaptive responses to dasatinib-treated lung squamous cell cancer cells harboring DDR2 mutations. Cancer Res. 2014;74:7217–7228. doi: 10.1158/0008-5472.CAN-14-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 38.Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio G, Pane F, Muller MC, Ernst T, Rosti G, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: Recommendations from an expert panel on behalf of European Leukemia Net. Blood. 2011;118:1208–1215. doi: 10.1182/blood-2010-12-326405. [DOI] [PubMed] [Google Scholar]
- 39.McConechy MK, Talhouk A, Li-Chang HH, Leung S, Huntsman DG, Gilks CB, McAlpine JN. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol Oncol. 2015;137:306–310. doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the analyzed results are included in this article. The original sequencing data were not submitted to a database, as a further study is underway. Datasets used and or analyzed during the current study are available from the corresponding author on reasonable request.