Abstract
Maize rayado fino virus (MRFV) is the type species of the genus Marafivirus in the family Tymoviridae. It infects maize (Zea mays), its natural host, to which it is transmitted by leafhoppers including Dalbulus maidis and Graminella nigrifrons in a persistent‐propagative manner. The MRFV monopartite RNA genome encodes a precursor polyprotein that is processed into replication‐associated proteins. The genome is encapsidated by two carboxy co‐terminal coat proteins, CP1 and CP2. Cloned MRFV can be readily transmitted to maize by vascular puncture inoculation (VPI), and such virus systems that can be used in maize are valuable to examine plant gene function by gene silencing. However, the efficacy of marafiviruses for virus‐induced gene silencing (VIGS) has not been investigated to date. To this end, MRFV genomic loci were tested for their potential to host foreign insertions without attenuating virus viability. This was done using infectious MRFV clones engineered to carry maize phytoene desaturase (PDS) gene fragments (ZmPDS) at various genomic regions. Several MRFV‐PDS constructs were generated and tested for infectivity and VIGS in maize. This culminated in identification of the helicase/polymerase (HEL/POL) junction as a viable insertion site that preserved virus infectivity, as well as several sites at which sequence insertion caused loss of virus infectivity. Transcripts of viable constructs, carrying PDS inserts in the HEL/POL junction, induced stable local and systemic MRFV symptoms similar to wild‐type infections, and triggered PDS VIGS initiating in veins and spreading into both inoculated and noninoculated leaves. These constructs were remarkably stable, retaining inserted sequences for at least four VPI passages while maintaining transmissibility by D. maidis. Our data thus identify the MRFV HEL/POL junction as an insertion site useful for gene silencing in maize.
Keywords: Dalbulus maidis, marafivirus, Zea mays
1. INTRODUCTION
Plant viruses have been engineered as vectors for gene expression or virus‐induced gene silencing (VIGS) for gene function studies in diverse plants (for recent reviews, see Cody & Scholthof, 2019; Hefferon, 2017). However, despite the ever‐increasing abundance of robust VIGS vectors for dicots, comparable tools for monocots have consistently lagged behind. This is the case particularly for maize (Zea mays), for which only a handful of viruses have been developed into VIGS vectors, most of them quite recently (reviewed in Kant & Dasgupta, 2019; Senthil‐Kumar & Mysore, 2011). These include brome mosaic virus (BMV) (Ding, Schneider, Chaluvadi, Mian, & Nelson, 2006), foxtail mosaic virus (FoMV) (Mei, Zhang, Kernodle, Hill, & Whitham, 2016), cucumber mosaic virus (CMV) (Wang et al., 2016), and barley stripe mosaic virus (BSMV) (Jarugula, Willie, & Stewart, 2018). Although these vector systems have offered timely tools for maize functional genomics, insert instability continues to be a problematic factor that limits silencing penetrance and longevity in plants. Thus, it is incumbent to continue to explore novel and established maize‐infecting viruses, until more robust and stable VIGS vector systems are developed for maize. Maize rayado fino virus (MRFV), a marafivirus, is a maize‐infecting virus whose genome has been sequenced (Hammond & Ramirez, 2001) and an infectious clone generated (Edwards, Weiland, Todd, & Stewart, 2015), thus offering a ready tool for vector development.
Viruses in the genus Marafivirus, along with two other genera (Tymovirus and Maculavirus), belong to the family Tymoviridae in the alphavirus superfamily (Gibbs et al., 2011; Martelli, Sabanadzovic, Sabanadzovic, Edwards, & Dreher, 2002). Marafivirus species, exemplified by MRFV, share close genomic similarities with tymoviruses, whose type species is Turnip yellow mosaic virus (TYMV) (Gibbs et al., 2011). MRFV is not readily transmissible by sap rub inoculation, although it has been efficiently experimentally transmitted using vascular puncture (Louie, 1995; Madriz‐Ordenana et al., 2000). MRFV is naturally transmitted in a persistent propagative manner by the maize leafhopper Dalbulus maidis, with virus replicating in both the plant host and insect vector (Rivera & Gamez, 1986). It has also been experimentally transmitted by several other leafhopper species including the black‐faced leafhopper, Graminella nigrifrons (Nault, Gingery, & Gordon, 1980). In maize, its natural host, MRFV causes a characteristic fine chlorotic striping and can markedly reduce crop yields in single or mixed infections with other pathogens (Gordon, Nault, Gordon, & Heady, 1985; Zambrano, Francis, & Redinbaugh, 2013). Since its first identification in Costa Rica and El Salvador (Gamez, 1969, 1973), MRFV has become an economically significant pathogen of maize in most of Central America.
The MRFV genome is monopartite, and its positive sense RNA has a 5’ cap and 3’ polyA tail (Edwards & Weiland, 2011). The MRFV genome, like that of other members of the family Tymoviridae, has a high cytidine content (Hammond & Ramirez, 2001). The genome encodes a precursor polyprotein that is post‐translationally cleaved into replication‐associated nonstructural proteins with canonical motifs of a methyltransferase (MTR), papain‐like protease (PRO), helicase (NTP/HEL), and polymerase (POL) (Hammond & Ramirez, 2001) and encodes structural proteins at the 3’ end. Most cleavage sites of the polyprotein have not been experimentally mapped. The MRFV HEL/POL and POL/CP1 junctions were predicted based on validated homologous positions in tymoviruses (Jakubiec, Drugeon, Camborde, & Jupin, 2007), with the POL/CP1 junction subsequently validated in another member of the genus Marafivirus, Oat blue dwarf virus (OBDV; Edwards & Weiland, 2014).
The genome is encapsidated in icosahedral particles in a ratio of 1:3 by two carboxy co‐terminal coat proteins: CP1, likely cleaved from the polyprotein; and CP2, likely expressed from a subgenomic RNA, as reported for OBDV (Edwards & Weiland, 2014). Synthesis of the subgenomic RNA is directed by the marafibox (Edwards et al., 2015), a 16‐nucleotide promoter that is conserved in marafiviruses and equivalent to the tymobox of tymoviruses. Interestingly, unexpected variability in genome organization between marafivirus species has been reported. Notably, a second small overlapping open reading frame not found in OBDV is found in the 3’ end in Citrus sudden death‐associated virus, another member of the genus (Maccheroni et al., 2005). Similarly, a second small overlapping open reading frame absent in OBDV is found in the 5’ end in MRFV, in which it encodes a P43 protein of unknown function that is not required for systemic movement or leafhopper transmission (Edwards, Weiland, Todd, Stewart, & Lu, 2016). Characterization of newly reported marafivirus species may shed more insights into the extent of the diversity of genome expression strategies within the Marafivirus genus.
Besides MRFV, full genome sequences have been determined for at least ten marafivirus species, including the better‐studied OBDV for which an infectious clone has also been reported (Edwards & Weiland, 2010; Edwards, Zhang, & Weiland, 1997). These offer novel genomic tools not only to better understand the biology of marafiviruses and their interactions with host plants and insect vectors, but for design of improved VIGS vectors for functional genomics. However, genomic loci amenable to insertion of heterologous sequences have not been determined for any member of the genus Marafivirus. Indeed, of all genera in the family Tymoviridae, a virus‐derived vector system has been previously described only for TYMV in the genus Tymovirus (Camborde, Tournier, Noizet, & Jupin, 2007; Pflieger et al., 2008; Yu et al., 2019). TYMV was developed as a VIGS vector system for Arabidopsis, based on insertion of a 76‐nt phytoene desaturase (PDS) inverted repeat sequence in the 3’ UTR immediately after the CP stop codon (Pflieger et al., 2008). The TYMV‐derived vector showed reduced symptom severity but maintained infectivity and systemic movement, resulting in VIGS in Arabidopsis (Pflieger et al., 2008).
In this study, we identified the MRFV helicase/polymerase junction as a site for generating a VIGS vector system for maize gene silencing exhibiting strong silencing phenotype penetrance and stability of inserted sequences. The insert remained fully intact for at least 60 days post‐inoculation (dpi) of the Silver Queen maize hybrid and over four serial passage infections.
2. MATERIALS AND METHODS
2.1. Virus strains, plasmids, and cloning procedures
The U.S. isolate of MRFV (MRFV‐US), first identified in maize leaf samples from Texas, was described previously (Bradfute et al., 1980), as well as an infectious MRFV‐US clone in the pGEM‐T Easy plasmid (Edwards et al., 2015). The binary vector pJL89 was used as a cloning vector for full length viral genomes (Crivelli, Ciuffo, Genre, Masenga, & Turina, 2011; Lindbo, 2007). Viral genomic cDNAs were cloned into SmaI/StuI‐cut pJL89 to position the viral cDNA precisely after the Cauliflower mosaic virus (35S) promoter transcription initiation site and immediately before the self‐cleaving Hepatitis delta virus ribozyme sequence (HDV Rz), thus ensuring in planta synthesis of viral transcripts with exact viral 5′‐ and 3′‐ends. The DNA fragments for cloning were obtained as synthetic gene blocks (Integrated DNA Technologies, Coralville, IA) or PCR amplified from cDNAs using PrimeSTAR HS DNA polymerase (TaKaRa). Oligonucleotide synthesis and sequencing services were provided by Eurofins Genomics and Genewiz. The constructs were built using the NEBuilder HiFi DNA Assembly Master Mix (NEB) and amplified in NEB Stable Competent Escherichia coli cells (C3040H) (NEB).
2.2. In vitro transcription of viral genomic RNAs
Viral genomic cDNA templates for in vitro transcription (IVT) were PCR‐amplified using the PrimeSTAR GXL Premix (TaKaRa) according to the manufacturer's protocol and cycling conditions. The forward primer (sm125: CGTACGCGTTAATACGACTCACTATAGTC CTCTGCCCCCTTCTTGC) was designed to incorporate the minimal sequence (underlined) of the T7 promoter. The reverse primer (sm127: TTTTTTTTTTTTTTTTTTTTTGGCCCACAGGTCTTAT GGCCGACC) was appended with a 21‐nt polyT tail (underlined) for generation of polyA‐tailed in vitro transcripts. The amplified DNA templates were cleaned using NEB Monarch PCR and DNA Cleanup Kit (NEB) and 1µg used for each IVT reaction. IVT was done using the NEB HiScribe T7 ARCA mRNA Kit (NEB), modified by omitting the polyA tailing reaction step for transcripts.
2.3. Plant inoculation, growth conditions, and virus detection
Inoculations were done using vascular puncture (VPI) with an engraving tool and 8‐pin tattoo needle as described previously (Edwards & Weiland, 2011; Louie, 1995). Maize seeds were sterilized in 30% bleach for 3 min and dried on a paper towel. Prior to VPI, seeds were soaked in sterile water for 2 hrs at 30°C. Seeds were inoculated on each side of the embryo with 1 µg of in vitro transcripts using 0.2‐mm gauge 8‐pin needle (needlejig.com, product 0815M1). Thus, a total of 2 µg of in vitro transcripts were used per seed. For serial passage assays, 200 mg of infected leaves were ground in 1ml of 0.01M potassium phosphate buffer (pH 7.0) and the extract used as inoculum (4 µl per seed) for VPI. The inoculated seeds were incubated on damp paper towels in a tray at 30°C for 2 days. Five seeds per pot were planted and grown in a chamber set at 24°C and 16‐hrs light. Virus was RT‐PCR assayed in ninth leaves 30 days post‐VPI and in last leaves preceding tassels (~16th leaf) 60 days post‐VPI.
2.4. Virus leafhopper transmission assays
D. maidis was obtained from Kings County, California, in 2010 and maintained on Early Sunglow maize (Schlessman Seed Co.) at 27°C (daytime) and 16°C (night) with a 15‐hrs light: 9‐hrs dark photoperiod. The MRFV constructs were acquired by 14 day post‐oviposition D. maidis insects, with a 3‐week acquisition access period from source maize plants (Silver Queen) infected by VPI. Virus‐exposed leafhoppers were then moved to transmission test plants. Each transmission replicate had 49 maize test plants (7‐day‐old Silver Queen) inoculated with 196 MRFV‐exposed leafhoppers allowed to fly freely within the cage of 49 plants (average of 4 leafhoppers/plant) for four days, after which leafhoppers were removed by aspiration. Any remaining insects were killed using Nuvan ProStrips (dichlorvos, AMVAC Chemical Corp.). Each transmission was repeated three times (three replicates). The infection rate was observed by scoring MRFV symptoms 7 and 14 days post‐infestation with viruliferous leafhoppers. The PDS insertions in MRFV‐symptomatic plants were confirmed by RT‐PCR with primers sm151 (CGTATGTGGACTTCTACTGCTT) and sm152 (GACGGAAGCGCAGTCTCTTA).
2.5. Total RNA isolation and gel blot analysis
High molecular weight (HMW) and low molecular weight (LMW) total RNA was isolated for gel blot analysis as described previously (Mlotshwa et al., 2005). For mRNA gel blot analysis, 5 µg of HMW RNA was run on formaldehyde denaturing agarose gels; and for siRNA gel blot analysis, 10 µg of LMW RNA was run on 20% polyacrylamide/7 M urea denaturing gels. The resolved RNA (HMW or LMW) was blotted onto Amersham hybond‐N+ membranes (GE Healthcare) and hybridized with RNA probes. PDS probes were derived from cDNA fragments corresponding to the maize PDS mRNA sequence (GenBank Accession # NM_001352010.1; nt 1493–1612 and 1613–1732 for siRNAs and mRNA probes, respectively). LSP probes were derived from cDNA fragments corresponding to the maize LSP mRNA sequence (NCBI sequence NM_001175829; nt 1217–1426 and 1427–1651 for siRNAs and mRNA probes, respectively). The respective cDNA fragments were PCR‐appended with a T7 promoter at the 3′‐ or 5′‐end to make RNA probes to detect mRNAs or antisense siRNAs, respectively. The [α‐32P]UTP‐labeled RNA probes were synthesized using the MAXIscript T7 in vitro transcription kit (Invitrogen) and hybridized to mRNA blots at 68°C, or to siRNA blots at 42°C in PerfectHyb Plus buffer (Sigma). The siRNA sizes were based on Decade molecular weight RNA markers (Invitrogen) labeled with gamma‐32P‐ATP.
2.6. Autoradiography, plant photography, and electron microscopy
RNA blots were exposed to a PhosphorImager screen and the radiolabeled bands captured using the Storm 860 PhosphorImager (Molecular Dynamics) and band intensity values relative to background quantitated using the ImageQuant software. The mean value for the healthy controls in each experiment was designated 1.00 and used as a baseline to accordingly compute relative accumulation level for each sample. Plant images were acquired using a Canon DS126431 digital camera (Canon). Virus particles were captured using the Hitachi H‐7500 transmission electron microscope (Hitachi, Ltd.) following negative staining with uranyl acetate.
3. RESULTS
3.1. MRFV insertion constructs and viability testing
As the only available precedent for insertion of foreign sequences into the genome of a Tymoviridae member, TYMV offered the only lead in our search for viable insertion sites in MRFV. Based on close genomic similarities between tymoviruses and marafiviruses, our test insertions into MRFV first targeted the 3’ UTR position (nucleotides 6211/6212) equivalent to that used for TYMV VIGS vector (Pflieger et al., 2008), followed by other selected genomic loci.
All experiments used the previously described infectious clone of the U.S. isolate of MRFV (Edwards et al., 2015). A 120‐nt maize PDS cDNA fragment, corresponding to nucleotides 1493 to 1612 in the NCBI sequence NM_001352010.1, was amplified and used as a test insert. This enabled us to simultaneously search for MRFV insertion sites that retained virus infectivity as well as test the potential of resulting constructs as VIGS vectors. The recombinant MRFV constructs, generated through Gibson assembly of the PDS insert and MRFV PCR fragments in StuI/SmaI‐cut pJL89 binary vector, are shown in Table 1, with sequences associated with each PDS insert at each locus shown in Figure 1 (a1–d1). The constructs served as templates to amplify viral genomic cDNAs appended with T7 minimal promoter at 5’‐ends and 21‐nt polyT tail at 3’‐ends, from which RNAs were in vitro transcribed for infectivity testing in maize using VPI of maize kernels as previously described (Edwards et al., 2015; Louie, 1995; Madriz‐Ordenana et al., 2000).
TABLE 1.
MRFV genomic loci targeted for viability testing and VIGS constructs generated
| Construct (same ID as insert) | PDS size/total insert size (nt) | Insert position in MRFV genome | Clone ID | Construct details (bp: fragment size; nt: fragment position in MRFV genome; pmol: fragment amount used in 10 µl Gibson reaction) | ||||
|---|---|---|---|---|---|---|---|---|
| SmaI/StuI‐cut pJL89 (4675bp) | Fragment 1 | Fragment 2 | Fragment 3 | Fragment 4 | ||||
| a1 | 120/183 | nt 127/128 (after the start codon of polyprotein) | pSM199‐223 | 0.06 pmol | 127 bp (nt 1–127) 0.1 pmol | 183 bp (a1 insert) 0.1 pmol | 3,105 bp (nt 128–3232) 0.05 pmol | 3,105 bp (nt 3233‐6337) 0.05 pmol |
| b1 | 120/201 | nt 3709/3710 (HEL/POL junction) | pSM163‐177 | 0.04 pmol | 3,709 bp (nt 1–3709) 0.05 pmol | 201 bp (b1 insert) 0.1 pmol | 2,628 bp (nt 3710–6337) 0.05 pmol | – |
| b2 | 231/312 | nt 3709/3710 (HEL/POL junction) | pSM326‐351 | 0.04 pmol | 3,709 bp (nt 1–3709) 0.05 pmol | 312 bp (b2 insert) 0.1 pmol | 2,628 bp (nt 3710–6337) 0.05 pmol | – |
| b3 | 210/291 (LSP) | nt 3709/3710 (HEL/POL junction) | pSM389‐392 | 0.04 pmol | 3,709 bp (nt 1–3709) 0.05 pmol | 291 bp (b2 insert) 0.1 pmol | 2,628 bp (nt 3710–6337) 0.05 pmol | – |
| c1 | 120/201 | nt 5494/5495 (POL/CP1 junction) | pSM153‐162 | 0.04 pmol | 5,494 bp (nt 1–5494) 0.05 pmol | 201 bp (c1 insert) 0.1 pmol | 843 bp (nt 5495–6337) 0.07 pmol | – |
| d1 | 120/120 | nt 6211/6212 (3’ UTR after CP2 stop codon) | pSM47‐55 | 0.1 pmol | 3,105 bp (nt 1–3105) 0.05 pmol | 3,106 bp (nt 3106–6211) 0.05 pmol | 120 bp (e1 insert) 0.2 pmol | 126 bp (nt 6212‐6337) 0.2 pmol |
FIGURE 1.
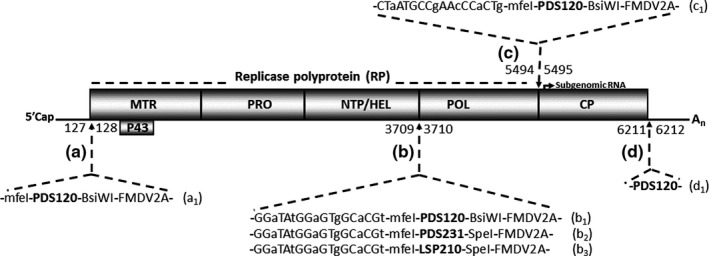
MRFV genome organization and loci tested for viability as insertion sites. The genome outline shows replication‐associated proteins (MTR, PRO, NTP/HEL, and POL) processed from the replicase polyprotein (RP) at predicted cleavage junctions. The genome is encapsidated by 2 carboxyl co‐terminal coat proteins: CP1, likely cleaved from the polyprotein; and CP2, likely expressed from a subgenomic RNA. Individual loci targeted for insertion of heterologous sequences are indicated, [(a)–(d)]. The numbers straddling the arrows indicate MRFV nucleotide junctions carrying insertions. Insertion cassettes are annotated as a1 to d1, as well as the constructs derived from each of the cassettes (see Table 1 first column). The sequence identity of each cassette is as indicated
The d1 construct‐derived clones (pSM47‐55), with a 120‐nt PDS insertion in the 3’ UTR immediately after the CP stop codon (nucleotides 6211/6212) (Figure 1d, Table 1), were VPI tested for infectivity. In vitro transcripts derived from these clones resulted in no symptomatic plants or RT‐PCR detectable virus in inoculated or systemic leaves (data not shown), suggesting lethality of insertions at this position in MRFV. Thus, unlike in TYMV, insertions at the CP2 stop codon/3’UTR junction likely disrupted functions critical for MRFV infectivity, possibly due to the larger insert size (120 nt) compared to the 76‐nt insert in TYMV.
Since no infection was observed from the d1 construct (clones pSM47‐55), we targeted gene junctions as they have proven to be good candidates for inserting foreign sequences in several virus systems (for recent reviews, see Cody & Scholthof, 2019; Hefferon, 2017). As most cleavage sites of the MRFV polyprotein have not been experimentally determined, we selected the predicted cleavage sites of the helicase/polymerase (HEL/POL; nt 3709/3710) (Figure 1b) and polymerase/coat protein1 (POL/CP1; nt 5494/5495) (Figure 1c) junctions for initial testing. The predictions were based on experimentally validated homologous positions in tymoviruses and OBDV for HEL/POL and POL/CP1 junctions, respectively (Edwards & Weiland, 2014; Jakubiec et al., 2007). Additionally, we tested insertions after the start codon of the polyprotein after the 5’ UTR (nucleotides 127/128) (Figure 1a). The insertion cassette was composed of the 120‐nt PDS fragment flanked by MfeI and BsiWI restriction sites, and a 51‐nt Foot‐and‐mouth disease virus (FMDV) 2A peptidase coding sequence (‐mfeI‐PDS120nt‐BsiWI‐FMDV2A‐), all in frame for no introduction of stop codons or frameshifts. To preserve polyprotein processing at carboxyl ends of the insertions, the FDMV 2A peptidase was included in lieu of duplicating native cleavage sites. Insertion cassettes between the HEL/POL and POL/CP1 junctions additionally included 18 nucleotide duplications of the 5’ end sequences of the POL and CP1 genes, respectively (Figure 1b: b1, b2, and b3; and Figure 1c: c1), to maintain native proteolytic cleavage at both junctions. For both duplicated sequences, degenerate codons were used to conserve amino acid sequence while reducing nucleotide sequence identity that might contribute to loss of inserts through recombination. Recombinant MRFV constructs and clones obtained from assembly of cassettes with MRFV PCR fragments in plasmid pJL89 are shown in Table 1.
For insertions in the POL/CP1 (5494/5495) (clones pSM153‐162) or after the start codon of the polyprotein (nucleotides 127/128) (clones pSM199‐223), no infectivity was RT‐PCR detected in inoculated or systemic leaves, indicating that disrupting these positions or the insert size was likely lethal to MRFV.
3.2. Viability of MRFV HEL/POL junction as an insertion site for nonviral sequences
For insertions at the MRFV HEL/POL junction, two clones (pSM165 and pSM167) of this construct (Figure 1b1; Table 1) were selected for testing. In vitro transcripts of the clones (pSM165 and pSM167), herein designated construct MRFV‐PDS120, were VPI tested for infectivity on maize in three replicates as shown in Table 2. Wild‐type MRFV (MRFV‐WT)‐inoculated and noninoculated plants were included as positive and negative controls respectively. MRFV‐PDS120 transcripts induced fine chlorotic striping symptoms similar to MRFV‐WT five dpi, thus indicating that MRFV‐PDS120 clones were infectious (Figure 2b), with VPI efficiency of 18% (9/51) to 40% (14/35) (Table 2). Similar to MRFV‐WT, MRFV‐PDS120 symptoms spread systemically into newly emerging noninoculated upper leaves, indicating that the insert did not compromise systemic virus movement. Interestingly, the onset of MRFV‐PDS120 symptoms triggered extensive visual photobleaching initiating from veins and spreading laterally in both local and systemic leaves (Figure 2b), suggesting intact PDS insert in MRFV‐PDS120 in both local and systemic infections.
TABLE 2.
VPI testing of MRFV‐PDS120 for infectivity and VIGS on maize
| Construct | Experiment 1 | Experiment 2 | Experiment 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MRFV‐PDS120 | MRFV‐WT | Healthy control | MRFV‐PDS120 | MRFV‐WT | Healthy control | MRFV‐PDS120 | MRFV‐WT | Healthy control | |
| Inoculated plants | 51 | 7 | 5 | 35 | 13 | 5 | 36 | 8 | 5 |
| MRFV symptomatic plants | 9 | 6 | 0 | 14 | 5 | 0 | 12 | 2 | 0 |
| PDS silencing plants | 9 | 0 | 0 | 14 | 0 | 0 | 12 | 0 | 0 |
FIGURE 2.
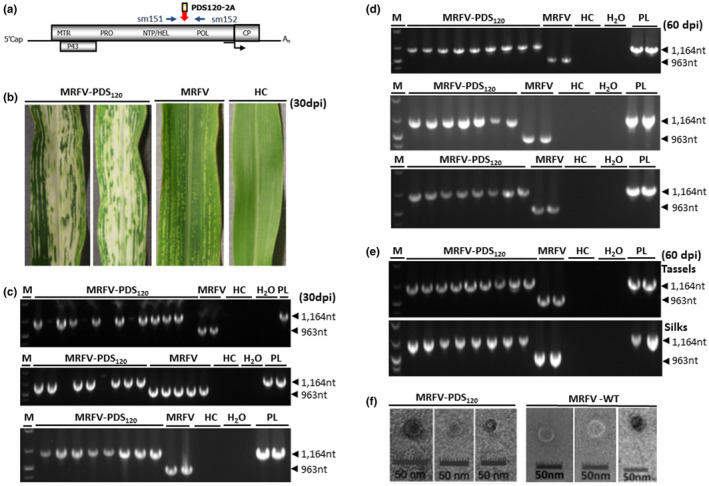
Viability of MRFV HEL/POL junction as insertion site. (a) Location of primers (sm151 and sm152) used for RT‐PCR detection of MRFV‐PDS120 and MRFV‐WT. The primers straddle the HEL/POL junction and amplify 1,164 bp in MRFV‐PDS120 and 963bp in wild‐type MRFV. (b) Virus symptoms and the chlorophyll photobleaching phenotype induced by MRFV‐PDS120 at 30 dpi, compared to leaves of plants inoculated with MRFV without insert and noninoculated leaves (HC). (c) RT‐PCR analysis of virus accumulation in systemic leaves at 30 days post‐inoculation. The three gels show RT‐PCR detection of MRFV‐PDS120 and MRFV‐WT in inoculated plants in three replicated experiments, with RNA from noninoculated plants (HC) and water (H2O) included as negative control templates; and the MRFV‐PDS120 plasmid (PL) serving as a positive control. Blank lanes in MRFV‐PDS120 inoculations represent nonsymptomatic plants. M: 100 bp DNA marker. (d) RT‐PCR analysis of insert retention in MRFV‐PDS120 at 60 days post‐inoculation. RT‐PCR was repeated at 60 dpi only for RT‐PCR positive (symptomatic/successfully inoculated) MRFV‐PDS120 plants in (c) above, with controls and DNA marker included as described above. This was done for all the three replicated experiments. (e) RT‐PCR assays for virus accumulation in tassels and silks for a subset of MRFV‐PDS120 symptomatic plants. Controls and DNA marker were included as described above. (f) Transmission electron micrographs of MRFV‐PDS120 (left panel) and MRFV‐WT particles (right panel) in maize (Silver Queen) extract
For molecular analysis of the integrity of the MRFV‐PDS120 insertion sequence in systemic leaves of symptomatic plants, we used RT‐PCR with MRFV‐specific primers sm151 and sm152 straddling the PDS insert and amplifying 963 bp in MRFV and 1,164 bp in MRFV‐PDS120 (Figure 2a). The expected 963 bp and 1,164 bp bands were amplified in MRFV‐WT and in MRFV‐PDS120, respectively, confirming the presence of intact insert in MRFV‐PDS120 30 dpi in ninth leaves (Figure 2c). The same set of plants was further RT‐PCR tested to determine the insert retention capacity of MRFV‐PDS120 at 60 days following initial inoculations with in vitro transcripts. All the analyzed plants still retained the inserts at 60 dpi in last leaves preceding tassels (~16th leaf) with no sign of insert loss (Figure 2d), including in tassels and silks for a subset of plants that produced them (Figure 2e). RT‐PCR assays also showed comparable detectability for MRFV‐PDS120 and MRFV‐WT in infected plants (Figure 2c,d,e). This correlated well with similar spatial and temporal symptom progression patterns of MRFV‐PDS120 and MRFV‐WT. The MRFV HEL/POL junction was thus identified as a viable site for insertion of heterologous sequences. The insertion did not affect the formation of icosahedral virus particles as confirmed using electron microscopy (Figure 2f), thus indicating maintenance of apparently normal MRFV virions for MRFV‐PDS120.
3.3. Northern blot analysis of the chlorophyll photobleaching phenotype induced by MRFV‐PDS120 in maize
Silencing induced by MRFV‐PDS120 was further assessed by Northern blotting to determine the accumulation of PDS mRNA and siRNAs. Northern blot analysis of total RNA isolated from leaf tissue 30 dpi showed reduced accumulation of PDS mRNAs in MRFV‐PDS120‐infected plants (Figure 3a, lanes 1–4) but not in MRFV‐WT‐infected and noninoculated plants (Figure 3a, lanes 5–6 and 7–8, respectively). The depletion of PDS mRNA in MRFV‐PDS120‐infected plants was associated with accumulation of predominantly 21‐nt PDS siRNAs (Figure 3a, lanes 1–4). PDS siRNAs were not detected in MRFV‐WT infected and healthy plants (Figure 3a, lanes 5–6 and 7–8, respectively). Overall, RNA analyses showed that the MRFV‐PDS120‐induced leaf photobleaching correlated with accumulation of PDS siRNAs and up to sixfold reduction in PDS mRNA levels, thus establishing VIGS of PDS mRNA by MRFV‐PDS120 as the molecular basis of the photobleaching phenotype. The marked depletion of PDS mRNAs at 30 dpi corroborates the extensive visual VIGS phenotype, both of which are consistent with stable maintenance of intact insert in MRFV‐PDS120 as well as retention of high levels of local and systemic infectivity similar to MRFV WT. In addition, RNA blot analysis showed reduction in PDS mRNA and accumulation of PDS siRNAs in MRFV‐PDS120‐infected tassels (Figure 3b, lanes 1–2) and silks (Figure 3b, lanes 3–4), an indication that VIGS also extends into reproductive tissue. This is consistent with detection of intact MRFV‐PDS120 in tassels and silks (Figure 2e).
FIGURE 3.
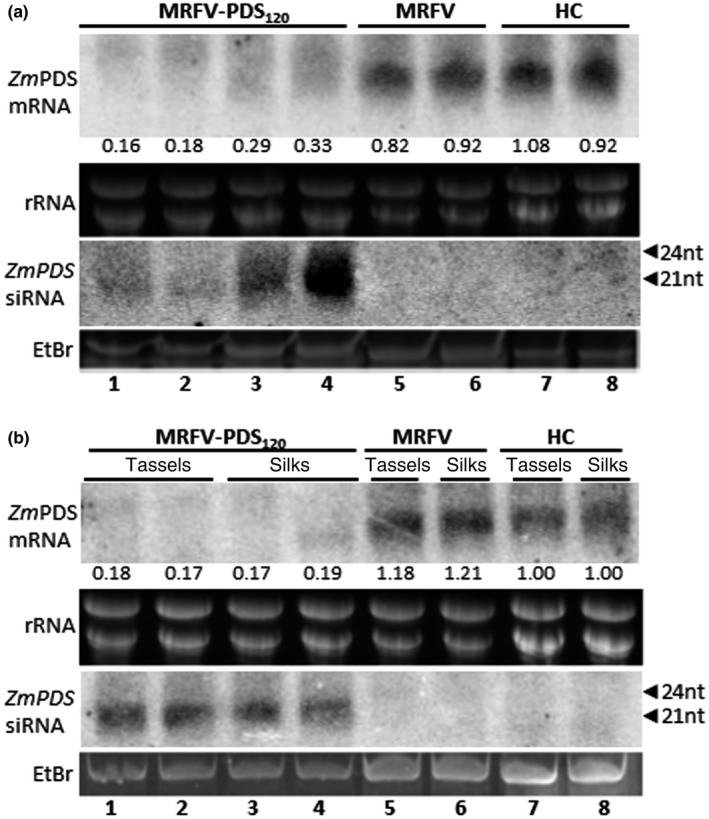
Northern blot analysis of photobleaching phenotype induced by MRFV‐PDS120. High and low molecular weight RNA blots were hybridized with PDS‐specific RNA probes to respectively detect PDS mRNAs or antisense siRNAs in RNA samples isolated from leaves 30 dpi (a), and from tassels and silks 60 dpi (b). For loading controls, rRNA and the major RNA species in low molecular weight RNA were ethidium bromide (EtBr)‐stained prior to blotting of gels onto membrane. The positions of 21‐ to 24‐nucleotide RNA size markers are indicated by arrowheads. (a) Levels of PDS mRNA and siRNAs from MRFV‐PDS120‐infected leaves is shown (lanes 1–4) compared to MRFV‐infected (lanes 5–6) and healthy plants (lanes 7–8). The relative level of PDS mRNA (determined using ImageQuant) is indicated for each sample normalized against the average of the two healthy control samples set at 1.00. (b) Levels of PDS mRNA and siRNAs from MRFV‐PDS120‐infected tassels is shown (lanes 1–2) compared to MRFV‐infected (Lane 5) and healthy tassels (Lane 7). Levels of PDS mRNA and siRNAs from MRFV‐PDS120‐infected silks are shown (lanes 3–4) compared to MRFV‐infected (Lane 6) and healthy silks (Lane 8). The relative levels of PDS mRNA were determined as described in (a), with band intensity values of healthy tassels (Lane 7) and silks (Lane 8) independently designated as 1.00 and samples from respective tissues normalized against these controls
3.4. Insert stability of MRFV‐PDS120 after VPI serial passages
Following initial plant inoculations with MRFV‐PDS120 in vitro transcripts, RT‐PCR analyses showed insert still intact in MRFV‐PDS120 at 30 and 60 dpi with no signs of reversions to MRFV‐WT (Figure 2c, d and e). Thus, we further tested the stability of MRFV‐PDS120 using serial passage inoculations. Plant extract prepared from each successive set of MRFV‐PDS120‐infected plants 20 dpi was used as inoculum in four serial passage inoculations using VPI. The numbers of symptomatic plants at each passage, reflecting successful inoculations, were 23/47 (49%), 24/53 (45%), 26/51 (51%) and 41/83 (49%) infected plants for passage 1 to 4 successively. All the infected plants continued to induce strong PDS VIGS at each of the four VPI passages of plant extract, indicating that MRFV‐PDS120 progeny still retained the insert after four passages. Representative VIGS phenotypes are shown in Figure 4a for each passage. The integrity of MRFV‐PDS120 in each of the infected plants at each passage was further evaluated at 15 and 60 dpi using RT‐PCR with primers sm151 and sm152 to ascertain insert stability post‐passaging. All the infected plants at each passage were individually RT‐PCR tested, of which representative results are shown in Figure 4b. Remarkably, MRFV‐PDS120 continued to stably maintain the insert as tested at 15 and 60 dpi after each of the four passages, with first sign of insert loss observed in only 1 of 41 infected fourth passage plants at 60 dpi (Figure 4b, psg 4 panel; Lane 8). Thus, in addition to retaining wild‐type MRFV levels of infectivity, the MRFV‐PDS120 recombinant virus shows remarkable stability of insert upon mechanical transmission using VPI.
FIGURE 4.
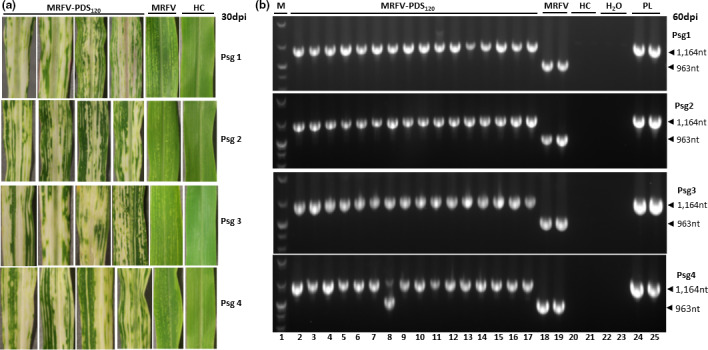
Testing of the stability of MRFV‐PDS120 using serial passage inoculations with crude plant sap. (a) MRFV‐PDS120 symptoms and chlorophyll photobleaching phenotype at 30 dpi for each passage, compared to MRFV‐WT and noninoculated plants (HC). (b) RT‐PCR analysis (primers sm151 and sm152) of the integrity of MRFV‐PDS120 at each of the four passages. All the MRFV‐PDS120‐infected plants obtained for each passage were RT‐PCR assayed both at 15 dpi (not shown) and at 60 dpi (representative gel shown here). Healthy plants (HC) and water (H2O) were included as negative controls, and MRFV‐PDS120 plasmid (PL) as a positive control. M: 100 bp DNA marker.
3.5. Transmissibility of MRFV‐PDS120 by the leafhopper D. maidis
To continue the characterization of MRFV‐PDS120, we investigated its transmissibility by the leafhopper D. maidis, which transmits MRFV to maize in a persistent‐propagative manner (Nault et al., 1980; Rivera & Gamez, 1986). The results of MRFV‐PDS120 transmissibility assays, done in three independent experiments of 49 plants each, are shown in Table 3 (row labeled first passage infections). Wild‐type MRFV was included as positive control. Test plants inoculated with MRFV‐WT began to show symptoms seven days post‐exposure to viruliferous D. maidis, and 19 of the 49 MRFV inoculated plants showed MRFV symptoms by 21 days post‐exposure to viruliferous D. maidis. The MRFV‐PDS120‐inoculated plants similarly developed MRFV symptoms as well as VIGS photobleaching 7 days after exposure to viruliferous D. maidis (Figure 5a, i, iii, and v), demonstrating that the recombinant virus was insect transmissible, with symptom onset similar to MRFV‐WT. The MRFV‐PDS120 transmission efficiency was variable between the three independent experiments, ranging from 5/49 (10%) to 10/49 (20%) plants compared to 19/49 (39%) for MRFV‐WT (Table 3).
TABLE 3.
Transmissibility of MRFV‐PDS120 by D. maidis and stability testing through passaging
| Experiment | 1 | 2 | 3 | |
|---|---|---|---|---|
| Construct | MRFV‐PDS120 | MRFV | MRFV‐PDS120 | MRFV‐PDS120 |
| First passage infections | 9/49 | 19/49 | 5/49 | 10/49 |
| Second passage infections | 11/49 | 17/49 | ||
| Third passage infections | 11/49 | 11/49 | ||
FIGURE 5.
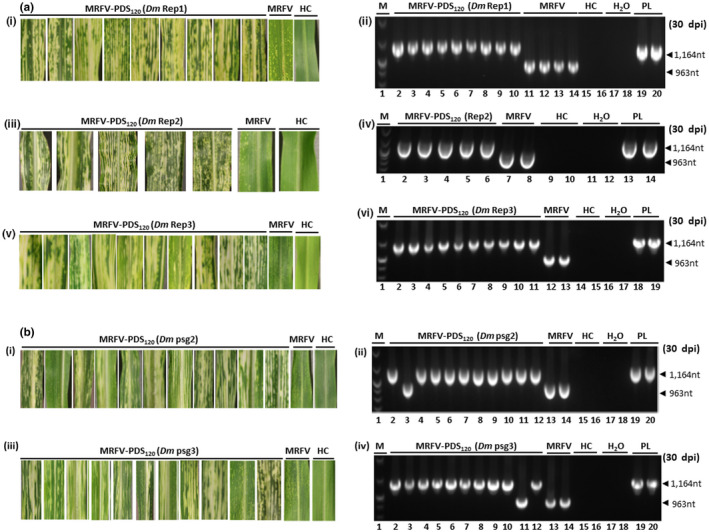
(a) Transmissibility of MRFV‐PDS120 by D. maidis (Dm) and RT‐PCR analysis (primers sm151 and 152) of insert stability. Replicated experiments showing virus symptoms and chlorophyll photobleaching phenotype induced by D. maidis‐transmitted MRFV‐PDS120 at 30 dpi compared to MRFV‐WT and noninoculated plants (i, iii and v); and corresponding RT‐PCR assays of MRFV‐PDS120 and MRFV‐WT in systemic leaves of infected plants (ii, iv and vi). Healthy plants (HC) and water (H2O) were included as negative controls, and the MRFV‐PDS120 plasmid (PL) as a positive control. M: 100 bp DNA marker. (b) Stability of MRFV‐PDS120 through D. maidis (Dm) passaging. (i) Virus symptoms and chlorophyll photo bleaching phenotype induced by MRFV‐PDS120 at 30 dpi after passage 2 acquisition and transmission by D. maidis, compared to MRFV‐WT and noninoculated plants, and (ii) corresponding RT‐PCR assays of MRFV‐PDS120 and MRFV‐WT in systemic leaves of infected plants. The controls and DNA marker were included as described above. (iii) Virus symptoms and chlorophyll photobleaching phenotype induced by MRFV‐PDS120 at 30 dpi after passage three acquisition and transmission by D. maidis, compared to MRFV‐WT and noninoculated plants, and (iv) corresponding RT‐PCR assays of MRFV‐PDS120 and MRFV‐WT in systemic leaves of infected plants. The controls were included as described above
Interestingly, the VIGS phenotype in D. maidis inoculated plants was still as visually strong as in VPI inoculated source plants, suggesting that the PDS insert was intact following insect transmission. This was further verified using RT‐PCR analysis with primers sm151 and sm152. The homogenous bands show that the insert was maintained intact in the hopper transmitted virus with no signs of insert loss (Figure 5a,ii,iv,vi). Overall, the data showed that MRFV‐PDS120 was transmissible by D. maidis and emerged from the propagative phase in the leafhopper with the insert still intact in 3 independent experiments.
3.6. Stability of MRFV‐PDS120 through leafhopper transmission
To fully investigate the effect of leafhopper transmission on the stability of MRFV‐PDS120 compared to VPI transmission, we carried out serial passage experiments using D. maidis. The nine first passage plants of the transmissibility test in Figure 5a were used as source plants for second passage MRFV‐PDS120 acquisition by D. maidis and transmission onto 49 healthy plants. Of the 49 plants, 11 developed MRFV symptoms and 10 showed photobleaching (Figure 5b,i). The 11 second passage plants were further RT‐PCR screened for insert stability using primers sm152 and sm153. The results confirmed the presence of the PDS insert in the 10 silencing plants as expected (Figure 5b,ii, lanes 2 and 4–12), and that the single MRFV symptomatic but nonsilenced plant had lost the PDS insert (Figure 5b,ii, Lane 3).
The ten silenced second passage plants, carrying intact insert confirmed by RT‐PCR, were further used as source plants for a third passage MRFV‐PDS120 acquisition by D. maidis and transmission onto 49 healthy plants. The third passage results recapitulated those of the second passage, with 11 plants developing MRFV symptoms, ten of which showed the photobleaching phenotype (Figure 5b,iii). RT‐PCR analysis of the 11 plants with primers sm151 and sm152 similarly showed the presence of the insert in the ten silenced plants (Figure 5b,iv, lanes 2–10; and Lane 12), and no insert in the single symptomatic but nonsilenced plant (Figure 5b,iv, Lane 11).
Thus, with D. maidis transmission, the first loss of insert was observed in one out of 11 plants at both second and third passage, whereas with VPI transmission the first insert loss was observed for the first time in 1 of 41 infected fourth passage plants at 60 dpi. Together, the results demonstrated that insertion loss was a rare and slow event.
3.7. Capacity of HEL/POL junction to hold larger inserts
The infectivity and unexpected stability of MRFV‐PDS120 prompted us to test the ability of the MRFV HEL/POL junction to host larger inserts. We inserted a 231 nt PDS fragment (NCBI sequence NM_001352010.1, nt 1382 to 1612), a size within the range shown to be optimal for PDS VIGS in maize (Mei et al., 2016; Wang et al., 2016). The MRFV‐PDS231 construct was built with the same insertion cassette as in MRFV‐PDS120, except for replacements of 120 nt PDS with 231nt PDS and the BsiWI site with SpeI (Figure 1b2). Thus, the total insertion cassette for MRFV‐PDS231 was 312 nt compared to the 201 nt insertion cassette in MRFV‐PDS120. In vitro transcripts derived from a subset of the MRFV‐PDS231 clones were VPI tested for infectivity, and two clones (pSM327 and pSM328; Table 1) were identified as infectious, inducing MRFV symptoms and PDS VIGS in local and systemic leaves (Figure 6b). RT‐PCR assays with primers sm151 and 152 (Figure 6a) amplified the expected 0.963 kb in MRFV and 1.275 kb in MRFV‐PDS231 in inoculated and systemic leaves of infected plants (Figure 6c), thus showing insert intact 30 dpi. Thus, the symptoms and VIGS pattern of MRFV‐PDS231 were similar to MRFV‐PDS120, with both retaining local and systemic infectivity similar to MRFV‐WT. Northern blot analysis, performed as described in Figure 3, showed accumulation of predominantly 21 nt PDS siRNAs and up to six‐fold reduction in PDS mRNA levels in MRFV‐PDS231 infected plants (Figure 6d, lanes 1–4), but not in MRFV‐WT infected or healthy plants (Figure 6d, lanes 5–6 and 7–8, respectively). The results thus showed that the MRFV HEL/POL junction tolerates inserts of the size range shown to be effective for VIGS in maize in other virus systems (Mei et al., 2016; Wang et al., 2016).
FIGURE 6.
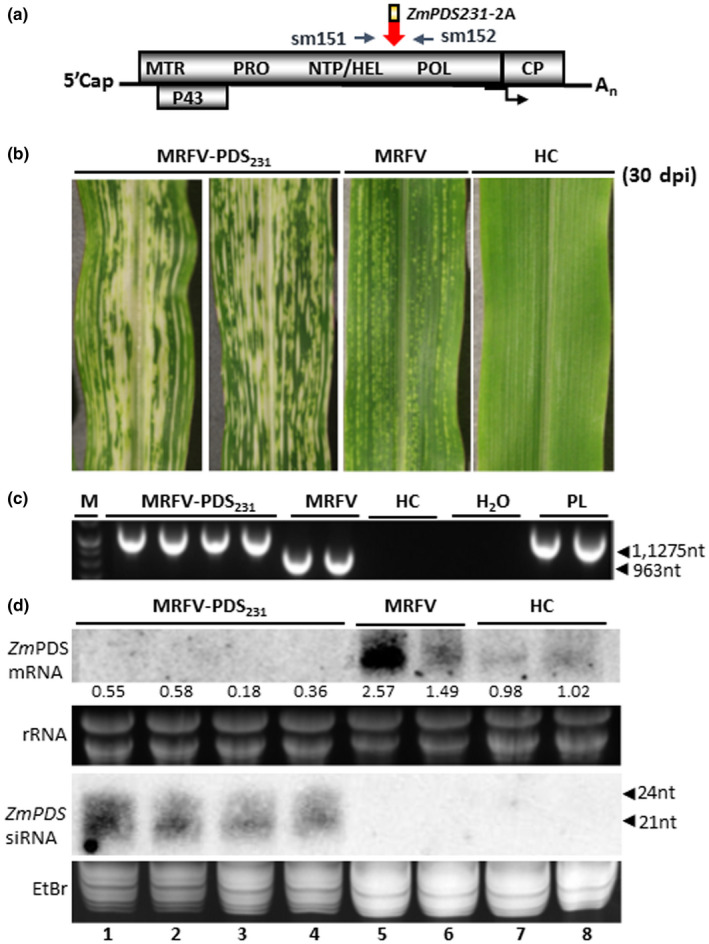
Capacity of HEL/POL junction to hold larger inserts. (a) Similar to MRFV‐PDS120, molecular assays for MRFV‐PDS231 used primers sm151 and 152 for RT‐PCR detection, amplifying 1,275 bp in MRFV‐PDS231 and 963bp in MRFV‐WT. (b) Virus symptoms and chlorophyll photobleaching phenotype induced by MRFV‐PDS231 at 30 dpi, compared to MRFV‐WT and noninoculated plants. (c) RT‐PCR analysis of virus accumulation in systemic leaves at 30 dpi. The gel shows RT‐PCR detection of MRFV‐PDS231 and MRFV‐WT in systemic leaves of inoculated plants. Healthy plants (HC) and water (H2O) were included as negative controls and the MRFV‐PDS231 plasmid (PL) as a positive control. M: 100 bp DNA marker. (d) Northern blot analysis of photobleaching phenotype induced by MRFV‐PDS231 was as described in Figure 3. Levels of PDS mRNA and siRNAs from MRFV‐PDS231‐infected leaves is shown (lanes 1–4) compared to MRFV‐infected (lanes 5–6) and healthy plants (lanes 7–8). The relative levels of PDS mRNA were determined as described in Figure 3
3.8. Potential of the MRFV VIGS system for wider applications
To evaluate the potential of the MRFV VIGS system for wider applications, we first examined its efficacy in silencing a different target gene besides PDS. A 210 nt fragment of ZmlspH (maize lemon white1, lw1) gene (NCBI sequence NM_001175829, nt 1217–1426) was used to replace the PDS insert in MRFV‐PDS231, resulting in MRFV‐LSP210 construct (clones pSM389‐392, Figure 1b3; Table 1). Because of its role in chloroplast development, silencing of ZmlspH results in a characteristic yellow photobleaching albino phenotype (Wang et al., 2016). VPI of Silver Queen kernels with in vitro transcripts of MRFV‐LSP210 clones pSM390 and pSM391 induced MRFV symptoms and LSP VIGS in inoculated and systemic leaves at least 5 dpi (Figure 7b). Virus accumulation was detected through RT‐PCR amplification of 0.963 kb in MRFV and 1.254 kb in MRFV‐LSP210 infected plants using primers sm151 and 152 (Figure 7a,c). Thus, the LSP insert was fully intact in MRFV‐LSP210 infected plants 30 dpi. MRFV‐LSP210 induced VIGS was further confirmed using Northern blot analysis of total RNA isolated 30 dpi. The LSP VIGS phenotype correlated with accumulation of predominantly 21 nt LSP siRNAs and reduced levels of LSP mRNA (Figure 7d, lanes 1–4). Overall, the MRFV‐LSP210 VIGS pattern and longevity was comparable to that described for MRFV‐PDS120/231.
FIGURE 7.
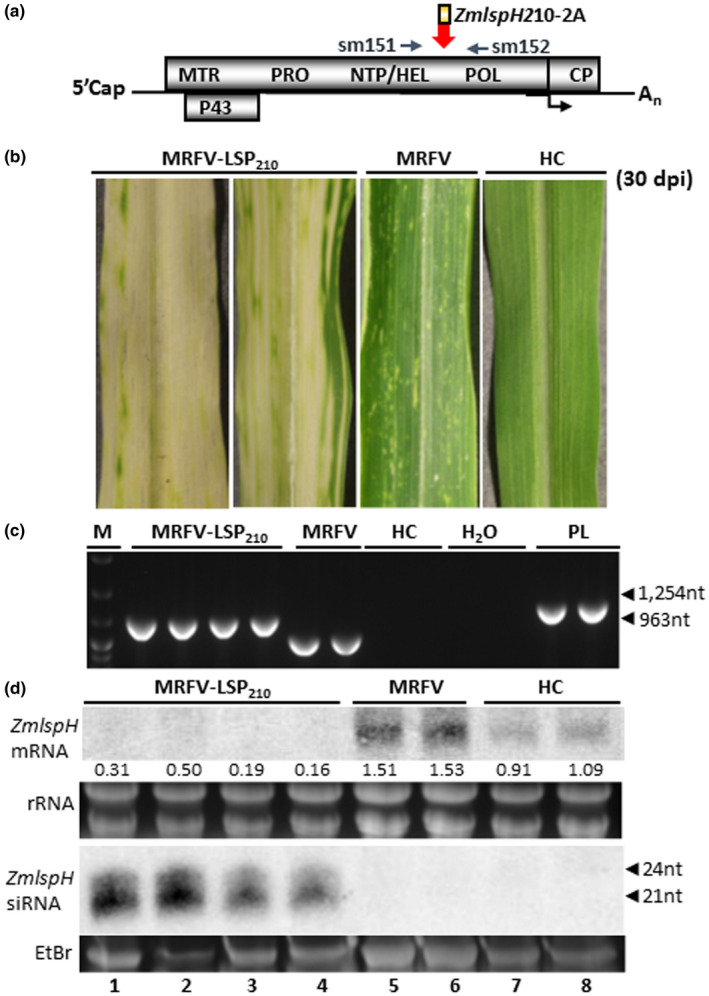
Efficacy of MRFV VIGS system in silencing ZmlspH gene. (a) Molecular assays for MRFV‐LSP210 used primers sm151 and 152 for RT‐PCR detection, amplifying 1,254 bp in MRFV‐LSP210 and 963bp in MRFV‐WT. (b) MRFV symptoms and yellow (albino) VIGS phenotype induced by MRFV‐LSP210 at 30 dpi, compared to plants inoculated with MRFV‐WT or noninoculated (HC). (c) RT‐PCR analysis of MRFV‐LSP210 and MRFV‐WT accumulation in systemic leaves 30 dpi. Noninoculated plants (HC) were included as negative controls; MRFV‐LSP210 plasmid (PL) served as a PCR‐positive control. M: 100 bp DNA marker. (d) Northern blot analysis of VIGS induced by MRFV‐LSP210. RNA blots were hybridized with LSP‐specific RNA probes to detect LSP mRNAs or antisense siRNAs. rRNA: EtBr‐stained ribosomal RNA used as loading control. For small RNA gel, the major low molecular weight RNA species was EtBr‐stained for loading control prior to bloating of gel onto membrane. Levels of LSP mRNA and siRNAs from MRFV‐LSP210‐infected plants (lanes 1–4) compared to MRFV‐infected (lanes 5–6) and healthy plants (lanes 7–8) are shown, with positions of 21‐ to 24‐nucleotide RNA size markers indicated by arrow heads. The relative levels of LSP mRNA were determined as described for PDS in Figure 3
Secondly, we also determined if the utility of the MRFV VIGS vector extends to other maize genotypes commonly used in genetic studies. Kernels of maize inbred lines B73, Mo17 and Va35 were VPI‐inoculated with MRFV‐PDS120 crude plant extract in three independent replicates, with MRFV‐WT included as positive control. MRFV‐PDS120 induced the typical MRFV chlorotic striping symptoms on all the three inbred lines five dpi (Figure 8a), with a total of 7/14, 6/21, and 24/29 MRFV‐PDS120 symptomatic B73, Mo17, and Va35 plants, respectively. The virus symptoms spread to emerging systemic leaves in which virus was RT‐PCR detected with primers sm151 and 152 (Figure 8b). MRFV‐PDS120‐infected plants showed onset of the PDS VIGS phenotype from seven to 15 dpi for all the three inbred lines (Figure 8a). The VIGS phenotype correlated with predominantly 21‐nt PDS siRNAs and about fivefold reduction in PDS mRNA levels (Figure 8c, lanes 1–2; 7–8; and 13–14) on Northern blots probed with same probes as used in Figure 3. Thus, the MRFV VIGS vector system shows potential for wider application in maize functional genomics.
FIGURE 8.
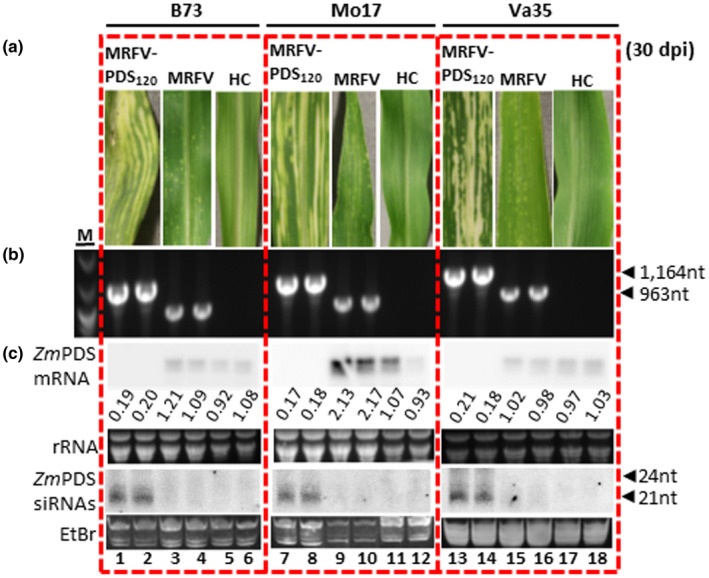
Utility of the MRFV VIGS system on different maize inbred lines. (a) MRFV symptoms and PDS photobleaching induced by MRFV‐PDS120 on B73, Mo17, and Va35 maize inbred lines compared to plants inoculated with MRFV‐WT or noninoculated healthy controls (HC). (b) RT‐PCR detection of MRFV‐PDS120 and MRFV‐WT in systemic leaves of B73, Mo17, and Va35 plants 30 dpi. Noninoculated plants (HC) and water (H2O) were included as negative controls; MRFV‐PDS120 plasmid (PL) served as a PCR‐positive control (not shown). M: 100 bp DNA marker. (c) Northern blot analysis of VIGS induced by MRFV‐PDS120 on B73, Mo17, and Va35 maize inbred lines. Blot hybridizations were as described for Figure 3. PDS mRNA and siRNA levels in MRFV‐PDS120‐infected plants are shown (lanes 1–2; 7–8; and 13–14) compared to MRFV infected (lanes 3–4; 9–10; and 15–16) and healthy plants (lanes 5–6; 11–12; and 17–18). The relative levels of PDS mRNA were determined as described in Figure 3
4. DISCUSSION
The first report of a VIGS vector for maize was based on BMV (Ding et al., 2006). Since then, several other maize‐infecting viruses have been engineered for VIGS, including FoMV (Mei et al., 2016), CMV (Wang et al., 2016), and BSMV (Jarugula et al., 2018). However, instability of inserts in these systems is considered a major constraint to silencing longevity and penetrance in infected plants, with some efforts to improve stability only yielding marginal gains (Ding et al., 2018).
Here, we report pioneering work identifying the MRFV HEL/POL junction as stable for insertion of heterologous sequences, culminating in highly stable VIGS vector that retains insert fully intact throughout the plant growth cycle and over four passage infections, with local and systemic infectivity similar to MRFV‐WT. Thus, our system circumvents a major limitation of maize VIGS vectors reported to date, all of which progressively lose insert after a few rounds of replication in infected plants, with concurrent fading of VIGS phenotype in systemic leaves attributed to insert instability.
However, while such explanations are certainly plausible for unstable VIGS vectors, some fading of the VIGS phenotype with plant growth should be expected regardless of the stability of a VIGS vector. This is because the virus construct is both the inducer and primary target of VIGS, with endogenous PDS transcripts being collateral damage. Thus, in longer‐growth‐span plants like maize, as the plant continues to gain biomass enabling it to mount stronger antiviral defenses, a reduction in virus titer means less viral replicative dsRNA trigger of silencing at the peak of the recovery phenomenon associated with VIGS (Ghoshal & Sanfacon, 2015), resulting in weaker targeting of endogenous PDS transcripts. Thus, progressive fading of VIGS, when seen in the recovery context, should be expected even for highly stable VIGS vectors. Indeed, despite the longevity and high penetrance of the MRFV‐PDS120 VIGS phenotype, in late leaves it was strong but not as potent as in early leaves despite 100% retention of insert throughout plant growth. Such observations are consistent with the impact of the host RNA‐dependent RNA polymerase‐mediated secondary silencing response in systemic leaves as a major antiviral arm of silencing. However, insert stability in MRFV‐PDS120 sustains VIGS phenotype over prolonged time frames, which would enable sufficient time to study the impact of VIGS knockdown of targeted genes.
Besides remarkable stability, the MRFV‐PDS system combines several attributes that likely explain its potency as a VIGS vector. Rapid establishment of VIGS phenotype in MRFV‐PDS, mostly initiating concomitantly with first onset of virus symptoms as early as 5 dpi, greatly extends the window for experimental analysis. With retention of wild‐type levels of systemic infectivity, MRFV‐PDS120 is as highly proliferative and invasive to newly emerging apical shoots and reproductive structures such as tassels and silks, thus increasing penetrance of the VIGS phenotype. MRFV being monopartite, the system has the simplicity of requiring in vitro transcripts of a single genomic RNA and is readily and robustly launched through VPI, resulting in similar patterns of infection and VIGS.
The simplicity of construct design is an added advantage of the MRFV‐PDS system. We initially perceived testing of the HEL/POL junction to be a long shot given the critical roles of helicase and polymerase genes in virus replication, perturbation of which should be lethal to the virus. However, the retention of both replication and systemic movement functions in MRFV‐PDS120, indistinguishable from MRFV‐WT shows the efficacy of our construct design, with duplication of only 6 amino acids of the POL N‐terminus and use of the FDMV 2A peptidase sufficient to preserve proteolytic cleavage and virus viability. With such design, the HEL/POL junction showed a higher threshold and permissibility for inserts than we expected, with sense oriented in‐frame insertions remarkably stable and sufficient for stable induction of VIGS. In this system, the 120‐nt PDS in a total insert context of 201 nt, showed more potent VIGS than larger fragments previously used for VIGS of maize genes in other virus systems (Mei et al., 2016; Wang et al., 2016). An increase in PDS fragment to 231 nt (312 nt total insertion) in MRFV‐PDS231 retained infectivity and VIGS phenotype indistinguishable from MRFV‐PDS120, but had no phenotypic silencing gains compared to the smaller insert construct MRFV‐PDS120. Thus, the HEL/POL junction can hold larger fragments within size ranges often used for VIGS in maize (Mei et al., 2016; Wang et al., 2016), with no notable increase in VIGS over that already potently induced by the smaller 120‐nt PDS fragment. The upper limit of stable insertion size was not determined.
With MRFV having a compact genome encapsidated in icosahedral particles, we expected MRFV‐PDS120 to be readily subject to genome size constraints on insert size and encapsidation. Packaging of the viral genome is expected to be required for systemic movement. However, our data indicated no such constraints on MRFV‐PDS120 encapsidation, as it stably maintained the insert and systemic movement at levels similar to MRFV‐WT, with virus particles confirmed using electron microscopy. The capacity of MRFV‐PDS120 to stably retain the PDS silencing insert was contrary to expected instability spurred by genome size constraints in viruses with icosahedral particles and compact genomes like MRFV. Thus, based on genomic similarities, the homologous HEL/POL junction in TYMV and other Tymoviridae members might be equally permissive to insertions and thus produce VIGS systems as robust as that described for MRFV in this work. This would greatly advance the functional genomics of plant hosts of viruses in the Tymoviridae family subsequent to this study and pioneering work on TYMV VIGS vector (Pflieger et al., 2008).
In summary, our work describes engineering of MRFV into a VIGS vector that is stable throughout the growth cycle of maize plants and over four serial passage infections. The stability of the MRFV VIGS system results in high VIGS penetrance and longevity in plants. The system has potential for wide application in maize functional genomics research.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with this work.
AUTHOR CONTRIBUTIONS
LRS led the project and LRS and SM designed the research and wrote the article with contributions of all the authors. SM performed most of the molecular experiments. JX performed insect vector transmission assays. KW, NK, and DM optimized and performed infection and stability assays and assisted in protocol development.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Michael C. Edwards and John J. Weiland (formerly at the United States Department of Agriculture, Agricultural Research Service Cereal Crops Research Unit, Fargo, ND 58102) for providing the infectious MRFV‐US clone. We thank Tea Meulia and Khwannarin Khemsom (Molecular & Cellular Imaging Center, Ohio State University) for assistance with electron microscopy and Jody Livesay for technical support. We thank Dr. Feng Qu (Ohio State University) and laboratory for assistance with radioisotopes for Northern blots. This research was sponsored by the Defense Advanced Research Projects Agency (DARPA) and was accomplished under cooperative agreement number HR0011‐17‐2‐0054. The views expressed are those of the author and should not be interpreted as representing the official views or policies of the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation hereon. This research was supported in part by the U.S. Department of Agriculture, Agricultural Research Service. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Mlotshwa S, Xu J, Willie K, Khatri N, Marty D, Stewart LR. Engineering Maize rayado fino virus for virus‐induced gene silencing. Plant Direct. 2020;4:1–15. 10.1002/pld3.224
REFERENCES
- Bradfute, O. E. , Nault, L. R. , Gordon, D. T. , Robertson, D. C. , Toler, R. W. , & Boothroyd, C. W. (1980). Identification of maize rayado fino virus in the United‐States. Plant Disease, 64, 50–53. [Google Scholar]
- Camborde, L. , Tournier, V. , Noizet, M. , & Jupin, I. (2007). A Turnip yellow mosaic virus infection system in Arabidopsis suspension cell culture. FEBS Letters, 581, 337–341. [DOI] [PubMed] [Google Scholar]
- Cody, W. B. , & Scholthof, H. B. (2019). Plant Virus Vectors 3.0: Transitioning into synthetic genomics. Annual Review of Phytopathology, 57, 211–230. [DOI] [PubMed] [Google Scholar]
- Crivelli, G. , Ciuffo, M. , Genre, A. , Masenga, V. , & Turina, M. (2011). Reverse genetic analysis of ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation. Journal of Virology, 85, 5091–5104. 10.1128/JVI.02565-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X. S. , Mannas, S. W. , Bishop, B. A. , Rao, X. L. , Lecoultre, M. , Kwon, S. , & Nelson, R. S. (2018). An improved brome mosaic virus silencing vector: Greater insert stability and more extensive VIGS. Plant Physiology, 176, 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X. S. , Schneider, W. L. , Chaluvadi, S. R. , Mian, M. A. R. , & Nelson, R. S. (2006). Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Molecular Plant‐Microbe Interactions, 19, 1229–1239. [DOI] [PubMed] [Google Scholar]
- Edwards, M. C. , & Weiland, J. J. (2010). First infectious clone of the propagatively transmitted Oat blue dwarf virus. Archives of Virology, 155, 463–470. [DOI] [PubMed] [Google Scholar]
- Edwards, M. C. , & Weiland, J. J. (2011). Presence of a polyA tail at the 3' end of maize rayado fino virus RNA. Archives of Virology, 156, 331–334. [DOI] [PubMed] [Google Scholar]
- Edwards, M. C. , & Weiland, J. J. (2014). Coat protein expression strategy of oat blue dwarf virus. Virology, 450, 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M. C. , Weiland, J. J. , Todd, J. , & Stewart, L. R. (2015). Infectious Maize rayado fino virus from Cloned cDNA. Phytopathology, 105, 833–839. [DOI] [PubMed] [Google Scholar]
- Edwards, M. C. , Weiland, J. J. , Todd, J. , Stewart, L. R. , & Lu, S. W. (2016). ORF43 of maize rayado fino virus is dispensable for systemic infection of maize and transmission by leafhoppers. Virus Genes, 52, 303–307. [DOI] [PubMed] [Google Scholar]
- Edwards, M. C. , Zhang, Z. J. , & Weiland, J. J. (1997). Oat blue dwarf marafivirus resembles the tymoviruses in sequence, genome organization, and expression strategy. Virology, 232, 217–229. [DOI] [PubMed] [Google Scholar]
- Gamez, R. (1969). A new leafhopper‐borned virus of corn in Central America. Plant Disease Reporter, 53, 929–932. [Google Scholar]
- Gamez, R. (1973). Transmission of Rayado fino virus of maize (Zea mays) by Dalbulus maidis. Annals of Applied Biology, 73, 285–292. [DOI] [PubMed] [Google Scholar]
- Ghoshal, B. , & Sanfacon, H. (2015). Symptom recovery in virus‐infected plants: Revisiting the role of RNA silencing mechanisms. Virology, 479, 167–179. 10.1016/j.virol.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Gibbs, A. J. , Haenni, A.‐L. , Jupin, I. , Martelli, G. P. , Edwards, M. C. , Koenig, R. , … Dreher, T. W. (2011). Family tymoviridae. Virus taxonomy. Oxford, UK: Elsevier. [Google Scholar]
- Gordon, D. T. , Nault, L. R. , Gordon, N. H. , & Heady, S. E. (1985). Serological detection of corn stunt spiroplasma and maize rayado fino virus in field‐collected Dalbulus spp. from Mexico. Plant Disease, 69, 108–111. [Google Scholar]
- Hammond, R. W. , & Ramirez, P. (2001). Molecular characterization of the genome of Maize rayado fino virus, the type member of the genus Marafivirus. Virology, 282, 338–347. [DOI] [PubMed] [Google Scholar]
- Hefferon, K. (2017). Plant virus expression vectors: A powerhouse for global health. Biomedicines, 5(4), 44 10.3390/biomedicines5030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec, A. , Drugeon, G. , Camborde, L. , & Jupin, I. (2007). Proteolytic processing of Turnip Yellow Mosaic Virus replication proteins and functional impact on infectivity. Journal of Virology, 81, 11402–11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarugula, S. , Willie, K. , & Stewart, L. R. (2018). Barley stripe mosaic virus (BSMV) as a virus‐induced gene silencing vector in maize seedlings. Virus Genes, 54, 616–620. [DOI] [PubMed] [Google Scholar]
- Kant, R. , & Dasgupta, I. (2019). Gene silencing approaches through virus‐based vectors: Speeding up functional genomics in monocots. Plant Molecular Biology, 100, 3–18. [DOI] [PubMed] [Google Scholar]
- Lindbo, J. A. (2007). TRBO: A high‐efficiency tobacco mosaic virus RNA‐Based overexpression vector. Plant Physiology, 145, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, R. (1995). Vascular puncture of maize kernels for the mechanical transmission of maize white line mosaic virus and other viruses of maize. Phytopathology, 85, 139–143. 10.1094/Phyto-85-139 [DOI] [Google Scholar]
- Maccheroni, W. , Alegria, M. C. , Greggio, C. C. , Piazza, J. P. , Kamla, R. F. , Zacharias, P. R. A. , … da Silva, A. C. R. (2005). Identification and genomic characterization of a new virus (Tymoviridae family) associated with citrus sudden death disease. Journal of Virology, 79, 3028–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madriz‐Ordenana, K. , Rojas‐Montero, R. , Lundsgaard, T. , Ramirez, P. , Thordal‐Christensen, H. , & Collinge, D. B. (2000). Mechanical transmission of maize rayado fino marafivirus (MRFV) to maize and barley by means of the vascular puncture technique. Plant Pathology, 49, 302–307. [Google Scholar]
- Martelli, G. P. , Sabanadzovic, S. , Sabanadzovic, N. A. G. , Edwards, M. C. , & Dreher, T. (2002). The family Tymoviridae. Archives of Virology, 147, 1837–1846. [DOI] [PubMed] [Google Scholar]
- Mei, Y. , Zhang, C. Q. , Kernodle, B. M. , Hill, J. H. , & Whitham, S. A. (2016). A Foxtail mosaic virus vector for virus‐induced gene silencing in maize. Plant Physiology, 171, 760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S. , Schauer, S. E. , Smith, T. H. , Mallory, A. C. , Herr, J. M. Jr , Roth, B. , … Vance, V. B. (2005). Ectopic DICER‐LIKE1 expression in P1/HC‐Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. The Plant Cell, 17, 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault, L. R. , Gingery, R. E. , & Gordon, D. T. (1980). Leafhopper transmission and host range of maize rayado fino virus. Phytopathology, 70, 709–712. [Google Scholar]
- Pflieger, S. , Blanchet, S. , Camborde, L. , Drugeon, G. , Rousseau, A. , Noizet, M. , … Jupin, I. (2008). Efficient virus‐induced gene silencing in Arabidopsis using a 'one‐step' TYMV‐derived vector. Plant Journal, 56, 678–690. [DOI] [PubMed] [Google Scholar]
- Rivera, C. , & Gamez, R. (1986). Multiplication of maize rayado fino virus in the leafhopper vector Dalbulus maidis. Intervirology, 25, 76–82. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. , & Mysore, K. S. (2011). New dimensions for VIGS in plant functional genomics. Trends in Plant Science, 16, 656–665. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Yang, X. X. , Wang, N. , Liu, X. D. , Nelson, R. S. , Li, W. M. , … Zhou, T. (2016). An efficient virus‐induced gene silencing vector for maize functional genomics research. Plant Journal, 86, 102–115. [DOI] [PubMed] [Google Scholar]
- Yu, J. , Gao, L. W. , Liu, W. S. , Song, L. X. , Xiao, D. , Liu, T. K. , … Zhang, C. W. (2019). Transcription coactivator ANGUSTIFOLIA3 (AN3) regulates leafy head formation in Chinese cabbage. Frontiers in Plant Science, 10, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano, J.‐L. , Francis, D. M. , & Redinbaugh, M. G. (2013). Identification of resistance to maize rayado fino virus in maize inbred lines. Plant Disease, 97, 1418–1423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


