Abstract
Background: Patients who suffered coronary heart disease (CHD) complicated with non-alcoholic fatty liver disease (NAFLD) were reported to have worse cardiac function and clinical outcomes than patients with CHD only. The mechanism was unclear. Previous study focused on the metabolism and showed it could be regulated by the microbiota. Few studies related to fungi. We aimed to investigate the characteristics of intestinal fungal microbiota in CHD patients complicated with NAFLD (CHD-NAFLD). Methods: 72 People were recruited and equally divided into three groups, including CHD patients (without NAFLD), CHD-NAFLD patients, and healthy controls (HCs). Fecal samples were collected. The Illumina sequencing of the internal transcribed spacer 3-4 rRNA was applied. Results: The BMI, uric acid and triglyceride in CHD-NAFLD patients increased compared with CHD patients. The abundance of Exophiala attenuata and Malassezia restricta in all CHD-NAFLD and CHD patients significantly reduced. The intestinal fungal microbiota in CHD-NAFLD patients showed an increase in the abundance of Preussia, Xylodon and Cladorrhinum, and a reduction in the abundance of Candida glabrata and Ganoderma. Among them, the abundance of Ganoderma was significantly lower than that in CHD patients. The ejection fraction was negatively correlated to the abundance of Xylodon. Uric acid was positively correlated with the abundance of Cladorrhinum and Preussia. Conclusions: These changes of intestinal fungal microbiota in CHD-NAFLD patients may be important factors affecting the degree of metabolic disorder. But there are few reports on these fungi. More studies are needed to confirm the effects of these fungi on human.
Keywords: Non-alcoholic fatty liver disease, coronary heart disease, intestinal microbiota
Introduction
Nonalcoholic fatty liver disease (NAFLD) was one of the most common chronic liver diseases worldwide [1,2]. However, the pathogenesis of NAFLD was still unclear. In recent years, it was believed that NAFLD tended to be caused by various factors including genetic differences, insulin resistance, intestinal microbial dysbiosis and lipid metabolism [3]. The intestinal microbiota was found to play an important role in the occurrence and development of NAFLD [4].
It was reported that NAFLD had a closely correlation with coronary atherosclerotic heart disease (CHD). The risk of cardiovascular disease was increased in NAFLD patients [5,6]. A observational study found that the incidence of atherosclerotic cardiovascular disease including CHD and ischemic stroke in CHD patients complicated with NALFD (CHD-NAFLD) was significantly higher than that in CHD patients [7]. The incidence and mortality of cardiovascular events in NAFLD patients significantly increased [8-13]. And the rate of coronary stenosis was higher in CHD-NAFLD patients than that in CHD patients without NAFLD [14] and the severity of CHD and cardiac function were also worse [15]. There were few studies on the mechanisms, especially from the perspective of intestinal microbiota.
A large number of recent studies also focused on the role of intestinal microbiota in CHD [16]. There was continuous evidence that intestinal microbiota was closely related to atherosclerosis [17]. Trimethylamine N-oxide (TMAO), formed by gut microbe-dependent metabolism, is a gut microbiota-derived metabolite that enhances both platelet responsiveness and in vivo thrombosis potential in animal models and could predict incident atherothrombotic event risks in human clinical studies. The drug TMAO inhibitor for CHD targeted on the intestinal bacterial microbiota had also made some progress [18].
Gut microbiota and metabolism played pivotal roles in the progression of CHD and NAFLD. Furthermore, fungal microbiota was an important component of the intestinal microbiota and some animal experiments showed that fungi also played a role in metabolic diseases [19,20]. However, current researches mainly focused on bacterial microbiota and there were few studies on fungal microbiota, coronary heart disease and NAFLD. Thus the characteristics of fungal microbiota in CHD patients, especially the characteristics of fungal microbiota in CHD-NAFLD patients have not been reported. This study was designed to investigate the characteristics and effects of intestinal fungal microbiota in CHD-NAFLD patients.
Materials and methods
Subject enrollment
Patients who were admitted to the Department of Gastroenterology or Cardiology in Peking University People’s Hospital from January to September in 2018 were recruited. They must meet: (1) No viral hepatitis, autoimmune liver disease and alcoholic hepatitis. No chronic gastrointestinal disease and previous abdominal surgery; (2) Left ventricular ejection fraction ≥40% and no heart failure; (3) Age between 18 and 80 years. Pregnant women or after an abortion would be also excluded in this study; (4) No antibiotics for nearly 2 weeks. No drinking alcohol, spicy food, yogurt and probiotics for nearly 1 week; (5) Normal stool frequency: 3 times/Day-3 times/week without diarrhea.
This study was approved by the Conjoint Health Research Ethics Board of Peking University People’s Hospital (No. 2018PHB033-01) and informed consent forms were obtained from all the participants. The study was carefully conducted complying with the Declaration of Helsinki.
People were divided into three groups, including CHD patients (without NAFLD), CHD-NAFLD patients and healthy controls (HCs). The overall CHD patients included CHD patients and CHD-NAFLD patients. CHD diagnosis was confirmed by coronary angiography and individuals that had ≥50% stenosis in single or multiple vessels were included. NAFLD diagnosis was confirmed based on the evidence of hepatic steatosis via imaging [21]. B-ultrasound is the preferred method for imaging diagnosis of NAFLD [22]. Considering that liver biopsy was an invasive procedure, the guidelines recommended patients with undiagnosed NAFLD or suspected coexisting chronic liver disease needed the biopsy [22]. No such patients were included in this study. Therefore, this study mainly used B-ultrasound for imaging diagnosis of NAFLD. All the healthy controls enrolled were free of NAFLD, CHD and had no clinically CHD evidence such as angina and abnormal electrocardiographic.
The CHD-NAFLD patients were 1:1 matched with CHD patients and HCs according to the gender and age (±5). All the patients would receive abdominal ultrasound and biochemical tests and the overall CHD patients has performed the coronary angiography examination in the Peking University People’s Hospital. Demographic data and clinical information were carefully collected.
Sampling and sequencing
Fresh feces of each subject were collected after admission to the hospital. All samples were collected in Stool Collection Tube with Stool Satilizer and stored in -80°C freezers before further analysis in 48 hours.
DNA was extracted from stool samples using the PSP® Spin Stool DNA Plus Kit protocol (Stratec, German). The full-length primer sequences, using standard IUPAC nucleotide nomenclature, to follow the protocol targeting this region are ITS V1-V2 Amplicon PCR Forward Primer = 5’-GGAAGTAAAAGTCGTAACAAGG, PCR Reverse Primer = 5’-GCTGCGTTCTTCATCGATGC [23]. Each PCR product of the appropriate size was purified and quantified. And then, they were added to a master pool of DNA, subsequently, a 2 × 250 paired-end sequencing was performed and base called using the MiSeq Reporter software and the MiSeq system. For the alignment, the software flash was used [24]. For the quality control, high-quality sequences (QC value ≥25) were retained by the software QC tools.
Sequencing data analysis
The main software used for sequence analysis is Vsearch v2.8.1 [25] and Usearch v10 (bit 32). The original data was merged using a double-ended sequence by Vsearch, followed by data quality control, excision of primers and barcodes. 4786644 sequences remained and 15848 sequences were removed. Then we used vsearch to remove the redundant sequences and sequences with <30 occurrences. There are 1575125143 base pairs in the 4786644 sequences with a minimum of 250 pairs and a maximum of 490 pairs (an average of 329 pairs). A total of 1889618 redundant sequences were removed and 29525 high quality sequences remained.
The chimera was removed by ESV non-cluster denoising [26] and Usearch v10 (balanced pattern) based on the reference sequence utax reference dataset 22.08.2016.fasta and a total of 1087 chimeric sequences were removed. 2497 non-chimera sequences were obtained. The Operational Taxonomic Unit (OTU) table was generated by Vsearch and the finally obtained sequence was clustered according to a certain threshold. The sequence of which the similarity is higher than 97% was defined as an OTU. In the 70 samples, a total of 4489199 reads (3584 OTUs) were obtained. Among these OTUs, 0 OTU appeared in all samples, 28 OTUs appeared in 90% of samples and 162 OTUs appeared in 50% of samples. All samples were equally sampled to 30,000 reads with Usearch V10, resulting in a total of 978664 reads (3584 OTUs). Among them, 0 OTU appeared in all samples, 15 OTUs appeared in 90% of samples and 104 OTUs appeared in 50% of samples.
Statistical analysis and visualization
The basic data were statistically analyzed using SPSSv21. Except for the special annotations, the measurement data were expressed as mean ± standard error (Mean ± SD). The data analysis between groups was analyzed by one-way ANOVA. P≤0.05 was considered statistically significant. The specific different statistical methods were described in the respective sections. Unless special annotations, the data was visualized by the ggplot2.
In the diversity analysis, Usearch v10 was used for alpha and beta diversity analysis. The beta diversity was based on the bray curtis distance. Data differences were evaluated using the adnois test.
In the difference analysis, we used the following methods: 1) Using the STAMP software [27], the two groups of independent samples were compared using the t-test. P≤0.05 was considered statistically significant. 2) Lefse (Linear Discriminant Analysis Effect Size) visualizes the abundance of the fungi with a difference of more than 2 times in abundance [28]. The method we used was the Kruskal-wallis method and the wilcoxon test. P≤0.05 was considered statistically significant and the corresponding fungi was included in the lefse analysis. Data visualization was achieved at the website (http://huttenhower.sph.harvard.edu [28]).
Indicator species analysis was performed on the genus and species levels using the indicspecies package, permutation = 999.
Correlation analysis was performed using the psych package and the stringr package, and the p value was corrected by the false discovery rate. Data visualization was performed using the pheatmap package. P≤0.05 was considered statistically significant and was labeled in the figure.
All 3584 OTU data were functionally annotated using the software FunGuild [29]. 1719 OTU data received functional annotations.
Results
Clinical characteristics
We have included three groups of 72 patients, 24 in each group. The basic information is shown. We could see that the ratio of male to female is 17/7 and the age and gender of the three groups of patients were matched. To be mentioned, though 72 patients were recruited, the microbiota information of two people in the 24 HCs was missed. So in the analysis of microbiota, 70 samples were used. The basic information is shown in (Table 1).
Table 1.
Clinical characteristics of the patients
| CHD-NAFLD | CHD | HC | (CHD-NAFLD+CHD) VS HC | CHD-NAFLD VS CHD | CHD-NAFLD VS HC | |
|---|---|---|---|---|---|---|
|
| ||||||
| (N = 24) | (N = 24) | (N = 22) | P | P | P | |
| Male/Female (N) | 17/7 | 17/7 | 15/7 | 0.822 | 1 | 0.845 |
| Age (Mean ± SD) | 63.54±7.21 | 63.50±7.70 | 63.83±7.22 | 0.622 | 0.985 | 0.67 |
| BMI | 27.74±2.72 | 24.46±5.80 | 24.84±4.22 | 0.229 | <0.001*** | 0.014* |
| HBP (N) | 18 | 17 | 11 | 0.026* | 0.745 | 0.04* |
| DM (N) | 11 | 6 | 9 | 0.659 | 0.131 | 0.736 |
| ALT (U/L) | 25.04±11.69 | 20.45±13.28 | 17.96±10.03 | 0.128 | 0.211 | 0.038* |
| AST (U/L) | 25.54±12.97 | 20.45±12.73 | 20.80±9.92 | 0.322 | 0.456 | 0.201 |
| UA (umol/L) | 405.21±103.08 | 371.33±112.13 | 328.04±76.40 | 0.032* | 0.282 | 0.01** |
| BUN (mmol/L) | 5.50±1.56 | 5.89±2.00 | 5.27±1.06 | 0.427 | 0.462 | 0.732 |
| HDL-C (mmol/L) | 1.04±0.34 | 1.03±0.24 | 1.08±0.26 | 0.441 | 0.879 | 0.581 |
| LDL-C (mmol/L) | 2.45±0.67 | 2.37±0.72 | 2.55±0.87 | 0.402 | 0.694 | 0.593 |
| TG (mmol/L) | 1.88±1.69 | 1.40±0.79 | 1.16±0.54 | 0.094 | 0.214 | 0.056 |
| Cre (umol/L) | 72.58±19.11 | 82.79±31.71 | 70.38±16.58 | 0.182 | 0.183 | 0.555 |
| EF (%) | 64.58±7,11 | 66.35±6.61 | 67.87±5.05 | 0.27 | 0.396 | 0.169 |
| NCA (N) | 1.78±0.85 | 1.63±1.10 | ||||
| HMI (N) | 6 | 5 | ||||
| Statin (N) | 24 | 24 | 12 | |||
BMI, Body mass index; HBP, High blood pressure; DM, diabetes mellitus; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; GGT, Glutamyl Transpeptidase; ALP, Alkaline phosphatase; UA, uric acid; BUN, Blood ureanitrogen, HDL-C, High-density lipoprotein cholesterol; LDL-C, Low-density lipoprotein cholesterol; TG, Triglyceride; Cre, creatinine; EF, ejection fractions; NCA, Narrowed coronary artery; HMI, History of myocardial infarction.
P≤0.05;
P≤0.01;
P≤0.001.
The levels of uric acid and triglyceride in CHD patients were higher than those in HCs. These clinical indexes in CHD-NAFLFD patients were further increased and the uric acid in CHD-NAFLD patients was significantly higher than that in HCs (P<0.05). The BMI of CHD patients was not significantly different from that of the HCs, but the BMI of CHD-NAFLFD patients was significantly higher than that of the HCs (P<0.0.5). These results indicated that the changes of BMI, uric acid and triglyceride in CHD-NAFLFD patients are higher than those in CHD patients.
In terms of cardiac function, the echocardiographic ejection fraction of CHD-NAFLD patients was lower than that of CHD patients. The number of narrowed coronary artery was higher than that of CHD patients. The narrowed coronary artery was defined as the coronary artery with more than 70% stenosis including left main coronary artery, left anterior descending artery, left circumflex artery and right coronary artery.
Diversity of the fecal fungal microbiota
We used Shannon index and chao1 index to assess the α-diversity of the fungal microbiota. Principal coordinate analysis (PCoA) was used for the β-diversity of the fungal microbiota.
The difference of Shannon index and chao1 index between the overall CHD patients and the HCs was analyzed and was not statistically different (Figure 1A and 1B). The PCOA analysis showed that there was no statistically significant difference in the composition pattern of the fungal microbiota between the overall CHD patients and the HCs (Figure 1C).
Figure 1.
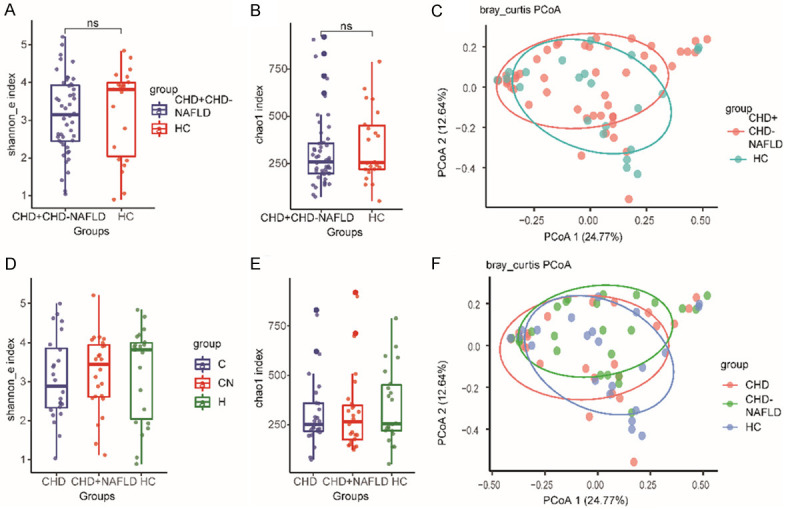
The diversity of the fecal fungal microbiota. A. The Shannon index in the overall CHD patients. B. The Chao1 index in the overall CHD patients. C. The PCoA analysis of the overall CHD patients. D. The Shannon index in CHD-NAFLD patients. E. The Chao1 index in CHD-NAFLD patients. F. The PCoA analysis of CHD-NAFLD patients. The “CHD+CHD-NAFLD” stood for the overall CHD patients. ns, not significant. 70 samples were used in each analysis. Kruskal-Wallis H test was used in the comparsion of Shannon index and Chao1 index. In the comparsion of PCoA analysis, adonis test was used.
The difference of Shannon index and chao1 index between CHD patients, CHD-NAFLD patients and the HCs was also not statistically different (Figure 1D and 1E). For the CHD-NAFLD patients, the PCoA analysis showed no significant difference with either CHD patients or HCs (Figure 1F). These results didn’t show a distinctive fungal composition in different groups.
The microbiota at phylum and genus level
Among all the identified OTUs, the Ascomycota and Basidiomycota were the two most abundant phylum (Figure 2A). For the CHD patients and CHD-NAFLD patients, the Ascomycota and Basidiomycota were also the dominant phylum (Figure 2C).
Figure 2.
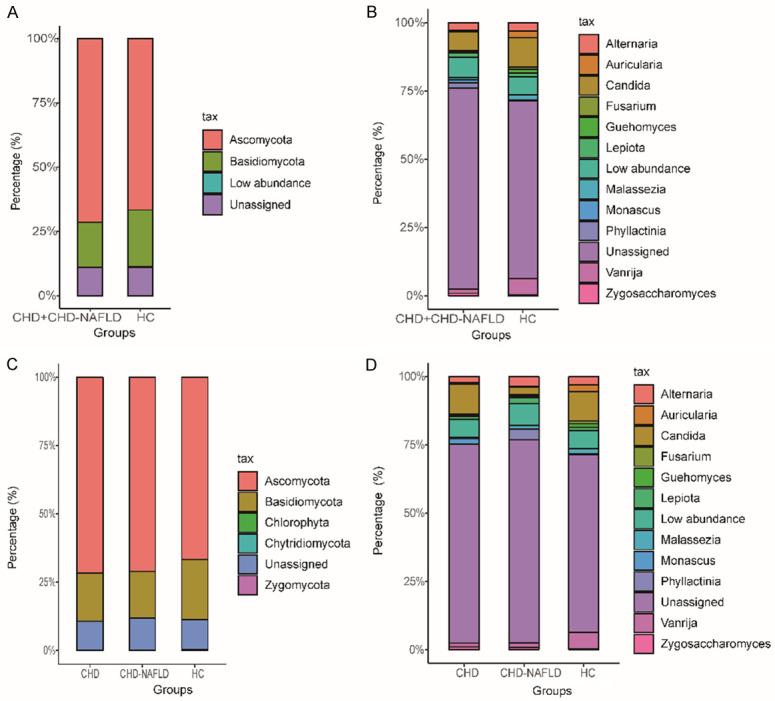
The composition of the fungi at the phylum and genus level. A. The fungi at the phylum level in the overall CHD patients. B. The fungi at the genus level in the overall CHD patients. The top 13 genus in abundance was listed. C. The fungi at the phylum level in the CHD-NAFLD and CHD patients. D. The fungi at the genus level in the CHD-NAFLD and CHD patients. The top 13 genus in abundance was listed. 70 samples were used in each analysis.
At the genus level, the composition of fungal microbiota of the overall CHD patients and HCs was analyzed (Figure 2B). Phyllactinia, Alternaria and Candida were the main genus of the fungal microbiota in the overall CHD patients and HCs.
For the CHD patients and CHD-NAFLD patients, Phyllactinia, Alternaria and Candida were also the main genus of the fungal microbiota (Figure 2D).
The characteristic of fungal microbiota of the overall CHD patients
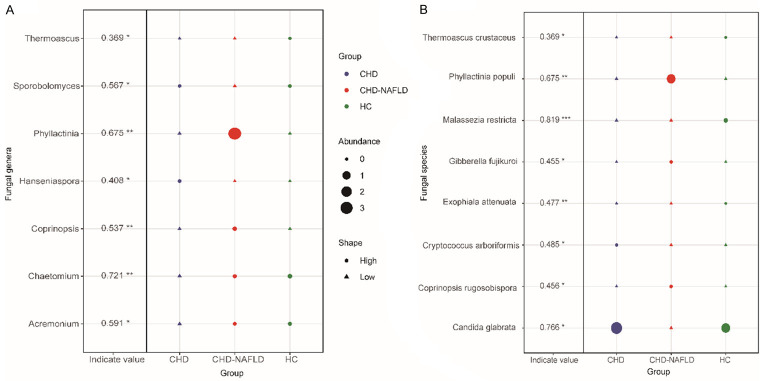
At the genus level, indicating species was used to find the characteristic of the fungal microbiota. We found that the abundance of Thermoascus in the overall CHD patients was lower than in HCs, which was the characteristic fungi of the overall CHD patients (Figure 3A).
Figure 3.
The specific microbiota at the genus and species level. A. The specific fungal microbiota at the genus level. B. The specific fungal microbiota at the species level. The R3.5.1 with indicspecies package was used. Permutation test was performed. The shape of the graph represents the comparison in enrichment (circle) or depletion (triangle) between three groups. The size of the graph indicates the relative abundance. *P≤0.05; **P≤0.01; ***P≤0.001. 70 samples were used in analysis.
At the species level, indicating species found that the abundance of Exophiala attenuata and Malassezia restricta in the overall CHD patients was lower than that in HCs, which was the characteristic of the fungal microbiota of the overall CHD patients (Figure 3B).
The characteristic of fungal microbiota of the CHD patients
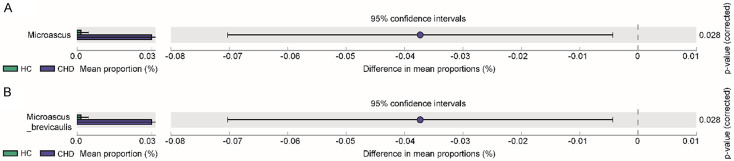
Compared with the HCs, the abundance of Microascus (P = 0.028), Microascus brevicaulis (P = 0.028) was significantly increased in the CHD patients (Figure 4).
Figure 4.
The comparison of fungal microbiota in CHD patients. A. The comparison of fungal microbiota between CHD patients and HCs at genus level. B. The comparison of fungal microbiota between CHD patients and HCs at species level. The t test and STAMP was used. 70 samples were used in each analysis.
The characteristic microbiota was analyzed using indicating species. We found that the indicating species in CHD patients was Chaetomium (P = 0.004) and Cryptococcus arboriformis (P = 0.038) (Figure 3B). Among the three groups, the Chaetomium and Cryptococcus arboriformis had the lowest abundance in the CHD patients. However, there were currently few reports on Chaetomium and Cryptococcus arboriformis.
The characteristic of fungal microbiota of the CHD-NAFLD patients
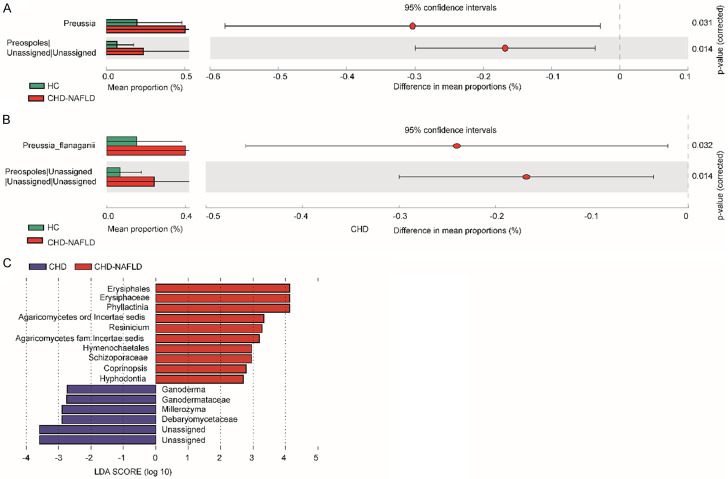
Compared with HCs, CHD-NAFLD patients had significantly higher abundance of Preussia (P = 0.031) (Figure 5). Compared with CHD patients, the abundance of Coprinopsis and Phyllactinia was significantly higher and the abundance of Ganoderma was relatively lower in CHD-NAFLD patients using the Lefse [28].
Figure 5.
The comparison of fungal microbiota in CHD-NAFLD patients. A. The comparison of fungal microbiota between CHD-NAFLD patients and HCs at genus level. B. The comparison of fungal microbiota between CHD-NAFLD patients and HCs at species level. C. The comparison of fungal microbiota between CHD-NAFLD patients and CHD patients using the Lefse. The t test and STAMP was used. 70 samples were used in analysis.
The indicating species analysis found that the indicating species of CHD-NAFLD patients was Candida glabrata (P = 0.025) (Figure 3B). Compared with the other two groups, the abundance of Candida glabrata was the lowest in CHD-NAFLD patients.
At present, there are few reports on fungi such as Preussia and Candida glabrata. Previous studies reported that Ganoderma had protective effects on atherosclerosis and NAFLD [19,20]. It was suggested that the reduction of abundance of Ganoderma in CHD-NAFLD patients might be an important factor affecting the degree of metabolic disorder.
Correlation analysis between clinical indexes and fungal microbiota at genus and species levels
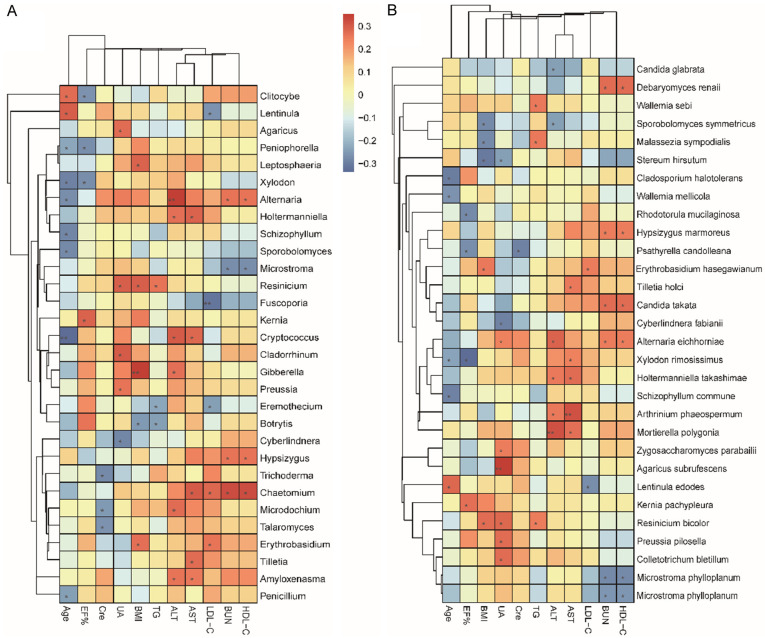
The fungal microbiota of all samples was included and Spearman’s correlation analysis was performed between fungal abundance and clinical indexes (Figure 6).
Figure 6.
The correlation analysis between clinical indexes and fungal microbiota. A. The correlation analysis between clinical indexes and fungal microbiota at the genus level. B. The correlation analysis between clinical indexes and fungal microbiota at the species level. The Spearman’s correlation analysis was used. R 3.5.1 software with pheatmap package was use for visualization. *P≤0.05; **P≤0.01; ***P≤0.001. 70 samples were used in analysis.
The ejection fraction of the overall CHD patients was lower than that of HCs. Correlation analysis found that the ejection fraction also negatively correlated with the abundance of Xylodon (P<0.05). The abundance of Xylodon in CHD-NAFLD patients was higher than that of CHD patients and HCs (the abundance of Xylodon in CHD-NAFLD, CHD, HCs: 0.0904, 0.0438, 0.0312).
Among the participants, UA increased in CHD-NAFLD patients. Correlation analysis showed that UA positively correlated with the abundance of Preussia, Cladorrhinum, Zygosaccharomyces parabailii and Alternaria eichhorniae. Compared with CHD patients and HCs, the abundance of Preussia and Cladorrhinum was the highest in CHD-NAFLD patients (the abundance of Preussia in CHD-NAFLD, CHD, HCs: 0.5233, 0.3372, 0.2468; the abundance of Cladorrhinum in CHD-NAFLD, CHD, HCs: 0.1598, 0.0796, 0.0807).
Function annotation of fungal microbiota
A total of 3,584 OTU data were annotated with the software FunGuild [29] and 1719 were functionally annotated. According to the nutrition method, the fungus can be divided into pathotroph, symbiotroph and saprotroph. And it was further subdivided into 12 categories including animal pathogens, arbuscular mycorrhizal fungi, ectomycorrhizal fungi, ericoid mycorrhizal fungi, foliar endophytes, lichenicolous fungi, lichenized fungi, mycoparasites, plantpathogens, undefined root endophytes, undefined saprotrophs and wood saprotrophs [29].
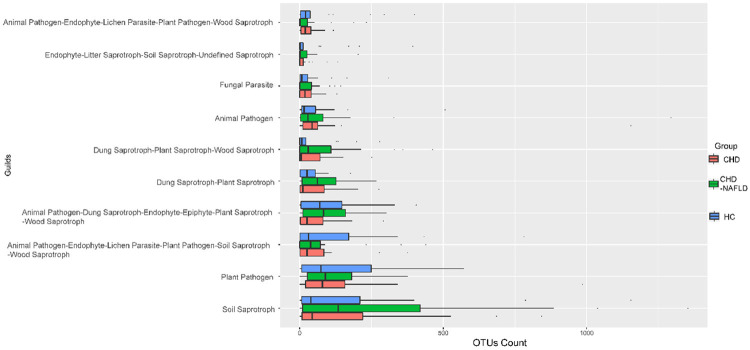
We compared the OTU levels under each function annotation. The animal pathogens increased in CHD patients compared with HCs and it was further increased in CHD-NAFLD patients, which was the characteristic of the disorder in fungal microbiota (Figure 7).
Figure 7.
Function annotation of fungal microbiota. The comparison of OTUs of function annotation in fungal microbiota between CHD patients, CHD-NAFLD patients and HCs. The software FunGuild was used. 70 samples were used in analysis.
Discussion
The change of intestinal microbiota was an important factor in the occurrence and progression of CHD and NAFLD. At present, most research focused on bacterial microbiota [17]. There was no research on the characteristics of fungal microbiota in CHD patients and CHD-NAFLD patients. Therefore, this study analyzed the characteristics of the microbiota of CHD-NAFLD patients from the perspective of fungal microbiota.
In this study, there was no significant difference in the α and β diversity of the microbiota between the overall CHD patients and HCs. At present, the diversity of the bacterial microbiota in the overall CHD patients and HCs was still controversial in previous studies and the differences in diversity reported by different studies were not consistent [30,31]. About the diversity of fungal microbiota, there was no previous reports.
There was no significant difference in the α and β diversity of the CHD-NAFLD patients compared with CHD patients and HCs, which suggested that the richness and diversity of the fungal microbiota had no significant difference between CHD patients, CHD-NAFLD patients and HCs. The diversity of gut microbiota represents the homeostasis of gut microbes and have been reported to be correlated with human health [32,33], but it was not positively correlated with the human health in all cases [34-37].
We firstly compared the overall CHD patients with HCs. In the overall CHD patients, we found that the abundance of Thermoascus and Malassezia restricta decreased compared with HCs, which was the characteristic for the overall CHD patients. There are few reports on Thermoascus. Some studies mentioned it was the pathogens of dialysis-related peritonitis [38] and its relationship with the human remained to be further studied.
The natural habitat of Malassezia was the skin of humans and other warm-blooded animals. It was classified into at least 14 species, 8 of which are isolated from human skin, including Malassezia restricta. Malassezia produced a variety of enzymes including lipases and phospholipases, which triggered an inflammatory response by releasing unsaturated free fatty acids in sebum and caused seborrheic dermatitis [39,40]. Whether Malassezia was clustered in seborrheic dermatitis remained controversial and the influence of Malassezia on lipid metabolism in human remained to be further studied [41]. Therefore, it was speculated that Malassezia restricta appeared in the intestinal microbiota and its abundance in the overall CHD patients reduced, which might be related to lipid metabolism disorder of the overall CHD patients.
The intestinal fungal microbiota in CHD-NAFLD patients showed an increase in the abundance of Preussia, Xylodon and Cladorrhinum and a reduction in the abundance of Candida glabrata and Ganoderma.
Previous studies reported that α-glucosidase inhibitors can be extracted from the products of Preussia minimoides [42], suggesting that the species in Preussia might involve in sugar metabolism and the specific mechanism remained to be confirmed. It was reported that Candida glabrata was present in plaques in patients with atherosclerosis [43]. However, it was not clear whether the presence of the fungi in the coronary atherosclerotic plaque was related to the severity of coronary artery disease. Some studies concluded that there was no correlation in it [44].
Ganoderma was reported to have a protective effect on atherosclerosis in a mouse model [45]. Some species of Ganoderma could improve the area of atherosclerotic plaque and was possibly through the regulation of macrophages and release of nitric oxide to protect atherosclerosis [20,46,47]. Some products of the Ganoderma also had a certain therapeutic effect on NAFLD mice [19] and hypoglycemic and hypolipidemic effects in diabetic mice [48]. The abundance of Ganoderma significantly reduced in CHD-NAFLD patients compared with CHD patients, which might aggravate the degree of metabolic disorder and the progression of atherosclerosis.
Correlation analysis showed that uric acid positively correlated with the abundance of Cladorrhinum and Preussia. The CHD-NAFLD showed an increase in both the UA and the abundance of Cladorrhinum and Preussia. It suggested that the abundance of Preussia and Cladorrhinum in CHD-NAFLD patients might be related to the disorder of purine metabolism and uric acid. Serum uric acid levels are closely related to cardiovascular risk factors such as hypertension and metabolic syndrome [49]. However, there are few reports on Preussia. Cladorrhinum was reported to be one of the pathogens of fungal keratitis [50]. The relationship between these two fungi and purine metabolism still needed further research.
In terms of cardiac function, the ejection fraction in CHD-NAFLD patients was the lowest compared with the CHD patients and HCs. Correlation analysis showed a negative correlation between ejection fraction and the abundance of Xylodon. The abundance of Xylodon increased in CHD-NAFLD patients. It suggested that the changes in abundance of Xylodon might be related to cardiac function. But the function of Xylodon in human was not reported yet and further research was needed.
Notably, this study has some limitations. Firstly, it is a correlation study and had no animal research on the function of the key differential fungi. And there are few previous reports about the metabolism function of the differential fungi. Thus, more studies are needed to confirm the function of the fungi. Secondly, considering that our study was a single center study, more multi-center study was needed to confirm the results. Thirdly, the study has a small sample size.
The changes of intestinal fungal microbiota in CHD-NAFLD patients may be important factors affecting the degree of metabolic disorder. But there are few reports on these fungi. More studies are needed to confirm the effects of these fungi on human.
Acknowledgements
The datasets generated during the current study are available in the Sequence Read Archive (SRA) [PRJNA541490] [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA541490]. This work was supported by the national key research and development program of China (2017YFC0908900), National Natural Science Foundation of China (81670499) and Beijing Municipal Science and Technology Project (Z171100000417022).
Disclosure of conflict of interest
None.
Abbreviations
- ALP
Alkaline phosphatase
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- BMI
Body mass index
- BUN
Blood urea nitrogen
- CHD
coronary atherosclerotic heart disease
- CHD-NAFLD
CHD patients complicated with NAFLD
- GGT
Glutamyl transpeptidase
- HCs
healthy controls
- HDL-C
High-density lipoprotein cholesterol
- LDL-C
Low-density lipoprotein cholesterol
- NAFLD
nonalcoholic fatty liver disease
- OTUs
Operational Taxonomic Units
- PCoA
Principal coordinate analysis
- PCR
Polymerase Chain Reaction
- TG
Triglyceride
- UA
Uric acid
References
- 1.Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, Landt CL, Harrison SA. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–31. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 2.Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 4.Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J, Liu Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia LS, Curzen NP, Calder PC, Byrne CD. Non-alcoholic fatty liver disease: a new and important cardiovascular risk factor? Eur Heart J. 2012;33:1190–1200. doi: 10.1093/eurheartj/ehr453. [DOI] [PubMed] [Google Scholar]
- 6.Gaggini M, Morelli M, Buzzigoli E, DeFronzo RA, Bugianesi E, Gastaldelli A. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients. 2013;5:1544–1560. doi: 10.3390/nu5051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamaguchi M, Kojima T, Takeda N, Nagata C, Takeda J, Sarui H, Kawahito Y, Yoshida N, Suetsugu A, Kato T, Okuda J, Ida K, Yoshikawa T. Nonalcoholic fatty liver disease is a novel predictor of cardiovascular disease. World J Gastroenterol. 2007;13:1579–1584. doi: 10.3748/wjg.v13.i10.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong MJ, Adams LA, Canbay A, Syn WK. Extrahepatic complications of nonalcoholic fatty liver disease. Hepatology. 2014;59:1174–1197. doi: 10.1002/hep.26717. [DOI] [PubMed] [Google Scholar]
- 9.Rafiq N, Bai C, Fang Y, Srishord M, McCullough A, Gramlich T, Younossi ZM. Long-term follow-up of patients with nonalcoholic fatty liver. Clin Gastroenterol Hepatol. 2009;7:234–238. doi: 10.1016/j.cgh.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Soderberg C, Stal P, Askling J, Glaumann H, Lindberg G, Marmur J, Hultcrantz R. Decreased survival of subjects with elevated liver function tests during a 28-year follow-up. Hepatology. 2010;51:595–602. doi: 10.1002/hep.23314. [DOI] [PubMed] [Google Scholar]
- 11.Ekstedt M, Franzen LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, Kechagias S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44:865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 12.Ekstedt M, Hagstrom H, Nasr P, Fredrikson M, Stal P, Kechagias S, Hultcrantz R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61:1547–1554. doi: 10.1002/hep.27368. [DOI] [PubMed] [Google Scholar]
- 13.Stepanova M, Rafiq N, Makhlouf H, Agrawal R, Kaur I, Younoszai Z, McCullough A, Goodman Z, Younossi ZM. Predictors of all-cause mortality and liver-related mortality in patients with non-alcoholic fatty liver disease (NAFLD) Dig Dis Sci. 2013;58:3017–3023. doi: 10.1007/s10620-013-2743-5. [DOI] [PubMed] [Google Scholar]
- 14.Wong WS, Wong LH, Yip WK, Lo OS, Limquiaco J, Chu CW, Chim ML, Yu CM, Yu J, Chan KL. Coronary artery disease and cardiovascular outcomes in patients with non-alcoholic fatty liver disease. Gut. 2011;60:1721. doi: 10.1136/gut.2011.242016. [DOI] [PubMed] [Google Scholar]
- 15.Alper AT, Hasdemir H, Sahin S, Ontürk E, Akyol A, Nurkalem Z, Cakmak N, Erdinler I, Gürkan K. The relationship between nonalcoholic fatty liver disease and the severity of coronary artery disease in patients with metabolic syndrome. Turk Kardiyol Dern Ars. 2008;36:376. [PubMed] [Google Scholar]
- 16.Sanduzzi Zamparelli M, Compare D, Coccoli P, Rocco A, Nardone OM, Marrone G, Gasbarrini A, Grieco A, Nardone G, Miele L. The metabolic role of gut microbiota in the development of nonalcoholic fatty liver disease and cardiovascular disease. Int J Mol Sci. 2016;17:1225. doi: 10.3390/ijms17081225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chacon MR, Lozano-Bartolome J, Portero-Otin M, Rodriguez MM, Xifra G, Puig J, Blasco G, Ricart W, Chaves FJ, Fernandez-Real JM. The gut mycobiome composition is linked to carotid atherosclerosis. Benef Microbes. 2018;9:185–198. doi: 10.3920/BM2017.0029. [DOI] [PubMed] [Google Scholar]
- 18.Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W, Gupta N, Skye SM, Cody DB, Levison BS, Barrington WT, Russell MW, Reed JM, Duzan A, Lang JM, Fu X, Li L, Myers AJ, Rachakonda S, DiDonato JA, Brown JM, Gogonea V, Lusis AJ, Garcia-Garcia JC, Hazen SL. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med. 2018;24:1407–1417. doi: 10.1038/s41591-018-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong D, Xie Z, Huang B, Zhu S, Wang G, Zhou H, Lin S, Lin Z, Yang B. Ganoderma lucidum polysaccharide peptide alleviates hepatoteatosis via modulating bile acid metabolism dependent on FXR-SHP/FGF. Cell Physiol Biochem. 2018;49:1163–1179. doi: 10.1159/000493297. [DOI] [PubMed] [Google Scholar]
- 20.Woo CW, Man RY, Siow YL, Choy PC, Wan EW, Lau CS, O K. Ganoderma lucidum inhibits inducible nitric oxide synthase expression in macrophages. Mol Cell Biochem. 2005;275:165–171. doi: 10.1007/s11010-005-1352-9. [DOI] [PubMed] [Google Scholar]
- 21.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American association for the study of liver diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 22.Wong VW, Chan WK, Chitturi S, Chawla Y, Dan YY, Duseja A, Fan J, Goh KL, Hamaguchi M, Hashimoto E, Kim SU, Lesmana LA, Lin YC, Liu CJ, Ni YH, Sollano J, Wong SK, Wong GL, Chan HL, Farrell G. Asia-pacific working party on non-alcoholic fatty liver disease guidelines 2017-part 1: definition, risk factors and assessment. J Gastroenterol Hepatol. 2018;33:70–85. doi: 10.1111/jgh.13857. [DOI] [PubMed] [Google Scholar]
- 23.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josse J, Sardy S, Wager S. denoiseR: a package for low rank matrix estimation. Cornell University. 2016 [Google Scholar]
- 27.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology. 2016;20:241–248. [Google Scholar]
- 30.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Li J, Guo J, Geng B, Ji W, Zhao Q, Li J, Liu X, Liu J, Guo Z, Cai W, Ma Y, Ren D, Miao J, Chen S, Zhang Z, Chen J, Zhong J, Liu W, Zou M, Li Y, Cai J. Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome. 2018;6:66. doi: 10.1186/s40168-018-0441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll IM, Tamar RK, Keku TO, Young-Hyo C, Packey CD, R Balfour S, Yehuda R. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Rob K. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huston MA, Aarssen LW, Austin MP, Cade BS, Fridley JD, Garnier E, Grime JP, Hodgson J, Lauenroth WK, Thompson K. No consistent effect of plant diversity on productivity. Science. 2000;289:1255. doi: 10.1126/science.289.5483.1255a. [DOI] [PubMed] [Google Scholar]
- 35.Fridley JD. The influence of species diversity on ecosystem productivity: how, where, and why? Oikos. 2001;93:514–526. [Google Scholar]
- 36.Cardinale BJ, Srivastava DS, J Emmett D, Wright JP, Downing AL, Mahesh S, Claire J. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature. 2006;443:989–992. doi: 10.1038/nature05202. [DOI] [PubMed] [Google Scholar]
- 37.Shade A. Diversity is the question, not the answer. ISME J. 2017;11:1–6. doi: 10.1038/ismej.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez E, Castillo A, Iturrieta I. Fungal peritonitis by Thermoascus crustaceus in a peritoneal dialysis patient from Chile. Rev Iberoam Micol. 2017;34:225–228. doi: 10.1016/j.riam.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol. 2016;55:494–504. doi: 10.1111/ijd.13116. [DOI] [PubMed] [Google Scholar]
- 40.Dawson TL Jr. Malassezia globosa and restricta: breakthrough understanding of the etiology and treatment of dandruff and seborrheic dermatitis through whole-genome analysis. J Investig Dermatol Symp Proc. 2007;12:15–19. doi: 10.1038/sj.jidsymp.5650049. [DOI] [PubMed] [Google Scholar]
- 41.Gupta AK, Kohli Y, Summerbell RC, Faergemann J. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med Mycol. 2001;39:243–251. doi: 10.1080/mmy.39.3.243.251. [DOI] [PubMed] [Google Scholar]
- 42.Rangel-Grimaldo M, Rivero-Cruz I, Madariaga-Mazon A, Figueroa M, Mata R. alpha-glucosidase inhibitors from Preussia minimoides double dagger. J Nat Prod. 2017;80:582–587. doi: 10.1021/acs.jnatprod.6b00574. [DOI] [PubMed] [Google Scholar]
- 43.Nurgeldiyeva MJ, Hojakuliyev BG, Muhammedov MB. Correlation of atherogenesis with an infection of Candida albicans. Int J Clin Exp Med. 2014;7:2137–2143. [PMC free article] [PubMed] [Google Scholar]
- 44.Masoumi O, Shahzadi M, Kordbacheh P, Zaini F, Mahmoudi S, Mahmoudi M, Bahreini H, Safara M, Mirhendi H. Detection of fungal elements in atherosclerotic plaques using mycological, pathological and molecular methods. Iran J Public Health. 2015;44:1121–1125. [PMC free article] [PubMed] [Google Scholar]
- 45.Meng J, Yang B. Protective effect of ganoderma (Lingzhi) on cardiovascular system. Adv Exp Med Biol. 2019;1182:181–199. doi: 10.1007/978-981-32-9421-9_7. [DOI] [PubMed] [Google Scholar]
- 46.Hsu PL, Lin YC, Ni H, Mo FE. Ganoderma triterpenoids exert antiatherogenic effects in mice by alleviating disturbed flow-induced oxidative stress and inflammation. Oxid Med Cell Longev. 2018;2018:3491703. doi: 10.1155/2018/3491703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andri Wihastuti T, Sargowo D, Heriansyah T, Eka Aziza Y, Puspitarini D, Nur Iwana A, Astrida Evitasari L. The reduction of aorta histopathological images through inhibition of reactive oxygen species formation in hypercholesterolemia rattus norvegicus treated with polysaccharide peptide of ganoderma lucidum. Iran J Basic Med Sci. 2015;18:514–519. [PMC free article] [PubMed] [Google Scholar]
- 48.Li F, Zhang Y, Zhong Z. Antihyperglycemic effect of ganoderma lucidum polysaccharides on streptozotocin-induced diabetic mice. Int J Mol Sci. 2011;12:6135–6145. doi: 10.3390/ijms12096135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ford ES, Li C, Cook S, Choi HK. Serum concentrations of uric acid and the metabolic syndrome among US children and adolescents. Circulation. 2007;115:2526–2532. doi: 10.1161/CIRCULATIONAHA.106.657627. [DOI] [PubMed] [Google Scholar]
- 50.Gajjar DU, Pal AK, Santos JM, Ghodadra BK, Vasavada AR. Severe pigmented keratitis caused by Cladorrhinum bulbillosum. Indian J Med Microbiol. 2011;29:434–437. doi: 10.4103/0255-0857.90191. [DOI] [PubMed] [Google Scholar]