Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the malignancies with the highest morality rate due to postoperative local invasion and distant metastasis. Although C-X-C motif chemokine receptor (CXCR) subunits have been reported as prognostic indicators in gastric cancer, the prognostic value of CXCR subunits in PDAC remains poorly understood. In the present study, the expression levels and biological functions of CXCR subunits were investigated using multiple publicly accessible bioinformatic platforms and databases. Survival analysis was used to evaluate the prognostic value of CXCR subunits in 112 early-stage PDAC cases by setting the median expression levels as the cut-off values. A nomogram was constructed to combine CXCR subunit expression levels and clinical data for prognosis prediction. Moreover, the association between CXCR subunit expression levels and tumor infiltration levels were detected in PDAC. The expression levels of CXCR subunits were elevated in PDAC tumor tissues. In the multivariate Cox proportional risk regression model, high CXCR2, CXCR4 and CXCR6 expression levels in early-stage PDAC were associated with a more favorable prognosis. Further, it was demonstrated that the differential expression levels of CXCR subunits in PDAC for combined survival analysis could contribute to risk stratification. The nomogram model demonstrated the contribution of CXCR subunits and clinical features in the prognosis of PDAC. Gene Set Enrichment Analysis suggested that CXCR subunits serve a role in immunomodulatory functions. The expression levels and somatic copy number alterations of CXCR subunits were associated with tumor infiltration levels in PDAC. CXCR subunits were associated with prognosis in patients with early-stage PDAC and may be potential drug targets for the treatment of pancreatic cancer.
Keywords: PDAC, CXCR, immune infiltration, survival analysis
Introduction
Pancreatic cancer is known to be one of the most lethal malignancies globally. Currently, due to the aggressiveness and lack of effective treatment of pancreatic cancer, it ranks as the fourth leading cause of cancer-associated death in the United States in 2017, with a 5-year survival rate of 6% (1). In China, pancreatic cancer ranks ninth and sixth in cancer morbidity and mortality, respectively, and it is estimated that 91,000 cases were diagnosed in 2015, of which 79,400 died from this disease (2). The major histological type of pancreatic cancer (>95% of cases) is pancreatic ductal adenocarcinoma (PDAC) (3). Since there is a lack of early clinical symptoms and effective biomarkers, most patients are diagnosed at an advanced stage and only 10% of patients are eligible for surgery (4).
The tumor microenvironment serves a key role in tumor self-monitoring and control of malignant transformation. The function of tumor infiltrating immune cells can be mediated by chemokine heterocomplexes of chemokine agonists and specific receptors that can activate multiple chemokine receptors modulating cell recruitment, localization and conversion of cellular invasive components at different tumor stages (5). In inflammatory types of cancer, patients with established T cell tumor infiltration have an improved prognosis and response to immunotherapy compared with patients with non-inflammatory tumors (6–9). High immune cell scores, determined by scoring tumor samples based on the total number of immune cells at the center of the tumor and on the edge of invasion, were significantly associated with an improved prognosis in patients with early-stage PDAC after pancreaticoduodenal resection (10). Lianyuan et al (11) reported that low levels of stromal tumor-infiltrating lymphocytes are a poor indicator of prognosis and liver metastasis in patients with PDAC after surgery (11). Poschke et al (12) analyzed infiltration of T-cells in tumor biopsy of resectable PDAC by immunohistochemistry and reported that adoptive T-cell therapy has significant beneficial therapeutic impact.
Chemokines are a family of chemotactic cytokines that regulate immune cell activation and migration under normal and inflammatory conditions (13). The binding of chemokines to 7 transmembrane binding receptors and multi-level conduction facilitates intracellular delivery of activation signals for cell migration (14). A recent study reported that chemokines and their receptors are involved in tumor growth, tumor cell invasion and distant metastasis (5). C-X-C motif chemokine receptor (CXCR) subunits belong to an important subfamily of chemokines. High expression levels of CXCR subunits were detected in tumor tissues from diverse tumor types, including lung adenocarcinoma, gastric cancer, colorectal cancer and hepatocellular carcinoma (15–18). Another recent study reported the prognostic values of CXCR subunits in gastric cancer (19); however, the potential prognostic values of CXCR subunits in PDAC remains unclear. The present study aimed to examine the potential regulation pathway of CXCR subunits in network enrichment analysis and the prognostic value of CXCR subunits in patients with early-stage PDAC.
Materials and methods
Expression levels of CXCR subunits in tissues and enrichment analyses
The expression levels of CXCR subunits in normal human tissues were analyzed in Genotype-Tissue Expression projects (GTEx) (gtexportal.org/). The comparison of CXCR subunit expression levels in pancreatic adenocarcinoma (PAAD) tumor and non-tumor tissues were performed using Gene Expression Profiling Interactive Analysis (GEPIA; gepia.cancer-pku.cn/) (20), which provides comprehensive analysis of the expression profiling data from The Cancer Genome Atlas (TCGA) and GTEx projects. The enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and annotation of Gene Ontology (GO) terms was performed using Database for Annotation, Visualization, and Integrated Discovery version 6.8 (david.ncifcrf.gov/home.jsp) (21).
In order to determine potential drug-target interaction profiles in CXCR subunits, the interactions between chemical compounds and proteins from the Search Tool for Interactions of Chemicals (STITCH) version 5.0 database (stitch.embl.de/) (22) were analyzed. To investigate the association between CXCR subunit mRNA expression levels, correlation analyses were performed using the Corrplot R package version 0.84 (cran.r-project.org/web/packages/corrplot/).
Survival analysis and prognostic model construction
The data on the expression levels of all CXCR subunits and the relevant clinical parameters were obtained from the UCSC Xena browser (xena.ucsc.edu/). Since the TCGA PAAD cohort contains a variety of histopathological types, inclusion and exclusion criteria of early-stage PDAC samples in TCGA were processed as described in previous studies (23,24). Patients who underwent pancreaticoduodenectomy and whose clinical pathology was diagnosed as American Joint Committee on Cancer 7th stage (25) I or II PDAC were enrolled in the survival analysis. Then, a total of 112 PDAC cases with complete survival data were analyzed in the study. The median expression levels of the CXCR subunit was used as the cut-off for classifying patients into high- and low-expression groups. Estimation of survival distribution and overall survival (OS) and disease-free survival (DFS) times were performed using Kaplan-Meier method and analyzed using the log-rank test. A Cox proportional hazard model was established and used to calculated hazard ratios (HRs) and 95% confidence intervals (CIs) to identify independent prognostic predictors. The association between the prognosis of patients with early-stage PDAC and CXCR subunit expression levels was identified using combined survival analysis.
The survivalROC R package (cran.r-project.org/web/packages/survivalROC/) version 1.0.3 was using to construct time-dependent receiver operating characteristic (ROC) curves and further evaluate the predictive accuracy of CXCR subunits in the clinical outcomes of patients with PDAC. All area under the ROC curve (AUC) values were calculated. Further, a prognostic nomogram model was constructed using the rms (26) R package, according to the selection of prognosis-associated variables in a univariate Cox proportional hazards regression model.
Gene set enrichment analysis (GSEA)
To investigate the potential mechanisms underlying the association of CXCR subunits between high- and low-expression groups and the prognosis of patients with PDAC, c2 (c2.all. version 7.0. symbols) was used for KEGG pathway analysis and c5 (c5.all.v7.0. symbols) was used for GO term analysis of the Molecular Signatures Database for GSEA (software.broadinstitute.org/gsea/index.jsp). P<0.05 was considered to indicate a statistically significant difference.
Comprehensive analysis of tumor-infiltrating immune cells
The correlation between 6 immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells) and CXCR subunit expression levels in patients with PAAD was identified using the Tumor IMmune Estimation Resource (TIMER) (27) and was analyzed using the purity-corrected partial Spearman method. The infiltration level for each somatic copy number alteration category was compared using the normal using two-sided Wilcoxon rank sum test, followed by Bonferroni's correction. The somatic copy number alteration categories were defined using GISTIC version 2.0 (28). Moreover, fractions of tumor-infiltrating immune cells in TCGA patients with PDAC were obtained from supplementary materials of a previously published literature (29). In addition, the sample IDs of all PDAC cases were matched with the sample IDs of the selected samples in the aforementioned steps for further analysis and other non-PDAC samples were excluded.
Statistical analysis
All statistical analyses were performed using SPSS version 24.0 (IBM Corp.) and R version 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/). The correlation of metric data was analyzed using Pearson's correlation coefficient. The comparison of CXCR subunit expression levels in PAAD tumor and non-tumor tissues were performed using a Student's t-test. P<0.05 was considered to indicate a statistically significant difference. All Kaplan-Meier survival curves were constructed using GraphPad Prism version 7.01 (GraphPad Software, Inc.).
Results
Expression levels of CXCR subunits in tumor and normal tissues and enrichment analysis
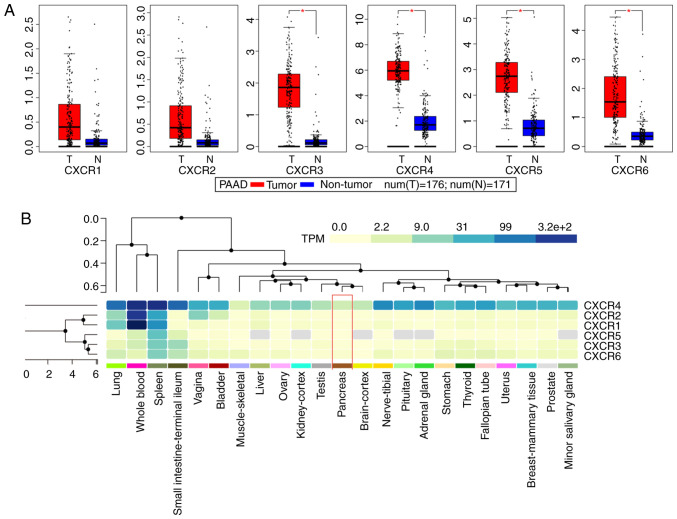
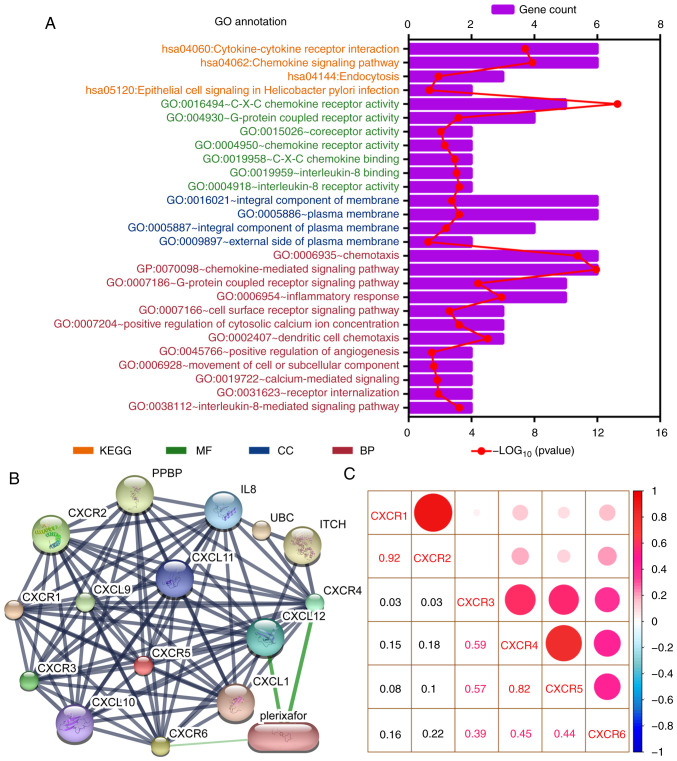
Public databases indicated that the expression levels of CXCR subunits (CXCR3, CXCR4, CXCR5 and CXCR6) were significantly elevated in patients with PAAD compared with healthy patients (Fig. 1A). The heatmap of the GTEx project showed that CXCR4 expression levels were slightly elevated in normal tissues, notably in whole blood and spleen (Fig. 1B). In normal pancreatic tissues, all CXCR subunits displayed a low expression levels. The enrichment analysis of CXCR subunits shows that the subunits may be involved in ‘cytokine-cytokine receptor interaction’, ‘C-X-C chemokine receptor activity’ and the ‘G-protein coupled receptor signaling pathway’ (Fig. 2A). Using the STITCH database, plerixafor was identified as a potential chemical drug that acts on CXCR4, CXCL12 and CXCR6 (Fig. 2B). Furthermore, correlation analysis of each two CXCR subunits expression levels in PDAC presented a relation with a positive trend (Fig. 2C). The aforementioned results indicated that CXCR subunit expression levels are low in normal tissues and highly expressed in pancreatic cancer tissues. These CXCR expression levels were positively correlated with each other in PDAC cancer and para-cancer tissues and could be novel targets for pancreatic cancer treatment.
Figure 1.
Expression analysis of CXCR subunits. (A) Expression levels of CXCR subunits in The Cancer Genome Atlas pancreatic tumor and non-tumor tissues. Comparisons between groups were performed using unpaired Student's t-test. (B) Heatmap analysis of CXCR subunits expression in Genotype-Tissue Expression normal tissues. Red box indicates normal pancreatic tissue expression and the color of the cell indicates the level of expression. *P<0.05. CXCR, C-X-C motif chemokine receptor; T, tumor; N, normal; TPM, transcripts per kilobase million.
Figure 2.
Enrichment and correlation analysis of CXCR subunits. (A) GO term and KEGG pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery. The vertical axis represents the KEGG pathways and GO terms, in which the CXCR subunits are enriched. The height of the histogram in the horizontal axis indicates the number of CXCR subunits enriched and the red dotted line represents the P-value of the enrichment analysis. (B) Drug target prediction using Search Tool for Interactions of Chemicals. Green edges indicate predictive drug association with target. (C) Correlation analysis for CXCR subunits in patients with early-stage pancreatic ductal adenocarcinoma. Red and blue represent positive and negative correlations, respectively. The size and color of the ball indicate the level of correlation coefficient. CXCR, C-X-C motif chemokine receptor; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
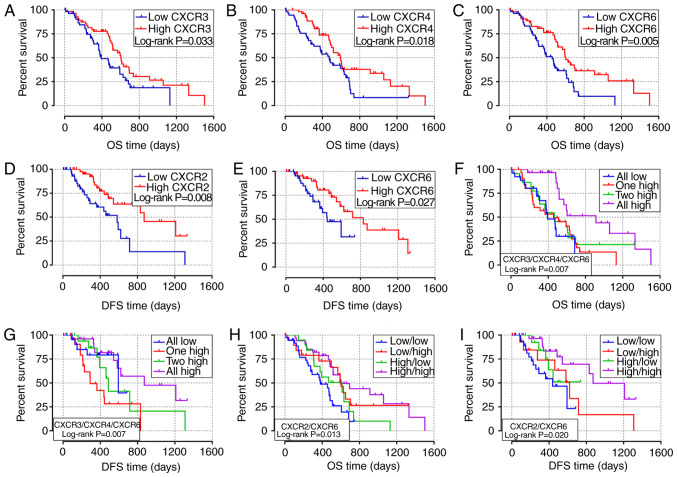
Survival analysis of CXCR subunit expression levels in patients with early-stage PDAC
The association between clinicopathological parameters and the prognosis of patients with early-stage PDAC in TCGA database is shown in Table SI. The survival curves of the association between CXCR subunit expression levels and prognosis of patients with PDAC are displayed in Fig. 3. In the univariate survival analysis, high expression level groups of CXCR3, CXCR4 and CXCR6 were associated with a more favorable OS time in patients with early-stage PDAC (P=0.033, 0.018 and 0.005, respectively; Fig. 3A-C). Compared with low expression levels of the CXCR subunits, PDAC patients with high CXCR3, CXCR4 and CXCR6 expression levels have longer OS times (HR=0.59, 0.56 and 0.49, respectively; Table I). Moreover, low CXCR2 and CXCR6 expression levels in patients with early-stage PDAC results in lower DFS time (P=0.008 and 0.027, respectively; Fig. 3D and E). The median DFS time of low CXCR2 and CXCR6 expression levels was 581 and 443 days, respectively, compared with 872 and 831 days in patients with high CXCR2 and CXCR6 expression levels, respectively (Table I). In the combined survival analysis, it was identified that all high expression levels of CXCR3, CXCR4 and CXCR6 had a significantly reduced risk of death and more favorable OS time in patients with early-stage PDAC (HR=0.29; P=0.007; Fig. 3F; Table II), compared with the combined all low expression levels of CXCR3, CXCR4 and CXCR6. Moreover, the joint effects of CXCR3, CXCR4 and CXCR6 were associated with DFS time of patients with PDAC, DFS time of different CXCR subunits joint expression levels were statistically different (Fig. 3G). In addition, it was demonstrated that combined all high expression levels of CXCR2 and CXCR6 were associated with higher OS and DFS time in patients with PDAC (HROS=0.36, POS=0.002, Fig. 3H; HRDFS=0.25, PDFS=0.003; Fig. 3-I; Table II) compared with all low expression levels of both CXCR2 and CXCR6.
Figure 3.
Survival analysis of different CXCR subunit expression levels in patients with early-stage pancreatic ductal adenocarcinoma. (A-C) Survival curves of OS time for high and low CXCR3, CXCR4 and CXCR6 expression level groups, respectively. (D and E) Survival curves of DFS time for high and low CXCR2 and CXCR6 expression levels groups, respectively. (F and G) Survival curves of OS and DFS times for different combination of CXCR3, CXCR4 and CXCR6 expression level groups. (H and I) Survival curves of OS and DFS for combination of CXCR2 and CXCR6 expression groups. CXCR, C-X-C motif chemokine receptor; OS, overall survival; DFS, disease-free survival.
Table I.
Survival analysis of the CXCR subunit expression levels in patients with early-stage PDAC.
| OS | DFS | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subunit | MST | P-value | HR crude (95% CI) | P-value | Coefficient Ba | HR adjusted (95% CI)b | P-value | MRT | P-value | HR crude (95%CI) | P-value | Coefficient Ba | HR adjusted (95% CI)b | P-value |
| CXCR1 | 0.654 | |||||||||||||
| Low | 511 | Ref. | 0.654 | Ref. | 0.290 | 593 | 0.466 | Ref. | 0.467 | Ref. | 0.124 | |||
| High | 518 | 0.90 (0.55-1.45) | −0.110 | 0.75 (0.44-1.28) | 716 | 0.78 (0.40-1.52) | −0.248 | 0.55 (0.25-1.18) | ||||||
| CXCR2 | 0.056 | |||||||||||||
| Low | 476 | Ref. | 0.058 | Ref. | 0.150 | 581 | 0.006 | Ref. | 0.008 | Ref. | 0.008 | |||
| High | 596 | 0.63 (0.39-1.02) | −0.469 | 0.66 (0.38-1.16) | 872 | 0.39 (0.20-0.78) | −0.939 | 0.32 (0.14-0.74) | ||||||
| CXCR3 | 0.033 | |||||||||||||
| Low | 393 | Ref. | 0.035 | Ref. | 0.071 | 716 | 0.500 | Ref. | 0.501 | Ref. | 0.292 | |||
| High | 603 | 0.59 (0.37-0.97) | −0.522 | 0.61 (0.36-1.04) | 620 | 0.78 (0.39-1.59) | −0.243 | 0.65 (0.30-1.45) | ||||||
| CXCR4 | 0.018 | |||||||||||||
| Low | 473 | Ref. | 0.019 | Ref. | 0.023 | 593 | 0.543 | Ref. | 0.544 | Ref. | 0.692 | |||
| High | 592 | 0.56 (0.34-0.91) | −0.586 | 0.51 (0.28-0.91) | 620 | 0.81 (0.41-1.59) | −0.208 | 0.84 (0.36-1.96) | ||||||
| CXCR5 | 0.231 | |||||||||||||
| Low | 593 | Ref. | 0.233 | Ref. | 0.405 | 461 | 0.057 | Ref. | 0.062 | Ref. | 0.412 | |||
| High | 517 | 0.74 (0.46-1.21) | −0.296 | 0.79 (0.45-1.38) | 872 | 0.50 (0.24-1.04) | −0.694 | 0.71 (0.31-1.62) | ||||||
| CXCR6 | 0.005 | |||||||||||||
| Low | 458 | Ref. | 0.005 | Ref. | 0.025 | 443 | 0.027 | Ref. | 0.030 | Ref. | 0.237 | |||
| High | 603 | 0.49 (0.30-0.80) | −0.715 | 0.53 (0.31-0.92) | 831 | 0.44 (0.21-0.93) | −0.819 | 0.60 (0.26-1.40) | ||||||
B, regression coefficient calculated using univariate Cox proportional hazards regression analysis.
Adjusted for pathological stage T and N stage, histological grade, radical resection and targeted molecular therapy. CXCR, C-X-C motif chemokine receptor; TCGA, The Cancer Genome Atlas; PDAC, pancreatic ductal adenocarcinoma; OS, overall survival time; DFS, disease-free survival time; MST, median survival time; MRT, median disease-free survival time; HR, hazard ratio; CI, confidence interval; Ref., reference.
Table II.
Combined survival analysis of the CXCR subunits in patients with early-stage pancreatic ductal adenocarcinoma.
| OS | DFS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | MST | P-value | HR crude (95% CI) | P-value | HR adjusted (95% CI)a | P-value | MRT | P-value | HR crude (95% CI) | P-value | HR adjusted (95% CI) a | P-value |
| CXCR3+4+6 | 0.007 | 0.007 | ||||||||||
| All low | 393 | Ref. | 0.011 | Ref. | 0.022 | 593 | Ref. | 0.013 | Ref. | 0.018 | ||
| 1 high | 458 | 0.84 (0.44-1.62) | 0.609 | 0.89 (0.43-1.83) | 0.751 | 291 | 3.03 (1.07-8.57) | 0.037 | 3.75 (1.15-12.18) | 0.028 | ||
| 2 high | 467 | 0.72 (0.35-1.46) | 0.360 | 0.69 (0.32-1.49) | 0.346 | 486 | 1.45 (0.46-4.54) | 0.527 | 1.05 (0.30-3.63) | 0.937 | ||
| All high | 913 | 0.29 (0.13-0.64) | 0.002 | 0.25 (0.10-0.65) | 0.004 | 872 | 0.75 (0.24-2.35) | 0.623 | 0.76 (0.19-3.05) | 0.696 | ||
| CXCR2+6 | 0.013 | 0.020 | ||||||||||
| Low/low | 381 | Ref. | 0.017 | Ref. | 0.087 | 439 | Ref. | 0.030 | Ref. | 0.057 | ||
| Low/high | 603 | 0.49 (0.24-0.99) | 0.048 | 0.84 (0.37-1.92) | 0.683 | 620 | 0.62 (0.24-1.61) | 0.324 | 1.38 (0.43-4.37) | 0.589 | ||
| High/low | 614 | 0.66 (0.34-1.28) | 0.219 | 1.08 (0.51-2.27) | 0.842 | NA | 0.52 (0.19-1.48) | 0.222 | 0.46 (0.14-1.51) | 0.199 | ||
| High/high | 596 | 0.36 (0.19-0.69) | 0.002 | 0.44 (0.22-0.91) | 0.026 | 872 | 0.25 (0.10-0.63) | 0.003 | 0.29 (0.10-0.85) | 0.024 | ||
Adjusted for pathological T and N stage, histological grade, radical resection and targeted molecular therapy. CXCR, C-X-C motif chemokine receptor; OS, overall survival time; DFS, disease-free survival time; MST, median survival time; MRT, median disease-free survival time; HR, hazard ratio; CI, confidence interval; NA, not available; Ref., reference.
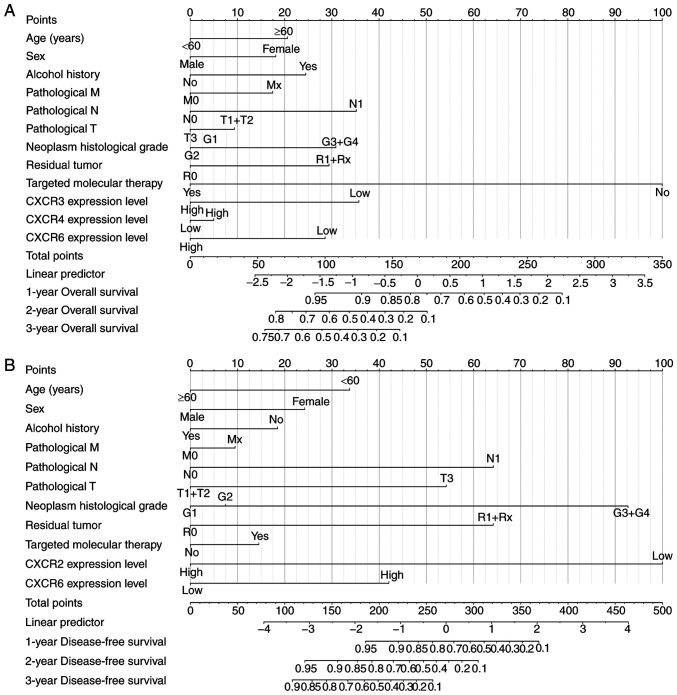
Prognostic model construction
Based on the results of univariate survival analysis, time-independent ROC curves were used to evaluate the prognostic ability of CXCR subunits in patients with PDAC. The AUCs of time-independent ROC curves were 0.283-0.461 in different survival time (Fig. S1A and B). The time-dependent ROC curves are univariate analyses that require a consideration of clinical case information to improve accuracy. Therefore, a prognosis nomogram for OS and DFS time of patients with PDAC was constructed to predict the prognostic risk of different CXCR subunits mRNA levels and clinical characteristics and analyze its contribution to prognosis prediction. In the OS time nomogram model, low CXCR3 and CXCR6 expression levels were adverse factors for long-term OS time (Fig. 4A), which is consistent with the aforementioned results. Moreover, low CXCR2 expression levels, high pathological stage, high neoplasm histological grade and non-radical resection were the primary factors affecting DFS time in patients with PDAC (Fig. 4B). The aforementioned results showed that combined CXCR subunits expression levels and clinical data can predict postoperative clinical outcomes of patients with PDAC for further improvement of treatment plans.
Figure 4.
Nomogram model constructed for CXCR subunits expression level for (A) overall survival and (B) disease-free survival in patients with early-stage pancreatic adenocarcinoma. To determine the value of each variable on the point axis, a vertical line is drawn on the total points axis according to the total value of variables and the predicted survival probability in the time-evaluated line. CXCR, C-X-C motif chemokine receptor.
GSEA report in patients with early-stage PDAC
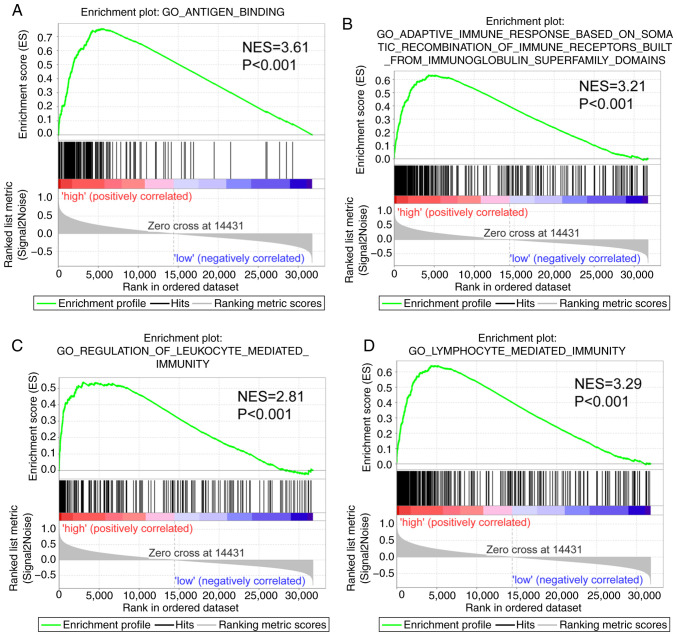
The GSEA report suggested that CXCR subunits in the genome-wide expression profile dataset of the TCGA PDAC cohort serves a role in ‘immune response’ and ‘antigen binding’ (Fig. 5). Thus, CXCR subunits are associated with the cellular immune antigen presentation process for tumor cell recognition. Furthermore, CXCR subunits function in the regulation of the immune cell activation and facilitate the adhesion and differentiation of immune cells (Fig. S2A-E), which may help immune cells to bind, recognize and destroy tumor cells to promote apoptosis.
Figure 5.
Gene set enrichment analysis in patients with early-stage PDAC. Gene Set Enrichment Analysis reports for high C-X-C motif chemokine receptor subunit expression levels using high- vs. low-risk groups. Enrichment plots for: (A) Antigen binding, (B) adaptive immune response based on the somatic recombination of immune receptors built from immunoglobulin superfamily domains, (C) regulation of leukocyte-mediated immunity and (D) lymphocyte-mediated immunity. These panels provide the genome-wide potential pathways and molecular mechanisms in patients with PDAC based on gene expression. NES, normalized enrichment score; PDAC, pancreatic ductal adenocarcinoma; GO, Gene Ontology.
Association of tumor infiltration levels with CXCR subunits expression levels in PDAC
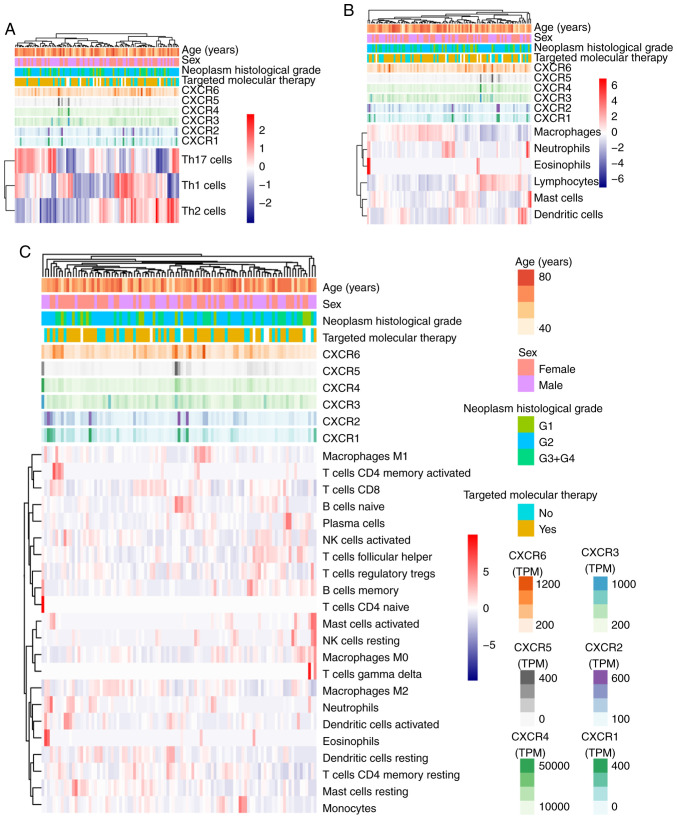
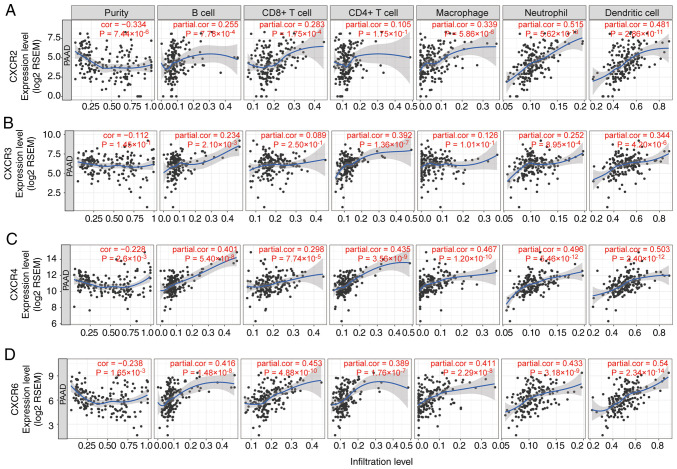
The association between tumor infiltration levels with CXCR subunit expression levels were detected using TIMER and TCGA. Using heatmap clustering analysis, it was demonstrated that Th1, Th2 and TH17 cell fractions were associated with CXCR subunit expression levels, and high CXCR1 and CXCR2 expression levels were associated with less Th1 cell fractions and more Th2 and TH17 cell fractions. High CXCR3, CXCR4 and CXCR5 expression levels were associated with more Th1 cell fractions, less Th2 and TH17 cell fractions (Fig. 6A). Similarly, a number of immune cells in PDAC tumor tissues were associated with CXCR subunits expression levels: High CXCR1 and CXCR2 expression levels were associated with more neutrophils, eosinophils and dendritic cell fractions; high CXCR3, CXCR4 and CXCR5 expression levels were associated with more lymphocyte cell fractions and fewer macrophages cell fractions (Fig. 6B and C). In the purity-corrected partial Spearman's correlation analysis, CXCR subunits (CXCR2, CXCR3, CXCR4 and CXCR6) were positively correlated with tumor infiltration levels of 6 immune infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells, in patients with PDAC, Spearman's correlation coefficient range from 0.234-0.54 (P<0.05, Fig. 7). A comparison of tumor infiltration levels among tumors with different somatic copy number alterations for CXCR subunits also showed a significant correlation, different copy number alterations of CXCR subunits were mainly different in the infiltration levels of CD4+ T cells (P<0.05, Fig. S3).
Figure 6.
Associations between CXCR subunit expression levels, clinical features and immune cell fractions in pancreatic ductal adenocarcinoma tumor tissues. (A) Helper cells. (B) Overall immune cells. (C) Immune cell subsets. Red to blue: High abundance of immune infiltration to low abundance. Various colored side boxes were used to characterize the age, sex, histological grade, targeted molecular therapy and CXCR subunit expression levels. Colors from light to dark indicate that the value of the continuity variable is from low to high. Multiple colors represent two or multiple categorical variables. CXCR, C-X-C motif chemokine receptor.
Figure 7.
Correlation of (A) CXCR2, (B) CXCR3, (C) CXCR4 and (D) CXCR6 expression levels with immune infiltration level in pancreatic cancer. The scatterplots show the purity-corrected partial Spearman's correlation and statistical significance. The abscissa and ordinate represent CXCR subunit expression levels of tumor tissues and tumor infiltration levels of immune cells, respectively. CXCR, C-X-C motif chemokine receptor; PAAD, pancreatic adenocarcinoma; RSEM, RNA-Seq by Expectation-Maximization; cor, correlation.
Discussion
In the present study, the differential expression levels of CXCR subunits in patients with PDAC were identified using data from TCGA. High CXCR3, CXCR4 and CXCR6 expression levels were associated with a favorable OS in patients with early-stage PDAC. In addition, a significant association was found between high CXCR2 and CXCR6 expression levels and favorable DFS in patients with PDAC. The prognostic ability of CXCR subunits was evaluated using time-independent ROC curves. A prediction model for the prognosis of patients with PDAC was then constructed using CXCR subunit expression levels and clinical features. Moreover, the results of GSEA analysis showed that CXCR subunit expression levels functioned in the regulation of immunity in PDAC. Heatmap clustering demonstrated that immune cell fragments in tumors were associated with CXCR expression levels. Expression level and somatic copy number alterations of CXCR subunits (CXCR2, CXCR3, CXCR4 and CXCR6) were positively correlated with tumor infiltration levels in pancreatic cancer. It was predicted that plerixafor was associated with CXCR6, CXCR4 and its ligand CXCL12, using drug target network analysis.
In the present results of tumor infiltration levels, CXCR subunits expression were positively associated with infiltration levels of tumor-infiltrating immune cells in PDAC tissues, indicating that immune cells were mediated by chemokines. In the tumor microenvironment, a variety of cells in the tumor release different chemokines, leading to the recruitment and activation of different immune cells, modulating the balance between tumor-promoting and anti-tumor responses (30). CXCR subunits are expressed to coordinate leukocyte trafficking under both physiological and pathological conditions (31).
Tumor-associated neutrophils serve a role in the early stages of cancer, advanced progression and therapeutic drug resistance (32–34). The inhibition of CXCR2 expression in pancreatic tumors prevents neutrophil accumulation, leading to tumor growth under T cell-dependent suppression. Activated and functionalized T cells infiltrate pancreatic cancer in the absence of neutrophils (35). Ijichi et al (36) found that tumor-stromal interactions via a CXCR2-dependent chemokine can regulate the progression of PDAC. Idorn et al (37) identified that CXCR2, as a candidate for chemokine receptor transduction, can improve the recruitment of transduced tumor ascites lymphocytes toward the ovarian cancer microenvironment. However, the function of CXCR2 may be different in immune cells to regulate the balance between antitumor and protumor responses. A recent study demonstrated that the CXC chemokine-receptor axis was associated with the invasion and migration of PDAC cells and that blockade of this axis prolonged survival and inhibited both tumor angiogenesis and PDAC microinvasion, following CXCR2 knockout in PKF mice (38). Moreover, high expression levels of CXCR2 were associated with a favorable DFS in patients with early-stage PDAC in the present study and the prognostic prediction model indicated that low CXCR2 expression level was a prognostic indicator for less favorable DFS.
A previous study reported that knockdown of CXCR2 diminished the DNA-damage response and CXCR2-binding chemokines reinforced growth arrest in senescent cells (39). In a further study, it was revealed that the overall effect of CXCR2 signaling was involved in the balance between tumor suppression and tumorigenicity, which induced senescence in benign lesions; however, this is reversed in more advanced tumors (40). It was suggested that the effect of CXCR2 signaling was determined by the stage and pathological status of the lesion, as well as its origin and genetic background. Studies performed in animal models require an understanding of the complex contribution of CXCR2 towards tumor progression in cell experiments. Intrinsic causes of the differences in results of CXCR2 towards tumor progression in vitro and in vivo experiments needs further verification to clarify the molecular mechanisms underpinning these differences.
In the present study, high expression levels of CXCR3 were associated with an improved OS in patients with early-stage PDAC. Studies have reported that agonists of CXCR3 promote the recruitment of NK cells, CD4+ and CD8+ T lymphocytes to the tumor microenvironment, where they exert potent antitumor activity (30,41). The present study also identified that CXCR3 expression levels were positively correlated to tumor infiltration levels in PDAC and that CXCR3 expression levels were elevated in tumor tissues, enhancing the antitumor activity of immune cells.
In the previous studies, CXCR4 was upregulated in cancer tissues and extensively expressed in pancreatic cancer cell lines (42,43). In the SDF-1α/CXCR4 axis, the migratory potential and invasion activity of pancreatic cancer cells were modulated by the induction of CXCR4 expression (42). Moreover, CXCR4 and its ligand CXCL12 are involved in the metastasis of different types of tumors, such as breast, prostate, lung and colorectal cancer (43). CXCR4 is important in the classical chemokine receptor response in adults, as well as neutrophil maturation (44). Maréchal et al (45) reported that patients with PAAD and high level of CXCR4 expression in tumor tissues had a shorter OS time compared with those with low CXCR4 expression levels, using immunohistochemistry. The group demonstrated that CXCR4 was expressed in 84.5% of PAAD tumor tissues, but CXCR4 expression levels were undetectable in 6 samples with a low proportion (<30%) of adenocarcinoma cells, indicating that CXCR4 expression levels are different in variable pathological types of PAAD. In contrast, Gebauer et al (46) reported that CXCR4 expression levels evaluated using immunohistochemistry in PAAD specimens were not associated with OS and DFS times. Gebauer et al (46) demonstrated that CXCR4 was expressed in 214 patients with PAAD (86.0%) and >90% patients received adjuvant gemcitabine chemotherapy, whereas Maréchal et al (45) reported only 17% of patients with PAAD to receive adjuvant chemotherapy. In the present study, high expression levels of CXCR4 were detected using RNA-sequencing of tumor tissues and were associated with longer OS in early-stage ductal adenocarcinoma type of PAAD. It was hypothesized that different types of histology and adjuvant therapy may lead to differences in CXCR subunit expression levels. The present results demonstrated that CXCR4 expression levels were positively correlated with tumor infiltration levels of PDAC. Studies have reported that CXCR4 knock-out mice are embryonically lethal due to multiple organ failure, such as hematopoiesis damage (47,48). In addition, the high levels of immune cell infiltration in the microenvironment recruited by the CXCL12/CXCR4 axis is positively correlated with a favorable prognosis in breast, ovarian and cervical cancer (49–51). More studies are required to evaluate the prognostic value of CXCR4 in patients with PDAC.
A positive correlation was also identified in the present study between CXCR6 and immune cell-infiltrate levels in pancreatic cancer. High expression levels of CXCR6 were associated with a favorable OS and DFS in patients with early-stage PDAC. However, a number of studies have been reported where high CXCR6 expression levels indicated a less favorable clinical outcome for patients with gastric cancer and clear cell renal cell carcinoma (52,53). In the present study, it was hypothesized that the prognostic value of CXCR6 was associated with the type and stage of the tumor. Late-stage PDAC is characterized by the matrix portion that forms a thick layer of connective tissue across the epithelial portion of the tumor, functioning as a physical barrier mediating chemotherapy resistance and hindering T-cell migration (54,55). Using the nomogram model to predict the clinical outcomes of patients with PDAC, it was demonstrated that the prognostic values of CXCR6 expression levels were different for OS and DFS. Based on the multivariate and combination survival analysis, high CXCR6 expression levels were associated with long-term OS and DFS of patients with PDAC in the present study. It was hypothesized that the prognostic value of CXCR may be associated with clinical information in the model, which needs further verification.
Using drug target network analysis, it was identified that plerixafor was associated with CXCR6, CXCR4 and its ligand CXCL12. Plerixafor is the only approved drug that targets CXCR4 and has been used in combination therapy clinical trials for other types of cancer, such as prostate and cervical cancer (56,57). Correlation analysis of the present study identified the association between tumor-associated immune cell-infiltrates levels, CXCR subunit expression levels and somatic copy number alterations, indicating that CXCR subunits are potential therapeutic targets for immunotherapy in PDAC. However, this needs further verification using in vitro and in vivo experiments to clarify the levels of tumor-associated immune cell infiltrates in PDAC tissues and associations between immunotherapy drugs and CXCR subunits in PDAC.
There are several limitations of the present study. Firstly, there was the lack of external and experimental validation of the results. Secondly, some important clinical baseline information and the postoperative adjuvant treatments in TCGA database were not available, such as chemotherapy and primary tumor location. Thirdly, the present study evaluated the association between CXCR subunits and tumor infiltration levels; however, no bioinformatic platforms were found for investigating the depth of PDAC cell infiltration. Moreover, an independent cohort is required to verify the constructed prognostic models and larger samples are needed for a more reliable prognostic assessment.
In conclusion, CXCR subunits are associated with infiltration levels of immune cells and the prognosis of patients with early-stage PDAC and these subunits may be potential drug targets for the treatment of pancreatic cancer.
Supplementary Material
Acknowledgements
Not available.
Glossary
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- CXCR
C-X-C motif chemokine receptor
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- STITCH
Search Tool for Interactions of Chemicals
- GTEx
Genotype-Tissue Expression
- PAAD
pancreatic adenocarcinoma
- TCGA
The Cancer Genome Atlas
- OS
overall survival
- DFS
disease-free survival
- HR
hazard ratio
- CI
confidence interval
- ROC
receiver operating characteristic
- AUC
area under curves
- GSEA
Gene Set Enrichment Analysis
- TIMER
Tumor IMmune Estimation Resource
Funding
This work was supported by The National Nature Science Foundation of China (grant. nos. 81160457 and 81460747).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
QW and LL designed the study. CY, QW, LR and JL performed data collection and integration. QW, CY, LR and JL interpreted the data. QW and CY wrote the manuscript. LL critically revised the manuscript and participated in the analysis and interpretation of the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by The Ethics Committee of the First Affiliated Hospital of Guangxi University of Chinese Medicine (Guangxi, China) (approval no. GXTCMU-KY-2-072).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka S. Molecular pathogenesis and targeted therapy of pancreatic cancer. Ann Surg Oncol. 2016;23(Suppl 2):S197–S205. doi: 10.1245/s10434-015-4463-x. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M, Cascinu S, Kleeff J, Labianca R, Löhr JM, Neoptolemos J, Real FX, Van Laethem JL, Heinemann V. Addressing the challenges of pancreatic cancer: Future directions for improving outcomes. Pancreatology. 2015;15:8–18. doi: 10.1016/j.pan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 5.D'Agostino G, Cecchinato V, Uguccioni M. Chemokine heterocomplexes and cancer: A novel chapter to be written in tumor immunity. Front Immunol. 2018;9:2185. doi: 10.3389/fimmu.2018.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss SA, Han SW, Lui K, Tchack J, Shapiro R, Berman R, Zhong J, Krogsgaard M, Osman I, Darvishian F. Immunologic heterogeneity of tumor-infiltrating lymphocyte composition in primary melanoma. Hum Pathol. 2016;57:116–125. doi: 10.1016/j.humpath.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 9.Becht E, Giraldo NA, Dieu-Nosjean MC, Sautès-Fridman C, Fridman WH. Cancer immune contexture and immunotherapy. Curr Opin Immunol. 2016;39:7–13. doi: 10.1016/j.coi.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Tahkola K, Mecklin JP, Wirta EV, Ahtiainen M, Helminen O, Böhm J, Kellokumpu I. High immune cell score predicts improved survival in pancreatic cancer. Virchows Arch. 2018;472:653–665. doi: 10.1007/s00428-018-2297-1. [DOI] [PubMed] [Google Scholar]
- 11.Lianyuan T, Dianrong X, Chunhui Y, Zhaolai M, Bin J. The predictive value and role of stromal tumor-infiltrating lymphocytes in pancreatic ductal adenocarcinoma (PDAC) Cancer Biol Ther. 2018;19:296–305. doi: 10.1080/15384047.2017.1416932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poschke I, Faryna M, Bergmann F, Flossdorf M, Lauenstein C, Hermes J, Hinz U, Hank T, Ehrenberg R, Volkmar M, et al. Identification of a tumor-reactive T-cell repertoire in the immune infiltrate of patients with resectable pancreatic ductal adenocarcinoma. Oncoimmunology. 2016;5:e1240859. doi: 10.1080/2162402X.2016.1240859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins-Green M, Petreaca M, Wang L. Chemokines and their receptors are key players in the orchestra that regulates wound healing. Adv Wound Care (New Rochelle) 2013;2:327–347. doi: 10.1089/wound.2012.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palomino DCT, Marti LC. Chemokines and immunity. Einstein (Sao Paulo) 2015;13:469–473. doi: 10.1590/S1679-45082015RB3438. (In English, Portuguese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saintigny P, Massarelli E, Lin S, Ahn YH, Chen Y, Goswami S, Erez B, O'Reilly MS, Liu D, Lee JJ, et al. CXCR2 expression in tumor cells is a poor prognostic factor and promotes invasion and metastasis in lung adenocarcinoma. Cancer Res. 2013;73:571–582. doi: 10.1158/0008-5472.CAN-12-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Liu H, Shen Z, Wang X, Zhang H, Qin J, Xu J, Sun Y, Qin X. The prognostic value of CXC-chemokine receptor 2 (CXCR2) in gastric cancer patients. BMC Cancer. 2015;15:766. doi: 10.1186/s12885-015-1793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, Zheng L, Li D, Chen G, Gu J, Chen J, Yao Q. CXCR4 overexpression is correlated with poor prognosis in colorectal cancer. Life Sci. 2018;208:333–340. doi: 10.1016/j.lfs.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 18.Gao Q, Zhao YJ, Wang XY, Qiu SJ, Shi YH, Sun J, Yi Y, Shi JY, Shi GM, Ding ZB, et al. CXCR6 upregulation contributes to a proinflammatory tumor microenvironment that drives metastasis and poor patient outcomes in hepatocellular carcinoma. Cancer Res. 2012;72:3546–3556. doi: 10.1158/0008-5472.CAN-11-4032. [DOI] [PubMed] [Google Scholar]
- 19.Yu C, Zhang Y. Characterization of the prognostic values of CXCR family in gastric cancer. Cytokine. 2019;123:154785. doi: 10.1016/j.cyto.2019.154785. [DOI] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Santos A, Von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao X, Huang K, Huang R, Liu X, Han C, Yu L, Yu T, Yang C, Wang X, Peng T. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther. 2017;10:4493–4506. doi: 10.2147/OTT.S142557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C, Yu T, Liu Z, Ye X, Liao X, Wang X, Han C, Zhu G, Qin W, Peng T. Cystatin F as a key family 2 cystatin subunit and prognostic biomarker for early-stage pancreatic ductal adenocarcinoma. Oncol Rep. 2019;42:79–90. doi: 10.3892/or.2019.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 26.Harrell FE., Jr Package ‘rms’: Regression modeling strategies. http://biostat.mc.vanderbilt.edu/wiki/Main/RmS 2016 Package Version 5.1-4. [Google Scholar]
- 27.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014;2:1125–1131. doi: 10.1158/2326-6066.CIR-14-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 32.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: Neutral no more. Nat Rev Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 33.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fridlender ZG, Albelda SM. Tumor-associated neutrophils: Friend or foe? Carcinogenesis. 2012;33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- 35.Chao T, Furth EE, Vonderheide RH. CXCR2-dependent accumulation of tumor-associated neutrophils regulates T-cell immunity in pancreatic ductal adenocarcinoma. Cancer Immunol Res. 2016;4:968–982. doi: 10.1158/2326-6066.CIR-16-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ijichi H, Chytil A, Gorska AE, Aakre ME, Bierie B, Tada M, Mohri D, Miyabayashi K, Asaoka Y, Maeda S, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J Clin Invest. 2011;121:4106–4117. doi: 10.1172/JCI42754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Idorn M, Olsen M, Halldórsdóttir HR, Skadborg SK, Pedersen M, Høgdall C, Høgdall E, Met Ö, Thor Straten P. Improved migration of tumor ascites lymphocytes to ovarian cancer microenvironment by CXCR2 transduction. Oncoimmunology. 2017;7:e1412029. doi: 10.1080/2162402X.2017.1412029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sano M, Ijichi H, Takahashi R, Miyabayashi K, Fujiwara H, Yamada T, Kato H, Nakatsuka T, Tanaka Y, Tateishi K, et al. Blocking CXCLs-CXCR2 axis in tumor-stromal interactions contributes to survival in a mouse model of pancreatic ductal adenocarcinoma through reduced cell invasion/migration and a shift of immune-inflammatory microenvironment. Oncogenesis. 2019;8:8. doi: 10.1038/s41389-018-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 40.Acosta JC, Jesús G. A role for CXCR2 in senescence, but what about in cancer? Cancer Res. 2009;69:2167–2170. doi: 10.1158/0008-5472.CAN-08-3772. [DOI] [PubMed] [Google Scholar]
- 41.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, Galon J. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Wang H, Cai J, Du S, Xin B, Wei W, Zhang T, Shen X. Artemin regulates CXCR4 expression to induce migration and invasion in pancreatic cancer cells through activation of NF-κB signaling. Exp Cell Res. 2018;365:12–23. doi: 10.1016/j.yexcr.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 44.Machado ID, Spatti M, Hastreiter A, Santin JR, Fock RA, Gil CD, Oliani SM, Perretti M, Farsky SH. Annexin A1 is a physiological modulator of neutrophil maturation and recirculation acting on the CXCR4/CXCL12 pathway. J Cell Physiol. 2016;231:2418–2427. doi: 10.1002/jcp.25346. [DOI] [PubMed] [Google Scholar]
- 45.Maréchal R, Demetter P, Nagy N, Berton A, Decaestecker C, Polus M, Closset J, Devière J, Salmon I, Van Laethem JL. High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer. 2009;100:1444–1451. doi: 10.1038/sj.bjc.6605020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gebauer F, Tachezy M, Effenberger K, von Loga K, Zander H, Marx A, Kaifi JT, Sauter G, Izbicki JR, Bockhorn M. Prognostic impact of CXCR4 and CXCR7 expression in pancreatic adenocarcinoma. J Surg Oncol. 2011;104:140–145. doi: 10.1002/jso.21957. [DOI] [PubMed] [Google Scholar]
- 47.Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt M, Böhm D, Von Törne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kölbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 50.Nedergaard BS, Ladekarl M, Nyengaard JR, Nielsen K. A comparative study of the cellular immune response in patients with stage IB cervical squamous cell carcinoma. Low numbers of several immune cell subtypes are strongly associated with relapse of disease within 5 years. Gynecol Oncol. 2008;108:106–111. doi: 10.1016/j.ygyno.2007.08.089. [DOI] [PubMed] [Google Scholar]
- 51.Milne K, Köbel M, Kalloger SE, Barnes RO, Gao D, Gilks CB, Watson PH, Nelson BH. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4:e6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin JJ, Dai FX, Long ZW, Cai H, Liu XW, Zhou Y, Hong Q, Dong QZ, Wang YN, Huang H. CXCR6 predicts poor prognosis in gastric cancer and promotes tumor metastasis through epithelial-mesenchymal transition. Oncol Rep. 2017;37:3279–3286. doi: 10.3892/or.2017.5598. [DOI] [PubMed] [Google Scholar]
- 53.Chang Y, Zhou L, Xu L, Fu Q, Yang Y, Lin Z, Xu J. High expression of CXC chemokine receptor 6 associates with poor prognosis in patients with clear cell renal cell carcinoma. Urol Oncol. 2017;35:675.e17–675.e24. doi: 10.1016/j.urolonc.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 54.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, Lolkema MP, Buchholz M, Olive KP, Gress TM, Tuveson DA. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 55.Hartmann N, Giese NA, Giese T, Poschke I, Offringa R, Werner J, Ryschich E. Prevailing role of contact guidance in intrastromal T-cell trapping in human pancreatic cancer. Clin Cancer Res. 2014;20:3422–3433. doi: 10.1158/1078-0432.CCR-13-2972. [DOI] [PubMed] [Google Scholar]
- 56.Chaudary N, Pintilie M, Jelveh S, Lindsay P, Hill RP, Milosevic M. Plerixafor improves primary tumor response and reduces metastases in cervical cancer treated with radio-chemotherapy. Clin Cancer Res. 2017;23:1242–1249. doi: 10.1158/1078-0432.CCR-16-1730. [DOI] [PubMed] [Google Scholar]
- 57.Conley-LaComb MK, Semaan L, Singareddy R, Li Y, Heath EI, Kim S, Cher ML, Chinni SR. Pharmacological targeting of CXCL12/CXCR4 signaling in prostate cancer bone metastasis. Mol Cancer. 2016;15:68. doi: 10.1186/s12943-016-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.