Abstract
Background: We aimed to explore the relationship between hypoxia-inducible factors-1α (HIF-1α) and lncRNA nuclear-enriched abundant transcript 1 (NEAT1), and their functions on hepatocellular carcinoma (HCC) under hypoxia. Methods: HIF-1α and NEAT1 levels in HCC tissues and corresponding non-tumor tissues were determined by qRT-PCR, and the correlations of their levels in HCC tissues were analyzed by Pearson test. The relationship between overall survival and the two genes (HIF-1α and NEAT1) for HCC patients was detected by log-rank test. Clinicopathological features of NEAT1 in HCC patients were collected. HIF-1α and NEAT1 levels in HCC cells were measured by qRT-PCR and Western blot, and their relationship was determined by co-immunoprecipitation (Co-IP) assay. Cell viability, migration and invasion were detected by CCK-8, scratch wound healing and transwell assay, respectively. The interaction of NEAT1 with HIF-1α in tumor development was determined by xenograft tumor assays in nude mice. Results: NEAT1 and HIF-1α were highly expressed and showed a positive relationship in HCC tissues, and specifically, higher NEAT1 expression was positively associated with advanced TNM stage and metastasis in HCC patients. Up-regulated NEAT1 or HIF-1α in HCC patients had poorer prognosis. NEAT1 was induced by HIF-1α and suppressed by siHIF-1α. NEAT1 overexpression further promoted development of HCC under hypoxia while promoting cell viability, migration and invasion and suppressing apoptosis, and such effects were reversed by down-regulating HIF-1α. NEAT1 overexpression promoted tumor growth, which was reversed by down-regulating HIF-1α. Conclusion: HIF-1α knockdown inhibits NEAT1 expression, which suppresses progression of HCC and improves its prognosis.
Keywords: HIF-1α, NEAT1, hepatocellular carcinoma, hypoxia, prognosis
Introduction
Hepatocellular carcinoma (HCC), which accounts for approximately 90% of primary liver cancers, is the main type of primary liver cancer and causes 700,000 deaths annually worldwide [1,2]. Meanwhile, HCC is also a major cause of cancer-related deaths in China [3]. Though some therapeutic options, such as systemic chemotherapy, targeted therapy, surgical resection, and liver transplant, have been adopted for the management of HCC [4], HCC has a high tolerance to the majority of anti-cancer treatments, and therefore the 5-year survival rate of HCC patients is still low [5]. Although efforts have been devoted to understanding basic cellular events in HCC, the exact mechanisms of liver carcinogenesis still remain unclear [6]. Thus, it is highly necessary to discover effective prognostic markers and potential molecular mechanism involved in HCC carcinogenesis so as to develop new targeted therapeutic methods for improving the prognosis of HCC patients.
Hypoxia, which can be commonly found in general solid tumors, is often involved in poor prognosis. It promotes cancer cell invasion and metastasis by activating related gene expressions via hypoxia-inducible factors (HIFs) [7]. Hypoxia-inducible factor 1 (HIF1), a heterodimeric transcription factor composed of an α subunit (HIF-1α) and a β subunit (HIF-1β), participates in hypoxic signaling pathways, and HIF-1α is found to be the most mature factor in studying the relationship between HIF-1 and tumor [8]. HIF-1α controls cellular and systemic homeostatic responses to oxygen availability [9]. Under hypoxia, it recognizes and binds to hypoxia response elements (HREs), activating the transcription of numerous genes to modulate pro-oncogenic events [10,11]. Pathogenesis of HCC is involved in tumor hypoxia and the activation of HIF [12]. In HCC, HIF-1α exerts a tumorigenic effect by promoting cell migration and invasion [13].
In recent years, multiple long non-coding RNAs (lncRNAs) have been reported to serve as important regulators in different cancers [14,15], and to be associated with the promotion or inhibition of typical cancer hallmarks, including continuous proliferation, surpassing apoptosis, drug resistance, invasion, and metastasis [16]. Structures and functions of lncRNA nuclear-enriched abundant transcript 1 (NEAT1) have been investigated widely for its significant role in numerous cancer-related pathological changes and metastasis [17]. However, the role of NEAT1 in HCC under hypoxia remains unclear.
The current study explored the expressions of HIF-1α and NEAT1, their correlations in HCC tissues and their relationships with prognosis for HCC patients. We also investigated the relationship between HIF-1α and NEAT1 and their roles in HCC progression in vitro under hypoxia and normoxia, and in tumor progression in vitro.
Materials and methods
Ethics statement
All patients signed the written informed consent before surgery, and Research Ethics Committee of Jiangsu Cancer Hospital (JCH20180504298) approved the current study according to the Declaration of Helsinki. In this study, all the animals were used in accordance with the Guidelines of the China Council on Animal Care and Use, and the experiments were approved by the Committee of Experimental Animals of Jiangsu Cancer Hospital (JCH2019060326). Efforts have been made to minimize pain or discomfort caused to the animals. The animal experiments were performed in Jiangsu Cancer Hospital.
Clinical specimens
Ninety-eight patients who were diagnosed as hepatocellular carcinoma (HCC) and received routine hepatic resection in Jiangsu Cancer Hospital from 02/16/2018 to 03/16/2019 were included in this study. The patients had no radiotherapy or chemotherapy history and were treated by surgery, and fresh tumor samples were obtained from their resected specimens. The clinicopathological characteristics (gender, age, tumor size, HBsAg, liver cirrhosis, histological differentiation, TNM stage, and metastasis) are displayed in Table 1. Tumors and adjacent fresh non-tumor tissues were frozen immediately in liquid nitrogen and stored at -80°C after the resection.
Table 1.
The relationship between NEAT1 and clinical characteristics of HCC patients
| Characteristics | N | Low NEAT1 expression (n=45) | High NEAT1 expression (n=53) | χ2 | P | |
|---|---|---|---|---|---|---|
| Gender | Male | 80 | 37 | 43 | 0.019 | 0.890 |
| Female | 18 | 8 | 10 | |||
| Age (years) | < 55 | 55 | 26 | 29 | 0.093 | 0.761 |
| ≥ 55 | 43 | 19 | 24 | |||
| Tumor size (cm) | < 5 | 56 | 27 | 29 | 0.277 | 0.598 |
| ≥ 5 | 42 | 18 | 24 | |||
| HBsAg | Negative | 10 | 5 | 5 | 0.075 | 0.785 |
| Positive | 88 | 40 | 48 | |||
| Liver cirrhosis | Absence | 22 | 10 | 12 | 0.002 | 0.960 |
| Presence | 76 | 35 | 41 | |||
| Histological differentiation | Well | 21 | 10 | 11 | 0.033 | 0.984 |
| Moderate | 35 | 16 | 19 | |||
| Poor | 42 | 19 | 23 | |||
| TNM stage | I + II | 46 | 27 | 19 | 5.699 | 0.017 |
| III + IV | 52 | 18 | 34 | |||
| Metastasis | No | 58 | 31 | 17 | 7.642 | 0.006 |
| Yes | 40 | 14 | 26 | |||
Prognosis analysis
Survival of HCC patients in relation to HIF-1α was analyzed by Kaplan-Meier and log-rank test based on data from The Cancer Genome Atlas (TCGA). Overall survival of NEAT1 in HCC patients was analyzed by Kaplan-Meier and log-rank test based on data from follow-up time (60 months).
Cell hypoxia culture
Two human liver cancer cell lines (Hep3B and SK-Hep1) were purchased from American Type Culture Collection and cultured in DMEM (12100, Solarbio, China) containing 10% fetal bovine serum (FBS; 11011-8611, Solarbio). For hypoxia treatment, the cells were cultured in an in vivo Hypoxia Work Station (Ruskinn Technology Ltd.) under normoxia (21% oxygen) or hypoxia (gas mixture of 1% oxygen, 5% CO2, and 94% N2).
Cell transfection
To construct plasmids that express NEAT1, the full-length human NEAT1 sequence was synthesized by RiboBio and subcloned into the pcDNA3.1 vector (V79020, ThermoFisher, USA). For HIF-1α knockdown, 100 pmol of HIF-1α siRNA (siHIF-1α) (siRNA; GenePharma, Shanghai, China) and siRNA negative control (NC) were respectively transfected into the cells. The empty vector was considered as a control (pc-Control) of pcDNA-NEAT1. NC served as a control of siHIF-1α. After seeded into 6-well plates, the cells were cultured for 24 h, and then transfected by Lipofectamine 2000 (11668, ThermoFisher, USA) following the instructions. The cells were collected after transfection for 48 h.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNAs were isolated from the collected tissues and cells by Trizol reagent (15596018, Invitrogen). PrimeScript™ RT Master Mix was used for RT-PCR (RR036B, Takara). Quantitative PCR was performed in 7300 real-time PCR system (Applied Biosystems, USA) using TB Green® Premix Ex Taq™ II (RR820Q, Takara). The conditions were set as follows: incubation at 95°C for 30 min, followed by amplification at 95°C for 5 sec and at 60°C for 34 sec for 40 cycles. Expressions of genes were normalized to that of GAPDH (Sangon Biotech) using 2-ΔΔCT method [18]. The sequences of primers used are displayed in Table 2.
Table 2.
Primer sequences used for quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
| Genes | Primer sequences (5’-3’) | |
|---|---|---|
| HIF-1α | Forward | GAACGTCGAAAAGAAAAGTCTCG |
| Reverse | CCTTATCAAGATGCGAACTCACA | |
| NEAT1 | Forward | CAGTTAGTTTATCAGTTCTCCCATCCA |
| Reverse | GTTGTTGTCGTCA CCTTTCAACTCT | |
| GAPDH | Forward | GCTTCGGCAGCACATATACTAAAAT |
| Reverse | CGCTTCACGAATTTGCGTGTCAT | |
Western blot
Total proteins were extracted from the cells using RIPA buffer (R0010, Solarbio). The supernatant from the total proteins was collected and the concentration was determined by BCA protein assay kit (PC0020, Solarbio). The supernatant (30 μg/lane) was loaded for SDS gel and transferred onto PVDF membranes (FFP32, Beyotime, China) by using an electroblotting apparatus. Non-specific bindings were blocked by incubating the membranes in 5% (w/v) skimmed milk in Tris-buffered saline containing 0.5% Tween-20 (w/v) buffer at 37°C for 1 h. The membranes were probed by specific primary antibodies [anti-HIF-1α (Rabbit, 1:1000, ab51608, Abcam, USA) and anti-GAPDH antibody (Mouse, 1:500, ab8245, Abcam, USA)] at 4°C overnight, and washed by TBS containing 0.1% Tween-20. The membranes were then incubated with HRP-conjugated secondary antibodies (Goat Anti-Mouse, 1:2000, ab205719, Abcam, USA; Goat Anti-Rabbit, 1:2000, ab205718, Abcam, USA) at 1:1000 dilution at 37°C for 1 h. After washing the membranes 3 times at an interval of 10 min, the signals were visualized by ECL detection kit (ECL, Premega, USA) and normalized to GAPDH.
Co-immunoprecipitation (Co-IP) assay
Co-IP assay was performed as described previously [19]. Briefly, the total proteins were extracted from the cell lines and the concentration was determined. The total proteins were incubated with anti-HIF-1α (ab51608, Abcam), or with control IgG antibodies and protein-G agarose beads (Beyotime) overnight at 4°C. The immunoprecipitates were boiled and then analyzed by Western blot.
Cell counting kit-8 (CCK-8) assay
Transfected cells (5×103 per well) were seeded in 96-well plates and cultured for 24 h. Then, CCK-8 solution (10 μL/well, CK04, Japan) was added into the plates for further incubation for 2 h. The absorbance was measured at 450 nm using a microplate reader (SpectraMax iD5, Molecular Devices, US).
Scratch wound healing assay
The cells were cultured in 6-well plates to reach 80-90% confluence, scratched by a 10 μl tip and further cultured for 24 h. The migration rate was calculated from photomicrographs under a 100× inverted microscope (Ts2r-FL, Nikon, Japan).
Invasion assays
The cells were cultured in Transwell chambers (8 mm pores, Corning Inc., Corning, USA) composed of Transwell®-precoated Matrigel™ membrane filter inserts. The 10% FBS was added into the lower chamber and served as the chemoattractant. After 24 h, the remaining cells were gently removed by cotton swabs, while those adhered to lower surface were fixed by 4% methanol for 30 min and stained by 0.1% crystal violet solution for 20 min at 37°C and photographed.
Flow cytometry assay
Cell apoptosis after treatment was determined by flow cytometry using an Annexin V-FITC Apoptosis Detection Kit (CA1020, Solarbio, China). Briefly, the cells were treated and cultured in 6-well plates. After washed by cold sterile PBS, the cells were counted, resuspended in 1X binding buffer, and incubated with 5 μL of Annexin V-FITC and blended gently for 10 min, followed by incubation with 5 μL of propidium iodide (PI) for 5 min in the dark. Then the cells were diluted with PBS to 500 μL. After that, the apoptotic rate was evaluated by flow cytometry in flow cytometer Accuri™ C6 (BD Biosciences) and the data were analyzed by Cell Quest software 3.3 (Becton-Dickinson).
Animal experiments in vivo
For tumorigenicity in vivo, 20 male BALB/c athymic nude mice (aged 6 weeks old) were randomly divided into four groups, with five mice in each group. SiHIF-1α, pc-NEAT1-transfected or SiHIF-1α, and pc-NEAT1 co-transfected Hep3B cells (1×107 cells in 100 μL) were injected subcutaneously into the left flanks of the nude mice. In the control group, tumor growth was measured every week and when the diameter reached 1 cm, the mice were sacrificed and tumors were weighted. The tumor volume was calculated according to the following equation: V = 0.5× D × d2 (V = volume; D = longitudinal diameter and d = latitudinal diameter). The tissue samples were paraffin-embedded and sectioned, and the expressions of HIF-1α protein and NEAT1 were determined by immunohistochemistry and qRT-PCR.
Immunohistochemistry
Immunohistochemistry was performed as previously described [6]. The tissues were sectioned into 4 μm thick, treated with antigen retrieval by heating ethylene diamine tetra acetic acid buffer in a microwave oven. Endogenous peroxidase activity was blocked by incubating the sections in 3% H2O2 for 30 min and then rinsed in PBS three times at an interval of 3 min. The sections were incubated with mouse monoclonal antibody against HIF-1α (ab1, 1:500, Abcam) at 4°C overnight and then incubated with secondary antibody (Goat Anti-Mouse, 1:2000, Abcam). The DAB Horseradish Peroxidase Color Development Kit (P0203, Beyotime) was used to develop color for targeting proteins. Next, the sections were redyed by Hematoxylin Staining Solution (C0107, Beyotime).
Statistical analysis
The data were shown as mean ± standard deviation (S.D.). Chi-square test was used to determine the correlation between NEAT1 and clinical features in liver cancer patients. Paired t-test was performed for paired samples. Correlation coefficients (r) were analyzed by Pearson correlation. Kaplan-Meier plots were analyzed by log-rank test. Comparisons among multiple groups were conducted by one-way ANOVA followed by Bonferroni-t post hoc test. Statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA), and P < 0.05 was considered to be of statistical significance.
Results
NEAT1 and HIF-1α were differentially overexpressed in HCC tissues and their overexpressions were associated with poorer prognosis
QRT-PCR results showed that NEAT1 and HIF-1α were obviously increased in HCC tissues compared with those in adjacent normal tissues (Figure 1A and 1B, P < 0.001). Pearson correlation analysis revealed that HIF-1α expression was positively correlated with NEAT1 expression in HCC specimens (Figure 1C, P < 0.05).
Figure 1.
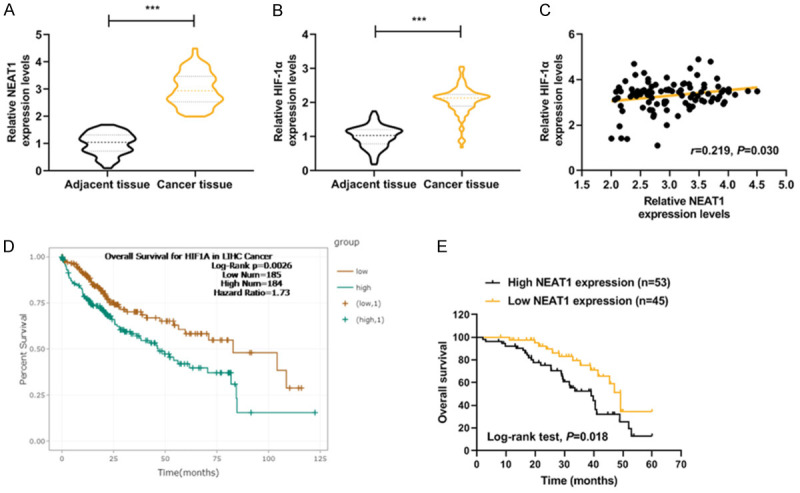
Correlations and relative expressions of HIF-1α and NEAT1 in HCC tissues and their relationships with overall survival of HCC patients. (A and B) HIF-1α (A) and NEAT1 (B) expressions were detected by qRT-PCR and normalized to GAPDH expression in 98 pairs of HCC tissues compared with adjacent nontumourous liver specimens. (C) Spearman’s rank correlation analysis was performed to analyze the correlations of HIF-1α and NEAT1 in HCC tissues. (D and E) Kaplan-Meier survival curve and log-rank test were used to evaluate whether HIF-1α and NEAT1 expression levels were associated with overall survival rate. ***P < 0.001 vs. Adjacent tissue. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; HCC, hepatocellular carcinoma.
Data on differential expression of HIF-1α in HCC cancer were collected from TCGA. The proportion of surviving HCC patients in the HIF-1α high group (n=184) was significantly lower than that in the HIF-1α low group (n=185), suggesting that HCC patients with low HIF-1α expression could have a better prognosis (Figure 1D, P=0.0026).
As shown in Table 1, HCC patients were divided into two groups, namely, low NEAT1-expression group (n=45) and high NEAT1-expression group (n=53), according to the cut-off value, which was defined as the median value of the NEAT1 level. Clinicopathological analysis showed that there were no statistically significant differences in gender, age, tumor size, HBsAg, and histological differentiation liver cirrhosis between the low NEAT1 and high NEAT1 groups (Table 1, P > 0.05); However, a high level of NEAT1 was found to be significantly associated with advanced tumor-node-metastasis (TNM) stage (Table 1, P=0.017) and metastasis of HCC (Table 1, P=0.006).
Kaplan-Meier plots showed that HCC patients in the high NEAT1-expression group (n=53) had lower overall survival rate (15%-25%) compared with the low NEAT1-expression group (n=45) (Figure 1E, P=0.018).
HIF-1α silencing reduced NEAT1 level in Hep3B and SK-Hep1 cells under hypoxia
SiHIF-1α significantly reduced the protein and mRNA levels of HIF-1α in Hep3B and SK-Hep1 cells (Figure 2A-F, P < 0.05 or P < 0.01 or P < 0.001). Nevertheless, the protein and mRNA levels of HIF-1α were highly expressed in Hep3B and SK-Hep1 cells under hypoxia (Figure 2A-F, P < 0.001), which, however, could be partially reversed by silencing HIF-1α (Figure 2A-F, P < 0.001).
Figure 2.
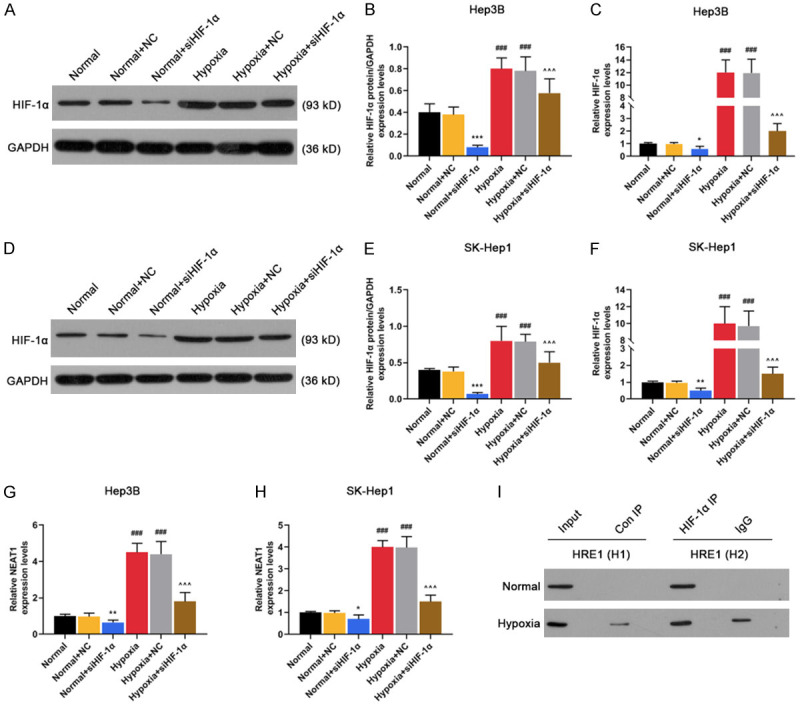
NEAT1 was induced by HIF-1α and HIF-1α interacted with the hypoxia response element (HRE) in the promoter region of NEAT1. A-F. The mRNA and protein levels of HIF-1α were detected in Normal (non-transfected control), Normal + NC (siRNA non-targeting control), Normal + siHIF-1α (siRNA-HIF-1α), Hypoxia, Hypoxia + NC, Hypoxia + siHIF-1α groups in Hep3 and SK-Hep1 cells by qRT-PCR and Western blot. G. The mRNA levels of NEAT1 were detected in Normal, Normal + NC, Normal + siHIF-1α, Hypoxia, Hypoxia + NC, Hypoxia + siHIF-1α groups in Hep3 cells by qRT-PCR. H. The mRNA levels of NEAT1 were detected in Normal, Normal + NC, Normal + siHIF-1α, Hypoxia, Hypoxia + NC, Hypoxia + siHIF-1α groups in SK-Hep1 cells by qRT-PCR. I. ChIP assays were performed to detect whether HIF-1α directly bound to the HRE of NEAT1 promoter. GAPDH served as an internal control. *P < 0.05 or **P < 0.01 or ***P < 0.001 vs. Normal + NC. ###P < 0.001 vs. Normal. ^^^P < 0.001 vs. Hypoxia + NC. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; NC, negative control.
Under hypoxia, NEAT1 was highly expressed in Hep3B (Figure 2G, P < 0.001) and SK-Hep1 cells (Figure 2H, P < 0.001), while this effect was partially reversed by HIF-1α silencing (Figure 2G and 2H, P < 0.001). In addition, we found that HIF-1α bound to hypoxia response elements (HRE) in gene promoters of NEAT1 in hypoxic Hep3B and SK-Hep1 cells (Figure 2I), suggesting that HIF-1α can regulate protein-coding genes of NEAT1.
HIF-1α silencing suppressed the development of HCC by negatively regulating NEAT1 level in Hep3B and SK-Hep1 cells under hypoxia
Hep3B and SK-Hep1 cells in the hypoxia, pc-control, pc-NEAT1, pc-NEAT1 + siHIF-1α, siHIF-1α groups were exposed to hypoxia, and we observed that hypoxia or NEAT1 overexpression significantly increased the level of NEAT1 in Hep3B (Figur 3A, P < 0.001) and SK-Hep1 cells (Figure 3B, P < 0.001), while HIF-1α silencing greatly reduced the protein level of NEAT1 (Figure 3A and 3B, P < 0.001).
Figure 3.
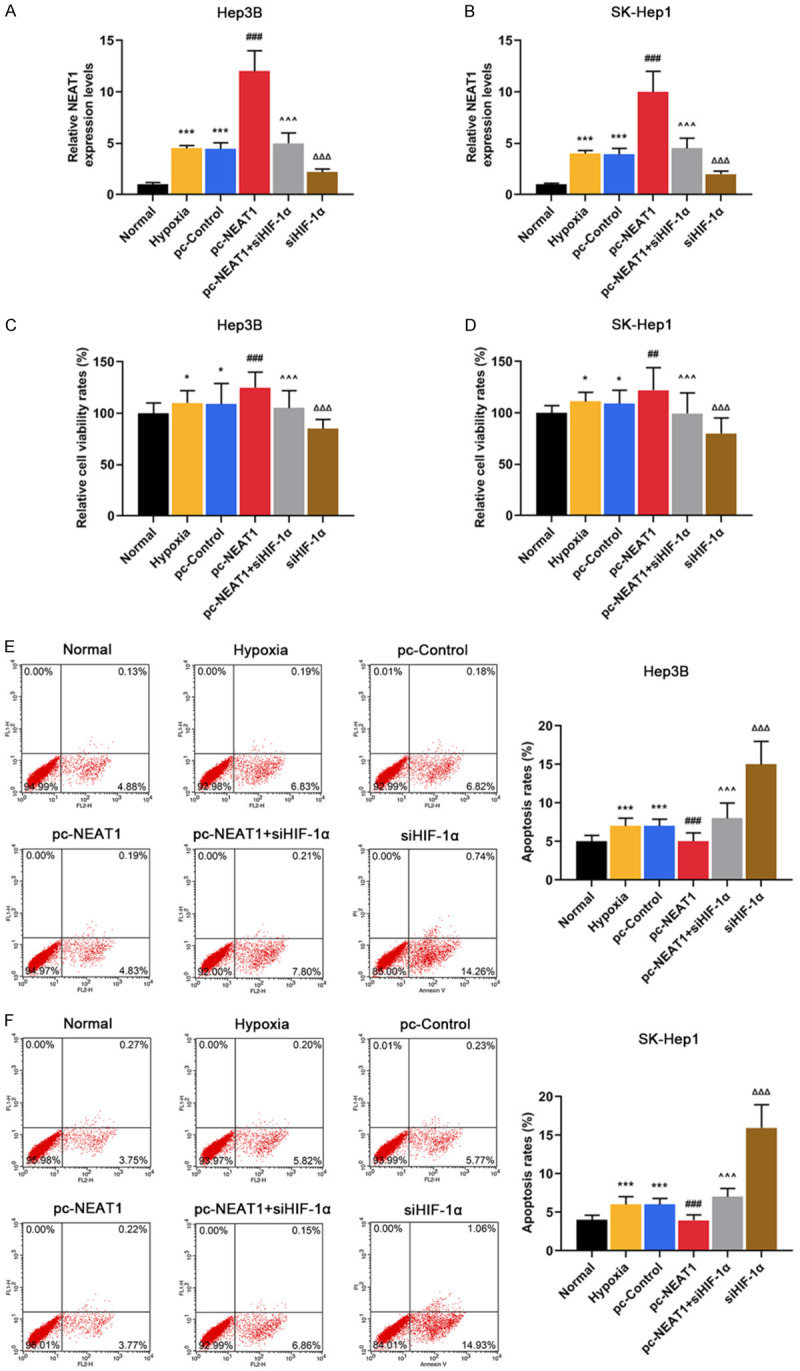
HIF-1α silencing suppressed the development of HCC by negatively regulating NEAT1 level in Hep3B and SK-Hep1 cells under hypoxia. (A and B) The mRNA levels of NEAT1 were detected in Normal (non-transfected and non-hypoxia control), Hypoxia, pc-Control (non-targeting and hypoxia control), pc-NEAT1 (NEAT1 transfected and hypoxia), pc-NEAT1 + siHIF-1α (NEAT1 and siRNA-HIF-1α co-transfected under hypoxia), siHIF-1α groups in Hep3 (A) and SK-Hep1 cells (B) by qRT-PCR. (C and D) The viability rates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1, pc-NEAT1 + siHIF-1α, siHIF-1α groups in Hep3 (C) and SK-Hep1 cells (D) by CCK-8. (E and F) The apoptosis rates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1, pc-NEAT1 + siHIF-1α, siHIF-1α groups in Hep3 (C) and SK-Hep1 cells (D) by flow cytometry assay. GAPDH served as an internal control. *P < 0.05 or ***P < 0.001 vs. Normal. ###P < 0.001 vs. pc-Control. ^^^P < 0.001 vs. pc-NEAT1. ΔΔΔP < 0.001 vs. pc-NEAT1 + siHIF-1α. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; CCK-8, cell counting kit-8.
NEAT1 overexpression further promoted the viability of Hep3B (Figure 3C, P < 0.001) and SK-Hep1 cells (Figure 3D, P < 0.01) under hypoxia. However, HIF-1α silencing significantly suppressed the cell viability under hypoxia (Figure 3C and 3D, P < 0.01), which was partially reversed NEAT1 overexpression (Figure 3C and 3D, P < 0.001). Moreover, the apoptosis rates of Hep3B (Figure 3E, P < 0.001) and SK-Hep1 cells (Figure 3F, P < 0.001) were slightly increased under hypoxia. HIF-1α silencing significantly increased the apoptosis rates of Hep3B (Figure 3E, P < 0.001) and SK-Hep1 cells (Figure 3F, P < 0.001) under hypoxia, which was partially reversed by NEAT1 overexpression (Figure 3E and 3F, P < 0.001).
As shown in Figure 4A-E, NEAT1 overexpression further promoted migration and invasion under hypoxia (P < 0.001), while HIF-1α silencing noticeably suppressed migration and invasion under hypoxia (P < 0.001), and the effects of HIF-1α silencing were partially reversed by NEAT1 overexpression (P < 0.001).
Figure 4.
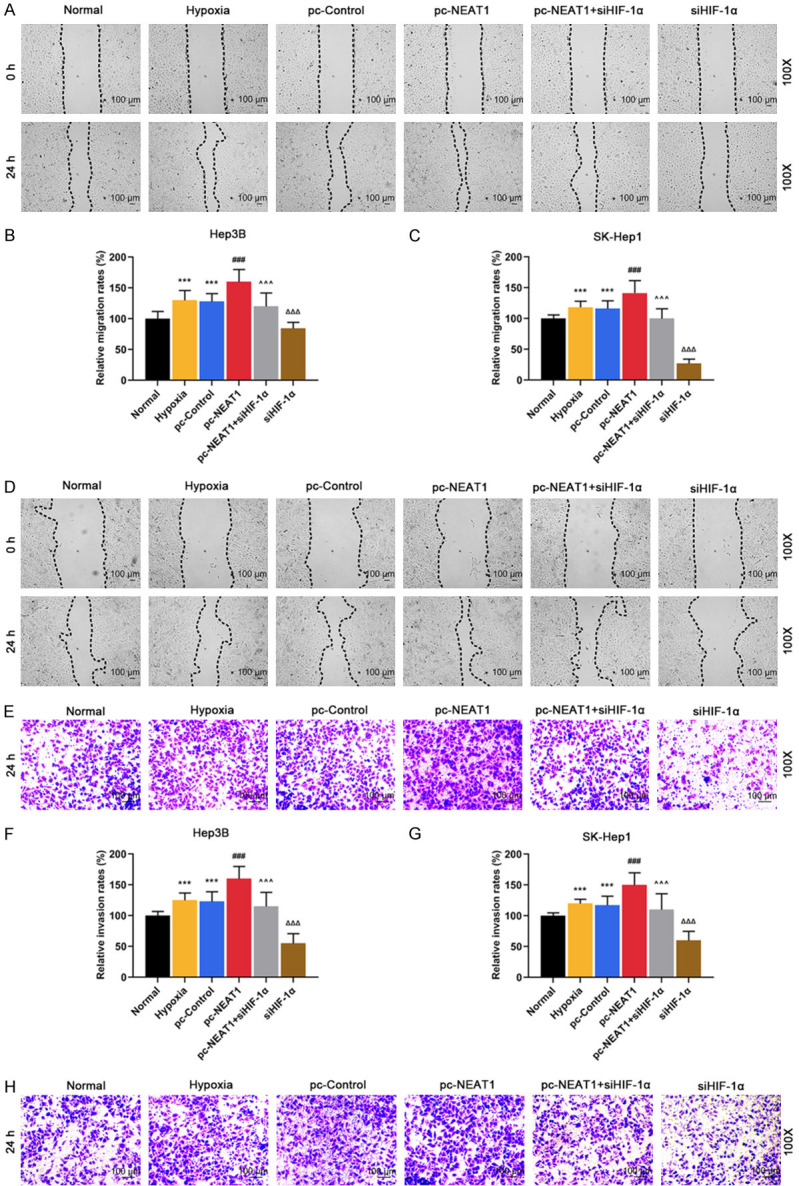
HIF-1α silencing suppressed the development of HCC by negatively regulating NEAT1 level in Hep3B and SK-Hep1 cells under hypoxia. (A, B) Migration rates were detected in Normal (non-transfected and non-hypoxia control), Hypoxia, pc-Control (non-targeting and hypoxia control), pc-NEAT1 (NEAT1 transfected and hypoxia), pc-NEAT1 + siHIF-1α (NEAT1 and siRNA-HIF-1α co-transfected under hypoxia), siHIF-1α groups in Hep3 (A and B) and SK-Hep1 cells (C and D) by scratch wound test. (E-H) Invasion rates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1, pc-NEAT1 + siHIF-1α, siHIF-1α groups in Hep3 (E and F) and SK-Hep1 cells (G and H) by transwell assay. ***P < 0.001 vs. Normal. ###P < 0.001 vs. pc-Control. ^^^P < 0.001 vs. pc-NEAT1. ΔΔΔP < 0.001 vs. pc-NEAT1 + siHIF-1α. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear-enriched abundant transcript 1.
Knocking down HIF-1α inhibited tumor growth in vivo through negatively regulating NEAT1 expression
To evaluate the effects of siHIF-1α and NEAT1 on the tumor growth of HCC cells in vivo, we established a xenograft model in which the Hep3B cells treated with siHIF-1α, or pc-NEAT1, or siHIF-1α and pc-NEAT1 were subcutaneously injected into the fland of athymic mice, respectively. The xenograft model was allowed to develop measurable tumors. The volume (Figure 5A and 5C, P < 0.001) and weight (Figure 5A and 5D, P < 0.001) of the tumors in the siHIF-1α group were significantly smaller and lighter than those in the control group, which, however, were reversed by pc-NEAT1 (Figure 5A, 5C and 5D, P < 0.001).
Figure 5.
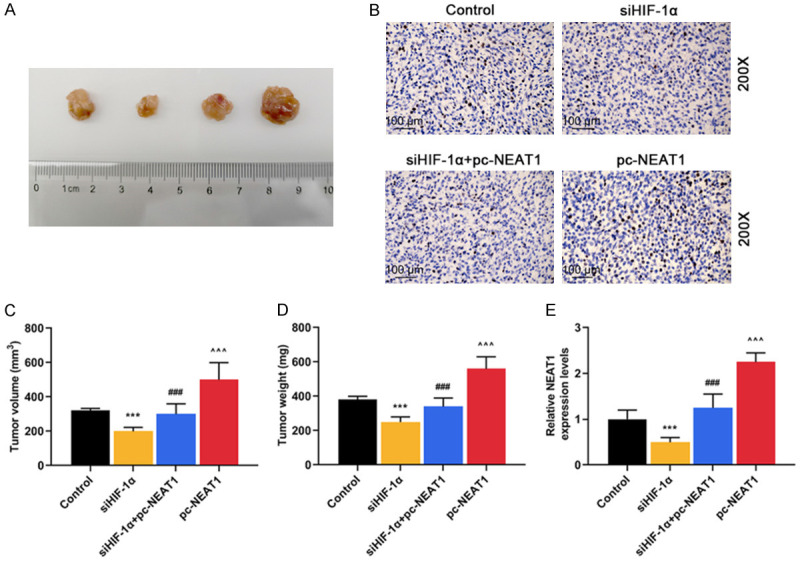
Knockdown of HIF-1α inhibited tumor growth in vivo through negatively regulating NEAT1 expression. A. Photographs of representative tumor diameter of nude mice. B. Immunohistochemistry assays were performed to detect HIF-1α protein expression in tumor tissues of Control (no injection of Hep3B cells), siHIF-1α (injection of Hep3B cells transfected with siRNA HIF-1α), siHIF-1α + pc-NEAT1 (injection of Hep3B cells transfected with siRNA HIF-1α and NEAT1) and pc-NEAT1 (injection of Hep3B cells transfected with NEAT1) groups. C. Tumor volumes were measured in tumor tissues of siHIF-1α, siHIF-1α + pc-NEAT1 and pc-NEAT1 groups. D. Tumor weights were measured in tumor tissues of siHIF-1α, siHIF-1α + pc-NEAT1 and pc-NEAT1 groups. E. The mRNA levels of NEAT1 were determined in tumor tissues of siHIF-1α, siHIF-1α + pc-NEAT1 and pc-NEAT1 groups by qRT-PCR. GAPDH served as an internal control. ***P < 0.001 vs. Control. ###P < 0.001 vs. siHIF-1α. ^^^P < 0.001 vs. siHIF-1α + pc-NEAT1. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
Next, qRT-PCR analysis of NEAT1 expression and immunostaining analysis of HIF-1α protein expression were performed in resected tumor tissues. As described in Figure 5E, the relative level of NEAT1 expression in the siHIF-1α group was significantly lower than that in the control group (P < 0.001), which was reversed by pc-NEAT1 (P < 0.001). As described in Figure 5B, siHIF-1α reduced the protein level of HIF-1α, while pc-NEAT1 did not affect the protein level of HIF-1α.
Collectively, our in vivo and in vitro data supported that NEAT1 can act as an oncogenic lncRNA in HCC, and its expression can be suppressed by HIF-1α knockdown.
Discussion
In the study, the data showed that NEAT1 and HIF-1α were highly expressed in HCC. Consistent with our findings, a recent study revealed that NEAT1 was obviously up-regulated in HCC tissues and cells [20]. We also found that higher expressions of HIF1α and NEAT1 were associated with poorer prognosis in HCC patients. Moreover, studies showed that in human HCC tissues, HIF1α is highly elevated and is associated with poorer prognosis [21,22]. Li G et al. reported that lncRNA plays an important role in tumorigenesis, subsequent prognosis and metastasis of HCC [23], which is consistent with our findings that higher NEAT1 expression was closely associated with advanced TNM stage, metastasis and prognosis in HCC patients. Liu Z et al. suggested that in HCC tissues NEAT1 overexpression is an independent risk factor associated with the prognosis of HCC patients [24]. Besides, our data directly confirmed that HIF-1α expression was positively correlated with NEAT1 expression in HCC tissues. Taken together, expressions of HIF-1α and NEAT1 have high predictive and diagnostic values and can serve as prognostic markers and treatment targets for HCC.
Hypoxia is a general symptom in solid cancers and is particularly common in HCC due to its rapid growth [25]. Hypoxia is closely related to tumor progression and negatively affects overall prognosis [26]. Our results demonstrated that HIF-1α and NEAT1 were highly expressed in HCC cells under hypoxia, suggesting that hypoxia caused poor prognosis to HCC patients possibly through overexpressing HIF-1α and NEAT1. Moreover, Co-IP results demonstrated that HIF-1α promoted NEAT1 transcription through directly binding to the HRE in the promoter region of NEAT1, which were consistent with the correlation of HIF-1α and NEAT1 in HCC tissues and the result that NEAT1 expression was inhibited by down-regulating the expression of HIF-1α. In view of this, we suggested that the overexpression of NEAT1 was induced partly by HIF-1α directly binding to the NEAT1 promoter in HCC cells under hypoxia. Therefore, we elucidated that it might be a promising therapeutic option of HCC to inhibit the expression of NEAT1 by HIF-1α silencing.
In the present study, the results demonstrated that NEAT1 further promoted the HCC cell development, evidenced by increased cell viability, migration and invasion, and suppressed apoptosis in HCC cells under hypoxia, while HIF-1α silencing further promoted the above phenomena. Consistently, previous study showed that down-regulating the expression of NEAT1 not only suppressed proliferation, migration, and invasion of HCC cells, but also promoted apoptosis of HCC cells [27]. We also found that the effects of NEAT1 overexpression in HCC cells under hypoxia were partially reversed by HIF-1α silencing. Migration refers to any directed cell movement within the body, and invasion of carcinomas is the penetration of tissue barriers [28]. The migration and invasion abilities of cancer cells enables diseases to spread through changing position within tissues or from the initial tumor [29]. Cancer cell migration and invasion into surrounding tissues and vasculature are the pivotal and initial events in cancer metastasis [29]. Frequent tumor metastasis will bring dismal outcome to HCC patients [30]. Thus, inhibition of HIF-1α might suppress HCC development and improve prognosis by negatively regulating the expression of NEAT1 under hypoxia. In addition, the results from xenograft tumor assays in nude mice further revealed that HIF-1α silencing suppressed HCC malignant progression in vitro through inhibiting the expression of NEAT1.
In brief, our results provide novel findings that improve the current understanding on HCC pathogenesis and provide a promising strategy for the diagnosis and treatment of HCC. However, HCC progression is a complex process involving the most gene networks and changes in signaling pathways, many of which still need to be illuminated [31]. For example, Ling Z A et al. [27] reported that NEAT1 promotes the development of HCC through interacting with several tumor-related genes such as SP1 and KAT2A; Mang Y et al. [32] found that NEAT1 promotes cell proliferation and invasion through modulating hnRNP A2 expression in HCC cells. These findings suggested that the mechanism of HIF-1α and NEAT1 in HCC under hypoxia may involve other downstream targets, yet more experiments are required to be performed in appropriate cells or animal models to confirm this hypothesis. However, the generate more data of some mechanistic studies should be further explore for supporting the observation and conclusion in this study.
Conclusion
Taken together, the study demonstrates that NEAT1 acts as an oncogene by promoting malignant development, and that higher expression of NEAT1 predicts a poorer prognosis of human HCC. The expression of NEAT1 could be suppressed by down-regulating the expression of HIF-1α.
Acknowledgements
This work was supported by the Young Talents Program of Jiangsu Cancer Hospital [Grant Number QL201801]; the National Natural Science Foundation of China [Grant Number 81872485]. Thanks for the financial supports.
Disclosure of conflict of interest
None.
References
- 1.Rawat D, Shrivastava S, Naik RA, Chhonker SK, Mehrotra A, Koiri RK. An overview of natural plant products in the treatment of hepatocellular carcinoma. Anticancer Agents Med Chem. 2018;18:1838–1859. doi: 10.2174/1871520618666180604085612. [DOI] [PubMed] [Google Scholar]
- 2.Pollicino T, Saitta C. Occult hepatitis B virus and hepatocellular carcinoma. World J Gastroenterol. 2014;37:439–440. doi: 10.3748/wjg.v20.i20.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu ZX, Huang JW, Liao MH, Zeng Y. Treatment strategy for hepatocellular carcinoma in China: radiofrequency ablation versus liver resection. Jpn J Clin Oncol. 2016;46:1075–1080. doi: 10.1093/jjco/hyw134. [DOI] [PubMed] [Google Scholar]
- 4.Shiani A, Narayanan S, Pena L, Friedman M. The role of diagnosis and treatment of underlying liver disease for the prognosis of primary liver cancer. Cancer Control. 2017;24:1073274817729240. doi: 10.1177/1073274817729240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin Cancer Res. 2010;16:5928–5935. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fallone F, Britton S, Nieto L, Salles B, Muller C. ATR controls cellular adaptation to hypoxia through positive regulation of hypoxia-inducible factor 1 (HIF-1) expression. Oncogene. 2013;32:4387–4396. doi: 10.1038/onc.2012.462. [DOI] [PubMed] [Google Scholar]
- 9.Snell CE, Turley H, McIntyre A, Li D, Masiero M, Schofield CJ, Gatter KC, Harris AL, Pezzella F. Proline-hydroxylated hypoxia-inducible factor 1alpha (HIF-1alpha) upregulation in human tumours. PLoS One. 2014;9:e88955. doi: 10.1371/journal.pone.0088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Zhao X, Zou H, Bai R, Yang K, Tian Z. Hypoxia Promotes gastric cancer malignancy partly through the HIF-1alpha dependent transcriptional activation of the long non-coding RNA GAPLINC. Front Physiol. 2016;7:420. doi: 10.3389/fphys.2016.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagaraju GP, Park W, Wen J, Mahaseth H, Landry J, Farris AB, Willingham F, Sullivan PS, Proia DA, El-Hariry I, Taliaferro-Smith L, Diaz R, El-Rayes BF. Antiangiogenic effects of ganetespib in colorectal cancer mediated through inhibition of HIF-1alpha and STAT-3. Angiogenesis. 2013;16:903–917. doi: 10.1007/s10456-013-9364-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Lou T. Hypoxia inducible factors in hepatocellular carcinoma. Oncotarget. 2017;8:46691–46703. doi: 10.18632/oncotarget.17358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng W, Xue T, Huang S, Shi Q, Tang C, Cui G, Yang G, Gong H, Guo H. HIF-1alpha promotes the migration and invasion of hepatocellular carcinoma cells via the IL-8-NF-kappaB axis. Cell Mol Biol Lett. 2018;23:26. doi: 10.1186/s11658-018-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, He J, Qian X, Xu P, Wang X, Li Z, Qian J, Yao J. The involvement of lncRNAs in the development and progression of pancreatic cancer. Cancer Biol Ther. 2017;18:927–936. doi: 10.1080/15384047.2017.1385682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Zhi X, Gao Y, Ta N, Jiang H, Zheng J. LncRNAs in pancreatic cancer. Oncotarget. 2016;7:57379–57390. doi: 10.18632/oncotarget.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. doi: 10.1007/978-981-10-5203-3_7. [DOI] [PubMed] [Google Scholar]
- 17.Fang L, Sun J, Pan Z, Song Y, Zhong L, Zhang Y, Liu Y, Zheng X, Huang P. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IkappaB. Am J Physiol Gastrointest Liver Physiol. 2017;313:G150–G156. doi: 10.1152/ajpgi.00426.2016. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression using different real-time quantitative PCR and 2-ΔΔCT method. Acta Agronomica Sinica. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Gao Y, Cui T, Yang T, Liu L, Li T, Chen J. Retinoic acid facilitates toll-like receptor 4 expression to improve intestinal barrier function through retinoic acid receptor beta. Cell Physiol Biochem. 2017;42:1390–1406. doi: 10.1159/000479203. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Zou Q, Song M, Chen J. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed Pharmacother. 2017;94:612–618. doi: 10.1016/j.biopha.2017.07.111. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Zhao X, Zhu D, Liu T, Liang X, Liu F, Zhang Y, Dong X, Sun B. HIF-1alpha promoted vasculogenic mimicry formation in hepatocellular carcinoma through LOXL2 up-regulation in hypoxic tumor microenvironment. J Exp Clin Cancer Res. 2017;36:60. doi: 10.1186/s13046-017-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SL, Liu LP, Jiang JX, Xiong ZF, He QJ, Wu C. The correlation of expression levels of HIF-1alpha and HIF-2alpha in hepatocellular carcinoma with capsular invasion, portal vein tumor thrombi and patients’ clinical outcome. Jpn J Clin Oncol. 2014;44:159–167. doi: 10.1093/jjco/hyt194. [DOI] [PubMed] [Google Scholar]
- 23.Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, He L, Miao R, Chen S, Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Chang Q, Yang F, Liu B, Yao HW, Bai ZG, Pu CS, Ma XM, Yang Y, Wang TT, Guo W, Zhou XN, Zhang ZT. Long non-coding RNA NEAT1 overexpression is associated with unfavorable prognosis in patients with hepatocellular carcinoma after hepatectomy: a Chinese population-based study. Eur J Surg Oncol. 2017;43:1697–1703. doi: 10.1016/j.ejso.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, Jiang K, Shen F, Wang Y, Zhang J, You Y. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci Rep. 2017;7:45029. doi: 10.1038/srep45029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling ZA, Xiong DD, Meng RM, Cen JM, Zhao N, Chen G, Li RL, Dang YW. LncRNA NEAT1 promotes deterioration of hepatocellular carcinoma based on in vitro experiments, data mining, and RT-qPCR analysis. Cell Physiol Biochem. 2018;48:540–555. doi: 10.1159/000491811. [DOI] [PubMed] [Google Scholar]
- 28.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschlager M, Dolznig H. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Duff D, Long A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal. 2017;35:250–255. doi: 10.1016/j.cellsig.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ, Ma WL. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:3076–3082. [PMC free article] [PubMed] [Google Scholar]
- 31.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239. e1224. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 32.Mang Y, Li L, Ran J, Zhang S, Liu J, Li L, Chen Y, Liu J, Gao Y, Ren G. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther. 2017;10:1003–1016. doi: 10.2147/OTT.S116319. [DOI] [PMC free article] [PubMed] [Google Scholar]


