Abstract
The discovery of new light-triggered prodrugs based on ruthenium (II) complexes is a promising approach for photoactivated chemotherapy (PACT). The light-mediated activation of “strained” Ru(II) polypyridyl complexes resulted in ligand release and produced a ligand-deficient metal center capable of forming covalent adducts with biomolecules such as DNA. Based on the strategy of exploiting structural distortion to activate photochemistry, biologically active small molecules were coordinated to a Ru(II) scaffold to create light-triggered dual-action agents. Thirteen new Ru(II) complexes with pyridyl-pyrazol(in)e ligands were synthesized, and their photochemical reactivity and anticancer properties were investigated. Isomeric bidentate ligands were investigated, where “regular” ligands (where the coordinated nitrogens in the heterocycles are linked by C–C atoms) were compared to “inverse” isomers (where the coordinated nitrogens in the heterocycles are linked by C–N atoms). Coordination of the regular 3-(pyrid-2-yl)-pyrazol(in)es to a Ru(II) bis-dimethylphenanthroline scaffold yielded photoresponsive compounds with promising photochemical and biological properties, in contrast to the inverse 1-(pyrid-2-yl)-pyrazolines. The introduction of a phenyl ring to the 1N-pyrazoline cycle increased the distortion in complexes and improved ligand release upon light irradiation (470 nm) up to 5-fold in aqueous media. Compounds 1–8, containing pyridyl-pyrazol(in)e ligands, were at least 20–80-fold more potent than the parent pyridyl-pyrazol(in)es, and exhibited biological activity in the dark, with half-maximal inhibitory concentration (IC50) values ranging from 0.2 to 7.6 μM in the HL60 cell line, with complete growth inhibition upon light irradiation. The diversification of coligands and introduction of a carboxylic acid into the Ru(II) complex resulted in compounds 9–12, with up to 146-fold improved phototoxicity indices compared with complexes 1–8.
1. Introduction
The investigation of new ruthenium-based compounds as alternatives to anticancer platinum drugs is a developing trend in modern medicinal inorganic chemistry.1−6 Based on the specific chemical features of ruthenium complexes, drug discovery efforts can be divided into two categories: (1) development of agents which are prone to ligand exchange (for example, NAMI-A and KP1339, which were in phase II clinical trials, and RAPTA-C, which is progressing toward clinical trials),7−10 and (2) investigation of kinetically inert ruthenium(II) polypyridyl complexes as either agents possessing anticancer potency without irradiation11−13 or prodrugs for photodynamic therapy (PDT; for example, TLD1433, now in phase II clinical trials) and photoactivated chemotherapy (PACT).2,6,14−16
The structural and functional diversification of bidentate ligands incorporated into octahedral ruthenium complexes has a crucial impact on both the photochemical features and the anticancer potency of compounds. The chemical modification of heteroleptic light-activated ruthenium-based prodrugs with bidentate ligands is usually focused on the optimization of polypyridine-type compounds14,17−22 or in some cases S-coordinating ligands.23,24 Recently, we investigated a class of bidentate ligands containing different five-membered ring heterocycles (indole, benzimidazole, benzoxazole, and benzothiazole).16 Using these systems, Ru(II) complexes were developed that rapidly release ligands in aqueous (aq) solutions, forming biologically active photoproducts. The compounds were highly effective in killing leukemic cells when irradiated, but the high toxicity of the compounds in the dark could be a limitation for their potential application as PACT agents.16 To continue the investigation of heteroleptic ruthenium complexes based on five-membered heterocycles, the purpose of this project was the coordination of pyridyl-pyrazoline (pyrazoline = 4,5-dihydropyrazole) and pyridyl-pyrazole ligands to Ru(II) scaffolds, aiming to identify new promising anticancer agents with potential for PDT or PACT.
Pyrazole and pyrazoline are important heterocycles contained in many bioactive agents that display a broad spectrum of biological activities, including antitumor properties.25 For example, celecoxib (a nonsteroidal anti-inflammatory drug, compound I, Chart 1) has shown promise in the prevention of cancer and has been used to reduce the number of adenomatous colorectal polyps in patients with the hereditary colon cancer susceptibility syndrome.26 Recently, encorafenib (II) has been approved for combination therapy for patients with unresectable or metastatic melanoma with a BRAF V600E or V600K mutation.27 The cytotoxic mechanisms of 3,5-diarylsubstituted pyrazolines have been associated with their inhibition of tubulin polymerization (compound III),28 mammalian cathepsin B and H (compounds IV),29 and replication protein A (compound V).30 Recently, one of us explored the hybridization of pyrazoline scaffold with other heterocycles (thiazolidine, indoline) as a direction for the enhancement of their biological properties. These pyrazoline-based conjugates possessed promising anticancer,31−36 antiviral,32 and antitrypanosomal37 activities in vitro. It is also known that the coordination of pyrazoles and pyrazolines to different metal centers is a successful approach to increase their anticancer activity. The cytotoxic effects of Pt(II),38 Pd(II),38 Cu(II),38,39 and Au(III)40 complexes (VI–VIII) with pyridyl-pyrazol(in)e ligands have been reported recently, with activity at micromolar concentrations. Ru(II) complexes with pyridyl-pyrazole ligand have been reported, and the arene complex IX exhibited selective cytotoxic activity against breast cancer cells in vitro.41 Complex X possessed light-mediated anticancer activity both in vitro and in vivo and has been identified as a potential candidate for PDT.42 Notably, the pyrazoline-thioamides coordinated to Ru(II) scaffolds XI possessed cytotoxic activity at sub-micromolar concentration (Chart 1).43
Chart 1. Small Molecules with Pyrazole (Red) and Pyrazoline (Blue) Scaffolds (I–V) and Metal Complexes Containing Pyrazol(in)e Ligands (VI–XI) That Exhibit Anticancer Activitya.
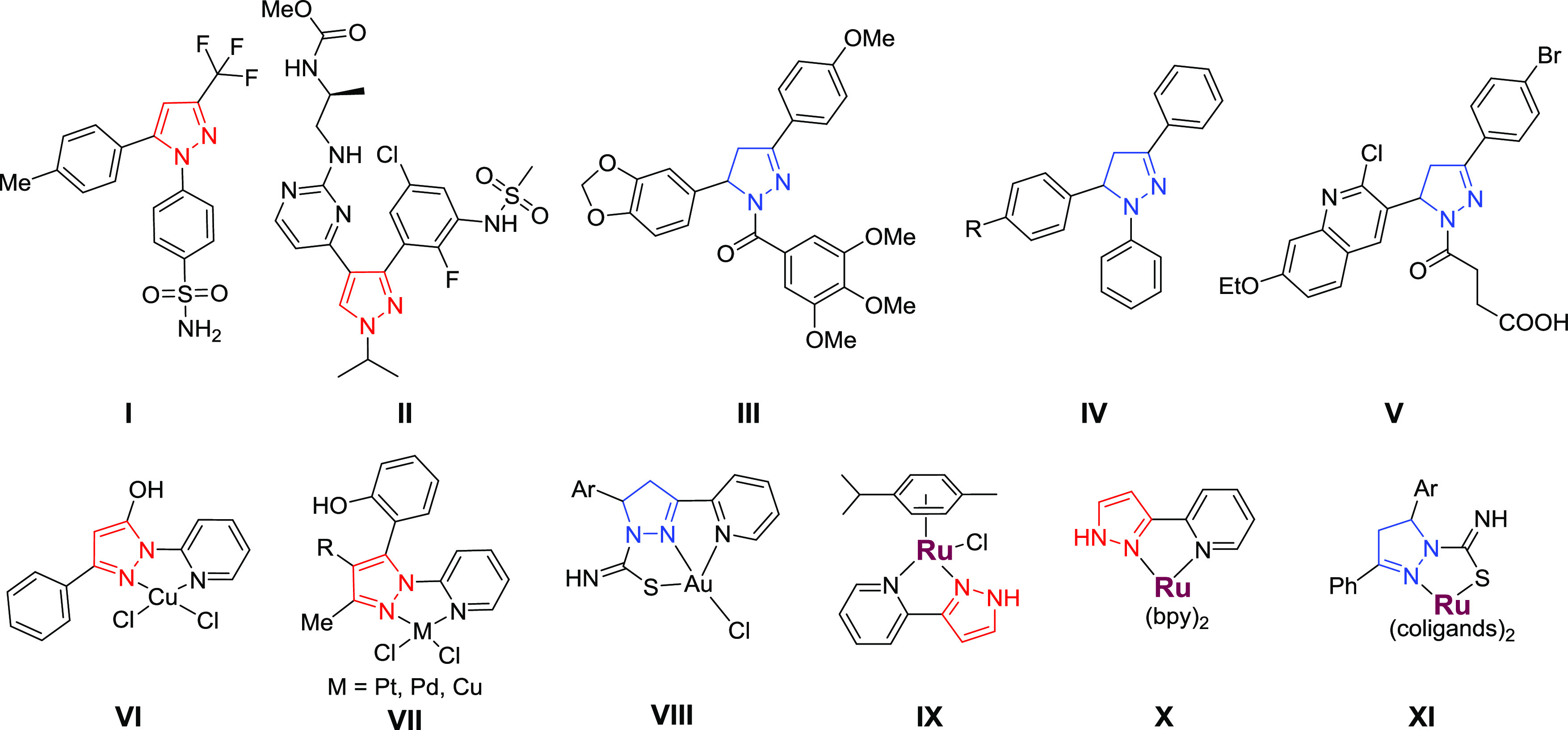
a Ru atoms are colored purple. The structures VI and VII contain C–N linkages (inverse ligands) and the structures VIII–X contain C–C linkages (regular ligands).
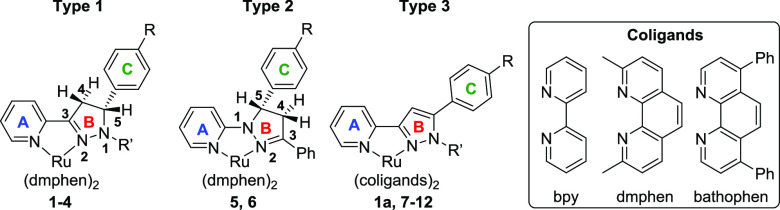
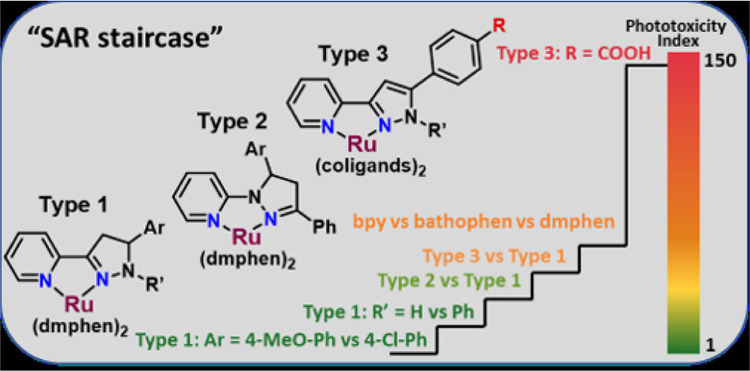
Coordination of ruthenium scaffolds containing ortho-substituted phenanthroline-type ligands ([Ru(dmphen)2]2+; dmphen is 2,9-dimethyl-1,10-phenanthroline) led to the identification of complexes with cytotoxicity toward cancer cells at micromolar concentrations.44−59 This scaffold exhibits an intrinsically distorted octahedral geometry, and when explored for PDT or PACT applications, some of the complexes exhibited sub-micromolar anticancer activity via light-mediated ligand release and covalent binding to DNA.16,17,60 In addition to steric factors, electronic features also can modulate photochemistry, and we have previously explored this through incorporation of additional nitrogens within coordinating heterocycles.61,62 Moreover, the placement of biaryl linkages in pyridyl triazoles, either at the regular position, between two carbons, or the inverse position, between a carbon and a nitrogen of the triazole, has been demonstrated to regulate the photochemistry of Ru(II) complexes.63,64 To test this strategy, we investigated analogous regular and inverse pyrazol(in)es, which we name by the same convention. Here, we report the design and synthesis of new [Ru(dmphen)2]2+ complexes based on regular 3-(pyrid-2-yl)-pyrazol(in)es (types 1 and 3, Chart 2) and inverse 1-(pyrid-2-yl)-pyrazolines (type 2), their photochemical properties in aqueous media, their in vitro anticancer activity, and the structure–activity relationships (SARs) that emerged for the tested compounds.
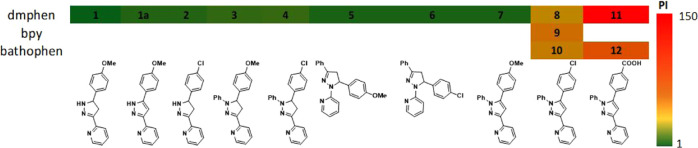
Chart 2. Ru(II) Complexes (1–12) with the Regular Pyridyl-pyrazolines (Type 1), Inverse Pyridyl-pyrazolines (Type 2), and Regular Pyridyl-pyrazoles (Type 3).
We have discovered that distorted heteroleptic Ru(II) complexes with pyridyl-pyrazol(in)e ligands are photoreactive and exhibited selective photoejection of the pyrazole-containing ligand when irradiated with blue light. Their photochemical properties and anticancer profile depended significantly on the type of pyrazol(in)e ligand (regular vs inverse). The coordination of inactive regular pyridyl-pyrazol(in)es to the Ru(II) scaffold resulted in compounds with sub-micromolar cytotoxicity without irradiation. This finding could be useful for further investigation of the complexes as traditional anticancer agents but precludes their application for PACT. Similarly, the inverse pyridyl-pyrazol(in)es were potent in the dark but were not light-active cytotoxins. However, the addition of the substituents to the aryl group appended to the 5-position of the five-membered ring heterocycle (cycle C, Chart 2) altered the key properties and rendered the regular pyridyl-pyrazol(in)es useful as PACT agents. Based on a rationale design theory, the inclusion of a carboxylic group into the pyridyl-pyrazole ligand (compound 11) radically reduced the dark cytotoxicity of Ru(II) complexes while simultaneously increasing the phototoxicity index (PI = 146). Considering the significant anticancer potential of pyrazol(in)e-based hybrids,25 these findings could be useful for the improvement of similar small-molecules’ cytotoxicity by coordination to the Ru(II) center, and/or further investigation of the potential light-activated, dual-action cytotoxic agents.
2. Results and Discussion
2.1. Chemistry
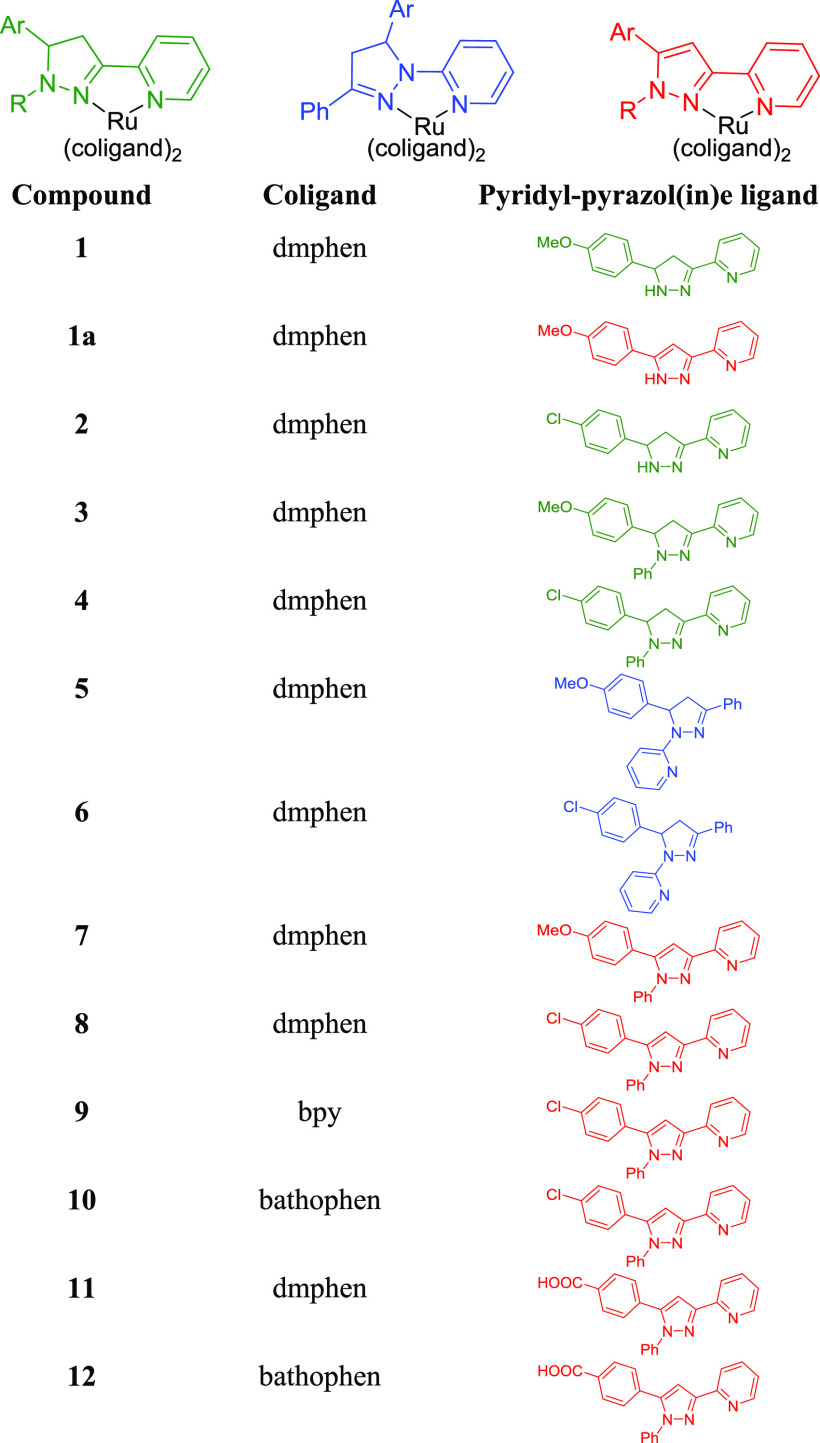
The regular 3-(pyrid-2-yl)-4,5-dihydropyrazoles were synthesized from 2-acetylpyridine via a Claisen–Schmidt condensation with aromatic aldehydes, followed by heterocyclization of chalcones to pyrazolines with hydrazine hydrate65 or phenylhydrazine.66 To expand the structure–activity relationships, we carried out the oxidation of the pyrazolines to pyrazoles67 and synthesized the inverse 1-(pyrid-2-yl)-pyrazoline ligands68 (Scheme S1). The Ru(II) complexes 1–12 (Chart 3) were synthesized from a racemic mixture of the Δ and Λ enantiomers of the Ru(II) starting materials, and form a mixture of enantiomers upon coordination of the pyridyl-pyrazol(in)e ligands. Compounds 1–8 and 11 were synthesized from Ru(dmphen)2Cl2, compound 9 from Ru(bpy)2Cl2 (bpy is 2,2′-bipyridine), and compounds 10 and 12 from Ru(bathophen)2Cl2 (bathophen is 4,7-diphenyl-1,10-phenanthroline). The purity of all compounds was determined by high-performance liquid chromatography (HPLC) (detection wavelength = 280 nm; Figures S33–S35). The Ru(II) complexes containing pyridyl-pyrazoline ligands (1–6) may exist as a mixture of four stereoisomers owing to the presence of two chiral centers (Figure S1), i.e., metal (Λ/Δ) and ligand (R/S). The 1H NMR spectra of 1–6 exhibited the characteristic patterns of two AMX systems for CH2–CH protons of the pyrazoline fragment and evidenced the presence of diastereomers in the racemic mixture. In contrast to compounds 3 and 4, there is no preference of one diastereomer for compounds 5 and 6. Interestingly, the HPLC chromatograms for 5 and 6 exhibited two peaks with similar UV/vis profiles (Figure S33), indicating the separation of different diastereomers under HPLC conditions.
Chart 3. Structures of Complexes Included in This Studya.
a The regular pyridyl-pyrazoline (green), inverse pyridyl-pyrazoline (blue), and pyridyl-pyrazole (red) ligands were combined with the indicated coligands to make bis heteroleptic Ru(II) complexes.
Upon coordination of the 3-(pyrid-2-yl)-pyrazoline ligand (L1, Scheme S1) to [Ru(dmphen)2], the oxidation of the pyrazoline cycle into pyrazole was observed. The oxidized product 1a was isolated and fully characterized by NMR, electrospray ionization mass spectrometry (ESI MS), and X-ray. Apparently, compound 1a may exist as an impurity of compound 1, and the oxidation of the pyrazoline cycle may occur not only under reaction conditions, but also upon storage when not protected from air. To minimize the impact of oxidation on compounds 1 and 2, rapid characterization was conducted by ESI MS and 1H NMR techniques. Compound 1 (as the PF6 salt) exhibited a [M]2+ ion at m/z 385.6 in the ESI MS spectrum. In contrast, the [M]2+ ion for compound 1a was detected at m/z 384.4 (Figures S36 and S37). The 1H NMR spectrum of 1 showed a minor impurity of oxidized form, but the HPLC trace of compound 1 (with Cl– counterions) exhibited two peaks (Figure S34). While the oxidation of compounds 1 and 2 under air condition is a significant disadvantage for their in-depth investigation and their potential advancement, we decided to include them for the studies reported below to determine if these structures were worth further efforts.
2.2. Crystallography
The structures of three complexes containing bidentate pyridyl-pyrazoline or pyrazole ligands, 1a, 4, and 8, were determined by X-ray crystallography and are contrasted in Figures 1 and S2–S5. Selected bond lengths and angles are listed in Table 1.
Figure 1.
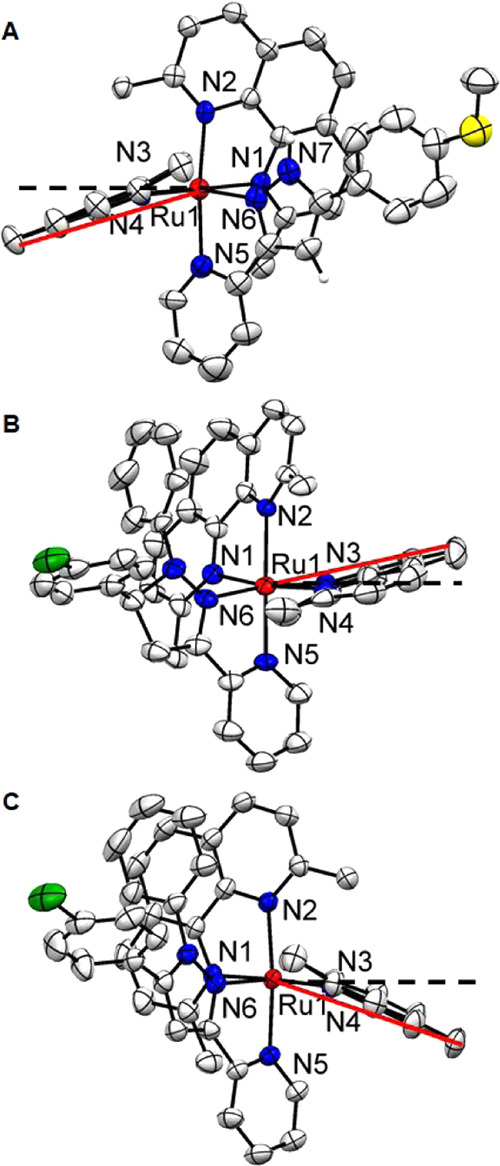
Ellipsoid plot of ruthenium complexes 1a (A), 4 (B), and 8 (C) at 50% probability with H atoms omitted for clarity. These side views highlight the distortion of the dmphen ligand. The black dashed line indicates the plane defined by the N3–Ru–N4 atoms, and the angle between red and black dash lines represents the ligand bend.
Table 1. Selected Bond Lengths (Å) and Bond Angles (deg) of 1a, 4, and 8.
| 1a | 4 | 8 | |
|---|---|---|---|
| Bond Lengths (Å) | |||
| Ru–N1 | 2.117(3) | 2.124(4) | 2.114(2) |
| Ru–N2 | 2.094(3) | 2.130(4) | 2.108(2) |
| Ru–N3 | 2.084(3) | 2.112(4) | 2.102(2) |
| Ru–N4 | 2.103(3) | 2.121(4) | 2.134(2) |
| Ru–N5 | 2.109(4) | 2.070(4) | 2.108(2) |
| Ru–N6 | 2.117(3) | 2.075(4) | 2.123(2) |
| Bond Angles (deg) | |||
| N1–Ru–N2 | 79.42(12) | 79.14(15) | 79.38(10) |
| N1–Ru–N3 | 102.41(12) | 93.16(16) | 100.63(9) |
| N1–Ru–N4 | 177.93(12) | 170.29(17) | 177.01(9) |
| N1–Ru–N5 | 96. 34(13) | 102.15(16) | 97. 5(1) |
| N1–Ru–N6 | 80.45(12) | 86.54(16) | 83.5(1) |
| N2–Ru–N3 | 93.81(12) | 85.41(15) | 92.99(9) |
| N2–Ru–N4 | 100.94(12) | 93.59(15) | 103.6(1) |
| N2–Ru–N5 | 173.62(13) | 178.71(15) | 174.12(9) |
| N2–Ru–N6 | 96.87(13) | 102.33(15) | 96.66(9) |
| N3–Ru–N4 | 79.62(12) | 79.74(18) | 79.42(9) |
| N3–Ru–N5 | 91.76(13) | 94.44(16) | 92.49(10) |
| N3–Ru–N6 | 169.30(13) | 172.04(15) | 170.07(9) |
| N4–Ru–N5 | 83.12(13) | 85.13(16) | 79.52(9) |
| N4–Ru–N6 | 97.48(13) | 101.41(17) | 95.98(9) |
| N5–Ru–N6 | 77.63(14) | 77.86(16) | 77.97(10) |
| dmphen (N1–N2) benda | 21.88 | –11.0 | 16.89 |
| dmphen (N3–N4) bendb | 19.58 | –10.64 | 22.06 |
Average angle (N3–Ru–C13/C14)—90°.
Average angle (N2–Ru–C27/C28)—90°.
All three complexes exhibited a distorted octahedral geometry due to the incorporation of two dmphen ligands. This resulted in the Ru–N(dmphen) bond lengthening to 2.100 Å (average value for 1a), 2.122 Å (average value for 4), and 2.115 Å (average value for 8) in comparison with 2.06 Å for analogous complexes with 2,2′-bipyridyl (bpy) coligands.69 The average bond length of Ru–N5 and Ru–N6 for complex 4 is about 2.07 Å, which is almost the same as that of the [Ru(bpy)2]2+ analogous complex and [Ru(bpy)3]2+.70 However, the average bond lengths for the pyridyl-pyrazole Ru–N5 and Ru–N6 for complexes 1a (2.113 Å) and 8 (2.116 Å) are equal to the length of Ru–N(dmphen) bonds and are longer than the analogous bonds for the pyridyl-pyrazoline ligand in complex 4 (Table 1) and the dipyridylphenazine ligand (dppz, 2.083 Å) coordinated to the [Ru(dmphen)2] scaffold.71
The bond angles between the trans-nitrogens of the two dmphen ligands (N1 and N4) are nonequivalent, with nearly a 10° deviation from the ideal 180° angle for complexes 1a and 4 (Table 1). Both dmphen ligands (Figure 1 and Table 1) for compounds 1a, 4, and 8 are considerably bent from the normal plane, exhibiting misdirected metal–ligand bonds.72 In addition, the directions of the dmphen bends are different. The dmphen ligand (N3–N4) is bent toward dmphen (N1–N2) for compound 4, while, in contrast, the dmphen ligand (N3–N4) bend for 1a and 8 is directed toward the pyridyl-pyrazole ligand (Figure 1). The significant distortion from the ideal octahedral geometry reflects the strain in the molecules. The nonequivalent bends of the dmphen ligands could cause a difference in the photoejection kinetics for Ru(II) complexes with pyridyl-pyrazoline and pyridyl-pyrazole ligands.
2.3. Photochemistry
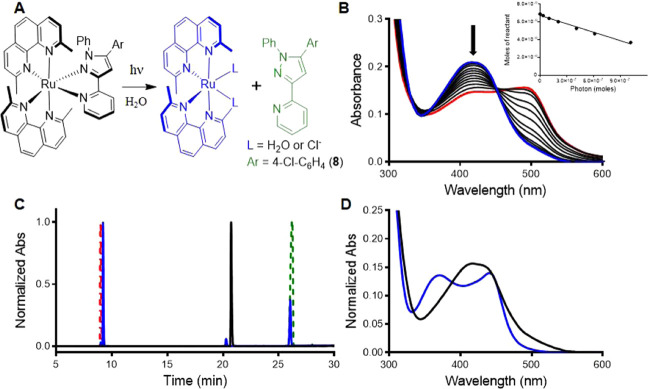
The strained Ru(II) complexes 1–8 and 11 all exhibited selective photoejection of the pyridyl-pyrazole or pyridyl-pyrazoline ligand when irradiated with 470 nm light, as shown in Figures 2 and S6–S14. The photochemical reactions were monitored by absorption spectroscopy, and the presence of an isosbestic point indicated the direct conversion to a single product (Figure 2A). The selective ejection of the pyridyl-pyrazoline/pyrazole ligands after irradiation of 4 and 8 in water was confirmed by HPLC through comparison with the starting complexes and ligands (Figure 2C,D). No dmphen was ejected under these conditions, excluding the possibility of the cytotoxic effect of dmphen in the cellular assay.73 The UV/vis spectra upon irradiation of 1–8 and 11 in water exhibited the absorbance peak at 495 nm (Figures 1B and S6–S14) corresponding to the [Ru(dmphen)2(H2O)2]2+ photoproduct.74 The photosubstitution quantum yields (ΦPS) for the complexes 1–8 and 11 with the Cl counterions were determined by an optical approach, as has been described previously.61 ΦPS values for complexes 3, 4, 7, and 8 were found to be up to 5-fold higher than for compounds 1 and 2. This variation in the photochemical reactivity suggests that introduction of a phenyl ring at the 1N of pyrazoline/pyrazole ligands increases the strain in the molecules (3, 4, 7, and 8) and improves the photochemical reactivity. Ru(II) complexes 5 and 6 with inverse ligands exhibited 2–4-fold lower ΦPS values than isomeric compounds 3 and 4 in both water and Opti-MEM. Considering that the same steric effect of the phenyl ring exists for both the regular and inverse pyridyl-ligand, the disparity in photoejection kinetics can potentially be explained by the different pKa values for bidentate ligands (Chart S1). The regular system has a lower pKa, which we have previously shown is associated with higher ΦPS values.61 Notably, the replacement of the electron-donating methoxy group (1, 3, 5, and 7) with an electron-withdrawing chlorine (2, 4, 6, and 8) had a minor effect on photoreactivity.
Figure 2.
Photochemical reactivity of compound 8. (A) Photoejection reaction scheme. (B) Photoejection of 8 (30 μM, in water) followed by UV/vis absorption over 0–60 min irradiation with 470 nm light. Inset: linear regression for moles of reactant vs moles of photons absorbed for complex 8. (C) Determination of the photoejection products by HPLC. Chromatogram of 8 before (black) and after 30 min irradiation (blue), in comparison with starting ligands dmphen (red) and pyridyl-pyrazole (green). (D) Absorption profile of 8 (black, retention time = 20.7 min) and the photochemical product [Ru(dmphen)2(L)2] (blue, retention time = 9.2 min); note that the presence of CH3CN in the HPLC experiment changes the absorption profile for the photoproduct.
2.4. DNA Damage
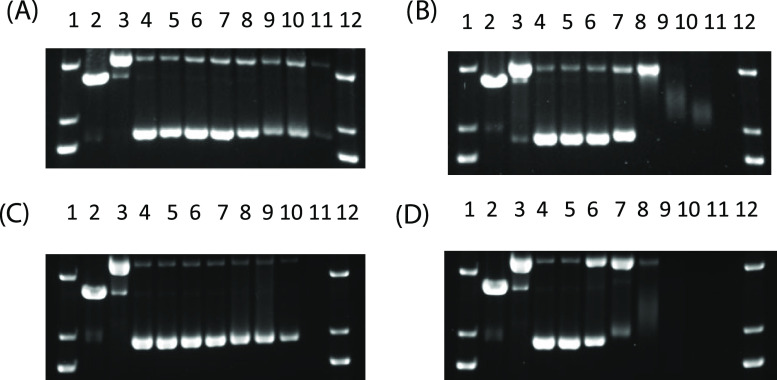
Due to the rapid photochemistry of the regular 3-(pyrid-2-yl)-1N-phenyl-pyrazol(in)e ligands in aqueous media, the interaction of compounds 4 and 8 with DNA was studied. DNA damage was assessed by gel electrophoresis using pUC19 plasmid DNA (Figure 3). Incubation of Ru(II) complexes 4 and 8 with plasmid DNA in the dark showed no interactions that caused structural perturbations at concentrations up to 125 μM (Figure 3A,C). In contrast, two types of DNA damage were observed upon irradiation with 470 nm light (40 J/cm2). Complexes 4 and 8 undergo ligand loss and covalent attachment to DNA at concentrations above 30 μM, as observed by the reduction of DNA mobility and loss of ethidium bromide (EtBr) staining.19 The presence of relaxed circular DNA (Figure 3B,D, lanes 6–8) indicated that complexes 4 and 8 also created single-strand breaks.
Figure 3.
Agarose gel electrophoresis showing the dose response of compounds 4 (A, B) and 8 (C, D) incubated with 40 μg/mL pUC19 DNA in the dark (A, C) and with irradiation (470 nm light, 40 J/cm2 (B, D)). Lanes 1 and 12, DNA ladder; lane 2, EcoRI; lane 3, Cu(OP)2; lane 4–11, 0–500 μM. EcoRI and Cu(OP)2 are controls for linear and relaxed circle DNA, respectively.
2.5. Cytotoxicity Studies
The anticancer activity of complexes 1–12 was determined against a leukemic cell line (HL60 human promyelocytic leukemia; Table 2). Cell death was determined after 72 h incubation with the complexes in the dark or following a 1 h incubation and subsequent light exposure (29.1 J/cm2) before the 72 h incubation. The initial SAR study for [Ru(dmphen)2]2+ complexes (1–8) with the pyridyl-pyrazol(in)e ligands was focused on: (a) the modification of the Ar substituent at the 5-position of the pyrazol(in)e moiety (cycle C, Chart 2); (b) the introduction of a phenyl moiety at the 1N-position of pyrazole (R′, see Chart 2); (c) regular and inverse pyridyl-pyrazoline isomerism, and (d) pyrazoline-pyrazole replacement.
Table 2. Photophysical and Photochemical Properties, Cytotoxicity Half-Maximal Inhibitory Concentration (IC50) Values (μM, HL60 Cell Line), and Heat Map for the Phototoxicity Indices for Ru(II) Complexes.
| ΦPS |
||||||
|---|---|---|---|---|---|---|
| compound | λmax (nm) | water | Opti-MEM | dark IC50, μMa | light IC50, μMa | phototoxicity index, PIb |
| 1 | 445 | 0.0011(3) | 0.0022(4) | 1.0 ± 0.1 | 1.6 ± 0.2 | 0.6 |
| 1a | 435 | 0.0062(4) | 0.0034(5) | 0.2 ± 0.1 | 0.2 ± 0.1 | 1 |
| 2 | 445 | 0.0009(3) | 0.0008(2) | 0.6 ± 0.05 | 0.5 ± 0.05 | 1.2 |
| 3 | 450 | 0.0045(5) | 0.0051(3) | 2.4 ± 0.2 | 1.5 ± 0.2 | 1.5 |
| 4 | 450 | 0.0024(3) | 0.0027(5) | 0.8 ± 0.2 | 0.5 ± 0.1 | 1.6 |
| 5 | 390 | 0.0014(3) | 0.0005(6) | 0.8 ± 0.2c | 1.3 ± 0.2 | 0.6 |
| 6 | 390 | 0.0009(3) | 0.0016(4) | 1.0 ± 0.2c | 1.4 ± 0.2 | 0.7 |
| 7 | 415 | 0.0026(4) | 0.0024(5) | 0.9 ± 0.1 | 0.9 ± 0.1 | 1 |
| 8 | 415 | 0.0032(6) | 0.0028(4) | 7.6 ± 0.3 | 1.5 ± 0.2 | 5 |
| 9 | 410 | n.r.d | n.r. | 44.7 ± 1.5 | 1.6 ± 0.3 | 28 |
| 10 | 425 | n.r. | n.r. | 8.5 ± 0.2 | 0.6 ± 0.2 | 14 |
| 11 | 415 | 0.0057(4) | 0.0061(6) | 116.7 ± 6.6 | 0.8 ± 0.3 | 146 |
| 12 | 425 | n.r. | n.r. | 29.3 ± 1.5 | 0.5 ± 0.2 | 59 |
Cytotoxicity of compounds evaluated as the average of three measurements. The IC50 value of cisplatin is 3.1 ± 0.3 μM.
The phototoxicity index (PI) is the ratio of the dark and light IC50 values.
Only 60% cell death was achieved at higher concentrations.
n.r. = no reaction.
Compounds 1–4, 7, and 8 containing the regular pyridyl-pyrazol(in)e ligands were at least 20–80-fold more potent than the parent pyridyl-pyrazol(in)e ligands (Figures 4A and S15–S19) and exhibited cytotoxicity IC50 values ranging from 0.2 to 7.6 μM. The activity of the complexes was sensitive to the modification of the Ar substituent at the 5-position of the pyrazoline moiety (cycle C), the introduction of a phenyl ring at the 1N-position (R′ = Ph), or oxidation of the pyrazoline moiety to the aromatic pyrazole (cycle B). The replacement of the electron-donating methoxy group with an electron-withdrawing chlorine had an ambiguous effect. Ruthenium complexes with chloro-substituted pyrazolines (2, 4) exhibited around 2-fold larger anticancer activities than complexes 1 and 3 with the MeO group. However, complex 7 with the p-MeO-phenyl-substituted pyrazole ligand was more active than 8. Despite only small reductions in the cytotoxic properties after introduction of the phenyl ring at the 1N-position (compounds 3, 4), it should be noted that compounds 1 and 2 (1NH-pyrazoline derivatives) exhibited total growth inhibition of cancer cells (0% viable cells) at lower concentrations than corresponding N-Ph-substituted analogous complexes 3 and 4 (Figure S16). The coordination of inverse pyridyl-pyrazoline ligands (type 2) to the [Ru(dmphen)2]2+ scaffold (complexes 5 and 6) did not result in significant improvements in anticancer activity compared to isomeric compounds 3 and 4. Notably, compound 1a exhibited the highest anticancer activity on the HL60 cell line with an IC50 value of 0.2 μM, making it 15-fold more potent than cisplatin.
Figure 4.
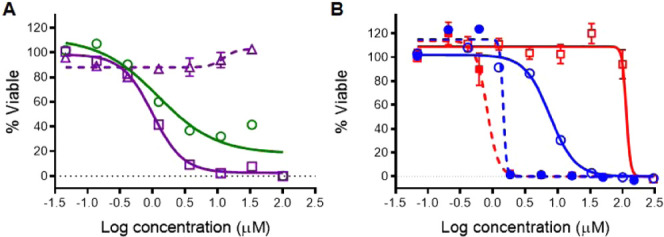
Cytotoxicity dose responses of ruthenium complexes 1, 3, 8, and 11 on the HL60 cells (n = 3): (A) compound 1 (□, solid purple line) in comparison with the parent pyridyl-pyrazoline ligand (Δ, dashed purple line) and complex 3 (○, solid green line); (B) light-mediated cytotoxicity for compounds 8 (○, blue) and 11 (□, red), indicating the improvement in the PI by introduction of a carboxylic acid group into the pyridyl-pyrazole ligand (solid line—dark conditions; dashed line—upon 1 min irradiation with >450 nm light, 29.1 J/cm2).
The photoreactive compounds 1–8 were tested for their anticancer activity upon light activation, but except for compound 8, did not exhibit significant improvements in cytotoxicity upon irradiation (see the phototoxicity index (PI) values in Table 2). However, light activation of 3–8 resulted in much steeper dose response curves, achieving essentially complete cell death (0% viable cells) at 10-fold lower concentrations than the same compound in the dark (Figures S16 and S17). This indicates a different mechanisms of action with regards to the cause of the cell death upon irradiation vs the nonirradiated cells.
Considering the low phototoxicity indices for compounds 1–8, the focus shifted to diversification of the coligands in the Ru(II) heteroleptic complexes. For this, bpy and bathophen coligands were incorporated, creating two unstrained Ru(II) complexes (9 and 10), analogous to complex 8 (Table 2). Introduction of the bpy coligands into heteroleptic complex 9 considerably reduced the dark cytotoxicity in comparison to complex 8 and improved the light-mediated potency, with PI = 28. Complex 10, with bathophen coligands, possessed the same range of activity as 8 under dark conditions, but showed sub-micromolar potency upon irradiation, with PI = 14.
Recently, we discovered that the introduction of a carboxylic acid-contained ligand (2,2′-biquinoline-4,4′-dicarboxylic acid) into Ru(II) complexes reduces the cytotoxicity and produces complexes that can be used for photocaging of small molecules.62 Therefore, we used the same approach to reduce the cytotoxicity under dark conditions for the complexes 8 and 10. Accordingly, the chlorine in the pyridyl-pyrazole ligand (type 3) was replaced with a carboxylic acid group and the complexes generated with dmphen (11) and bathophen (12) coligands. These +1 complexes were studied under dark conditions and possessed up to 15-fold lower cytotoxicity than the corresponding +2 charged complexes 8 and 10 (Figure 4B). Fortunately, incorporation of the carboxylic acid did not affect the light-mediated activity; sub-micromolar potency was exhibited by complex 11 upon irradiation, as the same ligand-deficient Ru(II) active species is produced as for complexes 1–8. This modification resulted in the largest phototoxicity index (146) for this group of compounds.
2.6. In-Cell Transcription and Translation Assay
To investigate the impact of the Ru(II) complexes on the essential biological processes, a cell-based transcription and translation assay was performed using the photoconvertible protein, Dendra2, as a reporter for protein synthesis.75 This allowed for a real-time report in live cells of any damage to the DNA, RNA, or the ribosome or inhibition of any essential components of the cellular machinery responsible for the processes of transcription and translation.
Dendra2 is a photoconvertible protein; upon irradiation, the chromophore within the protein undergoes a chemical reaction and switches from green to red fluorescence, while Dendra2 synthesized after this irradiation step will only show green fluorescence. Therefore, this assay provides a real-time observation of the newly synthesized protein, with ratiometric detection compared to the previously made protein, providing an assay for inhibition of protein synthesis that can be assessed in dose response and with kinetic information. Previously, we established that anticancer 8-hydroxyquinolines (HQs) coordinated with the [Ru(dmphen)2]2+ scaffold exhibit inhibition of protein production at sub-micromolar concentrations, as indicated by the reduction in the expression of Dendra2.76 Therefore, to narrow down the possible mechanisms of action causing the cytotoxicity, complexes 4 and 8 were tested for inhibition of protein synthesis (Figure 5A,B). Rapamycin was used as a positive control, with the results shown in Figure 5C. Rapamycin is an inhibitor of the mammalian target of rapamycin (mTOR) pathway, which is involved in regulating protein synthesis. An IC50 for inhibition of protein synthesis of 6.3 μM was observed for rapamycin. Complexes 4 and 8 had no effect that resulted in the reduction in Dendra2 production at concentrations up to 30 μM. These data indicate that inhibition of either transcription or translation is not the causative mechanism involved in the cytotoxicity of Ru(II) complexes with pyridyl-pyrazole ligands, in contrast to cytotoxic HQs coordinated with the [Ru(dmphen)2]2+ scaffold.76
Figure 5.
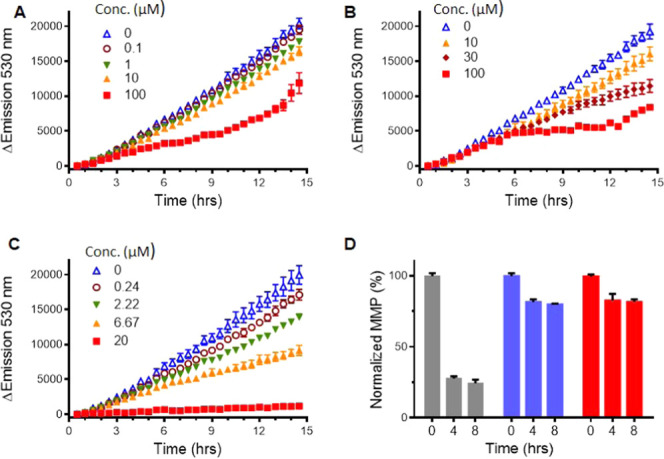
Inhibition of protein synthesis (A–C) and examination of mitochondria dysfunction (D). Emission of the photoconvertible protein, Dendra2, was monitored over time to report on protein production. (A) Complex 4 (0–100 μM); (B) complex 8 (0–100 μM); (C) rapamycin (0–20 μM). (D) Time-dependent inhibition of mitochondrial function with complex 2 upon dark (blue) and light (red) conditions compared with the control compound (carbonyl cyanide m-chlorophenyl hydrazone (CCCP), gray).
2.7. Mitochondria Dysfunction
Alternatively, the compounds could cause cell death through damage to the mitochondria. This mechanism of action has been previously established for a variety of anticancer Ru(II) complexes with dmphen coligands.44,51,55 To test this, the mitochondrial membrane potential was examined by fluorescent dye tetramethylrhodamine ethyl ester perchlorate (TMRE), which is excluded from active mitochondria but not dysfunctional mitochondria. A cyanide compound, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), was used as a positive control for mitochondria dysfunction. For this assay, A549 nonsmall cell lung cancer cells were treated with complexes 2, 4, and 8, at concentrations twice their IC50 values under both dark and light (1 min with Indigo LED, 29.1 J/cm2) conditions. The fluorescence intensity from TMRE was normalized, with the no-cells reading set as 0% and the no-treatment cell reading as 100%. Upon treatment of all three compounds, a rapid decrease of 20–30% of the mitochondrial membrane potential was observed (Figure 5D). In contrast, the positive control cyanide compound inhibits ∼80% of the mitochondria function. Thus, the loss of the mitochondrial membrane potential does not appear to be the cause of the cytotoxicity of these complexes.
3. Conclusions
Previous work using the [Ru(dmphen)2]2+ scaffold resulted in metal complexes that are photoreactive cytotoxins with potential utility for PACT.17 However, this framework has also shown potential to enhance the anticancer activity of small molecules, such as HQs,11,76 pyridyl-benzazoles,16 and modified phenanthrolines.44,50,55 To expand the repertoire of strained polypyridyl compounds with applications in PACT, we synthesized and investigated the photochemical and biological properties of a group of Ru(II) complexes with pyridyl-pyrazol(in)e ligands. The choice of new five-membered ligands was motivated by the biological potential of the free ligands and previous results for photoactive heteroleptic Ru(II) complexes with pyridyl-benzazole ligands.16 Recently, we established that the electronic features can be used to tune the photochemistry of Ru(II) complexes for biological applications; this was demonstrated for monodentate ligands, which act as leaving groups.61,62 Additionally, it is known that Ru(II) complexes that contain the pyridyl-1,2,3-triazole ligand63 or pyridyl-benzazole ligands16 are photolabile. Therefore, we hypothesized that pyridyl-pyrazol(in)es can be utilized as bidentate leaving groups for cytotoxic Ru(II) scaffolds. Fortunately, complexes 3, 4, 7, and 8, which include a phenyl moiety at the 1N-position of pyrazol(in)e, exhibited rapid and selective photoejection of the pyrazol(in)e-containing ligand when irradiated with >450 nm light. We established that compounds 1–4, 7, and 8, with regular pyridyl-pyrazol(in)e ligands, were at least 20–80-fold more potent than the parent pyridyl-pyrazol(in)es, and exhibited anticancer activity in the HL60 cell line, with IC50 values ranging from 0.2 to 7.6 μM. Light activation of 3, 4, 7, and 8 resulted in complete cell death at 10-fold lower concentrations than under dark conditions but did not show a significant improvement in the IC50 values. Complexes 5 and 6 with inverse pyridyl-pyrazoline ligands exhibited the same dark potency against the HL60 cell line as 1–4. In addition, these compounds exhibit slow photoejection, which diminishes their utility for PACT. Diversification of the coligands in the complex improved the phototoxicity indicies from PI = 5 up to 28 for compound 9. Finally, the introduction of a carboxylic acid in heteroleptic complexes 11 and 12 provided the largest PI values (146 and 59) by a remarkable reduction in the dark cytotoxicity. This points toward a rational design strategy utilizing pendent carboxylic acids for the creation of photoactive heteroleptic Ru (II) complex candidates for PACT.
4. Experimental Section
4.1. Materials and Instrumentation
All materials that were purchased from commercial sources were used without any further purification. All 1H NMR were obtained on a Varian mercury spectrometer (400 MHz), and chemical shifts are reported relative to the residual solvent peak of CD3CN (δ 1.94). Electrospray ionization (ESI) mass spectra were obtained on a Varian 1200L mass spectrometer at the Environmental Research Training Laboratory (ERTL) at the University of Kentucky. The UV/vis absorption spectra were obtained on a BMG Labtech FLUOstar Omega microplate reader. Light activation experiments were performed using a 470 nm LED array from Elixa (for photochemistry and DNA gels) or with >450 nm light using the Indigo LED (for cytotoxicity experiments). The Prism software package was used to analyze the data.
4.2. HPLC Analysis for Purity and Photoejection Products
The purity of each of the Ru(II) complexes 1–12 and photoejection products were analyzed using an Agilent 1100 series HPLC equipped with a model G1311A quaternary pump, G1315B UV diode array detector (detection wavelength of 280 nm), and ChemStation software version B.01.03. Chromatographic conditions were optimized on a Column Technologies Inc. A C18 120 Å column was used for compounds 4, 8, 10 and 12, and a Phonomenex Luna 5 μm C18(2) 100 Å was used for compounds 1–3, 6, 7, and 11. Both columns were fitted with a Phenomenex C18 guard column. Mobile phases consisted of 0.1% formic acid in dH2O and 0.1% formic acid in HPLC grade CH3CN. The samples of each Ru(II) complex were prepared at a final concentration of 10–100 μM in dH2O and protected from light (dark controls/purity analysis) or irradiated to determine the photoejection products.
4.3. Synthesis and Characterization of Ru(II) Complexes
4.3.1. General Procedure for the Synthesis of [Ru(dmphen)2L] Complexes 1–8 and 11
The starting material [Ru(dmphen)2Cl2] (120 mg, 0.2 mmol) and pyrazole-containing ligand (1.1 equiv) were added to 4 mL of ethylene glycol in a 15 mL pressure tube. The mixture was heated at 100–120 °C for 2 h (12 h for compound 1a) while protected from light. The dark orange solution was allowed to cool to room temperature and poured into 50 mL of dH2O. Addition of a saturated aq KPF6 solution (ca. 1 mL) produced a red or red-orange precipitate that was collected by vacuum filtration. The purification of the solid was carried out by flash chromatography (silica gel, loaded in 0.1% KNO3, 5%H2O in MeCN). A gradient was run, and the pure complex was eluted at 0.2% KNO3, 5–10% H2O in MeCN. The product fractions were concentrated under reduced pressure, and a saturated aq solution of KPF6 was added, followed by extraction of the complex into CH2Cl2. The solvent was removed under reduced pressure to give a solid.
4.3.1.1. Complex 1
Yield: 38 mg (18%). Ratio (%) of diastereomers (Λ)-(S)/(Λ)-(R) = 1:9. 1H NMR (CD3CN): δ 8.64–8.69 (m, 1H), 8.51–8.58 (m, 2H), 8.35–8.38 (m, 1H), 8.23–8.27 (m, 1H), 8.04–8.15 (m, 3H), 7.80 (d, J = 8.3 Hz, 0.9H), 7.51–7.75 (m, 4.2H), 7.44 (d, J = 7.9 Hz, 0.9H), 7.06 (d, J = 8.7 Hz, 0.2H), 6.82–6.90 (m, 2.2H), 6.76 (d, J = 8.7 Hz, 1.8H), 6.35 (d, J = 8.6 Hz, 1.8H), 6.21 (d, J = 3.3 Hz, 0.9H), 5.41 (d, J = 3.7 Hz, 0.1H), 4.76–4.83 (m, 0.9H), 4.27–4.35 (m, 0.1H), 3.74–3.82 (m, 3.9H), 3.49 (dd, J = 17.8, 11.5 Hz, 0.1H), 3.26–3.34 (m, 0.1H), 2.69–2.79 (m, 3.9H), 2.00 (s, 2.7H), 1.96 (s, 0.3H), 1.91 (s, 0.3H), 1.88 (s, 2.7H), 1.85–1.86 (m, 3H). Purity by HPLC = 98% (compound 1a was detected by HPLC due to oxidation of 1; less than 2% correspond to contaminants, see Figure S34). ESI MS calcd for C43H39N7ORu [M2+·PF6–]+ 916.19, [M]2+ 385.62; found 916.4 [M2+·PF6–]+, 385.6 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 445 (15.9).
4.3.1.2. Complex 1a
Yield: 95 mg (45%). 1H NMR (CD3CN): δ 10.70 (brs, 1H), 8.65–8.67 (m, 2H), 8.34 (t, J = 8.5 Hz, 2H), 8.24–8.28 (m, 2H), 8.12–8.14 (m, 2H), 7.86 (d, J = 7.7 Hz, 1H), 7.73–7.80 (m, 3H), 7.50 (d, J = 8.4 Hz, 1H), 7.41 (d, J = 8.3 Hz, 1H), 7.02–7.09 (m, 3H), 6.82–6.93 (m, 4H), 3.80 (s, 3H), 2.14 (s, 3H), 1.97 (s, 3H), 1.88 (s, 3H), 1.87 (s, 3H). Purity by HPLC = 99%. ESI MS calcd for C43H37N7ORu [M]2+ 384.61; found 384.5 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 435 (12.3).
4.3.1.3. Complex 2
Yield: 73 mg (28%). Ratio (%) of diastereomers (Λ)-(S)/(Λ)-(R) = 1:3. 1H NMR (CD3CN): δ 8.64–8.69 (m, 1H), 8.52–8.57 (m, 2H), 8.34–8.38 (m, 1H), 8.22–8.27 (m, 1H), 8.06–8.15 (m, 3H), 7.80 (d, J = 8.4 Hz, 0.75H), 7.50–7.75 (m, 5H), 7.44 (d, J = 7.8 Hz, 0.75H), 7.31 (d, J = 8.5 Hz, 0.5H), 7.24 (d, J = 8.4 Hz, 1.5H), 7.13 (d, J = 8.4 Hz, 0.5H), 6.83–6.86 (m, 2H), 6.39 (d, J = 8.4 Hz, 1H), 6.28 (d, J = 4.7 Hz, 0.75H), 5.45 (d, J = 4.5 Hz, 0.25H), 4.81–4.88 (m, 0.75H), 4.35–4.42 (m, 0.25H), 3.84 (dd, J = 17.9, 12.2 Hz, 0.75H), 3.59 (dd, J = 17.6, 11.3 Hz, 0.25H), 3.23–3.31 (m, 0.25H), 2.74–2.82 (m, 1.5H), 2.68 (s, 2.25H), 2.00 (s, 2.25H), 1.91 (s, 0.75H), 1.83–1.89 (m, 6H). Purity by HPLC = 95% (oxidized product was detected; less than 5% correspond to contaminants, see Figure S35). ESI MS calcd for C42H36ClN7Ru [M2+·PF6–]+ 920.14, [M]2+ 387.59; found 920.2 [M2+·PF6–]+, 387.5 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 445 (11.5).
4.3.1.4. Complex 3
Yield: 81 mg (42%). Ratio (%) of diastereomers (Λ)-(S)/(Λ)-(R) = 9:1. 1H NMR (CD3CN): δ 8.57–8.63 (m, 1H), 8.46–8.52 (m, 1H), 8.34 (d, J = 5.6 Hz, 0.9H), 8.11–8.16 (m, 2H), 7.88–8.03 (m, 3.8H), 7.61–7.82 (m, 5.1H), 7.31–7.39 (m, 2.7H), 7.13–7.21 (m, 0.3H), 7.07 (d, J = 8.3 Hz, 0.9H), 6.79–6.92 (m, 3.1H), 6.69 (d, J = 8.6 Hz, 0.2H), 6.22–6.39 (m, 1H), 5.86–6.14 (m, 3H), 5.59 (dd, J = 12.4, 10.0 Hz, 0.1H), 5.22 (dd, J = 15.0, 10.2 Hz, 0.9H), 4.32–4.44 (m, 1H), 3.88 (dd, J = 18.1, 10.1 Hz, 0.1H), 3.78 (dd, J = 17.8, 15.2 Hz, 0.9H), 3.74 (s, 2.7H), 3.65 (s, 0.3H), 2.91 (s, 0.3H), 2.84 (s, 2.7H), 2.44 (s, 2.7H), 2.40 (s, 0.3H), 1.53–1.54 (m, 5.4H), 1.26–1.28 (m, 0.6H). Purity by HPLC = 98%. ESI MS calcd for C49H43N7Ru [M2+·PF6–]+ 992.22, [M]2+ 423.63; found 992.4 [M2+·PF6–]+, 423.7 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 450 (11.0).
4.3.1.5. Complex 4
Yield: 88 mg (31%). Ratio (%) of diastereomers (Λ)-(S)/(Λ)-(R) = 3:1. 1H NMR (CD3CN): δ 8.63 (d, J = 8.3 Hz, 0.2H), 8.58 (d, J = 8.3 Hz, 0.8H), 8.51 (d, J = 8.3 Hz, 0.8H), 8.47 (d, J = 8.4 Hz, 0.2H), 8.34–8.36 (m, 1H), 8.13–8.18 (m, 2H), 7.88–8.03 (m, 4H), 7.61–7.81 (m, 5H), 7.31–7.49 (m, 4H), 7.12–7.22 (m, 1H), 7.07 (d, J = 8.3 Hz, 0.8H), 6.94–6.98 (m, 0.2H), 6.81–6.84 (m, 1H), 6.26–6.45 (m, 1H), 5.92–6.20 (m, 3H), 5.63 (dd, J = 12.7, 10.3 Hz, 0.2H), 5.27 (dd, J = 14.8, 10.6 Hz, 0.8H), 4.46 (dd, J = 18.0, 10.7 Hz, 0.8H), 4.40 (dd, J = 18.9, 12.6 Hz, 0.2H), 3.86 (dd, J = 18.9, 10.2 Hz, 0.2H), 3.80 (dd, J = 17.9, 14.7 Hz, 0.8H), 2.91 (s, 0.6H), 2.81 (s, 2.4H), 2.43 (s, 2.4H), 2.37 (s, 0.6H), 1.54 (brs, 4.8H), 1.28 (brs, 1.2H). Purity by HPLC = 96%. ESI MS calcd for C48H40ClN7Ru [M2+·PF6–]+ 996.17, [M]2+ 425.61; found 996.2 [M2+·PF6–]+, 425.5 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 450 (14.9).
4.3.1.6. Complex 5
Yield: 138 mg (56%). Ratio (%) of diastereomers (Λ)-(S)/(Λ)-(R) = 55:45. 1H NMR (CD3CN): δ 8.63 (d, J = 8.3 Hz, 0.45H), 8.46–8.54 (m, 2.55H), 8.04–8.19 (m, 2.55H), 7.81–7.97 (m, 2.45H), 7.72–7.75 (m, 1H), 7.60–7.63 (m, 1H), 7.55 (d, J = 8.4 Hz, 0.45H), 7.34–7.49 (m, 2.55H), 7.27–7.29 (m, 1H), 6.84–7.09 (m, 5H), 6.54–6.67 (m, 4H), 6.40–6.46 (m, 1.55H), 6.28 (t, J = 6.2 Hz, 0.45H), 5.49 (dd, J = 11.6, 7.7 Hz, 0.45H), 5.40 (dd, J = 12.2, 8.8 Hz, 0.55H), 3.95 (dd, J = 19.7, 12.2 Hz, 0.45H), 3.77–3.85 (m, 3.55H), 3.33 (s, 1.65H), 3.29 (dd, J = 19.6, 8.7 Hz, 0.55H), 3.19 (dd, J = 19.7, 7.8 Hz, 0.45H), 3.03 (s, 1.35H), 2.10 (s, 1.35H), 2.09 (s, 1.65), 1.87 (s, 1.65H), 1.77 (s, 1.35H), 1.54 (s, 1.35H), 1.41 (s, 1.65H). Purity by HPLC = 95%. ESI MS calcd for C49H43N7ORu [M2+·PF6–]+ 992.22, [M]2+ 423.63; found 992.3 [M2+·PF6–]+, 423.7 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 400 (10.6).
4.3.1.7. Complex 6
Yield: 64 mg (27%). Ratio (%) of diastereomers (Λ)-(S)/(Λ)-(R) = 3:2. 1H NMR (CD3CN): δ 8.48–8.63 (m, 3H), 8.06–8.20 (m, 2.6H), 7.83–7.97 (m, 2.4H), 7.72–7.75 (m, 1H), 7.29–7.63 (m, 7H), 6.85–7.11 (m, 3H), 6.29–6.69 (m, 6H), 5.53 (dd, J = 11.7, 8.1 Hz, 0.4H), 5.44 (dd, J = 12.3, 8.9 Hz, 0.6H), 3.99 (dd, J = 19.8, 12.4 Hz, 0.6H), 3.83 (dd, J = 19.7, 11.7 Hz, 0.4H), 3.34 (s, 1.8H), 3.29 (dd, J = 19.8, 9.1 Hz, 0.6H), 3.20 (dd, J = 19.6, 8.1 Hz, 0.4H), 3.00 (s, 1.2H), 2.13 (s, 1.2H), 2.07 (s, 1.8), 1.90 (s, 1.8H), 1.76 (s, 1.2H), 1.55 (s, 1.2H), 1.41 (s, 1.8H). Purity by HPLC = 91%. ESI MS calcd for C48H40ClN7Ru [M2+·PF6–]+ 996.17, [M]2+ 425.61; found 996.3 [M2+·PF6–]+, 425.6 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 400 (12.7).
4.3.1.8. Complex 7
Yield: 44 mg (22%). 1H NMR (CD3CN): δ 8.47 (d, J = 8.3 Hz, 1H), 8.39 (d, J = 8.3 Hz, 1H), 8.35 (d, J = 8.4 Hz, 1H), 7.99–8.12 (m, 4H), 7.95 (d, J = 8.7 Hz, 1H), 7.84 (td, J = 7.8, 1.3 Hz, 1H), 7.76 (d, J = 8.7 Hz, 1H), 7.65 (d, J = 8.4 Hz, 1H), 7.59 (d, J = 8.3 Hz, 1H), 7.49 (s, 1H), 7.41 (d, J = 8.3 Hz, 1H), 7.32 (d, J = 8.4 Hz, 1H), 7.15–7.16 (m, 1H), 6.95 (d, J = 8.9 Hz, 2H), 6.89 (ddd, J = 8.0, 5.9, 1.5 Hz, 2H), 6.78 (t, J = 7.0 Hz, 1H), 6.71 (d, J = 8.9 Hz, 2H), 6.15 (brs, 1H), 5.40 (brs, 1H), 3.68 (s, 3H), 2.59 (s, 3H), 2.13 (s, 3H), 1.73 (s, 3H), 1.41 (s, 3H). Purity by HPLC = 97%. ESI MS calcd for C49H41N7ORu [M2+·PF6–]+ 990.21, [M]2+ 422.62; found 990.2 [M2+·PF6–]+, 422.4 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 415 (12.8).
4.3.1.9. Complex 8
Yield: 58 mg (30%). 1H NMR (CD3CN): δ 8.47 (d, J = 8.3 Hz, 1H), 8.40 (d, J = 8.3 Hz, 1H), 8.35 (d, J = 8.4 Hz, 1H), 8.12 (d, J = 7.8 Hz, 1H), 8.04–8.07 (m, 2H), 8.00 (d, J = 8.7 Hz, 1H), 7.95 (d, J = 8.7 Hz, 1H), 7.85 (td, J = 7.8, 1.3 Hz, 1H), 7.76 (d, J = 8.7 Hz, 1H), 7.65 (d, J = 8.4 Hz, 1H), 7.59 (d, J = 8.3 Hz, 1H), 7.56 (s, 1H), 7.41 (d, J = 8.3 Hz, 1H), 7.32 (d, J = 8.4 Hz, 1H), 7.16–7.22 (m, 4H), 6.99 (ddd, J = 8.0, 5.9, 1.5 Hz, 2H), 6.88–6.92 (m, 1H), 6.79–6.82 (m, 2H), 6.16 (brs, 1H), 5.41 (brs, 1H), 2.58 (s, 3H), 2.13 (s, 3H), 1.96 (s, 3H), 1.73 (s, 3H). Purity by HPLC = 95%. ESI MS calcd for C48H38ClN7Ru [M2+·PF6–]+ 994.16, [M]2+ 424.60; found 994.1 [M2+·PF6–]+, 424.6 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 415 (12.2).
4.3.1.10. Complex 11
Yield: 20 mg (20%). 1H NMR (CD3CN): δ 8.46 (d, J = 8.3 Hz, 1H), 8.39 (d, J = 8.3 Hz, 1H), 8.35 (d, J = 8.4 Hz, 1H), 8.14 (d, J = 7.8 Hz, 1H), 7.99–8.06 (m, 3H), 7.94 (d, J = 8.7 Hz, 1H), 7.85 (t, J = 7.8 Hz, 1H), 7.75 (d, J = 8.6 Hz, 3H), 7.64 (d, J = 8.3 Hz, 1H), 7.62 (s, 1H), 7.59 (d, J = 8.3 Hz, 1H), 7.40 (d, J = 8.3 Hz, 1H), 7.31 (d, J = 8.3 Hz, 1H), 7.17 (d, J = 5.3 Hz, 1H), 7.09 (d, J = 8.0 Hz, 2H), 6.91 (t, J = 7.2 Hz, 1H), 6.78 (t, J = 6.4 Hz, 1H), 6.15 (brs, 1H), 5.40 (brs, 1H), 2.59 (s, 3H), 2.13 (s, 3H), 1.72 (s, 3H), 1.41 (s, 3H). Purity by HPLC = 95%. UV (CH3CN): λmax nm (ε × 10–3) 415 (11.1).
4.3.2. General Procedure for Synthesis of Unstrained Ru(II) Complexes 9, 10 and 12
Starting material [Ru(bpy)2Cl2] (for complex 9) or [Ru(bathophen)2Cl2] (for complexes 10 and 12, 0.2 mmol) and the pyridyl-pyrazole ligand (1.1 equiv) were added to 6 mL of EtOH/H2O mixture (1:1) in a 15 mL pressure tube. The mixture was heated at 90 °C for 2 h. The dark orange solution was allowed to cool to room temperature and poured into 50 mL of dH2O. Addition of a saturated aq KPF6 solution (ca. 1 mL) produced a red-orange precipitate that was collected by vacuum filtration. The purification of the solid was carried out by flash chromatography (silica gel, loaded in 0.1% KNO3, 5%H2O in MeCN). A gradient was run, and the pure complex was eluted at 0.2% KNO3, 5–10% H2O in MeCN. The product fractions were concentrated under reduced pressure, and a saturated aq solution of KPF6 was added, followed by extraction of the complex into CH2Cl2. The solvent was removed under reduced pressure to give a solid.
4.3.2.1. Complex 9
Yield: 122 mg (61%). 1H NMR (CD3CN): δ 8.42 (d, J = 8.0 Hz, 1H), 8.37 (d, J = 8.2 Hz, 2H), 8.29 (t, J = 7.7 Hz, 2H), 8.03–8.14 (m, 4H), 7.96 (ddd, J = 9.5, 8.0, 1.5 Hz, 1H), 7.86 (d, J = 6.5 Hz, 1H), 7.58–7.65 (m, 3H), 7.46–7.51 (m, 3H), 7.25–7.35 (m, 6H), 7.16–7.20 (m, 2H), 7.12 (d, J = 6.9 Hz, 1H), 7.02–7.08 (m, 2H), 6.87 (ddd, J = 7.2, 5.6, 1.3 Hz, 1H), 6.77 (t, J = 7.1 Hz, 1H), 6.20 (d, J = 7.2 Hz, 1H). Purity by HPLC = 99%. ESI MS calcd for C40H30ClN7Ru [M2+·PF6–]+ 890.09, [M]2+ 372.57; found 890.1 [M2+·PF6–]+, 372.5 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 410 (12.2).
4.3.2.2. Complex 10
Yield: 190 mg (68%). 1H NMR (CD3CN): δ 8.93 (d, J = 5.4 Hz, 1H), 8.47 (d, J = 5.4 Hz, 1H), 8.40 (d, J = 7.8 Hz, 1H), 8.06–8.21 (m, 4H), 7.99 (d, J = 9.4 Hz, 1H), 7.95 (d, J = 5.4 Hz, 1H), 7.83–7.86 (m, 2H), 7.45–7.76 (m, 25H), 7.16–7.27 (m, 5H), 7.04–7.07 (m, 2H), 6.92 (t, J = 7.5 Hz, 1H), 6.23 (t, J = 7.3 Hz, 1H), 5.77 (d, J = 7.8 Hz, 1H). Purity by HPLC = 98%. ESI MS calcd for C68H46ClN7Ru [M2+·PF6–]+ 1242.22, [M]2+ 548.63; found 1242.2 [M2+·PF6–]+, 548.6 [M]2+. UV (CH3CN): λmax nm (ε × 10–3) 425 (23.0).
4.3.2.3. Complex 12
Yield: 160 mg (57%). 1H NMR (CD3CN): δ 8.94 (d, J = 5.4 Hz, 1H), 8.47 (d, J = 5.4 Hz, 1H), 8.41 (d, J = 7.8 Hz, 1H), 8.07–8.20 (m, 4H), 8.00 (d, J = 9.4 Hz, 1H), 7.95 (d, J = 5.4 Hz, 1H), 7.82–7.86 (m, 4H), 7.45–7.78 (m, 25H), 7.24–7.30 (m, 3H), 7.04–7.08 (m, 2H), 6.94 (t, J = 7.4 Hz, 1H), 6.24 (t, J = 7.3 Hz, 1H), 5.79 (d, J = 7.9 Hz, 1H). Purity by HPLC = 96%. UV (CH3CN): λmax nm (ε × 10–3) 425 (24.1).
4.4. Crystallography
Single crystals of compounds 1a, 4, and 8 were grown from methylene chloride or acetone by vapor diffusion of diethyl ether, mounted in an inert oil, and transferred to the cold gas stream of the diffractometer. X-ray diffraction data were collected at 90.0(2) K on either a Nonius KappaCCD diffractometer using MoKα X-rays or on a Bruker-Nonius X8 Proteum diffractometer with graded-multilayer focused Cu Kα X-rays. Raw data were integrated, scaled, merged, and corrected for Lorentz-polarization effects using either the HKL-SMN package77 or the APEX2 package.78 Corrections for absorption were applied using SADABS79 and XABS2.80 The structures were solved by SHELXT81 and refined against F2 by weighted full-matrix least-squares using SHELXL-2014.82 For compound 8, the SQUEEZE routine83 was used to treat disordered solvent. Hydrogen atoms were placed at the calculated positions and refined using a riding model. Nonhydrogen atoms were refined with the anisotropic displacement parameters. Structures were checked using check CIF tools in Platon84 and by an R-tensor.85 Crystal data and relevant details of the structure determinations are summarized below.
4.4.1. Crystal Data (1a, CCDC 2006205)
C47H47F12N7O2P2Ru, Mr = 1132.92, monoclinic, C2/c, a = 25.3195(5) Å, α = 90°, b = 27.6183(5) Å, β = 116.666(1)°, c = 17.9680(3) Å, γ = 90°, V = 11228.3(4)Å3, Z = 8, ρ = 1.34 mg/m3, μ = 3.513 mm–1, F(000) = 4608, crystal size = 0.240 × 0.03 × 0.02 mm3, θ(max) = 68.450°, 73 439 reflections collected, 10 219 unique reflections (Rint = 0.0714), goodness of fit (GOF) = 1.029, R1 = 0.047 and wR2 = 0.1305 [I > 2σ(I)], R1 = 0.0681 and wR2 = 0.1442 (all indices), largest difference peak/hole = 1.009/–0.581 e/Å3.
4.4.2. Crystal Data (4, CCDC 1996034)
C41H40ClF12N7P2Ru, Mr = 1141.33, triclinic, P1̅, a = 10.8291(6) Å, α = 86.860(3)°, b = 11.0247(6) Å, β = 84.770(3)°, c = 21.5477(12) Å, γ = 79.645(2)°, V = 2518.2(2) Å3, Z = 4, ρ = 1.505 mg/m3, μ = 4.371 mm–1, F(000) = 1152, crystal size = 0.240 × 0.200 × 0.190 mm3, θ(max) = 68.460°, 32 007 reflections collected, 8942 unique reflections (Rint = 0.0535), GOF = 1.198, R1 = 0.0598 and wR2 = 0.1487 [I > 2σ(I)], R1 = 0.0620 and wR2 = 0.1501 (all indices), largest difference peak/hole = 0.999/–0.888 e/Å3.
4.4.3. Crystal Data (8, CCDC 1996035)
C99H84Cl2F24N14O2P4Ru2, Mr = 2354.72, monoclinic, C2/c, a = 28.6521(2) Å, α = 90°, b = 22.6211(2) Å, β = 128.0180(4)°, c = 21.4137(2) Å, γ = 90°, V = 10934.20(17) Å3, Z = 4, ρ = 1.430 mg/m3, μ = 0.477 mm–1, F(000) = 4760, crystal size = 0.350 × 0.300 × 0.210 mm3, θ(max) = 27.697°, 126 391 reflections collected, 12 653 unique reflections (Rint = 0.0357), GOF = 1.073, R1 = 0.0538 and wR2 = 0.1673 [I > 2σ(I)], R1 = 0.0669 and wR2 = 0.1785 (all indices), largest difference peak/hole = 1.347/–0.677 e/Å3.
4.5. Counterion Exchange
Compounds 1–12 were converted to Cl– salts by dissolving 5–20 mg of the product in 1–2 mL of methanol. The dissolved product was loaded onto an Amberlite IRA-410 chloride ion exchange column, eluted with methanol, and the solvent was removed in vacuo.
4.6. Photoejection Studies
Quantum yields for the complexes 1–8 and 11 with the Cl counterions were determined by an optical approach, as has been described previously.61 The Ru(II) complexes were analyzed in a 96-well plate at a final concentration of 25–35 μM and a path length of 0.5 cm. Scans were taken at set time points for 300 min. In all cases, the light source was a 470 nm LED array from Elixa. The photon flux of the lamp for irradiation in the plate was determined by a ferrioxalate actinometer (1.77 × 10–8 E/s).
The absorbance of complexes at a concentration of 25–35 μM at 470 nm was from 0.07 to 0.19 with photon absorption probability (F) from 0.14 to 0.36. Therefore, the moles of the photon absorbed have been calculated as the product of photons irradiated and photon absorption probability.
4.7. Cytotoxicity Assay
The HL60 cells were plated at 30 000 cells/well in Opti-MEM media with 1% fetal bovine serum (FBS) and Pen-Strep in 96-well plates. Compounds were serially diluted in opti-MEM with 1% FBS and Pen-Strep in a 96-well plate and then added to the cells. They were then irradiated with 29.1 J/cm2 light (>450 nm using the Indigo LED) for 1 min or kept in the dark. The cells were incubated with the compounds for 72 h followed by the addition of resazurin. The plates were incubated for 3 h and then read on a SpectraFluor Plus plate reader with an excitation filter of 535 nm and an emission of 595 nm.
4.8. DNA Gel Electrophoresis.
Compounds were mixed with 40 μg/mL pUC19 plasmid DNA in 10 mM potassium phosphate buffer, pH 7.4. To determine the effect of light, the samples were irradiated with light (470 nm) from a 200 W light source (LED array from Elixa) for total light doses of 40 J/cm2. The samples were then incubated for 12 h at room temperature in the dark. Single- and double-strand DNA break controls were prepared, and the DNA samples were resolved on agarose gels, as described previously.19 In brief, the samples were resolved on a 1% agarose gel prepared in tris-acetate buffer with 0.3 μg of plasmid/lane. The gels were stained with 0.5 μg/mL ethidium bromide in Tris-acetate buffer at room temperature for 40 min, destained with tris-acetate buffer, and imaged on a ChemiDoc MP system (Bio-Rad).
4.9. Dendra2 Transcription–Translation Assay
96-well plates were coated with matrigel followed by the addition of HEK T-Rex cells at a density of 30 000 cells/well and incubated with 1 μg/mL of tetracycline for 16 h. The media was removed and 50 μL of L-15 media containing 1 μg/mL tetracycline along with compound was added to each well and allowed to incubate for 1 h. The plates were then illuminated with a 405 nm LED flood array for 1 min and then read in kinetic mode on a SpectraFluor Plus (Tecan) set to 37 °C. The plates were read every 30 min for 15 h with excitation and emission wavelengths of 480 and 530 nm for newly translated Dendra2 and 535 and 595 nm for post-translated Dendra2.75
Acknowledgments
We gratefully acknowledge the National Institutes of Health (grant GM107586) for the support of this research. Crystallographic work was made possible by the National Science Foundation (NSF) MRI program, grants CHE-0319176 and CHE-1625732.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02079.
Additional schemes and figures for synthetic methods, characterization, photochemical and photobiological analyses (PDF)
Author Present Address
† (Yang Sun) Department of Medicine, Rheumatology, University of California, 513 Parnassus Avenue, San Francisco, California 94143, United States.
Author Contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Furrer J.; Süss-Fink G. Thiolato-bridged Dinuclear Arene Ruthenium Complexes and Their Potential as Anticancer Drugs. Coord. Chem. Rev. 2016, 309, 36–50. 10.1016/j.ccr.2015.10.007. [DOI] [Google Scholar]
- Mari C.; Pierroz V.; Ferrari S.; Gasser G. Combination of Ru(II) Complexes and Light: New Frontiers in Cancer Therapy. Chem. Sci. 2015, 6, 2660–2686. 10.1039/C4SC03759F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamo A.; Gaiddon C.; Schellens J. H.; Beijnen J. H.; Sava G. Approaching Tumour Therapy Beyond Platinum Drugs: Status of the Art and Perspectives of Ruthenium Drug Candidates. J. Inorg. Biochem. 2012, 106, 90–99. 10.1016/j.jinorgbio.2011.09.030. [DOI] [PubMed] [Google Scholar]
- Salassa L. Polypyridyl Metal Complexes with Biological Activity. Eur. J. Inorg. Chem. 2011, 2011, 4931–4947. 10.1002/ejic.201100376. [DOI] [Google Scholar]
- Zeng L.; Gupta P.; Chen Y.; Wang E.; Ji L.; Chao H.; Chen Z. S. The Development of Anticancer Ruthenium(II) Complexes: from Single Molecule Compounds to Nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. 10.1039/C7CS00195A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaro A.; Gasser G. Monomeric and Dimeric Coordinatively Saturated and Substitutionally Inert Ru(II) Polypyridyl Complexes as Anticancer Drug Candidates. Chem. Soc. Rev. 2017, 46, 7317–7337. 10.1039/c7cs00356k. [DOI] [PubMed] [Google Scholar]
- Rademaker-Lakhai J. M.; van den Bongard D.; Pluim D.; Beijnen J. H.; Schellens J. H. A Phase I and Pharmacological Study with Imidazolium-trans-DMSO-imidazole-tetrachlororuthenate, a Novel Ruthenium Anticancer Agent. Clin. Cancer Res. 2004, 10, 3717–3727. 10.1158/1078-0432.CCR-03-0746. [DOI] [PubMed] [Google Scholar]
- Trondl R.; Heffeter P.; Kowol C. R.; Jakupec M. A.; Berger W.; Keppler B. K. NKP-1339, the First Ruthenium-based Anticancer Drug on the Edge to Clinical Application. Chem. Sci. 2014, 5, 2925–2932. 10.1039/C3SC53243G. [DOI] [Google Scholar]
- Weiss A.; Berndsen R. H.; Dubois M.; Muller C.; Schibli R.; Griffioen A. W.; Dyson P. J.; Nowak-Sliwinska P. In Vivo Anti-tumor Activity of the Organometallic Ruthenium(II)-arene Complex [Ru(η6-p-cymene)Cl2(pta)] (RAPTA-C) in Human Ovarian and Colorectal Carcinomas. Chem. Sci. 2014, 5, 4742–4748. 10.1039/C4SC01255K. [DOI] [Google Scholar]
- Ang W. H.; Casini A.; Sava G.; Dyson P. J. Organometallic Ruthenium-based Antitumor Compounds with Novel Modes of Action. J. Organomet. Chem. 2011, 696, 989–998. 10.1016/j.jorganchem.2010.11.009. [DOI] [Google Scholar]
- Heidary D. K.; Howerton B. S.; Glazer E. C. Coordination of Hydroxyquinolines to a Ruthenium Bis-dimethyl-phenanthroline Scaffold Radically Improves Potency for Potential as Antineoplastic Agents. J. Med. Chem. 2014, 57, 8936–8946. 10.1021/jm501043s. [DOI] [PubMed] [Google Scholar]
- Pierroz V.; Joshi T.; Leonidova A.; Mari C.; Schur J.; Ott I.; Spiccia L.; Ferrari S.; Gasser G. Molecular and Cellular Characterization of the Biological Effects of Ruthenium(II) Complexes Incorporating 2-Pyridyl-2-pyrimidine-4-carboxylic Acid. J. Am. Chem. Soc. 2012, 134, 20376–20387. 10.1021/ja307288s. [DOI] [PubMed] [Google Scholar]
- Notaro A.; Jakubaszek M.; Rotthowe N.; Maschietto F.; Vinck R.; Felder P. S.; Goud B.; Tharaud M.; Ciofini I.; Bedioui F.; Winter R. F.; Gasser G. Increasing the Cytotoxicity of Ru(II) Polypyridyl Complexes by tuning the Electronic Structure of Dioxo Ligands. J. Am. Chem. Soc. 2020, 142, 6066–6084. 10.1021/jacs.9b12464. [DOI] [PubMed] [Google Scholar]
- Howerton B. S.; Heidary D. K.; Glazer E. C. Strained Ruthenium Complexes Are Potent Light-activated Anticancer Agents. J. Am. Chem. Soc. 2012, 134, 8324–8327. 10.1021/ja3009677. [DOI] [PubMed] [Google Scholar]
- Albani B. A.; Pena B.; Leed N. A.; de Paula N. A.; Pavani C.; Baptista M. S.; Dunbar K. R.; Turro C. Marked Improvement in Photoinduced Cell Death by a New Tris-heteroleptic Complex with Dual Action: Singlet Oxygen Sensitization and Ligand Dissociation. J. Am. Chem. Soc. 2014, 136, 17095–17101. 10.1021/ja508272h. [DOI] [PubMed] [Google Scholar]
- Havrylyuk D.; Heidary D. K.; Nease L.; Parkin S.; Glazer E. C. Photochemical Properties and Structure–Activity Relationships of RuII Complexes with Pyridylbenzazole Ligands as Promising Anticancer Agents. Eur. J. Inorg. Chem. 2017, 2017, 1687–1694. 10.1002/ejic.201601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidayatullah A. N.; Wachter E.; Heidary D. K.; Parkin S.; Glazer E. C. Photoactive Ru(II) Complexes With Dioxinophenanthroline Ligands Are Potent Cytotoxic Agents. Inorg. Chem. 2014, 53, 10030–10032. 10.1021/ic5017164. [DOI] [PubMed] [Google Scholar]
- Wachter E.; Glazer E. C. Mechanistic Study on the Photochemical ″Light Switch″ Behavior of [Ru(bpy)2dmdppz]2+. J. Phys. Chem. A 2014, 118, 10474–10486. 10.1021/jp504249a. [DOI] [PubMed] [Google Scholar]
- Wachter E.; Heidary D. K.; Howerton B. S.; Parkin S.; Glazer E. C. Light-Activated Ruthenium Complexes Photobind DNA and are Cytotoxic in the Photodynamic Therapy Window. Chem. Commun. 2012, 48, 9649–9651. 10.1039/c2cc33359g. [DOI] [PubMed] [Google Scholar]
- Wachter E.; Howerton B. S.; Hall E. C.; Parkin S.; Glazer E. C. A New Type of DNA ″Light-Switch″: a Dual Photochemical Sensor and Metalating Agent for Duplex and G-quadruplex DNA. Chem. Commun. 2014, 50, 311–313. 10.1039/C3CC47269H. [DOI] [PubMed] [Google Scholar]
- Kohler L.; Nease L.; Vo P.; Garofolo J.; Heidary D. K.; Thummel R. P.; Glazer E. C. Photochemical and Photobiological Activity of Ru(II) Homoleptic and Heteroleptic Complexes Containing Methylated Bipyridyl-type Ligands. Inorg. Chem. 2017, 56, 12214–12223. 10.1021/acs.inorgchem.7b01642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque J. III; Havrylyuk D.; Barrett P. C.; Sainuddin T.; McCain J.; Colón K.; Sparks W. T.; Bradner E.; Monro S.; Heidary D.; Cameron C. G.; Glazer E. C.; McFarland S. A. Strained, Photoejecting Ru(II) Complexes that are Cytotoxic Under Hypoxic Conditions. Photochem. Photobiol. 2020, 96, 327–339. 10.1111/php.13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello-Garibo J.-A.; Meijer M. S.; Bonnet S. To Cage or to Be Caged? The Cytotoxic Species in Ruthenium-Based Photoactivated Chemotherapy Is not Always the Metal. Chem. Commun. 2017, 53, 6768–6771. 10.1039/C7CC03469E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.; Ghrayche J. B.; Wei J.; Renfrew A. K. Photolabile Ruthenium(II)-Purine Complexes: Phototoxicity, DNA Binding, and Light-Triggered Drug Release. Eur. J. Inorg. Chem. 2017, 2017, 1679–1686. 10.1002/ejic.201601137. [DOI] [Google Scholar]
- Perez-Fernandez R.; Goya P.; Elguero J. A Review of Recent Progress (2002-2012) on the Biological Activities of Pyrazoles. ARKIVOC 2014, 2, 233–293. 10.3998/ark.5550190.p008.131. [DOI] [Google Scholar]
- Arber N.; Eagle C. J.; Spicak J.; Racz I.; Dite P.; Hajer J.; Zavoral M.; Lechuga M. J.; Gerletti P.; Tang J.; Rosenstein R. B.; Macdonald K.; Bhadra P.; Fowler R.; Wittes J.; Zauber A. G.; Solomon S. D.; Levin B. Celecoxib for the Prevention of Colorectal Adenomatous Polyps. New Engl. J. Med. 2006, 355, 885–895. 10.1056/NEJMoa061652. [DOI] [PubMed] [Google Scholar]
- https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-encorafenib-and-binimetinib-combination-unresectable-or-metastatic-melanoma-braf.
- Qin Y. J.; Li Y. J.; Jiang A. Q.; Yang M. R.; Zhu Q. Z.; Dong H.; Zhu H. L. Design, Synthesis and Biological Evaluation of Novel Pyrazoline-containing Derivatives as Potential Tubulin Assembling Inhibitors. Eur. J. Med. Chem. 2015, 94, 447–457. 10.1016/j.ejmech.2015.02.058. [DOI] [PubMed] [Google Scholar]
- Raghav N.; Singh M. SAR Studies of Differently Functionalized Chalcones Based Hydrazones and Their Cyclized Derivatives as Inhibitors of Mammalian Cathepsin B and Cathepsin H. Bioorg. Med. Chem. 2014, 22, 4233–4245. 10.1016/j.bmc.2014.05.037. [DOI] [PubMed] [Google Scholar]
- Shuck S. C.; Turchi J. J. Targeted Inhibition of Replication Protein A Reveals Cytotoxic Activity, Synergy with Chemotherapeutic DNA-damaging Agents, and Insight into Cellular Function. Cancer Res. 2010, 70, 3189–3198. 10.1158/0008-5472.CAN-09-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havrylyuk D.; Kovach N.; Zimenkovsky B.; Vasylenko O.; Lesyk R. Synthesis and Anticancer Activity of Isatin-based Pyrazolines and Thiazolidines Conjugates. Arch. Pharm. 2011, 344, 514–522. 10.1002/ardp.201100055. [DOI] [PubMed] [Google Scholar]
- Havrylyuk D.; Zimenkovsky B.; Vasylenko O.; Day C. W.; Smee D. F.; Grellier P.; Lesyk R. Synthesis and Biological Activity Evaluation of 5-pyrazoline Substituted 4-thiazolidinones. Eur. J. Med. Chem. 2013, 66, 228–237. 10.1016/j.ejmech.2013.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havrylyuk D.; Zimenkovsky B.; Vasylenko O.; Gzella A.; Lesyk R. Synthesis of New 4-thiazolidinone-, Pyrazoline-, and Isatin-based Conjugates with Promising Antitumor Activity. J. Med. Chem. 2012, 55, 8630–8641. 10.1021/jm300789g. [DOI] [PubMed] [Google Scholar]
- Havrylyuk D.; Zimenkovsky B.; Vasylenko O.; Zaprutko L.; Gzella A.; Lesyk R. Synthesis of Novel Thiazolone-based Compounds Containing Pyrazoline Moiety and Evaluation of Their Anticancer Activity. Eur. J. Med. Chem. 2009, 44, 1396–1404. 10.1016/j.ejmech.2008.09.032. [DOI] [PubMed] [Google Scholar]
- Havrylyuk D.; Roman O.; Lesyk R. Synthetic Approaches, Structure Activity Relationship and Biological Applications for Pharmacologically Attractive Pyrazole/pyrazoline-thiazolidine-based Hybrids. Eur. J. Med. Chem. 2016, 113, 145–166. 10.1016/j.ejmech.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdieiev S.; Gera L.; Havrylyuk D.; Hodges R. S.; Lesyk R.; Ribrag V.; Vassetzky Y.; Kavsan V. Bradykinin Antagonists and Thiazolidinone derivatives as New Potential Anti-cancer Compounds. Bioorg. Med. Chem. 2014, 22, 3815–3823. 10.1016/j.bmc.2014.06.046. [DOI] [PubMed] [Google Scholar]
- Havrylyuk D.; Zimenkovsky B.; Karpenko O.; Grellier P.; Lesyk R. Synthesis of Pyrazoline-thiazolidinone Hybrids with Trypanocidal Activity. Eur. J. Med. Chem. 2014, 85, 245–254. 10.1016/j.ejmech.2014.07.103. [DOI] [PubMed] [Google Scholar]
- Budzisz E.; Miernicka M.; Lorenz I.-P.; Mayer P.; Krajewska U.; Rozalski M. Synthesis and X-ray Structure of Platinum(II), Palladium(II) and Copper(II) Complexes with Pyridine-pyrazole Ligands: Influence of Ligands’ Structure on Cytotoxic Activity. Polyhedron 2009, 28, 637–645. 10.1016/j.poly.2008.12.013. [DOI] [Google Scholar]
- Kupcewicz B.; Ciolkowski M.; Karwowski B. T.; Rozalski M.; Krajewska U.; Lorenz I.-P.; Mayer P.; Budzisz E. Copper(II) Complexes with Pyrazole Derivatives – Synthesis, Crystal Structure, DFT Calculations and Cytotoxic Activity. J. Mol. Struct. 2013, 1052, 32–37. 10.1016/j.molstruc.2013.08.045. [DOI] [Google Scholar]
- Wang S.; Shao W.; Li H.; Liu C.; Wang K.; Zhang J. Synthesis, Characterization and Cytotoxicity of the Gold(III) Complexes of 4,5-dihydropyrazole-1-carbothioamide Derivatives. Eur. J. Med. Chem. 2011, 46, 1914–1918. 10.1016/j.ejmech.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Huang Y. C.; Haribabu J.; Chien C. M.; Sabapathi G.; Chou C. K.; Karvembu R.; Venuvanalingam P.; Ching W. M.; Tsai M. L.; Hsu S. C. N. Half-sandwich Ru(η(6)-p-cymene) Complexes Featuring Pyrazole Appended Ligands: Synthesis, DNA Binding and in vitro Cytotoxicity. J. Inorg. Biochem. 2019, 194, 74–84. 10.1016/j.jinorgbio.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Yang G. W.; Zhang X.; Li G. M.; Yang J.; Shen L.; Chen D. Y.; Li Q. Y.; Zou D. F. Photochemical Property of a Ru(II) Compound Based on 3-(2-pyridyl)pyrazole and 2,2’-bipyridine for Ablation of Cancer Cells. New J. Chem. 2018, 42, 5395–5402. 10.1039/C7NJ05034H. [DOI] [Google Scholar]
- Thota S.; Imran M.; Udugula M.; Yerra R.; Karki S. S.; Balzarini J.; De Clercq E. Synthesis, Spectroscopic Characterization and in vitro Antitumor Activities of Some Novel Mononuclear Ru(II) Complexes. Chin. Chem. Lett. 2012, 23, 466–469. 10.1016/j.cclet.2012.01.025. [DOI] [Google Scholar]
- Tang B.; Wan D.; Lai S.-H.; Yang H.-H.; Zhang C.; Wang X.-Z.; Zeng C.-C.; Liu Y.-J. Design, Synthesis and Evaluation of Anticancer Activity of Ruthenium (II) Polypyridyl Complexes. J. Inorg. Biochem. 2017, 173, 93–104. 10.1016/j.jinorgbio.2017.04.028. [DOI] [PubMed] [Google Scholar]
- Huang H.-L.; Li Z.-Z.; Liang Z.-H.; Liu Y.-J. Cell Cycle Arrest, Cytotoxicity, Apoptosis, DNA-Binding, Photocleavage, and Antioxidant Activity of Octahedral Ruthenium(II) Complexes. Eur. J. Inorg. Chem. 2011, 2011, 5538–5547. 10.1002/ejic.201100848. [DOI] [Google Scholar]
- Lai S.-H.; Jiang G.-B.; Yao J.-H.; Li W.; Han B.-J.; Zhang C.; Zeng C.-C.; Liu Y.-J. Cytotoxic Activity, DNA Damage, Cellular Uptake, Apoptosis and Western Blot Analysis of Ruthenium(II) Polypyridyl Complex Against Human Lung Decarcinoma A549 Cell. J. Inorg. Biochem. 2015, 152, 1–9. 10.1016/j.jinorgbio.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Liang Z.-H.; Li Z.-Z.; Huang H.-L.; Liu Y.-J. Ruthenium(II) Complexes: Synthesis, Cytotoxicity in vitro, Apoptosis, DNA-binding, Photocleavage, and Antioxidant Activity Studies. J. Coord. Chem. 2011, 64, 3342–3352. 10.1080/00958972.2011.619533. [DOI] [Google Scholar]
- Lin G.-J.; Jiang G.-B.; Xie Y.-Y.; Huang H.-L.; Liang Z.-H.; Liu Y.-J. Cytotoxicity, Apoptosis, Cell Cycle Arrest, Reactive Oxygen Species, Mitochondrial Membrane Potential, and Western Blotting Analysis of Ruthenium(II) Complexes. JBIC, J. Biol. Inorg. Chem. 2013, 18, 873–882. 10.1007/s00775-013-1032-2. [DOI] [PubMed] [Google Scholar]
- Liu Y.-J.; Zeng C.-H.; Yao J.-H.; Wu F.-H.; He L.-X.; Huang H.-L. Synthesis, Structure, DNA-Binding Properties, and Cytotoxicity of Ruthenium(II) Polypyridyl Complexes. Chem. Biodiversity 2010, 7, 1770–1783. 10.1002/cbdv.200900213. [DOI] [PubMed] [Google Scholar]
- Wan D.; Lai S.-H.; Zeng C.-C.; Zhang C.; Tang B.; Liu Y.-J. Ruthenium(II) Polypyridyl Complexes: Synthesis, Characterization and Anticancer Activity Studies on BEL-7402 Cells. J. Inorg. Biochem. 2017, 173, 1–11. 10.1016/j.jinorgbio.2017.04.026. [DOI] [PubMed] [Google Scholar]
- Wan D.; Tang B.; Wang Y.-J.; Guo B.-H.; Yin H.; Yi Q.-Y.; Liu Y.-J. Synthesis and Anticancer Properties of Ruthenium (II) Complexes as Potent Apoptosis Inducers Through Mitochondrial Disruption. Eur. J. Med. Chem. 2017, 139, 180–190. 10.1016/j.ejmech.2017.07.066. [DOI] [PubMed] [Google Scholar]
- Wang J.-Q.; Zhao Z.-Z.; Bo H.-B.; Chen Q.-Z. Synthesis, Characterization, and Antitumor Properties of Ruthenium(II) Anthraquinone Complexes. J. Coord. Chem. 2016, 69, 177–189. 10.1080/00958972.2015.1120291. [DOI] [Google Scholar]
- Xie Y.-Y.; Jiang G.-B.; Yao J.-H.; Lin G.-J.; Huang H.-L.; Wang X.-Z.; Liu Y.-J. DNA-binding, Antioxidant activity, and Bbioactivity Studies of Ruthenium(II) Complexes Containing Amino Substituents. J. Coord. Chem. 2013, 66, 2423–2433. 10.1080/00958972.2013.805214. [DOI] [Google Scholar]
- Xie Y.-Y.; Li Z.-Z.; Lin G.-J.; Huang H.-L.; Wang X.-Z.; Liang Z.-H.; Jiang G.-B.; Liu Y.-J. DNA Interaction, Cytotoxicity, Apoptotic Activity, Cell Cycle Arrest, Reactive Oxygen Species and Mitochondrial Membrane Potential Assay Induced by Ruthenium(II) Polypyridyl Complexes. Inorg. Chim. Acta 2013, 405, 228–234. 10.1016/j.ica.2013.06.002. [DOI] [Google Scholar]
- Zeng C.-C.; Lai S.-H.; Yao J.-H.; Zhang C.; Yin H.; Li W.; Han B.-J.; Liu Y.-J. The Induction of Apoptosis in HepG-2 Cells by Ruthenium(II) Complexes Through an Intrinsic ROS-mediated Mitochondrial Dysfunction Pathway. Eur. J. Med. Chem. 2016, 122, 118–126. 10.1016/j.ejmech.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Zeng C.-H.; Liu Y.-J.; Li Z.-Z.; Liang Z.-H.; Huang H.-L.; Wu F.-H. DNA-binding, Antioxidant Activity and in vitro Cytotoxicity Induced by Ruthenium(II) Complexes Containing Polypyridyl Ligands. Transition Met. Chem. 2010, 35, 731–736. 10.1007/s11243-010-9386-6. [DOI] [Google Scholar]
- Zhang C.; Han B.-J.; Zeng C.-C.; Lai S.-H.; Li W.; Tang B.; Wan D.; Jiang G.-B.; Liu Y.-J. Synthesis, Characterization, in vitro Cytotoxicity and Anticancer Effects of Ruthenium (II) Complexes on BEL-7402 Cells. J. Inorg. Biochem. 2016, 157, 62–72. 10.1016/j.jinorgbio.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Zeng C.-C.; Lai S.-H.; Xing D.-G.; Li W.; Han B.-J.; Liu Y.-J. Synthesis, Cytotoxicity in vitro, Apoptosis, Cell Cycle Arrest and Comet Assay of Asymmetry Ruthenium(II) Complexes. Polyhedron 2016, 106, 115–124. 10.1016/j.poly.2015.12.058. [DOI] [Google Scholar]
- Guo Q.-F.; Liu S.-H.; Liu Q.-H.; Xu H.-H.; Zhao J.-H.; Wu H.-F.; Li X.-Y.; Wang J.-W. Synthesis, Characterization, Photocleavage, Cytotoxicity in vitro, Apoptosis, and Cell Cycle Arrest of Ruthenium(II) Complexes. J. Coord. Chem. 2012, 65, 1781–1791. 10.1080/00958972.2012.680592. [DOI] [Google Scholar]
- Glazer E. C.; Heidary D. K.. Light-Activated Compounds. US9290528B1, 2016.
- Havrylyuk D.; Deshpande M.; Parkin S.; Glazer E. C. Ru(II) Complexes with Diazine Ligands: Electronic Modulation of the Coordinating Group Is Key to the Design of “Dual Action” Photoactivated Agents. Chem. Commun. 2018, 54, 12487–12490. 10.1039/C8CC05809A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havrylyuk D.; Stevens K.; Parkin S.; Glazer E. C. Toward Optimal Ru(II) Photocages: Balancing Photochemistry, Stability, and Biocompatibility Through Fine Tuning of Steric, Electronic, and Physiochemical Features. Inorg. Chem. 2020, 59, 1006–1013. 10.1021/acs.inorgchem.9b02065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. K. C.; Huff G. S.; Cubanski J. R.; Kennedy A. D. W.; McAdam C. J.; McMorran D. A.; Gordon K. C.; Crowley J. D. Comparison of Inverse and Regular 2-Pyridyl-1,2,3-triazole “Click” Complexes: Structures, Stability, Electrochemical, and Photophysical Properties. Inorg. Chem. 2015, 54, 1572–1587. 10.1021/ic502557w. [DOI] [PubMed] [Google Scholar]
- Scattergood P. A.; Khushnood U.; Tariq A.; Cooke D. J.; Rice C. R.; Elliott P. I. P. Photochemistry of [Ru(pytz)(btz)2]2+ and Characterization of a κ1-btz Ligand-Loss Intermediate. Inorg. Chem. 2016, 55, 7787–7796. 10.1021/acs.inorgchem.6b00782. [DOI] [PubMed] [Google Scholar]
- Mamolo M. G.; Zampieri D.; Falagiani V.; Vio L.; Banfi E. Synthesis and Antimycobacterial Activity of 5-aryl-1-isonicotinoyl-3-(pyridin-2-yl)-4,5-dihydro-1H-pyrazole Derivatives. Il Farmaco 2001, 56, 593–599. 10.1016/S0014-827X(01)01098-9. [DOI] [PubMed] [Google Scholar]
- Rurack K.; Bricks J. L. Towards Simple and Efficient Molecular Reporters: Combining Electron Transfer and Charge Transfer in Functional Dyes of Donor-acceptor-spacer-donor Constitution. ARKIVOC 2001, 11, 31–40. 10.3998/ark.5550190.0002.b03. [DOI] [Google Scholar]
- Li X.; Wang L.; Long L.; Xiao J.; Hu Y.; Li S. Synthesis and Biological Evaluation of 1,2,4-trisubstituted Imidazoles and 1,3,5-trisubstituted Pyrazoles as Inhibitors of Transforming Growth Factor β Type 1 Receptor (ALK5). Bioorg. Med. Chem. Lett. 2009, 19, 4868–4872. 10.1016/j.bmcl.2009.04.066. [DOI] [PubMed] [Google Scholar]
- Gong Z.-L.; Ge F.; Zhao B.-X. Novel Pyrazoline-based Selective Fluorescent Sensor for Zn2+ in Aqueous Media. Sens. Actuators, B 2011, 159, 148–153. 10.1016/j.snb.2011.06.064. [DOI] [Google Scholar]
- Wang P.; Onozawa-Komatsuzaki N.; Katoh R.; Himeda Y.; Sugihara H.; Arakawa H.; Kasuga K. Structure and Properties of Diastereoisomers of a Ruthenium(II) Complex Having a Pyridylpyrazoline Derivative as a Ligand. Chem. Lett. 2001, 30, 940–941. 10.1246/cl.2001.940. [DOI] [Google Scholar]
- Rillema D. P.; Jones D. S. Structure of Tris(2,2’-bipyridyl)ruthenium(II) Hexafluorophosphate, [Ru(bipy)3][PF6]2; X-ray Crystallographic Determination. J. Chem. Soc., Chem. Commun. 1979, 19, 849–851. 10.1039/C39790000849. [DOI] [Google Scholar]
- Greguric A.; Greguric I. D.; Hambley T. W.; Aldrich-Wright J. R.; Collins J. G. Minor Groove Intercalation of Δ-[Ru(Me2phen)2dppz]2+ to the Hexanucleotide d(GTCGAC)2. J. Chem. Soc., Dalton Trans. 2002, 849–855. 10.1039/b105689c. [DOI] [Google Scholar]
- Ashby M. T.; Alguindigue S. S.; Khan M. A. Kinetic Element Effect for Atropisomerization of an Organometallic Complex of the Misdirected Ligand 1,1‘-Biisoquinoline. Organometallics 2000, 19, 547–552. 10.1021/om990825l. [DOI] [Google Scholar]
- Azar D. F.; Audi H.; Farhat S.; El-Sibai M.; Abi-Habib R. J.; Khnayzer R. S. Phototoxicity of Strained Ru(II) Complexes: Is it the Metal Complex or the Dissociating Ligand?. Dalton Trans. 2017, 46, 11529–11532. 10.1039/C7DT02255G. [DOI] [PubMed] [Google Scholar]
- Collin J. P.; Sauvage J. P. Synthesis and Study of Mononuclear Ruthenium(II) Complexes of Sterically Hindering Diimine Chelates. Implications for the Catalytic Oxidation of Water to Molecular Oxygen. Inorg. Chem. 1986, 25, 135–141. 10.1021/ic00222a008. [DOI] [Google Scholar]
- Heidary D. K.; Fox A.; Richards C. I.; Glazer E. C. A High-Throughput Screening Assay Using a Photoconvertable Protein for Identifying Inhibitors of Transcription, Translation, or Proteasomal Degradation. SLAS Discovery 2017, 22, 399–407. 10.1177/2472555216684333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havrylyuk D.; Howerton B. S.; Nease L.; Parkin S.; Heidary D. K.; Glazer E. C. Structure-activity Relationships of Anticancer Ruthenium(II) Complexes with Substituted Hydroxyquinolines. Eur. J. Med. Chem. 2018, 156, 790–799. 10.1016/j.ejmech.2018.04.044. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997, 276, 307–326. 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- APEX2 . Programs for Data Collection and Data Reduction; Bruker-Nonius: Madison, WI. [Google Scholar]
- Krause L.; Herbst-Irmer R.; Sheldrick G. M.; Stalke D. Comparison of Silver and Molybdenum Microfocus X-ray Sources for Single-crystal Structure Determination. J. Appl. Crystallogr. 2015, 48, 3–10. 10.1107/S1600576714022985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin S.; Moezzi B.; Hope H. XABS2: an Empirical Absorption Correction Program. J. Appl. Crystallogr. 1995, 28, 53–56. 10.1107/S0021889894009428. [DOI] [Google Scholar]
- Sheldrick G. M. SHELXT – Integrated Space-group and Crystal-structure Determination. Acta Crystallogr., Sect. A: Found. Adv. 2015, 71, 3–8. 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. Structure Determination Revisited. Acta Crystallogr., Sect. A: Found. Adv. 2015, 71, S9. 10.1107/S2053273315099842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis P.; Spek A. L. BYPASS: an Effective Method for the Refinement of Crystal Structures Containing Disordered Solvent Regions. Acta Crystallogr., Sect. A: Found. Crystallogr. 1990, 46, 194–201. 10.1107/S0108767389011189. [DOI] [Google Scholar]
- Spek A. L. Structure Validation in Chemical Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65, 148–155. 10.1107/S090744490804362X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin S. Expansion of Scalar Validation Criteria to Three Dimensions: the R Tensor. Acta Crystallogr., Sect. A: Found. Crystallogr. 2000, 56, 157–162. 10.1107/S010876739901497X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.