Abstract
Studies characterizing stem cell lineages in different organs aim to understand which cells particular progenitors can give rise to and how this is controlled. Because the skin contains several resident stem cell populations and undergoes constant turnover, it is an ideal tissue in which to study this process. Furthermore, with the advent of two-photon microscopy techniques in combination with genetic tools for cell labeling, this question can be studied non-invasively by using live imaging. In this chapter, we describe an experimental approach that takes this technique one step further. We combine the Cre and Tet inducible genetic systems for single clone labeling and genetic manipulation in a specific stem cell population in the skin by using known drivers. Our system involves the use of gain and loss of function alleles activated only in a differentially labeled population to distinguish single clones. The same region within a tissue is imaged over time to document the fate and interactions of single clones with and without genetic modifications in the long term. By implementing a lineage tracing approach and documenting changes in cell behavior brought about by the genetic alterations allows both aspects to be linked. Because of the inherent flexibility of the approach, we expect it to have broad applications in studying stem cell function in the skin, but also in other tissues amenable to live imaging.
Keywords: Live imaging, stem cells, skin, lineage tracing, two-photon laser scanning fluorescent microscopy, clonal dynamics, inducible genetic mouse tools
1. Introduction
1.1. In vivo Lineage Tracing in the Skin
In vivo lineage tracing has been the gold standard for identifying and studying stem cells in various organs. In a classical lineage tracing experiment, cells are labeled to allow the fate of their descendants to be followed through time [1]. This experimental set-up has been widely used to characterize tissue-specific resident stem cell populations and their subsequent progeny. The visualization of stem cells through live imaging is a major step forward from static histological analysis and has significantly contributed to our understanding of stem cell behavior in their native in vivo environment. Studies carried out at a population-level provide information on the general aspects of stem cell behavior however; by averaging out individual behaviors, they bypass the opportunity to shed light on the inherent heterogeneity of the stem cell population [2]. Therefore, to gain a deeper understanding of the underlying mechanisms that regulate individual stem cell activity requires tools that allow us to document behavior at the individual level. Observing how the behavior of individual stem cells is altered when a particular gene is turned on or off compared to that of their neighbors allows a causative relationship between specific genes and cellular activity to be established. Therefore, the capacity to both trace and genetically manipulate single cells over time ultimately contributes to dissecting the underlying molecular heterogeneity that characterizes some stem cell populations. Furthermore, cellular interactions between cells with different genetic compositions can be studied by differentially labeling single genetically altered and “normal” cells within the same tissue. Experiments of this nature would also be useful to uncover the genetic drivers of aberrant stem cell activity in the context of wound healing and disease.
The skin is a well-studied and readily accessible organ that continuously self-regenerates thanks to the contribution of several resident stem cell populations located within self-contained compartments. These include the interfollicular epidermis (IFE), hair follicles (HF), and sebaceous and sweat glands [3]. As occurs in other adult epithelia, stem and progenitor cells in the skin constantly divide to replace the cells that are naturally lost through terminal differentiation and shedding. This process is crucial for the maintenance of organ homeostasis [4]. Due to the fast turnover of cells in the skin, conventional histological techniques are insufficient to provide a comprehensive understanding of complex mechanisms regulating stem cell activity [5]. Genomics tools provide a very detailed snapshot on the molecular heterogeneity in a cell population, but lack the temporal and spatial information required to put that information into the context of the three-dimensional tissue [6], a technical challenge that the field is currently tackling [7]. Live imaging together with powerful mouse genetic tools provides a unique opportunity for real-time single cell tracking in the intact adult skin. The approach has already provided unparalleled insight into the proliferation and differentiation dynamics of stem cells and their progeny [8, 9]. These genetic systems rely on the use of different promoters to drive the expression of a Cre recombinase in a specific stem cell population to activate expression of a fluorescent reporter. Labeled cells can be visualized in vivo in a noninvasive manner by using two-photon laser scanning fluorescent microscopy, which minimizes phototoxicity and photobleaching while increasing tissue imaging depth [10, 11]. Additionally, revisiting the same area in multiple imaging sessions enables the direct tracking of progenitor and descendant cell fate over time.
Studies that address how genetic changes impact stem cells on a population level lack the spatiotemporal resolution to distinguish variable cellular responses of singe cells. Because stem cell populations are heterogeneous, gaining a molecular understanding of the underlying mechanisms that regulate stem cell activity at the single cell level requires tools to genetically manipulate and monitor individual cells. In this chapter, we describe how live imaging can be combined with the use of two inducible systems, the Cre and Tet systems, to lineage trace cells whose gene expression is specifically altered. The method involves using the spatiotemporally regulated expression of Cre to generate single labeled clones within specific stem cell populations, whereas Tet drives the expression of gain- or loss-of- function alleles to genetically manipulate specific genes and signaling pathways in the labeled clones. In the example below, we propose a system in which expression of a single gene is modulated in fluorescently labeled skin stem cells whose long-term behavior can be recorded non-invasively at high resolution by two-photon microscopy. The activity of these cells is then compared to that of “normal” neighboring cells, which are also clonally marked but with a different fluorescent reporter. We use this design as proof of principle for a genetic system that can be expanded to modulate a wide assortment of signaling pathways and characterize their importance in stem cell dynamics in vivo.
1.2. Combining Cre-Mediated Recombination with Tetracycline-Controlled Transcriptional Activation
The P1 bacteriophage Cre-recombinase system efficiently mediates the recombination of palindromic LoxP recognition sites, resulting in the excision of the flanked DNA segment [12]. Cre activity is spatiotemporally controlled by using a cell population-specific promoter to control the expression of an inducible cre allele (CreER) in which the protein is fused to an estrogen receptor (ER) that sequesters it in the cytoplasm [13]. The chimeric CreER will translocate to the nucleus only in the presence of an ER ligand, most commonly, the estrogen analog Tamoxifen (see Note 1). Importantly, the number of cells that undergo Cre-mediated recombination increases with Tamoxifen treatment in a dose-dependent manner. Therefore, as a general rule, it is necessary to titrate the drug dosage to confine Cre mediated recombination to only few, spatially dispersed clones that are easily identifiable. The exact dose of Tamoxifen must be determined to account for variations in promoter activity, as well as tissue variations that may affect Cre-recombinase expression and activation to achieve clonal labeling. It is also necessary for mice to carry a Cre reporter allele that tracks recombinase activity for normal cells to be identified and tracked at the individual level.
Tetracycline(Tet)-inducible systems, derived from the E. coli tetracycline resistance operon, activate or repress genes under the control of the Tet Operon (tetO) sequence [14, 15]. Depending on whether the TetOFF or TetON system is used, two different transactivator fusion alleles can be used to drive expression of genes downstream of TetO sequences, the tTA (tetracycline transactivator protein) and rtTA (reverse tTA), respectively. Each has an opposite requirement of tetracycline or doxycycline, tetracycline’s commonly used analog. In the TetOFF system, doxycycline prevents tTA from binding to TetO sequences. Conversely, in the TetON system, doxycycline induces the binding of rtTA to TetO sequences and downstream gene expression is subsequently activated. Therefore, unlike the Cre-mediated recombination effects, which are irreversible, the activity of Tet-inducible systems can be modulated throughout the experiment by the administration or removal of doxycycline.
To label and monitor the behavior of normal and genetically altered cells within the same tissue we combine the two inducible systems to lineage trace the behavior of normal and genetically altered cells within the same tissue. In the method outlined in this chapter, we use the LoxP-STOP-LoxP (LSL) allele, in which a STOP cassette is placed between a ubiquitous promoter and the Tet transactivator of choice. This allele links the two inducible systems because Cre recombination is required for the excision of the STOP sequence to allow the expression of the tTA. Therefore, the activation of TetO-driven alleles is restricted to the cells in which Cre-recombination is activated. This system can be combined with any available Cre transgenic line and TetO alleles, enabling a wide range of manipulation possibilities. Similar genetic approaches have previously been used for the activation in non-sensory regions of the ear [16], and in the skin, to induce YAP expression [17] and drive oncogene expression [18]. The novelty of our approach relies on the simultaneous differential fluorescent labeling of single cells and its combined use with two-photon microscopy to perform lineage tracing studies in live mice.
2. Materials
2.1. Choice of Mouse Lines
Several inducible CreER reporter lines that target specific stem cell populations in the skin and other tissues are available from public vendors such as the Jackson Laboratory. The driver lines listed in Table 1 are three examples of CreER reporter lines that allow clonal labeling in unique stem cell populations in the epidermis (K14-CreER), isthmus/sebaceous gland (Lgr6-CreER) and hair follicle (Lgr5-CreER). Although there are several CreER reporter lines that can be used for lineage tracing, the presence of the mTom-LSL-mGFP allele ensures that cell membranes will be ubiquitously labeled by tdTomato (mT) expression. Because membrane labels can be used to visualize cellular morphology and tissue architecture, expression of tdTomato is necessary to determine spatial location of the clones within the epidermis. In cells where Cre recombinase is activated EGFP (mG) will replace tdTomato at the membrane to generate single “normal” clones (see Figure 1A). Cre-recombinase will also turn on expression of the tTA in single cells. The Tet transactivator drives expression of both a reporter TetO allele and the gain- or loss-of-function TetO allele of choice, thus labeling single altered clones. A model experimental timeline is shown in Figure 1B (see Note 2).
Table 1.
Useful publically available transgenic mouse lines
| Allele | Source | Reference |
|---|---|---|
| LSL-tTA | JAX Stock no. 008600 |
[19] |
| mTom-LSL-mGFP | JAX Stock no. 007676 |
[20] |
| TetO-H2BGFP | JAX Stock no. 005104 |
[21] |
| TetO-GeneofInterest | ||
| K14-CreER* | JAX Stock no. 005107 |
[22] |
| Lgr5-CreER* | JAX Stock no. 008875 |
[23] |
| Lgr6-CreER* | JAX Stock no. 016934 |
[24] |
Available Cre-reporters that can be used to target different stem cell populations in the skin
Figure 1.
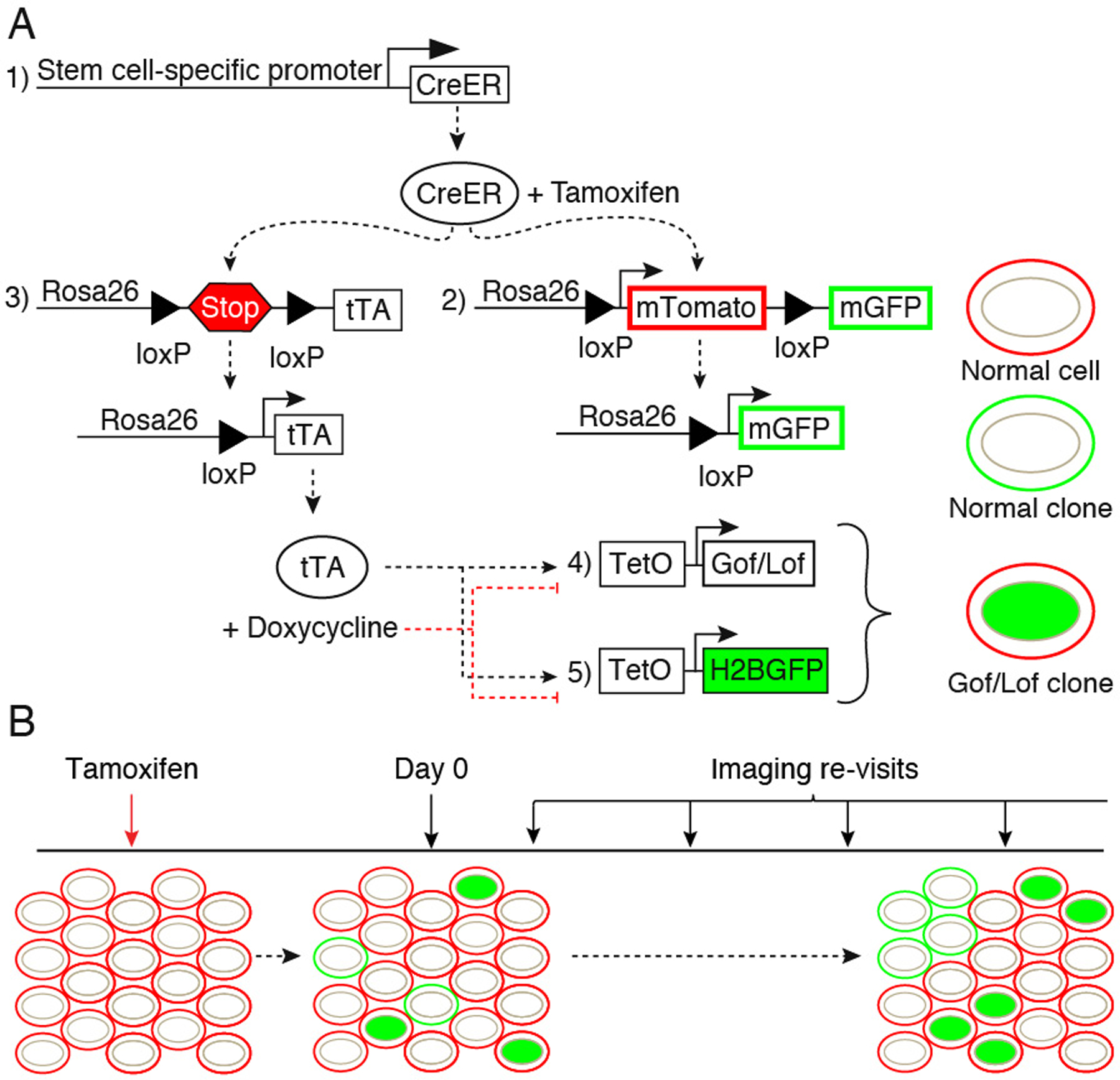
Genetic system and experimental timeline. To generate single genetically altered and normal clones, 5 alleles are used in our system. A stem cell-specific driver to control expression of an inducible Cre recombinase (1). Addition of Tamoxifen will enable the Cre-mediated excision of loxP-flanked sequences in two alleles: a reporter allele (2), in this case mT/mG, in which membrane-localized EGFP (mG) replaces the excised membrane-localized tdTomato (mT) in single clones, and the Tet transactivator allele (3), in this case LSL-tTA, where excision of a STOP cassette allows tTA expression. Expression of tTA in single clones will then enable activation of the TetO alleles: A gain- or loss-of-function (Gof/Lof) allele to alter a gene/pathway of interest (4) and a second reporter allele (5), in this case TetO-H2BGFP used to label the single “Lof/Gof clones” with nuclear EGFP. In this example, where the TetOFF system is used, doxycycline treatment will prevent binding of tTA to the TetO sequences, thus silencing TetO-driven gene expression (A). Before Tamoxifen treatment, all cells are uniformly labeled with membrane tdTomato (“normal cells”). After induction of Cre-mediated recombination, on the first imaging session taking place at “Day 0”, single clones labeled either with membrane EGFP instead of red (“normal clones”), or carrying a nuclear EGFP (“Gof/Lof clones”) are identified. Subsequent imaging sessions allow live tracking of clonal dynamics through time. The frequency of imaging revisits should be empirically determined according to experimental needs (B).
2.2. Systemic Induction by Intraperitoneal Injection of Tamoxifen
Tamoxifen (Sigma)
Corn oil
Scale
27G Insulin syringe (BD)
Preparation of Tamoxifen
Dissolve Tamoxifen in corn oil to a final concentration of 20 mg/ml by incubating in a 37 °C water bath. Vortex the solution frequently over the course of several hours. Tamoxifen is classified as carcinogenic and toxic, so always wear gloves when handling it. After Tamoxifen is in solution, wrap with foil and store in the dark at −20 °C.
2.3. Temporal Regulation of Transgene Expression by Doxycycline
Doxycyline (Sigma)
Drinking water
Amber water bottle
Preparation of Doxycycline
Dissolve Doxycycline in drinking water to a final concentration of 1 mg/ml by vortexing the solution for several minutes. This solution can be wrapped in foil and frozen in 5 ml aliquots at −20 °C.
2.4. Two-Photon Imaging
Mouse Preparation
Fluorescent reporter mice containing all alleles (see Table 1)
PBS, pH 7.2
Depilatory lotion (Nair)
Tattoo ink (Optional; Dr. Ph. Martin’s Black Star India Ink)
LubriFresh P.M. sterile eye ointment (MAJOR)
Ketamine (Midwest Veterinary Supply)
Xylazine (Midwest Veterinary Supply)
10 ml vial (GREER)
30G Insulin syringe for tattooing (Optional; BD)
27G Insulin syringe for IP injections (BD)
Scale
Small, electric hair clipper (Wahl)
Cotton swabs
Adhesive tape
Wall-mounted anesthesia machine (VetEquip)
Isoflurane vaporizer (VetEquip)
Anesthesia breathing circuit and nose cone (Braintree Scientific)
O2 gas flow regulator for E-cylinders (VetEquip)
O2 tank (E-cylinder; Airgas)
Imaging Equipment
Olympus FV1200MPE (Olympus) or equivalent multiphoton microscope with upright objective lens and motorized stage
Chameleon Vision II (Coherent) or equivalent Ti:sapphire tunable laser
25× objective (Olympus, XLPLN25XWMP2) or equivalent infrared optimized objective of desired magnification
Custom mounting stage with custom-made mounting spatulas [10] and micromanipulators (Edmund Optics, cat. nos. NT03–682 ×2 and NT36–347 ×2)
Heating pad (CryoLogic, BioTerm SmartStage no. SS12)
Glass coverslip (22 mm; Electron Microscopy Sciences)
Super glue (Loctite)
Bull’s-eye level (McMaster-Carr)
Thumb screw (ProTanium)
Vacuum grease (Dow Corning)
Purified water (Milli-Q, Millipore)
Preparation of Ketamine/Xylazine Solution
Combine 15 mg/ml ketamine (15 mg/ml) and xylazine (1 mg/ml) in PBS. Store solution in a sterile 10 ml vial at room temperature for up to 30 days.
2.5. Image Analysis
Fiji/ImageJ
3. Methods
3.1. Systemic Induction by Intraperitoneal Injection of Tamoxifen
Cre activation is induced with a single intraperitoneal (IP) injection of Tamoxifen (1μg/g in corn oil) at ~ postnatal day 50 (P50) to clonally label single epidermal cells.
Weigh the mouse to determine injection dose. For full induction, use approximately 75 mg Tamoxifen/kg body weight. Be sure to titrate the dose of Tamoxifen beforehand to determine the optimal dose for clonal induction using a particular Cre reporter.
Administer Tamoxifen via IP injection using an institution approved animal injection protocol.
Tamoxifen remains in the mouse’s system for ~3 days, and recombination events can occur during this time. Therefore, the first imaging time-point should be at least 3 days after Tamoxifen injection.
3.2. Temporal Regulation of Transgene Expression by Doxycycline
After the addition of Tamoxifen, cells that express tTA following recombination will also start to express H2BGFP (and any other transgenes under the control of the TetO element). To reverse the expression of all TetO alleles, the binding of tTA can be inhibited by treatment with doxycycline. This would lead to a reversal of both the genetic modifications and reporter expression, returning “Gof/Lof” clones to a “normal” state (see Note 3) Therefore, when TetO inhibition is desired, administer doxycycline (1 mg/ml in drinking water) at specified times.
Add the drinking water with dissolved Doxycycline to an amber water bottle and place in the mouse cage at selected time-points.
3.3. Two-Photon Imaging
On the fourth day after Tamoxifen induction, the first time-point in the time-course clonal analysis is collected by two-photon microscopy.
3.3.1. Preparing mouse for imaging
Weigh the mouse to determine injection dose of the ketamine (100 mg/kg)/xylazine (10 mg/kg) solution. Administer Tamoxifen via IP injection with a 27G insulin syringe using a institution approved animal injection protocol.
After the mouse is fully sedated, place the mouse on heating pad (see Note 4) on the stage. Apply eye ointment to keep eyes lubricated throughout imaging session and prevent damage. Next, use the electric clippers, appropriate for veterinary use in small animals, to shave the hair covering the area of skin to be imaged.
Apply depilatory cream to the shaved area using a cotton swab. Let the cream sit for 2 minutes, then use cotton swabs soaked in PBS to gently remove the cream.
Optional: Mark the area of the skin that is being imaged for later re-visiting by creating a small punctate tattoo. Dip a 30-gauge needle into an ink solution and quickly poke the skin of the anesthetized mouse. The pigment should be visible macroscopically.
3.3.2. Mounting mouse on imaging stage
Screw the skin-mounting spatula onto the micromanipulator and make sure it is level using a bull’s-eye level.
Place the mouse’s skin onto the spatula and ensure that the skin is completely flat.
Screw the coverslip spatula onto the second micromanipulator and make sure it is level using a bull’s-eye level. Lower the coverslip spatula using the micromanipulator towards the mounted skin.
Place a drop of super glue onto the edge of a round coverslip and gently press it onto the tip of the coverslip spatula to secure. After the super glue has dried, use the micromanipulator to lower the coverslip until it lightly presses down on the area of the skin to be imaged (see Figure 2B).
Figure 2.
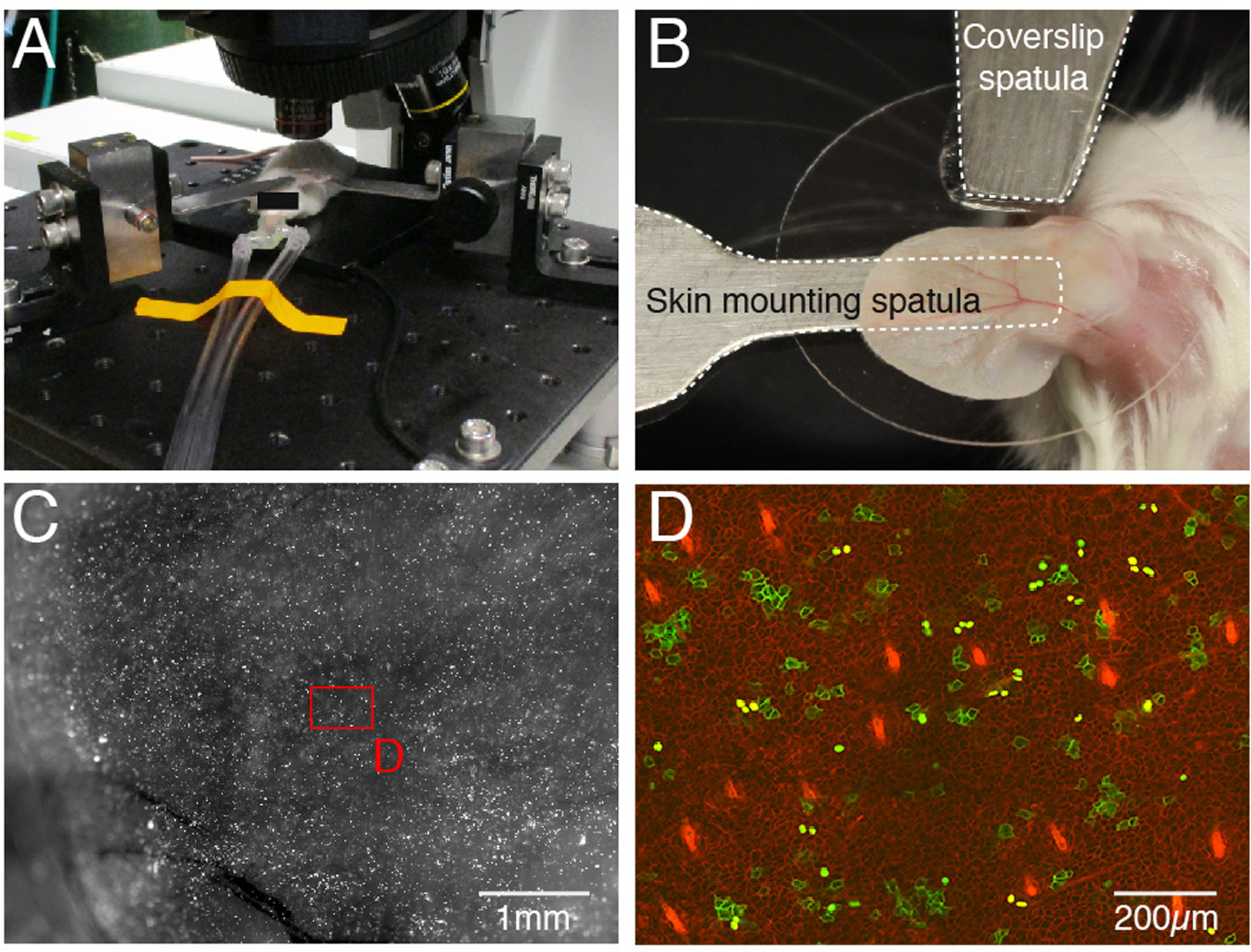
Mouse mounting. An anesthetized mouse on stage of confocal microscope. Anesthetic is delivered through nose cone (A). For ear mounting two spatulas are used, one to support the tissue form below (skin mounting spatula) and a second one to which the coverslip is glued placed on top (coverslip spatula). The ear is held firmly in place between the both spatulas (B). Ear after clonal induction at high magnification, shown is green channel (C). High magnification image of ear epidermis after clonal induction. Membrane EGFP (“normal clones”) and nuclear EGFP (“Gof/Lof clones”) are easily identifiable among red membrane-labeled cells (D).
3.3.3. Preparing stage for imaging
Move the stage from the preparation area to the imaging platform. Immediately plug in the outlet of the heating pad to the power supply.
Position the nose cone around the snout of the mouse and secure the connected tubing to the stage using a piece of adhesive tape (see Figure 2A).
Adjust the oxygen tank and vaporizer outlet to deliver 1 liter/min O2 and 1% isoflurane to the mouse throughout the imaging session.
If using a 25× water-immersion lens, deposit a drop of purified water to create a meniscus on the top of the coverslip.
Use the appropriate filters to view the fluorophores used on the mouse. Adjust the X-, Y-plane and Z-axis controllers to focus on the area of interest.
3.3.4. Two-photon image acquisition
Turn off all the lights in the room (see Note 5).
Use an Olympus FV1200MPE multiphoton microscope equipped with a Chameleon Vision II laser (or equivalent microscope and laser). Set-up to image with a 25× objective or an equivalent objective.
Tune the laser to a wavelength between 900 and 1000 nm. This is suitable to visualize both GFP and Tomato. However, 940 nm is optimal for green-yellow fluorophores (e.g., GFP and YFP) and 1040 nm is optimal for orange-red fluorophores (e.g., RFP, Tomato and mCherry).
Set the upper and lower limits of the Z stack. Normally, the epidermis is set as the start-point and the dermis just below the tip of the hair follicles are set as the endpoint. Set the step size between 1–3 μm.
Adjust the power increment to clearly visualize the cells of interest.
Acquire image stacks.
3.3.5. Performing re-visits
Prepare the mouse as detailed above to revisit the same area of the skin in the same mouse.
Use the tattoo and clusters of hair follicles as landmarks to find the same area.
Repeat Steps 1–20 to set-up and image the mouse.
3.4. Image Analysis
After image acquisition, cell fate of labeled clones is manually determined. Raw data from the imaging revisits is observed to document cell division and differentiation events. The first by quantifying the number of cells that a single clone identified at Day 0 gave rise to and the second by determining how many of those clones underwent terminal differentiation. Because the normal and Gof/Lof clones are uniquely labeled, the consequences of the genetic alteration(s) can be inferred from differences in the behavior of clones belonging to each population (see Figure 3).
Figure 3.
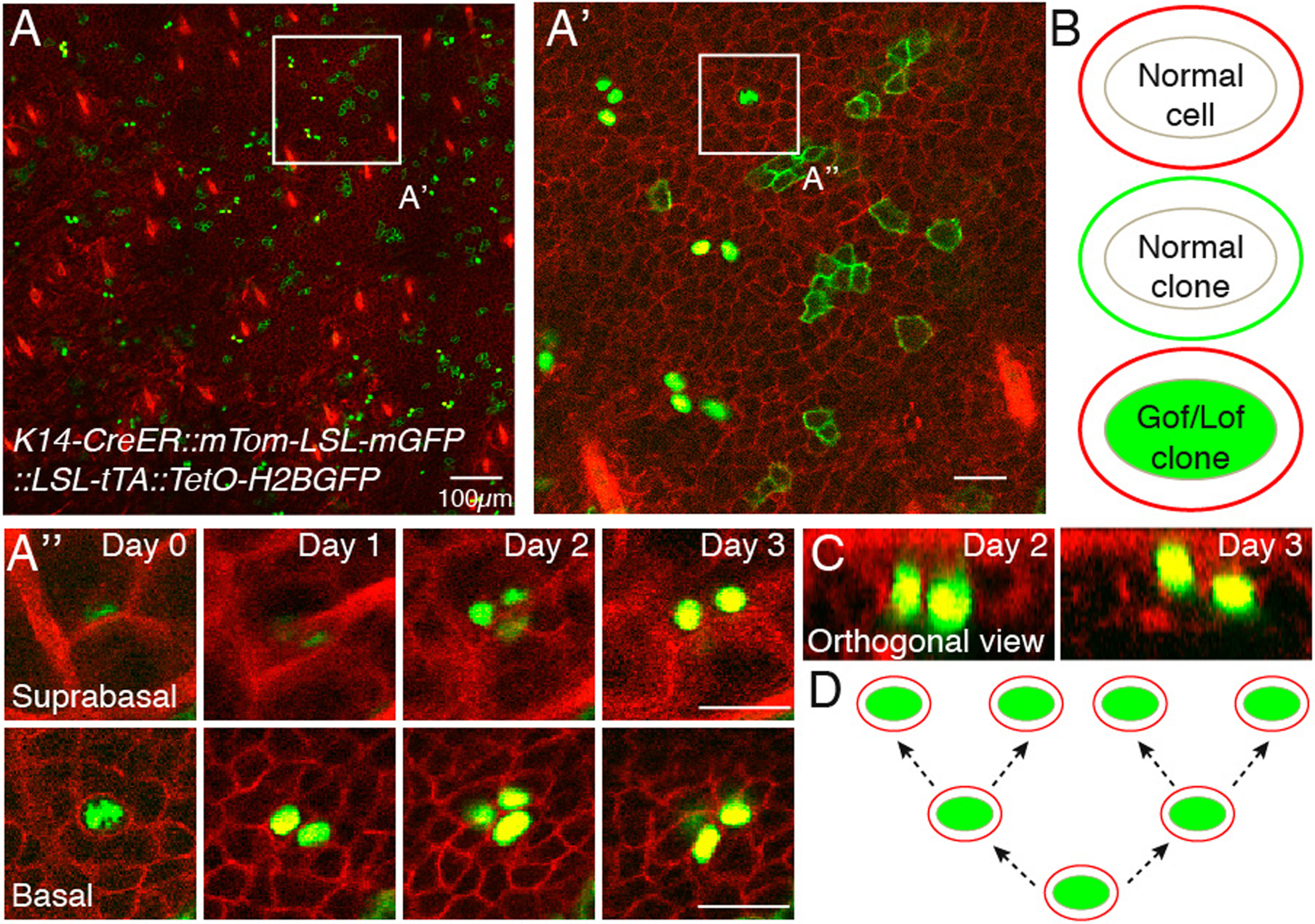
Image analysis. Example data set of a K14-CreER::mTom-LSL-mGFP::LSL-tTA::TetO-H2BGFP mouse acquired daily over 4 days. Overlay of green and red channels of entire imaged region of the skin at Day 0, basal layer is shown (A). Single labeled clones (membrane localized EGFP or nuclear EGFP) are distinguishable within the uniformly fluorescently labeled tissue (membrane tdTomato), with the hair follicle landmarks visible using the latter. Therefore, cells will be either “normal” (red membrane), “normal clones” (green membrane) or genetically modified (green nuclei with red membrane) (B). A region of interest is chosen for in-depth analysis of cell fate (A’). To determine the fate of single labeled clones, basal progenitor cells are looked at individually, along the entire Z stack on Day 0 (A”). For the chosen example, a single basal progenitor gives rise to 4 cells throughout the image period. Two of those can be seen in the suprabasal layer, indicating commitment to differentiation. The upward movement of cells can be more easily visualized by using orthogonal views of the acquired Z stack (C). Overall, in these panels, three division and two differentiation events are documented A possible lineage tree describing the division events (D). An accurate lineage tree cannot be determined with this experimental setup alone, but a label-dilution assay compatible with out system would enable this analysis (see Note 3). Scale bar 25μm unless otherwise indicated.
After image acquisition, use Fiji/ImageJ or a comparable image analysis software for clonal analysis. After importing the raw image files into the software, chose regions of interest within the first acquired time-point and select clones for analysis.
For each re-visit, document the fate of identified clones by quantifying each cell’s division events. The identity of the daughter cells can be determined by their location in the Z-axis because stem and progenitor cells will remain in the basal layer, while differentiating cells will migrate upwards.
Document these changes manually by comparing changes in cell number and location between time-points. A high resolution in the Z-axis and a tissue imaged as flat as possible will facilitate analysis.
4. Notes
It is well documented that unwanted Cre activity can occur even without treatment with Tamoxifen, known as “leakiness” [25]. This disadvantage must be considered when analyzing the data. If, at Day 0, clusters of clones instead of single ones are observed, it is likely that the recombination event precedes Tamoxifen induction. These cells should be disregarded from further analysis.
Mice used for live imaging should be bred into an albino background because pigment absorbs intensely in the infrared spectrum, which may be detrimental to the health of the tissue following prolonged exposure. Additionally, pigments produce strong auto-fluorescence that interferes with signal detection.
Administration of Doxycycline will inhibit expression of all TetO alleles, including the fluorescent nuclear label. Therefore, after Doxycyline administration, every round of cell division will dilute the nuclear label by half, which can be taken advantage of to carry out a label-dilution experiment [2, 9]. In this assay, the origin of the clones is determined based on the change in fluorescence level after every division. Quiescent cells retain their original level of fluorescence, whereas the levels of proliferative cells will fades over time.
Because mice are unable to regulate body temperature when under anesthesia, special care must be taken to prevent hypothermia during the experiment. Ensure that the anesthetized mice are placed on a heating pad at all times.
A lightproof curtain can be placed around the microscope to minimize light leakage.
References
- 1.Kretzschmar K, Watt FM (2012) Lineage tracing. Cell 148:33–45. doi: 10.1016/j.cell.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 2.Blanpain C, Simons BD (2013) Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol 14:489–502. doi: 10.1038/nrm3625 [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, Fuchs E (2009) Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10:207–217. doi: 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu Y-C, Li L, Fuchs E (2014) Emerging interactions between skin stem cells and their niches. Nat Med 20:847–856. doi: 10.1038/nm.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alcolea MP, Jones PH (2014) Lineage analysis of epidermal stem cells. Cold Spring Harb Perspect Med 4:a015206. doi: 10.1101/cshperspect.a015206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gawad C, Koh W, Quake SR (2016) Single-cell genome sequencing: current state of the science. Nat Rev Genet 17:175–188. doi: 10.1038/nrg.2015.16 [DOI] [PubMed] [Google Scholar]
- 7.Crosetto N, Bienko M, van Oudenaarden A (2015) Spatially resolved transcriptomics and beyond. Nat Rev Genet 16:57–66. doi: 10.1038/nrg3832 [DOI] [PubMed] [Google Scholar]
- 8.Rompolas P, Mesa KR, Greco V (2013) Spatial organization within a niche as a determinant of stem-cell fate. Nature 502:513–518. doi: 10.1038/nature12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V (2016) Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352:1471–1474. doi: 10.1126/science.aaf7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pineda CM, Park S, Mesa KR, Wolfel M, Gonzalez DG, Haberman AM, Rompolas P, Greco V (2015) Intravital imaging of hair follicle regeneration in the mouse. Nat Protoc 10:1116–1130. doi: 10.1038/nprot.2015.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, Rompolas P (2017) Two-photon microscopy for intracutaneous imaging of stem cell activity in mice. Exp Dermatol 26:379–383. doi: 10.1111/exd.13221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauer B, Henderson N (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85:5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzger D, Clifford J, Chiba H, Chambon P (1995) Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA 92:6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89:5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H (1995) Transcriptional activation by tetracyclines in mammalian cells. Science 268:1766–1769. [DOI] [PubMed] [Google Scholar]
- 16.Pan W, Jin Y, Stanger B, Kiernan AE (2010) Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proceedings of the National Academy of Sciences 107:15798–15803. doi: 10.1073/pnas.1003089107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, Camargo FD (2011) Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144:782–795. doi: 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grachtchouk M, Pero J, Yang SH, Ermilov AN, Michael LE, Wang A, Wilbert D, Patel RM, Ferris J, Diener J, Allen M, Lim S, Syu L-J, Verhaegen M, Dlugosz AA (2011) Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest 121:1768–1781. doi: 10.1172/JCI46307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Sharma K, Deng H-X, Siddique T, Grisotti G, Liu E, Roos RP (2008) Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol Dis 29:400–408. doi: 10.1016/j.nbd.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 20.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L (2007) A global double-fluorescent Cre reporter mouse. genesis 45:593–605. doi: 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 21.Tumbar T (2004) Defining the Epithelial Stem Cell Niche in Skin. Science 303:359–363. doi: 10.1126/science.1092436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasioukhin V, Degenstein L, Wise B, Fuchs E (1999) The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA 96:8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007. doi: 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- 24.Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, Stange DE, Toftgård R, Clevers H (2010) Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327:1385–1389. doi: 10.1126/science.1184733 [DOI] [PubMed] [Google Scholar]
- 25.Günschmann C, Chiticariu E, Garg B, Hiz MM, Mostmans Y, Wehner M, Scharfenberger L (2014) Transgenic mouse technology in skin biology: inducible gene knockout in mice. J Investig Dermatol 134:1–4. doi: 10.1038/jid.2014.213 [DOI] [PubMed] [Google Scholar]


