Abstract
PARP6 belongs to the mono-ADP-ribosyltransferase family and has been shown to be involved in the genesis and development of some tumours. However, the role of PARP6 in hepatocellular carcinoma (HCC) development remains to be fully elucidated. In the current study, we demonstrated that PARP6 was expressed at a low level in HCC cells and was negatively related to the degree of tumour differentiation. Additionally, silencing PARP6 led to an increase in the proliferation, invasion and migration ability of HCC cells in both in vitro and in vivo assays. Conversely, an elevation in the PARP6 expression level had the opposite effect. Through gene chip analysis combined with experimental verification, we confirmed that PARP6 can inhibit the expression of XRCC6 by inducing degradation and thus affect the Wnt/β-Catenin signalling pathway, which contributes to the suppression of HCC. Further mechanistic investigation demonstrated that the ubiquitin ligase HDM2 can interact with PARP6 and XRCC6, and mediated the regulatory effect of PARP6 on XRCC6 degradation. Taking together, PARP6 appears to inhibit HCC progression through the XRCC6/Wnt/β-catenin signal axis and could be used as a biomarker for the clinical monitoring of HCC development.
Keywords: PARP6, HCC, XRCC6, cell proliferation, metastasis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumours, and its mortality ranks third in the world [1]. Despite the decline in morbidity and mortality in recent years accompanying the improvement of early diagnosis techniques and surgical treatments, the long-term efficacy is not satisfactory due to the high rate of recurrence and metastasis [2]. The genesis and development of HCC is a complex process involving the interaction of many genetic and environmental factors, multi-gene participation, and multi-step regulation. Therefore, a key to developing new effective clinical treatments for HCC is actively exploring new molecules that affect the genesis and development of HCC and understanding the biological characteristics of these new molecules.
Poly(ADP-ribose) polymerases (PARP) are a class of multifunctional nuclear proteases involved in the process of poly(ADP)glycosylation [3]. These proteases are sensitive to DNA damage and can recognize and bind DNA strand gap or free DNA ends, thereby regulating a series of molecular events, such as intracellular DNA repair, replication, gene transcription, cell cycle progression, and cell death. It is now known that PARP family genes contribute to the genesis and development of various diseases, including tumours [3-5]. PARP6 is a new member of the PARP family and is located on chromosome 15q23 and consists of 630 amino acids. Since the discovery of PARP6, there have been only a small number of studies examining its role in malignant tumours. Our previous study in colorectal cancer found that PARP6 can regulate a member of the inhibitor of apoptosis family protein, surviving, and thus affect the progression of cancer cells [6,7]. In tandem with this study, others have shown that several molecules can affect the progression of breast cancer by interfering with PARP6 function and inhibiting the formation of the spindle in target cells [8]. Despite these advances, whether PARP6 is also involved in the genesis and development of HCC has not been determined.
In this study, we found that the expression level of PARP6 in HCC tissues is significantly lower than that in adjacent tissues, and this observation is closely related to the differentiation degree of HCC. In vitro and in vivo experiments confirmed that PARP6 can inhibit the proliferation, invasion and migration abilities of HCC cells. Mechanistic studies revealed that PARP6 can decay XRCC6 and affect the Wnt/β-catenin signalling pathway to inhibit the genesis and development of HCC.
Materials and methods
Patient information and clinical specimen
The tissue specimens and corresponding clinical data used in this study were randomly collected from the liver and gallbladder surgery of the Affiliated Hospital of Guilin Medical College from 2012 to 2015 and confirmed by pathologists, 50 cases of HCC and corresponding adjacent tissues (age 35-60 years old). The specimens were approved by the Medical Ethics Committee of the Affiliated Hospital of Guilin Medical College. The clinicopathological parameters include the gender, age, tumour location, tumour size and diameter, pathological type and stage, and liver metastasis of the patients. The tumours were staged by the TNM staging method revised by the Union for International Cancer Control (UICC) in 2002. The study protocol was approved by the Ethics Committee of the Guilin Medical College. Meanwhile, informed consent forms made according to the Declaration of Helsinki have been signed by the patients.
Immunohistochemical analysis (IHC)
The excised HCC and adjacent tissues were immersed and fixed in 4% paraformaldehyde, embedded in paraffin and cut into 4-mm-thick sections. Immunohistochemical staining was then performed according to the Envision two-step procedure [9]. The results were divided into four grades according to the degree of immunohistochemical staining: 0: <10% hepatoma cells stain positive; 1+: 11%-25% hepatoma cells stain positive; 2+: 26%-50% hepatoma cells stain positive; 3+: >50% hepatoma cells stain positive. Immunohistochemical analysis and scoring were performed simultaneously by two researchers.
Experimental reagent and antibody
Transfected liposomes and total RNA extraction reagent (TRIZOL) were purchased from Invitrogen (Grand Island, NY, USA). PARP6, XRCC6, Ku70, β-Catenin, p-EGFR, and MMP-7 antibodies were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). E-cadherin, N-cadherin, vimentin, and β-actin antibodies were purchased from Origine (Rockville, MD, USA). Other reagents were purchased from Sigma (St. Louis, MO, USA).
Culture and transfection of cells
We obtained HCC cells (HepG2, Hep3B, Sk-Hep-1, SMMC-7721, QGY-7701, LM-3) and normal liver cells (L02) from the Chinese Academy of Sciences cell bank (Shanghai, China). HepG2, Sk-Hep-1 and Hep3B cells were grown in Dulbecco’s modified Eagle medium (Thermo Fisher Scientific, South America), and SMMC-7721, LM-3, QGY-7701 and L02 cells were grown in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10% foetal bovine serum (Thermo Fisher Scientific) at 37°C and 5% CO2. The cells were revived every three to four months. The transfection of PARP6 plasmid and PARP6-siRNA were prepared and used according to a previously described protocol [15].
Cell proliferation test
The cell suspension of high PARP6 expression, downregulated PARP6 and negative control group were inoculated into a 96-well culture plate at a cell density of 1×104 cells/well. After a certain period of time, 10 μl of CCK8 solution was added, and the culture plate was placed in the incubator. After 1 to 4 hours, the absorbance at 450 nm was measured with a microplate reader and averaged.
Cloning formation experiment
The cells were inoculated in a six-well plate with a density of 500 cells/well. After three weeks of culture, the cells were fixed with 4% paraformaldehyde for 20 min and then stained with 1% crystal violet (G1062, Solarbio, Japan) overnight, and the number of cells per well was calculated after washing three times with PBS.
Cell invasion and migration experiments
The hepatoma cells of each group were collected, and the invasive ability of cancer cells was detected using a 24-well Transwell chamber with a pore size of 8 μm. A total of 1×105 cells were added to the upper chamber. The total volume was 200 μl. A total of 600 μl of culture solution was added to the lower chamber. After incubating at 37°C for 6 h, 12 h, and 24 h, the cells on the cover of the upper chamber at the basement membrane were wiped off with a cotton swab, stained with 1% crystal violet (G1062, Solarbio, Japan) and imaged. In addition, the Transwell cell membrane was not coated with Matrigel solution, and the cell migration experiment had the same steps as above.
Animal experiment
The study was carried out in accordance with the Declaration of Helsinki and the Guide to the Care and Use of Laboratory Animals adopted and promulgated by the National Institutes of Health. Male BALB/c nude mice 4-6 weeks old were purchased from Shanghai Slac Experimental Animal Co., Ltd. All animal experiments were approved by the Animal Care and Use Committee of Guilin Medical College. Stable transfected cells in the logarithmic growth phase were collected and injected subcutaneously into the right groin area of the animals. Five weeks after the inoculation, the animals were sacrificed. The tumour was excised, and the tumour weight was recorded. Tumour cells were injected into the animal through the tail vein to determine the ability of tumour metastasis. After 8 weeks, the lungs were removed after anesthetizing the animals to detect the number of metastases.
Western blot analysis
The cells of each experimental group were collected and added to the cell lysis buffer, the total protein was extracted, and the protein concentration was determined by BCA (Pierce, USA). Then, 30 μg of protein lysate was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis system (SDS-PAGE), and the specific antibody and peroxidase-labelled targeted secondary antibody were detected and displayed by chemiluminescence. The intensities of the bands were qualified by ImageJ (National Institutes of Health, Bethesda, MD, USA).
Gene chip analysis
The quality of total RNA was assessed using an Agilent 2100 Bioanalyzer and the NanoDrop ND-1000 assay. RNA expression was then analysed using the Affymetrix HU U133 plus 2.0 array according to the procedure. Raw data were analysed by normalization algorithm using robust multiarray average (RMA) in the expression control platform software (Affymetrix). Genes with significant changes in the PEA15 overexpression group and the control group were analysed by scatter diagram, and genes with up- or downregulated expression changes not less than 3-fold were selected. Cluster analysis was performed using Gene Cluster v3.0 software, and heat map visualization analysis was performed using Java TreeView v1.1.4r3 software.
Co-immunoprecipitation (Co-IP)
Cells were collected in cold PBS and lysed with RIPA buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 0.5 mM DTT, and protease inhibitor). The lysate was diluted 2-4-fold with dilution buffer (50 mM Tris (pH 7.4), 100 mM NaCl, 1 mM EDTA, 0.1% NP-40 and 10% glycerol, protease inhibitor). Then, 2-5 μg of antibodies was added to the diluted cell lysate and incubated overnight at 4°C. The next day, the protein complexes were isolated by magnetic Dynabeads Protein G for 2 h at 4°C with rotation. The bead-antibody-protein complexes were then washed 4 times with wash buffer (50 mM Tris (pH 7.4), 125 mM NaCl, 1 mM EDTA and 0.1% NP-40) and boiled for Western blot analysis.
Quantitative real time RT-PCR (qRT-PCR)
Total RNA was extracted from cells or tissues using TRIzol (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. cDNA synthesis was performed using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China). Real-time qRT-PCR analysis was performed using Platinum SYBR Green qPCR SuperMix-UDG kits (Life Technologies, Gaithersburg, MD, USA) or a TaqMan Probe Master Mix kit (Vazyme Biotech Co., Nanjing, China) according to the manufacturer’s protocol. The expression of genes was normalized to GAPDH. The primers used for amplification were as follows: PARP6 forward 5’-AGTTCTGGAATGATGACGACTCG-3’, reverse 5’-GTGGGTGTCGATACAGGTCAG-3’.
Statistical analysis
The results of this study were analysed with SPSS18.0 statistical software. The correlation between the expression level of PARP6 and clinical-pathological parameters was evaluated by chi-square test (x 2), and the quantitative data were evaluated by paired t-test. The patient survival rate and prognosis were estimated using the Kaplan-Meier method. Each group of experiments was repeated three times, and for the comparison of the mean of different sample data, P<0.05 indicates statistical difference.
Results
PARP6 expression in HCC tissue was lower and inversely correlated with tumour progression
We examined PARP6 expression using IHC in cancer and adjacent tissues from 50 cases of HCC selected randomly (Figure 1A). The results showed that the ratio of PARP6 in HCC tissue staining positive was significantly lower than that of corresponding adjacent tissue (Figure 1B). Additionally, we used qRT-PCR and Western blotting to examine the mRNA and protein expression of PARP6 in HCC and adjacent tissues. The results showed that the mRNA and protein expression levels of PARP6 in HCC tissues were significantly lower than those in adjacent tissues, which was consistent with the immunohistochemistry results (Figure 1C, 1D). Then, we referred to the Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) database to check the mRNA expression profile of PARP6 in normal liver tissues (n=109) and HCC tissues (n=369), further demonstrating that the expression of PARP6 was significantly downregulated in HCC tissues (Figure 1E).
Figure 1.

Expression of PARP6 was downregulated in HCC tissues. A. The expression of PARP6 in HCC and adjacent tissues of tumours was analysed by IHC (×400). B. The relative positive cells of PARP6 in 50 pairs of HCC and adjacent tissues of tumours. C. The protein expression of PARP6 was analysed by Western blot analysis in 9 pairs of HCC (T) and adjacent tissues (N). D. The mRNA expression of PARP6 was analysed by qRT-PCR in 98 pairs of HCC (T) and adjacent tissues (N). E. PARP6 expression was assessed using data from the TCGA databases. (**, P<0.01).
We divided the expression levels of PARP6 in HCC into a high expression group (IHC scores: 2+ and 3+, n=11) and a low expression group (IHC scores: 0 and 1+, n=39) in 50 cases of HCC. The results showed that the expression level of PARP6 was significantly correlated with clinical stage (P=0.013), TNM stage (P=0.002) and metastasis (P=0.042) but had no significant correlation with other clinicopathological features, including age, gender, tumour size, and tumour diameter (Table 1). The abovementioned results demonstrate that the expression level of PARP6 in HCC tissue was significantly reduced and was inversely associated with HCC progression in patients.
Table 1.
Relationship between PARP6 and clinicopathological parameters in 50 HCC patients
| Variables | All cases | PARP6 expression | χ 2 | P * | |
|---|---|---|---|---|---|
|
| |||||
| High (n=11) | Low (n=39) | ||||
| Age (years) | |||||
| ≥60 | 20 | 5 | 11 | ||
| <60 | 30 | 6 | 28 | 1.173 | 0.466 |
| Gender | |||||
| Male | 24 | 4 | 20 | ||
| Female | 26 | 7 | 19 | 0.765 | 0.501 |
| Tumor size (cm) | |||||
| <5 | 24 | 6 | 18 | ||
| ≥5 | 26 | 5 | 21 | 0.242 | 0.783 |
| AFP (ng/ml) | |||||
| <200 | 27 | 5 | 22 | ||
| >200 | 23 | 6 | 17 | 0.451 | 0.733 |
| Clinical stage | |||||
| I-II | 19 | 8 | 11 | ||
| III-IV | 31 | 3 | 28 | 7.219 | 0.013 |
| TNM stage | |||||
| I-II | 20 | 9 | 11 | ||
| III-IV | 30 | 2 | 28 | 10.276 | 0.002 |
| Metastasis | |||||
| Yes | 28 | 3 | 25 | ||
| No | 22 | 8 | 14 | 4.723 | 0.042 |
Probability, P, from χ2 test.
PARP6 may inhibit the proliferation of hepatoma cells in vitro and in vivo
We selected six HCC cell lines (HepG2, Hep3B, Sk-Hep-1, SMMC-7721, QGY-7701, LM-3) and one normal liver cell line (LO2) as a control and examined the difference in expression levels of PARP6 by Western blotting and qRT-PCR. The results showed that the expression levels of Sk-Hep-1 and QGY-7701 cells were higher, while the expression levels of HepG2, SMMC-7721 and LM-3 cells were relatively lower (P<0.05) (Figure 2A, 2B). Building on these observations, we constructed an over-expression PARP6 plasmid and several PARP6 siRNAs and introduced them into LM3 and SK-Hep-1 cell lines. The level of PARP6 expression was examined (Figure 2C, 2D), and we found that PARP6 siRNAs or plasmid were effective for PARP6 depletion or overexpression.
Figure 2.

Detection of PARP6 expression levels in HCC cells with PARP6 overexpression or depletion. A, B. The protein and mRNA expression of PARP6 were analysed by Western blot and qRT-PCR in different HCC and normal cell lines. C. Efficiency of four PARP6 siRNAs was detected by western blotting in HCC cells. D. The expression of PARP6 was detected by western blot after PARP6 plasmid transfection.
Subsequently, we examined proliferative capacity using the CCK8 method. Interestingly, the proliferation ability of LM3 cells after over-expression of PARP6 was significantly weakened compared with those in the normal group and empty vector group, while the proliferation ability of the Sk-Hep-1 cells was significantly enhanced after silencing of PARP6 (Figure 3A, 3B). In parallel, data from a clone formation assay showed that the colony formation ability of LM-3 cells was significantly lower than that of the normal and empty vector group when PARP6 was over-expressed (Figure 3C), while the clone formation of Sk-Hep-1 cells was significantly higher than that of the empty vector group when PARP6 was knocked down (Figure 3C). In addition, the in vivo animal experiment confirmed a significant reduction in tumour volume and tumour weight in nude mice after the upregulation of PARP6; conversely, tumour volume and tumour weight increased in nude mice after downregulation of PARP6 (Figure 3D-F). Taken together, the above data suggest that PARP6 can inhibit the proliferation of hepatoma cells both in vitro and in vivo.
Figure 3.
PARP6 inhibits the proliferation of HCC in vivo and in vitro. A, B. CCK-8 kit detection of cell proliferation after depletion or overexpression of PARP6. C. Plate cloning experiments were used to detect colony formation after depletion or overexpression of PARP6. D. Xenograft nude mouse model of tumorigenesis was used to confirm cell growth after depletion or overexpression of PARP6. E. The volume and weight of tumours were assessed in nude mice injected with PARP6-overexpressing LM3 and vector-transfected LM3. F. The volume and weight of tumours were assessed in nude mice injected with PARP6-silenced Sk-Hep-1 and control Sk-Hep-1. The data are expressed as the mean ± SD (*, P<0.05; **, P<0.01).
PARP6 can inhibit the invasion and migration of hepatoma cells in vitro and in vivo and participate in the regulation of EMT
We examined the effects of PARP6 on the invasion and migration of hepatoma cells using a Transwell chamber assay. The results showed that the invasion and migration of LM-3 cells were weakened when PARP6 was upregulated (Figure 4A, 4B) but enhanced invasion and migration was observed in the Sk-Hep-1 cells when PARP6 was knocked down (Figure 4C, 4D). Subsequently, we examined the effect of PARP6 expression on epithelial and mesenchymal protein levels in hepatoma cells by Western blotting. The results showed that overexpression of PARP6 in LM-3 cells led to increased levels of epithelial markers (E-cadherin) and decreased levels of mesenchymal markers (N-cadherin and vimentin) (Figure 4E). The depletion of PARP6 caused the opposite expression of these markers (Figure 4E). The above results suggest that PARP6 can control the invasion and migration of hepatoma cells, as well as the EMT in hepatoma cells.
Figure 4.
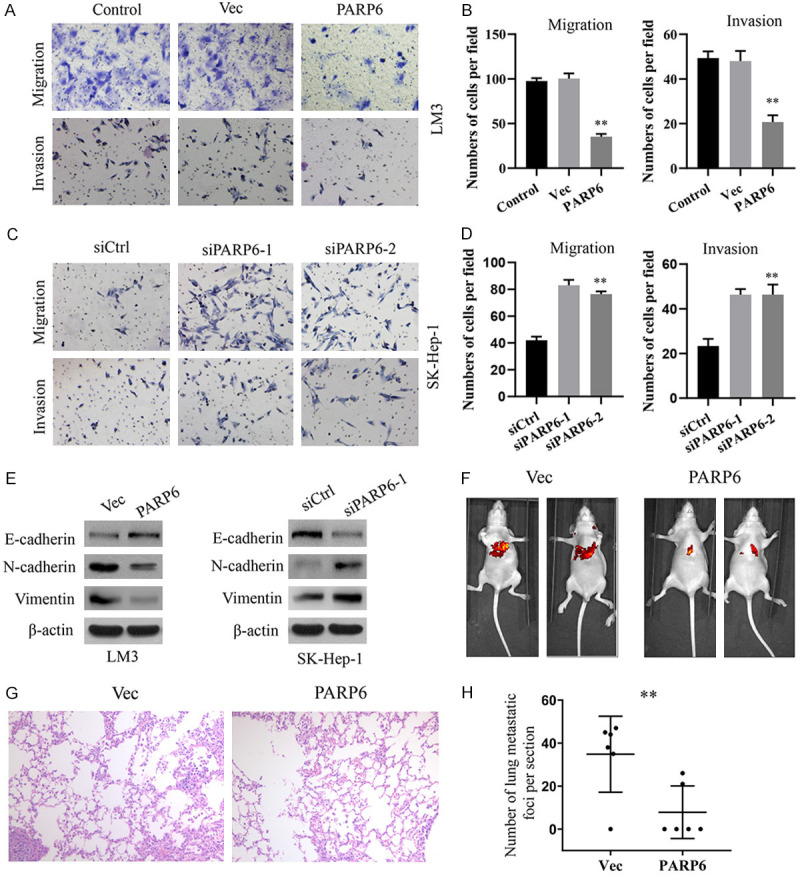
PARP6 inhibits the metastasis of HCC cells in vitro and in vivo. A, B. Migration and invasion were assessed by Transwell in LM3 cells overexpressing PARP6. C, D. Migration and invasion were assessed by Transwell in Sk-Hep-1 cells with PARP6 knockdown. E. Western blotting was used to analyse the expression of EMT markers in HCC cells with PARP6 overexpression or knockdown. F. Live imaging to observe lung metastasis. G, H. Representative images of the histological assessment of the lungs via H&E staining (×200). The metastatic lesion is indicated by an arrow. The number of metastatic lesions in the lung was significantly decreased in the mice injected with PARP6-overexpressing LM3 cells (*, P<0.05; **, P<0.01).
To verify whether PARP6 can affect HCC metastasis in vivo, we constructed a stably transfected LM-3 cell line that overexpressed PARP6 and injected intravenously into the tail vein of nude mice. The empty vector group was used as a negative control. After 60 days, the nude mice were sacrificed by cervical dislocation, and the lung metastases were observed and statistically recorded. The results showed that the incidence of lung metastasis and the number of lung metastases in nude mice overexpressing PARP6 were significantly lower than those in the empty vector group (Figure 4F-H), suggesting that PARP6 can inhibit lung metastasis of HCC.
PARP6 regulates the XRCC6/Wnt/β-Catenin signalling axis
To explore how PARP6 regulates the genesis and development of HCC, we carried out a global gene expression profile to examine the differential expression genes between PARP6-overexpressing LM-3 cells and normal LM-3 cells. Over 400 genes were identified to have significant changes (FC>3, Figure 5A). GO analysis revealed that the most significant differential genes are enriched in the Wnt/β-catenin signalling pathway (Figure 5B).
Figure 5.

PARP6 regulates the XRCC6/Wnt/β-Catenin signalling axis. A. Gene chip analysis were applied to examine the differential expression genes between PARP6-overexpressing LM-3 cells and vector-transfected LM-3 cells (FC>3). B. Gene microarray analysis revealed that these genes were involved in a variety of signalling pathways. C, D. Western blotting was used to analyse the protein levels of XRCC6, β-catenin, c-Myc, MMP-7 and cyclin D1 in PARP6-overexpressing LM-3 cells and PARP6-silenced SK-Hep-1 cells. E. Western blot was used to analyse the protein levels of XRCC6, β-Catenin, c-Myc, MMP-7 and Cyclin D1 in LM-3 cells transfected with PARP6 and XRCC6 plasmids (*, P<0.05; **, P<0.01).
XRCC6, a gene coding Ku70 protein, was reported to be involved in DNA recombination and repair [10]. Several studies revealed that XRCC6 was a regulator of the β-catenin/Wnt signalling pathway [11-13]. In addition, a member of the PARP family, PARP1, was reported to interact with Ku70 and regulate its function [14]. Hence, we assumed whether PARP6 suppresses HCC progression by regulating the XRCC6 and Wnt/β-Catenin signalling pathways. To validate this assumption, we used Western blotting to examine the expression levels of XRCC6 and related proteins of the Wnt/β-Catenin signalling pathway. The expression levels of XRCC6, β-Catenin and the downstream protein level of the Wnt/β-Catenin pathway (C-Myc, Cyclin D1, MMP-7) in LM-3 cells were significantly decreased after overexpression of PARP6 (Figure 5C). Conversely, silencing PARP6 had the opposite effect (Figure 5D). Then, we co-transfected over-expressed plasmids of PARP6 and XRCC6 into LM3 cells, and the results showed that XRCC6 over-expression restored the decreased levels of XRCC6, β-Catenin and the downstream protein level of the Wnt/β-catenin pathway induced by PARP6 overexpression (Figure 5E). Together, these data suggest that PARP6 regulates the XRCC6/Wnt/β-catenin signalling axis.
PARP6 degrades XRCC6 to regulate HCC progression
We then verified whether XRCC6 is a key factor of PARP6 in the regulation of the genesis and development of HCC. Overexpressed plasmids of PARP6 and XRCC6 were co-transfected into HCC cells, and the changes in phenotype were examined. Notably, XRCC6 overexpression restored the decreased proliferation, colony formation, invasion and migration induced by PARP6 overexpression in LM-3 cells (Figure 6A-D). As XRCC6 can activate the β-catenin/Wnt signalling pathway, a Wnt/β-catenin pathway inhibitor, XAV-939, was applied to inhibit the pathway activity. The results showed that XAV-939 treatment blocked the promoting effects of XRCC6 on cell proliferation, colony formation, invasion and migration (Figure 6A-D).
Figure 6.

PARP6 regulates HCC progression through XRCC6. A, B. The viability of PARP6-overexpressing LM3 cells was examined by CCK8 and colony formation assay after treatment with XRCC6 overexpression plasmid or XAV-939 with or without XRCC6 overexpression. C, D. The invasion and migration abilities were measured by Transwell assay in PARP6-overexpressing LM3 cells after treatment with XRCC6 overexpression plasmid or XAV-939 (*, P<0.05; **, P<0.01).
Previous studies reported that HDM2 can induce XRCC6 destabilization [15]. Therefore, we assumed whether PARP6 regulates the stability of the XRCC6 protein. We observed XRCC6 protein levels in the presence of cycloheximide (CHX), an inhibitor of protein translation. PARP6 overexpression resulted in considerably faster degradation of XRCC6 (Figure 7A). Co-IP experiments showed that PARP6 co-immunoprecipitated with XRCC6 reciprocally (Figure 7B). Moreover, treatment with MG132 (a proteasome inhibitor) apparently rescued the downregulation of XRCC6 protein levels caused by PARP6 overexpression (Figure 7C). These results indicated that XRCC6 was degraded by PARP6.
Figure 7.

PARP6 interacts with HDM2 to degrade XRCC6. A. LM3 cells were co-transfected with PARP6 and XRCC6 overexpression plasmids and treated with 50 lg/ml CHX before collection at designated time points. Relative quantification of XRCC6 protein levels was assessed at different time points. B. Reciprocal Co-immunoprecipitation of PARP6 and XRCC7 in LM3 cells. C. Before collection, the protein level was examined in PARP6-overexpressing LM3 cells with or without treatment with 10 μΜ MG132 for 4 h. D. Whole lysates were immunoprecipitates with anti-HDM2 beads, then products were blotted with anti-PARP6, and anti-XRCC6 antibodies. E. Western blotting was used to analyse the protein levels of XRCC6, PARP6 and HDM2 in LM3 cells treated with or without PARP6 overexpression plasmid, and in PARP6-overexpressed LM3 cells transfected with HDM2 siRNA, or Nutlin-3 (*, P<0.05; **, P<0.01).
Previous study showed that ubiquitin ligase activity of HDM2 (human homolog of murine double minutes 2) is required for XRCC6 destabilization [15], we came to examine whether HDM2 is involved in XRCC6 destabilization caused by PARP6. Notably, Co-IP experiments demonstrated that HDM2 antibody co-immunoprecipitated with XRCC6 and PARP6 (Figure 7D). Transfection with HDM2 siRNA or treatment with Nutlin-3, an Hdm2 inhibitor, can rescue the decreased XRCC6 level induced by PARP6 overexpression (Figure 7E) suggest that HDM2 is required for PARP6-induced XRCC6 destabilization.
Discussion
In this study, using qRT-PCR, IHC and Western blotting, we demonstrated that the level of PARP6 protein expression was significantly reduced in HCC tissues compared to normal adjacent liver tissues. In vitro and in vivo experiments showed that PARP6 inhibited proliferation, colony formation, invasion and migration in hepatoma cells. In addition, through analysing the data from gene chips combined with literature mining, we discovered that PARP6 influences the progression of HCC via the XRCC6/Wnt/β-catenin signalling pathway. Notably, we demonstrated for the first time that PARP6 suppressed XRCC6 through inducing its degradation.
To date, 17 members have been found in the PARP family, such as PARP1-4, 5α, 5β and 6-16, and many of them have been shown to directly impact the genesis and development of tumours [16]. PARP1 is the earliest and most well-recognized PARP member. Our previous studies have also shown that high levels of PARP1 expression in hepatoma cells can inhibit apoptosis [17]. Targeting PARP1 for synthetic lethality is a new strategy for breast cancer treatment [18]. PARP10 was originally identified as a Myc-interacting protein [19]. PARP10 can increase the migration and invasion of tumour cells through regulation of EMT in HCC [20]. In addition, PARP14 has also been shown to accelerate lymphomagenesis driven by persistent overexpression of the oncogene c-Myc [21], PARP14 is also upregulated in HCC and is required for tumour growth [5]. The function of PARP6 has been verified in colorectal cancer and breast cancer; however, the role of PARP6 in HCC is unknown. In our study, we demonstrated that PARP6 has antitumour effects, which manifest as suppressed proliferation and metastasis abilities.
XRCC is a family of DNA repair genes whose primary function is to repair single-strand breaks and DNA-based damage [10]. Aberrant expression of XRCC6 has been reported in several types of tumours, such as lung cancer [22], gastric cancer [23] and colorectal cancer [7]. Lim et al. demonstrated that XRCC6 expression was mediated by NF-κB and that overexpression of XRCC6 contributes to cell proliferation and carcinogenesis in gastric cancer [23]. Zhang et al. reported that XRCC6 regulates hepatocellular carcinoma cell proliferation and hepatic carcinogenesis by interacting with FOXO4 [24]. Wnt/β-catenin pathway activation has been observed in many malignancies and contributes to tumour occurrence and progression [25]. Studies revealed that XRCC6 is a regulator of the β-catenin/Wnt signalling pathway in several tumour types, such as colorectal cancer [26], and osteosarcoma [13]. In our study, overexpression of PARP6 downregulated XRCC6, β-catenin and downstream genes of the Wnt/β-catenin signalling pathway, and the inhibitory effects were reversed by XRCC6 overexpression, suggesting that PARP6 inhibits the Wnt/β-catenin signalling pathway via XRCC6, which is consistent with previous studies.
The mechanism by which PARP6 regulates XRCC6 is unclear. Several reports have revealed that PARP-1 can interact with XRCC6 [26,27]. These studies prompted us to examine whether PARP-6 can interact with XRCC6 in HCC cells. Interestingly, an IP assay demonstrated that both proteins can bind to each other. Previous studies reported that several molecules participated in the regulation of Ku70 stabilization. For example, Hdm2 (human homolog of murine double minute) has the ability to degrade Ku70 [15], and the deubiquitinating enzyme USP50 has a potential role in the regulation of Ku70 stability [28]. In terms of these proteins, we speculated whether PARP6 can decrease the amount of XRCC6 through inducing degradation. To evaluate this assumption, we treated cells with the protein synthesis inhibitor CHX, and the degradation rate of XRCC6 was enhanced by PARP6 overexpression. In addition, treating cells with a proteasome inhibitor, MG132, apparently rescued the downregulation of XRCC6 protein levels caused by PARP6 overexpression. suggesting that PARP6 is involved in mediating degradation of XRCC6. Indeed, PARP6 was reported to be a mono (ADP-ribose)-generating PARP, but its biological characterization was limited. A report in breast cancer showed that PARP6 directly ADP-ribosylates Chk1, leading to MPS formation, impaired cell growth, and induction of apoptosis [8]. In addition to ADP-ribosylation function, our study demonstrated a new function of regulating degradation of PARP6 for the first time. Considering that PARP6 might not modulate protein stability independently, we further identify whether the function of PARP6 in regulating degradation depends on the participation of other proteins. Hdm2 is defined as a RING-type E3 ubiquitin ligase with a prominent role in tumorigenesis, it has been shown that Hdm2 has an ability to facilitate XRCC6 degradation [15]. Notably, in our study, we found that HDM2 can interact with PARP6 and XRCC6, and inhibiting Hdm2 can abolish the inhibitory effects of PARP6 on XRCC6 stability. So we proposed that HDM2 mediated the regulation of PARP6 on XRCC6.
In the current work, we showed that PARP6 affects the progression of HCC by regulating the XRCC6/Wnt/β-catenin signal axis and demonstrated that PARP6 regulates XRCC6 through mediating its degradation. This study compensates for the lack of research on the function and mechanism of PARP6 and lays a foundation for future diagnosis and treatment of HCC.
Acknowledgements
This research was supported in part by The National Natural Science Foundation of China (No. 81871938, No. 81560393, No. 81702435), Guangxi Science Fund for Distinguished Young Scholars Program (2016GXNSFFA380003), The Natural Science Foundation of Jiangsu (BK20170264) and the China Postdoctoral Science Foundation (2018M630606). All participants provided written informed consent, and the study was approved by the ethics committee of the Affiliated Hospital of Guilin Medical College. All animal experiments complied with the Policy of the National Institutes of Health in Hiroshima Shudo University on the Care and Use of Laboratory Animals.
Disclosure of conflict of interest
None.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ, Wu F. Solitary large hepatocellular carcinoma: a specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann Surg. 2009;249:118–123. doi: 10.1097/SLA.0b013e3181904988. [DOI] [PubMed] [Google Scholar]
- 3.Posavec Marjanovic M, Crawford K, Ahel I. PARP, transcription and chromatin modeling. Semin Cell Dev Biol. 2017;63:102–113. doi: 10.1016/j.semcdb.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iansante V, Choy PM, Fung SW, Liu Y, Chai JG, Dyson J, Del Rio A, D’Santos C, Williams R, Chokshi S, Anders RA, Bubici C, Papa S. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat Commun. 2015;6:7882. doi: 10.1038/ncomms8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuncel H, Tanaka S, Oka S, Nakai S, Fukutomi R, Okamoto M, Ota T, Kaneko H, Tatsuka M, Shimamoto F. PARP6, a mono(ADP-ribosyl) transferase and a negative regulator of cell proliferation, is involved in colorectal cancer development. Int J Oncol. 2012;41:2079–2086. doi: 10.3892/ijo.2012.1652. [DOI] [PubMed] [Google Scholar]
- 7.Qi G, Kudo Y, Tang B, Liu T, Jin S, Liu J, Zuo X, Mi S, Shao W, Ma X, Tsunematsu T, Ishimaru N, Zeng S, Tatsuka M, Shimamoto F. PARP6 acts as a tumor suppressor via downregulating Survivin expression in colorectal cancer. Oncotarget. 2016;7:18812–18824. doi: 10.18632/oncotarget.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Grosskurth SE, Cheung T, Petteruti P, Zhang J, Wang X, Wang W, Gharahdaghi F, Wu J, Su N, Howard RT, Mayo M, Widzowski D, Scott DA, Johannes JW, Lamb ML, Lawson D, Dry JR, Lyne PD, Tate EW, Zinda M, Mikule K, Fawell SE, Reimer C, Chen H. Pharmacological inhibition of PARP6 triggers multipolar spindle formation and elicits therapeutic effects in breast cancer. Cancer Res. 2018;78:6691–6702. doi: 10.1158/0008-5472.CAN-18-1362. [DOI] [PubMed] [Google Scholar]
- 9.Pinato DJ, Tan TM, Toussi ST, Ramachandran R, Martin N, Meeran K, Ngo N, Dina R, Sharma R. An expression signature of the angiogenic response in gastrointestinal neuroendocrine tumours: correlation with tumour phenotype and survival outcomes. Br J Cancer. 2014;110:115–122. doi: 10.1038/bjc.2013.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rathaus M, Lerrer B, Cohen HY. DeubiKuitylation: a novel DUB enzymatic activity for the DNA repair protein, Ku70. Cell Cycle. 2009;8:1843–1852. doi: 10.4161/cc.8.12.8864. [DOI] [PubMed] [Google Scholar]
- 11.Tavana O, Puebla-Osorio N, Kim J, Sang M, Jang S, Zhu C. Ku70 functions in addition to nonhomologous end joining in pancreatic beta-cells: a connection to beta-catenin regulation. Diabetes. 2013;62:2429–2438. doi: 10.2337/db12-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang HW, Nam HY, Kim HJ, Moon SY, Kim MR, Lee M, Kim GC, Kim SW, Kim SY. Effect of beta-catenin silencing in overcoming radioresistance of head and neck cancer cells by antagonizing the effects of AMPK on Ku70/Ku80. Head Neck. 2016;38(Suppl 1):E1909–1917. doi: 10.1002/hed.24347. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B, Cheng D, Li S, Zhou S, Yang Q. High expression of XRCC6 promotes human osteosarcoma cell proliferation through the beta-catenin/wnt signaling pathway and is associated with poor prognosis. Int J Mol Sci. 2016;17:1188. doi: 10.3390/ijms17071188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paddock MN, Bauman AT, Higdon R, Kolker E, Takeda S, Scharenberg AM. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair (Amst) 2011;10:338–343. doi: 10.1016/j.dnarep.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gama V, Gomez JA, Mayo LD, Jackson MW, Danielpour D, Song K, Haas AL, Laughlin MJ, Matsuyama S. Hdm2 is a ubiquitin ligase of Ku70-Akt promotes cell survival by inhibiting Hdm2-dependent Ku70 destabilization. Cell Death Differ. 2009;16:758–769. doi: 10.1038/cdd.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupo B, Trusolino L. Inhibition of poly(ADP-ribosyl)ation in cancer: old and new paradigms revisited. Biochim Biophys Acta. 2014;1846:201–215. doi: 10.1016/j.bbcan.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Wei K, Zhang Q, Zeng S, Lin J, Qiao L, Liu L. Expression of cluster of differentiation-95 and relevant signaling molecules in liver cancer. Mol Med Rep. 2015;11:3375–3381. doi: 10.3892/mmr.2014.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali R, Al-Kawaz A, Toss MS, Green AR, Miligy IM, Mesquita KA, Seedhouse C, Mirza S, Band V, Rakha EA, Madhusudan S. Targeting PARP1 in XRCC1-deficient sporadic invasive breast cancer or preinvasive ductal carcinoma in situ induces synthetic lethality and chemoprevention. Cancer Res. 2018;78:6818–6827. doi: 10.1158/0008-5472.CAN-18-0633. [DOI] [PubMed] [Google Scholar]
- 19.Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, Mehraein Y, Mertsching J, Kraft R, Austen M, Luscher-Firzlaff J, Luscher B. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–1993. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Hu X, Wei L, Song D, Wang J, You L, Saiyin H, Li Z, Yu W, Yu L, Ding J, Wu J. PARP10 suppresses tumor metastasis through regulation of Aurora A activity. Oncogene. 2018;37:2921–2935. doi: 10.1038/s41388-018-0168-5. [DOI] [PubMed] [Google Scholar]
- 21.Cho SH, Ahn AK, Bhargava P, Lee CH, Eischen CM, McGuinness O, Boothby M. Glycolytic rate and lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc Natl Acad Sci U S A. 2011;108:15972–15977. doi: 10.1073/pnas.1017082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YC, Chen BS. A network-based biomarker approach for molecular investigation and diagnosis of lung cancer. BMC Med Genomics. 2011;4:2. doi: 10.1186/1755-8794-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim JW, Kim H, Kim KH. Expression of Ku70 and Ku80 mediated by NF-kappa B and cyclooxygenase-2 is related to proliferation of human gastric cancer cells. J Biol Chem. 2002;277:46093–46100. doi: 10.1074/jbc.M206603200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Zhang X, Shi W, Xu J, Fan H, Zhang S, Ni R. The DNA damage repair protein Ku70 regulates tumor cell and hepatic carcinogenesis by interacting with FOXO4. Pathol Res Pract. 2016;212:153–161. doi: 10.1016/j.prp.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Idogawa M, Masutani M, Shitashige M, Honda K, Tokino T, Shinomura Y, Imai K, Hirohashi S, Yamada T. Ku70 and poly(ADP-ribose) polymerase-1 competitively regulate beta-catenin and T-cell factor-4-mediated gene transactivation: possible linkage of DNA damage recognition and Wnt signaling. Cancer Res. 2007;67:911–918. doi: 10.1158/0008-5472.CAN-06-2360. [DOI] [PubMed] [Google Scholar]
- 27.Yang G, Liu C, Chen SH, Kassab MA, Hoff JD, Walter NG, Yu X. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 2018;46:3446–3457. doi: 10.1093/nar/gky088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai J, Wei J, Schrott V, Zhao J, Bullock G, Zhao Y. Induction of deubiquitinating enzyme USP50 during erythropoiesis and its potential role in the regulation of Ku70 stability. J Investig Med. 2018;66:1–6. doi: 10.1136/jim-2017-000622. [DOI] [PMC free article] [PubMed] [Google Scholar]



