Abstract
Background & Aims
Esophageal adenocarcinoma (EAC) develops from within Barrett’s esophagus (BE) concomitant with gastroesophageal reflux disease (GERD). Wound healing processes and cellular transitions, such as epithelial–mesenchymal transitions, may contribute to the development of BE and the eventual migratory escape of metastatic cancer cells. Herein, we attempt to identify the genes underlying esophageal cellular transitions and their potential regulation by the low pH environments observed in GERD and commonly encountered by escaping cancer cells.
Methods
Small interfering RNA library screening and high-content imaging analysis outlined changes in BE high-grade dysplasia (HGD) and EAC cell morphologies after gene silencing. Gene expression microarray data and low pH exposures studies modeling GERD-associated pulses (pH 4.0, 10 min) and tumor microenvironments (pH 6.0, constant) were used.
Results
Statistical analysis of small interfering RNA screening data defined 207 genes (Z-score >2.0), in 12 distinct morphologic clusters, whose suppression significantly altered BE-HGD cell morphology. The most significant genes in this list included KIF11, RRM2, NUBP2, P66BETA, DUX1, UBE3A, ITGB8, GAS1, GPS1, and PRC1. Guided by gene expression microarray study data, both pulsatile and constant low pH exposures were observed to suppress the expression of GPS1 and RRM2 in a nonoverlapping temporal manner in both BE-HGD and EAC cells, with no changes observed in squamous esophageal cells. Functional studies uncovered that GPS1 and RRM2 contributed to amoeboid and mesenchymal cellular transitions, respectively, as characterized by differential rates of cell motility, pseudopodia formation, and altered expression of the mesenchymal markers vimentin and E-cadherin.
Conclusions
Collectively, we have shown that low pH microenvironments associated with GERD, and tumor invasive edges, can modulate the expression of genes that triggered esophageal cellular transitions potentially critical to colonization and invasion.
Keywords: Barrett’s Esophagus, Esophageal Adenocarcinoma, Gastroesophageal Reflux Disease, Low pH, Tumor Microenvironment, Cellular Transitions, Metastasis, Cell Locomotion
Abbreviations used in this paper: A-T, A minus T; BE, Barrett’s esophagus; COP, constitutive photomorphogenesis; dNTP, deoxyribonucleotide; EAC, esophageal adenocarcinoma; ECM, extracellular matrix; GERD, gastroesophageal reflux disease; HCA, high-content imaging analysis; HGD, high-grade dysplasia; mRNA, messenger RNA; MTT, 3-[4, 5-dimethylthiazol-2-yl] 2, 5-diphenyltetrazolium bromide; PBS, phosphate-buffered saline; RNR, ribonucleotide reductase; siRNA, small interfering RNA
Graphical abstract
Summary.
Cellular transitions and microenvironmental conditions, such as reflux-associated and tumor-associated low pH, underlie Barrett’s esophagus and metastatic processes. Low pH–responsive genes supporting transitions of Barrett’s esophagus–associated dysplastic and adenocarcinoma cells were defined by small interfering RNA library screening and high-content analysis.
The prevalence of esophageal cancer has increased significantly both in Canada and in the United States (2.8%).1, 2, 3, 4 This increase, despite screening programs for preliminary lesions,3 has been attributed to the continued increase in the diagnosis of esophageal adenocarcinoma (EAC) over that of squamous cell carcinoma of the esophagus, whose rates have decreased.4 Furthermore, the poor 5-year survival rates for esophageal cancer in Canada (14%) and the United States (18%) add to the growing need to understand the etiology and develop targeted therapeutics to treat or manage this deadly disease.
EAC is derived from a precursor lesion known as Barrett’s esophagus (BE), which develops at the lower gastroesophageal junction as a consequence of ongoing gastroesophageal reflux disease (GERD). The origins of this metaplasia have been the subject of much debate with no significant consensus of understanding between trans-commitment of squamous cell lineages, cardia-resident stem cells, or stem cells deriving from the esophageal submucosal glands.5, 6, 7, 8, 9 However, recent evidence points toward BE stem cells that may have the potential to migrate from their niche and expand to replace the damaged epithelium through a process of wound healing analogous to epithelial–mesenchymal transitions.10, 11, 12 The development of high-grade dysplasia (HGD) within BE also is associated with localized remodeling of BE glandular structures, likely facilitated by similar cellular transitions. Thus, it may be hypothesized that inflammatory factors or refluxate constituents released upon damage to the esophageal surface epithelium supports the expanding population of BE stem cells and associated HGD through a poorly understood mechanism.
EAC is a highly metastatic cancer, whose invasive potential is partially facilitated by the lack of serosa surrounding the esophagus. Subsequent invasion of the tumor cells into surrounding lymph nodes correlates with poor patient prognosis.13,14 It is suggested that only a fraction of tumor cells acquire the capacity for invasion and dissemination through a separation from the primary tumor, migration, colonization, and metastatic growth at a distant site.15, 16, 17, 18 Dissemination of tumor cells involves the use of dynamic morphologic features to navigate the differing tissue geometries and escape the original tumor before migrating to distant sites. This movement is underlined by a sequence of locomotive cellular extensions and contractions, as well as adhering to and releasing from the local substrates.19 These hijacked developmental transitions such as epithelial–mesenchymal, mesenchymal–epithelial, and amoeboid transition have been found to confer cancer cells with useful attributes promoting invasive, locomotive, and migratory potential in addition to cancer stem cell–like properties necessary in surviving anoikis.20, 21, 22, 23, 24, 25 During the process of tumor escape, cancer cells are simultaneously reacting to, or following signals provided by, chemotactic or haptotactic gradients in the extracellular matrix (ECM) and cell adhesion molecules (integrins and cadherins) expressed on cell surfaces. Many of these signals are provided by the local and distal microenvironments such as gradients of vascular epithelial growth factor induced in response to hypoxia-inducible factor 1A–mediated oxygen-sensing transcriptional programs. Less commonly understood is the role of tissue-specific pH levels as a microenvironmental support for metastasis. However, low extracellular pH, sometimes termed acidosis, is observed in the hypoxic cores of solid tumors, where lengthy paths of diffusion exist. These paths are suggested to drive aggressive phenotypes through increased matrix metalloproteinase production, metastases, and invasion.26, 27, 28, 29 Furthermore, recent evidence has suggested that cancer cells also encounter acidified environments at tumor-stromal borders that correlate with greater levels of invasion.26
Thus, defining regulators of morphologic changes in BE and EAC cells that can support their expansion and metastasis may allow targeted therapies at either end of the esophageal adenocarcinoma sequence. Here, high-throughput small interfering RNA (siRNA) screening in BE-HGD cells, that reflect phenotypes shared by BE, HGD, and EAC, was used in conjunction with high-content imaging analysis (HCA) of cell morphology. This analysis uncovered gene-dependent regulation of epithelial–mesenchymal and amoeboid transitions in BE-HGD and EAC cells. Furthermore, we show that environmental exposures, reflecting GERD and the low pH associated with hypoxic tumor cores, suppress the expression of newly defined regulators of these transitions.
Results
High-Content Imaging of BE-HGD Cell Morphology After siRNA Library Screening
An siRNA screening protocol (Figure 1A) using a cell line representing Barrett’s-associated high-grade dysplasia, CP-D BE-HGD cells, a druggable genome siRNA library consisting of 6033 siRNA pools and HCA of cytoskeletal proteins (F-actin, α-tubulin, and Hoechst) was used as previously described.30 To reduce data dimensionality (8 images per well, 7000 wells, 4 channels), initial analysis and data transformations of screening images was performed using a singular analytic parameter measuring the difference between the overall area of the cell (F-actin) and that of tubulin staining (A minus T [A-T] parameter), thus reflecting extensions, contractions, or bundling associated with cellular transitions. Locally weighted polynomial regression normalization first was performed to account for cell shape changes as a consequence of alterations in cell density that occur as a result of gene loss-mediated cell death, growth inhibition, or regional variations (Figure 1B). Optimal library screening quality was then assessed using the A-T parameter with a Z factor greater than 0.5, defined through the performance, and separation, of negative and positive controls (Figure 1C).
Figure 1.
Screening metrics of high-content and high-throughput imaging analysis of esophageal cell morphology after siRNA druggable genome library screening. (A) Flow diagram of the siRNA library screening process in BE-HGD cells using measurement of morphologic parameters as determined through staining patterns, size, intensity, shape, location of F-actin, α-tubulin, and the nucleus (Hoechst). (B) Locally weighted polynomial regression (lowess) prenormalization of raw screening data, using the A-T parameter, which tracks overall cellular dynamics, controlled for changes in cell shape associated with variations in cell densities. Cell counts were determined by nuclear staining of images. Red dots and green dots represent negative and positive controls from screening data, respectively. (C) Plate normalized (to negative controls) values of A-T parameter showing significant separation of negative and positive controls throughout the screening plates. siNT, control nontargeting siRNA pool; siPOS, positive control siRNA pool targeting GATA6.
Statistical Z-score ranking of the normalized A-T parameter defined 207 siRNA targeted genes affecting cell morphology independent of cell density at a Z score >2 (Table 1). Notably, genes within the top 10 of this list whose suppression resulted in altered morphologies included KIF11, RRM2, NUBP2, P66BETA, DUX1, UBE3A, ITGB8, GAS1, GPS1, SERPINE, and PRC1. Gene Ontology analysis of the 207 genes ranked by their A-T Z-scores (Z > 2) highlighted an enrichment (13-fold) of genes associated with mitotic cell-cycle control and transitions specifically in negative regulation of mitotic cell cycle (Gene Ontology: 0045930) in keeping with the relationship between mitosis, cytokinesis, and changes in cell morphology as regulated by, for example, KIF11 and PRC1 (Table 2). Multiparameter analysis measuring discrete changes (N = 32 distinct morphologic parameters) in overall cell and nuclear polarity (level or roundness), nuclear positions, distances between membrane and nucleus, intensity of tubulin or F-actin staining, among many others, was used to project the overall functional effect of gene loss through changes in cell morphology. Hierarchical clustering of genes, as initially defined by A-T parameter (207 genes Z > 2), with multiple morphologic parameters next was used to study the relationships between changes in cell shape/morphology and specific gene sets (Figure 2A). This multiparameter HCA data resulted in 12 distinct gene–morphology clusters, as classified by the HCA software (Figure 2A and Table 3). Illustrative examples of the morphologic features associated with silencing of genes within these clusters are shown in Figure 2C and expanded in Figure 3, and show the scale and dynamics of changes in cell morphology.
Table 1.
siRNA-Targeted Genes Affecting the A-T Parameter as Ranked by Z Score
| Gene ID | Entrez gene | Z-score | Gene ID | Entrez gene | Z-Score | Gene ID | Entrez gene | Z-score |
|---|---|---|---|---|---|---|---|---|
| KIF11 | 3832 | 7.31 | XRCC2 | 7516 | 3.47 | CD24 | 934 | 3.00 |
| RRM2 | 6241 | 5.54 | WNT7B | 7477 | 3.36 | CSTF3 | 1479 | 2.99 |
| NUBP2 | 10101 | 5.06 | BRE | 9577 | 3.31 | TFRC | 7037 | 2.98 |
| P66BETA | 57459 | 4.91 | KCNQ4 | 9132 | 3.27 | TNNT2 | 7139 | 2.98 |
| DUX1 | 26584 | 4.83 | HOXD3 | 3232 | 3.24 | AQP8 | 343 | 2.97 |
| UBE3A | 7337 | 4.34 | TRIM32 | 22954 | 3.19 | HOXB13 | 10481 | 2.97 |
| ITGB8 | 3696 | 4.31 | CYLD | 1540 | 3.18 | HMGN4 | 10473 | 2.95 |
| GAS1 | 2619 | 4.02 | PLXNA1 | 5361 | 3.13 | RNF144 | 9781 | 2.93 |
| GPS1 | 2873 | 4.02 | CDC25B | 994 | 3.13 | FSCN2 | 25794 | 2.92 |
| SERPINI1 | 5274 | 3.98 | CPOX | 1371 | 3.13 | DPF2 | 5977 | 2.91 |
| PRC1 | 9055 | 3.92 | SERPINF1 | 5176 | 3.12 | ZFP64 | 55734 | 2.90 |
| RNF32 | 140545 | 3.88 | ACOX2 | 8309 | 3.11 | DNMT3B | 1789 | 2.89 |
| FBXO32 | 114907 | 3.73 | GBP1 | 2633 | 3.10 | FGA | 2243 | 2.89 |
| MTVR1 | 23625 | 3.66 | LHB | 3972 | 3.09 | NFKBIB | 4793 | 2.88 |
| HMGN1 | 3150 | 3.64 | RDH8 | 50700 | 3.08 | USP11 | 8237 | 2.86 |
| CASP8AP2 | 9994 | 3.62 | GABPA | 2551 | 3.08 | CD79A | 973 | 2.86 |
| PRDM8 | 56978 | 3.57 | TMEM4 | 10330 | 3.06 | SALL3 | 27164 | 2.85 |
| SPRY1 | 10252 | 3.54 | CRLF2 | 64109 | 3.04 | C9ORF100 | 84904 | 2.84 |
| NOL1 | 4839 | 3.52 | TM4SF4 | 7104 | 3.01 | AGL | 178 | 2.83 |
| RAG1 | 5896 | 3.48 | HOXA4 | 3201 | 3.00 | NR1D2 | 9975 | 2.83 |
| Gene ID | Entrez gene | Z-score | Gene ID | Entrez gene | Z-score | |||
|---|---|---|---|---|---|---|---|---|
| CDC6 | 990 | 2.82 | EPB42 | 2038 | 2.61 | |||
| DIABLO | 56616 | 2.77 | RING1 | 6015 | 2.60 | |||
| KRT18 | 3875 | 2.76 | HOXA7 | 3204 | 2.60 | |||
| BTG4 | 54766 | 2.76 | ACY1 | 95 | 2.58 | |||
| FRAG1 | 27315 | 2.74 | HSF2 | 3298 | 2.58 | |||
| AQP3 | 360 | 2.74 | PSMB4 | 5692 | 2.56 | |||
| NCOA7 | 135112 | 2.72 | MLL | 4297 | 2.55 | |||
| TCOF1 | 6949 | 2.72 | RSU1 | 6251 | 2.54 | |||
| DAXX | 1616 | 2.72 | TNC | 3371 | 2.54 | |||
| LGALS3BP | 3959 | 2.69 | ACADVL | 37 | 2.52 | |||
| IL27RA | 9466 | 2.68 | CDC42EP3 | 10602 | 2.50 | |||
| ARHGDIG | 398 | 2.68 | SPINK5 | 11005 | 2.50 | |||
| COL18A1 | 80781 | 2.68 | ABCC3 | 8714 | 2.50 | |||
| SERPINI2 | 5276 | 2.67 | XPMC2H | 57109 | 2.49 | |||
| TNFRSF13B | 23495 | 2.67 | CTGF | 1490 | 2.48 | |||
| MDS1 | 4197 | 2.66 | NGFRAP1 | 27018 | 2.47 | |||
| LRP8 | 7804 | 2.62 | NR4A1 | 3164 | 2.47 | |||
| SMARCA1 | 6594 | 2.62 | AXIN1 | 8312 | 2.46 | |||
| TNFSF11 | 8600 | 2.62 | CCNB3 | 85417 | 2.45 | |||
| DBI | 1622 | 2.61 | GPR135 | 64582 | 2.44 |
Table 2.
Over-represented Gene Ontologies Associated With Z Score–Ranked Screening Results
| PANTHER GO-Slim Biological process | GO code | Fold enrichment | Raw P valuea | FDR |
|---|---|---|---|---|
| Negative regulation of mitotic cell cycle | GO:0045930 | 13.82 | 2.97E-04 | 2.81E-02 |
| JAK-STAT cascade | GO:0007259 | 10.71 | 7.21E-04 | 4.63E-02 |
| STAT cascade | GO:0097696 | 10.71 | 7.21E-04 | 4.47E-02 |
| Mitotic cell-cycle phase transition | GO:0044772 | 8.78 | 3.64E-04 | 2.97E-02 |
| Regulation of cell-cycle process | GO:0010564 | 8.37 | 4.47E-04 | 3.21E-02 |
| Regulation of cell proliferation | GO:0042127 | 6.7 | 3.74E-04 | 2.92E-02 |
| Positive regulation of cellular process | GO:0048522 | 5.95 | 2.42E-04 | 2.56E-02 |
| Mitotic nuclear division | GO:0140014 | 4.61 | 2.03E-04 | 2.61E-02 |
| Mitotic cell cycle | GO:0000278 | 4.53 | 2.32E-04 | 2.78E-02 |
| Mitotic cell-cycle process | GO:1903047 | 4.53 | 2.32E-04 | 2.61E-02 |
FDR, false discovery rate; GO, Gene Ontology; JAK, Janus Kinase; PANTHER, Protein ANalysis THrough Evolutionary Relationships; STAT, Signal transducer and activator of transcription proteins.
Fisher exact test.
Figure 2.
Hierarchical clustering of high-content and high-throughput morphologic parameters with siRNA-targeted genes defines gene–morphology clusters. (A) Hierarchical clustering of both siRNA-targeted genes (side, vertical), at A-T parameter Z > 2 as the cut-off value, and associated cell morphologic parameters (n = 32, top horizontal) measured by In Cell Investigator suite, highlights 12 distinct morphologic clusters. (B) Z score bar chart of genes (A-T parameter Z ≤ 2) ordered by location within hierarchical clusters. (C) Zoomed screening images of cell shapes from selected siRNA-targeted genes (KIF11, GPS1, RRM2, PRC1, CDC6, CTGF, BTG4, NOL1, AXIN1, MYO9B, UBE3A, PPP1R10) clusters. Control nontargeting siRNA (siNT) and GATA6 represent negative and positive screening controls, respectively. Green and red pseudocolorization represent α-tubulin and F-actin staining, respectively. Nc, nucleus.
Table 3.
Gene Membership of Cluster From Hierarchical Clustering in Figure 2
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | Cluster 6 | Cluster 7 | Cluster 8 | Cluster 9 | Cluster 10 | Cluster 11 | Cluster 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SERPINI1 | NUBP2 | NGFRAP1 | CAMKIINALPHA | TMEM4 | APOC4 | BTG4a | NOL1a | LRP8 | NFKBIB | CPOX | LBP |
| WNT7B | GABPA | RRM2a | ACOX2 | ACADVL | TNNT2 | ARHGDIG | NCOA7 | CD79A | IL27RA | HMGN4 | TWIST1 |
| KIF11a | HOXB13 | P66BETA | AQP3 | CDC42EP3 | KCNQ4 | PME-1 | CD24 | SPINT2 | POLR2B | PAFAH1B3 | ACP2 |
| CASP8AP2 | ZFP64 | DUX1 | GAS1 | CDC6a | SUPT6H | SKP2 | DIABLO | LGALS3BP | GBP1 | PRDM8 | ZIC3 |
| DBI | HSF2 | ITGB8 | CCND1 | GLI2 | SRP72 | PSMB4 | ABCC3 | LHB | FBXO32 | RAG2 | ALPL |
| TNFSF11 | PHF1 | SPRY1 | RNF32 | DACH2 | RSU1 | GGT2 | CES1 | SPINK5 | AQP8 | FGD1 | CPT2 |
| FGA | GPS1a | HOXA7 | KRT18 | MTA1 | RDH8 | ACADSB | PLXNA1 | UNC5B | BIA2 | XPMC2H | |
| GSTT2 | XRCC2 | ACY1 | PRC1a | CTGFa | COL18A1 | THRAP5 | TCOF1 | CSTF3 | |||
| C9ORF100 | CCNB1 | C6ORF108 | AGL | DAXX | TRIM32 | DNMT3B | NR1D2 | PCOLCE | |||
| DPEP3 | TOP2A | HOXA4 | MTVR1 | LCHN | EPB42 | OLIG2 | |||||
| TAF6L | TNFRSF21 | MDS1 | CRLF2 | CTSC | AQP2 | DRPLA | |||||
| FSCN2 | MLL | SERPINF1 | HOXD3 | IFNA7 | ZNF74 | ||||||
| SALL3 | AVP | CSH1 | SERPINI2 | TNC | |||||||
| NRGN | FRAG1 | BRE | NCF1 | PPP1R10a | |||||||
| ZNF160 | PARC | CCNB3 | STAT5A | HSPB2 | |||||||
| ITGB1BP2 | SMURF2 | OAS3 | MMP25 | NRL | |||||||
| HMGN1 | AXIN1a | TCF12 | DELGEF | ||||||||
| GSPT1 | SMARCA1 | RASAL2 | UBE3Aa | ||||||||
| TFRC | RNF144 | FECH | USP11 | ||||||||
| GML | GPR135 | GJB2 | DPF2 | ||||||||
| PRDM13 | NOS2A | OXT | |||||||||
| GRAP | NR4A3 | CDC42 | |||||||||
| CYLD | APOC1 | IGHG3 | |||||||||
| MK-STYX | ILF2 | CPN2 | |||||||||
| XCL2 | P4HB | BUB3 | |||||||||
| TNFRSF13B | TM4SF4 | IGHG4 | |||||||||
| HLA-B | MYO9Ba | LOC94431 | |||||||||
| GPR142 | CTSK | NR5A2 | |||||||||
| CLTCL1 | FLJ10665 | ARPP-19 | |||||||||
| CMRF35 | CDYL | RAB38 | |||||||||
| RAG1 | SUFU | FGD6 | |||||||||
| RXRA | TFAP2BL1 | NPPA | |||||||||
| SCNN1A | |||||||||||
| SERPING1 | |||||||||||
| TLE4 | |||||||||||
| RING1 | |||||||||||
| NR4A1 | |||||||||||
| DNAI1 | |||||||||||
| NR2F6 | |||||||||||
| TIEG2 | |||||||||||
| APOA4 |
Figure 3.
Expanded screening images of BE-HGD cells after siRNA-mediated silencing of genes selected from preceding gene–morphology clustering. Images represent of each gene–morphology cluster as defined by hierarchical clustering of genes statistically significant at the Z ≤ 2 cut-off value in the A-T parameter as detailed in Figure 2. Green and red pseudocolorization represent α-tubulin and F-actin staining, respectively. Control nontargeting siRNA (siNT) and GATA6 represent negative and positive screening controls, respectively.
Gene Silencing-Dependent Changes in Cell Morphology Are Associated With Altered Cell Migration
The relationships between the cell phenotypes observed after siRNA-mediated gene suppression were examined through verification experiments, as well as their ability to migrate in scratch wound assays. The genes RRM2, ITGB8, GPS1, NOL1, MYO9B, and SPRY1 were selected to represent a cross-section of the differing morphologic clusters observed in preceding experiments. The silencing of these genes in BE-HGD cells, using alternate siRNA pool builds, resulted in similar changes in cell shape to that observed in the original screen (Figure 4A). HCA analysis of these images, using analogous morphologic parameters, resulted in similar levels of significant alteration to that observed in the original siRNA screening data (Table 4). Scratch-wound assays were next performed at 72 hours after transfection followed by measurement of cell migration into the scratched area at 96 hours. Silencing GPS1 and SPRY1 expression resulted in significant increases in the rate of scratch-wound closure (Figure 4B and C), which was not associated with changes in either cell viability (Figure 4D) or exact cell counts (Figure 4E). Suppression of MOY9B similarly resulted in significantly increased scratch-wound closure, despite a significant decrease in cell number and proliferative potential (Figure 4). Comparatively, silencing of RRM2 and ITGB8 resulted in significant effects on proliferative potential (Figure 4D and E), which consequently negatively affected scratch-wound assays. Thus, alterations in morphologic parameters translated into altered cell migration characteristics, validating the high-throughput imaging approaches used in this study.
Figure 4.
Changes in cellular morphology impacts cellular migration. (A) Changes in BE-HGD cell morphology as indicated by screening data were confirmed in independent experiments (n = 3) using an alternate siRNA format to that of the original screen (siOn-target plus). (B) Scratch-wound assays performed in confluent cells (n = 6) transfected with siRNAs targeting RRM2, ITGB8, GPS1, NOL1, MYO9B, and SPRY1. (C) Representative images of significantly affected wells. (D) Cell count and (E) cell viability after silencing of RRM2, ITGB8, GPS1, NOL1, MYO9B, and SPRY1. Negative, negative control growth media without fetal calf serum; Positive, positive control, epidermal growth factors in culture media. ∗P < .01, ∗∗P < .001, and ∗∗∗P < .0001. NS, not significant in Student t testing.
Table 4.
Summarized Table of HCA Parameter Results After Imaging of Separate Validation Experiments Using Alternate siRNA Pools in BE-HGD Cells
| Parameter | Resting | siNT | siRRM2 | siITGB8 | siCSN1 | siNOL1 | siMYO9B | siSPRY1 |
|---|---|---|---|---|---|---|---|---|
| Nuclear displacementa | 0.638 ± 0.058 | 0.68 ± 0.078 | 1.539 ± 0.066 | 0.899 ± 0.062 | 0.837 ± 0.049 | 0.974 ± 0.038 | 0.901 ± 0.086 | 0.92 ± 0.041 |
| Cell area (α-tubulin)b | 761.65 ± 103.71 | 839.14 ± 83.77 | 2282.11 ± 46.43 | 1034.36 ± 56.03 | 916.07 ± 29.99 | 1060.9 ± 53.87 | 963.23 ± 11.25 | 1098.4 ± 49.62 |
| Organelle area (F-actin)c | 604.89 ± 66.01 | 659.01 ± 55.14 | 1996.41 ± 65.51 | 799.54 ± 34.57 | 651.29 ± 7.94 | 810.17 ± 2.81 | 716.56 ± 10.94 | 865.19 ± 44.75 |
| Cell area – organelle aread | 156.75 ± 38.25 | 180.12 ± 29.75 | 285.70 ± 35.48 | 234.82 ± 21.51 | 264.78 ± 36.19 | 250.79± 1.47 | 246.67 ± 0.38 | 233.25 ± 35.14 |
| Cell counte | 3907.5 ± 720.62 | 3523.1 ± 654.06 | 472.5 ± 68.98 | 2043 ± 337.36 | 2566.667 ± 186.73 | 1850.17 ± 35.26 | 1526.83 ± 131.1 | 1739 ± 93.86 |
Distance between the nucleus and cell center of gravity divided by the gyration radius.
Tubulin area of the identified cell body.
Actin area of the identified cell body.
Area between the actin and tubulin.
Cell number calculated by nuclear stain.
Exposure of Esophageal Cells to Low pH Environments, Replicating GERD-Associated and Tumor Cores, Suppresses GPS1 and RRM2 Levels
Acidic microenvironments are commonly associated with hypoxic tumor cores, invasive tumor edges, and, obviously, GERD.9,12,26 Finally, acidic environments and exposures have been observed to induce epithelial–mesenchymal phenotypes in a variety of cell types including BE cells.12 Therefore, we next examined whether low pH exposure could alter the expression of genes that impact cellular morphologies, as defined earlier by HCA of siRNA library screening data. Using previously published gene expression microarray analysis data of EAC cells exposed to low pH,31 the expression of the 207 significant siRNA targets from the earlier-described screening data was investigated. GPS1 and RRM2 were selected through this investigation because genes with the highest cell morphology Z scores (within top 10) (Table 4) that were responsive to low pH and, as noted in preceding experiments, resulted in distinctive cell shapes when gene-silenced. This analysis suggested that both GPS1 and RRM2 expression may be suppressed by low pH exposure in SKGT4 EAC cells (Figure 5A). Subsequent validation in EAC cells exposed to continuous low pH 6.5, as used in the original gene expression microarray study,31 showed an immediate decrease in GPS1 messenger RNA (mRNA) levels that persisted over 2 hours, before returning to basal levels at 4 hours (Figure 5B). Compared with GPS1 levels, suppression of RRM2 mRNA in response to continuous low pH 6.5 exposure was delayed until 4 hours, and thus did not overlap with that of GPS1 (Figure 5B). Simulated GERD insults using pulsatile exposure of BE cells to pH 4.5 (10 min)32 followed by return to a normal pH of 7.4, similarly resulted in an immediate decrease in GPS1 mRNA levels that persisted for 5 hours (Figure 5C). Again, a decrease in RRM2 levels occurred in response to a pulsatile pH level of 4.5, which did not overlap with the initial loss of GPS1. Thus, both GPS1 and RRM2 levels may be suppressed in response to environmental conditions common to both hypoxic tumor cores and BE/EAC-associated GERD exposure.
Figure 5.

Low pH exposure, simulating GERD and tumor-associated environments, suppresses GPS1 and RRM2 expression in a temporal fashion. (A) Matrix heatmap of GPS1 and RRM2 expression in previously published gene expression microarray data of low pH-mediated signaling in SKGT4 cells. (B) Independent validation of GPS1 and RRM2 expression (n = 3) levels after continuous exposure to pH 6.5 in SKGT4 cells, (C) simulating a hypoxic tumor core with low pH over 4 hours or pulsatile exposure of BE-HGD cells simulating a reflux event (pH 4.5 for 10 min) followed by measurement over 8 hours. ∗P < .01, ∗∗P < .001, and ∗∗∗P < .0001 in Mann–Whitney nonparametric testing. Rel., relative.
GPS1 Suppression Promotes BE and EAC Cell Migration Through Epithelial–Amoeboid–Like Transition
GPS1, also known as COP9 signalosome complex 1 (CSN1) or constitutive photomorphogenesis (COP)S1, is a component of the COP9 signalosome regulating protein NEDDylation (the binding of a neural precursor cell expressed developmentally down-regulated protein 8 (NEDD8)), most notably, of cullin-RING ligases, thus controlling protein ubiquitination and impacting a diverse array of cellular events, including cell cycles, through ubiquitin-mediated protein turnover.33,34 β-catenin levels, controlled by cycles of ubiquitination, are centrally implicated in the metastatic phenotypes of many cancer types through catenin/cadherin complexes.35, 36, 37, 38, 39 Figure 6A shows the morphologic parameter data at the individual per cell level, highlighting the consistency of the overall shape change in GPS1-silenced cells. Under flow cytometric cell-cycle analysis, no expansion of GO/G1 or sub-G1 populations, or any notable global changes in cell-cycle distribution, was noted in GPS1-silenced CP-D BE-HGD cells when compared with the nontargeting siRNA-transfected cells (Figure 6B). However, silencing GPS1 expression in both BE-HGD and EAC cells resulted in stabilization of β-catenin protein expression in esophageal cells at 72 and 96 hours after siRNA transfection (Figure 6C and E). Interestingly, GPS1 levels were observed to accumulate significantly in control resting cells over 72 hours as cell culture vessel density increased and cells became nonmotile (Figure 6D and F).
Figure 6.
GPS1 silencing results in consistent cell phenotype and β-catenin accumulation without affecting the cell cycle. Individual cell values from (Ai) morphologic A-T and (Aii) organelle inclusion/cell intensity parameters in siGPS1-transfected (n = 1059 individual cells) and nontargeting siRNA (n = 2818) wells from HCA data from an independent validation study. (B) Flow cytometric analysis of cell-cycle dynamics after silencing of GPS1 shows no changes from nontargeting control transfected cells. (C and D) Expression levels of GPS1, β-catenin, and actin (loading control) by Western blot after siRNA-mediated silencing of GPS1 in (C) BE-HGD CP-D cells and (D) SKGT4 EAC cells over 96 hours. (E and F) Densitometry of proteins normalized to actin loading control levels from 3 replicate experiments. ∗P < .01, ∗∗P < .001, and ∗∗∗P < .0001 in Student t testing.
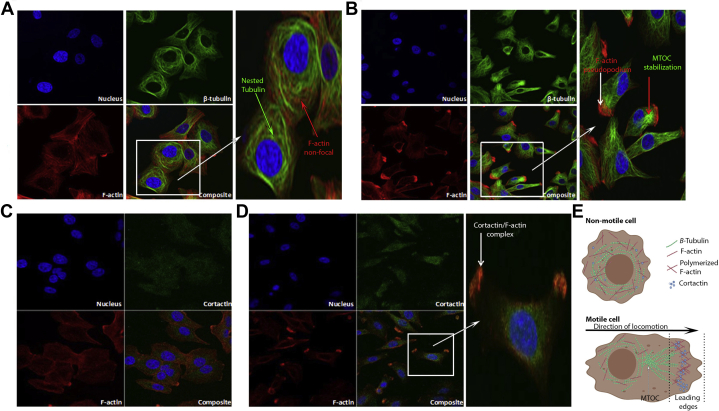
Cell motility is achieved through changes in actin- and tubulin-mediated structural alterations in cell morphology, leading to F-actin protrusions such as those observed in preceding validation experiments (Figure 4A). Confocal microscopy thus was performed upon resting and GPS1 silenced BE-HGD cells to further investigate actin, tubulin, and cortactin dynamics that underpin cellular locomotion and invasive phenotypes. Nested tubulin staining around the nucleus in addition to well-dispersed multiprocess actin filament staining was observed in nontargeting siRNA-transfected cells (Figure 7A). However, in GPS1-silenced cells, clear F-actin polymerization was observed in large foci that extended beyond the staining pattern of tubulin, which had retracted and stabilized to form a microtubule organizing center behind the F-actin–rich leading edge of the pseudopodium (Figure 7A). Next, the ability of these pseudopodia to facilitate locomotive and invasive phenotypes was validated further through localization of the actin–nucleation–promoting factor, cortactin in resting and GPS1-silenced cells. Cortactin (green) punctate cellular staining was dispersed throughout resting cells with no distinct colocalization with F-actin (red) (Figure 7C). Strikingly, colocalization was observed between F-actin and cortactin in the pseudopodia of GPS1-silenced cells identified by the pattern of the yellow punctate staining in the cellular protrusions (Figure 7D). Comparatively, the dispersed nonpolymerized F-actin (red) and cortactin (green) staining in the cytoplasmic/perinuclear region of GPS1-silenced cells did not form any yellow puncta indicative of colocalization. This finding further verifies that the pattern of staining specifically in the pseudopodia is indicative of interaction at the point where the leading locomotive, sensing, or invasive cellular edge had formed (Figure 7D). The overall pattern of F-actin, β-tubulin, and cortactin staining is suggestive of an amoeboid-like movement and transition of BE-HGD cells, representing a midway point between BE and EAC.
Figure 7.
(A and B) Cortactin-positive pseudopodia formation is increased in GPS1-silenced BE-HGD cells. Confocal imaging of nucleus, β-tubulin, and F-actin in (A) control nontargeting or (B) GPS1-silenced cells. Expanded panels show zoomed composite image of all 4 panels with arrows highlighting nested tubulin, microtubule organizing center (MTOC) formation, nonfocal F-actin, and polymerized F-actin in pseudopodia extensions. (C and D) Confocal imaging of F-actin and cortactin immunostaining in (C) control nontargeting siRNA-transfected and (D) GPS1-silenced cells. Expanded panel shows zoomed composite image of all 4 panels from GPS1-silenced cells; arrow shows co-localization of F-actin with cortactin in pseudopodia-like protrusions. (E) Graphic representation (created with BioRender.com) of nonmotile and motile cells where the MTOC is localized behind a leading-edge protrusion filled with polymerized F-actin and colocalized cortactin.
Amoeboid-like transitions are associated commonly with increased cellular motility. Live cell imaging of GPS1-silenced BE and EAC cell lines showed significantly increased motility over that of nontargeting siRNA-transfected cells. Cell tracking analysis performed in CP-D BE-HGD (Figure 8A–D) and SKGT4 EAC cells (Figure 8E–H) showed that GPS1-silenced cells showed significantly increased velocity (Figure 8C and G), increased accumulated distance (distance inclusive of changes in direction) (Figure 8D–H), and Euclidean distances (distance in a straight line), supportive of an amoeboid transition and motility in response to GPS1 suppression.
Figure 8.
GPS1 suppresses cellular locomotive velocity and accumulated distances in BE-HGD and EAC cells. Individual live cell tracking data in BE-HGD CP-D cells from live imaging of (A) control nontargeting (siNT) and (B) GPS1-silenced (siGPS1) cells. (C) Velocity and (D) accumulated distances of BE-HGD CP-D cells from live imaging of control transfected (siNT) or cells after GPS1 silencing. Individual cell tracking data of SKGT4 EAC cells transfected with (E) control nontargeting (siNT) or (F) GPS1 targeting (siGPS1) siRNAs. (G) Velocity and (H) accumulated distances of SKGT4 EAC cells after GPS1 silencing or nontargeting transfection. ∗∗∗∗P < .00001 in Student t testing.
Increased Expression of the RRM2B Subunit of the Ribonucleotide Reductase Holoenzyme After Low pH Exposure
The ribonucleotide reductase (RNR) enzyme that catalyzes the formation of ribonucleotides and deoxyribonucleotides is composed of 2 subunits formed through the association of the RRM1 subunit with either the RRM2 or RRM2B partner subunit.40 In normoxia, the RRM1/RRM2 variant is preferred to the RRM2B partnership that becomes predominant under hypoxic conditions where it sustains survival, maintains DNA replication of cancer cells, and avoids the accumulation of DNA damage.41 Constant low pH exposure of EAC cells, replicating hypoxic tumor cores and acidic invasive edges, was found to result in significant induction of RRM2B expression at the same time as RRM2 expression was reduced (4 hours post exposure) in replicated experiments (Figure 9B). Similarly, this RRM2 to RRM2B switching occurred in BE-HGD cells exposed to pulsatile pH 4.5 exposure (replicating GERD), reaching maximal levels of RRM2B induction at 4 hours postpulse (Figure 9C). Thus, GERD constituents such as gastric acid suppress RRM2 expression in a cell type–specific manner, with potential ramifications for bypassing DNA replication stress in BE/EAC cells types, and for processes such as wound healing that underlie metastasis and the development of BE.
Figure 9.

Suppression of RRM2 levels in response to low pH exposure is mitigated by induction of the RRM2B subunit. (A) Gene expression microarray (GEM) heatmap of RNR subunit proteins RRM1, RRM2, and RRM2B (no GEM probe on platform) expression in SKGT4 cells exposed to low pH 6.5 exposure over time (GATA6 and SOX2 are provided as validated comparators). (B and C) Independent validation of RRM1, RRM2, and RRM2B expression after exposure of EAC cells to (B) continuous (pH 6.5) simulating tumor hypoxic cores, or (C) BE-HGD cells to pulsatile low pH (pH 4.5, 10 min) simulating reflux exposure in replicate experiments (n = 3). ∗P < .01 and ∗∗∗P < .0001 in Mann–Whitney testing. FC, fold change; Rel., relative.
RRM2 Silencing Results in Epithelial–Mesenchymal Transition
The striking elongations or polarizations observed in RRM2-silenced BE-HGD cells (Figures 2 and 3) are specific to the RRM2 subunit with no such phenotypes observed in siRNA screening data of RRM1 and RRM2B knockdown (Figure 10). Individual cell level screening data of A-T morphologic and cell gyration radius parameters show the strength and significance of the observed changes in RRM2-silenced BE-HGD cell morphology throughout the entire cell population (Figure 11A and B). However, clear differences in cell number were observed between nontargeting siRNA control wells and siRRM2-transfected wells, indicative of changes in cell proliferation or apoptosis resulting from RRM2 silencing. Under flow cytometric cell-cycle analysis, RRM2 silencing resulted in decreased G0/G1 cell populations (55% ± 2.6%), increased S phase (10.2% ± 1.8%), and increased sub-G1 phase (31% ± 2.6%) cell populations in comparison with nontargeting siRNA-transfected EAC cells in flow cytometric analyses (G0/G1, 80.3% ± 2.2%; S phase, 2.4% ± 0.4%; sub-G1, 7.9% ± 1.7%) indicative of some cell-cycle arrest and apoptosis (Figure 11C). However, a significant surviving cell population showed obvious morphologic changes irrespective of cell-cycle arrest or apoptosis with significant elongation, centrally located nucleus, and apparent F-actin foci suggestive of epithelial–mesenchymal transition (Figures 10 and 11A). Tracking under live cell imaging highlighted distinct elongations and extensions directing BE-HGD cell movement in RRM2-silenced cells in addition to trailing and/or tethering of the hindmost foot/extension. No significant distances were traveled, however, by RRM2-silenced BE-HGD or EAC cells (Figure 11D and E). The epithelial–mesenchymal transition markers E-cadherin and vimentin were next examined in RRM2-silenced BE-HGD and EAC cells, showing decreased E-cadherin and increased vimentin expression when compared with nontargeting control-transfected cells at 96 hours, consistent with the development of a mesenchymal phenotype (Figure 11F). Thus, changes in RRM2 levels may underlie epithelial–mesenchymal phenotypes and transitions that have been shown to contribute to metastasis, genomic instabilities, and aneuploidy, common events in esophageal adenocarcinogenesis.42
Figure 10.
Polarized phenotype is specific to RRM2-silenced BE-HGD cells. Representative images (n = 8) of siRRM1-, siRRM2-, and siRRM2B-transfected wells highlighting the lack of cell polarizations in either RRM1- or RRM2B-silenced BE-HGD cells. Actin, red; tubulin, green; nucleus, blue.
Figure 11.
(A and B) Epithelial–mesenchymal transition occurs in a population of apoptotic-resistant cells after RRM2 silencing. Individual values from (A) morphologic A-T and (B) cell gyration radius parameters in siRRM2-transfected (n = 1059 individual BE-HGD cells) and nontargeting siRNA (n = 2818) wells (inset shows cell morphologies of siNT- or siRRM2-transfected cells). (C) Flow cytometric analysis of cell-cycle distribution in RRM2-silenced and nontargeting transfected (siNT) cells showing decreased cell proliferation (G0/G1), increased apoptosis (sub-G1), and increased S phase in RRM2-silenced BE-HGD cells. (D and E) Accumulated (D) distance and (E) velocity measured from live imaging of BE-HGD cells transfected with control (siNT) or RRM2 (siRRM2) targeting siRNA pools. (F) Expression of RRM2 E-cadherin, vimentin (epithelial-mesenchymal transition markers), and β-actin (loading control) proteins by Western blot in BE-HGD (CP-D) and EAC (SKGT4) cells at 96 hours after transfection with control (siNT) or RRM2 (siRRM2) targeting siRNA pools. All experiments, except screening data, were performed N = 3, representative blots are shown. ∗P < .01, ∗∗P < .001, ∗∗∗P < .0001 in Student t test.
Discussion
By using siRNA library screening, in conjunction with high content imaging analysis of cell morphology, this study uncovered genes whose expression levels selectively dictate modes of BE and EAC shape change and cell motility, through facilitating cellular transitions such as epithelial-mesenchymal and amoeboid. We further showed that exposure of esophageal cells to low pH, simulating GERD and tumor microenvironments, suppressed the expression of newly defined regulators of amoeboid and mesenchymal transitions, GPS1 and RRM2, respectively. Thus, microenvironmental conditions may support cellular transitions crucial in the metaplastic transformation observed during BE and in the metastasis of EAC cells in later stages of esophageal adenocarcinogenesis.
EAC is an aggressive cancer associated with distant metastasis in the lungs and liver. Numerous models propose native microenvironmental surfaces, such as collagen, muscle fibers, and both inner and outer endothelial surface as cancer disseminating roadways.22 Path-clearing alterations to normal and tumor tissue architectures, such as degradation of the ECM by epithelial–mesenchymal transition-associated cancer cell–secreted matrix metalloproteinases, also are suggested to create tracts on which cancer cells may navigate these roadways. Conversely, path-finding amoeboid-like movement associated with amoeboid transitions are suggested to allow cancer cells to navigate through ECM fibers independent of protease activity,20 adapting morphologically to pre-existing spaces. In a recent rodent model of esophageal reflux disease, involving surgical attachment of the jejunum to the esophagus, it was discovered that neoglandular epithelium distal from the surgical site was derived from jejunal stem cells rather than esophageal resident progenitors.10 Furthermore, progressive expansion of the neoglandular epithelium into the reflux-induced ulcer bed was observed. These findings suggested that invasive and wound healing processes, induced by ulceration, promoted the local expansion of the glandular lesion but also facilitated distal colonization away from the site of surgical attachment. Critically, epithelial to mesenchymal transition-mediated wound healing processes were observed to play a significant role in these colonizations. Thus, our identification of 207 genes underlying altered esophageal cell morphologies that may impact mesenchymal and amoeboid transitions in BE-HGD cells, or additional as yet undefined transitions, may be critical in understanding not only the metastatic process, but the mechanisms through which BE colonizes the lower esophagus.
Microenvironments of differing pH levels are encountered as cancer cells move through tissues. A decrease in extracellular pH is a common characteristic of hypoxic tumor cores26,43, 44, 45, 46 and was also recently observed at tumor–stromal boundaries where it facilitates tissue invasion.27 Furthermore, the reflux-mediated colonization of rat esophagus by jejunal cells, and subsequent in vitro evidence of a GERD-induced mesenchymal phenotype in BE cell lines, are suggestive of the ability of low pH exposure to promote BE stem cell colonization.10,12 In vivo low pH microenvironments associated with hypoxic conditions, GERD exposure, or at metastatic edges, are dynamic environments that may have both immediate and cumulative effects difficult to mimic in vitro. Cells undergoing dynamic transitions between epithelial, mesenchymal, ameboid, and other transitions necessary for colonization, likely encounter differing pH environments at differing points of transition. Its is likely that these pH challenges may result in transition-stage–dependent effects adding to the overall complexity of the relationships between microenvironment and cell morphology/transitions. In our study we selected pH challenges consistent with tissues exposures that would allow significant cell signaling effects but avoid levels of cell death in cell line culture models, thus allowing the investigation and exploration of these relationships. Furthermore, in esophageal cells and associated microenvironments, challenged by tissue and environmental pH changes, the genes underlying these processes are ill-defined. Thus, our study focused on genes such as GPS1 and RRM2, whose expression in esophageal cells was affected by low pH exposure, and whose suppression resulted in altered cellular transitions.
Silencing of GPS1 in esophageal CP-D BE-HGD cells resulted in an amoeboid-like morphology, a loss of cell polarity, significantly faster cellular locomotion, and cortactin-positive pseudopodia consistent with an amoeboid transition. This transition is associated with less-adhesive interactions with the basement membrane and, consequently, a faster rate of cell motility than that of cells undergoing EMT.20,21,23 GPS1 is a member of the COP9 signalosome central to the deneddylation of cullin-RING E3 ubiquitin ligases and thus the ubiquitinylation cycle of protein modification, activation, and turnover.33 Ubiquitination, and thus degradation of proteins such as β-catenin, can have wide-ranging cellular effects. The increased β-catenin levels observed in GPS1-silenced cells suggested possible alterations in ubiquitination pathways that might impact catenin/cadherin or WNT/catenin pathways. However, inhibition of other COP9 proteins CSN2 (COPS2), CSN3 (COPS3), or CSN5 (COPS5) did not result in any significant changes in morphology screening data. Thus, GPS1 was the only significant member of the COP9 and NEDD8 pathway whose suppression resulted in significant morphologic changes. The increased migration, velocity, and β-catenin levels in response to GPS1 loss, as we have observed, therefore is more consistent with COP9-independent GPS1 function. GPS1 is known to be a binding partner for a variety of kinases, such as ITPK1, mediating suppression of c-Jun phosphorylation,47 and its inhibition in NIH3T3 cells resulted in down-regulation of c-Jun N-terminal kinases 1 (JNK1) mRNA and protein expression.47 Thus, facilitation of increased cell motility may be through numerous mechanisms, and will form the basis of future studies.
RRM2 and RRM1 encode the traditional subunits of the RNR enzyme, which catalyzes the formation of ribonucleotides and dNTPs. Thus, the S-phase cell-cycle arrest, decreased rates of scratch-wound closure, and increased apoptosis observed in our study after RRM2 silencing may result in part from effects on dNTP synthesis rates. However, under hypoxic conditions, RRM1 may preferentially form the RNR enzyme through binding to RRM2B (p53-inducible ribonucleotide reductase) after loss of RRM2 expression.41 This RNR subunit switch is suggested to occur to preserve ongoing replication without DNA damage and was observed in a number of cancer cell lines including esophageal cells. In our study, both low pH exposure at levels modeling hypoxic tumor cores and pulsatile exposure modeling GERD similarly suppressed RRM2 expression and induced RRM2B mRNA in BE and EAC cells. A previous study in hepatocellular carcinoma cells observed that markers of epithelial–mesenchymal transition are increased after RRM2B suppression.48 However, no significant effects on cell morphology was noted in RRM2B-silenced esophageal cells in our study. The significant increase in cellular polarity, loss of CDH1/E-cadherin expression, and gain of vimentin expression in surviving RRM2-silenced esophageal cells is suggestive of further commitment toward a mesenchymal phenotype.
Exposure to low pH is one of many environmental and external factors encountered by cells during metastasis or colonization, but one of the most pertinent during early stages of esophageal adenocarcinogenesis. Thus, the multiparametric data set developed in our study of gene–morphology relationships in BE-HGD cells was used initially to investigate the ability of low pH challenge to modulate the expression of genes underlying mesenchymal and ameboid transitions in these cells. However, this data set also may be used to uncover relationships between cellular transitions and other factors underlying the development of BE and EAC such as genomic instability, polyploidy, inflammatory, immune regulatory, microbial, and metabolic factors. For example, the genes PRC1, KIF11, and CDC25B, with known roles in cytokinesis and mitosis, were uncovered to result in distinctive morphologic changes and multinucleated cells upon silencing in this study. Future studies may investigate these relationships through reanalysis of the HCA imaging data with particular focus on phenotypes underlying, for example, cytokinesis and mitotic control such as multinucleation. Furthermore, verification of the earlier-described findings in a wider array of esophageal cell lines, and more advanced models, may expand and further detail the relationships between low pH, cellular morphology, and locomotion in future studies. Nonetheless the identification of the changes outlined in BE-HGD cells, which still retain features of metaplastic change and the potential for formal malignant change, suggests that these findings have implications in the process of esophageal carcinogenesis.
The HCA imaging data set produced in our study and the identified genes provide a rich data source for reanalysis in future studies with a focus on different cellular parameters. In our initial study, we showed that cellular transitions may be triggered through GPS1 and RRM2 expression levels and that both of these genes are responsive to low environmental pH, critical throughout the metaplasia–dysplasia–adenocarcinoma sequence of the esophagus. Targeting amoeboid and mesenchymal transition processes mediated by GPS1 and RRM2 in future studies may provide avenues for the prevention of BE emergence and EAC metastasis. Thus, this data set provides the basis for future studies examining wide-ranging interactions between cellular transitions, locomotion, development, and colonization by BE, BE-HGD, and EAC cell types.
Materials and Methods
Cell Lines, Culture and Treatment
SK-GT-4 (11012007) cells were obtained from the European collection of authenticated cell culture collection and were cultured in RPMI-1640 without L-glutamine media with 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA), and added penicillin/streptomycin/L-glutamine (Gibco, Invitrogen). CP-D BE-HGD cells (CP-18821) were obtained from the American Type Culture Collection and maintained in bronchial epithelial base/growth media (Lonza, Mississauga, Canada) with bullet kit additives and 5% fetal bovine serum (Gibco, Invitrogen). Cells were fed every 2 days, and subcultured at 72 hours, before overconfluence, using trypsin-EDTA (Gibco) in Hank's balanced salt solution for detachment. Exposure of cells (5 × 105 in 6-well plates) to constitutive or pulsatile exposure was performed in triplicate independent experiments as follows: cell culture media was titrated to either pH 6.5 or pH 4.5 with hydrochloric acid under sterile conditions. Cell culture media in all wells was stored and replaced with base growth media without additives to wash remaining culture media away. The remaining media was removed by vacuum manifold before addition of base growth media with no additives at either pH 6.5 or pH 4.5. Pulsatile exposure was achieved by replacing media at pH 4.5 with the stored culture media from that specific well after 10 minutes of exposure to low pH followed by continuation of the experiment. RNA extractions were performed 2 hours before media changes and throughout the experiments to control for change in gene expression that might be owing to environmental changes unrelated to pH environment.
siRNA-Mediated Silencing
As previously described, siRNA siGenome-SMARTpool (Dharmacon, GE Healthcare, Horizon Discovery, Waterbeach, UK) were used for initial screening followed by siON-TARGETplus build (Dharmacon) in all other validation experiments as an alternate verification strategy.30 siRNAs were delivered to cells using Dharmafect transfection reagent (DF1-4; Dharmacon) in a reverse-transfection protocol with a cell culture media change at 24 hours after transfection. Validation of siRNA knockdown was achieved by Western blot and real-time reverse-transcription polymerase chain reaction for respective proteins (N = 3 biological replicates in 2 cell lines) and mRNAs (N = 3 technical and biological replicates) at 72–96 hours after transfection with siRNA.
Protein Expression Analysis
Lysis of cells for protein expression analysis was achieved through M-PER lysis reagent (Pierce, Thermofisher) following the manufacturer’s instructions, followed by protein quantitation by microbicinchoninic acid assay (Pierce, Thermofisher, Waltham, MA) with absorbance at 562 nm in a spectrophotometer (Tecan, Männedorf, Switzerland). Western blot was used to measure protein expression through 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by transfer to a polyvinylidene difluoride membrane in semidry conditions in a power blotter (Pierce, Thermofisher, Waltham, Massachusetts, US), as previously described.30 Polyvinylidene difluoride membranes were blocked with milk protein–based blocking solution. Primary antibodies anti-GPS1 (sc-365617; Santa Cruz Biotechnology, Dallas, TX), anti-RRM2 (ab712476; Abcam), anti–E-cadherin (Purified Mouse Anti–E-Cadherin, clone 36; Becton Dickinson, San Jose, CA), antivimentin (sc-66001; Santa Cruz Biotechnology), anti–β-catenin (Purified Mouse Anti–β-Catenin, cone 14; Becton Dickinson, Franklin Lakes, NJ), and anti–β-actin (Sigma, St. Louis, MO) were combined with horseradish–peroxidase–linked secondary anti-mouse (31430; Invitrogen, Thermofisher, Waltham, Massachusetts, US) and anti-rabbit (31460; Invitrogen, Thermofisher, Waltham, Massachusetts, US) antibodies as required. After incubations with either primary or secondary antibodies, membranes were washed (5 × 10 min) with phosphate buffered saline-tween 20 (PBST) followed by chemiluminescent detection with ECL Western Blot Substrate (Pierce, Thermofisher) and the LAS-3000 CCD lightbox imager (Fujifilm Life Sciences, Tokyo, Japan).
Gene Expression Analysis
Real-time reverse-transcription polymerase chain reaction analysis of gene expression was performed with TaqMan chemistry (Applied Biosystems, Foster City, CA; Thermofisher). RNA was extracted from cells with RNeasy columns (Qiagen, Hilden, Germany) and converted to complementary DNA with a reverse-transcription High-Capacity complementary DNA Reverse Transcription Kit (Applied Biosystems, Thermofisher) before polymerase chain reaction using a 7900HT and Quantstudio 3 thermocyclers with predesigned real-time primers and probes (Applied Biosystems). Fold changes were calculated using the ΔΔCt method of relative quantification with glyceraldehyde-3-phosphate dehydrogenase (Hs99999905_m1) as the denominator control gene. GPS1 (Hs00358804_m1), RRM1 (Hs01040698_m1), RRM2 (Hs00357247_g1), and RRM2B (Hs00968432_m1) primers also were used. Statistical significance was performed using GraphPad (Prism, San Diego, CA), and the nonparametric Mann–Whitney test for technical (N = 3) and biological (N = 3) replicates.
High-Throughput siRNA Library Screening
The siRNA library screen was performed as previously described30 using the BE-HGD cell line CP-D and the druggable genome library (siGenome format; Dharmacon) consisting of 6022 siRNA pools (4 individual gene-targeting siRNAs at 20 μmol/L) formatted in 76 library plates (96-well v-shaped bottom). Negative (n = 6) and positive controls (n = 6) consisting of nontargeting and GATA6-targeting siRNAs were placed within inner wells of the second column of 96-well screening plates. Automated dispensing (Thermofisher) was used to distribute the siRNA library over the screening plates followed by siRNA reverse transfection. At 96 hours after transfection the culture media was removed and replaced with phosphate-buffered saline (PBS) for 10 minutes followed by fixation of cells in 4% paraformaldehyde in PBS. The cells were permeabilized with 0.3% Triton X-100 (Sigma) in PBS followed by blocking in 3% bovine serum albumin in PBS for 30 minutes at room temperature. The primary mouse anti–α-tubulin in 3% bovine serum albumin–PBS was next added to the cells at a dilution of 1:2000 for 1 hour at room temperature, followed by 3 washes with 0.01% Tween 20 in PBS. The cells then were incubated in the dark for 30 minutes in a mixture of Alexa Fluor 488 anti-mouse secondary antibody (1:500), phalloidin–tetramethylrhodamine isothiocyanate (1:250), and Hoechst 33342 (1:5000) in the bovine serum albumin blocking agent. The cells were washed another 3 times with 0.01% Tween 20 in PBS before final storage in PBS and imaging (In Cell Analyzer 1000; GE Life Sciences, Piscataway, NJ) of 9 fields per well.
High Content and Confocal Imagining Analysis
The analysis of the acquired images then was performed with the Investigator software package (GE Healthcare) in conjunction with the R statistical programming language using CellHTS2 (https://bioconductor.org) and RNAither (https://bioconductor.org) platform for statistical analysis of high-throughput RNA interference screens. The measurement of the difference in the overall area occupied by F-actin (phalloidin) and tubulin was used in the initial statistical interpretation and transformations through normalization to the median of the nontargeting negative controls (n = 6) per plate. Plates with significant outliers (plates 22 and 75) were identified in the screen owing to cell infection and systemic error and thus removed from further analyses. Data were rescaled further through robust Z score normalization that substitutes outlier-insensitive median and median absolute deviation for means and SD as follows: Z-score = (x – median)/median absolute deviation. Changes in cell morphology dependent on cell density were controlled through locally weighted least squares regression transformation. Following these steps, Z-score ranking highlighted 207 gene-targeting siRNAs at a significant level Z-score >2.0. Multiparameter (n = 32) analysis then was performed on the images (9 fields per well) from these 207 gene-targeting siRNAs to further investigate the relationships between gene expression and morphologic function using In Cell Analyzer software (GE Life Sciences). Confocal imaging (Zeiss, Oberkochen, Germany) of F-actin, α-tubulin, and cortactin was performed as per the earlier-described staining protocol with the addition of cortactin antibody (sc-55579; Santa Cruz) at a 1:50 dilution.
Cell Viability and Apoptosis Assays
Cell viability was examined through flow cytometry, 3-[4, 5-dimethylthiazol-2-yl] 2, 5-diphenyltetrazolium bromide (MTT) and cell counting assays. In MTT assays the cells were plated into 96-well plates at a concentration of 2 × 105 cells/mL before siRNA reverse transfection as detailed earlier. After appropriate treatment times, 10 μL of MTT reagent was added to cells and incubated for 3 hours. Detergent reagent (100 μL) was added to lyse the cells and shaken after 4 hours to fully dissolve formazan crystals before measurement of absorbance at 570 nm on the VERSAmax Microplate Reader (Molecular Devices, Sunnyvale, CA). Flow cytometry assays were performed as follows: cells (1 × 106) were suspended in PBS and fixed by rapidly admixing with a Pasteur pipette into 10 mL ice-cold 70% ethanol for 2 hours followed by centrifugation for 5 minutes at 200 × g. After the ethanol was decanted, 50 μL DNA extraction buffer (0.2 mol/L phosphate citrate buffer, pH 7.8) was added to the tubes and incubated for 30 minutes at 37°C. The cells were centrifuged for 10 minutes at 1500 × g, and the DNA was stained for 15 minutes at room temperature in the dark, with 1 mL propidium iodide/Triton X-100/RNase A solution (Sigma). Propidium iodide–treated cells were analyzed immediately using a FACScan cytometer (FACS Calibur, Becton Dickinson). The percentage of apoptotic cells was calculated from the sub-G1 peak of the DNA histograms, and 10,000 events routinely were taken from each cell sample for calculation. Direct measurements of cell counts were achieved through Hoechst (33342) nuclear staining as detailed earlier, followed by nuclear counting in the HCA imager (In Cell Analyzer 1000; GE Life Sciences) with 9 fields per well.
Cell Migration and Time-Lapsed Imaging Assays
Cells seeded in a 96-well plate at a density of 1.2 × 104 cells were reverse-transfected with respective siRNAs as outlined earlier. Seventy-two hours after transfection sterile 200-μL pipette tips were used to create a linear scratch in cell monolayers. Images were captured using the In Cell 1000 automated microscope immediately after scratching (time = 0), and again at 96 hours after transfection (time = 24 h). The percentage of wound closure was calculated using the following formula: ([area at time 0 – area at time 24]/area at time 0) × 100. Replacement with serum-free media was used as a negative control, and epidermal growth factor (10 ng/mL) was used as a positive control for cell migration. The area of the wound was quantified by Java’s ImageJ software (National Institutes of Health, Bethesda, MD) using the polygon selection mode (http://rsb.info.nih.gov). The migration of cells into the wounds was expressed as a percentage of wound closure: % wound closure= ([At = 0 h - At = 24 h]/At = 0 h) × 100%, whereby At = 0 hours is the area measured immediately after wounding the cell monolayer, and At = 24 hours is the wound area. Significance of biological replicates (n = 3) was examined with the Student t test in in GraphPad. Time-lapsed cell imaging of cell nondirectional locomotion was achieved through transfection of 5 × 103 cells with respective siRNAs followed by time-lapsed white-light imaging at 72 hours after transfection throughout a 24-hour time period collecting 384 intervaled images until the end point at 96 hours. Manual cell tracking was performed using ImageJ with 18 individual cells tracked in each image stack file.
CRediT Authorship Contributions
Sinead Phipps (Conceptualization: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Equal); Catherine Garry (Data curation: Equal; Methodology: Equal; Resources: Equal); Sepehr Kamal (Investigation: Equal; Methodology: Equal); James Johnson (Methodology: Equal; Supervision: Equal); John Gilmer (Conceptualization: Equal; Project administration: Equal; Resources: Equal; Supervision: Equal; Writing – review & editing: Supporting); Aideen Long (Methodology: Equal; Project administration: Equal; Resources: Equal; Supervision: Equal); Dermot Kelleher (Conceptualization: Equal; Project administration: Equal; Resources: Equal; Supervision: Equal; Writing – review & editing: Supporting); Shane Duggan, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Resources: Equal; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Health Research Board (HRB) Ireland and the HRB PhD Scholars program grant HRB-TRA.2007.11 (S.P.D., C.E.G., S.M.P., D.K., A.L., J.G.), Imperial College London (S.P.D., D.K.), the University of British Columbia (D.K.), the Dr Barbara Hejdankova Cancer Research Fund (S.P.D., D.K.), and donations from the Chan family (S.P.D., D.K.) and Tiny Ventures, Ltd (S.P.D., D.K).
References
- 1.Otterstatter M.C., Brierley J.D., De P., Ellison L.F., Macintyre M., Marrett L.D., Semenciw R., Weir H.K. Esophageal cancer in Canada: trends according to morphology and anatomical location. Can J Gastroenterol. 2012;26:723–727. doi: 10.1155/2012/649108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thrift A.P., Whiteman D.C. The incidence of esophageal adenocarcinoma continues to rise: analysis of period and birth cohort effects on recent trends. Ann Oncol. 2012;23:3155–3162. doi: 10.1093/annonc/mds181. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen T., Thrift A.P., Yu X., Duan Z., El-Serag H.B. The annual risk of esophageal adenocarcinoma does not decrease over time in patients with Barrett's esophagus. Am J Gastroenterol. 2017;112:1049–1055. doi: 10.1038/ajg.2017.18. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Ouyang H., Yamamoto Y., Kumar P.A., Wei T.S., Dagher R., Vincent M., Lu X., Bellizzi A.M., Ho K.Y., Crum C.P., Xian W., McKeon F. Residual embryonic cells as precursors of a Barrett's-like metaplasia. Cell. 2011;145:1023–1035. doi: 10.1016/j.cell.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto Y., Wang X., Bertrand D., Kern F., Zhang T., Duleba M., Srivastava S., Khor C.C., Hu Y., Wilson L.H., Blaszyk H., Rolshud D., Teh M., Liu J., Howitt B.E., Vincent M., Crum C.P., Nagarajan N., Ho K.Y., McKeon F., Xian W. Mutational spectrum of Barrett's stem cells suggests paths to initiation of a precancerous lesion. Nat Commun. 2016;7:10380. doi: 10.1038/ncomms10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang M., Li H., Zhang Y., Yang Y., Lu R., Liu K., Lin S., Lan X., Wang H., Wu H., Zhu J., Zhou Z., Xu J., Lee D.K., Zhang L., Lee Y.C., Yuan J., Abrams J.A., Wang T.C., Sepulveda A.R., Wu Q., Chen H., Sun X., She J., Chen X., Que J. Transitional basal cells at the squamous-columnar junction generate Barrett's oesophagus. Nature. 2017;550:529–533. doi: 10.1038/nature24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu K., Zhao T., Wang J., Chen Y., Zhang R., Lan X., Que J. Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett. 2019;458:21–28. doi: 10.1016/j.canlet.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Que J., Garman K.S., Souza R.F., Spechler S.J. Pathogenesis and cells of origin of Barrett's esophagus. Gastroenterology. 2019;157:349–364 e1. doi: 10.1053/j.gastro.2019.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agoston A.T., Pham T.H., Odze R.D., Wang D.H., Das K.M., Spechler S.J., Souza R.F. Columnar-lined esophagus develops via wound repair in a surgical model of reflux esophagitis. Cell Mol Gastroenterol Hepatol. 2018;6:389–404. doi: 10.1016/j.jcmgh.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D.H., Clemons N.J., Miyashita T., Dupuy A.J., Zhang W., Szczepny A., Corcoran-Schwartz I.M., Wilburn D.L., Montgomery E.A., Wang J.S., Jenkins N.A., Copeland N.A., Harmon J.W., Phillips W.A., Watkins D.N. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett's metaplasia. Gastroenterology. 2010;138:1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q., Agoston A.T., Pham T.H., Zhang W., Zhang X., Huo X., Peng S., Bajpai M., Das K., Odze R.D., Spechler S.J., Souza R.F. Acidic bile salts induce epithelial to mesenchymal transition via VEGF signaling in non-neoplastic Barrett's cells. Gastroenterology. 2019;156:130–144 e10. doi: 10.1053/j.gastro.2018.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson M., Rosato E.L., Chojnacki K.A., Chervoneva I., Kairys J.C., Cohn H.E., Rosato F.E., Sr, Berger A.C. Prognostic significance of lymph node metastases and ratio in esophageal cancer. J Surg Res. 2008;146:11–15. doi: 10.1016/j.jss.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aurello P., D'Angelo F., Nigri G., Bellagamba R., Cicchini C., Ruzzetti R., Ramacciato G. Comparison between site N-category and number N-category for nodal staging in carcinoma of the gastroesophageal junction: our experience and literature review. Am Surg. 2006;72:118–123. [PubMed] [Google Scholar]
- 15.Wang W., Goswami S., Lapidus K., Wells A.L., Wyckoff J.B., Sahai E., Singer R.H., Segall J.E., Condeelis J.S. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 16.Klein C.A. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 17.Klein C.A., Stoecklein N.H. Lessons from an aggressive cancer: evolutionary dynamics in esophageal carcinoma. Cancer Res. 2009;69:5285–5288. doi: 10.1158/0008-5472.CAN-08-4586. [DOI] [PubMed] [Google Scholar]
- 18.Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rorth P. Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev Cell. 2011;20:9–18. doi: 10.1016/j.devcel.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y.J., Le Berre M., Lautenschlaeger F., Maiuri P., Callan-Jones A., Heuze M., Takaki T., Voituriez R., Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Wolf K., Mazo I., Leung H., Engelke K., von Andrian U.H., Deryugina E.I., Strongin A.Y., Brocker E.B., Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul C.D., Mistriotis P., Konstantopoulos K. Cancer cell motility: lessons from migration in confined spaces. Nat Rev Cancer. 2017;17:131–1140. doi: 10.1038/nrc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankova K., Rosel D., Novotny M., Brabek J. The molecular mechanisms of transition between mesenchymal and amoeboid invasiveness in tumor cells. Cell Mol Life Sci. 2010;67:63–71. doi: 10.1007/s00018-009-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benoit M.E., Clarke E.V., Morgado P., Fraser D.A., Tenner A.J. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J Immunol. 2012;188:5682–5693. doi: 10.4049/jimmunol.1103760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark A.G., Vignjevic D.M. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol. 2015;36:13–22. doi: 10.1016/j.ceb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Rohani N., Hao L., Alexis M.S., Joughin B.A., Krismer K., Moufarrej M.N., Soltis A.R., Lauffenburger D.A., Yaffe M.B., Burge C.B., Bhatia S.N., Gertler F.B. Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. 2019;79:1952–1966. doi: 10.1158/0008-5472.CAN-18-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brisson L., Reshkin S.J., Gore J., Roger S. pH regulators in invadosomal functioning: proton delivery for matrix tasting. Eur J Cell Biol. 2012;91:847–860. doi: 10.1016/j.ejcb.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Paradise R.K., Whitfield M.J., Lauffenburger D.A., Van Vliet K.J. Directional cell migration in an extracellular pH gradient: a model study with an engineered cell line and primary microvascular endothelial cells. Exp Cell Res. 2013;319:487–497. doi: 10.1016/j.yexcr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Robey I.F., Baggett B.K., Kirkpatrick N.D., Roe D.J., Dosescu J., Sloane B.F., Hashim A.I., Morse D.L., Raghunand N., Gatenby R.A., Gillies R.J. Bicarbonate increases tumor pH and inhibits spontaneous metastases. Cancer Res. 2009;69:2260–2268. doi: 10.1158/0008-5472.CAN-07-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duggan S.P., Garry C., Behan F.M., Phipps S., Kudo H., Kirca M., Zaheer A., McGarrigle S., Reynolds J.V., Goldin R., Kalloger S.E., Schaeffer D.F., Long A., Strid J., Kelleher D. siRNA library screening identifies a druggable immune-signature driving esophageal adenocarcinoma cell growth. Cell Mol Gastroenterol Hepatol. 2018;5:569–590. doi: 10.1016/j.jcmgh.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duggan S.P., Gallagher W.M., Fox E.J., Abdel-Latif M.M., Reynolds J.V., Kelleher D. Low pH induces co-ordinate regulation of gene expression in oesophageal cells. Carcinogenesis. 2006;27:319–327. doi: 10.1093/carcin/bgi211. [DOI] [PubMed] [Google Scholar]
- 32.Duggan S.P., Behan F.M., Kirca M., Zaheer A., McGarrigle S.A., Reynolds J.V., Vaz G.M., Senge M.O., Kelleher D. The characterization of an intestine-like genomic signature maintained during Barrett's-associated adenocarcinogenesis reveals an NR5A2-mediated promotion of cancer cell survival. Sci Rep. 2016;6:32638. doi: 10.1038/srep32638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubiel D., Rockel B., Naumann M., Dubiel W. Diversity of COP9 signalosome structures and functional consequences. FEBS Lett. 2015;589:2507–2513. doi: 10.1016/j.febslet.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Chamovitz D.A. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009;10:352–358. doi: 10.1038/embor.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damsky W.E., Curley D.P., Santhanakrishnan M., Rosenbaum L.E., Platt J.T., Gould Rothberg B.E., Taketo M.M., Dankort D., Rimm D.L., McMahon M., Bosenberg M. Beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi J., Yu Y., Akilli Ozturk O., Holland J.D., Besser D., Fritzmann J., Wulf-Goldenberg A., Eckert K., Fichtner I., Birchmeier W. New Wnt/beta-catenin target genes promote experimental metastasis and migration of colorectal cancer cells through different signals. Gut. 2016;65:1690–1701. doi: 10.1136/gutjnl-2014-307900. [DOI] [PubMed] [Google Scholar]
- 37.Bian Y.S., Osterheld M.C., Bosman F.T., Fontolliet C., Benhattar J. Nuclear accumulation of beta-catenin is a common and early event during neoplastic progression of Barrett esophagus. Am J Clin Pathol. 2000;114:583–590. doi: 10.1309/3QLC-5MF1-JYXU-A5XX. [DOI] [PubMed] [Google Scholar]
- 38.Beavon I.R. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur J Cancer. 2000;36:1607–1620. doi: 10.1016/s0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura E., Sugihara H., Bamba M., Hattori T. Dynamic alteration of the E-cadherin/catenin complex during cell differentiation and invasion of undifferentiated-type gastric carcinomas. J Pathol. 2005;205:349–358. doi: 10.1002/path.1718. [DOI] [PubMed] [Google Scholar]
- 40.Aye Y., Li M., Long M.J., Weiss R.S. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 41.Foskolou I.P., Jorgensen C., Leszczynska K.B., Olcina M.M., Tarhonskaya H., Haisma B., D'Angiolella V., Myers W.K., Domene C., Flashman E., Hammond E.M. Ribonucleotide reductase requires subunit switching in hypoxia to maintain DNA replication. Mol Cell. 2017;66:206–220 e9. doi: 10.1016/j.molcel.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comaills V., Kabeche L., Morris R., Buisson R., Yu M., Madden M.W., LiCausi J.A., Boukhali M., Tajima K., Pan S., Aceto N., Sil S., Zheng Y., Sundaresan T., Yae T., Jordan N.V., Miyamoto D.T., Ting D.T., Ramaswamy S., Haas W., Zou L., Haber D.A., Maheswaran S. Genomic instability is induced by persistent proliferation of cells undergoing epithelial-to-mesenchymal transition. Cell Rep. 2016;17:2632–2647. doi: 10.1016/j.celrep.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baron A., Migita T., Tang D., Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53. doi: 10.1002/jcb.10708. [DOI] [PubMed] [Google Scholar]
- 44.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 45.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 46.Burbridge M.F., West D.C., Atassi G., Tucker G.C. The effect of extracellular pH on angiogenesis in vitro. Angiogenesis. 1999;3:281–288. doi: 10.1023/a:1009092511894. [DOI] [PubMed] [Google Scholar]
- 47.Tsuge T., Menon S., Tong Y., Wei N. CSN1 inhibits c-Jun phosphorylation and down-regulates ectopic expression of JNK1. Protein Cell. 2011;2:423–432. doi: 10.1007/s13238-011-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian H., Ge C., Li H., Zhao F., Hou H., Chen T., Jiang G., Xie H., Cui Y., Yao M., Li J. Ribonucleotide reductase M2B inhibits cell migration and spreading by early growth response protein 1-mediated phosphatase and tensin homolog/Akt1 pathway in hepatocellular carcinoma. Hepatology. 2014;59:1459–1470. doi: 10.1002/hep.26929. [DOI] [PubMed] [Google Scholar]