Abstract
Metabolite carriers of the mitochondrial inner membrane are crucial for cellular physiology since mitochondria contribute essential metabolic reactions and synthesize the majority of the cellular ATP. Like almost all mitochondrial proteins, carriers have to be imported into mitochondria from the cytosol. Carrier precursors utilize a specialized translocation pathway dedicated to the biogenesis of carriers and related proteins, the carrier translocase of the inner membrane (TIM22) pathway. After recognition and import through the mitochondrial outer membrane via the translocase of the outer membrane (TOM) complex, carrier precursors are ushered through the intermembrane space by hexameric TIM chaperones and ultimately integrated into the inner membrane by the TIM22 carrier translocase. Recent advances have shed light on the mechanisms of TOM translocase and TIM chaperone function, uncovered an unexpected versatility of the machineries, and revealed novel components and functional crosstalk of the human TIM22 translocase.
Keywords: mitochondrial carrier, metabolite transport, mitochondrial pyruvate carrier, sideroflexin, TOM, TIM chaperones, TIM22, protein translocation, mitochondrial biogenesis
1. Introduction
The mitochondrial inner membrane separates two aqueous compartments, the matrix and the intermembrane space, that differ in their protein and metabolite composition and host distinct metabolic pathways. The inner membrane is also the site of oxidative phosphorylation, and its integrity is crucial to maintain the electrochemical membrane potential that fuels ATP synthesis as well as mitochondrial biogenesis and function. Therefore, metabolite transport into or out of the matrix relies on carrier proteins that facilitate diffusion of specific substrates across the membrane or use the membrane potential to transport metabolites.
Most mitochondrial metabolite carriers belong to the mitochondrial carrier family (MCF, in humans SLC25 for solute carrier family 25). It comprises more than 50 members in humans and over 30 in yeast, and includes the most abundant inner membrane proteins [1,2,3,4]. MCF substrates range from nucleotides and amino acids to cofactors, intermediates of oxidative metabolism, and inorganic ions. Thus, they perform crucial functions in mitochondrial metabolism, and mutations in carrier genes are associated with a variety of human pathologies [5]. The mitochondrial pyruvate carrier (MPC) belongs to an unrelated protein family and functions as a hetero-dimer that requires both subunits for carrier activity [6,7]. The sideroflexin family constitutes a third metabolite carrier family, members of which have recently been discovered to function as serine transporters in one-carbon metabolism [8,9,10].
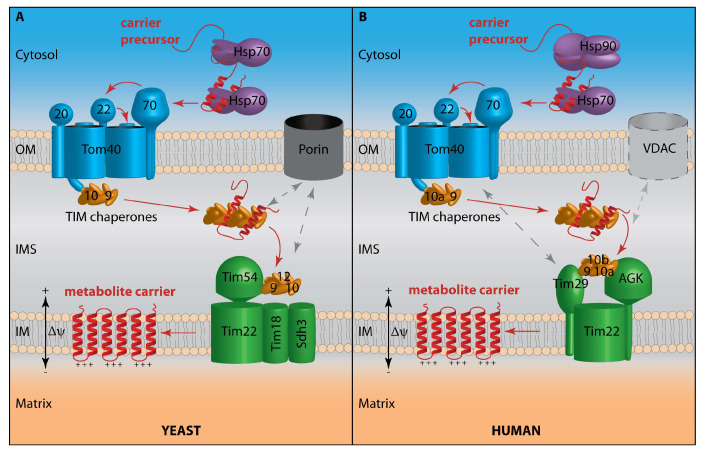
Like the vast majority of mitochondrial proteins, carriers are encoded in the nuclear genome and synthesized by cytosolic ribosomes. Therefore, they have to be specifically recognized and imported into the correct mitochondrial compartment. A multitude of protein translocases cooperates in the biogenesis of proteins destined for the different mitochondrial compartments [11,12,13,14]. Most precursors of mitochondrial inner membrane proteins are imported by the presequence translocase of the inner membrane (TIM23). However, carrier precursors generally lack a presequence, although a few contain an N-terminal extension that can improve solubility and translocation across the outer membrane [15,16,17]. Instead, carriers are targeted to mitochondria by internal signals. The internal targeting signals are not well defined, and carriers apparently contain several such motifs with different properties. For their biogenesis, metabolite carriers utilize a specialized import pathway involving the carrier translocase of the inner membrane (TIM22) [12,13,17,18,19,20] (Figure 1). This pathway can be divided into consecutive, biochemically defined stages: Stages I and II take place in the cytosol and on the mitochondrial surface, leading to import of the carrier precursor by the translocase of the outer membrane (TOM). In the intermembrane space (IMS), the precursor is bound by small TIM chaperones (stage III) and handed over to the TIM22 translocase (stage IV) which integrates the carrier into the inner membrane (stage V). The membrane potential across the inner membrane provides the driving force for membrane integration: Positively charged carrier sequences are subject to an electrophoretic force that pulls them into the matrix.
Figure 1.
The carrier pathway in the yeast S. cerevisiae (A) and in humans (B) handles the recognition, translocation and membrane integration of mitochondrial metabolite carriers into the inner membrane. Carrier precursors are bound by chaperones in the cytosol and recognized at the translocase of the outer membrane (TOM) by the Tom70 receptor. After their transfer through the outer membrane, they are bound in the intermembrane space by the hexameric TIM chaperones, Tim9-Tim10 in yeast (A) or Tim9-Tim10a in humans (B). The TIM chaperones guide the precursor through the aqueous compartment to the membrane-bound TIM chaperone complex consisting of Tim9-Tim10-Tim12 in yeast (A) or Tim9-Tim10a-Tim10b in humans (B). Substrate transfer to the carrier translocase of the inner membrane (TIM22) is aided by interactions with outer membrane proteins (dashed arrows) involving the metabolite channel porin/VDAC in yeast (A), or the TOM complex in humans (B). In humans, VDAC was found in association with TIM22 components (B) and, thus, might participate in carrier biogenesis similarly to porin. The TIM22 carrier translocase integrates the precursors into the inner membrane in a membrane potential-dependent manner. OM, outer membrane; IMS, intermembrane space; IM, inner membrane; ∆ψ, membrane potential; Hsp70, Hsp90, cytosolic ATP-dependent chaperones; Tom40, pore-forming component of the TOM complex; Tom20, Tom22, Tom70, receptors of the TOM complex; porin/VDAC, voltage-dependent anion channel; Tim22, core component of the TIM22 translocase; Tim18, Sdh3 (succinate dehydrogenase 3), Tim54, auxiliary subunits of the yeast TIM22 translocase; Tim29, AGK (acylglycerol kinase), auxiliary subunits of the human TIM22 translocase.
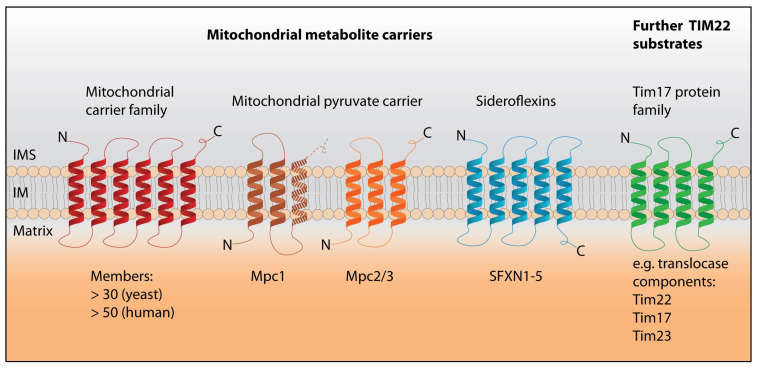
Mitochondrial carriers of the MCF/SLC25 family are the eponymous substrates of the TIM22 carrier translocase pathway [12,13,17,19] (Figure 1). The best studied carrier is the ADP/ATP carrier AAC (yeast)/ANT (human; adenine nucleotide translocator) that has been employed both as a model substrate to study carrier biogenesis and for the analysis of MCF structure and transport mechanism [4,17,18,21,22,23]. Mitochondrial carriers of the MCF/SLC25 family have a tripartite organization, with three homologous repeats consisting of two transmembrane segments each. They uniformly possess six transmembrane segments and expose both their N- and C-termini to the intermembrane space (Figure 2) [4,12,17,19,24]. Mitochondrial carriers transport substrates by enabling alternate access of the substrate(s) to the matrix and the intermembrane space while maintaining membrane impermeability for non-substrates [4,25,26,27,28]. Divergent members of the MCF with a differing number of TM segments are localized in the outer membrane and have acquired functions distinct from metabolite transport [4,29,30].
Figure 2.
Substrates of the TIM22 carrier import pathway. Mitochondrial carriers of the mitochondrial carrier family (MCF)/SLC25 family (red), the components of the mitochondrial pyruvate carrier (brown, orange), as well as sideroflexins (blue) are imported into mitochondria via the TIM22 carrier pathway. MCF/SLC25 proteins have a uniform topology with 6 transmembrane segments [4,12,17,19,24]. In contrast, Mpc2/Mpc3 has only 3 TM segments, and Mpc1 has 2 or 3 TM segments [6,7,35,36,37]. The third unique family of metabolite carriers, the sideroflexins, has 5 TM domains with the N-terminus in the intermembrane space (IMS) [8,38,39]. Aside from metabolite carriers, the TIM22 pathway also imports the members of the Tim17 protein family including the translocase components Tim17, Tim22 and Tim23 (green). IMS, intermembrane space; IM, inner membrane.
Interestingly, the components of the heterodimeric mitochondrial pyruvate carrier (MPC) have recently been discovered as further substrates of the TIM22 pathway (Figure 1) [31,32]. In contrast to the classical mitochondrial carriers, they are related to sugar transporters of the eukaryotic sugars will eventually be exported transporter (SWEET) and prokaryotic semiSWEET families [33,34]. SWEET transporters possess seven TM segments that are arranged into two triple-helix bundles connected by another α-helix. SemiSWEETs instead consist of one triple-helix bundle and assemble to dimers, forming a six TM functional unit like the SWEETs. MPC subunits MPC2 (mammals) as well as Mpc2 and Mpc3 (yeast) have three TM segments (Figure 2). For MPC1/Mpc1, the topology is not entirely clear: It has been suggested that they have only two TM segments with both termini in the matrix, or alternatively that they share the same topology as Mpc2/Mpc3 [6,7,35,36,37] (Figure 2).
Additionally, recent studies indicate that the sideroflexins, with five transmembrane segments and the N-terminus in the intermembrane space (IMS), also depend on the TIM22 carrier pathway for their biogenesis [38,39] (Figure 2). Thus, the mitochondrial pyruvate carrier components and the sideroflexins with their unique topologies have challenged long-held views of the structural requirements for TIM22 substrates.
2. Carrier Recognition at the TOM Complex
Due to their hydrophobic nature, mitochondrial carrier precursors in the cytosol are bound by chaperones to prevent their aggregation (stage I). Since carrier import takes place post-translationally, the soluble stage can be distinguished from stage II that consists in precursor targeting to the translocase of the outer membrane (Figure 1, Table 1) [13,14,17,19]. The TOM complex is the main entry gate by which almost all precursor proteins destined to the different mitochondrial compartments gain access to the organelle [12,14,40,41]. It forms dimers in vivo and consists of the Tom40 β-barrel pore and six α-helical membrane proteins: Tom5, Tom6, Tom7 as well as the receptors Tom22, Tom20 and Tom70 [42,43,44]. Aggregation of the highly hydrophobic carrier precursors in the cytosol is prevented by molecular chaperones. In yeast, carriers are chaperoned mainly by Hsp70, whereas in mammalian cells both Hsp70 and Hsp90 participate in carrier biogenesis, along with several co-chaperones [14,45,46,47,48,49]. Recognition of precursors at the TOM complex is mediated by the receptors Tom20 and Tom70. They can functionally substitute for each other sufficiently well for single deletions to be viable, however Tom20 preferentially recognizes precursors that contain a presequence, while Tom70 preferentially binds precursors with internal targeting sequences including carrier proteins such as the ADP/ATP carrier or the phosphate carrier [12,13,49,50,51,52,53,54,55,56,57,58,59,60]. Tom70 not only interacts with the precursor, but also with the associated chaperone(s) via tetratricopeptide repeats (TPR) that bind the C-termini of Hsp70 or Hsp90 chaperones [14,46,49,61]. Moreover, one tripartite carrier precursor of the MCF/SLC25 family can recruit three Tom70 dimers, with each of the repeats participating in the interaction [21]. Thus, the interactions of Tom70 with the precursor-chaperone complex likely contribute to prevention of its aggregation. Import of carriers can be stalled at stage II by depletion of ATP (Table 1). ATP binding to Hsp70 triggers substrate release from Hsp70, the carrier precursor is handed over to the central receptor Tom22, and individual helix-loop-helix modules are threaded into the Tom40 pore in a hairpin-like conformation [21,62]. It is currently unclear how mitochondrial pyruvate carrier precursors with their distinct topology are handled by the TOM complex, although it is tempting to speculate that at least the two C-terminal TM segments of Mpc2/Mpc3 may be recognized in a fashion similar to classical carriers. Interestingly, even during translocation through the TOM complex, carriers follow a different route than presequence precursors, involving the distal regions of the Tom40 dimer and the N-terminal extension of Tom40 [42,44]. It was proposed that translocation through TOM is aided by interaction of positively charged regions in the precursors with the negatively charged inner surface of the Tom40 β-barrel [63], which is consistent with the previously reported head-first insertion of helix-loop-helix modules by their positively charged loops [21].
Table 1.
Stages of carrier biogenesis via the TIM22 carrier import pathway.
| TIM22 Carrier Import Pathway | Characteristics of Individual Stages in Carrier Biogenesis |
|---|---|
| Stage I | The carrier precursor is bound to cytosolic chaperones upon its synthesis, forming a soluble complex not associated with mitochondria. |
| Stage II | The precursor-chaperone complex is recognized by the Tom70 receptor of the TOM complex and can be arrested on the mitochondrial surface by ATP depletion. ATP binding triggers dissociation of Hsp70 chaperones and progression of the precursor. |
| Stage III IIIa: | The carrier precursor is translocated through the TOM complex into the IMS and concomitantly bound by soluble TIM chaperones (mainly Tim9-Tim10 in yeast, Tim9-Tim10a in humans). |
| IIIb: | The carrier precursor is handed over to the TIM22-bound TIM chaperones (Tim9-Tim10-Tim12 in yeast, Tim9-Tim10a-Tim10b in humans), resulting in tethering to the inner membrane. |
| Stage IV | In the presence of a low membrane potential, the carrier precursor is transferred to TIM22 and inserted into the inner membrane as a docked precursor. |
| Stage V | Formation of the mature, inner-membrane integrated carrier and release from TIM22 requires the presence of a higher membrane potential. |
The efficiency of carrier recognition at the TOM complex is subject to metabolic regulation. Tom70 is phosphorylated by protein kinase A specifically during non-respiratory growth of yeast on glucose, resulting in an impaired interaction with Hsp70 and concomitantly reduced carrier import [64]. Thus, the efficiency of carrier biogenesis can be adjusted to the metabolic requirements of respiratory versus non-respiratory growth.
3. En Route through the Intermembrane Space
Unlike the TIM23 translocase that imports presequence proteins into the matrix or the inner membrane, the translocation of carrier precursors through the TOM complex is apparently not tightly coupled to integration into the inner membrane by the TIM22 carrier translocase [12,17,65,66]. Instead, once a carrier precursor has traversed the Tom40 pore and reached the IMS, it is bound by small TIM chaperones to prevent aggregation during its transit through the aqueous IMS environment to the inner membrane (stage III) [67,68,69,70,71,72,73] (Figure 1, Table 1). Translocation through the TOM complex is coupled to TIM chaperone binding [71]. The small TIM chaperones form ring-like hetero-hexameric complexes that bind carriers in an extended conformation [73,74,75]. The predominant TIM chaperone complex consists of alternating Tim9 and Tim10 subunits (Tim9 and Tim10a in humans) and is required for carrier import; the alternative Tim8-Tim13 complex (Tim8a or Tim8b and Tim13 in humans) has partially redundant substrate specificity [73,75,76,77,78,79]. Mutations in Tim8a cause the deafness–dystonia syndrome called Mohr–Tranebjærg syndrome [80], however, novel evidence suggests that the underlying molecular mechanism reflects a new function of Tim8a in cytochrome c oxidase maturation rather than a defective TIM22 carrier pathway [79]. The small TIM chaperones interact with the N-terminal extension of Tom40 that participates in carrier translocation, so they are ideally positioned to receive their cargo from the TOM complex [21,42,44,71]. Stage III of carrier import can be further subdivided, where stage IIIa denotes carriers bound to soluble TIM chaperones that may be associated with TOM or soluble in the IMS (Table 1) [17,19]. A recent comprehensive study demonstrated that an MCF carrier with six TM segments is bound by two TIM chaperone hexamers, and the precursor is chaperoned by interacting with a conserved hydrophobic cleft between the two tentacle-like α-helices of the small TIM proteins [73]. This conserved substrate binding region is also required for chaperoning of the structurally unrelated mitochondrial pyruvate carrier [31]. The soluble TIM complexes transfer carrier precursors to a separate TIM chaperone complex that is associated with the TIM22 carrier translocase of the inner membrane. This membrane-bound TIM hexamer consists of Tim9/Tim10/Tim12 (yeast) or Tim9/Tim10a/Tim10b (human) [67,81,82,83,84,85,86] (Figure 1). Carrier precursors bound to the membrane-associated TIM chaperones represent stage IIIb of the carrier import pathway where the precursor is tethered to the inner membrane, but not yet inserted (Table 1) [17,19]. Since this is the last step that is independent of the membrane potential ∆ψ, precursors accumulate in stage III upon dissipation of the membrane potential. The dependence of carrier import on TIM chaperones also allows to experimentally distinguish this import pathway into the inner membrane from the presequence pathway where import into a protease-protected environment depends on ∆ψ due to the coupling of TOM and TIM23 [65,66].
Unexpectedly, transport of carriers to the TIM22 carrier translocase is also aided by porin/voltage-dependent anion channel (VDAC), the major metabolite channel of the outer membrane (Figure 1) [87,88,89,90]. In addition to interacting with TIM chaperones as well as carrier precursors, porin recruits the TIM22 translocase and thereby brings the outer and inner membrane in close proximity, which may enhance carrier biogenesis [88,89]. Since porin, unlike the protein translocases, is significantly upregulated upon respiratory growth [91], this novel physiological role may support the higher levels of protein import required for metabolic remodeling of the mitochondria. While a contribution of VDACs to carrier biogenesis in human mitochondria has not been studied directly, they were found to interact with TIM22 [92,93].
4. Membrane Integration by the TIM22 Carrier Translocase
The TIM22 translocase consists of the Tim22 protein, several auxiliary subunits that have roles in assembly and stabilization of the complex, and TIM chaperones. In yeast, the additional subunits comprise Tim54, Sdh3, which also interacts with Sdh4 as part of the succinate dehydrogenase complex, and Tim18, a homolog of Sdh4 [12,17,19,20,94,95,96,97,98,99,100] (Figure 1A). Until recently, the only known membrane integral TIM22 component in humans was Tim22 itself, however, several studies have discovered two new subunits (Figure 1B). The metazoan-specific subunit Tim29, which like Tim54 is exposed to the IMS and can be crosslinked to TIM chaperones, is required for TIM22 assembly and efficient import of some substrates [92,101]. Moreover, Tim29 interacts with Tom40, indicating that there may be coupling between TOM and TIM22 in human cells [92], in contrast to yeast. The most recently identified subunit of human TIM22 is acylglycerol kinase (AGK) that phosphorylates glycerides to generate lysophosphatidic acid or phosphatidic acid [102,103,104]. AGK has a dual role in human mitochondria: It is required for TIM22 stability and carrier import independently of its lipid kinase activity, and loss of AGK results in TCA (tricarboxylic acid) cycle defects; however, kinase deficiency causes aberrant mitochondrial ultrastructure and concomitantly reduced respiration [103,104]. Mutations in AGK cause the mitochondrial disease Sengers syndrome [105,106], and its novel role as part of the TIM22 translocase appears to account for the disease phenotype [103]. Aside from AGK, mutations in Tim22 that impair carrier import were also recently reported to result in human pathology with neuromuscular defects [32,107]. Interestingly, human TIM22 interacts with the mitochondrial contact site and cristae organizing system (MICOS) [93], an inner membrane protein complex that is crucial for native cristae architecture and that forms contact sites between the two mitochondrial membranes [108,109,110]. Upon MICOS disruption, carrier import is specifically impaired, indicating that MICOS-mediated membrane contact sites might support efficient carrier biogenesis in human cells [93].
The TIM22 complex is a voltage-gated preprotein translocase that is thought to insert one helix—matrix loop—helix repeat at a time in a hairpin conformation into the inner membrane [19,20,86,97]. At least a low membrane potential is required for transfer of a precursor from the TIM chaperones to TIM22. Precursors can be trapped experimentally at this stage IV by reducing the membrane potential with ionophores (Table 1) [20]. In the presence of a higher membrane potential as well as of an internal targeting sequence, TIM22 integrates the protein into the inner membrane by an unknown mechanism involving lateral release, and the carrier reaches the mature stage V [20].
The modular topology of mitochondrial carriers of the MCF/SLC25 family—with repeats of helix-loop-helix domains, the termini facing the IMS and positively charged loops in the matrix—was long assumed to be a requirement for substrates of the TIM22 carrier pathway. This pathway also imports members of the Tim17 protein family that includes Tim22 itself as well as the TIM23 translocase components Tim23 and Tim17, all of which have four TM segments but otherwise share the topology of classical carriers [73,75,76,77,111,112,113] (Figure 2). As mentioned, both TOM and TIM22 translocases act on paired helices during import of classical carriers, including the dicarboxylate carrier, the ADP/ATP carrier and the phosphate carrier, as well as during import of Tim22-related proteins [21,70,77,114,115,116], in contrast to the linear import of other mitochondrial proteins. Until recently, multi-spanning inner membrane proteins apart from the MCF and Tim17 families were thought to be imported into mitochondria by the other translocases of the inner membrane: By the TIM23 translocase [11,12,13,65,66,117], by the oxidase assembly (OXA) translocase that is responsible for the membrane integration of mitochondrially encoded proteins [118,119] or by a combination of both machineries [120,121,122]. Moreover, truncated variants of carrier proteins are no longer recognized as substrates of the TIM22 pathway and instead are imported by TIM23 or remain in the intermembrane space [115,116]. However, recent work has identified the mitochondrial pyruvate carrier proteins with their divergent topology as substrates of the TIM22 pathway [31,32]. What is more, the unrelated sideroflexins also rely on the TIM22 carrier translocase for their biogenesis [38,39]. Of the MPC components, at least Mpc2 and Mpc3 have unpaired TM helices and none of the MPC proteins possess more than three TM segments [6,7,35,36,37], while the sideroflexins also have an uneven number of TM segments and yet another topology [8,39] (Figure 2). Thus, multiple recent studies have revealed a surprising versatility of the TIM22 pathway. The mechanistic differences in the handling of substrates with 4 or 6 TM segments (classical TIM22 substrates) versus 2/3 (MPC subunits) or 5 TM segments (sideroflexins) are still unclear. The C-terminal helix-loop-helix domain of Mpc2 and Mpc3 might conceivably be treated similarly as a typical carrier repeat, since they share the topology and positively charged matrix loop. In addition, MPC subunits were reported to have an N-terminal α-helix whose function is unknown [37]. For the human TIM22 translocase a differential requirement of the auxiliary subunits for different substrate classes has been reported: AGK is required for efficient carrier import and dispensable for the import of Tim22 and related proteins [103,104], whereas the opposite is the case for Tim29 [92]. Interestingly, sideroflexins, like classical carriers, rely on AGK, while Tim29 is dispensable [38]. It will be very interesting to learn how TIM22 adapts its function to import this range of structurally distinct substrates.
5. Perspectives
The biogenesis of mitochondrial metabolite carriers is still not fully understood despite the fact that the TIM22 import pathway has been under scientific investigation for decades. The recent discovery of two human TIM22 components and the very limited insight into the mechanism of membrane integration by the carrier translocase exemplify how much fundamental information is still lacking. Novel findings indicate that carrier import may benefit from contact sites between the outer and inner membranes after all. Moreover, the TIM22 pathway has turned out to be unexpectedly versatile regarding its substrate requirements. Finally, it seems likely that the biogenesis of proteins with such a central role in mitochondrial physiology is regulated at different steps. While there is precedent for this notion from yeast, the human TIM22 pathway still awaits characterization of regulatory factors. Thus, the biogenesis of mitochondrial metabolite carriers remains an exciting field of study that is expected to generate important insights into mitochondrial physiology.
Acknowledgments
We thank Nils Wiedemann and Nikolaus Pfanner for discussion and helpful suggestions. Work included in this study has been performed in partial fulfillment of the requirements for the doctoral thesis of P.H.
Funding
This work was supported by the Excellence Initiative/Strategy of the German Federal and State Governments (EXC 2189 CIBSS Project ID 390939984).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Palmieri F., Monné M. Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochim. Biophys. Acta. 2016;1863:2362–2378. doi: 10.1016/j.bbamcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Taylor E.B. Functional Properties of the Mitochondrial Carrier System. Trends Cell Biol. 2017;27:633–644. doi: 10.1016/j.tcb.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogunbona O.B., Claypool S.M. Emerging Roles in the Biogenesis of Cytochrome c Oxidase for Members of the Mitochondrial Carrier Family. Front. Cell Dev. Biol. 2019;7:3. doi: 10.3389/fcell.2019.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruprecht J.J., Kunji E.R.S. The SLC25 Mitochondrial Carrier Family: Structure and Mechanism. Trends Biochem. Sci. 2020;45:244–258. doi: 10.1016/j.tibs.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmieri F., Scarcia P., Monné M. Diseases caused by mutations in mitochondrial carrier genes SLC25: A review. Biomolecules. 2020;10:655. doi: 10.3390/biom10040655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.-C., Cox J.E., Cardon C.M., Van Vranken J.G., Dephoure N., et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herzig S., Raemy E., Montessuit S., Veuthey J.-L., Zamboni N., Westermann B., Kunji E.R.S., Martinou J.-C. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 8.Kory N., Wyant G.A., Prakash G., Uit de Bos J., Bottanelli F., Pacold M.E., Chan S.H., Lewis C.A., Wang T., Keys H.R., et al. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science. 2018;362:eaat9528. doi: 10.1126/science.aat9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzi A., Glerum M., Koller R., Mertens W., Spycher S. The mitochondrial tricarboxylate carrier. J. Bioenerg. Biomembr. 1993;25:515–524. doi: 10.1007/BF01108408. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham C.N., Rutter J. 20,000 picometers under the OMM: Diving into the vastness of mitochondrial metabolite transport. EMBO Rep. 2020;21:e50071. doi: 10.15252/embr.202050071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neupert W. A Perspective on Transport of Proteins into Mitochondria: A Myriad of Open Questions. J. Mol. Biol. 2015;427:1135–1158. doi: 10.1016/j.jmb.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Wiedemann N., Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 13.Hansen K.G., Herrmann J.M. Transport of Proteins into Mitochondria. Protein J. 2019;38:330–342. doi: 10.1007/s10930-019-09819-6. [DOI] [PubMed] [Google Scholar]
- 14.Becker T., Song J., Pfanner N. Versatility of Preprotein Transfer from the Cytosol to Mitochondria. Trends Cell Biol. 2019;29:534–548. doi: 10.1016/j.tcb.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Zara V., Palmieri F., Mahlke K., Pfanner N. The cleavable presequence is not essential for import and assembly of the phosphate carrier of mammalian mitochondria but enhances the specificity and efficiency of import. J. Biol. Chem. 1992;267:12077–12081. [PubMed] [Google Scholar]
- 16.Zara V., Dolce V., Capobianco L., Ferramosca A., Papatheodourou P., Rassow J., Palmieri F. Biogenesis of eel liver citrate carrier (CIC): Negative charges can substitute for positive charges in the presequence. J. Mol. Biol. 2007;365:958–967. doi: 10.1016/j.jmb.2006.10.077. [DOI] [PubMed] [Google Scholar]
- 17.Ferramosca A., Zara V. Biogenesis of mitochondrial carrier proteins: Molecular mechanisms of import into mitochondria. Biochim. Biophys. Acta Mol. Cell Res. 2013;1833:494–502. doi: 10.1016/j.bbamcr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Ryan M.T., Müller H., Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- 19.Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 20.Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H.E., Kühlbrandt W., Wagner R., Truscott K.N., Pfanner N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003;299:1747–1751. doi: 10.1126/science.1080945. [DOI] [PubMed] [Google Scholar]
- 21.Wiedemann N., Pfanner N., Ryan M.T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta Biomembr. 2008;1778:1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Ruprecht J.J., King M.S., Zögg T., Aleksandrova A.A., Pardon E., Crichton P.G., Steyaert J., Kunji E.R.S. The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier. Cell. 2019;176:435–447.e15. doi: 10.1016/j.cell.2018.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmieri F. Mitochondrial carrier proteins. FEBS Lett. 1994;346:48–54. doi: 10.1016/0014-5793(94)00329-7. [DOI] [PubMed] [Google Scholar]
- 25.Klingenberg M. The ADP, ATP shuttle of the mitochondrion. Trends Biochem. Sci. 1979;4:249–252. doi: 10.1016/0968-0004(79)90215-9. [DOI] [Google Scholar]
- 26.Indiveri C., Tonazzi A., Palmieri F. The reconstituted carnitine carrier from rat liver mitochondria: Evidence for a transport mechanism different from that of the other mitochondrial translocators. Biochim. Biophys. Acta. 1994;1189:65–73. doi: 10.1016/0005-2736(94)90281-X. [DOI] [PubMed] [Google Scholar]
- 27.Monné M., Palmieri F. Antiporters of the Mitochondrial Carrier Family. Curr. Top. Membr. 2014;73:289–320. doi: 10.1016/B978-0-12-800223-0.00008-6. [DOI] [PubMed] [Google Scholar]
- 28.Ruprecht J.J., Kunji E.R.S. Structural changes in the transport cycle of the mitochondrial ADP/ATP carrier. Curr. Opin. Struct. Biol. 2019;57:135–144. doi: 10.1016/j.sbi.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coonrod E.M., Karren M.A., Shaw J.M. Ugo1p is a multipass transmembrane protein with a single carrier domain required for mitochondrial fusion. Traffic. 2007;8:500–511. doi: 10.1111/j.1600-0854.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 30.Hoppins S.C., Horner J., Song C., McCaffery J.M., Nunnari J. Mitochondrial outer and inner membrane fusion requires a modified carrier protein. J. Cell Biol. 2009;184:569–581. doi: 10.1083/jcb.200809099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rampelt H., Sucec I., Bersch B., Horten P., Perschil I., Martinou J.-C., van der Laan M., Wiedemann N., Schanda P., Pfanner N. The mitochondrial carrier pathway transports non-canonical substrates with an odd number of transmembrane segments. BMC Biol. 2020;18:2. doi: 10.1186/s12915-019-0733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomkale R., Cruz-Zaragoza L.D., Suppanz I., Guiard B., Montoya J., Callegari S., Pacheu-Grau D., Warscheid B., Rehling P. Defining the Substrate Spectrum of the TIM22 Complex Identifies Pyruvate Carrier Subunits as Unconventional Cargos. Curr. Biol. 2020;30:1119–1127.e5. doi: 10.1016/j.cub.2020.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y., Tao Y., Cheung L.S., Fan C., Chen L.-Q., Xu S., Perry K., Frommer W.B., Feng L. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature. 2014;515:448–452. doi: 10.1038/nature13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng L., Frommer W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015;40:480–486. doi: 10.1016/j.tibs.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Bender T., Pena G., Martinou J.-C. Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. EMBO J. 2015;34:911–924. doi: 10.15252/embj.201490197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanderperre B., Cermakova K., Escoffier J., Kaba M., Bender T., Nef S., Martinou J.-C. MPC1-like Is a Placental Mammal-specific Mitochondrial Pyruvate Carrier Subunit Expressed in Postmeiotic Male Germ Cells. J. Biol. Chem. 2016;291:16448–16461. doi: 10.1074/jbc.M116.733840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavoulari S., Thangaratnarajah C., Mavridou V., Harbour M.E., Martinou J.-C., Kunji E.R.S. The yeast mitochondrial pyruvate carrier is a hetero-dimer in its functional state. EMBO J. 2019;38:e100785. doi: 10.15252/embj.2018100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson T.D., Hock D., Palmer C.S., Kang Y., Fujihara K.M., Clemons N.J., Thorburn D.R., Stroud D., Stojanovski D. The TIM22 complex regulates mitochondrial one-carbon metabolism by mediating the import of sideroflexins. bioRxiv. 2020 doi: 10.1101/2020.02.06.937920. [DOI] [Google Scholar]
- 39.Acoba M.G., Alpergin E.S.S., Renuse S., Fernández-del-Río L., Lu Y.-W., Clarke C.F., Pandey A., Wolfgang M.J., Claypool S.M. The mitochondrial carrier SFXN1 is critical for complex III integrity and cellular metabolism. bioRxiv. 2020 doi: 10.1101/2020.06.18.157495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Endo T., Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim. Biophys. Acta. 2010;1803:706–714. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Kang Y., Fielden L.F., Stojanovski D. Mitochondrial protein transport in health and disease. Semin. Cell Dev. Biol. 2018;76:142–153. doi: 10.1016/j.semcdb.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 42.Shiota T., Imai K., Qiu J., Hewitt V.L., Tan K., Shen H.-H., Sakiyama N., Fukasawa Y., Hayat S., Kamiya M., et al. Molecular architecture of the active mitochondrial protein gate. Science. 2015;349:1544–1548. doi: 10.1126/science.aac6428. [DOI] [PubMed] [Google Scholar]
- 43.Bausewein T., Mills D.J., Langer J.D., Nitschke B., Nussberger S., Kühlbrandt W. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell. 2017;170:693–700.e7. doi: 10.1016/j.cell.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Araiso Y., Tsutsumi A., Qiu J., Imai K., Shiota T., Song J., Lindau C., Wenz L.-S., Sakaue H., Yunoki K., et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature. 2019;575:395–401. doi: 10.1038/s41586-019-1680-7. [DOI] [PubMed] [Google Scholar]
- 45.Komiya T., Rospert S., Schatz G., Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young J.C., Hoogenraad N.J., Hartl F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/S0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 47.Bhangoo M.K., Tzankov S., Fan A.C.Y., Dejgaard K., Thomas D.Y., Young J.C. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell. 2007;18:3414–3428. doi: 10.1091/mbc.e07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opaliński Ł., Song J., Priesnitz C., Wenz L.-S., Oeljeklaus S., Warscheid B., Pfanner N., Becker T. Recruitment of Cytosolic J-Proteins by TOM Receptors Promotes Mitochondrial Protein Biogenesis. Cell Rep. 2018;25:2036–2043.e5. doi: 10.1016/j.celrep.2018.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avendaño-Monsalve M.C., Ponce-Rojas J.C., Funes S. From cytosol to mitochondria: The beginning of a protein journey. Biol. Chem. 2020;401:645–661. doi: 10.1515/hsz-2020-0110. [DOI] [PubMed] [Google Scholar]
- 50.Hines V., Brandt A., Griffiths G., Horstmann H., Brütsch H., Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Söllner T., Pfaller R., Griffiths G., Pfanner N., Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- 52.Steger H.F., Söllner T., Kiebler M., Dietmeier K.A., Pfaller R., Trülzsch K.S., Tropschug M., Neupert W., Pfanner N. Import of ADP/ATP carrier into mitochondria: Two receptors act in parallel. J. Cell Biol. 1990;111:2353–2363. doi: 10.1083/jcb.111.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brix J., Dietmeier K., Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 54.Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- 55.Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/S0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 56.Saitoh T., Igura M., Obita T., Ose T., Kojima R., Maenaka K., Endo T., Kohda D. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 2007;26:4777–4787. doi: 10.1038/sj.emboj.7601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamano K., Yatsukawa Y.-I., Esaki M., Hobbs A.E.A., Jensen R.E., Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto H., Fukui K., Takahashi H., Kitamura S., Shiota T., Terao K., Uchida M., Esaki M., Nishikawa S.I., Yoshihisa T., et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 2009;284:31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto H., Itoh N., Kawano S., Yatsukawa Y.I., Momose T., Makio T., Matsunaga M., Yokota M., Esaki M., Shodai T., et al. Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria. Proc. Natl. Acad. Sci. USA. 2011;108:91–96. doi: 10.1073/pnas.1014918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Backes S., Hess S., Boos F., Woellhaf M.W., Gödel S., Jung M., Mühlhaus T., Herrmann J.M. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 2018;217:1369–1382. doi: 10.1083/jcb.201708044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 62.van Wilpe S., Ryan M.T., Hill K., Maarse A.C., Meisinger C., Brix J., Dekker P.J., Moczko M., Wagner R., Meijer M., et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- 63.Tucker K., Park E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2019;26:1158–1166. doi: 10.1038/s41594-019-0339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt O., Harbauer A.B., Rao S., Eyrich B., Zahedi R.P., Stojanovski D., Schönfisch B., Guiard B., Sickmann A., Pfanner N., et al. Regulation of Mitochondrial Protein Import by Cytosolic Kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 65.Moulin C., Caumont-Sarcos A., Ieva R. Mitochondrial presequence import: Multiple regulatory knobs fine-tune mitochondrial biogenesis and homeostasis. Biochim. Biophys. Acta Mol. Cell. Res. 2019;1866:930–944. doi: 10.1016/j.bbamcr.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Callegari S., Cruz-Zaragoza L.D., Rehling P. From TOM to the TIM23 complex – handing over of a precursor. Biol. Chem. 2020 doi: 10.1515/hsz-2020-0101. [DOI] [PubMed] [Google Scholar]
- 67.Sirrenberg C., Endres M., Fölsch H., Stuart R.A., Neupert W., Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 68.Koehler C.M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R.J., Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 69.Koehler C.M., Merchant S., Oppliger W., Schmid K., Jarosch E., Dolfini L., Junne T., Schatz G., Tokatlidis K. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Endres M., Neupert W., Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Truscott K.N., Wiedemann N., Rehling P., Müller H., Meisinger C., Pfanner N., Guiard B. Mitochondrial Import of the ADP/ATP Carrier: The Essential TIM Complex of the Intermembrane Space Is Required for Precursor Release from the TOM Complex. Mol. Cell. Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Curran S.P., Leuenberger D., Oppliger W., Koehler C.M. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinhäupl K., Lindau C., Hessel A., Wang Y., Schütze C., Jores T., Melchionda L., Schönfisch B., Kalbacher H., Bersch B., et al. Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space. Cell. 2018;175:1365–1379.e25. doi: 10.1016/j.cell.2018.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Webb C.T., Gorman M.A., Lazarou M., Ryan M.T., Gulbis J.M. Crystal Structure of the Mitochondrial Chaperone TIM9•10 Reveals a Six-Bladed α-Propeller. Mol. Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 75.Beverly K.N., Sawaya M.R., Schmid E., Koehler C.M. The Tim8–Tim13 Complex Has Multiple Substrate Binding Sites and Binds Cooperatively to Tim23. J. Mol. Biol. 2008;382:1144–1156. doi: 10.1016/j.jmb.2008.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paschen S.A., Rothbauer U., Káldi K., Bauer M.F., Neupert W., Brunner M. The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 2000;19:6392–6400. doi: 10.1093/emboj/19.23.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Curran S.P., Leuenberger D., Schmidt E., Koehler C.M. The role of the Tim8p–Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 2002;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis A.J., Alder N.N., Jensen R.E., Johnson A.E. The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochondrial protein import. Mol. Biol. Cell. 2007;18:475–486. doi: 10.1091/mbc.e06-06-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang Y., Anderson A.J., Jackson T.D., Palmer C.S., De Souza D.P., Fujihara K.M., Stait T., Frazier A.E., Clemons N.J., Tull D., et al. Function of hTim8a in complex IV assembly in neuronal cells provides insight into pathomechanism underlying Mohr-Tranebjærg syndrome. eLife. 2019;8:e48828. doi: 10.7554/eLife.48828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koehler C.M., Leuenberger D., Merchant S., Renold A., Junne T., Schatz G. Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl. Acad. Sci. USA. 1999;96:2141–2146. doi: 10.1073/pnas.96.5.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adam A., Endres M., Sirrenberg C., Lottspeich F., Neupert W., Brunner M. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 1999;18:313–319. doi: 10.1093/emboj/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mühlenbein N., Hofmann S., Rothbauer U., Bauer M.F. Organization and Function of the Small Tim Complexes Acting along the Import Pathway of Metabolite Carriers into Mammalian Mitochondria. J. Biol. Chem. 2004;279:13540–13546. doi: 10.1074/jbc.M312485200. [DOI] [PubMed] [Google Scholar]
- 83.Gebert N., Chacinska A., Wagner K., Guiard B., Koehler C.M., Rehling P., Pfanner N., Wiedemann N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008;9:548–554. doi: 10.1038/embor.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lionaki E., de Marcos Lousa C., Baud C., Vougioukalaki M., Panayotou G., Tokatlidis K. The essential function of Tim12 in vivo is ensured by the assembly interactions of its C-terminal domain. J. Biol. Chem. 2008;283:15747–15753. doi: 10.1074/jbc.M800350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weinhäupl K., Wang Y., Hessel A., Brennich M., Lindorff-Larsen K., Schanda P. Architecture and subunit dynamics of the mitochondrial TIM9·10·12 chaperone. bioRxiv. 2020 doi: 10.1101/2020.03.13.990150. [DOI] [PubMed] [Google Scholar]
- 86.Qi L., Wang Q., Guan Z., Wu Y., Cao J., Zhang X., Yan C., Yin P. Cryo-EM structure of the human mitochondrial translocase TIM22 complex. bioRxiv. 2019 doi: 10.1101/869289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mertins B., Psakis G., Essen L.-O. Voltage-dependent anion channels: The wizard of the mitochondrial outer membrane. Biol. Chem. 2014;395:1435–1442. doi: 10.1515/hsz-2014-0203. [DOI] [PubMed] [Google Scholar]
- 88.Ellenrieder L., Dieterle M.P., Doan K.N., Mårtensson C.U., Floerchinger A., Campo M.L., Pfanner N., Becker T. Dual Role of Mitochondrial Porin in Metabolite Transport across the Outer Membrane and Protein Transfer to the Inner Membrane. Mol. Cell. 2019;73:1056–1065.e7. doi: 10.1016/j.molcel.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 89.Grevel A., Becker T. Porins as helpers in mitochondrial protein translocation. Biol. Chem. 2020;401:699–708. doi: 10.1515/hsz-2019-0438. [DOI] [PubMed] [Google Scholar]
- 90.Becker T., Wagner R. Mitochondrial Outer Membrane Channels: Emerging Diversity in Transport Processes. Bioessays. 2018;40:e1800013. doi: 10.1002/bies.201800013. [DOI] [PubMed] [Google Scholar]
- 91.Morgenstern M., Stiller S.B., Lübbert P., Peikert C.D., Dannenmaier S., Drepper F., Weill U., Höß P., Feuerstein R., Gebert M., et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang Y., Baker M.J., Liem M., Louber J., McKenzie M., Atukorala I., Ang C.-S., Keerthikumar S., Mathivanan S., Stojanovski D. Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. eLife. 2016;5:e17463. doi: 10.7554/eLife.17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Callegari S., Müller T., Schulz C., Lenz C., Jans D.C., Wissel M., Opazo F., Rizzoli S.O., Jakobs S., Urlaub H., et al. MICOS-TIM22 Association Promotes Carrier Import into Human Mitochondria. J. Mol. Biol. 2019;431:2835–2851. doi: 10.1016/j.jmb.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 94.Sirrenberg C., Bauer M.F., Guiard B., Neupert W., Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 95.Koehler C.M., Murphy M.P., Bally N.A., Leuenberger D., Oppliger W., Dolfini L., Junne T., Schatz G., Or E. Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol. Cell. Biol. 2000;20:1187–1193. doi: 10.1128/MCB.20.4.1187-1193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kerscher O., Sepuri N.B., Jensen R.E. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell. 2000;11:103–116. doi: 10.1091/mbc.11.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kovermann P., Truscott K.N., Guiard B., Rehling P., Sepuri N.B., Müller H., Jensen R.E., Wagner R., Pfanner N. Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel. Mol. Cell. 2002;9:363–373. doi: 10.1016/S1097-2765(02)00446-X. [DOI] [PubMed] [Google Scholar]
- 98.Hwang D.K., Claypool S.M., Leuenberger D., Tienson H.L., Koehler C.M. Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J. Cell Biol. 2007;178:1161–1175. doi: 10.1083/jcb.200706195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wagner K., Gebert N., Guiard B., Brandner K., Truscott K.N., Wiedemann N., Pfanner N., Rehling P. The Assembly Pathway of the Mitochondrial Carrier Translocase Involves Four Preprotein Translocases. Mol. Cell. Biol. 2008;28:4251–4260. doi: 10.1128/MCB.02216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gebert N., Gebert M., Oeljeklaus S., von der Malsburg K., Stroud D.A., Kulawiak B., Wirth C., Zahedi R.P., Dolezal P., Wiese S., et al. Dual Function of Sdh3 in the Respiratory Chain and TIM22 Protein Translocase of the Mitochondrial Inner Membrane. Mol. Cell. 2011;44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 101.Callegari S., Richter F., Chojnacka K., Jans D.C., Lorenzi I., Pacheu-Grau D., Jakobs S., Lenz C., Urlaub H., Dudek J., et al. TIM29 is a subunit of the human carrier translocase required for protein transport. FEBS Lett. 2016;590:4147–4158. doi: 10.1002/1873-3468.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bektas M., Payne S.G., Liu H., Goparaju S., Milstien S., Spiegel S. A novel acylglycerol kinase that produces lysophosphatidic acid modulates cross talk with EGFR in prostate cancer cells. J. Cell Biol. 2005;169:801–811. doi: 10.1083/jcb.200407123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang Y., Stroud D.A., Baker M.J., De Souza D.P., Frazier A.E., Liem M., Tull D., Mathivanan S., McConville M.J., Thorburn D.R., et al. Sengers Syndrome-Associated Mitochondrial Acylglycerol Kinase Is a Subunit of the Human TIM22 Protein Import Complex. Mol. Cell. 2017;67:457–470.e5. doi: 10.1016/j.molcel.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 104.Vukotic M., Nolte H., König T., Saita S., Ananjew M., Krüger M., Tatsuta T., Langer T. Acylglycerol Kinase Mutated in Sengers Syndrome Is a Subunit of the TIM22 Protein Translocase in Mitochondria. Mol. Cell. 2017;67:471–483.e7. doi: 10.1016/j.molcel.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 105.Mayr J.A., Haack T.B., Graf E., Zimmermann F.A., Wieland T., Haberberger B., Superti-Furga A., Kirschner J., Steinmann B., Baumgartner M.R., et al. Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am. J. Hum. Genet. 2012;90:314–320. doi: 10.1016/j.ajhg.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haghighi A., Haack T.B., Atiq M., Mottaghi H., Haghighi-Kakhki H., Bashir R.A., Ahting U., Feichtinger R.G., Mayr J.A., Rötig A., et al. Sengers syndrome: Six novel AGK mutations in seven new families and review of the phenotypic and mutational spectrum of 29 patients. Orphanet J. Rare Dis. 2014;9:119. doi: 10.1186/s13023-014-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pacheu-Grau D., Callegari S., Emperador S., Thompson K., Aich A., Topol S.E., Spencer E.G., McFarland R., Ruiz-Pesini E., Torkamani A., et al. Mutations of the mitochondrial carrier translocase channel subunit TIM22 cause early-onset mitochondrial myopathy. Hum. Mol. Genet. 2018;27:4125–4144. doi: 10.1093/hmg/ddy305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rampelt H., Zerbes R.M., van der Laan M., Pfanner N. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta. 2017;1864:737–746. doi: 10.1016/j.bbamcr.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 109.Wollweber F., von der Malsburg K., van der Laan M. Mitochondrial contact site and cristae organizing system: A central player in membrane shaping and crosstalk. Biochim. Biophys. Acta. 2017;1864:1481–1489. doi: 10.1016/j.bbamcr.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 110.Kozjak-Pavlovic V. The MICOS complex of human mitochondria. Cell Tissue Res. 2017;367:83–93. doi: 10.1007/s00441-016-2433-7. [DOI] [PubMed] [Google Scholar]
- 111.Káldi K., Bauer M.F., Sirrenberg C., Neupert W., Brunner M. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J. 1998;17:1569–1576. doi: 10.1093/emboj/17.6.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davis A.J., Ryan K.R., Jensen R.E. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol. Biol. Cell. 1998;9:2577–2593. doi: 10.1091/mbc.9.9.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Žárský V., Doležal P. Evolution of the Tim17 protein family. Biol. Direct. 2016;11:54. doi: 10.1186/s13062-016-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brandner K., Rehling P., Truscott K.N. The carboxyl-terminal third of the dicarboxylate carrier is crucial for productive association with the inner membrane twin-pore translocase. J. Biol. Chem. 2005;280:6215–6221. doi: 10.1074/jbc.M412269200. [DOI] [PubMed] [Google Scholar]
- 115.Vergnolle M.A.S., Sawney H., Junne T., Dolfini L., Tokatlidis K. A cryptic matrix targeting signal of the yeast ADP/ATP carrier normally inserted by the TIM22 complex is recognized by the TIM23 machinery. Biochem. J. 2005;385:173–180. doi: 10.1042/BJ20040650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yamano K., Ishikawa D., Esaki M., Endo T. The phosphate carrier has an ability to be sorted to either the TIM22 pathway or the TIM23 pathway for its import into yeast mitochondria. J. Biol. Chem. 2005;280:10011–10017. doi: 10.1074/jbc.M413264200. [DOI] [PubMed] [Google Scholar]
- 117.Reinhold R., Kruger V., Meinecke M., Schulz C., Schmidt B., Grunau S.D., Guiard B., Wiedemann N., van der Laan M., Wagner R., et al. The Channel-Forming Sym1 Protein Is Transported by the TIM23 Complex in a Presequence-Independent Manner. Mol. Cell. Biol. 2012;32:5009–5021. doi: 10.1128/MCB.00843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hell K., Neupert W., Stuart R.A. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bonnefoy N., Fiumera H.L., Dujardin G., Fox T.D. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bohnert M., Rehling P., Guiard B., Herrmann J.M., Pfanner N., van der Laan M. Cooperation of Stop-Transfer and Conservative Sorting Mechanisms in Mitochondrial Protein Transport. Curr. Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 121.Stiller S.B., Höpker J., Oeljeklaus S., Schütze C., Schrempp S.G., Vent-Schmidt J., Horvath S.E., Frazier A.E., Gebert N., van der Laan M., et al. Mitochondrial OXA Translocase Plays a Major Role in Biogenesis of Inner-Membrane Proteins. Cell Metab. 2016;23:901–908. doi: 10.1016/j.cmet.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park K., Botelho S.C., Hong J., Osterberg M., Kim H. Dissecting Stop Transfer versus Conservative Sorting Pathways for Mitochondrial Inner Membrane Proteins in Vivo. J. Biol. Chem. 2013;288:1521–1532. doi: 10.1074/jbc.M112.409748. [DOI] [PMC free article] [PubMed] [Google Scholar]