Abstract
Exosomes (EXOs) are naturally occurring nanosized lipid bilayers that can be efficiently used as a drug delivery system to carry small pharmaceutical, biological molecules and pass major biological barriers such as the blood-brain barrier. It was hypothesized that EXOs derived from human endometrial stem cells (hEnSCs-EXOs) can be utilized as a drug carrier to enhance tumor-targeting drugs, especially for those have low solubility and limited oral bioactivity. In this study, atorvastatin (Ato) loaded EXOs (AtoEXOs) was prepared and characterized for its physical and biological activities in tumor growth suppression of 3 D glioblastoma model. The AtoEXOs were obtained in different methods to maximize drug encapsulation efficacy. The characterization of AtoEXOs was performed for its size, stability, drug release, and in vitro anti-tumor efficacy evaluated comprising inhibition of proliferation, apoptosis induction of tumor cells. Expression of apoptotic genes by Real time PCR, Annexin V/PI, tunnel assay was studied after 72 h exposing U87 cells where encapsulated in matrigel in different concentrations of AtoEXOs (5, 10 μM). The results showed that the prepared AtoEXOs possessed diameter ranging from 30–150 nm, satisfying stability and sustainable Ato release rate. The AtoEXOs was up taken by U87 and generated significant apoptotic effects while this inhibited tumor growth of U87 cells. Altogether, produced AtoEXOs formulation due to its therapeutic efficacy has the potential to be an adaptable approach to treat glioblastoma brain tumors.
Keywords: Atorvastatin loaded exosome, Glioblastoma, Apoptotic effects, anti-Glioblastoma carrier
Highlights
-
•
Isolated exosome for endometrial stem cells show sizes between 30 and 150 nm in spherical-shaped, and atorvastatin could be efficiently loaded into exosome.
-
•
Atorvastatin loaded exosome (AtoEXO) were stable during 30 days' incubation and released relatively slowly from exosome.
-
•
U87 cells untaken AtoEXO in short time efficiently.
-
•
AtoEXO had anti-proliferative impact on U87 cells which proved by attenuated cell density and viable cell as well as decreasing expression of Ki-67 protein.
-
•
Efficient activity of AtoEXO to control tumor cell growth proved through up regulation of pro-apoptotic and apoptosis pathways including caspase-3, 8 and 9.
1. Introduction
Glioblastoma (GBM) is highly aggressive and most common form of primary brain tumor that has a dire prognosis with high mortality [1–3]. Generally, the GBM microenvironment generates under secretion of several pro-inflammatory cytokines into a hypoxic condition. In this process, pro-inflammatory mediators including interleukin-1β (IL-1β), interleukin-6 (IL-6), and also interleukin-8 (IL-8) play key roles in the GBM microenvironment [1,2], through activation of their related downstream targets such as the JAK-STAT, PI3K-AKT, and mitogen-activated protein kinase (MAPK) signaling pathways. During GBM progression, the tumor cell proliferation, invasion and also tumor angiogenesis arouse a dreadful disorder [[3], [4], [5]]. Meanwhile, conventional therapeutic treatments including radiotherapy and chemotherapy are strongly restricted in treating GBM due to their toxicity to normal brain tissues that have further consequences impact on brain damage and decreased quality of life in patients [2,6,7]. Besides, direct application of stem cells as a well-known therapy is restricted due to immunological barriers and rejection, as well as ethical problems of their preparation and concerns for tumor formation at the site of transplantation [8,9]. The development of a novel cell-free medical approach with tumor-suppressive effects is of great importance. Innovative methods of drug delivery to prevent systemic toxicity of cancer drugs as well as targeted drug delivery to the tumor site are among the new approaches that are being considered as proper approaches in cancer therapy [10,11].
Recently the stem cell products as bioactive drug carriers are developed to overcome stem cell therapy restrictions [12]. Exosome (EXO) as a drug carrier can be a good cell-free material in the field of GBM treatment due to being natural product of the body which leads to low immune responses. Implantation feature of the EXOs in transport of their cargoes to distant targets introduce it as a desirable carrier. The EXOs are small nanometer vesicles that originate from the cell membrane and release into the microenvironment of the cells as contain a selective set of ligands, receptors, and enzymatic substances [[13], [14], [15]]. Moreover, EXOs could drive stimulatory, inhibitory, genetic re-programming messages to the target cells through binding and internalizing into the cell membrane [[16], [17], [18]]. More interestingly, the EXOs are good carriers for delivering medicinal compounds including anti-cancer drugs that inhibit inflammatory and angiogenic pathways by enzyme-reductase (hydroxymethylglutaryl coenzyme A) inhibitors [[19], [20], [21]]. Recently, application of EXOs is developing in regenerative medicine due to their advantages, including non-immune rejection, homing effect, easy control of their concentration and stability for sustain release of drugs [13,14,[22], [23], [24]]. Besides, statins have been shown pleiotropic effects within the specified concentrations and could hinder pathways associated with inflammation [[25], [26], [27]]. The exact molecular mechanisms of statin effects that can be useful for tumor avoidance and/or treatment including apoptosis induction, proliferation inhibition are not well cleared [28]. Increasing investigations, both in vitro and in vivo, have shown inhibitory effects of statins especially atorvastatin (Ato) on tumor cells such as glioblastoma [[29], [30], [31], [32]], melanoma cell line [30], breast cancer cell lines [33,34], prostate tumor [35]. Additionally, the anti-cancer ability of Ato have also examined in some clinical trials [[36], [37], [38]]. It's suggested that the apoptotic effects of statin can be made through several mechanisms consist of upregulation of pro-apoptotic protein expression (e.g., Bax, Bim), while downregulation of anti-apoptotic proteins (e.g., Bcl-2) [39,40] and activation of caspase proteins (caspase 3, 8, 9) [[41], [42], [43]]. The EXOs generally are isolated from variety of cell sources such as mesenchymal stem cells, progenitor stem cells, adipose derived stem cells and endometrial stem cells. In particular, EnSC would be used for proposed study due to its immunomodulatory function, ease acquisition and minor donor site morbidity which has made it as a good candidate for clinical and therapeutic applications. Thus, we have taking into account that by isolation of EnSC-derived EXOs and their loading with Ato (AtoEXOs) could restrict tumor cell growth, pro-apoptosis and apoptosis pathways as well as necrosis in GBM model in vitro.
In this study, EXOs isolated from human EnSC (hEnSC) were used as a nano-carrier to load chemotherapeutic drug Ato for GBM treatment. The physical and biological characteristics of isolated EXOs were evaluated through scanning electron microscopy (SEM), size distribution analysis, western blot as well as drug loading release and stability measurement during extended time of incubation. The 3 D spheroid tumor model was produced using hanging drop of U87 cells in matrigel for tumor growth suppression study in different concentrations of AtoEXOs and free Ato in vitro. The anti-proliferative effects of AtoEXOs were proved by microscopic observation of U87 cell morphology, MTT assay and down expression of Ki-67 protein. The annexin/PI flowcytometry and tunnel staining of U87 cells display highest level of apoptosis for treated cells with AtoEXOs in both utilized 5 and 10 μM. The real time PCR was up-regulated apoptosis-related molecules including caspase 3, caspase 8 and caspase 9 in highest levels for AtoEXOs conditions.
2. Materials and methods
2.1. Materials
Atorvastatin ([R-(R*, R*)-2-(4-Fluorophenyl)-ß, d-dihydroxy-5-(1-methylethyl)-3-Phenyl-4-(Phenylamino) Carbonyl]-1-H-Pyr-role-1-heptanoic acid), tween 20, MTT (3-(4,5 Diamethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide) were obtained from Sigma Co. (St. Louis, USA). A lipophilic fluorescent dyes, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindo-carbocyanine perchlorate (DIL), trizol solution were purchased from Invitrogen (Carlsbad, CA, USA). Matrigel matrix from Corning, UK, tunnel assay kit from Abcam, UK and cDNA synthesis kit from Thermo Scientific Fermentas, USA were obtained.
2.2. Cultured cells
Human derived endometrial stem cells (hEnSCs) were isolated from donors using laparoscopy procedure according previously described protocol [44]. The harvested cells were sub-cultured in 100 mm culture dish in a 37 °C and 5% CO2 incubator for 2 weeks. The U87 cell line was purchased from the Cell Bank of the Pasteur Institute (Tehran, Iran). These cells were all grown and maintained Dulbecco's Modified Eagle Medium/F12 (DMEM/F12) containing 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin were purchased from Scotland (Gibco, Scotland). The FBS-exosome depleted was obtained from Capricorn Scientific GmbH (Germany). Medium of cells was changed every 2~3 days.
2.3. EXO isolation and characterization
The hEnSCs at 70–80% confluency were grown in DMEM/F12 + 10% FBS-EXO depleted +1% pen/strep. After 24 h, hEnSCs supernatant was collect to apply for EXO isolation by differential centrifugation (300×g/10 min, 2000×g/10 min, 20000×g/30 min) that proceed with ultracentrifugation at 120000×g/120 min/4 °C (Beckman Coulter X-14R centrifuge). The hEnSCs EXO characterization assays including size distribution, morphology evaluation and CD63 immunoblotting was performed according to our recently published protocol [14].
2.4. Atorvastatin loading into EXO (AtoEXO)
Ato was dissolved in DMSO and passed 0.2 μm filter. The Ato solution was diluted with culture medium at 10 μM and then 100 μg extracted EXOs were added to Ato solution. Four approaches for Ato incorporation into EXOs were evaluated: freeze-thaw cycles (Method I), sonication (Method II), diffusion (Method III) and conventional suspension (Method VI). For Method I, EX and Ato containing solution were exposed several times to the freeze-thawing cycle. To do this, the solution was exposed tree times rapidly to freezing and thawing at -80 °C and 37 °C, respectively. Method II, contents of the EXO and Ato solution were place in ice bath and sonicated with voltage 500 V, frequency of 2 kHz and 20% power. During sonication operation, the pulse cycle was set for 4 s run and 2 s pause for 2 min. For Method III, sample incubation was performed by addition of 0.1% tween-20 and incubation with gentle shaking on rotary shaker for 18 h at room temperature. Method VI, loading Ato in EXO without addition of tween-20 and just incubation. Indirect method was used to evaluate Ato loading into EXOs. The tubes containing EXOs and Ato were ultra-centrifuged for at 12000 g and 120 min. The absorbance of supernatant was at 246 nm and the difference between absorbance of samples just before addition of EXOs and supernatant correlated with the amount of loaded Ato in EXO using calibration curve.
2.5. AtoEXO characterization
The various characterization methods were applied to evaluate the quality of AtoEXO Nanoformulation.
2.5.1. Size distribution analysis
Hydrodynamic diameter of AtoEXO was studied by dynamic light scattering (DLS) assessments using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) as claimed by company.
2.5.2. Morphology of EXOs
To visualize AtoEXO morphology, scanning electron microscopy (SEM) were utilized. AtoEXO pellets were vortexed then were re-suspended in phosphate-buffered saline (PBS). The AtoEXO suspension 10 μL was fixed in 2.5% paraformaldehyde. The process followed by sample dehydration with 75% ethanol, drying and finally covering with a thin layer of gold layer to analysis under SEM (QUANTA SEM system; FEI Company, Hillsboro, OR, USA).
2.5.3. Immunoblotting of EXOs
The effective immunoblotting of CD63 as a specific CD marker of EXO was performed on isolated EXOs and AtoEXOs. In detail, briefly, 12% SDS-PAGE was prepared for exosomal total proteins that extracted using RIPA buffer (Radioimmuno Percipitation Assay). After that proteins were transferred to nitrocellulose membrane, multi-steps including blocking with (5% milk and 0.05% tween-20 in PBS), incubation with primary anti-CD63 monoclonal antibody (Santa Cruz Biotechnology, Dallas, Texas, USA) for 2 h. Then, samples were washed in PBS and incubated with secondary horseradish peroxidase (HRP)-conjugated antibody (SinaClon, Tehran, Iran) for 2 h. The CD63 bands related to naive EXO and AtoEXO were detected using DAB solution.
2.6. Release profile of Ato
Time-courses for the diffusion of Ato from EXOs were measured as follows. Harvested AtoEXOs were placed in 10 mL PBS and mixed on a rotary shaker at 4 °C and the concentration of Ato remaining in the solution was analyzed at prescribed time points. In brief, AtoEXO solution was centrifuged at 120000 g and 120 min. Then, supernatant 1 mL was used for UV-measurement at 246 nm and equal volume fresh PBS was added to EXO solution and mixing of rotary was continued until 168 h. The quantity of released Ato was normalized per initial degree of loading.
2.7. EXO size stability
The size stability of AtoEXOs was measured through size distribution measurement. For this, 50 μL AtoEXOs was suspended in 1 mL PBS and was shacked gently at physiological temperature until 30 days. The changes in size of AtoEXOs was measured using size distribution analysis and averaged.
2.8. Cellular uptake of AtoEXOs
The feasibility of cellular uptake of AtoEXOs within the intercellular filamentous structures of U87 cells were carry out using fluorescent labeling process. Briefly, 1, 1′-Dioctadecyl-3, 3, 3′, 3′-tetramethylindocarbocyanine perchlorate (DiI) dye solution was added to AtoEXOs suspension and incubated for 1 h under gentle shaking in physiological temperature. The nuclei of U87 cells were stained with DAPI (4′,6‐diamidino‐2‐phenylindole; light blue) (Sigma‐Aldrich) at 1 μM. The non-diffused DiI or DAPI from samples were removed through centrifugation and washing with PBS.
2.9. Production of glioblastoma model and 3 D encapsulation
In brief, the U87 cell line was cultured in T25 culture flasks containing 5 mL of pre-warmed cell culture media (DMEM/F12 + 10% FBS + 1% pen/strep) at 37 °C, 5% CO2, 95% humidity. The U87 cells at 80% confluency were removed from T25 cell culture flask by trypsinization and suspension of cells were centrifuged at 300×g/10 min. The acquired U87 cells at density of 1 × 104 cells was placed on the surface of well-plate in hanging-drop method and incubated at physiological condition for 48 h to provide desirable U87 spheroid. The culture media was added and incubated for 48 h to provide desirable U87 spheroid. Then, prepared U87 spheroid was removed from surface of well via sterilized blade. Separately, 200 μL matrigel (Corning, Matrigel, UK) was poured on surface of well and after 30 min incubation, the prepared U87 spheroid was placed on its surface and in following additional 200 μL matrigel poured on prepared matrix surface which resulted U87 spheroid encapsulation in 3 D model of matrigel.
2.10. MTT evaluation of U87 cells
The U87 spheroid in 3D model of matrigel was incubated in DMEM/F12 medium supplemented with concentrated free at 5, 10 μM and Ato loaded EXOs in same concentration of Ato was poured on wells until 72 h. The MTT assay was used to measure anti-proliferation by MTT Kit (thiazolyl blue tetrazolium bromide; Sigma) on U87 spheroids as a glioblastoma tumor model. At the indicated time points, the cells were released from matrigel through incubation of specimens in collagenase A (1mg/mL) at 4 °C within 20 min. After washing cells with culture media, MTT solution 5 mg/mL was added into each well followed by incubation for 4 h. The medium was removed and insoluble purple formazan product was dissolved by dimethyl sulfoxide (DMSO) for 20 min. The absorbance was measured by a microplate reader at 570 nm (EL340 Bio-Tek Instruments, Hopkinton, MA).
2.11. Ki-67 expression of U87 cells
The U87 cells were seeded in 6-well plates at a density of 2 × 104 cells/cm2 and cultured for 72 h. The cells were fixed 4% paraformaldehyde for 10 min and in 70% ethanol at room temperature. After that, the cells were divided in two parts. The first part of cells suspended in cold PBS containing fluorescein isothiocyanate conjugate-labeled anti-Ki-67 antibody (catalog No. 556026; BD Biosciences) and incubated at 4 °C for 30 min. Then, treated cells were washed via PBS and were centrifuged to remove non-reacted fluorophore molecules. The fluorescence of labeled cells was measured using a BD Accuri U87 flow cytometer (BD Biosciences, CA, USA).
2.12. Annexin-PI assay
Likewise the Ki-67 expression test, theU87 cells were seeded well plates at a density of 2 × 104 cells/cm2 and in following treated by Atro and AtoEXOs substrates. After 72 h incubation, annexin-PI assay was used to detect the feasible percentage of U87 cells which induced in apoptosis phase. The second part of harvested U87 cells from were washed with PBS and stained with diluted Annexin V–FITC and propidium iodide (PI), fluorophore in binding buffer for 30 min in the dark. Finally, U87 cells were washed with PBS and subjected to flow cytometry technique using flow cytometer.
2.13. TUNEL labeling for apoptosis
The after fixation process of cell from matrigel, harvested cells were washed with 10 mM tris hydrochloride buffer and permeabilized with protein kinase 2% at 37 °C for 60 min. The cells were poured in TUNEL reaction mixture and incubated in physiological condition for 60 min, rinsed with PBS. The stained cells were observed by a light microscope at × 100 magnification for detecting TUNEL-positive cells.
2.14. Quantitative real-time polymerase chain reaction
RNA extraction and real-time polymerase chain reaction (RT-PCR) analysis was performed in order to define the expression level of initiative and executive apoptosis genes. For this, trizol solution (Invitrogen, USA) was used to extract total RNA from U87 cells for each experimental conditions according to the manufacturer's instructions. Afterward, cDNA was synthesized by Revert Aid first-strand cDNA synthesis kit (Thermo Scientific Fermentas, USA), according to the manufacturer's instructions. Primers for real-time PCR were designed using Beacon Designer 7 software. The primer sequences information presented in Table 1. Quantitative RT-PCR (qRT-PCR) was performed on cDNAs using Rotor-Gene Q 2plex HRM platform real-time PCR system (Corbett Life Science). The relative expression levels of caspase 3, 8 and 9 were evaluated in comparison with GAPDH as an endogenous control gene. Each 15 μL reaction volume was contained 7.5 μL of the concentrated 2 × SYBR green master mix (Invitrogen, USA), 1 μL of cDNA, and 0.6 μL (10 ppm) of each pair of oligonucleotide primers. The RT-PCR reactions were done in duplicate. The PCR cycling began with an initial step of 95 °C for 10 min followed by 35 cycles of 95 °C for 20 s, 57 °C for 20 s and 72 °C for 15 s; then a melting curve was analyzed. Rotor-gene Q sequence detection system determined the threshold cycle (CT) values. Comparative threshold cycle (2−ΔΔCT) method was used in order to analyze the data.
Table 1.
Forward and reverse primers of genes used for RT-PCR experiments.
| Gene | Primer sequence | Tm (°C) |
|---|---|---|
| Caspase 3 | FAAAAGCACTGGAATGACATCTCG RGAAACATCACGCATCAATTCCAC | 59 |
| Caspase 8 | FACTGGATGATGACATGAACCTGC R CCTCCCCTTTGCTGAATTCTTC | 60 |
| Caspase 9 | FCATTTCATGGTGGAGGTGAAG RGGGAACTGCAGGTGGCTG |
61 |
2.15. Statistical analysis
All experiments were carried out in triplicate for each condition. The data are shown as means ± standard deviation (SD). Statistical analysis was done by Minitab 18 software (Minitab, Inc., State College, USA). Significant differences are expressed as *P < 0.05, **P < 0.01, and ns P > 0.05 in bar graphs.
3. Results
3.1. AtoEXO characterization
The harvested EXO and AtoEXO analyzed by particle size measurement, SEM, and western blot observations (Fig. 1). The obtained EXOs and AtoEXO possessed a spheroid-shape morphology with uniform and narrow size distribution in diameters of just about 30–150 nm which indicate appearance of all EXOs were consistent with previous studies [14,45] (Fig. 1A and B). Besides, western blotting analysis confirmed that hEnSCs vesicles and isolated AtoEXO nanoparticles were positive for CD63 protein (Fig. 1C). The CD63 expression was highly expressed in isolated EXOs as well as AtoEXO nanoformulation. Meanwhile, expression of CD63 marker was not distinguishable in the supernatant of the isolated AtoEXO condition (Fig. 1C). The results proved that extracellular vesicles from hEnSCs were indeed EXOs and these could be utilized as effective nano-biocarrier for the loading of Ato in it. These results are consistent with previous reports that EXOs derived from other cell sources [13,14,[22], [23], [24]].
Fig. 1.
Characterization of atorvastatin loaded exosome (AtoEXO). (A) Size distribution of prepared AtoEXO, (B) SEM microphotograph of AtoEXO and (C) western blot of isolated AtoEXO, EXO and supernatant media of AtoEXO (Supernatant).
3.2. Nanoformulation of Ato in AtoEXO production
Drug encapsulation efficiency is an essential parameter for developing successful formulations and evaluating drug delivery potential of prepared biocarriers [46]. Therefore, we assessed the Ato encapsulation efficiency in EXOs. The loading capacity of Ato in EXOs in different conditions evaluated using UV–vis spectroscopy at 246 nm. Meanwhile, Ato encapsulation efficiency was lowest amount for freeze/thaw method about 10% (Fig. 2A). The Ato encapsulation was found to be highest at diffusion condition which sample was incubated in presences of tween at about 28% (Fig. 2A). The sonication and normal incubation methods had also moderate encapsulation efficacy at about 20% (Fig. 2A). It is expected that tween 20 promotes Ato compound diffusion and permeabilization from EXO transmembrane likewise its effects on cell membrane. This compound could remove cholesterol binds at exosomal membrane and create pores in exosomal lipid bilayer membrane which allows superior diffusion of Ato into the EXOs [47]. It is reported that using detergent tween helps in the extraction of drug from the EXOs by dissociating drug-EXO interaction [21]. The low release profile of loaded Ato in EXOs suggest that most of existed Ato inside of EXOs could not associated with their membranes (Fig. 2B). The average size of EXO is depended on cell type and is ranged from 30 to 200 nm [[48], [49], [50]]. The size of AtoEXO was slightly comparable with EXOs obtained from cells treated with naive EXO vehicle (Fig. 1A and B). Moreover, modulation of sustainable drug delivery is recognized as an influential factor for tumor treatment [[51], [52], [53]]. The Ato release profile from the EXO sample shown in Fig. 2B. The Ato was continuously released within 168 h from AtoEXO. The controlled release of Ato from AtoEXO with slightly fast release was about 50% in 48 h and this continuous controlled release was constant until 168 h at 75%. These results demonstrate the advantage of using developed AtoEXO in cancer treatment due to the increasing time of drug release by an initial, slightly fast release and then a sustained release phase. The structure and size stability of Ato loaded EXOs at -20 °C over time was also investigated until 30 days. The size of EXOs shows quite stable form and did not change during extended time of incubation until 30 days which prove integration of Ato with EXOs (Fig. 2C). This could be related to hydrophobic property of Ato and lipid layer of EXOs. The uniform and stable size of AtoEXO demonstrates that Ato encapsulation and its hydrophobic characteristics resulted in feasibility of AtoEXO [[54], [55], [56]].
Fig. 2.
The evaluation of produced AtoEXOs nanoformulation. (A) Permeation of Ato in EXOs, (B) controlled release of encapsulated Ato in EXOs until 168 h incubation (C) size stability of AtoEXO at -20 °C and (D) cellular uptake of AtoEXO in U87 cells.
In following AtoEXO uptake by U87 cells was evaluated to elaborate applicability of developed AtoEXO for growth tumor suppression. The uptake of AtoEXO observed in the recipient U87 cells after 1 h incubation which indicate intracellular accumulation of AtoEXO in cancer cell line (Fig. 2D).
3.3. MTT assay
To evaluate effect of EXO loaded with Ato on cell proliferation in glioblastoma models, we performed dose-response study as well as with/without EXO carrier conditions for seeded U87 cell line. Fig. 3 shows the result of MTT assay and morphology of U87 cells. As can be seen, U87 cell viability increased in control group (non-treated condition) by increasing time of culture. Most of glioblastoma cells remained viable after treatments in lower concentration of Ato content. An interesting point from Fig. 3 is that increasing Ato content at 10 μM decreases the U87 cell viability in a time-depended manner even for initial 24 h. More interestingly, incorporation of Ato in EXOs decreased significantly cell viability and it was continued during extended time of incubation until 72 h (Fig. 3A and B). Moreover, cellular morphology changed from spindle shape to spheroid-shaped by incorporation of high concentration of free Ato and AtoEXO substrates. Recent studies demonstrate that Ato affects mitochondrial functions which has an important role in cell survival [57,58]. Increasing Ato content had a positive effect on decrease of U87 cell viability. The Ato had an important role as a cofactor in cellular physiology and function [57,58]. Its positive effect on the mitochondrial function of different kind of cells at adjacent concentrations has been reported [59]. Results showed that 5 and 10 μM concentration of Ato when combined with EXOs had synergistic cytotoxicity on glioblastoma cells, via inducing apoptosis and interfering with the cell cycle.
Fig. 3.
Cell viability measurement (A) and microphotographs (B) of U87 spheroids in matrigel treated with free Ato at 5 and 10 μM (Ato 5 and Ato 10 respectively) and also AtoEXO at 5 and 10 μM (AtoEXO 5 and AtoEXO 10 respectively) at 72 h.
3.4. Ki-67 protein expression
Degree of Ki-67 expression is highly responsible for cell proliferation in tumor cells and may more accurately help to define high-risk patients [60,61]. Therefore, anti-proliferative effects of AtoEXO was evaluated by flowcytometry for Ki-67 expression [62]. The U87 cells in control condition (non-treated cells) was expressed Ki-67 more 90% which shows proliferation potential of U87 cells in 3 D matrigel (Fig. 4A and B). The utilized free Ato and AtoEXO were able to inhibit U87 cell proliferation as evidenced by a significant reduction in Ki-67 labeling index (Fig. 4A and B). Interestingly, AtoEXO showed more significant impact on cellular proliferation and Ki-67 expression index was lower than 11% for these conditions (Fig. 4A and B). Meanwhile, Ki-67 index was more than 21% for direct application of Ato in different concentrations (Fig. 4A and B). However, there was no significant difference between doses studied as far as inhibition of cell proliferation is concerned. The obtained results for Ki-67 index for experimental conditions were consistent with MTT assay and cellular growth inhibition (Fig. 3). The results proved that utilization of AtoEXO was highly effective in U87 cell growth suppression and could utilize this produced substrate for GBM tumor suppression.
Fig. 4.
Ki-67 expression of U87 cells harvested from 3D matrigels after 72 h treatment with Ato and AtoEXO in different concentrations. (A) Flowcytometry assay to evaluate the effects of the Ato and AtoEXO. (B) Quantitative amount of Ki-67 expression of U87 cells. The U87 cells were harvested, stained with Ki-67-FITC (FL-I) and analyzed by the flow cytometry. The statistical difference in the Ki-67 level was compared between among all groups.
3.5. Annexin/PI and cell cycle
It is well known that collapse of mitochondrial membrane is mainly induces cell apoptosis (programmed cell death) and mitochondrial membrane disruption results in releasing apoptogenic proteins which causes cell death [63]. In this study, we compared U87 cell apoptosis and necrosis that are induced by releasing of Ato from EXOs. The non-treated condition possessed lowest level of apoptosis and necrosis compared with free Ato and AtoEXO conditions. The non-treated U87 cells with AtoEXO in matirgels shows total apoptosis 2.94% with early and late apoptosis 1.33% and 1.61% respectively. The viability of cells was significantly decreased in all experimental conditions treated with free Ato and AtoEXO which indicate effectiveness of Ato and EXOs to induce cellular apoptosis and decrease cell viability (Fig. 5) as obtained consistent results with MTT assay and Ki-67 measurement (Fig. 3, Fig. 4). The AtoEXO 10 μL displays highest level of apoptosis at 30.37% including early and late apoptosis 4.31% and 26.6% respectively (Fig. 5). Meanwhile, treatment of U87 cells with Ato 5 and 10 μL induces necrosis in 24.1% and 23.4% of the cells, respectively which was comparable with AtoEXO 5 μL at 19.5% of the cells. The significant higher late apoptotic effect of AtoEXO can be related to long term and a moderate release of Ato as shown in Fig. 5.
Fig. 5.
Flowcytometry assay to evaluate the effects of the free Ato and AtoEXO on the U87 cell apoptosis induction. (A) shows the histograms (B) (B) Quantitative amounts of necrotic, apoptotic and live cells. U87 cells were harvested from 2 D culture dish, free Ato and AtoEXO conditions after 72 h incubation and stained with Annexin V-FITC (FL-I) and PI for analyzing by the flow cytometry.
3.6. Apoptosis protein analysis
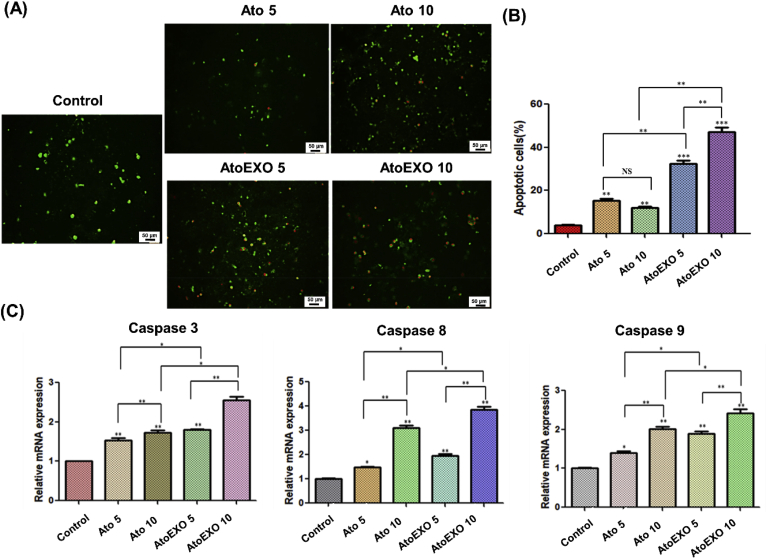
Apoptosis is a main mechanism regarding the cell fate and possibility for tumor progression, since relative amounts of anti- and pro survival signals will be controlled whether the cell undergoes cell death and plays a critical role in cancer initiation and progression or not [[64], [65], [66]]. We have measured several family of proteins includes apoptosis-related molecules including tunnel as well as caspase 3, caspase 8 and caspase 9 for 3 D cellular tumor spheroid which were exposed to AtoEXO for 72 h. Results showed that significantly altered the expression mRNA levels of these genes in U87 cell line (Fig. 6 A and B). The TUNEL assay (TUNEL Assay Kit, ab66110) was utilized to determine if the administration of different concentration of free Ato and AtoEXO enhances the apoptosis of the cells. The number of apoptotic-positive cells was counted in 5 high-power fields ( × 400 magnification) and the mean percentage of apoptotic cells was reported. The proportion of apoptotic-positive cells in the AtoEXO 5 and AtoEXO 10 groups was significantly higher than that in the free Ato 5 and 10 as well as control group respectively (Fig. 6 A). The proportion of apoptotic-positive cells between free drug (Ato 5 and 10 groups) and the control group was also significant (p < 0.05). The pro-apoptotic gene caspase 8 and 9 as an extrinsic and intrinsic cascades [67], respectively and another marker of apoptosis caspase 3 were significantly upregulated in all concentrations used free Ato and AtoEXO conditions (Fig. 6B). The expression degree of these genes was in maximum level for AtoEXO 10 condition compared with other experimental conditions (Fig. 6B). It is most likely that free Ato and AtoEXO induce apoptosis by elevating their upstream signal where cytochrome C interacts with caspases signaling pathways [68]. The indirect delivery of anti-cancer drug using EXO in order to induce the apoptotic process demonstrates more efficient activity to control cancer cell growth through up regulation of pro-apoptotic and apoptosis pathways (Fig. 6A and B). Caspases are key factors in the execution of apoptosis. Mitochondrial pathways and death receptor-mediated pathways are two main apoptosis pathways, when caspase 8 protease is activated, it activates caspase 3 to trigger apoptosis [69,70]. Therefore, both pathways converge at activation of the effector caspases. Besides, the cleavage of caspase 9 was indicative of apoptotic cell death. Whereas, there is some crosstalk between the activated caspase 8 can also trigger activation of caspase 9 [68,70]. Therefore, we could demonstrate anti-cancer toxicity of AtoEXOs in U87 cancer cell line via the caspase cascade of pro-apoptotic and apoptosis pathways. Hence, AtoEXOs represent a potentially biocompatible and applicable nano-carrier for sustainable drug delivery. These finding support the potential of AtoEXOs for targeted brain drug delivery in the treatment of brain cancer.
Fig. 6.
TUNEL staining of U87 cells in 3D matrigel model and (B) quantifying degrees of apoptotic cells (C) Expression level of caspase 3, caspase 8, for caspase 9 for U87 cells treated by Ato and AtoEXO in different concentrations. TUNEL assay was performed to quantify the apoptotic cells in different groups. The quantification of the apoptotic cells was determined and shown in the diagram. The proportion of apoptotic-positive cells significantly increased in AtoEXO 5 and 10 samples when compared with those of other experimental groups.
4. Conclusion
In summary, the present study demonstrates feasibility of EXO isolation from hEnSCs which this could be used as a bioactive drug nanocarrier to load Ato with high loading efficiency for inhibition of glioblastoma growth. The AtoEXOs showed uniform size distribution, stable structure and controlled release of Ato during extended time of incubation. Besides, the AtoEXO could be taken up by U87 cells and induce glial cell death. The AtoEXO inhibited the glial cell line growth and proliferation by minimizing cell density and viable cell as well as decreasing expression of Ki-67 protein. Moreover, upregulation of apoptosis genes proves that AtoEXO would provide a promising sustainable drug delivery vehicle for tumor-targeted treatment in further study.
Author declaration
I wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
I confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of authors.
I confirm that all of authors have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing I confirm that I have followed the regulations of our institutions concerning intellectual property.
All authors understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. I confirm that all of authors have provided a current, correct email address which is accessible by the Corresponding Author and which has been configured to accept email from authors.
Declaration of competing interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgment
The authors with to thank Kashan University of Medical Sciences, Iran for supporting the study (Grant No. 9527).
Contributor Information
Mehdi Khanmohammadi, Email: mehdi.khanmohammadi84@gmail.com.
Jafar Ai, Email: Jafar_ai@tums.ac.ir.
References
- 1.Lichtor T., Libermann T.A. Coexpression of interleukin-1β and interleukin-6 in human brain tumors. Neurosurgery. 1994;34(4):669–673. doi: 10.1227/00006123-199404000-00015. [DOI] [PubMed] [Google Scholar]
- 2.Parsons D.W., Jones S., Zhang X., Lin J.C.-H., Leary R.J., Angenendt P. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiesner S.M., Freese A., Ohlfest J.R. Emerging concepts in glioma biology: implications for clinical protocols and rational treatment strategies. Neurosurg. Focus. 2005;19(4):1–6. doi: 10.3171/foc.2005.19.4.4. [DOI] [PubMed] [Google Scholar]
- 4.Kanu O.O., Hughes B., Di C., Lin N., Fu J., Bigner D.D. Glioblastoma multiforme oncogenomics and signaling pathways. Clin. Med. Oncol. 2009;3 doi: 10.4137/cmo.s1008. CMO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao H., LeBrun D.G., Yang J., Zhu V.F., Li M. Deregulated signaling pathways in glioblastoma multiforme: molecular mechanisms and therapeutic targets. Canc. Invest. 2012;30(1):48–56. doi: 10.3109/07357907.2011.630050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pashaki A.S., Hamed E.A., Mohamadian K., Abassi M., Safaei A.M., Torkaman T. Efficacy of high dose radiotherapy in post-operative treatment of glioblastoma multiform--a single institution report. Asian Pac. J. Cancer Prev. APJCP. 2014;15(6):2793–2796. doi: 10.7314/apjcp.2014.15.6.2793. [DOI] [PubMed] [Google Scholar]
- 7.Souhami L., Seiferheld W., Brachman D., Podgorsak E.B., Werner-Wasik M., Lustig R. Randomized comparison of stereotactic radiosurgery followed by conventional radiotherapy with carmustine to conventional radiotherapy with carmustine for patients with glioblastoma multiforme: report of Radiation Therapy Oncology Group 93-05 protocol. Int. J. Radiat. Oncol. Biol. Phys. 2004;60(3):853–860. doi: 10.1016/j.ijrobp.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Strauer B.E., Kornowski R. Stem cell therapy in perspective. Circulation. 2003;107(7):929–934. doi: 10.1161/01.cir.0000057525.13182.24. [DOI] [PubMed] [Google Scholar]
- 9.Trounson A., McDonald C. Stem cell therapies in clinical trials: progress and challenges. Cell stem cell. 2015;17(1):11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Juillerat-Jeanneret L. The targeted delivery of cancer drugs across the blood–brain barrier: chemical modifications of drugs or drug-nanoparticles? Drug Discov. Today. 2008;13(23–24):1099–1106. doi: 10.1016/j.drudis.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Ren J., Shen S., Wang D., Xi Z., Guo L., Pang Z. The targeted delivery of anticancer drugs to brain glioma by PEGylated oxidized multi-walled carbon nanotubes modified with angiopep-2. Biomaterials. 2012;33(11):3324–3333. doi: 10.1016/j.biomaterials.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 12.Tan A., De La Peña H., Seifalian A.M. The application of exosomes as a nanoscale cancer vaccine. Int. J. Nanomed. 2010;5:889. doi: 10.2147/IJN.S13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nooshabadi V.T., Mardpour S., Yousefi‐Ahmadipour A., Allahverdi A., Izadpanah M., Daneshimehr F. The extracellular vesicles‐derived from mesenchymal stromal cells: a new therapeutic option in regenerative medicine. J. Cell. Biochem. 2018;119(10):8048–8073. doi: 10.1002/jcb.26726. [DOI] [PubMed] [Google Scholar]
- 14.Verdi Javad, Ebrahimi-Barough Somayeh, Mowla Javad, Atlasi Mohammad Ali, Mazoochi Tahereh, Valipour Elahe, Shafiei Shilan, Ai Jafar, Banafshe Hamid Reza. Endometrial mesenchymal stem cell-derived exosome promote endothelial cell angiogenesis in a dose dependent manner: a new perspective on regenerative medicine and cell-free therapy. Arch. Neurosci. 2019;6(4) doi: 10.5812/ans.94041. [DOI] [Google Scholar]
- 15.Johnsen K.B., Gudbergsson J.M., Skov M.N., Pilgaard L., Moos T., Duroux M. A comprehensive overview of exosomes as drug delivery vehicles—endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta Rev. Canc. 2014;1846(1):75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Tran T.-H., Mattheolabakis G., Aldawsari H., Amiji M. Exosomes as nanocarriers for immunotherapy of cancer and inflammatory diseases. Clin. Immunol. 2015;160(1):46–58. doi: 10.1016/j.clim.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Shafei S., Khanmohammadi M., Heidari R., Ghanbari H., Nooshabadi V.T., Farzamfar S. Exosome loaded alginate hydrogel promotes tissue regeneration in full‐thickness skin wounds: an in‐vivo study. J. Biomed. Mater. Res. 2019;108(3):545–556. doi: 10.1002/jbm.a.36835. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Mark M.T. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang T., Martin P., Fogarty B., Brown A., Schurman K., Phipps R. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharmaceut. Res. 2015;32(6):2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun D., Zhuang X., Xiang X., Liu Y., Zhang S., Liu C. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 2010;18(9):1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munagala R., Aqil F., Jeyabalan J., Gupta R.C. Bovine milk-derived exosomes for drug delivery. Canc. Lett. 2016;371(1):48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batrakova E.V., Kim M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Contr. Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascucci L., Coccè V., Bonomi A., Ami D., Ceccarelli P., Ciusani E. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J. Contr. Release. 2014;192:262–270. doi: 10.1016/j.jconrel.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 24.Wei H., Chen J., Wang S., Fu F., Zhu X., Wu C. A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int. J. Nanomed. 2019;14:8603. doi: 10.2147/IJN.S218988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanae M., Tsubaki M., Satou T., Itoh T., Imano M., Yamazoe Y. Statin-induced apoptosis via the suppression of ERK1/2 and Akt activation by inhibition of the geranylgeranyl-pyrophosphate biosynthesis in glioblastoma. J. Exp. Clin. Canc. Res. 2011;30(1):74. doi: 10.1186/1756-9966-30-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C.-Y., Liu P.-Y., Liao J.K. Pleiotropic effects of statin therapy: molecular mechanisms and clinical results. Trends Mol. Med. 2008;14(1):37–44. doi: 10.1016/j.molmed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson P.D., Panza G., Zaleski A., Taylor B. Statin-associated side effects. J. Am. Coll. Cardiol. 2016;67(20):2395–2410. doi: 10.1016/j.jacc.2016.02.071. [DOI] [PubMed] [Google Scholar]
- 28.Hindler K., Cleeland C.S., Rivera E., Collard C.D. The role of statins in cancer therapy. Oncol. 2006;11(3):306–315. doi: 10.1634/theoncologist.11-3-306. [DOI] [PubMed] [Google Scholar]
- 29.Song H.-H.G., Park K.M., Gerecht S. Hydrogels to model 3D in vitro microenvironment of tumor vascularization. Adv. Drug Deliv. Rev. 2014;79:19–29. doi: 10.1016/j.addr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayat N., Ebrahimi-Barough S., Norouzi-Javidan A., Saberi H., Tajerian R., Ardakan M.M.M. Apoptotic effect of atorvastatin in glioblastoma spheroids tumor cultured in fibrin gel. Biomed. Pharmacother. 2016;84:1959–1966. doi: 10.1016/j.biopha.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Yongjun Y., Shuyun H., Lei C., Xiangrong C., Zhilin Y., Yiquan K. Atorvastatin suppresses glioma invasion and migration by reducing microglial MT1-MMP expression. J. Neuroimmunol. 2013;260(1–2):1–8. doi: 10.1016/j.jneuroim.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Peng P., Wei W., Long C., Li J. Atorvastatin augments temozolomide's efficacy in glioblastoma via prenylation-dependent inhibition of Ras signaling. Biochem. Biophys. Res. Commun. 2017;489(3):293–298. doi: 10.1016/j.bbrc.2017.05.147. [DOI] [PubMed] [Google Scholar]
- 33.Xu P., Yu H., Zhang Z., Meng Q., Sun H., Chen X. Hydrogen-bonded and reduction-responsive micelles loading atorvastatin for therapy of breast cancer metastasis. Biomaterials. 2014;35(26):7574–7587. doi: 10.1016/j.biomaterials.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 34.Seeger H., Wallwiener D., Mueck A. Statins can inhibit proliferation of human breast cancer cells in vitro. Exp. Clin. Endocrinol. Diabetes. 2003;111(1):47–48. doi: 10.1055/s-2003-37501. [DOI] [PubMed] [Google Scholar]
- 35.Peng X., Li W., Yuan L., Mehta R.G., Kopelovich L., McCormick D.L. Inhibition of proliferation and induction of autophagy by atorvastatin in PC3 prostate cancer cells correlate with downregulation of Bcl2 and upregulation of miR-182 and p21. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0070442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shojaei S., Alizadeh J., Thliveris J., Koleini N., Kardami E., Hatch G.M. Statins: a new approach to combat temozolomide chemoresistance in glioblastoma. J. Invest. Med. 2018;66(8):1083–1087. doi: 10.1136/jim-2018-000874. [DOI] [PubMed] [Google Scholar]
- 37.Altwairgi A.K., Alghareeb W., AlNajjar F., Alsaeed E., Balbaid A., Alhussain H., Aldanan S., Orz Y., Lari A., Alsharm A. Phase II study of atorvastatin in combination with radiotherapy and temozolomide in patients with glioblastoma (ART): interim analysis report. Ann. Oncol. 2016;27:103–113. [Google Scholar]
- 38.Happold C., Gorlia T., Nabors L.B., Erridge S.C., Reardon D.A., Hicking C. Do statins, ACE inhibitors or sartans improve outcome in primary glioblastoma? J. Neuro Oncol. 2018;138(1):163–171. doi: 10.1007/s11060-018-2786-8. [DOI] [PubMed] [Google Scholar]
- 39.Spampanato C., De Maria S., Sarnataro M., Giordano E., Zanfardino M., Baiano S. Simvastatin inhibits cancer cell growth by inducing apoptosis correlated to activation of Bax and down-regulation of BCL-2 gene expression. Int. J. Oncol. 2012;40(4):935–941. doi: 10.3892/ijo.2011.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fromigue O., Hay E., Modrowski D., Bouvet S., Jacquel A., Auberger P. RhoA GTPase inactivation by statins induces osteosarcoma cell apoptosis by inhibiting p42/p44-MAPKs-Bcl-2 signaling independently of BMP-2 and cell differentiation. Cell Death Differ. 2006;13(11):1845. doi: 10.1038/sj.cdd.4401873. [DOI] [PubMed] [Google Scholar]
- 41.Vallianou G.N., Kostantinou A., Kougias M., Kazazis C. Statins and cancer. Anti-cancer agents in medicinal chemistry (formerly current medicinal chemistry-anti-cancer agents) 2014;14(5):706–712. doi: 10.2174/1871520613666131129105035. [DOI] [PubMed] [Google Scholar]
- 42.Kim W., Yoon J.-H., Kim J.-R., Jang I.-J., Bang Y.-J., Kim Y.-J. Synergistic anti-tumor efficacy of lovastatin and protein kinase C-beta inhibitor in hepatocellular carcinoma. Canc. Chemother. Pharmacol. 2009;64(3):497–507. doi: 10.1007/s00280-008-0897-1. [DOI] [PubMed] [Google Scholar]
- 43.Follet J., Rémy L., Hesry V., Simon B., Gillet D., Auvray P. Adaptation to statins restricts human tumour growth in Nude mice. BMC Canc. 2011;11(1):491. doi: 10.1186/1471-2407-11-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavakol S., Azedi F., Hoveizi E., Ai J., Joghataei M.T. Human endometrial stem cell isolation from endometrium and menstrual blood. Bio Protoc. 2018;8(2) doi: 10.21769/BioProtoc.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng Y.H., Rome S., Jalabert A., Forterre A., Singh H., Hincks C.L. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PloS One. 2013;8(3) doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalani A., Chaturvedi P., Kamat P.K., Maldonado C., Bauer P., Joshua I.G. Curcumin-loaded embryonic stem cell exosomes restored neurovascular unit following ischemia-reperfusion injury. Int. J. Biochem. Cell Biol. 2016;79:360–369. doi: 10.1016/j.biocel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J. Contr. Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turchinovich A., Weiz L., Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 2012;37(11):460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Kastelowitz N., Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem. 2014;15(7):923–928. doi: 10.1002/cbic.201400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Jong O.G., Verhaar M.C., Chen Y., Vader P., Gremmels H., Posthuma G. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles. 2012;1(1) doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J.H., Yeo Y. Controlled drug release from pharmaceutical nanocarriers. Chem. Eng. Sci. 2015;125:75–84. doi: 10.1016/j.ces.2014.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh M., Kundu S., Reddy A., Sreekanth V., Motiani R.K., Sengupta S. Injectable small molecule hydrogel as a potential nanocarrier for localized and sustained in vivo delivery of doxorubicin. Nanoscale. 2014;6(21):12849–12855. doi: 10.1039/c4nr04064c. [DOI] [PubMed] [Google Scholar]
- 53.Chen T., Zhao T., Wei D., Wei Y., Li Y., Zhang H. Core–shell nanocarriers with ZnO quantum dots-conjugated Au nanoparticle for tumor-targeted drug delivery. Carbohydr. Polym. 2013;92(2):1124–1132. doi: 10.1016/j.carbpol.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 54.Kim M.-S., Jin S.-J., Kim J.-S., Park H.J., Song H.-S., Neubert R.H. Preparation, characterization and in vivo evaluation of amorphous atorvastatin calcium nanoparticles using supercritical antisolvent (SAS) process. Eur. J. Pharm. Biopharm. 2008;69(2):454–465. doi: 10.1016/j.ejpb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Rajith B., Ravindran A. BSA nanoparticle loaded atorvastatin calcium-a new facet for an old drug. PloS One. 2014;9(2) doi: 10.1371/journal.pone.0086317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mekhail G.M., Kamel A.O., Awad G.A., Mortada N.D. Anticancer effect of atorvastatin nanostructured polymeric micelles based on stearyl-grafted chitosan. Int. J. Biol. Macromol. 2012;51(4):351–363. doi: 10.1016/j.ijbiomac.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Yamanaka R., Tabata S., Shindo Y., Hotta K., Suzuki K., Soga T. Mitochondrial Mg2+ homeostasis decides cellular energy metabolism and vulnerability to stress. Sci. Rep. 2016;6 doi: 10.1038/srep30027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lea A.P., McTavish D. Atorvastatin. Drugs. 1997;53(5):828–847. doi: 10.2165/00003495-199753050-00011. [DOI] [PubMed] [Google Scholar]
- 59.Song T., Liu J., Tao X., Deng J. Protection effect of atorvastatin in cerebral ischemia-reperfusion injury rats by blocking the mitochondrial permeability transition pore. Genet. Mol. Res. 2014;13(4):10632–10642. doi: 10.4238/2014.December.18.5. [DOI] [PubMed] [Google Scholar]
- 60.Urruticoechea A., Smith I.E., Dowsett M. Proliferation marker Ki-67 in early breast cancer. J. Clin. Oncol. 2005;23(28):7212–7220. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- 61.Surov A., Meyer H.J., Wienke A. Associations between apparent diffusion coefficient (ADC) and KI 67 in different tumors: a meta-analysis. Part 1: ADCmean. Oncotarget. 2017;8(43) doi: 10.18632/oncotarget.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruna J., Brell M., Ferrer I., Gimenez‐Bonafe P., Tortosa A. Ki‐67 proliferative index predicts clinical outcome in patients with atypical or anaplastic meningioma. Neuropathology. 2007;27(2):114–120. doi: 10.1111/j.1440-1789.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- 63.Green D.R. Apoptotic pathways: ten minutes to dead. Cell. 2005;121(5):671–674. doi: 10.1016/j.cell.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 64.Debatin K.-M. Apoptosis pathways in cancer and cancer therapy. Canc. Immunol. Immunother. 2004;53(3):153–159. doi: 10.1007/s00262-003-0474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stegh A.H., Kesari S., Mahoney J.E., Jenq H.T., Forloney K.L., Protopopov A. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105(31):10703–10708. doi: 10.1073/pnas.0712034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu S., Zhou Y., Yang G., Tian H., Geng Y., Hu Y. Sulforaphane-cysteine induces apoptosis by sustained activation of ERK1/2 and caspase 3 in human glioblastoma U373MG and U87MG cells. Oncol. Rep. 2017;37(5):2829–2838. doi: 10.3892/or.2017.5562. [DOI] [PubMed] [Google Scholar]
- 67.Cosford N.D.P., VAMOS M.D. Google Patents; 2018. Inhibitor of Apoptosis Protein (IAP) Antagonists. [Google Scholar]
- 68.Bao X-m, Wu C-f, Lu G-p. Atorvastatin inhibits homocysteine-induced dysfunction and apoptosis in endothelial progenitor cells. Acta Pharmacol. Sin. 2010;31(4):476. doi: 10.1038/aps.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spierings D., McStay G., Saleh M., Bender C., Chipuk J., Maurer U. Connected to death: the (unexpurgated) mitochondrial pathway of apoptosis. Science. 2005;310(5745):66–67. doi: 10.1126/science.1117105. [DOI] [PubMed] [Google Scholar]
- 70.Lemasters J.J., Qian T., He L., Kim J.-S., Elmore S.P., Cascio W.E. Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxidants Redox Signal. 2002;4(5):769–781. doi: 10.1089/152308602760598918. [DOI] [PubMed] [Google Scholar]