Abstract
Purpose
microRNAs (miRNAs) mediate the pathological mechanisms of diabetic retinopathy. In this study, we compared miRNA expression profiles in the vitreous between patients with proliferative diabetic retinopathy (PDR) and patients with a macular hole as non-diabetic controls, and between PDR patients treated with anti-vascular endothelial growth factor (VEGF) therapy and untreated PDR patients.
Methods
Vitreous samples of non-diabetic and PDR patients were screened for miRNAs with quantitative polymerase chain reaction (qPCR) panels. miRNA candidates were validated in vitreous samples of a second, independent cohort. In addition, the effect of anti-VEGF therapy was investigated in the vitreous of a third study population consisting of PDR patients who had not received anti-VEGF therapy and PDR patients who had received preoperative anti-VEGF therapy.
Results
During screening, seven miRNAs were found to be significantly higher in the vitreous of PDR patients, whereas two miRNAs were found to be significantly lower compared with non-diabetic controls. Validating the expression of these miRNAs in a second cohort resulted in the identification of six miRNAs that were expressed at significantly higher rates in the vitreous of PDR patients: hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-326, and hsa-miR-362-5p. Among these six miRNAs, hsa-miR-23b-3p levels were lower in the anti-VEGF-treated group of PDR patients compared with untreated PDR patients.
Conclusions
In this study, we identified six miRNAs that are expressed more highly in PDR patients and one miRNA that is expressed at a lower levels in anti-VEGF-treated PDR patients.
Translational Relevance
miRNAs identified in the vitreous of PDR patients may improve our understanding of the mechanisms leading to PDR.
Keywords: proliferative diabetic retinopathy, microRNA, vitreous humor, anti-VEGF
Introduction
Diabetic retinopathy (DR) is a vision-threatening complication of diabetes and an important cause of blindness worldwide.1 It is a multistage disease that develops slowly and can be divided clinically into a non-proliferative or a proliferative state. The latter is referred to as proliferative diabetic retinopathy (PDR), which is characterized by retinal neovascularization and ultimately vision loss.2,3 Even though photocoagulation or anti-vascular endothelial growth factor (VEGF) therapy offers treatment options, treatment success is quite variable.4,5 Furthermore, the mechanisms driving DR are still not completely understood, so a deeper understanding of them has the potential to lead to new and improved therapies.6
The expression of microRNAs (miRNAs) is altered in DR and is anticipated to contribute to disease progression.7–9 miRNAs are small, non-coding RNAs, 19 to 22 nucleotides in length, that regulate gene expression by inhibiting messenger RNA translation.10,11 In addition, they are highly stable in body fluids,12 as they either are actively secreted from cells or passively accumulate due to cellular processes such as apoptosis.13–15 Once in a body fluid, miRNAs can be taken up by cells and consecutively modulate their gene expression.14 The expression of vitreal miRNAs is also changed in various vitreoretinal diseases,16–22 suggesting that vitreal miRNAs may influence disease progression.
Which miRNAs are differently expressed in the human vitreous during PDR is still largely unexplored. Furthermore, correctly measuring and interpreting miRNA expression in the vitreous is challenging due to the lack of valid internal controls.23
In this study, we therefore aimed to identify miRNAs that are differently expressed in the vitreous of PDR patients compared with non-diabetic control patients. Furthermore, we investigated whether or not preoperative anti-VEGF therapy in PDR was associated with an altered miRNA expression profile.
Materials and Methods
Study Population
Vitreous samples were obtained from the University of Newcastle upon Tyne. A favorable ethical opinion for the collection of the samples was obtained from the National Health Service (NHS) research ethics committee (South East Coast-Surrey Research Ethics Committee reference 12/LO/0130), and the collection was carried out at Sunderland Eye Infirmary under the sponsorship of the City Hospitals Sunderland NHS foundation trust. After the aims of the study were explained, written consent was obtained from all patients, and the study was conducted according to the Declaration of Helsinki. Consecutive patients undergoing vitrectomy for the complications of either a macular hole or PDR were recruited by one surgeon from 2015 to 2018. Eyes with previous vitrectomy surgery were excluded. Eyes with active PDR were pretreated with bevacizumab, ranibizumab, or aflibercept 2 to 5 days prior to vitrectomy. All clinical variables were determined by a single experienced retinal specialist.
Patient characteristics can be found in Table 1. A screening cohort was used initially for a screening purposes; it consisted of 10 vitreous samples from patients with macular holes as controls and 10 samples from PDR patients. There was no statistically significant difference in age or gender ratio between the control and PDR patients.
Table 1.
Patient Characteristics
| Gender | Diabetes Mellitus | ||||
|---|---|---|---|---|---|
| Age (y), | Duration (y), | HbA1c (mmol/mol), | |||
| Cohort | mean ± SD | Female, n (%) | Male, n (%) | mean ± SD | mean ± SD |
| Screening cohort | |||||
| Controls (n = 10) | 66.0 ± 5.2 | 5 (50) | 5 (50) | — | — |
| PDR (n = 10) | 64.7 ± 6.1 | 5 (50) | 5 (50) | 26.7 ± 14.6 | 96.3 ± 22.6 |
| P | 0.614 | 1.000 | |||
| Confirmation cohort | |||||
| Controls (n = 17) | 71.1 ± 3.9 | 12 (71) | 5 (29) | — | — |
| PDR (n = 11) | 69.1 ± 9.0 | 4 (36) | 7 (64) | 20.1 ± 7.2 | 74.9 ± 10.9 |
| P | 0.417 | 0.079 | |||
| Anti-VEGF cohort | |||||
| Untreated PDR (n = 16) | 59.1 ± 13.4 | 8 (50) | 8 (50) | 21.7 ± 10.0 | 78.2 ± 20.8 |
| Treated PDR (n = 17) | 50.6 ± 13.3 | 6 (35) | 11 (65) | 22.5 ± 9.2 | 79.0 ± 22.0 |
| P | 0.076 | 0.409 | 0.817 | 0.918 | |
| Bevacizumab + ranibizumab (n = 11)a | 47.1 ± 10.6 | 4 (36) | 7 (64) | 19.5 ± 7.4 | 84.7 ± 20.0 |
| Aflibercept (n = 6) | 57.0 ± 16.3 | 2 (33) | 4 (67) | 27.8 ± 10.6 | 69.5 ± 23.7 |
| P | 0.149 | 0.908 | 0.076 | 0.190 | |
P < 0.05 was considered to be statistically significant.
This group includes 10 patients pretreated with bevacizumab and one patient pretreated with ranibizumab.
A second cohort, the confirmation cohort, consisted of 28 patients, including 17 control and 11 PDR patients, and was used for confirmation of the identified miRNAs. There was no statistically significant difference in age or gender ratio between control and PDR patients. The HbA1c value for one patient could not be retrieved.
Finally, a third cohort of patients, the anti-VEGF cohort, was used to compare untreated and anti-VEGF-treated patients. Of 33 PDR patients, 17 had received preoperative anti-VEGF treatment (PDR(+)) in the form of bevacizumab (n = 10), ranibizumab (n = 1), or aflibercept (n = 6), and 16 of them had not received any anti-VEGF therapy and were referred to as PDR(–). The samples of 8 of the 16 patients without anti-VEGF treatment were also used in the confirmation cohort. There were no statistically significant differences in age, gender ratio, duration of diabetes, or HbA1c concentration between patients with or without anti-VEGF therapy. The HbA1c value of one patient in each group is not known. In addition, no statistically significant differences were found between the two different pretreatment groups (bevacizumab + ranibizumab vs. aflibercept) (Table 1).
RNA Isolation
RNA was isolated using either the miRCury RNA Isolation Kit (#300112; Exiqon, Vedbaek, Denmark) for the vitreous samples of the screening study group or the miRNeasy Serum/Plasma Advanced Kit (#217204; Qiagen, Hilden, Germany) for the vitreous samples of the remaining two study groups. The difference in isolation kits is a consequence of the integration of Exiqon into Qiagen in October 2017, when the miRCury Kit of Exiqon became no longer available. Vitreous samples were thawed on ice and centrifuged (14,000×g, 15 minutes, 4°C); 200 µl of clear supernatant was then transferred to a fresh tube, and RNA was isolated according to the respective manufacturer's instructions. The isolation step was performed with various spike-ins, including UniSP2 (2 fmol), UniSP4 (0.02 fmol), UniSP5 (0.0002 fmol), UniSP6 (0.15 fmol), and cel-miR-39-3p (0.002 fmol), all of which are synthetic RNAs (#203203; Exiqon) that served as exogenous controls, as well as 10 ng of glycogen (Invitrogen, Carlsbad, CA, USA). RNA was eluted with 50 µl of RNase free water and stored as aliquots of 8 µl at –80°C until further use.
miRNA qPCR
To identify miRNAs of interest, miRNA expression was measured in the screening cohort using the Human microRNA PCR Panel I (V4.M, #203617; Exiqon). We chose quantitative polymerase chain reaction (qPCR) panels, because they offer a focused search and are more sensitive than hybridization panels,24,25 thus increasing the chance of detecting miRNAs expressed at potentially low abundance in the vitreous.
In subsequent experiments, the expression of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, and hsa-miR-199a-5p was measured using the Qiagen miRCURY SYBR Green system. Because hsa-miR-223-3p, hsa-miR-326, hsa-miR-362-5p, and hsa-miR-662 could not be detected with this system, their expression was measured using TaqMan Advanced miRNA Assays (#A28007 and #4444557; Thermo Fisher Scientific, Waltham, MA, USA). All reactions were run in duplicate.
In the confirmation cohort, the expression of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, and hsa-miR-199a-5p was measured with the Qiagen system for 17 control and 11 PDR samples, and the expression of hsa-miR-223-3p, hsa-miR-326, hsa-miR-362-5p, and hsa-miR-662 was measured with the TaqMan assays for 17 control and 8 PDR samples. In the anti-VEGF cohort, the expression of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, and hsa-miR-185-5p was measured with the Qiagen system for 16 PDR(–) and 17 PDR(+) samples, and the expression of hsa-miR-326 and hsa-miR-362-5p was measured with the TaqMan assays for 13 PDR(–) and 16 PDR(+) samples. The difference in samples sizes within the two cohorts can be explained by vitreous being used up while measuring hsa-miR-223-3p, hsa-miR-326, hsa-miR-362-5p, and hsa-miR-662 with the Qiagen miRCURY system. To check for blood or hemolysis as a source of miRNA expression in our samples, we additionally measured the expression of hsa-miR-451 and hsa-miR-144, two miRNAs indicative of hemolysis.26
All experiments were performed following the respective manufacturer's instructions and run on an ABI ViiA7 real-time PCR system (Applied Biosystems, Foster City, CA, USA). As RNA concentration is frequently below 2 ng/µl in body fluids and thus cannot be reliably measured with a spectrophotometer,27 using a constant input volume of RNA is recommended. The array was run with 16 µl input RNA, and the SYBR Green and TaqMan assays were run with 4 µl and 2 µl input RNA, respectively. A detailed overview of experiment parameters, kits, and primers used can be found in Supplementary Table S1.
PCR Data Analysis
Raw qPCR data were analyzed using QuantStudio Real-Time PCR System software (Applied Biosystems). Ct values were converted to arbitrary absolute amounts (2−Ct × 1E12) and expressed as fold change compared with controls.
In the screening cohort, miRNAs expressed in fewer than 70% of samples in both the control group and the PDR group were not considered representative for a difference in controls and PDR and were omitted from further analysis (Supplementary Table S2).
Normalization
In the screening cohort, different normalization methods (i.e., global mean,28 UniSp3 IPC, UniSp2, UniSp4, UniSp5, UniSp6, cel-miR-39-3p, and snoU6) were compared. snoU6 was expressed in only 8 out of 20 samples and could therefore not be used for normalization. Ct values of all selected miRNAs under investigation were converted to arbitrary absolute amounts (2–Ct × 1E12). The coefficient of variation (CV), equal to (SD/mean) × 100%, of each miRNA under each method of normalization was then calculated separately for the controls-only sample set (Supplementary Fig. S1A) and the complete sample set (Supplementary Fig. S1B). No normalization, global normalization, and normalization by UniSp3 IPC gave the lowest CV, and there were no significant differences between no normalization and global normalization; all other normalization methods resulted in significantly higher CVs compared with that of no normalization. Because global normalization or normalization by UniSP3 IPC could not be applied in the confirmation and anti-VEGF cohorts, we opted to use no normalization in all cohorts.
miRNA Target Analysis
Validated targets were extracted from miRTarBase 7.0.29 Predicted targets were extracted from miRWalk 3.0.30 To limit the number of validated and predicted targets, we focused on genes with elevated protein levels in the vitreous of PDR patients according to Klaassen et al.31
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8.0.1 for Mac (GraphPad Software, La Jolla, CA, USA; www.graphpad.com). For the screening analysis, a two-tailed Mann–Whitney U test was used to detect a statistically significant difference between the groups. For further qPCR experiments, a one-tailed Mann–Whitney test was used to confirm a statistically significant difference if not indicated otherwise. P < 0.05 was considered statistically significant.
Results
Identification of miRNAs Differently Expressed in the Vitreous of PDR Patients and Controls
To identify miRNAs differently expressed in the vitreous of PDR patients compared with patients with a macular hole (MH) as non-diabetic controls, we measured the expression of multiple miRNAs with a qPCR panel. In the vitreous of control patients, 24 miRNAs were regularly expressed, being defined as miRNAs that are detectable in at least 70% of the samples of one group. In the vitreous samples of PDR patients, 26 miRNAs were regularly expressed (Supplementary Table S2).
Of these miRNAs, hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-223-3p, hsa-miR-362-5p, and hsa-miR-662 expression levels were significantly higher in the vitreous of PDR patients as compared to controls, whereas hsa-miR-199a-5p and hsa-miR-326 levels were significantly lower (Fig. 1). Two miRNAs related to hemolysis, hsa-miR-451 and hsa-miR-144, did not show elevated levels in the vitreous of PDR patients as compared to controls.
Figure 1.
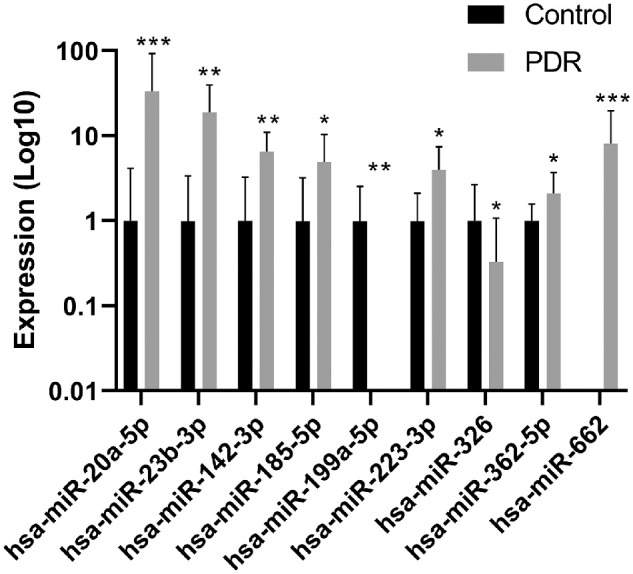
Expression levels of miRNAs in the vitreous that were significantly different between control and PDR patients. The expression levels of miRNAs in the screening cohort consisted of control and PDR samples (control, n = 10; PDR, n = 10). The expression levels are shown as the means of absolute amounts with standard deviations as compared with controls on a log10 scale. The expression levels of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-199a-5p, hsa-miR-223-3p, hsa-miR-326, and hsa-miR-362-5p are relative to the control group. The expression levels of hsa-miR-662 were calculated as absolute amounts (2−Ct × 1E12). *P < 0.05, **P < 0.01, and ***P < 0.001.
Confirmation of Different miRNA Expression in a Cohort of PDR Patients and Controls
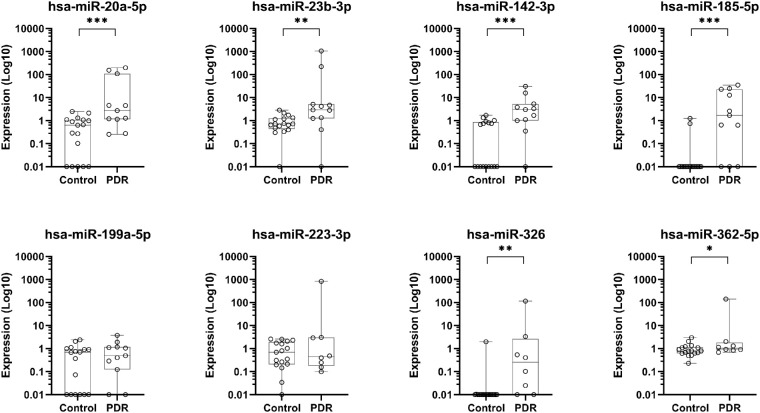
To verify our screening results, we measured the expression of these eight miRNAs in a second, independent cohort of PDR patients and non-diabetic MH controls. The expression of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, and hsa-miR-199a-5p was measured with Qiagen reagents, and the expression of hsa-miR-223-3p, hsa-miR-326, hsa-miR-362-5p, and hsa-miR-662 was measured with TaqMan reagents. The levels of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-326, and hsa-miR-362-5p were significantly higher in the vitreous of PDR patients compared with controls, whereas the expression levels of hsa-miR-199a-5p and hsa-miR-223-3p were not significantly different (Fig. 2). The expression of miR-662 could not be detected.
Figure 2.
Expression levels of miRNAs in the vitreous of the confirmation cohort. The expression levels of miRNAs with a significant difference in the screening cohort were measured in control and PDR samples of a second, larger cohort. The expression levels are shown as absolute amounts on a log10 scale relative to the control group. Box plots indicate median, minimum, and maximum and first and third quartiles. Symbols on the x-axis represent individuals without detectable expression. *P < 0.05, **P < 0.01, and ***P < 0.001.
Anti-VEGF Treatment is Associated with a Lower Expression of hsa-miR-23b-3p in the Vitreous of PDR Patients
To investigate the effect of anti-VEGF treatment on the miRNAs that had significantly higher levels in PDR patients (hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-326, and hsa-miR-362-5p), we measured their expression in the vitreous of a third study group consisting of patients with PDR who had or had not received anti-VEGF treatment prior to surgery.
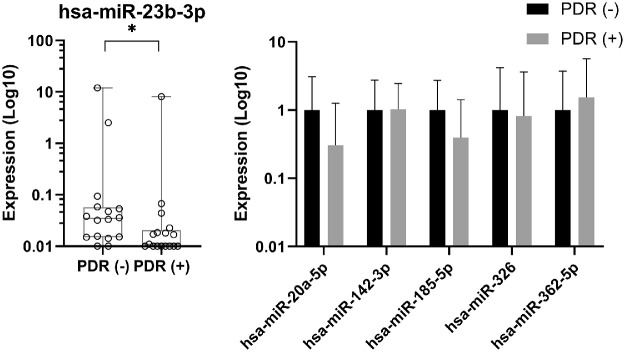
The level of hsa-miR-23b-3p was significantly lower (3.5-fold, P = 0.02) in the vitreous of patients with anti-VEGF therapy compared with PDR patients without anti-VEGF treatment; the expression levels of the other miRNAs were not significantly different (Figs. 3A, 3B). There were no significant differences in miRNA expression between patients with type 1 diabetes and those with type 2 diabetes (data not shown).
Figure 3.
Expression levels of miRNAs in the vitreous with and without anti-VEGF therapy. The expression levels of miRNAs were measured in the vitreous of patients with PDR who had not received anti-VEGF treatment, PDR(–) and in the vitreous of patients who had received anti-VEGF treatment, PDR(+). For hsa-miR-23b-3p, the expression levels are shown as absolute amounts on a log10 scale relative to the PDR(–) group. Box plots indicate median, minimum, and maximum and first and third quartiles. (A) The symbol on the x-axis represents one individual without detectable expression. (B) For hsa-miR-20a-5p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-326, and hsa-miR-362-5p, the expression levels are shown as the means of absolute amounts with standard deviations as compared with the PDR(–) group on a log10 scale. *P = 0.03.
miRNAs of Interest Are Related to Proteins That Are Increased in Vitreous of PDR Patients
To estimate possible functions of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-326, and hsa-miR-362-5p, we analyzed validated and predicted targets of these miRNAs. Within these targets, we focused on proteins that are differently expressed in the vitreous of PDR patients31 and are associated with DR.
All six miRNAs are predicted to target proteins that are elevated in DR,31 such as bone morphogenetic protein 2 or angiopoietin 1 (Table 2), or are associated with angiogenesis and wound healing responses, major processes involved in PDR (VEGF, angiopoietin-2, or connective tissue growth factor [CTGF]). VEGF-A is a confirmed target protein for hsa-miR-20a-5p and hsa-miR-185-5p.
Table 2.
Validated and Predicted Gene Targets of miRNAs of Interest
| miRNA | Validated Targets | Predicted Targets |
|---|---|---|
| hsa-miR-20a-5p | BMP2, PDGFB, THBS1, VEGFA | ANGPT1, ICAM1, IGFBP3, NRG1, UBB |
| hsa-miR-23b-3p | — | ADIPOQ, ANGPT2, BMP2, CTGF, HGF1, NRG1, PDGFB, THBS1, VEGFA |
| hsa-miR-142-3p | — | ANGPT2, HGF, IGFBP3, NRG1, VEGFA |
| hsa-miR-185-5p | VEGFA | ADIPOQ, ANGPT2, BMP2, ICAM1, NOV, NRG1, PDGFB |
| hsa-miR-326 | NOTCH1, NOTCH2 | CTGF, GDF15, HGF, IGFBP2, IGFBP3, MMP2, MMP9, NGFR, NOTCH3, NRG1, PDGFA, PDGFB, PDGFRB, TIMP2, VEGFA |
| hsa-miR-362-5p | — | ADIPOQ, ANGPT1, ANGPT2, MMP2, NGFR, NRG1, PDGFB, PDGFRA, PDGFRB, THBS1, TIMP2 |
Discussion
In the present study, our aims were to identify differently expressed miRNAs in the vitreous of PDR patients compared to the vitreous of non-diabetic MH patients and to determine whether the expression of these miRNAs was different after preoperative anti-VEGF treatment. We chose patients with MH as controls, because sufficient numbers of patients undergoing vitrectomy are available, this condition has the least retinovascular pathology compared to other eye conditions, and the patients are non-diabetic.
The Change of Vitreal miRNA Expression in PDR Patients
We found 24 miRNAs regularly expressed in the vitreous of control patients and 26 miRNAs regularly expressed in PDR patients. Of these miRNAs, we determined that the expression of hsa-miR-20a-5p, hsa-miR-20a-23b-3p, hsa-miR-20a-142-3p, hsa-miR-20a-185-5p, hsa-miR-20a-326, and hsa-miR-20a-362-5p was higher in the vitreous of PDR patients. This difference in miRNA expression also seems to be independent of diabetes type. Furthermore, the expression of hsa-miR-23b-3p was lower in patients who had received preoperative anti-VEGF treatment compared with untreated PDR patients.
Targets of Increased Vitreal miRNAs in PDR
Because we compared PDR patients to non-diabetic controls, the differences observed may be related to the diabetic state, DR mechanisms in general, or specifically to PDR. However, all six miRNAs with elevated expression in PDR patients can possibly target angiogenic or profibrotic proteins that are increased in the vitreous during PDR,31 including VEGF-A, angiopoietin 2, platelet-derived growth factor B (PDGF-B), and CTGF, which implies that these miRNAs may be actively involved in regulating angiogenesis and wound healing responses specific to PDR. The increase in miRNA expression might be a compensatory mechanism to decrease VEGF-A, angiopoietin 2, PDGF-B, or other protein levels. On the other hand, protein levels may be regulated by miRNAs in a different fashion. One example is the interaction between miR-20a-5p and VEGF, where miR-20a-5p targets VEGF but also stimulates its expression.32 In contrast, VEGF itself is able to increase the expression of miR-20a-5p.33 This indicates that the interplay between miRNAs and their targets is complex, and further research on the role of hsa-miR-20a-5p, miR-20a-23b-3p, miR-20a-142-3p, miR-20a-185-5p, miR-20a-326, and miR-20a-362-5p in PDR is necessary.
Association of Anti-VEGF Therapy with the Vitreal miRNA Expression in PDR
Single preoperative anti-VEGF therapy 2 to 5 days before surgery is a recognized treatment in PDR,34 particularly prior to vitrectomy in patients with active neovascularization.35 Here, we observed that such anti-VEGF therapy was associated with a lower expression of hsa-miR-23b-3p in PDR as compared with untreated PDR patients. It should be noted in this context that selection bias may be involved in the composition of these two groups; nevertheless, as VEGF increases the expression of hsa-miR-23b-3p,36 anti-VEGF therapy may result in a decrease of hsa-miR-23b-3p expression. Anti-VEGF therapy, however, may also influence the expression of miRNAs in ways other than we have investigated. Additionally, different anti-VEGF agents may also influence the expression of different miRNAs.
Comparison with Other Studies Investigating Vitreal miRNAs in PDR
Thus far, to the best of our knowledge, no other studies have performed a large-scale miRNA inventory in PDR patients as compared to controls with groups of more than four patients. The team of Hirota et al.19 was the first to publish differentially expressed miRNAs in the vitreous of PDR patients as compared with control patients with a macular hole, with four patients in each group. No overlap in miRNAs with our study was found, nor did we find a significant difference in expression levels for miR-200b, a single miRNA that was investigated by another group.20
The lack of overlap between our study and these two studies may be explained by differences in screening platforms, normalization methods, group sizes, patient characteristics, and whether or not the results were confirmed in subsequent experiments. However, this lack of overlap also emphasizes the importance of interpreting results carefully and highlights the need for standardized approaches, such as a standardized method of normalization.
Normalization of Vitreal miRNAs
One major challenge when investigating miRNAs in body fluids is to reliably normalize their expression for correct interpretation. The purpose of normalization is to compensate for experimentally induced technical variation, so sample variance after an effective normalization strategy should be lower than when normalization is not performed. We established that normalization by global mean or UniSp6 as a spike-in did not significantly improve sample variance compared with no normalization, whereas other normalization strategies or spike-ins significantly increased sample variance. For this reason, we chose not to normalize our data. In addition, we found snoU6, that is commonly used to normalize miRNA expression,16,20 to be only sparsely expressed in our screening cohort, which raises the question of whether or not snoU6 is suitable for miRNA normalization in the vitreous.
Limitations
One limitation of the study is the use of panels with predefined miRNAs for screening. Other methods such as next-generation sequencing may result in the identification of additional miRNAs. In addition, the miRNA profile was examined only in the vitreous of patients with end-stage PDR and compared to non-diabetic controls. As indicated, the differences observed may therefore be related to the diabetic state, DR mechanisms in general, or specifically to PDR. By investigating miRNA profiles in the vitreous of patients with diabetes without DR, earlier stages of DR and early PDR may shed further light on miRNA expression over the course of DR, further illuminating the role of miRNAs in the progression of DR. Finally, because preoperative anti-VEGF treatment was indicated by the treating physician, selection bias may have introduced other relevant differences between the groups of PDR patients with or without anti-VEGF patients.
Conclusions
We found the expression of hsa-miR-20a-5p, hsa-miR-23b-3p, hsa-miR-142-3p, hsa-miR-185-5p, hsa-miR-326, and hsa-miR-362-5p to be elevated in the vitreous of PDR patients compared with controls. These miRNAs are, therefore, interesting new candidates to study in order to better understand the pathophysiology of DR, particularly PDR. Furthermore, anti-VEGF therapy was associated with lower levels of hsa-miR-23b-3p in PDR patients, which may suggest the involvement of a regulatory mechanism.
Future research should clarify how these miRNAs influence the course of DR and PDR and if they can serve either as biomarkers or as new therapeutic targets.
Supplementary Material
Acknowledgments
This research was funded by grants from Deutsche Diabetes Gesellschaft; Jan Kornelis de Cock Stichting; Stichting Blinden-Penning, which contributed through UitZicht (UitZicht 2017-30); Stichting Blindenhulp; Nederlandse Vereniging tot Verbetering van het Lot der Blinden; and the Rotterdamse Stichting Blindenbelangen (B20170068). This study was published with the help of the Edmond en Marianne Blaauw Fonds voor Oogheelkunde. The funding organizations had no role in the design or conduct of this research. They provided unrestricted grants.
Disclosure: J. Friedrich, None; D.H.W. Steel, None; R.O. Schlingemann, None; M.J. Koss, None; H.-P. Hammes, None; G. Krenning, None; I. Klaassen, None
References
- 1. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010; 376: 124–136. [DOI] [PubMed] [Google Scholar]
- 2. Solomon SD, Chew E, Duh EJ, et al.. Diabetic retinopathy: a position statement by the American Diabetes Association. Diabetes Care. 2017; 40: 412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wong TY, Sun J, Kawasaki R, et al.. Guidelines on diabetic eye care: the International Council of Ophthalmology recommendations for screening, follow-up, referral, and treatment based on resource settings. Ophthalmology. 2018; 125: 1608–1622. [DOI] [PubMed] [Google Scholar]
- 4. Maturi RK, Bleau L, Saunders J, Mubasher M, Stewart MW. A 12-month, single-masked, randomized controlled study of eyes with persistent diabetic macular edema after multiple anti-VEGF injections to assess the efficacy of the dexamethasone-delayed delivery system as an adjunct to bevacizumab compared with continued bevacizumab monotherapy. Retina. 2015; 35: 1604–1614. [DOI] [PubMed] [Google Scholar]
- 5. Stewart MW. Treatment of diabetic retinopathy: recent advances and unresolved challenges. World J Diabetes. 2016; 7: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kusuhara S, Fukushima Y, Ogura S, Inoue N, Uemura A. Pathophysiology of diabetic retinopathy: the old and the new. Diabetes Metab J. 2018; 42: 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mortuza R, Feng B, Chakrabarti S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia. 2014; 57: 1037–1046. [DOI] [PubMed] [Google Scholar]
- 8. Fulzele S, El-Sherbini A, Ahmad S, et al.. MicroRNA-146b-3p regulates retinal inflammation by suppressing adenosine deaminase-2 in diabetes. Biomed Res Int. 2015; 2015: 846501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaidonis G, Gillies MC, Abhary S, et al.. A single-nucleotide polymorphism in the microRNA-146a gene is associated with diabetic nephropathy and sight-threatening diabetic retinopathy in Caucasian patients. Acta Diabetol. 2016; 53: 643–650. [DOI] [PubMed] [Google Scholar]
- 10. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014; 15: 509–524. [DOI] [PubMed] [Google Scholar]
- 11. Garcia-Lopez J, Brieno-Enriquez MA, Del Mazo J. MicroRNA biogenesis and variability. Biomol Concepts. 2013; 4: 367–380. [DOI] [PubMed] [Google Scholar]
- 12. Gong Q, Su G. Roles of miRNAs and long noncoding RNAs in the progression of diabetic retinopathy. Biosci Rep. 2017; 37: BSR20171157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang H, Gong F, Zhang S, Zhang CY, Zen K, Chen X. The origin, function, and diagnostic potential of extracellular microRNAs in human body fluids. Wiley Interdiscip Rev RNA. 2014; 5: 285–300. [DOI] [PubMed] [Google Scholar]
- 14. Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Mol Oncol. 2017; 11: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao ZY, Chen WB, Shao SS, et al.. Role of exosome-associated microRNA in diagnostic and therapeutic applications to metabolic disorders. J Zhejiang Univ Sci B. 2018; 19: 183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Usui-Ouchi A, Ouchi Y, Kiyokawa M, Sakuma T, Ito R, Ebihara N. Upregulation of Mir-21 levels in the vitreous humor is associated with development of proliferative vitreoretinal disease. PLoS One. 2016; 11: e0158043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ragusa M, Caltabiano R, Russo A, et al.. MicroRNAs in vitreus humor from patients with ocular diseases. Mol Vis. 2013; 19: 430–440. [PMC free article] [PubMed] [Google Scholar]
- 18. Menard C, Rezende FA, Miloudi K, et al.. MicroRNA signatures in vitreous humour and plasma of patients with exudative AMD. Oncotarget. 2016; 7: 19171–19184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirota K, Keino H, Inoue M, Ishida H, Hirakata A. Comparisons of microRNA expression profiles in vitreous humor between eyes with macular hole and eyes with proliferative diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 2015; 253: 335–342. [DOI] [PubMed] [Google Scholar]
- 20. Gomaa AR, Elsayed ET, Moftah RF. MicroRNA-200b expression in the vitreous humor of patients with proliferative diabetic retinopathy. Ophthalmic Res. 2017; 58: 168–175. [DOI] [PubMed] [Google Scholar]
- 21. Russo A, Ragusa M, Barbagallo C, et al.. miRNAs in the vitreous humor of patients affected by idiopathic epiretinal membrane and macular hole. PLoS One. 2017; 12: e0174297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kakkassery V, Schroers R, Coupland SE, et al.. Vitreous microRNA levels as diagnostic biomarkers for vitreoretinal lymphoma. Blood. 2017; 129: 3130–3133. [DOI] [PubMed] [Google Scholar]
- 23. Sheinerman KS, Umansky SR. Circulating cell-free microRNA as biomarkers for screening, diagnosis and monitoring of neurodegenerative diseases and other neurologic pathologies. Front Cell Neurosci. 2013; 7: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mestdagh P, Hartmann N, Baeriswyl L, et al.. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014; 11: 809–815. [DOI] [PubMed] [Google Scholar]
- 25. Andreasen D, Fog JU, Biggs W, et al.. Improved microRNA quantification in total RNA from clinical samples. Methods. 2010; 50: S6–S9. [DOI] [PubMed] [Google Scholar]
- 26. Blondal T, Jensby Nielsen S, Baker A, et al.. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013; 59: S1–S6. [DOI] [PubMed] [Google Scholar]
- 27. Gautam A, Kumar R, Dimitrov G, Hoke A, Hammamieh R, Jett M. Identification of extracellular miRNA in archived serum samples by next-generation sequencing from RNA extracted using multiple methods. Mol Biol Rep. 2016; 43: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hellemans J, Vandesompele J. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol Biol. 2014; 1160: 19–26. [DOI] [PubMed] [Google Scholar]
- 29. Chou CH, Shrestha S, Yang CD, et al.. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018; 46: D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018; 13: e0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klaassen I, de Vries EW, Vogels IMC, et al.. Identification of proteins associated with clinical and pathological features of proliferative diabetic retinopathy in vitreous and fibrovascular membranes. PLoS One. 2017; 12: e0187304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luengo-Gil G, Gonzalez-Billalabeitia E, Perez-Henarejos SA, et al.. Angiogenic role of miR-20a in breast cancer. PLoS One. 2018; 13: e0194638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng HT, Liu HL, Zhai BB, et al.. Vascular endothelial growth factor suppresses TNFSF15 production in endothelial cells by stimulating miR-31 and miR-20a expression via activation of Akt and Erk signals. FEBS Open Bio. 2017; 7: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Y, Singh RP. The role of anti-vascular endothelial growth factor (anti-VEGF) in the management of proliferative diabetic retinopathy. Drugs Context. 2018; 7: 212532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith JM, Steel DH. Anti-vascular endothelial growth factor for prevention of postoperative vitreous cavity haemorrhage after vitrectomy for proliferative diabetic retinopathy. Cochrane Database Syst Rev. 2015; 8: CD008214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsu MY, Hung YC, Hwang DK, et al.. Detection of aqueous VEGF concentrations before and after intravitreal injection of anti-VEGF antibody using low-volume sampling paper-based ELISA. Sci Rep. 2016; 6: 34631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.