Abstract
Purpose
Over 9.5 million Latinos could be affected by cataracts by 2050. However, no known cataract genetic risk alleles have been identified in Latinos. Moreover, no mitochondrial genome-wide association studies (MiWAS) have been conducted on cataracts in a Latino cohort despite the association between mitochondrial dysfunction and cataracts. Our purpose was to identify a mitochondrial DNA variant that associated with cataracts in a large-scale Latino population.
Methods
We conducted an MiWAS to identify mitochondrial single-nucleotide polymorphisms that modify cataract risk in nearly 3500 individuals enrolled in the Los Angles Latino Eye Study cohort, the largest Latino-specific cohort with comprehensive cataract data. Our analytic strategy for MiWAS included logistic regression on cataract occurrence while controlling for mitochondrial genetic ancestry, age, and biological sex.
Results
We found that MitoG228A (rs41323649) alternative allele carriers experienced a five times greater risk for cataracts compared with reference allele carriers. Alternative allele carriers also developed cataracts earlier in life compared with reference allele carriers. Intracohort cross-validation with 10-fold resampling and five repeats showed that the effect of MitoG228A remained significant.
Conclusions
MitoG228A increased risk for cataracts five-fold in approximately 3500 Latinos. To the best of our knowledge, this is the first cataract MiWAS on a large-scale Latino population. This association needs to be validated in an independent cohort.
Translational Relevance
Our discovery hypothesis-generating study suggest MitoG228A has potential to be used as a risk factor in the clinic and as a target for therapeutics. With validation via an independent cohort, MitoG228A could be used to estimate cataract risk for a Latino to reduce complications later in life.
Keywords: genome-wide, genetic epidemiology, ophthalmology
Introduction
Cataract is the leading cause of blindness and affects nearly half of Latinos over the age of 75 years.1 Strategies that delay cataract treatment just by 10% are projected to save the United States $660 million in Medicare expenses per year.2 However, the number of Latinos diagnosed with cataracts is projected to grow from nearly 2 million to over 9.5 million by 2050. No other racial/ethnic group is projected to experience such a drastic prevalence shift for cataracts in the United States.3
Cataracts are principally treated by surgical removal of the lens opacity. Although cataract surgeries are generally successful with few postoperative complications, uncorrected visual impairment and significant surgical costs are problematic for Latinos. Indeed, 67% of Latinos reported uncorrected refractive error in the Los Angeles Latino Eye Study (LALES) despite successful cataract removal.4 These cataract surgeries have comprised over 10% of Medicare's total budget,5 and Latinos are 50% more likely to undergo complex cataract surgery through Medicare compared with individuals who self-reported as white.6 In addition, Latinos with annual income under $20,000 are at a 2.6 times greater risk for an unmet need for cataracts. Those who only undergo one eye examination per 5 years are almost four times more likely to suffer from complications associated with unmet cataract surgery.1 Hence, identifying Latinos who are at high risk for cataracts could decrease medical costs and protect individuals from residual visual impairment.
One possible therapeutic target for cataracts is the mitochondrion, a cellular organelle with its own genome that encodes proteins involved in oxidative phosphorylation and peptides that modify cellular metabolism. Dysfunction of mitochondria is the greatest source of reactive oxygen species (ROS) and has been observed in cataract patients and experimental models. For instance, cataract patients’ mitochondrial DNA (mtDNA) showed high levels of oxidative stress, as measured by a quantitative polymerase chain reaction assay that quantifies strand breaks and base modification.7 Quenching oxidative stress via overexpression of mitochondrial catalase has improved lifespan and delayed cataract formation in mice, which suggests lens are especially prone to oxidative damage.8 In fact, scavenging ROS via overexpression of superoxide dismutase in whole lens prevented damage caused by ROS,9 and promoting mitochondrial dysfunction has induced damage in lens in vitro and in vivo via protein oxidation.10 Moreover, during the aging process in mice, most cataracts colocalized with mitochondria.11 Given the role of mitochondria in lens physiology and age-related damage, we hypothesized that mitochondrial genetic variation predisposes individuals to lens opacities.
Several single-nucleotide polymorphisms (SNPs) have been associated with cataracts that affect genes involved in fructose metabolism, nervous system development, cytoskeleton integrity, and protein ubiquination.12 Gene-gene interaction scans have also revealed cataract loci involved in immunity, DNA replication, and cell signaling.13 These risk alleles, however, were observed in cohorts of mainly European ancestry and may not be generalized to all racial/ethnic groups, including Latinos. No mitochondrial genome-wide association studies (MiWAS) have been conducted on cataracts despite the ubiquitous role of mitochondria in eye physiology and the ethnic-specificity of mtDNA. To the best of our knowledge, we conducted the first MiWAS on cataracts in a large-scale Latino cohort.
Methods
Study Sample
We examined patients in the LALES cohort who self-identify as Latinos living in La Puente, California, and who are at least age 40 years. We studied this population because they represent the demographic and socioeconomic status of individuals from Mexico who reside in Los Angeles County, California, and the United States. Complete details of the examinations have been previously described.14 All study participants provided informed consent, and study procedures were approved by the Los Angeles County/University of Southern California Medical Center Institutional Review.
Genotyping and Quality Control
LALES participants were genotyped using either the Illumina OmniExpress BeadChip (∼730k markers; Illumina, Inc., San Diego, CA) for 4122 samples or the Illumina Hispanic/SOL BeadChip (∼2.5 million markers; Illumina, Inc.) for 716 samples. Genotyping occurred at the Los Angeles Biomedical Research Institute, and variants were called by using Illumina GenomeStudio (v2011.1; Illumina, Inc.). These two datasets were harmonized by filtering all SNPs at a genotype call rate of at least 90% and nuclear SNP Hardy-Weinberg equilibrium P values less than 5E−8. Note that the mitochondrial genome does not follow Hardy-Weinberg equilibrium and were not subjected to the same thresholds as nuclear SNPs. For downstream nuclear SNP analyses, variants with alleles not matched to the correct forward strand in the hg19 genome assembly were removed. These methods retained 697,553 and 1,608,650 markers, as well as 3642 and 706 individuals, respectively, from the OmniExpress and SOL arrays. A total of 120 mitochondrial single-nucleotide polymorphisms (mtSNPs) were retained when merging the two datasets together for 3822 individuals.
Measurement of Phenotypes
Lens were examined at the slit lamp after patients’ eyes were dilated with 1% tropicamide and 2.5% phenylephrine by the examiner. The Lens Opacities Classification System II (LOCS II) was used to assign opacities into separate groups, including 5 nuclear (N0, NI, NII, NIII, NIV), 5 posterior subcapsular (P0, PI, PII, PIII, PIV), and 7 cortical (C0, Ctr, CI, CII, CIII, CIV, CV) grades of increasing severity, according to photographic standards. Individuals were classified as having a cataract if the following criteria were met: opacity in at least one eye of (1) any gradable nuclear (≥ NII), cortical (≥ CII), or PSC (≥ PII) lens opacity; or (2) too advanced to grade; or (3) history of lens surgery. Cataract classifications were reproduced by a separate examiner who graded 50 participants independently, and classification accuracy was measured for agreement using proportionally weighted K statistics, which revealed moderate to good intergrader agreement, results of which have been previously reported.15
Additional markers include blood pressure, glycosylated hemoglobin, and lipids. Blood pressure was measured by random zero sphygmomanometer while the participant was in a relaxed, upright, sitting position. Systolic and diastolic blood pressure was obtained after averaging two consecutive measurements. Nonfasting glycosylated hemoglobin was measured by the DCA 2000+ System (Bayer HealthCare, Tarrytown, NY). Nonfasting total cholesterol, high-density lipoprotein (HCL), low-density lipoprotein (LDL), and triglycerides were measured using the Cholestech Analyzer (Cholestech LDX System; Alere, Waltham, MA).
Statistics
Our analytic strategy for MiWAS included logistic regression on cataract occurrence while controlling for genetic ancestry, age, and biological sex. Minor allele frequency and missing individual SNP thresholds were set at 0.01 and 0.1, respectively. We performed MiWAS using PLINK2.0 (Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4) and corrected for statistical significance using the Benjamini-Hochberg false discovery rate (FDR; i.e., q value). There is no standard statistical significance threshold for mitochondrial genome association analyses; some groups considered mtSNPs statistically significant if under an FDR of 0.2, whereas other groups included adaptive permutation tests.16,17 We set the FDR at 0.2 because of the limited number of mtSNPs that passed the 1% minor allele frequency threshold, previous genome-wide association study (GWAS) approaches on LALES, and our objective to include relatively rare variants.18 After identifying one statistically significant mtSNP (i.e., MitoG228A), we implemented an intracohort cross-validation strategy because an independent Latino-specific cohort with cataract data were not available. We performed 10-fold cross-validation with five repeats, modeling cataracts as a function of MitoG228A.
We examined genetic ancestry two ways. First, mitochondrial genetic ancestry was controlled in the MiWAS model by using principal components calculated from binary-coded mtSNPs, as previously described for mtSNPs.19 These principal components were derived using singular value-decomposition of the mitochondrial genetic data matrix that outputs eigenvectors that closely approximate the matrix with a minimal number of values (prcomp function in R (version 3.5.1, R Foundation, Vienna, Austria, 2018)).20 Second, nuclear genetic ancestry was examined using maximum likelihood estimation. We combined our samples with the 1000 Genomes reference population using all overlapping nuclear SNPs and pruned to 12,980 variants to facilitate downstream computation. We then calculated the proportions of the ancestral populations using the ADMIXTURE (v.1.3.0, http://software.genetics.ucla.edu/admixture/download.html), software, as previously described.21 After findings that MitoG228A was significantly associated with cataracts, we examined the effect of the mtSNP on a variety of metabolic markers and types of cataracts. Linear regression for metabolic markers was performed using the lm command in R. Multinomial regression was performed for type of cataracts using the multinom command in R; the effect of MitoG228A was modeled as a function of cataract type.
Results
Study Sample
Descriptive statistics of the samples, including genotypic and phenotypic data, are presented in the Table. A total of 3819 individuals were assessed during the MiWAS stage, whereas a total of 3531 individuals were assessed at the blood biomarker stage. The overall mean age of the studied sample was 54.3 years (SD, 10.5). Approximately 59% of the sample were women. A total of 1577 cases of cataracts were reported.
Table.
Descriptives of LALES Cohort
| Measure | No Cataracts | Cataracts |
|---|---|---|
| Sample size | 1951 | 1577 |
| Female | 56.8% | 62.1% |
| Age (years), mean (SD) | 49.2 (7.5) | 60.5 (10.3) |
MitoG22A Increases Cataract Risk in Latinos
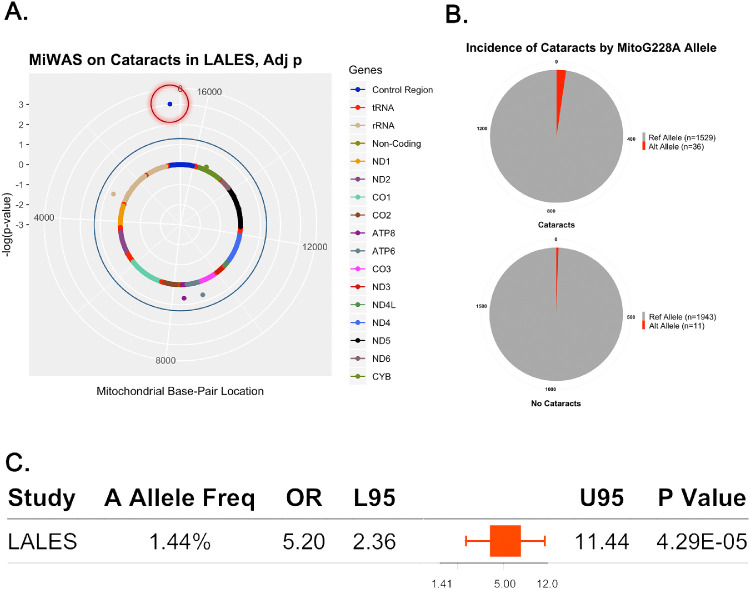
We conducted an MiWAS to examine the effects of 120 mtSNPs on cataracts in individuals who self-report as Latino. After filtering by a minor allele frequency of 1%, only 25 mtSNPs remained. Of the 25 mtSNPs, 21 variants were subjected to statistical adjustment. Genomic inflation was not observed based on the median χ2 statistic (λ = 1). MitoG228A (rs41323649) was the only statistically significant mtSNP that was associated with cataracts (odds ratio [OR], 5.20; P = 4.29e-05; q value = 9.01e-03; mitochondrial genetic ancestry corrected; Figs. 1A, 1C).
Figure 1.
MitoG228A alternative allele carriers are at greater risk for cataracts. (A) Mitochondrial solar plot illustrating that MitoG228A (red circle) significantly associates with cataracts. Dots that do not pass the blue outer rim (FDR 0.05) are not considered statistically significant. (B) Pie graph illustrating the number of alternative MitoG228A carriers with and without cataracts. (C) Forest plot representing the effects of MitoG228A.
After filtering thresholds were applied, 36 participants with the MitoG228A alternative allele had an occurrence of cataracts, whereas 11 participants with the MitoG228A alternative allele did not have an occurrence of cataracts (Fig. 1B). Modeling MitoG228A as a function of cataract type showed that the alternative allele had no significant association with cortical, nuclear, posterior subscapular, or mixed cataracts. The effects of the 21 mtSNPs are included in Supplementary Table S1.
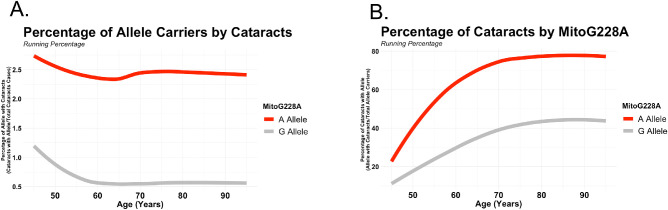
The frequency of cataract cases with MitoG228A was nearly five times greater across age (Fig. 2A), and nearly 80% of MitoG228A carriers developed cataracts late in life (Fig. 2B). We did not observe a statistically significant difference in overall age between MitoG228A reference and alternative allele carriers (G allele = 54.2 years; A allele = 56.9 years) within the cohort. We also did not observe a significant overall age difference between MitoG228A reference and alternative allele carriers with cataracts (G allele = 60.2 years; A allele = 60.5 years). However, we observed a statistically significant difference in age between MitoG228A alternative allele carriers with and without cataracts (A allele with cataracts: 60.2 years; A allele without cataracts: 45.4 years; P < 0.001), which suggests that the 11 alternative allele are not at an age when cataracts are particularly likely to develop.
Figure 2.
MitoG228A alternative allele frequency of cataracts by age. (A) Red line illustrates the frequency of alternative allele carriers in cataracts cases over time. (B) Red line illustrates the frequency of cataracts in alternative allele carriers over time.
Validating the effect of MitoG228A on cataract risk via an independent cohort is crucial, but there is currently no robust Latino-specific cohort with cataract data. Therefore we performed an intracohort cross-validation strategy by implementing a 10-fold cross-validation with five repeats on cataract as a function of MitoG228A (generalized linear model). The final reported model from this sample included an OR for MitoG228A of 4.36 (P = 1.88E-05).
MitoG228A Carriers are not Genetically Admixed
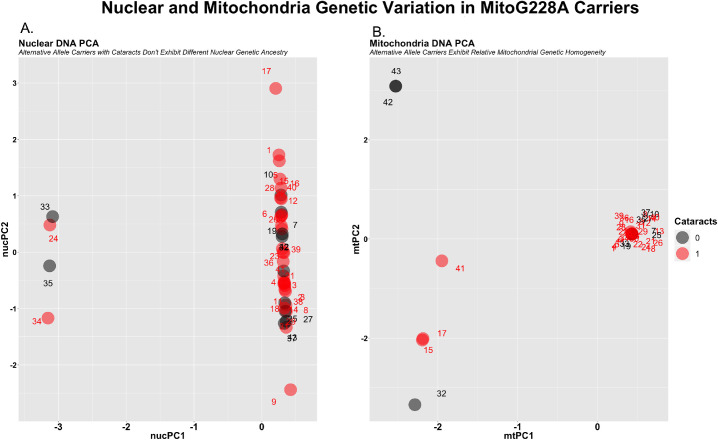
We examined nuclear and mitochondrial genetic ancestry in alternative MitoG228A carriers. The purpose of this analysis is to (1) identify nuclear genetic admixture that segregate with MitoG228A alternative allele carriers with and without cataracts, and (2) to examine the degree of mitochondrial genetic admixture in MitoG228A alternative allele carriers with and without cataracts.
Both nuclear and mtDNA principal-component analysis (PCA) revealed that (1) nuclear genetic admixture does not confound the effects of MitoG228A, and (2) mitochondrial genetic admixture is also relatively homogenous between MitoG228A carriers. Most MitoG228A alternative allele carriers have nearly the same mtDNA profile regardless of cataracts, as shown by one singular tight cluster (Fig. 3A; x = 0.5, y = 0.1). Similarly, most MitoG228A alternative allele carriers have minimal nuclear DNA admixture, as illustrated by a linear cluster (Fig. 3B; x = 0.5; y = –1:2). These findings suggest that the effect of MitoG228A on cataracts is not confounded by nuclear or mitochondrial genetic ancestry.
Figure 3.
MitoG228A alternative allele carriers are relatively genetically homogenous. (A) Illustrating nuclear DNA principal components for MitoG228A alternative allele carriers with and without cataracts (red, black). (B) Illustrating mitochondria DNA principal components for MitoG228A alternative allele carriers with and without cataracts.
To completely rule out the possibility of nuclear genetic admixture confounding effects of MitoG228A, we examined the effect of MitoG228A on cataracts while controlling for nuclear genetic ancestry (i.e., that derived from nuclear DNA PCA). The effect of the alternative while controlling nuclear genetic ancestry (OR, 4.7; P = 1.87e-04) was similar to its effects while controlling for mitochondrial genetic ancestry (OR, 5.20; P = 4.29e-05). Therefore the effects of MitoG228A appear independent from genetic admixture.
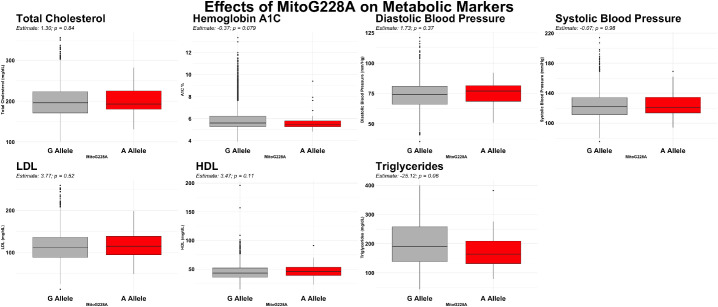
MitoG228A does not Significantly Associate with Metabolic Dysfunction
Because metabolic dysfunction has been associated with cataracts,22 we examined the effect of MitoG228A on total cholesterol, hemoglobin A1C, blood pressure, LDL, HDL, and triglycerides. Alternative allele carriers did not exhibit any statistically significant metabolic differences compared with reference allele carriers (Fig. 4).
Figure 4.
Box plots illustrating the effects of MitoG228A on total cholesterol, hemoglobin A1c, diastolic blood pressure, systolic blood pressure, LDL, HDL, and triglycerides. All analyses were not statistically significant: hemoglobin A1c (P = 0.079), HDL (P = 0.11), and triglycerides (P = 0.06).
Discussion
We identified a novel cataract genetic target for Latinos in the form of the mtDNA SNP MitoG228A by using an approach called MiWAS. We showed that Latino MitoG228A alternative allele carriers had a five times greater risk for cataracts compared with Latino reference allele carriers. The hazardous effect of MitoG228A is not confounded by nuclear or mitochondrial genetic ancestry, as controlling for both mitochondrial and nuclear genetic ancestry did not affect risk. The effects of MitoG228A on cataracts also appears independent from metabolic dysfunction; MitoG228A had no significant effect on metabolic markers, although hemoglobin A1C, HDL, and triglycerides were borderline significant. As a result, the precise effect of MitoG228A on cataracts is worth exploring experimentally.
MitoG228A occurs in the mitochondrial displacement-loop (D-loop), a 1.2 kb noncoding region involved in mitochondrial transcription and genome replication. Previous reports showed that mutations in the D-loop are nearly 17 times greater than mutations in nuclear DNA.23 Polymorphisms in this region could theoretically modify mitochondrial transcription and mitochondrial biogenesis, although the precise effects of D-loop mtSNPs are not well understood.
In addition to the potential effects of MitoG228A on the D-loop, MitoG228A might also affect peptides derived from mitochondrial small open reading frames (smORFs). Protein characterization has historically only considered open reading frames greater than approximately 100 nucleotides as viable protein-encoding genes. Nevertheless, emerging proteomic and genetic evidence has revealed thousands of peptides encoded by mitochondrial, nuclear, and human microbiome smORFs.24–27 Because we observed minimal effects of MitoG228A on global metabolic markers, the effects of the mtSNP might not be completely due to mitochondrial dysfunction. Rather, the specific effect of MitoG228A might be mediated through altered mitochondrial smORF peptides. Humanin, small humanin-like peptides (SHLPs), MOTS-c, and emerging new mitochondrial-derived peptides have highly specific and profound effects on various systems and organs.28–34 Yen et al.27 reported that an mtSNP in the humanin smORF was associated with lower circulating levels of humanin and worse cognitive function. Moreover, the mitochondrial-derived peptides SHLP2 and humanin protected against a model of macular degeneration in vitro.35,36 Hence the experimental effects of MitoG228A on mitochondrial biology, and specifically mitochondrial-derived peptides could reveal therapeutic targets for cataracts.
MitoG228A occurs in 12 haplogroups, but only one of the 12 haplogroups, B2a4a, is Latino-specific (i.e., derived from the Sinaloa state of Mexico).37 No associations between haplogroup B2a4a and any phenotype has been reported to the best of our knowledge. B2a4a is estimated to be roughly 9 ka years according to the molecular clock proposed by Soares et al.38,39 MitoG228A is the only B2a4a determining variant, and one subclade from the haplogroup, B2a4a1, exists and includes three additional variants (A3663G, G10685A, and T16325C). None of these three variants were included on the LALES genome arrays. It is possible that the effects we observed of MitoG228A are mediated or modified by A3663G, G10685A, and T16325C, which are worth exploring in future analyses.
The identification of MitoG228A suggests that MiWAS is an underexplored technique that could yield vital genetic therapeutic targets for ethnic-specific diseases. Most GWAS reports have excluded the mitochondrial genome because it does not follow Hardy-Weinberg principle. However, MiWAS has been used by additional groups to find novel mitochondrial genetic targets for metabolic and chronic diseases.40–42 Furthermore, medical genetics research has mostly analyzed populations of European descent, although recent efforts have since expanded studies to include additional ethnicities.43 Separate studies on Latino-specific cohorts have identified several novel medical genetic targets.44 Our study was the first to examine the effect of mitochondrial genetic variation on cataracts in a large-scale Latino population.
Conclusions
These findings are discovery hypothesis generating with clinical potential in the form of precision-based medicine. With further validation via an independent cohort, clinicians might gauge one's risk for cataracts early in life by genotyping MitoG228A. Moreover, with experimental follow-up, therapeutics designed to target MitoG228A could have potential to slow down cataract development. Given the effects of the D-loop on mitochondrial biology, it is possible that MitoG228A affects a series of mitochondrial-derived peptides that have been shown to modify eye disease, peptides of which could represent druggable targets. Overall, our findings suggest that MitoG228A could be a cataracts genetic target for Latinos that has translational relevance as a diagnostic and drug target.
Supplementary Material
Acknowledgments
Supported by NIH Grants (PC; R01AG061834, R56AG062693, P01AG034906, R01EY027363, U54CA233465), NIA T32 AG000037 (BM), and the NEI Grant U10-EY-11753 (RV).
Disclosure: B. Miller, None; M. Torres, None; X. Jiang, None; R. McKean-Cowdin, None; D. Nousome, None; S.-J. Kim, None; H.H. Mehta, None; K. Yen, None; P. Cohen, CohBar Inc. (C, I); R. Varma, None
References
- 1. Richter GM, Chung J, Azen SP, Varma R, Los Angeles Latino Eye Study G. Prevalence of visually significant cataract and factors associated with unmet need for cataract surgery: Los Angeles Latino Eye Study. Ophthalmology. 2009; 116: 2327–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oostra TD, Mauger TF.. Cost effect of surgeon and patient discretion in regard to cataract surgery. Clin Ophthalmol. 2018; 12: 2563–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. National Eye Institute. Cataract data and statistics. 2019. Available at: https://www.nei.nih.gov/eyedata/cataract. Accessed September 25, 2019.
- 4. Baranano AE, Wu J, Mazhar K, Azen SP, Varma R; Los Angeles Latino Eye Study Group. Visual acuity outcomes after cataract extraction in adult Latinos. The Los Angeles Latino Eye Study. Ophthalmology. 2008; 115: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellwein LB, Urato CJ.. Use of eye care and associated charges among the Medicare population: 1991-1998. Arch Ophthalmol. 2002; 120: 804–811. [DOI] [PubMed] [Google Scholar]
- 6. Mahr MA, Hodge DO, Erie JC. Racial/ethnic differences in rates of complex cataract surgery among United States Medicare beneficiaries. J Cataract Refract Surg. 2018; 44: 140–143. [DOI] [PubMed] [Google Scholar]
- 7. Erol Tinaztepe O, Ay M, Eser E. Nuclear and mitochondrial DNA of age-related cataract patients are susceptible to oxidative damage. Curr Eye Res. 2017; 42: 583–588. [DOI] [PubMed] [Google Scholar]
- 8. Schriner SE, Linford NJ, Martin GM, et al.. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005; 308: 1909–1911. [DOI] [PubMed] [Google Scholar]
- 9. Lin D, Barnett M, Grauer L, et al.. Expression of superoxide dismutase in whole lens prevents cataract formation. Mol Vis. 2005; 11: 853–858. [PubMed] [Google Scholar]
- 10. Nita M, Grzybowski A.. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxid Med Cell Longev. 2016; 2016: 3164734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pendergrass W, Penn P, Possin D, Wolf N. Accumulation of DNA, nuclear and mitochondrial debris, and ROS at sites of age-related cortical cataract in mice. Invest Ophthalmol Vis Sci. 2005; 46: 4661–4670. [DOI] [PubMed] [Google Scholar]
- 12. Ritchie MD, Verma SS, Hall MA, et al.. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol Vis. 2014; 20: 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- 13. Hall MA, Verma SS, Wallace J, et al.. Biology-driven gene-gene interaction analysis of age-related cataract in the eMERGE Network. Genet Epidemiol. 2015; 39: 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varma R, Paz SH, Azen SP, et al.. The Los Angeles Latino Eye Study: design, methods, and baseline data. Ophthalmology. 2004; 111: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 15. Varma R, Richter GM, Torres M, et al.. Four-year incidence and progression of lens opacities: the Los Angeles Latino Eye Study. Am J Ophthalmol. 2010; 149: 728–734.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giorgi EE, Li Y, Caberto CP, et al.. No association between the mitochondrial genome and prostate cancer risk: the multiethnic cohort. Cancer Epidemiol Biomarkers Prev. 2016; 25: 1001–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lakatos A, Derbeneva O, Younes D, et al.. Association between mitochondrial DNA variations and Alzheimer's disease in the ADNI cohort. Neurobiol Aging. 2010; 31: 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao X, Gauderman WJ, Liu Y, et al.. A genome-wide association study of central corneal thickness in Latinos. Invest Ophthalmol Vis Sci. 2013; 54: 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller B, Arpawong TE, Jiao H, et al.. Comparing the utility of mitochondrial and nuclear DNA to adjust for genetic ancestry in association studies. Cells. 2019; 8:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Z, Castello A.. Principal components analysis in clinical studies. Ann Transl Med. 2017; 5: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009; 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sabanayagam C, Wang JJ, Mitchell P, et al.. Metabolic syndrome components and age-related cataract: the Singapore Malay Eye Study. Invest Ophthalmol Vis Sci. 2011; 52: 2397–2404. [DOI] [PubMed] [Google Scholar]
- 23. Lievre A, Blons H, Houllier AM, et al.. Clinicopathological significance of mitochondrial D-loop mutations in head and neck carcinoma. Br J Cancer. 2006; 94: 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ikonen M, Liu B, Hashimoto Y, et al.. Interaction between the Alzheimer's survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc Natl Acad Sci USA. 2003; 100: 13042–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H, Xiao L, Zhang L, et al.. FSPP: a tool for genome-wide prediction of smORF-encoded peptides and their functions. Front Genet. 2018; 9: 96.29675032 [Google Scholar]
- 26. Saghatelian A, Couso JP.. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol. 2015; 11: 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yen K, Wan J, Mehta HH, et al.. Humanin prevents age-related cognitive decline in mice and is associated with improved cognitive age in humans. Sci Rep. 2018; 8: 14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiao J, Howard L, Wan J, et al.. Low circulating levels of the mitochondrial-peptide hormone SHLP2: novel biomarker for prostate cancer risk. Oncotarget. 2017; 8: 94900–94909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Widmer RJ, Flammer AJ, Herrmann J, et al.. Circulating humanin levels are associated with preserved coronary endothelial function. Am J Physiol Heart Circ Physiol. 2013; 304: H393–H397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oh YK, Bachar AR, Zacharias DG, et al.. Humanin preserves endothelial function and prevents atherosclerotic plaque progression in hypercholesterolemic ApoE deficient mice. Atherosclerosis. 2011; 219: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee C, Kim KH, Cohen P. MOTS-c: a novel mitochondrial-derived peptide regulating muscle and fat metabolism. Free Radic Biol Med. 2016; 100: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim SJ, Xiao J, Wan J, Cohen P, Yen K. Mitochondrially derived peptides as novel regulators of metabolism. J Physiol. 2017; 595: 6613–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim SJ, Miller B, Mehta HH, et al.. The mitochondrial-derived peptide MOTS-c is a regulator of plasma metabolites and enhances insulin sensitivity. Physiol Rep. 2019; 7: e14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim SJ, Mehta HH, Wan J, et al.. Mitochondrial peptides modulate mitochondrial function during cellular senescence. Aging (Albany NY). 2018; 10: 1239–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nashine S, Cohen P, Nesburn AB, Kuppermann BD, Kenney MC. Characterizing the protective effects of SHLP2, a mitochondrial-derived peptide, in macular degeneration. Sci Rep. 2018; 8: 15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sreekumar PG, Ishikawa K, Spee C, et al.. The mitochondrial-derived peptide humanin protects RPE cells from oxidative stress, senescence, and mitochondrial dysfunction. Invest Ophthalmol Vis Sci. 2016; 57: 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Auken K, Jaffery J, Chan J, Muller HM, Sternberg PW. Semi-automated curation of protein subcellular localization: a text mining-based approach to Gene Ontology (GO) Cellular Component curation. BMC Bioinformatics. 2009; 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Achilli A, Perego UA, Lancioni H, et al.. Reconciling migration models to the Americas with the variation of North American native mitogenomes. Proc Natl Acad Sci USA. 2013; 110: 14308–14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soares P, Ermini L, Thomson N, et al.. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009; 84: 740–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kraja AT, Liu C, Fetterman JL, et al.. Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am J Hum Genet. 2019; 104: 112–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu C, Yang Q, Hwang SJ, et al.. Association of genetic variation in the mitochondrial genome with blood pressure and metabolic traits. Hypertension. 2012; 60: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klarin D, Lynch J, Aragam K, et al.. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat Med. 2019; 25: 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019; 177: 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goodarzi MO, Langefeld CD, Xiang AH, et al.. Insulin sensitivity and insulin clearance are heritable and have strong genetic correlation in Mexican Americans. Obesity (Silver Spring). 2014; 22: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.