Abstract
Vitamin D has a potential anticarcinogenic role, possibly through regulation of cell proliferation and differentiation, stimulation of apoptosis, immune modulation and regulation of estrogen receptor levels. Because breast cancer (BC) risk varies among individuals exposed to similar risk factors, we hypothesize that genetic variants in the vitamin D pathway genes are associated with BC risk. To test this hypothesis, we performed a larger meta-analysis using 14 published GWAS datasets in the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) Study. We assessed associations between 2,994 (237 genotyped in the DRIVE study and 2,757 imputed from the 1000 Genomes Project) single nucleotide polymorphisms (SNPs) in 33 vitamin D pathway genes and BC risk. In unconditional logistic regression analysis, we found 11 noteworthy SNPs to be associated with BC risk after multiple comparison correction by the Bayesian false-discovery probability method (<0.80). In stepwise logistic regression analysis, with adjustment for age, principal components and previously published SNPs in the same study populations, we identified three independent SNPs (SNAI1 rs1047920 C>T, AMDHD1 rs11826 C>T and CUBN rs3914238 C>T) to be associated with BC risk (P = 0.0014, 0.0020 and 0.0022, respectively). Additional expression quantitative trait loci analysis revealed that the rs73276407 A allele, in a high LD with the rs1047920 T allele, was associated with decreased SNAI1 mRNA expression levels, while the rs11826 T allele was significantly associated with elevated AMDHD1 mRNA expression levels. Once replicated by other investigators and additional mechanistic studies, these genetic variants may serve as new biomarkers for susceptibility to BC.
Keywords: Breast cancer susceptibility, single nucleotide polymorphism, Vitamin D, expression quantitative trait loci analysis
Introduction
Breast cancer (BC) is the most common malignancy and the second leading cause of cancer deaths in women in the United States [1]. In 2019, there were about 268,600 cases estimated to be diagnosed with BC in the United States, accounting for 15.2% of all new cancer cases, while an estimated 41,760 individuals died from BC, accounting for 6.9% of all cancer deaths [2]. Although the mortality of BC has declined due to early detection and advanced treatments, the incidence rate is still rising each year (https://seer.cancer.gov/statfacts/html/breast.html). Therefore, it is urgent to search for additional risk factors to identify individuals who are at high risk of BC for early screening and prevention that will reduce the incidence and mortality of BC.
There are several risk factors for developing female BC, including aging, unhealthy lifestyle, estrogen status, germ-line mutations and family history [3]. Genetic variation, such as single nucleotide polymorphisms (SNPs) as reported by genome-wide association studies (GWASs) is believed to contribute to BC risk. However, the risk contribution of individual SNPs is considered small [4] as identified in the hypothesis-free GWASs, which have always focused on the most important SNPs with a rigorous P value after the most stringent multiple test correction. Furthermore, few of the GWAS-identified SNPs are found to be functionally annotated. In the post-GWAS era, hypothesis-driven and combined analyses of all published GWASs are performed to identify cancer-risk associated functional SNPs in a combination of pathway analysis, meta-analysis, and functional analysis. With such a hypothesis-driven approach, investigators can focus on SNPs with potential biological functions by using available genotyping data from previously published GWAS datasets with the hope to be able to identify truly cancer-risk associated functional variants.
Vitamin D is a fat-soluble steroid hormone obtained from both dietary sources and sun exposure to ultraviolet B radiation, and once present in the body, it regulates the expression of genes in many types of tissue [5-7]. In addition, vitamin D may have a potential anticarcinogenic role, by regulating cell proliferation and differentiation, apoptosis, immune modulation and estrogen receptor levels [8,9]. Therefore, the vitamin D pathway plays a role in regulating cell growth and immune function, relevant to tumor progression. For example, studies have demonstrated that the vitamin D pathway has an effect on T cell function, monocyte/macrophage differentiation and cytokine production [10-12]. Other studies have found that vitamin D may affect the pathogenesis, prognosis and survival of BC at the cellular level [13,14]. Many epidemiological studies have also attempted to determine associations of vitamin D levels with risk and mortality of various types of cancer [15-17]. In a Brazilian study of postmenopausal BC patients, low vitamin D levels were found to be a risk factor for individuals negative for estrogen receptor with a higher rate of cell proliferation and positive axillary lymph node [18]. However, few studies have comprehensively investigated the effect of genetic variation in vitamin D pathway genes on BC risk.
Considering the importance of the vitamin D pathway in cancer development, we hypothesize that genetic variants in vitamin D pathway genes are associated with BC risk, and we tested this hypothesis in a larger meta-analysis of 53,107 BC case-control study subjects with genotyping datasets from 14 previously published GWAS datasets in the DRIVE study.
Materials and methods
Study subjects
The subjects in this case-control meta-analysis were from 14 out of 17 previously published BC GWASs from the DRIVE study (phs001265.v1.p1), which is different from previously used by others named the DRIVE-Genome-Wide Association meta-analysis (phs001263.v1.p1); and the details of the specific differences between the two studies have been previously described [19]. The DRIVE study we used was one of five projects funded by the NCI’s Genetic Associations and Mechanisms in Oncology (GAME-ON) in 2010. We removed three studies out of the 17 GWASs: one was “Women of African Ancestry Breast Cancer Study (WAABCS)”, because it was on African ancestry study with a relatively small study population, and the other two were “The Sister Study (SISTER)” and “the Two Sister Study (2 SISTER)”, because they had different research designs from others and used cases’ sisters as controls. As a result, all the subjects of European ancestry in 14 GWAS studies were included in the final analysis, including 28,758 BC cases and 24,349 controls, whose characteristics are presented in Supplementary Table 1.
These 14 GWASs included Breast Oncology Galicia Network (BREOGAN); Copenhagen General Population Study (CGPS); Cancer Prevention Study-II Nutrition Cohort (CPSII); European Prospective Investigation Into Cancer and Nutrition (EPIC); Melbourne Collaborative Cohort Study (MCCS); Multiethnic Cohort (MEC); Nashville Breast Health Study (NBHS); Nurses’ Health Study (NHS); Nurses’ Health Study 2 (NHS2); NCI Polish Breast Cancer Study (PBCS); The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO); Study of Epidemiology and Risk factors in Cancer Heredity (SEARCH); Swedish Mammography Cohort (SMC); and Women’s Health Initiative (WHI). Illumina Infinium OncoArray-500k BeadChip genotyping platforms were used for these GWAS datasets, and information on both sex and age at interview was obtained for all the subjects. For the cases, there were three other variables including age at diagnosis, estrogen receptor status, and tumor histology type; for age variables, we used the age at diagnosis for the cases and the age at interview for the controls. A written informed consent was obtained from subjects by each of the original studies that were approved by the Institutional Review Boards of the Participating institutions.
Identification of vitamin D pathway genes and their SNP extraction
We searched for candidate genes in the vitamin D pathway according to the online datasets “PathCards” (http://pathcards.genecards.org/) and the databases of KEGG, BIOCARTA, REACTOME, Canonical pathways and Gene Ontology (GO) in the “Molecular Signatures Database v7.0 (MsigDB)” (http://software.broadinstitute.org/gsea/msigdb/search.jsp) used by the keyword “vitamin D”, and previously published studies (GWASs and vitamin D levels)[20]. In total, we identified 33 genes as candidate genes after excluding some duplicate genes (Supplementary Table 2).
We performed quality control before imputation with the following stringent criteria: (1) the minor allelic frequency (MAF) ≥1%, (2) genotyping success rate ≥95%, (3) missing rate ≤10% and (4) Hardy-Weinberg equilibrium (HWE) P≥1 × 10-6. SNPs located in the 33 candidate genes and their ± 500 kb flanking regions were extracted for imputation with the reference panel from the 1000 Genomes Project data (phase 3) by using IMPUTE2 software [21]. After quality control, SNPs within 2-kb up- and down-stream of genes in the vitamin D pathway were extracted for further analysis, and SNPs for the final meta-analysis with the following criteria: (1) Imputed SNPs with an information score ≥0.80; (2) MAF ≥5%; (3) a genotyping call rate ≥95%; and (4) HWE P≥10-6.
Statistical analysis
Principal components (PCs) analysis was performed for each study separately and their combined dataset by using Genome-wide Complex Trait Analysis (GCTA) [22]. Through univariate logistic regression analysis, the associations between the top 20 PCs and BC risk were evaluated. In further SNP association analysis, significant PCs together with age as covariates were adjusted for in the final models. For each SNP, we calculated odds ratios (ORs) and 95% confidence intervals (CIs) with adjustment for covariates (age and PCs with significant associations) by unconditional logistic regression. We also adjusted for the four SNPs we previously published from the DRIVE Study to identify additional independent SNPs. A meta-analysis was further performed using the results of an additive model of the 14 studies with the inverse variance method. If the Cochran’s Q test P-value >0.1 and I 2≤50%, a fixed-effects model was used; otherwise a random-effects model was employed. The results were first corrected for multiple comparisons by false discovery rate (FDR). Because many SNPs under investigation were in high LD as a result of imputations, the Bayesian false-discovery probability (BFDP) approach was also used in substitution for the false discovery rate, with a cut-off value of 0.8 was used to reduce the probability of false-positive findings for multiple comparisons, as recommended [23]. We applied a prior probability of 0.1 to detect an upper bound of 3.0 for an association with variant genotypes or minor alleles of the SNPs. The number of risk genotypes (NRGs) derived from the independent SNPs as a genetic score was subsequently used to assess the joint effects of these SNPs. Additionally, Haploview v4.2 [24] was employed to construct a Manhattan plot, and LocusZoom [25] was applied to produce regional association plots for significant SNPs. Other statistical analyses, if not specifically mentioned otherwise, were performed with SAS software Version 9.4 (SAS Institute, Cary, NC), R (version 3.5.0) and PLINK (version 1.90).
Functional analysis
Potential functions of independent SNPs were predicted by RegulomeDB (http://www.regulomedb.org/) and HaploReg [26] (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php). In addition, the expression quantitative trait locus (eQTL) analysis was performed to assess the associations between genotypes of independent SNPs and mRNA expression levels of their corresponding genes by using the genotyping and expression data from the lymphoblastoid cell lines of 373 European descendants available in the 1000 Genomes Project [27]. We also used data the Genotype-Tissue Expression (GTEx) Project to assess the correlations between identified SNPs and their corresponding mRNA expression levels in whole blood and breast tissues (https://gtexportal.org/home/) [28].
Results
Single locus analysis
The results of the top 20 PCs are shown in Supplementary Table 3. The workflow of the present analysis is showed in Figure 1. In total, 2,994 SNPs, including 237 genotyped SNPs and 2,757 imputed SNPs, were included in the meta-analysis of the 14 studies, and the distribution of information score for these SNPs is showed in Supplementary Figure 3, of which 304 SNPs were significantly associated with BC risk at P<0.05 in an additive model, and 11 SNPs remained significant after multiple comparison corrections with BFDP <0.8 (Figure 2). To further identify independent SNPs associated with BC risk, stepwise logistic regression analyses were performed to assess the effects of 11 significant SNPs with adjustment for age, significant PCs and four previously published risk-associated SNPs in the DRIVE study. As a result, three independent SNPs (i.e., SNAI1 rs1047920 C>T, AMDHD1 rs11826 C>T and CUBN rs3914238 C>T) remained statistically significantly associated with BC risk (P = 0.0014, 0.0020 and 0.0022, respectively) and used for further analysis (Table 1).
Figure 1.
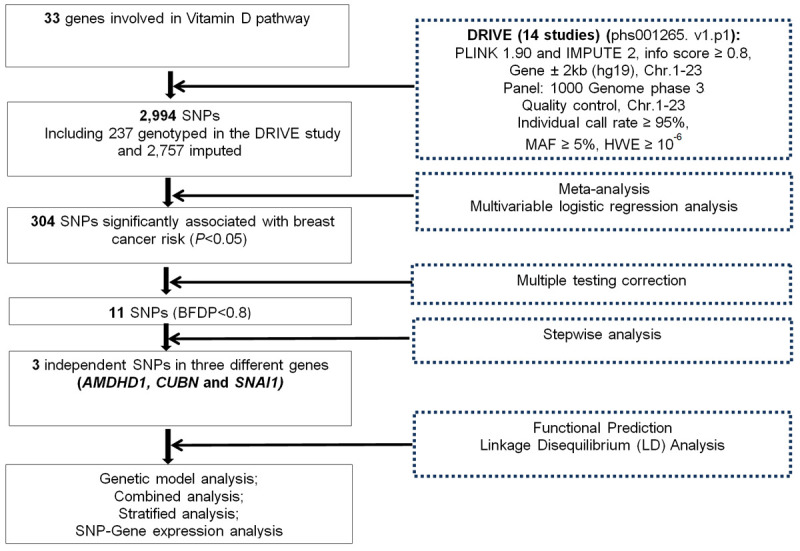
The workflow of present study.
Figure 2.

Manhattan Plot of the 2,994 SNPs of Vitamin D pathway Genes in the DRIVE study. The x-axis represents each chromosome. The y-axis represents the association P values (-log10 transformed) with breast cancer risk. The red horizontal line indicates P value equal to 0.05 and the blue horizontal line represents BFDP value equal to 0.8. Abbreviations: AMDHD1, amidohydrolase domain containing 1; BFDP, Bayesian false-discovery probability; CUBN, cubilin; DRIVE, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; SNAI1, snail family transcriptional repressor 1.
Table 1.
Three genetic variants as independent BC risk predictors obtained from stepwise logistic regression analysis in the DRIVE study
| SNP1 | Location | MAF | Category2 | Frequency | OR (95% CI)1 | P 1 |
|---|---|---|---|---|---|---|
| SNAI1 rs1047920_T | 20q13.13 | 0.07 | CC/CT/TT | 45730/6957/276 | 0.93 (0.88-0.97) | 0.0014 |
| AMDHD1 rs11826_T | 12q23.1 | 0.24 | CC/CT/TT | 30973/18993/2997 | 0.96 (0.93-0.98) | 0.0020 |
| CUBN rs3914238_T | 10p13 | 0.38 | CC/CT/TT | 20752/24693/7518 | 1.04 (1.01-1.07) | 0.0022 |
Stepwise logistic regression analysis included age, PC1, PC3, PC4, PC5, PC6, PC8, PC10, PC11, PC14, PC16, four previously published risk-associated SNPs (rs1323697, rs1264308, rs141308737 and rs1469412) in the same study by Jie Ge (PMID: 31026346) and 11 SNPs (rs3914238, rs1047920, rs7913144, rs7898138, rs11424438, rs2271464, rs77090490, rs4141977, rs11826, rs1800629 and rs1800628).
The most left-hand side “category” was used as the reference.
Note: there were 20 PCs in the combined datasets as listed in Supplementary Table 3, of which ten remained significant and were adjusted in the final stepwise logistic regression analysis. Abbreviations: AMDHD1, amidohydrolase domain containing 1; BC, breast cancer; CI, confidence interval; CUBN, cubilin; DRIVE, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; SNAI1, snail family transcriptional repressor 1.
The locations of these three SNPs and their associations with BC risk are presented in Supplementary Table 4. There was no heterogeneity found among the 14 GWASs for the effects of these three independent SNPs. The forest plots of three independent SNPs by the meta-analysis are presented in Supplementary Figure 1. The results showed that two SNPs were associated with a significantly decreased risk of BC (SNAI1 rs1047920 C>T: OR = 0.92, 95% CI = 0.88-0.97, P = 6.14 × 10-4; and AMDHD1 rs11826 C>T: OR = 0.95, 95% CI = 0.92-0.98, P = 2.77 × 10-4), while the other SNP was associated with a significantly increased BC risk (CUBN rs3914238 C>T: OR = 1.05, 95% CI = 1.02-1.07, P = 3.16 × 10-4). Regional association plots of the three independent SNPs in the 100 kb up- and down-stream regions are summarized in Supplementary Figure 2. As shown in Table 2, the effects of the SNAI1 rs1047920 T, AMDHD1 rs11826 T and CUBN rs3914238 T alleles on BC risk were statistically significant (trend test in univariate analysis: P = 0.001, 0.002 and 0.002, respectively; trend test in multivariate analysis: P = 0.001, 0.002 and 0.003, respectively).
Table 2.
Associations between three independent SNPs in the vitamin D pathway genes and BC risk in the DRIVE study
| Genotype | N Control/N Case | Univariate analysis | Multivariate analysis1 | ||
|---|---|---|---|---|---|
|
|
|
||||
| OR (95% CI) | P | OR (95% CI) | P | ||
| SNAI1 rs1047920 C>T | |||||
| CC | 20787/24943 | 1.00 | 1.00 | ||
| CT | 3324/3633 | 0.91 (0.87-0.96) | 0.0003 | 0.91 (0.87-0.96) | 0.0003 |
| TT | 124/152 | 1.02 (0.81-1.30) | 0.861 | 1.02 (0.80-1.30) | 0.869 |
| Trend test | 0.001 | 0.001 | |||
| CT+TT | 3448/3785 | 0.92 (0.87-0.96) | 0.0004 | 0.91 (0.87-0.96) | 0.0004 |
| AMDHD1 rs11826 C>T | |||||
| CC | 14033/16940 | 1.00 | 1.00 | ||
| CT | 8761/10232 | 0.97 (0.93-1.00) | 0.074 | 0.97 (0.93-1.00) | 0.069 |
| TT | 1441/1556 | 0.89 (0.83-0.96) | 0.004 | 0.89 (0.82-0.96) | 0.002 |
| Trend test | 0.002 | 0.002 | |||
| CT+TT | 10202/11788 | 0.96 (0.93-0.99) | 0.013 | 0.96 (0.92-0.99) | 0.011 |
| CUBN rs3914238 C>T | |||||
| CC | 9676/11076 | 1.00 | 1.00 | ||
| CT | 11183/13510 | 1.06 (1.02-1.10) | 0.004 | 1.05 (1.02-1.09) | 0.006 |
| TT | 3376/4142 | 1.07 (1.02-1.13) | 0.010 | 1.07 (1.01-1.13) | 0.014 |
| Trend test | 0.002 | 0.003 | |||
| CT+TT | 14559/17652 | 1.06 (1.02-1.10) | 0.001 | 1.06 (1.02-1.10) | 0.002 |
| Number of combined risk genotypes2 | |||||
| 0 | 596/628 | 1.00 | 1.00 | ||
| 1 | 5128/5640 | 1.04 (0.93-1.18) | 0.475 | 1.05 (0.93-1.18) | 0.451 |
| 2 | 11284/13489 | 1.14 (1.01-1.27) | 0.031 | 1.14 (1.01-1.28) | 0.028 |
| 3 | 7232/8976 | 1.18 (1.05-1.32) | 0.006 | 1.18 (1.05-1.33) | 0.005 |
| Trend test | <0.0001 | <0.0001 | |||
| 0-2 | 17008/19757 | 1.00 | 1.00 | ||
| 3 | 7232/8976 | 1.07 (1.03-1.11) | 0.0005 | 1.07 (1.03-1.11) | 0.0005 |
Abbreviations: AMDHD1, amidohydrolase domain containing 1; BC: breast cancer; CI: confidence interval; CUBN, cubilin; DRIVE: Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; OR: odds ratio; SNAI1, snail family transcriptional repressor 1; SNP: single nucleotide polymorphism.
Adjusted for age, PC1, PC3, PC4, PC5, PC6, PC8, PC10, PC11, PC14 and PC16.
Risk genotypes were rs1047920 CC, rs11826 CC and rs3914238 CT+TT.
Combined genotype analyses of the three independent SNPs
To assess the joint effect of the three independent SNPs on BC risk, we further combined risk genotypes of SNAI1 rs1047920 CC, AMDHD1 rs11826 CC and CUBN rs3914238 CT+TT into a genetic score as the NRG, which categorized all the individuals into four groups with 0 to 3 risk genotypes. The trend test indicated that the increased NRG was significantly associated with BC risk (P<0.0001, Table 2). According to the effect values and the frequency of each group, we further dichotomized all the individuals into two groups: low-risk (0-2 NRG) and high-risk (3 NRG). We found that the high-risk group had a higher BC risk (in multivariate analysis: OR = 1.07, 95% CI = 1.03-1.11, P = 0.0005, Table 2).
Stratified analysis of combined risk genotypes on BC risk
To further evaluate the interaction between genotypes and age, we stratified the analysis by age. As shown in Table 3, we found that the risk was more evident in the subgroup of age >60 (in univariate analysis: OR = 1.10, 95% CI = 1.04-1.16, P = 0.0005; in multivariate analysis: OR = 1.10, 95% CI = 1.04-1.15, P = 0.0007) compared with that for the subgroup of age ≥60. Further subgroup analysis (ER status and histological type) also revealed that the risk was more evident in the subgroup aged >60 than in the subgroup of age ≥60 among individuals with ER+ and invasive tumor. Both ER status and histological type had any effect on BC risk associated with the genotypes, nor heterogeneity was found between these strata (all P>0.05) (Table 3).
Table 3.
Stratified analysis for associations between the combined risk genotypes and BC in the DRIVE study
| Characteristics | NRG 0-2 | NRG 3 | Univariate analysis | Multivariate analysis1 | Pinter 2 | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| Control | Case | Control | Case | OR (95% CI) | P | OR (95% CI) | P | ||
| Age | |||||||||
| ≤60 | 8866 | 8541 | 3852 | 3872 | 1.04 (0.99-1.10) | 0.120 | 1.04 (0.99-1.10) | 0.156 | 0.163 |
| >60 | 8142 | 11216 | 3380 | 5104 | 1.10 (1.04-1.16) | 0.0005 | 1.10 (1.04-1.15) | 0.0007 | |
| ER+ vs. control | |||||||||
| ≤60 | 8866 | 5270 | 3852 | 2322 | 1.01 (0.95-1.08) | 0.656 | 1.02 (0.96-1.09) | 0.470 | 0.135 |
| >60 | 8142 | 7925 | 3380 | 3570 | 1.09 (1.03-1.15) | 0.005 | 1.09 (1.03-1.15) | 0.004 | |
| ER- vs. control | |||||||||
| ≤60 | 8866 | 1283 | 3852 | 634 | 1.14 (1.03-1.26) | 0.014 | 1.09 (0.98-1.21) | 0.118 | 0.655 |
| >60 | 8142 | 1238 | 3380 | 551 | 1.07 (0.96-1.20) | 0.207 | 1.05 (0.94-1.17) | 0.356 | |
| Invasiveness vs. control | |||||||||
| ≤60 | 8866 | 7620 | 3852 | 3454 | 1.04 (0.99-1.10) | 0.132 | 1.04 (0.99-1.10) | 0.140 | 0.213 |
| >60 | 8142 | 10212 | 3380 | 4634 | 1.09 (1.04-1.15) | 0.001 | 1.09 (1.04-1.15) | 0.001 | |
| In-situ vs. control | |||||||||
| ≤60 | 8866 | 747 | 3852 | 343 | 1.06 (0.93-1.21) | 0.416 | 1.04 (0.91-1.19) | 0.555 | 0.223 |
| >60 | 8142 | 807 | 3380 | 393 | 1.17 (1.03-1.33) | 0.014 | 1.16 (1.02-1.32) | 0.020 | |
Adjusted for PC1, PC3, PC4, PC5, PC6, PC8, PC10, PC11, PC14 and PC16.
Pinter: P value for interaction analysis between age and NRG.
Abbreviations: BC: breast cancer; CI: confidence interval; DRIVE: Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; ER: estrogen receptor; NRG: number of risk genotypes; OR: odds ratio.
LD analysis and functional prediction of the three independent SNPs
Functional prediction by RegulomeDB showed that SNAI1 rs1047920 C>T, AMDHD1 rs11826 C>T and CUBN rs3914238 C>T had RegulomeDB scores of 4, 6 and no data, respectively. We further searched for SNPs in high-LD (r2≥0.7) with these three independent SNPs and made functional prediction by using HaploReg. We found that SNAI1 rs1047920 C>T located in the 3’-UTR may change the motifs of BDP1 and LF-A1, which is also located in DNase I hypersensitive sites and that AMDHD1 rs11826 C>T is located in the 3’-UTR with the selected eQTL for 5 hits, whereas CUBN rs3914238 C>T may change the motifs of Hand1 and PU.1 (Supplementary Table 5). We further assessed potential functions of these three independent SNPs using data from the ENCODE Project. The results revealed that SNAI1 rs1047920 C>T is located at H3K27AC acetylation enriched region (Supplementary Figure 4).
Correlation analysis
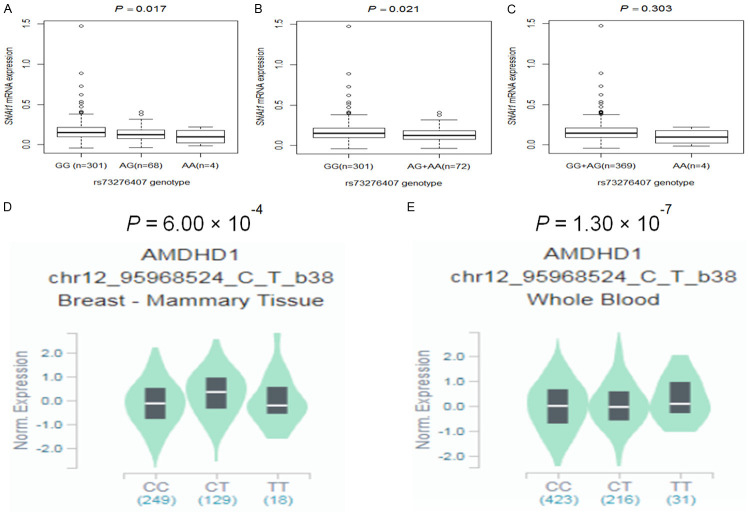
We further performed correlation analysis between genotypes of the three independent SNPs and their corresponding mRNA expression levels in the publically available RNA-seq data of lymphoblastoid cell lines generated from 373 European descendants in the 1000 Genomes Project. We found that the rs1047920 C>T SNP was not significantly associated with levels of mRNA expression of SNAI1 in all additive, dominant and recessive genetic models (P = 0.093, P = 0.102, and P = 0.491, respectively; Supplementary Figure 5A-C) but that another SNP (rs73276407 G>A) with a high LD (r2 = 0.71) of rs1047920 C>T was significantly associated with decreased levels of mRNA expression of SNAI1 in additive and dominant models (P = 0.017 and P = 0.021, respectively; Figure 3A and 3B), but not for the recessive model (P = 0.303, Figure 3C). We also found that rs11826 C>T was not significantly associated with levels of mRNA expression of AMDHD1 in all additive, dominant and recessive genetic models (P = 0.894, P = 0.815, and P = 0.861, respectively; Supplementary Figure 5D-F), while rs3914238 C>T was also not significantly associated with levels of mRNA expression of CUBN in all additive, dominant and recessive genetic models (P = 0.889, P = 0.864, and P = 0.608, respectively; Supplementary Figure 5G-I).
Figure 3.
The expression quantitative trait loci (eQTLs) analysis for SNAI1 rs73276407 (high LD with SNAI1 rs1047920, r2 = 0.71) and AMDHD1 rs11826. Correlation between SNAI1 mRNA expression and rs73276407 genotype in 373 Europeans from the 1000 Genomes Project in the (A) additive model, (B) dominant model and (C) recessive model; Correlation between AMDHD1 mRNA expression and rs11826 genotype in the GTEx Project (D) Breast tissue, (E) Whole Blood. Abbreviations: AMDHD1, amidohydrolase domain containing 1; GTEx, Genotype-Tissue Expression; LD, Linkage disequilibrium; SNAI1, snail family transcriptional repressor 1.
To gain more evidence on the correlations between the independent SNPs and mRNA expression levels, we further evaluated eQTL using data from the GTEx Project (http://www.gtexportal.org/home). The results showed that rs11826 C>T had a positive correlation with AMDHD1 mRNA expression levels in breast tissue (P = 6.00 × 10-4, Figure 3D) and the whole blood (P = 1.30 × 10-7, Figure 3E). However, there were no significant associations between rs1047920 genotypes and SNAI1 mRNA expression levels in the whole blood (P = 0.670; Supplementary Figure 5J) or breast tissue (P = 0.900; Supplementary Figure 5K) from GTEx, nor there were any significant associations between rs3914238 genotypes and CUBN mRNA expression levels in the whole blood (P = 0.350; Supplementary Figure 5L) or breast tissue (P = 0.170; Supplementary Figure 5M) from GTEx.
Discussion
To investigate whether genetic variants in the vitamin D pathway genes contribute to BC risk, we assessed associations between 2,994 SNPs in 33 vitamin D pathway genes and BC risk by using a large-scale meta-analysis of 14 GWASs in the DRIVE study. We identified three potential susceptibility variants (i.e., SNAI1 rs1047920 at 20q13.13, AMDHD1 rs11826 at 12q23.1, and CUBN rs3914238 at 10p13) that were independently or jointly associated with BC risk. Individuals with a higher NRG of these three genetic variants had a higher BC risk. Further eQTL analysis showed that the SNAI1 rs73276407 A allele, in a high LD with the rs1047920 T allele, was associated with decreased SNAI1 mRNA expression levels in lymphoblastoid cell lines, while the AMDHD1 rs11826 T allele was found to be significantly associated with elevated AMDHD1 mRNA expression levels in the whole blood cells and breast tissues. These findings suggest that SNPs in vitamin D pathway genes may play important biological roles in the development of BC, possibly by influencing their gene expression.
Vitamin D may play a controlling role in normal breast cell growth and has the ability to stop cancer cell growth in the breast. The bioactive form of vitamin D is 1,25-dihydroxyvitamin D that is largely considered a chemoprevention agent for its anti-cancer effect [29]. Several experimental studies have shown that 1,25-dihydroxyvitamin D can induce cell differentiation and apoptosis as well as inhibit angiogenesis and cell proliferation in normal and malignant breast cells [30,31]. In the present study, we showed that genetic variants in some genes of the vitamin D pathway were associated with BC risk; however, their exact biological mechanisms need to be further investigated. Furthermore, the inconsistence observed for the correlations between genotypes of the risk-associated SNPs and mRNA expression levels of their corresponding genes in different human tissues (i.e., blood cells and breast tissues) remain to be resolved in more rigorously designed experimental and mechanistic studies.
SNAI1, also known as SLUGH2, SNA, SNAH, SNAIL or SNAIL1, encodes SNAIL that is a transcription factor and regulates the epithelial to mesenchymal transition by activating N-cadherin and repressing E-cadherin during embryonic development and cell migration [32,33]. Few studies have investigated the roles of SNAI1 in BC susceptibility. One study found that in the immune cells from the peripheral blood of BC patients, the expression of SNAI1 was significantly higher in stage I patients than that in other higher stages [34], suggesting the involvement of this gene in early carcinogenesis. Another study showed that breast tumor tissue had lower levels of SNAl1 protein product than normal breast tissue [35], further suggesting the involvement of this gene in breast carcinogenesis. One GWAS study has indicated that MYT1 shares the same location of 20q13.13 as SNAl1 [36], but the previously reported rs6062356 SNP in MYT1 was not in LD with SNAI1 rs1047920 (Supplementary Figure 6A), nor MYT1 has any effect on the function of vitamin D. However, we found in the present study that the SNAI1 rs73276407 A allele, in a high LD with the rs1047920 T allele, might down-regulate the expression of SNAI1, suggesting that SNAI1 may play an oncogenic role in BC risk. Nevertheless, how SNAI1 rs73276407 C>T influences BC risk needs additional experimental investigation.
AMDHD1 is also known as HMFT1272, and there were few studies focusing on the roles of AMDHD1 in oncogenesis, especially in BC. In one study on quantitative analysis of gene expression, AMDHD1 was found to be overexpressed in adrenal adenoma, compared with adrenal cancer [37]. Data from one GWAS of esophageal squamous cell carcinoma showed a statistically significant association between SNPs in AMDHD1 (other than the ones identified in the present study) and cancer risk [38]. Although the GWAS study indicated that LINC02452 shared the same location at 12q23.1 as AMDHD1 [36], AMDHD1 rs11826 has no LD with the previously published LINC02452 rs77034926 (Supplementary Figure 6B) and lack of functional analysis. In the present study, we found that the rs11826 T allele might up-regulate the expression of AMDHD1, suggesting that AMDHD1 may play a protective role in BC risk, but we did not have additional experimental data to explain how AMDHD1 rs11826 C>T influenced BC risk.
CUBN encodes cubilin that is a large protein with three types of domain, an N-terminal stretch, 27 CUB domains and eight epidermal growth-factor-like repeats [39]. One study showed that mutations of CUBN caused glycosylation, endoplasmic reticulum retention, and abrogation of surface expression [40]. A meta-exome-wide association study of 33,985 Europeans found that a novel CUBN mutation was associated with albuminuria levels, with a 3.5-fold increase in the effect in type 2 diabetes compared with non-diabetic individuals [41], and CUBN variants have also been associated with albuminuria [42,43]. A genome-wide colorectal cancer association study has identified a promising cancer risk-associated intronic CUBN rs10904849 SNP [44]. However, there is no report on an association between SNPs in CUBN and BC risk. One BC GWAS study indicated that FRMD4A shared the same location 10p13 as CUBN [36], but the previously reported FRMD4A rs10906522 is not in LD with CUBN rs3914238 (Supplementary Figure 6C). In the present study, we did not find any evidence for an association between CUBN rs3914238 and its mRNA expression levels. Therefore, further experimental studies are needed to explain biological mechanisms underlying the observed associations between CUBN rs3914238 and BC risk.
The present study has several limitations that should be mentioned. First, some known risk factors, such as unhealthy lifestyle, physical activities, and incomplete information on estrogen status prevented us from complete adjustment in the analysis. Second, the biological mechanisms by which these three independent SNPs may influence BC risk remain unclear. Third, the risk-predicting model was built in non-Hispanic white, which may not be generalizable to the general population. Finally, our study was based on a signaling pathway and did not adjust the previously published BC risk associated SNPs, but only adjust previously published SNPs from the DRIVE study.
In summary, we have comprehensively analyzed SNPs in the vitamin D pathway genes for their associations with BC risk using genotyping data from 14 previously published GWASs among 53,107 subjects of European descendants, and we identified three independent BC susceptibility loci in the vitamin D pathway genes (i.e., SNAI1 rs1047920, AMDHD1 rs11826, and CUBN rs3914238). Once replicated by other investigators, these genetic variants may serve as new biomarkers for predicting BC risk.
Acknowledgements
DRIVE (dbGaP Study Accession: phs001265.v1.p1): The genotyping and phenotype data obtained from the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) breast-cancer case control samples was supported by U19 CA148065 and X01 HG007491 and by Cancer Research UK (C1287/A16563). Genotyping was conducted by the Centre for Inherited Disease Research (CIDR), University of Cambridge, Centre for Cancer Genetic Epidemiology, and the National Cancer Institute. The following studies provided germline DNA for breast cancer cases and controls: the Two Sister Study (2SISTER), Breast Oncology Galicia Network (BREOGAN), Copenhagen General Population Study (CGPS), Cancer Prevention Study 2 (CPSII), The European Prospective Investigation into Cancer and Nutrition (EPIC), Melbourne Collaborative Cohort Study (MCCS), Multi-ethnic Cohort (MEC), Nashville Breast Health Study (NBHS), Nurses’ Health Study (NHS), Nurses’ Health Study 2 (NHS2), Polish Breast Cancer Study (PBCS), Prostate Lung Colorectal and Ovarian Cancer Screening Trial (PLCO), Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH), The Sister Study (SISTER), Swedish Mammographic Cohort (SMC), Women of African Ancestry Breast Cancer Study (WAABCS), Women’s Health Initiative (WHI). GTEx: The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: GTEx Analysis V7 (dbGaP Accession phs000424.v7.p2). Qing-yi Wei was supported by the P30 Cancer Center Support Grant from the Duke Cancer Institute (Grant ID: NIH CA014236). Hai-jiao Wang and Ling-ling Zhao were supported by funds from The First Hospital of Jilin University in China.
Disclosure of conflict of interest
None.
Abbreviations
- AMDHD1
amidohydrolase domain containing 1
- BC
breast cancer
- BFDP
Bayesian false-discovery probability
- BREOGAN
Breast Oncology Galicia Network
- CGPS
Copenhagen General Population Study
- CI
confidence interval
- CPSII
Cancer Prevention Study-II Nutrition Cohort
- CUBN
cubilin
- DRIVE
Discovery, Biology, and Risk of Inherited Variants in Breast Cancer
- EPIC
European Prospective Investigation Into Cancer and Nutrition
- eQTL
expression quantitative trait loci
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MCCS
Melbourne Collaborative Cohort Study
- MEC
Multiethnic Cohort
- NBHS
Nashville Breast Health Study
- NHS
Nurses’ Health Study
- NHS2
Nurses’ Health Study 2
- NRGs
number of risk genotypes
- OR
odds ratio
- PBCS
NCI Polish Breast Cancer Study
- PCs
principal components
- PLCO
The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial
- SEARCH
Study of Epidemiology and Risk factors in Cancer Heredity
- SMC
Swedish Mammography Cohort
- SNAI1
snail family transcriptional repressor 1
- SNP
single nucleotide polymorphism
- WHI
Women’s Health Initiative
Supporting Information
References
- 1.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387–1397. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiovitz S, Korde LA. Genetics of breast cancer: a topic in evolution. Ann Oncol. 2015;26:1291–1299. doi: 10.1093/annonc/mdv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014;14:342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 6.Dawson-Hughes B, Heaney RP, Holick MF, Lips P, Meunier PJ, Vieth R. Estimates of optimal vitamin D status. Osteoporos Int. 2005;16:713–716. doi: 10.1007/s00198-005-1867-7. [DOI] [PubMed] [Google Scholar]
- 7.Shao T, Klein P, Grossbard ML. Vitamin D and breast cancer. Oncologist. 2012;17:36–45. doi: 10.1634/theoncologist.2011-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 9.Krishnan AV, Swami S, Feldman D. Vitamin D and breast cancer: inhibition of estrogen synthesis and signaling. J Steroid Biochem Mol Biol. 2010;121:343–348. doi: 10.1016/j.jsbmb.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C. Vitamin D: modulator of the immune system. Curr Opin Pharmacol. 2010;10:482–496. doi: 10.1016/j.coph.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Shirvani SS, Nouri M, Sakhinia E, Babaloo Z, Mohammadzaeh A, Alipour S, Jadideslam G, Khabbazi A. The molecular and clinical evidence of vitamin D signaling as a modulator of the immune system: role in Behcet’s disease. Immunol Lett. 2019;210:10–19. doi: 10.1016/j.imlet.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Provvedini DM, Tsoukas CD, Deftos LJ, Manolagas SC. 1,25-dihydroxyvitamin D3 receptors in human leukocytes. Science. 1983;221:1181–1183. doi: 10.1126/science.6310748. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin PJ, Ennis M, Pritchard KI, Koo J, Hood N. Prognostic effects of 25-hydroxyvitamin D levels in early breast cancer. J. Clin. Oncol. 2009;27:3757–3763. doi: 10.1200/JCO.2008.20.0725. [DOI] [PubMed] [Google Scholar]
- 14.Neuhouser ML, Sorensen B, Hollis BW, Ambs A, Ulrich CM, McTiernan A, Bernstein L, Wayne S, Gilliland F, Baumgartner K, Baumgartner R, Ballard-Barbash R. Vitamin D insufficiency in a multiethnic cohort of breast cancer survivors. Am J Clin Nutr. 2008;88:133–139. doi: 10.1093/ajcn/88.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn J, Peters U, Albanes D, Purdue MP, Abnet CC, Chatterjee N, Horst RL, Hollis BW, Huang WY, Shikany JM, Hayes RB. Serum vitamin D concentration and prostate cancer risk: a nested case-control study. J Natl Cancer Inst. 2008;100:796–804. doi: 10.1093/jnci/djn152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Rohan TE. Vitamin D, calcium, and breast cancer risk: a review. Cancer Epidemiol Biomarkers Prev. 2006;15:1427–1437. doi: 10.1158/1055-9965.EPI-06-0075. [DOI] [PubMed] [Google Scholar]
- 17.Weinstein SJ, Yu K, Horst RL, Ashby J, Virtamo J, Albanes D. Serum 25-hydroxyvitamin D and risks of colon and rectal cancer in Finnish men. Am J Epidemiol. 2011;173:499–508. doi: 10.1093/aje/kwq398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Sousa Almeida-Filho B, De Luca Vespoli H, Pessoa EC, Machado M, Nahas-Neto J, Nahas EAP. Vitamin D deficiency is associated with poor breast cancer prognostic features in postmenopausal women. J Steroid Biochem Mol Biol. 2017;174:284–289. doi: 10.1016/j.jsbmb.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Ge J, Liu H, Qian D, Wang X, Moorman PG, Luo S, Hwang S, Wei Q. Genetic variants of genes in the NER pathway associated with risk of breast cancer: a large-scale analysis of 14 published GWAS datasets in the DRIVE study. Int J Cancer. 2019;145:1270–1279. doi: 10.1002/ijc.32371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang X, O’Reilly PF, Aschard H, Hsu YH, Richards JB, Dupuis J, Ingelsson E, Karasik D, Pilz S, Berry D, Kestenbaum B, Zheng J, Luan J, Sofianopoulou E, Streeten EA, Albanes D, Lutsey PL, Yao L, Tang W, Econs MJ, Wallaschofski H, Volzke H, Zhou A, Power C, McCarthy MI, Michos ED, Boerwinkle E, Weinstein SJ, Freedman ND, Huang WY, Van Schoor NM, van der Velde N, Groot L, Enneman A, Cupples LA, Booth SL, Vasan RS, Liu CT, Zhou Y, Ripatti S, Ohlsson C, Vandenput L, Lorentzon M, Eriksson JG, Shea MK, Houston DK, Kritchevsky SB, Liu Y, Lohman KK, Ferrucci L, Peacock M, Gieger C, Beekman M, Slagboom E, Deelen J, Heemst DV, Kleber ME, Marz W, de Boer IH, Wood AC, Rotter JI, Rich SS, Robinson-Cohen C, den Heijer M, Jarvelin MR, Cavadino A, Joshi PK, Wilson JF, Hayward C, Lind L, Michaelsson K, Trompet S, Zillikens MC, Uitterlinden AG, Rivadeneira F, Broer L, Zgaga L, Campbell H, Theodoratou E, Farrington SM, Timofeeva M, Dunlop MG, Valdes AM, Tikkanen E, Lehtimaki T, Lyytikainen LP, Kahonen M, Raitakari OT, Mikkila V, Ikram MA, Sattar N, Jukema JW, Wareham NJ, Langenberg C, Forouhi NG, Gundersen TE, Khaw KT, Butterworth AS, Danesh J, Spector T, Wang TJ, Hypponen E, Kraft P, Kiel DP. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat Commun. 2018;9:260. doi: 10.1038/s41467-017-02662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Methods Mol Biol. 2013;1019:215–236. doi: 10.1007/978-1-62703-447-0_9. [DOI] [PubMed] [Google Scholar]
- 23.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson G. Human genetics. GTEx detects genetic effects. Science. 2015;348:640–641. doi: 10.1126/science.aab3002. [DOI] [PubMed] [Google Scholar]
- 29.Wulaningsih W, Sagoo HK, Hamza M, Melvin J, Holmberg L, Garmo H, Malmstrom H, Lambe M, Hammar N, Walldius G, Jungner I, Van Hemelrijck M. Serum calcium and the risk of breast cancer: findings from the Swedish AMORIS study and a meta-analysis of prospective studies. Int J Mol Sci. 2016;17:1487. doi: 10.3390/ijms17091487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantell DJ, Owens PE, Bundred NJ, Mawer EB, Canfield AE. 1 alpha,25-dihydroxyvitamin D(3) inhibits angiogenesis in vitro and in vivo. Circ Res. 2000;87:214–220. doi: 10.1161/01.res.87.3.214. [DOI] [PubMed] [Google Scholar]
- 31.Colston KW, Berger U, Coombes RC. Possible role for vitamin D in controlling breast cancer cell proliferation. Lancet. 1989;1:188–191. doi: 10.1016/s0140-6736(89)91204-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Liu R, Zhu W, Chu H, Yu H, Wei P, Wu X, Zhu H, Gao H, Liang J, Li G, Yang W. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571:127–131. doi: 10.1038/s41586-019-1340-y. [DOI] [PubMed] [Google Scholar]
- 33.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 34.Naik A, Al-Zeheimi N, Bakheit CS, Al Riyami M, Al Jarrah A, Al Moundhri MS, Al Habsi Z, Basheer M, Adham SA. Neuropilin-1 associated molecules in the blood distinguish poor prognosis breast cancer: a cross-sectional study. Sci Rep. 2017;7:3301. doi: 10.1038/s41598-017-03280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Itoh T, Wu K, Zhou D, Yang C. Transcriptional regulation of aromatase expression in human breast tissue. J Steroid Biochem Mol Biol. 2002;83:93–99. doi: 10.1016/s0960-0760(02)00276-5. [DOI] [PubMed] [Google Scholar]
- 36.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemacon A, Soucy P, Glubb D, Rostamianfar A, Bolla MK, Wang Q, Tyrer J, Dicks E, Lee A, Wang Z, Allen J, Keeman R, Eilber U, French JD, Qing Chen X, Fachal L, McCue K, McCart Reed AE, Ghoussaini M, Carroll JS, Jiang X, Finucane H, Adams M, Adank MA, Ahsan H, Aittomaki K, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Arun B, Auer PL, Bacot F, Barrdahl M, Baynes C, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Bernstein L, Blomqvist C, Bogdanova NV, Bojesen SE, Bonanni B, Borresen-Dale AL, Brand JS, Brauch H, Brennan P, Brenner H, Brinton L, Broberg P, Brock IW, Broeks A, Brooks-Wilson A, Brucker SY, Bruning T, Burwinkel B, Butterbach K, Cai Q, Cai H, Caldes T, Canzian F, Carracedo A, Carter BD, Castelao JE, Chan TL, David Cheng TY, Seng Chia K, Choi JY, Christiansen H, Clarke CL, Collee M, Conroy DM, Cordina-Duverger E, Cornelissen S, Cox DG, Cox A, Cross SS, Cunningham JM, Czene K, Daly MB, Devilee P, Doheny KF, Dork T, Dos-Santos-Silva I, Dumont M, Durcan L, Dwek M, Eccles DM, Ekici AB, Eliassen AH, Ellberg C, Elvira M, Engel C, Eriksson M, Fasching PA, Figueroa J, Flesch-Janys D, Fletcher O, Flyger H, Fritschi L, Gaborieau V, Gabrielson M, Gago-Dominguez M, Gao YT, Gapstur SM, Garcia-Saenz JA, Gaudet MM, Georgoulias V, Giles GG, Glendon G, Goldberg MS, Goldgar DE, Gonzalez-Neira A, Grenaker Alnaes GI, Grip M, Gronwald J, Grundy A, Guenel P, Haeberle L, Hahnen E, Haiman CA, Hakansson N, Hamann U, Hamel N, Hankinson S, Harrington P, Hart SN, Hartikainen JM, Hartman M, Hein A, Heyworth J, Hicks B, Hillemanns P, Ho DN, Hollestelle A, Hooning MJ, Hoover RN, Hopper JL, Hou MF, Hsiung CN, Huang G, Humphreys K, Ishiguro J, Ito H, Iwasaki M, Iwata H, Jakubowska A, Janni W, John EM, Johnson N, Jones K, Jones M, Jukkola-Vuorinen A, Kaaks R, Kabisch M, Kaczmarek K, Kang D, Kasuga Y, Kerin MJ, Khan S, Khusnutdinova E, Kiiski JI, Kim SW, Knight JA, Kosma VM, Kristensen VN, Kruger U, Kwong A, Lambrechts D, Le Marchand L, Lee E, Lee MH, Lee JW, Neng Lee C, Lejbkowicz F, Li J, Lilyquist J, Lindblom A, Lissowska J, Lo WY, Loibl S, Long J, Lophatananon A, Lubinski J, Luccarini C, Lux MP, Ma ESK, MacInnis RJ, Maishman T, Makalic E, Malone KE, Kostovska IM, Mannermaa A, Manoukian S, Manson JE, Margolin S, Mariapun S, Martinez ME, Matsuo K, Mavroudis D, McKay J, McLean C, Meijers-Heijboer H, Meindl A, Menendez P, Menon U, Meyer J, Miao H, Miller N, Taib NAM, Muir K, Mulligan AM, Mulot C, Neuhausen SL, Nevanlinna H, Neven P, Nielsen SF, Noh DY, Nordestgaard BG, Norman A, Olopade OI, Olson JE, Olsson H, Olswold C, Orr N, Pankratz VS, Park SK, Park-Simon TW, Lloyd R, Perez JIA, Peterlongo P, Peto J, Phillips KA, Pinchev M, Plaseska-Karanfilska D, Prentice R, Presneau N, Prokofyeva D, Pugh E, Pylkas K, Rack B, Radice P, Rahman N, Rennert G, Rennert HS, Rhenius V, Romero A, Romm J, Ruddy KJ, Rudiger T, Rudolph A, Ruebner M, Rutgers EJT, Saloustros E, Sandler DP, Sangrajrang S, Sawyer EJ, Schmidt DF, Schmutzler RK, Schneeweiss A, Schoemaker MJ, Schumacher F, Schurmann P, Scott RJ, Scott C, Seal S, Seynaeve C, Shah M, Sharma P, Shen CY, Sheng G, Sherman ME, Shrubsole MJ, Shu XO, Smeets A, Sohn C, Southey MC, Spinelli JJ, Stegmaier C, Stewart-Brown S, Stone J, Stram DO, Surowy H, Swerdlow A, Tamimi R, Taylor JA, Tengstrom M, Teo SH, Beth Terry M, Tessier DC, Thanasitthichai S, Thone K, Tollenaar R, Tomlinson I, Tong L, Torres D, Truong T, Tseng CC, Tsugane S, Ulmer HU, Ursin G, Untch M, Vachon C, van Asperen CJ, Van Den Berg D, van den Ouweland AMW, van der Kolk L, van der Luijt RB, Vincent D, Vollenweider J, Waisfisz Q, Wang-Gohrke S, Weinberg CR, Wendt C, Whittemore AS, Wildiers H, Willett W, Winqvist R, Wolk A, Wu AH, Xia L, Yamaji T, Yang XR, Har Yip C, Yoo KY, Yu JC, Zheng W, Zheng Y, Zhu B, Ziogas A, Ziv E, Lakhani SR, Antoniou AC, Droit A, Andrulis IL, Amos CI, Couch FJ, Pharoah PDP, Chang-Claude J, Hall P, Hunter DJ, Milne RL, Garcia-Closas M, Schmidt MK, Chanock SJ, Dunning AM, Edwards SL, Bader GD, Chenevix-Trench G, Simard J, Kraft P, Easton DF. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assie G, Guillaud-Bataille M, Ragazzon B, Bertagna X, Bertherat J, Clauser E. The pathophysiology, diagnosis and prognosis of adrenocortical tumors revisited by transcriptome analyses. Trends Endocrinol Metab. 2010;21:325–334. doi: 10.1016/j.tem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Hyland PL, Zhang H, Yang Q, Yang HH, Hu N, Lin SW, Su H, Wang L, Wang C, Ding T, Fan JH, Qiao YL, Sung H, Wheeler W, Giffen C, Burdett L, Wang Z, Lee MP, Chanock SJ, Dawsey SM, Freedman ND, Abnet CC, Goldstein AM, Yu K, Taylor PR. Pathway, in silico and tissue-specific expression quantitative analyses of oesophageal squamous cell carcinoma genomewide association studies data. Int J Epidemiol. 2016;45:206–220. doi: 10.1093/ije/dyv294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moestrup SK, Kozyraki R, Kristiansen M, Kaysen JH, Rasmussen HH, Brault D, Pontillon F, Goda FO, Christensen EI, Hammond TG, Verroust PJ. The intrinsic factor-vitamin B12 receptor and target of teratogenic antibodies is a megalin-binding peripheral membrane protein with homology to developmental proteins. J Biol Chem. 1998;273:5235–5242. doi: 10.1074/jbc.273.9.5235. [DOI] [PubMed] [Google Scholar]
- 40.Udagawa T, Harita Y, Miura K, Mitsui J, Ode KL, Morishita S, Urae S, Kanda S, Kajiho Y, Tsurumi H, Ueda HR, Tsuji S, Saito A, Oka A. Amnionless-mediated glycosylation is crucial for cell surface targeting of cubilin in renal and intestinal cells. Sci Rep. 2018;8:2351. doi: 10.1038/s41598-018-20731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahluwalia TS, Schulz CA, Waage J, Skaaby T, Sandholm N, van Zuydam N, Charmet R, Bork-Jensen J, Almgren P, Thuesen BH, Bedin M, Brandslund I, Christensen CK, Linneberg A, Ahlqvist E, Groop PH, Hadjadj S, Tregouet DA, Jorgensen ME, Grarup N, Pedersen O, Simons M, Groop L, Orho-Melander M, McCarthy MI, Melander O, Rossing P, Kilpelainen TO, Hansen T. A novel rare CUBN variant and three additional genes identified in Europeans with and without diabetes: results from an exome-wide association study of albuminuria. Diabetologia. 2019;62:292–305. doi: 10.1007/s00125-018-4783-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bedin M, Boyer O, Servais A, Li Y, Villoing-Gaude L, Tete MJ, Cambier A, Hogan J, Baudouin V, Krid S, Bensman A, Lammens F, Louillet F, Ranchin B, Vigneau C, Bouteau I, Isnard-Bagnis C, Mache CJ, Schafer T, Pape L, Godel M, Huber TB, Benz M, Klaus G, Hansen M, Latta K, Gribouval O, Moriniere V, Tournant C, Grohmann M, Kuhn E, Wagner T, Bole-Feysot C, Jabot-Hanin F, Nitschke P, Ahluwalia TS, Kottgen A, Andersen CBF, Bergmann C, Antignac C, Simons M. Human C-terminal CUBN variants associate with chronic proteinuria and normal renal function. J Clin Invest. 2020;130:335–344. doi: 10.1172/JCI129937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boger CA, Chen MH, Tin A, Olden M, Kottgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tonjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarod K, Skorpen F, Syvanen AC, Illig T, Baumert J, Koenig W, Kramer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Volzke H, Stumvoll M, Magi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlov J, Hallan S, Navis G, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH. CUBN is a gene locus for albuminuria. J Am Soc Nephrol. 2011;22:555–570. doi: 10.1681/ASN.2010060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Tassan NA, Whiffin N, Hosking FJ, Palles C, Farrington SM, Dobbins SE, Harris R, Gorman M, Tenesa A, Meyer BF, Wakil SM, Kinnersley B, Campbell H, Martin L, Smith CG, Idziaszczyk S, Barclay E, Maughan TS, Kaplan R, Kerr R, Kerr D, Buchanan DD, Win AK, Hopper J, Jenkins M, Lindor NM, Newcomb PA, Gallinger S, Conti D, Schumacher F, Casey G, Dunlop MG, Tomlinson IP, Cheadle JP, Houlston RS. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep. 2015;5:10442. doi: 10.1038/srep10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.